Abstract
The primary oncogenic event in ∼85% of Ewing sarcomas is a chromosomal translocation that generates a fusion oncogene encoding an aberrant transcription factor. The exact genomic breakpoints within the translocated genes, EWSR1 and FLI1, vary; however, in EWSR1, breakpoints typically occur within introns 7 or 8. We previously found that in Ewing sarcoma cells harboring EWSR1 intron 8 breakpoints, the RNA-binding protein HNRNPH1 facilitates a splicing event that excludes EWSR1 exon 8 from the EWS–FLI1 pre-mRNA to generate an in-frame mRNA. Here, we show that the processing of distinct EWS–FLI1 pre-mRNAs by HNRNPH1, but not other homologous family members, resembles alternative splicing of transcript variants of EWSR1. We demonstrate that HNRNPH1 recruitment is driven by guanine-rich sequences within EWSR1 exon 8 that have the potential to fold into RNA G-quadruplex structures. Critically, we demonstrate that an RNA mimetic of one of these G-quadruplexes modulates HNRNPH1 binding and induces a decrease in the growth of an EWSR1 exon 8 fusion-positive Ewing sarcoma cell line. Finally, we show that EWSR1 exon 8 fusion-positive cell lines are more sensitive to treatment with the pan-quadruplex binding molecule, pyridostatin (PDS), than EWSR1 exon 8 fusion-negative lines. Also, the treatment of EWSR1 exon 8 fusion-positive cells with PDS decreases EWS–FLI1 transcriptional activity, reversing the transcriptional deregulation driven by EWS–FLI1. Our findings illustrate that modulation of the alternative splicing of EWS–FLI1 pre-mRNA is a novel strategy for future therapeutics against the EWSR1 exon 8 containing fusion oncogenes present in a third of Ewing sarcoma.
Keywords: HNRNPH1, G-quadruplex, splicing, Ewing sarcoma, EWS–FLI1, EWSR1
INTRODUCTION
Members of the RNA-binding protein family heterogeneous nuclear ribonucleoproteins (hnRNPs) are linked to the development of distinct cancers and neurodegenerative diseases, making these proteins attractive unexploited therapeutic targets (Han et al. 2013; Geuens et al. 2016). However, because hnRNPs are involved in numerous pathways of nucleic acid metabolism, it is essential that any modulation of their function not disrupt transcriptome-wide RNA processing (Han et al. 2005). To this end, a more appropriate approach is to selectively target disease-specific hnRNP interactions, which may limit off-target inhibition of the global mRNA processing machinery. Compounds that block hnRNP interactions are currently underexplored but are not unprecedented. For example, the anti-tumor natural product camptothecin was found to inhibit hnRNP A1-topoisomerase 1 interactions (Manita et al. 2011), while a synthetic phenanthrene-based tylophorine derivative was determined to bind to hnRNP A2/B1 and reduce lung adenocarcinoma growth and metastasis (Chen et al. 2014). Thus, understanding the molecular mechanisms of hnRNPs cellular functions may help identify new therapeutic strategies to combat cancers and neurodegenerative diseases.
A subgroup of hnRNPs which includes HNRNPH1, HNRNPH2, and HNRNPF, regulate alternative splicing by binding to poly(G) tracts (Caputi and Zahler 2001; Black 2003; Han et al. 2005; Wang et al. 2007). There is precedent that poly(G) tract sequences have the potential to fold into stable secondary structures known as RNA G-quadruplexes and sequester hnRNP H/F proteins (Decorsiere et al. 2011; Fisette et al. 2012; Smith et al. 2014; Conlon et al. 2016; Huang et al. 2017). The mechanism by which these proteins regulate splicing is multifaceted since the hnRNP H/F proteins can mediate both exon inclusion and exclusion (Katz et al. 2010). For instance, when HNRNPH1 binds intronic elements in transcripts, such as BCLX (Garneau et al. 2005; Wang et al. 2007), GRIN1 (Han et al. 2005), and PLP1 (Wang et al. 2007), exon inclusion is enhanced. In contrast, when HNRNPH1 interacts with exonic elements in transcripts, such as α/β-tropomyosin (Chen et al. 1999; Expert-Bezançon et al. 2004; Crawford and Patton 2006) and human immunodeficiency virus type 1 (Jacquenet et al. 2001; Domsic et al. 2003), exon exclusion is enhanced. Furthermore, exon inclusion and exclusion for a given pre-mRNA may be independently regulated by HNRNPH1 as seen with different reported splice variants of ERBB2 (HER2) in breast cancer (Gautrey et al. 2015).
Ewing sarcoma (EWS) is a cancer of soft tissues and bones that occurs most frequently in children and young adults (Hawkins et al. 2011). We recently determined that HNRNPH1 is required to alternatively splice EWSR1 exon 8 fusion transcripts in cell lines representing a subgroup of EWS (Grohar et al. 2016). Briefly, the pathology in ∼85% of EWS is caused by a translocation involving chromosomes 11 and 22, which results in a fusion between the 5′ end of the EWSR1 (Ewing sarcoma breakpoint region 1) and the 3′ end of the FLI1 (Friend leukemia virus integration site 1) genes (Delattre et al. 1992; May et al. 1993). The generated EWS–FLI1 fusion oncogene encodes an aberrant transcription factor upon which EWS cells are dependent for proliferation and survival (May et al. 1993; Bailly et al. 1994). In about a third of EWS cases, the tumors that harbor EWS–FLI1 fusions must exclude EWSR1 exon 8 from the chimeric pre-mRNA to generate an in-frame transcript (Hawkins et al. 2011). Our previous study demonstrated that to facilitate the exclusion of EWSR1 exon 8, HNRNPH1 binds to G-rich regions within the fusion pre-mRNA transcripts (Grohar et al. 2016). However, the molecular mechanism that promotes HNRNPH1 binding and processing of EWS–FLI1, as well as whether HNRNPH1 plays a role in regulating the splicing of EWSR1 necessitates further elucidation.
Herein, we demonstrate using biochemical and cell-based studies that HNRNPH1 binds to a G-rich region at the 3′ end of EWSR1 exon 8 that can fold into RNA G-quadruplexes. Our analysis suggests HNRNPH1 may regulate the expression of two rare EWSR1 transcripts, facilitating the removal of all, or part, of EWSR1 exon 8. In contrast, HNRNPH1-mediated splicing becomes dominant and pivotal for the processing of EWS–FLI1 pre-mRNAs in EWS cells with EWSR1 intron 8 breakpoints. We also show that a single-stranded RNA oligomer can mimic one of the G-rich HNRNPH1 binding sites in EWSR1 exon 8 and, upon transfection, decrease EWS–FLI1 mRNA and protein expression in an EWS cell line harboring an EWSR1 intron 8 breakpoint. Furthermore, we can phenocopy these biological effects with a quadruplex-specific compound, pyridostatin (PDS), which blocks the interaction between HNRNPH1 and a G-rich region within EWSR1 exon 8. Critically, PDS treatment also leads to the reversal of transcriptional deregulation driven by EWS–FLI1. To the best of our knowledge, this is the first demonstration of the targeting of a fusion pre-mRNA transcript using a tool compound that modulates alternative splicing and provides a foundation for the direct targeting of the fusion transcript expressed in approximately one-third of Ewing sarcomas.
RESULTS AND DISCUSSION
The processing of distinct EWS–FLI1 pre-mRNAs by HNRNPH1 resembles alternative splicing of transcript variants of EWSR1
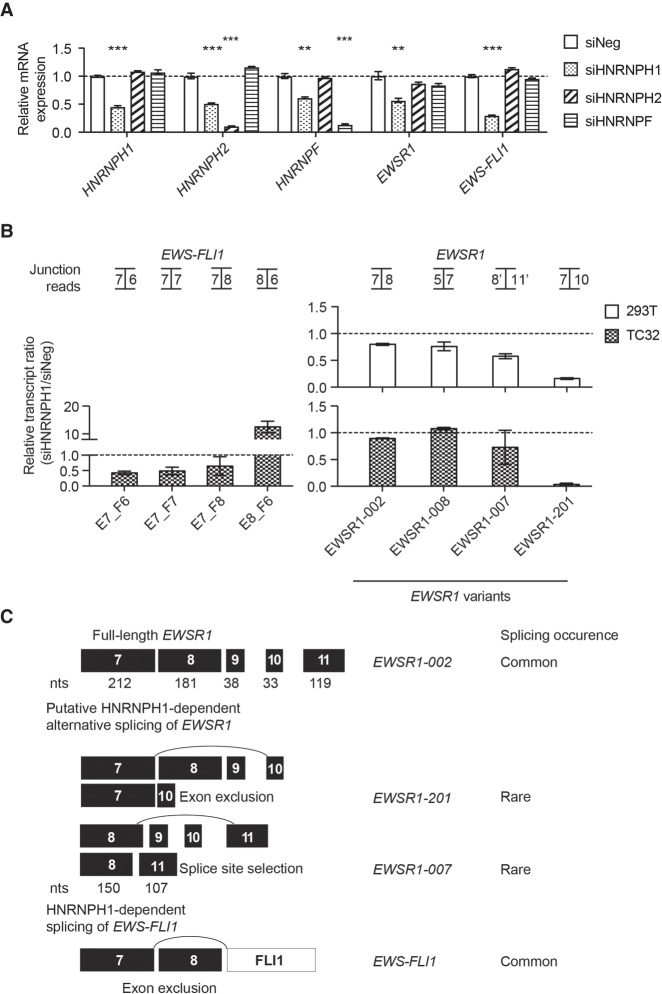
HNRNPH1 mediates the exclusion of EWSR1 exon 8 from EWS–FLI1 pre-mRNAs in EWS cells that express fusion transcripts containing this exon (Grohar et al. 2016). However, removal of EWSR1 exon 8 is atypical because the inclusion of exon 8 is crucial to express the dominant full-length, protein-coding transcripts of EWSR1. To better understand the possible functions of HNRNPH1 in the processing of EWS–FLI1 and EWSR1 pre-mRNAs, we performed silencing studies of HNRNPH1 in four different EWS cells (TC32 and SKNMC—EWSR1 intron 8 breakpoints, TC71 and RD-ES—EWSR1 intron 7 breakpoints) and a non-EWS line, HEK-293T. As expected, silencing of HNRNPH1 reduces EWS–FLI1 mRNA levels in cell lines expressing EWSR1 exon 8 containing fusion pre-mRNAs (TC32 and SKNMC; Fig. 1A; Supplemental Fig. S1A), but not in cell lines that harbor EWSR1 intron 7 breakpoints (Supplemental Fig. S1A). qPCR analysis also showed reduced EWSR1 mRNA levels upon depletion of HNRNPH1 in all cell lines tested (Fig. 1A; Supplemental Fig. S1A). An examination of HNRNPH2 and HNRNPF mRNA levels upon silencing of HNRNPH1 showed reduced mRNA expression suggesting that HNRNPH1 may function in the regulation of the expression of these other family members. Critically, however, the silencing of HNRNPH2 or HNRNPF directly had no significant effects on HNRNPH1 expression, nor EWSR1 or EWS–FLI1 mRNA levels (Fig. 1A). See Supplemental Figure S1B for immunoblot analysis of the siRNAs targeting the HNRNPH/F genes. As our findings also indicate a function for HNRNPH1 in the expression of EWSR1, consistent with its role in the regulation of RNA processing and metabolism (Uren et al. 2016), we next analyzed the effects of silencing HNRNPH1 on the expression of EWSR1 in more detail.
FIGURE 1.
HNRNPH1-mediated processing of EWSR1 exon 8 containing pre-mRNAs. (A) qPCR assessment of EWS–FLI1, EWSR1, HNRNPH1, HNRNPH2, and HNRNPF mRNA expression in HNRNPH1, HNRNPH2, or HNRNPF silenced TC32 EWS cells (48 h). Data are normalized to the geomean of reference genes, ACTB, RPL27, and NACA, and expressed relative to siNeg-transfected cells (mean ± SEM, n = 3). (**) P < 0.01; (***) P < 0.001 compared to siNeg-transfected cells. For additional experimental details, refer to Supplemental Table S7. (B) Expression of selected EWSR1 and EWS–FLI1 exon–exon junctions from targeted RNA-seq data analyzed using splicing junction analysis for HNRNPH1-silenced HEK-293T cells (top graph) and TC32 EWS cells (bottom graph). Data are expressed relative to siNeg-transfected cells (48 h, mean ± SEM, n = 3). (C) Schematic representation of putative HNRNPH1-dependent regulation of EWSR1 exon 8 containing pre-mRNAs.
There are over twenty reported transcript variants of EWSR1 (Supplemental Table S6). To assess the putative function of HNRNPH1 in the generation of one or more EWSR1 transcript variants, we used an enriched RNA-seq approach. Briefly, we harvested RNA from control and HNRNPH1-silenced HEK-293T and TC32 EWS cells, generated cDNA libraries, and enriched for EWSR1 sequences using a Chr. 22 fosmid clone containing the entire EWSR1 locus. Alignment of paired-end reads to EWSR1 transcripts (human assembly GRCh37), mapped RNA-seq reads to 20 out of the 21 reported splice variants (Supplemental Table S6; Supplemental Fig. S1C). To filter out the likelihood for EWSR1 transcript artifacts versus lowly expressed EWSR1 transcripts, we retained only those transcripts that represent at least 1% of the per-component (IsoPct) expression level. This analysis resulted in the detection of eight highly expressed and ten rare transcripts in both cell lines (Supplemental Fig. S1D,E), in which eight out of the 18 transcripts were protein-coding variants (Supplemental Fig. S1F). Consistently, depletion of HNRNPH1 was associated with a loss of EWS–FLI1 transcripts that skip only EWSR1 exon 8 and a substantial increase in an out-of-frame transcript that retains exon 8 in TC32 EWS cells (Fig. 1B). Analysis of read counts across exon junctions revealed that depletion of HNRNPH1 in HEK-293T and TC32 EWS cells was associated with loss of a rare transcript—EWSR1-201—that excludes both exon 8 and 9 (Fig. 1B). Also, silencing of HNRNPH1 had a minor effect on another rare variant that uses an alternative donor site in EWSR1 exon 8 (150 nt) and an alternative acceptor site in exon 11 (EWSR1-007). According to RNA expression data deposited in GTEx, EWSR1-007 is more readily expressed in skeletal muscle and different compartments of the brain where HNRNPH1 levels are decreased compared to other tissues such as skin (Supplemental Figs. S2, S3). Together with the targeted RNA-seq results, we hypothesize that HNRNPH1 interacts directly at the same binding site in EWSR1 exon 8 to mediate alternative splicing of EWSR1 and EWS–FLI1 transcripts (Fig. 1C).
Identification and validation of the rG1 oligomer to investigate HNRNPH1 binding specificity
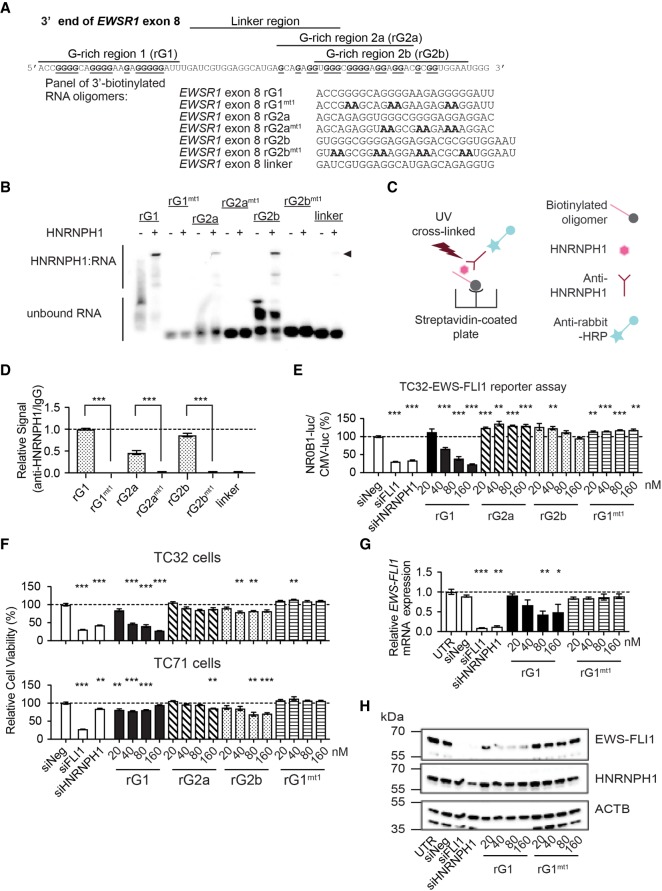
We previously reported that HNRNPH1 could bind oligonucleotides corresponding to G-rich sequences located at the 3′ end of EWSR1 exon 8 (Grohar et al. 2016). To determine the HNRNPH1 recognition sequence required for binding, we used chemiluminescent electrophoretic mobility shift assays (EMSA) and a series of 3′biotinylated single-stranded RNA (ssRNA) oligomers corresponding to different regions within the 3′ end of EWSR1 exon 8 (Fig. 2A; Supplemental Table S1). We observed mobility shifts for rG1, rG2a, and rG2b in the presence of purified HNRNPH1. Significantly, no mobility shifts were seen upon G to A nucleotide mutations (rG1mt1, rG2amt1, and rG2bmt1) (Fig. 2B). To confirm HNRNPH1 binding, we performed an antibody-based RNA binding assay with these biotinylated oligomers and purified HNRNPH1, as illustrated in Figure 2C. We saw an enrichment of HNRNPH1 binding in the order of rG1 > rG2b > rG2a (Fig. 2D). As expected, mutant RNAs (rG1mt1, rG2amt1, and rG2bmt1) exhibited significantly diminished enrichment of HNRNPH1 binding. Notably, we observed little to no HNRNPH1 binding of the linker sequence using either binding assay.
FIGURE 2.
Identification of the HNRNPH1 binding site, rG1, within the 3′ end of EWSR1 exon 8. (A) The sequence of the 3′ end of EWSR1 exon 8 (top) and the sequences of a series of 3′-biotinylated RNA oligomers corresponding to this exon (bottom). (B) Chemiluminescent EMSA gel for 3′-biotinylated EWSR1 exon 8 RNA oligomers shown in (A) in the absence or presence of purified HNRNPH1 protein. Arrowhead indicates mobility shift for each labeled probe. (C) Schematic representation of the antibody-based RNA oligomer binding assay. (D) The fold-enrichment for HNRNPH1-bound RNA oligomers determined by comparison of chemiluminescent signals from pull-down performed using HNRNPH1 antibody and an IgG isotype control. Data are shown as the mean ± SEM of three technical replicates. (***) P < 0.001. Recombinant, purified HNRNPH1 was used as the protein source. (E) Ratio of the TC32-NR0B1-luc and TC32-CMV-luc reporter signals 72 h posttransfection of the stated siRNA (20 nM) or synthetic RNA oligomer at the concentrations indicated. Data are normalized to control, siNeg (mean normalized, ±SEM, n = 4). (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 compared to siNeg. (F) The relative viability of EWS cell lines TC32 (top panel) and TC71 (bottom panel) 72 h posttransfection of the stated siRNA (20 nM) or synthetic RNA oligomer at the concentrations indicated. Data are normalized to control, siNeg (mean normalized, ±SEM, n = 4). (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 compared to siNeg. (G) qPCR assessment of EWS–FLI1 mRNA expression in siRNA or synthetic RNA oligomer transfected-TC32 EWS cells (48 h). Data are normalized to the reference gene NACA and expressed relative to untreated cells (mean ± SEM, n = 3). (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 compared to untreated cells. For additional experimental details, refer to Supplemental Table S7. (H) Immunoblot analysis of whole-cell lysates prepared from siRNA or synthetic RNA oligomer transfected TC32 EWS cells using antibodies against the proteins indicated (48 h). (UTR) Untreated cells.
Next, we tested whether RNA mimetic of the three EWSR1 G-rich regions (rG1, rG2b, and rG2a) that bind HNRNPH1 could compete with the EWS–FLI1 pre-mRNA for protein binding in cells. We first used a reporter assay of the transcriptional activity of EWS–FLI1 (Grohar et al. 2011). This assay makes use of TC32 EWS cells that express a luciferase reporter protein via either a CMV promoter (TC32-CMV-luc) or a promoter of the EWS–FLI1 regulated gene NR0B1 (TC32-NR0B1-luc). We transfected each reporter line with increasing concentrations of the rG1, rG2a, rG2b, or rG1mt1 ssRNA oligomers used for the binding studies and compared their effects on the readout of EWS–FLI1 transcriptional activity observed following transfection of siFLI1 or siHNRNPH1. As concentrations of rG1 increase, we observed a selective decrease in the TC32-NR0B1-luc reporter relative to the TC32-CMV-luc reporter, not seen for the rG2a, rG2b, or rG1mt1 oligomers (Fig. 2E). Importantly, transfection of the rG1 ssRNA oligomer at the higher concentrations (80 and 160 nM) recapitulated the effects of depleting EWS–FLI1 or HNRNPH1 by RNAi. Loss of EWS–FLI1 transcriptional activity should alter the viability of EWS cells. We thus assessed the effect of the rG1, rG2a, rG2b, or rG1mt1 ssRNA oligomers on the viability of TC32 and TC71 cells (Fig. 2F). Increasing concentrations of rG1 ssRNA oligomer altered the viability of TC32 cells with minimal decrease in TC71 cells, supporting a selective growth inhibition of EWS cells harboring EWSR1 exon 8 fusions.
To confirm that the changes in transcriptional activity and cell viability mediated by rG1 result from disruption of the expression of EWS–FLI1 in TC32 cells, we evaluated fusion transcript and protein levels 48 h posttransfection of the rG1 or rG1mt1 ssRNA oligomers. qPCR analysis of EWS–FLI1 mRNA levels showed a significant reduction by ∼50% at 80 and 160 nM rG1 compared to untreated control (Fig. 2G). Also, at rG1 concentrations ≥40 nM, immunoblot analysis revealed substantial depletion of EWS–FLI1 protein levels (Fig. 2H). In contrast, transfections of the negative control oligomer, rG1mt1, showed no significant effects on EWS–FLI1 mRNA or protein.
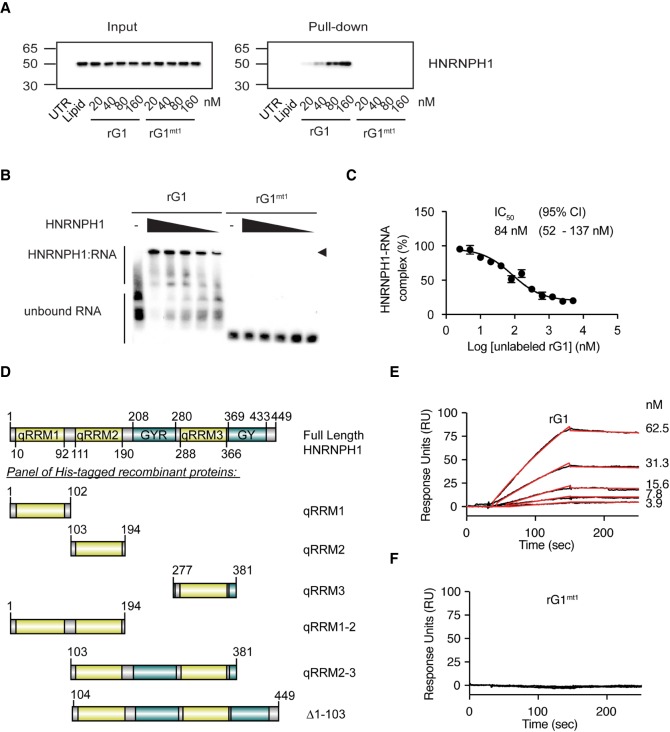
To establish in cells that the rG1 ssRNA oligomer interacts with HNRNPH1, we transfected TC32 cells with varying concentrations of the biotinylated rG1 or control rG1mt1 RNA oligomers for 24 h. We then prepared cell lysates, UV crosslinked and used streptavidin beads to pull-down protein associated with the biotinylated rG1 or the rG1mt1 RNA oligomers. Western blot analysis using an anti-HNRNPH1 antibody showed a concentration-dependent enrichment for HNRNPH1 binding to the rG1 oligomer not observed for the mutant RNA (Fig. 3A). These results suggest that the mode of action for rG1 is through direct interaction with HNRNPH1. We further verified rG1 specificity for HNRNPH1 using orthogonal in vitro assays. Using chemiluminescent EMSA, we observed that the intensity of the rG1:HNRNPH1 shifted band diminished as we decreased the concentration of purified HNRNPH1 (Fig. 3B). Furthermore, we performed antibody-based RNA binding assays at constant concentrations of purified HNRNPH1 and biotinylated rG1 with varying concentrations of nonbiotinylated rG1 as the RNA competitor. Nonbiotinylated rG1 was able to compete with labeled RNA with an IC50 of 84 nM (Fig. 3C), confirming HNRNPH1 binding specificity.
FIGURE 3.
Specificity of the HNRNPH1 and rG1 oligomer interaction. (A) TC32 cells were transfected with siRNA or synthetic ssRNA oligomer for 24 h and followed by streptavidin pull-down. Interaction between HNRNPH1 and rG1/rG1mt1 was confirmed by immunoblotting for HNRNPH1. (UTR) Untreated cells. (B) Chemiluminescent EMSA gel for 3′-biotinylated EWSR1 exon 8 rG1 and rG1mt1 at varying concentrations of purified HNRNPH1. Wedges indicate decreasing amounts of purified HNRNPH1 (20, 10, 5, 2.5, and 1.25 nM). Arrowhead indicates mobility shift for each labeled probe. (C) The fold-enrichment for HNRNPH1-bound rG1 at varying concentrations of nonbiotinylated rG1 determined by comparison of chemiluminescent signals from pull-down performed using HNRNPH1 antibody and an IgG isotype control. Data are shown as the mean ± SEM of three technical replicates. (D) A schematic representation of the protein domains of HNRNPH1 and a panel of recombinant His-tagged HNRNPH1 domains used in this study. All domain annotations are adapted from Van Dusen et al. (2010). Schematic diagrams were constructed using Domain Graph (DOG), version 2.0 (Ren et al. 2009). (E) The binding of full-length HNRNPH1 protein to a rG1 RNA target surface by SPR is shown. Black lines represent the binding responses for HNRNPH1 at the indicated concentrations on each sensorgram. HNRNPH1 was exposed to the surface for 2 min (association phase) followed by 10 min flow of running buffer (dissociation phase). Data were fit globally to a 1:1 binding model and the respective fits are shown in red. The resulting parameter values are provided in Table 1. (F) The binding of full-length HNRNPH1 protein to a rG1mt1 RNA target surface by SPR is shown. Black lines represent the binding responses for HNRNPH1 at 15.6, 31.3, 62.5, and 125 nM. HNRNPH1 was exposed to the surface as described in E; however, no binding was observed.
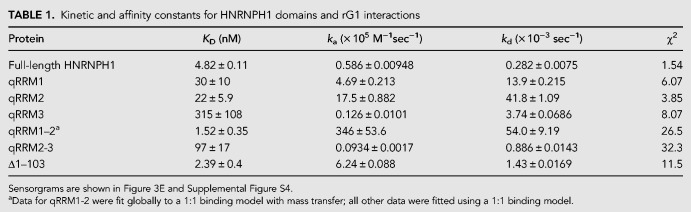
The HNRNPH1 protein includes three RNA binding domains known as quasi-RNA recognition motifs (qRRMs), a glycine-tyrosine-arginine-rich (GYR) domain, and a carboxy-terminal glycine-rich (GY) domain (Fig. 3D). To quantify the binding affinity of HNRNPH1 and its different domains for rG1, we performed SPR experiments with full-length protein and a series of recombinant proteins corresponding to different regions of HNRNPH1 (Fig. 3D). Table 1 summarizes the rate and binding constants for complexes of the different HNRNPH1 recombinant proteins with rG1 (sensorgrams are shown in Supplemental Fig. S4) compared to full-length HNRNPH1. The binding data for the full-length HNRNPH1 and rG1 interaction were fitted to a Langmuir (1:1 binding) model, yielding an equilibrium dissociation constant (KD) of 4.8 nM (Table 1; Fig. 3E). The mutated RNA oligomer, rG1mt1, exhibited no binding at all HNRNPH1 concentrations tested (Fig. 3F). Out of the three individual qRRM domains, qRRM1 and qRRM2 showed the highest affinity for rG1 with KD values of 30 and 22 nM, respectively (Table 1). qRRM3 displayed a 10-fold weaker affinity (KD = 315 nM). Precise KD values were more challenging to determine for proteins consisting of multiple domains of HNRNPH1 based on the model fit statistics. Nevertheless, a protein consisting of both the qRRM1 and qRRM2 domains showed comparable binding to full-length protein to rG1 (KD of 1.5 nM). We also detected weaker binding for the protein consisting of the qRRM2, GYR, and qRRM3 domains (KD of 97 nM), while a protein lacking the qRRM1 domain of HNRNPH1 (Δ1–103 = qRRM2, GYR, qRRM3, and GY domains) displayed high-affinity interactions for rG1 with a KD of 2.4 nM. The SPR results show that the three individual qRRM domains and the multidomain recombinant proteins all bind to rG1. Overall, our in vitro and cell-based data revealed that HNRNPH1 displays enhanced binding to the rG1 sequence within EWSR1 exon 8 in comparison to other G-rich sequences within the 3′ end of this exon. More importantly, we conclude this rG1 sequence is most likely the biologically relevant HNRNPH1 binding site within EWSR1 exon 8.
TABLE 1.
Kinetic and affinity constants for HNRNPH1 domains and rG1 interactions
The rG1 oligomer forms RNA G-quadruplexes in vitro
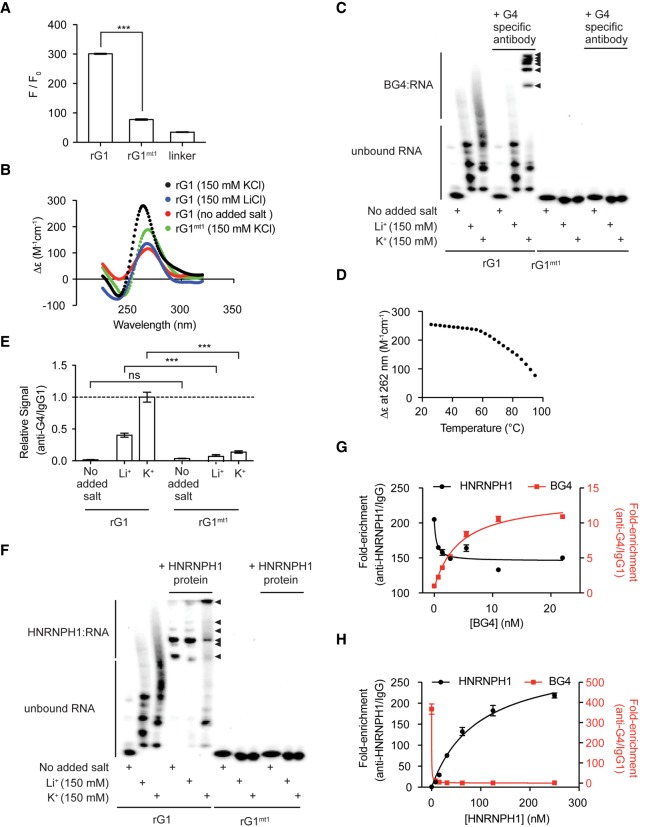
DNA and RNA sequences containing G-rich nucleotides have the potential to self-associate and form quadruplexes (Gellert et al. 1962; Arnott et al. 1974; Zimmerman et al. 1975; Howard et al. 1977). Quadruplexes are stable structures formed by Hoogsteen hydrogen bonding of G nucleotides around a central cation (Williamson 1994; Davis 2004). Since rG1 contains three successive G-runs in its sequence, we assessed the computational likelihood of rG1 forming stable quadruplexes using the quadruplex-forming G-rich sequences (QGRS) Mapper (Kikin et al. 2006). QGRS mapper predicted 53 quadruplexes in the rG1 sequence, in which the highest scoring QGRS comprised of repeats of two guanines and is consistent with a noncanonical, two-quartet quadruplex structure (Qin et al. 2015). To confirm RNA G-quadruplex formation, we utilized thioflavin T (ThT), a probe that is known to fluoresce in the presence of quadruplex structures (Xu et al. 2016). The rG1 oligomer generated ThT fluorescence while the rG1mt1 had a significantly lower signal than the nonmutated version of the oligomer (Fig. 4A). The linker region was utilized as a negative control and displayed a comparable fluorescence signal as rG1mt1. Our data suggest that rG1 forms RNA G-quadruplexes consistent with QGRS Mapper prediction.
FIGURE 4.
HNRNPH1 directly binds to unstructured G-tracts and RNA G-quadruplex structures within EWSR1 exon 8 rG1. (A) Fluorescence intensity enhancement (F/F0) of 6 µM thioflavin T with 2 µM RNA at the excitation and emission wavelengths of 440/487 nm in a buffer containing 20 mM Tris-HCl, pH 7.5, 150 mM KCl at 25°C for the indicated RNA oligomers. (***) P < 0.001. (B) Smoothened circular dichroism (CD) spectra for the 3′-biotinylated EWSR1 exon 8 rG1 or rG1mt1 oligomer (10 µM) at 25°C in a buffer containing 20 mM Tris-HCl, pH 7.5 with either no added salt, 150 mM LiCl, or 150 mM KCl. (C) Chemiluminescent EMSA gel for biotinylated EWSR1 exon 8 rG1 and rG1mt1 incubated in the absence and presence of the BG4-antibody under different salt conditions at 25°C in 20 mM Tris-HCl, pH 7.5. Arrowheads indicate mobility shifts for each labeled probe. (D) Smoothened CD melt curve of EWSR1 exon 8 rG1 (25 µM) in a buffer containing 20 mM Tris-HCl pH 7.5 and 150 mM KCl. (E) Fold-enrichment determined using a quadruplex-specific antibody for biotinylated EWSR1 exon 8 rG1 and rG1mt1 under different salt conditions at 25°C in 20 mM Tris-HCl (pH 7.5), which was determined by a change in the chemiluminescent signal using BG4-antibody and IgG1 isotype control. Data are shown as the mean ± SEM of three technical replicates. ns, not significant; (***) P < 0.001. (F) Chemiluminescent EMSA gel for biotinylated EWSR1 exon 8 rG1 and rG1mt1 incubated in the absence and presence of purified HNRNPH1 under different salt conditions at 25°C in 20 mM Tris HCl, pH 7.5. Arrowheads indicate mobility shifts for each labeled probe. (G) Fold-enrichment plots for rG1–HNRNPH1 (black) and rG1–BG4 (red) immunocomplexes obtained at 30 nM HNRNPH1 and varying concentrations of BG4-antibody. Data are shown as the mean ± SEM of three technical replicates. (H) Fold-enrichment plots for rG1–HNRNPH1 (black) and rG1–BG4 (red) immunocomplexes obtained at 0.5 nM BG4-antibody and varying concentrations of HNRNPH1. Data are shown as the mean ± SEM of three technical replicates.
Next, we evaluated the effects metal cations have on rG1 folding and stability. It is known that monovalent cations found in the central cavity of quadruplexes impact stability, formation, and polymorphism (Bhattacharyya et al. 2016). Hence, the stability of rG1 was evaluated using CD spectroscopy in the presence of lithium (Li+), potassium (K+), or no added cations. The CD spectra for rG1 had a positive peak at 262 nm and a negative peak at 240 nm under K+ conditions (Fig. 4B), characteristic of parallel-stranded RNA G-quadruplexes (Vorlíčková et al. 2012; Randazzo et al. 2013). Furthermore, the intensity of the peak at 262 nm was higher in the presence of K+ than with either Li+ or no salt added, confirming that K+ induces a more folded state. Also, the spectra for rG1mt1 differed from rG1 under K+ conditions, having a lower intensity in the positive peak at 262 nm and the negative peak slightly shifts to 245 nm. Overall, our CD results for rG1 in the presence of K+ are consistent with findings for RNA G-quadruplexes reported in VEGF, TRF2, BCL-2, and NRAS transcripts (Xu et al. 2015).
To directly confirm quadruplex formation in the presence of metal cations, we then compared the folded state for rG1 in the presence and absence of a quadruplex-specific antibody (BG4-antibody) using chemiluminescent EMSA. In the RNA only samples, we observed multiple bands for rG1 under Li+ and K+ conditions, suggesting metal ions induce various structural conformations and/or multimerization of rG1 (Fig. 4C). Addition of the BG4-antibody to rG1 led to several mobility shifts in the presence of K+ only, which corroborates quadruplex formation. The multiple bands observed upon the addition of the BG4-antibody indicate that various quadruplex structures could interact with this antibody. Given that there are 53 putative QGRS found in the rG1, it is plausible that this structure may be polymorphic. Since CD spectra of rG1 display characteristics of a parallel-stranded G-quadruplex, all guanines in a quartet must adopt the same glycosidic bond angle and the groove dimensions must remain identical (Tran et al. 2013). These distinct features allow for the possible formation of tetramolecular quadruplexes, which can resist high temperatures (e.g., 95°C–100°C) in the presence of potassium. Therefore, we tested the thermal stability of rG1 by CD melting experiments (Fig. 4D). We observed no distinct melting temperature transition, possibly confirming tetramolecular interactions.
We continued to assess BG4-antibody binding using the antibody-based RNA binding assay because this assay can capture both tight-binding and transient interactions through UV-crosslinking. Our results show that the binding affinity of rG1 for BG4-antibody was salt-dependent (Fig. 4E). The BG4-antibody binding was enriched by six- and 14-fold in the presence of Li+ and K+, respectively, compared to no salt. Overall, these salt-dependent studies demonstrate that potassium ions induced the most stable quadruplex formation for rG1.
HNRNPH1 interacts with both rG1 unstructured G-tracts and G-quadruplexes in vitro
To determine if the formation of rG1 quadruplexes affect HNRNPH1 recognition, we evaluated HNRNPH1 binding to rG1 under different metal cation conditions. Using EMSA, we observed distinct mobility shift patterns for rG1 in the presence of purified HNRNPH1 under each condition (Fig. 4F). There are three overall shifted bands with the highest signal intensity, which may indicate different conformations for the HNRNPH1–RNA complexes (Fried 1989). In contrast to the no salt and Li+ conditions, we detected unbound RNA after the addition of HNRNPH1 in the presence of K+. It is plausible that HNRNPH1 binding is modulated in the presence of K+ because rG1 has the potential to adopt multiple conformations and multimerizes. Moreover, we observed a dominant supershifted band for one of the HNRNPH1–RNA complexes under the K+ condition, compared to the no added salt and Li+ condition. This result suggests that multimerization of HNRNPH1 itself may be another reason for alteration of its binding specificity. Heterodimers and homodimers between members of the hnRNP H/F family have been implicated in modulating RNA binding in other studies (Alkan et al. 2006). Overall, these studies reveal that HNRNPH1 can interact with rG1 under conditions where the RNA is unfolded or stably folded into G-quadruplexes.
To further assess HNRNPH1 substrate specificity, we performed antibody-based binding assays containing both purified HNRNPH1 and BG4-antibody in the presence of rG1. To conduct these experiments, we first evaluated the apparent dissociation constant (KDapp) for each immunocomplex. We determined KDapp constants for the rG1–HNRNPH1 and rG1–BG4 immunocomplexes of 30 ± 2 nM and 0.5 ± 0.05 nM, respectively (Supplemental Fig. S5A,B). Then, we performed protein binding assays at the KDapp for each immunocomplex while varying the concentration of the other protein. As the concentration of BG4-antibody increased, there was a partial loss of HNRNPH1 binding to rG1 (Fig. 4G). However, as the concentration of HNRNPH1 increased, there was a complete loss of BG4-antibody binding to rG1 (Fig. 4H). The loss of protein binding (BG4 or HNRNPH1) to rG1 in these assays is most likely due to substrate sequestration that prevents the other protein from binding to RNA. We conclude that the differences in the RNA binding patterns observed are because the BG4-antibody solely recognizes quadruplexes, while HNRNPH1 binding is both structural- and sequence-dependent.
Insights into global recognition and selectivity of exonic RNA elements by HNRNPH1
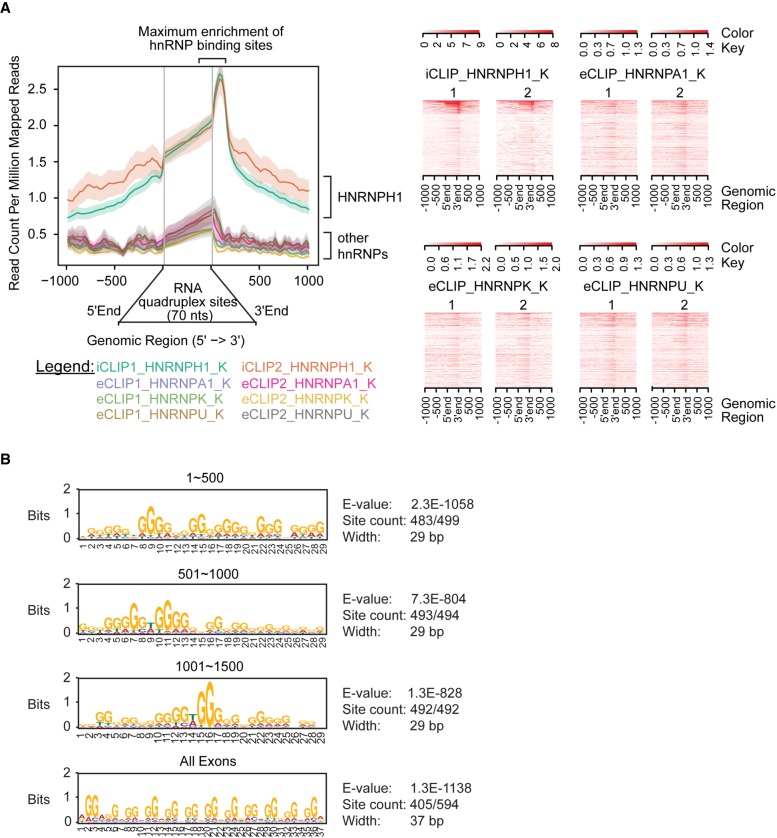
To investigate the broader context of RNA structural dependency for HNRNPH1 recognition within exonic elements, we analyzed the reported transcriptome-wide profiles for RNA G-quadruplexes and RNA targets of HNRNPH1. First, the RNA G-quadruplex sequencing data set GSE77282 was used to identify quadruplexes within polyadenylated-enriched RNA transcriptome-wide (Kwok et al. 2016). The RNA targets and binding sites of HNRNPH1 were identified from individual-nucleotide UV crosslinking and immunoprecipitation (iCLIP) or RNA immunoprecipitation (RIP) data sets that utilized total RNA (Uren et al. 2016). We then generated an enrichment profile of HNRNPH1 binding sites across different genomic regions and overlapping sequences that form RNA G-quadruplexes using ngs.plot (Shen et al. 2014). This analysis showed a correlation of 3346 RNA G-quadruplex sites with nearby sequences bound by HNRNPH1 (Fig. 5A; Supplemental Fig. S6A). Also, the absolute peak numbers for HNRNPH1 were four- to fivefold higher than other hnRNP family proteins (HNRNPA1, HNRNPK, and HNRNPU), suggesting that a subgroup of RNA targets may depend on structural binding specificity for HNRNPH1. These trends were confirmed using another RNA G-quadruplex sequencing data set GSE83617 that utilized polyadenylated-enriched RNA (Supplemental Fig. S6B; Guo and Bartel 2016). Our analysis is consistent with previous studies showing that the hnRNP H/F family of proteins regulates alternative splicing by binding to poly(G) tracts that fold into RNA G-quadruplexes (Decorsiere et al. 2011; Fisette et al. 2012; Smith et al. 2014; Conlon et al. 2016; Huang et al. 2017). Out of the 3346 RNA G-quadruplex sites that correlated with HNRNPH1 binding sites, 549 sites resided in exonic coding regions, and 313 sites were neither within the first nor the last exon. Critically, the EWSR1 gene ranked 1486 out of 3346 genes, and the HNRNPH1 binding site mapped to G-rich regions at the 3′ end of EWSR1 exon 8 (last 70 nt), consistent with our current findings.
FIGURE 5.
A subgroup of RNA G-quadruplex regions is enriched near HNRNPH1 binding sites in the human genome. (A) Coverage plots (left) and read density heat maps (right) represent binding read count for the indicated hnRNPs near RNA G-quadruplex sites (±1 Kb). The RNA targets and binding sites for each hnRNP were obtained from iCLIP or eCLIP data sets (Uren et al. 2016; Van Nostrand et al. 2016). The RNA G-quadruplex sites were obtained from the sequencing data set GSE77282 (Kwok et al. 2016). (B) Overrepresented sequence motifs at HNRNPH1 binding sites that overlap with either the highest-ranked 1500 RNA guanine quadruplex regions (binned into sets of 500) or all exonic regions containing potential RNA guanine quadruplexes.
To examine sequence context of this identified HNRNPH1–RNA quadruplex interactome, we used multiple EM for motif elicitation (MEME) analysis to predict HNRNPH1 binding sites in the exonic RNA G-quadruplex regions compared to all RNA G-quadruplex regions. Briefly, we ranked all RNA G-quadruplex regions by HNRNPH1 binding (iCLIP-Seq read counts) and subsequently performed a motif search analysis of all exonic sequences and the highest-ranked 1500 RNA G-quadruplex regions overall, binned into sets of 500 (Fig. 5B). Most of the first 500 regions (483 out of 499) contain a motif comprising four repeats of three to five guanines (G3-5)4 (Fig. 5B, top panel), which is consistent with the consensus sequence for canonical RNA G-quadruplex structures (G3L1-7)3G3 (Huppert et al. 2008). These results indicate a strong RNA G-quadruplex signal associated with the binding of HNRNPH1. Interestingly, most of the exonic regions (405 out of 549) contain repeats of two guanines with a consensus sequence that is consistent with noncanonical, two-quartet G-quadruplexes G2NXG2NYG2NZG2 (Fig. 5B, bottom panel; Qin et al. 2015). Thus, two-quartet G-quadruplex configurations, as predicted for rG1 using QGRS mapper, are overrepresented in these exonic HNRNPH1 binding regions, which may indicate sequence motifs associated with exon inclusion or exclusion.
Pyridostatin binds to rG1 oligomer and displaces HNRNPH1
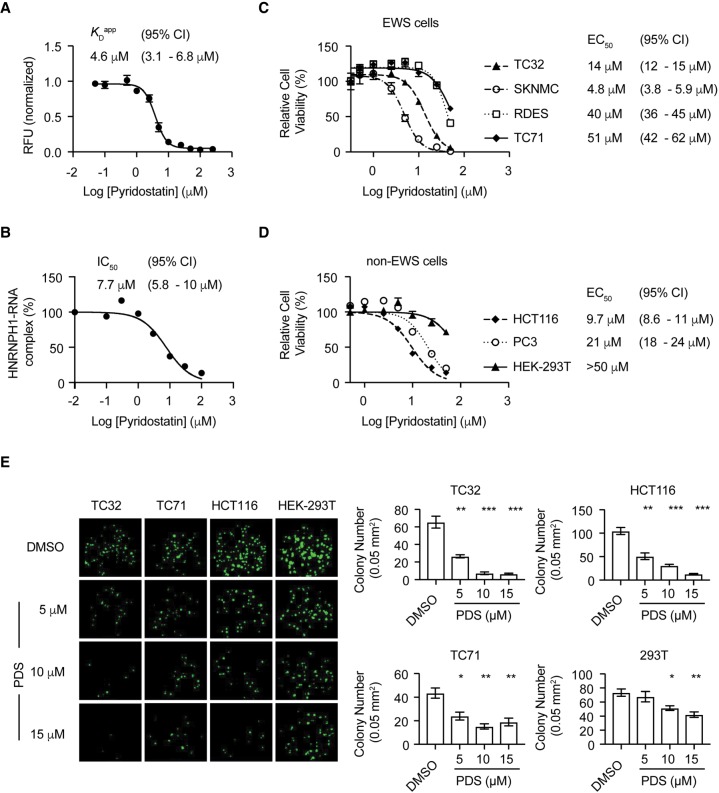
The selective chemical targeting of the HNRNPH1 and EWS–FLI1 pre-mRNA interaction may provide a rational treatment strategy for a subgroup of Ewing sarcoma patients. Since there are no known inhibitors of HNRNPH1 function to date, we pursued an alternative approach and selected to assess the quadruplex-binding compound pyridostatin (PDS). PDS is a synthetic molecule that stabilizes both RNA and DNA quadruplexes (Mela et al. 2012; Biffi et al. 2014) and is known to quench the fluorescence of fluorophore-labeled quadruplexes by proximal ligand binding at individual G-tetrads (Le et al. 2015). Hence, we first performed fluorescence titration binding assays at varying concentrations of PDS with a 5′-Alexa Fluor 647 labeled rG1 oligomer. This compound resulted in a dose-dependent loss of fluorescence emission for the 5′-Alexa Fluor 647 labeled rG1 oligomer, resulting in a KDapp of 4.6 µM (Fig. 6A). We subsequently performed an antibody-based RNA binding assay using the rG1 oligomer and HNRNPH1 from TC32 cell lysates at varying concentrations of PDS. We observed a PDS-induced loss of the HNRNPH1–rG1 complex, generating an IC50 for PDS of 7.7 µM (Fig. 6B). Our results indicate that a small molecule, such as PDS, can bind to rG1 and block its interaction with HNRNPH1. Since PDS is known to have a planar aromatic scaffold that can facilitate pi-stacking interactions with the G-tetrad ends (Le et al. 2015), the loss of HNRNPH1 binding could be attributed to gain of an unfavorable RNA conformation and/or limited substrate accessibility to allow HNRNPH1 to interact with the G-tetrad ends directly. Irrespective of the major driver of HNRNPH1 displacement, this mechanism of action differs from another G-quadruplex binding molecule, TMPyP4. TMPyP4 can destabilize and unfold RNA quadruplex structures (Morris et al. 2012; Zamiri et al. 2014), and previous studies have shown that TMPyP4 enhances HNRNPH1 binding to sequences containing G-quadruplexes within p53 RNA (Decorsiere et al. 2011). However, it is not atypical for PDS to displace nucleic acid-binding proteins as studies by Rodriguez and colleagues showed PDS could dissociate the shelterin component POT1 (protection of telomeres 1) and telomeric DNA complex (Rodriguez et al. 2008). To guide our understanding of how molecules like PDS modulate protein substrate recognition will require future study of the structure of the binary and ternary complexes comprising HNRNPH1, RNA substrate, and G-quadruplex stabilizers.
FIGURE 6.
PDS interacts with rG1 and selectivity inhibits the growth of EWS cell lines harboring EWSR1 exon 8 fusion transcripts. (A) PDS-induced fluorescence quenching of 5′-Alexa Fluor 647 labeled EWSR1 exon 8 rG1 oligomer. (B) The fold-enrichment for HNRNPH1-bound rG1 at varying concentrations of PDS determined by comparison of chemiluminescent signals from pull-down performed using HNRNPH1 antibody and an IgG isotype control. HNRNPH1 within TC32 whole cell lysates was used as the protein source. (C) Relative viability of EWS cell lines (TC32, SKNMC, RD-ES, and TC71) 72 h posttreatment at varying concentrations of pyridostatin normalized to control, 0.1% DMSO (mean normalized, ±SEM, n = 6). (D) Relative viability of non-EWS cell lines (HCT116, PC3, and HEK-293T) 72 h posttreatment at varying concentrations of pyridostatin normalized to control, 0.1% DMSO (mean normalized, ±SEM, n = 6). (E) Colony formation assay using cells pretreated with indicated concentrations of pyridostatin for 72 h and then grown for 7 d in compound-free medium. We imaged and counted colonies using a Celigo microwell image cytometer (mean normalized, ±SEM, n = 4). Colonies on plate are pseudocolored in green. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 compared to control, 0.1% DMSO. (CI) confidence interval, (UTR) untreated cells.
PDS modulates EWS–FLI1 transcriptional activity and inhibits viability in EWS cell lines harboring EWSR1 exon 8 fusion transcripts
To understand the effect of modulating quadruplex-specific interactions in cells, we assessed the viability of four EWS and three non-EWS cell lines treated with PDS. We observed that relatively low EC50 concentrations of PDS inhibited the growth of the EWSR1 exon 8 fusion-positive EWS cell lines TC32 (14 μM) and SKNMC (4.8 μM) compared to the EWS cell lines with different breakpoints in EWSR1 (>40 µM) (Fig. 6C). The differences in the sensitivity of TC32 and SKNMC cells may be attributed to differences in cell line origin and distinctive molecular characteristics (May et al. 2013). DNA G-quadruplexes have been assessed as potential molecular targets in both HCT116 and PC3 cells (McLuckie et al. 2013; Lu et al. 2016; Chilka et al. 2019) and we found the PDS EC50 values of 10 and 20 μM for these cell lines (Fig. 6D) to be comparable to that seen in the EWS cells harboring EWSR1 exon 8 fusions (TC32 and SKNMC). In contrast, the EC50 values observed for RD-ES and TC71 cells were closer to the higher concentrations of PDS treatment needed to inhibit the growth of HEK-293T cells (EC50 > 50 μM) (Fig. 6D). To our knowledge, there is no experimental evidence establishing that targeting DNA or RNA G-quadruplexes in this cell line alters the growth of this cell line.
We also examined more prolonged effects of PDS treatment with a clonogenic endpoint (Fig. 6E). Treatment of TC32, TC71, HCT116, and HEK-293T cells with PDS for 72 h decreased the number of colony-forming cells versus control in all cell lines to varying percentages up to the highest PDS concentration tested. Treatment at the lowest PDS concentration (5 μM) reduced the number of colony-forming cells for TC32 and HCT116 to ∼40%–50% versus controls, and this reduction was comparable to that observed at the highest PDS treatment (15 μM) for TC71 and HEK-293T cells. Moreover, treatment with 15 μM PDS drastically reduced the number of colony-forming cells to ∼10% for TC32 and HCT116. These results corroborate the selective growth inhibition of cell lines that have links to specific G-quadruplex molecular targets.
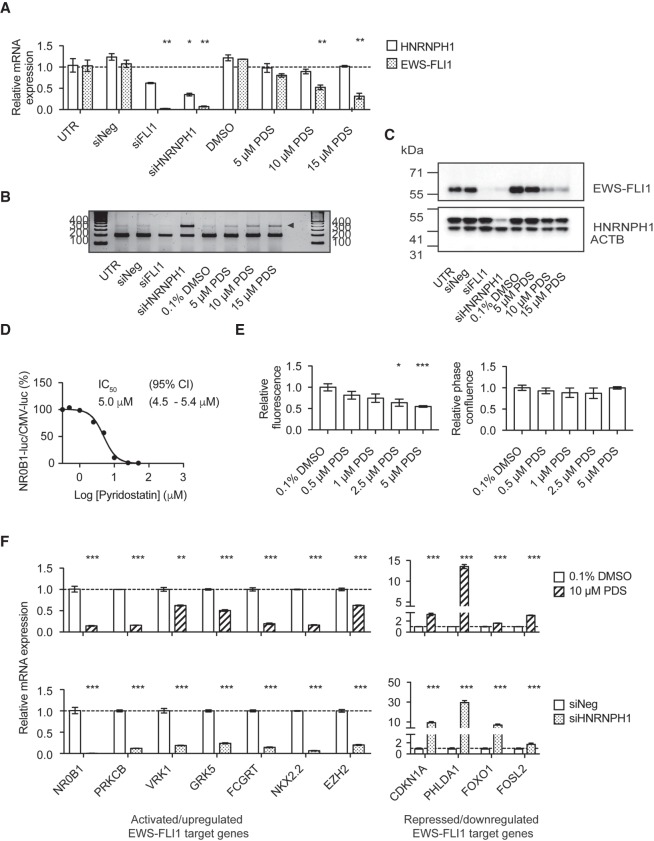
To further evaluate the effect of PDS treatment in EWS cells harboring EWSR1 exon 8 fusions, we compared mRNA and protein levels in PDS-treated TC32 cells with those transfected with siHNRNPH1 or siFLI1. Analysis of qPCR data shows a dose-dependent decrease in EWS–FLI1 mRNA levels following PDS treatment that phenocopied the silencing of HNRNPH1 and FLI1 (Fig. 7A). Using PCR splicing analysis, we also observe an additional PCR product for PDS-treated samples compared to control of a size consistent with the retention of EWSR1 exon 8 (Fig. 7B). Sanger sequencing confirmed retention of EWSR1 exon 8 following PDS treatment, and these results are comparable with those observed for HNRNPH1 silencing (Fig. 7B; Supplemental Fig. S7A). The inclusion of EWSR1 exon 8 results in the generation of a premature stop codon and leads to a loss of full-length EWS–FLI1 protein levels (Supplemental Fig. S7A,C).
FIGURE 7.
In TC32 cells, PDS modulates EWS–FLI1 mRNA processing, decreases EWS–FLI1 protein levels, and restores mRNA expression of EWS–FLI1 deregulated targets. (A) qPCR assessment of HNRNPH1 and EWS–FLI1 mRNA expression in PDS-treated or siRNA transfected TC32 EWS cells (48 h). Data are normalized to the geomean of reference genes, ACTB and RPL27, and expressed relative to untreated cells (mean ± SEM, n = 3). (*) P < 0.05; (**) P < 0.01 compared to untreated cells. For additional experimental details, refer to Supplemental Table S7. (B) PCR analysis of the splicing of EWS–FLI1 using primers corresponding to EWSR1 exon 7 forward and FLI1 exon 7 reverse primer pair for PDS-treated or siRNA transfected TC32 EWS cells (48 h). Arrowhead indicates additional PCR product band. (C) Immunoblot analysis of whole-cell lysates prepared from PDS-treated or siRNA transfected TC32 cells using antibodies against the proteins indicated (48 h). (D) Ratio of the TC32-NR0B1-luc and TC32-CMV-luc reporter signals 48 h posttreatment at varying concentrations of pyridostatin normalized to control, 0.1% DMSO (mean normalized, ±SEM, n = 6). (E) Relative fluorescence units at the excitation and emission wavelengths of 585/635 nm (top) and phase confluence (bottom) of TC32-NR0B1-mCherry cells at varying concentrations of pyridostatin normalized to control, 0.1% DMSO (48 h, mean normalized, ±SEM, n = 6). (*) P < 0.05; (***) P < 0.001 compared to control. (F) qPCR assessment of EWS–FLI1-regulated genes in PDS-treated (top) or HNRNPH1-silenced (bottom) TC32 EWS cells (72 h). Data normalized to ACTB are expressed relative to control, 0.1% DMSO or siNeg-transfected cells (mean ± SEM, n = 3). (**) P < 0.01; (***) P < 0.001 compared to control. For additional experimental details, refer to Supplemental Table S7. (CI) confidence interval, (UTR) untreated cells.
We expect lower EWS–FLI1 levels to affect the expression of genes that are typically deregulated by EWS–FLI1; thus, we performed two cell-based reporter assays to examine EWS–FLI1 transcriptional activity. First, we treated TC32-CMV-luc and TC32-NR0B1-luc reporter cells with PDS, and we assessed luciferase activity 48 h posttreatment. PDS-treatment induces a selective decrease in the TC32-NR0B1-luc reporter relative to the TC32-CMV-luc reporter with an IC50 of 5 µM (Fig. 7D). Next, we quantified EWS–FLI1 activity based on the change in fluorescence of a mCherry fluorescent reporter gene inserted at the 3′ end of the NR0B1 gene by CRISPR–Cas9 gene editing (TC32-NR0B1-mCherry). TC32-NR0B1-mCherry cells were treated with PDS at 0–5 µM to monitor changes in EWS–FLI1 activity. At these concentrations, fluorescence signals decreased in a dose-dependent manner with minimal effects on cell confluence (Fig. 7E). Both cell-based reporter assays demonstrate PDS can modulate EWS–FLI1 transcriptional activity.
To further investigate changes in EWS–FLI1 activity upon PDS-treatment in TC32 cells, we extended our mRNA expression studies to examine 11 genes that are deregulated by EWS–FLI1. Analysis of qPCR data 48 and 72 h posttreatment shows the mRNA levels of EWS–FLI1 target genes are significantly altered after 72 h, which is analogous to the observed trends for HNRNPH1 silencing (Fig. 7F; Supplemental Fig. S7B). These results demonstrate that PDS treatment in TC32 cells can reverse the expression of genes deregulated by EWS–FLI1.
Conclusion
Recent studies have shown an emerging role of HNRNPH1 in atypical splicing in hepatocellular carcinoma (Chettouh et al. 2013), glioblastoma (LeFave et al. 2011), breast cancer (Gautrey et al. 2015), African American prostate cancer (Jia et al. 2006) and a subgroup of Ewing sarcoma (Grohar et al. 2016). Thus, understanding the molecular basis for HNRNPH1 recognition in processing oncogenic fusion and cancer-related transcripts is imperative to develop novel RNA-binding small molecules and therapeutics. In this study, we have rigorously dissected RNA sequence and structural elements within EWSR1 exon 8 that led to identification and validation of the recruitment of HNRNPH1 to the rG1 binding site within this exon. Our studies show that HNRNPH1 can interact with both rG1 unstructured G-tracts and G-quadruplexes in vitro, though the conformation of the HNRNPH1–RNA complexes differs under conditions that can modulate RNA structure. A bioinformatic enrichment profile of HNRNPH1 binding sites and overlapping RNA G-quadruplex sites revealed a subgroup of exonic HNRNPH1 binding sites that contain sequences with two-quartet G-quadruplex configurations. However, it is still unclear if the mode of HNRNPH1-mediated exon exclusion is strictly dependent on RNA structure or sequence, as our studies suggest that both features may be mechanistically important. Future studies will focus on investigating this gap in knowledge as it will provide better insights into whether hnRNP H/F bind G-quadruplexes or prevent G-quadruplex formation. Furthermore, insights into the sequence and structural features of rG1 supplied information that allowed us to exploit a breakpoint-dependent vulnerability in a subgroup of Ewing sarcoma and modulate EWS–FLI1 pre-mRNA processing by both a synthetic RNA substrate mimic and a generic quadruplex-binding molecule, pyridostatin. Our findings confirm that HNRNPH1-mediated exon exclusion can be targetable through alteration of HNRNPH1 substrate recognition for a specific binding site.
The existence of RNA quadruplexes in vivo is currently controversial because current methods used to examine these structures may predispose experimental outcomes to lean toward a folded or unfolded RNA structure. For example, the use of quadruplex-specific antibodies in immunofluorescence studies can be beneficial to detect these distinct structural motifs in cells (Biffi et al. 2013, 2014; Henderson et al. 2014). However, it is possible that antibody specificity may induce the formation of quadruplexes not typically folded in a cellular context. Another example includes the use of chemical probes that can help modify nucleic acid substituents that are not directly participating in quadruplex-specific interactions (Guo and Bartel 2016). However, in vivo chemical modifications could nonspecifically alter the structure and destabilize the quadruplex depending on the concentration of the chemical reagent. Captivatingly, the processing of both EWSR1 and EWS–FLI1 transcripts, via HNRNPH1 recognition, provides a thought-provoking model system to address the ongoing debate of quadruplex cellular relevance. The exclusion of all or part of EWSR1 exon 8 within wild-type EWSR1 is rare; however, once an exon 8-containing segment of EWSR1 fuses to FLI1, this alternative splicing event becomes pivotal and supports the recruitment of HNRNPH1. Thus, future work will focus on understanding additional cis- and trans-acting elements that alter alternative splicing decisions for EWSR1 and its fusion transcripts. Overall, our study provides a glimpse into the molecular mechanisms of HNRNPH1 that orchestrate transcript fate and provides a foundation for a novel approach to target fusion oncogenes.
MATERIALS AND METHODS
Materials and reagents
All single-stranded RNA oligomers were purchased from Integrated DNA Technologies or Dharmacon. Supplemental Table S1 details the sequences and modifications of these oligomers used in each experiment. Control (siNegative [siNeg]) and gene-specific siRNAs (siFLI1, siHNRNPH1, siHNRNPH2, and siHNRNPF), and PCR primers are detailed in Supplemental Tables S2–S4. Antibodies against: HNRNPH1 (A300-511A, Bethyl Laboratories, Inc.), HNRNPH2 (ab181171, Abcam), HNRNPF (ab50982, Abcam), FLI1 (ab15289, Abcam), ACTB (sc-47778, Santa Cruz Biotechnology), BG4 (Ab00174-1.1, Labscoop), anti-Flag (sc-166355, Santa Cruz), and anti-6X His-tag (ab9108, Abcam) were used to analyze protein expression using standard immunoblotting and/or for the detection of protein–RNA complexes. See Supplemental Figure S1B for validation of HNRNPH/F siRNAs and antibodies. Purified full-length HNRNPH1 was purchased from OriGene Technologies. We designed DNA sequences for the expression of truncated HNRNPH1 proteins qRRM1, qRRM2, qRRM3, qRRM1-2, and qRRM2-3 (Supplemental Table S5) and the recombinant proteins were expressed and purified (see Supplemental Methods) by the Protein Expression Laboratory, NCI Frederick. The purified HNRNPH1 proteins Δ1–103 and qRRM2–3 were purchased from Creative BioMart. Pyridostatin (cat# S7444) was obtained from Selleckchem. The 96-well black plates, polystyrene (cat# 237105) and streptavidin-coated (cat# 15119), were purchased from ThermoFisher Scientific. The 384-well black, polystyrene plates (cat# 09-761-86) were purchased from Fisher Scientific. The 96-well white tissue culture-treated, CulturPlate-96 plates (cat# 6005680) were purchased from PerkinElmer.
Cell culture and transfection and compound treatments
All cell lines were grown in RPMI-1640 (Invitrogen) supplemented with 10% FBS (Invitrogen). The EWS cell lines TC32 (EWSR1 intron 8 breakpoint) and TC71 (EWSR1 intron 7 breakpoint) were gifted from Tim Triche (The Saban Research Hospital, Children's Hospital of Los Angeles) and we obtained the SKNMC cell line (EWSR1 intron 8 breakpoint) from Javed Khan (Genetics Branch, CCR, NCI). We purchased the RD-ES (EWSR1 intron 7 breakpoint), HEK-293T (human embryonic kidney), PC3 (human prostate cancer), and HCT116 (human colorectal cancer) cell lines from ATCC. The Pediatric Oncology Branch, CCR supplied the TC32-NR0B1-luc and TC32-CMV-luc reporter lines (Grohar et al. 2011). The generation of mCherry-NR0B1 tagged TC32 cells is described in Supplemental Methods. We confirmed the identity of cell lines by short tandem repeat analysis (ATCC; November 2015 and February 2018) and in the case of EWS cell lines expression of the EWS–FLI1 fusion transcript. We checked that cell lines were mycoplasma free twice a year.
For gene expression, splicing, and protein analysis, the following number of cells per well of a six-well plate were plated in 2 mL of growth medium: TC32, 1.35 × 105 cells per well; SKNMC, 2.7 × 105 cells per well; TC71, 1.8 × 105 cells per well; RD-ES, 1.8 × 105 cells per well; and HEK-293T, 1.8 × 105 cells per well. For RNAi-based studies, cells were transfected with 20 nM siRNA complexed with Lipofectamine RNAi-Max (Invitrogen) as follows: TC32, 4.5 µL/well; SKNMC, 4 µL/well; TC71 and RD-ES, 3.5 µL/well; and HEK-293T, 5 µL/well. For the synthetic RNA transfection studies, ssRNA oligomers were first heated and cooled to enable self-assembly and formation of stable tertiary structures and then transfected into the cells. We heated 50 µM of ssRNA oligomer in a buffer containing 20 mM Tris-HCl (pH 7.5) and 150 mM KCl to 95°C for 3 min and then cooled the RNA slowly to room temperature for 1.5 h. The RNA was diluted and complexed with 4.5 µL/well Lipofectamine RNAi-Max (final concentrations of 20, 40, 80, or 160 nM). Freshly passaged cells were then added to preplated transfection complexes. For compound treatment studies, cells were grown overnight and treated with 5, 10, or 15 µM pyridostatin (0.1% final DMSO). Cells were harvested 48 and/or 72 h posttransfection or -treatment. For reporter assays of EWS–FLI1 transcriptional activity or assessment of cell viability, cells were plated in a 96-well plate (cell viability and luciferase assays—6000 cells per well; mCherry assay—4000 cells per well), grown overnight, and then treated at varying concentrations of pyridostatin (0 to 50 µM; 0.1% DMSO) for 24–72 h.
For clonogenic assays, cells plated in a six-well plate were grown overnight and followed by 72 h of pyridostatin treatment (0, 5, 10, or 15 µM; 0.1% DMSO). After compound treatment, we collected each sample and plated 200–250 cells per well of a 12-well plate. We then incubated cells for 7 d in compound-free medium, after which we fixed colonies with 100% ethanol and stained with crystal violet (0.04% crystal violet, 2% methanol in PBS). Each experiment was performed in biological quadruplicates. The number of colonies was determined using a Celigo microwell image cytometer and software (Nexcelom Bioscience).
For the RNA pull-down experiments, transfected TC32 cells were irradiated, lysed with RNA immunoprecipitation (RIP) buffer (20 mM Tris HCl pH 7.5, 150 mM KCl, 0.5% Triton X-100, 1 mM PMSF, protease inhibitor cocktail and RNase inhibitor), sonicated three times for 5 sec, and centrifuged at 20,000g at 4°C for 30 min. Then, 80 µg of cellular extract was incubated overnight at 4°C with 25 µL Dynabeads M-280 Streptavidin (ThermoFisher Scientific) that were blocked in RIP buffer containing 0.5 mg/mL of BSA (ThermoFisher Scientific) and 0.05 mg/mL of tRNA (Thermo Fisher Scientific) for 4 h at 4°C. Beads were washed four times with RIP buffer. Bound proteins were eluted by heating the samples at 95°C for 5 min in 40 µL SDS-PAGE sample buffer. Twenty microliters of each sample was then loaded into a 3%–8% polyacrylamide gel (ThermoFisher Scientific), and a western blot against HNRNPH1 using anti-HNRNPH1 primary and anti-rabbit HRP secondary was performed.
Gene expression and splicing analysis
For PCR-based gene expression and splicing analysis, RNA was purified using Maxwell 16 LEV simplyRNA Kits (Promega) and reverse-transcribed using iScript Reverse Transcription Supermix (Bio-Rad). The synthesized first-strand cDNA was subsequently assayed by quantitative reverse transcription PCR (qPCR) using Fast SYBR Green Master Mix and StepOne Plus Real-time PCR system (Applied Biosystems). The expression of ACTB, RPL27, and/or NACA was used for normalization. Additional experimental details regarding each qPCR data set are provided in Supplemental Table S7. To detect gene splice variants, we conducted parallel analysis, synthesizing the first-strand cDNA using Platinum PCR SuperMix High Fidelity (Invitrogen). For Sanger sequencing, PCR products were further amplified using T7-forward and T3-reverse tagged primers, and after gel electrophoresis, the amplified products isolated utilizing an x-tractra Gel Extractor tool (LabGadget).
For RNA-sequencing, we collected poly(A)-selected RNA from three biological replicates, and sequencing libraries were created using a TruSeq Stranded mRNA LT Sample Preparation Kit as described by manufacturer's instructions (Illumina). To investigate differential expression of EWSR1 and EWS–FLI1, we incorporated a capture step using fosmid clones (CHORI) of full genomic coverage for EWSR1 (WIBR2-3436F12; GRCh37/hg19 chr22:29662533-29698996) or EWS–FLI1 (WIBR2-3436F12 and WIBR2-1520I3; GRCh37/hg19 chr11:128640224–128682737). Capture bait was prepared by Klenow extension of biotinylated random hexamers priming on a purified fosmid DNA template. The prepared bait was bound to streptavidin-coated Dynabeads C1 (Invitrogen) and hybridized to denatured, pooled libraries for 48 h. The enriched library was recovered after a series of stringency washes by PCR using primers homologous to library end-sequences (Forward: 5′-AATGATACGGCGACCACC-3′, Reverse: 5′-CAAGCAGAAGACGGCATA-3′). Libraries were quantified by qPCR (Kapa Biosystems) and sequenced on a NextSeq 500 Sequencer (Illumina).
Chemiluminescent electrophoretic mobility shift assays (EMSA)
Gel shift assays were performed using the LightShift chemiluminescent RNA EMSA kit (ThermoFisher Scientific). Briefly, 125 nM biotinylated RNA oligomer was annealed in a buffer containing 20 mM Tris-HCl (pH 7.5) and 150 mM KCl by heating to 95°C for 3 min and cooling slowly to room temperature for 1.5 h. In a total volume of 20 µL, 6.25 nM of annealed biotinylated RNA oligomer was incubated in the presence and absence of 0.01 µg of purified HNRNPH1. The binding buffer contained 2 µg tRNA, 10 mM HEPES (pH 7.3), 20 mM KCl, 1 mM MgCl2, and 1 mM DTT. For the salt dependency assays, the binding buffer was modified to contain 2 µg tRNA and 20 mM Tris-HCl (pH 7.5) with and without 150 mM LiCl or KCl. After 30-min incubation at room temperature, the samples were resolved on a 6% nondenaturing polyacrylamide gel (ThermoFisher Scientific) at 4°C, electrotransferred onto a 0.45 µm Biodyne B nylon membrane (ThermoFisher Scientific) at 400 mA for 30 min at 4°C, and crosslinked to the membrane (120 mJ/cm2, Stratalinker). The blots were then developed using the Chemiluminescent Nucleic Acid Detection Module (ThermoFisher Scientific).
Antibody-based RNA binding assays
Each biotinylated RNA oligomer (3 µM) was annealed in a buffer containing 20 mM Tris-HCl (pH 7.5) and 150 mM KCl by heating to 95°C for 3 min and cooling slowly to room temperature for 1.5 h. The RNA sample was then diluted to 20 nM in binding buffer (SuperBlock T20 PBS Blocking Buffer, ThermoFisher Scientific) and 100 µL aliquots were added in triplicate to a streptavidin-coated 96-well plate that was prewashed three times with wash buffer (Gibco PBS buffer, pH 7.4, containing 0.1% Tween-20). Subsequently, the plate was incubated with gentle shaking for 1 h at room temperature and then washed three times with wash buffer to remove unbound RNA. We then added 100 µL aliquots of 3 µg/mL TC32 whole cell lysate or 0.35 µg/mL purified HNRNPH1 in the binding buffer to the respective wells, incubated for 30 min, and crosslinked the samples by UV light irradiation (250 mJ/cm2, Stratalinker). Unbound protein was removed by washing three times with wash buffer, and the plate was then incubated with antibodies against HNRNPH1 (100 µL per well, 0.125 µg/mL) or rabbit IgG (cat # 02-6102, Invitrogen) as the negative control for 30 min at room temperature. The plate was washed three times with wash buffer and bound protein–RNA complexes were detected using 100 µL HRP-conjugated anti-rabbit IgG (1:200 dilution, cat# 7074, Cell Signaling Technology) per well (20 min, room temperature). The signal was developed using chemiluminescent reagents (SuperSignal West Dura, ThermoFisher Scientific), incubated at 37°C for 5 min, and luminescence was read on an EnSight Multimode plate reader (PerkinElmer). For compound treatment studies, varying concentrations of pyridostatin (0 to 100 µM; 1% DMSO) were added to the wells at the same step as the HNRNPH1 protein source. Data were normalized to DMSO only control and plotted against pyridostatin concentration. The IC50 value was determined using a dose-response inhibition model with variable slope (GraphPad Prism software).
To examine RNA salt-dependency and structural conformations, we modified the above procedure as follows. To evaluate salt-dependency, we changed the binding buffer to 20 mM Tris-HCl (pH 7.5) with and without 150 mM LiCl or KCl and the wash buffer to 20 mM Tris-HCl, pH 7.5, containing 0.1% Tween-20. BG4-antibody (100 µL per well, 3 ng/mL) or mouse IgG1 (cat# 02-6100, Invitrogen) as negative control were added to the respective wells following the removal of unbound RNA, incubated for 1 h and crosslinked by UV light irradiation (250 mJ/cm2, Stratalinker). The plate was washed three times with wash buffer, and RNA G-quadruplexes were detected using 100 µL HRP-conjugated anti-mouse IgG (1:200 dilution, cat# 7076, Cell Signaling Technology) per well (20 min, room temperature). Detection of the developed signal was as described above.
Circular dichroism spectroscopy
RNA oligomers were prepared at a concentration of 10 or 25 µM in 20 mM Tris-HCl (pH 7.5) in the presence and absence of either 150 mM LiCl or KCl. The RNA oligomers were folded by heating to 95°C for 3 min followed by slow cooling to room temperature for 1.5 h. CD spectra were recorded on an Aviv Biomedical Inc. Model 420 CD spectrometer at 25°C. The scans were performed in the wavelength range of 200–320 nm using a 3 sec averaging time, 1 nm step size, and 1 nm bandwidth. The melting curves were obtained by recording the change of molar circular dichroism or delta epsilon (Δε) at 262 nm in a range of temperatures from 25°C to 97°C with 1°C intervals. CD spectra shown were averaged from three individual spectra after subtraction of the buffer solution and then smoothened. A 1 mm path length quartz cuvette was used in all experiments.
Thioflavin T binding assays
In a black polystyrene 96 well-plate, annealed RNA oligomers (2.5 µM) were mixed with 6 µM Thioflavin T in 20 mM Tris-HCl (pH 7.5), 150 mM KCl in triplicate. The fluorescence emission was collected at 487 nm with excitation at 440 nm at room temperature using an EnSight Multimode plate reader (PerkinElmer).
Fluorescence intensity assay
In a black polystyrene 384 well-plate, pyridostatin was diluted to final concentrations ranging from 0 to 250 µM in triplicate in 10 mM Tris, 100 mM KCl (5% final DMSO). After shaking for 5 min, annealed 5′-Alexa Fluor 647-labeled RNA oligomer was added to a final concentration of 100 nM and the samples equilibrated for 30 min with shaking. The fluorescence intensity was then measured on a Synergy Mx microplate reader (BioTek) at an excitation wavelength of 649 nm and an emission wavelength of 670 nm. The fluorescence intensity was then normalized to the values obtained for RNA incubated with a DMSO control and was plotted against pyridostatin concentration. The apparent dissociation constants were determined using a single site model to fit the curve (GraphPad Prism software).
Surface plasmon resonance (SPR)
Surface plasmon resonance (SPR) was conducted on the Biacore 3000 platform (Biocore/GE Healthcare). All experiments were performed at 25°C in a SPR running buffer containing 10 mM Tris (pH 7.4), 100 mM KCl, 0.01% Tween-20, and 3% DMSO. A flow rate of 5 µL/min was used to build the Sensor chip. Streptavidin (ThermoFisher) at a concentration of 200 µg/mL in 10 mM sodium acetate (pH 5) was immobilized to ∼3000 response units (RU) in each flow cell using EDC/NHS coupling to a CM5 chip (GE Healthcare). The surface was then blocked with ethanolamine (50 µL, 1 M, pH 8.4) followed by fast injections at 50 µL/min of 1M NaCl, 50 mM NaOH (25 µL) over each flow cell. Next, 250 nM 5′-biotinylated RNA oligomer was annealed in the SPR running buffer by heating at 95°C for 5 min, followed by cooling to room temperature for over 1 h. The RNA was immobilized on one flow cell of the Sensor chip at a flow rate of 5 µL/min to a density of ∼250 RU. Serial dilutions of either full-length HNRNPH1 protein or its His-tagged domains were injected at a flow rate of 50 µL/min for 2 min. Each protein concentration was assessed twice, and binding data were fit to the Langmuir (1:1 binding) model or 1:1 binding with mass transfer model as indicated. Rate and binding constants for each RNA–protein complexes were determined using BIAevaluation software (GE Healthcare).
Phenotypic and functional assays
Cell viability and luciferase reporter expression were assayed on an EnSight Multimode plate reader (PerkinElmer) using CellTiter Glo and OneGlo assay systems, respectively (Promega) following the manufacturer's instructions. We used an IncuCyte ZOOM (Accela) system to monitor cell confluence and mCherry fluorescence (excitation: 585 nm, emission: 635 nm), taking measurements every 4 h for 2 d.
RNA-sequencing mapping and transcript variant analysis
RNA-seq reads were aligned to the hg19 reference genome using TopHat (Kim et al. 2013). Gene expression values were calculated as Reads Per Kilobase of transcript per Million mapped reads (RPKM) using Cufflinks (Trapnell et al. 2012) and the UCSC reference at the gene level. The non-EWS cell line HEK-293T was validated as a suitable model for assessing EWSR1 variants by comparison of RNA expression data deposited in the Genotype-Tissue Expression (GTEx) portal (http://www.gtexportal.org/).
RSEM tools were utilized for the analysis of the expression level of protein-coding EWSR1 transcript variants from targeted RNA-seq data. Reads counts across the signature junction of each transcript variant were extracted and calculated using an in-house splicing junction analysis tool developed in Python (v 3.6.0).
Bioinformatic analysis of RNA G-quadruplexes and hnRNP binding sites
We collected RNA G-quadruplex sequencing data from recently published studies (Guo and Bartel 2016; Kwok et al. 2016). The RNA targets and binding sites of HNRNPH1 were identified from iCLIP and RIP (Uren et al. 2016). The RNA targets and binding sites for the other HNRNP family proteins (A1, K, and U) were collected from eCLIP data (ENCODE) (Van Nostrand et al. 2016). The ngs.plot program was used to generate read density heat maps and the read coverage profiles of HNRNPH1 binding sites nearby each quadruplex region (Shen et al. 2014). The highest-ranked 1500 regions and all exonic regions that contained overlapping RNA G-quadruplexes and HNRNPH1 binding sites were submitted for motif discovery analysis by the meme-chip software (http://meme-suite.org/tools/meme-chip) using default parameters. The most significant motifs and E-values are reported.
Statistical analysis
Statistical analysis was conducted using a Student's t-test in GraphPad Prism 7. Multiple comparisons were corrected using the Bonferroni–Dunn method or two-stage linear set-up procedure of Benjamini, Krieger, and Yekutieli. P < 0.05 was considered significant. Concentration curves, IC50 values, and confidence intervals were generated using GraphPad Prism 7.
DATA DEPOSITION
The data sets containing enriched EWSR1 sequencing reads are available in the NCBI Gene Expression Omnibus repository (accession GSE119974).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Maggie Cam (CCR Collaborative Bioinformatics Resource), Thomas Meyer (CCR Collaborative Bioinformatics Resource), Yuelin Jack Zhu (Genetics Branch, CCR, NCI), and Sven Bilke (Genetics Branch, CCR, NCI) for their support with bioinformatic analyses; and Dr. Paul Meltzer (Genetics Branch, CCR, NCI) for advice on the study of enriched-RNA-seq data. We thank Mrs. Jane Jones (Protein Expression Laboratory, CCR, NCI) for expression and purification of recombinant HNRNPH1 proteins. Also, we would like to thank Dr. Sergey G. Tarasov for his technical assistance (Biophysics Resource, Structural Biophysics Laboratory, CCR, NCI). The CCR Genomics Core, CCR, NCI performed the Sanger sequencing. This work was supported by the Intramural Research Program of the National Cancer Institute (NCI), Center for Cancer Research (CCR); project numbers ZIA BC 011704 (N.J.C.) and ZIA BC 011585 (J.S.S.) and under contract no. HHSN261200800001E. Also, C.N. received funding support through a National Cancer Institute Director's Innovation Award, and N.A. was funded by the NCI Molecular Target and Drug Discovery Fellowship program (ZIJ BC 011133).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.072454.119.
Freely available online through the RNA Open Access option.
REFERENCES
- Alkan SA, Martincic K, Milcarek C. 2006. The hnRNPs F and H2 bind to similar sequences to influence gene expression. Biochem J 393: 361–371. 10.1042/BJ20050538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnott S, Chandrasekaran R, Marttila CM. 1974. Structures for polyinosinic acid and polyguanylic acid. Biochem J 141: 537–543. 10.1042/bj1410537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly RA, Bosselut R, Zucman J, Cormier F, Delattre O, Roussel M, Thomas G, Ghysdael J. 1994. DNA-binding and transcriptional activation properties of the EWS-FLI-1 fusion protein resulting from the t(11;22) translocation in Ewing sarcoma. Mol Cell Biol 14: 3230–3241. 10.1128/MCB.14.5.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya D, Mirihana Arachchilage G, Basu S. 2016. Metal cations in G-quadruplex folding and stability. Front Chem 4: 38 10.3389/fchem.2016.00038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G, Tannahill D, McCafferty J, Balasubramanian S. 2013. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem 5: 182–186. 10.1038/nchem.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biffi G, Di Antonio M, Tannahill D, Balasubramanian S. 2014. Visualization and selective chemical targeting of RNA G-quadruplex structures in the cytoplasm of human cells. Nat Chem 6: 75–80. 10.1038/nchem.1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336. 10.1146/annurev.biochem.72.121801.161720 [DOI] [PubMed] [Google Scholar]
- Caputi M, Zahler AM. 2001. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J Biol Chem 276: 43850–43859. 10.1074/jbc.M102861200 [DOI] [PubMed] [Google Scholar]
- Chen CD, Kobayashi R, Helfman DM. 1999. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat β-tropomyosin gene. Genes Dev 13: 593–606. 10.1101/gad.13.5.593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Yang SC, Lee KH, Yang X, Wei LY, Chow LP, Wang TC, Hong TM, Lin JC, Kuan C, et al. 2014. The antitumor agent PBT-1 directly targets HSP90 and hnRNP A2/B1 and inhibits lung adenocarcinoma growth and metastasis. J Med Chem 57: 677–685. 10.1021/jm401686b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chettouh H, Fartoux L, Aoudjehane L, Wendum D, Clapéron A, Chrétien Y, Rey C, Scatton O, Soubrane O, Conti F, et al. 2013. Mitogenic insulin receptor-A is overexpressed in human hepatocellular carcinoma due to EGFR-mediated dysregulation of RNA splicing factors. Cancer Res 73: 3974–3986. 10.1158/0008-5472.CAN-12-3824 [DOI] [PubMed] [Google Scholar]
- Chilka P, Desai N, Datta B. 2019. Small molecule fluorescent probes for G-quadruplex visualization as potential cancer theranostic agents. Molecules 24: 752 10.3390/molecules24040752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon EG, Lu L, Sharma A, Yamazaki T, Tang T, Shneider NA, Manley JL. 2016. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. Elife 5: e17820 10.7554/eLife.17820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford JB, Patton JG. 2006. Activation of α-tropomyosin exon 2 is regulated by the SR protein 9G8 and heterogeneous nuclear ribonucleoproteins H and F. Mol Cell Biol 26: 8791–8802. 10.1128/MCB.01677-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JT. 2004. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew Chem Int Ed Engl 43: 668–698. 10.1002/anie.200300589 [DOI] [PubMed] [Google Scholar]
- Decorsiere A, Cayrel A, Vagner S, Millevoi S. 2011. Essential role for the interaction between hnRNP H/F and a G quadruplex in maintaining p53 pre-mRNA 3'-end processing and function during DNA damage. Genes Dev 25: 220–225. 10.1101/gad.607011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delattre O, Zucman J, Plougastel B, Desmaze C, Melot T, Peter M, Kovar H, Joubert I, de Jong P, Rouleau G, et al. 1992. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature 359: 162–165. 10.1038/359162a0 [DOI] [PubMed] [Google Scholar]
- Domsic JK, Wang Y, Mayeda A, Krainer AR, Stoltzfus CM. 2003. Human immunodeficiency virus type 1 hnRNP A/B-dependent exonic splicing silencer ESSV antagonizes binding of U2AF65 to viral polypyrimidine tracts. Mol Cell Biol 23: 8762–8772. 10.1128/MCB.23.23.8762-8772.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expert-Bezançon A, Sureau A, Durosay P, Salesse R, Groeneveld H, Lecaer JP, Marie J. 2004. hnRNP A1 and the SR proteins ASF/SF2 and SC35 have antagonistic functions in splicing of β-tropomyosin exon 6B. J Biol Chem 279: 38249–38259. 10.1074/jbc.M405377200 [DOI] [PubMed] [Google Scholar]
- Fisette JF, Montagna DR, Mihailescu MR, Wolfe MS. 2012. A G-rich element forms a G-quadruplex and regulates BACE1 mRNA alternative splicing. J Neurochem 121: 763–773. 10.1111/j.1471-4159.2012.07680.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried MG. 1989. Measurement of protein-DNA interaction parameters by electrophoresis mobility shift assay. Electrophoresis 10: 366–376. 10.1002/elps.1150100515 [DOI] [PubMed] [Google Scholar]
- Garneau D, Revil T, Fisette JF, Chabot B. 2005. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J Biol Chem 280: 22641–22650. 10.1074/jbc.M501070200 [DOI] [PubMed] [Google Scholar]
- Gautrey H, Jackson C, Dittrich AL, Browell D, Lennard T, Tyson-Capper A. 2015. SRSF3 and hnRNP H1 regulate a splicing hotspot of HER2 in breast cancer cells. RNA Biol 12: 1139–1151. 10.1080/15476286.2015.1076610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M, Lipsett MN, Davies DR. 1962. Helix formation by guanylic acid. Proc Natl Acad Sci 48: 2013–2018. 10.1073/pnas.48.12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuens T, Bouhy D, Timmerman V. 2016. The hnRNP family: insights into their role in health and disease. Hum Genet 135: 851–867. 10.1007/s00439-016-1683-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohar PJ, Woldemichael GM, Griffin LB, Mendoza A, Chen QR, Yeung C, Currier DG, Davis S, Khanna C, Khan J, et al. 2011. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. J Natl Cancer Inst 103: 962–978. 10.1093/jnci/djr156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohar PJ, Kim S, Rangel Rivera GO, Sen N, Haddock S, Harlow ML, Maloney NK, Zhu J, O'Neill M, Jones TL, et al. 2016. Functional genomic screening reveals splicing of the EWS-FLI1 fusion transcript as a vulnerability in Ewing sarcoma. Cell Rep 14: 598–610. 10.1016/j.celrep.2015.12.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Bartel DP. 2016. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 353: aaf5371 10.1126/science.aaf5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Yeo G, An P, Burge CB, Grabowski PJ. 2005. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol 3: e158 10.1371/journal.pbio.0030158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han N, Li W, Zhang M. 2013. The function of the RNA-binding protein hnRNP in cancer metastasis. J Cancer Res Ther 9: S129–S134. 10.4103/0973-1482.122506 [DOI] [PubMed] [Google Scholar]
- Hawkins DS, Bolling T, Dubois S, Hogendoorn PCW, Jürgens H, Paulussen M, Randall RL, Lessnick SL. 2011. Ewing sarcoma. In Principles and practice of pediatric oncology (ed. Pizzo PA, Poplack DG), pp. 987–1014. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- Henderson A, Wu YL, Huang YC, Chavez EA, Platt J, Johnson FB, Brosh RM, Sen D, Lansdorp PM. 2014. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res 42: 860–869. 10.1093/nar/gkt957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard FB, Frazier J, Miles HT. 1977. Stable and metastable forms of poly(G). Biopolymers 16: 791–809. 10.1002/bip.1977.360160407 [DOI] [PubMed] [Google Scholar]
- Huang H, Zhang J, Harvey SE, Hu X, Cheng C. 2017. RNA G-quadruplex secondary structure promotes alternative splicing via the RNA-binding protein hnRNPF. Genes Dev 31: 2296–2309. 10.1101/gad.305862.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huppert JL, Bugaut A, Kumari S, Balasubramanian S. 2008. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res 36: 6260–6268. 10.1093/nar/gkn511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquenet S, Méreau A, Bilodeau PS, Damier L, Stoltzfus CM, Branlant C. 2001. A second exon splicing silencer within human immunodeficiency virus type 1 tat exon 2 represses splicing of Tat mRNA and binds protein hnRNP H. J Biol Chem 276: 40464–40475. 10.1074/jbc.M104070200 [DOI] [PubMed] [Google Scholar]
- Jia DW, Davis R, Moparty K, Haque S, Crawford BE, Srivastav SK, Abdel-Mageed A. 2006. hnRNPH1, a differentially expressed African American prostate cancer gene induces mitogenic response via a ligand-independent activation of androgen receptor. J Urol 175: 385. [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015. 10.1038/nmeth.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikin O, D'Antonio L, Bagga PS. 2006. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res 34: W676–W682. 10.1093/nar/gkl253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok CK, Marsico G, Sahakyan AB, Chambers VS, Balasubramanian S. 2016. rG4-seq reveals widespread formation of G-quadruplex structures in the human transcriptome. Nat Methods 13: 841–844. 10.1038/nmeth.3965 [DOI] [PubMed] [Google Scholar]
- Le DD, Di Antonio M, Chan LKM, Balasubramanian S. 2015. G-quadruplex ligands exhibit differential G-tetrad selectivity. Chem Commun 51: 8048–8050. 10.1039/C5CC02252E [DOI] [PubMed] [Google Scholar]
- LeFave CV, Squatrito M, Vorlova S, Rocco GL, Brennan CW, Holland EC, Pan YX, Cartegni L. 2011. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. EMBO J 30: 4084–4097. 10.1038/emboj.2011.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YJ, Hu DP, Zhang K, Wong WL, Chow CF. 2016. New pyridinium-based fluorescent dyes: a comparison of symmetry and side-group effects on G-quadruplex DNA binding selectivity and application in live cell imaging. Biosens Bioelectron 81: 373–381. 10.1016/j.bios.2016.03.012 [DOI] [PubMed] [Google Scholar]
- Manita D, Toba Y, Takakusagi Y, Matsumoto Y, Kusayanagi T, Takakusagi K, Tsukuda S, Takada K, Kanai Y, Kamisuki S, et al. 2011. Camptothecin (CPT) directly binds to human heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) and inhibits the hnRNP A1/topoisomerase I interaction. Bioorg Med Chem 19: 7690–7697. 10.1016/j.bmc.2011.09.059 [DOI] [PubMed] [Google Scholar]
- May WA, Lessnick SL, Braun BS, Klemsz M, Lewis BC, Lunsford LB, Hromas R, Denny CT. 1993. The Ewing's sarcoma EWS/FLI-1 fusion gene encodes a more potent transcriptional activator and is a more powerful transforming gene than FLI-1. Mol Cell Biol 13: 7393–7398. 10.1128/MCB.13.12.7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May WA, Grigoryan RS, Keshelava N, Cabral DJ, Christensen LL, Jenabi J, Ji L, Triche TJ, Lawlor ER, Reynolds CP. 2013. Characterization and drug resistance patterns of Ewing's sarcoma family tumor cell lines. PLoS ONE 8: e80060 10.1371/journal.pone.0080060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLuckie KI, Di Antonio M, Zecchini H, Xian J, Caldas C, Krippendorff BF, Tannahill D, Lowe C, Balasubramanian S. 2013. G-quadruplex DNA as a molecular target for induced synthetic lethality in cancer cells. J Am Chem Soc 135: 9640–9643. 10.1021/ja404868t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mela I, Kranaster R, Henderson RM, Balasubramanian S, Edwardson JM. 2012. Demonstration of ligand decoration, and ligand-induced perturbation, of G-quadruplexes in a plasmid using atomic force microscopy. Biochemistry 51: 578–585. 10.1021/bi201600g [DOI] [PubMed] [Google Scholar]
- Morris MJ, Wingate KL, Silwal J, Leeper TC, Basu S. 2012. The porphyrin TmPyP4 unfolds the extremely stable G-quadruplex in MT3-MMP mRNA and alleviates its repressive effect to enhance translation in eukaryotic cells. Nucleic Acids Res 40: 4137–4145. 10.1093/nar/gkr1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M, Chen Z, Luo Q, Wen Y, Zhang N, Jiang H, Yang H. 2015. Two-quartet G-quadruplexes formed by DNA sequences containing four contiguous GG runs. J Phys Chem B 119: 3706–3713. 10.1021/jp512914t [DOI] [PubMed] [Google Scholar]
- Randazzo A, Spada GP, da Silva MW. 2013. Circular dichroism of quadruplex structures. Top Curr Chem 330: 67–86. 10.1007/128_2012_331 [DOI] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. 2009. DOG 1.0: illustrator of protein domain structures. Cell Res 19: 271–273. 10.1038/cr.2009.6 [DOI] [PubMed] [Google Scholar]
- Rodriguez R, Müller S, Yeoman JA, Trentesaux C, Riou JF, Balasubramanian S. 2008. A novel small molecule that alters shelterin integrity and triggers a DNA-damage response at telomeres. J Am Chem Soc 130: 15758–15759. 10.1021/ja805615w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E. 2014. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15: 284 10.1186/1471-2164-15-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LD, Dickinson RL, Lucas CM, Cousins A, Malygin AA, Weldon C, Perrett AJ, Bottrill AR, Searle MS, Burley GA, et al. 2014. A targeted oligonucleotide enhancer of SMN2 exon 7 splicing forms competing quadruplex and protein complexes in functional conditions. Cell Rep 9: 193–205. 10.1016/j.celrep.2014.08.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PL, De Cian A, Gros J, Moriyama R, Mergny JL. 2013. Tetramolecular quadruplex stability and assembly. Top Curr Chem 330: 243–273. 10.1007/128_2012_334 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren PJ, Bahrami-Samani E, de Araujo PR, Vogel C, Qiao M, Burns SC, Smith AD, Penalva LO. 2016. High-throughput analyses of hnRNP H1 dissects its multi-functional aspect. RNA Biol 13: 400–411. 10.1080/15476286.2015.1138030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dusen CM, Yee L, McNally LM, McNally MT. 2010. A glycine-rich domain of hnRNP H/F promotes nucleocytoplasmic shuttling and nuclear import through an interaction with transportin 1. Mol Cell Biol 30: 2552–2562. 10.1128/MCB.00230-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514. 10.1038/nmeth.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorlíčková M, Kejnovská I, Sagi J, Renčiuk D, Bednářová K, Motlová J, Kypr J. 2012. Circular dichroism and guanine quadruplexes. Methods 57: 64–75. 10.1016/j.ymeth.2012.03.011 [DOI] [PubMed] [Google Scholar]
- Wang E, Dimova N, Cambi F. 2007. PLP/DM20 ratio is regulated by hnRNPH and F and a novel G-rich enhancer in oligodendrocytes. Nucleic Acids Res 35: 4164–4178. 10.1093/nar/gkm387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson JR. 1994. G-quartet structures in telomeric DNA. Annu Rev Biophys Biomol Struct 23: 703–730. 10.1146/annurev.bb.23.060194.003415 [DOI] [PubMed] [Google Scholar]
- Xu S, Li Q, Xiang J, Yang Q, Sun H, Guan A, Wang L, Liu Y, Yu L, Shi Y, et al. 2015. Directly lighting up RNA G-quadruplexes from test tubes to living human cells. Nucleic Acids Res 43: 9575–9586. 10.1093/nar/gkv1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Li Q, Xiang J, Yang Q, Sun H, Guan A, Wang L, Liu Y, Yu L, Shi Y, et al. 2016. Thioflavin T as an efficient fluorescence sensor for selective recognition of RNA G-quadruplexes. Sci Rep 6: 24793 10.1038/srep24793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamiri B, Reddy K, Macgregor RB Jr, Pearson CE. 2014. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J Biol Chem 289: 4653–4659. 10.1074/jbc.C113.502336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman SB, Cohen GH, Davies DR. 1975. X-ray fiber diffraction and model-building study of polyguanylic acid and polyinosinic acid. J Mol Biol 92: 181–192. 10.1016/0022-2836(75)90222-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.