Summary
Cucumber mosaic virus (CMV) is a highly prevalent viral pathogen causing substantial damage to the bulb and cut‐flower production of Lilium spp. Here, we performed an Illumina RNA sequencing (RNA‐Seq) study on the leaf tissues of a virus‐resistant species Lilium regale inoculated with mock control and CMV. A total of 1346 differentially expressed genes (DEGs) were identified in the leaves of L. regale upon CMV inoculation, which contained 34 up‐regulated and 40 down‐regulated DEGs that encode putative transcription factors (TFs). One up‐regulated TF, LrNAC35, belonging to the NAM/ATAF/CUC (NAC) superfamily, was selected for further functional characterization. Aside from CMV, lily mottle virus and lily symptomless virus infections provoked a striking increase in LrNAC35 transcripts in both resistant and susceptible Lilium species. The treatments with low temperature and several stress‐related hormones activated LrNAC35 expression, contrary to its reduced expression under salt stress. Ectopic overexpression of LrNAC35 in petunia (Petunia hybrida) resulted in reduced susceptibility to CMV and Tobacco mosaic virus infections, and enhanced accumulation of lignin in the cell walls. Four lignin biosynthetic genes, including PhC4H, Ph4CL, PhHCT and PhCCR, were found to be up‐regulated in CMV‐infected petunia lines overexpressing LrNAC35. In vivo promoter‐binding tests showed that LrNAC35 specifically regulated the expression of Ph4CL. Taken together, our results suggest a positive role of transcriptome‐derived LrNAC35 in transcriptional modulation of host defence against viral attack.
Keywords: Cucumber mosaic virus, lignin, Lilium regale, NAC transcription factor, petunia, Tobacco mosaic virus, transcriptome
Introduction
Viral pathogens are the major constraining factors for crop growth and production worldwide. They occur ubiquitously in various environmental conditions. Disease symptoms resulting from virus invasion often include chlorotic or necrotic leaves, shortened internodes, stunted upright growth and weakened vitality (Galvez et al., 2014; Rojas et al., 2007). Plants have evolved multiple defensive mechanisms against the threats posed by massive virus proliferation. The recognition of foreign virions by plants firstly stimulates the establishment of a basal defence system, which involves the participation of reactive oxygen species, cell wall structural components, and antiviral proteins and compounds (He et al., 2007; van Loon et al., 2006). If viruses break through the basal guard, a second‐line host defence called the innate immune response is activated (Liu et al., 2004).
Numerous virus‐responsive genes have essential roles in the innate immunity system, such as the resistance (R) gene (Kang et al., 2005), tobacco N gene (Marathe et al., 2002; Whitham et al., 1996), and the genes associated with RNA silencing (Agius et al., 2012; Sun et al., 2016a) and translation suppression (Zorzatto et al., 2015). The transcription of these genes is potentially controlled by specific transcription factors (TFs), of which many members of various families, including MYB (Yang and Klessig, 1996), zinc finger (Guo et al., 2004), WRKY (Park et al., 2006), AP2/ERF (Huang et al., 2016), bZIP (Gaguancela et al., 2016), bHLH (Aparicio and Pallás, 2017) and NAC (Huang et al., 2017), have been revealed to be implicated in the response to viral stimuli. In particular, NAC domain‐containing proteins comprise a large TF family in plants and play essential roles in plant growth, development and stress responses (Puranik et al., 2012). Approximately 105 and 75 NAC proteins are present in Arabidopsis and Oryza sativa genomes, respectively. Based on sequence similarity, NAC proteins from both species are classified into two large groups and 18 subgroups (Ooka et al., 2003). These proteins share five highly conserved subdomains (A to E) within the N‐terminal region or a divergent C‐terminal domain (Hegedus et al., 2003; Kikuchi et al., 2000).
Multiple studies have demonstrated the critical functions of NAC TFs in abiotic stress resistance, such as drought (Tran et al., 2004), salt (Balazadeh et al., 2010) and low temperature (Hao et al., 2011), as well as their disease resistance to fungal and bacterial pathogens, including Botrytis cinerea, Pseudomonas syringae and Alternaria brassicicola (Wang et al., 2009). In addition, a number of studies have also reported the involvement of NAC TFs in plant response to virus infection. For instance, GRAB1 and GRAB2 interact with wheat dwarf geminivirus (WDV) RepA protein, and their expression in wheat cells restrains DNA replication of WDV (Xie et al., 1999). SlNAC1 overexpression causes an enhanced replication of tomato leaf curl virus by specifically binding to geminivirus replication enhancer (REn) protein (Selth et al., 2005). The mutant of rim1‐1, a novel NAC gene, shows reduced susceptibility to rice dwarf virus, whereas its overexpression enhances virus multiplication (Yoshii et al., 2009). Transgenic Arabidopsis plants overexpressing NAC TF gene ATAF2 exhibit decreased proliferation of tobacco mosaic virus (TMV) and up‐regulation of some defence‐associated marker genes (Wang et al., 2009). The interaction between NAC protein TIP and turnip crinkle virus coat protein interferes with the basal defence against virus invasion in Arabidopsis (Donze et al., 2014). Another NAC protein, NAC083, interacts with mungbean yellow mosaic India virus replication initiator protein in Arabidopsis, but its biological role in virus resistance has not yet been examined (Suyal et al., 2014). A recent study shows that six NAC genes differentially respond to tomato yellow leaf curl virus (TYLCV) infection in tomato plants, and among them virus‐induced gene silencing (VIGS) of SlNAC61 leads to increased TYLCV accumulation (Huang et al., 2017). These findings indicate that NAC TFs have a conserved biological role in virus resistance mostly through the interaction with viral proteins. However, little is known about the transcriptional regulation of virus‐responsive genes by NAC TFs in plants.
Lily belongs to the genus Lilium of the family Liliaceae. It is a summer‐blooming perennial bulbous plant with various food, aesthetic, medicinal and economic values (Wang et al., 2016). The fleshy bulb scales are rich in nutrients and antioxidants (Jin et al., 2012). Viral infection is a frequently occurring disease in lily plants, which affects bulb and cut‐flower production (Ram et al., 2000). More than 20 virus species have been reported to be capable of infecting Lilium and of these, Cucumber mosaic virus (CMV, genus Cucumovirus), Lily mottle virus (LMoV; genus Potyvirus) and Lily symptomless virus (LSV, genus Carlavirus) are most prevalent (Chinestra et al., 2010). To cultivate new lily varieties with desirable antiviral characteristics, conventional crossbreeding or molecular genetic manipulation cannot be inseparable from the excellent wild germplasm resources. A representative lily species native to China, Lilium regale, enjoys a high reputation because of its broad‐spectrum resistance to abiotic stresses, fungi and viruses (Li et al., 2014; Rao et al., 2013). A couple of genes have been identified from L. regale, and one example is that LrP5CS overexpression confers increased tolerance of transgenic Arabidopsis plants to osmotic, drought and high salinity stresses (Wei et al., 2016). With respect to virus resistance, our recent research shows the lowest viral disease incidence of L. regale among ten species tested under natural infection in the field (Sun et al., 2016b). L. regale's outstanding antiviral performance is also supported by our mechanical inoculation experiments in which CMV, LMoV and LSV could not elicit some visible symptoms, such as vein clearing, leaf mosaic, leaf curling, necrotic spots or hypersensitive response (HR), in L. regale initially. Only mild leaf curling symptom appeared in CMV‐inoculated L. regale leaves at the late stage of infection.
Considering the recalcitrance of lily plants to stable transformation, we employed petunia as a heterologous expression model system for studies of antiviral machinery in lilies. In previous work, we constructed a CMV‐induced L. regale cDNA library based on suppression subtractive hybridization (SSH), from which a gene termed LrABCF1 was functionally determined to modulate the resistance to CMV and tobacco rattle virus (TRV) in transgenic petunia (Sun et al., 2016b). However, SSH analysis can only generate a relatively small amount of differentially expressed transcripts. In this study, we used a high‐throughput Illumina RNA sequencing (RNA‐Seq) approach to further investigate the molecular basis of CMV resistance in L. regale. A NAC TF gene, annotated as LrNAC35, was selected from up‐regulated TFs based on transcriptome data and validated for its crucial role in virus (CMV and TMV) resistance by ectopic overexpression in petunia.
Results
Sampling, RNA sequencing and de novo assembly
To investigate the host transcriptome response of L. regale to CMV infection, the second broad true leaves of seed‐grown plantlets were used for virus inoculation (Fig. 1A). Viral gene expression tests showed a moderate increase in transcripts of CMV‐1a, ‐2a and ‐coat protein (CP) during an initial 24 h of infection, followed by a rather sharp rise at 48 and 72 h post‐inoculation (hpi) (Fig. 1B). The sampling time of L. regale leaves for RNA extraction and sequencing, representing a surge of virus replication, was thus determined to be 48 hpi.
Figure 1.
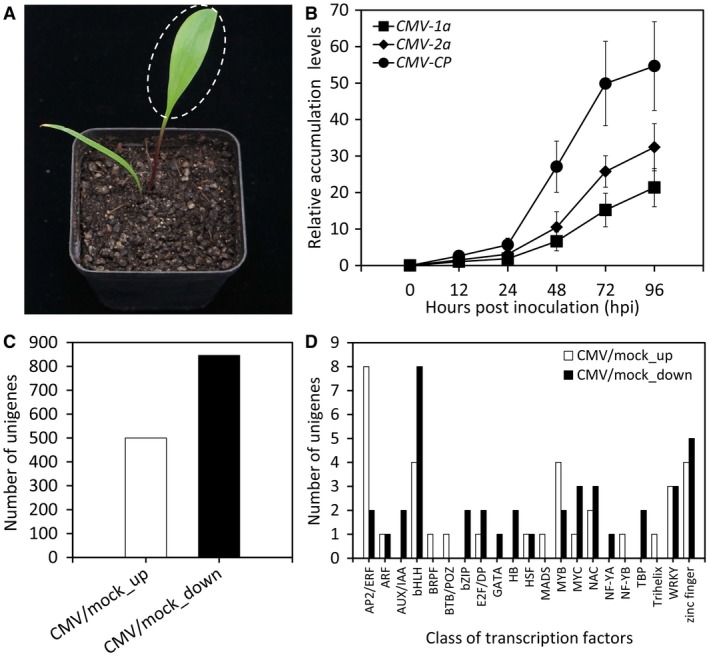
Differentially expressed transcripts in CMV‐infected Lilium regale leaves. (A) Representative growth phenotypes of two‐leaf‐stage L. regale seedlings propagated from seeds at 4 weeks post‐germination. The second newly sprouted leaves used for CMV inoculation and further RNA‐Seq are marked in a dashed circle. (B) qRT‐PCR analysis of CMV accumulation levels (CMV‐1a, ‐2a and ‐CP) in the inoculated leaves of L. regale seedlings at various hours post‐inoculation (hpi). Expression levels were standardized to LrGAPDH. Error bars represent standard error (SE) of the mean from three biological replicates. Number of differentially expressed genes (C) and transcription factors (D) in CMV‐infected L. regale leaves by comparison with mock. Up, up‐regulated; down, down‐regulated.
Illumina paired‐end sequencing in total generated 154 921 638 and 154 400 476 raw reads for the mock control and CMV‐infected L. regale libraries. Correspondingly, 151 810 642 and 150 595 506 clean data were obtained after trimming off the adaptor sequences, and ambiguous and low‐quality reads. De novo assembly resulted in a total of 115 826 unigenes. The maximum, minimum and mean lengths of the assembled unigenes are 11 418, 224 and 615.8 bp, respectively (Table 1). The sequences and functional annotation of all assembled unigenes are shown in Fig. S1 and Table S1 (see Supporting information). Their size distribution revealed that 75 601 (65.3%) unigenes ranged from 200 to 499 bp, 22 496 (19.4%) from 500 to 999 bp and 17 729 (15.3%) were more than 1000 bp (Fig. S2, see Supporting information). To corroborate the accuracy of RNA‐Seq data, nine unigenes, comprising one unknown and eight annotated transcripts, were randomly selected and analysed via quantitative real‐time PCR (qRT‐PCR). The qRT‐PCR results for the unigenes tested were consistent with the transcriptome data (Fig. S3, see Supporting information). The reliability of differential gene expression identified by RNA‐Seq was accordingly validated. Principal component analysis (PCA) of transcriptome data showed that principal components 1 and 2 explained 60.8% and 21% of the variance, respectively, and CMV‐infected samples were significantly different from the mock control (Fig. S4, see Supporting information).
Table 1.
Summary of CMV‐infected Lilium regale transcriptome sequencing dataset
| Item | Number |
|---|---|
| Raw reads from mock‐inoculated samples | 154 921 638 |
| Raw reads from CMV‐inoculated samples | 154 400 476 |
| Clean reads from mock‐inoculated samples | 151 810 642 |
| Clean reads from CMV‐inoculated samples | 150 595 506 |
| Total assembled unigenes | 115 826 |
| Total assembled bases | 71 330 030 |
| Maximum length (bp) | 11 418 |
| Minimum length (bp) | 224 |
| Average length (bp) | 615.8 |
| Unigenes against NR database | 43 064 |
| Unigenes against Swiss‐Prot database | 36 468 |
| Unigenes against COG database | 32 211 |
| Unigenes against KEGG database | 14 641 |
| Annotated unigenes | 47 307 |
| Unannotated unigenes | 68 519 |
Detection of differentially expressed genes during CMV infection
Comparative transcriptome analysis of mock control and CMV‐infected L. regale leaves generated 1346 differentially expressed genes (DEGs). Of all DEGs, 500 unigenes were significantly up‐regulated in CMV‐infected leaves, while 846 unigenes were down‐regulated (Fig. 1C and Table S2, see Supporting information). Gene Ontology (GO) enrichment analysis showed that 663, 418 and 158 DEGs were assigned to three functional categories: biological process, cellular component and molecular function, respectively. The largest number of enriched genes fell into the metabolic process, response to stimulus and single‐organism process (Fig. S5, see Supporting information). We further implemented a pathway‐based categorization of DEGs by searching against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and found that 219 DEGs were mapped to 58 biochemical pathways. Among these mapped pathways, the metabolic pathway contained the largest number of DEGs (36), followed by secondary metabolites biosynthesis (28) and plant–pathogen interaction (22) pathways (Table S3, see Supporting information).
Candidate transcriptional regulators involved in response to CMV
To understand the transcriptional modulation mechanism of CMV defence in L. regale, 74 DEGs encoding putative TFs, of which 34 were up‐regulated and 40 were down‐regulated, were obtained based on gene annotation. These TFs could be classified into 21 TF families, with a considerable number of genes grouped into the bHLH, AP2/ERF, zinc finger, MYB, WRKY and NAC families (Fig. 1D and Table S4, see Supporting information). Fifty‐six DEGs covering the nine TF families that are most probably correlated with the virus response of L. regale plants were identified (Table 2). Next, a time‐course expression profile analysis of selected candidate TF genes was conducted. The results show that CMV inoculation leads to a continuous increase in transcript abundances of LrERF61, LrTINY, LrCPC, LrNAC35, LrNAC100, LrWRKY28, LrWRKY48 and LrDOF5.6, and reduction in LrIAA17, LrRF2a and LrNAC48 expression levels throughout the infection process up to 72 hpi. An increase followed by a drop occurred when examining the expression of LrbHLH100, LrbHLH18, LrMYB98, LrMYC4 and LrZFP28 (Fig. 2).
Table 2.
Putative transcription factors associated with the defence response of Lilium regale to CMV infection
| Gene ID | Annotation | RPKM | Log2 (CMV/mock) | |
|---|---|---|---|---|
| Mock | CMV | |||
| AP2/ERF family | ||||
| Unigene0000389 | Ethylene‐responsive transcription factor ERF061‐like | 0.362 | 2.026 | 2.486 |
| Unigene0000390 | Ethylene‐responsive transcription factor ERF061‐like | 0.371 | 2.501 | 2.752 |
| Unigene0014641 | Ethylene‐responsive transcription factor ERF061‐like | 0.169 | 0.957 | 2.503 |
| Unigene0052014 | Ethylene‐responsive transcription factor ERF061‐like | 0.140 | 0.580 | 2.047 |
| Unigene0052016 | Ethylene‐responsive transcription factor ERF061‐like | 0.401 | 1.947 | 2.281 |
| Unigene0072729 | Ethylene‐responsive transcription factor ERF061‐like | 0.001 | 0.8321 | 9.701 |
| Unigene0073936 | Ethylene‐responsive transcription factor TINY‐like | 0.001 | 0.7314 | 9.515 |
| Unigene0089226 | Ethylene‐responsive transcription factor ERF061‐like | 0.038 | 1.575 | 5.377 |
| Unigene0020351 | Ethylene‐responsive transcription factor PLT2‐like | 6.224 | 0.983 | −2.662 |
| Unigene0037049 | Ethylene‐responsive transcription factor ERF012‐like | 0.644 | 0.001 | −9.331 |
| AUX/IAA family | ||||
| Unigene0090221 | AUX/IAA transcriptional regulator family protein | 0.979 | 0.203 | −2.272 |
| Unigene0108320 | Auxin‐responsive protein IAA17‐like | 129.788 | 48.931 | −1.407 |
| bHLH family | ||||
| Unigene0015592 | Transcription factor ABA‐inducible bHLH‐type‐like | 0.001 | 0.495 | 8.951 |
| Unigene0038912 | Transcription factor bHLH100‐like | 1.341 | 6.279 | 2.227 |
| Unigene0038913 | Transcription factor bHLH100‐like | 0.727 | 4.409 | 2.600 |
| Unigene0111737 | Transcription factor bHLH18‐like | 4.817 | 25.620 | 2.411 |
| Unigene0009308 | Transcription factor bHLH75‐like | 1.880 | 0.045 | −5.395 |
| Unigene0029409 | Transcription factor PAR1‐like | 0.414 | 0.050 | −3.064 |
| Unigene0031740 | Transcription factor bHLH137‐like | 1.508 | 0.020 | −6.255 |
| Unigene0038757 | Transcription factor bHLH35‐like | 0.832 | 0.094 | −3.141 |
| Unigene0042758 | BHLH family transcriptional factor | 6.980 | 1.631 | −2.098 |
| Unigene0051700 | Transcription factor bHLH35‐like | 0.223 | 0.001 | −7.802 |
| Unigene0059321 | Transcription factor bHLH30‐like | 1.232 | 0.431 | −1.514 |
| Unigene0011609 | BHLH family protein | 1.397 | 0.134 | −3.379 |
| bZIP family | ||||
| Unigene0010955 | Transcription factor RF2a‐like | 5.961 | 0.724 | −3.041 |
| Unigene0027495 | Bzip‐like transcription factor‐like protein | 1.748 | 0.459 | −1.928 |
| MYB family | ||||
| Unigene0003524 | Transcription factor MYB98‐like | 0.139 | 2.429 | 4.123 |
| Unigene0006408 | Transcription factor RL9 | 0.122 | 0.408 | 1.736 |
| Unigene0007577 | MYB‐related protein Zm1‐like | 0.226 | 1.360 | 2.592 |
| Unigene0054070 | Transcription factor CPC‐like | 0.604 | 6.966 | 3.527 |
| Unigene0003522 | Transcription factor MYB98‐like | 0.335 | 0.001 | −8.386 |
| Unigene0097039 | Transcription factor R2R3‐MYB | 1.876 | 0.135 | −3.800 |
| MYC family | ||||
| Unigene0053370 | Transcription factor MYC4‐like | 0.001 | 3.317 | 11.696 |
| Unigene0076682 | Transcription factor ICE1‐like | 0.956 | 0.051 | −4.237 |
| Unigene0105852 | Transcription factor MYC2‐like | 5.815 | 0.096 | −5.917 |
| Unigene0111554 | Transcription factor MYC4‐like | 5.449 | 0.796 | −2.776 |
| NAC family | ||||
| Unigene0012070 | NAC domain‐containing protein 35‐like | 5.611 | 38.013 | 2.760 |
| Unigene0025824 | NAC domain‐containing protein 100‐like | 0.088 | 0.558 | 2.656 |
| Unigene0050073 | NAC domain‐containing protein 90‐like | 1.611 | 0.112 | −3.718 |
| Unigene0093619 | NAC domain‐containing protein 21/22‐like | 3.284 | 0.446 | −2.881 |
| Unigene0112474 | NAC domain‐containing protein 48‐like | 8.865 | 2.983 | −1.571 |
| WRKY family | ||||
| Unigene0001022 | WRKY transcription factor 28 | 0.385 | 1.892 | 2.296 |
| Unigene0039394 | WRKY transcription factor 28 | 1.759 | 12.578 | 2.838 |
| Unigene0104357 | WRKY transcription factor 48 | 14.561 | 34.974 | 1.264 |
| Unigene0042962 | WRKY family protein | 5.930 | 2.080 | −1.512 |
| Unigene0051891 | WRKY transcription factor 53 | 4.355 | 0.411 | −3.404 |
| Unigene0078584 | WRKY transcription factor 51 | 0.665 | 0.001 | −9.377 |
| Zinc finger family | ||||
| Unigene0071735 | Zinc finger protein WIP3‐like | 0.429 | 1.637 | 1.932 |
| Unigene0076256 | Zinc finger protein RICESLEEPER 2‐like | 0.723 | 2.298 | 1.668 |
| Unigene0109816 | Zinc finger protein ZAT11 | 0.001 | 1.402 | 10.453 |
| Unigene0111263 | Dof zinc finger protein DOF5.6‐like | 0.240 | 2.770 | 3.530 |
| Unigene0018640 | Zinc finger protein 7 | 1.513 | 0.121 | −3.645 |
| Unigene0092120 | Zinc finger CCCH domain‐containing protein 28‐like | 1.094 | 0.158 | −2.792 |
| Unigene0093967 | Zinc finger protein ZNFphex133 | 0.713 | 0.151 | −2.239 |
| Unigene0027578 | RING‐H2 zinc finger protein ATL79‐like | 32.849 | 12.804 | −1.359 |
| Unigene0060238 | RING‐H2 zinc finger protein ATL3‐like | 1.615 | 0.010 | −7.357 |
Figure 2.
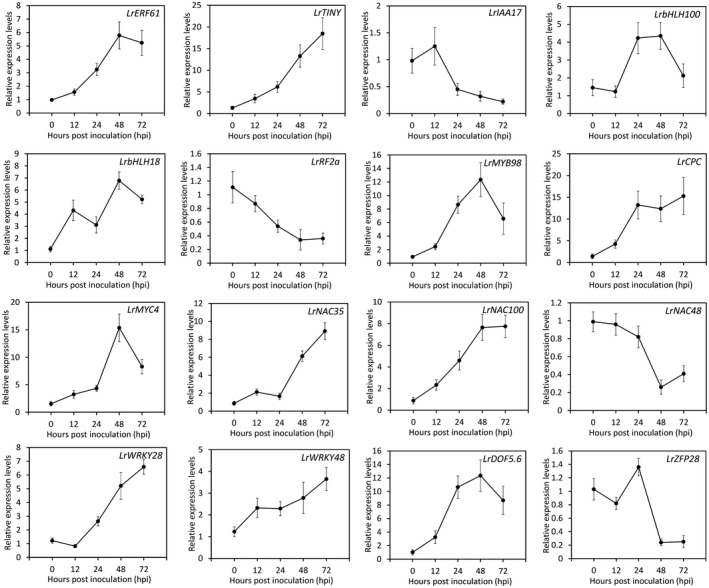
Expression of candidate transcription factors (TFs) associated with Lilium regale defence against CMV infection. Sixteen unigenes, classified into nine families, were chosen from up‐ or down‐regulated TFs and evaluated by qRT‐PCR at given time points. Transcript abundances were normalized to LrActin. SE of the mean from three biological replicates is shown as error bars.
Identification of LrNAC35
We selected LrNAC35 (GenBank accession no. MK805884) from the expression‐quantified TF genes above for further functional characterization. The full length of LrNAC35 cDNA includes a complete coding region encoding 358 amino acids (Fig. S6, see Supporting information), possessing five subdomains A to E. Alignment and the phylogenetic tree showed that LrNAC35 has high homology to NAC49 and NAC75 from O. sativa, and NAC35 proteins from Petunia hybrida, Arabidopsis thaliana, Triticum aestivum, Solanum lycopersicum and Zea mays (Fig. 3A,B).
Figure 3.
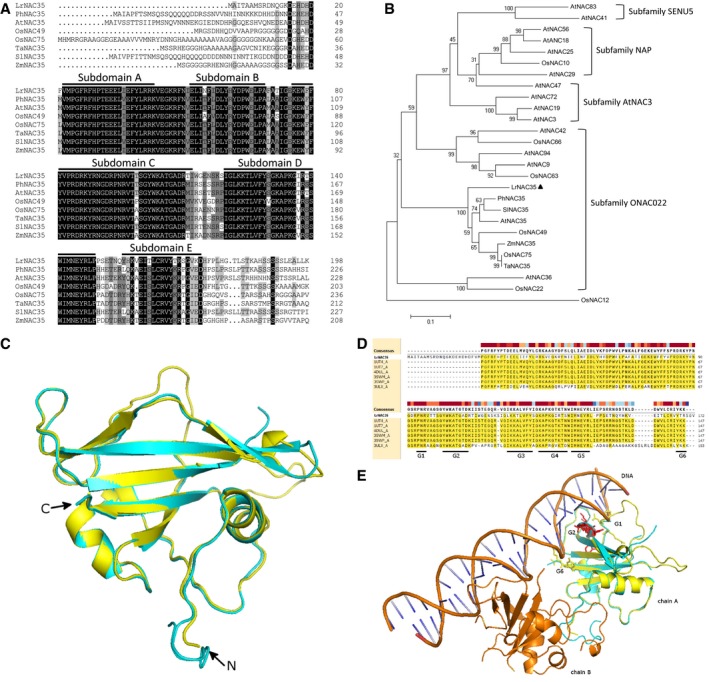
Sequence analysis of LrNAC35 from Lilium regale. (A) Alignment of conserved regions of deduced LrNAC35 amino acid sequence with similar proteins, including Petunia hybrida PhNAC35 (GBRU01060495), Arabidopsis thaliana AtNAC35 (AtLOV1, AT2G02450), Oryza sativa OsNAC49 (AJO53625) and OsNAC75 (XP_015631974), Triticum aestivum TaNAC35 (CDM85391), Solanum lycopersicum SlNAC35 (XP_004230395) and Zea mays ZmNAC35 (PWZ31007). Solid lines indicate the conserved subdomains A to E. (B) Phylogenetic analysis of LrNAC35 with the aligned proteins above and other similar proteins including AtNAC3 (At1g02220), AtNAC9 (At1g26870), AtNAC18 (At1g52880), AtNAC19 (At1g52890), AtNAC25 (At1g61110), AtNAC29 (At1g69490), AtNAC36 (At2g17040), AtNAC41 (At2g33480), AtNAC42 (At2g43000), AtNAC47 (At3g04070), AtNAC56 (At3g15510), AtNAC72 (At4g27410), AtNAC83 (At5g13180), AtNAC94 (At5g39820), OsNAC10 (XP_015645677), OsNAC22 (XP_015630174), OsNAC63 (XP_015649454) and OsNAC66 (XP_015628846). LrNAC35 is highlighted by a black triangle. OsNAC12 (EEC79300) belonging to subfamily TERN of NAC proteins served as the out‐group. Boot‐strap values are expressed as a percentage of 1000 replicates and shown at branch nodes. (C) Superimposition of LrNAC35 (cyan) with AtNAC19 (3SWM chain A, yellow). (D) Amino acid sequence alignment of LrNAC35 with the identified homologous protein templates. G1–G6 represents the identified DNA‐interacting residue groups in AtNAC19. (E) The potential DNA‐binding interaction of LrNAC35 (cyan) in superimposition with chain A (yellow) of AtNAC19 (3SWM). The spatial position of monomer chain B (orange) is also displayed. The side chains of the corresponding G1, G2 (red) and G6 residues in LrNAC35 are shown as sticks.
To investigate the protein structural features of LrNAC35, particularly its DNA‐binding potential, homology‐based protein structural modelling was performed for the NAC DNA‐binding domain. A combination of multiple homologous protein structures (chain A of Protein Data Banks (PDBs) 1UT4, 1UT7, 4DUL, 3SWM, SWP and 3ULX) was identified and used as a template for protein modelling. As shown in Fig. 3C, LrNAC35 harbours a conserved NAC domain (N‐terminal) and a divergent transcription regulatory domain (C‐terminal). The NAC domain of LrNAC35 adopts the typical NAC protein fold, which mainly consists of beta‐sheets. Based on the report on the structure of Arabidopsis AtNAC19 bound with DNA (PDB 3SWM) (Welner et al., 2012), six residue groups (G1–G6) have been suggested to have DNA‐binding potential (Fig. 3D). The spatial positions of the corresponding G1, G2 and G6 in LrNAC35 were displayed in superimposition with the chain A of AtNAC19 (PDB 3SWM). The protein model of LrNAC35 resembles the spatial coordination of AtNAC19. The side chains of the residues in G1, G2 and G6 are in close proximity to the bound DNA molecule. Each of these three residue groups inserts into the adjacent grooves of the DNA, similar as that observed for AtNAC19 (Fig. 3E).
Biotic, abiotic stresses and hormone treatments alter LrNAC35 expression
To assess the expression patterns of LrNAC35 during multiple plant–virus interactions, additional virus infection tests using CMV, LMoV and LSV were carried out in five wild Lilium species, including virus‐resistant species L. regale, L. pumilum and L. duchartrei, and susceptible species L. brownii and L. tigrinum (Sun et al., 2016b). The overall expression levels of LrNAC35 increased dramatically in all species tested when challenged with three viruses, and in particular L. regale displayed the maximum abundances at 48 or 72 hpi (Fig. 4A–C). CMV infection resulted in significantly higher transcript levels of LrNAC35 in L. regale, L. pumilum not L. duchartrei than in two susceptible species at 48 or 72 hpi (Fig. 4A), while only L. regale exhibited higher LrNAC35 expression than the susceptible ones at 24 or 48 hpi with LMoV (Fig. 4B). A different expression profile was observed after inoculation with LSV, showing that LrNAC35 transcripts in the susceptible species L. brownii surpassed those in the resistant species L. pumilum or L. duchartrei at 24 or 48 hpi (Fig. 4C).
Figure 4.
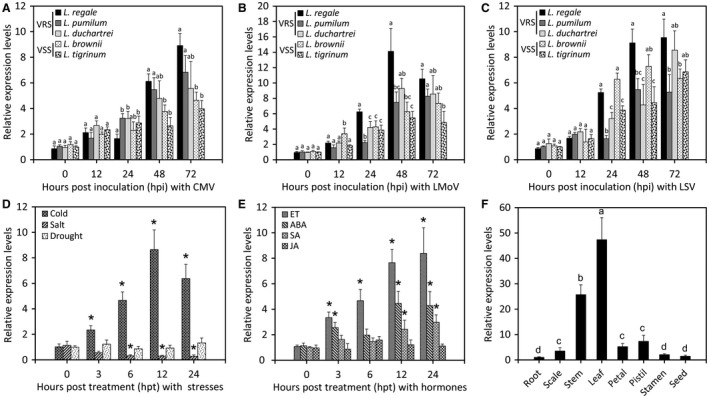
Expression of LrNAC35 in lily leaves under virus infection, abiotic stress and hormone treatments or in different tissues. qRT‐PCR analysis of LrNAC35 transcript levels in the leaves of five wild Lilium species at different hours post‐inoculation (hpi) with CMV (A), LMoV (B) and LSV (C). VRS, virus‐resistant species; VSS, virus‐susceptible species. qRT‐PCR analysis of LrNAC35 transcript levels in L. regale leaves treated with abiotic stressors (4 °C, 150 mM NaCl and dehydration) (D) and stress‐related hormones (15 μL/L ethylene (ET), 50 μM abscisic acid (ABA), 100 μM salicylic acid (SA) and 100 μM jasmonic acid (JA)) (E) at indicated time points. (F) Tissue‐specific expression of LrNAC35 in various tissues of L. regale plants by qRT‐PCR. Six‐week‐old plantlets of bulb‐propagated wild Lilium species were used for virus inoculation, abiotic stress and hormone treatments. Different tissues of L. regale propagated from bulbs were collected at 12 or 16 weeks post‐germination. LrActin was used as an internal control. Error bars represent SE of the mean from three biological replicates. Asterisks or different letters at the top of columns indicate statistical significance as determined by Student's t‐test or one‐way ANOVA test at P < 0.05, respectively.
For treatment with abiotic stresses and plant growth regulators, LrNAC35 transcripts increased markedly at low temperature but reduced under high salinity (Fig. 4D). A pronounced elevation in transcript levels of LrNAC35 occurred following treatments with ethylene (ET) and abscisic acid (ABA), while a moderate and delayed up‐regulation was observed upon exposure to salicylic acid (SA) (Fig. 4E). No significant change in the expression of LrNAC35 was found under dehydration and jasmonic acid (JA) treatments (Fig. 4D,E). Furthermore, LrNAC35 was constitutively expressed in stems and leaves with higher abundances than in roots, scales, seeds and floral tissues (Fig. 4F).
Ectopic overexpression of LrNAC35 alters flowering time in petunia
To elucidate the possible function of LrNAC35, we performed a genetic transformation assay in petunia, which is a common model platform for molecular research of floral crops. LrNAC35 was ectopically overexpressed under the control of the CaMV 35S promoter in petunia. Three transgenic lines (3‐1, 5‐6 and 12‐3) with LrNAC35 overexpression showed delayed flowering phenotypes compared with wild‐type (WT) plants (Fig. 5A,C). A dual expression analysis using semiquantitative and quantitative methods confirmed the substantial transcription of LrNAC35 in transgenic lines (Fig. 5B).
Figure 5.
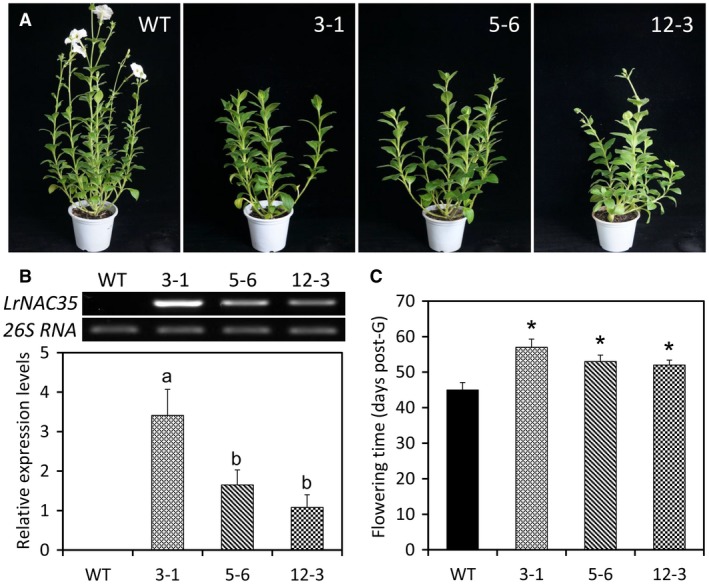
Impact of LrNAC35 overexpression on flowering time of transgenic petunia plants. (A) Representative growth phenotypes of wild‐type (WT) and LrNAC35‐overexpressing lines (3‐1, 5‐6 and 12‐3) at 8 weeks post‐germination. (B) Semiquantitative RT‐PCR and qRT‐PCR analyses of LrNAC35 transcript abundances in the leaves of WT and transgenic petunia lines overexpressing LrNAC35. 26S rRNA was used as a normalization control. Error bars represent SE of the mean from three biological replicates. Significance of difference was evaluated using one‐way ANOVA test (P < 0.05) and shown as various letters. (C) Flowering time of transgenic petunia plants overexpressing LrNAC35 post‐germination (post‐G) compared to WT plants. Five independent plants for each line were used for the analysis of flowering time. Asterisks denote significant difference based on calculation by Student's t‐test at P < 0.05.
LrNAC35 affects susceptibility to CMV
In view of LrNAC35's prominent induction when viruses invade, the function of LrNAC35 in the defence response against CMV was investigated. Both WT and transgenic lines were inoculated with CMV. At 18 days post‐inoculation (dpi), LrNAC35‐overexpressing transgenic petunia lines showed milder leaf distortion and necrosis than WT plants (Fig. 6A). Accumulation of CMV‐CP at both transcript and protein levels was reduced in systemically infected leaves of overexpression lines compared with the control at 8 or 12 dpi (Fig. 6B,C). In the inoculated leaves, CMV infection elicited a higher level of HR‐like cell death in WT lines than that in transgenic lines overexpressing LrNAC35, where relatively smaller lesion areas were observed (Fig. 6D,E). Transgenic lines generated significantly lower levels of electrolyte leakage than the WT control after CMV inoculation (Fig. 6F). There was a notable increase in Klason lignin content in the CMV‐inoculated leaves of three overexpression lines compared to WT lines (Fig. 6G).
Figure 6.
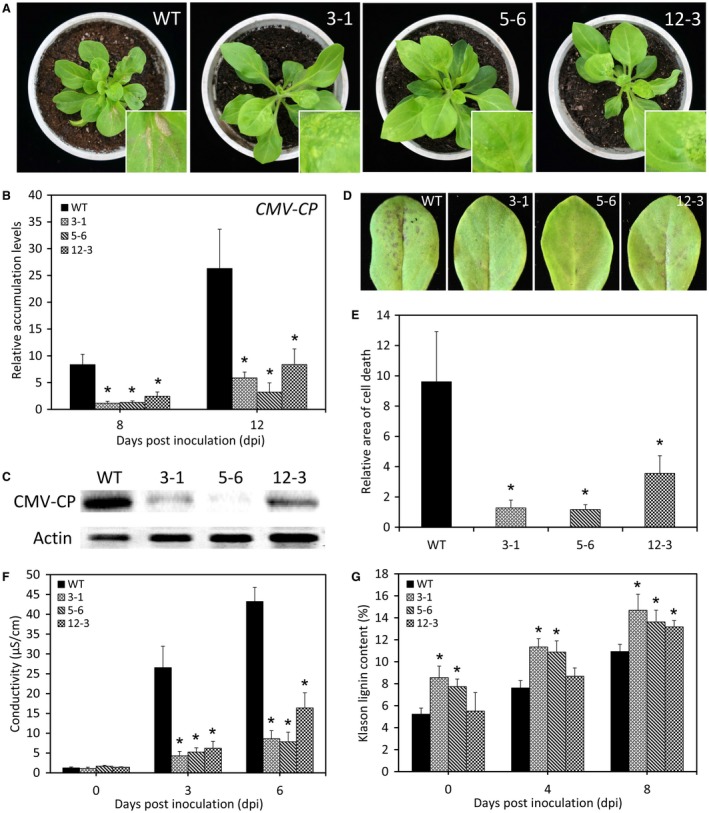
Increased resistance to CMV infection in transgenic petunia plants overexpressing LrNAC35. (A) Disease symptoms of wild‐type (WT) and LrNAC35‐overexpressing lines (3‐1, 5‐6 and 12‐3) at 18 days post‐inoculation (dpi) with CMV. The magnified views of symptoms in systemically infected leaves with CMV are indicated as the insets. Four‐leaf‐stage seedlings were used for inoculation. qRT‐PCR (B) and western blot (C) analyses of CMV coat protein (CMV‐CP) transcript and its protein levels in the uppermost leaves of WT and overexpression lines at 8 or 12 dpi with CMV, and the leaf samples at 12 dpi were used for western blots. 26S rRNA and actin were used as a reference gene and protein, respectively. Disease symptoms (D) and relative cell death area (E) of inoculated leaves of WT and LrNAC35‐overexpressing lines (3‐1, 5‐6 and 12‐3) at 6 dpi with CMV. (F) Electrolyte leakage (conductivity) in the inoculated leaves of WT and transgenic lines challenged with CMV at intervals. (G) Klason lignin content in CMV‐inoculated leaves of WT and transgenic lines at different time points. Error bars represent SE of the mean from three biological replicates. Asterisks indicate significant difference as evaluated by Student's t‐test at P < 0.05.
For functional validation of LrNAC35, we selected a petunia orthologue of LrNAC35, designated PhNAC35, to perform TRV‐based virus‐induced gene silencing (VIGS) assay. Petunia leaves infiltrated with TRV empty vector or TRV‐PhNAC35 were subsequently inoculated with CMV, and the mock control was used to differentiate the symptoms triggered by TRV vector and CMV. PhNAC35 transcripts were markedly down‐regulated in TRV‐PhNAC35‐infected petunia leaves. We observed more severe symptoms in CMV‐infected petunia plants with PhNAC35 silencing. CMV‐CP transcript and protein levels were significantly higher in PhNAC35‐silenced petunia leaves than those in non‐silenced ones (Fig. S7A–D, see Supporting information), suggesting that PhNAC35 silencing caused a compromised resistance to CMV.
LrNAC35 modulates expression of lignin biosynthesis‐related genes
Lignification creates a non‐degradable mechanical barrier to hinder pathogen spread within the host tissues (Moura et al., 2010). To verify if LrNAC35 can regulate lignin biosynthesis, the expression of the core genes in the lignin biosynthetic pathway were examined. Transgenic petunia lines with LrNAC35 overexpression showed higher transcript abundances of cinnamate 4‐hydroxylase (PhC4H), 4‐coumarate: CoA ligase (Ph4CL), hydroxycinnamoyl CoA: quinate/shikimate hydroxycinnamoyl transferase (PhHCT) and cinnamoyl‐CoA reductase (PhCCR) than WT plants at 0 and 48 hpi with CMV. The expression levels of the other structural genes, such as phenylalanine ammonia‐lyase (PhPAL), coumaroyl‐quinate/shikimate 3‐hydroxylase (PhC3H), caffeoyl CoA O‐methyltransferase (PhCCoAOMT), ferulate 5‐hydroxylase (PhF5H), caffeic acid O‐methyltransferase (PhCOMT) and cinnamyl alcohol dehydrogenase (PhCAD), remained unchanged between WT and overexpression lines during CMV infection (Fig. 7A).
Figure 7.
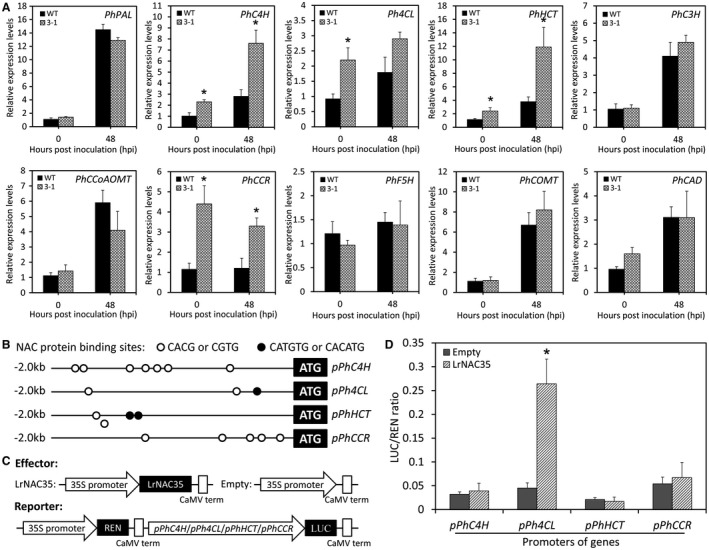
Involvement of LrNAC35 in the lignin biosynthesis pathway. (A) qRT‐PCR analysis of lignin biosynthesis‐associated genes, including PhPAL, PhC4H, Ph4CL, PhHCT, PhC3H, PhCCoAOMT, PhCCR, PhF5H, PhCOMT and PhCAD, in the leaves of wild‐type (WT) and LrNAC35‐overexpressing transgenic petunia line (3‐1) at 0 or 48 h post‐inoculation (hpi) with CMV. Expression levels were standardized to 26S rRNA. (B) Graphic representation of petunia PhC4H, Ph4CL, PhHCT and PhCCR promoters (pPhC4H, pPh4CL, pPhHCT and pPhCCR) with 2.0 kb region upstream of their coding sequences. Hollow and black circles denote two different NAC protein binding sites. (C) Schematic diagrams of the effector and reporter constructs for dual luciferase assay. REN, Renilla luciferase; LUC, firefly luciferase. (D) Dual luciferase assay of the pPhC4H, pPh4CL, pPhHCT and pPhCCR. The activation was expressed as a LUC/REN ratio. Error bars indicate SE of the mean from three biological replicates. Statistical significance was determined using Student's t‐test (P < 0.05) and shown as asterisks.
To investigate the possible transactivation of PhC4H, Ph4CL, PhHCT and PhCCR promoters by LrNAC35, the promoter binding sites of these genes were predicted. Two types of NAC protein binding sites with the core CACG and CATGTG (or CGTG and CACATG in the opposite strand, respectively) elements were found in the 2.0 kb promoter regions upstream of their coding sequences (Fig. 7B). Based on the effector and reporter constructs (Fig. 7C), a dual luciferase assay was performed to examine the potential interaction of LrNAC35 with PhC4H, Ph4CL, PhHCT and PhCCR promoters in petunia leaves. The co‐expression of 35S::LrNAC35 with pPh4CL::LUC led to a 5.9‐fold rise in firefly luciferase (LUC) activity, whereas no significant change in LUC activity was detected for the pPhC4H/pPhHCT/pPhCCR::LUC constructs (Fig. 7D).
LrNAC35 affects susceptibility to TMV
To determine whether LrNAC35 participates in the response to other viruses, a TMV vector expressing green fluorescent protein (TMV‐GFP) was used to inoculate the transgenic and WT petunia lines. Under UV irradiation, the LrNAC35‐overexpressing lines had visibly smaller sizes of fluorescent foci than the WT lines at 6 dpi (Fig. 8A,B). This fluorescence variation was consistent with the measured transcript levels of GFP, which were decreased significantly in transgenic petunia lines compared to WT lines (Fig. 8C). At 4 and 6 dpi, the petunia plants overexpressing LrNAC35 accumulated lower abundances of TMV‐CP transcripts than the WT plants (Fig. 8D).
Figure 8.
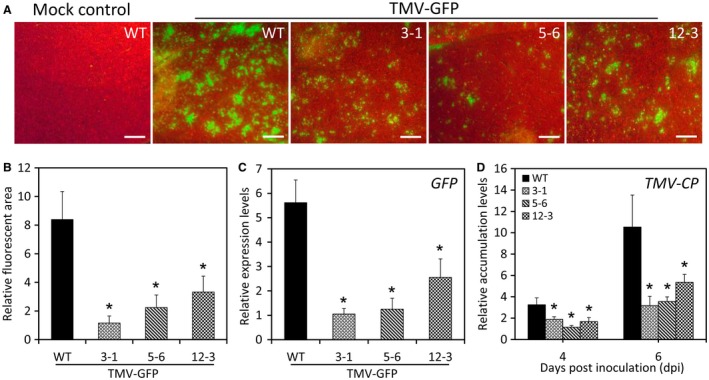
Enhanced resistance to TMV infection in LrNAC35‐overexpressing transgenic petunia plants. GFP fluorescent foci (A) and relative fluorescent area (B) in the inoculated leaves of wild‐type (WT) and transgenic petunia lines (3‐1, 5‐6 and 12‐3) at 6 days post‐inoculation (dpi) with Agrobacterium bearing no TMV vector (mock control) or TMV‐GFP. Photographs were taken under UV light. Scale bars = 2.0 mm. qRT‐PCR analysis of transcripts of GFP (C) and TMV‐CP (D) encoding TMV coat protein in the leaves of WT and transgenic plants at 4 or 6 dpi with TMV‐GFP, and the samples at 6 dpi were used for assessment of GFP expression. Error bars represent SE of the mean from three biological replicates. Asterisks denote significant difference as calculated by Student's t‐test at P < 0.05.
To further determine the impact of PhNAC35 silencing on the defence response to TMV, TRV empty vector‐ and TRV‐PhNAC35‐infected petunia leaves were challenged with TMV‐GFP. A clearly enlarged fluorescent area and increased accumulation of TMV‐CP were found in PhNAC35‐silenced petunia leaves compared to empty vector control (Fig. S7E,F, see Supporting information).
Discussion
The wild lily species L. regale displays exceptional resistance to various biotic stress factors and has drawn extensive attention from lily breeders and plant biologists. Interspecific hybridization between L. regale and L. rubellum (Niimi et al., 1996) or L. nobilissimum (Obata et al., 2000) has been performed with attempts to introduce resistance traits of L. regale to other species sensitive to fungi or viruses. The L. regale × L. nobilissimum hybrids show some resistance to fungal disease, including Fusarium and Botrytis. Analogously, Lim et al. (2003) found that L. regale is highly resistant to Fusarium oxysporum. In recent reports, differentially expressed transcripts in L. regale response to F. oxysporum have been identified using the SSH method (Rao et al., 2013). Transcriptome‐wide identification has also been performed to characterize the microRNAs (Gao et al., 2017) and genes (Cui et al., 2018a) with significantly changed expression in Botrytis elliptica‐infected L. regale, and two TFs, LrWRKY4 and LrWRKY12, have been characterized as important regulators of resistance to B. cinerea (Cui et al., 2018b). In this study, we employed a comparative transcriptome approach to dissect the antiviral molecular mechanism in L. regale.
We screened the DEGs encoding putative TFs in CMV‐infected L. regale leaves. One NAC TF gene, LrNAC35, was identified to be consistently and significantly up‐regulated upon virus infection. Protein modelling analysis revealed that three residue groups (G1, G2 and G6) of LrNAC35 within the NAC domain seem to be associated with potential DNA binding (Fig. 3E). As reported by Welner et al. (2012), G2 has been suggested to be related to DNA binding specificity recognition, whilst G1 and G6 may affect the binding affinity. The results also showed a great resemblance with AtNAC19 in DNA binding, suggesting that the NAC domain is highly conserved in LrNAC35 in comparison with other NAC proteins. LrNAC35 may adopt a similar DNA‐binding mechanism with AtNAC19.
LrNAC35 belongs to the ONAC022 subgroup of the NAC family in plants (Fig. 3B). Several members of the ONAC022 subgroup have been linked with responses to fungal or viral stimuli. For example, AtNAC94 transcripts accumulate more at fungus‐infected sites with hypersensitive cell death in Arabidopsis (Michel et al., 2006). Arabidopsis rosette leaves infected with cabbage leaf curl virus display elevated abundances of AtNAC36 and AtNAC42 (Ascencio‐Ibánez et al., 2008). Our RNA‐Seq and qRT‐PCR analyses based on CMV infection tests showed that the up‐regulation of LrNAC35 is probably associated with CMV resistance in L. regale (Table 2 and Fig. 4A).
LrNAC35 transcripts also increased significantly in all resistant or susceptible lily species infected with two other common lily viruses, LMoV and LSV. It is particularly interesting that the expression of LrNAC35 appears not to be in complete accord with virus‐resistant levels of five species when challenged with LSV (Fig. 4C). This may be explained by the potential genetic variations in LrNAC35 across different lily species. This hypothesis is supported by several previous studies in which plant sensitivity to viral diseases has been associated with a single‐nucleotide or amino acid polymorphism in the defensive gene or protein (Lee et al., 2010; Ling et al., 2009; Wang et al., 2011). Thus, we performed sequence analysis of LrNAC35 by amplifying its complete coding sequences and translating the nucleotides to amino acids. Sequence alignment verified that L. brownii and L. tigrinum had four and one amino acid variations in the amino acid sequences of LrNAC35, respectively, compared to three resistant species (Fig. S8A, see Supporting information). L. brownii also exhibited the most polymorphic nucleotide sites (Fig. S8B, see Supporting information). This could perhaps explain the discrepancy between higher LrNAC35 transcript levels and lower resistance levels in L. brownii during LSV infection.
Apart from virus‐caused induction, increased expression of LrNAC35 under low temperature treatment may suggest the involvement of LrNAC35 in L. regale cold tolerance (Fig. 4D). This notion is supported by the report that a mutant of AtLOV1, an Arabidopsis homologue of LrNAC35, exhibits enhanced hypersensitivity to cold, while the AtLOV1 gain‐of‐function allele displays reduced hypersensitivity (Yoo et al., 2007). An opposite expression of LrNAC35 was observed in L. regale leaves during salt stress (Fig. 4D). The biological role of LrNAC35 in response to high salinity awaits further investigation.
Moreover, we also measured the effects of plant hormone treatments, including ET, ABA, SA and JA, on LrNAC35 transcripts. These plant hormones have all been shown to affect virus resistance in different plants (Alazem and Lin, 2015). In the present study, expression analysis demonstrated that LrNAC35 was significantly up‐regulated in ET‐, ABA‐ and SA‐treated L. regale leaves (Fig. 4E), providing further support that these hormones may be involved in virus resistance in plants. The crucial involvement of LrNAC35 in L. regale response to viruses may be attributed to the interplay between LrNAC35 and these hormones. Future studies will examine the hormone variation in WT and transgenic plants overexpressing LrNAC35.
We verified the enhanced resistance to CMV conferred by LrNAC35 overexpression in transgenic petunia plants (Fig. 6A–F). We also detected reduced fluorescence signals and TMV‐CP expression in transgenic lines infiltrated with an artificially modified vector, TMV‐GFP (Fig. 8), but no significant difference in disease symptoms was observed between TMV‐GFP‐infected transgenic and WT lines (data not shown). It was speculated that the GFP tag may have repressed the full manifestation of visual symptoms caused by TMV infection. In addition, LrNAC35's function in antiviral defence was validated through the TRV‐VIGS assay of a petunia orthologous gene PhNAC35 (Fig. S7, see Supporting information). We observed lessened symptoms of HR‐like necrotic lesions and reduced electrolyte leakage in systemically infected or inoculated leaves of overexpression plants with CMV compared to WT lines. Earlier studies have indicated that virus‐induced HR is correlated with increased lignin, which is a major constituent of the plant secondary cell wall (Kimmins and Wuddah, 1977). Lignin deposition perhaps acts as a physical barrier in preventing the spread of viruses (Candela et al., 1994; Nicholson and Hammerschmidt, 1992).
There have been a large number of publications on the role of lignification or lignin biosynthetic genes in plant responses to fungal penetration (Moura et al., 2010; Wang et al., 2018; Zeyen et al., 1995). However, the reports on lignin's function in response to viruses are fewer and sometimes controversial. Tobacco necrosis virus and tomato spotted wilt virus infections increase lignin content in dwarf bean (Kimmins and Wuddah, 1977) and petunia leaves (Quecini et al., 2007), respectively, but Kofalvi and Nassuth (1995) found no significant change in wheat streak mosaic virus‐infected wheat leaves. Application of aminooxyacetate, a competitive inhibitor of phenylalanine ammonia‐lyase upstream of the lignin biosynthesis pathway, to tobacco leaves enlarges the size of TMV‐induced necrotic lesions (Massala et al., 1980). A significant discovery is that synthetic or natural lignin could inhibit the replication and cytopathogenicity of human immunodeficiency virus infection (Nagashima et al., 1992).
Indeed, we detected higher lignin content in LrNAC35‐overexpressing plants than WT plants inoculated with CMV (Fig. 6G). It appears likely that lignin is an essential intermediate in transcriptional modulation of antiviral defence by LrNAC35. This hypothesis is further confirmed by the altered abundances of several lignin biosynthesis‐related genes in LrNAC35‐overexpressing lines, and a particularly direct activation of Ph4CL promoter by LrNAC35 (Fig. 7A,D). However, the specific DNA binding sites recognized by LrNAC35 remain enigmatic. To date, multiple cis‐acting elements have been clearly identified in the target gene promoters of NAC members. For example, a complete NAC recognition sequence containing the core elements CATGT or CACG was determined in Arabidopsis (Tran et al., 2004). AtNAC19, AtNAC55 and AtNAC72 physically bind to the CATGTG motif of the ERD1 promoter (He et al., 2005). Two NAC proteins, SND1 and VND7, specifically bind to a secondary wall NAC binding element (SNBE) (Zhong et al., 2010). The binding sequence of ATAF2, an NAC TF, harbours an imperfect palindrome CAAATNNNATTTG (Huh et al., 2012). In a recent study, ThNAC13 was reported to bind to NACRS and CBNAC elements with the core sequence CGT(A/G) and GCTT, respectively (Wang et al., 2017). In the present study, we only identified the CACG and CATGTG motifs in the promoter regions of Ph4CL (Fig. 7B). The unanswered question is whether LrNAC35 could bind to these two motifs or to other unrevealed ones. Thus, a deletion analysis of the Ph4CL promoter is required to identify the exact DNA binding sites of LrNAC35 in the subsequent studies.
Given the impact of LrNAC35 overexpression on several lignin biosynthesis‐related genes, we searched the corresponding orthologous unigenes against our RNA‐Seq data in CMV‐infected L. regale, and two 4CL and three CCR homologues were identified. Although all these homologues displayed a false discovery rate (FDR) > 0.05, their transcript abundances were increased by 2.0‐ to 4.8‐fold upon CMV infection (Table S5, see Supporting information). We found no orthologous genes of PhC4H and PhHCT in our transcriptome data, probably due to a very large genome size in Lilium (Siljak‐Yakovlev et al., 2003) and an incomplete coverage through the Illumina RNA‐Seq approach.
In summary, our results suggest that LrNAC35 plays an important role in antiviral defence by mediating the biosynthesis of lignin. The identification of LrNAC35, arising from a comprehensive transcriptome sequencing effort, provides a valuable genetic solution to reduce the susceptibility of lilies to viral disease. To find more promising candidate genes, functional screening of massive transcriptome data requires a rapid and efficient approach. For instance, high‐throughput VIGS is an attractive tool for permitting the quick knockdown of gene expression in plants, and a CMV‐based VIGS system has been established in L. leichtlinii (Tasaki et al., 2016). Using the VIGS method to unravel the functions of candidate genes in response to CMV or other viruses in Lilium, not in heterologous model plants, should be pursued in future work.
Experimental Procedures
Plant materials and growth conditions
Wild lily species L. regale seeds were planted in small pots filled with sterile soil mix (peat:perlite:vermiculite = 3:1:1 v/v/v) and germinated in a growth chamber at 22/20 °C day/night temperature with a 16/8 h light/dark photoperiod. The second newly sprouted leaves with relatively large sizes were inoculated with CMV for subsequent viral detection, transcriptome assay and expression analysis of candidate genes. Uppermost young leaves of 6‐week‐old plantlets of bulb‐propagated L. regale, L. pumilum, L. duchartrei, L. brownii and L. tigrinum were used for assessment of LrNAC35 expression upon CMV, LMoV and LSV inoculation, abiotic stress and hormone treatments. These lily plants were cultivated in the germplasm resource nursery under 25/22 °C day/night temperature and natural photoperiods. Tissue‐specific expression analysis of LrNAC35 was carried out using rootlets, outer bulb scales, top stems, young leaves, floral tissues at anthesis (petals, pistils and stamens) and mature seeds collected from L. regale plants at 12 or 16 weeks after bulb germination. To study the LrNAC35's function in resistance response to CMV, we selected petunia (P. hybrida ‘Mitchell Diploid’) as material for genetic transformation experiments. Petunia seeds were sown in a 36‐well tray, and four‐leaf‐stage seedlings were then transferred to pots under identical chamber conditions to those described above. The leaves three to eight from terminal were collected as explant source for genetic transformation.
Inoculation assay
Viral inoculum of CMV strain LS (CMV‐LS, subgroup II) was obtained from the infected leaves of Lilium Oriental hybrids cultivar Sorbonne, while both strains of LMoV and LSV were isolated from the wild species L. davidii. Leaf tissues infected with three types of viruses were separately homogenized with kieselguhr in 100 mM phosphate buffer adjusted at pH 7.0 (1:6 w/v) to prepare infectious sap. The sap from leaves of virus‐free plants was used as mock control. The virus preparations were rub‐inoculated onto healthy young leaves of wild lily species, WT and transgenic petunia plants through the mechanical method (Hull, 2009). An agroinjection method with TMV‐GFP plasmids was used to inoculate petunia (Dorokhov et al., 2006). For functional analysis of LrNAC35 in petunia, virus symptom development was evaluated during the post‐inoculation growth periods. Accumulation levels of CMV coat protein (CMV‐CP), GFP and TMV‐CP in the infected leaves were tested via qRT‐PCR (Sun et al., 2017) or western blot (Jeon et al., 2017). GFP fluorescence was monitored using a Blak‐Ray long‐wave ultraviolet lamp (UV products, Upland, CA, USA; Model B 100AP) and photographed using a Canon EOS 40D digital camera. The cell death and GFP fluorescence at the infection sites were quantified by measuring pixel sizes of the necrotic and fluorescent spots using Photoshop CS6. The calculation of relative cell death and fluorescence area was based on the quantitative data, and the lowest relative data was set to 1.0.
RNA extraction, library construction and sequencing
Lilium regale leaves from 20 seedlings at 48 hpi with CMV or mock were randomly divided into two groups serving as two independent biological replicates. Ten seedlings for each replicate were pooled to prepare RNA samples. Total RNA was isolated using the modified cetyltrimethylammonium bromide (CTAB) method (Li et al., 2011) and purified with RNase‐free DNase I (Promega, Madison, WI, USA) to avoid genomic DNA contamination. The quality and quantity of RNA was assessed using a NanoDrop ND‐2000c spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Generation of cDNA libraries and sequencing projects was carried out using Illumina protocols at Gene Denovo Bio‐Technology Co., Ltd (Guangzhou, China). Briefly, mRNA harbouring poly (A) was enriched through magnetic beads with oligo(dT) and randomly chopped into small fragments. These pieces were used as templates to synthesize cDNAs. The resulting cDNAs were subjected to purification and the 3’‐end repair process and then ligated with sequencing adaptors. The ligation products were size‐selected, PCR‐amplified and sequenced on an Illumina HiSeq2000 platform. The raw transcriptome reads were submitted to the NCBI Sequence Read Archive (SRA) database under accession no. SRP193127.
RNA‐Seq data processing
Raw data/reads from the sequencing machine were yielded through base calling. Before assembly, raw reads were handled by removing reads harbouring adaptor sequences, more than 10% of unknown nucleotides (N) and more than 40% of low‐quality (Q value ≤ 10) bases using a Perl script tool. The generated high‐quality clean reads were assembled de novo using Trinity software (v. 2.1.1) with k‐mer size parameter set to 25 by default (Grabherr et al., 2011). Clean data were mapped back onto the assembled transcriptome sequences, and the reads for each unigene were counted based on the mapping results. Trinity combined reads with certain length of overlap to create longer fragments not containing N or contigs. Next, these contigs were processed using sequence clustering software TGICL (v. 2.1) (Pertea et al., 2003) to form sequences longer than 200 bp, which were defined as unigenes.
A homology search of all unigenes against the public Non‐Redundant (NR) (released on July 24, 2015, http://blast.ncbi.nlm.nih.gov/), Swiss‐Prot (released on July 24, 2015, http://www.expasy.ch/sprot), Clusters of Orthologous Groups (COG) (released on July 24, 2015, http://www.ncbi.nlm.nih.gov/COG) and KEGG (released on July 27, 2015, https://www.genome.jp/kegg) databases with a cut‐off e value of 1 × 10−5 was performed for functional annotation, sequence orientation and coding region prediction, using the standalone BLAST (v. 2.2.29). Transcript levels of unigenes were calculated and normalized to reads per kilobase exon model per million mapped reads (RPKM). PCA was conducted using the gmodels package of statistical program R (http://www.r-project.org/). Significant DEGs between mock‐ and CMV‐inoculated L. regale samples were identified with a threshold for FDR ≤ 0.05 and fold change ≥ 2.0 in multiple comparisons using edgeR package (v. 3.12) (Chen et al., 2014) with an R environment wrapper (v. 3.2.1). Based on NR annotation results, the GO annotation of unigenes was carried out using Blast2GO (v. 2.3.5) (Conesa et al., 2005) and then the WEGO tool (http://wego.genomics.org.cn/cgi-bin/wego/index.pl) (Ye et al., 2006) was used to perform GO functional categorization of unigenes. To identify the active metabolic pathways in CMV‐infected L. regale leaves, a BLAST search against the KEGG database was employed to perform the mapping of DEGs to reference canonical pathways (Wixon and Kell, 2000). A formula was used for calculation of P values (Lu et al., 2012), which was gone through FDR correction. KEGG pathways with FDR ≤ 0.05 were defined as significantly enriched.
Semiquantitative RT‐PCR and qRT‐PCR
Total RNA from lily and petunia tissues was extracted using the modified CTAB method and TRIzol reagent (Invitrogen, Carlsbad, CA, USA), respectively. After genomic DNA elimination, 2–5 μg of RNA samples was reversely transcribed to first‐strand cDNA using a PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, Kyoto, Japan). PCR amplification was performed using Premix Taq DNA polymerase (TaKaRa, Kyoto, Japan), and the abundances of products were analysed by electrophoresis on 1% agarose gel stained with GelRed (Biotium, Hayward, CA, USA). qRT‐PCR was conducted using the SYBR Green PCR Master Mix (2×) (Applied Biosystems, Foster City, CA, USA) in a LightCycler480 Real‐Time PCR System (Roche Diagnostic, Basel, Switzerland). Glyceraldehyde‐3‐phosphate dehydrogenase (LrGAPDH) and LrActin served as internal controls in Lilium species, and the reference gene in petunia was 26S ribosomal RNA. Data were calculated using the 2–△△CT method (Livak and Schmittgen, 2001). A set of oligonucleotide primers used for analyses of virus accumulation, RNA‐Seq validation and gene expression are listed in Table S6 (see Supporting information). The unigene sequences used for expression or functional analysis in this study are provided in Fig. S9 (see Supporting information).
Isolation and sequence analysis of LrNAC35
A 1366‐bp cDNA sequence of LrNAC35 containing a complete coding region was selected from up‐regulated TFs in CMV‐infected L. regale leaves. To amplify the coding sequences of LrNAC35 from various Lilium species, the forward primer 5′‐ATGGCAATTACCGCAGCCATGAG‐3′ and reverse primer 5′‐TCACTCCCATAGCTTGTCTGGAT‐3′ were used. Its deduced amino acids were obtained by the ExPASy translated tool (http://web.expasy.org/translate/). Proteins homologous to LrNAC35 were found by a BLAST search. The conserved motifs were identified based on comparative NAC domains analysis between O. sativa and Arabidopsis (Ooka et al., 2003). Multiple protein alignments were performed using DNAMAN (v. 5.2.2) (Wang, 2015). A phylogenetic tree was constructed using MEGA (v. 4.0.2) (Tamura et al., 2007). Homologous templates for protein modelling were identified from the RCSB Protein Data Bank (https://www.rcsb.org/). The modelling process was carried out using the Modeller server (v. 9.20) (Webb and Sali, 2014) based on sequence alignment performed in Chimera (v. 1.12) (Pettersen et al., 2004). A combination of multiple protein structures (chain A of PDBs: 1UT4, 1UT7, 4DUL, 3SWM and 3SWP) was used for the homologous modelling of LrNAC35. Only the NAC domains were used and modelled. The best model was chosen based on the lowest Discrete Optimized Protein Energy (DOPE) values and GA 341 score of 1, which suggest that these models are reliable. The final model was validated by Ramachandran plot analysis using PROCHECK (http://www.ebi.ac.uk/thornton-srv/software/PROCHECK). Molecular visualizations were performed using PyMOL (v. 1.3r1. Schrodinger, LLC, Cambridge, MA, USA).
Abiotic stress and hormone treatments
To determine the impacts of abiotic stresses and stress‐related hormones on transcript levels of LrNAC35, c. 20‐cm long stems from the top were cut from 6‐week‐old L. regale plantlets and used for the follow‐up experiments. For the cold treatment, the plants were placed into a large glass container with fresh deionized water at 4 °C in cold storage. For the salinity and dehydration treatments, the seedlings were exposed to a NaCl solution with a concentration of 150 mM and no water. For treatments with hormones, the stems were immersed in water without addition and treated with continuous 15 μL/L ET in a sealed chamber, or in water supplemented with 50 μM ABA, 100 μM SA and 100 μM JA. Terminal upper leaves were excised from three individual plants at five time points (0, 3, 6, 12 and 24 h) after the treatments.
Plasmid construct and petunia plant transformation
The full‐length of the LrNAC35 open reading frame (ORF) sequence was PCR‐amplified using the forward primer 5′‐ATGGTACCATGGCAATTACCGCAGCCAT‐3′ bearing a KpnI restriction site, and reverse primer 5′‐ATGTCGACTCACTCCCATAGCTTGTCTG‐3′ bearing a SalI restriction site. To generate the overexpression construct, the amplified DNA fragment was inserted into the corresponding position of a modified pCAMBIA1300 vector downstream of the CaMV 35S promoter in the sense orientation. Agrobacterium tumefaciens LBA4404 was transformed with the recombinant plasmid by electroporation. Agrobacterium‐mediated genetic transformation and regeneration of ‘Mitchell Diploid’ leaf discs were carried out according to a previously described protocol (Sun et al., 2016a). After a continuous hygromycin selection on MS plates and cultivation process in soil mixtures, the T 2 lines 3‐1, 5‐6 and 12‐3, verified as homozygotes, were finally chosen for phenotype and further antiviral analyses of LrNAC35.
Western blot assay
The CMV‐infected leaves were homogenized in liquid nitrogen, and proteins were prepared using a Plant Total Protein Extraction Kit (Sigma‐Aldrich, St Louis, MO, USA). Equal amounts of samples were resolved by 10% sodium dodecyl sulphate‐polyacrylamide gel electrophoresis. The fractionated proteins were transferred to a polyvinylidene fluoride membrane (0.45 μm) on a Mini Trans‐Blot Cell (Bio‐Rad, Richmond, CA, USA). The primary antibody anti‐CMV‐CP was applied to probe the blots, which were then incubated with goat anti‐rabbit IgG secondary antibody conjugated to horseradish peroxidase. The antigen–antibody complexes were visualized using an ECL western blot detection kit (Pierce, Waltham, MA, USA). The actin antibody was used for examination of reference protein.
VIGS assay
A 310‐bp fragment of PhNAC35, the petunia homologue of LrNAC35, was PCR‐amplified and cloned into TRV plasmid. The generated TRV‐PhNAC35 was electrotransformed into A. tumefaciens GV3101. The transformed bacteria were cultured, centrifuged and resuspended in the infiltration buffer as previously described (Sun et al., 2017). The bacterial mixture containing TRV1 and TRV2 plasmids in a 1:1 ratio was used to infiltrate four‐leaf‐stage petunia seedlings. For virus inoculation, mock control and CMV or TMV‐GFP were inoculated onto the leaves of petunia seedlings at 5 dpi with TRV empty vector and TRV‐PhNAC35. PCR primers to sequence beyond the inserted fragment for silencing were used to examine expression levels of PhNAC35 in VIGS assay (Table S6, see Supporting information).
Electrolyte leakage assay
Leaf discs, 8 mm in diameter, were prepared from the leaves of WT and LrNAC35‐overexpressing transgenic petunia lines at intervals after CMV inoculation using a hole punch, and thoroughly immersed in 20 mL of deionized water with gentle shaking at room temperature. Conductivity was measured using an Orion conductivity meter (Thermo Scientific, Waltham, MA, USA).
Measurement of Klason lignin content
The CMV‐inoculated leaves of WT and LrNAC35‐overexpressing transgenic petunia plants were harvested, ground into a fine powder in liquid nitrogen and sequentially extracted using a method previously described (Wang et al., 2012). Sulphuric acid (72%) was added to the methanol extract upon air‐dry treatment for hydrolysis reaction. The hydrolysate was diluted with distilled water to adjust the acid concentration to 4% and boiled under reflux for 1 h. Lignin obtained as an insoluble solid residue was filtered, washed with hot water and freeze dried. Determination of Klason lignin content, expressed as a weight percentage of dried cell wall residues, was conducted using the Klason technique (Dence, 1992).
Dual luciferase assay
The dual luciferase assay was performed using a previously reported method (Chen et al., 2017). The full‐length ORF of LrNAC35 was PCR‐amplified and introduced into pGreenII62‐SK vector, with the recombinant construct being used as effector. The complete promoter regions of PhC4H, Ph4CL, PhHCT and PhCCR were amplified using the primers listed in Table S6 (see Supporting information). Next, these promoter sequences were ligated into pGreenII0800‐LUC vector to generate reporter plasmids, where the promoters drive a firefly luciferase (LUC) gene and the CaMV 35 promoter drives a Renilla luciferase (REN) gene. The A. tumefaciens GV3101 cells transformed with the effector and reporter plasmids were co‐infiltrated into the petunia plants at the four‐leaf stage. The LUC and REN enzyme activities were tested by a Tecan Infinite M200 luminometer (Männedorf, Switzerland) and represented as the LUC/REN ratio.
Supporting information
Fig. S1 Sequences of all assembled unigenes in Lilium regale.
Fig. S2 Length distribution of unigenes in Lilium regale.
Fig. S3 Validation of RNA‐Seq data by qRT‐PCR. Nine unigenes were randomly selected for expression analysis in mock‐ and CMV‐inoculated Lilium regale leaves at 48 h post‐inoculation (hpi). LrActin was used as a reference gene. Error bars indicate standard error (SE) of the mean from three biological replicates. Significance of difference was calculated using Student's t test (P < 0.05) and is shown as asterisks. Unigene0000074, cytochrome P450 86B1; Unigene0004763, lecithine‐cholesterol acyltransferase 4; Unigene0007097, endonuclease/exonuclease/phosphatase family protein; Unigene0011570, auxin‐induced 15A; Unigene0013897, unknown protein; Unigene0017759, mannose‐specific lectin 3; Unigene0063829, argonaute 1; Unigene0073681, ABC transporter G family member 11; Unigene0079669, geranylgeranyl diphosphate synthetase.
Fig. S4 Principal component analysis of Lilium regale transcriptome data.
Fig. S5 GO functional classification of differentially expressed genes in Lilium regale.
Fig. S6 Full‐length cDNA sequence of LrNAC35 and its deduced amino acids. LrNAC35 cDNA sequence harbours a 1077‐bp open reading frame region encoding a polypeptide of 358 amino acids. The italic bold type and bold type in squares denote the start and stop codons, respectively. The type shaded in grey indicate the conserved region of LrNAC35 protein containing subdomains A to E.
Fig. S7 Reduced resistance to CMV and TMV infections in petunia plants with PhNAC35‐VIGS silencing. (A) Disease symptoms of TRV empty vector‐ and TRV‐PhNAC35‐infected petunia plants at 14 days post‐inoculation (dpi) with mock control and CMV. (B) qRT‐PCR analysis of PhNAC35 expression levels in uppermost leaves of TRV empty vector‐ and TRV‐PhNAC35‐infected petunia plants at 14 dpi with mock control and CMV. qRT‐PCR (C) and western blot (D) analyses of CMV coat protein (CMV‐CP) transcript and its protein levels in the uppermost leaves of TRV constructs‐infected petunia plants at 14 dpi with CMV. 26S rRNA and actin were used as a reference gene and protein, respectively. GFP fluorescent foci (E) and qRT‐PCR analysis of transcripts of TMV‐CP (F) encoding TMV coat protein in the leaves of TRV constructs‐inoculated petunia plants at 6 dpi with TMV‐GFP. Four‐leaf‐stage petunia seedlings were used for VIGS assay, and the seedlings at 5 dpi with TRV constructs were thereafter inoculated with mock control, CMV and TMV‐GFP. Error bars represent SE of the mean from three biological replicates. Asterisks indicate significant difference as determined by Student's t‐test at P < 0.05.
Fig. S8 Sequence analysis of LrNAC35 from five Lilium species. Alignment of LrNAC35 amino acid sequences (A) and nucleotide sequences (B) from L. regale, L. pumilum, L. duchartrei, L. brownii and L. tigrinum. Identical amino acids and nucleotides are shaded in black, while similar ones are shaded in grey.
Fig. S9 Sequences of unigenes used for qRT‐PCR or functional analysis.
Table S1 Functional annotation of all assembled unigenes in Lilium regale.
Table S2 Differentially expressed genes in Lilium regale.
Table S3 KEGG pathway annotation of differentially expressed genes in Lilium regale.
Table S4 Differentially expressed transcription factors in Lilium regale.
Table S5 Unigenes associated with lignin synthesis exhibiting increased expression in Lilium regale.
Table S6 Primers used for qRT‐PCR, gene promoter and fragment amplification.
Acknowledgements
We are grateful to Yule Liu for kindly providing the TMV‐GFP vector. We thank Pilong Liu and Zhaowei Wu for experimental assistance in western blot and dual luciferase assay. We are appreciative of Lihang Xie's advice on operation of laboratory instruments. We appreciate Ayla Norris's kind help for editing this manuscript. We also appreciate the invaluable and helpful comments from anonymous reviewers for improving this manuscript. This study was funded by the Doctoral Scientific Start‐Up Foundation of Northwest A&F University (grant no. Z109021711). The authors declare that no competing interests exist.
References
- Agius, C. , Eamens, A.L. , Millar, A.A. , Watson, J.M. and Wang, M.B. (2012) RNA silencing and antiviral defense in plants In: Antiviral Resistance in Plants ( Watson, J.M. and Wang, M.B. eds.), pp. 17–38. Totowa, NJ, USA: Humana Press. [DOI] [PubMed] [Google Scholar]
- Alazem, M. and Lin, N.S. (2015) Roles of plant hormones in the regulation of host–virus interactions. Mol. Plant Pathol. 16, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio, F. and Pallás, V. (2017) The coat protein of Alfalfa mosaic virus interacts and interferes with the transcriptional activity of the bHLH transcription factor ILR3 promoting salicylic acid‐dependent defence signalling response. Mol. Plant Pathol. 18, 173–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascencio‐Ibánez, J.T. , Sozzani, R. , Lee, T.J. , Chu, T.M. , Wolfinger, R.D. , Cella, R. and Hanley‐Bowdoin, L. (2008) Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148, 436–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazadeh, S. , Siddiqui, H. , Allu, A.D. , Matallana‐Ramirez, L.P. , Caldana, C. , Mehrnia, M. , Zanor, M.I. , Köhler, B. and Mueller‐Roeber, B. (2010) A gene regulatory network controlled by the NAC transcription factor ANAC092/AtNAC2/ORE1 during salt‐promoted senescence. Plant J. 62, 250–264. [DOI] [PubMed] [Google Scholar]
- Candela, M.E. , Muñoz, R. , Alcázar, M.D. and Espín, A. (1994) Isoperoxidase involvement in the resistance of Capsicum annuum to infection by Cucumber mosaic virus . J. Plant Physiol. 143, 213–217. [Google Scholar]
- Chen, Y. , Lun, A.T. and Smyth, G.K. (2014) Differential expression analysis of complex RNA‐seq experiments using edgeR In: Statistical Analysis of Next Generation Sequencing Data ( Datta, S. and Nettleton, D. , eds.), pp. 51–74. Cham, Switzerland: Springer. [Google Scholar]
- Chen, K. , Liu, H. , Lou, Q. and Liu, Y. (2017) Ectopic expression of the grape hyacinth (Muscari armeniacum) R2R3‐MYB transcription factor gene, MaAN2, induces anthocyanin accumulation in tobacco. Front. Plant Sci. 8, 965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinestra, S.C. , Facchinetti, C. , Curvetto, N.R. and Marinangeli, P.A. (2010) Detection and frequency of lily viruses in Argentina. Plant Dis. 94, 1188–1194. [DOI] [PubMed] [Google Scholar]
- Conesa, A. , Götz, S. , García‐Gómez, J.M. , Terol, J. , Talón, M. and Robles, M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21, 3674–3676. [DOI] [PubMed] [Google Scholar]
- Cui, Q. , Liu, Q. , Gao, X. , Yan, X. and Jia, G. (2018a) Transcriptome‐based identification of genes related to resistance against Botrytis elliptica in Lilium regale . Can. J. Plant Sci. 98, 1058–1071. [Google Scholar]
- Cui, Q. , Yan, X. , Gao, X. , Zhang, D. , He, H. and Jia, G. (2018b) Analysis of WRKY transcription factors and characterization of two Botrytis cinerea‐responsive LrWRKY genes from Lilium regale . Plant Physiol. Bioch. 127, 525–536. [DOI] [PubMed] [Google Scholar]
- Dence, C.W. (1992) The determination of lignin In: Methods in Lignin Chemistry ( Lin, S.Y. and Dence, C.W. , eds.), pp. 33–61. Berlin, Germany: Springer. [Google Scholar]
- Donze, T. , Qu, F. , Twigg, P. and Morris, T.J. (2014) Turnip crinkle virus coat protein inhibits the basal immune response to virus invasion in Arabidopsis by binding to the NAC transcription factor TIP. Virology, 449, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorokhov, Y.L. , Frolova, O.Y. , Skurat, E.V. , Ivanov, P.A. , Gasanova, T.V. , Sheveleva, A.A. , Ravin, N.V. , Mäkinen, K.M. , Klimyuk, V.I. and Skryabin, K.G. (2006) A novel function for a ubiquitous plant enzyme pectin methylesterase: the enhancer of RNA silencing. FEBS Lett. 580, 3872–3878. [DOI] [PubMed] [Google Scholar]
- Gaguancela, O.A. , Zúñiga, L.P. , Arias, A.V. , Halterman, D. , Flores, F.J. , Johansen, I.E. , Wang, A. , Yamaji, Y. and Verchot, J. (2016) The IRE1/bZIP60 pathway and bax inhibitor 1 suppress systemic accumulation of potyviruses and potexviruses in Arabidopsis and Nicotiana benthamiana plants. Mol. Plant–Microbe Interact. 29, 750–766. [DOI] [PubMed] [Google Scholar]
- Galvez, L.C. , Banerjee, J. , Pinar, H. and Mitra, A. (2014) Engineered plant virus resistance. Plant Sci. 228, 11–25. [DOI] [PubMed] [Google Scholar]
- Gao, X. , Cui, Q. , Cao, Q. , Liu, Q. , He, H. , Zhang, D. and Jia, G. (2017) Transcriptome‐wide analysis of Botrytis elliptica responsive microRNAs and their targets in Lilium regale Wilson by high‐throughput sequencing and degradome analysis. Front. Plant Sci. 8, 753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr, M.G. , Haas, B.J. , Yassour, M. , Levin, J.Z. , Thompson, D.A. , Amit, I. , Adiconis, X. , Fan, L. , Raychowdhury, R. and Zeng, Q. (2011) Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X. , Carroll, J.W.N. , MacDonald, M.R. , Goff, S.P. and Gao, G. (2004) The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J. Virol. 78, 12781–12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, Y.J. , Wei, W. , Song, Q.X. , Chen, H.W. , Zhang, Y.Q. , Wang, F. , Zou, H.F. , Lei, G. , Tian, A.G. and Zhang, W.K. (2011) Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 68, 302–313. [DOI] [PubMed] [Google Scholar]
- He, X.J. , Mu, R.L. , Cao, W.H. , Zhang, Z.G. , Zhang, J.S. and Chen, S.Y. (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916. [DOI] [PubMed] [Google Scholar]
- He, P. , Shan, L. and Sheen, J. (2007) Elicitation and suppression of microbe‐associated molecular pattern‐triggered immunity in plant–microbe interactions. Cell. Microbiol. 9, 1385–1396. [DOI] [PubMed] [Google Scholar]
- Hegedus, D. , Yu, M. , Baldwin, D. , Gruber, M. , Sharpe, A. , Parkin, I. , Whitwill, S. and Lydiate, D. (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol. Biol. 53, 383–397. [DOI] [PubMed] [Google Scholar]
- Huang, Y. , Zhang, B. , Sun, S. , Xing, G. , Wang, F. , Li, M. , Tian, Y. and Xiong, A. (2016) AP2/ERF transcription factors involved in response to Tomato yellow leaf curl virus in tomato. Plant Genome, 9. doi:10.3835/plantgenome2015.09.0082. [DOI] [PubMed] [Google Scholar]
- Huang, Y. , Li, T. , Xu, Z. , Wang, F. and Xiong, A. (2017) Six NAC transcription factors involved in response to TYLCV infection in resistant and susceptible tomato cultivars. Plant Physiol. Biochem. 120, 61–74. [DOI] [PubMed] [Google Scholar]
- Huh, S.U. , Lee, S.B. , Kim, H.H. and Paek, K.H. (2012) ATAF2, a NAC transcription factor, binds to the promoter and regulates NIT2 gene expression involved in auxin biosynthesis. Mol. Cells, 34, 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, R. (2009) Mechanical inoculation of plant viruses. Curr. Protoc. Microbiol. 13, 16B. 6. 1–16B. 6. 4. [DOI] [PubMed] [Google Scholar]
- Jeon, E.J. , Tadamura, K. , Murakami, T. , Inaba, J.I. , Kim, B.M. , Sato, M. , Atsumi, G. , Kuchitsu, K. , Masuta, C. and Nakahara, K.S. (2017) rgs‐CaM detects and counteracts viral RNA silencing suppressors in plant immune priming. J. Virol. 91, e00761–00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, L. , Zhang, Y. , Yan, L. , Guo, Y. and Niu, L. (2012) Phenolic compounds and antioxidant activity of bulb extracts of six Lilium species native to China. Molecules, 17, 9361–9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, B.C. , Yeam, I. and Jahn, M.M. (2005) Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43, 581–621. [DOI] [PubMed] [Google Scholar]
- Kikuchi, K. , Ueguchi‐Tanaka, M. , Yoshida, K. , Nagato, Y. , Matsusoka, M. and Hirano, H. (2000) Molecular analysis of the NAC gene family in rice. Mol. Gen. Genet. 262, 1047–1051. [DOI] [PubMed] [Google Scholar]
- Kimmins, W. and Wuddah, D. (1977) Hypersensitive resistance: determination of lignin in leaves with a localized virus infection. Phytopathology, 67, 1012. [Google Scholar]
- Kofalvi, S. and Nassuth, A. (1995) Influence of Wheat streak mosaic virus infection on phenylpropanoid metabolism and the accumulation of phenolics and lignin in wheat. Physiol. Mol. Plant Pathol. 47, 365–377. [Google Scholar]
- Lee, J.H. , Muhsin, M. , Atienza, G.A. , Kwak, D.Y. , Kim, S.M. , De Leon, T.B. , Angeles, E.R. , Coloquio, E. , Kondoh, H. and Satoh, K. (2010) Single nucleotide polymorphisms in a gene for translation initiation factor (eIF4G) of rice (Oryza sativa) associated with resistance to Rice tungro spherical virus . Mol. Plant–Microbe Interact. 23, 29–38. [DOI] [PubMed] [Google Scholar]
- Li, X. , Wang, C. , Sun, H. and Li, T. (2011) Establishment of the total RNA extraction system for lily bulbs with abundant polysaccharides. Afr. J. Biotechnol. 10, 17907–17915. [Google Scholar]
- Li, H. , Liu, D. , He, H. , Zhang, N. , Ge, F. and Chen, C. (2014) Molecular cloning of a 14‐3‐3 protein gene from Lilium regale Wilson and overexpression of this gene in tobacco increased resistance to pathogenic fungi. Sci. Hortic. 168, 9–16. [Google Scholar]
- Lim, J. , Rhee, H. , Kim, Y. , Lim, K. and Van Tuyl, J. (2003) Resistance to Fusarium oxysporum f. sp. lilii in Lilium . Acta Hort. 620, 311–318. [Google Scholar]
- Ling, K.S. , Harris, K.R. , Meyer, J.D. , Levi, A. , Guner, N. , Wehner, T.C. , Bendahmane, A. and Havey, M.J. (2009) Non‐synonymous single nucleotide polymorphisms in the watermelon eIF4E gene are closely associated with resistance to Zucchini yellow mosaic virus . Theor. Appl. Genet. 120, 191–200. [DOI] [PubMed] [Google Scholar]
- Liu, Y. , Schiff, M. and Dinesh‐Kumar, S. (2004) Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COI1 and CTR1 in N‐mediated resistance to Tobacco mosaic virus . Plant J. 38, 800–809. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- van Loon, L.C. , Rep, M. and Pieterse, C.M. (2006) Significance of inducible defense‐related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. [DOI] [PubMed] [Google Scholar]
- Lu, J. , Du, Z.X. , Kong, J. , Chen, L.N. , Qiu, Y.H. , Li, G.F. , Meng, X.H. and Zhu, S.F. (2012) Transcriptome analysis of Nicotiana tabacum infected by Cucumber mosaic virus during systemic symptom development. PLoS One, 7, e43447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marathe, R. , Anandalakshmi, R. , Liu, Y. and Dinesh‐Kumar, S. (2002) The Tobacco mosaic virus resistance gene, N, Mol. Plant Pathol. 3, 167–172. [DOI] [PubMed] [Google Scholar]
- Massala, R. , Legrand, M. and Fritig, B. (1980) Effect of α‐aminooxyacetate, a competitive inhibitor of phenylalanine ammonia‐lyase, on the hypersensitive resistance of tobacco to Tobacco mosaic virus . Physiol. Plant Pathol. 16, 213–226. [Google Scholar]
- Michel, K. , Abderhalden, O. , Bruggmann, R. and Dudler, R. (2006) Transcriptional changes in powdery mildew infected wheat and Arabidopsis leaves undergoing syringolin‐triggered hypersensitive cell death at infection sites. Plant Mol. Biol. 62, 561–578. [DOI] [PubMed] [Google Scholar]
- Moura, J.C.M.S. , Bonine, C.A.V. de Oliveira Fernandes Viana, J. , Dornelas, M.C. and Mazzafera, P., (2010) Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 52, 360–376. [DOI] [PubMed] [Google Scholar]
- Nagashima, H. , Murakami, T. , Yamamoto, N. , Naoe, T. , Kawazoe, Y. , Konno, K. and Sakagami, H. (1992) Lignified materials as medicinal resources. V. Anti‐HIV (Human immunodeficiency virus) activity of some synthetic lignins. Chem. Pharm. Bull. 40, 2102–2105. [DOI] [PubMed] [Google Scholar]
- Nicholson, R.L. and Hammerschmidt, R. (1992) Phenolic compounds and their role in disease resistance. Annu. Rev. Phytopathol. 30, 369–389. [Google Scholar]
- Niimi, Y. , Nakano, M. and Maki, K. (1996) Production of interspecific hybrids between Lilium regale and L. rubellum via ovule culture. J. Jpn. Soc. Hortic. Sci. 64, 919–925. [Google Scholar]
- Obata, Y. , Niimi, Y. , Nakano, M. , Okazaki, K. and Miyajima, I. (2000) Interspecific hybrids between Lilium nobilissimum and L. regale produced via ovules‐with‐placental‐tissue culture. Sci. Hortic. 84, 191–204. [Google Scholar]
- Ooka, H. , Satoh, K. , Doi, K. , Nagata, T. , Otomo, Y. , Murakami, K. , Matsubara, K. , Osato, N. , Kawai, J. and Carninci, P. (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana . DNA Res. 10, 239–247. [DOI] [PubMed] [Google Scholar]
- Park, C.J. , Shin, Y.C. , Lee, B.J. , Kim, K.J. , Kim, J.K. and Paek, K.H. (2006) A hot pepper gene encoding WRKY transcription factor is induced during hypersensitive response to Tobacco mosaic virus and Xanthomonas campestris . Planta, 223, 168–179. [DOI] [PubMed] [Google Scholar]
- Pertea, G. , Huang, X. , Liang, F. , Antonescu, V. , Sultana, R. , Karamycheva, S. , Lee, Y. , White, J. , Cheung, F. and Parvizi, B. (2003) TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics, 19, 651–652. [DOI] [PubMed] [Google Scholar]
- Pettersen, E.F. , Goddard, T.D. , Huang, C.C. , Couch, G.S. , Greenblatt, D.M. , Meng, E.C. and Ferrin, T.E. (2004) UCSF Chimera‐a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- Puranik, S. , Sahu, P.P. , Srivastava, P.S. and Prasad, M. (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. [DOI] [PubMed] [Google Scholar]
- Quecini, V. , Lopes, M.L. , Pacheco, F.T. and Ongarelli, M.d.G. (2007) Tomato spotted wilt virus triggers specific and shared defense mechanisms in hypersensitive and susceptible Solanaceae hosts. Physiol. Mol. Plant Pathol. 70, 189–197. [Google Scholar]
- Ram, R. , Sharmaj, A. , Dhyani, D. , Sood, A. and Zaidi, A. (2000) Viral diseases of lilies & their management–a review. Bharatiya Vaigyanik evam Audyogik Anusandhan Patrika, 8, 63–67. [Google Scholar]
- Rao, J. , Liu, D. , Zhang, N. , He, H. , Ge, F. and Chen, C. (2013) Identification of genes differentially expressed in a resistant reaction to Fusarium oxysporum in Lilium regale by SSH. IERI Procedia, 5, 95–101. [Google Scholar]
- Rojas, M. , Kon, T. , Natwick, E. , Polston, J. , Akad, F. and Gilbertson, R. (2007) First report of Tomato yellow leaf curl virus associated with tomato yellow leaf curl disease in California. Plant Dis. 91, 1056. [DOI] [PubMed] [Google Scholar]
- Selth, L.A. , Dogra, S.C. , Rasheed, M.S. , Healy, H. , Randles, J.W. and Rezaian, M.A. (2005) A NAC domain protein interacts with Tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell, 17, 311–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siljak‐Yakovlev, S. , Peccenini, S. , Muratovic, E. , Zoldos, V. , Robin, O. and Valles, J. (2003) Chromosomal differentiation and genome size in three European mountain Lilium species. Plant Syst. Evol. 236, 165–173. [Google Scholar]
- Sun, D. , Nandety, R.S. , Zhang, Y. , Reid, M.S. , Niu, L. and Jiang, C. (2016a) A petunia ethylene‐responsive element binding factor, PhERF2, plays an important role in antiviral RNA silencing. J. Exp. Bot. 67, 3353–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, D. , Zhang, X. , Li, S. , Jiang, C. , Zhang, Y. and Niu, L. (2016b) LrABCF1, a GCN‐type ATP‐binding cassette transporter from Lilium regale, is involved in defense responses against viral and fungal pathogens. Planta, 244, 1185–1199. [DOI] [PubMed] [Google Scholar]
- Sun, D. , Li, S. , Niu, L. , Reid, M.S. , Zhang, Y. and Jiang, C. (2017) PhOBF1, a petunia ocs element binding factor, plays an important role in antiviral RNA silencing. J. Exp. Bot. 68, 915–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suyal, G. , Rana, V.S. , Mukherjee, S.K. , Wajid, S. and Choudhury, N.R. (2014) Arabidopsis thaliana NAC083 protein interacts with Mungbean yellow mosaic India virus (MYMIV) Rep protein. Virus Genes, 48, 486–493. [DOI] [PubMed] [Google Scholar]
- Tamura, K. , Dudley, J. , Nei, M. and Kumar, S. (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599. [DOI] [PubMed] [Google Scholar]
- Tasaki, K. , Terada, H. , Masuta, C. and Yamagishi, M. (2016) Virus‐induced gene silencing (VIGS) in Lilium leichtlinii using the Cucumber mosaic virus vector. Plant Biotechnol. 33, 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, L.S.P. , Nakashima, K. , Sakuma, Y. , Simpson, S.D. , Fujita, Y. , Maruyama, K. , Fujita, M. , Seki, M. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2004) Isolation and functional analysis of Arabidopsis stress‐inducible NAC transcription factors that bind to a drought‐responsive cis‐element in the early responsive to dehydration stress 1 promoter. Plant Cell, 16, 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. (2015) The molecular detection of Corynespora cassiicola on cucumber by PCR assay using DNAman software and NCBI In: International Conference on Computer and Computing Technologies in Agriculture ( Li, D.L. and Li, Z.B. , eds.), pp. 248–258. Cham, Switzerland: Springer. [Google Scholar]
- Wang, X.E. , Basnayake, B.V.S. , Zhang, H. , Li, G. , Li, W. , Virk, N. , Mengiste, T. and Song, F. (2009) The Arabidopsis ATAF1, a NAC transcription factor, is a negative regulator of defense responses against necrotrophic fungal and bacterial pathogens. Mol. Plant–Microbe Interact. 22, 1227–1238. [DOI] [PubMed] [Google Scholar]
- Wang, D. , Ma, Y. , Yang, Y. , Liu, N. , Li, C. , Song, Y. and Zhi, H. (2011) Fine mapping and analyses of R SC8 resistance candidate genes to Soybean mosaic virus in soybean. Theor. Appl. Genet. 122, 555–565. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Wang, W. , Jin, S. , Wang, J. , Wang, B. and Hou, B. (2012) Overexpression of a putative poplar glycosyltransferase gene, PtGT1, in tobacco increases lignin content and causes early flowering. J. Exp. Bot. 63, 2799–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Ma, S. , Li, W. , Wang, Q. , Cao, H. , Gu, J. and Lu, Y. (2016) Genetic variability and diversity of the main resources of lily assessed via phenotypic characters, pollen morphology, and ISSR markers. Genet. Mol. Res. 15, gmr.15027638. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Li, Z. , Lu, M. and Wang, Y. (2017) ThNAC13, a NAC transcription factor from Tamarix hispida, confers salt and osmotic stress tolerance to transgenic tamarix and Arabidopsis . Front. Plant Sci. 8, 635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Zhu, X. , Wang, K. , Lu, C. , Luo, M. , Shan, T. and Zhang, Z. (2018) A wheat caffeic acid 3‐O‐methyltransferase TaCOMT‐3D positively contributes to both resistance to sharp eyespot disease and stem mechanical strength. Sci. Rep. 8, 6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, B. and Sali, A. (2014) Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 47, 5.6. 1–5.6. 32. [DOI] [PubMed] [Google Scholar]
- Wei, C. , Cui, Q. , Zhang, X. , Zhao, Y. and Jia, G. (2016) Three P5CS genes including a novel one from Lilium regale play distinct roles in osmotic, drought and salt stress tolerance. J. Plant Biol. 59, 456–466. [Google Scholar]
- Welner, D.H. , Lindemose, S. , Grossmann, J.G. , Møllegaard, N.E. , Olsen, A.N. , Helgstrand, C. , Skriver, K. and Leggio, L.L. (2012) DNA binding by the plant‐specific NAC transcription factors in crystal and solution: a firm link to WRKY and GCM transcription factors. Biochem. J. 444, 395–404. [DOI] [PubMed] [Google Scholar]
- Whitham, S. , McCormick, S. and Baker, B. (1996) The N gene of tobacco confers resistance to Tobacco mosaic virus in transgenic tomato. Proc. Natl. Acad. Sci. USA, 93, 8776–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wixon, J. and Kell, D. (2000) Website review: the Kyoto encyclopedia of genes and genomes‐KEGG. http://www.genome.ad.jp/kegg. Yeast, 17, 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Q. , Sanz‐Burgos, A.P. , Guo, H. , García, J.A. and Gutiérrez, C. (1999) GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol. Biol. 39, 647–656. [DOI] [PubMed] [Google Scholar]
- Yang, Y. and Klessig, D.F. (1996) Isolation and characterization of a Tobacco mosaic virus‐inducible myb oncogene homolog from tobacco. Proc. Natl. Acad. Sci. USA, 93, 14972–14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J. , Fang, L. , Zheng, H. , Zhang, Y. , Chen, J. , Zhang, Z. , Wang, J. , Li, S. , Li, R. and Bolund, L. (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34, W293–W297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo, S.Y. , Kim, Y. , Kim, S.Y. , Lee, J.S. and Ahn, J.H. (2007) Control of flowering time and cold response by a NAC‐domain protein in Arabidopsis . PLoS One, 2, e642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii, M. , Shimizu, T. , Yamazaki, M. , Higashi, T. , Miyao, A. , Hirochika, H. and Omura, T. (2009) Disruption of a novel gene for a NAC‐domain protein in rice confers resistance to Rice dwarf virus . Plant J. 57, 615–625. [DOI] [PubMed] [Google Scholar]
- Zeyen, R. , Bushnell, W. , Carver, T. , Robbins, M. , Boyles, D. and Vance, C. (1995) Inhibiting phenylalanine ammonia lyase and cinnamyl‐alcohol dehydrogenase suppresses Mla1 (HR) but not mlo5 (non‐HR) barley powdery mildew resistances. Physiol. Mol. Plant Pathol. 47, 119–140. [Google Scholar]
- Zhong, R. , Lee, C. and Ye, Z. (2010) Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis . Mol. Plant, 3, 1087–1103. [DOI] [PubMed] [Google Scholar]
- Zorzatto, C. , Machado, J.P.B. , Lopes, K.V. , Nascimento, K.J. , Pereira, W.A. , Brustolini, O.J. , Reis, P.A. , Calil, I.P. , Deguchi, M. and Sachetto‐Martins, G. (2015) NIK1‐mediated translation suppression functions as a plant antiviral immunity mechanism. Nature, 520, 679–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Sequences of all assembled unigenes in Lilium regale.
Fig. S2 Length distribution of unigenes in Lilium regale.
Fig. S3 Validation of RNA‐Seq data by qRT‐PCR. Nine unigenes were randomly selected for expression analysis in mock‐ and CMV‐inoculated Lilium regale leaves at 48 h post‐inoculation (hpi). LrActin was used as a reference gene. Error bars indicate standard error (SE) of the mean from three biological replicates. Significance of difference was calculated using Student's t test (P < 0.05) and is shown as asterisks. Unigene0000074, cytochrome P450 86B1; Unigene0004763, lecithine‐cholesterol acyltransferase 4; Unigene0007097, endonuclease/exonuclease/phosphatase family protein; Unigene0011570, auxin‐induced 15A; Unigene0013897, unknown protein; Unigene0017759, mannose‐specific lectin 3; Unigene0063829, argonaute 1; Unigene0073681, ABC transporter G family member 11; Unigene0079669, geranylgeranyl diphosphate synthetase.
Fig. S4 Principal component analysis of Lilium regale transcriptome data.
Fig. S5 GO functional classification of differentially expressed genes in Lilium regale.
Fig. S6 Full‐length cDNA sequence of LrNAC35 and its deduced amino acids. LrNAC35 cDNA sequence harbours a 1077‐bp open reading frame region encoding a polypeptide of 358 amino acids. The italic bold type and bold type in squares denote the start and stop codons, respectively. The type shaded in grey indicate the conserved region of LrNAC35 protein containing subdomains A to E.
Fig. S7 Reduced resistance to CMV and TMV infections in petunia plants with PhNAC35‐VIGS silencing. (A) Disease symptoms of TRV empty vector‐ and TRV‐PhNAC35‐infected petunia plants at 14 days post‐inoculation (dpi) with mock control and CMV. (B) qRT‐PCR analysis of PhNAC35 expression levels in uppermost leaves of TRV empty vector‐ and TRV‐PhNAC35‐infected petunia plants at 14 dpi with mock control and CMV. qRT‐PCR (C) and western blot (D) analyses of CMV coat protein (CMV‐CP) transcript and its protein levels in the uppermost leaves of TRV constructs‐infected petunia plants at 14 dpi with CMV. 26S rRNA and actin were used as a reference gene and protein, respectively. GFP fluorescent foci (E) and qRT‐PCR analysis of transcripts of TMV‐CP (F) encoding TMV coat protein in the leaves of TRV constructs‐inoculated petunia plants at 6 dpi with TMV‐GFP. Four‐leaf‐stage petunia seedlings were used for VIGS assay, and the seedlings at 5 dpi with TRV constructs were thereafter inoculated with mock control, CMV and TMV‐GFP. Error bars represent SE of the mean from three biological replicates. Asterisks indicate significant difference as determined by Student's t‐test at P < 0.05.
Fig. S8 Sequence analysis of LrNAC35 from five Lilium species. Alignment of LrNAC35 amino acid sequences (A) and nucleotide sequences (B) from L. regale, L. pumilum, L. duchartrei, L. brownii and L. tigrinum. Identical amino acids and nucleotides are shaded in black, while similar ones are shaded in grey.
Fig. S9 Sequences of unigenes used for qRT‐PCR or functional analysis.
Table S1 Functional annotation of all assembled unigenes in Lilium regale.
Table S2 Differentially expressed genes in Lilium regale.
Table S3 KEGG pathway annotation of differentially expressed genes in Lilium regale.
Table S4 Differentially expressed transcription factors in Lilium regale.
Table S5 Unigenes associated with lignin synthesis exhibiting increased expression in Lilium regale.
Table S6 Primers used for qRT‐PCR, gene promoter and fragment amplification.


