Pentatricopeptide repeat protein DEK40 functions in processing of cox3, nad2, and nad5 transcripts and is required for mitochondrial function in maize.
Keywords: Kernel development, maize, mitochondrion, pentatricopeptide repeat protein, RNA editing
Abstract
Pentatricopeptide repeat (PPR) proteins are one of the largest protein families, which consists of >400 members in most species. However, the molecular functions of many PPR proteins are still uncharacterized. Here, we isolated a maize mutant, defective kernel 40 (dek40). Positional cloning, and genetic and molecular analyses revealed that DEK40 encodes a new E+ subgroup PPR protein that is localized in the mitochondrion. DEK40 recognizes and directly binds to cox3, nad2, and nad5 transcripts and functions in their processing. In the dek40 mutant, abolishment of the C-to-U editing of cox3-314, nad2-26, and nad5-1916 leads to accumulated reactive oxygen species and promoted programmed cell death in endosperm cells due to the dysfunction of mitochondrial complexes I and IV. Furthermore, RNA sequencing analysis showed that gene expression in some pathways, such as glutathione metabolism and starch biosynthesis, was altered in the dek40 mutant compared with the wild-type control, which might be involved in abnormal development of the maize mutant kernels. Thus, our results provide solid evidence on the molecular mechanism underlying RNA editing by DEK40, and extend our understanding of PPR-E+ type protein in editing functions and kernel development in maize.
Introduction
RNA editing mediated by organellar genes at the transcript level is an essential step in the production of functional proteins (Castandet and Araya, 2011). Most RNA editing events are conversions of cytidines (C) to uridines (U) in mitochondria or plastids (Ichinose and Sugita, 2017). These RNA modification processes are mostly regulated by nuclear-encoded factors (Takenaka et al., 2013b; Ichinose and Sugita, 2017). Interestingly, many RNA-binding proteins, such as pentatricopeptide repeat (PPR) proteins, function in RNA post-transcriptional processes in organelles (Fujii and Small, 2011; Barkan and Small, 2014).
PPR proteins are characterized by tandem repeats of a highly degenerate 35 amino acid motif (Small and Peeters, 2000). They are found in all eukaryotes, but their numbers are greatly increased in land plants (Schmitz-Linneweber and Small, 2008; Takenaka et al., 2014). Based on the structure of their PPR motifs, PPR proteins are divided into two categories, the P subfamily and the PLS subfamily (Lurin et al., 2004). The PLS subfamily proteins are further divided into four subgroups, PLS, E, E+, and DYW, according to their C-terminal motifs (Lurin et al., 2004). The PPR motif contains two antiparallel α-helices (helix A and helix B), which interact to form a superhelix as a helix–turn–helix motif (Small and Peeters, 2000; Yin et al., 2013). RNA substrates of PPR proteins can be identified in a motif sequence-specific manner. Positions 6 and 1′ (position 1 in the next motif) or positions 4′ and 34′ in the PPR motif are responsible for RNA binding and nucleotide recognition (Barkan et al., 2012; Takenaka et al., 2013a; Yin et al., 2013). All three types of PPR proteins, the E, E+, and DYW subgroups, are involved in mitochondrial or plastid RNA editing (Fujii and Small, 2011; Barkan and Small, 2014; Li et al., 2014; Sun et al., 2015; Yang et al., 2017; Li et al., 2019). Defects in these proteins lead to phenotypes associated with organelle dysfunction, such as embryo developmental arrest, embryo lethality, albino plants, cytoplasmic male sterility, and poor growth (Saha et al., 2007; Schmitz-Linneweber and Small, 2008).
The plant respiratory chain is composed of four main complexes and an ATP synthase (Dudkina et al., 2006). These four main complexes comprise complex I (NADH-ubiquinone oxidoreductase), complex II (succinate-ubiquinone oxidoreductase), complex III (ubiquinol-cytochrome c oxidoreductase), and complex IV (cytochrome c-O2 oxidoreductase). In the mitochondrial electron transport chain (ETC), complexes I and III can catalyze reactive oxygen species (ROS) formation under physiological and pathological conditions (Blokhina and Fagerstedt, 2010; Gill and Tuteja, 2010). Impaired complex I activity in the mitochondrial ETC results in redox imbalance and increased ROS accumulation (Liu et al., 2010; Xie et al., 2016; Lee et al., 2017). The defective cytochrome c pathway in the ppr40 mutant enhanced ROS accumulation (Zsigmond et al., 2008). ROS not only are important signaling molecules that regulate and coordinate vital processes including growth, the cell cycle, programmed cell death (PCD), abiotic stress responses, defense, systemic signaling, and development, but they also control protein stability and gene expression (Laloi et al., 2004; Gechev et al., 2006). Besides the cytochrome oxidase (COX) pathway, the alternative oxidase (AOX) pathway represents an alternative electron transport pathway in mitochondrial ETC (Vanlerberghe, 2013). When electron transport in the cytochrome c pathway is blocked, mitochondrial dysfunction signaling up-regulates the expression of AOX genes, which regulate the ROS levels initiated by mitochondrial retrograde signaling (Zsigmond et al., 2008).
In this study, DEK40, a new maize E+ type PPR protein was identified, which directly binds to cox3, nad2, and nad5 transcripts. Its dysfunction abolished the C-to-U editing of cox3-314, nad2-26, and nad5-1916, impaired mitochondrial complex I and IV activity, and promoted ROS accumulation and PCD of endosperm cells. Gene expression profile analysis demonstrated that some biosynthesis and metabolism pathways were altered in the dek40 mutant, for example glutathione metabolism and starch biosynthesis pathways, which might be involved in abnormal development of the maize kernels.
Materials and methods
Plant materials
The maize dek40-reference (dek40-ref) mutant was isolated from an ethyl methanesulfonate (EMS) population (in the B73 genetic background). The second allele dek40-1 was obtained from the MEMD website (Maize EMS-induced Mutant Database, http://www.elabcaas.cn/memd/; last accessed 8 September 2019) (Lu et al., 2018). The dek40-ref allele was maintained in heterozygotes and the dek40-ref mutant was backcrossed to B73 to generate BC3F1; BC3F2 seeds were used for further experiments. The dek40-ref heterozygotes were used as male parents in crosses with four inbred lines, Mo17, V3b1, Chang7-2, and S162. The wild-type (WT) plants used in this study were siblings of the mutant from self-pollinated segregating ears. Arabidopsis (Arabidopsis thaliana) line MT-GK (CS16263) used for subcellular localization analysis of DEK40 was obtained from the Arabidopsis Biological Resource Center.
Histological analysis
Kernels at different days after pollination (DAP) were cut longitudinally into three equal parts. The central part containing the intact embryo was fixed with formalin–acetic acid–alcohol (FAA) solution (50 ml of 100% ethanol, 10 ml of glacial acetic acid, 5 ml of 37% formaldehyde, and 35 ml of ddH2O) for 12–14 h at 4 °C. The fixed material was dehydrated in a graded ethanol series (50, 70, 85, 95, and 100%), which was subsequently replaced with xylene, and infiltrated with paraffin. The samples were embedded and cut into 12 μm sections using an RM2235 microtome (Leica, Wetzlar, Germany). The sections were stained with 0.1% (w/v) Toluidine Blue O (Sigma-Aldrich, USA). Finally, photographs were taken using a microscope (Olympus BX51).
TEM and SEM
For TEM analysis, immature WT and dek40-ref endosperm at 11 and 13 DAP was collected from the same ear, cut into small pieces, and fixed with 2.5% glutaraldehyde and 1% osmium tetroxide at 4 °C for 12 h. The samples were subsequently washed five times with phosphate-buffered saline (PBS; pH 7.2). After dehydration in an ethanol gradient, they were transferred to propylene epoxide and gradually infiltrated with acrylic resin. Ultrathin sections of the samples were cut with a diamond knife and photomicrographs were taken using a transmission electron microscope (JEM-1400Plus) at Shandong Agricultural University.
For SEM analysis, mature WT and dek40-ref seeds were harvested from the same ear and cut longitudinally with a knife. The samples were vacuum-dried, spray-coated with gold, and observed using a scanning electron microscope (JSM-6610LV) at Shandong Agricultural University.
Map-based cloning
The Dek40 locus was mapped using 702 mutant individuals from an F2 mapping population of the cross between dek40-ref heterozygotes and the S162 inbred line. For preliminary mapping, >100 polymorphic simple sequence repeat (SSR) markers evenly distributed over the whole genome were selected to analyze both parents and F2 mutant kernels. For fine mapping, molecular markers were developed based on single nucleotide polymorphisms (SNPs), SSRs, and Indels. The molecular markers narrowed the Dek40 locus down to a 300 kb region. Based on candidate gene annotation in this region, DNA fragments were amplified from 10 dek40-ref and WT kernels using KOD DNA polymerase (Toyobo) and sequenced. The primer sequences are listed in Supplementary Table S4 at JXB online.
Phylogenetic analysis
All the sequences were aligned with the MAFFT program (Katoh et al., 2002). The phylogenetic trees were reconstructed by the maximum likelihood (ML) method implemented in IQ-TREE, with the best-fit model automatically selected by ModelFinder. Support for the inferred ML tree was obtained by ultrafast bootstrap approximation (UFBoot) with 10 000 replicates (Nguyen et al., 2015; Kalyaanamoorthy et al., 2017). Figtree was used for visualization (Figtree v1.4.4 visualization; http://tree.bio.ed.ac.uk/software/figtree/; last accessed 8 September 2019).
DNA isolation and RNA extraction
Genomic DNA (gDNA) for each sample was isolated from mature maize kernels without pericarp using the GMO Crop Extraction & Amplification kit (Tiangen, Beijing, China) following the manufacturer’s instructions. Total RNA of developing kernels was extracted using a cetyltrimethylammonium bromide (CTAB)-based method (Gambino et al., 2008). Total RNA of other samples was extracted using the Trizol reagent (Ambion; Cat. No. 15596-026).
Reverse transcription-PCR (RT–PCR) and quantitative RT–PCR (qRT–PCR)
Total RNA was treated with gDNA buffer to remove residual DNA contamination and reverse transcribed to cDNA using a FastQuant RT Kit (Tiangen). RT–PCR was used to examine RNA editing, and the products were introduced into the pEAZY-Blunt3 Cloning vector (Tiangen) and directly sequenced. Amplification of mitochondrial transcripts was performed using primers as described previously (Liu et al., 2013).
qRT–PCR was performed with cDNA dilutions using SuperReal PreMix Plus (Tiangen) with a LightCycler 96 (Roche Diagnostics) in a 20 µl reaction volume with 45 cycles. The RNA levels were normalized to the maize ZmActin gene (GRMZM2G126010) with three biological replicates, and three technical replicates each. The primers used for qRT–PCR are listed in Supplementary Table S4.
Subcellular localization
The full-length coding sequence of Dek40 without the termination codon was amplified with specific primers and cloned into the pROKII-GFP vector to generate a green fluorescent protein (GFP) fusion product. The modified subcellular localization assays were conducted according to the method described by Yoo et al. (2007). Briefly, 0.5–1 mm leaf strips were cut from etiolated leaves at 14 d after sowing and digested with 1.5% (w/v) cellulase R10 and 0.5% (w/v) macerozyme R10 (Yakult Pharmaceutical Ind. Co., Ltd, Japan). The digestion continued for 5 h at room temperature with 50 rpm shaking in the dark. The DEK40–GFP fusion products were transfected into maize protoplasts by polyethylene glycol/calcium-mediated transformation (Yoo et al., 2007). The protoplasts were incubated for 14–18 h in the dark at 23 °C. After incubation, the protoplasts were treated with 500 nM of the mitochondrion-selective dye Mito Tracker Red CMX-Ros (Invitrogen) for 30 min at 37 °C, and washed with W5 solution three times. DEK40–GFP and Mito Tracker Red CMX-Ros were excited at 488 nm and 561 nm, and emissions were collected at 505–550 nm and 575–615 nm, respectively. Images were captured using a Leica TCS SP5 II laser scanning confocal microscope. The subcellular localization of DEK40–red fluorescent protein (RFP) in the Arabidopsis marker line for mitochondria (MT-GK) was performed as described previously (Zhang et al., 2018). Primer sequences are listed in Supplementary Table S4.
Measurement of ROS in endosperms
Developing WT and dek40-ref mutant kernels were isolated from self-pollinated segregating ears at 11, 13, and 15 DAP, and used to measure ROS in the endosperms. For 3,3'-diaminobenzidine (DAB) staining to detect hydrogen peroxide (H2O2), the kernels were cut longitudinally into two equal parts and placed in 1 mg ml−1 DAB (Sigma-Aldrich) in 50 mM Tris–HCl buffer (pH 5.0) in the dark for 12 h. For nitroblue tetrazolium chloride (NBT) staining to detect superoxides, the kernels were cut, immersed in a 0.5 mg ml−1 NBT (Sigma-Aldrich) solution in 10 mM potassium phosphate buffer (pH 7.6), and incubated in complete darkness at room temperature for 2 h. After each assay, the kernels were fixed in 70% (v/v) ethanol at room temperature. Photographs were taken using an Olympus DP72 microscope. The relative DAB and NBT staining intensities were analyzed with the Image J software.
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assays
The TUNEL assay was performed as described previously (Wang et al., 2014). Kernels were fixed in formaldehyde solution overnight at 4 °C, embedded in paraffin after dehydration, and sectioned at 12 µm thickness. The sections were dewaxed in xylene, rehydrated in an ethanol series, treated with 20 μg ml−1 Proteinase K (Gibco, Carlsbad, CA, USA) in Proteinase K buffer (10 mM Tris–HCl and 5 mM EDTA, pH 7.5) for 15 min at 37 °C, and washed with PBS solution twice. The samples were incubated in a mixture of fluorescein-labeled deoxynucleotides and TdT (TUNEL mix) at 37 °C for 1 h. After incubation, the slides were washed with PBS solution, treated with 10 μg ml–1 propidium iodide (PI) for 15 min at 37 °C, and washed again with PBS solution. The samples were observed using a Leica TCS SP5 II laser scanning confocal microscope.
RNA EMSAs
The full-length coding sequence of Dek40 was amplified with specific primers and cloned into the pGEX-4T-1 vector to generate recombinant DEK40–glutathione S-transferase (GST). The DEK40–GST construct was expressed in the Escherichia coli BL21 (DE3) cell line. DEK40 tagged with GST was purified using glutathione Sepharose 4B (GE Healthcare). Two RNA probes for cox3-314, nad2-26, and nad5-1916 were synthesized and labeled with biotin at the 3′ end. The LightShift Chemiluminescent RNA EMSA Kit (Thermo Scientific; catalog no. 20158) was used for binding reactions and labeled complex detection following the manufacturer’s instructions. Competition experiments were performed with different concentrations of unlabeled probe. The primer sequences are listed in Supplementary Table S4.
Isolation and analysis of mitochondrias complexes
Mitochondria for blue native PAGE (BN-PAGE) and complex activity analysis were isolated from kernels without pericarp of self-pollinated segregating ears at 15 DAP as described previously (Chen et al., 2017). The enriched mitochondria were resuspended in 40 μl of sample buffer [50 mM Bis-Tris, 6 N HCl, 50 mM NaCl, 10% (w/v) glycerol, 0.001% Ponceau S] with 20% n-dodecyl-β-d-maltoside added to a final concentration of 1%, and incubated for 30 min on ice. The samples were centrifuged at 20 000 g for 30 min at 4 °C and loaded on native PAGE Novex 4–16% Bis-Tris gels (Invitrogen, BN1002BOX). The electrophoresis was performed following the manufacturer’s protocol. The gels were stained with Coomassie Blue R-250 (Sigma-Aldrich). The activities of mitochondrial complexes were determined by colorimetry as described previously (Luo et al., 2008; Muhling et al., 2010; Gadicherla et al., 2012).
RNA sequencing (RNA-seq) and data analysis
Total RNAs were extracted from the endosperm of WT and dek40-ref at 10 DAP using the CTAB-based method. Two biological repeats (8–10 individuals per pool) of WT (WT-A, WT-B) and dek40-ref (dek-A, dek-B) were isolated from two different segregating ears, and used for RNA-seq. High-throughput sequencing was performed on the Illumina HiSeq4000 platform at the Beijing Genomics Institute (BGI; Shenzhen, China). After sequencing, reads of low quality, adaptor polluted, and with a high content of unknown bases (N) were filtered to obtain clean reads, and these clean reads were then mapped to the reference genome (http://www.maizegdb.org/; last accessed 8 September 2019, Zea mays.AGPv3) using HISAT (Kim et al., 2015). For gene expression analysis, clean reads were mapped to the reference using Bowtie2 (Langmead and Salzberg, 2012) and then the gene expression level was calculated with RSEM (Li and Dewey, 2011). We determined differentially expressed genes (DEGs) as described {[fold change ≥2.00 and –log10 (1–probability) ≥0.8] and [fold change ≥2.00 and false discovery rate (FDR) ≤0.001]}. RNA-seq data are available from the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; last accessed 8 September 2019) under the series entry GEO: GSE122962.
Results
Genetic and phenotypic characterization of the dek40 mutant
A defective kernel mutant was isolated from an EMS mutant library of inbred line B73 and designated dek40-ref. Self-pollinated dek40-ref heterozygotes produced ~25% defective kernels (defective kernel number/total kernel number=292/1211, P=0.48). To confirm this segregation, we crossed dek40-ref heterozygotes to each of four maize inbred lines, S162, V3b1, Mo17, and Chang7-2. Among these monohybrid crosses, ~50% (106/209) of the first generation (F1) had phenotype-segregating ears, which matched the 1:1 segregation ratio. Moreover, the ratio of WT to mutant phenotype kernels in the second-generation (F2) offspring of the segregating ears was 3:1 in all four genetic backgrounds (Supplementary Table S1). Thus, a monogenic recessive mutation was responsible for the phenotype of dek40-ref.
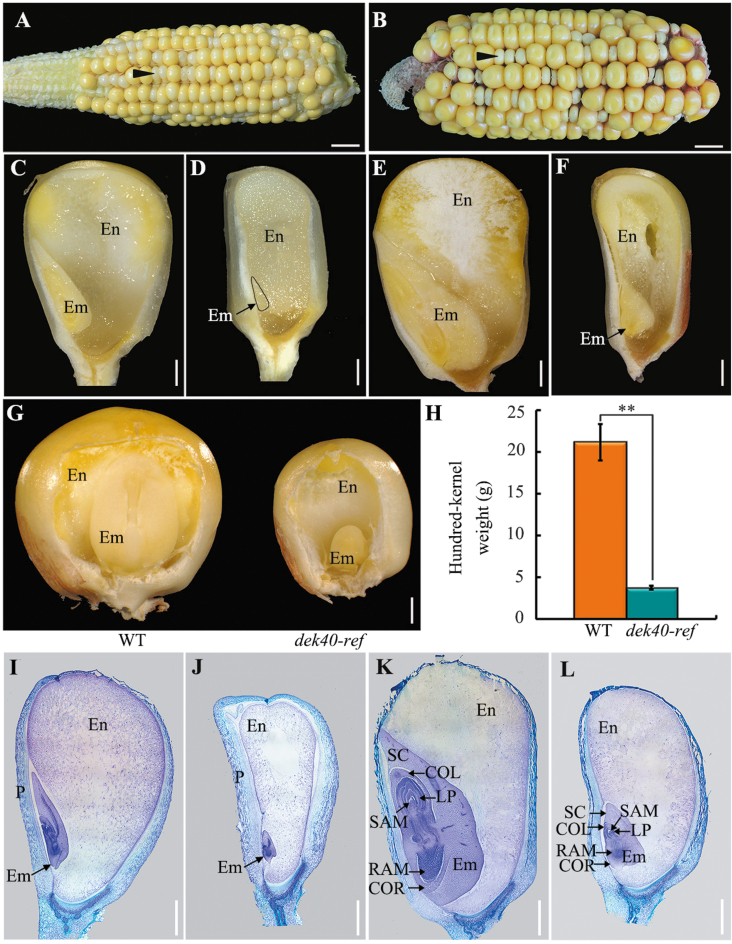
A germination test showed that dek40-ref kernels could not geminate, indicating embryo lethality. Therefore, the dek40-ref allele was maintained with heterozygotes. On self-pollinated heterozygous ears at 15 and 25 DAP, the dek40-ref mutant kernels displayed small, white, and translucent phenotypes (Fig. 1A, B). Longitudinal dissection of WT and mutant sibling kernels from segregated ears at 15 and 25 DAP showed that kernels with the WT phenotype contained starch-filled endosperm and normal embryos, while dek40-ref kernels had a soft texture and abnormal embryos (Fig. 1C–F). Moreover, there was a cavity in the central region of the dek40-ref mutant endosperm at 25 DAP (Fig. 1F). In the frontal view, both the mutant kernel and embryo were smaller than in the WT (Fig. 1G). The 100-kernel weight of dek40-ref was nearly 82.4% less than that of the WT (Fig. 1H), indicating that dek40-ref kernel development was severely defective.
Fig. 1.
Phenotypic characterization of dek40-ref kernels. (A and B) The self-pollinated dek40-ref heterozygote ears at 15 DAP (A) and 25 DAP (B). The arrowhead indicates one of the dek40-ref kernels. Scale bars=10 mm. (C–F) Developmental comparisons of dissected WT and dek40-ref kernels at 15 and 25 DAP. (C and E) WT kernels at 15 and 25 DAP. (D and F) dek40-ref kernels at 15 and 25 DAP. Scale bars=1 mm. (G) The front view of mature WT and dek40-ref kernels whose partial pericarp was removed to display an embryo. Scale bars=1 mm. (H) Comparison of the 100-kernel weight of randomly selected mature WT and dek40-ref kernels. (I–L) Histological analysis of WT and dek40-ref kernels at 15 and 25 DAP. (I and K) WT at 15 and 25 DAP. (J and L) dek40-ref kernels at 15 and 25 DAP. Scale bars=1 mm. En, endosperm; Em, embryo; P, pericarp; LP, leaf primordia; RAM, root apical meristem; SAM, shoot apical meristem; SC, scutellum; COL, coleoptile; COR, coleorhiza.
Due to the dramatic dek40-ref kernel defects, we analyzed embryo and endosperm development of dek40-ref and WT kernels at 7–25 DAP from the same segregating ear using histological sections (Fig. 1I–L; Supplementary Fig. S1). The mutant kernels exhibited developmental delay compared with WT kernels. At 15 DAP, the WT pericarp was filled with endosperm, and the embryo had developed an obvious scutellum, coleoptile, two leaf primordia, and both shoot and root apical meristems (Fig. 1I). However, the mutant kernel embryo had only a coleoptile and an empty space between the pericarp and the endosperm (Fig. 1J). At 25 DAP, the WT embryos were larger and produced five or six leaf primordia (Fig. 1K); in contrast, the mutant kernel embryos remained small and had a shoot apical meristem-like structure with one or two tiny leaf primordia (Fig. 1L). These results indicated that embryo and endosperm development was arrested in dek40-ref kernels.
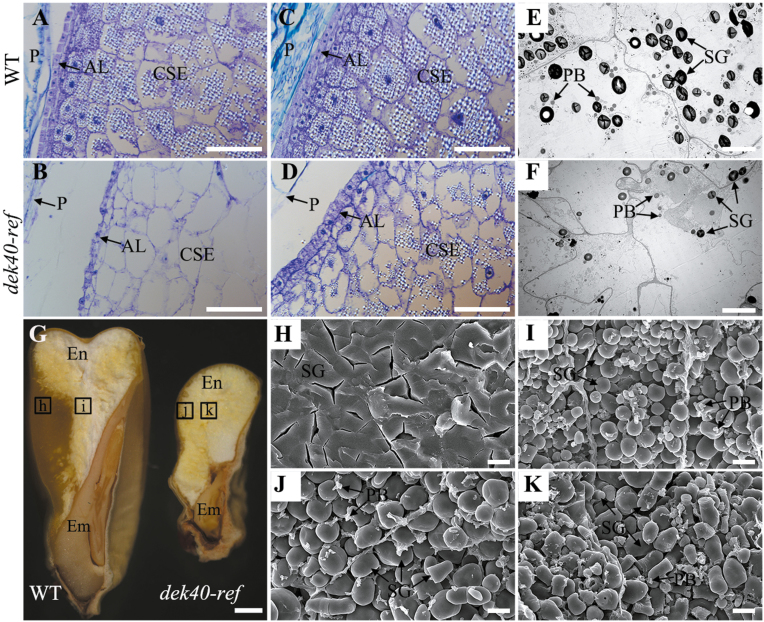
Further analysis showed an obvious difference in starch accumulation between mutant and WT endosperm cells. As shown in Fig. 2A–F, WT kernels accumulated many starch granules, while mutant kernels contained fewer starch granules. Longitudinal dissection of WT and dek40-ref dry seeds showed that the ratio of hard to soft endosperm was decreased in dek40-ref (Fig. 2G). SEM analysis revealed that the starch granules in the peripheral hard endosperm cells were tightly packed in the WT (Fig. 2H), but loosely packed in dek40-ref (Fig. 2J). The central starchy endosperm cells were filled with irregular polyhedrons and large starch granules in the dek40-ref endosperm (Fig. 2I, K). In addition, protein body accumulation appeared to be reduced in the central endosperm cells of dek40-ref compared with those in the WT (Fig. 2I, K).
Fig. 2.
Comparisons of endosperm in the WT and dek40-ref at different developmental stages. (A and B) Histological section analysis of 13 DAP endosperm of WT (A) and dek40-ref (B) kernels. Scale bars=100 μm. (C and D) Histological section analysis of 15 DAP endosperm of the WT (C) and dek40-ref (D) kernels. Scale bars=100 μm. (E and F) TEM images of 13 DAP endosperms of the WT and dek40-ref kernels. Scale bars=10 μm. (G) Longitudinal dissection of WT and dek40-ref mature kernels. Scale bars=1 mm. (H–K) SEM images of WT and dek40-ref mature endosperm in (G). (H and I) SEM images of WT endosperm in the areas h and i indicated in (G). Scale bars=10 μm. (J and K) SEM images of dek40-ref endosperm of in the areas j and k indicated in (G). Scale bars=10 μm. En, endosperm; Em, embryo; P, pericarp; AL, aleurone layer; CSE, central starch endosperm; PB, protein body; SG, starch grain.
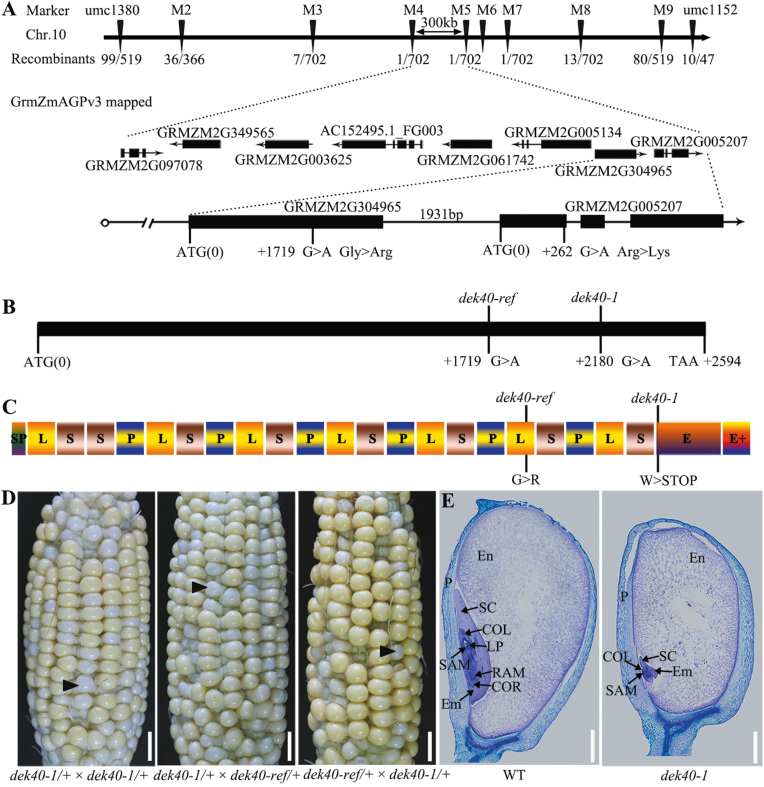
Positional cloning of Dek40
To identify the locus responsible for the dek40-ref mutant phenotype, an F2 mapping population from a cross between dek40-ref heterozygotes and the WT inbred line S162 were used for map-based cloning of the candidate gene. Through analysis of 48 mutant individuals, the Dek40 gene was mapped between the SSR markers umc1380 and umc1152 on the short arm of chromosome 10 (Fig. 3A). To narrow down the location of Dek40, we selected eight polymorphic markers (Supplementary Table S4) to screen 702 mutant individuals. Finally, the Dek40 locus was mapped to a 300 kb region between molecular markers M4 and M5. Eight candidate genes were identified in this region (Fig. 3A). DNA sequence comparison of these candidate genes between WT and mutant alleles revealed two SNPs in two adjacent genes. One was a putative PPR gene whose SNP mutation (G to A) at base pair +1719 resulted in an amino acid replacement (Gly to Arg) (Fig. 3A, B), and the other was a putative WRKY transcription factor and its SNP transition (G to A) at base pair +262 caused an Arg to Lys substitution (Fig. 3A).
Fig. 3.
Map-based cloning and identification of Dek40. (A) Fine mapping of the Dek40 locus. The Dek40 locus was mapped to a 300 kb region by markers M4 and M5 on chromosome 10, which contains eight candidate genes. The molecular markers and number of recombinants are indicated. The structure and mutation site of the two candidate genes are indicated. (B) Gene structure of Dek40 and mutation sites in the Dek40 gene. (C) Schematic diagram of DEK40 protein with a total of 21 PPR domains (P, L, and S), E, and E+ motifs. The mutation sites of amino acid change are indicated. SP, signal peptide. (D) Heterozygous dek40-ref and dek40-1 were used in an allelism test of Dek40. The black arrow indicates the mutant kernel. Scale bars=1 cm. (E) Histological analysis of WT and dek40-1 kernels at 15 DAP. Scale bars=1 mm. En, endosperm; Em, embryo; P, pericarp; LP, leaf primordia; RAM, root apical meristem; SAM, shoot apical meristem; SC, scutellum; COL, coleoptile; COR, coleorhiza.
To determine which mutation was responsible for the dek40-ref phenotype, we examined co-segregation between the mutation sites and the dek40-ref mutant phenotypes by PCR and DNA sequencing in 158 WT phenotype individuals that were segregated from the self-progenies of dek40-ref heterozygotes. Only the self-progenies of the individuals with a heterozygote mutation (GA) in the PPR gene were segregated (Supplementary Table S2), suggesting that the SNP mutation (G to A) at base pair +1719 in the gene GRMZM2G304965 was the locus responsible for the dek40-ref phenotypes. Thus, the gene was named Dek40.
To confirm that the candidate PPR gene was the gene responsible for the dek40-ref phenotypes, we identified another mutant allele of this gene designated dek40-1. dek40-1 had an SNP mutation (G to A) at base pair +2180, resulting in a premature stop codon in the mature transcript (Fig. 3B, C). The self-progenies of dek40-1 heterozygotes produced similar defective kernels to dek40-ref in a recessive pattern (Fig. 3D). Crosses between dek40-ref and dek40-1 heterozygotes produced defective kernels, with ears segregating with normal and defective kernels in an expected 3:1 ratio, demonstrating that they were allelic (Fig. 3D). The results were further confirmed by PCR sequencing genotyping (Supplementary Table S3). Additionally, histological analysis of WT and dek40-1 kernels at 15 DAP showed that embryo and endosperm development were severely defective in dek40-1 kernels (Fig. 3E). Thus, the Dek40 gene encoding a PPR protein was responsible for the defective kernel mutant phenotype.
Dek40 encodes a mitochondrion-targeted PPR-E+ subclass protein
Genomic DNA structure analysis revealed that Dek40 contained only one 2595 bp long exon (Fig. 3A). It encoded a novel PPR protein of 864 amino acids with 21 PRR motifs, an E domain and an E+ domain (Fig. 3C; Supplementary Fig. S2). The mutation sites in the dek40-ref and dek40-1 mutants were in the 17th PPR repeat and E domain, respectively (Fig. 3C; Supplementary Fig. S2). The premature stop mutation in dek40-1 resulted in a truncated protein without E and E+ domains (Fig. 3C; Supplementary Fig. S2). These results suggested that DEK40 belongs to the PPR-E+ subclass, and its E and E+ domains are required for its function.
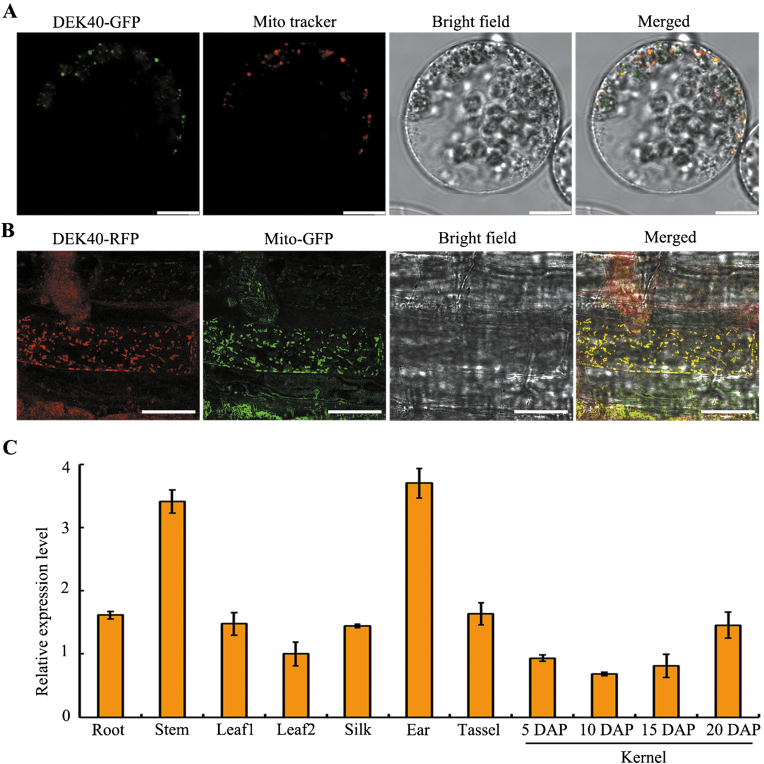
Previous studies have shown that most PPRs are targeted to either plastids or mitochondria (Lurin et al., 2004; Sosso et al., 2012a; Colcombet et al., 2013; Li et al., 2014). To determine the subcellular localization of DEK40, the fused construct p35S:DEK40-GFP was generated and transfected into maize protoplasts. As shown in Fig. 4A, the GFP signal was co-localized with the Mito tracker for mitochondria. To confirm this localization, the construct p35S:DEK40-RFP was transformed into the Arabidopsis marker line MT-GK with GFP-specific expression in mitochondria. The RFP signal was co-localized with the GFP signal in mitochondria of root cells (Fig. 4B), suggesting that the DEK40 protein is targeted to the mitochondria in maize. A gene expression analysis showed that Dek40 was constitutively expressed in all examined tissues (Fig. 4C).
Fig. 4.
Subcellular localization of DEK40 and expression pattern of Dek40. (A) Fluorescence signals of transiently expressed DEK40–GFP in maize protoplasts. Mitochondria are marked by Mito Tracker (red). Scale bars=10 µm. (B) Subcellular localization of DEK40 protein in the Arabidopsis marker line of mitochondria. The T1 transgenic line roots were used for imaging by confocal microscopy. The fluorescence signals of expressed DEK40–RFP in the Arabidopsis marker line are shown in red. The Arabidopsis marker line (MT-GK) has a specially expressed GFP in mitochondria, which is marked as Mito-GFP. Scale bars=25 µm. (C) The expression profiles of Dek40 in a variety of organs and maize kernels at different developmental stages. Leaf1, juvenile leaves; Leaf2, mature leaves; DAP, days after pollination. The RNA levels were normalized to that of the maize ZmActin gene (GRMZM2G126010). Values are means of three technical replicates and three biological repeats. Error bars represent the SD.
DEK40 is required for C-to-U RNA editing of multiple mitochondrial transcripts
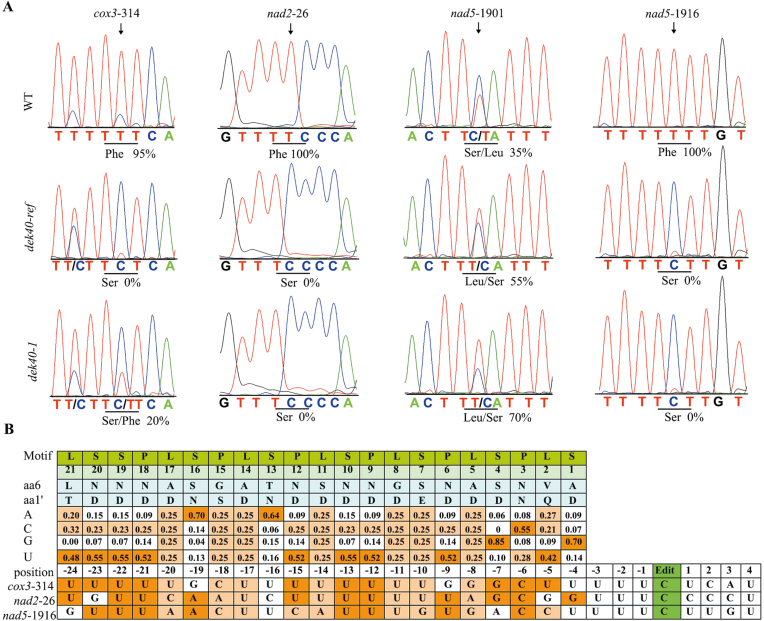
To investigate the function of Dek40, we used gene-specific primers to examine 35 protein-coding mitochondrial genes as described previously (Liu et al., 2013). The results showed that the editing of three C-to-U editing sites, cox3-314, nad2-26, and nad5-1916, in dek40-ref was completely abolished (Fig. 5A). Normally, the cox3-314 RNA editing site is 95% C-to U-edited, and the nad2-26 and nad5-1916 editing sites are completely edited. Surprisingly, the editing ratio of nad5-1901 was increased in dek40-ref (Fig. 5A). We then analyzed these editing sites in the dek40-1 mutant. RNA editing at the two editing sites nad2-26 and nad5-1916 was also completely abolished (Fig. 5A). At the cox3-314 site, the RNA editing ratio was reduced to 20% (Fig. 5A). At nad5-1901, the editing ratio was increased to 70% (Fig. 5A). These data suggested that the DEK40 protein is necessary for RNA editing at cox3-314, nad2-26, nad5-1901, and nad5-1916 in maize.
Fig. 5.
DEK40 is required for cox3-314, nad2-26, nad5-1901, and nad5-1916 C-to-U editing in maize mitochondria. (A) Analysis of RNA editing in cox3, nad2, and nad5 transcripts of developing kernels in the WT, dek40-ref, and dek40-1. The arrow marks the editing site. The amino acid encoded is indicated on the bottom, and the editing efficiency is presented under each target site. (B) Prediction and alignment of DEK40 to editing sites in the mitochondrial targets. Prediction of DEK40 target sites of the amino acids at positions 6 and 1' with the coding rule. The probability of each nucleotide binding a DEK40 PPR element is given in the bottom panel, and nucleotides matching the DEK40 RNA recognition site sequences are indicated.
The defective editing at cox3-314, nad2-26, nad5-1901, and nad5-1916 in dek40-ref and dek40-1 resulted in an amino acid replacement compared with the WT (Fig. 5A). The DEK40 target sites of the amino acids at positions 6 and 1′ were analyzed using a previously described method (Barkan et al., 2012; Takenaka et al., 2013a). The sequences around the cox3-314, nad2-26, and nad5-1916 RNA editing sites were highly conserved with putative recognition sites (Fig. 5B), indicating that DEK40 is important for RNA editing at these four sites in mitochondria.
DEK40 recognizes and directly bind to cox3, nad2, and nad5 transcripts
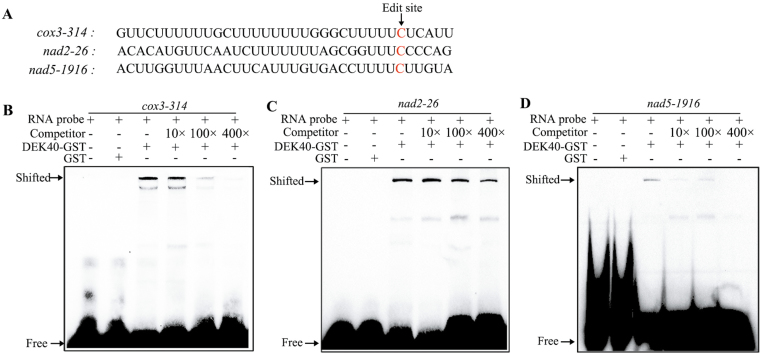
In the dek40-ref mutant, the C-to-U editing at cox3-314, nad2-26, and nad5-1916 was completely abolished, implying that the three transcripts may be targets of DEK40. To investigate whether DEK40 directly binds to these transcripts, an RNA EMSA was carried out. Because the amino acids at positions 6 and 1′ are essential in PPR protein target sites (Barkan et al., 2012; Takenaka et al., 2013a), we predicted the DEK40 recognition sequences (Fig. 5B). Three RNA probes (base pairs –29 upstream to +4 downstream of the RNA editing site of cox3, nad2, and nad5) that included 35 nucleotides surrounding the three RNA editing sites were designed and biotin labeled (Fig. 6A). Shifted bands were detected when DEK40–GST was incubated with the RNA probes, but were not identified when the RNA probes were incubated with GST (Fig. 6B–D). The competition assay further showed that the shifted bands distinctly decreased as the competitor concentration increased and the labeled RNA probes increased (Fig. 6B–D). Thus, DEK40 can directly bind cox3, nad2, and nad5 transcripts and is necessary for RNA editing at cox3-314, nad2-26, and nad5-1916 in maize.
Fig. 6.
DEK40 recognizes and directly binds cox3, nad2, and nad5 transcripts. (A) Nucleotide sequence of RNA probes. Edited sites are indicated in red. (B) RNA EMSAs indicated that DEK40 directly binds to cox3 transcripts that surround the cox3 edited site. Unlabeled probe was used as a competitor, and GST was used as a negative control. Black arrows show the shifted bound and free RNA probe. (C) RNA EMSAs indicated that DEK40 directly binds to nad2 transcripts that surround the nad2 edited site. Unlabeled probe was used as a competitor, and GST was used as a negative control. The shifted bound and free RNA probesw are indicated by black arrows. (D) RNA EMSAs indicated that DEK40 directly binds to nad5 transcripts that surround the nad5 edited site. Unlabeled probe was used as a competitor, and GST was used as a negative control. The shifted bound and free RNA probes are indicated by black arrows.
The activities of mitochondrial complexes I and IV are reduced in the dek40-ref mutant
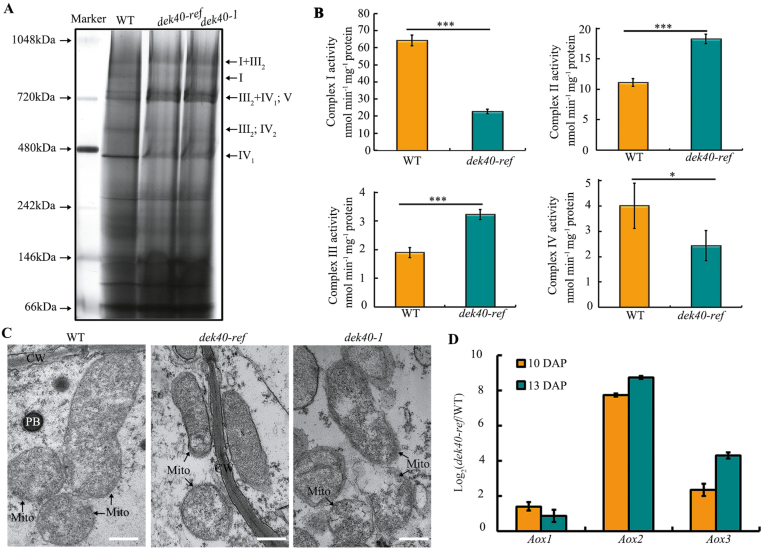
The nad2 and nad5 genes encode complex I subunits NAD2 and NAD5, respectively. The cox3 gene encodes subunit III of the COX that is part of complex IV of the ETC. Thus, the editing defects at cox3-314, nad2-26, nad5-1901, and nad5-1916 in dek40-ref might affect complex I and IV activity. Mitochondrial complexes were isolated from 15 DAP kernels without pericarp and separated by BN-PAGE. Analyses revealed that the abundances of complexes I and IV decreased dramatically in the dek40-ref and dek40-1 mutants compared with the WT (Fig. 7A). We then investigated the activities of mitochondrial complexes I, II, III, and IV in dek40-ref kernels. The activities of complexes I and IV decreased dramatically, but those of complexes II and III increased noticeably in the dek40-ref mutant (Fig. 7B). The results suggested that the editing defects at cox3-314, nad2-26, nad5-1901, and nad5-1916 in the mutant kernels affected complex I and IV assembly and led to decreased activities of these complexes in the mitochondrial respiratory chain.
Fig. 7.
Comparison of mitochondrial complexes, ultrastructure, and alternative respiratory pathways between the WT and mutants. (A) The BN-PAGE analyses of mitochondrial complexes. The BN-PAGE gels were stained with Coomassie Brilliant Blue (CBB). (B) Activity analyses of four mitochondrial complexes. Values are means of three biological replicates. Error bars represent the SD. Significant differences are indicated. *P<0.05, **P<0.01, ***P<0.001 (Student’s t-test). (C) TEM analysis of mitochondrial ultrastructure in 11 DAP endosperm of WT, dek40-ref, and dek40-1. Mito, mitochondria; PB, protein body; CW, cell wall. Scale bars=500 nm. (D) qRT–PCR analysis of Aox gene expression in endosperm at 10 and 13 DAP. Values and bars represent the mean and the SD of three biological replicates, respectively. The RNA levels were normalized to that of the maize ZmActin gene (GRMZM2G126010).
Mitochondrial ultrastructure was altered and alternative respiratory pathway activities were increased in the dek40 mutant
To determine the functions of Dek40 in mitochondria, we analyzed the structure of mitochondria in dek40-ref and dek40-1 mutant endosperm at 11 DAP. The WT had normal mitochondria with densely folded inner membranes and distinct cristae (Fig. 7C), whereas the mitochondria of dek40-ref and dek40-1 had large internal spaces and lacked obvious membrane structures (Fig. 7C), implying that DEK40 is required for mitochondrial structure and function during kernel development.
Besides classical ETCs, plants have evolved an alternative respiratory pathway. The alternative oxidase (Aox) genes are usually used as markers for the alternative respiratory pathway in the mitochondrial ETC. There are three Aox genes, Aox1 (AC233960.1_FG002), Aox2 (GRMZM2G125669), and Aox3 (GRMZM2G074743), in the maize genome. Because the dek40 mutant appeared to reduce mitochondrial complex I and IV activity, we examined Aox transcript accumulation in dek40-ref by qRT–PCR. The results showed that Aox gene transcript levels, especially that of Aox2, were noticeably higher in the dek40-ref mutant than in the WT (Fig. 7D). These results indicated that the respiration function of dek40-ref mitochondria is severely impaired because of the decrease in complex I and IV activity, which induces the alternative respiratory pathway in the dek40-ref mutant.
ROS were overproduced and PCD was initiated early in dek40-ref endosperm
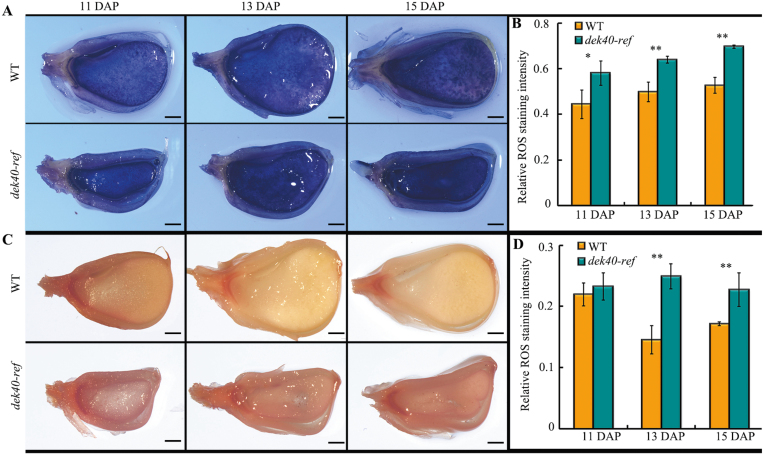
ROS production is mainly caused by the mitochondrial ETC complexes I and III (Gill and Tuteja, 2010). Thus, we analyzed the H2O2 and O2− levels in dek40-ref and the WT at 11, 13, and 15 DAP by DAB and NBT staining, respectively. The endosperm of dek40-ref accumulated much more ROS than that of the WT (Fig. 8A–D). Furthermore, ROS accumulation was found near the cavity in the dek40-ref endosperm (Fig. 8A, C).
Fig. 8.
Abnormal accumulation of ROS in dek40-ref endosperm. (A) NBT staining for the detection of superoxide (O2−) in endosperm of the WT and dek40-ref at 11, 13, and 15 DAP. Scale bars=1 mm. (B) Relative NBT staining intensity in endosperm of the WT and dek40-ref at the indicated developmental stage. (C) DAB staining for the detection of hydrogen peroxide (H2O2) in endosperm of the WT and dek40-ref at 11, 13, and 15 DAP. Scale bars=1 mm. (D) Relative DAB staining intensity in endosperm of the WT and dek40-ref at the indicated developmental stages. Values and bars represent the mean and the SD of more than five biological replicates, respectively. Significant differences are indicated. *P<0.05, **P<0.01 (Student’s t-test).
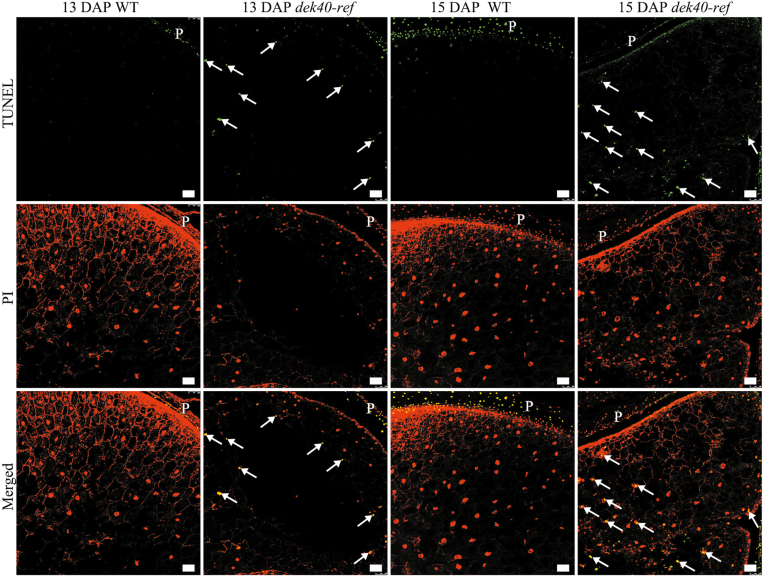
To understand the cavity formation phenotype, we performed TUNEL assays on the WT and dek40-ref using longitudinal sections of 11, 13, and 15 DAP kernels. No TUNEL-positive signal was detected at 11 DAP in WT or dek40-ref endosperm nuclei (Supplementary Fig. S3). Interestingly, strong TUNEL-positive signals started to be detected at 13 DAP in dek40-ref endosperm nuclei, but no TUNEL-positive signal was detected in WT endosperm at the same stage (Fig. 9). At 15 DAP, WT endosperm showed weak TUNEL-positive signals close to the pericarp, whereas the dek40-ref mutant showed strong TUNEL-positive signals in the central region of the starchy endosperm near the cavity (Fig. 9), implying that nuclear DNA fragmentation started earlier in dek40-ref endosperm cells than in the WT. The cavity in the central region of the endosperm might be caused by abnormal PCD in dek40-ref.
Fig. 9.
Earlier PCD in dek40-ref endosperm. Nuclear DNA fragmentation in WT and dek40-ref endosperm at 13 and 15 DAP was detected by TUNEL assay. Green indicates the TUNEL signals. Red is PI showing the nuclei. Yellow is a merge of TUNEL-detected nuclei and PI-detected nuclei. The white arrowheads show examples of a positive signal. P, pericarp. Scale bars=100 μm.
The cysteine proteinase and metacaspases are involved in PCD (Suarez et al., 2004; Takahashi et al., 2015). To verify the initiation of PCD in dek40-ref mutant endosperm, we examined the expression levels of cysteine proteinase and metacaspase genes. Their expression levels were increased in the dek40 mutant at 11, 13, and 15 DAP (Supplementary Fig. S4), suggesting that dek40-ref endosperm PCD started earlier than in the WT.
Genes were differentially expressed between the WT and dek40-ref
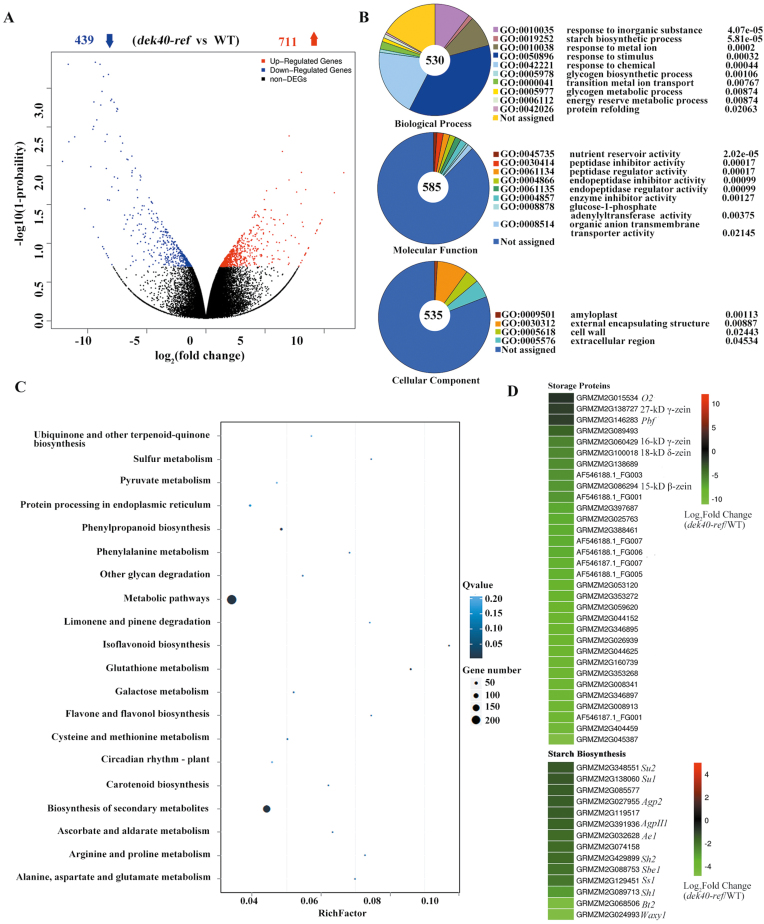
To explain the developmental defects at the gene expression level, RNA-seq analysis was performed with total RNA isolated from WT and dek40-ref mutant endosperms at 10 DAP. In total, 1150 genes showed significantly altered expression between the WT and dek40-ref mutant (Fig. 10A). To identify metabolic pathways in which the DEGs were enriched, we performed Gene Ontology (GO) classification and pathway analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (Fig. 10B, C). The largest category was biological process, with 10 subcategories, namely: response to inorganic substance; starch biosynthetic process; response to metal ion; response to stimulus; response to chemical; glycogen biosynthetic process; transition metal ion transport; glycogen metabolic process; energy reserve metabolic process; and protein refolding. The second largest was molecular function with: nutrient reservoir activity; peptidase inhibitor activity; peptidase regulator activity; endopeptidase inhibitor activity; endopeptidase regulator activity; enzyme inhibitor activity; glucose-1-phosphate adenylyltransferase activity; organic anion transmembrane transporter activity; and cellular component (Fig. 10B). A total of 739 DEGs were classified into 120 pathways, 22 of which were significantly enriched (P<0.05). Among the 22 pathways, the extremely significant pathways were biosynthesis of secondary metabolites, followed by glutathione metabolism pathways, isoflavonoid biosynthesis, and phenylpropanoid biosynthesis (Fig. 10C). Notably, many genes functioning in nutrient storage activity and starch biosynthesis were markedly down-regulated in dek40-ref (Fig. 10D).
Fig. 10.
Identification of differentially expressed genes (DEGs) between the WT and dek40-ref by RNA-seq. (A) The number of DEGs between the dek40-ref mutant and WT. Fold change ≥2; –log10(1–probability) ≥0.8. Blue and red arrows represent the number of down- and up-regulated genes in dek40-ref compared with the WT, respectively. (B) The most significantly related GO terms of the functional annotated DEGs, P<0.05. (C) Pathway functional enrichment of DEGs. The x-axis represents the enrichment factor and the y-axis represents the pathway name. (D) Heat maps of differentially expressed genes involved in storage protein and starch biosynthesis.
Discussion
The E+ PPR protein DEK40 is essential for post-transcriptional processing of cox3, nad2, and nad5 transcripts in maize
PPR proteins play an essential role in post-transcriptional processes in mitochondria and chloroplasts, including RNA editing, splicing and cleavage, and translation (Schmitz-Linneweber and Small, 2008). So far, several Dek genes have been reported in previous studies; to investigate the relationships between the DEK40 and other DEK proteins, the phylogenetic tree was constructed (Supplementary Fig. S5). DEK40 is more closely related to DEK36, DEK39, and DEK10 (Supplementary Fig. S5), and they all belong to the PLS-type PPR protein subfamily. They are involved in RNA editing in mitochondria and maize kernel development (Qi et al., 2017; Wang et al., 2017; Li et al., 2018), suggesting that their functions are conserved in maize. In this study, we identified the E+ subgroup PPR protein DEK40 localized to mitochondria in maize (Fig. 4A, B). Dek40 loss of function caused kernel defects and embryo lethality in maize (Fig. 3D). The dek40-ref mutant has a nucleotide change resulting in an amino acid replacement (Gly to Arg) at the 21st amino acid of the 17th PPR motif (L motif) (Fig. 3B, C; Supplementary Fig. S2). The amino acid alignment of maize DEK40 with similar PPR proteins in other plant species showed that the Gly residue was highly conserved (Supplementary Fig. S2). Previous studies have shown that the Gly residue is highly conserved in L motifs (Lurin et al., 2004; Wang et al., 2017). Therefore, the Gly residue in the α-helices of the L motif is required for PPR protein function. The single point mutation in dek40-1 led to a premature stop codon in Dek40, resulting in the loss of the E and E+ domains in DEK40. The studies on EMP5, EMP9, and MEF3 revealed that the E domain but not the E+ domain is essential for RNA editing (Verbitskiy et al., 2012; Liu et al., 2013; Yang et al., 2017), which in combination with our results suggests that the E domain is an important functional domain in E/E+ subgroup PPR proteins.
Bioinformatics and structural analyses indicated that RNA substrates of PPR proteins could be identified in a motif sequence-specific manner (Barkan et al., 2012; Takenaka et al., 2013a; Yin et al., 2013). However, to our knowledge, very few studies have shown direct evidence for the recognition and binding between PPR proteins and their targets. In our study, we confirmed the binding capacity between DEK40 and target RNAs (Fig. 6B–D), which provides very important information for the illumination of the functional mechanism of PPR proteins. Editing at the cox3-314, nad2-26, and nad5-1916 sites in dek40-ref was completely abolished, but the editing ratio at nad5-1901 was increased (Fig. 5A), suggesting that DEK40 is involved in multiple editing events. MEF13, the Arabidopsis ortholog of DEK40, is responsible for editing at eight target sites (ccmFc-50, nad2-59, nad5-1665, nad5-1916, nad7-213, cox3-314, nad4-158, and ccmFc-415) (Glass et al., 2015). However, we did not find RNA editing at ccmFc-50, nad7-213, and nad5-1665 in WT or dek40-ref mitochondria in maize. Sequence alignment showed that the maize nad2-26, cox3-314, and nad5-1916 editing positions are consistent with Arabidopsis nad2-53, cox3-314, and nad5-1916 transcripts, respectively. Interestingly, the nad5-1901 editing ratio was increased in dek40-ref, which was not be found in Arabidopsis mef13. Taken together, DEK40 has divergent functions in regulating mitochondrial RNA metabolism between maize and Arabidopsis.
Post-transcriptional processing of cox3, nad2, and nad5 transcripts is required for mitochondrial functions in maize
In maize, 22 proteins in the ETC are encoded by mitochondrial genes, which include nine subunits (NAD1, 2, 3, 4, 4L, 5, 6, 7, and 9) of complex I, one subunit (cob) of complex III, three subunits (COX 1, 2, and 3) of complex IV, four subunits (CCMB, C, FN, and FC) of cytochrome c, and five subunits (ATP 1, 4, 6, 8, and 9) of complex V (Clifton et al., 2004). Here, we found that editing defects at cox3-314, nad2-26, nad5-1901, and nad5-1916 in the dek40 mutant resulted in an amino acid change compared with the WT (Fig. 5A). nad2 and nad5 encode complex I subunits NAD2 and NAD5, respectively, while cox3 encodes complex IV subunit COX3. The abundance and activities of complexes I and IV were dramatically decreased in the dek40 mutant, but the activities of complexes II and III were noticeably increased (Fig. 7A, B). These changes might lead to dysfunction of the mitochondria and damage of the mitochondrial ultrastructure (Fig. 7C, D). NAD2 and NAD5 are essential for the assembly and activity of complex I. The splicing efficiency of the mitochondrial nad2 intron was affected by EMP16, EMP10, DEK37, PPR-SMR1, or EMP12 (Xiu et al., 2016; Cai et al., 2017; Dai et al., 2018; Chen et al., 2019; Sun et al., 2019). Loss of PPR78 led to a reduction in the steady-state level of the mitochondrial nad5 mature mRNA (Zhang et al., 2017). These mutations reduce the assembly and activity of complex I. COX3 is also critical for mitochondrial functions. For example, RNA editing defects in cox3 resulted in reduced COX activity and inhibition of the mitochondrial respiratory pathway (Tang et al., 2010). Thus, we suggest that the NAD2-9 (Phe), NAD5-639 (Phe). and COX3-105 (Phe) amino acids correctly encoded at the editing site are required for mitochondrial function in maize, and the editing at cox3-314, nad2-26, nad5-1901, and nad5-1916 is critical to mitochondrial functions.
Mitochondrial dysfunction affects kernel development
As well as the classical oxidative phosphorylation system, the alternative respiratory pathways play an important role in plant respiratory electron transport in mitochondria (Eubel et al., 2004). Reduced assembly and activity of mitochondrial complexes increases the AOX protein content and AOX transcript levels (Zsigmond et al., 2008; Sosso et al., 2012b; Xie et al., 2016; Xiu et al., 2016; Ren et al., 2017). In the dek40 mutant, AOX transcript levels were increased and more ROS were generated than in the WT, implying that mitochondrial dysfunction leads to cellular redox imbalance and causes ROS overproduction.
The ROS generated in the ETC play a key role in triggering plant PCD (Wu et al., 2015). In dek40-ref, complex I was defective, but ROS accumulation was increased in the endosperm (Fig. 8A–D), which might explain why PCD began earlier in dek40-ref endosperm than in the WT (Fig. 9). This is probably caused by ROS overproduction through mitochondrial complex III in dek40-ref and is consistent with a previous study in which the ROS generated from complex III play a major role in triggering mod1 cell death (Wu et al., 2015). Generally, PCD starts in the central region of the endosperm and then spreads to the periphery (Young et al., 1997; Young and Gallie, 2000). In sh2 endosperm cells, PCD occurs earlier in the central endosperm and progresses outwardly more rapidly than in WT endosperm, which interferes with starch and storage protein synthesis (Young et al., 1997). In fl4, accelerated PCD may stop the development of the central starchy endosperm cells (Wang et al., 2014). Similarly, the dek40-ref mutant showed a large cavity in the central starchy endosperm that might be caused by early PCD.
In brief, we propose that DEK40 is essential for post-transcriptional processing of cox3, nad2, and nad5 transcripts and kernel development in maize. Abnormal DEK40 could not edit its target sites in cox3, nad2, and nad5 transcripts, which disrupted the assembly of mitochondrial complexes and led to damage to the mitochondrial ultrastructure. Increased accumulation of ROS in the mutants promoted PCD through the regulation of gene expression and caused defective maize kernels.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Analysis of paraffin sections of WT and dek40-ref kernels at different developmental stages.
Fig. S2. Amino acid alignment of maize DEK40 with homologous PPR proteins in other plant species.
Fig. S3. Nuclear DNA fragmentation was detected in WT and dek40-ref endosperm at 11 DAP by TUNEL assay.
Fig. S4. The expression levels of PCD-associated genes.
Fig. S5. Phylogenetic relationships between DEK40 and other maize DEK proteins.
Table S1. Genetic analysis of the kernels in the F2 population of the segregating ears.
Table S2. Co-segregation analysis of the mutant sites and phenotypes of progenies from wild-type phenotype kernels segregated from self-pollinated dek40-ref heterozygous ears.
Table S3. Genotypes of kernels in the crosses between dek40-ref and dek40-1 heterozygote ears were identified by PCR sequencing.
Table S4. Primer sequences used in this study.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (91735301 and 91535109), the National Plant Transgenic Program (2016ZX08003-003), Taishan Scholars Project (ts201712024), funds of Shandong ‘Double Tops’ Program (SYL2017YSTD03), and a project (dxkt201707) from the State Key Laboratory of Crop Biology. The authors declare no competing financial interests.
Author contributions
XYZ and XSZ conceived the project; RCR, YMW, YJZ, and LLW performed the experiments; RCR and LZ conducted bioinformatics analyses; XL and CZ isolated the dek40 mutants; and XYZ, RCR, and XSZ wrote the paper.
References
- Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I. 2012. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genetics 8, e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Small I. 2014. Pentatricopeptide repeat proteins in plants. Annual Review of Plant Biology 65, 415–442. [DOI] [PubMed] [Google Scholar]
- Blokhina O, Fagerstedt KV. 2010. Reactive oxygen species and nitric oxide in plant mitochondria: origin and redundant regulatory systems. Physiologia Plantarum 138, 447–462. [DOI] [PubMed] [Google Scholar]
- Cai M, Li S, Sun F, Sun Q, Zhao H, Ren X, Zhao Y, Tan BC, Zhang Z, Qiu F. 2017. Emp10 encodes a mitochondrial PPR protein that affects the cis-splicing of nad2 intron 1 and seed development in maize. The Plant Journal 91, 132–144. [DOI] [PubMed] [Google Scholar]
- Castandet B, Araya A. 2011. RNA editing in plant organelles. Why make it easy? Biochemistry 76, 924–931. [DOI] [PubMed] [Google Scholar]
- Chen X, Feng F, Qi W, Xu L, Yao D, Wang Q, Song R. 2017. Dek35 encodes a PPR protein that affects cis-splicing of mitochondrial nad4 intron 1 and seed development in maize. Molecular Plant 10, 427–441. [DOI] [PubMed] [Google Scholar]
- Chen Z, Wang HC, Shen J, Sun F, Wang M, Xu C, Tan BC. 2019. PPR-SMR1 is required for the splicing of multiple mitochondrial introns and interacts with Zm-mCSF1 and is essential for seed development in maize. Journal of Experimental Botany 70, 5245–5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton SW, Minx P, Fauron CM, et al. 2004. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiology 136, 3486–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colcombet J, Lopez-Obando M, Heurtevin L, Bernard C, Martin K, Berthomé R, Lurin C. 2013. Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biology 10, 1557–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Luan S, Chen X, Wang Q, Feng Y, Zhu C, Qi W, Song R. 2018. Maize Dek37 encodes a P-type PPR protein that affects cis-splicing of mitochondrial nad2 Intron 1 and seed development. Genetics 208, 1069–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkina NV, Heinemeyer J, Sunderhaus S, Boekema EJ, Braun HP. 2006. Respiratory chain supercomplexes in the plant mitochondrial membrane. Trends in Plant Science 11, 232–240. [DOI] [PubMed] [Google Scholar]
- Eubel H, Heinemeyer J, Sunderhaus S, Braun HP. 2004. Respiratory chain supercomplexes in plant mitochondria. Plant Physiology and Biochemistry 42, 937–942. [DOI] [PubMed] [Google Scholar]
- Fujii S, Small I. 2011. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytologist 191, 37–47. [DOI] [PubMed] [Google Scholar]
- Gadicherla AK, Stowe DF, Antholine WE, Yang M, Camara AK. 2012. Damage to mitochondrial complex I during cardiac ischemia reperfusion injury is reduced indirectly by anti-anginal drug ranolazine. Biochimica et Biophysica Acta 1817, 419–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambino G, Perrone I, Gribaudo I. 2008. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochemical Analysis 19, 520–525. [DOI] [PubMed] [Google Scholar]
- Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C. 2006. Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28, 1091–1101. [DOI] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. 2010. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiology and Biochemistry 48, 909–930. [DOI] [PubMed] [Google Scholar]
- Glass F, Härtel B, Zehrmann A, Verbitskiy D, Takenaka M. 2015. MEF13 requires MORF3 and MORF8 for RNA editing at eight targets in mitochondrial mRNAs in Arabidopsis thaliana. Molecular Plant 8, 1466–1477. [DOI] [PubMed] [Google Scholar]
- Ichinose M, Sugita M. 2017. RNA editing and its molecular mechanism in plant organelles. Genes 8, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14, 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nature Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi C, Apel K, Danon A. 2004. Reactive oxygen signalling: the latest news. Current Opinion in Plant Biology 7, 323–328. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Han JH, Park YI, Colas des Francs-Small C, Small I, Kang H. 2017. The mitochondrial pentatricopeptide repeat protein PPR19 is involved in the stabilization of NADH dehydrogenase 1 transcripts and is crucial for mitochondrial function and Arabidopsis thaliana development. New Phytologist 215, 202–216. [DOI] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gu W, Sun S, Chen Z, Chen J, Song W, Zhao H, Lai J. 2018. Defective Kernel 39 encodes a PPR protein required for seed development in maize. Journal of Integrative Plant Biology 60, 45–64. [DOI] [PubMed] [Google Scholar]
- Li XJ, Zhang YF, Hou M, et al. 2014. Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). The Plant Journal 79, 797–809. [DOI] [PubMed] [Google Scholar]
- Li XL, Huang WL, Yang HH, Jiang RC, Sun F, Wang HC, Zhao J, Xu CH, Tan BC. 2019. EMP18 functions in mitochondrial atp6 and cox2 transcript editing and is essential to seed development in maize. New Phytologist 221, 896–907. [DOI] [PubMed] [Google Scholar]
- Liu Y, He J, Chen Z, Ren X, Hong X, Gong Z. 2010. ABA overly-sensitive 5 (ABO5), encoding a pentatricopeptide repeat protein required for cis-splicing of mitochondrial nad2 intron 3, is involved in the abscisic acid response in Arabidopsis. The Plant Journal 63, 749–765. [DOI] [PubMed] [Google Scholar]
- Liu YJ, Xiu ZH, Meeley R, Tan BC. 2013. Empty pericarp5 encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize. The Plant Cell 25, 868–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Liu J, Ren W, et al. 2018. Gene-indexed mutations in maize. Molecular Plant 11, 496–504. [DOI] [PubMed] [Google Scholar]
- Luo C, Long J, Liu J. 2008. An improved spectrophotometric method for a more specific and accurate assay of mitochondrial complex III activity. Clinica Chimica Acta 395, 38–41. [DOI] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, et al. 2004. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. The Plant Cell 16, 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühling J, Tiefenbach M, López-Barneo J, et al. 2010. Mitochondrial complex II participates in normoxic and hypoxic regulation of α-keto acids in the murine heart. Journal of Molecular and Cellular Cardiology 49, 950–961. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Tian Z, Lu L, Chen X, Chen X, Zhang W, Song R. 2017. Editing of mitochondrial transcripts nad3 and cox2 by Dek10 is essential for mitochondrial function and maize plant development. Genetics 205, 1489–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Pan Z, Zhao H, Zhao J, Cai M, Li J, Zhang Z, Qiu F. 2017. EMPTY PERICARP11 serves as a factor for splicing of mitochondrial nad1 intron and is required to ensure proper seed development in maize. Journal of Experimental Botany 68, 4571–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha D, Prasad AM, Srinivasan R. 2007. Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiology and Biochemistry 45, 521–534. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. 2008. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends in Plant Science 13, 663–670. [DOI] [PubMed] [Google Scholar]
- Small ID, Peeters N. 2000. The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends in Biochemical Sciences 25, 46–47. [DOI] [PubMed] [Google Scholar]
- Sosso D, Canut M, Gendrot G, Dedieu A, Chambrier P, Barkan A, Consonni G, Rogowsky PM. 2012a PPR8522 encodes a chloroplast-targeted pentatricopeptide repeat protein necessary for maize embryogenesis and vegetative development. Journal of Experimental Botany 63, 5843–5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosso D, Mbelo S, Vernoud V, et al. 2012. b PPR2263, a DYW-subgroup pentatricopeptide repeat protein, is required for mitochondrial nad5 and cob transcript editing, mitochondrion biogenesis, and maize growth. The Plant Cell 24, 676–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez MF, Filonova LH, Smertenko A, Savenkov EI, Clapham DH, von Arnold S, Zhivotovsky B, Bozhkov PV. 2004. Metacaspase-dependent programmed cell death is essential for plant embryogenesis. Current Biology 14, R339–R340. [DOI] [PubMed] [Google Scholar]
- Sun F, Wang X, Bonnard G, Shen Y, Xiu Z, Li X, Gao D, Zhang Z, Tan BC. 2015. Empty pericarp7 encodes a mitochondrial E-subgroup pentatricopeptide repeat protein that is required for ccmFN editing, mitochondrial function and seed development in maize. The Plant Journal 84, 283–295. [DOI] [PubMed] [Google Scholar]
- Sun F, Xiu Z, Jiang R, Liu Y, Zhang X, Yang YZ, Li X, Zhang X, Wang Y, Tan BC. 2019. The mitochondrial pentatricopeptide repeat protein EMP12 is involved in the splicing of three nad2 introns and seed development in maize. Journal of Experimental Botany 70, 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Yamauchi T, Rajhi I, Nishizawa NK, Nakazono M. 2015. Transcript profiles in cortical cells of maize primary root during ethylene-induced lysigenous aerenchyma formation under aerobic conditions. Annals of Botany 115, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Verbitskiy D, Zehrmann A, Härtel B, Bayer-Császár E, Glass F, Brennicke A. 2014. RNA editing in plant mitochondria—connecting RNA target sequences and acting proteins. Mitochondrion 19, 191–197. [DOI] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Brennicke A, Graichen K. 2013a Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS One 8, e65343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Verbitskiy D, Härtel B, Brennicke A. 2013b RNA editing in plants and its evolution. Annual Review of Genetics 47, 335–352. [DOI] [PubMed] [Google Scholar]
- Tang J, Kobayashi K, Suzuki M, Matsumoto S, Muranaka T. 2010. The mitochondrial PPR protein LOVASTATIN INSENSITIVE 1 plays regulatory roles in cytosolic and plastidial isoprenoid biosynthesis through RNA editing. The Plant Journal 61, 456–466. [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC. 2013. Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. International Journal of Molecular Sciences 14, 6805–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbitskiy D, Merwe JA, Zehrmann A, Härtel B, Takenaka M. 2012. The E-class PPR protein MEF3 of Arabidopsis thaliana can also function in mitochondrial RNA editing with an additional DYW domain. Plant & Cell Physiology 53, 358–367. [DOI] [PubMed] [Google Scholar]
- Wang G, Qi W, Wu Q, Yao D, Zhang J, Zhu J, Wang G, Wang G, Tang Y, Song R. 2014. Identification and characterization of maize floury4 as a novel semidominant opaque mutant that disrupts protein body assembly. Plant Physiology 165, 582–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhong M, Shuai B, Song J, Zhang J, Han L, Ling H, Tang Y, Wang G, Song R. 2017. E+ subgroup PPR protein defective kernel 36 is required for multiple mitochondrial transcripts editing and seed development in maize and Arabidopsis. New Phytologist 214, 1563–1578. [DOI] [PubMed] [Google Scholar]
- Wu J, Sun Y, Zhao Y, et al. 2015. Deficient plastidic fatty acid synthesis triggers cell death by modulating mitochondrial reactive oxygen species. Cell Research 25, 621–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie T, Chen D, Wu J, Huang X, Wang Y, Tang K, Li J, Sun M, Peng X. 2016. Growing slowly 1 locus encodes a PLS-type PPR protein required for RNA editing and plant development in Arabidopsis. Journal of Experimental Botany 67, 5687–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiu Z, Sun F, Shen Y, Zhang X, Jiang R, Bonnard G, Zhang J, Tan BC. 2016. EMPTY PERICARP16 is required for mitochondrial nad2 intron 4 cis-splicing, complex I assembly and seed development in maize. The Plant Journal 85, 507–519. [DOI] [PubMed] [Google Scholar]
- Yang YZ, Ding S, Wang HC, Sun F, Huang WL, Song S, Xu C, Tan BC. 2017. The pentatricopeptide repeat protein EMP9 is required for mitochondrial ccmB and rps4 transcript editing, mitochondrial complex biogenesis and seed development in maize. New Phytologist 214, 782–795. [DOI] [PubMed] [Google Scholar]
- Yin P, Li Q, Yan C, et al. 2013. Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR. 2000. Regulation of programmed cell death in maize endosperm by abscisic acid. Plant Molecular Biology 42, 397–414. [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR, DeMason DA. 1997. Ethylene-mediated programmed cell death during maize endosperm development of wild-type and shrunken2 genotypes. Plant Physiology 115, 737–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang TT, Liu H, Shi Y, Wang M, Bie XM, Li XG, Zhang XS. 2018. Thioredoxin-mediated ROS homeostasis explains natural variation in plant regeneration. Plant Physiology 176, 2231–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YF, Suzuki M, Sun F, Tan BC. 2017. The mitochondrion-targeted PENTATRICOPEPTIDE REPEAT78 protein is required for nad5 mature mRNA stability and seed development in maize. Molecular Plant 10, 1321–1333. [DOI] [PubMed] [Google Scholar]
- Zsigmond L, Rigó G, Szarka A, Székely G, Otvös K, Darula Z, Medzihradszky KF, Koncz C, Koncz Z, Szabados L. 2008. Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport. Plant Physiology 146, 1721–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.