Significance
Short tandem repeats (STRs) and variable number tandem repeats (VNTRs) are among the most mutable regions of our genome but are frequently underascertained in studies of disease and evolution. Using long-read sequence data from apes and humans, we present a sequence-based evolutionary framework for ∼20,000 phased STRs and VNTRs. We identify 1,584 tandem repeats that are specifically expanded in human lineage. We show that VNTRs originate by short interspersed nuclear element–VNTR–Alu retrotransposition or accumulate near genes in subtelomeric regions. We identify associations with expanded tandem repeats and genes differentially spliced or expressed between human and chimpanzee brains. We identify 52 loci with long, uninterrupted repeats (≥40 pure tandem repeats) as candidates for genetically unstable regions associated with disease.
Keywords: tandem repeat, STR, VNTR, tandem repeat expansion, genome instability
Abstract
Short tandem repeats (STRs) and variable number tandem repeats (VNTRs) are important sources of natural and disease-causing variation, yet they have been problematic to resolve in reference genomes and genotype with short-read technology. We created a framework to model the evolution and instability of STRs and VNTRs in apes. We phased and assembled 3 ape genomes (chimpanzee, gorilla, and orangutan) using long-read and 10x Genomics linked-read sequence data for 21,442 human tandem repeats discovered in 6 haplotype-resolved assemblies of Yoruban, Chinese, and Puerto Rican origin. We define a set of 1,584 STRs/VNTRs expanded specifically in humans, including large tandem repeats affecting coding and noncoding portions of genes (e.g., MUC3A, CACNA1C). We show that short interspersed nuclear element–VNTR–Alu (SVA) retrotransposition is the main mechanism for distributing GC-rich human-specific tandem repeat expansions throughout the genome but with a bias against genes. In contrast, we observe that VNTRs not originating from retrotransposons have a propensity to cluster near genes, especially in the subtelomere. Using tissue-specific expression from human and chimpanzee brains, we identify genes where transcript isoform usage differs significantly, likely caused by cryptic splicing variation within VNTRs. Using single-cell expression from cerebral organoids, we observe a strong effect for genes associated with transcription profiles analogous to intermediate progenitor cells. Finally, we compare the sequence composition of some of the largest human-specific repeat expansions and identify 52 STRs/VNTRs with at least 40 uninterrupted pure tracts as candidates for genetically unstable regions associated with disease.
Short tandem repeats (STRs) and variable number tandem repeats (VNTRs), also referred to as micro- and minisatellites (1, 2), are operationally defined as tandemly repeating units of DNA of 1 to 6 and ≥7 bp in length, respectively (3). The mutation rates among these tandem repeats can be several orders of magnitude higher than the unique portions of the genome, ranging from 10−6 to 10−2 nucleotides per generation in STRs (4, 5). The mutation rate for a given locus can vary widely, while the longest and purest tandem repeat tract often defines the most unstable STRs and VNTRs (6–8). As a result, STRs/VNTRs have long been recognized among the most polymorphic markers of genomes. They are also an important source of genomic instability associated with several human disorders, including repeat expansion disorders, due to their tendency to expand through replication slippage, DNA repair, or nonallelic homologous recombination (9). Tandem repeats can also harbor cryptic disease-causing variation in the form of single-nucleotide variants (SNVs) (10) or short insertions and deletions (indels) (11, 12), emphasizing the importance of accurately predicting both their size and sequence composition.
Despite their established importance in population genetics and disease association, tandem repeats, particularly VNTRs, are among the least characterized forms of genetic variation in the human genome (13, 14). Their repetitive nature and sometimes extreme GC content make them particularly challenging to sequence and assemble with standard whole-genome shotgun sequencing assembly strategies, including next generation sequencing approaches that depend on bridge amplification (15). Their large size, often many kilobases in length, and their inherent instability during clonal propagation through Escherichia coli vectors have limited their accurate representation in the human reference genome, which was largely dependent on hierarchical bacterial artificial chromosome (BAC) clone sequence and assembly (16, 17). As a result, recent surveys of human genomes using orthogonal single-molecule long-read sequencing technologies (18, 19) have shown that the length and number of these repeats have been systematically underestimated. Because their length and purity are critical to determining their mutability and inherent instability, accurate haplotype resolution of larger alleles is important (20).
The goal of this study is 2-fold: 1) produce a high-quality set of haplotype-resolved tandem repeat loci in the human genome with a specific emphasis on those that are misrepresented in the current human reference genome and 2) generate an evolutionary framework for their origin by establishing the likely ape ancestral state of each allele. As a starting point, we leveraged the haplotype-resolved structural variants (SVs) from 3 diverse individuals generated as part of the Human Genome Structural Variation Consortium (HGSVC) (20). Next, we generated haplotype-resolved sequences for the homologous loci in nonhuman primates (NHPs) using a combination of PacBio long reads and 10x Genomics linked reads from individuals of 3 species: chimpanzee, gorilla, and orangutan (21). This comparative analysis allowed us to delineate human-specific tandem repeat expansions and further investigate their potential effects on gene expression and splicing using single-cell and tissue-specific RNA sequencing (RNA-seq) of human and NHP brains. This study represents a step toward accurately sequencing and characterizing the most abundant SV class in the human genome (19), including the identification of candidate loci that may contribute to disease through repeat instability.
Materials and Methods
Genome Sequence Data.
NHP single-molecule, real-time (SMRT) PacBio sequencing data were generated previously from western chimpanzee (Clint) and Sumatran orangutan (Susie) lymphocyte cell lines as described previously (21). We also generated long-read sequence data (for the western lowland reference gorilla Kamilah) using the same PacBio RS II platform (P6v2-C4v2 chemistry; 6-h movies) and the same protocol as the 2 other NHP samples, with the following exception: DNA was sheared at the 45 kbp setting for size selection at 20 kbp or 50 kbp for size selection at 30 kbp. Kamilah was sequenced to a depth of 67-fold sequence coverage. Human SMRT sequence data for human samples CHM13 and Yoruban NA19240 were generated similarly (Washington University, St. Louis) as part of the Human Reference Genome Sequencing Consortium. The 3 children samples were sequenced across multiple sequencing and linking technologies as part of the HGSVC (20); these represent daughters of families of Puerto Rican (HG00733), Yoruban (NA19240), and Han Chinese (HG00514) ancestries. The 10x Genomics chromium (CHRO) data were generated from the same 3 individuals that we had PacBio data for, namely orangutan, gorilla, and chimpanzee (Washington University, St. Louis) as part of the Primate Reference Genome project. The CHRO libraries were generated to approximate coverage of 70-fold (Clint), 52-fold (Kamilah), and 81-fold (Susie), with average molecule lengths of 166,454, 105,984, and 178,993 bp, respectively.
Discovery of Tandem Repeats in Human Haplotypes.
Using SVGain (https://github.com/mchaisso/hgsvg/blob/master/sv/analysis/bin_analysis/SVGain.snakefile), we selected regions of the genome that contained SVs of variable sizes across the 6 human haplotypes from the HGSVC (20). Briefly, the pipeline divides the assembled genomes into bins and identifies regions that have a net gain (i.e., insertion) relative to the human reference of ≥50 bp. The sequence of each SV was padded by 2 kbp and analyzed by RepeatMasker (v.4.0.3) (22) and Tandem Repeats Finder (TRF v.4.07b) (23) to identify repeat motifs from 1 bp to 2 kbp. Ultimately, our set of STRs and VNTRs were projected onto GRCh38.
Phasing of the Tandem Repeat Loci in NHP Genomes.
The 10x Genomics linked-read data were generated for 3 NHPs and processed through the Long Ranger (v.2.2.2) pipeline using the human GRCh38 p12 primary assembly (i.e., the same reference as the one used for the HGSVC analysis) and FreeBayes (24) for SNV calling. The final variant call format (VCF) file contained phased SNV genotypes for each of the 3 NHP samples, with heterozygous phased SNVs being the most informative for long-read partitioning. The binary alignment map and the phased VCF files from 10x Genomics data were analyzed in combination using PhasedSV (20) (https://github.com/mchaisso/phasedsv). SI Appendix has more details.
Comparative Analyses.
Following the sequence annotation with TRF and RepeatMasker, the tandem repeat copy numbers were compared between the 7 human and 6 NHP haplotypes, with the goal of identifying human-specific expansions (HSEs) of tandem repeats. SI Appendix has more details.
We also identified ab initio human repeats; these consisted of loci with tandem repeats content in all human samples and no tandem repeat content in any of the NHPs’ homologous loci. Due to the filters used above for both ab initio and HSE, we expect to observe a set of tandem repeats that are differently expanded in humans while also being ab initio, which leads to some STRs/VNTRs being classified in both categories. Thus, we took into account this redundancy between the 2 categories to avoid double counting in the downstream analyses.
A subset of the tandem repeats is expected to differ in both length and sequence composition from the human genome reference. To estimate the number of reference collapses or misassemblies in GRCh38, we counted STRs/VNTRs with ≥2-fold as many tandem copies in the shortest human haplotype than in GRCh38 (i.e., collapsed regions) and repeat unit sequence with ≤90% sequence identity between the human haplotypes and the reference (i.e., misassembled). Lastly, a tandem repeat locus was classified as polymorphic if its standard deviation of copy numbers across the human haplotypes was ≥10th percentile.
Differential Gene Expression and Splicing Analyses.
Single-cell RNA-seq data from cerebral organoid models of chimpanzee and human were recently used to identify gene expression differences in a cell type-specific manner (25). We used the sets of genes that had higher expression in human (i.e., human up-regulated) or chimpanzee (i.e., chimpanzee up-regulated) organoid models for the following 4 brain cell types: excitatory neurons (n = 912 genes), inhibitory neurons (n = 386), intermediate progenitor cells (IPCs; n = 758), and radial glial (RG; n = 877) (25). We identified overlaps between these genes and the human tandem repeat expansions. Next, we assessed whether the tandem repeat-overlapping genes had a cell-type specificity within the human brain using primary tissue single-cell RNA-seq of 4,261 cells originating from human cortex and medial ganglionic eminence regions (26). Overall, our analysis aimed to identify tandem repeat overrepresentations within these sets of species- and cell type-specific genes. We also conducted differential splicing analyses using bulk RNA-seq data from chimpanzee and human brain tissues (SI Appendix).
Statistical Enrichment Analyses.
Permutation tests.
Tandem repeats were defined as genic when overlapping with the open reading frame (ORF) of a RefSeq gene. We calculated the significance of the observed overlap between our tandem repeat regions and those from various functionally relevant gene sets. The regioneR package (27) was used to calculate the significance of the overlap between utilizing the region coordinates from both sets. The enrichment test is carried out by function overlapPermTest, which conducts permutations of the query regions (i.e., tandem repeat coordinates in our case) over the available GRCh38 coordinate space while inherently controlling for the size as well as the relative distance between regions. We did not allow permutation over the regions of GRCh38 where we did not have sequence information (i.e., the acrocentric arms 13p, 14p, 15p, 21p, and 22p and centromeres). We also did not allow the query regions to be shuffled across chromosomes, thereby preserving the chromosome label of each region. A total of 100,000 permutations were carried out for each enrichment analysis, corresponding to a minimum empirical P value = 0.00001.
Multiple regression analyses.
To formally test for association between tandem repeats and biologically relevant genes or genomic regions, we constructed different generalized linear models (SI Appendix). While in each generalized linear model we tried to control for potential confounding effects that may lead to a false positive association, we cannot exclude entirely the possibility of underlying occult confounding effects as contributors to this.
Pathway enrichment.
We conducted Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway-level enrichment for genes that overlapped with our set of human-specific repeat expansions using the overrepresentation analysis module of WebGestalt (28). Briefly, this approach utilizes a hypergeometric model to calculate the significance of the intersection between 2 gene sets.
Validation Experiments.
We performed 3 independent sequence validation experiments. 1) BAC sequencing: we aligned 199 NHP BAC insert sequences (21) to haplotype-resolved tandem repeat sequences from our 3 NHP genomes. 2) Macaque assembly comparisons: we used the most recent macaque genome long-read assembly (Macaca mulatta; GenBank accession no. GCA_003339765.3) to extract sequence from the regions homologous to the human ab initio and expanded tandem repeats. 3) Validation with orthogonal long-read sequence datasets: we carried out an additional orthogonal validation for STRs and VNTRs from the CHM13 continuous long-read (CLR) assembly using orthogonal high-fidelity (HiFi) circular consensus sequencing data and ultralong Oxford Nanopore Technologies (UL-ONT) sequence reads generated from the same source cell line (CHM13) (29). SI Appendix has more details.
Data Access.
All long-read human and NHP assemblies of the STRs/VNTRs presented in this study were aligned against the GRCh38 and deposited in Zenodo (accession no. 10.5281/zenodo.3401477).
Results
Sequence Characterization of Human Haplotype-Resolved Tandem Repeats.
Analyzing the SVs identified by the HGSVC (20) (Fig. 1), we classify a total of 21,442 as either human STRs (7,036; motif length ≤ 6 bp) or VNTRs (14,406; motif length ≥ 7 bp). After requiring that each STR/VNTR is flanked by ≥100 bp of unique sequence on both ends and completely resolved in at least 2 human haplotypes, we obtain a high-confidence set of 17,494 STRs/VNTRs (Dataset S1), of which 5,729 are STRs and 11,765 are VNTRs; 85% of these loci are copy number polymorphic (Fig. 2 and Dataset S2) and measure 246 bp on average longer than the remaining 15% that are invariable. Introns show the greatest variation in tandem repeat length, while protein-coding sequence, predictably, is the least variable (Fig. 2). Additionally, 56% (n = 9,794) map to the transcribed portion of protein-encoding genes. Specifically, 9,230 STRs/VNTRs overlap introns, which is 1.3-fold higher than null expectation (empirical P value < 1 × 10−5); 289 overlap exons (1.7%), and 104 and 195 map within the 5′ and 3′ untranslated region (UTR) respectively (1.7%).
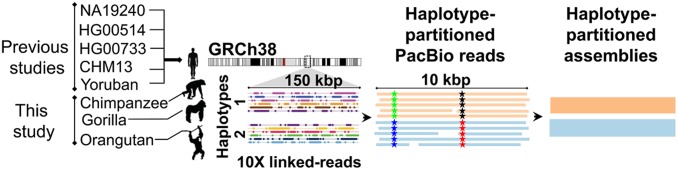
Fig. 1.
Phasing and assembly of STRs/VNTRs. The targeted phasing of tandem repeat sequences in 3 NHPs: chimpanzee, gorilla, and orangutan. 10x Genomics linked reads from each of the great apes were mapped to the human genome reference (GRCh38) followed by the identification of SNVs and phasing of their genotypes using Long Ranger. Next, the phased SNV genotypes were used to partition the PacBio reads of each individual into the 2 parental haplotypes followed by the assembly of 2 haplotype-partitioned contigs per locus. Materials and Methods has more details.
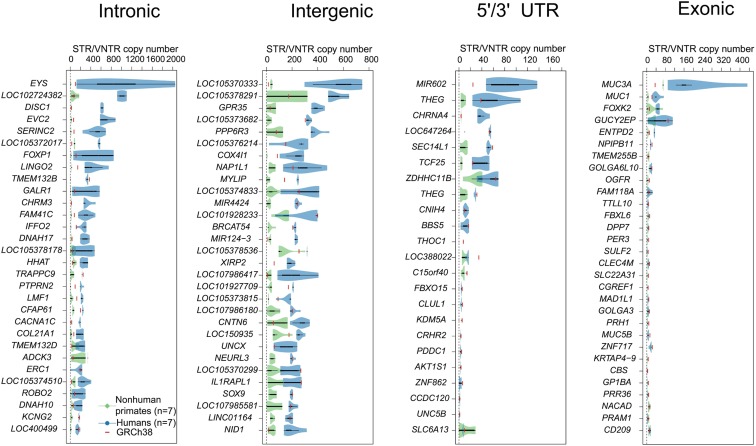
Fig. 2.
Human lineage-specific expansion. The largest human vs. NHP STR/VNTR copy number differences. The top 30 ab initio (no evidence of tandem repeats in other ape genomes) and HSE loci for intronic, intergenic, UTR, and exonic regions, if available, are shown in green (NHP) and blue (human) violin plots, while the solid red lines represent the number of tandem repeat copies for each locus in the human genome reference (GRCh38). GRCh38 carries a significantly shorter allele in 73, 57, 9, and 13% of the intronic, intergenic, 5′/3′ UTR, and exonic loci, respectively. In the case of exonic STRs/VNTRs, we selected additional protein-coding loci from the high-quality STR/VNTR set.
We first compared this set of STRs/VNTRs with the human reference genome (GRCh38) in an effort to identify potential collapses or misassemblies. GRCh38 is on average 111 bp shorter per locus for 6,221 STRs (or the equivalent of 46 tandem repeats) when compared with the HGSVC human haplotypes. Similarly, the reference genome is on average 74 bp shorter or the equivalent of 4 tandem repeat units for 11,279 VNTRs (Datasets S3 and S4), consistent with the previous observation of a systematic underrepresentation of larger alleles in the current reference genome (13). We identified 281 loci (1.6%) that are completely devoid of tandem repeats in GRCh38. Interestingly, 250 (89%) of these map inside retrotransposons (e.g., short interspersed nuclear element–VNTR–Alu [SVA]), which are polymorphically inserted or deleted in the human population, and thus, their absence from GRCh38 is likely the result of normal allelic variation. A relatively small number of simple tandem repeat loci (n = 25) was unaffiliated with mobile elements, and they were present in all human haplotypes but missing from the human reference genome (Dataset S3). For example, a pentameric CCCTC repeat, mapping within the intron and <100 bp away from a splice site of the mutation intolerant gene MGAT5B (probability of loss-of-function intolerance [pLI] = 0.98), ranges from 144 to 188 copies in human haplotypes but is completely absent from GRCh38.
NHP STR/VNTR Characterization.
We assembled the homologous STR/VNTR regions in the genome of NHPs at a single-haplotype resolution by partitioning the PacBio long reads using phasing data obtained from 10x Genomics linked reads generated for chimpanzee, gorilla, and orangutan (Materials and Methods, Fig. 1, and Dataset S5). Of the 17,494 STRs/VNTRs, 16,712 (95.5%) phased successfully in ≥2 NHP haplotypes (Dataset S6). Interestingly, when we compared these haplotype-resolved VNTRs with the previous haplotype-unaware diploid genome assemblies generated from the same individuals, namely panTro6 from Clint the chimpanzee and ponAbe3 from Susie the orangutan (21), we readily identified loci that were longer. In the chimpanzee genome, we identified 2,822 loci where the STR/VNTR alleles were longer in both haplotypes (i.e., h0 and h1) by an average of 103 bp compared with the corresponding sequences in panTro6. We also identified 2,925 loci where the panTro6 assembly contained a larger allele than both phased alleles in chimpanzee by an average of 64 bp. Similarly, for orangutan, we observed 3,337 loci where the STR/VNTR alleles were longer in both h0 and h1 compared with ponAbe3 by an average of 91 bp. The inverse set contained 2,544 STRs/VNTRs, which were longer in the ponAbe3 compared with both assembled haplotypes by 66 bp. These results demonstrate that haplotype-aware assembly of STR/VNTR loci is critical even when long-read sequence data are used; otherwise, diploid assembly will favor the smaller allele or a hybrid that is smaller than both alleles.
To assess the accuracy of our call set, we sequenced 151 STRs/VNTRs from large-insert BAC clones isolated from the same individuals (80 chimpanzee, 12 gorilla, and 59 orangutan) and compared the sequence identity and length concordance of these with the haplotype-resolved assemblies. Overall, the sequences matched with an average sequence identity of 97.5% (Dataset S7). Length concordance, however, showed greater variability, with the BACs tending toward smaller repeat lengths, which would appear as a deletion relative to the assemblies. Overall, BACs contained severalfold more deleted than inserted tandem repeat DNA bases relative to the assemblies; for instance, after controlling for nontandem repeat SVs, we observed a total of 7,116 bp of deletions and 1,324 bp of insertions in the chimpanzee BACs, 416 bp of deletions and 38 bp of insertions in the gorilla BACs, and 347 bp of deletions and 210 bp of insertions in the orangutan BACs. After requiring ≥99% length and sequence concordance (Materials and Methods) between BACs and our assemblies, the validation rate for all 151 loci was 77.8% across the 3 NHPs. We consider this validation rate a lower bound due to the clonal instability of longer VNTRs and STRs (29). One VNTR locus that failed to validate, for example, contained a 1.2-kbp insertion of an E. coli mobile element encoding for insertion sequence 3 (IS3)-like element IS2 family transposase that inserted precisely into the expanded VNTR locus (SI Appendix, Fig. S1), highlighting potential propagation artifacts.
Given the limitations of BACs in validating PacBio assemblies of tandem repeats, we used 2 additional orthogonal technologies for length concordance and sequence identity validations (Materials and Methods): PacBio’s recent HiFi sequencing reads and UL-ONT reads. While the data assembled for our study originated from PacBio’s CLR sequencing, HiFi circular consensus sequence and UL-ONT data were recently generated from the same CHM13 hydatidiform cell line (29). This allowed us to compare sequence length and compositional accuracy for 1 human haplotype using 3 different sequencing platforms. Of the 17,494 STRs/VNTRs, 17,132 (98%) assembled successfully in both HiFi and CLR assemblies of CHM13. Of the assembled loci, similar proportions of 98.2% for HiFi and 98.0% for CLR were validated by UL-ONT (i.e., differed in length by ≤5%) (Materials and Methods). Next, we identified all of the STR/VNTR loci that showed discrepancy in length between HiFi and CLR (i.e., >1% length discordance) and compared each with the UL-ONT reads to determine which of the 2 assemblies was more consistent. This set consisted of 513 discordant STRs/VNTRs (3% of the call set), and higher length concordance with UL-ONT was observed in the case of HiFi (455 or 89%) than CLR (402 or 78%) (Dataset S8). Lastly, we determined the sequence accuracy of our CLR data via a direct comparison of sequence between CLR and individual high-quality HiFi reads (error of less than 1 in every 1,000 bases) (Materials and Methods). Ultimately, of the 17,132 regions with available assemblies in both HiFi and CLR, 15,251 (89%) were both length and sequence concordant by ≥99%. The 11% of the STR/VNTR regions that did not validate by HiFi clustered primarily near the centromere and the pericentromeric regions; ∼4.1-fold as many loci situated ≤1 Mbp of the centromere as expected by random chance (χ2 P value = 7.3 × 10−15).
Human-Specific Expansions.
Next, we classified the human STRs/VNTRs into 2 categories. Human ab initio tandem repeats (or simply ab initio) are tandem repeats where there is no evidence of tandem repeats in the orthologous location in the 3 NHP genomes. HSEs, in contrast, are those where there is an expansion in length in the human haplotypes compared with the NHP haplotypes (Materials and Methods). We classify 1,151 HSEs and 640 ab initio STRs/VNTRs, of which 207 belong to both categories, for a total of 1,584 nonredundant loci (Dataset S9). VNTRs, in particular, predominate, representing 71% of loci expanded in the human lineage (70% ab initio and 72% HSE), and these map preferentially to intronic regions (57% for ab initio and 59% for HSE; an enrichment ratio of 1.46 compared with the null expectation; empirical P value < 1 × 10−5). Both of the categories were overall particularly GC rich (Fig. 3E).
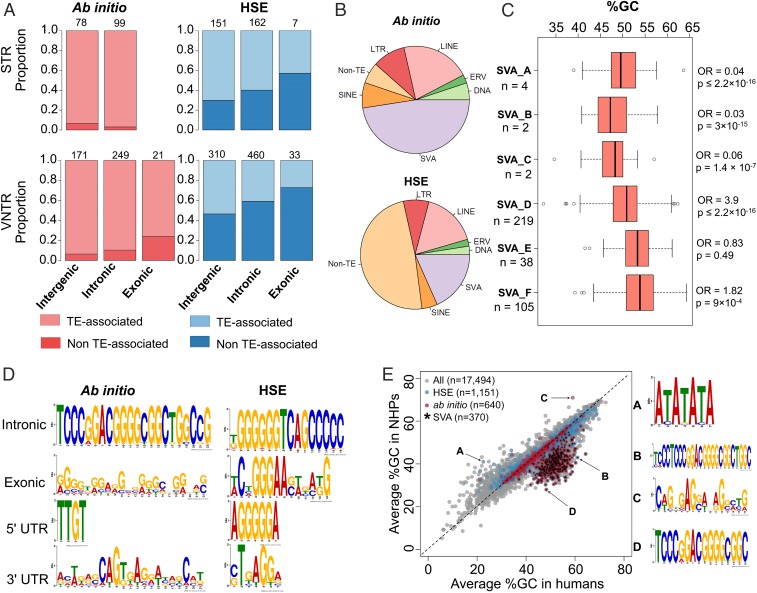
Fig. 3.
Sequence properties of human-expanded STRs/VNTRs. Ab initio and HSE tandem repeats have distinct sequence composition. (A) The relative abundance of STRs and VNTRs was broken down by position relative to a gene (i.e., intergenic, intronic, and protein coding). The lighter color corresponds to transposable element (TE)-associated tandem repeats, while the darker color corresponds to simple (i.e., non–TE-associated) tandem repeats. (B) Pie charts for the ab initio (Upper) and HSE (Lower) tandem repeats. The labels correspond to short interspersed nuclear element (SINE), long interspersed nuclear element (LINE), long terminal repeat (LTR), endogenous retrovirus (ERV), and transposons (DNA). (C) Boxplots of the percentage of GC content for all HSE and ab initio tandem repeats that are SVA elements. The total counts for the SVA elements are shown by each of the 6 subfamilies (A to F), and the ORs and P values on right-hand side were calculated using Fisher’s exact test on the observed SVA counts in our call set compared with their relative abundance in GRCh38 (i.e., 1,128 SVA_A, 848 SVA_B, 501 SVA_C, 1,546 SVA_D, 701 SVA_E, and 1,026 SVA_F). An OR > 1 represents an enrichment. The boxplots use the GC content from all of the assembled human sequences. (D) A multiple expectation maximization for motif elicitation (MEME) analysis using the sequences categorized by functional annotation with respect to the gene body. (E) A comparison of average percentage of GC content for all STR/VNTR loci in our call set. The different characters in the scatterplot correspond to A = HSE tandem repeats that are not SVA associated, B = HSE tandem repeats that are associated with SVAs, C = ab initio tandem repeats that are not associated with SVAs, and D = ab initio tandem repeats that are associated with SVA elements. The MEME motifs are shown for each of the 4 categories.
Since we are comparing a relatively small number of individuals, where not all haplotypes are always fully resolved, we quantified to what extent missing human and NHP haplotype data might confound our classification of HSE and ab initio loci. We considered 6 different nonmissing thresholds (i.e., ≥1, ≥2, …, ≥6 assembled samples) for both human and NHP haplotypes separately. For each of the 6 thresholds, we observed the following HSE and ab initio proportions: 9.2, 9.2, 9.1, 9.1, 8.9, and 9.2% (SI Appendix, Fig. S2). These proportions of repeat expansions were not significantly different from each other (χ2 P value > 0.05), suggesting that missing sequence data introduced minimal bias in the discovery of ab initio and HSE loci.
As a final test of potential false positives and evolutionary misclassification, we compared a set of 1,584 HSE and ab initio tandem repeat expansions with the latest long read-based assembly of the macaque, which serves as a primate outgroup to the ape lineages (Materials and Methods). We successfully mapped 1,407 loci (88.8%) uniquely to macaque, with the majority of those that failed to map (97%) originating from large retrotransposable elements or highly divergent sequence between macaque and human. From the set of 1,036 HSEs with available homologous sequence, 98.5% remain classified as HSEs, with only 15 loci failing to validate (i.e., the macaque genome contains a larger tandem repeat than the largest NHP haplotype-resolved sequence). Of the 551 human ab initio loci with available macaque homologous sequence, 115 showed the presence of a tandem repeat at the orthologous location, which was counted as evidence for false positives, resulting in a validation rate of 79%. We note, however, that nearly all macaque tandem repeats are more divergent than expected (<95% sequence identity), raising the possibility of homoplasy at these particular positions.
Among VNTRs, we find that most HSE and ab initio events (91.7%) associate with retrotransposons in contrast to STRs, where this association is less pronounced (50.4%) (Fig. 3A). This 1.82-fold retrotransposon enrichment for VNTRs over STRs is significant (Fisher’s exact test P value < 2.2 × 10−16). Predictably, SVA retrotransposons (Fig. 3B) represent the most abundant substrate of ab initio (47%) compared with HSE (18%) elements. Because different classes of SVA have been active at different time points during ape evolution, we considered enrichment by SVA subtype (A through F) (30) across the combined set of ab initio and HSE events (n = 1,584) and observe the greatest enrichment for the most abundant subfamilies SVA-D (odds ratio [OR] = 3.9, P value < 2.2 × 10−16) and SVA-F (OR = 1.8, P value = 9 × 10−4) (Fig. 3C). The SVA-D subfamily is estimated to have emerged ∼9 to 10 million years ago, shortly after Asian and African apes diverged, while SVA-F is human specific (30–32). We searched for specific sequence motifs associated with human lineage repeats using MEME. Consistent with their SVA origin, intronic HSE and ab initio VNTRs are enriched in high-GC composition sequence motifs and in particular, long stretches of homopolymer guanines and cytosines (Fig. 3D), resulting in the highest differential GC content of all tandem repeats (Fig. 3E). While far fewer in number, non-SVA HSE events are almost exclusively AT rich (Fig. 3E).
We previously reported a strong enrichment for VNTRs within subtelomeric chromosomal regions based on a sequence-resolved SV analysis from 15 PacBio genomes (19). We reassessed this effect for the genome-wide distribution of human haplotype-resolved VNTRs (SI Appendix, Fig. S3) after adjusting for retrotransposable element origin, size of tandem repeat, and all pairwise interactions. While we continue to observe a strong subtelomeric enrichment for VNTRs relative to STRs (OR = 4.53 [3.6 to 5.7], P value < 2.2 × 10−16), this effect is driven almost exclusively by VNTRs not associated with SVA elements (Materials and Methods). This association is further supported by the observation of a strong subtelomeric depletion for STRs/VNTRs originating from retrotransposons, such as SVA (OR = 0.45 [0.36 to 0.56], P value = 6.6 × 10−12). We also tested whether STRs or VNTRs were differentially enriched with respect to genes. After adjusting for the subtelomeric distribution bias and other covariates (Materials and Methods), we observe a small but significant gene enrichment for VNTRs over STRs (OR = 2.23 [1.11 to 4.48], P value = 0.02) and a significant gene depletion for all retrotransposon-associated tandem repeats (OR = 0.57 [0.38 to 0.85], P value = 0.006). These results suggest 2 distinct evolutionary trajectories: VNTRs not associated with retrotransposons have a propensity to cluster near genes within the subtelomeric regions of chromosomes, while retrotransposons, primarily SVAs, serve as a vector to distribute them more uniformly across the genome with a bias against genic regions.
Gene and Differential Expression Analyses.
Numerous studies have implicated expanding STRs and VNTRs as regulators of transcription (12, 33, 34). Using the KEGG pathways, we initially conducted an overrepresentation analysis using the combined set of genes overlapping with our ab initio and HSE tandem repeats. Two pathways, in particular, show a striking enrichment by this analysis: dopaminergic synapse (enrichment ratio = 4.0, false discovery rate (FDR)-adjusted P value = 6.7 × 10−4) and the glutamatergic synapse (enrichment ratio = 4.02, FDR P value = 0.0013) (SI Appendix, Fig. S4). Consistent with this observation, the tissue-specific enrichment analyses suggest that the brain is particularly enriched for ab initio and HSE genes (SI Appendix, Fig. S5). Next, we performed a more detailed overrepresentation analysis between our STR/VNTR calls and differentially expressed genes between the human and chimpanzee brains at the single-cell level. Briefly, we used the gene expression differences estimated from single-cell RNA-seq data from human and chimpanzee cerebral organoid models (25) for 4 different cell types: RGs, excitatory neurons, inhibitory neurons, and IPCs. These measurements represent spatiotemporal proxies of gene expression in the primate brain at a single-cell resolution.
We observe 3 interesting and potentially related trends. First, we find a significant overrepresentation of subtelomeric STRs/VNTRs in human up-regulated RG genes (OR = 1.78 [1.33 to 2.38], P value = 0.0001) (SI Appendix, Fig. S6 and Dataset S10). Conversely, there is a depletion observed between human down-regulated RG genes and subtelomeric STRs/VNTRs (OR = 0.58 [0.38 to 0.88], P value = 0.01). Second, we observe an overrepresentation of VNTRs that overlap with gene enhancers (OR = 8.63 [1.73 to 157], P value = 0.038) as defined by the GeneHancer database (35) in human up-regulated excitatory neuronal genes. In contrast, human down-regulated inhibitory neuronal genes are enriched for ab initio repeat expansions (OR = 3.76 [1.05 to 10.5], P value = 0.02). Third, human up-regulated genes in IPCs are significantly overrepresented for subtelomeric tandem STRs/VNTRs (OR = 2.21 [1.59 to 3.08], P value = 2.5 × 10−6). Overall, our results suggest that tandem repeats have likely impacted gene expression in progenitor and neuronal cells differently depending on their cell type and evolutionary trajectory. Subtelomeric tandem repeats (primarily devoid of retrotransposable elements and enriched for VNTRs) are associated with the up-regulation of genes in human RG cells and excitatory neurons, especially tandem repeats associated with enhancers. In contrast, human ab initio repeat expansions (associated with SVA retrotransposon) are associated with the down-regulation of genes in inhibitory neuronal cells.
These genes and human lineage-specific tandem repeats represent candidates for future investigation and functional testing (Dataset S11). For example, one of the most differentially expressed RG genes, VPS53, overlaps a subtelomeric VNTR and has a 1.61-fold higher expression in human compared with chimpanzee RG cells (P value < 2.2 × 10−16). Furthermore, VPS53 shows a modest enrichment in RG compared with other cells types (adjusted P value = 0.0076) (Dataset S11). The most human up-regulated gene with an enhancer-overlapping VNTR is the autism-implicated gene TRIO (1.41-fold increase relative to chimpanzee, P value = 5.7 × 10−14); TRIO is particularly enriched in excitatory neurons (P value = 2 ×10−5) compared with other human brain cell types. The gene SVIL contains an SVA-mediated ab initio repeat in its intron, and it shows a 1.73-fold reduction in expression in human relative to chimpanzee excitatory neurons (adjusted P value = 0.0022). Interestingly, this gene is also depleted in human RG cells compared with other human brain cells (adjusted P value = 1 × 10−6). TBC1D22A is a human up-regulated gene in IPCs (ratio = 1.63-fold, P value = 6.22 × 10−15), and it contains a human-expanded VNTR in its intron. This gene is broadly expressed in other human cell types.
Differential Gene Splicing Analysis.
Since expanded tandem repeats are known to alter transcript splicing patterns (36), we tested for an association between expanded intronic STRs/VNTRs and the genes that we identified as differentially spliced between human and chimpanzee striatum and cortex using tissue-specific RNA-seq datasets. A tandem repeat expansion is considered to overlap with a differentially spliced transcript only if the STR/VNTR occurs in an intron located in between a pair of differentially spliced exons. Among the 656 intronic HSE tandem repeats, we observe a small fraction overlapping with differentially spliced transcripts in cortex (20 or 3%) and striatum (11 or 1.7%). Similarly, of the 378 intronic ab initio events, 12 (3.2%) and 6 (1.6%) overlap differentially spliced transcripts in cortex and striatum, respectively. These proportions are not significantly different from expectation (i.e., the null expectations are 2.6 and 1.8% for each brain tissue). Next, we further investigated whether a more direct link could be established between the expanded STR/VNTR sequences and the altered splicing at that locus. Using SpliceAI (37), we searched specifically for splice donor and acceptor loss or gain mutations occurring within the tandem repeat sequence. The SpliceAI scores predict a high likelihood for cryptic splice variants within 2 HSE VNTRs (overlapping genes PTPRN2 and NTRK2) and 2 ab initio VNTRs (overlapping genes GPR176 and PIGQ). Interestingly, NTRK2 has a high mutational intolerance score (pLI > 0.99), while GPR176 is differentially spliced in the cortex but not in the striatum, suggesting potential tissue specificity for the differential splicing (SI Appendix, Fig. S7).
Disease-Associated Genes and Candidates for Instability.
We tested for association between STRs/VNTRs and genes that overlap disease-causing CNVs and are enriched for de novo point mutations in autism spectrum disorder (38). We observe a nominal overrepresentation of enhancer-overlapping STRs/VNTRs (OR = 1.67 [1.0 to 2.65], P value = 0.038) as well as a nominal depletion for retrotransposable STRs/VNTRs (OR = 0.72 [0.53 to 1.0], P value = 0.048). When we expand our test for enrichment to all Simons Foundation Autism Research Initiative (SFARI) autism genes, we observe a more significant depletion for retrotransposable STRs/VNTRs (OR = 0.76 [0.67 to 0.87], P value = 3.4 × 10−5), suggesting that these highly conserved genes are biased in their proximity to expanded STRs/VNTRs. Next, we tested for any STR/VNTR subset overrepresentation within tandem repeats that occur ≤2 kbp of genome-wide association study (GWAS) single-nucleotide polymorphisms (SNPs; n = 1,719) (Dataset S12); we observed a significant overrepresentation of enhancer-overlapping STRs/VNTRs relative to STRs/VNTRs that do not overlap enhancers (OR = 4.34 [1.77 to 10.2], P value = 0.001), which decreased sharply to OR = 1.8 over a distance of 10 kbp (SI Appendix, Fig. S8). However, after controlling for the inherent enrichment for gene enhancers near GWAS SNPs (39), we no longer observed a significant enrichment for enhancer-overlapping tandem repeats; specifically, 12.9% of all published GWAS SNPs (n = 104,746) overlapped enhancers, which is more than expected by random chance (OR = 1.47 [1.42 to 1.52], P value < 2.2 × 10−16) using 1,000 permutations of the enhancer coordinates. A comparable 12.7% of all GWAS-overlapping STRs/VNTRs also overlapped enhancers. Hence, the association between GWAS-overlapping STRs/VNTRs and enhancers is driven by the inherent bias in GWAS markers to localize in proximity to gene enhancers.
We selected 38 tandem repeat loci that are known to be involved in genomic instability and disease and investigated their sequence composition in more detail (Dataset S13). Some of these loci have been studied previously, and their patterns of evolutionary and human variation are better understood (e.g., FMR1 and HTT), while others have been only recently discovered, such as the disease-causing CAG triplet repeat loci (XYLT1) (40) (SI Appendix, Fig. S9). In general, we faithfully reconstruct the sequence composition of the disease-associated repeats in both humans and NHPs. For example, we assembled normal alleles of the trinucleotide repeat locus responsible for fragile X syndrome (41), which maps to the promoter of FMR1 (SI Appendix, Fig. S9A). Most of the human alleles contain 1 or 2 AGG interruptions occurring with a periodicity of once every 9 or 10 CGG repeats, while NHPs carry up to 4 AGG interruptions and concomitantly carry shorter uninterrupted CGG tracts. This is consistent with previous comparative studies (42). Similarly, the largest representation of the protein-encoding MUC1 locus (SI Appendix, Fig. S9B) contains up to 72 tandem repeat copies of a 60-mer, consistent with known patterns of human variation. This is in contrast to the largest NHP haplotype, which contains only 19 copies of the same motif, and thus, we classify this locus as an HSE (Materials and Methods). With this level of resolution, it should be possible to readily identify a 1-bp insertion in one of the copies of this 60-mer that disrupts the ORF leading to medullary cystic kidney disease type I(11). We assembled a disease-associated VNTR, located in the protein-coding portion of MUC21, and it is composed of 27 to 31 copies of a 45-mer in the human haplotypes and 19 to 37 copies of the same 45-mer in NHP haplotypes (SI Appendix, Fig. S9M). A 4-bp deletion in this VNTR has been shown to increase disease risk for diffuse panbronchiolitis (43).We also assembled a VNTR (2) that has been associated with type I diabetes (44). This locus is situated in the promoter region of INS and consists of 41 to 144 tandem copies of a 14-bp motif in our human haplotypes and 3 to 21 copies of a 15-bp motif in the NHP haplotypes (SI Appendix, Fig. S9N). Both 14- and 15-bp VNTR motifs have been previously reported in humans (45). While most normal alleles associated with genomic instability and disease were sequence resolved, there were some notable exceptions. Only 5 of the potential 12 haplotypes corresponding to the lipoprotein A (LPA) Kringle-IV VNTR were assembled, and most of these were at the lower range of the variable ∼5.5-kbp tandem repeat motifs, each containing multiple exons of the LPA gene (46, 47). Sequence analysis suggests that most other haplotypes are only partially resolved or collapsed (SI Appendix, Fig. S9L).
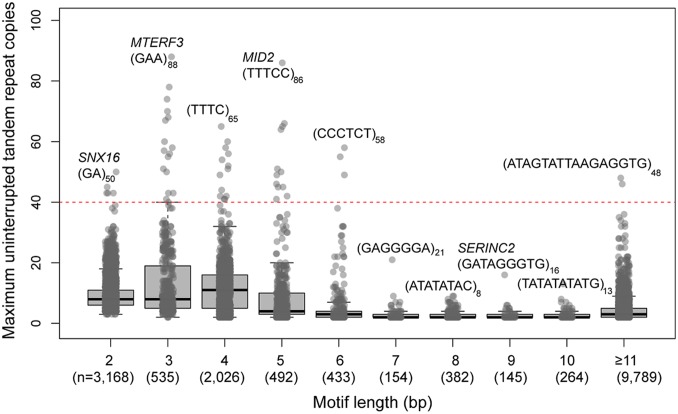
Since long tracts of uninterrupted repeats are a hallmark of genomic instability for many disease-associated loci (7, 48), we specifically searched for tandem repeats that contained a long pure tract (LPT) in excess of 39 pure repeat units (Fig. 4). We identified 52 loci based on our examination of 6 human haplotypes (Dataset S14). Almost all are STRs (50 or 96%), and most (31 or 60%) map within the intron of genes. We classify 31% (n = 16) as either ab initio or HSEs, representing a 3.4-fold enrichment over the null expectation of 9% (χ2 P value = 0.01). The human reference genome (GRCh38) is shorter for 41 (79%) of these loci, with 6 loci showing ≥2-fold difference in length between the newly assembled haplotypes and the reference assembly. Eleven of these outliers map within the introns of genes highly intolerant to mutation (pLI ≥ 0.90). The largest LPT region in terms of copy number units maps within MTERF3 (88 tandem copies of a GAA triplet repeat) (Fig. 4), while the overall largest LPT in terms of length is located in the intron of CLCN5 (46 copies of a 26-mer repeat unit). CLCN5 is a mutationally intolerant gene (online mendelian inheritance in man [OMIM] identification no. 300008, pLI = 0.989) associated with various X-linked recessive disorders, including Dent disease (phenotype OMIM no. 300009), hypophosphatemic rickets (phenotype OMIM no. 300554), Nephrolithiasis type I (phenotype OMIM no. 310468), and proteinuria with hypercalciuric nephrocalcinosis (phenotype OMIM no. 308990). We consider expansion of these uninterrupted tracts as candidates for future investigations into genome instability associated with disease, including missing heritability (40).
Fig. 4.
The longest pure repeat tract length. Distributions are shown using boxplots for motif sizes of ≥2 bp. The motif sequence and the corresponding number of tandem repeats are shown for the longest pure repeat tract, and the gene name is shown for intronic STRs/VNTRs only. The dotted horizontal line corresponds to the 40-tandem repeat copies threshold used to identify longest pure tracks. The numbers (n) in parentheses on the x axis correspond to the total numbers of tandem repeats observed for a given motif size. Motifs ≥11 bp were binned into 1 boxplot.
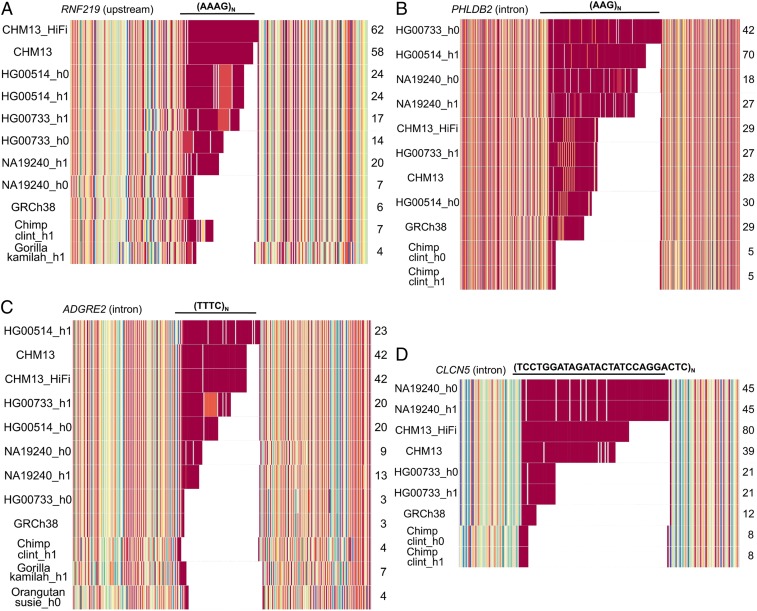
We further characterized the structure and sequence composition of these repeat loci by developing a k-mer compositional visualization for human and NHP haplotypes (Materials and Methods, Fig. 5, and SI Appendix, Fig. S10). The analysis revealed some interesting properties regarding the structure and mutability of these loci. First, the overall longest alleles did not always carry the longest tract of pure repeats due to the presence of multiple interruptions (e.g., PHLDB2) (Fig. 5B). Second, the pattern of interruptions often differs between species and even between human haplotypes (e.g., ADGRE2) (Fig. 5C). Third, secondary tandem repeat loci embedded within the primary repeat are not uncommon and often differ by a mutation of a single base pair of the original motif, which becomes subsequently expanded [e.g., RNF219, where the tetranucleotide (AAAG)n in some haplotypes becomes (AAGG)n or simply a run of homopolymer adenines] (Fig. 5A). Lastly, we observe 1 locus that contains a higher-order tandem repeat reminiscent of patterns observed for centromeric satellites on a smaller scale (SI Appendix, Fig. S10 A and Q).
Fig. 5.
STR/VNTR sequence composition plots. The 4 loci represent STRs/VNTRs with ≥40 tandem repeat copies. The sequences from each human and NHP haplotype were colored according to their k-mer abundance (Materials and Methods). For the CHM13 sample, sequences from both CLR and HiFi assemblies have been included labeled as “CHM13” and “CHM13_HiFi,” respectively, which provide a replicate measure of sequence accuracy. (A) An STR located upstream of RNF219 is composed of 7 to 62 uninterrupted tandem copies of an AAAG expansion in humans. Three human haplotypes contain clustered AAGG interruptions, while 1 chimpanzee haplotype contains a clustered interruption of AG repeats. (B) A human-specific STR expansion is located in the intron of PHLDB2 and is composed of 18 to 70 uninterrupted repeats of AAG. Periodic interruptions of AGG exist in 3 human haplotypes and GRCh38. (C) An STR located in the intron of ADGRE2 contains 3 to 42 uninterrupted tandem repeat copies of TTTC. A cluster of a continuous tract of pure TC repeats interrupts the tetranucleotide repeat in 1 of the Puerto Rican haplotypes. (D) A human-specific VNTR expansion is located in the intron of CLCN5 and is composed of 21 to 80 tandem repeat copies of a 26-bp motif. A single interruption by a 30-bp motif that contains 3 additional adenines in position 23 occurs in the Puerto Rican and Yoruban haplotypes (gray bars).
Discussion
In this study, we have developed a framework to study the evolution and mutability of tandem repeats in human and NHP genomes. Such regions are frequently misassembled or incomplete in the reference genome. As a source of human genetic variation, these elements are often overlooked in disease association studies. We characterized 17,494 haplotype-resolved human STRs/VNTRs with respect to their orthologous alleles in 3 primate outgroups using a uniform strategy that involves phasing long reads with 10x Genomics data followed by sequence assembly of the locus. The analysis allows us to readily identify HSEs as well as investigate the origin of one of the most mutable classes of human genetic variation (49). Our analysis of the apes suggests 2 distinct evolutionary trajectories. The first involves the dispersal of particularly GC-rich minisatellites throughout the genome via retrotransposition. SVA element insertions, in particular, frequently lead to the formation of ab initio VNTRs with a bias against genic regions (Fig. 3). The second trajectory involves the emergence and expansion of both STRs and VNTRs, particularly within the introns of genes. Such events accumulate in the last 5 Mbp of ape chromosomes, likely as a result of increased double-strand breakage and male meiotic recombination (19).
Because differences in VNTR and STR lengths have been associated with gene expression and transcript splicing differences, we searched for potential association based on available chimpanzee and human transcriptomic datasets (25). In general, we find that human-expanded STRs/VNTRs mapping to known gene enhancers associate with human up-regulated genes, especially in excitatory neurons, supporting the idea that repeat copy number may modulate enhancer activity (50). The same proportion of ∼12.7% GWAS-associated STRs/VNTRs overlapping known gene enhancers is frequently found to correspond to GWAS loci alone. For instance, we identify a previously described enhancer-overlapping CACNA1C VNTR (a human-specific enhancer) located near the GWAS SNP rs117888112 (12). In addition, we identify 1,719 other GWAS STR/VNTR associations, 121 of which are located nearby HSEs and 29 near ab initio repeat expansions (4 of these are shown in SI Appendix, Fig. S11). One such locus is rs407203, previously associated with esophageal and gastric cancer and located ≤2 kbp from an MUC1 HSE VNTR (SI Appendix, Fig. S9B and Dataset S12). Another is GWAS SNP rs2526882, which has been recently associated with schizophrenia (51) (Dataset S12). This finding suggests that regions of recent and rapid evolutionary change frequently correspond to sites of disease association. These loci represent candidates for experimental follow-up, such as more detailed linkage disequilibrium analysis between GWAS SNPs and the sequence content of the STR/VNTR alleles or experimental evaluation of the effects of the expanded VNTR on splicing or expression.
Other disease-associated tandem repeat loci are tagged by GWAS SNPs and have more complex effects than simple gene expression differences. One such example includes the CTG repeat expansion in the intron of TCF4, which when expanded, is known to cause Fuch’s endothelial corneal dystrophy (FECD) (52). This triplet repeat is successfully assembled in all human and NHP haplotypes. The first report of a GWAS signal associated with FECD originally suggested an intronic SNP in TCF4 (namely rs613872-G) as the most strongly associated risk allele (i.e., OR = 5.47) (53). Two years later, it was shown that the size of a CTG repeat located ∼43 kbp downstream from that GWAS SNP was highly predictive of disease status (54). Thus, the original risk SNP was simply tagging the pathogenic CTG repeat locus. The hyperexpanded allele of this triplet repeat was recently shown to sequester splicing factors and increase RNA foci accumulation in the nucleus (55).
Similarly, the well-known amyotrophic lateral sclerosis (ALS)-associated gene C9orf72 harbors 2 genome-wide significant SNPs (rs10122902 and rs3849942) (56), both of which are in the same linkage disequilibrium block as the hexanucleotide repeat CCGGGG. This repeat, which has been shown to expand in up to half of familial ALS cases, causes the loss of one C9orf72 isoform while increasing repeat-containing RNA foci in the nucleus (57, 58). This VNTR was resolved in all 6 of the human haplotypes and 5 of the 6 NHP haplotypes, with lengths ranging from 30 to 90 bp and from 48 to 60 bp, respectively. It is interesting that normal-sized alleles were not properly represented in the original versions of the human genome. These examples stress the importance of high-quality sequencing of these loci and obtaining population-level STR/VNTR genotypes in large and diverse cohorts in order to more finely map additional GWAS SNPs.
Sequence compositional analysis (Fig. 5 and SI Appendix, Fig. S10) shows considerable evolutionary turnover in the structure and length of tandem repeats between and within species. A common feature of most tandem repeats is the presence of interruptions that disrupt the longest tract of pure repeats. These interruptions typically differ by a single-base pair mutation of the major motif and can lead themselves to the formation of secondary VNTRs/STRs or higher-order structures embedded within or immediately adjacent to the repeat. Remarkably, these structures have the potential to vary dramatically between human haplotypes. It is interesting that tracts of uninterrupted repeats (≥40 units) are relatively rare in the 6 human haplotypes, with only 52 (0.3%) of such loci being identified. Almost all (96%) of these longer pure alleles are restricted to the STRs (≤6-bp motifs), suggesting that VNTRs are inherently more heterogeneous, possibly as a result of frequent interallelic recombination during meiosis, which leads to truncated or scrambled motifs (49). Indeed, VNTRs were relatively depleted among the long tracts of uninterrupted repeats (OR = 0.038 [0.025 to 0.055], P value < 2.2 × 10−16). The variance in the number of tandem repeat copies was ∼115 times higher in the case of loci with ≥40 compared with <40 uninterrupted repeats (Wilcoxon rank sum test, P value < 2.2 × 10−16), indicating that the longer the uninterrupted tracts, the more variable their size, consistent with their increased instability. The loss of interruptions and the concomitant increase in tract length are important considerations for mutability, slipped strand structure formation, and disease risk (7, 59). The loss of AGG interruptions, for example, associates with smaller unstable premutations, leading to fragile X syndrome (7, 60). Similarly, loss of CAA interruptions in the Huntington (HTT) CAG repeat increases instability, leading to an association between shorter alleles and an earlier age of onset (48). We predict that the loci with the longest pure tracts of repeats that we have identified will be among the most unstable and represent candidates for disease association.
As part of this study, we have haplotype resolved the sequence structure of many clinically relevant STRs/VNTRs in the normal population (SI Appendix, Fig. S9). Based on our validation experiments, the level of accuracy will not only be sufficient to delineate pathogenic contractions or expansions but also has the potential to distinguish internal variants that have often taken decades to deduce (11). For example, we have successfully sequenced full-length VNTR alleles corresponding to MUC1, MUC21, and ABCA7, where single-base pair insertions or indels of the tandem repeat are associated with medullary cystic kidney disease type I(11), panbronchiolitis (43), and increased Alzheimer disease risk (36), respectively. An important experiment going forward will be to determine how accurately and at what level of accuracy such disease-associated variants can be routinely detected in both long- and short-read datasets. As a test of feasibility, we selected 687 of the most human polymorphic and largest VNTRs and used our sequence-resolved alleles to generate all possible 30-bp k-mers. Each k-mer was then mapped against GRCh38 using an edit distance of 2 (61) to account for polymorphisms and potential sequencing errors. We were able to identify uniquely mappable k-mers for 681 (99.1%) of the loci, suggesting that nearly all of these large VNTRs may be at least partially traversable by short genome sequencing reads and allowing smaller internal variants to be deduced as long as a sufficient number of full-length alleles has been resolved to accurately represent their composition.
Despite these advances, there are several limitations. First, not all VNTR loci are sequence resolved. Most notable was the poor recovery of the LPA VNTR, where copy number variation of the protein-coding region is associated with coronary heart disease risk (47). The multiple-exon ∼5.5- to 5.6-kbp repeat of LPA is one of the largest VNTRs in the human population and will likely require longer and/or more accurate sequence reads to fully sequence resolve (62, 63). Second, we have only analyzed 6 haplotypes from 3 humans, and a much larger number of individuals will be required to fully understand the diversity of these hypervariable loci. A dedicated effort that systematically haplotype resolves such loci will serve to identify additional candidate loci for genomic instability and disease association. Third, additional resources are needed to improve STR/VNTR association analyses. These include the development of genotyping methods and resources, where tissue and/or single-cell transcript data from a larger population of individuals are generated for expression quantitative trait loci (eQTL) analysis. We recently demonstrated that genotyping of sequence-resolved SVs (including STRs/VNTRs) may be used to uncover eQTL associations (19). Of the various SV classes, VNTRs, however, showed the lowest genotyping accuracy and, as a result, are a major source of false negatives (19). Hence, the improvement of tandem repeat genotyping methods against short-read genomes will likely require the cataloguing of additional haplotype-resolved STRs/VNTRs sequences in order to accurately discover and assign variation.
The availability of long-read sequence data is providing some of our first glimpses of these more complex forms of genetic variation at the whole-genome level (19, 20) as well as revealing systematic biases in reference genomes that guide variant and genetic association studies. In the case of the human reference, for example, we identified 279 tandem repeat loci that are expanded in most human genomes representing potential reference collapses or polymorphisms (Dataset S3). Similarly, when we compare our haplotype-resolved primate assemblies with previous whole-genome assemblies that were generated without consideration of phase (21), we identify 3,113 and 3,496 STRs and VNTRs, respectively, that are larger in both haplotype-resolved assemblies, despite being generated from the same NHP individual. These findings emphasize the importance of haplotype phasing and partitioning of long reads prior to assembly in order to fully resolve STR/VNTR sequence structures (20) in human and other genomes.
Supplementary Material
Acknowledgments
We thank M. Sorensen and K. Munson for technical assistance in generating sequencing data and T. Brown for manuscript editorial assistance. The authors thank the 3 anonymous reviewers for their thoughtful and constructive comments. This work was supported, in part, by US NIH Grants HG002385 (to E.E.E.) and HG010169 (to E.E.E.). A.S. was supported by NIH Genome Training Grant T32 HG000035-23. M.R.V. was supported by National Library of Medicine Big Data Training Grant for Genomics and Neuroscience 5T32LM012419-04. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Competing interest statement: E.E.E. is on the scientific advisory board of DNAnexus, Inc.
This article is a PNAS Direct Submission.
Data deposition: All long-read human and nonhuman primate assemblies of the short tandem repeats/variable number tandem repeats presented in this study were aligned against the GRCh38 and deposited in Zenodo (https://zenodo.org/record/3401477).
1A complete list of the Human Genome Structural Variation Consortium can be found in the SI Appendix.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912175116/-/DCSupplemental.
Contributor Information
Collaborators: Mark J.P. Chaisson, Ashley D. Sanders, Xuefang Zhao, Ankit Malhotra, David Porubsky, Tobias Rausch, Eugene J. Gardner, Oscar L. Rodriguez, Li Guo, Ryan L. Collins, Xian Fan, Jia Wen, Robert E. Handsaker, Susan Fairley, Zev N. Kronenberg, Xiangmeng Kong, Fereydoun Hormozdiari, Dillon Lee, Aaron M. Wenger, Alex R. Hastie, Danny Antaki, Thomas Anantharaman, Peter A. Audano, Harrison Brand, Stuart Cantsilieris, Han Cao, Eliza Cerveira, Chong Chen, Xintong Chen, Chen-Shan Chin, Zechen Chong, Nelson T. Chuang, Christine C. Lambert, Deanna M. Church, Laura Clarke, Andrew Farrell, Joey Flores, Timur Galeey, David U. Gorkin, Madhusudan Gujral, Victor Guryev, William Haynes Heaton, Jonas Korlach, Sushant Kumar, Jee Young Kwon, Ernest T. Lam, Jong Eun Lee, Joyce Lee, Wan-Ping Lee, Sau Peng Lee, Shantao Li, Patrick Marks, Karine Viaud-Martinez, Sascha Meiers, Katherine M. Munson, Fabio C.P. Navarro, Bradley J. Nelson, Conor Nodzak, Amina Noor, Sofia Kyriazopoulou-Panagiotopoulou, Andy W.C. Pang, Yunjiang Qiu, Gabriel Rosanio, Mallory Ryan, Adrian Stütz, Diana C.J. Spierings, Alistair Ward, AnneMarie E. Welch, Ming Xiao, Wei Xu, Chengsheng Zhang, Qihui Zhu, Xiangqun Zheng-Bradley, Ernesto Lowy, Sergei Yakneen, Steven McCarroll, Goo Jun, Li Ding, Chong Lek Koh, Bing Ren, Paul Flicek, Ken Chen, Mark B. Gerstein, Pui-Yan Kwok, Peter M. Lansdorp, Gabor T. Marth, Jonathan Sebat, Xinghua Shi, Ali Bashir, Kai Ye, Scott E. Devine, Michael E. Talkowski, Ryan E. Mills, Tobias Marschall, Jan O. Korbel, Evan E. Eichler, and Charles Lee
References
- 1.Sueoka N., Correlation between base composition of deoxyribonucleic acid and amino acid composition of protein. Proc. Natl. Acad. Sci. U.S.A. 47, 1141–1149 (1961). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeffreys A. J., Wilson V., Thein S. L., Hypervariable ‘minisatellite’ regions in human DNA. Nature 314, 67–73 (1985). [DOI] [PubMed] [Google Scholar]
- 3.Tautz D., Notes on the definition and nomenclature of tandemly repetitive DNA sequences. EXS 67, 21–28 (1993). [DOI] [PubMed] [Google Scholar]
- 4.Chakraborty R., Kimmel M., Stivers D. N., Davison L. J., Deka R., Relative mutation rates at di-, tri-, and tetranucleotide microsatellite loci. Proc. Natl. Acad. Sci. U.S.A. 94, 1041–1046 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stead J. D., Jeffreys A. J., Structural analysis of insulin minisatellite alleles reveals unusually large differences in diversity between Africans and non-Africans. Am. J. Hum. Genet. 71, 1273–1284 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richards R. I., Sutherland G. R., Dynamic mutations: A new class of mutations causing human disease. Cell 70, 709–712 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Eichler E. E., et al. , Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat. Genet. 8, 88–94 (1994). [DOI] [PubMed] [Google Scholar]
- 8.Berg I., Neumann R., Cederberg H., Rannug U., Jeffreys A. J., Two modes of germline instability at human minisatellite MS1 (locus D1S7): Complex rearrangements and paradoxical hyperdeletion. Am. J. Hum. Genet. 72, 1436–1447 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gatchel J. R., Zoghbi H. Y., Diseases of unstable repeat expansion: Mechanisms and common principles. Nat. Rev. Genet. 6, 743–755 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Coassin S., et al. , A comprehensive map of single-base polymorphisms in the hypervariable LPA kringle IV type 2 copy number variation region. J. Lipid Res. 60, 186–199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirby A., et al. , Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat. Genet. 45, 299–303 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song J. H. T., Lowe C. B., Kingsley D. M., Characterization of a human-specific tandem repeat associated with bipolar disorder and schizophrenia. Am. J. Hum. Genet. 103, 421–430 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chaisson M. J., et al. , Resolving the complexity of the human genome using single-molecule sequencing. Nature 517, 608–611 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaisson M. J., Wilson R. K., Eichler E. E., Genetic variation and the de novo assembly of human genomes. Nat. Rev. Genet. 16, 627–640 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fedurco M., Romieu A., Williams S., Lawrence I., Turcatti G., BTA, a novel reagent for DNA attachment on glass and efficient generation of solid-phase amplified DNA colonies. Nucleic Acids Res. 34, e22 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanger F., Nicklen S., Coulson A. R., DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U.S.A. 74, 5463–5467 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lander E. S., et al. ; International Human Genome Sequencing Consortium , Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). Erratum in: Nature 411, 720 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Eid J., et al. , Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Audano P. A., et al. , Characterizing the major structural variant alleles of the human genome. Cell 176, 663–675.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chaisson M. J. P., et al. , Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 10, 1784 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kronenberg Z. N., et al. , High-resolution comparative analysis of great ape genomes. Science 360, eaar6343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tempel S., Using and understanding RepeatMasker. Methods Mol. Biol. 859, 29–51 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Benson G., Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrison E., Marth G., Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 (17 July 2012).
- 25.Pollen A. A., et al. , Establishing cerebral organoids as models of human-specific brain evolution. Cell 176, 743–756.e17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowakowski T. J., et al. , Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gel B., et al. , regioneR: An R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32, 289–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang B., Kirov S., Snoddy J., WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–W748 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vollger M. R., et al. , Improved assembly and variant detection of a haploid human genome using single-molecule, high-fidelity long reads. bioRXiv:10.1101/635037 (10 May 2019). [DOI] [PMC free article] [PubMed]
- 30.Wang H., et al. , SVA elements: A hominid-specific retroposon family. J. Mol. Biol. 354, 994–1007 (2005). [DOI] [PubMed] [Google Scholar]
- 31.Hancks D. C., Ewing A. D., Chen J. E., Tokunaga K., Kazazian H. H. Jr, Exon-trapping mediated by the human retrotransposon SVA. Genome Res. 19, 1983–1991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Damert A., et al. , 5′-Transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 19, 1992–2008 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kõks G., et al. , Genetic interaction between two VNTRs in the SLC6A4 gene regulates nicotine dependence in Vietnamese men. Front. Pharmacol. 9, 1398 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bellizzi D., et al. , A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics 85, 258–263 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Fishilevich S., et al. , GeneHancer: Genome-wide integration of enhancers and target genes in GeneCards. Database 2017, bax028 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Roeck A., et al. ; BELNEU Consortium , An intronic VNTR affects splicing of ABCA7 and increases risk of Alzheimer’s disease. Acta Neuropathol. 135, 827–837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaganathan K., et al. , Predicting splicing from primary sequence with deep learning. Cell 176, 535–548.e24 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Coe B. P., et al. , Neurodevelopmental disease genes implicated by de novo mutation and copy number variation morbidity. Nat. Genet. 51, 106–116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y. I., et al. , RNA splicing is a primary link between genetic variation and disease. Science 352, 600–604 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LaCroix A. J., et al. ; University of Washington Center for Mendelian Genomics , GGC repeat expansion and exon 1 methylation of XYLT1 is a common pathogenic variant in Baratela-Scott syndrome. Am. J. Hum. Genet. 104, 35–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu Y. H., et al. , Variation of the CGG repeat at the fragile X site results in genetic instability: Resolution of the Sherman paradox. Cell 67, 1047–1058 (1991). [DOI] [PubMed] [Google Scholar]
- 42.Eichler E. E., Hammond H. A., Macpherson J. N., Ward P. A., Nelson D. L., Population survey of the human FMR1 CGG repeat substructure suggests biased polarity for the loss of AGG interruptions. Hum. Mol. Genet. 4, 2199–2208 (1995). [DOI] [PubMed] [Google Scholar]
- 43.Hijikata M., et al. , Molecular cloning of two novel mucin-like genes in the disease-susceptibility locus for diffuse panbronchiolitis. Hum. Genet. 129, 117–128 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Pugliese A., et al. , The insulin gene is transcribed in the human thymus and transcription levels correlated with allelic variation at the INS VNTR-IDDM2 susceptibility locus for type 1 diabetes. Nat. Genet. 15, 293–297 (1997). [DOI] [PubMed] [Google Scholar]
- 45.Bell G. I., Selby M. J., Rutter W. J., The highly polymorphic region near the human insulin gene is composed of simple tandemly repeating sequences. Nature 295, 31–35 (1982). [DOI] [PubMed] [Google Scholar]
- 46.Nordestgaard B. G., et al. ; European Atherosclerosis Society Consensus Panel , Lipoprotein(a) as a cardiovascular risk factor: Current status. Eur. Heart J. 31, 2844–2853 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lackner C., Cohen J. C., Hobbs H. H., Molecular definition of the extreme size polymorphism in apolipoprotein(a). Hum. Mol. Genet. 2, 933–940 (1993). [DOI] [PubMed] [Google Scholar]
- 48.Wright G. E. B., et al. , Length of uninterrupted CAG, independent of polyglutamine size, results in increased somatic instability, hastening onset of Huntington disease. Am. J. Hum. Genet. 104, 1116–1126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bois P., Jeffreys A. J., Minisatellite instability and germline mutation. Cell. Mol. Life Sci. 55, 1636–1648 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Usdin K., The biological effects of simple tandem repeats: Lessons from the repeat expansion diseases. Genome Res. 18, 1011–1019 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z., et al. , Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat. Genet. 49, 1576–1583 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Vasanth S., et al. , Expansion of CTG18.1 trinucleotide repeat in TCF4 is a potent driver of Fuchs’ corneal dystrophy. Invest. Ophthalmol. Vis. Sci. 56, 4531–4536 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baratz K. H., et al. , E2-2 protein and Fuchs’s corneal dystrophy. N. Engl. J. Med. 363, 1016–1024 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Wieben E. D., et al. , A common trinucleotide repeat expansion within the transcription factor 4 (TCF4, E2-2) gene predicts Fuchs corneal dystrophy. PLoS One 7, e49083 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu J., et al. , Oligonucleotides targeting TCF4 triplet repeat expansion inhibit RNA foci and mis-splicing in Fuchs’ dystrophy. Hum. Mol. Genet. 27, 1015–1026 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Es M. A., et al. , Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat. Genet. 41, 1083–1087 (2009). [DOI] [PubMed] [Google Scholar]
- 57.DeJesus-Hernandez M., et al. , Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Renton A. E., et al. ; ITALSGEN Consortium , A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pearson C. E., Sinden R. R., Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry 35, 5041–5053 (1996). [DOI] [PubMed] [Google Scholar]
- 60.Nolin S. L., et al. , Fragile X full mutation expansions are inhibited by one or more AGG interruptions in premutation carriers. Genet. Med. 17, 358–364 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Hach F., et al. , mrsFAST-Ultra: A compact, SNP-aware mapper for high performance sequencing applications. Nucleic Acids Res. 42, W494–W500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jain M., et al. , Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36, 338–345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wenger A. M., et al. , Highly-accurate long-read sequencing improves variant detection and assembly of a human genome. bioRXiv:10.1101519025 (13 January 2019). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.