Abstract
Heat shock proteins (HSPs) are a large family of ubiquitously expressed proteins with diverse functions, including protein assembly and folding/unfolding. These proteins have been associated with the progression of various gastrointestinal tumours. Dysregulation of cellular redox has also been associated with gastrointestinal carcinogenesis, however, a link between HSPs and dysregulation of cellular redox in carcinogenesis remains unclear. In this study, we analysed mRNA co-expression and methylation patterns, as well as performed survival analysis and gene set enrichment analysis, on gastrointestinal cancer data sets (oesophageal, stomach and colorectal carcinomas) to determine whether HSP activity and cellular redox dysregulation are linked. A widespread relationship between HSPs and cellular redox was identified, with specific combinatorial co-expression patterns demonstrated to significantly alter patient survival outcomes. This comprehensive analysis provides the foundation for future studies aimed at deciphering the mechanisms of cooperativity between HSPs and redox regulatory enzymes, which may be a target for future therapeutic intervention for gastrointestinal tumours.
Keywords: Heat shock proteins (HSPs), Redox regulators, Gene expression analysis, Gastrointestinal carcinomas
Graphical abstract
1. Introduction
Heat shock proteins (HSPs) are a highly conserved, ubiquitously expressed family of proteins, that act as molecular chaperones to correctly fold a new polypeptide chain, or support the refolding of damaged proteins [1]. HSPs have been implicated in a variety of cellular processes, including cell cycle regulation, organelle trafficking, and signal transduction [2]. Interestingly, experimental evidence indicates that stress-induced activation of HSPs endows cells with a survival advantage by mechanism(s) that, on the one hand, activate pro-survival networks, and on the other, inhibit apoptotic execution [3]. These attributes and the association with pro-inflammatory signalling lend credence to the involvement of HSPs in promoting carcinogenesis and its progression [4], [5]. Along similar lines, the differential expression of various HSP family members and dysregulation of redox homeostasis have been individually implicated in tumorigenesis, particularly involving the gastrointestinal (GI) tract [6], [7]. However, the interplay between HSPs and redox regulation in the setting of GI cancers is less widely understood.
HSPs are divided into five families, including the DNAJ (HSP40) family, the HSPB (small heat shock protein family), the HSP90/HSPC family, chaperonins (HSPD/E and CCT) and the HSP70 superfamily, containing the HSPA (HSP70) and HSPH (HSP110) subfamilies [8]. These families mainly differ in their functions and cellular localisations. The DNAJ/HSP40 family primarily function by stimulating the ATPase activity of HSP70s [9], while the HSP70 family has been implicated in most aspects of protein folding, import, recovery from aggregation and drug resistance in cancer [10]. HSP90 is involved in many cellular processes, including DNA repair [11], and has been implicated in a variety of neoplasia [12]. The HSPB family, in particular HSP27, has been shown to be associated with several human cancers, as it promotes epithelial to mesenchymal transition (EMT) [13], [14], blocks apoptotic signalling [15], and triggers activation of oncogenic transcription factors such as NF-κB [13] and mitogen activated protein kinases (MAPK) [16], independent of their ATP hydrolysis function [17]. Chaperonins, or CCT (chaperonin containing TCP-1) proteins, are ring complexes encompassing a central cavity; this allows protein folding in a shielded environment. They can be divided into two groups: Group 1 (HSPD), that are present in the mitochondria, and Group 2, that are localized to the cytosol [18].
An imbalance in intracellular redox conditions can promote or inhibit tumour growth [19]. This is dependent on a tight balance between the different reactive oxygen species present within the cell [20]. Increased superoxide (O2-) production, favouring a pro-oxidant milieu, has been demonstrated to act as a pro-survival signal in some cancers [21], [22], [23], while increased peroxides, particularly hydrogen peroxide (H2O2), can initiate death signalling, leading to apoptosis [24], [25]. There is a close relationship between intracellular redox and HSPs. Protein folding, specifically, disulfide bond formation, is an oxidative process and generates ROS, primarily in the form of H2O2 [26], [27]. Dysregulation of protein folding can cause oxidative stress and stimulate the unfolded protein response (UPR), which is dependent on HSP proteins [28]. Furthermore, ROS-mediated DNA oxidation and lipid peroxidation can result in the induction of HSP synthesis [29]. For example, a redox-regulated HSP family member, HSP33, functions as a cellular protective agent against oxidative stress in prokaryotes; a coordinated zinc ion of HSP33 is released from the protein following the oxidation of cysteine residues by H2O2 to activate the protein [30]. Mammalian HSP1 has been reported to exhibit a similar oxidative stress response to prokaryotic HSP33. Two conserved cysteine residues near the DNA-binding domain are crucial for the formation of higher order homomultimers, required for target gene transcription and protection from oxidative stress [31]. These redox-mediated reversible reactions are critical for the activity of HSP33 and HSP1 and demonstrate the importance of cellular redox state on HSP function.
Datasets describing molecular and clinical features of patient tumours have been made publicly available through consortia such as The Cancer Genome Atlas (TCGA), accessible through websites such as cBioPortal [32], [33]. These datasets contain large numbers of patient samples across many cancer types, linking mutation, copy-number alterations, transcriptomic, proteomic and methylomic data with clinical attributes, including survival outcomes, age, gender and therapeutic interventions. These large datasets have provided the foundation for studies that have resulted in the successful identification of biomarkers, development of in vivo models and the identification of drug candidates [34], [35], [36]. The generation of a gene signature in response to different drugs using only public databases enabled the repurposing of niclosamide ethanolamine, previously used as an anti-helminthic, as an anti-tumour agent against hepatocellular carcinoma (HCC); this molecule disrupted the interaction between cell division cycle 37 and HSP90, and was effective at reducing tumour burden in vivo [34]. Similarly, an in silico approach using public datasets identified protein tyrosine kinase PTK7 as a potential oncogenic driver in non-small cell lung cancer (NSCLC); these findings were validated in vitro and in vivo, where PTK7 was found to regulate MKK7-JNK signalling, indicating its utility as a potential therapeutic target [35].
In this study, we identified a range of previously unreported genes in GI cancer patients – specifically, oesophageal, stomach and colorectal cancers – that resulted in significantly altered survival outcomes when differentially expressed. We have also identified new combinations of HSP and redox genes that, when differentially expressed, predicted overall and disease-free survival in the selected cohorts. These genes may represent clinically relevant targets for use as biomarkers in monitoring disease progression and severity, as well as offering potential interest for the development of new cancer therapeutics.
2. Methods
2.1. Data collection
Gastrointestinal carcinoma datasets were obtained from the relevant TCGA studies via cBioPortal [33] in November 2017, specifically the oesophageal carcinoma [37], stomach adenocarcinoma [38] and colorectal adenocarcinoma data sets [39]. Based on the families set out in the guidelines for the nomenclature for HSPs, we examined 96 HSP genes [8] and 30 cancer-associated genes involved in redox generation and detoxification [40] in a range of analyses, including co-expression of HSP and redox regulator genes, correlation of gene methylation and expression at different cancer stages, survival, and enrichment of other gene families in susceptible populations. For all analyses, the three cancer types were analysed separately. Analysis was facilitated via Perl scripts developed in-house.
2.2. Co-expression and methylation analysis
For co-expression analyses, Spearman's ρ values were calculated using mRNA expression z-scores (calculated from RNA Seq V2 RSEM data, compared to the expression distribution of each gene in tumours that are diploid for that gene, as provided by cBioPortal) for all gene combinations, using all patient data. Spearman's ρ was also used to assess correlation between methylation β-values (from HM450 data) and mRNA expression z-scores for all genes, separating patient data by tumour stage; positive ρ values indicate that increased gene methylation can be correlated to increased gene expression, while conversely, a negative ρ values that increased gene methylation can be correlated to decreased gene expression.
2.3. Survival analysis
Survival analysis was performed for selected patient groups meeting specific criteria. Patients were categorized as “high” or “low” expressers of genes, using an absolute mRNA expression z-score cutoff of 0.75; such groups required a minimum of 10 patients before further analysis was performed. The log-rank test was used to assess whether differences in survival were statistically significant; comparisons yielding p-values less than 0.05 were selected as statistically significant. Patient groups with single gene high/low phenotypes were assessed, as well as groups with two gene high/low phenotypes.
2.4. Gene set enrichment analysis
For patient groups with phenotypes affording significant differences in survival, gene set enrichment analysis (GSEA) was performed using the GSEA software [41], [42]. The lists of patients corresponding to each patient group for comparison were manually provided to GSEA to create phenotypes for analysis. GO gene sets [43] were used in the analysis. Gene sets affording p-values less than 0.05 and q-values (false discovery rates) of less than 0.25 were selected as being significantly altered between the patient groups. The remaining gene sets were then ranked by the GSEA normalised enrichment score (NES), and up to the top 350 ranked gene sets were analysed using REVIGO [44] to further summarize the biological importance of the identified gene sets. NESs for each gene set were supplied to REVIGO as a measure of the relative significance of each gene set. The human database was used and allowed similarity of output gene sets was set to tiny (0.4). The SimRel semantic similarity measure was used [45].
3. Results
3.1. Analysis of co-expression patterns of heat shock proteins and redox regulator genes in GI carcinomas
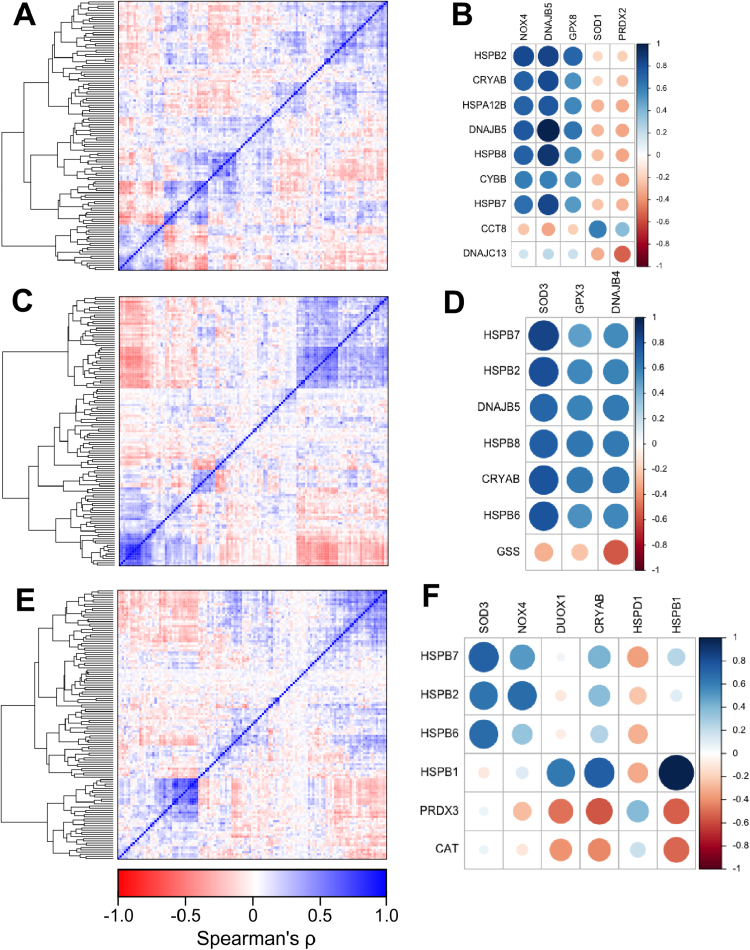
To determine co-expression patterns of the 126 HSP and redox regulator genes, we calculated Spearman's rank correlation coefficient (ρ) for all gene combinations for the stomach adenocarcinoma, colorectal adenocarcinoma and oesophageal carcinoma datasets. Hierarchical clustering revealed numerous HSPs and redox regulators that were strongly positively and negatively correlated in all carcinoma datasets.
The colorectal dataset showed a strong positive correlation in gene expression between NADPH oxidase 4 (NOX4) and numerous members of the small HSP family, including HSPB2, HSPB5 (CRYAB), HSPB7 and HSPB8, as well as members of the HSP40 family DNAJB5 and DNAJC5B. NOX4 also has a strong positive correlation with HSPA12B, a member of the HSP70 family (Fig. 1A & B). Other strong positive correlations in gene expression were identified between superoxide dismutase 1 (SOD1) with CCT8 and glutathione peroxidase 8 (GPX8) with HSPB2. Peroxiredoxin 2 (PRDX2) gene expression was negatively correlated with DNAJC13.
Fig. 1.
Co-expression analysis of HSPs and redox regulators in gastrointestinal tumours. Heatmaps of Spearman's rank correlation coefficient (ρ) for all HSP and redox genes (A, C,E) and selected gene combinations (B, D,F) in the following TCGA datasets: colorectal carcinoma (A, B), stomach adenocarcinoma (C, D), oesophageal carcinoma (E, F). In all panels, blue indicates negative ρ, red indicates positive ρ. In panels B, D and F, circle size indicates magnitude of ρ.
In the stomach adenocarcinoma dataset, strong positive correlations in gene expression were identified between members of the small HSP family, including HSPB2, HSPB5 (CRYAB), HSPB6, HSPB7 and HSPB8, as well as the HSP40 family member DNAJB5, with superoxide dismutase (SOD3) (Fig. 1C & D). Glutathione peroxidase 3 (GPX3) gene expression was strongly positive correlated with HSPB8. The only redox/HSP combination that exhibited a noteworthy negative correlation at the level of gene expression was between glutathione synthetase (GSS) and DNAJB4.
Strong positive correlations in the expression of SOD3 with the expression of HSPB2, HSPB6 and HSPB7 were found in the oesophageal carcinoma dataset (Fig. 1E & F). Gene expression of the NADPH oxidase family members NOX4 and DUOX1 was positively correlated with expression of HSPB1 and HSPB2. Negative correlations were found between PRDX3 with both HSPB5 and HSPD1, as well as between CAT and HSPB1.
3.2. Survival analysis of patient groups exhibiting differential expression of HSP and redox genes
We then set to determine if there were novel predictors of overall and disease-free survival based on combinations of HSP or redox genes, which exhibited either high or low expression. Single gene and two-gene survival analyses were performed.
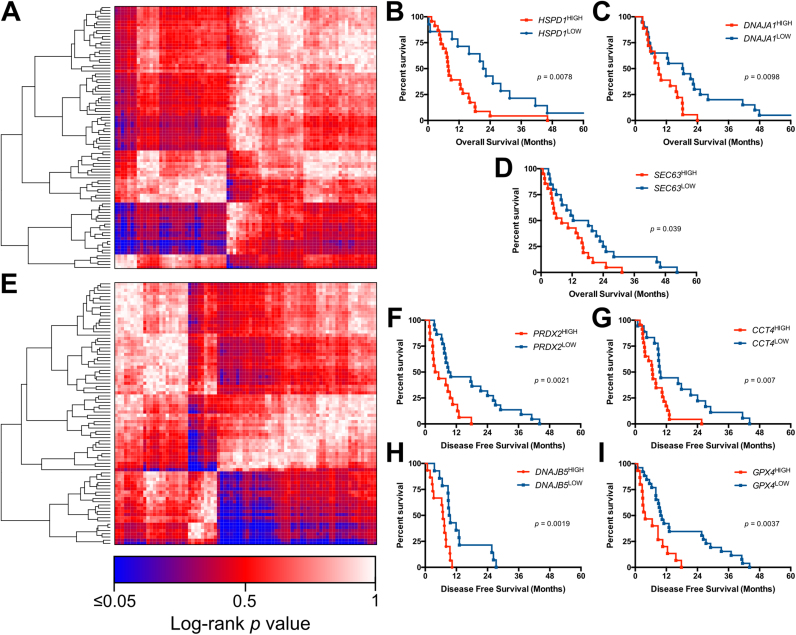
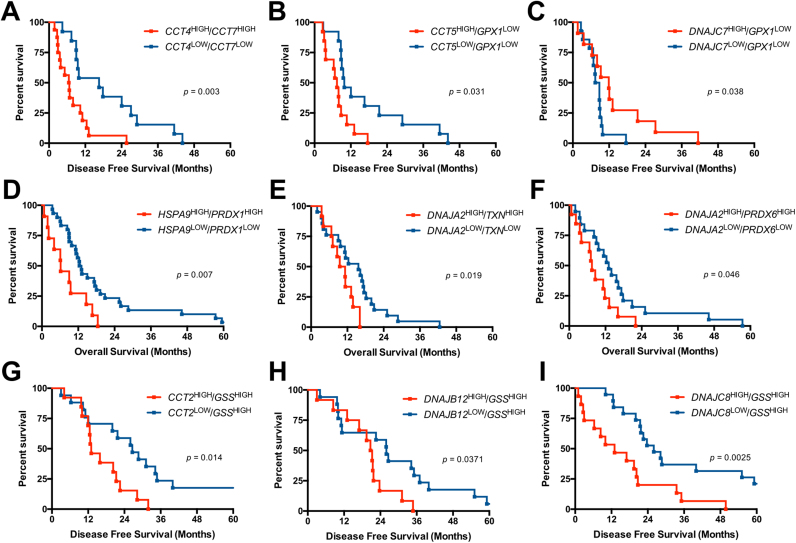
In the oesophageal carcinoma dataset, patients exhibiting high expression of HSPD1, DNAJA1 and SEC63 all resulted in significantly lower overall survival when compared to patients with low expression (Fig. 2A-D). High expression of PRDX2 and GPX4 reduced the disease-free survival significantly compared to patients with lower mRNA levels of these genes. Furthermore, patients with higher expression of DNAJB5 and CCT4 had significantly reduced disease-free survival rates (Fig. 2E-I). Two-gene survival analysis indicated that patients with CCT2high/GSS high, DNAJB12 high/GSS high and DNAJC8 high/GSS high phenotypes all exhibited poorer disease-free survival, compared to patients with CCT2low/GSShigh, DNAJB12low/GSShigh and DNAJC8low/GSShigh expression (Fig. 5A–C).
Fig. 2.
Single gene survival analysis of HSPs and redox regulators in oesophageal carcinoma. A) Heatmap of log-rank p-values for overall survival differences of HSPs and redox regulators in oesophageal carcinoma. Blue indicates significant (p < 0.05) survival differences, Red indicates non-significant (p > 0.05) survival differences. Overall survival analysis for B) HSPD1high vs HSPD1low C) DNAJA1high vs DNAJA1low D) SEC63high vs SEC63low. E) Heatmap of log-rank p-values for disease-free survival differences of HSPs and redox regulators in oesophageal carcinoma. Overall disease-free survival analysis for F) PRDX2high vs PRDX2low G) CCT4high vs CCT4low H) DNAJB5high vs DNAJB5low I) GPX4high vs GPX4low.
Fig. 5.
Combinatorial gene survival analysis of HSPs and redox regulators in gastrointestinal cancers. Disease-free survival analysis for A) CCT4/CCT7high vs CCT4/CCT7low, B) CCT5/GPX1high vs CCT5/GPX1low C) DNAJC7/GPX1high vs DNAJC7/GPX1low in oesophageal carcinoma dataset. Overall survival analysis for D) HSPA9/PRDX1high vs HSPA9/PRDX1low E) DNAJA2/TXNhigh vs DNAJA2/TXNlow and F) DNAJA2/PRDX6high vs DNAJA2/PRDX6low in stomach adenocarcinoma dataset. Disease-free survival analysis for G) CCT2high/GSShigh vs CCT2low/GSSlow H) DNAJB12high/GSShigh vs DNAJB12low/GSSlow I) DNAJC8high/GSShigh vs DNAJC8low/GSSlow in colorectal carcinoma dataset.
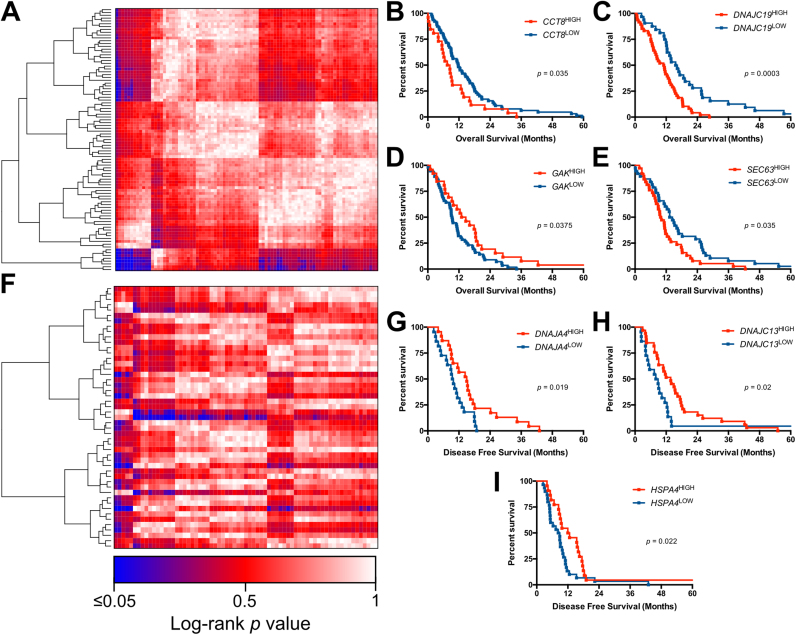
The stomach adenocarcinoma dataset revealed that overall survival was significantly altered as a result of differential expression of four HSPs - CCT8, DNAJC19, GAK and SEC63 (Fig. 3B-E). Low expression of CCT8, DNAJC19 and SEC63 significantly increased overall survival, whereas low expression of GAK significantly decreased overall survival (Fig. 3D). Significantly poorer disease-free survival rates were found in patients with high DNAJA4, DNAJC13 and HSPA4 expression compared to patients with low expression (Fig. 3G-I). Two-gene survival analysis revealed significantly poorer survival in patients with CCT5high/GPX1low expression compared to CCT5low/GPX1low expression. Conversely, patients with DNAJC7high/GPX1low expression had a significantly better disease free survival rate than those with DNAJC7low/GPX1low expression. High expression of both CCT4 and CCT7 resulted in significantly poorer survival than those with low expression of CCT4 and CCT7 (Fig. 5D–F).
Fig. 3.
Single gene survival analysis of HSPs and redox regulators in stomach adenocarcinoma. A) Heatmap of log-rank p-values for overall survival differences of HSPs and redox regulators in stomach adenocarcinoma. Blue indicates significant (p < 0.05) survival differences, Red indicates non-significant (p > 0.05) survival differences. Overall survival analysis for B) CCT8high vs CCT8low C) DNAJC19high vs DNAJAC19low D) GAKhigh vs GAKlow E) SEC63high vs SEC63low. F) Heatmap of log-rank p-values for disease-free survival differences of HSPs and redox regulators in stomach adenocarcinoma. Disease-free survival analysis for G) DNAJA4high vs DNAJA4low H) DNAJC13high vs DNAJC13low I) HSPA4high vs HSPA4low
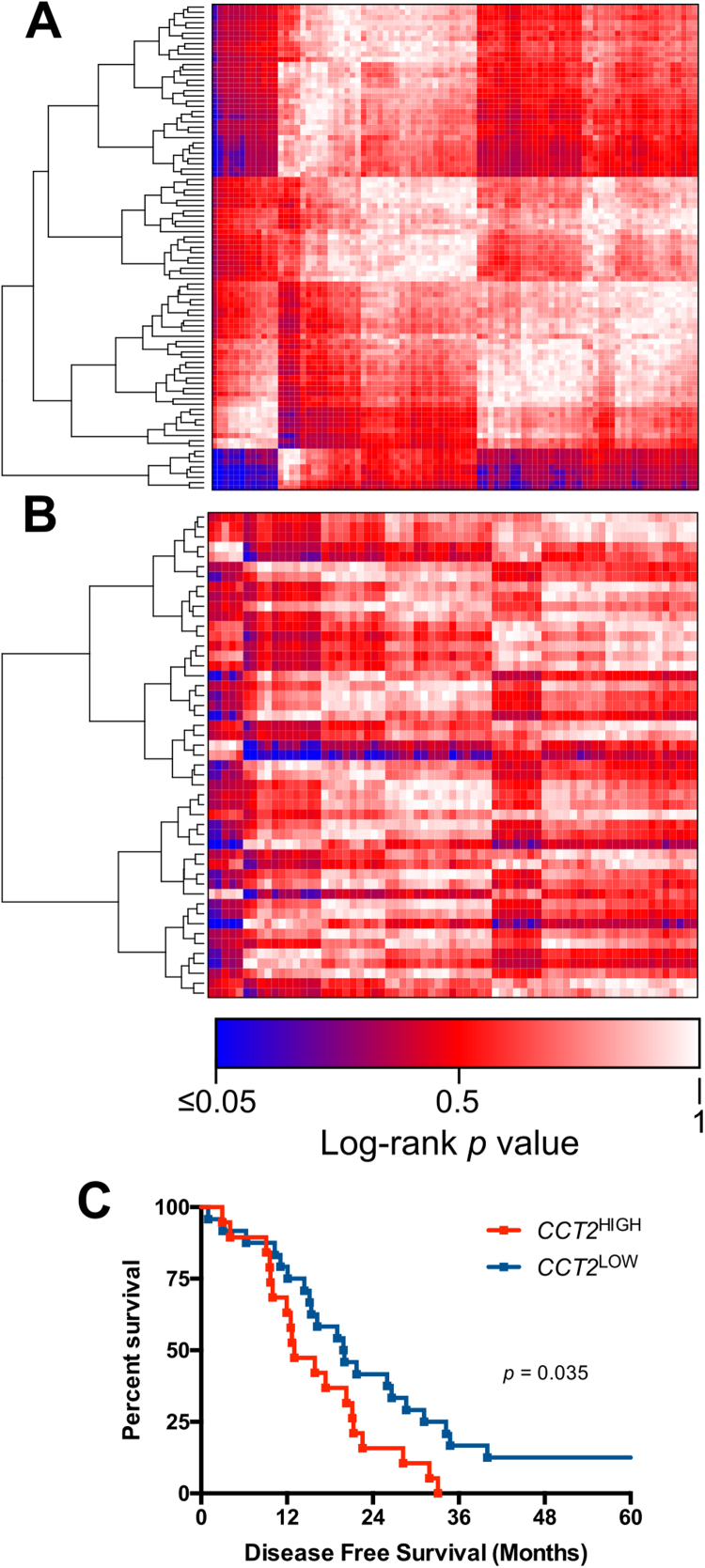
The colorectal adenocarcinoma dataset revealed that the increased expression of CCT2 resulted in a significantly decreased disease-free survival (Fig. 4C). When two-gene survival analysis was performed combinations of HSP and redox regulators, patients with HSPA9high/PRDX1high, DNAJA2high/TXNhigh and DNAJA2high/PRDX6high genotypes all had significantly decreased overall survival rates when compared to the same combinations with low expression (Fig. 5G–I).
Fig. 4.
Single gene survival analysis of HSPs and redox regulators in colorectal carcinoma. A) Heatmap of log-rank p-values for overall survival differences of HSPs and redox regulators in colorectal carcinoma. B) Heatmap of log-rank p-values for disease-free survival differences of HSPs and redox regulators in colorectal carcinoma. Blue indicates significant (p < 0.05) survival differences, Red indicates non-significant (p > 0.05) survival differences. C) Disease-free survival analysis for CCT2high vs CCT2low.
3.3. Correlations between gene methylation and expression at different pathological stages
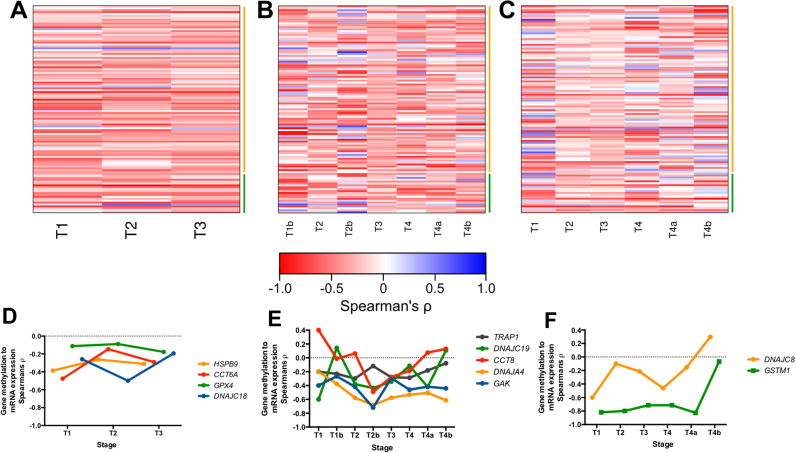
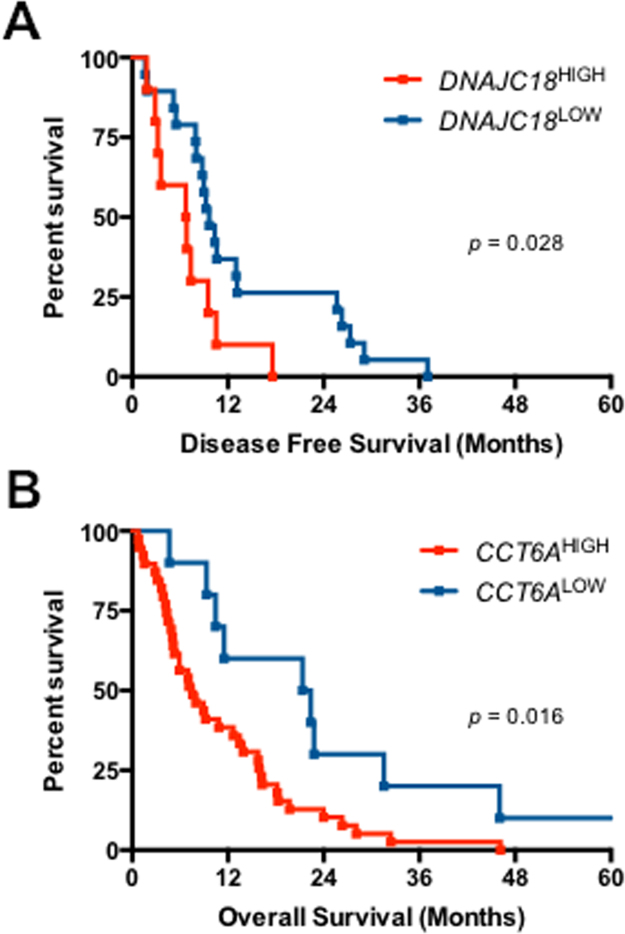
We set to determine Spearman correlation coefficients between gene methylation and expression for 126 genes in our HSP and redox regulator set, and to draw connection between this, pathological stage and survival (Fig. 6A-C). The majority of genes in oesophageal carcinoma unexpectedly exhibited a negative correlation between methylation and mRNA expression at stages T1, T2 and T3 (Fig. 6D). HSPB9 showed a negative correlation across these stages (Fig. 6D). GPX4 demonstrated a weak negative correlation that remained mainly unchanged across different pathological stages. CCT6A exhibited the strongest negative correlation at T1, which decreased with tumour stage progression. DNAJC18 showed a strong negative correlation at T2, which progressed to a weak negative correlation in stage T3. The loss of a strong negative correlation during the progression of tumours suggests that there may be decreased methylation and increased mRNA expression during the more progressed stages. This is consistent with survival data for DNAJC18 (Fig. S4A) and CCT6A (Fig. S4B), both of which, when highly expressed, result in significantly poorer overall patient survival compared to patients with low expression. Conversely, consistently low negative correlations between stages in HSPB9 suggest no loss of methylation and low mRNA expression, consistent with low levels of HSPB9 reflecting significantly poorer patient survival.
Fig. 6.
Correlation of methylation and gene expression of HSPs and redox regulators in gastrointestinal cancers. Heatmap of Spearman's rank correlation coefficient (ρ) for TCGA datasets of A) oesophageal carcinoma B) stomach adenocarcinoma and C) colorectal carcinoma for various tumour stages (T1-T4b). Blue indicates negative Spearman's coefficient, Red indicates positive Spearman's coefficient. Green line indicates redox regulator genes, yellow line indicates HSP genes. D) Line graph plotting gene methylation to mRNA expression through tumour stages T1-T3 of HSPB9, CCT6A, GPX4 and DNAJC18 for oesophageal carcinoma dataset. E) Line graph plotting gene methylation to mRNA expression through tumour stages T1-T4b of TRAP1, DNAJC19, CCT8, DNAJA4 and GAK for stomach adenocarcinoma dataset. F) Line graph plotting gene methylation to mRNA expression through tumour stages T1-T4b of DNAJC8, and GSTM1 for colorectal carcinoma dataset.
When the analysis was performed on the stomach adenocarcinoma data, a strong negative correlation between gene expression and methylation was observed for DNAJA4. This correlation was stronger in the later stages (T2, T2b, T3, T4, T4a and T4b) than in T1 (Fig. 6E). The survival analysis revealed that patients with DNAJA4low had a significantly poorer disease free survival than those with the DNAJA4high phenotype, suggesting that increased gene methylation of DNAJA4 may predict poorer survival. A similar trend of a strong negative correlation was seen in GAK in all stages (T1-T4b). Again, patients with a GAKlow phenotype had a significantly poorer overall survival than those with a GAKhigh phenotype, suggesting that the methylation status, corresponding to overall gene expression, plays an important role in overall patient survival.
In the colorectal dataset, two genes were identified where methylation may play an important role in gene expression at different tumour stages. GSTM1 exhibited a very strong negative correlation between methylation and gene expression in stages T1 through to T4a, however, this correlation was nearly completely lost in stage T4b (Fig. 6F). Methylation of DNAJC8 was strong negatively correlated in stage T1, suggesting gene silencing through methylation, although in T4a and T4b, this correlation was reversed, suggesting that methylation was correlated with an increase in gene expression.
3.4. Gene set enrichment analysis in patient groups associated with significantly altered survival
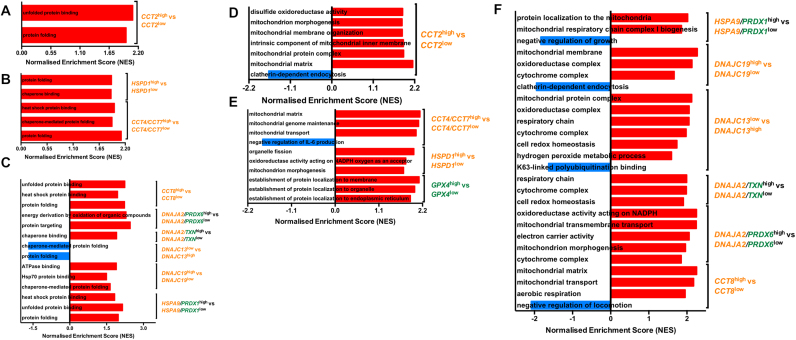
To determine whether the genotypes associated with differing disease-free or overall survival outcomes were also broadly associated with other biological processes and molecular functions, we performed gene set enrichment analysis (GSEA). The Gene Ontology (GO) gene sets were used for this purpose. Across the three gastrointestinal tumour types, grouped individuals with high expression of HSP family members were, unsurprisingly, enriched in gene sets covering processes involved in canonical HSP function, for example, protein folding, chaperone-mediated protein folding and heat shock protein binding (Fig. 7A–C).
Fig. 7.
Gene set enrichment analysis (GSEA) of HSPs and redox regulators in gastrointestinal cancers. Bar graphs of normalised enrichment scores (NES) from GSEA of various HSPhigh vs HSPlow phenotypes in A) colorectal carcinoma B) oesophageal adenocarcinoma and C) stomach adenocarcinoma for validation of HSP related processes. Bar graphs of normalised enrichment scores (NES) from GSEA of various redox regulatorshigh/low/HSPhigh/low vs redox regulatorshigh/low/HSPlow/high phenotypes in D) colorectal carcinoma E) oesophageal carcinoma and F) stomach adenocarcinoma. Heat shock protein genes indicated by yellow writing, redox regulators indicated by green writing.
The colorectal adenocarcinoma dataset, which had only one statistically significant survival phenotype (CCT2high vs CCT2low), was primarily enriched in GO gene sets involved in mitochondrial transport and organisation. (Fig. 7D). CCT2low individuals were enriched for gene sets involved in clathrin-mediated endocytosis compared to CCT2high individuals.
GSEA on the oesophageal carcinoma phenotypes associated with differential survival outcomes were enriched in GO terms associated with mitochondrial matrix, transport and genome maintenance (CCT4/CCT7high vs CCT4/CCT7low), mitochondrial morphogenesis and oxidoreductase activity (HSPD1high vs HSPD1low) and protein localisation to the membrane, organelles and ER (GPX4high vs GPX4low). The CCT4/CCT7low vs CCT4/CCT7high phenotypes were enriched for gene sets involved in the negative regulation of IL-6 production (Fig. 7E).
In the stomach adenocarcinoma dataset, GO gene sets for mitochondrial membrane transport and structure, as well as mitochondrial respiration components, were significantly enriched. Gene sets involved in hydrogen peroxide metabolic processes and cell redox homeostasis were enriched in patients with a DNAJC13low vs DNAJC13high phenotypes. HSPA9high/PRDX1high individuals were significantly enriched in genes mediating mitochondrial respiratory chain complex I biogenesis and protein localisation to the mitochondria, while conversely, HSPA9low/PRDX1low individuals were significantly enriched for gene sets involved in the negative regulation of growth. Gene sets for mitochondrial matrix, transport and aerobic respiration were all enriched in the CCT8high individuals, while genes mediating negative regulation for locomotion were enriched in grouped individuals demonstrating a CCT8low phenotype (Fig. 7F).
4. Discussion
We analysed publicly available TCGA datasets from three gastrointestinal tumours utilising a list of 96 HSP family genes and 30 genes that have been implicated in cellular redox functions. We analysed gene correlations, correlations between methylation status in tumour stages with gene expression, and overall and disease free survival in patients. We then selected the genotypes that demonstrated significant differences in patient survival and performed gene set enrichment analysis to reveal up or downregulated gene sets associated with these phenotypes. The colorectal and oesophageal dataset revealed positive gene correlations between NOX4 and a variety of HSP family members. NOX4 has been previously shown to mediate the expression of HSP27 mediated by TGFβ [46]. This is in line with previous reports that have demonstrated co-localisation of NOX4 with HSP27 and HSP70 in vascular smooth muscle cells [47] and myoblasts [48] respectively. In all datasets, members of the superoxide dismutase family, which are responsible for the production of H2O2 through the dismutation of O2-, have a positive correlation with many HSPs. This is consistent with previous observations that have demonstrated the induction of a variety of HSPs, including HSP30, HSP90 and HSP70, through H2O2 [49], [50], [51]. HSP70 itself directly interacts with SOD2 and may chaperone the enzyme to the mitochondria, although the exact mechanism remains unclear [52]. Interestingly, enzymes involved in ROS detoxification, such as catalase and peroxiredoxins, exhibited a negative correlation with HSPs. This aligns with the observation that HSP gene expression is induced by H2O2. Higher levels of intracellular hydrogen peroxide, as a result of decreased catalase or peroxidredoxin expression, may facilitate cellular redox conditions permissible for the induction of high levels of HSP family members. Strong positive correlations were shown between various members of the small HSP family and redox-related genes. It has previously been shown that HSP27 plays roles in the pathological state of cancer cells in response to oxidative stress through modulation of the ROS-glutathione pathway [53]. HSP27 has been shown to act as a protectant against the oxidation of proteins [53]. Overexpression of HSP27 has also been shown to increase extracellular O2- production to produce a pro-oxidant state beneficial for cell survival [53], [54]. Furthermore, phosphorylation-mediated downregulation of HSP27 was shown to sensitize cancer cells to TRAIL-mediated apoptosis upon exposure to a small molecule [55] that signals through extracellular H2O2 production [56]. These observations align with results demonstrated in all these gastrointestinal cancer datasets.
Gene methylation is widely known to influence gene expression [57]. We observed a strong negative correlation between DNAJA4 gene methylation and expression in stomach adenocarcinoma, which suggested that methylation of DNAJA4 may suppress gene expression. This is consistent with reports that DNAJA4 expression is strongly influenced by its methylation status [58]. Our data also revealed that patients with low DNAJA4 expression had poorer survival outcomes. Together, this data suggests that gene methylation of DNAJA4 may suppress its expression, leading to poorer patient outcomes. DNAJA4 has been shown to promote metastasis and angiogenesis in melanoma [59], but may act as a tumour suppressor in stomach cancer. DNAJA4 and its methylation status may be a new biomarker for stomach adenocarcinoma, although this may not be applicable to other carcinomas. Gene expression of GAK (cyclin G-associated kinase) was also found to be negatively correlated in stomach cancer, and patients with a GAKlow phenotype had poorer overall survival. While GAK has been shown to promote oncogenesis in some cancers [60], in stomach cancer it appears that higher expression of GAK may be beneficial. GAK has been shown to be essential for clathrin-mediated endocytosis [61], which, if impaired, can lead to prolonged cell signalling and higher rates of proliferation [62]. In the colorectal dataset, GSTM1 demonstrated a high negative correlation in all stages prior to stage T4a, whereas in stage T4b this was completely lost, suggesting that low levels of GSTM1 are associated with the onset and progression of colorectal cancer. This has been confirmed in different populations, where the GSTM1 null variant was correlated with increased colorectal cancer risk [63], [64]. The mechanisms of GSTM1 gene methylation may provide interesting insights into the onset of colorectal cancer.
Our gene set enrichment data implicated many biological processes that are crucial for tumorigenesis in these cancer types surveyed, many being involved in mitochondrial functions, an organelle crucial in the progression of cancer [65]. In the colorectal dataset, the CCT2high phenotype, which displayed poorer survival, was enriched in gene sets involved in mitochondrial organisation and membrane processes. While CCT2 overexpression has previously been shown to adversely affect colorectal patient survival [66], its role in mitochondrial membrane organisation is less clearly understood. A recent study in colorectal cancer identified CCT2 as a synthetic lethal partner to mutant KRAS, which was found to be dependent on mitochondrial translation for viability [67]. Similarly, in oesophageal cancer, patients with HSPD1high and CCT4high/CCT7high phenotypes displayed enrichment for gene sets involved in mitochondrial processes, such as organelle fission, transport and genome maintenance, which were associated with poorer survival. HSPD1 was identified as an enriched gene in the ESCC4 subtype of oesophageal carcinoma, which also has the poorest survival outcomes [68]. HSPD1 has been previously shown to be critical for mitochondrial protein folding [69]. It also functions as a protectant against pharmacologically-induced oxidative stress [70]; therefore, combination therapy that initiates oxidative stress-induced apoptosis, such as myrtucommulone while simultaneously inhibiting HSP60 may result in better outcomes [71]. Glutathione peroxidase family member GPX4 protects against lipid peroxidation and ferroptosis, and is predominantly expressed in the mitochondria of the testis [72], [73]. Overexpression of GPX4 in oesophageal cancer patients correlates with the upregulation of gene sets involved in protein localisation to the ER and membrane, a process that, if dysregulated, can contribute to carcinogenesis [74]. GPX4 overexpression may represent a novel biomarker and a potential therapeutic target for the treatment of oesophageal cancer.
The stomach adenocarcinoma dataset revealed many combinations of HSPs and redox regulators that influenced patient survival. These were also associated with many biological processes involved in mitochondrial transport, metabolism and cellular redox homeostasis. The overexpression of DNAJC19, which encodes for the protein TIM14, has yet to be shown to influence oncogenesis or patient survival in stomach cancer. It functions as a translocase on the inner mitochondrial membrane [75], and GSEA on DNAJC19high patients revealed enrichment of gene sets for mitochondrial membrane and oxidoreductase activity. Conversely, DNAJC13 overexpression may benefit patient survival outcomes. The primary role of DNAJC13 is in early endosome transportation, [76] and in ERBB2 positive breast cancers has been implicated in the trafficking of epidermal growth factor receptor (EGFR) to the plasma membrane [77]. In this context, DNAJC13 is seen as a potential therapeutic target, whereas the situation is reversed in stomach cancer; the enriched gene sets for the DNAJC13low phenotype primarily included cell redox homeostasis and those involved in mitochondrial complexes and metabolism. Therefore, DNAJC13 depletion may facilitate mitochondrial complex function and pro-oxidant state necessary to promote tumorigenesis. CCT8high individuals were found to have a significantly poorer overall survival, and interestingly, the GSEA revealed that CCT8low individuals were enriched for gene sets involved in the negative regulation of locomotion. This is consistent with the reduction in cell migration and invasion observed in glioma cells following siRNA-mediated knockdown of CCT8 [78].
This study has uncovered a variety of novel HSP and redox genes that may influence patient outcomes and survival in gastrointestinal cancers. This high-throughput method of data extraction and analysis will become more valuable as more patient data becomes available. This study also sets a foundation for the analysis and development of multi-gene signatures that can predict patient survival, and exponentially expand our list of biomarkers for the onset and progression of a variety of cancers.
Acknowledgements
S.Ö.G.P is supported an Australian Government Research Training Program Scholarship, the Rotary Club of Belmont, Australian Rotary Health Research Fund and Curtin University School of Pharmacy and Biomedical Sciences. M.A. is supported by a Curtin Research Fellowship, and a recipient of a Raine Priming Grant (Raine Medical Research Foundation).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.11.018.
Appendix A. Supplementary material
Data S1. Correlation co-efficients for paired gene co-expression in all colorectal adenocarcinoma (COAD), oesophageal carcinoma (ESCA) and stomach adenocarcinoma (STAD) sets, ordered as depicted in Figs. 1A, 1C, and 1E.
Data S2. Correlation co-efficients between methylation and gene expression for patients at progressing stages of colorectal adenocarcinoma (COAD), oesophageal carcinoma (ESCA) and stomach adenocarcinoma sets (STAD), ordered as depicted in Figs. 5A, 5B and 5C.
Data S3. p-values for log-rank tests on overall (os) and disease-free survival (dfs) in high- vs low-expressing colorectal adenomcarcinoma (crc), oesophageal carcinoma (eso) and stomach adenocarcinoma sets (stom), ordered as depicted in Figs. 2A, 2E, 3A, 3F, 4A and 4B.
Fig. S4.
A) Disease-free survival analysis for DNAJC18high vs DNAJC18low patients in the esophageal carcinoma dataset. B) Overall survival analysis for CCT6Ahigh vs CCT6Alow patients in the oesophageal carcinoma dataset.
References
- 1.Whitley D., Goldberg S.P., Jordan W.D. Heat shock proteins: a review of the molecular chaperones. J. Vasc. Surg. 1999;29:748–751. doi: 10.1016/s0741-5214(99)70329-0. [DOI] [PubMed] [Google Scholar]
- 2.Li Z., Srivastava P. Heat-shock proteins. Curr. Protoc. Immunol. 2004 doi: 10.1002/0471142735.ima01ts58. (Appendix 1, Appendix 1T) [DOI] [PubMed] [Google Scholar]
- 3.Romanucci M., Bastow T., Della Salda L. Heat shock proteins in animal neoplasms and human tumours--a comparison. Cell Stress Chaperon. 2008;13:253–262. doi: 10.1007/s12192-008-0030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calderwood S.K., Gong J. Heat shock proteins promote cancer: it's a protection racket. Trends Biochem. Sci. 2016;41:311–323. doi: 10.1016/j.tibs.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lianos G.D. The role of heat shock proteins in cancer. Cancer Lett. 2015;360:114–118. doi: 10.1016/j.canlet.2015.02.026. [DOI] [PubMed] [Google Scholar]
- 6.Bhattacharyya A., Chattopadhyay R., Mitra S., Crowe S.E. Oxidative stress: an essential factor in the pathogenesis of gastrointestinal mucosal diseases. Physiol. Rev. 2014;94:329–354. doi: 10.1152/physrev.00040.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dudeja V., Vickers S.M., Saluja A.K. The role of heat shock proteins in gastrointestinal diseases. Gut. 2009;58:1000–1009. doi: 10.1136/gut.2007.140194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kampinga H.H. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperon-. 2009;14:105–111. doi: 10.1007/s12192-008-0068-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu X.B., Shao Y.M., Miao S., Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol. Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radons J. The human HSP70 family of chaperones: where do we stand? Cell Stress Chaperon-. 2016;21:379–404. doi: 10.1007/s12192-016-0676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schopf F.H., Biebl M.M., Buchner J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017;18:345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 12.Whitesell L., Lindquist S.L. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 13.Wei L. Hsp27 participates in the maintenance of breast cancer stem cells through regulation of epithelial-mesenchymal transition and nuclear factor-kappaB. Breast Cancer Res. 2011;13:R101. doi: 10.1186/bcr3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cordonnier T. Hsp27 regulates EGF/beta-catenin mediated epithelial to mesenchymal transition in prostate cancer. Int. J. Cancer. 2015;136:E496–E507. doi: 10.1002/ijc.29122. [DOI] [PubMed] [Google Scholar]
- 15.Concannon C.G., Gorman A.M., Samali A. On the role of Hsp27 in regulating apoptosis. Apoptosis. 2003;8:61–70. doi: 10.1023/a:1021601103096. [DOI] [PubMed] [Google Scholar]
- 16.Cuenda A., Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta. 2007;1773:1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Arrigo A.P., Gibert B. HspB1, HspB5 and HspB4 in human cancers: potent oncogenic role of some of their client proteins. Cancers. 2014;6:333–365. doi: 10.3390/cancers6010333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith H.L., Li W., Cheetham M.E. Molecular chaperones and neuronal proteostasis. Semin. Cell Dev. Biol. 2015;40:142–152. doi: 10.1016/j.semcdb.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sosa V. Oxidative stress and cancer: an overview. Ageing Res. Rev. 2013;12:376–390. doi: 10.1016/j.arr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Pervaiz S., Clement M.V. Superoxide anion: oncogenic reactive oxygen species? Int. J. Biochem. Cell Biol. 2007;39:1297–1304. doi: 10.1016/j.biocel.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Chen Z.X., Pervaiz S. Bcl-2 induces pro-oxidant state by engaging mitochondrial respiration in tumor cells. Cell Death Differ. 2007;14:1617–1627. doi: 10.1038/sj.cdd.4402165. [DOI] [PubMed] [Google Scholar]
- 22.Clement M.V., Hirpara J.L., Pervaiz S. Decrease in intracellular superoxide sensitizes Bcl-2-overexpressing tumor cells to receptor and drug-induced apoptosis independent of the mitochondria. Cell Death Differ. 2003;10:1273–1285. doi: 10.1038/sj.cdd.4401302. [DOI] [PubMed] [Google Scholar]
- 23.Pervaiz S., Cao J., Chao O.S., Chin Y.Y., Clement M.V. Activation of the RacGTPase inhibits apoptosis in human tumor cells. Oncogene. 2001;20:6263–6268. doi: 10.1038/sj.onc.1204840. [DOI] [PubMed] [Google Scholar]
- 24.Hug H. Reactive oxygen intermediates are involved in the induction of CD95 ligand mRNA expression by cytostatic drugs in hepatoma cells. J. Biol. Chem. 1997;272:28191–28193. doi: 10.1074/jbc.272.45.28191. [DOI] [PubMed] [Google Scholar]
- 25.Ahmad K.A., Clement M.V., Hanif I.M., Pervaiz S. Resveratrol inhibits drug-induced apoptosis in human leukemia cells by creating an intracellular milieu nonpermissive for death execution. Cancer Res. 2004;64:1452–1459. doi: 10.1158/0008-5472.can-03-2414. [DOI] [PubMed] [Google Scholar]
- 26.Zito E. ERO1: a protein disulfide oxidase and H2O2 producer. Free Radic. Biol. Med. 2015;83:299–304. doi: 10.1016/j.freeradbiomed.2015.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Gross E. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc. Natl. Acad. Sci. USA. 2006;103:299–304. doi: 10.1073/pnas.0506448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chong W.C., Shastri M.D., Eri R. Endoplasmic reticulum stress and oxidative stress: a vicious nexus implicated in bowel disease pathophysiology. Int. J. Mol. Sci. 2017;18 doi: 10.3390/ijms18040771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minisini M.P., Kantengwa S., Polla B.S. DNA damage and stress protein synthesis induced by oxidative stress proceed independently in the human premonocytic line U937. Mutat. Res. 1994;315:169–179. doi: 10.1016/0921-8777(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 30.Jakob U., Muse W., Eser M., Bardwell J.C. Chaperone activity with a redox switch. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 31.Ahn S.G., Thiele D.J. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 2003;17:516–528. doi: 10.1101/gad.1044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao J. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6 doi: 10.1126/scisignal.2004088. (pl1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerami E. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen B. Computational discovery of niclosamide ethanolamine, a repurposed drug candidate that reduces growth of hepatocellular carcinoma cells in vitro and in mice by inhibiting cell division cycle 37 signaling. Gastroenterology. 2017;152:2022–2036. doi: 10.1053/j.gastro.2017.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen R. A meta-analysis of lung cancer gene expression identifies PTK7 as a survival gene in lung adenocarcinoma. Cancer Res. 2014;74:2892–2902. doi: 10.1158/0008-5472.CAN-13-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butte A.J., Ito S. Translational bioinformatics: data-driven drug discovery and development. Clin. Pharmacol. Ther. 2012;91:949–952. doi: 10.1038/clpt.2012.55. [DOI] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Research N. Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pohl S.O., Agostino M., Dharmarajan A., Pervaiz S. Cross talk between cellular redox state and the antiapoptotic protein Bcl-2. Antioxid. Redox Signal. 2018 doi: 10.1089/ars.2017.7414. [DOI] [PubMed] [Google Scholar]
- 41.Subramanian A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mootha V.K. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 43.Ashburner M. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Supek F., Bosnjak M., Skunca N., Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlicker A., Domingues F.S., Rahnenfuhrer J., Lengauer T. A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinform. 2006;7:302. doi: 10.1186/1471-2105-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez I. Hic-5 mediates TGFbeta-induced adhesion in vascular smooth muscle cells by a Nox4-dependent mechanism. Arterioscler. Thromb. Vasc. Biol. 2015;35:1198–1206. doi: 10.1161/ATVBAHA.114.305185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gil Lorenzo A.F., Bocanegra V., Benardon M.E., Cacciamani V., Valles P.G. Hsp70 regulation on Nox4/p22phox and cytoskeletal integrity as an effect of losartan in vascular smooth muscle cells. Cell Stress Chaperon-. 2014;19:115–134. doi: 10.1007/s12192-013-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Desai L.P. Negative regulation of NADPH oxidase 4 by hydrogen peroxide-inducible clone 5 (Hic-5) protein. J. Biol. Chem. 2014;289:18270–18278. doi: 10.1074/jbc.M114.562249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muller M., Gauley J., Heikkila J.J. Hydrogen peroxide induces heat shock protein and proto-oncogene mRNA accumulation in Xenopus laevis A6 kidney epithelial cells. Can. J. Physiol. Pharmacol. 2004;82:523–529. doi: 10.1139/y04-059. [DOI] [PubMed] [Google Scholar]
- 50.Courgeon A.M., Rollet E., Becker J., Maisonhaute C., Best-Belpomme M. Hydrogen peroxide (H2O2) induces actin and some heat-shock proteins in Drosophila cells. Eur. J. Biochem. 1988;171:163–170. doi: 10.1111/j.1432-1033.1988.tb13772.x. [DOI] [PubMed] [Google Scholar]
- 51.Motoyama S. Hydrogen peroxide induces midzonal heat shock protein 72 and apoptosis in sinusoidal endothelial cells of hypoxic rat liver. Crit. Care Med. 2000;28:1509–1514. doi: 10.1097/00003246-200005000-00042. [DOI] [PubMed] [Google Scholar]
- 52.Afolayan A.J. Inducible HSP70 regulates superoxide dismutase-2 and mitochondrial oxidative stress in the endothelial cells from developing lungs. Am. J. Physiol. Lung Cell Mol. Physiol. 2014;306:L351–L360. doi: 10.1152/ajplung.00264.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arrigo A.P. Hsp27: novel regulator of intracellular redox state. IUBMB Life. 2001;52:303–307. doi: 10.1080/152165401317291156. [DOI] [PubMed] [Google Scholar]
- 54.Souren J.E., Van Aken H., Van Wijk R. Enhancement of superoxide production and protection against heat shock by HSP27 in fibroblasts. Biochem. Biophys. Res. Commun. 1996;227:816–821. doi: 10.1006/bbrc.1996.1590. [DOI] [PubMed] [Google Scholar]
- 55.Mellier G., Liu D., Bellot G., Holme A.L., Pervaiz S. Small molecule sensitization to TRAIL is mediated via nuclear localization, phosphorylation and inhibition of chaperone activity of Hsp27. Cell Death Dis. 2013;4:e890. doi: 10.1038/cddis.2013.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shenoy K., Wu Y., Pervaiz S. LY303511 enhances TRAIL sensitivity of SHEP-1 neuroblastoma cells via hydrogen peroxide-mediated mitogen-activated protein kinase activation and up-regulation of death receptors. Cancer Res. 2009;69:1941–1950. doi: 10.1158/0008-5472.CAN-08-1996. [DOI] [PubMed] [Google Scholar]
- 57.Baylin S.B. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005;2(Suppl 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 58.Esteve P.O. Binding of 14-3-3 reader proteins to phosphorylated DNMT1 facilitates aberrant DNA methylation and gene expression. Nucleic Acids Res. 2016;44:1642–1656. doi: 10.1093/nar/gkv1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pencheva N. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151:1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ray M.R. Cyclin G-associated kinase: a novel androgen receptor-interacting transcriptional coactivator that is overexpressed in hormone refractory prostate cancer. Int. J. Cancer. 2006;118:1108–1119. doi: 10.1002/ijc.21469. [DOI] [PubMed] [Google Scholar]
- 61.Lee D.W., Zhao X., Yim Y.I., Eisenberg E., Greene L.E. Essential role of cyclin-G-associated kinase (Auxilin-2) in developing and mature mice. Mol. Biol. Cell. 2008;19:2766–2776. doi: 10.1091/mbc.E07-11-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Floyd S., De Camilli P. Endocytosis proteins and cancer: a potential link? Trends Cell Biol. 1998;8:299–301. doi: 10.1016/s0962-8924(98)01316-6. [DOI] [PubMed] [Google Scholar]
- 63.Li J., Xu W., Liu F., Huang S., He M. GSTM1 polymorphism contribute to colorectal cancer in Asian populations: a prospective meta-analysis. Sci. Rep. 2015;5:12514. doi: 10.1038/srep12514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Khabaz M.N. GSTM1 gene polymorphism and the risk of colorectal cancer in a Saudi Arabian population. Genet Mol. Res. 2016;15 doi: 10.4238/gmr.15017551. [DOI] [PubMed] [Google Scholar]
- 65.Vyas S., Zaganjor E., Haigis M.C. Mitochondria and cancer. Cell. 2016;166:555–566. doi: 10.1016/j.cell.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coghlin C. Characterization and over-expression of chaperonin t-complex proteins in colorectal cancer. J. Pathol. 2006;210:351–357. doi: 10.1002/path.2056. [DOI] [PubMed] [Google Scholar]
- 67.Martin T.D. A role for mitochondrial translation in promotion of viability in K-Ras mutant cells. Cell Rep. 2017;20:427–438. doi: 10.1016/j.celrep.2017.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiong T. An esophageal squamous cell carcinoma classification system that reveals potential targets for therapy. Oncotarget. 2017;8:49851–49860. doi: 10.18632/oncotarget.17989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ostermann J., Horwich A.L., Neupert W., Hartl F.U. Protein folding in mitochondria requires complex formation with hsp60 and ATP hydrolysis. Nature. 1989;341:125–130. doi: 10.1038/341125a0. [DOI] [PubMed] [Google Scholar]
- 70.Sarangi U. Hsp60 chaperonin acts as barrier to pharmacologically induced oxidative stress mediated apoptosis in tumor cells with differential stress response. Drug Target Insights. 2013;7:35–51. doi: 10.4137/DTI.S12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiechmann K. Mitochondrial chaperonin HSP60 Is the apoptosis-related target for myrtucommulone. Cell Chem. Biol. 2017;24:614–623. doi: 10.1016/j.chembiol.2017.04.008. (e616) [DOI] [PubMed] [Google Scholar]
- 72.Hambright W.S., Fonseca R.S., Chen L., Na R., Ran Q. Ablation of ferroptosis regulator glutathione peroxidase 4 in forebrain neurons promotes cognitive impairment and neurodegeneration. Redox Biol. 2017;12:8–17. doi: 10.1016/j.redox.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Imai H. Depletion of selenoprotein GPx4 in spermatocytes causes male infertility in mice. J. Biol. Chem. 2009;284:32522–32532. doi: 10.1074/jbc.M109.016139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hung M.C., Link W. Protein localization in disease and therapy. J. Cell Sci. 2011;124:3381–3392. doi: 10.1242/jcs.089110. [DOI] [PubMed] [Google Scholar]
- 75.Mokranjac D., Sichting M., Neupert W., Hell K. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 2003;22:4945–4956. doi: 10.1093/emboj/cdg485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fujibayashi A. Human RME-8 is involved in membrane trafficking through early endosomes. Cell Struct. Funct. 2008;33:35–50. doi: 10.1247/csf.07045. [DOI] [PubMed] [Google Scholar]
- 77.Girard M., McPherson P.S. RME-8 regulates trafficking of the epidermal growth factor receptor. FEBS Lett. 2008;582:961–966. doi: 10.1016/j.febslet.2008.02.042. [DOI] [PubMed] [Google Scholar]
- 78.Qiu X. Overexpression of CCT8 and its significance for tumor cell proliferation, migration and invasion in glioma. Pathol. Res. Pract. 2015;211:717–725. doi: 10.1016/j.prp.2015.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Correlation co-efficients for paired gene co-expression in all colorectal adenocarcinoma (COAD), oesophageal carcinoma (ESCA) and stomach adenocarcinoma (STAD) sets, ordered as depicted in Figs. 1A, 1C, and 1E.
Data S2. Correlation co-efficients between methylation and gene expression for patients at progressing stages of colorectal adenocarcinoma (COAD), oesophageal carcinoma (ESCA) and stomach adenocarcinoma sets (STAD), ordered as depicted in Figs. 5A, 5B and 5C.
Data S3. p-values for log-rank tests on overall (os) and disease-free survival (dfs) in high- vs low-expressing colorectal adenomcarcinoma (crc), oesophageal carcinoma (eso) and stomach adenocarcinoma sets (stom), ordered as depicted in Figs. 2A, 2E, 3A, 3F, 4A and 4B.