Abstract
The diagnosis of Autism Spectrum Disorder (ASD) is a subjective process requiring clinical expertise in neurodevelopmental disorders. Since such expertise is not available at many clinics, automated diagnosis using machine learning (ML) algorithms would be of great value to both clinicians and the imaging community to increase the diagnoses’ availability and reproducibility while reducing subjectivity. This research systematically compares the performance of classifiers using over 900 subjects from the IMPAC database [1], using the database’s derived anatomical and functional features to diagnose a subject as autistic or healthy. In total 12 classifiers are compared from 3 categories including: 6 nonlinear shallow ML models, 3 linear shallow models, and 3 deep learning models. When evaluated with an AUC ROC performance metric, results include: (1) amongst the shallow learning methods, linear models outperformed nonlinear models, agreeing with [2]. (2) Deep learning models outperformed shallow ML models. (3) The best model was a dense feedforward network, achieving 0.80 AUC which compares to the recently reported 0.79±0.01 AUC average of the top 10 methods from the IMPAC challenge [3]. These results demonstrate that even when using features derived from imaging data, deep learning methods can provide additional predictive accuracy over classical methods.
Keywords: autism spectrum disorder, deep learning, machine learning, MRI, neuroimaging
1. INTRODUCTION
Autism spectrum disorder (ASD) is a common psychiatric disorder characterized by social and communication deficits and a restricted pattern of interests [4]. It is known that individuals with ASD have altered neuroanatomy and connectivity, though the full extent of these relationships has not been fully elucidated. Currently, the diagnosis of ASD is a subjective process that requires an expert in neurodevelopmental disorders who may be unavailable at many clinics. Noninvasive imaging captures structural and functional aspects of brain development that are promising for an automated machine learning (ML) based diagnosis. Such automated approaches would reduce subjectivity and increase reproducibility and availability of the diagnosis. Existing literature on automated diagnostics are limited in two ways. First, in these studies, just one category of predictive model is typically proposed and fully optimized; making comparisons to comparable methods biased. Second, they often depend on access to raw image data. Sharing patient images can be problematic due to concerns for patient identifiability. However preprocessed data, such as volumetry and functional connectivity are more easily shared. The Paris IMPAC Autism Challenge [1] is one such sharable dataset containing the derived features from structural MRI (sMRI) and resting state functional MRI (rs-fMRI).
While in many problem domains, such as real world object recognition, deep learning outperforms shallow learning, this increase in performance has been attributed to the replacement of hand-engineered features with learned features. To date there is limited research aimed at understanding how deep learning methods compare in performance to shallow ML methods on datasets with pre-derived features. Elucidating the comparative performance of model categories for such large sharable datasets would be of great significance to guide the image analysis community.
In order to perform a fair comparison, in this study each model is similarly hyperparameter optimized using a random search-based approach. Identical randomly chosen cross-validation splits are used to train each model, ensuring similar training opportunities for each model.
The primary contribution of this work is four-fold. First the study provides a systematic comparison of 3 broad categories of methods: linear and nonlinear shallow ML models and deep learning models, and assesses their relative performance. Second the study examines the relative performance of anatomical features, functional features and their combination and provides evidence of their level of complementarity. Third evidence for the effective level of granularity for deriving regional features from whole brain parcellations is obtained by comparing 7 atlases. Fourth the relative performance of 12 individual classifiers is compared and recommendations for ASD diagnosis is made for a specific winning deep learning model, which achieves greater performance than all other tested models.
2. MATERIALS AND METHODS
2.1. Materials
This study uses 915 subjects from the IMPAC dataset [1] that received both sMRI and rs-fMRI and were identified by the IMPAC organizers as having satisfactory images. The focus of this study is the comparison of two-category classifiers for diagnosing ASD or healthy control. The IMPAC dataset includes the clinical diagnosis (the classifier target) for which there were 418 ASD patients and 497 healthy control subjects. Structural MRI (sMRI) and resting state functional MRI (rs-fMRI) were acquired for each subject. Figure 1 illustrates how the features were derived from the MRI. From the sMRI, 207 features were extracted, including volumes of cortical and subcortical structures, cortical thickness, and area per region of interest (ROI) defined by the Desikan-Killiany gyral atlas [5]. From the rs-fMRI, functional connectivity matrices were derived. For this derivation, the rs-fMRI was first parcellated into ROIs using seven different atlases including: atlases (1–3) The BASC Atlas, whose regions are defined by k-means clustering of stable coherent groups [6] for k=64, 122, and 197 ROIs, atlas (4) the Craddock atlas, which defines 249 ROIs by coherence of local graph connectivity [7], atlas (5) the Harvard-Oxford Anatomical atlas, which defines 69 ROIs using anatomical features, atlas (6) the MSDL atlas, which has 39 ROIs defined by correlations of spontaneous activity [8], and atlas (7) the Power atlas [9], which is defined by local graph-connectivity into 264 ROIs. The rs-fMRI time signals from each region were converted into a connectivity matrix by projection into tangent space, a procedure which captures connectivity aspects from both the correlation and partial correlation [10]. Clinical data including patient gender and age were also collected.
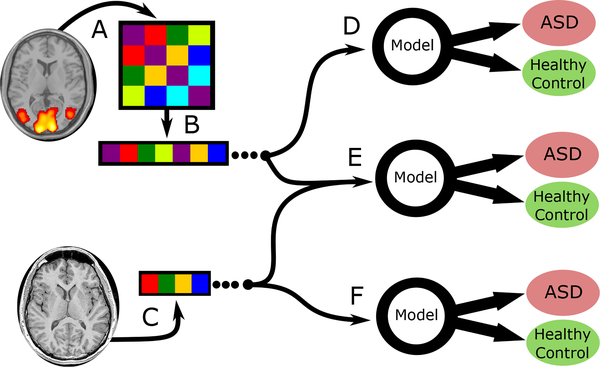
Fig. 1.
Combinations of derived features used by the predictive models tested in this study. Image data (rs-fMRI and sMRI) and was gathered externally by IMPAC organizers. (A) The rs-fMRI was transformed into a symmetric connectivity matrix for each atlas. (B) Upper triangular elements of matrix were flattened into a 1D vector. (C) The sMRI was transformed into a vector of cortical/subcortical ROI volumes and cortical thickness features. In (D) the connectivity matrix vector is used as the sole input for the predictive model, in (E) both anatomical and connectivity derived feature vectors are appended and used, while in (F) the anatomical features are used as the sole input for the predictive model.
2.2. Data partitioning
Subjects were randomly partitioned with 80% assigned to a training set and 20% to a test set with the split having matching proportions of diagnosis (ASD/healthy) and gender (male/female). The test subjects were set aside and not used during training or model selection. The training set was further split into validation and training folds using a 3-fold stratified cross validation approach. To ensure fair subsequent model comparison, the same splits were used for all tested machine learning models.
2.3. Model construction
Systematic testing of a broad array of 12 machine learning classifiers was conducted. This included 6 nonlinear shallow machine learning methods: a naїve Bayes classifier, a support vector machine with a Gaussian kernel, a random forest classifier, an extremely randomized trees classifier, adaptive boosting, and gradient boosting with decision tree base models; 3 linear shallow models; a support vector machine with a linear kernel, logistic regression with ridge regularization, logistic regression with lasso regularization; and 3 deep learning approaches: a fully connected dense feedforward (FeedFWD) network, and a long-short term memory (LSTM) based recurrent neural network classifier (RNN), and the BrainNetCNN [11]. Classical models were constructed using the scikit-learn and XGBoost pakages, while the deep learning models used the keras, tensorflow, and caffe packages. The LSTM classifier uses an LSTM network followed by a dense feedforward layer for classification like [12] which can succeed even on non-sequential fixed vector data, as suggested by [13]. The BrainNetCNN classifier is a graph-convolutional network classifier [11]. The models were trained on an NVIDIA Tesla p100.
2.4. Random search
To optimize each ML model, a random search was conducted over the model’s hyperparameter space. A total of 50 random points in hyperparameter space were sampled for each model. To illustrate, consider the examples of simple and complex dense FeedFWD networks that were tested are illustrated in Table 1, in the left and middle columns respectively. In detail, for the models tested these hyperparameter points were randomly chosen from the following dimensions and ranges: Random Forest: number of estimators [50, 5000], max nodes [5, 50]; Extremely Random Trees: number of estimators [50, 5000], max nodes [5, 50]; Adaptive Boosting: number of estimators [20, 2000], learning rate [0.1, 0.9]; Gradient Boosting: number of estimators [5, 5000], max depth [1, 10], subsampling fraction per tree [0.2, 0.8], fraction of columns per tree [0.2, 1], learning rate [0.01, 1]; SVM with Gaussian Kernel: C [0.0001, 10000], maximum iterations: [10000, 100000], gamma [0.01, 100]; SVM with Linear Kernel: C [0.0001, 10000], maximum iterations: [10000, 100000]; logistic regression with lasso regularization: C [0.0001, 10000], maximum iterations [1000, 100000]; logistic regression with ridge regularization: C [0.0001, 10000], maximum iterations [1000, 100000]; dense FeedFWD network: number of hidden layers [1, 3], layer width [16, 256]; dropout fraction [0.1, 0.6], L2 regularization coefficient [0.0001, 0.02], LSTM: number of hidden layers [1, 3], layer width [16, 256], dropout fraction [0.1, 0.6], L2 regularization coefficient [0.0001, 0.02]; BrainNetCNN: number of hidden layers [0, 2], layer width [16, 64], dropout fraction [0.1, 0.6], ReLU slope for x<1 [0.1, 0.5]. Deep learning models used the leaky ReLU activation function, early stopping, the Nesterov ADAM optimizer, a batch size of 128, and the binary cross-entropy loss function.
Table 1.
Examples of dense FeedFWD network architectures tested in the random search. Hyperparameters shown include the regularization coefficient, # of layers, # of neurons/layer, and dropout fraction. Left column illustrates a simple network. Middle shows a complex network. Right column shows the architecture of the highest performing network.
| Simple Dense Network | Complex Dense Network | Highest Performing Dense Network |
|---|---|---|
| L2 Regularization: 2.3e-4 | L2 Regularization: 2.3e-4 | L2 Regularization: 1.1e-4 |
| Dense 16 neurons | Dense 128 neurons | Dense 64 neurons |
| Dropout: 53% removed | Dropout 18% removed | Dropout 13% removed |
| Dense 16 neurons | Dense 128 neurons | Dense 64 neurons |
| Dense 1 neuron | Dropout 18% removed | Dense 1 neuron |
| Dense 64 neurons | ||
| Dropout 18% removed | ||
| Dense 42 neurons | ||
| Dense 1 neuron |
Each of our 12 models types was trained on 15 different feature sets, for a total of 180 model type × feature set combinations. The feature sets contain measures of anatomical volume and functional connectivity from the IMPAC dataset. These feature sets included: (1) 207 measures of regional volume and thickness, (2–8) functional connectivity measured between regions defined by one of the 7 atlases described in the materials section above, (9–15) the union of the anatomical with one of the functional feature sets. The model with the highest average area under ROC curve over the cross validation folds was selected as the best model for each model type × feature set combination. This model was then trained on all training data and evaluated on the held out test set not used in training.
3. RESULTS
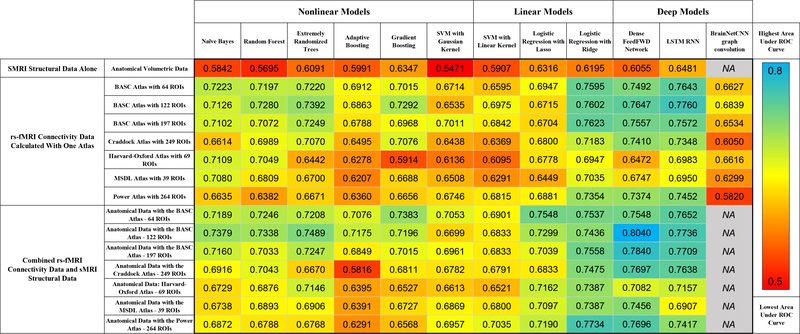
The results are summarized in Figure 2, which shows the area under the ROC curve of different machine learning models predicting ASD vs healthy control on the test data that was held out from training.
Fig. 2.
Performance of classifiers predicting the diagnosis of ASD versus healthy control. Performance is measured as the area under the ROC curve on held-out test data. Cooler colors indicate superior performance.
3.1. The importance of feature set combination
Comparing the 15 feature sets (rows of Figure 2), it can be observed that the anatomical features yielded the lowest prediction accuracy by area under the ROC curve, while the rs-fMRI functional connectivity features alone yielded models with higher predictive power than anatomical features. For functional connectivity data alone, the BASC atlas with any number of parcellations and the Power atlas generated models with more predictive power than other atlases. However the combination of anatomical and functional features yielded models with even higher predictive power, suggesting their complementarity. The best performing models used the anatomical features with connectivity features from the Power atlas, Craddock atlas, or BASC atlas. Models trained on the Harvard-Oxford atlas connectivity data and volumetric data are notably lower performing, and models trained on the MSDL atlas were slightly better than those trained on the Harvard-Oxford atlas.
3.2. The importance of model type
The most accurate shallow machine learning algorithm was logistic regression with ridge regularization, which had a maximum ROC AUC of 0.773. Of the nonlinear methods, the extremely randomized trees performed the highest, and the adaptive boosting methods performed the lowest. Overall, deep learning outperformed shallow learning, and the highest performing shallow linear methods outperformed the highest performing shallow nonlinear methods.
As shown in the columns towards the right of Figure 2, the deep learning methods performed higher than the other categories of models. The most successful deep learning algorithms were the dense FeedFWD network which achieved an ROC AUC of 0.804 and LSTM, which achieved an ROC AUC of 0.776. The BrainNetCNN model is only defined for functional connectivity input features, but on those features it performed lower than the other deep learning models with a performance similar to the linear models. Like the shallow methods, the deep learning methods performed best when using the combination of functional and anatomical features. The highest overall performance was the dense FeedFWD network, whose architecture is shown in Table 1 right column, using the rs-fMRI connectivity data with the BASC atlas with 122 ROIs and the sMRI volumetric data combined, achieving an AUC of 0.804. Other permutations using the BASC atlases, Craddock atlas, and Power atlas as training data for the dense FeedFWD network also performed well.
4. DISCUSSION AND CONCLUSIONS
Mensch et al. [14] reported high performance using deep learning networks for decoding brain activity to predict of the class of psychological stimuli presented in neuroimaging studies. This study focused on the classification of ASD versus healthy control and also demonstrated high performance using deep learning, adding to the evidence that deep learning is effective at classification from multidimensional neuroimaging data. The highest performing model in this study was a dense FeedFWD network which achieved 0.80 AUC, which is quite similar to the 0.79±0.01 AUC average of the top 10 methods recently reported from the IMPAC challenge [3]. Classification of ASD has been reported by Parisot et al. [15] and Meenashki et al. [16]. These methods both employ novel convolutional neural networks to achieve state-of-the-art performance of 70.4% and 73.3% accuracy respectively on the open source ABIDE dataset for ASD [17], but both depend on the raw imaging data.
The results of this study indicate multiple conclusions: First, the results show that deep learning is still a valuable tool that is able to extract additional predictive power over shallow methods even when provided pre-extracted feature sets. Second, the subset of atlases that performed better is informative for ASD diagnosis. Across many different machine learning modalities, the functional BASC atlas, derived using a k-means clustering approach, performed very well. Its 122 and 197 ROI versions performed better than the 64 ROI version. This suggests the scale or granularity of neuroimaging-detectable changes in functional connectivity in ASD. Also, this suggests that k-means clustering and other graph-based clustering methods such as the Power and Craddock atlases, may be more suited to accurately elucidate functional connectivity changes in ASD than other parcellation methods. Third, the uniformly poor performance observed when models use purely anatomical features suggests that the deficits in ASD are reflected more by changes in functional connectivity than by changes in volume and cortical thickness. This finding is in agreement with results of previous studies [16]. However, the fact that in general, combining anatomical features with functional connectivity features tended to improve model performance across model categories, supports the notion that the information is complementary and should be combined to maximize predictive accuracy.
In summary, this study provides insights into the comparative performance of three categories of widely used machine learning models, including both linear and nonlinear shallow models as well as deep learning models for the important task of automating diagnosis for Autism Spectrum Disorder. It provides insights into the combination of anatomical and functional features that are most useful for diagnosis of ASD and demonstrates that their combination is most appropriate. The study also demonstrates that a finer level of granularity in whole brain parcellation with roughly 120 ROIs outperforms coarser parcellations. Lastly the study shows that a dense FeedFWD network outperforms other models even when features are pre-extracted from MRI and attains highly accurate diagnosis compared to previously published methods. In the future we aim to continue to improve upon automated classification performance in ASD and other neuropathologies.
5. REFERENCES
- [1].Toro R, Traut N, Beggatio A, Heuer K, and Varoquaux G et al. , “IMPAC: Imaging-psychiatry challenge: predicting autism, a data challenge on autism spectrum disorder detection,” Online Challenge, 2018. [Google Scholar]
- [2].Dadi K, Rahim M, Abraham A, Chyzhyk D, and Milham M et al. , “Benchmarking functional connectome-based predictive models for resting-state fMRI,” HAL-Inria, 2018. [DOI] [PubMed] [Google Scholar]
- [3].Varoquaux G, “MRI biomarkers extraction, teachings from an autism-prediction challenge,” MICCAI Conference, Sept 20, 2018. [Google Scholar]
- [4].Stanfield AC, McIntosh AM, Spencer MD, Philip R, Gaur S, and Lawrie SM, “Towards a neuroanatomy of autism: A systematic review and meta-analysis of structural magnetic resonance imaging studies,” European Psychiatry, vol. 23, no. 4, pp. 289–299, June 2008. [DOI] [PubMed] [Google Scholar]
- [5].Desikan RS, Sgonne F, Fischl B, Quinn BT, and Dickerson BC et al. , “An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest,” NeuroImage, vol. 31, no. 3, pp. 968–980, 2006. [DOI] [PubMed] [Google Scholar]
- [6].Bellec P, Rosa-Neto P, Lyttelton OC, Benali H, and Evans AC, “Multi-level bootstrap analysis of stable clusters in resting-state fMRI,” NeuroImage, vol. 51, no. 3, pp. 1126–1139, 2010. [DOI] [PubMed] [Google Scholar]
- [7].Craddock RC, James GA, Holtzheimer PE, Hu XP, and Mayberg HS, “A whole brain fMRI atlas generated via spatially constrained spectral clustering,” Human Brain Mapping, vol. 33, no. 8, pp. 10.1002/hbm.21333, August 2012, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Varoquaux G, Gramfort A, Pedregosa F, Michel V, and Thirion B, “Multi-subject dictionary learning to segment an atlas of brain spontaneous activity,” in Information Processing in Medical Imaging, Kaufbeuren, Germany, July 2011, ekely Gabor Sz, Horst Hahn, vol.´ 6801 of Lecture Notes in Computer Science, pp. 562–573, Springer. [DOI] [PubMed] [Google Scholar]
- [9].Power JD, Cohen AL, Nelson SM, Wig GS, and Barnes KA et al. , “Functional network organization of the human brain,” Neuron, vol. 72, no. 4, pp. 665–678, November 2011, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Varoquaux G, Baronnet F, Kleinschmidt A, Fillard P, and Thirion B, “Detection of brain functional-connectivity difference in post-stroke patients using group-level covariance modeling,” in Medical Image Computing and Computer Assisted Intervention, 2010. [DOI] [PubMed] [Google Scholar]
- [11].Kawahara J, Brown CJ, Miller SP, Booth BG, and Chau V et al. , “BrainNetCNN: Convolutional neural networks for brain networks; towards predicting neurodevelopment,” NeuroImage, vol. 146, pp. 1038–1049, 2017. [DOI] [PubMed] [Google Scholar]
- [12].Yan W, Zhang H, Sui J, and Shen D, “Deep chronnectome learning via full bidirectional long short-term memory networks for mci diagnosis,” arXiv.org, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Karpathy A, “The unreasonable effectiveness of recurrent neural networks,” http://karpathy.github.io/2015/05/21/rnn-effectiveness, 2015. [Google Scholar]
- [14].Mensch A, Mairal J, Thirion B, and Varoquaux G, “Extracting universal representations of cognition across brain-imaging studies,” European Psychiatry, September 2018. [Google Scholar]
- [15].Parisot S, Ktena SI, Ferrante E, Lee M, and Guerrero R et al. , “Disease prediction using graph convolutional networks: Application to autism spectrum disorder and Alzheimer’s disease,” Medical image analysis, vol. 48, pp. 117–130, 2018. [DOI] [PubMed] [Google Scholar]
- [16].Meenakshi K, Jamison K, Kuceyeski A, and Sabuncu MR, “3D convolutional neural networks for classification of functional connectomes,” arXiv.org, 2018. [Google Scholar]
- [17].Di Martino A, Yan CG, Li Q, Denio E, and Castellanos FX et al. , “The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism,” Mol. Psychiatry, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]