Abstract
Background
The adaptation of organisms to changing environments is self-evident, with the adaptive evolution of organisms to environmental changes being a fundamental problem in evolutionary biology. Bees can pollinate in various environments and climates and play important roles in maintaining the ecological balance of the earth.
Results
We performed an analysis of 462 Apis cerana (A. cerana) specimens from 31 populations in 11 regions and obtained 39 representative morphological features. We selected 8 A. cerana samples from each population and performed 2b-RAD simplified genome sequencing. A total of 11,506 high-quality single nucleotide polymorphism (SNP) loci were obtained. For these SNPs, the minor allele frequency (MAF) was > 1%, the average number of unique labels for each sample was 49,055, and the average depth was 72.61x. The ratios of the unique labels of all samples were 64.27–86.33%.
Conclusions
Using 39 morphological characteristics as the data set, we proposed a method for the rapid classification of A. cerana. Using genomics to assess population structure and genetic diversity, we found that A. cerana has a large genetic difference at the ecotype level. A comparison of A. cerana in North China revealed that some physical obstacles, especially the overurbanization of the plains, have isolated the populations of this species. We identified several migration events in North China and Central China. By comparing the differences in the environmental changes in different regions, we found that A. cerana has strong potential for climate change and provides a theoretical basis for investigating and protecting A. cerana.
Keywords: Apis cerana, Apis mellifera, Genomics, Population genetics, Genetic differentiation, Population structure, Standard morphometrics
Background
Adaptation to a changing environment is a fundamental problem in evolutionary biology. Understanding the impact of the environment on biological genetic diversity can not only clarify the course of evolution but can also provide information on how to protect the ecological environment [1, 2]. Bees have unique ecological value, and one-third of human food is derived from insect pollination, which is critical for maintaining the dynamics of the ecosystem. Bees are important mediators of pollination, bringing ecological value of more than 200 billion US dollars per year [3, 4].
Apis contains 10 different species, most of which are distributed throughout Asia [5]. A. cerana is an Asian honeybee species with a wide geographical distribution. Previously, A. cerana was divided into five or six local categories based on a comprehensive study of mitochondrial DNA and microsatellite DNA, and the A. cerana species in China belonged to a subspecies [6, 7]. A. cerana was defined as one of the most important pollinators in China, domesticating the region for more than 2000 years [8]. The various climate types and complex terrains in China have promoted bee differentiation. Previous studies on the differentiation of A. cerana in China have classified A. cerana into different ecological types only by morphometric measurements. However, the results were variable due to different measurement indicators [9, 10]. It is generally accepted that there are nine ecotypes of A. cerana in China, including Hainan, the Yunnan-Guizhou Plateau, Tibet, Aba, Changbai Mountain, southern Yunnan, North China, South China, and Central China [11]. However, these results were mainly based on the classification method of A. mellifera, and the conclusions were obtained from no more than 12 traits; therefore, the unique features of A. cerana were likely missing from the analysis [12]. A subsequent study used the mitochondrial tRNALeu-COII region to classify Apis mellifera (A. mellifera) [6, 13–17]. As more investigators began to study the genome, our understanding of A. cerana continued to improve until 2015, when the reference genome was published [18]. Recently, Chen et al. conducted a comprehensive genomics study of A. cerana in most parts of China, which paralleled the simple hierarchical division and proposed that physical barriers (such as islands and mountains), rather than physical distances, lead to population differentiation. In other words, these barriers are the main obstacles leading to population exchange [19]. Other studies have reported that A. cerana, even in the same region in China, can be highly variable [17, 20–23], which has prompted us to study this species.
A. cerana uses small-scale nectar plants effectively. This species also has a strong harvesting ability, a long honey-collecting period and adaptability, strong disease resistance, and low consumption of feed, which are highly suitable for changing landscapes. The number of A. cerana has fallen sharply in recent decades [24], with climate change being one of the main threats to this species [25]. On the one hand, A. cerana has unique economic and ecological values in agriculture in China. In recent years, due to the continuous increase of China’s population, A. cerana has helped to maintain a balance between demand and production by contributing to high yields of grain. On the other hand, A. cerana is at the center of research in evolutionary biology, which attempts to understand how the population responds to climate change or environmental insults, thereby providing basic knowledge of the evolution and adaptation of A. cerana.
In this study, we collected samples from regions of different topography, environment, and climate and carried out a comprehensive morphometric determination, with the A. cerana genome [18] serving as the basic resource. The accuracy of the division of A. cerana can also be used to determine the genetic variation of the adaptive and economic shape, thereby providing an in-depth understanding of the genetic structure of A. cerana as a whole. We conducted a more comprehensive measurement of North China and analyzed the effects of topographical and environmental changes, including human activities, by combining morphological characteristics and genomic differences. We identified genetic variations, proposed a rapid classification method, explored the population structure and factors affecting differentiation, and screened the A. cerana genes that adapted to the local environment, which provides evidence of genetic diversity for future research investigating A. cerana.
Results
Population structure
The morphological measurements of the collected A. cerana samples included: (1) Bee right front wing: length of bee right forewing cubital vein a (a), length of bee right forewing cubital vein b (b), width of bee right forewing (FB), length of bee right forewing (FL), angle A4, B4, D7, E9, G18, J10, J16, K19, L13, N23, O26 of bee forewing (A4, B4, D7, E9, G18, J10, J16, K19, L13, N23, O26), bee index number of cubital vein (Ci); (2) Bee right hind wing: Bee branches midrib (NT), Number of bee hindwing hamule (Nh); (3) Bee tergum: bee length of tergum 3,4 (T3, T4), bee width of tomentum on tergum 5 (T5); (4) Bee sternum: (bee length of sternum 7 (L7), bee width of sternum 7 (T7), bee length of sternum 4 (S4), bee distance of bee wax mirror on sternum 4 (WD), bee length of bee wax mirror on sternum 4 (WL), bee width of bee wax mirror on sternum 4 (WT); (5) Bee hind leg (length of femur of bee hind leg (Fe), length of bee hind leg basitarsus (ML), width of bee hind leg basitarsus (MT), length of tibia of bee hind leg (Ti); (6) Color of the bee: bee pigment of scutellum B,K area (B, K), bee pigment of tergum 2,3,4 (P2, P3, P4), bee pigment of clypeus (Pc), bee pigment of labrum (PL), and bee pigment of scutellum Sc area (Sc) (Additional file 2: Table S1). The abbreviations corresponding to these 39 traits and other names are located in Additional file 3: Table S2. The results showed significant differences between the different regions (P < 0.01).
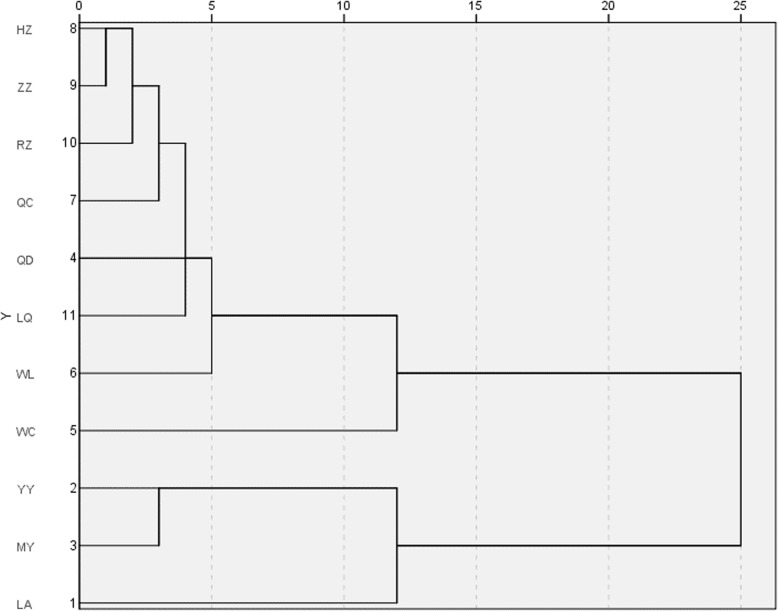
We divided 462 A. cerana from 31 populations collected from 11 regions in China (Mengyin (MY), Yiyuan (YY), Rizhao (RZ), Qingdao (QD), Linqu (LQ) and Zaozhuang (ZZ) Chongqing (WL), Sichuan (QC), Anhui (LA), Shanxi (HZ) and Hainan (WC)) and used the average of the morphological data of A. cerana from the 11 regions for the cluster analysis. Figure 1 shows the A. cerana interregional relationships. At a threshold of 12, A. cerana from the 11 regions was divided into 4 groups, in which YY and MY were grouped together, LA and WC were in separate groups, and the remaining seven regions were clustered into one large group. According to the Chinese recognized ecotypes of A. cerana, YY, MY, LA, QD, ZZ, RZ, and LQ were from the North China ecological type; WC was sampled from the Hainan ecological type; and HZ, WL, and QC were from the Central China ecological type [11] (National Commission for Animal Genetic Resources 2011). Although ZZ, RZ, QD, and LQ were from North China, they were clustered into a group associated with Central China ecological type according to our measurements.
Fig. 1.
Cluster analysis based on 39 morphological indicators of A. cerana from 11 regions
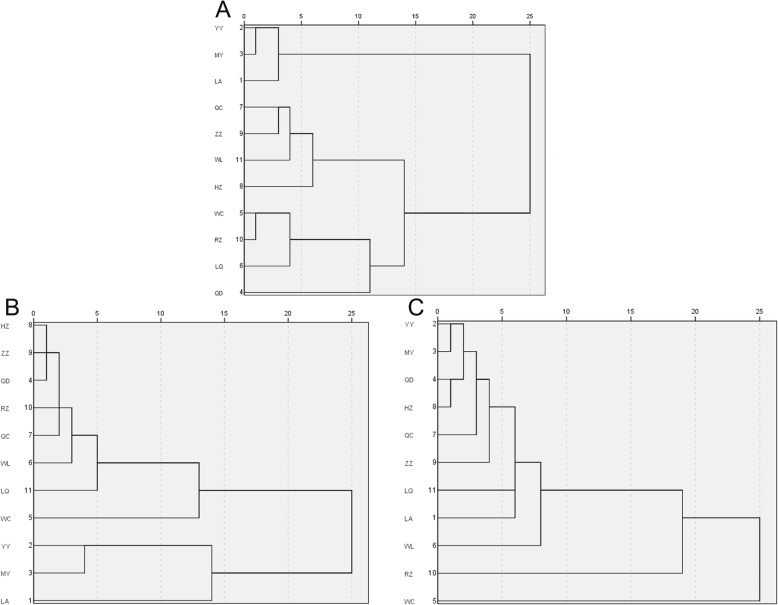
We clustered the 39 traits into three categories—body size, color type, and angle of right forewing—for cluster analysis (Fig. 2). We found differences in the clustering results, and different trait categories revealed different characteristics for the bees in each area. According to the color classification, we divided A. cerana from the 11 regions into three groups as follows: YY, MY, and LA formed the first group; ZZ, LQ, QC, and HZ fell into the second group; and WC, RZ, WL, and QD formed the third group. This finding was notably different from the results we had used to classify all the trait data. In particular, RZ, LQ, QD, and WC were relatively close in the four regions of the third group. We found that these regions were located in coastal areas, which seemed to be related to the color of the bees. According to the results of body size clustering, there was a significant difference in the size of the Hainan Bee (WC) compared to other bees, whereas clustering according to the angle of right forewing showed no difference between the 12 indicators of wing geometry and the total 39 traits.
Fig. 2.
a Cluster analysis of color using eight indicators, including B, K, P2, P3, P4, Pc, PL, and Sc. b Cluster analysis of angle of right forewings using 12 indicators, including A4, B4, D7, E9, G18, J10, J16, K19, L13, N23, Nh, and O26. c Cluster analysis of body size traits using 19 indicators, including a, b, Ci, FB, Fe, FL, L7, ML, MT, NT, S4, T3, T4, T5, T7, Ti, WD, WL, and WT
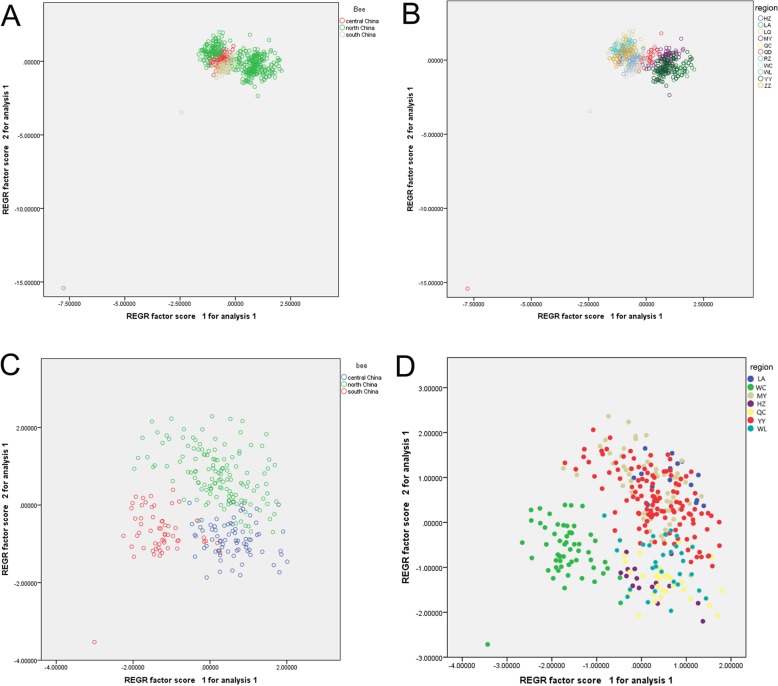
We performed principal component analysis using individual values and selected the first two factors with cumulative variability values of 57.4%. To facilitate the observations, we divided the collected bees according to the A. cerana ecological type of the sampling area. The northern ecological type was divided into two regions, the larger region and the smaller region that overlapped with the central ecological type. The central and south ecological types were nearby, but the division was clear (4A). When the bees were divided according to the sampling region, the bees in the 11 regions grouped together more obviously. The distribution pattern was consistent with the morphometric measurement of 39 traits. RZ, ZZ, LQ, and QD, which belong to North China, are close to central China (Fig. 1, 3a). We attempted to remove them and observe them again. The regions are clearly divided by the scatter plot of principal factor 1 and principal factor 2 (Fig. 3c, Fig. 3d) and divided A. cerana into three categories. In general, the population distance is consistent with the geographical distance, but there is a contradiction between the North China region and its corresponding ecological type.
Fig. 3.
Principal component analysis of the population structure in different regions of A. cerana. a Scatter plot of the main component 1 to 2 (PCA1 vs PCA2) from North China, Central China, and Hainan. b Scatter plot of the main component 1 to 2 (PCA1 vs. PCA2) from the 11 regions of LA, WC, LQ, MY, QD, RZ, HZ, QC, YY, ZZ, and WL (c) After removing the regions of RZ, ZZ, LQ, and QD, the RZ, ZZ, LQ, and QD regions were removed according to the sampling region in the scatter plot (d) of the main factor 1 and main factor 2. The main factor 1 and main factor 2 were passed according to the sampling region in the scatter plot
Population genetic structure analysis
Population structure
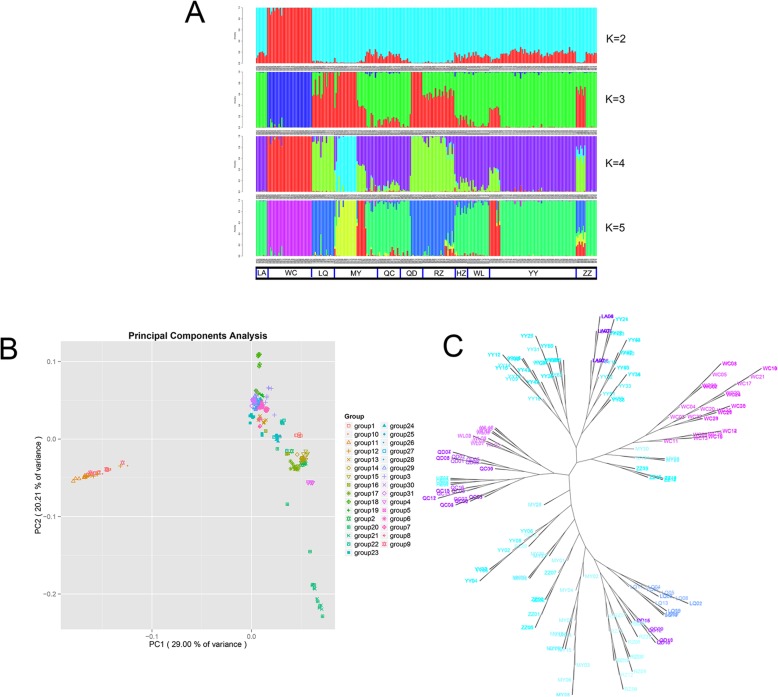
We used STRUCTURE 2.3.4 for cluster analysis [26] (Fig. 4a). When the K value was 2, A. cerana in the WC region was divided into one class, and the other regions formed a large group with clear boundaries between regions. The results of the principal component analysis (PCA) scatter plot of principal factor 1 (variation 29%) and principal factor 2 (variation 20.21%) further supported the patterns (Fig. 4b) [27]. When the K value was 3 or 4, the large clusters were split. The central region (HZ, WL, QC) formed its unique pedigree composition with LA, and most of YY (YY1-YY7) formed a cluster. LQ and RZ formed a cluster; MY, ZZ, and QD showed different degrees of pedigree, and approximately one-half of the MY area (MY1 and MY2) showed a special lineage with clear differences in the area. When the K value was 5, the differentiation of MY, ZZ, and QD was further emphasized. MY was split into three areas, ZZ was split into two areas, and RZ was split into two areas, showing a highly mixed state. These results were further supported by the phylogenetic tree of the SNP data, as analyzed with Tessal 5.0 [28] (Fig. 4c). WC did not split as a whole, and most of YY and LA were close. A small region of YY was split with MY, ZZ, MY, and QD were highly split. HZ, WL, and QC were clustered into one class. LQ, RZ, and part of QD and ZZ were clustered into a large cluster, which is consistent with the results of STRUCTURE at K values ranging from 3 to 5. ΔK was determined by the Harvester method [29], where the best K value was 3, and all regions were divided into 4 genetic components; blue region (WC), green region (LA, YY, HZ, WL, QC), red region (LQ, RZ), and a mixed area of red and green (MY, QD, ZZ).
Fig. 4.
Population structure analysis results. a Stacked diagram generated using STRUCTURE. b Scatter plot of principal components 1 and 2 (PCA1 vs. PCA2) from 31 populations collected from 11 regions. Groups 1–31 correspond to LQ1, LQ2, QD1, QD2, WL1, WL2, QC1, QC2, WC1, WC2, WC3, WC4, HZ1, RZ1, RZ2, RZ3, ZZ1, ZZ2, LA, MY1, MY2, MY3, MY4, YY1, YY2, YY3, YY4, YY5, YY6, YY7, YY8 (c) Individual bee are adjacent to phylogenetic trees
We studied the environmental characteristics of the bee sampling sites because they play important roles in the differentiation of A. cerana (Table 1). We divided the bees collected from the measurement areas into island type (WC), mountain type (LA, YY, HZ, WL, QC, MY), and plain type (QD, ZZ, LQ, RZ), which was consistent with the results of STRUCTURE. We found that climate can differentiate mountain-type bees, such as those from central and northern regions (Fig. 3c).
Table 1.
Type of climate between different regions
| Region | Altitude | Climate type |
|---|---|---|
| MY | 314.675 | Temperate monsoon climate |
| YY | 309.875 | Temperate monsoon climate |
| RZ | 148.8 | Temperate monsoon climate |
| QD | 137.9 | Temperate monsoon climate |
| ZZ | 221.75 | Temperate monsoon climate |
| LQ | 168 | Temperate monsoon climate |
| WL | 1221 | Subtropical monsoon climate |
| QC | 908 | Subtropical monsoon climate |
| LA | 337 | Temperate monsoon climate |
| HZ | 1260 | Subtropical monsoon climate |
| WC | 28.475 | Tropical monsson climate |
Genetic differentiation
Prior to genetic diversity analysis, we performed secondary filtering of the SNP data. SNP loci that could be classified in < 80% of individuals were excluded, and SNP loci with minor allele frequency (MAF) < 0.01 were excluded. Finally, only biallelic genes (−min-allele 2 - max-allele 2) and SNP loci with P-values < 0.01 according to the Hardy-Weinberg equilibrium test were filtered out. Finally, 2737 high-quality SNPs were used for the analysis (Additional file 12). The F-statistics (include Fis, Fst, and Fit; Additional file 4: Table S3) of each SNP locus in all individuals were statistically analyzed using Genepop software (ver. 4.2.2) [26]. To balance the sample size, we selected a similar number of bees from each region. The paired Fst between populations was calculated to quantify the genetic differentiation (Table 2). The Fst ranged from 0.041 (HZ, QC) to 0.257 (AH and WC), with an overall mean of 0.132, indicating moderate genetic differentiation. The states (0.05 < 0.132 < 0.15) in which WC, LA, and LQ were highly differentiated (average Fst, > 0.15) were consistent with the overall heterogeneity of the population structure and gene flow. Compared with A. mellifera (average Fst, 0.10) [30], A. cerana was more highly differentiated at the level of genetic differentiation (average Fst, 0.132). The Fst difference between the different regions of the bee was large, showing rich species diversity at the ecotype level [30, 31]. A. cerana (SX, QC, WL) in the central mountainous region had the lowest Fst distribution (average Fst, 0.103), and WC had the highest differentiation state (average Fst, 0.216). The region of North China (LA, YY, MY, QD, LQ, RZ, ZZ) showed variable differentiation states, with most being in a low differentiation state, although LA and LQ were in a highly differentiated state. We found that the mountainous regions (YY, HZ, WL, MY, QC) were higher than the other regions. Given the lower average Fst of 0.166, we calculated the Fst (Additional file 5: Table S4) and gene flow (Additional file 6: Table S5) among all groups. In the central mountainous region (HZ, WL, QC), the lowest Fst average value and the highest gene flow mean indicated that the genetic composition of the central mountainous region was similar, and the degree of differentiation was low, whereas A. cerana in the plain region showed a high degree of differentiation.
Table 2.
Paired Fst genetic distances between A. cerana populations
| Pop. | LA | WC | LQ | MY | QC | QD | RZ | HZ | WL | YY | ZZ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| WC | 0.257 | ||||||||||
| LQ | 0.204 | 0.252 | |||||||||
| MY | 0.139 | 0.194 | 0.127 | ||||||||
| QC | 0.126 | 0.185 | 0.128 | 0.061 | |||||||
| QD | 0.167 | 0.227 | 0.143 | 0.103 | 0.093 | ||||||
| RZ | 0.178 | 0.245 | 0.156 | 0.115 | 0.11 | 0.122 | |||||
| HZ | 0.139 | 0.192 | 0.131 | 0.069 | 0.041 | 0.093 | 0.118 | ||||
| WL | 0.147 | 0.198 | 0.142 | 0.076 | 0.058 | 0.107 | 0.126 | 0.062 | |||
| YY | 0.149 | 0.202 | 0.142 | 0.078 | 0.064 | 0.105 | 0.121 | 0.075 | 0.081 | ||
| ZZ | 0.15 | 0.204 | 0.141 | 0.081 | 0.079 | 0.11 | 0.12 | 0.083 | 0.092 | 0.087 | |
| Ave | 0.166 | 0.216 | 0.157 | 0.104 | 0.095 | 0.127 | 0.141 | 0.1 | 0.109 | 0.11 | 0.115 |
The polymorphism information content (Pic), effective allele number (Ne), nucleic acid diversity (Pi), heterozygosity (Ho), and expected heterozygosity (He) of each SNP locus in the population were separately calculated (Table 3). The number of effective alleles in the region was 1.46 (accounting for 73.05%). Pic measured the degree of variation of the population DNA (the overall Pic average was 0.236). Among these regions, the QC region (QC1, QC2) has the lowest Pic at 0.209, whereas the average Pic of the QD region (QD1, QD2) was the highest at 0.274. The more isolated populations had lower genetic polymorphisms (YY, HZ, WL, QC) with an average Pic of 0.214, which is consistent with the distribution of Pi. Nucleotide diversity is an indicator of diversity within or between populations. The Pi distribution of YY, HZ, WL, and QC was similar with average Pi values of 0.281, 0.275, 0.253, and 0.266, respectively, which are lower than the average (the average Pi = 0.306). Furthermore, the WC region surpassed the mean polymorphic information (Pi = 0.321 > 0.306), and there was high differentiation (mean Fst = 0.216) and low genetic variation (low polymorphism, Pic = 0.242 < 0.25). At the same time, Fis < 0 indicated that the Wenchang area was in a closed island, and it had a large surface area, which caused A. cerana in the WC area to have more biodiversity and heterozygosity than other areas. There were significant differences among heterozygosity (He), variance (Pic), and diversity (Pi) (P < 0.01) in MY, among which MY2 and MY3 had higher heterozygosity, variation, and diversity, and MY1 and MY4 had lower heterozygosity, variation, and diversity. A comparison of the STRUCTURE stacking map with a K value of 3 (Fig. 4a) revealed that the MY area was divided into three categories, whereas MY1 and MY2 belonged to one category. MY3 was in the mixed area, belonged to the second category, and MY4 belonged to the third category with a slight inbreeding phenomenon (Fis = 0.05 > 0). In addition, a large gene flow was observed between MY1 and MY2 (Nm = 18.1). MY2 had higher heterozygosity (He = 0.364), whereas MY1 remained relatively stable (He = 0.28), suggesting that the genes flowed from MY1 to MY2. Overall, we found that Ho was greater than He at the population level and that the inbreeding coefficient was generally less than zero. We also calculated all sampled groups with an average observed heterozygosity (Ho) of 0.3368, which was higher than the expected heterozygosity (He) of 0.2865. The QD region (QD1, QD2) has the highest Ho, He and Pic (0.34, 0.43, and 0.274, respectively) indicated that the population heterozygosity was high, which could be affected by the combined effects of selection, mutation and genetic drift.
Table 3.
Genetic diversity information of 31 A. cerana groups
| Group | Ne | He | Ho | PIC | Fis | Pi |
|---|---|---|---|---|---|---|
| YY1 | 1.512 | 0.309 | 0.388 | 0.251 | −0.253 | 0.331 |
| YY2 | 1.385 | 0.248 | 0.284 | 0.207 | −0.112 | 0.264 |
| YY3 | 1.397 | 0.257 | 0.288 | 0.215 | −0.133 | 0.274 |
| YY4 | 1.396 | 0.255 | 0.289 | 0.213 | −0.114 | 0.272 |
| YY5 | 1.417 | 0.27 | 0.301 | 0.226 | −0.017 | 0.294 |
| YY6 | 1.394 | 0.254 | 0.29 | 0.213 | −0.033 | 0.271 |
| YY7 | 1.415 | 0.264 | 0.31 | 0.219 | −0.055 | 0.282 |
| YY8 | 1.387 | 0.251 | 0.28 | 0.21 | −0.1 | 0.267 |
| LA | 1.523 | 0.319 | 0.379 | 0.26 | −0.198 | 0.341 |
| WL1 | 1.408 | 0.258 | 0.298 | 0.214 | −0.113 | 0.275 |
| WL2 | 1.407 | 0.258 | 0.296 | 0.214 | −0.092 | 0.275 |
| HZ | 1.356 | 0.237 | 0.246 | 0.201 | 0.004 | 0.253 |
| ZZ1 | 1.46 | 0.286 | 0.333 | 0.236 | −0.136 | 0.308 |
| ZZ2 | 1.543 | 0.324 | 0.418 | 0.261 | −0.243 | 0.346 |
| MY1 | 1.449 | 0.28 | 0.317 | 0.231 | −0.131 | 0.3 |
| MY2 | 1.611 | 0.364 | 0.432 | 0.293 | −0.196 | 0.389 |
| MY3 | 1.569 | 0.333 | 0.446 | 0.268 | −0.19 | 0.36 |
| MY4 | 1.345 | 0.232 | 0.239 | 0.197 | 0.005 | 0.247 |
| LQ1 | 1.545 | 0.322 | 0.413 | 0.26 | −0.239 | 0.345 |
| LQ2 | 1.541 | 0.325 | 0.393 | 0.263 | −0.158 | 0.347 |
| RZ1 | 1.47 | 0.292 | 0.358 | 0.24 | −0.178 | 0.311 |
| RZ2 | 1.505 | 0.308 | 0.365 | 0.251 | −0.129 | 0.328 |
| RZ3 | 1.402 | 0.261 | 0.268 | 0.218 | 0.005 | 0.278 |
| QD1 | 1.518 | 0.316 | 0.396 | 0.257 | −0.213 | 0.337 |
| QD2 | 1.613 | 0.362 | 0.459 | 0.291 | −0.224 | 0.386 |
| WC1 | 1.452 | 0.281 | 0.333 | 0.231 | −0.127 | 0.3 |
| WC2 | 1.509 | 0.308 | 0.37 | 0.251 | −0.172 | 0.329 |
| WC3 | 1.484 | 0.295 | 0.346 | 0.241 | −0.145 | 0.315 |
| WC4 | 1.519 | 0.316 | 0.367 | 0.257 | −0.139 | 0.338 |
| QC1 | 1.425 | 0.27 | 0.307 | 0.224 | −0.085 | 0.288 |
| QC2 | 1.338 | 0.228 | 0.233 | 0.194 | 0.015 | 0.243 |
| Average | 1.461 | 0.287 | 0.337 | 0.236 | N/A | 0.306 |
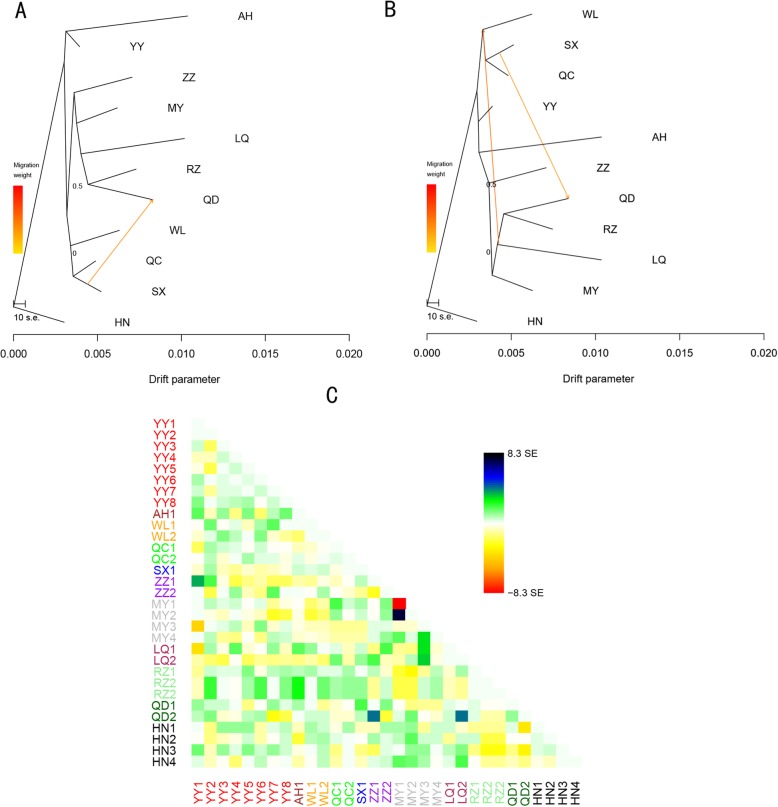
Treemix was used to construct the maximum likelihood tree with 1 and 2 migration events between regions in our study [32] using the graph with the highest probability of 100 independent runs in the species tree model (Fig. 5a, b). Modern populations shared common ancestors through shared branch points. The results of the tree model were consistent with the population state and phylogenetic tree (Fig. 4c) at a K value of 3 in the STRUCTURE stacking map (Fig. 4a, b), which largely demonstrated the known relationships between different groups. This result also explained the correlation between populations. The QD, RZ, and LQ regions of A. cerana evolved from MY and had a migratory relationship with the central region (WL, SX, QC). The first migration showed that the bloodline of QC was 18.1% for HZ (w = 18.1%). The second migration showed that the ancestors of HZ, QC, and WL were the ancestors of QD, RZ, and LQ (w = 26.5%). We evaluated the residual model using the 31 populations without migration to reveal the nonstrict hybrid events. All reported migration edges had P-values < 0.01. The three-population test was performed based on the “f3 structure” (Additional file 7: Table S6) [33] to further understand the characteristics of the mixed population. We calculated the corresponding f3 statistic for all possible combinations of the three mixed populations. The negative values indicated that there was mixing between the populations. The results indicated that only MY1 and MY2 showed a clear mixed signal. Compared to other regions, the WC region had the highest f3 statistic. We used all sampled groups information to generate a population maximum likelihood tree (Additional file 1: Figure S1), and the residual model was used to obtain a more intuitive understanding of the mixing between the populations (Fig. 5c) [32]. We found that MY and LQ, QD and ZZ, LQ had mixed events, which reveal to some extent the phenomenon of genetic structure divergence in the MY, QD and ZZ regions (Fig. 4a, K = 3–5).
Fig. 5.
a Maximum likelihood tree with two migration event. The different regions are colored according to the geographical location, and the scale shows 10 times the average standard error of the sample covariance matrix W. b Maximum likelihood tree with two migration events. c Residual map of the residual fit of the maximum likelihood tree calculated using 31 populations without migration. We divided the mean standard error between all pairs of residual covariances i and j between each pair of populations. We then plotted the scaled residual. The color is described in the palette on the right. A positive residual value indicates that the groups are more closely related to each other
Mantel test between A. cerana groups
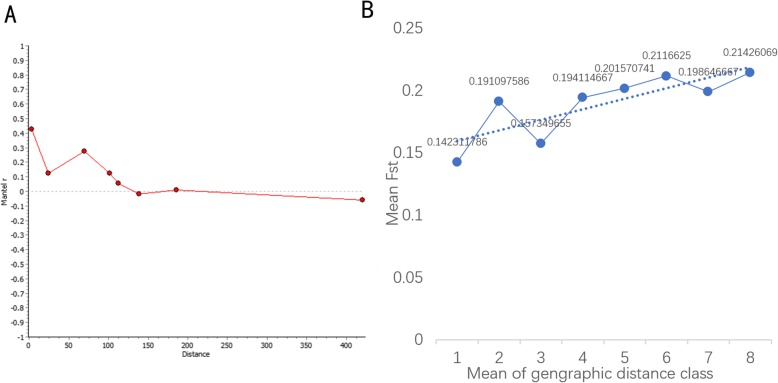
The difference between the genetic distance and geographical distance calculated by the paired Fst is one of the most common methods for assessing the spatial communication of the population structure [34–36]. We used ade4 within R [37], and the physical distance and genetic distance Fst were tested by Mantel [34, 36]. We observed a significant correlation between geographical distance and genetic distance (P = 0.008). We also used PASSaGE for repeated tests and obtained the same result (P = 0.0093). There was a significant correlation between physical distance and genetic distance, and the correlation degree was r = 0.34493. We calculated this value for A. cerana in North China. The results showed that there was a weak correlation between geographic distance and genetic distance (P = 0.051), and the correlation degree was reduced to r = 0.273. This finding may be related to the high plains in North China and low altitude.
North China has a rich terrain and belongs to the temperate monsoon climate. To further understand the changes in the correlation between genetic distance and geographical distance, we separately calculated the Fst and Manchester correlation of A. cerana in North China (Additional file 8: Table S7, Fig. 6a) [38, 39]. The most important parameter in the correlation diagram is that it should capture a continuous distribution within the geospatial space [40], and we divided A. cerana from North China into eight segments (Additional file 9: Table S8). When the observation sample was the same for each segment (0–6.23 Km), the population similarity was higher (r = 0.42; P < 0.01) in the first segment. When the distance was 156.4 Km, the population similarity decreased to 0 and then changed to a negative value, indicating that the population is not genetically similar. To make it easier to observe the Manchester correlation graph, we simultaneously calculated the Fst value at each node and created an interference graph (Fig. 6b). The results show that the spatial structure is not strong, and the mixed terrain of the North China Plain and the mountain could be the cause of fluctuation. In general, when the geographical distance increases, the genetic similarity of A. cerana in North China fluctuates and shows a linear decreasing trend (lower correlation and higher Fst value).
Fig. 6.
Manchester correlation diagram (a) and interference diagram (b), the latter by the average Fst in each distance class
Candidate genes under natural selection
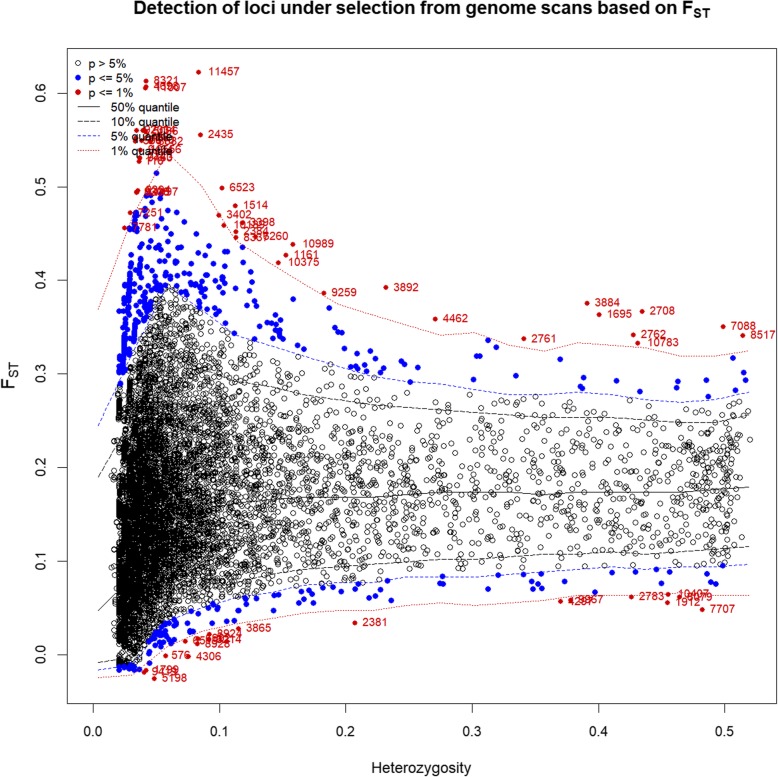
We examined divergent SNP loci in 243 A. cerana according to the Arlequin stratification method and identified 121 candidate SNPs at a significance level of 1% (Fig. 7). A list of these SNP loci, together with the A. cerana genomic information obtained from the alignment with A. mellifera and Drosophila genomes, is shown in Additional file 10: Table 9.
Fig. 7.
Identifies the different SNP loci assumed under the selection of the direction of the Fst outlier method. Use The hierarchical model Arlequin 3.5. was used. Fst: site-specific genetic differences between populations; heterozygosity: a measure of heterozygosity at each locus. Significant sites are indicated by red dots (P < 0.01)
According to Gene Ontology (GO) analysis and the related candidate genes in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, 20 significantly enriched GO terms and three significantly enriched KEGG pathways were selected (Table 4), in which the most significant was the Wnt signaling pathway. The Wnt pathway is highly conserved among different species, where it plays an important role in development, such as in the formation of Drosophila fins [41, 42]. In A. mellifera, the Hippo signaling pathway is generally considered to play an important role in cold climate adaptation [43], and a similar pathway has been identified in A. cerana, which is indicative of convergent evolution between the two species [19]. Metabolic pathways underlie how organisms respond to environmental stress, and studies have shown that parasitic infections, pesticides, and malnutrition can affect metabolic pathways in bees and reflect the adaptability of bees to changing environments [44].
Table 4.
Significantly enriched KEGG pathways
| Category | Term | count | P-Value | Benjamini | Fisher Exact |
|---|---|---|---|---|---|
| KEGG_PATHWAY | Wnt signaling pathway | 4 | 1.90E-02 | 3.30E-01 | 2.80E-03 |
| KEGG_PATHWAY | Hippo signaling pathway-fly | 3 | 5.40E-02 | 4.40E-01 | 6.90E-03 |
| KEGG_PATHWAY | Metabolic pathways | 5 | 8.90E-02 | 5.20E-01 | 8.00E-02 |
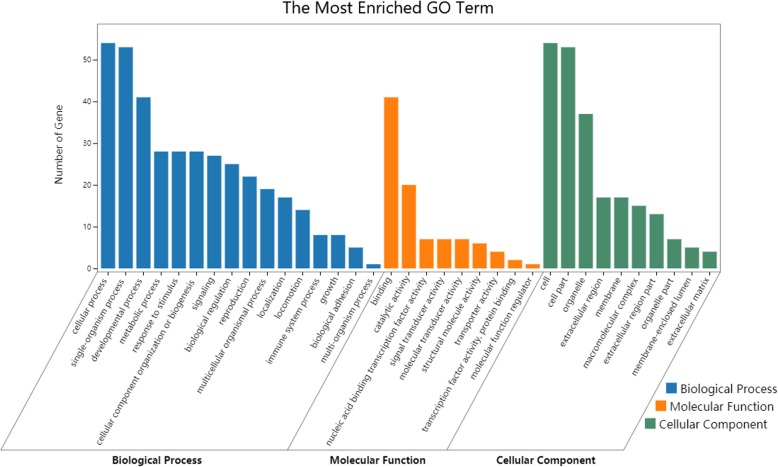
For the enriched GO terms, adaptive terms, such as “response to stimulus”, “signal transduction”, “locomotion”, and “biological regulation”, were significant (Additional file 11: Table S10), indicating the importance of the processing of signals and responses when bees adapt to different environments. We also identified key enriched GO terms (Fig. 8), “developmental process”, “metabolic process”, “biological regulation”, “immune system process”, “catalytic activity”, and other genes related to growth and development that are involved in natural selection.
Fig. 8.
GO terms enriched by abnormal genes. The asterisks represent significant terms
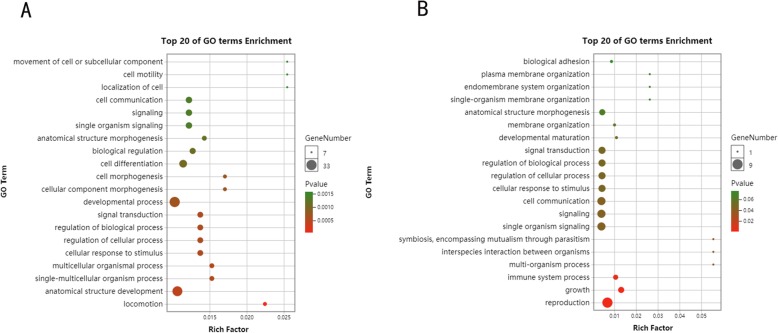
We further analyzed the genes under balanced selection and directed selection. Among the genes corresponding to a total of 121 abnormal SNP loci, there were 30 genes under balanced selection and 91 genes under directed selection. We separately performed enrichment analysis of the genes under balanced selection and directed selection. Under balanced selection, the GO terms that were enriched in response to the stimuli, such as “locomotion”, “signal transduction”, and “biological regulation”, we found that directional selection under natural selection allowed the adaptive evolution of organisms (Fig. 9). However, the enriched GO terms “symbiosis, encompassing mutualism through parasitism”, “signal transduction”, “interspecies interaction between organisms”, and “information processing” indicated that balanced selection maintained the diversity of alleles between bees. Therefore, collaboration, communication, and other activities are important for the survival of bee populations. For instance, collaboration is critical for parasite identification, and foraging behaviors, highly diverse symbiosis, and genes involved in communication are conducive to interactions among bees and their ability to cope with changing environments.
Fig. 9.
The left image shows the top 20 GO terms under directional selection, and the right image shows the top 20 GO terms under balanced selection. Rich Factor: Ratio of enriched gene number to total gene number in each GO term. Dot size were decided with enrichment gene number. Dot color were decided with enrichment p-value
Discussion
The evolution of species and the generation of diversity are mostly due to environmental changes. To understand the relationships between genetic differentiation and the environment, this study collected A. cerana from 11 regions in China and analyzed the morphological measurements. To obtain as much information as possible about local A. cerana, we include as many collection points as possible in one collection area. The samples were mainly collected from frame beehives, semiartificial beehives, including tree barrel hives and wall hole hives, and natural beehives. The domestication process of A. cerana is rather slow, and domestic bees and wild bees often undergo mutual transformation [11]. To ensure the stability of A. cerana society, all frame of artificial and semiartificial beehives have to mimic their natural beehives. Therefore, in this study, we believe that different beehives have negligible effect on A. cerana behavior. We used 39 morphological indicators to classify A. cerana and classified the different morphological indicators, such as color, angle of right forewing, and body length, to obtain more comprehensive classification results. Overall, the classification of the angle of right forewing was consistent with the classification that included all traits. Francoy et al. used wing morphology to quickly identify similarities among Africanized bees [45]. We believe that the wing morphology can also be used to rapidly identify species.
The phenotypes of ZZ, RZ, QD, and LQ in North China were similar to those in the central region, which seems counterintuitive. We performed a genome-level analysis, and the STRUCTURE results showed that there was considerable genetic diversity in ZZ, RZ, QD, and MY. At K = 5, MY showed three different pedigree types. The two migrations of the Treemix population phylogenetic tree revealed signs of migration for QD, LQ, RZ and the central region, while the residual plot showed signs of mixing between ZZ and QD. Previous studies on Italian honeybees (A. mellifera) showed that human-mediated gene flow led to the management of honey bee colonies High genetic diversity [46, 47], and the MY region may be the result of human action.
Genetic variation is critical for the survival of bees in changing climates. For bees, high genetic diversity at the population level can increase the adaptability of colonies, as bee colonies with higher diversity exhibit a more stable ethnic balance [48, 49], are more productive [50, 51] and are more resistant to disease [52]. We calculated the Fst value of population differentiation, which showed a large difference between WC and the inland region (average Fst = 0.216). The Fst value was previously calculated for A. cerana at the subspecies level between the more isolated populations (ecological type), for which the average Fst differentiation was 0.162 in the range of 0.099 to 0.228 [19]. We calculated the Fst between MY, YY, LA and WL, HZ, and QC between 0.6 and 0.9, but the morphological data show that it belongs to a different ecological type. Therefore, we believe that the Fst range of A. cerana should be between 0.06 and 0.228, while the Fst range for the A. mellifera subspecies of the same lineage was 0.05 to 0.15 [30]. Clearly, relative to A. mellifera, A. cerana has a richer genetic diversity.
Although ZZ, RZ, QD, and LQ may have undergone mixed population events, they are currently in a state of equilibrium. Furthermore, different mixed regions have gradually formed relatively independent ecological types. Previous studies have suggested that bees migrate rapidly in areas without strong physical barriers, which can promote gene flow and reduce genetic differentiation. The rapid spread of bees in Australia and the Americas can better illustrate the potential for the rapid migration of bees [53–55]. The results of the Mantel test showed that geographical distance had little effect on population differentiation, which is consistent with the results of studies using microsatellite markers [56, 57]; this finding allows us to focus our interest on other environmental variables. Previous studies have suggested that islands and mountains can form physical barriers that, in turn, cause group isolation. Therefore, the rapid migration of honeybees without a strong physical barrier can promote gene flow and reduce genetic differentiation. In addition, population growth can promote gene flow between populations [19], but our research shows that gene flow in the mountains is more pronounced than in plain areas (Additional file 6: Table S5). In China, various human activities, such as the loss of agricultural land and urbanization, led to the loss of habitat of A. cerana, especially in the vast plains of North China, which are particularly conducive to urban construction. However, complex terrains are more resistant to human activities, which could be the reason for the higher degree of differentiation of A. cerana in ZZ, RZ, QD, and LQ. We believe that the isolation of A. cerana by human urban agglomerations is more stringent than the physical isolation caused by mountains.
Researchers believe that the survival of a species through climate change, habitat loss, and ecosystem changes is due to their physiological tolerance limits and resilience, ecological characteristics (e.g., behavior, heat tolerance), and genetic diversity [58]. A. cerana shows the potential of using all three strategies. Research has described a method to identify the performance of bee populations based on outliers [59, 60]. In this study, we screened 121 SNP loci, and functional enrichment analysis highlighted several processes such as “neurology”, “biological regulation”, “behavior and growth”, and “interspecies interactions between organisms”, suggesting that genes associated with these loci may experience selective stress as the population spreads through various unwelcome habitats. Sensory transduction refers to the conversion of input stimuli into signals received by the brain. Specific changes in these genes may require adaptation to different food sources or other resources in the new environment. Bees exhibit a range of different behaviors in a social environment, and hundreds of genes are involved in the brain function and physiological behavior of bees [61]. The social behavior of bees helps the population maintain the homeostasis of the nest and counteract changes in the external environment [62, 63]. Furthermore, chemical signals coordinate the behavior and physiology of colony members, and changes in protein coding sequences may be related to the evolution of chemical communication systems found in bees [64]. Genetic diversity is an adaptive evolution of biological individuals to changing environments. The ability to adapt to changing environmental variables strongly depends on sufficient genetic variation, the indirect impact of population size on evolution, and the balance between environmental rates [65]. We further analyzed 91 loci under balanced selection and directed selection. The results of the enrichment analysis showed that equilibrium selection plays an important role in maintaining the diversity of alleles and increasing the stability of the population. Directed selection leads to the unilateral adaptive evolution of the population. Taken together, the results of our study indicated rich genetic diversity in A. cerana.
In summary, as a wide-range species, A. cerana shows good potential for climate change, but the effects of stress, such as pesticides, pollutants, pathogens, parasites, and limited flower resources caused by human activities, are posing a larger threat to A. cerana in China [66, 67]. In this study, we provided new phenotypic and genomic insights for further climate adaptation studies that can enhance our understanding of bee health and breeding management.
Conclusions
Our study utilized the wide-range A. cerana species, as well as morphological characteristics and genomic methods, to provide insights into the differentiation and adaptation of the species in China. We proposed a method for the rapid identification of the morphology of the wing and found some historical migration between the North and Central regions. A. cerana in China exhibits high genetic diversity, and physical barriers, rather than distance, are the driving factors for the differentiation of this highly migratory species. Human activities, such as urbanization, have a great impact on the differentiation of A. cerana. We screened for abnormal SNP loci and obtained related genes, performed enrichment analysis and gene ontology categories to identify candidate genes. The results of this study may help to elucidate the evolution of A. cerana in different environments and promote our understanding of how bee populations will respond to increasing climate change and how to protect bees from current and future challenges.
Methods
Sample collection
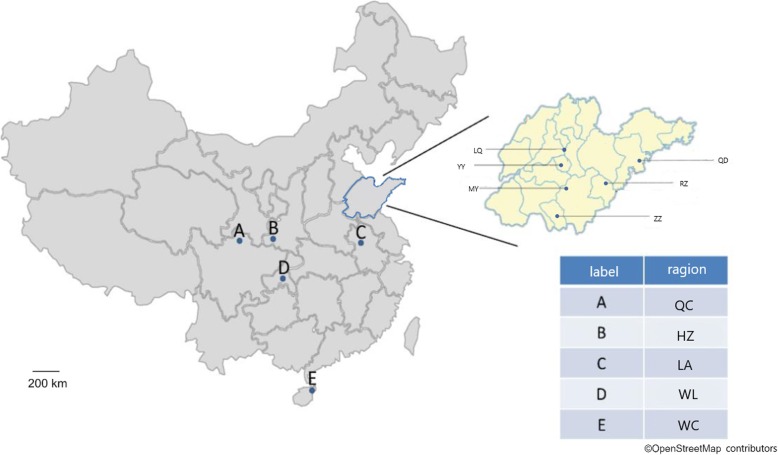
Samples were collected from six regions, Mengyin (MY), Yiyuan (YY), Rizhao (RZ), Qingdao (QD), Linqu (LQ) and Zaozhuang (ZZ), in Shandong (SD) Province and A. cerana germplasm conservation areas or traditional breeding sites in 11 provinces and municipalities, such as Chongqing (WL), Sichuan (QC), Anhui (LA), Shanxi (HZ) and Hainan (WC) (Fig. 10). To obtain as much information as possible about local A. cerana, we include as many collection points as possible in one collection area. The samples were mainly collected from frame beehives, semiartificial beehives, including tree barrel hives and wall hole hives, and natural beehives (Table 5). All frame beehives and semiartificial beehives are designed to mimic natural beehives. At least one sample group of ~ 30–50 adult worker bees at each sampling point, after being soaked in absolute ethanol and placed in a sealed grinding bottle, were brought back and stored in a − 20 °C freezer.
Fig. 10.
The position of the sample in China, where the blue area represents the dense sampling site of the representative area of North China f10:2 (Shandong). Sampling positions in Shandong Province were specially highlighted as yellow magnified area map. Base layer map date©OpenStreetMap contributors (page: https://www.openstreetmap.org/copyright)
Table 5.
Geographical information of the A. cerana sampling location
| Province | Region | Longitude | Latitude | Nest/Hive type | Number |
|---|---|---|---|---|---|
| Shandong | MY | 117.4954~118.0072 | 35.3608~35.5471 | natural beehive | 60 |
| YY | 118.2056~118.3009 | 36.0034~36.0501 | frame beehive | 120 | |
| RZ | 119.2493~119.3662 | 35.6367~35.6802 | frame beehive | 45 | |
| QD | 119.7948~120.5137 | 35.8845~36.2050 | frame beehive | 30 | |
| ZZ | 117.6521~117.6927 | 35.0355~35.0431 | natural beehives | 30 | |
| LQ | 118.3106 | 36.1846 | frame beehive | 30 | |
| Chongqing | WL | 107.7061 | 29.3941 | semi-artificial hive | 30 |
| Sichuan | QC | 105.1457 | 32.3455 | semi-artificial hive | 30 |
| Anhui | LA | 117.0127 | 31.408 | frame beehive | 15 |
| Shanxi | HZ | 116.4401 | 33.156 | semi-artificial hive | 15 |
| Hainan | WC | 110.7254~110.7805 | 19.5541~19.6845 | semi-artificial hive | 60 |
Dissection and collection of morphometric data
This test was carried out in accordance with the indicator project of the honeybee morphology researcher [68, 69] and the color standards of A. cerana [70]. First, a picture of the entire body, which included the color pattern of the bee specimen, was taken, and tweezers were used to remove the front and rear wings from the chest. The sternal remnants were removed by tearing the connective tissue between the wings. The cut sternum was cleaned with a soft-bristle brush, and the residual tissue was removed and fixed to the assay plate with scotch tape. Using a specific measuring microscope and its matching measurement software ImageView 3.7, morphological measurements were performed after adjusting the scale. The morphological markers were as follows: front and rear wing length; angle of right forewing (A4, B4, D7, E9, K19, G18, J10, L13, J16, N23, O26); elbow pulse length; and color aspect measurement of the lip base, upper lip, small scutellum SC area, K area, B area, second back board, third back board, and fourth back board (a total of 39 index). To avoid bias and improve accuracy, each sample was evaluated for pigmentation by three different observers, and the highest number of each score was recorded and used for subsequent analysis.
Sequencing, read mapping, and quality control
A. cerana was randomly selected from each of the sampled sites for sequencing, and a tag sequencing library of A. cerana samples was constructed by the 2b-RAD technique. All samples were constructed using standard 5′-NNN-3′ linkers. To obtain high quality reads, both the library and quality-controlled libraries were subjected to paired-end sequencing data filtering on the HiSeq X Ten platform, and SOAP2 was used to compare the sequencing data to the genome [71, 72] using maximum likelihood (maximum likelihood, ML) perform site typing and SNP detection, as well as VCF tools, to carry out strict filtering to obtain the most informative SNPs [60]. The detected SNPs were filtered out as follows: (1) except for sites in which less than 80% of the individuals could be typed, (2) sites with an MAF below 0.05, (3) one or four allelic sites were excluded, (4) exclude more than one SNP locus in the tag, (5) eliminate loci with a genotype deletion ratio greater than 0.2, (7) exclude sites with a sample deletion ratio of 0.2 or higher, and (8) filter out the Hardy-Weinberg equilibrium test for p-values less than 0.05. A total of 11,504 SNP loci were obtained for all samples (Table 6).
Table 6.
Number of A.cerana SNPs detected in each functional area
| SNP | Number |
|---|---|
| 3_prime_UTR_variant | 154 |
| 5_prime_UTR_premature_start_codon_gain_variant | 29 |
| 5_prime_UTR_variant | 167 |
| downstream_gene_variant | 606 |
| intergenic_region | 3048 |
| intragenic_variant | 24 |
| intron_variant | 5181 |
| missense_variant | 223 |
| non_coding_exon_variant | 5 |
| splice_acceptor_variant&intron_variant | 1 |
| splice_donor_variant&intron_variant | 1 |
| splice_region_variant | 4 |
| splice_region_variant&intron_variant | 23 |
| splice_region_variant&synonymous_variant | 10 |
| start_lost | 1 |
| stop_gained | 2 |
| synonymous_variant | 872 |
| upstream_gene_variant | 1153 |
| total | 11,504 |
Morphometric analysis
We used SPSS 22 to analyze the morphological data of 462 A. cerana. First, we calculated the mean and coefficient of variation between the regions. ANOVA was used to determine which variables best distinguish the differences between the different regions, and principal component analysis (PCA) was performed. Then, we used the first two main factors of PCA (cumulative variability reached 57.4) and constructed different scatter plots. Finally, we use the hierarchical cluster function in SPSS 22, select Ward’s method to perform hierarchical clustering, and combine the most similar samples according to the degree of closeness between the different indicators. The sequential aggregation method classified all samples to obtain an intuitive understanding of the population structure.
Population genetics analyses
Prior to population genetic analysis, VCF tools were used to rigorously obtain the most informative SNPs [60]. The inbreeding coefficient (F), expected heterozygosity (He), and observed heterozygosity (Ho) were calculated using PLINK2 [73]. The ratio of the minor allele frequency (MAF) to the polymorphic SNP (Pic) was calculated using a custom Perl script. The Fis, Fst, and Fit values of the SNP locus in 243 samples were statistically analyzed using Genepop software (version 4.2.2) [26]. The Mantel test of the Fst matrix and distance matrix was performed using the ade4 package in R [37]. We used TreeMix [32] to infer the history of the population division and mixture, allowing for three mixed events [33]. The method constructed a differentiation tree of the population and then identified the potential events of the population mixture from the residual covariance matrix.
Bayesian population structure and principal component analysis
A model-based Bayesian clustering method was used to characterize the different genetic clustering patterns between the different regions with STRUCTURE 2.3.4 [74]. The genome of each bee was located in a predetermined set K, and the variable ancestor ratio was determined by the allele frequency of the population. This approach allowed the use of mixed bees to characterize ancestral populations [74]. We used a hybrid model and applied a putative number of clusters K from one to ten for 11,506 SNPs to run on STRUCTURE. The analysis was performed without prior knowledge of the demographic identity by simulating 50,000 preburning steps and 100,000 iterations of the MCMC algorithm per run. Ten independent runs were performed for each K to estimate the most reliable different genetic clusters using the probability of posterior probability (LnP(N/K)) [75] and the temporary amount DK of each K partition. The posterior probability variation with respect to K between the different runs was designated as the method for determining the true K value [76]. The most likely value for K was based on the network software STRUCTURE HARVESTER [29] and Evanno’s ΔK method [76], which was based on the average log likelihood, Ln P(D). Using TASSEL v5.0 [28] and SPSS 22 to perform PCA of the genetic and morphological traits of different individuals, respectively, enabled us to visualize the correlations between individual bees in individuals/regions on a multidimensional scale.
Correlations between environmental variables and genetic diversity
The Mantel test was performed using the Fst matrix and distance matrix with the ade4 software package in R [37]. To describe the possible changes in the correlation between the genetic distance and geographical distance, we used PASSaGE 2.0 for Manchester correlation analysis [77], according to the distance. The rank divided the distance matrix into submatrices. The populations within the boundary geographical distance described by each submatrix corresponded to populations with different genetic distances. The average Fst of the set “distance level” was calculated separately to generate a polyline “interference graph” that was combined with the Manchester correlation map for a more intuitive view.
Detecting SNP loci under selection based on Fst outlier tests
A coalescence-based simulation was used to detect deviating SNP sites. With these methods, we expected to detect low-level differentiation sites under balanced selection (neutral loci) and high-level differentiation sites under directed selection (differentiation loci). The layering method we used was a modification of the FDIST method that was performed with Arlequin ver 3.5.2.2 [78, 79].. We used a hierarchical island model with 50,000 simulations to calculate the relationship between the Fst and heterozygosity. A locus with an Fst value higher than the 0.99 limit of the neutral distribution was considered to be a putative outlier under divergent selection [59]. The remaining loci with nonsignificant Fst values were considered to be neutral SNPs. All procedures reduced bias and maintained highly differentiated loci between ecotypes individuals. We chose Fst values that were higher than the expected neutral distribution as the directional selection sites and Fst values that were lower than the expected neutral distribution as the balanced selection sites [60].
Gene ontology analysis
SNP loci were screened using high-quality SNP loci [59, 80], and the reference sequence was obtained from NCBI (https://www.ncbi.nlm.nih.gov/assembly/GCF_001442555.1). A 50-base sequence upstream and downstream of the extraction site was sequenced and aligned with the A. mellifera sequence to obtain the most recent genes. Enrichment analysis was based on their orthologs in Drosophila melanogaster. The genes were assigned to GO and KEGG pathways. The functional enrichment of gene IDs was performed using GeneMania and g: Profiler [81, 82] with the online analysis site DAVID for KEGG pathway exploration [83] and Benjamini to correct the P-values to identify clusters of significantly enriched terms.
Supplementary information
Additional file 1: Figure S1. Maximum likelihood tree with one migration event.
Additional file 2: Table S1. The 39 morphological measurements of A. cerana.
Additional file 3: Table S2. Abbreviation table.
Additional file 4: Table S3. The Fis, Fst, and Fit of each SNP locus.
Additional file 5: Table S4. The Fst table between all groups.
Additional file 6: Table S5. The gene flow table between all group.
Additional file 7: Table S6. Three population tests for admixture.
Additional file 8: Table S7. The Fst value table and distance table in North China.
Additional file 9: Table S8. Mantel test table.
Additional file 10: Table S9. Details of 121 candidate SNP loci.
Additional file 11: Table S10. GO term enrichment result.
Additional file 12. SNP data after filtering.
Acknowledgments
The author would like to thank all the members of the Animal Protection Laboratory, School of Animal Science and Technology, Shandong Agricultural University, who collected the whole sample of our experiment. We are grateful to the livestock department and friends of apiculture in the sampling area for their help in sample collection on site/for providing samples. We would also thank Qingdao OE Biotechnology Company Limited for their help.
Abbreviations
- a
Length of bee right forewing cubital vein a
- A. cerana
Apis cerana
- A. mellifera
Apis mellifera
- A4
Angle A4 of bee forwing
- B
Bee pigment of scutellum B area
- b
Length of bee right forewing cubita lvein b
- B4
Angle B4 of bee forwing
- Ci
Bee index number of cubital veins
- D7
Angle D7 of bee forwing
- E9
Angle E9 of bee forwing
- FB
Width of bee right forewing
- Fe
Length of femur of bee hindleg
- FL
Length of bee right forewing
- Fst
F-statistics, Genetic differences among population.
- G18
Angle G18 of bee forwing
- GO
Gene ontology
- He
Expected heterozygosity
- Ho
Heterozygosity
- HZ
Shanxi
- J10
Angle J10 of bee forwing
- J16
Angle J16 of bee forwing
- K
Bee pigment of scutellum Karea
- K19
Angle K19 of bee forwing
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- L13
Angle L13 of bee forwing
- L7
Bee length of sternum 7
- LA
Anhui
- LQ
Linqu
- MAF
Minor allele frequency.
- ML
Length of bee hind leg metabasitarsus
- MT
Width of bee hind leg metabasitarsus
- MY
Mingyin
- N23
Angle N23 of bee forwing
- Ne
Effective allele number
- Nh
Number of bee hindwing hamuli
- NT
Bee branches midrib
- O26
Angle O26 of bee forwing
- P2
Bee pigment of tergum 2
- P3
Bee pigment of tergum 3
- P4
Bee pigment of tergum 4
- Pc
Bee pigment of clypeus
- PCA
Principal component analysis
- Pi
Nucleic acid diversity
- PIC
Polymorphism information content
- PL
Bee pigment of labrum
- QC
Sichuan
- QD
Qingdao
- RZ
Rizhao
- S4
Bee length ofsternum 4
- Sc
Bee pigment of scutellum Sc area
- SNP
Single nucleotide polymorphism.
- T3
Bee length of tergum 3
- T4
Bee length of tergum 4
- T5
Bee width of tomentum 1 ontergum 5
- T7
Bee width of sternum 7
- Ti
Length of tibia of bee hind leg
- WC
Hainan
- WD
Bee distance of bee wax mirror on sternum 4
- WL
Bee length of bee wax mirror on sternum 4
- WL
Chongqing
- WT
Bee width of bee wax mirror on sternum 4
- YY
Yiyuan
- ZZ
Zaozhuang
Authors’ contributions
WG, LD and LY designed and coordinated the research. LY and FY collected the samples. LY and FY conducted library construction and sequencing. WG, LY and CT analyzed the data. WG, LY and CT drafted the paper. All authors read and approved the final manuscript.
Funding
This work was financially supported by Funds of Shandong Province Modern Agricultural Industry Technology System Bee Industry Innovation Team Breeding and Resource Protection Post Expert Project (No. SDAIT-24-02), Shandong Province Agricultural Variety Project (No. 2016LZGC039), Shandong “Double Tops” Program (No. SYL2017YSTD12). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The raw genome 2b-RAD tag-sequencing datasets generated during the current study are publicly available under NCBI Bioproject: PRJNA579872. All other data analysed in the present study are included in this published article (and its Supplementary Information files).
Ethics approval and consent to participate
All animal experiments were performed in accordance with the “Guidelines for Experimental Animals” of the Ministry of Science and Technology (Beijing, China), and approved by the Institutional Animal Care and Use Ethics Committee of Shandong Agricultural University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Li Yancan and Chao Tianle contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12864-019-6246-4.
References
- 1.Skelly DK, Joseph LN, Possingham HP, Freidenburg LK, Farrugia TJ, Kinnison MT, et al. Evolutionary Responses to Climate Change. Conserv Biol. 2007;21:1353–1355. doi: 10.1111/j.1523-1739.2007.00764.x. [DOI] [PubMed] [Google Scholar]
- 2.Balanyà J, Huey RB, Gilchrist GW, Serra L. The chromosomal polymorphism of Drosophila subobscura: a microevolutionary weapon to monitor global change. Heredity. 2009;103:364. doi: 10.1038/hdy.2009.86. [DOI] [PubMed] [Google Scholar]
- 3.Wallberg A, Han F, Wellhagen G, Dahle B, Kawata M, Haddad N, et al. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat Genet. 2014;46:1081. doi: 10.1038/ng.3077. [DOI] [PubMed] [Google Scholar]
- 4.Gallai N, Salles J-M, Settele J, Vaissière BE. Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol Econ. 2009;68:810–821. doi: 10.1016/j.ecolecon.2008.06.014. [DOI] [Google Scholar]
- 5.Arias MC, Sheppard WS. Phylogenetic relationships of honey bees (Hymenoptera:Apinae:Apini) inferred from nuclear and mitochondrial DNA sequence data. Mol Phylogenet Evol. 2005;37:25–35. doi: 10.1016/j.ympev.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 6.Smith DR, Villafuerte L, Otis G, Palmer MR. Biogeography of Apis cerana F. and A. nigrocincta Smith: insights from mtDNA studies. Apidologie. 2000;31:265–279. doi: 10.1051/apido:2000121. [DOI] [Google Scholar]
- 7.Radloff SE, Hepburn C, Hepburn HR, Fuchs S, Hadisoesilo S, Tan K, et al. Population structure and classification of Apis cerana. Apidologie. 2010;41:589–601. doi: 10.1051/apido/2010008. [DOI] [Google Scholar]
- 8.Hou C, Li B, Luo Y, Deng S, Diao Q. First detection of Apis mellifera filamentous virus in Apis cerana cerana in China. J Invertebr Pathol. 2016;138:112–115. doi: 10.1016/j.jip.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Gong Y. Taxonomy and evolution of honeybees (in Chinese) Fuzhou: Fujian Science and Technology Press; 2000. [Google Scholar]
- 10.Yang G. Chinese honeybee (in Chinese) Beijing: China Agricultural Science and Technology Press; 2001. [Google Scholar]
- 11.The National Animal Genetic Resources Committee . Animal genetic resources in China bees. Beijing: China Agriculture Press; 2011. [Google Scholar]
- 12.Meixner MD, Pinto MA, Bouga M, Kryger P, Ivanova E, Fuchs S. Standard methods for characterising subspecies and ecotypes of Apis mellifera. J Apic Res. 2013;52:1–28. doi: 10.3896/IBRA.1.52.4.05. [DOI] [Google Scholar]
- 13.de La Rúa P, Simon UE, Tilde AC, Moritz RFA, Fuchs S. MtDNA variation in Apis cerana populations from the Philippines. Heredity. 2000;84:124–130. doi: 10.1046/j.1365-2540.2000.00646.x. [DOI] [PubMed] [Google Scholar]
- 14.Tan K, Meixner MD, Fuchs S, Zhang X, He S, Kandemir I, et al. Geographic distribution of the eastern honeybee, Apis cerana (Hymenoptera: Apidae), across ecological zones in China: morphological and molecular analyses. Syst Biodivers. 2006;4:473–482. doi: 10.1017/S1477200006002015. [DOI] [Google Scholar]
- 15.Tan K, Warrit N, Smith DR. Mitochondrial DNA diversity of Chinese Apis cerana. Apidologie. 2007;38:238–246. doi: 10.1051/apido:2007008. [DOI] [Google Scholar]
- 16.Tan K, Qu Y, Wang Z, Liu Z, Engel MS. Haplotype diversity and genetic similarity among populations of the eastern honey bee from Himalaya-Southwest China and Nepal (Hymenoptera: Apidae) Apidologie. 2016;47:197–205. doi: 10.1007/s13592-015-0390-x. [DOI] [Google Scholar]
- 17.Zhao W, Tan K, Zhou D, Wang M, Cheng C, Yu Z, et al. Phylogeographic analysis of Apis cerana populations on Hainan Island and southern mainland China, based on mitochondrial DNA sequences. Apidologie. 2014;45:21–33. doi: 10.1007/s13592-013-0223-8. [DOI] [Google Scholar]
- 18.Park D, Jung JW, Choi B-S, Jayakodi M, Lee J, Lim J, et al. Uncovering the novel characteristics of Asian honey bee, Apis cerana, by whole genome sequencing. BMC Genomics. 2015;16:1. doi: 10.1186/1471-2164-16-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C, Wang H, Liu Z, Chen X, Tang J, Meng F, et al. Population genomics provide insights into the evolution and adaptation of the eastern honey bee (Apis cerana) Mol Biol Evol. 2018;35:2260–2271. doi: 10.1093/molbev/msy130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, Zhu X, Zhou S, Wu X, Zhou B. Genetic differentiation between Apis cerana cerana populations from Damen Island and adjacent mainland in China. Acta Ecol Sin. 2013;33:122–126. doi: 10.1016/j.chnaes.2013.02.001. [DOI] [Google Scholar]
- 21.Yin L, Ji T. Genetic diversity of the honeybee Apis cerana in Yunnan, China, based on mitochondrial DNA. Genet Mol Res. 2013;12:2002–2009. doi: 10.4238/2013.June.20.1. [DOI] [PubMed] [Google Scholar]
- 22.Liu F, Shi T, Huang S, Yu L, Bi S. Genetic structure of mount Huang honey bee (Apis cerana) populations: evidence from microsatellite polymorphism. Hereditas. 2016;153:8. doi: 10.1186/s41065-016-0010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao L, Gu P, Lin Y. Genetic diversity of Apis cerana cerana based on mitochondrial DNA in Lishui, Zhejiang, China. J Zhejiang Univ Agric Life Sci. 2017;43:425–430. [Google Scholar]
- 24.Theisen-Jones H, Bienefeld K. The Asian honey bee (Apis cerana) is significantly in decline. Bee World. 2016;93:90–97. doi: 10.1080/0005772X.2017.1284973. [DOI] [Google Scholar]
- 25.Thomann M, Imbert E, Devaux C, Cheptou P-O. Flowering plants under global pollinator decline. Trends Plant Sci. 2013;18:353–359. doi: 10.1016/j.tplants.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 26.Rousset F. genepop’007: a complete re-implementation of the genepop software for windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- 27.Patterson N, Price AL, Reich D. Population structure and Eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 29.Earl DA. vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- 30.Harpur BA, Kent CF, Molodtsova D, Lebon JMD, Alqarni AS, Owayss AA, et al. Population genomics of the honey bee reveals strong signatures of positive selection on worker traits. Proc Natl Acad Sci. 2014;111:2614 LP–2612619. doi: 10.1073/pnas.1315506111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cridland JM, Tsutsui ND, Ramírez SR. The complex demographic history and evolutionary origin of the Western honey bee, Apis Mellifera. Genome Biol Evol. 2017;9:457–472. doi: 10.1093/gbe/evx009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diniz-Filho JAF, Soares TN, Lima JS, Dobrovolski R, Landeiro VL, de Campos Telles MP, et al. Mantel test in population genetics. Genet Mol Biol. 2013;36:475–485. doi: 10.1590/S1415-47572013000400002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mantel N. The Detection of Disease Clustering and a Generalized Regression Approach. Cancer Res. 1967;27(2 Part 1):209 LP–209220. [PubMed] [Google Scholar]
- 36.Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting F (ST) Nat Rev Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dray S, Dufour A-B. The ade4 package: implementing the duality diagram for ecologists. J Stat Softw. 2015;22:4.
- 38.Oden NL, Sokal RR. Directional autocorrelation: an extension of spatial Correlograms to two dimensions. Syst Zool. 2006;35:608. doi: 10.2307/2413120. [DOI] [Google Scholar]
- 39.Legendre P, Legendre L. Numerical Ecology. 3. Amsterdam: Elsevier; 2012. p. 990. [Google Scholar]
- 40.SOKAL ROBERT R., ODEN NEAL L. Spatial autocorrelation in biology: 1. Methodology. Biological Journal of the Linnean Society. 1978;10(2):199–228. doi: 10.1111/j.1095-8312.1978.tb00013.x. [DOI] [Google Scholar]
- 41.Mlodzik M. Planar polarity in the Drosophila eye: a multifaceted view of signaling specificity and cross-talk. EMBO J. 1999;18:6873–6879. doi: 10.1093/emboj/18.24.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adler PN, Lee H. Frizzled signaling and cell–cell interactions in planar polarity. Curr Opin Cell Biol. 2001;13:635–640. doi: 10.1016/S0955-0674(00)00263-5. [DOI] [PubMed] [Google Scholar]
- 43.Chen C, Liu Z, Pan Q, Chen X, Wang H, Guo H, et al. Genomic analyses reveal demographic history and temperate adaptation of the newly discovered honey bee subspecies Apis mellifera sinisxinyuan n. ssp. Mol Biol Evol. 2016;33:1337–1348. doi: 10.1093/molbev/msw017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Broadrup RL, Mayack C, Schick SJ, Eppley EJ, White HK, Macherone A. Honey bee (Apis mellifera) exposomes and dysregulated metabolic pathways associated with Nosema ceranae infection. PLoS One. 2019;14:e0213249. doi: 10.1371/journal.pone.0213249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Francoy TM, Wittmann D, Drauschke M, Müller S, Steinhage V, Bezerra-Laure MAF, et al. Identification of Africanized honey bees through wing morphometrics: two fast and efficient procedures. Apidologie. 2008;39:488–494. doi: 10.1051/apido:2008028. [DOI] [Google Scholar]
- 46.HARPUR BA, MINAEI S, KENT CF, ZAYED A. Management increases genetic diversity of honey bees via admixture. Mol Ecol. 2012;21:4414–4421. doi: 10.1111/j.1365-294X.2012.05614.x. [DOI] [PubMed] [Google Scholar]
- 47.Harpur BA, Minaei S, Kent CF, Zayed A. Admixture increases diversity in managed honey bees: reply to De la Rúa et al. (2013) Mol Ecol. 2013;22:3211–3215. doi: 10.1111/mec.12332. [DOI] [PubMed] [Google Scholar]
- 48.Jones JC, Myerscough MR, Graham S, Oldroyd BP. Honey Bee Nest Thermoregulation: Diversity Promotes Stability. Science (80-) 2004;305:402 LP–402404. doi: 10.1126/science.1096340. [DOI] [PubMed] [Google Scholar]
- 49.Oldroyd BP, Fewell JH. Genetic diversity promotes homeostasis in insect colonies. Trends Ecol Evol. 2007;22:408–413. doi: 10.1016/j.tree.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 50.Oldroyd BP, Rinderer TE, Harbo JR, Buco SM. Effects of Intracolonial genetic diversity on honey bee (Hymenoptera: Apidae) Colony performance. Ann Entomol Soc Am. 1992;85:335–343. doi: 10.1093/aesa/85.3.335. [DOI] [Google Scholar]
- 51.Mattila HR, Seeley TD. Genetic Diversity in Honey Bee Colonies Enhances Productivity and Fitness. Science (80- ) 2007;317:362 LP–362364. doi: 10.1126/science.1143046. [DOI] [PubMed] [Google Scholar]
- 52.R. TD. Genetic diversity within honeybee colonies prevents severe infections and promotes colony growth. Proc R Soc Lond Ser B Biol Sci. 2003;270:99–103. doi: 10.1098/rspb.2002.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gloag R, Ding G, Christie JR, Buchmann G, Beekman M, Oldroyd BP. An invasive social insect overcomes genetic load at the sex locus. Nat Ecol Amp Evol 2016;1:11. doi: 10.1038/s41559-016-0011. [DOI] [PubMed]
- 54.Winston ML. The biology and Management of Africanized Honey Bees. Annu Rev Entomol. 1992;37:173–193. doi: 10.1146/annurev.en.37.010192.001133. [DOI] [Google Scholar]
- 55.Scott Schneider Stanley, DeGrandi-Hoffman Gloria, Smith Deborah Roan. THEAFRICANHONEYBEE: Factors Contributing to a Successful Biological Invasion. Annual Review of Entomology. 2004;49(1):351–376. doi: 10.1146/annurev.ento.49.061802.123359. [DOI] [PubMed] [Google Scholar]
- 56.Yin L, Ji T, Liu M, Bao W, Chen G. Genetic diversity and relationship between genetic distance and geographical distance of 6 Apis cerana cerana populations in China. Res J Anim Sci. 2008;2:183–187. [Google Scholar]
- 57.Ji T, Yin L, Liu M, Chen G. Genetic diversity and genetic differentiation of six geographic populations of Apis cerana cerana (Hymenoptera: Apidae) in East China. Acta Entomol Sin. 2009;52:413–419. [Google Scholar]
- 58.Williams SE, Shoo LP, Isaac JL, Hoffmann AA, Langham G. Towards an integrated framework for assessing the vulnerability of species to climate change. PLoS Biol. 2008;6:e325. doi: 10.1371/journal.pbio.0060325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Achere V, Favre JM, Besnard G, Jeandroz S. Genomic organization of molecular differentiation in Norway spruce (Picea abies) Mol Ecol. 2005;14:3191–3201. doi: 10.1111/j.1365-294X.2005.02646.x. [DOI] [PubMed] [Google Scholar]
- 60.Eimanifar A, Brooks SA, Bustamante T, Ellis JD. Population genomics and morphometric assignment of western honey bees (Apis mellifera L.) in the Republic of South Africa. BMC Genomics. 2018;19:615. doi: 10.1186/s12864-018-4998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ben-Shahar Y, Leung H-T, Pak WL, Sokolowski MB, Robinson GE. cGMP-dependent changes in phototaxis: a possible role for the foraging gene in honey bee division of labor. J Exp Biol. 2003;206(Pt 14):2507–2515. doi: 10.1242/jeb.00442. [DOI] [PubMed] [Google Scholar]
- 62.Seeley TD. Honeybee ecology: a study of adaptation in social life. Princeton: Princeton University Press; 1985. [Google Scholar]
- 63.Schmickl T, Crailsheim K. Inner nest homeostasis in a changing environment with special emphasis on honey bee brood nursing and pollen supply. Apidologie. 2004;35:249–263. doi: 10.1051/apido:2004019. [DOI] [Google Scholar]
- 64.Woodard SH, Fischman BJ, Venkat A, Hudson ME, Varala K, Cameron SA, et al. Genes involved in convergent evolution of eusociality in bees. Proc Natl Acad Sci. 2011;108:7472 LP–7477477. doi: 10.1073/pnas.1103457108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Burger R, Lynch M. Evolution and Extinction in a Changing Environment: A Quantitative-Genetic Analysis. Evolution (N Y) 2006;49:151. doi: 10.1111/j.1558-5646.1995.tb05967.x. [DOI] [PubMed] [Google Scholar]
- 66.Goulson D, Nicholls E, Botías C, Rotheray EL. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science (80- ) 2015;347:1255957. doi: 10.1126/science.1255957. [DOI] [PubMed] [Google Scholar]
- 67.Klein S, Cabirol A, Devaud JM, Barron AB, Lihoreau M. Why bees are so vulnerable to environmental stressors. Trends Ecol Evol. 2017;32:268–278. doi: 10.1016/j.tree.2016.12.009. [DOI] [PubMed] [Google Scholar]
- 68.Liu X. Honey bee morphological identification index. Apiculture China. 1978;4(9):15–17. [Google Scholar]
- 69.Tan K, Zhang X, He SY, Zhou DY. Morphology and biogeography research of Chinese Apis cerana. J Yunnan Agric Univ. 2005;20(3):410–414. [Google Scholar]
- 70.Wang GZ, Shi W, Zhang XL, Lv LP, Ding GL. Criterion about tergum pigmentation of Apis cerana. Apiculture China. 2006;57(5):7–8. [Google Scholar]
- 71.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 72.Fu Xiaoteng, Dou Jinzhuang, Mao Junxia, Su Hailin, Jiao Wenqian, Zhang Lingling, Hu Xiaoli, Huang Xiaoting, Wang Shi, Bao Zhenmin. RADtyping: An Integrated Package for Accurate De Novo Codominant and Dominant RAD Genotyping in Mapping Populations. PLoS ONE. 2013;8(11):e79960. doi: 10.1371/journal.pone.0079960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 77.Rosenberg MS, Anderson CD. PASSaGE: pattern analysis, spatial statistics and geographic exegesis. Version 2. Methods Ecol Evol. 2011;2:229–232. doi: 10.1111/j.2041-210X.2010.00081.x. [DOI] [Google Scholar]
- 78.Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond Ser B Biol Sci. 1996;263:1619–1626. doi: 10.1098/rspb.1996.0237. [DOI] [Google Scholar]
- 79.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 80.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(suppl_2):W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Reimand J, Arak T, Adler P, Kolberg L, Reisberg S, Peterson H, et al. G:profiler—a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44:W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Maximum likelihood tree with one migration event.
Additional file 2: Table S1. The 39 morphological measurements of A. cerana.
Additional file 3: Table S2. Abbreviation table.
Additional file 4: Table S3. The Fis, Fst, and Fit of each SNP locus.
Additional file 5: Table S4. The Fst table between all groups.
Additional file 6: Table S5. The gene flow table between all group.
Additional file 7: Table S6. Three population tests for admixture.
Additional file 8: Table S7. The Fst value table and distance table in North China.
Additional file 9: Table S8. Mantel test table.
Additional file 10: Table S9. Details of 121 candidate SNP loci.
Additional file 11: Table S10. GO term enrichment result.
Additional file 12. SNP data after filtering.
Data Availability Statement
The raw genome 2b-RAD tag-sequencing datasets generated during the current study are publicly available under NCBI Bioproject: PRJNA579872. All other data analysed in the present study are included in this published article (and its Supplementary Information files).