Abstract
Background
Acquired resistance to next-generation ALK tyrosine kinase inhibitors (TKIs) is often driven by secondary ALK mutations. Here we investigated utility of plasma genotyping for identifying ALK resistance mutations at relapse on next-generation ALK TKIs.
Methods
We analyzed 106 plasma specimens from 84 patients with advanced ALK-positive lung cancer treated with second- and third-generation ALK TKIs using a commercially available next-generation sequencing (NGS) platform (Guardant360). Tumor biopsies from TKI-resistant lesions underwent targeted NGS to identify ALK mutations.
Results
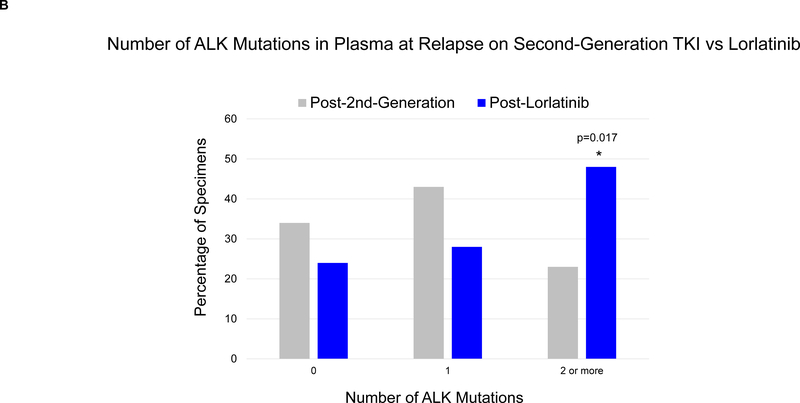
By genotyping plasma, we detected an ALK mutation in 46 (66%) of 70 patients relapsing on a second-generation ALK TKI. When post-alectinib plasma and tumor specimens were compared, there was no difference in frequency of ALK mutations (67% vs 63%), but plasma specimens were more likely to harbor ≥2 ALK mutations (24% vs 2%, p=0.004). Among 29 patients relapsing on lorlatinib, plasma genotyping detected an ALK mutation in 22 (76%), including 14 (48%) with ≥2 ALK mutations. The most frequent combinations of ALK mutations were G1202R/L1196M and D1203N/1171N. Detection of ≥2 ALK mutations was significantly more common in patients relapsing on lorlatinib compared to second-generation ALK TKIs (48% vs 23%, p=0.017). Among 15 patients who received lorlatinib after a second-generation TKI, serial plasma analysis demonstrated that 8 (53%) acquired ≥1 new ALK mutations on lorlatinib.
Conclusions
ALK resistance mutations increase with each successive generation of ALK TKI and may be underestimated by tumor genotyping. Sequential treatment with increasingly potent ALK TKIs may promote acquisition of ALK resistance mutations leading to treatment-refractory compound ALK mutations.
Keywords: ALK, Lung Cancer, Liquid Biopsy, Resistance, Lorlatinib, Alectinib
INTRODUCTION
Anaplastic lymphoma kinase (ALK)-rearranged non-small cell lung cancers (NSCLCs) are driven by a constitutively active fusion protein that confers marked sensitivity to ALK tyrosine kinase inhibitors (TKIs).1 Recent studies exploring the activity of second-generation ALK TKIs (ceritinib, alectinib, and brigatinib) in treatment-naïve patients have positioned these more potent ALK TKIs as the preferred first-line treatment for advanced ALK-positive NSCLC.2–4 However, virtually all patients will eventually develop resistance to therapy. In patients relapsing on second-generation ALK TKIs, molecular analysis of repeat biopsies from resistant disease sites suggests that secondary mutations in the ALK kinase domain contribute to 50–60% of treatment relapses.5–7
Lorlatinib is a third-generation ALK TKI that was specifically designed to overcome ALK kinase domain mutations.7,8 In the registrational phase 2 study, responses to lorlatinib were seen in approximately 70% of patients whose tumors harbored an ALK kinase domain mutation prior to lorlatinib.9 However, even ALK-positive NSCLCs that initially respond to lorlatinib will become resistant, often due to the development of compound ALK mutations (i.e., two or more ALK mutations located on the same allele).10 Thus, ALK mutations are a key driver of resistance to both second- and third-generation ALK TKIs. Interestingly, due to the distinct chemical structures of different ALK TKIs, a subset of lorlatinib-resistant compound ALK mutations may be sensitive to treatment with earlier generation ALK TKIs.11,12 The identification of ALK mutations can therefore inform selection of ALK TKIs at multiple points in the disease course.
Plasma genotyping is a promising strategy for analyzing TKI resistance in oncogene-driven NSCLCs.13,14 As plasma contains an amalgam of tumor-derived DNA from multiple metastatic sites, genotyping plasma may be more informative than biopsy of a single disease site. Several recent studies suggest a potential role for circulating tumor DNA (ctDNA) analysis in management of ALK-positive NSCLC.15,16 For example, two studies demonstrated that the Guardant360 assay reliably detects ALK fusions and kinase domain mutations in patients with ALK TKI-resistant disease.9,16 In another study using a different plasma assay, our group explored the role of longitudinal plasma genotyping in monitoring the evolution of resistance to ALK TKIs.15 These three studies primarily analyzed plasma from lorlatinib-naïve patients. As the genetic alterations that drive on-target resistance increase in complexity after exposure to lorlatinib,10 additional studies are needed to establish the utility of plasma genotyping for characterizing ALK resistance mutations across the spectrum of next-generation ALK TKIs, particularly lorlatinib.
Here we analyzed over 100 plasma samples from patients with ALK-positive NSCLC relapsing on next-generation ALK TKIs and show that ALK resistance mutations increase with each successive generation of ALK TKIs.
PATIENTS AND METHODS
Data Collection
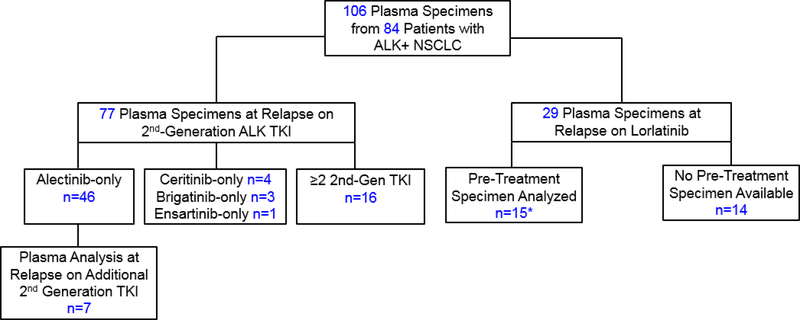
Between March 2016 and March 2019, we analyzed 106 plasma specimens from 84 patients with ALK-positive NSCLC who were relapsing on a second-generation ALK TKI or the third-generation ALK TKI lorlatinib (Figure 1). Medical records were retrospectively reviewed to extract data on clinicopathologic and molecular features. Data were updated as of March 15, 2019. This study was approved by the Massachusetts General Hospital Institutional Review Board. Patient studies were conducted according to the Declaration of Helsinki, the Belmont Report, and the U.S. Common Rule.
Figure 1. Study Population.
Serial analysis of plasma specimens was performed for 22 patients (7 patients who received a second-generation TKI followed by second-generation TKI, and 15 patients who received a second-generation TKI followed by lorlatinib, indicated by asterisk). Among 16 patients who received ≥2 second-generation ALK TKIs before plasma analysis, 9 received alectinib and ceritinib, 4 received alectinib and brigatinib, 2 received alectinib, ceritinib, and brigatinib, and 1 received alectinib, brigatinib, and ensartinib. *Included in both second-generation and lorlatinib cohorts; 2nd-Gen=second-generation; TKI: tyrosine kinase inhibitor.
Molecular Testing
All plasma specimens were genotyped using the CLIA-certified, Guardant360 NGS assay as previously described.17 The majority of plasma specimens (n= 99, 93%) were genotyped after November 2016 using a 73-gene Guardant360 assay. This iteration of the Guardant360 assay employs a panel that covers six fusions (including ALK), all exons of 19 genes, critical exons of 73 genes (including ALK), and insertions/deletions and copy number variations in select genes (23 genes and 18 genes, respectively). The remaining seven specimens (7%) were analyzed with an earlier 70-gene version of the Guardant360 assay that encompassed fewer insertions/deletions. Both versions of the Guardant360 assay use hybridization capture probes targeting the conserved ALK breakpoint in intron 19 and a variety of breakpoints in EML4 and other upstream ALK fusion partners to detect ALK rearrangements. For a subset of patients (n=22), contemporaneous tissue specimens were analyzed using SNaPshot NGS (n=17),18 Foundation One (n=2),19 DFCI Oncopanel (n=1),20 and MSK Impact (n=1)21 as previously described. All patients included in this study provided consent for molecular testing.
We conducted a tissue-plasma concordance analysis to evaluate the performance characteristics of the Guardant360 plasma assay. Twenty-two patients underwent paired tissue and plasma genotyping (Supplementary Table 1). Using tissue as the reference standard, the sensitivity of plasma genotyping for detecting tissue-identified ALK mutations was 90%, confirming that plasma genotyping can reliably detect ALK resistance mutations in patients relapsing on ALK TKIs. However, due to intratumor heterogeneity specificity was 48%, as plasma genotyping detected additional ALK mutations not identified by genotyping a single disease site.
Statistical Analysis
Fisher’s exact test was used to compare ALK mutation frequency between specimen and treatment groups. All p-values were based on a two-sided hypothesis and computed using Stata 12.1.
RESULTS
Study Population
We analyzed 106 plasma specimens from 84 patients with metastatic ALK-positive NSCLC (Figure 1). Baseline characteristics of the study cohort are described in Supplementary Table 2. The dataset included 77 plasma specimens from 70 patients relapsing on a second-generation ALK TKI. Fifty-four (77%) of these patients had only been exposed to one second-generation ALK TKI prior to analysis. In addition, we analyzed plasma from 29 patients relapsing on lorlatinib, fifteen of whom underwent plasma analysis after failure of a second-generation ALK TKI prior to initiating lorlatinib. These 15 patients were also included in the second-generation TKI group.
Resistance to Second-Generation ALK Inhibitors
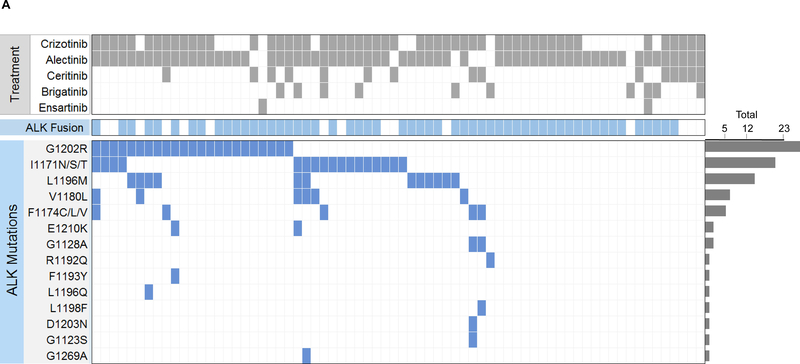
We first evaluated the spectrum of ALK resistance mutations in plasma at progression on second-generation ALK TKIs. We detected an ALK mutation in plasma from 46 (66%) of 70 patients relapsing on a second-generation ALK TKI (Figure 2A). An ALK fusion was detected in plasma from 19 (79%) of the 24 patients who did not have ALK mutations in plasma. The most frequently observed ALK mutation was G1202R, detected in 23 (33%) specimens. ALK I1171X and L1196M were also frequently seen (n=17, 24% and n=12, 17%, respectively). Sixteen (23%) plasma specimens contained ≥2 ALK mutations. Nineteen patients were relapsing only in the brain or thoracic cavity at the time of plasma analysis. As sensitivity of plasma genotyping may be lower when relapse is confined to these sites,14,22 we were particularly interested in ctDNA findings from these 19 patients. An ALK mutation and/or ALK fusion was detected in plasma from 17 (89%) of the 19 patients. The remaining patient’s plasma analysis failed to identify an ALK fusion or ALK mutation in the setting of brain-only relapse.
Figure 2. ALK Mutations at Relapse on Second-Generation ALK TKIs.
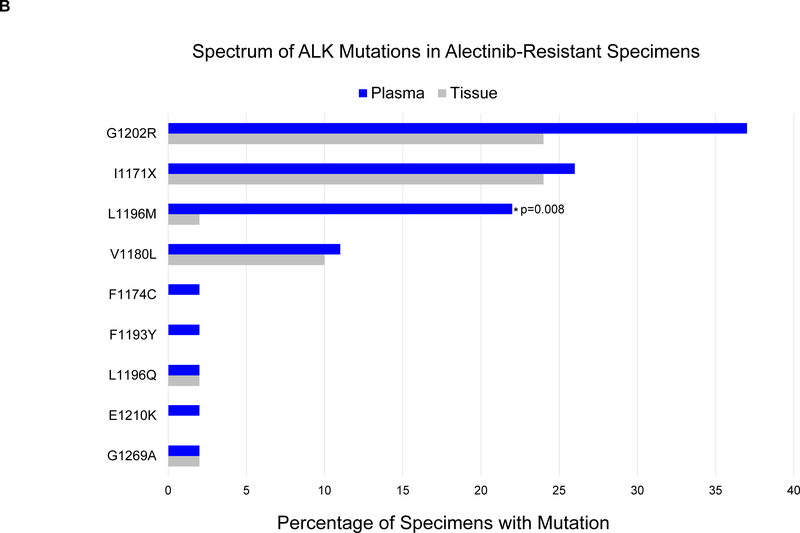
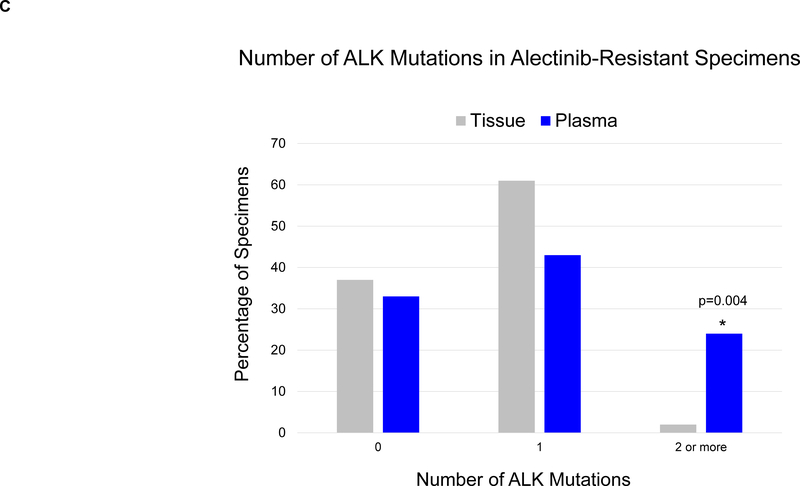
(A) The grid depicts ALK mutations detected in plasma from patients relapsing on second-generation ALK inhibitors. Gray boxes indicate ALK inhibitors received prior to plasma collection. Blue boxes indicate detection of an ALK fusion or ALK mutation in plasma. Bar graphs quantify the number of plasma specimens with each ALK mutation. (B) Bar graphs indicate the percentage of post-alectinib plasma or tissue specimens with each ALK mutation. Apart from L1196M (indicated by asterisk), there was no significant difference in frequency of specific ALK mutations in plasma vs tissue. (C) Bar graphs indicate the number of specimens (tissue vs plasma) harboring 0,1, or ≥2 ALK mutations at relapse on alectinib. Detection of ≥2 ALK mutations was significantly more common (asterisk) in plasma than tissue.
Spectrum of ALK Mutations in Plasma vs Tissue at Alectinib Relapse
To determine whether the frequency and spectrum of ALK mutations are consistent in tissue and plasma, we compared plasma genotyping results to tissue analysis results for patients seen at our institution who underwent tissue biopsy at progression on alectinib. We limited the analysis to patients who received alectinib as their only next-generation TKI as these patients constituted the majority of cases in our plasma cohort. In total, we analyzed 46 plasma and 41 tissue specimens. Twelve patients underwent paired plasma and tissue genotyping at progression on alectinib and were included in both cohorts.
Of the 46 patients who underwent plasma genotyping at relapse on alectinib, thirty-one (67%) had ALK mutations in plasma, including G1202R (n=17, 37%), I1171X (n=12, 26%), L1196M (n=10, 22%), and V1180L (n=5, 11%). Eleven (24%) plasma specimens contained ≥2 ALK mutations (Figure 2A, Supplementary Figure 1). By tumor genotyping, ALK mutations were identified in 26 (63%) of 41 alectinib-resistant tissue biopsies (Supplementary Figure 1). The most common tissue ALK mutations were G1202R (n=10, 24%), I1171X (n=10, 24%), and V1180L (n=4, 10%) (Figure 2B). ALK L1196M was notably less prevalent in tissue than plasma (2% vs 22%, p=0.008). As ALK L1196M is a gatekeeper mutation that confers resistance to crizotinib but can be overcome by all next-generation ALK TKIs, it is likely that the mutation emerged during prior treatment with crizotinib and was not the primary driver of failure of a second-generation ALK TKI. Thus, the decreased frequency of ALK L1196M in tissue relative to plasma may reflect targeting of truly resistant disease sites for tumor biopsies. There was no significant difference in the frequency of other ALK mutations (Figure 2B). Tumor genotyping was also significantly less likely than plasma genotyping to identify ≥2 ALK mutations at relapse on alectinib (2% vs 24%, p=0.004, Figure 2C). Overall, our findings suggest that the proportion of patients relapsing on alectinib due to secondary ALK mutations is similar based on tissue or plasma genotyping, but plasma can identify a subset of alectinib-resistant cancers harboring multiple ALK mutations.
Plasma ALK Mutations in Patients Exposed to Multiple Second-Generation ALK TKIs
We analyzed plasma from 23 patients who had been exposed to ≥2 second-generation ALK TKIs (Supplementary Table 3). For 16 of these patients, the plasma analysis was conducted after failure of multiple second-generation ALK TKIs (Figure 2A). Eight (50%) patients had an ALK mutation in plasma at relapse, three (19%) of whom had ≥2 ALK mutations. In comparison, thirty-five (69%) patients had an ALK mutation in plasma after exposure to only one second-generation TKI, including 13 (25%) patients with ≥ 2 ALK mutations (Figure 2A). Thus, detection of ≥2 ALK mutations in plasma was similar in patients exposed to one versus multiple second-generation TKIs (25% vs 19%, p=0.743).
The remaining seven patients underwent plasma analysis twice during the study period with each timepoint representing progression on a different second-generation ALK TKI (Supplementary Table 4). MGH9220 developed an alectinib-resistant (but ceritinib-sensitive) ALK V1180L mutation at relapse on alectinib. At progression after 7 months on ceritinib, ALK V1180L cleared but ALK G1202R was detected. Six patients received alectinib followed by brigatinib. In three cases (all with ALK mutations), the number and composition of ALK mutations did not change at subsequent relapse on brigatinib. For two of these patients, the best response to brigatinib was disease stabilization lasting four months. The other patient (MGH989) had primary progression on brigatinib. Three patients developed an additional ALK mutation after 4–6 months of treatment with brigatinib, including two patients who did not have detectable ALK mutations in plasma at alectinib relapse Overall, among six patients who were treated with alectinib followed by brigatinib, detection of ≥2 ALK mutations in plasma at relapse on brigatinib was seen in two cases, one of whom acquired all mutations prior to initiating brigatinib.
Resistance to the Third-Generation ALK Inhibitor Lorlatinib
Lorlatinib is a potent third-generation ALK TKI with clinical activity against the broadest range of ALK kinase domain mutations.7,9 However, lorlatinib cannot overcome all ALK-dependent resistance mechanisms.23 Our group recently identified lorlatinib-resistant compound ALK mutations in biopsy specimens from patients relapsing on lorlatinib.10 Based on these findings, we hypothesized that plasma from patients relapsing on lorlatinib would be more likely to contain ≥2 ALK mutations than plasma from patients relapsing on a second-generation TKI.
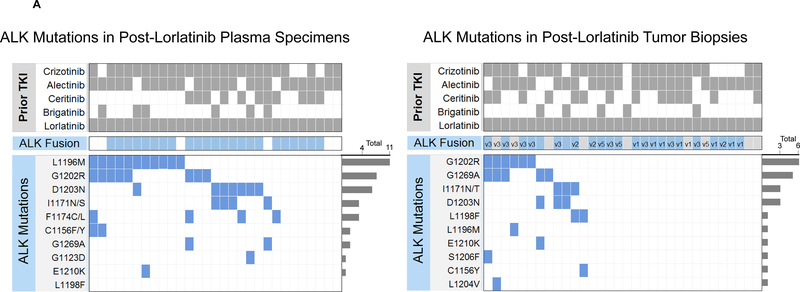
We detected ALK mutations in 22 (76%) plasma specimens from 29 patients at progression on lorlatinib, all of whom had received at least one prior second-generation ALK TKI (Figure 3A, left). An ALK fusion was detected in five of the 7 plasma specimens that lacked an ALK mutation. Six patients in the post-lorlatinib cohort were relapsing only in the brain or thoracic cavity at the time of plasma analysis. An ALK fusion and/or ALK mutation was detected in plasma from all six patients. In the group of 29 post-lorlatinib specimens, the following ALK mutations were recurrently seen: L1196M (38%), G1202R (28%), D1203N (24%), F1174C/L (14%), and I1171X (14%). Fourteen (48%) plasma specimens contained ≥2 ALK mutations. Detection of ≥2 ALK mutations in plasma was twice as common at relapse on lorlatinib than at relapse on a second-generation ALK TKI (Figure 3B, 48% vs 23%, p=0.017). Thus, while exposure to multiple second-generation ALK TKIs does not appear to predispose to developing multiple ALK mutations in plasma, these findings suggest that exposure to a more potent ALK TKI like lorlatinib may select for multiple ALK mutations.
Figure 3. ALK Mutations at Relapse on Lorlatinib.
(A) Grids depict ALK mutations detected in plasma (left) and tumor biopsies (right) from patients relapsing on lorlatinib. Dark grey boxes indicate ALK inhibitors received prior to specimen collection. Blue boxes indicate detection of an ALK kinase domain mutation or fusion. Light gray boxes in fusion row indicate that a specimen was not assessed for presence of ALK fusion. ALK fusion variants (based on current or prior testing) are indicated in the fusion row for tissue specimens when known. Bar graphs quantify the number of specimens with each ALK mutation. (B) Bar graphs indicate percentage of specimens harboring 0, 1, or ≥2 ALK mutations at relapse on a second-generation ALK TKI vs lorlatinib. Detection of ≥2 ALK mutations was more common at relapse on lorlatinib (asterisk). (C) Bar graphs indicate the percentage of plasma vs tissue specimens harboring 0, 1, ≥2 ALK mutations at relapse on lorlatinib. Compared to plasma, tissue specimens were significantly less likely to have ALK mutations (asterisk).
Spectrum of ALK Mutations in Plasma vs Tissue at Lorlatinib Relapse
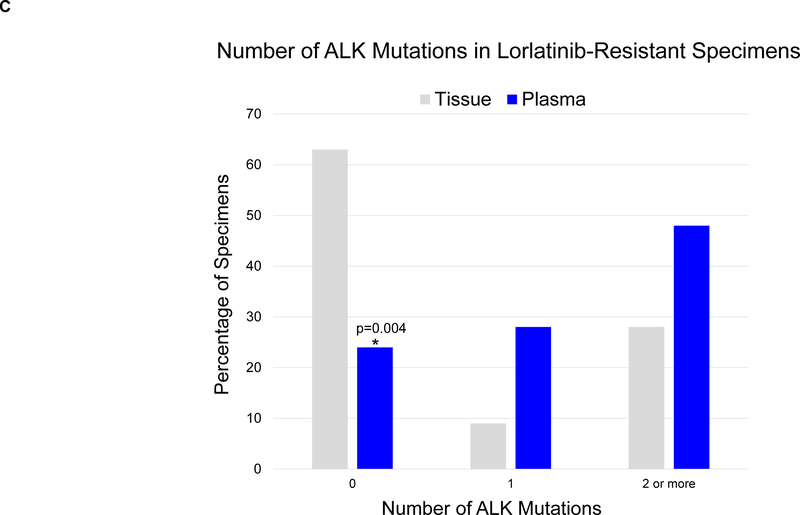
We compared ALK mutations in plasma to those detected in tumor biopsies from 32 patients relapsing on lorlatinib (Figure 3A, right), seven of whom were included in both cohorts. For six of the 32 patients who underwent tissue analysis, findings from genotyping post-lorlatinib tumor specimens were reported in an earlier analysis.10 Twelve (38%) tumor biopsies harbored ALK mutations, including nine with ≥2 ALK mutations. The most common tissue ALK mutations were G1202R (19%), G1269A (16%), D1203N (9%), and I1171X (9%). Three biopsies contained both ALK G1269A and G1202R. ALK G1202R/L1196M and D1203N/I1171X mutations were identified in one and two biopsies, respectively. Compared to plasma, tissue genotyping was half as likely to detect ALK mutations at relapse (38% vs 76%, p=0.004). The frequency of detecting ≥2 ALK mutations was also higher in plasma than in tissue (48% vs 28%, Figure 3C), but the comparison (p=0.12) was not strictly significant due to low power.
Allelic Configuration of ALK Mutations
We analyzed lorlatinib-resistant plasma specimens with ≥2 ALK mutations to determine whether the mutations occurred in cis (i.e., compound mutations). Five post-lorlatinib specimens contained ALK mutations located close enough to determine allelic configuration (Supplementary Figure 2), all of which were confirmed to represent compound mutations (Supplementary Table 5). Notably, MGH953’s plasma contained three distinct compound ALK mutations: G1202R/L1196M, L1196M/F1174L, and L1196M/F1174C. As previously reported, the patient underwent sampling of an enlarging pleural effusion at lorlatinib relapse which demonstrated ALK G1202R/L1196M in cis but did not detect the other two compound mutations.10 The remaining four patients did not have paired tumor biopsies. Of note, we performed a similar analysis on plasma specimens from patients relapsing on second-generation TKIs. Among six second-generation TKI-resistant cases where plasma ALK mutations were proximal enough to assess allelic configuration, three represented compound mutations while the remaining three had ALK mutations located in trans (Supplementary Table 5). While the numbers are small, these data suggest that less potent second-generation ALK TKIs are less likely than lorlatinib to select for resistant compound ALK mutations.
Dynamic Changes in Plasma ALK Mutations During Sequential ALK TKI Therapy
Analyses of longitudinal tissue biopsies suggest that compound ALK mutations can result from stepwise acquisition of ALK mutations during sequential treatment with second-generation ALK TKIs followed by lorlatinib.10,11 To evaluate whether ALK mutations accumulate on lorlatinib, we analyzed serial plasma samples from 15 patients at relapse on a second-generation ALK TKI and then again at progression on lorlatinib (Table 1). Thirteen patients had ALK mutations prior to initiating lorlatinib. Neither of the two patients who did not have a pretreatment ALK mutation developed new ALK mutations after treatment with lorlatinib.
Table 1.
ALK Mutations in Pre- and Post-Lorlatinib Plasma Specimens
| Patient | Pre-Lorlatinib Plasma ALK Mutations | Post-Lorlatinib Plasma ALK Mutations |
|---|---|---|
| MGH9200 | L1196M | L1196M*, G1202R* |
| MGH9035 | G1202R, L1196M, V1180L | G1202R*, L1196M*, C1156Y |
| MGH9118 | D1203N | D1203N, G1123D |
| MGH9158 | L1196Q^, L1196M^, G1202R^ | L1196M |
| MGH087 | G1202R | G1202R |
| MGH9185 | G1202R | I1171N, D1203N |
| MGH9027 | I1171N | I1171N, D1203N |
| MGH059 | E1210K*, I1171N, V1180L, L1196M* | E1210K*, L1196M* |
| MGH9213 | L1196M | L1196M*, D1203N* |
| MGH990 | G1202R, I1171N | I1171N, D1203N |
| MGH9212 | I1171S, F1174L | I1171S, G1269A |
| MGH919 | G1202R*, L1196M* | G1202R*, L1196M* |
| MGH9176 | G1202R | G1202R |
| MGH915 | None | None |
| MGH964 | None | None |
Blue=mutation only seen in pre-lorlatinib specimen; Red=mutation only seen in post-lorlatinib specimen
ALK mutations occurred in cis
ALK mutations occurred in trans
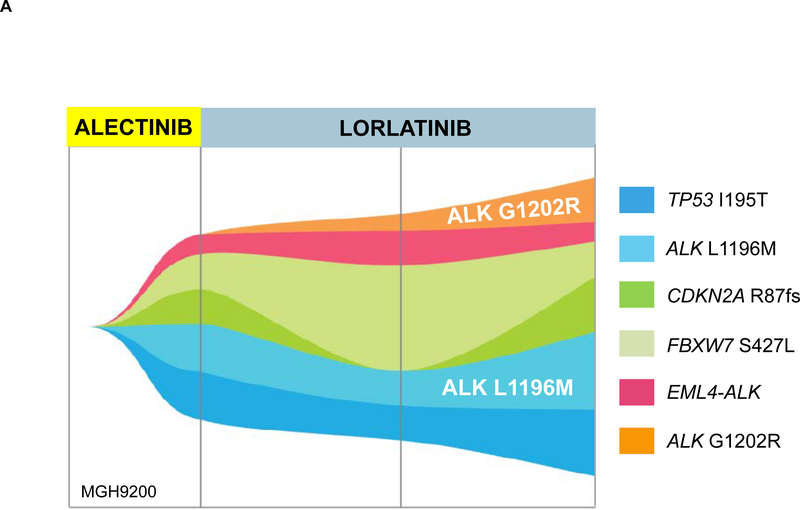
Among the 13 patients with pre-lorlatinib ALK mutations, eight developed at least one new ALK mutation (Table 1). In two cases (MGH9200 and MGH9213) where a new ALK mutation was acquired and the allelic relationship between post-lorlatinib ALK mutations could be assessed, the two ALK mutations resided on the same allele, consistent with generation of a compound mutation (G1202R/L1196M as seen in Figure 4A and D1203N/L1196M). Interestingly, four of the 8 patients acquired ALK D1203N during treatment with lorlatinib (Table 1). ALK D1203N was significantly more common at relapse on lorlatinib than at relapse on a second-generation ALK TKI (24% vs 4%, p=0.005). Among the 13 patients with pre-lorlatinib ALK mutations, we also observed “loss” of one or more ALK resistance mutations in six patients. For example, MGH9035 cleared ALK V1180L from plasma during treatment with lorlatinib and gained ALK C1156Y while retaining ALK G1202R and ALK L1196M (Figure 4B). The most frequently “lost” mutation was ALK G1202R in three cases, suggesting suppression of the G1202R-mutant clone by lorlatinib. These dynamic changes were also reflected by changes in the allelic frequency of ALK mutations in post-lorlatinib vs pre-lorlatinib plasma specimens (Supplementary Figure 3). Taken together, these results suggest that in the presence of a pre-existing ALK resistance mutation, lorlatinib treatment can foster acquisition of additional ALK resistance mutations, generating highly refractory compound ALK mutations.
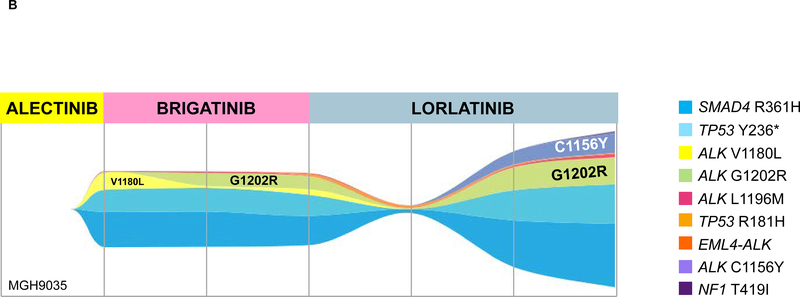
Figure 4. Evolution of ALK Mutations During Treatment with Sequential Second- and Third-Generation ALK TKIs.
(A) ALK L1196M was detected in MGH9200’s plasma at relapse on alectinib. After failure of lorlatinib, the mutation persisted and ALK G1202R emerged. (B) MGH9035’s clonal evolution plot depicts acquisition and “loss” of ALK mutations in plasma during sequential treatment with alectinib, brigatinib, and lorlatinib.
DISCUSSION
In this manuscript, we analyzed over 100 plasma specimens from patients with advanced ALK-positive NSCLC who were relapsing on next-generation ALK TKIs. We compared plasma versus tissue genotyping results, and also compared ALK mutation profiles after failure of second- versus third-generation ALK TKIs. Our results demonstrate that the number of ALK resistance mutations increases with each successive generation of ALK TKI. Under the selective pressure of lorlatinib, these mutations are often in cis and lead to generation of treatment-refractory compound ALK mutations.
In this study, plasma genotyping consistently detected more ALK mutations than tumor genotyping. For example, among patients relapsing on alectinib, we identified ≥2 ALK mutations in plasma specimens from 24% of cases. In contrast, we detected ≥2 ALK mutations in only 2% of alectinib-resistant tumor biopsies. Similarly, in lorlatinib-resistant patients, we detected ≥2 ALK mutations in 48% and 28% of cases using plasma and tumor genotyping, respectively. An analysis of allelic configuration among the subset of cases where the location of ALK mutations allowed for assessing allelic relationships demonstrated that all post-lorlatinib specimens with ≥ 2 ALK mutations had compound mutations whereas only half of those analyzed at relapse on a second-generation ALK TKI represented compound mutations. Although we expected that compound mutations would be more prevalent after exposure to lorlatinib, it was surprising to find that compound mutations can develop at relapse on second-generation ALK TKIs. Nevertheless, as many patients relapsing on second-generation ALK TKIs will not have compound mutations, the discordance between the frequency of detecting ≥2 ALK mutations with plasma vs tissue testing may in some cases reflect the underlying spatial heterogeneity of resistant disease, i.e. independent ALK-mutant clones located at different anatomical sites.24,25 In support of this, in our concordance study with paired plasma and tissue samples, six of 22 patients had additional ALK mutations in plasma that were not detected in a single-site tumor biopsy (Supplementary Table 1). In addition, as mentioned above, four of 11 plasma specimens evaluated for allelic configuration of plasma-detected ALK mutations had ALK mutations on separate alleles, possibly reflecting distinct resistant clones (Supplementary Table 5). These findings highlight the potential advantage of using plasma genotyping to dissect the complex and heterogeneous ALK resistance landscape.
In addition to capturing spatial heterogeneity of resistance at higher resolution than tissue biopsy, plasma genotyping offers a non-invasive strategy for assessing temporal evolution of resistance. In this study, we evaluated serial plasma samples from 15 patients with ALK-positive NSCLC treated with sequential next generation ALK TKIs. Plasma genotyping revealed that exposure to distinct ALK TKIs fuels highly dynamic shifts in clonal/subclonal populations. For example, both MGH990 and MGH9185 relapsed on second-generation TKIs due to an acquired ALK G1202R mutation. After initial response and then relapse on lorlatinib, ALK G1202R was no longer detectable in the plasma of either patient, and instead ALK I1171N and D1203N were identified at comparable allelic frequencies in both cases. The extent of genomic space between these two particular ALK mutations precluded us from establishing whether they resided on the same allele or different alleles. However, in the five post-lorlatinib cases where we were able to assess the allelic context of plasma ALK mutations, we confirmed that the ALK mutations were on the same allele. Taken together with our previous study of lorlatinib-resistant tumor specimens,10 these findings support the notion that stepwise acquisition of ALK mutations occurs as patients move from second- to third-generation ALK TKIs, culminating in the development of lorlatinib-resistant compound ALK mutations.
To date, several lorlatinib-resistant compound ALK mutations have been identified in patients relapsing after sequential next-generation TKIs. Remarkably, some lorlatinib-resistant compound ALK mutations may re-sensitize tumors to earlier-generation ALK TKIs.10,11,23 ALK L1198F-containing compound mutations have been shown to re-sensitize to crizotinib,11 but ALK L1198F was not detected in the plasma of any of our lorlatinib-resistant cases. For one patient relapsing on lorlatinib, plasma genotyping detected ALK G1269A and I1171S (Figure 3A). Based on in vitro models, a G1269A/I1171S compound mutation may re-sensitize to ceritinib or brigatinib.23 Outside of the compound mutations above, the remaining lorlatinib-resistant compound ALK mutations are likely refractory to currently available ALK TKIs.
The most common compound ALK mutation identified in plasma in this study was L1196M/G1202R, which we previously demonstrated confers high level resistance to first-, second-, and third-generation ALK TKIs.10 Interestingly, ALK D1203N, which resides at the solvent front of the kinase domain beside G1202R, was also frequently detected in post-lorlatinib cases harboring ≥2 plasma ALK mutations, including four instances where it was shown to be newly-acquired on lorlatinib. In three lorlatinib-resistant cases, ALK D1203N in exon 23 co-occurred with the exon 22 mutation I1171N. The allele frequencies of the two ALK mutations suggest that the mutations were likely in cis, but the allelic configuration could not be confirmed as the two exons are separated by approximately 1600 base pairs. Given the limitations of DNA-based plasma sequencing approaches in the setting of significant inter-exon distance, there is rationale for developing platforms for analyzing circulating gene transcripts. Early studies demonstrate feasibility of detecting ALK fusions and ALK resistance mutations in tumor-derived RNA contained in exosomes or platelets.26,27 Overall, our results highlight the growing complexity of on-target mechanisms of resistance in patients treated with increasingly potent ALK TKIs. While this work identifies potential new targets for drug discovery, including G1202R/L1196M and D1203N/I1171N compound mutations, the multiplicity of ALK mutations emerging after sequential next-generation TKIs suggests that alternative strategies such as allosteric inhibitors and ALK degraders should be explored.28
Our study has several important limitations. First, most patients had been exposed to other ALK TKIs (e.g., crizotinib) prior to initial plasma analysis, but plasma specimens were not available at earlier timepoints. In theory, ALK mutations may have developed on prior therapies, and this may have led us to overestimate the impact of lorlatinib on the accumulation of ALK mutations. To overcome this limitation, we examined serial plasma specimens from patients on sequential next generation ALK inhibitors, including pre- and post-lorlatinib specimens when available. Second, during the study period, we attempted to collect and analyze post-lorlatinib specimens from all patients seen at our institution. However, it is possible that patients with ALK mutations were inadvertently overrepresented given the small size of the post-lorlatinib dataset. As mentioned above, we could not establish the allelic configuration of ALK mutations in over one-half of plasma samples harboring ≥2 ALK mutations due to the location of the mutations within the large region of genomic DNA encompassed by exons 22–23 of ALK. Furthermore, although we have made the novel observation that ALK D1203N and ALK I1171N commonly co-occur at relapse on lorlatinib, we have not characterized the potential functional significance of co-localization of these ALK mutations on the same allele. Finally, as secondary ALK kinase domain mutations represent a major class of resistance to ALK TKIs, we focused our analysis on ALK mutations. However, 30–40% of patients relapsing on second- and third-generation ALK inhibitors do not acquire ALK resistance mutations suggesting that their cancers are driven by ALK-independent mechanisms. As these ALK-independent resistance mechanisms cause refractoriness to further ALK inhibition, it is essential for plasma genotyping platforms to comprehensively assess for genetic drivers of both ALK-independent and ALK-dependent resistance. Future studies characterizing potential ALK-independent resistance mechanisms will be critical to developing effective therapeutic strategies for patients who have become TKI-refractory.
In summary, the current treatment paradigm for advanced ALK-positive NSCLC involves sequential treatment with increasingly potent second- and third-generation ALK TKIs. While patients can derive significant clinical benefit from this approach, this work demonstrates that the selective pressure of sequential TKIs fosters the stepwise acquisition of secondary ALK resistance mutations, leading to a diverse array of compound ALK mutations, most of which are refractory to currently available ALK TKIs. As the initial ALK mutation provides the substrate for generating compound mutations, a more effective treatment strategy may be to move lorlatinib, which has shown the broadest range of activity against single ALK resistance mutations, into the first-line setting. A phase 3 clinical trial evaluating lorlatinib as first-line therapy in advanced ALK-positive NSCLC is ongoing and will help inform the optimal sequencing of next-generation ALK TKIs.
Supplementary Material
Translational Relevance.
Management of advanced ALK-positive lung cancer involves sequential treatment with a second-generation ALK inhibitor followed by lorlatinib. Early analyses of tumor biopsies suggest that ALK mutations acquired at initial relapse provide the substrate for generating compound ALK mutations during treatment with lorlatinib. As a composite of tumor DNA from multiple lesions, genotyping plasma may provide deeper insights into the dynamic and complex nature of ALK-dependent resistance than genotyping a single tumor. Here, through analysis of over 100 plasma specimens, we demonstrate that accumulation of ALK mutations during sequential treatment with next-generation ALK inhibitors promotes formation of refractory compound mutations. Our study highlights the potential for plasma genotyping to refine current understanding of the evolution of on-target resistance to ALK inhibitors and provides a blueprint for the rational design of fourth generation ALK inhibitors aimed at overcoming compound ALK mutations.
Acknowledgments
Conflict of Interest Statement/Disclosures:
IDJ has received honoraria from Foundation Medicine, consulting fees from Boehringer Ingelheim, and research support from Novartis, Pfizer, and Guardant Health. JJL has received honoraria from Chugai and Boehringer Ingelheim and institutional research support from Loxo. RJN and RBL are employees of Guardant Health. AFF has served consulting/advisory role for PharmaMar, Abbvie, Loxo, Stemcentrx, Genentech/Roche, Bayer, AstraZeneca, and Boehringer Ingelheim and research funding from PharmaMar, Abbvie, AstraZeneca, Bristol-Myers Squibb, Merck, Loxo, Ignyta, Amgen, Genentech/Roche, Bayer, and Novartis. ANH has received research grants/funding from Pfizer, Novartis, Amgen, Relay Therapeutics, and Roche/Genentech. JFG has served as a compensated consultant or received honoraria from Bristol-Myers Squibb, Genentech, Ariad/Takeda, Loxo, Pfizer, Incyte, Novartis, Merck, Agios, Amgen, Jounce, Regeneron, Oncorus, Jounce, Array, and Clovis Oncology, and research funding from Novartis, Genentech/Roche, and Ariad/Takeda, and institutional research support from Tesaro, Moderna, Blueprint, BMS, Jounce, Array, Adaptimmune, Novartis, Genentech/Roche, Alexo and Merck. ATS has served as a compensated consultant or received honoraria from Achilles, ARIAD, Bayer, Blueprint Medicines, Chugai, Daiichi Sankyo, EMD Serono, Foundation Medicine, Genentech/Roche, Guardant, Ignyta, KSQ Therapeutics, LOXO, Natera, Novartis, Pfizer, Servier, Taiho Pharmaceutical, Takeda, and TP Therapeutics, has received research (institutional) funding from Daiichi Sankyo, Ignyta, Novartis, Pfizer, Roche/Genentech, and TP Therapeutics; and has received travel support from Pfizer and Genentech/Roche. The remaining authors have no disclosures to report.
Funding:
This work was support by a Conquer Cancer/Amgen Young Investigator Award (I.D.J), an institutional research grant from American Cancer Society (I.D.J.), a National Cancer Institute Career Development Award (K12CA087723–16 to I.D.J.), a grant from the National Cancer Institute (R01CA164273 to A.T.S.), by Be a Piece of the Solution, and by Targeting a Cure for Lung Cancer Research Fund at MGH.
References
- 1.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. [DOI] [PubMed] [Google Scholar]
- 2.Soria JC, Tan DS, Chiari R, et al. First-line ceritinib versus platinum-based chemotherapy in advanced ALK-rearranged non-small-cell lung cancer (ASCEND-4): a randomised, open-label, phase 3 study. Lancet. 2017;389(10072):917–929. [DOI] [PubMed] [Google Scholar]
- 3.Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2017. [DOI] [PubMed] [Google Scholar]
- 4.Camidge DR, Kim HR, Ahn MJ, et al. Brigatinib versus Crizotinib in ALK-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2018;379(21):2027–2039. [DOI] [PubMed] [Google Scholar]
- 5.McCoach CE, Le AT, Gowan K, et al. Resistance Mechanisms to Targeted Therapies in ROS1(+) and ALK(+) Non-small Cell Lung Cancer. Clin Cancer Res. 2018;24(14):3334–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin JJ, Yeap BY, Ferris LA, et al. Long-term efficacy and outcomes with sequential crizotinib followed by alectinib in ALK+ NSCLC. In:2018.
- 7.Gainor JF, Dardaei L, Yoda S, et al. Molecular Mechanisms of Resistance to First- and Second-Generation ALK Inhibitors in ALK-Rearranged Lung Cancer. Cancer Discov. 2016;6(10):1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shaw AT, Felip E, Bauer TM, et al. Lorlatinib in non-small-cell lung cancer with ALK or ROS1 rearrangement: an international, multicentre, open-label, single-arm first-in-man phase 1 trial. Lancet Oncol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaw AT, Solomon BJ, Besse B, et al. ALK Resistance Mutations and Efficacy of Lorlatinib in Advanced Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer. J Clin Oncol. 2019:JCO1802236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoda S, Lin JJ, Lawrence MS, et al. Sequential ALK Inhibitors Can Select for Lorlatinib-Resistant Compound ALK Mutations in ALK-Positive Lung Cancer. Cancer Discov. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shaw AT, Friboulet L, Leshchiner I, et al. Resensitization to Crizotinib by the Lorlatinib ALK Resistance Mutation L1198F. N Engl J Med. 2016;374(1):54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katayama R, Friboulet L, Koike S, et al. Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib. Clin Cancer Res. 2014;20(22):5686–5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oxnard GR, Hu Y, Mileham KF, et al. Assessment of Resistance Mechanisms and Clinical Implications in Patients With EGFR T790M-Positive Lung Cancer and Acquired Resistance to Osimertinib. JAMA Oncol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aggarwal C, Thompson JC, Black TA, et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non-Small Cell Lung Cancer. JAMA Oncol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dagogo-Jack I, Brannon AR, Ferris LA, et al. Tracking the Evolution of Resistance to ALK Tyrosine Kinase Inhibitors through Longitudinal Analysis of Circulating Tumor DNA. JCO Precis Oncol. 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCoach CE, Blakely CM, Banks KC, et al. Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non-Small Cell Lung Cancer. Clin Cancer Res. 2018;24(12):2758–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odegaard JI, Vincent JJ, Mortimer S, et al. Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin Cancer Res. 2018;24(15):3539–3549. [DOI] [PubMed] [Google Scholar]
- 18.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20(12):1479–1484. [DOI] [PubMed] [Google Scholar]
- 19.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1(19):e87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17(3):251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol. 2016;2(8):1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okada K, Araki M, Sakashita T, et al. Prediction of ALK mutations mediating ALK-TKIs resistance and drug re-purposing to overcome the resistance. EBioMedicine. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goyal L, Saha SK, Liu LY, et al. Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion-Positive Cholangiocarcinoma. Cancer Discov. 2017;7(3):252–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russo M, Siravegna G, Blaszkowsky LS, et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov. 2016;6(2):147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brinkmann K, Enderle D, Flinspach C, Meyer L, Skog J, Noerholm M. Exosome liquid biopsies of NSCLC patients for longitudinal monitoring of ALK fusions and resistance mutations. In:2018.
- 27.Nilsson RJ, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget. 2016;7(1):1066–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang CH, Lee DH, Lee CO, Du Ha J, Park CH, Hwang JY. Induced protein degradation of anaplastic lymphoma kinase (ALK) by proteolysis targeting chimera (PROTAC). Biochem Biophys Res Commun. 2018;505(2):542–547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.