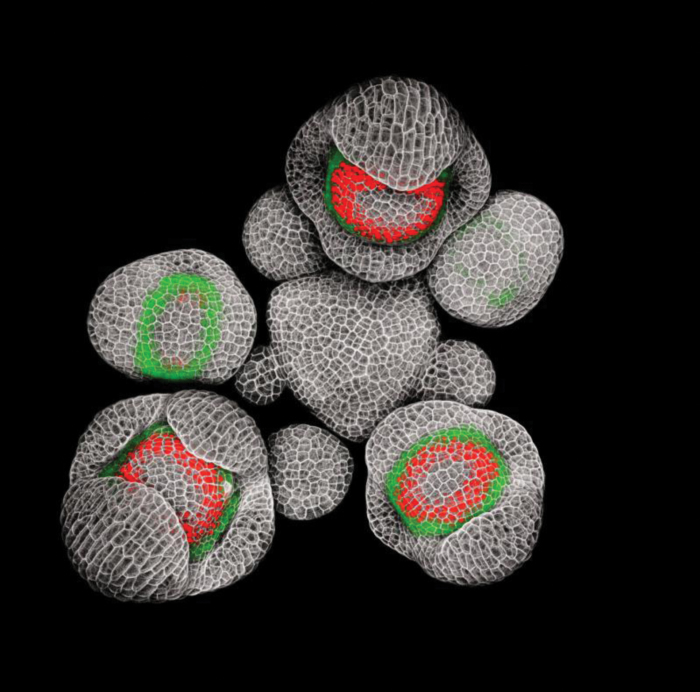
When asked to provide a picture for the cover of the Flowering Newsletter, I picked this image of an Arabidopsis thaliana inflorescence expressing fluorescent reporters for two key regulators of flower development: APETALA3 (AP3), which promotes petal and stamen identity, and SUPERMAN (SUP), which encodes a transcriptional repressor that defines the boundary between stamens and pistil (Fig. 1). The choice was easy: it was an important breakthrough in my research on the role of SUP in the separation of stamens in whorl 3 and carpels in whorl 4; and among the images of flowers I have taken with a confocal microscope, it is also one my favourites aesthetically. The image won awards at the 2015 Nikon Small World and FASEB BioArt competitions and is published in Prunet et al. (2017).
Fig. 1.
AP3 and SUP expression in young Arabidopsis flower buds. Arabidopsis inflorescence expressing gAP3-GFP (green) and gSUP-3xVenusN7 (red) fluorescent reporters. Cell walls were stained with propidium iodide (grey). Siliques and older flower buds were removed, and the inflorescence was prepared and imaged on a Zeiss LSM780 with a 20× water-dipping lens as described in Prunet (2017) and Prunet et al. (2016). Background noise was digitally removed for aesthetic reasons.
The molecular mechanisms underlying the determination of floral organ identity have been extensively studied over the last three decades, from the description of mutants with floral organ homeosis (Bowman et al., 1989, 1991; Irish and Sussex, 1990) to the characterization of the corresponding genes, most of which encode transcription factors of the MADS-box family (Yanofsky et al., 1990; Jack et al., 1992; Mandel et al., 1992; Goto and Meyerowitz, 1994), and the identification of their targets (Kaufmann et al., 2009, 2010; Wuest et al., 2012; Ó’Maoiléidigh et al., 2013). Floral organ identity is determined by the combinatorial action of four classes of MADS-box transcription factors [class A, AP1; class B, AP3 and PISTILLATA (PI); class C, AGAMOUS (AG); and class E, SEPALLATAs (SEPs)], which form different protein quartets in each whorl (reviewed in Prunet and Jack, 2014). For instance, quartets composed of class B, C, and E transcription factors orchestrate stamen development in whorl 3, while quartets composed of class C and E transcription factors alone determine carpel identity in whorl 4. These quartets recruit different transcription co-regulators and histone modification factors to regulate the transcription of their targets (Smaczniak et al., 2012). While the genetic networks downstream of these quartets have been partially deciphered (reviewed in Stewart et al., 2016), questions remain about how boundaries between floral whorls are established.
Mutations in SUP disrupt the boundary between whorls 3 and 4, with the formation of numerous extra stamens, usually at the expense of carpels, which are reduced or missing in most alleles (Schultz et al., 1991; Bowman et al., 1992). This phenotype is associated with the expansion of the expression of AP3 and PI towards the center of the flower (Bowman et al., 1992; Goto and Meyerowitz, 1994), but does not result from a simple homeotic conversion of carpels into stamens: the overall number of floral organs is increased in sup compared to the wild type, indicating an excess of cell proliferation in sup flowers. Two models have been proposed for the developmental origin of the extra stamens in sup flowers. It was first suggested that these extra stamens form in whorl 4, due to the ectopic expression of class B genes, and that the increase in floral organ number comes from delayed termination of the floral stem cells (Schultz et al., 1991; Bowman et al., 1992). However, when the SUP gene was identified, in situ hybridization experiments suggested that SUP was co-expressed with AP3 and PI in the inner part of whorl 3, but not expressed in whorl 4, casting doubts on the fact that SUP might function to prevent ectopic expression of class B genes in the fourth whorl (Sakai et al., 1995). Instead, SUP was proposed to control the balance of cell proliferation between whorls 3 and 4. According to this new model, extra stamens arise from whorl 3 cells that over-proliferate, while reduced proliferation in whorl 4 results in a loss of carpel tissue (Sakai et al., 1995, 2000). For more than 25 years after the isolation of the sup mutant it had not been possible to discriminate between these two models. This was mostly due to limitations in the techniques that were used at the time, such as in situ hybridizations or GUS reporter lines, which lack sufficient cellular resolution and cannot not be used on live tissues. The image I have chosen helped solve this question.
I first became interested in SUP during my PhD with Christophe Trehin and Ioan Negrutiu at École Normale Supérieure de Lyon. I was studying three different mutants with a minor delay in the termination of floral stem cells that was manifesting through a slight increase in the number of carpels and the occasional formation of extra organs inside the gynoecium (Prunet et al., 2008). This phenotype was correlated with a decrease in the expression of AG—which acts as the main switch to terminate floral stem cells (Lenhard et al., 2001; Lohmann et al., 2001)—in the center of the flower meristem (Prunet et al., 2008). However, the combination of these three mutations resulted in a spectacular phenotype, with the formation of an indeterminate spiral of stamens at the center of the flower. This phenotype is also observed when combining the sup-1 mutation with the moderate loss-of-function allele ag-4 (Prunet et al., 2008). While SUP initially appeared at the margin of the genetic networks I was studying, I started to increasingly suspect that it was involved in the timely termination of floral stem cells.
When I started my postdoc in Tom Jack’s lab at Dartmouth College, I decided to investigate the function of SUP using a live confocal imaging approach—a technique that allows us to monitor the expression of multiple genes in live tissue with good cellular resolution. Our data supported the model in which extra stamens in sup mutant flowers arise from whorl 4 rather than whorl 3. We observed a prolonged expression of the stem cell marker CLAVATA3 and stem cell activator WUSCHEL in sup flowers compared to the wild type, suggesting that the increase in floral organ number resulted from delayed termination of the floral stem cells rather than from an over-proliferation of cells in whorl 3 (Prunet et al., 2017). Time-lapse experiments also demonstrated that a ring of cells in whorl 4, adjacent to the boundary with whorl 3, starts expressing AP3 ectopically at the transition between whorl 4 and 5 in sup mutant flowers, thus confirming that extra stamens form in the fourth whorl in sup (Prunet et al., 2017). Our data also seemed to point at a mostly non cell-autonomous effect of SUP, which, based on hard-to-interpret in situ hybridizations, was believed to be expressed in whorl 3, and not in whorl 4 (Sakai et al., 1995). We generated a translational fluorescent reporter for SUP to have a closer look at the SUP expression pattern. It turned out to be a slow and painful process—it took 4 years and some pretty acrobatic cloning by Tom—but we finally obtained a fluorescent SUP reporter just as I moved from Dartmouth to Elliot Meyerowitz’s lab at Caltech. This image of an Arabidopsis inflorescence expressing two translational reporters for AP3 (fused with a single GFP) and SUP (fused with three Venus proteins and a nuclear localization signal) was one of the first images I took at Caltech; it was also the first time I managed to separate signals from GFP and YFP, which have partially overlapping emission spectra. But most importantly, this image clearly showed that contrary to what was previously thought, SUP is expressed on both sides of the boundary between whorls 3 and 4, not just in whorl 3. SUP and AP3 are expressed along two opposite gradients that only partially overlap in whorl 3, and whorl 4 cells that express SUP in wild-type flowers at stage 5 ectopically express AP3 instead in sup mutant flowers, indicating that SUP prevents AP3 expression in whorl 4 in a cell-autonomous manner (Prunet et al., 2017).
Independently of the scientific significance of this image, I love it for aesthetic reasons. One of the reasons why I studied biology in the first place is that of all sciences, it leaves the most room for artistic expression: observational drawing is an integral part of the learning process. This science-meets-art aspect—for which the term SciArt has been coined—has long been an important driver for my work. I chose to study development for my PhD because of the rich imaging possibilities this field offers. I later based my postdoc research on a confocal imaging approach for the power of the technique to solve developmental questions but also for the beauty of the images that can be generated. I admit to spending more time on the microscope than strictly required to answer my initial scientific questions, trying to get aesthetically perfect images (and I consider myself lucky to work with Elliot, who has been very supportive of that). But then, as Samuel H. Scudder noticed once he decided to draw his fish (‘At last a happy thought struck me—I would draw the fish; and now with surprise I began to discover new features in the creature. Just then the Professor returned. ‘That is right’, said he; ‘a pencil is one of the best of eyes’; Scudder, 1974), carefully crafted images often bring to our attention interesting biological details that we would not have suspected otherwise.
Acknowledgements
I would like to thank Tom Jack and Elliot Meyerowitz for their support. Funding in Tom’s lab was supported by the National Science Foundation Grant IOS-0926347; and funding in the Elliot’s laboratory was provided by the Howard Hughes Medical Institute, the National Institutes of Health Grant R01 GM104244, and the Gordon and Betty Moore Foundation through Grant GBMF3406.
References
- Bowman JL, Sakai H, Jack T, Weigel D, Mayer U, Meyerowitz EM. 1992. SUPERMAN, a regulator of floral homeotic genes in Arabidopsis. Development 114, 599–615. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. 1989. Genes directing flower development in Arabidopsis. The Plant Cell 1, 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Smyth DR, Meyerowitz EM. 1991. Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20. [DOI] [PubMed] [Google Scholar]
- Goto K, Meyerowitz EM. 1994. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes & Development 8, 1548–1560. [DOI] [PubMed] [Google Scholar]
- Irish VF, Sussex IM. 1990. Function of the apetala-1 gene during Arabidopsis floral development. The Plant Cell 2, 741–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack T, Brockman LL, Meyerowitz EM. 1992. The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68, 683–697. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Muiño JM, Jauregui R, Airoldi CA, Smaczniak C, Krajewski P, Angenent GC. 2009. Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biology 7, e1000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muiño JM et al. . 2010. Orchestration of floral initiation by APETALA1. Science 328, 85–89. [DOI] [PubMed] [Google Scholar]
- Lenhard M, Bohnert A, Jürgens G, Laux T. 2001. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105, 805–814. [DOI] [PubMed] [Google Scholar]
- Lohmann JU, Hong RL, Hobe M, Busch MA, Parcy F, Simon R, Weigel D. 2001. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793–803. [DOI] [PubMed] [Google Scholar]
- Mandel MA, Gustafson-Brown C, Savidge B, Yanofsky MF. 1992. Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273–277. [DOI] [PubMed] [Google Scholar]
- Ó’Maoiléidigh DS, Wuest SE, Rae L et al. . 2013. Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. The Plant Cell 25, 2482–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N. 2017. Live confocal imaging of developing Arabidopsis flowers. Journal of Visualized Experiments 122, e55156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N, Jack TP. 2014. Flower development in Arabidopsis: there is more to it than learning your ABCs. Methods in Molecular Biology 1110, 3–33. [DOI] [PubMed] [Google Scholar]
- Prunet N, Jack TP, Meyerowitz EM. 2016. Live confocal imaging of Arabidopsis flower buds. Developmental Biology 419, 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N, Morel P, Thierry AM, Eshed Y, Bowman JL, Negrutiu I, Trehin C. 2008. REBELOTE, SQUINT, and ULTRAPETALA1 function redundantly in the temporal regulation of floral meristem termination in Arabidopsis thaliana. The Plant Cell 20, 901–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunet N, Yang W, Das P, Meyerowitz EM, Jack TP. 2017. SUPERMAN prevents class B gene expression and promotes stem cell termination in the fourth whorl of Arabidopsis thaliana flowers. Proceedings of the National Academy of Sciences, USA 114, 7166–7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Krizek BA, Jacobsen SE, Meyerowitz EM. 2000. Regulation of SUP expression identifies multiple regulators involved in arabidopsis floral meristem development. The Plant Cell 12, 1607–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Medrano LJ, Meyerowitz EM. 1995. Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378, 199–203. [DOI] [PubMed] [Google Scholar]
- Schultz EA, Pickett FB, Haughn GW. 1991. The FLO10 gene product regulates the expression domain of homeotic genes AP3 and PI in Arabidopsis flowers. The Plant Cell 3, 1221–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scudder SH. 1974. In the laboratory with Agassiz. Every Saturday 16, 369–370. [Google Scholar]
- Smaczniak C, Immink RG, Muino JM et al. . 2012. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proceedings of the National Academy of Sciences, USA 109, 1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart D, Graciet E, Wellmer F. 2016. Molecular and regulatory mechanisms controlling floral organ development. The FEBS Journal 283, 1823–1830. [DOI] [PubMed] [Google Scholar]
- Wuest SE, O’Maoileidigh DS, Rae L, Kwasniewska K, Raganelli A, Hanczaryk K, Lohan AJ, Loftus B, Graciet E, Wellmer F. 2012. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proceedings of the National Academy of Sciences, USA 109, 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. 1990. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. [DOI] [PubMed] [Google Scholar]