Key Points
Autoreactive T cells drive the lethal disease in Malt1PD mice.
Malt1PD mice display B cell hyperactivation to environmental Ags.
IPEX-like disease occurs in Malt1PD mice despite in vitro Treg suppressive activity.
Abstract
The paracaspase Malt1 is a key regulator of canonical NF-κB activation downstream of multiple receptors in both immune and nonimmune cells. Genetic disruption of Malt1 protease function in mice and MALT1 mutations in humans results in reduced regulatory T cells and a progressive multiorgan inflammatory pathology. In this study, we evaluated the altered immune homeostasis and autoimmune disease in Malt1 protease-deficient (Malt1PD) mice and the Ags driving disease manifestations. Our data indicate that B cell activation and IgG1/IgE production is triggered by microbial and dietary Ags preferentially in lymphoid organs draining mucosal barriers, likely as a result of dysregulated mucosal immune homeostasis. Conversely, the disease was driven by a polyclonal T cell population directed against self-antigens. Characterization of the Malt1PD T cell compartment revealed expansion of T effector memory cells and concomitant loss of a CD4+ T cell population that phenotypically resembles anergic T cells. Therefore, we propose that the compromised regulatory T cell compartment in Malt1PD animals prevents the efficient maintenance of anergy and supports the progressive expansion of pathogenic, IFN-γ–producing T cells. Overall, our data revealed a crucial role of the Malt1 protease for the maintenance of intestinal and systemic immune homeostasis, which might provide insights into the mechanisms underlying IPEX-related diseases associated with mutations in MALT1.
Introduction
Primary immunodeficiencies represent a large group of genetic disorders in which affected individuals suffer from dysregulated immune responses often resulting in autoimmune diseases (1–3). As an example, mutations in the transcription factor FOXP3, a master regulator of regulatory T cells (Tregs), result in absence or dysfunction of Tregs and a lethal, systemic syndrome called immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) in both humans (4) and mice (5). Patients frequently demonstrate additional clinical manifestations, including the failure to thrive as a result of the enteropathy and deregulated humoral responses associated with food allergies and a broad variety of autoantibodies (6–9). Beyond FOXP3, defects in multiple other genes involved in immune homeostasis can give rise to IPEX-like diseases with diverse sets of clinical manifestations that may overlap with IPEX syndrome depending on the cellular and signaling pathways affected by the genetic defect (9). One such case is loss of function mutations in MALT1, which cause IPEX-like autoimmune manifestations in humans and are fatal unless treated with hematopoietic stem cell transplantation (10–14). The MALT1 paracaspase is a ubiquitously expressed key regulator of canonical NF-κB activation downstream of multiple receptors such as the TCR, BCR, FcγRs, several C-type lectin receptors, or G protein–coupled receptors (15–19). Together with BCL10 and various CARD molecules and upon activation, MALT1 forms the “CBM” signalosome (20), which acts as a scaffold for recruitment of effector proteins that ultimately trigger NF-κB–dependent signaling (21–23). In addition, MALT1 has a protease function that profoundly reshapes the cellular response to activation by cleavage of a growing family of substrates (24, 25). To study the relative contribution of the scaffolding and protease functions of MALT1, multiple Malt1 knockout (Malt1−/−) as well as Malt1 protease-deficient (Malt1PD) mouse lines have been generated (26–31). In Malt1PD animals, a catalytically inactive form of Malt1 retains the capacity to activate canonical NF-κB responses, but these are reduced or altered as a consequence of the defective cleavage of Malt1 substrates (29, 32, 33). Both Malt1−/− and Malt1PD mice show reduced Treg frequencies and defective Ab responses to T-dependent and T-independent type 2 Ags (26–29, 34). In contrast to Malt1−/− mice, which display a generally immunodeficient phenotype and are viable, Malt1PD animals develop a spontaneous and lethal IPEX-like disease associated with lymphadenopathy, elevated serum IgG1 and IgE levels, expansion of effector T cells (Teffs), and lymphocytic infiltrates in various tissues (26–29). Treg deficiency is the leading cause of disease development in Malt1PD mice because the adoptive transfer of wild-type (WT) Tregs early after birth prevents the appearance of disease symptoms (26). Although Malt1−/− mice do not develop a lethal pathologic condition, these animals still display subclinical disease features reflected by milder lymphadenopathy and limited immune infiltrates in various organs (29). Human MALT1 mutations described so far result in unstable or absent MALT1 protein but paradoxically cause IPEX-like phenotypes similar to those observed in the Malt1PD mice (10–14). In both MALT1-deficient patients and the Malt1PD mouse model, the immune dysregulation caused by partial Treg deficiency seems to drive lymphocyte effector functions despite profound defects in adaptive immunity. These parallels prompted us to further dissect the underlying causes of the disease developing in the Malt1PD mouse model.
The analysis of Malt1PD mouse lines has revealed some of the underlying causes of the IPEX-like disease (26–29). Although agreeing on most observations, some differences were observed between different Malt1PD lines, such as neuropathological symptoms like hind limb paralysis (26, 27, 29). This was likely caused by differences in the design of the lines or alternatively by the environmental factors related to different housing conditions. Therefore, several questions remained to understand the pathways driving distinct disease manifestations at specific anatomical locations and their link to environmental cues. In this study, we assessed the relative contribution of T and B lymphocytes to disease development and describe how Malt1 protease deficiency disrupts mucosal immunity and independently results in a systemic, ultimately lethal autoimmune disease. We found that environmental Ags and commensal-derived pathogen-associated molecular pattern molecules drive hyper IgG1 and IgE in Malt1PD animals via joint action of the BCR and the pattern recognition receptor pathways. Finally, we show that Malt1PD Tregs retained partial in vitro suppressive function and seemed to counteract increased inflammatory signals in vivo by upregulation of effector molecules and clonal expansion at specific anatomical sites. Therefore, we propose that the disease driven by Malt1 protease dysfunction in mice is a combination of a lethal, T cell–driven genuinely autoimmune disease and an independent, B cell–driven hyperreaction to environmental Ags.
Materials and Methods
Mice
Malt1PD (B6-Malt1tm1[C472A]Npa) mice on a C57BL/6 genetic background have been described previously (29). For experiments comparing Malt1PD to WT mice, cohoused WT littermates were used as controls. Germ-free (GF) Malt1PD and WT littermate animals were bred and housed in flexible film isolators at the Clean Mouse Facility of the University of Bern. The following parent lines were used to set up internal breedings with Malt1PD heterozygous animals: B6.Cg-Foxp3tm2(EGFP)Tch/J (stock no. 006772; purchased from The Jackson Laboratory) (35), Traj18−/− (B6-Ja281tm1810_0.2Arte) mice (kindly provided by M. Kronenberg) (36), B10.BR-Tg(IghelMD4)4Ccg (37, 38), and B10.BR-Tg(TcrHEL3A9)1MmD (both lines were kindly provided by J. Moreno) (39, 40). Of note, the resulting MD4 BCR transgenic (tg) × Malt1PD and 3A9 TCRtg × Malt1PD animals are on a mixed B10.BR:C57BL/6 genetic background. All experiments using these animals included the corresponding littermate controls of the following genotypes: MD4 BCRtg or 3A9 TCRtg, Malt1PD and WT. We observed no differences in disease kinetics or severity between pure B6-Malt1PD animals and B6:B10.BR–mixed background Malt1PD animals. B6.SJL-PtprcaPepcb/BoyJ CD45.1 WT animals (stock no. 002014; The Jackson Laboratory) were purchased from Taconic Europe. All animal studies were performed in accordance with the animal experimentation laws and guidelines laid down by the Swiss Federal and Cantonal Authorities. Health of Malt1PD mice and bone marrow (BM) chimeras was assessed by visual checks twice weekly. Animals were euthanized upon body weight loss of >20% or appearance of clinical symptoms such as hind limb paralysis, eye inflammation, or hunched posture. Mixed groups of male and female animals were used at the age of 8–12 wk if not stated otherwise.
Flow cytometry
Stainings of cell suspensions from spleen and lymph node (LN) were prepared by passing tissues through a 70-μm sieve followed by RBC lysis using ACK hypotonic solution. Cells were washed once in FACS buffer (PBS containing 2% FCS, 0.05% NaN3), blocked with mouse Fc block (clone 2.4G2; BD Biosciences), and stained for 30 min at 4°C with the indicated combination of fluorochrome-conjugated Abs. Intracellular stainings were performed using the Foxp3 Fix/Perm Buffer Set (BioLegend). Cells were acquired using a BD FACSFortessa flow cytometer, and data were analyzed using the FlowJo software. All populations were pregated on singlets and live lymphocytes based on their side scatter versus forward light scatter profiles and using appropriate live/dead stains. From live lymphocytes, cells were further gated on CD4 or CD8 for T cells and on CD19 and B220 for B cells as indicated on graphs, followed by assessment of the specified activation markers. In case of CD4+ T cells, populations were further divided based on Foxp3 or Neuropilin1 (Nrp1). Cells isolated from nonlymphoid tissues were additionally gated on CD45 or CD45.2. Cells originating from BM chimeras were gated on CD45.1 versus CD45.2 to differentiate between donor and recipient origin as specified.
T cell stimulation ex vivo
Pooled cells isolated from spleen of three Malt1PD × Foxp3EGFP or WT littermates were enriched for T cells using the EasySep Mouse T Cell Isolation Kit (no. 19851; STEMCELL Research). For cytokine detection, CD4+Foxp3−Nrp1− naive T cells, CD4+Foxp3+ Tregs, and CD4+Foxp3−Nrp1+ T cells were sorted using a BD FACSAria and were stimulated ex vivo with 10 ng/ml PMA and 1 μM ionomycin for 4 h prior to assessment of cytokine concentrations in culture media by multiplex ELISA (Meso Scale Diagnostics). For detection of intracellular IFN-γ by flow cytometry, cells were isolated from spleen, cervical/mandibular LN (cLN), or mesenteric LN (mLN), FACS sorted, and stimulated with 10 ng/ml PMA and 1 μM ionomycin in presence of 10 μg/ml brefeldin A (all Sigma-Aldrich) for 4 h at 37°C.
Treg suppression assay in vitro
BM was harvested from a WT mouse, and cells were cultured in presence of rmGM-CSF to induce differentiation into dendritic cells as described previously (41). At day 7 cells were harvested and cultured with a mix of Foxp3EGFP+CD4+CD25+ Tregs sorted from WT × Foxp3EGFP or Malt1PD × Foxp3EGFP mice and cell trace violet (no. C34557; Thermo Fisher Scientific)–labeled WT CD4+CD25− Teffs in presence of 1 μg/ml anti-CD3 Ab (clone 1.45-2C11). A total of 3,000 nonirradiated dendritic cells per well were added to 50,000 Teffs together with different numbers of Tregs to yield Treg/Teff ratios between 1:1 and 1:20. Proliferation of Teffs was assessed after 5 d of culture by flow cytometry, and the percentage of suppression was calculated as follows: 100 − [(% dividing cells − % spontaneously dividing cells)/(% max dividing cells − % spontaneously dividing cells)×100].
TCR repertoire sequencing
Foxp3EGFP+CD4+ Tregs were sorted from spleens, mLNs, and cLNs of WT × Foxp3EGFP or Malt1PD × Foxp3EGFP mice using a BD FACSAria. Per organ and genotype, three replicate samples were generated of which each contained pooled cells from two WT × Foxp3EGFP or two Malt1PD × Foxp3EGFP mice. Prior to the sort, cells were enriched for CD3+ T cells using the EasySep Mouse T Cell Isolation Kit (no. 19851; STEMCELL Research), and Foxp3EGFP+ cells were sorted directly into RLT lysis buffer for subsequent isolation of RNA using the RNeasy Micro Kit (no. 74004; Qiagen). Quality and quantity of RNA were tested with the Agilent 2100 Bioanalyzer using the RNA 6000 Pico Chip and reagents (no. 5067-1513; Agilent). 5′RACE-based PCR amplification of TCRβ V region VDJ sequences and Illumina library generation was done using the Smarter TCRα/β Profiling Kit following the manufacturer’s instructions (no. 634404; Takara Biotech). Briefly, all libraries were subject to two rounds of seminested PCR amplification, and amplified libraries were purified using the Agencourt AMPure XP PCR Purification Kit (no. A63881; Beckman Coulter). Libraries were subjected to one double-size selection as per the user manual; beads were pelleted using a Magnetic Separator - PCR Strip (Takara Bio). Library validation was performed on an Agilent 2100 Bioanalyzer with the DNA 1000 Kit (no. 5067-1505; Agilent). Libraries were pooled, supplemented with 10% PhiX Control v3 (Illumina), and sequenced at a final concentration of 12 pM using the 600-cycle MiSeq Reagent Kit v3 (Illumina) to 2 × 301 base paired-end reads. Data were analyzed using the MiXCR software package (42). All statistical analysis was performed using R Statistical Software (i.e., “ineq” package to calculate the Gini coefficient and “ggplot2” for generating figures [43]). All sequences are available at the European Nucleotide Archive (http://www.ebi.ac.uk/ena/data/view/PRJEB34151; accession number: PRJEB34151).
Depletion of CD4 and CD8 T cells using mAbs
Four-week-old WT and Malt1PD animals on a C57BL/6 background were treated with mAbs against CD4 (clone GK1-5; 10 mg/kg bodyweight i.p. twice per wk) or CD8 (clone 2-43; 10 mg/kg bodyweight i.p. twice per wk) or respective isotype control Ab (rat IgG2b, κ) for around 5 wk. Successful depletion of the corresponding subset was confirmed by FACS on blood cells.
Serum Ig subtype analysis, ELISpot, and ELISA
Serum preparation.
Blood was collected in BD Microtainer tubes (SST tubes), allowed to clot for 30 min at room temperature, and centrifuged at 14,000 rpm for 2 min to separate the serum from clotted components.
IgG1 and IgE detection from serum.
IgG1 and IgE in mouse serum were quantified using the following two kits and according to the manufacturer’s protocol: Merck Millipore Mouse Ig Isotyping Magnetic Bead Panel (catalog no. MGAMMAG-300K and MGAMMAG-300E).
ELISpot for total IgG and IgE.
Single-cell suspensions were prepared from cLN, axillary LN, brachial LN, mLN, and inguinal LN as well as from Peyer patches as described above (Flow cytometry). BM was prepared by flushing cells from femur and tibia using a 27-gauge needle and passage of the cells through a 70-μm cell strainer. IgG- and IgE-secreting cells were detected using the Mouse IgG ELISpot BASIC Kit (no. 3825-2H; Mabtech) following the manufacturer’s protocol. For detection of IgE-secreting cells, the anti-IgG Ab set was replaced by the following: IgE (clone R35-72) for coating and IgE-Biotin (clone R35-118) for detection (both BD).
Anti-cecal or food-derived Ag ELISA.
Fresh cecal content was collected, pooled from four WT mice, weighed, and carefully resuspended in PBS at 0.01 g/ml. The obtained suspension was centrifuged at 400 × g for 5 min to remove larger particles from bacteria. Bacteria were lysed by physical disruption through sonication (interval, three times 30 s at 50% on ice), and the protein concentration was quantified using Bradford protein assay (Pierce BCA; Thermo Fisher Scientific). A total of 0.5 μg of protein in 50 μl of PBS was used for coating of 96-well half-area plates (polystyrene; Costar) at 4°C overnight. To isolate food proteins from mouse chow pellets, 9 g of food was dissolved in 40 ml of PBS and shaken for 4 h at 37°C prior to centrifugation of the obtained suspension at 3000 × g for 10 min to remove larger particles. The supernatant was recentrifuged at 3000 × g for 5 min, and the protein concentration of the resulting suspension was determined using the BCA assay. A total of 5 μg of protein in 50 μl of PBS was used for coating of 96-well half-area plates. All plates were washed four times with PBS + 0.05% Tween 20, blocked for 1 h (10% FBS in PBS), and subsequently probed with diluted serum (1:5 to 1:20,000) from WT or Malt1PD animals for 16 h at 4°C. Cecal content– or food Ag–binding Igs were detected using biotin-conjugated goat anti-mouse IgG1 (no. 1070-08; 1/10 000; Southern Biotech) or rat anti-mouse IgE (no. 553419; 1/250; BD) and avidin-conjugated alkaline phosphatase (ExtrAvidin, no. E2636; Sigma-Aldrich) plus phosphatase substrate tablets (no. N2770; Sigma-Aldrich). The reaction was stopped by the addition of 4 M NaOH prior to spectrophotometric analysis at 405 nm (PerkinElmer).
TNP-LPS immunization and ELISA for anti-TNP IgM and IgG
WT (C57BL/6), Malt1PD mice were immunized i.v. with 30 μg of TNP-LPS (no. T-6769; Sigma-Aldrich) in PBS; total volume: 200 μl/mouse (n = 5 mice per group). Animals were sacrificed on days 6 and 12 postimmunization. For detection of anti-TNP IgM and IgG in serum, plates were coated with TNP-BSA (Biocat), probed with diluted serum, and developed using the following Abs: anti-ms IgM-Biotin (no. 1020-08; Southern Biotech) and anti-ms IgG-Biotin (no. 1030-08; Southern Biotech) in combination with avidin-conjugated alkaline phosphatase (ExtrAvidin, no. E2636; Sigma-Aldrich) plus phosphatase substrate tablets (no. N2770; Sigma-Aldrich). This reaction was stopped by the addition of 50 μl of 4 M NaOH prior to spectrophotometric analysis at 405 nm (Perkin Elmer).
Isolation of colonic lamina propria cells and intraepithelial lymphocytes
Lamina propria (LP) cells and intraepithelial lymphocytes (IELs) were purified using the Mouse Lamina Propria Dissociation Kit (no. 130-097-410; Miltenyi Biotec) according to the manufacturer’s recommendations. Briefly, colons were removed from naive mice, cleaned from feces, opened longitudinally, and cut into small pieces. Epithelial cells were stripped by incubating for 20 min in a 37°C water bath in cell dissociation solution (20 ml of 1× HBSS (w/o) containing 5 mM EDTA, 5% FBS, 1 mM DTT) under continuous rotation. The supernatant containing IELs and epithelial cells was kept for further processing. Colonic tissue was then incubated in a digestion mixture containing 5 ml of HBSS (w), 10% FBS, collagenase D (1 mg/ml), DNAse I (0.1 mg/ml), and Dispase (0.1 U/ml) per intestine at 37°C for 45 min. Digested tissue was passed through a 70-μm sieve and washed before lymphocytes were separated using a 30/37/70% Percoll gradient (no. 17089101; GE Healthcare). Similarly, IELs were purified from the supernatant obtained after step 1 using a 30/37/70% Percoll gradient. Cells were resuspended in complete RPMI 1640 supplemented with 10% FCS, 1% HEPES, 50 mM 2-ME, 1% sodium pyruvate, and penicillin and streptomycin (all from Life Technologies) or FACS buffer for further analysis.
Generation of BM chimeras
Donor BM cells were isolated by flushing cells from femur and tibia using a 27-gauge needle. Cells were passed through a 70-μm cell strainer, and RBCs were lysed subsequently using ACK hypotonic solution. A total of 4 × 106 BM cells derived from female CD45.1 WT mice (B6.SJL-PtprcaPepcb/BoyJ; Taconic) were injected i.v. into sublethally irradiated (two times 450 rad, 4 h apart) CD45.2+ WT (WT→WT) or CD45.2+ Malt1PD (WT → Malt1PD) recipient animals. Control Malt1PD→Malt1PD BM chimeras were made of sublethally irradiated CD45.2+ Malt1PD recipients receiving CD45.2+ Malt1PD BM cells, and thus lacked congenic markers. The average age of the recipient animals on the day of irradiation was 7.3 wk.
WT:Malt1PD mixed BM chimeras were set up by injecting a 1:1 mix of 5 × 106 WT (CD45.1) BM cells and 5 × 106 Malt1PD (CD45.2) BM cells into sublethally irradiated CD45.1 WT recipients.
Acute dextran sodium sulfate–induced colitis in BM chimeras
WT→WT and WT → Malt1PD BM chimeras were allowed to reconstitute for 9 wk with regular body weight assessment before induction of dextran sodium sulfate (DSS) colitis. Acute colitis was induced by the supply of 4% (weight per volume) DSS with a molecular mass of 36–50 kDa (MP Biomedicals, Irvine, CA) in drinking water. Animals were treated with DSS for 5 d (day 0–5) followed by regular drinking water (day 6–11). Animals were scored daily (disease activity index) and sacrificed on day 11. The disease activity index (0–10) was calculated based on body weight loss (0–4), diarrhea (0–3), and bloody stool (blood in the colon at the day of sacrifice; 0–3).
Statistics
Bar graphs in the figures represent average values + SEM unless indicated otherwise. Statistical significance between groups was calculated using a two-tailed unpaired Student t test or ordinary one-way ANOVA (Dunnett multiple comparisons) using GraphPad Prism (GraphPad Software) and is indicated in the graphs as follows: *p < 0.05, **p < 0.01, ***p < 0.001, p < 0.0001. Nonsignificant differences were not indicated.
Results
Excessive Ig production at barrier sites is driven by commensal Ags
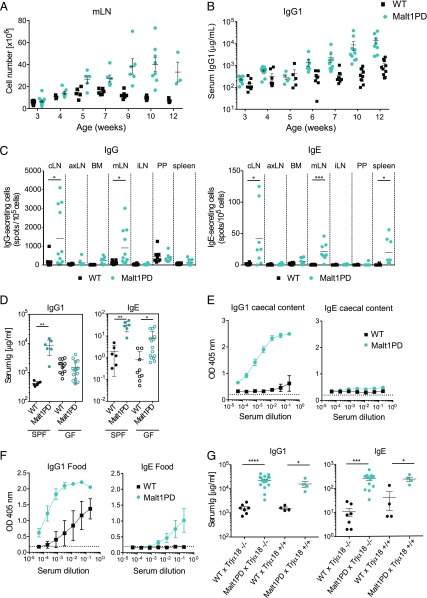
We and others previously described a lethal inflammatory syndrome in Malt1PD mice that leads to death of the animals by 10–20 wk of age (26–29). To understand the kinetics of disease development, we analyzed spleen and LN cellularity, frequency of T and B cell subsets, and serum Ig levels in naive Malt1PD and WT animals starting from 3 wk of age. In line with the previously reported reduction in thymic and peripheral Tregs in adult Malt1PD animals (29), we observed a 2–3-fold lower percentage of Foxp3+ Tregs in spleens and peripheral LNs of Malt1PD animals (Supplemental Fig. 1A). The expansion of Ag-experienced CD44hiCD8+ Teffs as well as the increased cellularity in mLN and pooled cLN progressively appeared at around 7–9 wk of age as a potential consequence of the reduction in the Treg compartment (Supplemental Fig. 1B, 1C). Compared with WT animals, adult Malt1PD mice display significantly higher serum levels of IgG1 and IgE (26, 29). The kinetics of serum IgG1 and IgE elevation in Malt1PD mice correlated with the increased cellularity in mLN and cLN (Fig. 1A, 1B, Supplemental Fig. 1C, 1D), and both IgG- and IgE-secreting plasma cells were mainly detected in mLN and cLN of Malt1PD animals (Fig. 1C). This indicated that formation of IgG1 and IgE Abs occurred in the LN draining sites exposed to environmental Ags such as the mouth, the eyes, and the intestinal tract, likely as a consequence of uncontrolled B cell responses to these Ags.
FIGURE 1.
Excessive IgG1 and IgE production at barrier sites is driven by environmental Ags. (A) Counts of total live cells isolated from mLN from WT or Malt1PD animals at different ages. Each dot represents a mouse. (B) Age-related evolution of serum IgG1 concentration in WT versus Malt1PD mice. The results are expressed as the mean ± SEM; each dot represents a mouse. (C) IgG- and IgE-secreting cells among live cells from different organs detected by ELISpot (age of the animals = 8–12 wk). Pooled data from two independent experiments are shown; lines indicate the mean. (D) IgG1 and IgE serum concentration in SPF and GF Malt1PD animals and WT littermates. The results are expressed as the mean ± SEM. (A–D) Each dot represents a mouse. (E and F) Proteins isolated from the cecal content of WT mice (E) or food pellets (F) were coated onto an ELISA plate and probed for serum IgG1 or IgE from 8–12-wk-old Malt1PD animals and WT littermates. The results are expressed as the mean ± SEM (n = 3). (G) Serum concentration of IgG1 and IgE in WT × Trjα18−/−, Malt1PD × Trjα18−/−, WT × Trjα18+/+, and Malt1PD × Trjα18+/+ animals determined at the age of 14 wk. The results are expressed as the mean ± SEM; each dot represents a mouse. All data are representative of at least two independent experiments if not indicated otherwise. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. axLN, axillary LN; brLN, brachial LN; iLN, inguinal LN; PP, Peyer patches.
To test whether microbiota-derived Ags drive excessive Ab formation, Malt1PD animals and WT littermate controls were rederived to GF conditions, and their serum Ig levels were analyzed at the age of 8–12 wk. GF Malt1PD mice still displayed the typical clinical symptoms, but these tended to appear with a faster kinetic compared with Malt1PD animals housed under specific pathogen–free (SPF) conditions (data not shown). Strikingly, Malt1PD mice displayed no elevation in serum IgG1 in GF conditions, and IgE levels were reduced ∼5-fold compared with those observed in Malt1PD animals housed under SPF conditions (Fig. 1D). These findings suggested that IgG1 and IgE were directed against commensal bacterial Ags. Because IgE was still partially elevated in GF Malt1PD mice, nonmicrobial Ags might also drive hyper IgE. Therefore, the reactivity of IgG1 and IgE to bacterial or food-derived proteins was tested by analysis of the binding of serum Igs from SPF Malt1PD and WT animals to proteins isolated from either cecal content of WT animals or from standard food pellets. As hypothesized, Malt1PD serum showed significant IgG1 binding to cecal Ags that was not detectable in WT serum (Fig. 1E). Binding of IgE to cecal contents was absent in WT and very low in Malt1PD serum. When testing binding to food-derived proteins, serum IgG1 from Malt1PD mice and, to a lesser extent, from WT mice reacted against food Ags (Fig. 1F). Binding of IgE to food-derived Ags was observed from Malt1PD serum but not from WT (Fig. 1F). Overall, these data demonstrate that the elevated serum IgG1 and IgE observed in our Malt1PD animals reflect excessive B cell responses to both commensal bacteria-derived and dietary Ags at barrier sites.
Because humoral responses requiring conventional T cell help or BCR cross-linking are defective in Malt1PD animals (26–29), we tested whether IgG1 and IgE Abs were produced via alternative pathways in these animals. Apart from conventional CD4 T cells, invariant NKT (iNKT) helper cells are an additional source of B cell help, specifically for B cells reactive against glycolipids, including those of microbial origin (44–46). To clarify the relevance of iNKT cell help in mediating B cell activation and Ab production in Malt1PD animals, we generated Malt1PD mice deficient in iNKT cells via an intercross with Traj18−/− mice (36). We observed no difference in serum Ab levels compared with Malt1PD × Traj18+/+ littermate controls (Fig. 1G). Therefore, iNKT cells are not required to drive elevated IgG1/IgE in Malt1PD mice. Based on these results, we thus hypothesized that B cells could be induced to produce IgG1 and IgE in Malt1PD in response to environmental Ags via joint activation of BCR and MALT1-independent pattern recognition receptor pathways such as TLRs (47). In this line, Malt1PD animals readily produced both IgM and IgG Abs upon immunization with TNP-LPS (Supplemental Fig. 2). Thus, hyper IgG1 and IgE in Malt1PD animals could be induced by an opening of the mucosal barrier and increased presence of microbial products. Luminal Ags could drive B cell activation via BCR-dependent recognition of commensal and food-derived Ags in conjunction with activation of pattern recognition receptors.
Deficiency of the Malt1 protease in nonhematopoietic cells is insufficient to disrupt intestinal barrier integrity
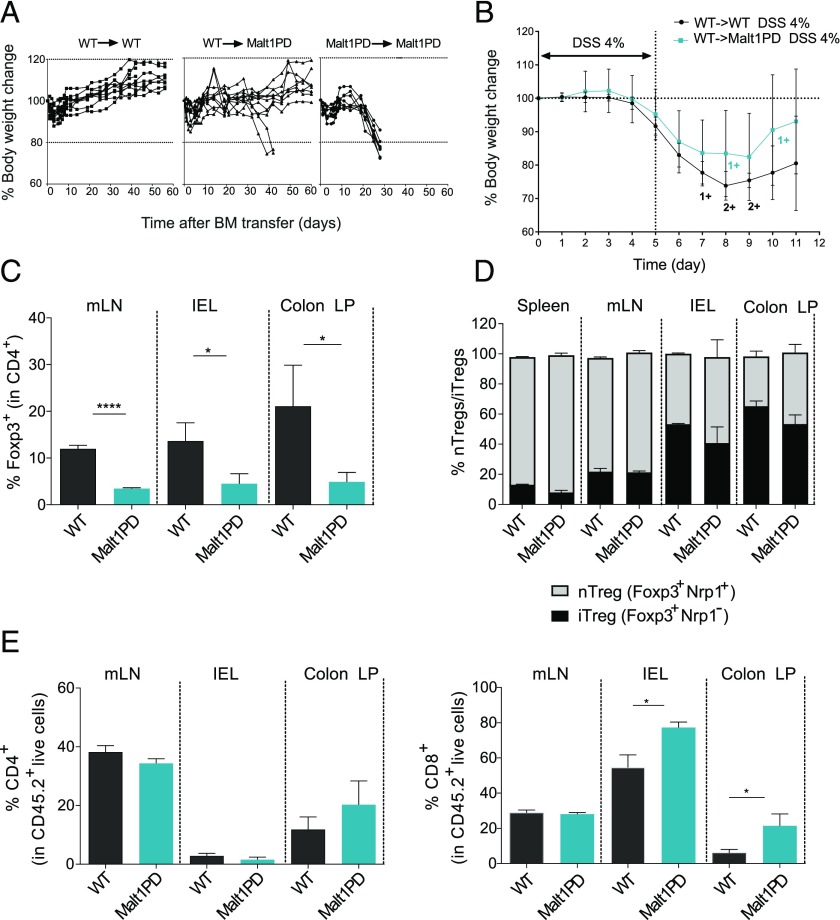
The excessive humoral response to cecal and food-derived Ags observed in Malt1PD animals indicated that a leaky intestinal barrier may have provided increased access of luminal Ags to the intestinal immune system. Impaired barrier function could either result from increased intestinal inflammation (48–50) or be a direct consequence of defective Malt1 protease signaling in the epithelial cells. Indeed, increasing evidence supports a vital role for the CBM complex in both endothelial and epithelial cells downstream of multiple receptors (17, 18, 51–54). To assess whether Malt1 protease signaling in nonimmune cells influenced epithelial barrier integrity in the absence of Malt1 protease function in the immune compartment, we generated BM chimeras. To this end, 7–8-wk-old sublethally irradiated Malt1PD (WT→Malt1PD) or WT control (WT→WT) recipients were reconstituted with WT BM. As a control, Malt1PD mice were reconstituted with Malt1PD BM. As expected, all the Malt1PD→Malt1PD BM chimeras rapidly developed clinical symptoms and wasting disease and accordingly had to be euthanized by 3–4 wk following BM reconstitution (Fig. 2A, right panel). Malt1PD mice are known to display different degrees of subclinical immune alterations already at the age of 7–8 wk (Fig. 1, Supplemental Fig. 1). Remarkably, when replacing the Malt1PD immune compartment with WT BM, the Malt1PD pathologic condition could be rescued in most animals. Indeed, 7 out of 10 WT→Malt1PD animals survived without apparent disease for up to 9 wk after reconstitution (Fig. 2A, middle panel). However, three animals demonstrated disease symptoms, including eye inflammation, hunched posture, and body weight loss 40 d post BM transfer and had to be euthanized according to local animal welfare regulations. The survival of 70% of the WT→Malt1PD BM chimeras suggested that the immune compartment is the main component driving the Malt1PD inflammatory disease. We hypothesized that the lack of protection observed in the remaining animals may reflect an excessively advanced subclinical pathologic condition that could not be completely rescued by the sublethal irradiation and the WT BM transfer process. However, it remains possible that the nonhematopoietic compartment contributed to disease progression in these animals.
FIGURE 2.
Deficiency of the Malt1 protease in nonhematopoietic cells is insufficient to disrupt intestinal barrier integrity. (A) Body weight of WT→WT, Malt1PD→WT, or control Malt1PD→Malt1PD BM chimeras over time. Each line represents a mouse. (B) Acute DSS colitis was induced in WT (WT→WT) and Malt1PD (Malt1PD→WT) BM chimeras from (A) 9 wk after reconstitution by addition of 4% DSS to the drinking water for 5 d, followed by pure drinking water until the day of analysis (day 11). Body weight during the course of acute DSS colitis (day 0–day 5) and recovery phase (day 6–day 11). The results are expressed as the mean ± 95% confidence interval, n = 7–10. 1+ and 2+ stand for one or two animals that died at the indicated time point. (C) Frequency of Foxp3+ Tregs in CD4+ T cells from mLN, IELs, and colon LP of naive Malt1PD animals and WT littermates determined by flow cytometry. The results are expressed as the mean ± SEM (n = 3). (D) Ratio of nTregs versus iTregs in CD4+Foxp3+ cells from the spleen, mLN, IEL, and LP of Malt1PD mice and WT littermates, assessed by flow cytometry. The results are expressed as the mean ± SEM (n = 3–5). (E) Frequency of CD4+ (left) and CD8+ (right) T cells in mLN, IEL, and colon LP determined by flow cytometry. The results are expressed as the mean ± SEM (n = 3). All data are representative of at least two independent experiments. *p < 0.05, ****p < 0.0001.
To address the contribution of the nonimmune compartment to barrier integrity, we next challenged barrier function in the surviving WT→WT and WT→Malt1PD BM chimeras. To this end, colitis was induced experimentally by oral administration of 4% DSS for five consecutive days in drinking water, followed by 6 d of regular drinking water to allow for barrier regeneration. Interestingly, Malt1PD animals reconstituted with WT BM (WT→Malt1PD) demonstrated a similar degree of disease compared with the WT→WT chimeras. We observed no significant difference, neither in body weight loss at any time point (Fig. 2B) nor in colon length and disease score at termination (day 11) between the two groups (Supplemental Fig. 3A, 3B). These results demonstrated that Malt1 protease deficiency in nonhematopoietic cells alone is not sufficient to disrupt intestinal barrier integrity and to confer increased sensitivity to DSS-induced colitis.
Because Treg deficiency was identified as the major driver of imbalanced systemic immune responses in Malt1PD mice (26–29), we determined Treg frequency in intestinal tissues such as the colon LP, IELs, and the mLN of naive Malt1PD animals and observed drastically reduced frequencies of total Foxp3+ Tregs at all sites (Fig. 2C). We further evaluated the proportions of inducible Tregs (iTregs) and natural, thymus-derived Tregs (nTregs) based on their differential expression of Nrp1 (55–57). Compared with WT, there was no difference in the ratios of nTregs (Foxp3+Nrp1+) to iTregs (Foxp3+Nrp1−) within Malt1PD Tregs (Fig. 2D). When characterizing the local immune alterations, we found increased proportions of CD8+ T cells within the LP and among IELs (Fig. 2E). Taken together, our data suggested that the main factor driving the excessive humoral response to commensal and dietary Ags is the disrupted intestinal immune homeostasis rather than a primary leak in the intestine due to an intrinsic role of Malt1 in intestinal epithelial cells. However, because of the limitation of our studies with BM chimeras, we cannot exclude the possibility that nonhematopoietic cells also contributed to the progression of the disease occurring upon systemic disruption of Malt1 protease in both immune and nonimmune cells.
Elevated IgG1 and IgE levels and B cells are not the key drivers of the Malt1PD inflammatory disease
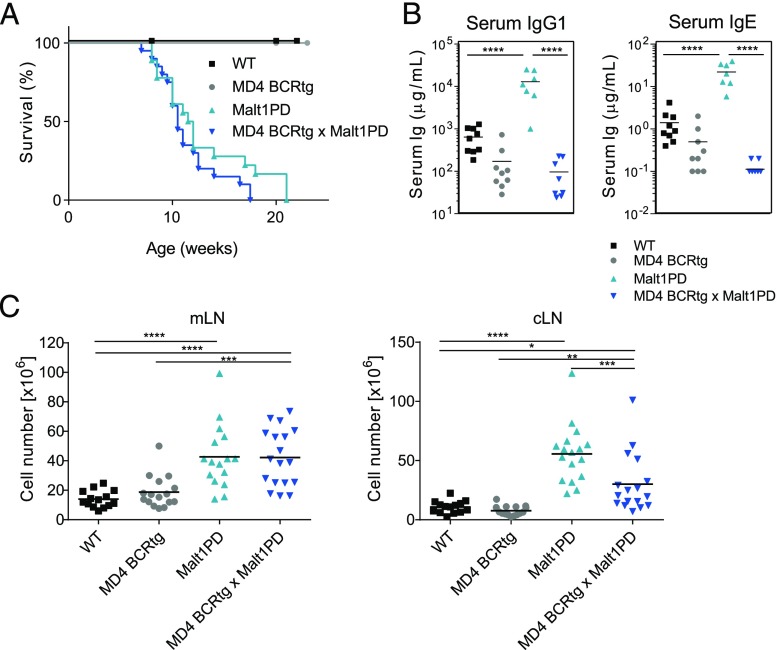
The above data indicated that alterations in the Malt1PD immune compartment are the major driver of the Malt1PD inflammatory disease, in line with previous reports demonstrating that the combined absence of T and B cells in Rag-deficient Malt1PD mice prevents the disease (27). Because B cells and excessive (auto-) Ab production are critical to numerous autoimmune diseases, we investigated the relevance of the B cell compartment as mediator of the Malt1PD pathologic condition. To this end, we crossed the Malt1PD line to MD4 transgenic mice expressing the hen-egg lysozyme (HEL)–specific BCR (MD4 BCRtg mice) (40). The resulting offspring harbored a Malt1PD and monoclonal IgD/IgM–restricted B cell population but retained polyclonal Malt1PD T cells. Both MD4 BCRtg × Malt1PD and littermate Malt1PD mice with a polyclonal B cell repertoire displayed the typical features of the progressive, lethal Malt1PD disease, such as lymphadenopathy and hind limb paralysis. Accordingly, all animals had to be euthanized because of wasting disease (Fig. 3A). Production of IgG1 or IgE subtype Abs by both MD4 BCRtg or MD4 BCRtg × Malt1PD B cells was similar and very limited because of the monoclonal B cell population (37) (Fig. 3B). MD4 BCRtg × Malt1PD mice, as well as Malt1PD controls, showed higher cell counts in mLNs and cLNs as compared with WT or MD4 BCRtg animals. Although the cLN cellularity was reduced in MD4 BCRtg × Malt1PD mice when compared with Malt1PD littermates, these data suggested that the clonality of the B cell population had only little impact on the expansion of other immune cell subsets in Malt1PD animals (Fig. 3C). Thus, the pathologic condition did not require polyclonal B cells or the induction of hyper IgG1 and IgE. Together, these data indicate that the lethal manifestations of disease in Malt1PD mice occurred independently from B cells reacting to commensal, dietary, or self-antigens.
FIGURE 3.
B lymphocytes and IgE/IgG1 elevation are irrelevant for the development and fatal course of the Malt1PD pathologic condition. (A) Kaplan–Meier survival analysis of WT, MD4 BCRtg, Malt1PD, and MD4 BCRtg × Malt1PD mice (animals were euthanized upon loss of ≥20% body weight or appearance of clinical symptoms). Pooled data from three independent experiments. (B) Serum concentration of IgG1 and IgE in animals of strains described in (A) determined at the age of 8–12 wk. Data are representative of at least two independent experiments. (C) Counts of total live cells isolated from mLN (left) or cLN (right) of WT, MD4 BCRtg, Malt1PD, and MD4 BCRtg × Malt1PD animals at 8–12 wk of age. Pooled data from three independent experiments. (A–C) Each dot represents a mouse; lines indicate the mean. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, one-way ANOVA.
T lymphocytes drive the Malt1PD inflammatory disease
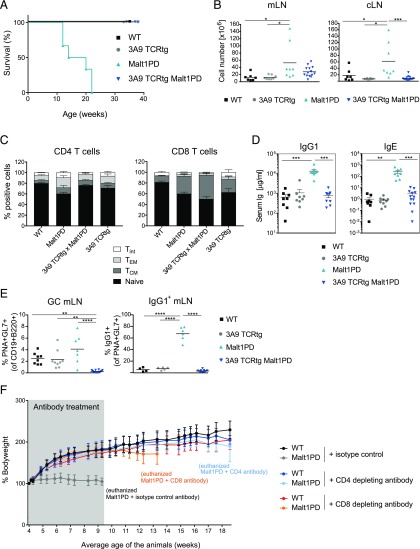
Because B cell reactivity did not drive the lethality of Malt1PD mice, yet when on a Rag−/− background Malt1PD mice were protected (27), T cells were the most likely candidates mediating disease. To evaluate this, we crossed Malt1PD mice to 3A9 CD4 TCRtg animals that harbor a TCR specific for HEL (39) and lack a polyclonal T cell compartment. In contrast to Malt1PD and to MD4 BCRtg × Malt1PD animals, 3A9 TCRtg × Malt1PD animals reached >40 wk of age without displaying clinical symptoms such as body weight loss, hunched posture, eye inflammation, or hind limb paralysis (Fig. 4A and data not shown). Further analysis revealed normal size of cLNs and a trend for reduced mLN cellularity compared with Malt1PD mice (Fig. 4B). Similar to Malt1PD mice, 3A9 TCRtg × Malt1PD animals displayed reduced Treg numbers (data not shown), yet this did not unleash self-reactive T cells. Consistent with the lack of disease symptoms, the distribution of naive, T effector memory (TEM) and central memory CD4 and CD8 T cells were similar between 3A9 TCRtg and 3A9 TCRtg × Malt1PD, as shown for mLN (Fig. 4C). In addition, serum IgG1 and IgE levels in 3A9 TCRtg × Malt1PD animals were normal (Fig. 4D), and there was no increase in GL7+PNA+ germinal center (GC) B cells or IgG1-skewed GC B cells in mLNs (Fig. 4E). Overall, the absence of a polyclonal T cell compartment abrogated the immune alterations and clinical manifestations typical of the Malt1PD pathologic condition, including the activation of B cells and hyperproduction of IgG1 and IgE.
FIGURE 4.
T lymphocytes are the key drivers of the Malt1PD inflammatory disease. (A) Kaplan–Meier survival analysis of WT, 3A9 TCRtg, Malt1PD, and 3A9 TCRtg × Malt1PD animals (mice were euthanized upon loss of ≥20% body weight or appearance of clinical symptoms). Data are representative of at least two independent experiments. (B) Counts of total live cells isolated from mLN (left) and cLN (right) of WT, 3A9 TCRtg, Malt1PD, and 3A9 TCRtg × Malt1PD animals at 8–12 wk of age. Each dot represents a mouse; lines indicate the mean, pooled data from three independent experiments (n = 7–14). (C) Proportion of CD62L+CD44− naive, CD62L+CD44+ central memory (TCM), CD62L−CD44+ TEM or CD62L−CD44− intermediate (Tint) T cells within CD4+ (left) or CD8+ (right) T cells detected by flow cytometry within cells isolated from mLN of WT, Malt1PD, 3A9 TCRtg, and 3A9 TCRtg × Malt1PD animals. The results are expressed as the mean ± SEM, pooled data from three independent experiments (n = 7–14). (D) Serum concentration of IgG1 and IgE in animals of strains described in (A) determined at the age of 8–12 wk. Data are representative of at least two independent experiments. Each dot represents one mouse; results are expressed as the mean ± SEM. (E) Frequency of PNA+GL-7+ GC B cells within CD19+B220+ total B cells (left) and IgG1+ GC B cells within total GC B cells (right) detected by flow cytometry within cells isolated from mLN of WT, 3A9 TCRtg, Malt1PD, and 3A9 TCRtg × Malt1PD animals. Each dot represents a mouse; lines indicate the mean, pooled data from three independent experiments (n = 7–14). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, one-way ANOVA. (F) Bodyweight measurement in C57BL6 WT and Malt1PD animals treated with CD4- or CD8-depleting Abs or the respective isotype control Ab two times per week from 4 wk to ∼9 wk of age (average). At ∼9 wk of age, all Malt1PD isotype control Ab-treated animals had been euthanized because of the appearance of severe clinical symptoms. Four and nine weeks after treatment stop, the CD8- and CD4-depleting Ab-treated groups, respectively, were euthanized because of severe clinical symptoms.
To substantiate the relevance of the T cells and evaluate the contribution of CD4 and CD8 T cells to the overall Malt1PD pathologic condition, we next treated C57BL/6 Malt1PD mice with either CD4- or CD8-depleting Abs. To this end, 4-wk-old Malt1PD animals were treated with depleting CD4 or CD8 mAbs or a relevant isotype control Ab for around 5 wk. Successful depletion of the corresponding subset was confirmed by FACS on blood cells (data not shown). In line with our results from the 3A9 TCRtg animal studies, Malt1PD animals did not show any clinical signs of disease during the treatment with either CD4 or CD8-depleting Abs, and they gained bodyweight to the same extent as isotype control–treated WT animals (Fig. 4F). As expected, isotype control–treated Malt1PD mice failed to gain weight. Interestingly, upon cessation of the treatment, both CD4- and CD8-depleted groups developed clear symptoms of the disease, albeit with distinct kinetics. Although CD8-depleted Malt1PD mice started to lose weight 5 wk after cessation of the treatment, CD4-depleted animals started to display the first clinical signs only 8 wk after treatment stop. Taken together, these data confirm that T cells are the major drivers of disease and indicate that both CD4 and CD8 T cells contribute to the Malt1PD pathologic condition.
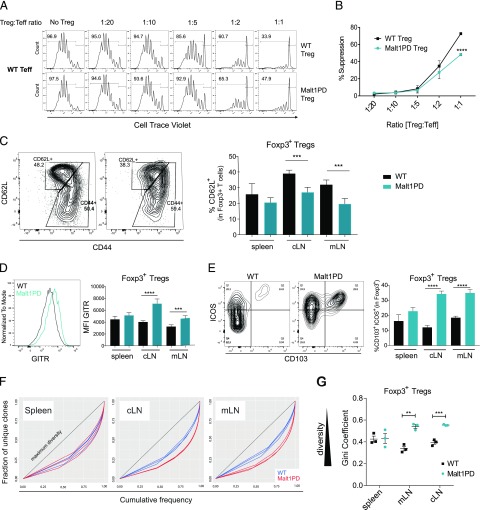
Malt1PD Tregs retain in vitro functional capacity and demonstrate an effector phenotype in vivo
Prior research confirmed that the Malt1PD mouse pathological symptoms can be largely rescued via the transfer of WT Tregs early after birth (26), suggesting that the reduction in Tregs is the root cause of pathogenic cell expansion (29, 58). It remained unknown, however, whether Tregs are functionally normal in a Malt1PD situation. To explore the phenotype and function of Malt1PD Tregs ex vivo, we crossed the Foxp3EGFP reporter mouse (35) to Malt1PD mice. In an in vitro suppression assay using WT conventional T cells as effectors, Malt1PD Tregs isolated from pooled spleen and LNs retained substantial suppressive capacity compared with WT Tregs, which demonstrated superior function only at high Treg to Teff ratios (Fig. 5A, 5B). Malt1PD and WT Tregs were also able to inhibit the proliferation of Malt1PD Teffs (Supplemental Fig. 4A). Although Malt1PD Teffs proliferated less extensively than WT effector cells, and consequently were more efficiently suppressed, these data suggested that Malt1PD Teffs are not intrinsically resistant to Treg suppression.
FIGURE 5.
Malt1PD Tregs display a tissue-specific activation pattern and retain partial suppressive capacity. (A) Increasing numbers of WT or Malt1PD CD4+Foxp3EGFP+CD25+ Tregs were added to 105 WT CD4+ Foxp3EGFP−CD25− T cells in presence of nonirradiated BM-derived dendritic cells and an anti-CD3 Ab. The proliferation of Teffs, as measured by dilution of cell trace violet, was assessed after 5 d by flow cytometry. Teffs and Tregs were sorted from pooled LNs and spleen. Data are representative of four independent experiments. (B) Percent suppression relative to Treg/Teff ratios. The results are expressed as the mean ± SEM (n = 3). Statistical differences were determined using two-way ANOVA. (C–E) Expression of activation markers on WT or Malt1PD Foxp3EGFP+ CD4+ Tregs assessed by flow cytometry. The results are expressed as the mean ± SEM (n = 5). Data are representative of three independent experiments. (C) Dot plots showing expression of CD62L versus CD44 in WT or Malt1PD Foxp3EGFP+ Tregs from cLNs. Bar graph displays the proportion of CD62L+ Tregs assessed in the indicated organs. (D) Histogram showing expression of GITR in WT or Malt1PD Foxp3EGFP+ Tregs from cLN. Bar graph displays GITR mean fluorescence intensity on Foxp3EGFP+ Tregs assessed in the indicated organs. (E) Dot plots showing expression of ICOS versus CD103 in WT or Malt1PD EGFP+ Tregs from cLNs. Bar graph displays % CD103+ICOS+ within Foxp3EGFP+ Tregs assessed in indicated organs. (F and G) WT or Malt1PD CD4+ Foxp3EGFP+ Tregs were FACS sorted and subjected to deep sequencing after 5′RACE-based amplification of TCRβ sequences. (F) Lorenz curve visualization of TCRβ sequence diversity in WT or Malt1PD Foxp3EGFP+ Tregs isolated from spleen (left), cLN (middle), and mLN (right). (G) Evaluation of Treg TCRβ sequence diversity based on the Gini coefficient. **p < 0.01, ***p < 0.001, ****p < 0.0001.
Further phenotyping by FACS revealed an LN-specific activation pattern for Tregs in Malt1PD animals, as exemplified by reduced proportions of naive CD62L+ Tregs (Fig. 5C) and increased expression of GITR (Fig. 5D), CD103, and ICOS (Fig. 5E) specifically in cLN and mLN. Another important determinant of T cell function is the diversity of the TCR repertoire. Whether a restricted Treg TCR diversity compromises Treg suppressive capacity in vivo is still a subject of debate (59–63). Given the severe impairment of thymic Treg development in Malt1PD animals (26–29), we evaluated whether Malt1PD Tregs in peripheral lymphoid organs displayed a somewhat restricted TCR repertoire. To test this, cDNA was generated from sorted splenic and LN-derived CD4+Foxp3EGFP+ Tregs isolated from Malt1PD × Foxp3EGFP animals or control WT × Foxp3EGFP littermates, and amplified TCR-Vβ sequences were subjected to deep sequencing. The diversity of TCR sequences was evaluated and visualized using the Gini coefficient and Lorenz curve as previously described (43, 64). In such a representation, a straight diagonal line indicates maximum diversity (i.e., all clonotypes are present at the same frequency, whereas repertoire skewing is shown by curves deviating below the diagonal, meaning individual clones occur more abundantly). When comparing clonotypes from WT to Malt1PD Tregs in the spleen we found equal measures of diversity in both sample sets, implying that the thymic Treg output in Malt1PD animals did not significantly impact the Treg TCR repertoire diversity (Fig. 5F, 5G). In contrast, Malt1PD Tregs in cLN and mLN demonstrated significantly less diverse TCR repertoires as indicated by greater deviation from the diagonal (Fig. 5F) and lower Gini coefficients (Fig. 5G) compared with WT Tregs in those LNs. Interestingly, these are the anatomical locations of increased cellularity and IgG1 and IgE Ab production. Consistent with the increased activation and expansion of T and B effector cells in cLN and mLN of Malt1PD animals, Tregs displayed an effector phenotype and restricted TCR repertoires in enlarged LNs potentially reflecting Ag-induced activation and clonal expansion in response to the local inflammation. In summary, although Malt1PD Tregs displayed some in vitro suppressive activity, variable TCR diversity, and an effector phenotype, they were not able to prevent the expansion of pathogenic T cells in vivo.
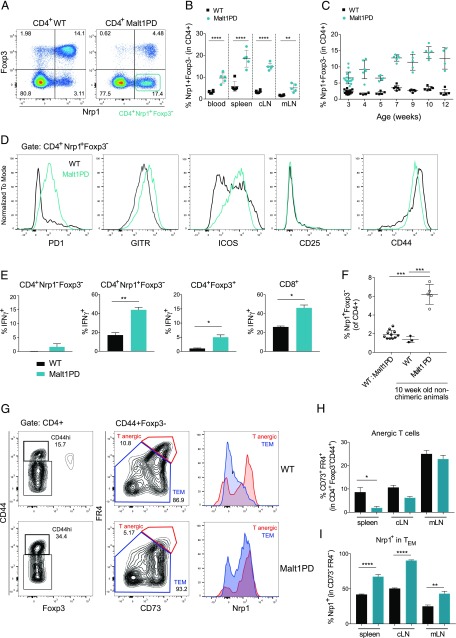
Lack of Tregs favors progressive expansion of CD4+ Nrp1+ Foxp3− T cells
When characterizing the distribution of nTreg versus iTreg based on the expression of Nrp1 (55–57), we noticed a population of CD4+ Nrp1+Foxp3− T cells (Fig. 6A), which was found at an elevated frequency in all lymphoid organs of adult Malt1PD animals (Fig. 6B). These cells appeared early after weaning and expanded progressively over time (Fig. 6C). Expression of CD44 and ICOS marked both WT and Malt1PD CD4+Nrp1+Foxp3− T cells as Ag-experienced, activated T cells, although only low levels of CD25 were detected (Fig. 6D). Malt1PD CD4+Nrp1+Foxp3− T cells additionally expressed high levels of GITR and PD1, concordant with a Teff/memory T cell phenotype. When we determined the in vitro cytokine profile upon PMA/ionomycin stimulation, CD4+Nrp1+Foxp3− T cells turned out to be the major producers of various proinflammatory cytokines in both WT and Malt1PD mice compared with naive CD4+Nrp1−Foxp3− T cells (>95% CD44−CD62L+) or CD4+Foxp3+ Tregs, (Supplemental Fig. 4B). IFN-γ, however, was more readily produced by Malt1PD CD4+Nrp1+Foxp3− T cells and CD8+ T cells than by the corresponding WT subsets (Fig. 6E). These data thus indicated Th1-skewed T cell responses in Malt1PD animals. Consistently, previous reports demonstrated a crucial role for IFN-γ+ cells in mediating the observed Malt1PD neuropathology (27). Interestingly, CD4+Nrp1+Foxp3− T cells did not expand in 1:1 (WT/Malt1PD) mixed BM chimeras (Fig. 6F) and in 3A9 TCRtg × Malt1PD animals (Supplemental Fig. 4C), consistent with their disease-free phenotype. In these settings, either the presence of WT Tregs or the lack of polyclonal conventional T cells seemed to prevent the expansion of CD4+Nrp1+Foxp3− T cells. Therefore, IFN-γhiNrp1+Foxp3− CD4+ Teffs were identified as an expanded subset in Malt1PD mice, which are potentially involved in disease development.
FIGURE 6.
Lack of Tregs favors progressive expansion of CD4+Nrp1+Foxp3− T cells. (A–H) Flow cytometry–based characterization of CD4+Nrp1+Foxp3− T cells in WT or Malt1PD cells isolated from blood, spleen, cLN, or mLN. (A) Dot plot displaying Foxp3 versus Nrp1 pregated on CD4+ T cells in cLN. (B) Frequency of CD4+Nrp1+Foxp3− T cells in 8–12-wk-old WT or Malt1PD animals detected in the indicated organs. The results are expressed as the mean ± SEM; each dot represents a mouse. (C) Frequency of Nrp1+Foxp3− T cells among CD4+ T cells in cLNs of 3–12-wk-old WT or Malt1PD animals. (D) Histograms showing expression of PD1, GITR, ICOS, CD25, and CD44 in WT or Malt1PD Nrp1+Foxp3− among CD4+ T cells T cells from cLNs. (E) Frequency of IFN-γ+ cells within populations of CD4+Nrp1−Foxp3− naive T cells, CD4+Nrp1+Foxp3− T cells, CD4+Foxp3+ Tregs, and CD8+ T cells after 4 h PMA/Ionomycin stimulation of cLN cells isolated from WT or Malt1PD animals. The results are expressed as the mean ± SEM (n = 5). Data are representative of at least two independent experiments. (F) Frequency of Nrp1+Foxp3− T cells among CD4+ T cells T cells in blood of 1:1 (WT/Malt1PD) mixed BM chimeras 9 wk after reconstitution compared with 10–12-wk-old nonchimeric WT or Malt1PD animals. (G) Dot plots showing the gating on anergic T cells in spleens of WT (upper panel) and Malt1PD (lower panel) animals at the age of 8–12 wk. CD4 T cells were gated on CD44+Foxp3− cells, which were further divided into CD73+FR4+ anergic T cells (Tanergic) and CD73lowFR4low TEM. Histograms display expression of Nrp1 in Tanergic versus TEM. (H and I) Frequency of anergic T cells (H) and Nrp1+ cells within TEM (I) in indicated organs of 8–12-wk-old WT or Malt1PD animals. The results are expressed as the mean ± SEM (n = 5). Data are representative of at least two independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
The source of the Nrp1+Foxp3− CD4+ Teff subset is not well understood. These may originate from either Foxp3+Nrp1+ nTregs losing Foxp3 expression, be derived from Foxp3+Nrp1− iTregs losing Foxp3 and acquiring Nrp1, or differentiate from Foxp3−Nrp1+ or Foxp3−Nrp1− CD4+ T cell precursors. Identifying the exact source of these cells deserves to be further explored using appropriate tracing or reporter mouse strains. However, we were intrigued by a similar population of CD4+Foxp3−CD44hi T cells expressing high levels of Nrp1, folate receptor 4 (FR4), and CD73, which was recently suggested to identify a population of anergic T cells endowed with a high degree of plasticity and the capacity to convert either into Foxp3+ Tregs or autoreactive TEM cells (65). Of particular note, abrogation of the Treg compartment upon transfer of these anergic cells into T cell–deficient recipients led to the rapid expansion of IFN-γ–producing pathogenic CD4+Foxp3−CD44hiFR4−CD73− TEM cells and systemic activation of autoantibody-secreting B cells (65). Although FR4−CD73− TEM cells were mostly Nrp1−, more than 80% of FR4hiCD73hi anergic T cells expressed high levels of Nrp1, thus resembling the CD4+Nrp1+Foxp3− T cell population expanding in Malt1PD mice. Based on these parallels, we evaluated if the reduced Treg frequency in Malt1PD mice was associated with reduced maintenance of anergic cells. In support of this, Malt1PD animals harbored less (CD4+Foxp3−CD44hi) FR4hiCD73hi anergic T cells than WT mice in spleen and cLNs (Fig. 6G, 6H). The reduction in anergic cells was paralleled by the expansion of CD4+FR4−CD73− TEM expressing Nrp1 (Fig. 6I). Reduced anergic and increased TEM markers were also observed on T cells isolated from blood or kidneys of Malt1PD animals (data not shown), indicating that this phenotype was not restricted to specific anatomical sites or lymphoid organs. Overall, based on these findings, we propose that the lack of Tregs in Malt1PD mice impairs the maintenance of T cell anergy, leading to their progressive conversion into pathogenic CD4+ TEM retaining Nrp1 expression.
In summary, our data determine the Treg deficiency in Malt1PD animals as the primary cause of both disturbed mucosal immunity driven by commensal-derived factors, and a systemic, T cell–driven IPEX-like disease very similar to the pathologies characterizing human MALT1 mutant patients.
Discussion
Our group and others have extensively described the IPEX-like syndrome that is consequent to the disruption of Malt1 protease function in mice (26–29), a disease with many similarities to the conditions affecting human patients bearing mutations in the MALT1-encoding gene (11–14, 56). In this study, we further dissected the triggers and the relative contribution of T and B cells to immune dysregulation and disease development.
Our data revealed that the excessive humoral immune response in Malt1PD mice, reflected by elevated serum IgG1 and IgE, was mainly directed against microbial and food Ags. This was supported by absence of hyper IgG1 and significant reduction of IgE in Malt1PD animals when housed under GF conditions, which also suggested that Ab responses are not primarily directed against systemic autoantigens. By studying MD4 BCRtg × Malt1PD mice, which are characterized by a monoclonal B cell repertoire and defective IgG1/IgE production, we demonstrated that B cells and elevated IgG1/IgE responses in Malt1PD mice are not the drivers of the most severe pathological symptoms. These findings contrast with a B cell–dependent disease component observed in Foxp3-deficient scurfy mice and human IPEX patients, which develop a broad set of autoantibodies (66–69). The lack of systemic autoantibodies in SPF Malt1PD mice per se can be explained by the fact that Malt1PD B cell responses to BCR-driven T-dependent and T-independent type 2 Ags are impaired (Refs. 26–29 and K. Martin, G. Cvijetic, V. Ottaviani, R. Touil, Y. Kolb, D. Zotos, F. Bornancin, D. Tarlinton, and T. Calzascia, manuscript in preparation). Given the intrinsic defect in mounting BCR-driven B cell responses in Malt1PD mice, we propose that activation of B cells by Malt1-independent pathways such as through pattern recognition receptors like TLRs is involved in the generation of elevated serum Igs in these animals. The complete normalization of IgG1 and partial reduction of IgE in GF animals indicated that commensal-derived signals were required for B cell activation to both microbial and food-derived Ags. The restricted presence of IgG- and IgE-secreting B cells in LNs draining barrier sites such as the eyes, the oral mucosa, and the intestine further supports the localized activation of B cells by environmental Ags. Also, given the crucial role of Tregs in the establishment of oral tolerance to dietary Ags (70–72) and the low percentages of both nTregs and iTregs in mucosal tissues of Malt1PD animals, we suggest that impaired oral tolerance results in food-reactive IgG1 and IgE in Malt1PD animals. Based on these data, we propose that Malt1PD B cells produce IgG1 and IgE Abs to environmental Ags as a result of impaired oral tolerance and upon activation through a combination of BCR triggering and microbial-derived innate signals that activate pattern recognition receptors.
Consistent with a disruption of intestinal immune homeostasis and barrier function, Malt1PD mice have previously been reported to display exacerbated disease in the DSS-induced colitis model (18). This, together with the hyper reactivity of Malt1PD mice to food and other Ags from the intestinal lumen, prompted us to investigate whether Malt1 signaling in epithelial cells could render intestinal tight junctions more permissive for Ags present in the intestinal lumen. However, although CBM complex activation plays a role in both endothelial and epithelial cells (17, 19, 52, 73, 74), we could not demonstrate a dominant role for Malt1 signaling in the nonhematopoietic compartment in driving the exacerbation of DSS-induced colitis. Instead, we confirmed the hematopoietic compartment as major driver of the Malt1PD mouse pathologic condition (26–29) and propose that, rather than defective Malt1 signaling in epithelial cells, the disrupted intestinal immune homeostasis consequent to the global reduction of nTregs and iTregs in lymphoid organs and in the local gut environment is the main factor driving the loss of the intestinal barrier and the consequent induction of Abs against commensal Ags. In this context, an inflammatory microenvironment can indirectly affect mucosal homeostasis by altering epithelial intercellular junctions and thus allow for increased accessibility of microbiota- and food-derived Ags (75). Although our data indicate that Malt1 protease deficiency restricted to nonhematopoietic cells is not sufficient on its own to disrupt the intestinal barrier, our BM chimera studies do not rule out a potential contribution of nonimmune cells to the progression of the disease that occurs in mice characterized by a systemic Malt1 protease deficiency.
In contrast to our observations, Jaworski et al. reported the reactivity of serum IgG1 and IgE to glandular stomach tissue Ags and suggested autoimmune gastritis to be the leading cause of death in their Malt1PD colony (26). We were unable to confirm the reactivity of serum IgG1 or IgE to glandular stomach tissue in our Malt1PD strain. This discrepancy may relate to differences in the approaches used for the detection of Abs or to differences in animal housing conditions, including variable hygiene levels and the microbiome. Supporting the relevance of the microbiome, we observed that GF Malt1PD mice display a faster and more severe appearance of clinical symptoms, suggesting that microbiota-derived factors can influence the disease kinetic and its clinical manifestations (76–78). As an example, commensals were reported to protect hosts from overt intestinal inflammation by the production of IL-10 family cytokines (79). With this, early life exposure to microbes may also influence susceptibility to and the outcome of specific autoimmune manifestations (80).
The limited contribution of the B cell compartment to the disease in our Malt1PD colony led us to identify T cells as the drivers of the severe clinical symptoms. In absence of polyclonal T cells, 3A9 TCRtg × Malt1PD animals displayed no signs of overt inflammatory disease and no clinical symptoms. Interestingly, the hyper IgG1/IgE was also absent in 3A9 TCRtg × Malt1PD animals, suggesting that a T cell–dependent component was required for humoral responses to environmental Ags. Because B cell help from conventional CD4 T cells is nonfunctional in Malt1PD animals, we evaluated whether iNKT helper cells may drive B cell responses to microbial Ags of glycolipid nature in Malt1PD mice (18, 45, 81). However, we found no relevance for these cells for B cell activation. Overall, our data strongly indicate that B cell activation is secondary to the T cell–mediated inflammatory environment and likely driven by bacterial triggers as a consequence of the opening of the intestinal barrier. In contrast to the systemic autoimmune disease in Foxp3-deficient individuals, the more restricted and specific immune alterations and clinical manifestations characterizing the IPEX-like syndrome in Malt1PD animals likely reflect the lack of systemic autoantibody responses and the suboptimal properties of T cells consequent to the disruption of the catalytic function of Malt1.
An additional learning derived from our studies is that Malt1PD Tregs, albeit reduced, retain substantial suppressive capacity in vitro. These cells also displayed an activated phenotype and reduced TCR diversity specifically in LNs affected by the pathologic condition. These data suggest that Tregs at these locations may clonally expand to suppress and oppose the local effector cell activation. Consistently, previous reports suggest that distinct sets of Treg TCRs found in diverse locations confer tissue-specific immunity (60, 82, 83), and expansion of specific TCR sequences in tissue-draining LNs is characteristic of the activated effector Treg population (84). Despite the in vitro suppressive activity, variable TCR diversity, and effector phenotype in vivo, Malt1PD Tregs were unable to prevent the expansion of pathogenic T cells. Although the reduced frequency of Tregs in Malt1PD mice may on its own be sufficient to drive the inflammatory disease, the absence of Malt1 protease function in Tregs may also impact additional Treg immunoregulatory mechanisms critical for the maintenance of immune homeostasis in vivo. To this point, four recent studies provided critical novel insights into the functional consequences of the disruption of the CBM complex in Foxp3+ Tregs via the genetic deletion of CARD11, BCL10, or MALT1 or via the genetic inactivation of the MALT1 protease (85–88). In line with our findings, Cheng et al. reported that Malt1PD Tregs display an overall normal in vitro suppression activity, whereas Rosenbaum et al. suggested a partial reduction in in vitro suppressive activity. Importantly, Cheng et al. also demonstrated that despite the overall normal suppressive activity in vitro, Malt1PD Tregs displayed impaired in vivo suppressive activity in an adoptive transfer colitis model. Consistent with this, both groups observed that the genetic inactivation of the Malt1 protease function selectively in Foxp3+ Tregs is sufficient to drive a severe IPEX-like inflammatory disease. In line with this, Demeyer et al. (86) reported that mice characterized by a T cell–restricted Malt1 protease deficiency develop an IPEX-like syndrome similar to the one reported in mice with a systemic Malt1 protease defect. Additional data involving the use of Malt1PD Tregs or WT Tregs treated with a MALT1 inhibitor suggest that inhibition of the Malt1 protease function results in defective expression of CTLA4, IL-10, and TGF-β (85, 88). Overall, these studies indicate that CBM components and the MALT1 protease function are critically required to maintain the optimal suppressive function and identity of Tregs in vivo. These results highlight that, despite the normal or partially reduced suppressive activity of Malt1PD Tregs observed in in vitro suppression assays, these in vitro systems do not recapitulate the multitude of functional properties and suppressive mechanisms required by Tregs to maintain immune homeostasis in vivo. In summary, these novel reports complement our work and indicate that besides the reduced frequency of Tregs in our Malt1PD animals, defects in their functional properties in vivo also contribute to the expansion of pathogenic T cells in Malt1PD mice.
A limitation of our system is the use of TCRtg (and BCRtg) animals instead of specific T cell (and B cell) knockout strains to dissect the role of B versus T cells. Nevertheless, our data clearly mark T cells as the drivers of disrupted intestinal immune homeostasis and the systemic inflammation. Consistently, prior research identified IFN-γ–producing cells as the subset responsible for wasting disease, the destruction of Purkinje cells, and the associated neurological disease, but not for lymphadenopathy (27). Although innate B cells, intestinal IL-C1, or myeloid cells have been shown to produce IFN-γ in response to cytokines or pathogens (89, 90), IFN-γ is predominantly released by T and NK cells (91). As NK cells and other cell subsets capable of producing IFN-γ are still present in 3A9 TCRtg × Malt1PD animals, we suggest that conventional αβ T lymphocytes are the main drivers of the Malt1PD pathologic condition. This is supported also by the clear delay in disease progression observed upon depletion of either CD8 or CD4 T cells in Malt1PD mice. Interestingly, CD4+ helper T cells rather than CD8+ Teffs are the driver of the systemic autoimmunity in Foxp3-deficient scurfy mice, and a mixed Th1/Th2 response caused the death of these animals (92). In analogy to these results, we previously noticed a mixed Th1/Th2 cytokine expression upon in vitro stimulation of Malt1PD CD4+ T cells (Ref. 29 and T. Calzascia and R. Touil, unpublished observations). Based on our findings, we propose that both CD4+ Foxp3− T cells expressing Nrp1 and CD8+ T cells contribute to the pathologic condition in Malt1PD animals. However, at this stage we have been unable to formally demonstrate the pathogenicity of either cell subset via adoptive transfer experiments into Rag2−/− recipient animals. This may reflect the reduced functionality of Malt1PD T cells, limiting their expansion and activity after transfer. In the same line, Malt1PD CD4+ T cells largely failed to induce T cell–mediated colitis upon adoptive transfer into Rag2-deficient recipient animals (26). The expansion of CD4+Nrp1+Foxp3− T cells was also reported in association with an IPEX-like disease occurring in CD4-specific TGF-βRII−/− animals, which are also characterized by reduced Tregs (55). CD4+Nrp1+Foxp3− T cells in those mice were described as activated/memory T cells that could not convert into Tregs upon thymic injection (18). Also, expression of Nrp1 was recently described on a CD4+Foxp3−CD44+FR4+CD73+ T cell population that the authors associated with anergic T cells (65). Interestingly, these cells displayed the capacity to generate Foxp3+ Tregs upon adoptive transfer into T cell–deficient hosts. However, upon elimination of Tregs, anergic T cells rapidly converted into autoreactive IFN-γ–secreting TEM cells driving systemic B cell activation. Based on these similarities we propose that the expansion of CD4+Nrp1+Foxp3− T cells in Malt1PD mice reflects the progressive expansion of a subset of ex-anergic T cells that differentiate into pathogenic effector cells as a consequence of the Treg reduction. Although such a hypothesis is supported by the reduced frequency of CD4+Foxp3−CD44+FR4+CD73+ anergic T cells in Malt1PD mice, additional work is required to demonstrate the pathogenicity of FR4−CD73−Nrp1+ TEM cells and their direct relationship to anergic cells. Alternative sources of CD4+Foxp3−Nrp1+ T cells may include nTregs or iTregs losing Foxp3 expression or even CD4+Foxp3−Nrp1− subsets. Dedicated studies using appropriate tracing or reporter mouse lines will be required to address the exact origin of these cells. Overall, this work establishes the Malt1PD pathologic condition as a truly T cell–driven autoimmune disease. Nevertheless, both the activation of autoreactive T cells resulting in systemic disease and the induction of B cells by environmental Ags as a result of mucosal immune imbalance have their roots in the defective regulatory compartment characterizing Malt1PD animals.
In light of the great interest in pharmacological MALT1 protease inhibition for the treatment of multiple autoimmune diseases and lymphoid malignancies (15, 93), it is an important goal to understand the contribution of the MALT1 protease function to the maintenance of both intestinal and systemic immune homeostasis. Given the progressive disease in Malt1PD animals, it remains to be tested whether long-term treatment using such inhibitors is safe. Short-term use of the pharmacological inhibitors of the MALT1 protease, such as MI-2 and mepazine, proved beneficial for the outcome of DSS-induced colitis in mice due to reduced proinflammatory cytokine production by myeloid cells in the colon and did not provoke major toxicities (94–96). Similarly, treatment with mepazine attenuated the onset and progression of experimental autoimmune encephalomyelitis and was safe in mice when given over a period of 17 d (97). Despite the protective effects reported for these inhibitors, their potency is limited and their selectivity is questionable (23, 98). Therefore, novel potent and more selective pharmacological inhibitors are required to evaluate whether durable MALT1 inhibition in adult individuals will be safe or impact immune homeostatic mechanisms that will result in a progressive IPEX-like disease similar to that observed in Malt1PD mice.
Supplementary Material
Acknowledgments
We thank Mitchell Kronenberg for providing the Traj18 knockout animals, Dirk Brenner for the support provided in establishing and monitoring the Malt1PD mouse line in Toronto, Ulrike Naumann for the sequencing of T cell samples and analysis of the raw data using MiXCR, Elisabetta Traggiai and Vanessa Cornacchione for input on the set up of the anti-cecal ELISA, and Ursula Junker Walker and Diethilde Theil for help in assessing the phenotype of TCRtg and BCRtg Malt1PD animals.
The sequences presented in this article have been submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/data/view/PRJEB34151) under accession number PRJEB34151.
The online version of this article contains supplemental material.
- BM
- bone marrow
- cLN
- cervical/mandibular LN
- DSS
- dextran sodium sulfate
- FR4
- folate receptor 4
- GC
- germinal center
- GF
- germ-free
- IEL
- intraepithelial lymphocyte
- iNKT
- invariant NKT
- IPEX
- immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome
- iTreg
- inducible Treg
- LN
- lymph node
- LP
- lamina propria
- Malt1PD
- Malt1 protease-deficient
- Nrp1
- Neuropilin1
- nTreg
- natural, thymus-derived Treg
- SPF
- specific pathogen–free
- Teff
- effector T cell
- TEM
- T effector memory
- Treg
- regulatory T cell
- WT
- wild-type.
Disclosures
K. Martin, R.T., Y.K., G.C., L.I., F.D., D.B., A.G., S.N., M.B., T.J., N.Z., P.S., F.B., and T.C. are or were employees and/or shareholders of Novartis Pharma AG. The other authors have no financial conflicts of interest.
References
- 1.Azizi G., Yazdani R., Rae W., Abolhassani H., Rojas M., Aghamohammadi A., Anaya J. M. 2018. Monogenic polyautoimmunity in primary immunodeficiency diseases. Autoimmun. Rev. 17: 1028–1039. [DOI] [PubMed] [Google Scholar]
- 2.Notarangelo L. D., Gambineri E., Badolato R. 2006. Immunodeficiencies with autoimmune consequences. Adv. Immunol. 89: 321–370. [DOI] [PubMed] [Google Scholar]
- 3.Casanova J. L., Holland S. M., Notarangelo L. D. 2012. Inborn errors of human JAKs and STATs. Immunity 36: 515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett C. L., Christie J., Ramsdell F., Brunkow M. E., Ferguson P. J., Whitesell L., Kelly T. E., Saulsbury F. T., Chance P. F., Ochs H. D. 2001. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27: 20–21. [DOI] [PubMed] [Google Scholar]
- 5.Godfrey V. L., Wilkinson J. E., Russell L. B. 1991. X-linked lymphoreticular disease in the scurfy (sf) mutant mouse. Am. J. Pathol. 138: 1379–1387. [PMC free article] [PubMed] [Google Scholar]
- 6.Bacchetta R., Barzaghi F., Roncarolo M. G. 2018. From IPEX syndrome to FOXP3 mutation: a lesson on immune dysregulation. Ann. N. Y. Acad. Sci. 1417: 5–22. [DOI] [PubMed] [Google Scholar]
- 7.Gambineri E., Ciullini Mannurita S., Hagin D., Vignoli M., Anover-Sombke S., DeBoer S., Segundo G. R. S., Allenspach E. J., Favre C., Ochs H. D., Torgerson T. R. 2018. Clinical, immunological, and molecular heterogeneity of 173 patients with the phenotype of immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) syndrome. Front. Immunol. 9: 2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsuda M., Torgerson T. R., Selmi C., Gambineri E., Carneiro-Sampaio M., Mannurita S. C., Leung P. S., Norman G. L., Gershwin M. E. 2010. The spectrum of autoantibodies in IPEX syndrome is broad and includes anti-mitochondrial autoantibodies. J. Autoimmun. 35: 265–268. [DOI] [PubMed] [Google Scholar]
- 9.Verbsky J. W., Chatila T. A. 2013. Immune dysregulation, polyendocrinopathy, enteropathy, X-linked (IPEX) and IPEX-related disorders: an evolving web of heritable autoimmune diseases. Curr. Opin. Pediatr. 25: 708–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKinnon M. L., Rozmus J., Fung S. Y., Hirschfeld A. F., Del Bel K. L., Thomas L., Marr N., Martin S. D., Marwaha A. K., Priatel J. J., et al. 2014. Combined immunodeficiency associated with homozygous MALT1 mutations. J. Allergy Clin. Immunol. 133: 1458–1462, 1462.e1–7. [DOI] [PubMed] [Google Scholar]
- 11.Punwani D., Wang H., Chan A. Y., Cowan M. J., Mallott J., Sunderam U., Mollenauer M., Srinivasan R., Brenner S. E., Mulder A., et al. 2015. Combined immunodeficiency due to MALT1 mutations, treated by hematopoietic cell transplantation. J. Clin. Immunol. 35: 135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rozmus J., McDonald R., Fung S. Y., Del Bel K. L., Roden J., Senger C., Schultz K. R., McKinnon M. L., Davis J., Turvey S. E. 2016. Successful clinical treatment and functional immunological normalization of human MALT1 deficiency following hematopoietic stem cell transplantation. Clin. Immunol. 168: 1–5. [DOI] [PubMed] [Google Scholar]
- 13.Jabara H. H., Ohsumi T., Chou J., Massaad M. J., Benson H., Megarbane A., Chouery E., Mikhael R., Gorka O., Gewies A., et al. 2013. A homozygous mucosa-associated lymphoid tissue 1 (MALT1) mutation in a family with combined immunodeficiency. J. Allergy Clin. Immunol. 132: 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charbit-Henrion F., Jeverica A. K., Bègue B., Markelj G., Parlato M., Avčin S. L., Callebaut I., Bras M., Parisot M., Jazbec J., et al. GENIUS Group 2017. Deficiency in mucosa-associated lymphoid tissue lymphoma translocation 1: a novel cause of IPEX-like syndrome. J. Pediatr. Gastroenterol. Nutr. 64: 378–384. [DOI] [PubMed] [Google Scholar]
- 15.Jaworski M., Thome M. 2016. The paracaspase MALT1: biological function and potential for therapeutic inhibition. Cell. Mol. Life Sci. 73: 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosebeck S., Rehman A. O., Lucas P. C., McAllister-Lucas L. M. 2011. From MALT lymphoma to the CBM signalosome: three decades of discovery. Cell Cycle 10: 2485–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klei L. R., Hu D., Panek R., Alfano D. N., Bridwell R. E., Bailey K. M., Oravecz-Wilson K. I., Concel V. J., Hess E. M., Van Beek M., et al. 2016. MALT1 protease activation triggers acute disruption of endothelial barrier integrity via CYLD cleavage. Cell Rep. 17: 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borthakur A., Bhattacharyya S., Alrefai W. A., Tobacman J. K., Ramaswamy K., Dudeja P. K. 2010. Platelet-activating factor-induced NF-kappaB activation and IL-8 production in intestinal epithelial cells are Bcl10-dependent. Inflamm. Bowel Dis. 16: 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zotti T., Polvere I., Voccola S., Vito P., Stilo R. 2018. CARD14/CARMA2 signaling and its role in inflammatory skin disorders. Front. Immunol. 9: 2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staal J., Driege Y., Haegman M., Borghi A., Hulpiau P., Lievens L., Gul I. S., Sundararaman S., Gonçalves A., Dhondt I., et al. 2018. Ancient origin of the CARD-coiled coil/Bcl10/MALT1-like paracaspase signaling complex indicates unknown critical functions. Front. Immunol. 9: 1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wegener E., Krappmann D. 2007. CARD-Bcl10-Malt1 signalosomes: missing link to NF-kappaB. Sci. STKE 2007: pe21. [DOI] [PubMed] [Google Scholar]
- 22.Lu H. Y., Bauman B. M., Arjunaraja S., Dorjbal B., Milner J. D., Snow A. L., Turvey S. E. 2018. The CBM-opathies-A rapidly expanding spectrum of human inborn errors of immunity caused by mutations in the CARD11-BCL10-MALT1 complex. Front. Immunol. 9: 2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bardet M., Unterreiner A., Malinverni C., Lafossas F., Vedrine C., Boesch D., Kolb Y., Kaiser D., Glück A., Schneider M. A., et al. 2018. The T-cell fingerprint of MALT1 paracaspase revealed by selective inhibition. Immunol. Cell Biol. 96: 81–99. [DOI] [PubMed] [Google Scholar]
- 24.Coornaert B., Baens M., Heyninck K., Bekaert T., Haegman M., Staal J., Sun L., Chen Z. J., Marynen P., Beyaert R. 2008. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat. Immunol. 9: 263–271. [DOI] [PubMed] [Google Scholar]
- 25.Rebeaud F., Hailfinger S., Posevitz-Fejfar A., Tapernoux M., Moser R., Rueda D., Gaide O., Guzzardi M., Iancu E. M., Rufer N., et al. 2008. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat. Immunol. 9: 272–281. [DOI] [PubMed] [Google Scholar]
- 26.Jaworski M., Marsland B. J., Gehrig J., Held W., Favre S., Luther S. A., Perroud M., Golshayan D., Gaide O., Thome M. 2014. Malt1 protease inactivation efficiently dampens immune responses but causes spontaneous autoimmunity. EMBO J. 33: 2765–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gewies A., Gorka O., Bergmann H., Pechloff K., Petermann F., Jeltsch K. M., Rudelius M., Kriegsmann M., Weichert W., Horsch M., et al. 2014. Uncoupling Malt1 threshold function from paracaspase activity results in destructive autoimmune inflammation. Cell Rep. 9: 1292–1305. [DOI] [PubMed] [Google Scholar]
- 28.Yu J. W., Hoffman S., Beal A. M., Dykon A., Ringenberg M. A., Hughes A. C., Dare L., Anderson A. D., Finger J., Kasparcova V., et al. 2015. MALT1 protease activity is required for innate and adaptive immune responses. PLoS One 10: e0127083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bornancin F., Renner F., Touil R., Sic H., Kolb Y., Touil-Allaoui I., Rush J. S., Smith P. A., Bigaud M., Junker-Walker U., et al. 2015. Deficiency of MALT1 paracaspase activity results in unbalanced regulatory and effector T and B cell responses leading to multiorgan inflammation. J. Immunol. 194: 3723–3734. [DOI] [PubMed] [Google Scholar]
- 30.Ruland J., Duncan G. S., Wakeham A., Mak T. W. 2003. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity 19: 749–758. [DOI] [PubMed] [Google Scholar]
- 31.Ruefli-Brasse A. A., French D. M., Dixit V. M. 2003. Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science 302: 1581–1584. [DOI] [PubMed] [Google Scholar]
- 32.Hachmann J., Salvesen G. S. 2016. The paracaspase MALT1. Biochimie 122: 324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Afonina I. S., Elton L., Carpentier I., Beyaert R. 2015. MALT1--a universal soldier: multiple strategies to ensure NF-κB activation and target gene expression. FEBS J. 282: 3286–3297. [DOI] [PubMed] [Google Scholar]
- 34.Brüstle A., Brenner D., Knobbe-Thomsen C. B., Cox M., Lang P. A., Lang K. S., Mak T. W. 2017. MALT1 is an intrinsic regulator of regulatory T cells. Cell Death Differ. 24: 1214–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haribhai D., Lin W., Relland L. M., Truong N., Williams C. B., Chatila T. A. 2007. Regulatory T cells dynamically control the primary immune response to foreign antigen. J. Immunol. 178: 2961–2972. [DOI] [PubMed] [Google Scholar]
- 36.Chandra S., Zhao M., Budelsky A., de Mingo Pulido A., Day J., Fu Z., Siegel L., Smith D., Kronenberg M. 2015. A new mouse strain for the analysis of invariant NKT cell function. Nat. Immunol. 16: 799–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodnow C. C., Crosbie J., Adelstein S., Lavoie T. B., Smith-Gill S. J., Brink R. A., Pritchard-Briscoe H., Wotherspoon J. S., Loblay R. H., Raphael K., et al. 1988. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature 334: 676–682. [DOI] [PubMed] [Google Scholar]
- 38.Rodríguez-Pinto D., Moreno J. 2005. B cells can prime naive CD4+ T cells in vivo in the absence of other professional antigen-presenting cells in a CD154-CD40-dependent manner. Eur. J. Immunol. 35: 1097–1105. [DOI] [PubMed] [Google Scholar]
- 39.Ho W. Y., Cooke M. P., Goodnow C. C., Davis M. M. 1994. Resting and anergic B cells are defective in CD28-dependent costimulation of naive CD4+ T cells. J. Exp. Med. 179: 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nashar T. O., Drake J. R. 2005. The pathway of antigen uptake and processing dictates MHC class II-mediated B cell survival and activation. J. Immunol. 174: 1306–1316. [DOI] [PubMed] [Google Scholar]
- 41.Müller P., Martin K., Theurich S., Schreiner J., Savic S., Terszowski G., Lardinois D., Heinzelmann-Schwarz V. A., Schlaak M., Kvasnicka H. M., et al. 2014. Microtubule-depolymerizing agents used in antibody-drug conjugates induce antitumor immunity by stimulation of dendritic cells. Cancer Immunol. Res. 2: 741–755. [DOI] [PubMed] [Google Scholar]
- 42.Bolotin D. A., Poslavsky S., Mitrophanov I., Shugay M., Mamedov I. Z., Putintseva E. V., Chudakov D. M. 2015. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12: 380–381. [DOI] [PubMed] [Google Scholar]
- 43.Shifrut E., Baruch K., Gal H., Ndifon W., Deczkowska A., Schwartz M., Friedman N. 2013. CD4(+) T cell-receptor repertoire diversity is compromised in the spleen but not in the bone marrow of aged mice due to private and sporadic clonal expansions. Front. Immunol. 4: 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang P. P., Barral P., Fitch J., Pratama A., Ma C. S., Kallies A., Hogan J. J., Cerundolo V., Tangye S. G., Bittman R., et al. 2011. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat. Immunol. 13: 35–43. [DOI] [PubMed] [Google Scholar]
- 45.Vinuesa C. G., Chang P. P. 2013. Innate B cell helpers reveal novel types of antibody responses. Nat. Immunol. 14: 119–126. [DOI] [PubMed] [Google Scholar]
- 46.Bai L., Deng S., Reboulet R., Mathew R., Teyton L., Savage P. B., Bendelac A. 2013. Natural killer T (NKT)-B-cell interactions promote prolonged antibody responses and long-term memory to pneumococcal capsular polysaccharides. Proc. Natl. Acad. Sci. USA 110: 16097–16102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green N. M., Marshak-Rothstein A. 2011. Toll-like receptor driven B cell activation in the induction of systemic autoimmunity. Semin. Immunol. 23: 106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beaurepaire C., Smyth D., McKay D. M. 2009. Interferon-gamma regulation of intestinal epithelial permeability. J. Interferon Cytokine Res. 29: 133–144. [DOI] [PubMed] [Google Scholar]
- 49.Wang F., Graham W. V., Wang Y., Witkowski E. D., Schwarz B. T., Turner J. R. 2005. Interferon-gamma and tumor necrosis factor-alpha synergize to induce intestinal epithelial barrier dysfunction by up-regulating myosin light chain kinase expression. Am. J. Pathol. 166: 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang F., Schwarz B. T., Graham W. V., Wang Y., Su L., Clayburgh D. R., Abraham C., Turner J. R. 2006. IFN-gamma-induced TNFR2 expression is required for TNF-dependent intestinal epithelial barrier dysfunction. Gastroenterology 131: 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Israël L., Bardet M., Huppertz A., Mercado N., Ginster S., Unterreiner A., Schlierf A., Goetschy J. F., Zerwes H. G., Roth L., et al. 2018. A CARD10-dependent tonic signalosome activates MALT1 paracaspase and regulates IL-17/TNF-α-driven keratinocyte inflammation. J. Invest. Dermatol. 138: 2075–2079. [DOI] [PubMed] [Google Scholar]
- 52.Martin D., Galisteo R., Gutkind J. S. 2009. CXCL8/IL8 stimulates vascular endothelial growth factor (VEGF) expression and the autocrine activation of VEGFR2 in endothelial cells by activating NFkappaB through the CBM (Carma3/Bcl10/Malt1) complex. J. Biol. Chem. 284: 6038–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McAllister-Lucas L. M., Jin X., Gu S., Siu K., McDonnell S., Ruland J., Delekta P. C., Van Beek M., Lucas P. C. 2010. The CARMA3-Bcl10-MALT1 signalosome promotes angiotensin II-dependent vascular inflammation and atherogenesis. J. Biol. Chem. 285: 25880–25884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Delekta P. C., Apel I. J., Gu S., Siu K., Hattori Y., McAllister-Lucas L. M., Lucas P. C. 2010. Thrombin-dependent NF-{kappa}B activation and monocyte/endothelial adhesion are mediated by the CARMA3·Bcl10·MALT1 signalosome. J. Biol. Chem. 285: 41432–41442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss J. M., Bilate A. M., Gobert M., Ding Y., Curotto de Lafaille M. A., Parkhurst C. N., Xiong H., Dolpady J., Frey A. B., Ruocco M. G., et al. 2012. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J. Exp. Med. 209: 1723–1742, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Singh K., Hjort M., Thorvaldson L., Sandler S. 2015. Concomitant analysis of Helios and Neuropilin-1 as a marker to detect thymic derived regulatory T cells in naïve mice. Sci. Rep. 5: 7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yadav M., Louvet C., Davini D., Gardner J. M., Martinez-Llordella M., Bailey-Bucktrout S., Anthony B. A., Sverdrup F. M., Head R., Kuster D. J., et al. 2012. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J. Exp. Med. 209: 1713–1722, S1–S19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baens M., Stirparo R., Lampi Y., Verbeke D., Vandepoel R., Cools J., Marynen P., de Bock C. E., Bornschein S. 2018. Malt1 self-cleavage is critical for regulatory T cell homeostasis and anti-tumor immunity in mice. Eur. J. Immunol. 48: 1728–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Föhse L., Suffner J., Suhre K., Wahl B., Lindner C., Lee C. W., Schmitz S., Haas J. D., Lamprecht S., Koenecke C., et al. 2011. High TCR diversity ensures optimal function and homeostasis of Foxp3+ regulatory T cells. Eur. J. Immunol. 41: 3101–3113. [DOI] [PubMed] [Google Scholar]
- 60.Levine A. G., Hemmers S., Baptista A. P., Schizas M., Faire M. B., Moltedo B., Konopacki C., Schmidt-Supprian M., Germain R. N., Treuting P. M., Rudensky A. Y. 2017. Suppression of lethal autoimmunity by regulatory T cells with a single TCR specificity. J. Exp. Med. 214: 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu A., Dee M. J., Adeegbe D., Dwyer C. J., Altman N. H., Malek T. R. 2017. The lower limit of regulatory CD4+ Foxp3+ TCRβ repertoire diversity required to control autoimmunity. J. Immunol. 198: 3127–3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kern J., Drutel R., Leanhart S., Bogacz M., Pacholczyk R. 2014. Reduction of T cell receptor diversity in NOD mice prevents development of type 1 diabetes but not Sjögren’s syndrome. PLoS One 9: e112467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishio J., Baba M., Atarashi K., Tanoue T., Negishi H., Yanai H., Habu S., Hori S., Honda K., Taniguchi T. 2015. Requirement of full TCR repertoire for regulatory T cells to maintain intestinal homeostasis. Proc. Natl. Acad. Sci. USA 112: 12770–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas P. G., Handel A., Doherty P. C., La Gruta N. L. 2013. Ecological analysis of antigen-specific CTL repertoires defines the relationship between naive and immune T-cell populations. Proc. Natl. Acad. Sci. USA 110: 1839–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kalekar L. A., Schmiel S. E., Nandiwada S. L., Lam W. Y., Barsness L. O., Zhang N., Stritesky G. L., Malhotra D., Pauken K. E., Linehan J. L., et al. 2016. CD4(+) T cell anergy prevents autoimmunity and generates regulatory T cell precursors. Nat. Immunol. 17: 304–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aschermann S., Lehmann C. H., Mihai S., Schett G., Dudziak D., Nimmerjahn F. 2013. B cells are critical for autoimmune pathology in Scurfy mice. Proc. Natl. Acad. Sci. USA 110: 19042–19047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leonardo S. M., Josephson J. A., Hartog N. L., Gauld S. B. 2010. Altered B cell development and anergy in the absence of Foxp3. J. Immunol. 185: 2147–2156. [DOI] [PubMed] [Google Scholar]
- 68.McGinness J. L., Bivens M. M., Greer K. E., Patterson J. W., Saulsbury F. T. 2006. Immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) associated with pemphigoid nodularis: a case report and review of the literature. J. Am. Acad. Dermatol. 55: 143–148. [DOI] [PubMed] [Google Scholar]
- 69.Chen C. A., Chung W. C., Chiou Y. Y., Yang Y. J., Lin Y. C., Ochs H. D., Shieh C. C. 2016. Quantitative analysis of tissue inflammation and responses to treatment in immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome, and review of literature. J. Microbiol. Immunol. Infect. 49: 775–782. [DOI] [PubMed] [Google Scholar]
- 70.Josefowicz S. Z., Niec R. E., Kim H. Y., Treuting P., Chinen T., Zheng Y., Umetsu D. T., Rudensky A. Y. 2012. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature 482: 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wawrzyniak M., O’Mahony L., Akdis M. 2017. Role of regulatory cells in oral tolerance. Allergy Asthma Immunol. Res. 9: 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hadis U., Wahl B., Schulz O., Hardtke-Wolenski M., Schippers A., Wagner N., Müller W., Sparwasser T., Förster R., Pabst O. 2011. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity 34: 237–246. [DOI] [PubMed] [Google Scholar]
- 73.Israel L., Mellett M. 2018. Clinical and genetic heterogeneity of CARD14 mutations in psoriatic skin disease. Front. Immunol. 9: 2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ekambaram P., Lee J. L., Hubel N. E., Hu D., Yerneni S., Campbell P. G., Pollock N., Klei L. R., Concel V. J., Delekta P. C., et al. 2018. The CARMA3-Bcl10-MALT1 signalosome drives NFκB activation and promotes aggressiveness in angiotensin II receptor-positive breast cancer. Cancer Res. 78: 1225–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luissint A. C., Parkos C. A., Nusrat A. 2016. Inflammation and the intestinal barrier: leukocyte-epithelial cell interactions, cell junction remodeling, and mucosal repair. Gastroenterology 151: 616–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Belkaid Y., Harrison O. J. 2017. Homeostatic immunity and the microbiota. Immunity 46: 562–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gensollen T., Iyer S. S., Kasper D. L., Blumberg R. S. 2016. How colonization by microbiota in early life shapes the immune system. Science 352: 539–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lathrop S. K., Bloom S. M., Rao S. M., Nutsch K., Lio C. W., Santacruz N., Peterson D. A., Stappenbeck T. S., Hsieh C. S. 2011. Peripheral education of the immune system by colonic commensal microbiota. Nature 478: 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fung T. C., Bessman N. J., Hepworth M. R., Kumar N., Shibata N., Kobuley D., Wang K., Ziegler C. G. K., Goc J., Shima T., et al. 2016. Lymphoid-tissue-resident commensal bacteria promote members of the IL-10 cytokine family to establish mutualism. Immunity 44: 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Arrieta M. C., Stiemsma L. T., Amenyogbe N., Brown E. M., Finlay B. 2014. The intestinal microbiome in early life: health and disease. Front. Immunol. 5: 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Vinuesa C. G., Cook M. C., Ball J., Drew M., Sunners Y., Cascalho M., Wabl M., Klaus G. G., MacLennan I. C. 2000. Germinal centers without T cells. J. Exp. Med. 191: 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sprouse M. L., Scavuzzo M. A., Blum S., Shevchenko I., Lee T., Makedonas G., Borowiak M., Bettini M. L., Bettini M. 2018. High self-reactivity drives T-bet and potentiates Treg function in tissue-specific autoimmunity. JCI Insight 3: e97322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lathrop S. K., Santacruz N. A., Pham D., Luo J., Hsieh C. S. 2008. Antigen-specific peripheral shaping of the natural regulatory T cell population. J. Exp. Med. 205: 3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bergot A. S., Chaara W., Ruggiero E., Mariotti-Ferrandiz E., Dulauroy S., Schmidt M., von Kalle C., Six A., Klatzmann D. 2015. TCR sequences and tissue distribution discriminate the subsets of naïve and activated/memory Treg cells in mice. Eur. J. Immunol. 45: 1524–1534. [DOI] [PubMed] [Google Scholar]
- 85.Cheng L., Deng N., Yang N., Zhao X., Lin X. 2019. Malt1 protease is critical in maintaining function of regulatory T cells and may Be a therapeutic target for antitumor immunity. J. Immunol. 202: 3008–3019. [DOI] [PubMed] [Google Scholar]
- 86.Demeyer A., Skordos I., Driege Y., Kreike M., Hochepied T., Baens M., Staal J., Beyaert R. 2019. MALT1 proteolytic activity suppresses autoimmunity in a T cell intrinsic manner. Front. Immunol. 10: 1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Di Pilato M., Kim E. Y., Cadilha B. L., Prüßmann J. N., Nasrallah M. N., Seruggia D., Usmani S. M., Misale S., Zappulli V., Carrizosa E., et al. 2019. Targeting the CBM complex causes Treg cells to prime tumours for immune checkpoint therapy. Nature 570: 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rosenbaum M., Gewies A., Pechloff K., Heuser C., Engleitner T., Gehring T., Hartjes L., Krebs S., Krappmann D., Kriegsmann M., et al. 2019. Bcl10-controlled Malt1 paracaspase activity is key for the immune suppressive function of regulatory T cells. Nat. Commun. 10: 2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fuchs A., Vermi W., Lee J. S., Lonardi S., Gilfillan S., Newberry R. D., Cella M., Colonna M. 2013. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity 38: 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bao Y., Liu X., Han C., Xu S., Xie B., Zhang Q., Gu Y., Hou J., Qian L., Qian C., et al. 2014. Identification of IFN-γ-producing innate B cells. Cell Res. 24: 161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schoenborn J. R., Wilson C. B. 2007. Regulation of interferon-gamma during innate and adaptive immune responses. Adv. Immunol. 96: 41–101. [DOI] [PubMed] [Google Scholar]
- 92.Blair P. J., Bultman S. J., Haas J. C., Rouse B. T., Wilkinson J. E., Godfrey V. L. 1994. CD4+CD8- T cells are the effector cells in disease pathogenesis in the scurfy (sf) mouse. J. Immunol. 153: 3764–3774. [PubMed] [Google Scholar]
- 93.Juilland M., Thome M. 2016. Role of the CARMA1/BCL10/MALT1 complex in lymphoid malignancies. Curr. Opin. Hematol. 23: 402–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee K. W., Kim M., Lee C. H. 2018. Treatment of dextran sulfate sodium-induced colitis with mucosa-associated lymphoid tissue lymphoma translocation 1 inhibitor MI-2 is associated with restoration of gut immune function and the microbiota. Infect. Immun. 86: e00091-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu W., Guo W., Hang N., Yang Y., Wu X., Shen Y., Cao J., Sun Y., Xu Q. 2016. MALT1 inhibitors prevent the development of DSS-induced experimental colitis in mice via inhibiting NF-κB and NLRP3 inflammasome activation. Oncotarget 7: 30536–30549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Monajemi M., Pang Y. C. F., Bjornson S., Menzies S. C., van Rooijen N., Sly L. M. 2018. Malt1 blocks IL-1β production by macrophages in vitro and limits dextran sodium sulfate-induced intestinal inflammation in vivo. J. Leukoc. Biol. 104: 557–572. [DOI] [PubMed] [Google Scholar]
- 97.Mc Guire C., Elton L., Wieghofer P., Staal J., Voet S., Demeyer A., Nagel D., Krappmann D., Prinz M., Beyaert R., van Loo G. 2014. Pharmacological inhibition of MALT1 protease activity protects mice in a mouse model of multiple sclerosis. J. Neuroinflammation 11: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Unterreiner A., Stoehr N., Huppertz C., Calzascia T., Farady C. J., Bornancin F. 2017. Selective MALT1 paracaspase inhibition does not block TNF-α production downstream of TLR4 in myeloid cells. Immunol. Lett. 192: 48–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.