Abstract
Spinal muscular atrophy (SMA) is a neuromuscular disease caused by deletions or mutations in survival motor neuron 1 (SMN1). The molecular mechanisms underlying motor neuron degeneration in SMA remain elusive, as global cellular dysfunction obscures the identification and characterization of disease-relevant pathways and potential therapeutic targets. Recent reports have implicated microRNA (miRNA) dysregulation as a potential contributor to the pathological mechanism in SMA. To characterize miRNAs that are differentially regulated in SMA, we profiled miRNA levels in SMA induced pluripotent stem cell (iPSC)-derived motor neurons. From this array, miR-23a downregulation was identified selectively in SMA motor neurons, consistent with previous reports where miR-23a functioned in neuroprotective and muscle atrophy-antagonizing roles. Reintroduction of miR-23a expression in SMA patient iPSC-derived motor neurons protected against degeneration, suggesting a potential miR-23a-specific disease-modifying effect. To assess this activity in vivo, miR-23a was expressed using a self-complementary adeno-associated virus serotype 9 (scAAV9) viral vector in the Smn2B/− SMA mouse model. scAAV9-miR-23a significantly reduced the pathology in SMA mice, including increased motor neuron size, reduced neuromuscular junction pathology, increased muscle fiber area, and extended survival. These experiments demonstrate that miR-23a is a novel protective modifier of SMA, warranting further characterization of miRNA dysfunction in SMA.
Introduction
Spinal muscular atrophy (SMA) is a pediatric neuromuscular disease and a leading genetic cause of infantile mortality. SMA is caused by the homozygous deletion or mutation of survival motor neuron 1 (SMN1), resulting in insufficient levels of the SMN protein (1). Humans possess an additional copy of the gene, termed SMN2. In contrast to SMN1, approximately 10% of SMN2 transcripts encode the full-length SMN protein, while 90% of SMN2 transcripts encode a truncated, rapidly degraded protein termed SMN∆7 (2). Although extremely low levels of SMN result in systemic defects that affect multiple organ systems, motor neuron pathology results in the most severe clinical outcomes including muscle wasting, paralysis, and respiratory failure (3). Thus, motor neurons remain a focal point of SMA research and therapeutic design. An increasing number of reports have implicated dysregulated molecular pathways in SMA motor neuron degeneration, including RNA metabolism (4,5), cytoskeletal dynamics (6–9), endocytosis (10–12), apoptosis (13–15), and ubiquitin-proteasome system (16,17). The specific mechanism of motor neuron degeneration, however, remains incompletely understood. Despite continued clinical advancements, including the SMN2 splice-switching oligonucleotide Spinraza and the progression of several SMN-inducing experimental therapies, the identification of novel disease-related mechanisms is warranted from a biological perspective and with the goal of continuing to improvement SMA therapeutics (18–21).
microRNAs (miRNAs) represent an important class of disease-relevant non-protein coding RNAs that have been implicated in a variety genetic diseases, including Duchenne muscular dystrophy (22), Alzheimer's disease (23), amyotrophic lateral sclerosis (24,25), Parkinson's disease (26,27), and SMA (28–32). miRNAs are approximately 22 nucleotide RNAs that posttranscriptionally silence their target mRNAs. A miRNA complexes with an RNA-induced silencing complex and associates with its target mRNAs via full or partial complementarity in the target 3′ UTR; mRNA decay or translational repression then ensues. A single miRNA can regulate multiple transcripts, fine-tuning the expression levels of networks of developmentally regulated transcripts (33). Therefore, the identification and characterization of differentially expressed miRNAs in SMA should reveal gene populations that are improperly regulated in the disease state.
Recent reports have suggested that miRNA dysregulation contributes to the pathogenic mechanism of SMA. Altered levels of miR-9 and miR-132 suggest potential dysregulation in subsets of genes involved in neuronal outgrowth and neuritogenesis, yet their contributions to disease pathology remain unclear (28,34–36). Manipulation of several differentially regulated miRNAs can modify pathology in a variety of in vitro disease contexts of SMA. Correction of dysregulated miR-375 and miR-431 in motor neurons reduced caspase-3 activity and rescued neurite outgrowth defects in cell culture models, respectively (30,31). Astrocyte-expressed miRNAs are also implicated in SMA; astrocytes derived from human SMA patient induced pluripotent stem cells (iPSCs) produce high levels of miR-146a that contribute to motor neuron cell death (31). Despite the identification of multiple dysregulated miRNAs in SMA, there are limited data that support the hypothesis that correction of miRNA disruption can modify the SMA phenotype in vivo. An exception was the demonstration that upregulation of miR-183, which silences mTOR transcripts, modifies the SMA phenotype and extends survival in severe SMA mice by an average of 3 days (29). To validate the potential therapeutic effects of miRNAs in vivo, further characterization of differentially regulated miRNAs is needed.
To identify miRNA expression differences between wild-type and SMA iPSC-derived motor neurons, we employed a non-biased polymerase chain reaction (PCR) array. miR-23a was identified as a potential candidate after observing significant reduction of miR-23a expression in SMA motor neurons. We hypothesized that miR-23a was a putative modifier based on reports that miR-23a exhibits neuroprotective effects through PTEN and Apaf-1 suppression as well as skeletal muscle atrophy-inhibiting activity through Atrogin1 and MuRF1 suppression (37–40). Transfection of synthetic miR-23a into iPSC-derived SMA motor neurons reduced motor neuron death following exposure to SMA astrocyte-conditioned media. In the Smn2B/− SMA mouse, administration of miR-23a using self-complementary adeno-associated virus serotype 9 (scAAV9) significantly reduced disease severity, including reduced loss of motor neuron soma area, reduced neuromuscular junction (NMJ) defects, increased muscle fiber area, and extended survival. These results demonstrate that modulation of differentially regulated miRNA can significantly lessen the severity of SMA and confirm miR-23a as a novel protective modifier of SMA.
Results
miR-23a is downregulated in SMA patient iPSC-derived motor neurons
Altered miRNA profiles in motor neurons from SMA have been reported; therefore, we investigated whether SMA iPSC-derived motor neurons also exhibit differential miRNA expression (41–46). iPSCs derived from fibroblast lines taken from a Type I SMA patient, a Type II SMA patient, and two healthy individuals were differentiated into motor neurons cultures lacking astrocytes using a previously published protocol (47). We used a miRNA PCR array assessing approximately 80 of the most abundantly expressed miRNAs in human tissue to identify dysregulated miRNAs from motor neurons harvested at 28 days post differentiation (Supplementary Material, Table S1). We discovered that a subset of 16 miRNAs was significantly decreased greater than 2-fold in the SMA iPSC-derived motor neurons compared to control motor neurons (Fig. 1A, Table 1). Of these miRNAs, miR-7, -9, -19a/b, -22, -23a, and -142 can exhibit neuroprotective properties or regulate axonal development (34,36,37,48–52). From this list, miR-23a also can also suppress skeletal muscle atrophy (40,53). The multifunctional roles of miR-23a suggest that its activity might provide protection against motor neuron degeneration and skeletal muscle atrophy in the context of SMA, providing the initial justification to focus upon this miRNA. To validate expression changes from the miRNA array, we confirmed that miR-23a was significantly downregulated using quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) in the SMA iPSC-derived motor neurons compared to control iPSC-derived motor neurons (Fig. 1B).
Figure 1.
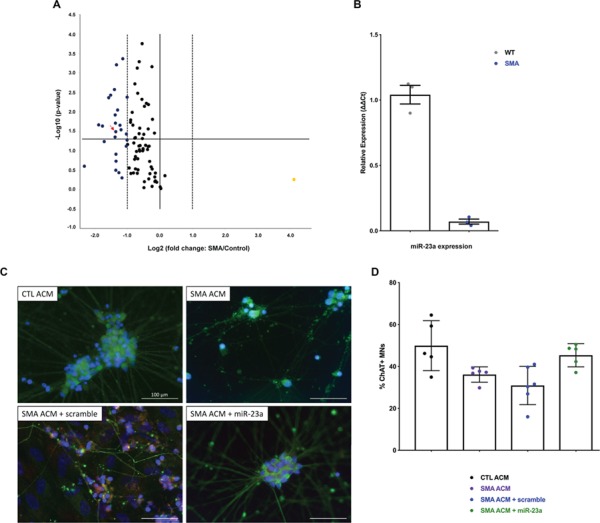
miR-23a is downregulated in SMA patient iPSC-derived motor neurons. Motor neurons were differentiated from control and SMA patient-derived iPSCs and analyzed for miR expression. (A) Volcano plot of miRNA PCR array revealing 16 significantly downregulated miRNAs in SMA vs. control iPSC-derived motor neurons at 28 days of differentiation. Horizontal line denotes a P-value 0.05. Dashed vertical lines indicate 2-fold change. Arrow indicates miR-23a. (B) qRT-PCR comparing miR-23a levels in wild-type and SMA motor neurons, compared using Student's t-test. (C) Representative images of immunohistochemical staining of iPSC-derived SMA motor neurons cultured with control astrocyte-conditioned media (CTL ACM), SMA astrocyte-conditioned media (SMA ACM), SMA astrocyte-conditioned media and transfected with miR scramble (SMA ACM + miR scramble), or SMA astrocyte-conditioned media and transfected with synthetic miR-23a (SMA ACM + miR-23a). Cells were stained with ChAT (red) to label motor neurons, Tuj1 (green) to label neurons, and Hoescht (blue) to label nuclei. Images represent merged channels. Images were taken at 40× (scale bar = 100 μm). (D) Survival of iPSC-derived SMA motor neurons cultured with CTL ACM, SMA ACM, SMA ACM + scramble, or SMA ACM + miR-23a at 28 days of differentiation. Groups were compared using one-way ANOVA and Newman-Keuls multiple comparisons test. Scatter plot is shown as mean ± SEM (*P < 0.05, **P < 0.01, ***P < 0.001, significance not indicated P>0.05).
Table 1.
Dysregulated miRNAs in SMA vs. control iPSC-derived motor neurons
| miRNAs underexpressed in SMA vs. control Mature ID | Fold regulation | P-value |
|---|---|---|
| hsa-miR-142-5p | −2.6 | 0.044536 |
| hsa-miR-9-5p | −2.52 | 0.000622 |
| hsa-miR-27b-3p | −2.04 | 0.037263 |
| hsa-miR-32-5p | −3.69 | 0.021736 |
| hsa-miR-142-3p | −2.98 | 0.004351 |
| hsa-miR-146a-5p | −2.63 | 0.002685 |
| hsa-miR-29b-3p | −2.29 | 0.028975 |
| hsa-miR-19a-3p | −2.86 | 0.003742 |
| hsa-miR-18a-5p | −2.21 | 0.000434 |
| hsa-miR-23a-3p | −2.56 | 0.032432 |
| hsa-miR-19b-3p | −2.58 | 0.019569 |
| hsa-miR-17-5p | −2.01 | 0.00422 |
| hsa-miR-27a-3p | −2.46 | 0.009028 |
| hsa-miR-22-3p | −2.59 | 0.012084 |
| hsa-miR-29c-3p | −2.34 | 0.022198 |
| hsa-miR-7-5p | −3.4 | 0.023218 |
Although SMA iPSC-derived motor neurons do not exhibit selective loss when cultured in isolation, SMA iPSC-derived motor neurons exhibit enhanced degeneration when cultured in the presence of SMA iPSC-derived astrocytes and/or SMA astrocyte-conditioned media (ACM) (32). With this as an experimental backdrop, we then asked whether increasing expression of miR-23a would prevent ACM-induced motor neuron pathology in vitro in SMA motor neurons. SMA iPSC-derived motor neurons were treated with synthetic miR-23a (or a scrambled control) in the presence or absence of iPSC-derived ACM. As anticipated, SMA iPSC-derived motor neurons showed selective loss in the presence of SMA ACM compared to the control ACM treated motor neurons. In contrast, addition of miR-23a mitigated the SMA ACM-induced motor neuron loss (Fig. 1C and D, Supplementary Material, Fig. S1). Together, these data suggest that miR-23a deficiency in SMA iPSC-derived motor neurons is functionally relevant to disease development and partially contributes to motor neuron pathology in vitro.
scAAV9-miR-23a increases motor neuron soma size in SMA model mice
Several SMA mouse models exist, and it has become increasingly clear that milder models represent more appropriate contexts to examine SMN-independent modalities, such as miR-23a (10,54–56). Therefore, we utilized the Smn2B/− SMA mouse model in the congenic C57BL/6 background (57,58). Unlike the commonly used severe SMN∆7 SMA mouse model, the less severe Smn2B/− model mice display ambulatory activity for several weeks and have a longer lifespan but maintain important histological and cellular hallmarks of SMA disease pathology such as motor neuron death, NMJ denervation, and muscle fiber atrophy.
To express miR-23a in the Smn2B/− mouse model, an scAAV9 vector was used. AAV9 transduces disease-relevant tissues including motor neurons, astrocytes, and skeletal muscle, as demonstrated in reports that AAV9-mediated SMN replacement rescues severe SMA mice (59–62). The constitutively active chicken β-actin (CBA) promoter drives expression of miR-23a within the 3′ UTR of a Green fluorescent protein (GFP) reporter. Mice were injected with 1 × 1011 vector genomes of scAAV9-miR-23a on postnatal day 1 (P1).
Smn2B/− mice exhibit progressive motor neuron death during mid and late symptomatic stages of SMA disease progression, as evidenced by a continual decrease in the number of motor neuron soma in the anterior horn in the lumbar spinal cord (57). To determine if miR-23a overexpression can reduce the loss of motor neuron soma, the L3–L5 lumbar spinal cord was analyzed at P21, a late symptomatic disease stage in the Smn2B/− SMA model (Fig. 2A, Supplementary Material, Fig. S2). As expected, the number of motor neuron somas was significantly reduced in untreated Smn2B/− mice compared to unaffected mice. Delivery of scAAV9-miR-23a, however, resulted in motor neuron soma numbers that were not significantly different from unaffected mice and untreated SMA mice (Fig. 2B). scAAV9-miR-23a treatment resulted in a small, but significant, increase in the area and perimeter of motor neuron soma in the L3–L5 lumbar region compared to untreated Smn2B/− mice (Fig. 2C and D). Taken together, these results highlight a subtle neuroprotective effect of miR-23a on motor neurons in an in vivo SMA context.
Figure 2.
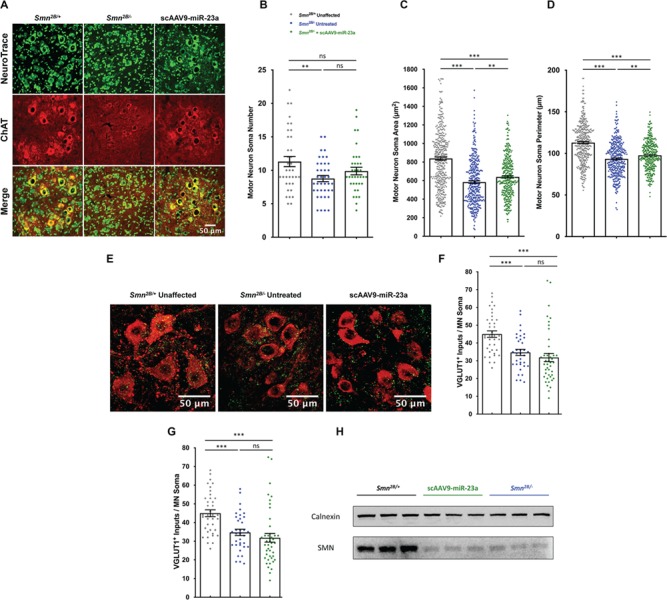
scAAV9-miR-23a improves motor neuron soma size and perimeter. L3–L5 spinal cords were dissected from mice harvested at P21, cross sectioned, and stained (n = 3 mice). (A) Representative images of L3–L5 cross sections immunohistochemically labeled with NeuroTrace Nissl (green) to label Nissl substance and ChAT (red) to label motor neurons. Images were taken with 40× objective lens (scale bar = 50 μm). (B) Scatter plot representing number of ChAT-positive motor neuron soma in the L3–L5 spinal cord. (C) Scatter plot representing cross-sectional area of ChAT-positive motor neuron soma in the L3–L5 spinal cord. (D) Scatter plot representing perimeter of ChAT-positive motor neuron soma in the L3–L5 spinal cord. (E) Representative images of L3–L5 cross sections immunohistochemically labeled with VGLUT1 (green) to label proprioceptive inputs and ChAT (red). Images were taken with 40× objective lens (scale bar = 50 μm). (F) Scatter plot representing quantification of VGLUT1+ proprioceptive synaptic inputs on motor neuron soma. (G) Western blot of SMN in spinal cord whole tissue lysates. Experimental groups were compared using one-way ANOVA with Newman-Keuls multiple comparisons test. Scatter plots are shown as mean ± SEM (**P < 0.01, ***P < 0.001, nsP > 0.05).
Disrupted sensory-motor connectivity, evidenced by the loss of excitatory glutamatergic proprioceptive synapses onto motor neurons, is an additional defect in the CNS of SMA mouse models (63,64). To determine if scAAV9-miR-23a treatment reduced defects in proprioceptive circuitry, the number of VGLUT+ proprioceptive synapses on motor neuron soma was determined (Fig. 2E). In contrast to the protective effect observed with motor neuron somas, scAAV9-miR-23a treatment did not prevent the loss of VGLUT+ proprioceptive synapses compared to untreated Smn2B/− mice (Fig. 2F).
miR-23a has been shown to exert neuroprotective effects through SMN-independent mechanisms. Consistent with this, SMN levels were not increased following delivery of scAAV9-miR-23a in the spinal cord (Fig. 2G). A potential mechanism of the protective benefit of miR-23a on SMA motor neurons could be attributed to miR-23a-induced PTEN suppression (39). In this regard, recent reports have demonstrated that shRNA-mediated PTEN suppression reduced disease severity in severe SMA mice (65,66). We did not observe reduction of PTEN levels or increase in phosphorylation of Akt following scAAV9-miR-23a treatment (Supplementary S-Table 1). Collectively, these data demonstrate that miR-23a protection from motor neuron loss in vitro translates to the in vivo context, further supporting the neuroprotective role of miR-23a; however, PTEN or Akt phosphorylation is likely not involved.
scAAV9-miR-23a reduces NMJ pathology in SMA mice
Disruption of NMJ architecture is a well-defined hallmark in patients and mouse models of SMA, characterized by developmental delays, denervation, and decreased soma size (67,68). To determine whether the neuroprotective effect of scAAV9-miR-23a extends to improved NMJ integrity, we characterized the NMJ phenotype of the transverse abdominis (TVA) muscle at P17, considered to be mid symptomatic disease stage. In Smn2B/− mice, the TVA muscle is vulnerable to denervation, which can be corrected by scAAV9-mediated delivery of SMA genetic modifiers (69). Delivery of scAAV9-miR-23a significantly improved gross NMJ architecture compared the untreated cohort (Fig. 3A) and significantly increased the number of innervated synapses in the TVA muscle compared to untreated controls (Fig. 3B). scAAV9-miR-23a treatment also increased endplate area compared to untreated controls in these TVA samples (Fig. 3C). scAAV9-miR-23a improved NMJ innervation and soma size compared to untreated mice, suggesting that reintroduction of miR-23a improves pre- and postsynaptic NMJ pathology.
Figure 3.
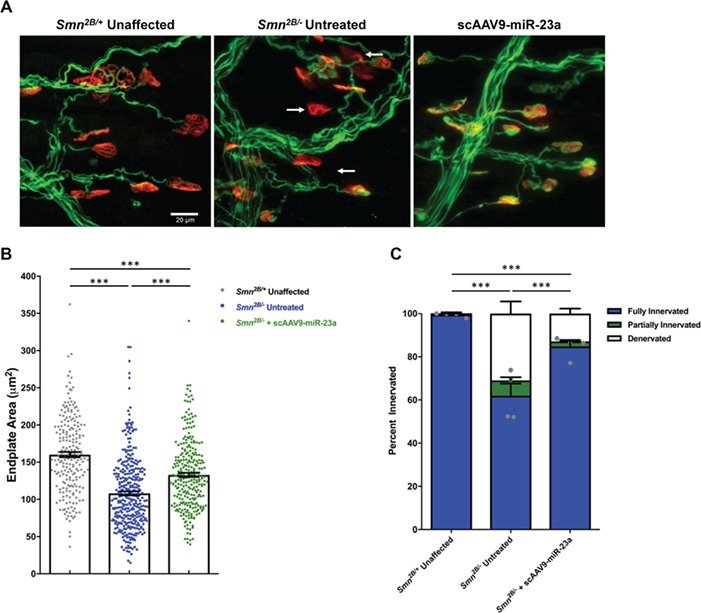
scAAV9-miR-23a reduces neuromuscular junction defects in Smn2B/− mice. Transverse abdominus muscles were dissected from mice harvested on P17, fixed, and stained (n = 5 mice). (A) Representative images of neuromuscular junctions stained with neurofilament (green) to label motor axons, synaptophysin (green) to label presynaptic terminals, and α-bungarotoxin (red) to label endplates. Arrows point toward fully denervated endplates. Images were taken with 40× objective lens (scale bar = 20 μm). (B) Bar graph representing average percentages of fully innervated, partially innervated, and denervated motor endplates. Statistical analysis was performed comparing degree of fully innervated endplates, represented by grey dots. (C) Scatter plot representing quantification of motor endplate area. Experimental groups were compared using one-way ANOVA with Newman-Keuls multiple comparisons test. Scatter plots are shown as mean ± SEM (***P < 0.001).
scAAV9-miR-23a improves muscle fiber area and reduces atrogene levels in SMA model mice
Skeletal muscle atrophy is, in part, regulated by the ubiquitin-mediated degradation of muscle-specific proteins. Degradation is coordinated by the E3 ubiquitin ligases Atrogin1 and MuRF1, referred to as atrogenes (70,71). Consequently, deletion of Atrogin1 or MuRF1 prevents muscle atrophy following denervation (70,72). miR-23a can downregulate Atrogin1 and MuRF1, protecting skeletal muscle from atrophy inducing stimuli (40). Consistent with this, we sought to characterize the influence of miR-23a on skeletal muscle atrophy in Smn2B/− mice. To determine whether skeletal muscle atrophy in SMA is associated with decreased expression of miR-23a, miR-23a-3p transcripts were quantitated in hindlimb skeletal muscle of Smn2B/− mice. A small, but significant, downregulation of miR-23a-3p was observed in SMA muscle compared to unaffected controls (Fig. 4A). To determine whether miR-23a downregulation resulted in decreased expression of Atrogin1 and MuRF1, we first analyzed transduction via GFP expression in skeletal muscle. GFP expression was observed in treated samples, demonstrating efficient transduction of scAAV9-miR-23a (Supplementary Material, Fig. S4) and a ~1.5 fold-increase in miR-23a-3p following scAAV9-miR-23a delivery to Smn2B/− mice (Fig. 4A). The increase in miR-23a following treatment was inversely correlated with transcript levels of both Atrogin1 and MuRF1, which were decreased in scAAV9-miR-23a-treated compared to untreated Smn2B/− mice (Fig. 4B).
Figure 4.
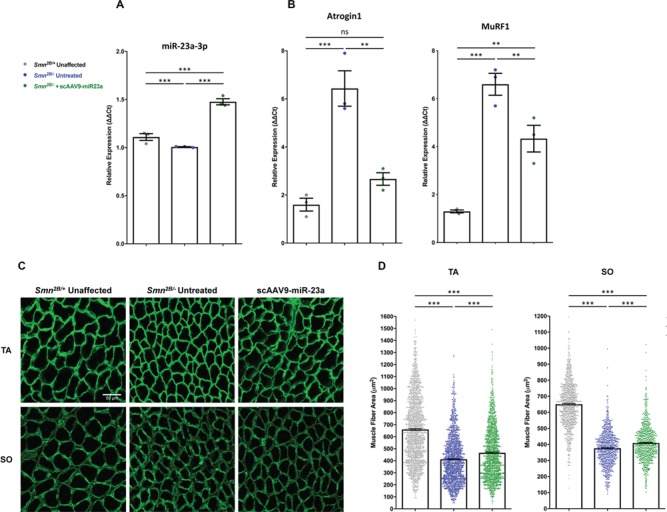
scAAV9-miR-23a decreases atrogene transcript levels and increases muscle fiber area and in Smn2B/− mice. Histological and molecular analyses were conducted on muscle harvested from mice at P17. (A) Quantification of relative transcript levels of miR-23a-3p in hindlimb skeletal muscle via qRT-PCR (n = 3 mice per group). (B) Quantification of relative transcript levels of MuRF1 and Atrogin1 in hindlimb skeletal muscle via qRT-PCR (n = 3 mice per group). (C) Representative images of cross sections of TA and SO. Sections were stained with laminin (green) to label muscle fiber boundaries. Images were taken with 40× objective lens (scale bar = 50 μm). (D) Scatter plot representing cross-sectional area of TA and SO muscle fibers (n = 4 mice per group; n > 350 fibers per TA; n > 200 fibers per SO). Experimental groups were compared using one-way ANOVA with Newman-Keuls multiple comparisons test. Scatter plots are shown as mean ± SEM (**P < 0.0 1, ***P < 0.001, nsP > 0.05).
To determine if suppression of atrophy-inducing genes following scAAV9-miR-23a treatment reduced skeletal muscle atrophy in Smn2B/− mice, we measured muscle fiber cross-sectional area in the tibialis anterior (TA) and soleus (SO) muscles, which are predominantly composed of fast-twitch and slow-twitch fibers, respectively. We evaluated muscle fibers at P17, a mid-symptomatic stage, by staining against laminin (Fig. 4C). Delivery of scAAV9-miR-23a significantly increased the mean fiber cross-sectional area of both muscles (Fig. 4D). Additionally, scAAV9-miR-23a induced a subtle shift in the size distribution of muscle fibers toward that of the unaffected littermates (Supplementary Material, Fig. S5). Despite reduction in skeletal muscle atrophy and reduced levels of atrophy-inducing genes, scAAV9-miR-23a treatment did not increase grip strength at mid and late disease stages compared to untreated littermates (Supplementary Material, Fig. S6). Taken together, these data demonstrate that miR-23a protects against muscle atrophy but does not significantly improve muscle function.
scAAV9-miR-23a extends survival in SMA model mice
Finally, we assessed whether the observed protective effects following scAAV9-miR-23a treatment conferred an increase in the lifespan of Smn2B/− mice. Delivery of 1 × 1011 viral particles through both intravenous (IV) and intracerebral ventricular (ICV) routes of administration significantly extended survival compared to untreated Smn2B/− mice (Fig. 5A). No significant increase in weight gain was observed with either treatment group compared to untreated controls (Fig. 5B). The absence of increased weight gain, despite prolonged survival, is consistent with reports of modifiers in SMA mice (54).
Figure 5.
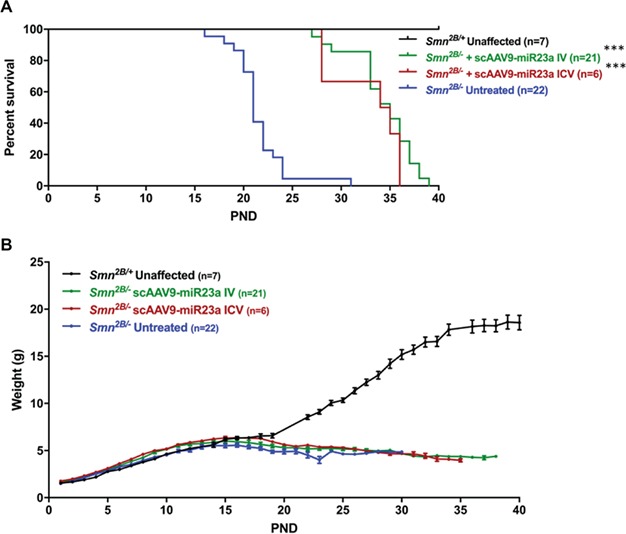
scAAV9-miR-23a extends survival in Smn2B/− mice. (A) Kaplan-Meier curve of Smn2B/− mice treated with scAAV9-miR-23a via IV or ICV injection. Difference in survival between treatment groups and untreated Smn2B/− controls was calculated using the log-rank Mantel-Cox test. (B) Weight gain of Smn2B/− mice treated with scAAV9-miR-23a via IV or ICV injection (***P < 0.001).
Discussion
miRNAs contribute to a multitude of neurodegenerative diseases and are also essential for motor neuron development, warranting investigation into the role of miRNA dysregulation in SMA (22–32,34,35). To date, several perturbations in miRNAs have been identified in various experimental SMA contexts, but investigation into the in vivo disease-modifying effects following correction of these defects has been limited. We sought to identify novel differentially regulated miRNAs using SMA patient iPSC-derived motor neurons to determine if reversal of miRNA dysregulation reduces disease pathology in SMA mice. Our nonbiased profiling of miRNA levels in patient iPSC-derived motor neurons identified several differentially regulated miRNAs. Here, we publish our findings concerning reduction of miR-23a expression on SMA disease development. Expression of miR-23a is associated with neuroprotection and prevention of muscle fiber atrophy, suggesting that miR-23a is a putative protective modifier of SMA (37–40). Reintroduction of miR-23a confers subtle protection against degeneration of motor neurons in patient iPSC-derived motor neurons, and scAAV9-medaited gene therapy of miR-23a slightly reduced motor neuron soma defects, improved NMJ size and innervation, decreased muscle fiber atrophy, and improved survival in Smn2B/− SMA mice. Taken together, these results support a modifying role of miR-23a in SMA.
miRNAs regulate gene cascades and often target multiple genes, making the analysis of their targets complicated due to the overlapping functionality in vivo. In an attempt to dissect the molecular pathways involved in the protective activity we observed in the SMA mice, we investigated whether previously known targets of miR-23a were altered in the SMA context. PTEN, Atrogin1, and MuRF1 were examined in this study; however, we were not able to detect suppression of PTEN in the CNS of Smn2B/− mice by western blot. We cannot rule out the possibility that PTEN suppression plays a role in the neuroprotective effects of miR-23a and that the level of sensitivity for this assay was below the level of functionality. We detected a reduction in Atrogin1 and MuRF1 transcripts in skeletal muscle of SMA mice following scAAV9-miR-23a treatment; however, this observation might be a result, rather than a cause, of improvements in muscle pathology following enhancement of NMJ architecture. Thus, the specific mechanism by which miR-23a acts as a phenotypic modifier in the context of SMA requires further verification. Bioinformatic analysis using miRNA prediction software reveals 866 predicted targets of miR-23a-3p, linking a potentially large network of genes to SMA pathology (73). In order to further characterize the specific molecular mechanism by which miR-23a provides phenotypic improvements, a carefully designed high throughput assay should be employed to determine the molecular response downstream of miR-23a upregulation in SMA models.
There are several plausible explanations for the downregulation of miR-23a in SMA tissues. Gemin3 and Gemin4, components of the SMN complex, are also components of miRNA processing complex that includes Argonaute-2 (74). This suggests potential overlapping functions of the SMN complex in mRNA and miRNA processing, where the SMN complex may regulate the subpopulations of Gemin3 and Gemin4 that are accessible for miRNA processing. Thus, disruption of the SMN complex may alter the levels or the availability of Gemin3 and Gemin4 for microRNA ribonucleoprotein (miRNP) processing, leading to improper maturation of a general subset of miRNAs. The results of our miRNA profile are consistent with this notion, in which miR-23a is one of many dysregulated miRNAs. In addition, low levels of SMN lead to significant shifts in transcriptional regulation (41–43,45,75). miR-23a and miR-27a, which are processed from the same pre-miRNA transcript, are downregulated in our miRNA profile. This suggests that the observed downregulation is a transcriptional event, consistent with other reports that the precursor transcript is downregulated following injury in neurons and during atrophy-inducing conditions in skeletal muscle (51,76). Thus, miR-23a downregulation is potentially due to defects in miRNA maturation and transcriptional downregulation of its pre-miRNA transcript.
Recent advances in oligonucleotide therapeutics have enabled targeting of miRNAs in human patients, and therapeutic correction of miRNA defects in SMA represents a potential SMN-independent approach to SMA therapy (77). This work contributes to the growing body of evidence that correction of miRNA dysregulation can reduce disease severity of SMA in vivo and provides the proof of principle that a single miRNA can improve multiple aspects of disease pathology. While these results suggest potential therapeutic promise of miR-23a, the feasibility of a translational application of miR-23a is limited by its oncogenic potential in certain tissues (78,79). Additionally, since miRNAs regulate a multitude of targets, the specificity for any miRNA therapy would need to be addressed. Toward this end, elucidation of the specific cellular mechanisms of miR-23a modification in the SMA context is necessary to determine the tissue-specific requirements for its activity. Further experimentation is warranted to identify additional disease-modifying miRNAs to better understand the pathways involved in SMA pathogenesis as well as potentially opening the door to new molecular therapeutic targets.
Materials and Methods
miRNA profiling array from iPSC-derived motor neurons
iPSCs were derived from parent fibroblast lines from one SMA Type I patient (GM09677, Coriell), one SMA Type II patient (GM03813, Coriell), and two unaffected individuals (GM03814 and GM02183, Coriell). (80,81). iPSCs were grown as embryoid bodies and cultured in motor neuron progenitor cell medium for 14 days followed by dissociation and platedown in motor neuron maturation medium for an additional 14 days to generate highly purified cultures of motor neurons lacking astrocytes as previously described (32,47). Mature motor neurons were collected at 28 days of differentiation using TrypLE (Thermo Fisher) and processed for miRNA extraction using the mirVana PARIS isolation kit (Life Technologies™). RNA was reverse transcribed using miScript II RT (QIAGEN), and the resulting cDNA was used for real-time PCR using the SYBR green human miFinder miRNA PCR Array system (catalog MIHS-001Z; QIAGEN) following manufacturer's protocols. Data were imported into the associated online analysis software (https://dataanalysis.qiagen.com/mirna/arrayanalysis.php). Validation of miR-23a downregulation was performed using miRCURY™ LNA Universal RT microRNA cDNA synthesis kit (Exiqon), SYBR Green master mix, Universal RT (Exiqon), and LNA PCR primer set for miR-23a and normalized to miR-103a-3p (Exiqon, Woburn, MA, USA). Relative abundance was calculated using the ΔΔCt method.
Immunocytochemistry and cell counts of miR-23a-treated iPSC-derived motor neurons
To induce motor neuron loss, 250 μl of ACM from SMA iPSC-derived astrocytes was added to motor neurons grown in isolation (32). ACM from healthy iPSCs was used as a control. Day 28 SMA iPSC-derived motor neurons were treated for 48 hours with 40 nM mirVana miR-23a oligonucleotide or random miRNA sequence (mirVana negative control 1; Ambion) in 1 μM EndoPorter (Gene Tools, LLC) transfection reagent and then fixed in 4% paraformaldehyde for 20 minutes at room temperature and rinsed with Phosphate buffered saline (PBS). Cells were blocked in donkey serum and permeabilized in 0.2% Triton X as well as subsequently incubated in primary antibodies rabbit anti-Tuj1 (1:1000; catalog MRB-435P; Covance) and goat anti-choline acetyltransferase (anti-ChAT) (1:100; catalog AB144P; MiliporeSigma) overnight at 4°C. Cells were then labeled with secondary antibodies Donkey anti-Goat Alexa Fluor-546 (1:1000; Catalog A11056; Invitrogen) and Donkey anti-Rabbit Alexa Fluor-488 (1:1000; Catalog A21206; Invitrogen). Hoechst nuclear dye labeled the cell nuclei. Representative images were taken using a fluorescence microscope (20× objective; Nikon E400; Nikon Corporation).
Production and purification of AAV9-miR-23a vector
HEK 293T (ATCC® CRL-3216™) cells were cultured in three 10-floor cell factories. When approximately 85% confluent, cells were transfected at a 1:1:1 molar ratio of plasmids Rep2Cap9 (a gift from James Wilson, University of Pennsylvania, Philadelphia, PA, USA), pHelper, and scAAV-CBA-GFP-miR-23a using 25-kDa polyethyleneimine as the transfection reagent. Forty-eight hours posttransfection, cells were harvested and lysed by 5 rapid freeze-thaw cycles, treated with DNase and protease, and subjected to three rounds of CsCl density gradient ultracentrifugation. Titers were determined after each round of centrifugation using quantitative polymerase chain reaction (qPCR) following a previously described protocol (82). CsCl was removed from final fractions by dialysis in PBS, and virus was subsequently stored at 4°C.
Animal procedures and delivery of therapeutics
Smn2B/− mice of the C57/BL6 background were a gift from Dr. Rashmi Kothary at the University of Ottawa, Canada. Animals of both sexes were used in all experiments. Treatment groups were assigned at random. Genotyping was performed by PCR assay following tail biopsy on P0. Virus was administered systemically using IV or ICV injection, performed on P1. IV injections utilized the superficial facial vein, and ICV injections were performed as previously described (62,83). ICV injection was utilized for the characterization of the effects of scAAV9-miR-23a on motor neuron pathology in order to increase viral transduction in motor neurons; for all other histological analyses, IV delivery was utilized. Grip-strength assays were performed using BioSeb Model BP32025 (Vitrolles) by placing all four paws on the grid. Mice were given two trials, one for training and the second for data collection. Mice were fed low-fat stock diets (Harlan Teklad 8640).
Immunohistochemistry of in vivo motor neuron soma
Three animals from both control cohorts and the ICV-treated AAV9-miR-23a cohort were randomly selected at P21, euthanized, and perfused in 4% formaldehyde in PBS. The L3–L5 region was dissected, cryoprotected in 30% sucrose overnight, embedded in optimal cutting temperature (OCT) media, and cryosectioned at 16 μm thickness. Every 10th section was collected and stained with ChAT primary antibody (1:100; catalog AB144P; MilliporeSigma), Donkey anti-Goat Alexa Fluor-594 secondary antibody (1:250; catalog 705-585-003; Jackson ImmunoResearch Laboratories, Inc.), and NeuroTrace Green Fluorescent Nissl (Thermo Fisher). VGLUT+ proprioceptive synapses were stained with VGLUT1 primary antibody (1:500; catalog 135 316; Synaptic Systems) and Donkey anti-Chicken Alexa Fluor-647 secondary antibody (1:400; Catalog AP194SA6, MilliporeSigma). Soma counts, soma area, soma perimeter, and VGLUT+ synaptic inputs were quantified manually using ImageJ software (National Institutes of Health [NIH]) on fluorescence micrographs (40× objective; Leica DM5500 B, Leica Microsystems Inc.). Representative images were taken using a laser scanning confocal microscope (40× objective; Leica TCS SP8, Leica Microsystems, Inc.).
Immunoblot
Hindlimb skeletal muscle and spinal cord tissue were harvested at P17. Tissue was lysed using JLB buffer (50 mm Tris, pH = 8.0, 150 mm NaCl, 10% glycerol, 20 mm NaH2PO4, 50 mm NaF, 2 mm EDTA, 5 mm NaVO3) supplied with cOmplete™ Mini Protease Inhibitor Cocktail (Roche). Following SDS-PAGE, protein was transferred to Immobilon®-P transfer membrane (MilliporeSigma) and probed with the following primary antibodies: anti-SMN (1:10,000; 610647, BD Transduction Laboratories™), anti-Calnexin (1:2000; catalog C4731, MilliporeSigma), anti-PTEN (1:1000; 9552S, Cell Signaling Technology®), anti-GFP (1:2000; A11122, Life Technologies™), anti-Akt (1:1000; 9272S, Cell Signaling Technology®), and anti-Phospho-Akt (Ser473) (1:1000; 4058S, Cell Signaling Technology®). Horseradish peroxidase-conjugated secondary antibodies were used (Jackson ImmunoResearch Laboratories, Inc.). Immunoblots were visualized using a BioSpectrum® 815 Imaging System (UVP, LLC).
Immunohistochemistry of NMJs
Four animals from both control cohorts and the IV-treated AAV9-miR-23a cohort were randomly selected and harvested at P17. Transverse abdominus muscles were dissected and fixed in 4% formaldehyde in PBS. Muscles were stained using antineurofilament-heavy (1:4000; catalog CPCA-NF-L, EnCor) and antisynaptophysin primary antibodies (1:200; catalog YE269, Life Technologies™), and Donkey anti-Chicken Alexa Fluor-647 (1:400; Catalog AP194SA6, MilliporeSigma) and Donkey anti-Rabbit Alexa Fluor-647 (1:200; Catalog AP187SA6, MilliporeSigma) secondary antibodies. Acetylcholine receptors were labeled with Alexa Fluor 594-conjugated α-bungarotoxin (Life Technologies™). Muscles were whole mounted using Citiflour AF1 mounting media (Ted Pella, Inc.). NMJ analysis was performed on 4 randomly selected fields of view per mouse (20× objective; Leica DM5500 B, Leica Microsystems Inc.). Endplate area was measured using ImageJ software (NIH). Representative images were taken using a laser scanning confocal microscope (20× objective; Leica TCS SP8, Leica Microsystems).
Immunohistochemistry of skeletal muscle
Four animals from both control cohorts and the IV-treated AAV9-miR-23a cohort were randomly selected and harvested at P17. TA and SO muscles were dissected and flash frozen in O.C.T. media (Tissue-Tek) using liquid nitrogen-cooled isopentane. Frozen muscle was cryosectioned at 18 μm per section. Muscle fibers were labeled using antilaminin primary antibody (1:300; catalog L9393, MilliporeSigma) and Donkey anti-Rabbit Alexa Fluor-647 secondary antibody (1:200; Catalog AP187SA6, MilliporeSigma). Samples were mounted using Citiflour AFI mounting media (Ted Pella, Inc.). Imaging was performed using a laser scanning confocal microscope (20× objective; Leica TCS SP8, Leica Microsystems, Inc.). Numbers of fibers analyzed are as follows: TA (Smn2B/+ n = 1631, scAAV9-miR-23a n = 1543, Smn2B/− n = 1506) and SO (Smn2B/+ n = 939, scAAV9-miR-23a n = 832, Smn2B/− n = 864). Fibers were blinded and analyzed manually using ImageJ software (NIH).
RNA isolation from mouse tissue and qRT-PCR
For in vivo analyses, total RNA was isolated from hindlimb skeletal muscle tissue using miRCURY™ RNA Isolation Kit (product 300111, Exiqon). The miRCURY LNA™ PCR Assay Starter Kit (product 339320, QIAGEN) was used to measure relative levels of the mature miR-23a-3p (catalog YP00204772, QIAGEN). Primers to detect miR-103-5p were included in the kit as the recommended normalizer. qPCR was performed using an Applied Biosystems 7500 instrument. For analysis of MuRF1 and Atrogin1 levels, RNA was reverse transcribed using Supescript III (Invitrogen). Relative levels of Atrogin1 and MuRF1 were determined by reference to GAPDH using previously published primers (53). qPCR was performed using a BioRad 89252 instrument. ∆∆Ct values were calculated to determine the fold difference between cohorts, and samples from four biological replicates each were run using three technical replicates. Plates were performed in triplicate. Data points represent averages per experimental replicate.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism version 8.0.1 for Mac OS X (Graphpad Software, San Diego, CA, USA) or Microsoft Excel version 15.19.1 (Microsoft Corporation). Data were analyzed using one-way analysis of variance (ANOVA) with Newman-Keuls post-hoc tests for comparisons between more than two conditions, Student's t-test for comparisons between two conditions, and log-rank Mantel-Cox test for survival comparisons. Changes were considered statistically significant when P < 0.05.
Study Approval
All animal procedures were carried out in accordance with procedures approved by NIH and MU animal Care and Use Committee.
Supplementary Material
Acknowledgement
The authors would like to thank Zachary Lorson and Elinor Stanley for technical assistance with the mouse colony.
Conflict of Interest Statement. C.L.L. is the cofounder and Chief Scientific Officer of Shift Pharmaceuticals, LLC.
Funding
This work was supported by the National Institute of Health [R21NS106490 to C.L.L.] and a research grant from the MU College of Veterinary Medicine to K.A.K. K.A.K. is supported by the University of Missouri Life Sciences Fellowship; C.E.S. is supported by a National Institutes of Health training grant [T32 GM008396]; and J.M. is supported by NIH PREP R25GM064120.
References
- 1. Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M. et al. (1995) Identification and characterization of a spinal muscular atrophy-determining gene. Cell, 80, 155–165. [DOI] [PubMed] [Google Scholar]
- 2. Lorson C.L., Hahnen E., Androphy E.J. and Wirth B. (1999) A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA, 96, 6307–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Crawford T.O. and Pardo C.A. (1996) The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis, 3, 97–110. [DOI] [PubMed] [Google Scholar]
- 4. Coady T.H. and Lorson C.L. (2011) SMN in spinal muscular atrophy and snRNP biogenesis. Wiley Interdiscip Rev RNA, 2, 546–564. [DOI] [PubMed] [Google Scholar]
- 5. Fallini C., Bassell G.J. and Rossoll W. (2012) Spinal muscular atrophy: the role of SMN in axonal mRNA regulation. Brain Res, 1462, 81–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Giesemann T., Rathke-Hartlieb S., Rothkegel M., Bartsch J.W., Buchmeier S., Jockusch B.M. and Jockusch H. (1999) A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with smn in nuclear gems. J Biol Chem, 274, 37908–37914. [DOI] [PubMed] [Google Scholar]
- 7. Bowerman M., Shafey D. and Kothary R. (2007) Smn depletion alters profilin II expression and leads to upregulation of the RhoA/ROCK pathway and defects in neuronal integrity. J Mol Neurosci, 32, 120–131. [DOI] [PubMed] [Google Scholar]
- 8. Oprea G.E., Krober S., McWhorter M.L., Rossoll W., Muller S., Krawczak M., Bassell G.J., Beattie C.E. and Wirth B. (2008) Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science, 320, 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Donlin-Asp P.G., Fallini C., Campos J., Chou C.C., Merritt M.E., Phan H.C., Bassell G.J. and Rossoll W. (2017) The survival of motor neuron protein acts as a molecular chaperone for mRNP assembly. Cell Rep, 18, 1660–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hosseinibarkooie S., Peters M., Torres-Benito L., Rastetter R.H., Hupperich K., Hoffmann A., Mendoza-Ferreira N., Kaczmarek A., Janzen E., Milbradt J. et al. (2016) The power of human protective modifiers: PLS3 and CORO1C unravel impaired endocytosis in spinal muscular atrophy and rescue SMA phenotype. Am J Hum Genet, 99, 647–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Riessland M., Kaczmarek A., Schneider S., Swoboda K.J., Lohr H., Bradler C., Grysko V., Dimitriadi M., Hosseinibarkooie S., Torres-Benito L. et al. (2017) Neurocalcin delta suppression protects against spinal muscular atrophy in humans and across species by restoring impaired endocytosis. Am J Hum Genet, 100, 297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janzen E., Mendoza-Ferreira N., Hosseinibarkooie S., Schneider S., Hupperich K., Tschanz T., Grysko V., Riessland M., Hammerschmidt M., Rigo F. et al. (2018) CHP1 reduction ameliorates spinal muscular atrophy pathology by restoring calcineurin activity and endocytosis. Brain, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tsai M.S., Chiu Y.T., Wang S.H., Hsieh-Li H.M., Lian W.C. and Li H. (2006) Abolishing Bax-dependent apoptosis shows beneficial effects on spinal muscular atrophy model mice. Mol Ther, 13, 1149–1155. [DOI] [PubMed] [Google Scholar]
- 14. Trulzsch B., Garnett C., Davies K. and Wood M. (2007) Knockdown of SMN by RNA interference induces apoptosis in differentiated P19 neural stem cells. Brain Res, 1183, 1–9. [DOI] [PubMed] [Google Scholar]
- 15. Sareen D., Ebert A.D., Heins B.M., McGivern J.V., Ornelas L. and Svendsen C.N. (2012) Inhibition of apoptosis blocks human motor neuron cell death in a stem cell model of spinal muscular atrophy. PloS one, 7, e39113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wishart T.M., Mutsaers C.A., Riessland M., Reimer M.M., Hunter G., Hannam M.L., Eaton S.L., Fuller H.R., Roche S.L., Somers E. et al. (2014) Dysregulation of ubiquitin homeostasis and beta-catenin signaling promote spinal muscular atrophy. J Clin Invest, 124, 1821–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Powis R.A., Karyka E., Boyd P., Come J., Jones R.A., Zheng Y., Szunyogova E., Groen E.J., Hunter G., Thomson D. et al. (2016) Systemic restoration of UBA1 ameliorates disease in spinal muscular atrophy. JCI Insight, 1, e87908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Finkel R.S., Mercuri E., Darras B.T., Connolly A.M., Kuntz N.L., Kirschner J., Chiriboga C.A., Saito K., Servais L., Tizzano E. et al. (2017) Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N Engl J Med, 377, 1723–1732. [DOI] [PubMed] [Google Scholar]
- 19. Mercuri E., Darras B.T., Chiriboga C.A., Day J.W., Campbell C., Connolly A.M., Iannaccone S.T., Kirschner J., Kuntz N.L., Saito K. et al. (2018) Nusinersen versus sham control in later-onset spinal muscular atrophy. N Engl J Med, 378, 625–635. [DOI] [PubMed] [Google Scholar]
- 20. Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K. et al. (2017) Single-dose gene-replacement therapy for spinal muscular atrophy. N Engl J Med, 377, 1713–1722. [DOI] [PubMed] [Google Scholar]
- 21. Parente V. and Corti S. (2018) Advances in spinal muscular atrophy therapeutics. Ther Adv Neurol Disord, 11, 1756285618754501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Coenen-Stass A.M.L., Wood M.J.A. and Roberts T.C. (2017) Biomarker potential of extracellular miRNAs in Duchenne muscular dystrophy. Trends Mol Med, 23, 989–1001. [DOI] [PubMed] [Google Scholar]
- 23. Iranifar E., Seresht B.M., Momeni F., Fadaei E., Mehr M.H., Ebrahimi Z., Rahmati M., Kharazinejad E. and Mirzaei H. (2018) Exosomes and microRNAs: new potential therapeutic candidates in Alzheimer disease therapy. J Cell Physiol, in press. [DOI] [PubMed] [Google Scholar]
- 24. Di Pietro L., Lattanzi W. and Bernardini C. (2018) Skeletal muscle MICRORNAS as key players in the pathogenesis of amyotrophic lateral sclerosis. Int J Mol Sci, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rinchetti P., Rizzuti M., Faravelli I. and Corti S. (2018) MicroRNA metabolism and dysregulation in amyotrophic lateral sclerosis. Mol Neurobiol, 55, 2617–2630. [DOI] [PubMed] [Google Scholar]
- 26. Martinez B. and Peplow P.V. (2017) MicroRNAs in Parkinson's disease and emerging therapeutic targets. Neural Regen Res, 12, 1945–1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leggio L., Vivarelli S., L'Episcopo F., Tirolo C., Caniglia S., Testa N., Marchetti B. and Iraci N. (2017) microRNAs in Parkinson's disease: from pathogenesis to novel diagnostic and therapeutic approaches. Int J Mol Sci, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Catapano F., Zaharieva I., Scoto M., Marrosu E., Morgan J., Muntoni F. and Zhou H. (2016) Altered levels of microRNA-9, -206, and -132 in spinal muscular atrophy and their response to antisense oligonucleotide therapy. Mol Ther Nucleic Acids, 5, e331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kye M.J., Niederst E.D., Wertz M.H., Goncalves Ido C., Akten B., Dover K.Z., Peters M., Riessland M., Neveu P., Wirth B. et al. (2014) SMN regulates axonal local translation via miR-183/mTOR pathway. Hum Mol Genet, 23, 6318–6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bhinge A., Namboori S.C., Bithell A., Soldati C., Buckley N.J. and Stanton L.W. (2016) MiR-375 is essential for human spinal motor neuron development and may be involved in motor neuron degeneration. Stem Cells, 34, 124–134. [DOI] [PubMed] [Google Scholar]
- 31. Wertz M.H., Winden K., Neveu P., Ng S.Y., Ercan E. and Sahin M. (2016) Cell-type-specific miR-431 dysregulation in a motor neuron model of spinal muscular atrophy. Hum Mol Genet, 25, 2168–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sison S.L., Patitucci T.N., Seminary E.R., Villalon E., Lorson C.L. and Ebert A.D. (2017) Astrocyte-produced miR-146a as a mediator of motor neuron loss in spinal muscular atrophy. Hum Mol Genet, 26, 3409–3420. [DOI] [PubMed] [Google Scholar]
- 33. Zhou S., Ding F. and Gu X. (2016) Non-coding RNAs as emerging regulators of neural injury responses and regeneration. Neurosci Bull, 32, 253–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Coolen M., Katz S. and Bally-Cuif L. (2013) miR-9: a versatile regulator of neurogenesis. Front Cell Neurosci, 7, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang L.T., Chiou S.S., Liao Y.M., Jong Y.J. and Hsu S.H. (2014) Survival of motor neuron protein downregulates miR-9 expression in patients with spinal muscular atrophy. Kaohsiung J Med Sci, 30, 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Clovis Y.M., Enard W., Marinaro F., Huttner W.B. and De Pietri Tonelli D. (2012) Convergent repression of Foxp2 3'UTR by miR-9 and miR-132 in embryonic mouse neocortex: implications for radial migration of neurons. Development, 139, 3332–3342. [DOI] [PubMed] [Google Scholar]
- 37. Chen Q., Xu J., Li L., Li H., Mao S., Zhang F., Zen K., Zhang C.Y. and Zhang Q. (2014) MicroRNA-23a/b and microRNA-27a/b suppress Apaf-1 protein and alleviate hypoxia-induced neuronal apoptosis. Cell Death Dis, 5, e1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ning K., Pei L., Liao M., Liu B., Zhang Y., Jiang W., Mielke J.G., Li L., Chen Y., El-Hayek Y.H. et al. (2004) Dual neuroprotective signaling mediated by downregulating two distinct phosphatase activities of PTEN. J Neurosci, 24, 4052–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin S.T., Huang Y., Zhang L., Heng M.Y., Ptacek L.J. and Fu Y.H. (2013) MicroRNA-23a promotes myelination in the central nervous system. Proc Natl Acad Sci USA, 110, 17468–17473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wada S., Kato Y., Okutsu M., Miyaki S., Suzuki K., Yan Z., Schiaffino S., Asahara H., Ushida T. and Akimoto T. (2011) Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J Biol Chem, 286, 38456–38465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Z., Pinto A.M., Wan L., Wang W., Berg M.G., Oliva I., Singh L.N., Dengler C., Wei Z. and Dreyfuss G. (2013) Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy. Proc Natl Acad Sci USA, 110, 19348–19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maeda M., Harris A.W., Kingham B.F., Lumpkin C.J., Opdenaker L.M., McCahan S.M., Wang W. and Butchbach M.E. (2014) Transcriptome profiling of spinal muscular atrophy motor neurons derived from mouse embryonic stem cells. PloS one, 9, e106818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saal L., Briese M., Kneitz S., Glinka M. and Sendtner M. (2014) Subcellular transcriptome alterations in a cell culture model of spinal muscular atrophy point to widespread defects in axonal growth and presynaptic differentiation. RNA (New York, N.Y.), 20, 1789–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Custer S.K., Gilson T.D., Li H., Todd A.G., Astroski J.W., Lin H., Liu Y. and Androphy E.J. (2016) Altered mRNA splicing in SMN-depleted motor neuron-like cells. PloS one, 11, e0163954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Murray L.M., Beauvais A., Gibeault S., Courtney N.L. and Kothary R. (2015) Transcriptional profiling of differentially vulnerable motor neurons at pre-symptomatic stage in the Smn (2b/-) mouse model of spinal muscular atrophy. Acta Neuropathol Commun, 3, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baumer D., Lee S., Nicholson G., Davies J.L., Parkinson N.J., Murray L.M., Gillingwater T.H., Ansorge O., Davies K.E. and Talbot K. (2009) Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet, 5, e1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maury Y., Come J., Piskorowski R.A., Salah-Mohellibi N., Chevaleyre V., Peschanski M., Martinat C. and Nedelec S. (2015) Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat Biotechnol, 33, 89–96. [DOI] [PubMed] [Google Scholar]
- 48. Li S., Lv X., Zhai K., Xu R., Zhang Y., Zhao S., Qin X., Yin L. and Lou J. (2016) MicroRNA-7 inhibits neuronal apoptosis in a cellular Parkinson's disease model by targeting Bax and Sirt2. Am J Transl Res, 8, 993–1004. [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Y., Ueno Y., Liu X.S., Buller B., Wang X., Chopp M. and Zhang Z.G. (2013) The MicroRNA-17-92 cluster enhances axonal outgrowth in embryonic cortical neurons. J Neurosci, 33, 6885–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang C.P., Zhang Z.H., Zhang L.H. and Rui H.C. (2016) Neuroprotective role of MicroRNA-22 in a 6-hydroxydopamine-induced cell model of Parkinson's disease via regulation of its target gene TRPM7. J Mol Neurosci, 60, 445–452. [DOI] [PubMed] [Google Scholar]
- 51. Sabirzhanov B., Zhao Z., Stoica B.A., Loane D.J., Wu J., Borroto C., Dorsey S.G. and Faden A.I. (2014) Downregulation of miR-23a and miR-27a following experimental traumatic brain injury induces neuronal cell death through activation of proapoptotic Bcl-2 proteins. J Neurosci, 34, 10055–10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu D.M., Wen X., Han X.R., Wang S., Wang Y.J., Shen M., Fan S.H., Zhuang J., Zhang Z.F., Shan Q. et al. (2018) MiR-142-3p enhances cell viability and inhibits apoptosis by targeting CDKN1B and TIMP3 following sciatic nerve injury. Cell Physiol Biochem, 46, 2347–2357. [DOI] [PubMed] [Google Scholar]
- 53. Zhang A., Li M., Wang B., Klein J.D., Price S.R. and Wang X.H. (2018) miRNA-23a/27a attenuates muscle atrophy and renal fibrosis through muscle-kidney crosstalk. J Cachexia Sarcopenia Muscle, 9, 755–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kaifer K.A., Villalón E., Osman E.Y., Glascock J.J., Arnold L.L., Cornelison D.D.W. and Lorson C.L. (2017) Plastin-3 extends survival and reduces severity in mouse models of spinal muscular atrophy. JCI Insight, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Feng Z., Ling K.K., Zhao X., Zhou C., Karp G., Welch E.M., Naryshkin N., Ratni H., Chen K.S., Metzger F. et al. (2016) Pharmacologically induced mouse model of adult spinal muscular atrophy to evaluate effectiveness of therapeutics after disease onset. Hum Mol Genet, 25, 964–975. [DOI] [PubMed] [Google Scholar]
- 56. Gray K.M., Kaifer K.A., Baillat D., Wen Y., Bonacci T.R., Ebert A.D., Raimer A.C., Spring A.M., Have S.T., Glascock J.J. et al. (2018) Self-oligomerization regulates stability of survival motor neuron protein isoforms by sequestering an SCF (Slmb) degron. Mol Biol Cell, 29, 96–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Eshraghi M., McFall E., Gibeault S. and Kothary R. (2016) Effect of genetic background on the phenotype of theSmn2B/-mouse model of spinal muscular atrophy. Human Molecular Genetics, in press., ddw278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bowerman M., Murray L.M., Boyer J.G., Anderson C.L. and Kothary R. (2012) Fasudil improves survival and promotes skeletal muscle development in a mouse model of spinal muscular atrophy. BMC Med, 10, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Foust K.D., Nurre E., Montgomery C.L., Hernandez A., Chan C.M. and Kaspar B.K. (2009) Intravascular AAV9 preferentially targets neonatal neurons and adult astrocytes. Nat Biotechnol, 27, 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Passini M.A., Bu J., Roskelley E.M., Richards A.M., Sardi S.P., O'Riordan C.R., Klinger K.W., Shihabuddin L.S. and Cheng S.H. (2010) CNS-targeted gene therapy improves survival and motor function in a mouse model of spinal muscular atrophy. J Clin Invest, 120, 1253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dominguez E., Marais T., Chatauret N., Benkhelifa-Ziyyat S., Duque S., Ravassard P., Carcenac R., Astord S., Pereira de Moura A., Voit T. et al. (2011) Intravenous scAAV9 delivery of a codon-optimized SMN1 sequence rescues SMA mice. Hum Mol Genet, 20, 681–693. [DOI] [PubMed] [Google Scholar]
- 62. Glascock J.J., Shababi M., Wetz M.J., Krogman M.M. and Lorson C.L. (2012) Direct central nervous system delivery provides enhanced protection following vector mediated gene replacement in a severe model of spinal muscular atrophy. Biochem Biophys Res Commun, 417, 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ling K.K., Lin M.Y., Zingg B., Feng Z. and Ko C.P. (2010) Synaptic defects in the spinal and neuromuscular circuitry in a mouse model of spinal muscular atrophy. PloS one, 5, e15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mentis G.Z., Blivis D., Liu W., Drobac E., Crowder M.E., Kong L., Alvarez F.J., Sumner C.J. and O'Donovan M.J. (2011) Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron, 69, 453–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ning K., Drepper C., Valori C.F., Ahsan M., Wyles M., Higginbottom A., Herrmann T., Shaw P., Azzouz M. and Sendtner M. (2010) PTEN depletion rescues axonal growth defect and improves survival in SMN-deficient motor neurons. Hum Mol Genet, 19, 3159–3168. [DOI] [PubMed] [Google Scholar]
- 66. Little D., Valori C.F., Mutsaers C.A., Bennett E.J., Wyles M., Sharrack B., Shaw P.J., Gillingwater T.H., Azzouz M. and Ning K. (2015) PTEN depletion decreases disease severity and modestly prolongs survival in a mouse model of spinal muscular atrophy. Mol Ther, 23, 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kong L., Wang X., Choe D.W., Polley M., Burnett B.G., Bosch-Marce M., Griffin J.W., Rich M.M. and Sumner C.J. (2009) Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J Neurosci, 29, 842–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kariya S., Park G.H., Maeno-Hikichi Y., Leykekhman O., Lutz C., Arkovitz M.S., Landmesser L.T. and Monani U.R. (2008) Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum Mol Genet, 17, 2552–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kline R.A., Kaifer K.A., Osman E.Y., Carella F., Tiberi A., Ross J., Pennetta G., Lorson C.L. and Murray L.M. (2017) Comparison of independent screens on differentially vulnerable motor neurons reveals alpha-synuclein as a common modifier in motor neuron diseases. PLoS Genet, 13, e1006680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bodine S.C., Latres E., Baumhueter S., Lai V.K., Nunez L., Clarke B.A., Poueymirou W.T., Panaro F.J., Na E., Dharmarajan K. et al. (2001) Identification of ubiquitin ligases required for skeletal muscle atrophy. Science, 294, 1704–1708. [DOI] [PubMed] [Google Scholar]
- 71. Gomes M.D., Lecker S.H., Jagoe R.T., Navon A. and Goldberg A.L. (2001) Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc Natl Acad Sci USA, 98, 14440–14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gomes A.V., Waddell D.S., Siu R., Stein M., Dewey S., Furlow J.D. and Bodine S.C. (2012) Upregulation of proteasome activity in muscle RING finger 1-null mice following denervation. Faseb j, 26, 2986–2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wong N. and Wang X. (2015) miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic acids research, 43, D146–D152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mourelatos Z., Dostie J., Paushkin S., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M. and Dreyfuss G. (2002) miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev, 16, 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ng S.Y., Soh B.S., Rodriguez-Muela N., Hendrickson D.G., Price F., Rinn J.L. and Rubin L.L. (2015) Genome-wide RNA-Seq of human motor neurons implicates selective ER stress activation in spinal muscular atrophy. Cell Stem Cell, 17, 569–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hudson M.B., Woodworth-Hobbs M.E., Zheng B., Rahnert J.A., Blount M.A., Gooch J.L., Searles C.D. and Price S.R. (2014) miR-23a is decreased during muscle atrophy by a mechanism that includes calcineurin signaling and exosome-mediated export. Am J Physiol Cell Physiol, 306, C551–C558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rupaimoole R. and Slack F.J. (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov, 16, 203–222. [DOI] [PubMed] [Google Scholar]
- 78. Tian K., Di R. and Wang L. (2015) MicroRNA-23a enhances migration and invasion through PTEN in osteosarcoma. Cancer Gene Ther, 22, 351–359. [DOI] [PubMed] [Google Scholar]
- 79. Han Z., Zhou X., Li S., Qin Y., Chen Y. and Liu H. (2017) Inhibition of miR-23a increases the sensitivity of lung cancer stem cells to erlotinib through PTEN/PI3K/Akt pathway. Oncol Rep, 38, 3064–3070. [DOI] [PubMed] [Google Scholar]
- 80. Ebert A.D., Yu J., Rose F.F. Jr., Mattis V.B., Lorson C.L., Thomson J.A. and Svendsen C.N. (2009) Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature, 457, 277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Induced pluripotent stem cells from patients with Huntington's disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell, 11, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mayginnes J.P., Reed S.E., Berg H.G., Staley E.M., Pintel D.J. and Tullis G.E. (2006) Quantitation of encapsidated recombinant adeno-associated virus DNA in crude cell lysates and tissue culture medium by quantitative, real-time PCR. J Virol Methods, 137, 193–204. [DOI] [PubMed] [Google Scholar]
- 83. Glascock J.J., Osman E.Y., Coady T.H., Rose F.F., Shababi M. and Lorson C.L. (2011) Delivery of therapeutic agents through intracerebroventricular (ICV) and intravenous (IV) injection in mice. J Vis Exp, 56, 2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


