Abstract
Differential gene isoform expression is a ubiquitous mechanism to enhance proteome diversity and maintain cell homeostasis. Mechanisms such as splicing that drive gene isoform variability are highly dynamic and responsive to changes in cell signaling pathways. Wnt/β-catenin signaling has profound effects on cell activity and cell fate and is known to modify several splicing events by altering the expression of individual splicing factors. However, a global assessment of how extensively Wnt signaling regulates splicing and other mechanisms that determine mRNA isoform composition in cancer is lacking. We used deep time-resolved RNA-seq in two independent in vivo Wnt-addicted tumor models during treatment with the potent Wnt inhibitor ETC-159 and examined Wnt regulated splicing events and splicing regulators. We found 1025 genes that underwent Wnt regulated variable exon usage leading to isoform expression changes. This was accompanied by extensive Wnt regulated changes in the expression of splicing regulators. Many of these Wnt regulated events were conserved in multiple human cancers, and many were linked to previously defined cancer-associated splicing quantitative trait loci. This suggests that the Wnt regulated splicing events are components of fundamental oncogenic processes. These findings demonstrate the wide-ranging effects of Wnt signaling on the isoform composition of the cell and provides an extensive resource of expression changes of splicing regulators and gene isoforms regulated by Wnt signaling.
Keywords: Wnt signaling, alternative splicing, cancer
INTRODUCTION
Greater than 90% of all multiexon human protein-coding genes produce multiple isoforms, generating increased functional diversity (Pan et al. 2008; Wang et al. 2008). Some of these isoform expression changes can alter non-protein-coding regions, whereas others can modify protein domains, leading to either complete or partial change of function (Kelemen et al. 2013). Although differential gene isoform expression is prevalent and essential in normal physiology, dysregulated isoform expression can result in disease and is often observed in cancer. These events can be brought about via the aberrant regulation of multiple mechanisms, including alternative promoter usage, alternative splicing, or alternative polyadenylation. Moreover, aberrant splicing is the target of emerging therapeutic strategies in cancer (for reviews, see Lee and Abdel-Wahab 2016; Jyotsana and Heuser 2018). Isoform diversity is controlled by tissue-specific factors that can be regulated by extracellular signals (Srebrow and Kornblihtt 2006; Ortis et al. 2010; Buljan et al. 2012; Martinez et al. 2012) and cancer-associated alterations in isoform composition can also be caused by dysregulated cell signaling pathways (Anczuków and Krainer 2016; Sveen et al. 2016).
Wnt signaling has potent and diverse effects on organismal development and the physiology of adult tissues. This pathway maintains homeostasis of a number of organs including the gut, skin, hair, and taste buds (Mah et al. 2016; Gaillard et al. 2017; Veltri et al. 2018). In its simplest form (Supplemental Fig. S1), Wnt signaling regulates the abundance of β-catenin, which then regulates the expression of tissue-specific target genes by binding to its cognate TCF/LEF transcription factors (Arce et al. 2006; Archbold et al. 2012; Cadigan and Waterman 2012). However, Wnt signaling can also regulate diverse downstream processes independent of β-catenin (Kohn and Moon 2005; Acebron and Niehrs 2016; Daulat and Borg 2017). If the mechanisms that tightly regulate these pathways are interrupted or perturbed, it can lead to diseases such as cancer (for review, see Nusse and Clevers 2017). Indeed, dysregulated Wnt signaling significantly modifies the gene expression pattern of cancer cells (Madan et al. 2016, 2018a).
Wnt-addicted cancers are an ideal model system to examine Wnt regulated variable exon usage leading to splicing changes. Mutations in RNF43 or RSPO3 that cause an increase in the abundance of Wnt receptors at the cell surface can cause Wnt addiction in cancers (for review, see Madan and Virshup 2015). Treatment of mice bearing Wnt-addicted cancer xenografts with orally administered PORCN inhibitors leads to a rapid cessation of Wnt signaling, followed by widespread changes in Wnt regulated gene expression, culminating in the suppression of cell proliferation and induction of cellular differentiation (Ho and Keller 2015; Madan et al. 2016, 2018a). Our group recently reported that >75% of the cell transcriptome in pancreatic and colorectal cancer is altered after inhibition of Wnt signaling in vivo (Madan et al. 2016, 2018a). Previous work has shown that dysregulation of the Wnt/β-catenin signaling pathway, examined particularly in colorectal cancers, can alter the gene expression of certain splicing regulators such as SRPK1, SRSF1, and SRSF3, affecting the splicing pattern of select genes such as CD44 and SLC39A14 (Gonçalves et al. 2008; Thorsen et al. 2011; Bordonaro 2013). Although the gene expression changes driven by Wnt signaling in multiple cell types have been explored by RNA sequencing (Madan et al. 2016, 2018a; Zimmerli et al. 2018; Michels et al. 2019), the same level of inquiry has not yet been extended to changes in gene isoform expression.
Here we report that Wnt signaling modifies the expression of numerous isoform expression regulators such as splicing factors, including many that were previously not known to be associated with Wnt signaling. Importantly Wnt signaling exerts a strong influence over the differential variable exon usage in more than 1000 genes, leading to isoform expression changes. Cis-regulatory sites that can influence the Wnt regulated differential exon usage were identified using splicing-associated quantitative trait loci (sQTLs). Some of these cis-regulatory sites can act as binding sites for transcription factors and function at the DNA level, whereas other cis-regulatory sites function only in RNA molecules by allowing the binding of regulatory proteins such as polyadenylation and splicing factors to promote or suppress the inclusion of the variable exon. This work highlights the considerable impact of Wnt signaling on differential gene isoform expression and provides a resource of Wnt regulated splicing events and splicing regulators.
RESULTS
Wnt signaling regulates the expression of multiple splicing regulators
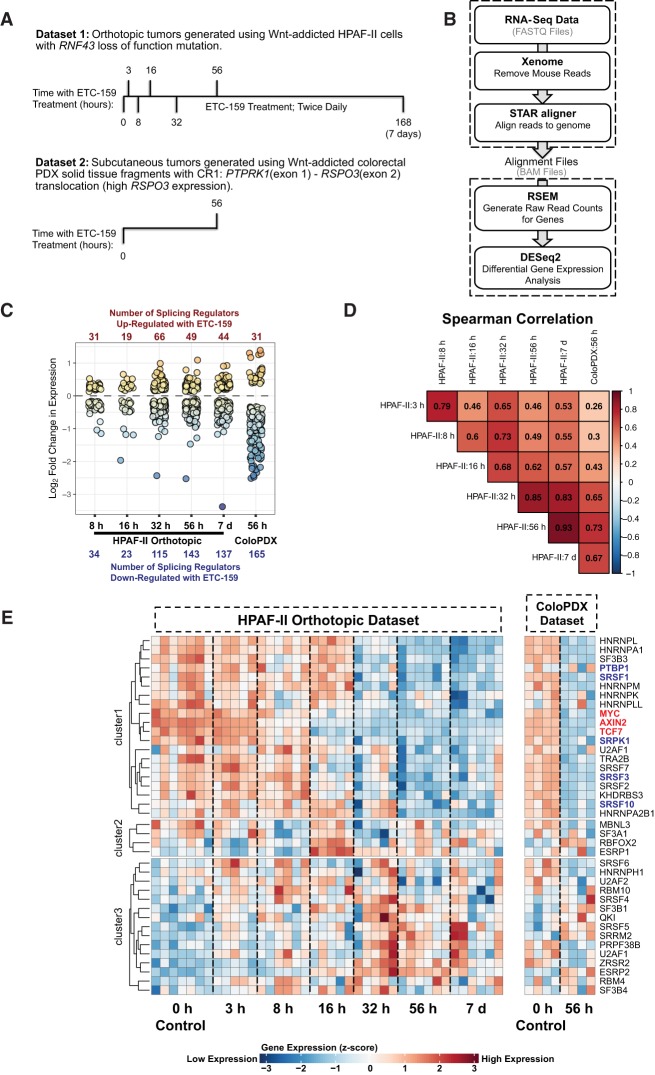
To ascertain the effects of Wnt signaling on the expression of gene isoforms and splicing regulators, we analyzed our large RNA-seq data sets from two physiologically relevant in vivo models of Wnt-addicted cancer (Madan et al. 2016, 2018a). The first data set is from an RNF43-mutant pancreatic cancer (HPAF-II) orthotopic xenograft model in which gene expression changes were assessed with five to seven replicates at each of seven time points over 7 d after the start of Wnt inhibition using ETC-159 treatment (Madan et al. 2018a). ETC-159 is a potent inhibitor of Wnt secretion with an IC50 of 2.9 nM (Madan et al. 2016). No significant off-target effects were identified during the evaluation of ETC-159 prior to its approval for human clinical trials. Additionally, the toxicities seen in both mice and humans treated with ETC-159 are shared with other PORCN inhibitors and are directly explainable by the mechanism of action of the drugs (Madan et al. 2018b). The time points were chosen in order to distinguish between the early and direct effects of Wnt signaling inhibition (3, 8, and 16 h) against the late and indirect consequences of Wnt signaling inhibition (32 h, 56 h, and 7 d). Changes in variable exon usage were also assessed in a second system with a RSPO3-mutant colorectal cancer patient-derived xenograft (PDX) sequenced 56 h after the start of ETC-159 treatment (Madan et al. 2016). A schematic illustration of the two experimental systems is shown in Figure 1A.
FIGURE 1.
Wnt signaling regulates the expression of multiple splicing regulators involved in cancer biology. (A) Overview of the two independent in vivo Wnt-addicted tumor models that were used to detect Wnt regulated differential gene and isoform expression. (B) Workflow used to assess Wnt regulated differential gene expression for 402 splicing regulators using the two in vivo tumor models. (C) Summary of the magnitude of gene expression change (measured as log2 fold changes) for the splicing regulators genes differentially expressed (relative to the untreated control) in the ETC-159-treated HPAF-II orthotopic model and colorectal PDX (ColoPDX) model. Note that the 3 h ETC-159-treated time point in the HPAF-II orthotopic model did not have any statistically significant differentially expressed splicing regulator genes. The numbers of up-regulated and down-regulated genes are indicated in red and blue, respectively. (D) Spearman correlation between the differential expression patterns of the splicing regulators at each time point using the log2 fold changes from 304 differentially expressed splicing regulators. (E) Heatmap of the scaled differential gene expression values (z-scores) of selected cancer-associated splicing regulators in response to Wnt signaling inhibition (with ETC-159 treatment). Three well-established Wnt target genes (AXIN2, MYC, and TCF7) have been included and highlighted in red for reference. The splicing regulators that are highlighted in blue (PTBP1, SRSF1, SRPK1, SRSF3, and SRSF10) were found in previous studies to be regulated by Wnt signaling. The heatmap was clustered into three sets via k-means clustering followed by hierarchical clustering to sort the genes within the sets.
Splicing regulators are proteins that regulate which exons are included or excluded in an mRNA transcript and consequently determine the exact isoforms that are produced from a specific gene. Proteins recognized to influence changes in isoform expression include splicing factors, kinases such as SRPK1 (Colwill et al. 1996), and chromatin modifiers (Kornblihtt 2006; Zhou et al. 2014). To determine which splicing regulators are Wnt regulated, we assessed the differential gene expression (Fig. 1B) of 402 human genes annotated as involved in RNA splicing (GO:0008380) in the Amigo gene ontology database (Ashburner et al. 2000; Carbon et al. 2009; The Gene Ontology Consortium 2017) following ETC-159 treatment. These 402 genes include splicing factors, transcription factors, chromatin remodelers, and 3′ end processing factors. Seventy-six percent (304) of these genes were differentially expressed (adjusted P-value ≤0.05) after inhibition of Wnt signaling in the least one of the two ETC-159-treated Wnt-addicted cancer RNA-seq data sets, with the majority of these expression changes occurring after 32 h of Wnt signaling inhibition (Fig. 1C). The magnitude of the changes in the differentially expressed splicing regulator genes was higher in the colorectal patient-derived xenograft than in the HPAF-II pancreatic cancer cell line implanted orthotopically, consistent with the higher dynamic range seen with other genes in our earlier study (Madan et al. 2018a). Nonetheless, there was a high degree of correlation in the expression changes (measured in terms of log2 fold changes of ETC-159-treated samples relative to the untreated controls) seen in the colorectal PDX (ColoPDX) data set and the later time points (32 h to 7 d) of the HPAF-II orthotopic data set (Fig. 1D). Among the 402 splicing regulators that we tested, the expression of 148 regulators changed consistently in the ColoPDXs and at least one of the time points in the HPAF-II orthotopic xenografts. Correlation between these putative cell-type agnostic Wnt regulated splicing regulators in the ColoPDXs and each of the five other time points in the HPAF-II orthotopic xenografts is shown in Supplemental Figure S4.
Many of these splicing regulators have been implicated in cancer (for reviews, see Anczuków and Krainer 2016; Sveen et al. 2016). A small number have previously been established as Wnt target genes—for example, SRSF1, SRPK1, SRSF3, and PTBP1 (Gonçalves et al. 2008; Thorsen et al. 2011; Bordonaro 2013). We also assessed the effect of Wnt signaling inhibition by ETC-159 on a range of cancer-associated splicing regulators (Fig. 1E; Chen et al. 2015b; Gabriel et al. 2015; Tian et al. 2015; Anczuków and Krainer 2016; Sveen et al. 2016; Zhou et al. 2017; Matsumoto et al. 2018; Sakuma et al. 2018a,b). We found that multiple such regulators, such as HNRNPA1, HNRNPA2B1, and HNRNPM, that have not previously been linked to Wnt signaling were affected by the suppression of Wnt signaling.
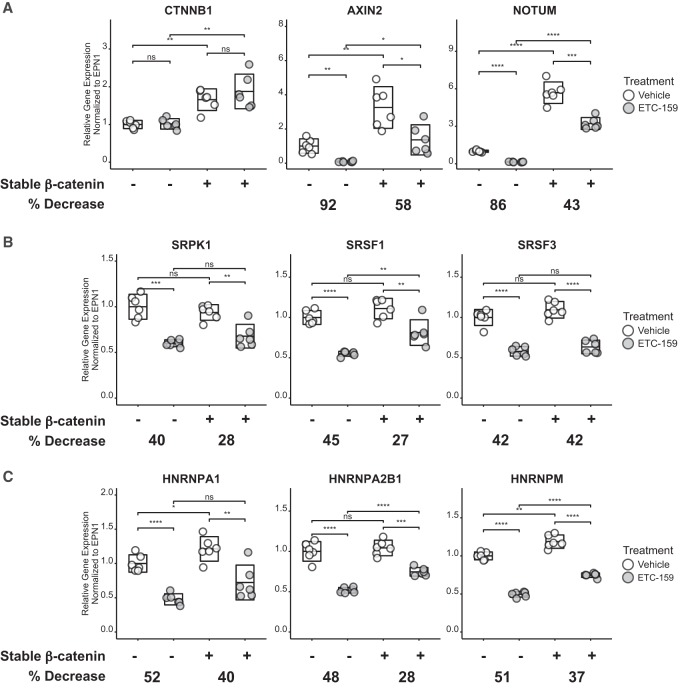
Selected gene expression changes were further validated in vitro by qPCR using control HPAF-II cells or the HPAF-II cells expressing stabilized β-catenin (Fig. 2). As shown in Figure 2A, ETC-159 treatment of HPAF-II cells suppresses the expression of core Wnt target genes AXIN2 and NOTUM by 92% and 86%, respectively. In contrast, in cells with stabilized β-catenin, the effect of ETC-159 was attenuated, as AXIN2 and NOTUM expression were reduced by only 58% and 43%, respectively. Stabilized β-catenin also attenuated the decrease in the expression of SRPK1 and SRSF1, genes that have been previously associated with Wnt signaling. Conversely, the ETC-159-induced decrease in SRSF3 was not attenuated by stabilized β-catenin (Fig. 2B). Inhibition of Wnt signaling suppressed the expression of HNRNPA1, HNRNPA2B1, and HNRNPM in HPAF-II cells and the ability of ETC-159 to suppress the expression of these genes was also attenuated in the cells with the stabilized β-catenin (Fig. 2C). Confirming that selected splicing factors were regulated by β-catenin signaling and not altered by an off-target effect of ETC-159, β-catenin knockdown with two independent siRNAs similarly decreased the expression of these genes (Supplemental Fig. S5A). Taken together we observed that the expression of many of these genes was largely regulated by the Wnt/β-catenin pathway. In our hands, SRSF3 expression was dependent on ETC-159 but not β-catenin, suggesting it could be regulated by previously described Wnt-dependent but β-catenin-independent pathways (Acebron et al. 2014; Koch et al. 2015).
FIGURE 2.
In vitro validation of the Wnt regulated differential gene expression of (A) β-catenin (CTNNB1) and two well-known Wnt target genes (AXIN2 and NOTUM) (note that both the endogenous and mutant CTNNB1 gene are detected by the primers), (B) three splicing regulators that were found to be regulated by Wnt signaling in previous studies (SRPK1, SRSF1, and SRSF3), and (C) three cancer-associated splicing regulators that were previously not associated with Wnt signaling (HNRNPA1, HNRNPA2B1, and HNRNPM). ETC-159 was administered to Wnt-addicted HPAF-II cells with or without the stabilized β-catenin. The percentage decrease in expression upon ETC-159 treatment was calculated. The boxplot shows the average relative gene expression (center line) and the 95% confidence interval (length of the box). A two-tailed t-test was used to determine significance between highlighted comparisons. For statistical tests, (ns) P > 0.05, (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001, (****) P ≤ 0.0001.
These findings demonstrate that Wnt signaling can directly or indirectly regulate the expression of a wide range of splicing regulators, many of which have been implicated in cancer. The complete differential expression analysis for the 402 splicing regulators and three Wnt target genes (AXIN2, MYC, and TCF7) is available in Supplemental Table 1.
Wnt signaling broadly regulates variable exon usage leading to isoform expression changes
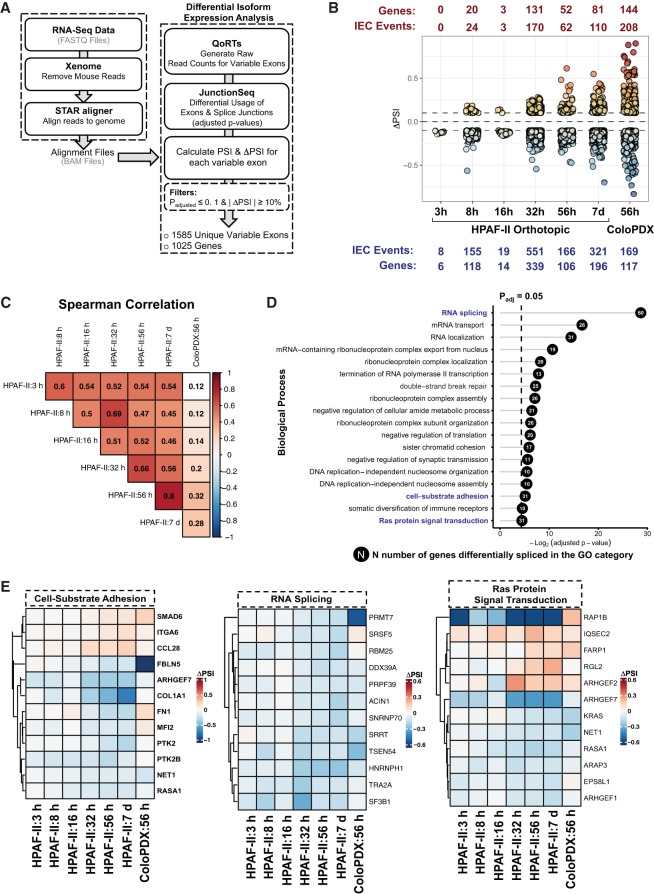
We next asked how many variable exon usage events leading to isoform expression changes were regulated by Wnt signaling. Differential exon expression was determined using the workflow defined in Figure 3A (see Materials and Methods). Briefly, we determined the statistical significance (adjusted P-value ≤0.1) and magnitude of relative exon inclusion (defined by the change in percent-spliced-in values |ΔPSI| ≥ 10%) for each variable exon (defined as differentially expressed exonic parts in the JunctionSeq algorithm) in ETC-159-treated samples relative to untreated controls (see Materials and Methods). This analysis identified 1025 unique genes with differential exon usage induced by Wnt signaling inhibition, accounting for 1585 variable exons. We considered isoform changes detected in either RNA-seq data set that passed this filter as events that were regulated by Wnt signaling. Validating our approach, we identified previously characterized Wnt regulated splicing changes including isoform changes that stem from the differential expression of mutually exclusive exons in SLC39A14 and exon skipping events in CD44 (Supplemental Figs. S6A,B, respectively; Wielenga et al. 1999; Thorsen et al. 2011). Details about the 1585 Wnt regulated variable exon usage events is presented in Supplemental Table 2.
FIGURE 3.
Wnt signaling regulates the differential isoform expression of genes involved in important biological processes. (A) Workflow with the different algorithms that were used to determine differential exon usage leading to isoform expression changes. (B) Summary of the magnitude of isoform expression change (IEC) events (measured as the change in percent-spliced in values [ΔPSI]) for the differentially expressed variable exons and their associated genes in both tumor models at indicated time points after the start of treatment with ETC-159. (C) Spearman correlation of the magnitude of variable exon usage (ΔPSI) between the various differentially expressed variable exons in each time point, shown as a heatmap. (D) Gene Ontology (GO) categories of genes that underwent isoform expression changes in at least one time point in either of the tumor models used, sorted by adjusted P-value. The numbers within the circles refer to the number of genes that underwent isoform changes in each of the categories. (E) Heatmaps corresponding to three GO categories (found in D and highlighted in blue) represent the Wnt regulated differential exon usage for a subset of genes. Each row represents a gene with a Wnt regulated variable exon. The ΔPSI for each variable exon measures the relative inclusion of the variable exon in the ETC-159-treated time points relative to the untreated control. Hierarchical clustering was applied to each of the heatmaps.
Wnts can regulate signaling through both transcriptional and posttranscriptional mechanisms. One method of posttranscriptional regulation of downstream signaling includes the rapid inhibition of GSK3 kinase activity (Taelman et al. 2010; Acebron et al. 2014). Consistent with such rapid posttranscriptional regulation, a few differential exon usage events were detected as early as 3 h following treatment with ETC-159 in HPAF-II orthotopic cancers (Fig. 3B). These events occurred prior to any statistically significant changes in the expression of core Wnt target genes and the 402 splicing regulators that we examined (Fig. 1C–E). We speculate that these early isoform expression changes may occur due to nontranscriptional Wnt regulated changes in cellular signaling involving kinases such as GSK3 and CK1 (Taelman et al. 2010; Acebron et al. 2014). This is consistent with a report by Shinde et al. (2017), who showed that GSK3 can phosphorylate and affect the activity of splicing factors such as RBM8A, SRSF9, and PSF in mouse embryonic stem cells. Indeed, we identified some overlap between splicing events in these distinct tissue types (Supplemental Fig. S7).
The number of differential exon usage events in the ETC-159-treated samples (measured in terms of ΔPSI values relative to the untreated controls) reached a peak at 32 h following the start of therapy (Fig. 3B). Most of the variability in exon usage events was seen at early time points (3 h to 32 h of ETC-159 treatment). The variability in these splicing events was reduced in the later time points as the correlation of the magnitude of change in exon inclusion (measured in ΔPSI) peaked between 56 h and 7 d ETC-159 treatment at 0.8 (Fig. 3C). This may be driven by the finding that most of the variability in splicing regulator gene expression was seen at the early time points, which then became more consistent (with a correlation of 0.93) at the later time points (Fig. 3C). Despite the inherent differences between the two tumor types used in the analysis, we found 35 differential exon usage events from 30 genes were shared (significant exon usage events [adjusted P-value ≤ 0.1] with the same direction of relative exon inclusion [ΔPSI] and significant magnitude of differential exon usage [|ΔPSI| ≥ 0.1]) between the pancreatic and colorectal cancers. The complete list of these genes and differential exon usage events is shown in Supplemental Figure S8. This highlights that a number of the Wnt regulated splicing events were shared between the pancreatic and colorectal cancers.
Gene ontology (GO) analysis revealed that the genes that underwent Wnt regulated differential exon usage were involved in a number of important biological processes that are implicated in tumor progression, such as RNA splicing, cell-substrate adhesion, and Ras signaling (Fig. 3D; Supplemental Fig. S9). Examples of genes with alternative exon usage that comprise these categories are shown in Figure 3E. Some of the genes with differentially expressed exons, such as SMAD6, DDX39A, FBLN5, and KRAS, have strong and consistent differential exon usage patterns in both tumor types. This is consistent with Wnt signaling broadly modulating the expression of gene isoforms involved in biological processes associated with cancer.
Wnt regulated splicing events found in multiple tumor types
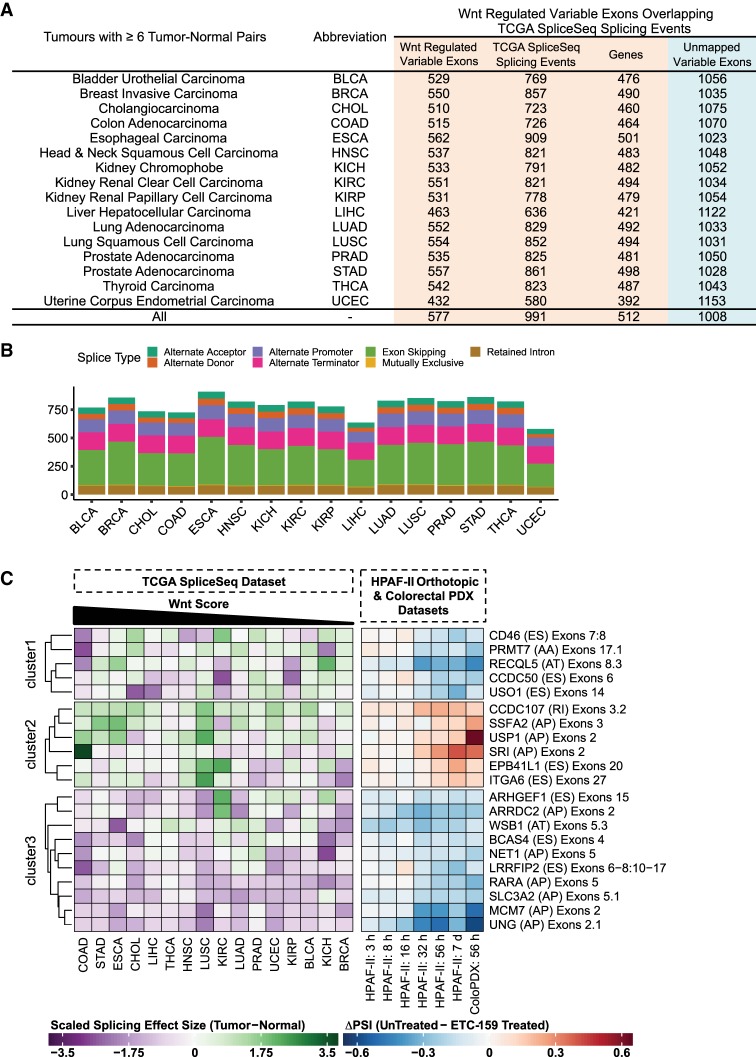
Our analysis in Wnt-addicted pancreatic and colorectal cancer models identified several genes with Wnt regulated variable exon usage. We next asked if these variable exons were significantly dysregulated in other human cancers, which would suggest a more fundamental role in cancer progression. A number of studies have examined isoform expression alterations in different cancer types using The Cancer Genome Atlas (TCGA) RNA-seq data sets (Danan-Gotthold et al. 2015; Sebestyén et al. 2016). We looked at the cancer-associated splicing events described in the TCGA SpliceSeq database (https://bioinformatics.mdanderson.org/TCGASpliceSeq/) (Ryan et al. 2016). This database contains mRNA splicing information (including alternative promoter usage events and alternative termination or polyadenylation events) for a broad range of cancers, derived using TCGA RNA-seq data sets. We determined the differential splicing (ΔPSI = PSItumor − PSImatched normal) of cancer types in which data from at least six tumor-normal pairs was available. We found 991 cancer-associated splicing events that overlapped with 577 Wnt regulated variable exons from 512 genes (Fig. 4A). Detailed splicing information for the 991 unique splicing events found in 16 different cancer types can be found in Supplemental Table 4. The most frequent types of Wnt regulated splicing events detected in multiple cancers were exon skipping, alternative promoter usage and alternative polyadenylation events (Fig. 4B). This suggests that these shared variable exon changes represent core events undergoing positive selection in cancer.
FIGURE 4.
Wnt regulated isoform expression changes are found in multiple cancer types. (A) Tabulation of the number of Wnt regulated variable exons that overlap with splicing events described in the TCGA SpliceSeq database for each tumor type. Only tumors with at least six tumor-normal pairs were considered for the analysis. The unmapped Wnt regulated variable exons did not overlap with any significant splicing events in the TCGA SpliceSeq database for the tumors tested. (B) Frequency plot of the different types of splicing events (n = 991) in the TCGA SpliceSeq database that overlapped with Wnt regulated variable exons. Plot shows the distribution of the splicing events for each tumor type. (C) Heatmap of a subset of the splicing events (described in the TCGA SpliceSeq database) that overlap with Wnt regulated variable exons. We define these as Wnt regulated splicing events. Each row is annotated with gene name, (splice type), and affected exons. The left panel shows the effect size (Hedges’ g-estimate) of the splicing change (ΔPSI) seen in the 16 cancer types relative to the matched normals. The effect sizes were scaled by row (divided by the root mean square) but not centered. The tumor types were sorted according to the Wnt score (Materials and Methods). The higher the Wnt score, the greater the relative influence of Wnt/β-catenin signaling on the tumor. The right panel shows the magnitude of splicing change (ΔPSI) for different time points in the HPAF-II orthotopic and the colorectal PDX (ColoPDX) models. Both panels refer to the same set of splicing events. The different splice types include (AA) alternative acceptor, (AD) alternative donor, (AP) alternative promoter, (AT) alternative terminator, (ES) exon skipping , (RI) retained intron. The heatmap was clustered into three sets via k-means clustering followed by hierarchical clustering to sort the genes within the sets.
Splicing effect sizes for each splicing event in each cancer, based on the ΔPSI values, indicate the relative inclusion of an exon related to the splicing event. Figure 4C describes some of these Wnt regulated splicing events that were also detected in different cancers. The two heatmaps compare the same splicing events with each row representing a particular splicing event (or expression of variable exons). The right panel shows Wnt regulated splicing events (measured in ΔPSI values) identified using our HPAF-II orthotopic xenografts and ColoPDXs xenograft models treated with ETC-159. The left panel compares these Wnt regulated splicing in different cancer types from the TCGA SpliceSeq data set. The tumor samples from the TCGA data set are arranged according to their Wnt score, determined based on the level of expression of Wnt target genes in paired normal versus tumor samples. For example, our analysis identified that the inclusion of SRI exon 2 was Wnt regulated in HPAF-II and ColoPDX models (right panel). Comparing across additional tumor types from TCGA SpliceSeq data, we observe that the Wnt-high cancers such as colorectal cancer (COAD) had higher expression of SRI exon 2, providing independent validation that this event is Wnt regulated.
Another example, uracil-DNA glycosylase (UNG) undergoes alternative promoter usage with variable exon 2.1 (schematic available in Supplemental Fig. S6C). The inclusion of this exon is higher in the ETC-159-treated samples than the untreated controls in both colorectal cancer PDX and HPAF-II tumor models, indicating that Wnt signaling suppresses the use of the promoter linked to this variable exon. From the TCGA SpliceSeq data, we find that the inclusion of this variable exon is similarly suppressed in the majority of tumors relative to the matched control tissue, regardless of the magnitude of Wnt signaling (Wnt score) in these tumors. This suggests that the shifting of the promoter preference for the gene, UNG, away from variable exon 2.1 could be a potential oncogenic process that is relevant in many cancers. It remains an open question of whether Wnt signaling drives this shift in promoter preference in all cancers or if diverse mechanisms converge on the same event in different tumor types.
sQTLs define cis-regulatory loci for Wnt regulated splicing events
Multiple proteins can regulate differential exon usage events in a combinatorial and coordinated manner (for review, see Lee and Rio 2015). Splicing regulators can influence exon inclusion by binding to cis-regulatory sites (such as splice enhancers and silencers), but the precise location of these sites is difficult to determine as they can be at highly variable locations within or outside exons. Thus, it becomes challenging to define splicing regulators and cis-regulatory sites for specific variable exons when the number of differentially expressed or dysregulated splicing regulator genes is large (Fig. 1C).
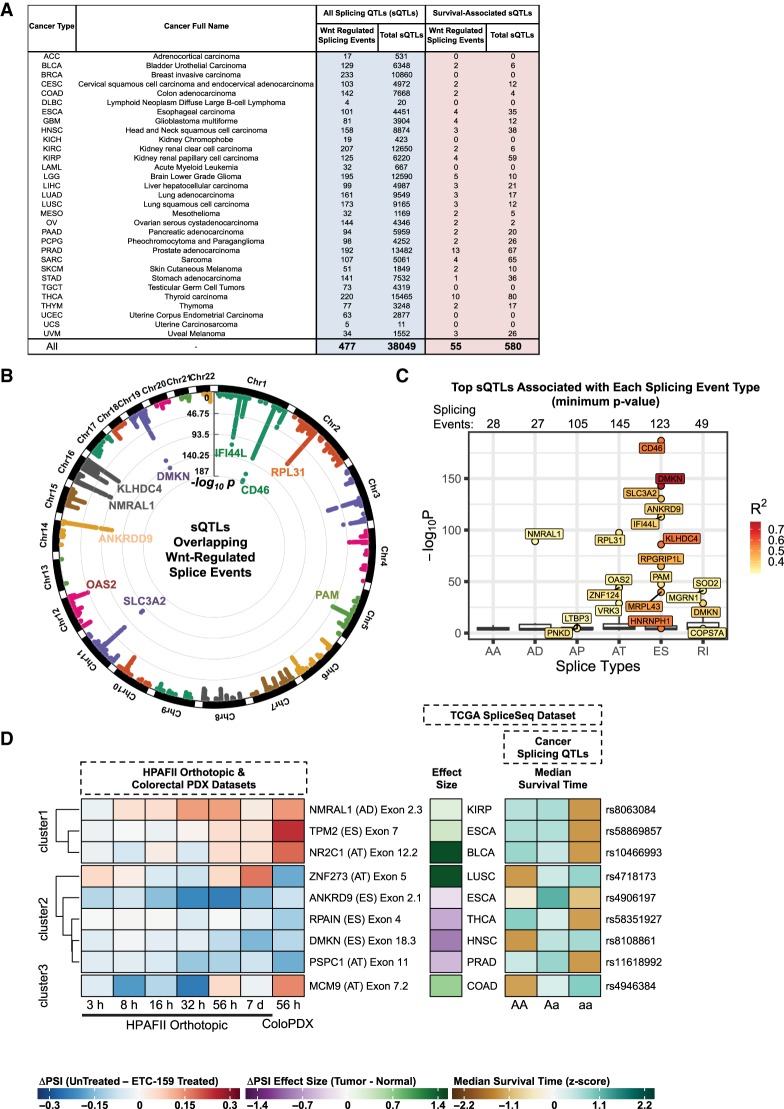
As an approach to define cis-regulatory elements that affect the inclusion of Wnt regulated variable exons, we used the sQTLs from the CancerSplicingQTL database (www.cancersplicingqtl-hust.com) (Tian et al. 2019). These sQTLs correlate genetic loci (marked by SNPs) to the relative inclusion of the variable exons described in the splicing events from the TCGA SpliceSeq database for specific cancer types. We wanted to identify the sQTLs that were linked to the 991 Wnt regulated splicing events. A total of 477 Wnt regulated splicing events detected in our system were mapped to 38,049 sQTLs from the CancerSplicingQTL database (Fig. 5A; Supplemental Table 5). There are multiple sQTLs per event because of multiple tightly haplotype-linked SNPs in the database. The genome-wide distribution of genomic loci for all of the 38,049 sQTLs is summarized in Figure 5B. The top sQTLs for each of the 477 splicing events (sQTLs with the minimum P-value) are summarized in Figure 5C. Wnt regulated exon skipping events, such as CD46 exon 8, have high confidence cis-regulatory sites involved in the inclusion of the spliced exon. In Figure 5C, this is measured in terms of the statistical significance of the sQTLs (P-value) and the R2 values (which quantify the extent of splicing, using normalized PSI values, that is explained by the genetic variation [sQTLs]).
FIGURE 5.
Splicing-associated quantitative trait loci (sQTLs) associated with Wnt regulated splicing events highlight potential cis-regulatory regions that can impact patient prognosis. (A) Tabulation of the total number of sQTLs and survival-associated sQTLs linked to Wnt regulated splicing events in each cancer type. (B) Genome-wide overview of all the 38,049 sQTLs associated with Wnt regulated splicing events. (C) Top sQTLs associated with each of the 477 Wnt regulated splicing events. Because of linkage within a haplotype, each splicing event can have multiple sQTLs that pass the significance filter. In such cases, the sQTLs were sorted based on the P-value and only the most significant (smallest P-value) sQTL was chosen to represent the splicing event. The splicing events were then segregated by splice type. Only sQTLs with R2 ≥ 0.3 are labeled. Exon skipping (ES), alternative termination (AT), and alternative promoter (AP) events had the largest number of splicing events with associated sQTLs (145, 123, and 105, respectively), whereas retained intron (RI), alternative acceptor (AA), and alternative donor (AD) events had fewer splicing events with associated sQTLs (49, 28, and 27, respectively). (D) Heatmap of a subset of the sQTLs that correlate with Wnt regulated splicing events and associated with patient survival in certain cancer types. The left panel shows the ΔPSI values for the splicing events in the two in vivo cancer models in response to Wnt signaling inhibition with ETC-159. Positive ΔPSI values (in red) indicate that Wnt signaling promotes the inclusion of the spliced exon. The middle panel shows the effect size (Hedges’ g estimate) of the splicing change (ΔPSI) in the TCGA SpliceSeq data set for the annotated tumor type. A positive effect size (in green) indicates that the spliced exon is included more in the tumor relative to its matched normal. The right panel shows the median survival time (z-score) associated with the sQTL. Positive median survival times (in green) indicate better patient prognosis for the annotated tumor. Each column represents a different haplotype for the various sQTLs. The heatmap was clustered into three sets via k-means clustering followed by hierarchical clustering to sort the genes within the sets.
This analysis suggests potential binding sites for the Wnt regulated splicing regulators described in Figure 1. To overlap the binding sites of trans-acting factors to the sQTL sites, we used the ReMap database (Chèneby et al. 2018) (which contains genomic binding sites for transcriptional regulators derived from ChIP-seq data that could affect alternative promoter usage events) and POSTAR2 database (Zhu et al. 2019) (which contains binding sites for RNA-binding proteins such as splicing factors derived from CLIP-seq data that could affect alternative splicing events). A list of the different trans-acting factors with binding sites that overlap sQTLs associated with the 477 Wnt regulated splicing events is detailed in Supplemental Table 5.
The CancerSplicingQTL database also contains sQTLs that are associated with patient prognosis for specific cancers. Fifty-five Wnt regulated splicing events were associated with 580 unique sQTLs that were also linked to patient survival (Fig. 6A). Some of these splicing events are highlighted in Figure 5D. A list of sQTLs that were associated with Wnt regulated splicing events and linked to patient survival (for cancer types, as shown in Fig. 5A, where splicing data was available) is shown in Supplemental Table 6. An example of a Wnt regulated splicing event that is also linked to patient survival for a particular cancer type is NR2C1 (Nuclear Receptor Subfamily 2 Group C Member 1). Wnt signaling promotes the alternative termination (or polyadenylation) of NR2C1 at variable exon 12.2. The inclusion of this variable exon is enriched in bladder cancer samples relative to matched normal tissue. The inclusion of this variable exon is also linked to a specific sQTL (rs10466993), in which changes in the haplotype of this sQTL are associated with changes in the relative inclusion of the variable exon and the prognosis for patients with bladder cancer. This underscores the significance of this Wnt regulated splicing event in specific cancer types and the effect of DNA sequence variation in regulating this process.
FIGURE 6.
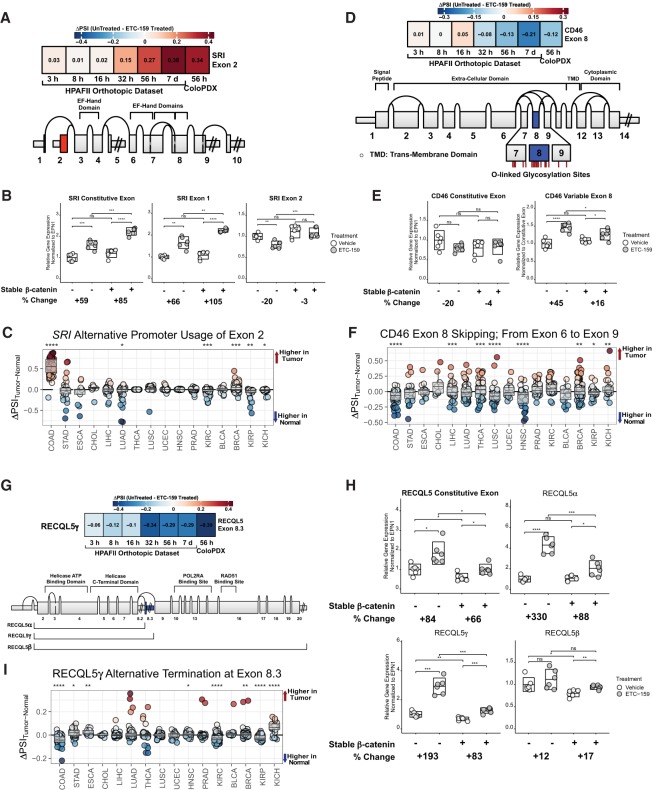
Wnt signaling (A–C) drives the alternative promoter usage of SRI (with preference to the promoter proximal to exon 2), (D–F) suppresses the inclusion of CD46 exon 8, and (G–I) inhibits the alternative polyadenylation of RECQL5 with transcription terminating at exon 8.3, in the two independent in vivo tumor models descibed in Figure 1A. (A,D,G) Each of the plots includes a schematic illustration of the exon structure of genes with Wnt regulated differential exon usage, together with the heatmaps indicating the magnitude of variable exon usage in both the HPAF-II orthotopic and ColoPDX tumor models. A positive value (in red) indicates that Wnt signaling promotes the inclusion of the exon, whereas a negative value (in blue) indicates that Wnt signaling suppresses the inclusion of the exon. (B,E,H) In vitro qPCR validation is also shown for the Wnt regulated (B) change in promoter preference of SRI in favor of the promoter proximal to exon 2, (E) skipping of CD46 exon 8, and (H) inhibition of the gene expression of the truncated RECQL5α/γ isoforms. ETC-159 was administered to Wnt-addicted HPAF-II cells with or without the stabilized β-catenin. The percentage change in expression upon ETC-159 treatment was calculated. The boxplot shows the average relative gene expression (center line) and the 95% confidence interval (length of the box). A two-tailed t-test was used to determine significance between comparisons. For statistical tests, (ns) P > 0.05, (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001, (****) P ≤ 0.0001. (C,F,I) The differential inclusion of Wnt regulated exons (measured as ΔPSI) in multiple tumors from the TCGA SpliceSeq database were assessed for the Wnt regulated (C) alternative promoter usage of SRI (with preference for the promoter proximal to exon 2), (F) skipping of exon 8 of CD46, and (I) inhibition of the expression of RECQL5γ isoform with transcription terminating at exon 8.3. A positive ΔPSI value indicates that the variable exon is included more in the tumors relative to the matched normals. The tumor types are sorted by the Wnt score (see Materials and Methods) with colorectal cancer (COAD) having the highest Wnt score. The higher the Wnt score, the stronger the relative influence of Wnt/β-catenin signaling in the cancer. A two-tailed t-test was used with H0: average ΔPSI value = 0, for each cancer type. For statistical tests, (ns) P > 0.05, (*) P ≤ 0.05, (**) P ≤ 0.01, (***) P ≤ 0.001, (****) P ≤ 0.0001.
Wnt signaling regulates isoform expression changes in SRI, CD46, and RECQL5
Wnt signaling induces isoform expression changes in genes that are strongly linked to cancer (Fig. 1E). We highlight and experimentally validate three such cancer-associated genes with Wnt regulated variable exon usage (Fig. 6). These changes in isoform expression include the alternative promoter usage of sorcin (SRI), the skipping of exon 8 in CD46, and alternative polyadenylation in RECQL5. The cis-regulatory sites for these variable exon usage events, determined using sQTLs, and the cognate transcriptional and RNA-binding proteins for these sites are highlighted in Supplemental Figure S10.
Sorcin (SRI) is a cytoplasmic calcium-binding protein involved in intracellular calcium homeostasis and is implicated in resistance to a variety of anticancer drugs (Maddalena et al. 2011; Zheng et al. 2012; Hu et al. 2013; Colotti et al. 2014; Gong et al. 2014; Liu et al. 2014; Yamagishi et al. 2014). Expression of the long form of SRI, with transcription originating at exon 2, is suppressed with ETC-159 treatment in vivo in both tumor models, suggesting that Wnt signaling strongly induces the inclusion of SRI exon 2 (Fig. 6A). Protein sequences found in the long form of SRI but not in the short form (with transcription originating at exon 1) have been linked to variable protein binding affinities to target proteins, including the formation of homodimers (Ilari et al. 2015). To test if the Wnt regulated variable exon use can be reproduced in vitro, we compared the ETC-159-mediated changes in control HPAF-II cells and HPAF-II cells expressing a stabilized nondegradable form of β-catenin. In the control HPAF-II, the expression of SRI transcripts containing exon 2 is reduced by 20% with ETC-159 treatment (Fig. 6B). This ETC-159-mediated effect is abolished in cells expressing stabilized β-catenin. This suggests that the inclusion of SRI exon 2 is regulated by Wnt/β-catenin signaling. Moreover, the knockdown of β-catenin in the HPAF-II cells reduced the expression of SRI transcripts containing exon 2 by an average of ∼30% (Supplemental Fig. S5B). Additionally, the inclusion of SRI exon 2 was strongly induced in colorectal cancers relative to matched normal tissues with an average ΔPSI of ∼54% (Fig. 6C). Much smaller (average ΔPSI ∼ 3%), but statistically significant, inclusion of SRI exon 2 was also seen in breast cancers (BRCA).
CD46 is a transmembrane protein involved in immune regulation (Ni Choileain et al. 2017; Arbore et al. 2018; Hansen et al. 2018) and is a potential target for cancer therapy (Su et al. 2018). This gene has a cluster of variable exons spanning exon 7 to exon 9 (Tang et al. 2016). These regions encode multiple O-linked glycosylation sites in the protein. Wnt signaling induced the skipping of CD46 exon 8 in both HPAF-II xenografts and ColoPDX models (Fig. 6D), which can lead to the expression of a hypoglycosylated form of the protein. In vitro analysis in HPAF-II cells showed that the proportion of CD46 exon 8 containing transcripts in the cell increased by 45% in the cells treated with ETC-159 (Fig. 6E). However, when the cells with stabilized β-catenin were treated with ETC-159, the proportion of CD46 exon 8 containing transcripts in the cell increased only by 16%. This shows that the inclusion of CD46 exon 8 is mediated, at least in part, by Wnt/β-catenin signaling. Moreover, the knockdown of β-catenin in HPAF-II cells led to an increase in the proportion of CD46 transcripts containing exon 8 (Supplemental Fig. S5B). The skipping of CD46 exon 8 was observed in various human cancers including colorectal cancer (COAD; average |ΔPSI| ∼ 15%), head and neck cancer (HNSC; average |ΔPSI| ∼ 13%), and lung cancer (LUSC; average |ΔPSI| ∼ 12%) (Fig. 6F). To a lesser extent (all with |ΔPSI| < 6%), papillary kidney cancers (KIRPs), liver cancers (LIHCs), and thyroid cancers (THCAs) also showed statistically significant exclusion of CD46 exon 8.
RECQL5 is a DNA helicase that binds to RNA polymerase II to regulate its transcriptional activity and maintain genomic stability (Islam et al. 2010; Tadokoro et al. 2012; Popuri et al. 2013; Hosono et al. 2014; Saponaro et al. 2014). RECQL5 can also support the growth and proliferation of cancers by alleviating the cellular stress induced by multiple cycles of replication (Arora et al. 2016; Patterson et al. 2016). There are three well-known isoforms for RECQL5. RECQL5β is the full-length and fully functional isoform, whereas RECQL5α/γ, generated via alternative polyadenylation events, are shorter variants with the helicase domains but without the domains required for binding to RNA polymerase II or RAD51 (Hu et al. 2007; Schwendener et al. 2010). From the differential exon usage analysis in both in vivo xenograft models, we find that the Wnt signaling represses the expression of the shorter, functionally compromised variant—especially RECQL5γ (Fig. 6G). In vitro assessment of RECQL5γ expression confirmed it to be highly sensitive to Wnt signaling inhibition (Fig. 6H). In the control HPAF-II cells, treatment with ETC-159 led to a twofold increase in the expression of the RECQL5γ isoform, whereas in cells with stabilized β-catenin, the ETC-159-mediated increase in the expression of this isoform was markedly blunted, indicating that it is partly regulated by Wnt/β-catenin signaling. A similar effect was observed for the other truncated isoform, RECQL5α, while Wnt signaling does not appear to have a strong influence over the expression of the full-length isoform, RECQL5β (Fig. 6H). The expression of RECQL5γ (with transcription ending at exon 8.3) was reduced in colorectal cancers (average |ΔPSI| ∼ 4%) as well as clear cell and papillary kidney cancers (KIRC and KIRP; each with average |ΔPSI| of ∼2%) (Fig. 6I).
DISCUSSION
Our study demonstrates that Wnt signaling has a profound impact on the expression of gene isoforms in cancer. We found that variable exon usage is regulated by Wnt signaling in 1025 genes. Some of these genes are involved in important biological processes that are commonly dysregulated in cancers (Fig. 3D,E). These Wnt regulated changes in gene isoform expression were due in part to broad changes in the expression of splicing regulators, because 76% (304) of the tested factors showed significant differential expression in at least one of the two tumor models we used. This is consistent with the finding that Wnt signaling affects the expression of 75% of the cellular transcriptome (Madan et al. 2018a) including some splicing regulators. This analysis also identified a large number of splicing regulators that were previously not linked to Wnt signaling. These findings are broadly applicable, because many of these Wnt regulated splicing regulators were previously associated with different cancers (Chen et al. 2015b; Gabriel et al. 2015; Tian et al. 2015; Anczuków and Krainer 2016; Sveen et al. 2016; Zhou et al. 2017; Matsumoto et al. 2018; Sakuma et al. 2018a,b). Wnt signaling was found to promote the gene expression of some of these cancer-associated splicing factors such as HNRNPA1, HNRNPA2B1, and HNRNPM. Interestingly, MYC, a well-known Wnt target gene, was previously found to promote the expression of HNRNPA1 (David et al. 2010), which strengthens its association with Wnt signaling. As such, the prooncogenic effects of some of these splicing factors could be attenuated with the inhibition of Wnt signaling.
Additionally, 577 out of 1585 Wnt-induced exon usage changes identified here were also dysregulated in multiple types of human cancers, suggesting the convergence of diverse mechanisms to promote prooncogenic splicing (Fig. 4A). Mechanistically, our data also identified potential cis-regulatory elements controlling the inclusion of Wnt regulated exons using 38,039 splicing QTLs associated with 477 Wnt regulated splicing events (Fig. 5A). These cis-regulatory sites may serve as binding sites for Wnt regulated transcriptional regulators and RNA-binding proteins involved in differential exon usage leading to isoform changes.
Different splicing events that were regulated by Wnt signaling were seen in many different cancer types (Fig. 4C). One reason for the commonality of these splicing events, even in cancers that are generally not known to be associated with Wnt signaling, is that Wnt signaling is only one of a number of pathways that might regulate the splicing event. Another reason that could account for the presence of shared splicing events is that there are specific downstream components of the Wnt signaling pathway, such as GSK3 and MYC, that are also regulated by other signaling networks. Modulating the activity of GSK3 protein, for example, was previously shown to lead to changes in phosphorylation and activity of proteins including splicing factors (Taelman et al. 2010; Acebron et al. 2014; Shinde et al. 2017). GSK3 is a key kinase downstream of Wnt as well as multiple other signaling pathways. Wnt signaling suppresses GSK3 activity by several proposed mechanisms (Wu and Pan 2010; Metcalfe and Bienz 2011), whereas growth factor signaling can suppress GSK3 via site-specific phosphorylation, catalyzed by kinases such as AKT or S6K1 (Cross et al. 1995; Zhang et al. 2006). This highlights one potential mechanism whereby Wnt and other pathways can regulate similar exon usage and splicing events.
Confirming the results of our RNA-seq analysis, we further validated three Wnt regulated isoform changes—namely, the alternative promoter usage of sorcin (SRI), the exon skipping events seen in CD46, and the alternative polyadenylation events present in RECQL5 that showed a consistent variable exon usage pattern in both tumor models. We report how β-catenin-dependent Wnt signaling drives a strong preference for the SRI promoter proximal to exon 2. This preference for exon 2 was particularly apparent in colorectal cancers relative to normal colon tissue (Fig. 6C). Colorectal cancers have elevated Wnt signaling because of the high incidence of loss-of-function mutations in APC, gain-of-function mutations in β-catenin, chromosomal translocation in R-spondins, or RNF43 loss-of-function mutations (Sparks et al. 1998; Rowan et al. 2000; Madan and Virshup 2015). This suggests that the Wnt-induced increase in SRI transcripts containing exon 2 may be a useful biomarker for colorectal cancer. SRI attenuates the buildup of calcium in the cytoplasm and has been implicated in the drug resistance phenotype in many cancers (Maddalena et al. 2011; Zheng et al. 2012; Hu et al. 2013; Colotti et al. 2014; Gong et al. 2014; Liu et al. 2014; Yamagishi et al. 2014). Reducing cytoplasmic calcium accumulation and protecting against ER stress requires SRI to form homodimers to bind and inhibit the ryanodine receptor (Maddalena et al. 2011). Structural analysis of SRI by Illari and colleagues suggest that the oligomerization of SRI requires the amino-terminal region of SRI that originates in the differentially expressed exon 2 (Ilari et al. 2015). This region containing the hexapeptide (GYYPGG) may affect the binding affinity of SRI to other proteins. Therefore, our data suggest that Wnt signaling can potentially generate different isoforms of SRI protein with known and important functional differences.
Wnt signaling was also found to suppress the inclusion of exon 8 of CD46, which corresponds to a heavily glycosylated serine, threonine, and proline-rich (STP) region at its extracellular domain. CD46 is involved in immune regulation through complement inactivation and acts as a costimulatory receptor to promote T-cell proliferation and function (Yamamoto et al. 2013; Arbore et al. 2018; Hansen et al. 2018). Although CD46 was shown to play an oncogenic role in cancers by suppressing complement activity (Buettner et al. 2007), the role of its STP region is not well-understood. The loss of exons within the STP region has been observed in multiple cancers, including colorectal cancers, suggesting that the reduced glycosylation of CD46 could potentially promote oncogenesis in certain cancers.
The final Wnt regulated isoform change that we examined was the alternative polyadenylation of RECQL5. Full-length RECQL5 protein maintains genome stability and prevents transcriptional and replication-induced stress based on its ability to act as a helicase as well as to bind to proteins such as RNA polymerase II and RAD51 (Islam et al. 2010; Tadokoro et al. 2012; Popuri et al. 2013; Saponaro et al. 2014; Chen et al. 2015a). The RECQL5 transcript undergoes alternative polyadenylation to produce three protein isoforms: RECQL5β is the full-length isoform and RECQL5α/γ are truncated at the carboxy-terminal end and lack the binding sites for RNA polymerase II and RAD51 (Shimamoto et al. 2000). Wnt signaling favored the generation of the full-length and functional RECQL5β isoform over the truncated isoforms. The truncated RECQL5 isoforms cannot bind to RNA Pol II and RAD51; hence, their ability to protect cancer cells from replication stress could be compromised. Thus, the prooncogenic functions of RECQL5 can be inhibited through two potential mechanisms. One approach is to directly inhibit the enzymatic activity of RECQL5β. The other approach is to switch the isoform composition of RECQL5 to its truncated variants (RECQL5α/γ) by suppressing Wnt signaling in Wnt-driven tumors.
This work identifies a broad repertoire of variable isoforms and splicing regulators that are regulated by Wnt signaling, which will facilitate future functional characterization of these events in normal physiology and cancer. Here we took advantage of a PORCN inhibitor, ETC-159, which allowed the suppression of Wnt signaling induced by all Wnt ligands in physiologically relevant in vivo cancer models. Confirming the on-target effect of ETC-159 on splicing and splicing regulators, knockdown of β-catenin had a similar effect. Future work on Wnt regulated isoform expression changes is required to elucidate the effect of individual Wnt ligands and the combinatorial effect of different Wnts on the expression of variable exons. Likewise, β-catenin gain-of-function mutations or APC loss-of-function mutations that are prevalent in colorectal cancers may regulate subsets of these exon usage and splicing events. It is also likely that the cross talk of other cell signaling pathways with Wnt signaling can alter exon usage and splicing patterns to shape the cancer proteome. These new insights into the regulation of gene isoform expression could illuminate new therapeutic strategies and mechanisms to combat diseases such as cancer.
MATERIALS AND METHODS
RNA-seq data sets used to identify Wnt regulated differential gene and isoform expression
To determine Wnt regulated differential isoform expression, we used RNA-seq data sets from two independent in vivo cancer models (shown in Fig. 1A). The first was generated using a Wnt-sensitive (due to RNF43 loss of function mutation) pancreatic cancer cell line (HPAF-II) that was orthotopically injected into mice and treated with ETC-159 in a time course dependent manner, as described in (Madan et al. 2018a). The corresponding data is available from NCBI GEO as GSE118041, GSE118231, GSE118190, and GSE118179. The second was derived from subcutaneous tumors, generated using a Wnt-sensitive (due to RSPO3 chromosomal translocation leading to its enhanced expression) colorectal cancer patient-derived xenograft, treated with the PORCN inhibitor, ETC-159, as described in (Madan et al. 2016). The corresponding RNA-seq data is available from NCBI GEO as GSE69687.
Detection of Wnt regulated differential gene and isoform expression using the two in vivo Wnt-addicted cancer models
Raw sequence reads for the in vivo data sets were filtered using Xenome (Conway et al. 2012) to remove reads originating from the mouse. The filtered reads were aligned to the human genome (GRCh38; Ensembl version 79) using STAR (v2.5.2) (Dobin et al. 2013). The read counts were quantified from the aligned bam files using RSEM (v1.2.31) (Li and Dewey 2011), and DESeq2 (Love et al. 2014) was used to detect differential gene expression (Fig. 1B). Detailed instruction on the methods used to detect differential gene expression using the RNA-seq data sets can be found in Madan et al. (2018a).
To detect differential gene isoform expression, QoRTs (v1.1.8) (Hartley and Mullikin 2015) was first used to generate a “flattened” gene annotation file, which, together with the aligned bam files, was used to generate read counts for each variable exon. Differential expression analysis was performed for each variable exon with JunctionSeq (v1.4.0) (Hartley and Mullikin 2016). JunctionSeq is based on the statistical framework that underpins a widely used splicing analysis algorithm, DEXSeq. JunctionSeq allows us to better evaluate novel splice junctions, and it has a clear visualization of the results. The JunctionSeq algorithm outputs P-values that were derived from statistical tests of the relative usage of the variable exons in the ETC-159-treated samples relative to the untreated controls. A Benjamini–Hochberg multiple testing correction was applied to generate the adjusted P-values. Percent spliced in (PSI) values were also determined for each variable exon using the pipeline described by Schafer et al. (2015). ΔPSI values were calculated by subtracting the PSI value of the untreated (Wnt-high) sample with the ETC-159-treated (Wnt-low) samples. A positive ΔPSI value would indicate that Wnt signaling promotes the inclusion of the variable exon. Variable exons with an adjusted P-value ≤ 0.1 and |ΔPSI| ≥ 10% in at least one time point in either of the two in vivo cancer model data sets were determined to be regulated by Wnt signaling. The threshold used to determine differentially expressed variable exons includes both the ΔPSI metric that measures the magnitude of the splicing change and the adjusted P-value that provides an evaluation of the statistical significance of the splicing change. The workflow for the detection of these differentially expressed variable exons is shown in Figure 3A.
Identification of 402 genes associated with regulation of isoform composition in cells
To determine if Wnt signaling can affect the expression of genes that code for regulatory proteins that affect differential isoform expression in a cell, we examined a set of 402 unique splicing regulators extracted from the gene ontology category of “RNA Splicing” (GO:0008380) in the Amigo gene ontology database (Ashburner et al. 2000; Carbon et al. 2009; The Gene Ontology Consortium 2017). These 402 regulators are composed of splicing factors, chromatin modifiers, and transcriptional coactivators that can affect alternative promoter usage. The differential gene expression analysis for this set was performed using the two in vivo Wnt-addicted cancer models as described above.
In vitro qPCR validation of differential Wnt regulated gene and isoform expression
Wnt-sensitive human pancreatic cancer cells (HPAF-II) were stably transduced with the β-catenin gene with four mutations (S33A, S37A, T41A, and S45A). These sites correspond to amino acid residues that are phosphorylated by CK1 and GSK3 (Supplemental Fig. S1). Phosphorylation of β-catenin at these residues triggers its degradation. The mutations to β-catenin prevent its degradation and HPAF-II cells with the mutant β-catenin have constitutively active β-catenin signaling. HPAF-II cells with or without the stabilized β-catenin were treated with 100 nM ETC-159 (PORCN inhibitor), to suppress Wnt signaling for 3 d. RNA was extracted from the cells using the RNeasy MiniKit (QIAGEN) according to the manufacturer's protocol. Primers used for RT-qPCR are listed in Supplemental Figure S2. Note that the primer set used to detect the β-catenin gene (CTNNB1) can detect both the endogenous and the transduced mutant variants of the gene.
Gene ontology analysis on the 1025 genes that underwent Wnt regulated variable exon usage
Gene ontology analysis was performed on the 1025 genes that were found to undergo Wnt regulated differential exon usage leading to isoform expression changes (Fig. 2A). The analysis was performed using clusterProfiler (v3.2.14) (Yu et al. 2012) in R (v3.3.1). The background genes encompassed all expressed protein-coding genes from the human transcriptome. GO categories that had at least 70% overlapping genes were considered redundant, and only the GO category with the smallest adjusted P-value was retained.
Integration of Wnt regulated variable exon usage with splicing events from the TCGA SpliceSeq data
To determine if the Wnt regulated variable exon expression that we detected from the two in vivo cancer model RNA-seq data sets was also differentially expressed in other cancer types, we overlapped the Wnt regulated variable exon usage with splicing events described in the TCGA SpliceSeq database (https://bioinformatics.mdanderson.org/TCGASpliceSeq/) (Ryan et al. 2016). TCGA SpliceSeq database contains splicing information for genes in tumor and normal samples derived from RNA-seq data from The Cancer Genome Atlas (TCGA). Note that “splicing” information from this database includes alternative promoter usage and alternative termination (alternative polyadenylation) events. We classify the splicing events described in this database that contain Wnt regulated variable exons as Wnt regulated isoform expression changes.
To determine the prevalence of these Wnt regulated isoform expression changes in other cancer types, we extracted the PSI values from 16 different tumors with at least six tumor-normal pairs available in the TCGA SpliceSeq database. Only the tumor-normal pairs were used for downstream analyses. ΔPSI values were calculated by subtracting the PSI values from the tumors with the PSI values from the paired normals. A positive ΔPSI value would indicate that the variable exon from the Wnt regulated isoform expression changes is included more in the tumors than in the matched normals. Splicing events for rectal adenocarcinomas (READs) were combined into the events described for colorectal adenocarcinomas (COADs).
Sorting TCGA cancer types by relative magnitude of Wnt signaling
To sort the 16 different cancer types in terms of the relative level of Wnt signaling effect, we devised a Wnt score based on the differential expression of 10 well-established Wnt target genes (SOX9, DKK1, CCND1, MYC, LGR5, SP5, AXIN2, TCF7, RNF43, and BMP4) in the tumors relative to paired normal tissue (Supplemental Fig. S3).
To perform this differential gene expression analysis on TCGA RNA-seq data sets, raw read counts for 16 different cancer types (see Fig. 4A for a list of the different cancer types used) with at least six tumor-normal pairs were downloaded from Broad Institute GDAC Firehose (https://gdac.broadinstitute.org/). To maintain consistency with our data sets we used the TCGA samples that were sequenced using the Illumina HiSeq platform. The raw reads from tumor samples with paired normals were used in the DESeq2 pipeline (Love et al. 2014) to determine the differential expression of the genes in the tumors relative to the paired normals. The results of the differential expression analysis for all 16 tumor types can be found in Supplemental Table 3.
Wnt Score Calculation:
Gene Score = LFC × −log10(Padjusted), for each gene,
Scaling Factor= max(Gene Score),
Scaled Gene Score = Gene Score/Scaling Factor, for each of the 10 genes,
Wnt score = average(Scaled Gene Score),
where LFC is the log2 fold change (tumors vs. matched normals).
The larger the Wnt score, the stronger the relative influence of Wnt/β-catenin signaling in the tumors. As shown in Supplemental Figure S3 and in line with our expectation, the cancer types with the highest Wnt score include colorectal cancers (COADs), stomach cancers (STADs), esophageal cancers (ESCAs), cholangiocarcinomas (CHOLs), and liver cancers (LIHCs), all of which are known to have mutationally activated Wnt/β-catenin signaling.
Integration of Wnt regulated splicing events with cancer-associated splicing quantitative trait loci to determine putative cis-regulatory sites
sQTLs correlate gene loci (marked by single-nucleotide polymorphisms [SNPs]) with splicing events described in the TCGA SpliceSeq database in multiple tumors (Ryan et al. 2016). All the sQTLs associated with Wnt regulated splicing events were extracted, together with the survival-associated sQTLs from the CancerSplicingQTL database (http://www.cancersplicingqtl-hust.com) (Tian et al. 2019).
Because these sQTLs can mark the genetic location of potential cis-regulatory sites that mediate the inclusion of the Wnt regulated variable exon, we identified some trans-acting factors with binding sites that overlap the sQTLs. Protein binding data for transcriptional regulators (including transcription factors) (from the ReMap database [Chèneby et al. 2018]) and RNA-binding proteins (including splicing factors) (from the POSTAR2 database [Zhu et al. 2019]) were used to determine if these cis-regulatory sites (sQTLs) overlapped with binding sites for these trans-acting proteins. Protein binding data for corresponding sQTLs is available in Supplemental Table 5. The protein binding data is shown for the alternative promoter usage of SRI (Supplemental Fig. S10A), the exon skipping event seen in CD46 (Supplemental Fig. S10B), and the alternative polyadenylation events for RECQL5 (Supplemental Fig. S10C) in its genomic contex
DATA DEPOSITION
The RNA-seq data for the HPAF-II orthotopic tumor model is available at NCBI GEO GSE118041, GSE118231, GSE118190, and GSE118179. The RNA-seq data for the ColoPDX tumor model is available in NCBI GEO GSE69687. Detailed results for the Wnt regulated gene expression for splicing regulators and Wnt regulated splicing events can be found in the Supplemental Tables.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
COMPETING INTEREST STATEMENT
B.M. and D.M.V. have a financial interest in ETC-159.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Virshup lab and the Duke-NUS scientific community for their helpful comments and discussion. This research is supported in part by NMRC/STAR/0017/2013 from the National Research Foundation Singapore and administered by the Singapore Ministry of Health's National Medical Research Council (to D.M.V.). E.P. acknowledges the support of the MRC London Institute of Medical Sciences, Imperial College, London. B.M. acknowledges the support of the Singapore Ministry of Health's National Medical Research Council Open Fund–Independent Research Grant NMRC/OFIRG/0055/2017.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.071506.119.
REFERENCES
- Acebron SP, Niehrs C. 2016. β-catenin-independent roles of Wnt/LRP6 signaling. Trends Cell Biol 26: 956–967. 10.1016/j.tcb.2016.07.009 [DOI] [PubMed] [Google Scholar]
- Acebron SP, Karaulanov E, Berger BS, Huang Y-L, Niehrs C. 2014. Mitotic Wnt signaling promotes protein stabilization and regulates cell size. Mol Cell 54: 663–674. 10.1016/j.molcel.2014.04.014 [DOI] [PubMed] [Google Scholar]
- Anczuków O, Krainer AR. 2016. Splicing-factor alterations in cancers. RNA 22: 1285–1301. 10.1261/rna.057919.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbore G, West EE, Rahman J, Le Friec G, Niyonzima N, Pirooznia M, Tunc I, Pavlidis P, Powell N, Li Y, et al. 2018. Complement receptor CD46 co-stimulates optimal human CD8+ T cell effector function via fatty acid metabolism. Nat Commun 9: 4186 10.1038/s41467-018-06706-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arce L, Yokoyama NN, Waterman ML. 2006. Diversity of LEF/TCF action in development and disease. Oncogene 25: 7492–7504. 10.1038/sj.onc.1210056 [DOI] [PubMed] [Google Scholar]
- Archbold HC, Yang YX, Chen L, Cadigan KM. 2012. How do they do Wnt they do?: regulation of transcription by the Wnt/β-catenin pathway. Acta Physiol (Oxf) 204: 74–109. 10.1111/j.1748-1716.2011.02293.x [DOI] [PubMed] [Google Scholar]
- Arora A, Abdel-Fatah TMA, Agarwal D, Doherty R, Croteau DL, Moseley PM, Hameed K, Green A, Aleskandarany MA, Rakha EA, et al. 2016. Clinicopathological and prognostic significance of RECQL5 helicase expression in breast cancers. Carcinogenesis 37: 63–71. 10.1093/carcin/bgv163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordonaro M. 2013. Crosstalk between Wnt signaling and RNA processing in colorectal cancer. J Cancer 4: 96–103. 10.7150/jca.5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner R, Huang M, Gritsko T, Karras J, Enkemann S, Mesa T, Nam S, Yu H, Jove R. 2007. Activated signal transducers and activators of transcription 3 signaling induces CD46 expression and protects human cancer cells from complement-dependent cytotoxicity. Mol Cancer Res 5: 823–832. 10.1158/1541-7786.MCR-06-0352 [DOI] [PubMed] [Google Scholar]
- Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, Bateman A, Babu MM. 2012. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol Cell 46: 871–883. 10.1016/j.molcel.2012.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan KM, Waterman ML. 2012. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol 4: a007906 10.1101/cshperspect.a007906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, AmiGO Hub Web Presence Working Group. 2009. AmiGO: online access to ontology and annotation data. Bioinformatics 25: 288–289. 10.1093/bioinformatics/btn615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E, Ahn JS, Sykes DB, Breyfogle LJ, Godfrey AL, Nangalia J, Ko A, DeAngelo DJ, Green AR, Mullally A. 2015a. RECQL5 suppresses oncogenic JAK2-induced replication stress and genomic instability. Cell Rep 13: 2345–2352. 10.1016/j.celrep.2015.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Du H, Liu B, Zou L, Chen W, Yang Y, Zhu Y, Gong Y, Tian J, Li F, et al. 2015b. The associations between RNA splicing complex gene SF3A1 polymorphisms and colorectal cancer risk in a Chinese population. PLoS One 10: e0130377 10.1371/journal.pone.0130377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chèneby J, Gheorghe M, Artufel M, Mathelier A, Ballester B. 2018. ReMap 2018: an updated atlas of regulatory regions from an integrative analysis of DNA-binding ChIP-seq experiments. Nucleic Acids Res 46: D267–D275. 10.1093/nar/gkx1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colotti G, Poser E, Fiorillo A, Genovese I, Chiarini V, Ilari A. 2014. Sorcin, a calcium binding protein involved in the multidrug resistance mechanisms in cancer cells. Molecules 19: 13976–13989. 10.3390/molecules190913976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K, Feng LL, Yeakley JM, Gish GD, Cáceres JF, Pawson T, Fu XD. 1996. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J Biol Chem 271: 24569–24575. 10.1074/jbc.271.40.24569 [DOI] [PubMed] [Google Scholar]
- Conway T, Wazny J, Bromage A, Tymms M, Sooraj D, Williams ED, Beresford-Smith B. 2012. Xenome—a tool for classifying reads from xenograft samples. Bioinformatics 28: i172–i178. 10.1093/bioinformatics/bts236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. 1995. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378: 785–789. 10.1038/378785a0 [DOI] [PubMed] [Google Scholar]
- Danan-Gotthold M, Golan-Gerstl R, Eisenberg E, Meir K, Karni R, Levanon EY. 2015. Identification of recurrent regulated alternative splicing events across human solid tumors. Nucleic Acids Res 43: 5130–5144. 10.1093/nar/gkv210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daulat AM, Borg J-P. 2017. Wnt/Planar cell polarity signaling: new opportunities for cancer treatment. Trends Cancer 3: 113–125. 10.1016/j.trecan.2017.01.001 [DOI] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. 2010. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463: 364–368. 10.1038/nature08697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel M, Delforge Y, Deward A, Habraken Y, Hennuy B, Piette J, Klinck R, Chabot B, Colige A, Lambert C. 2015. Role of the splicing factor SRSF4 in cisplatin-induced modifications of pre-mRNA splicing and apoptosis. BMC Cancer 15: 227 10.1186/s12885-015-1259-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard D, Bowles SG, Salcedo E, Xu M, Millar SE, Barlow LA. 2017. β-catenin is required for taste bud cell renewal and behavioral taste perception in adult mice. PLoS Genet 13: e1006990 10.1371/journal.pgen.1006990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. 2017. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 45: D331–D338. 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves V, Matos P, Jordan P. 2008. The β-catenin/TCF4 pathway modifies alternative splicing through modulation of SRp20 expression. RNA 14: 2538–2549. 10.1261/rna.1253408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Sun P, Chu H, Zhu H, Sun D, Chen J. 2014. Overexpression of sorcin in multidrug-resistant human breast cancer. Oncol Lett 8: 2393–2398. 10.3892/ol.2014.2543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Slater J, Biltoft M, Bundgaard BB, Møller BK, Höllsberg P. 2018. CD46 is a potent co-stimulatory receptor for expansion of human IFN-γ-producing CD8+ T cells. Immunol Lett 200: 26–32. 10.1016/j.imlet.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley SW, Mullikin JC. 2015. QoRTs: a comprehensive toolset for quality control and data processing of RNA-Seq experiments. BMC Bioinformatics 16: 224 10.1186/s12859-015-0670-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley SW, Mullikin JC. 2016. Detection and visualization of differential splicing in RNA-Seq data with JunctionSeq. Nucleic Acids Res 44: e127 10.1093/nar/gkw501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SY, Keller TH. 2015. The use of porcupine inhibitors to target Wnt-driven cancers. Bioorg Med Chem Lett 25: 5472–5476. 10.1016/j.bmcl.2015.10.032 [DOI] [PubMed] [Google Scholar]
- Hosono Y, Abe T, Ishiai M, Islam MN, Arakawa H, Wang W, Takeda S, Ishii Y, Takata M, Seki M, et al. 2014. Tumor suppressor RecQL5 controls recombination induced by DNA crosslinking agents. Biochim Biophys Acta 1843: 1002–1012. 10.1016/j.bbamcr.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. 2007. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev 21: 3073–3084. 10.1101/gad.1609107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Cheng X, Li S, Zhou Y, Wang J, Cheng T, Yang M, Xiong D. 2013. Inhibition of sorcin reverses multidrug resistance of K562/A02 cells and MCF-7/A02 cells via regulating apoptosis-related proteins. Cancer Chemother Pharmacol 72: 789–798. 10.1007/s00280-013-2254-2 [DOI] [PubMed] [Google Scholar]
- Ilari A, Fiorillo A, Poser E, Lalioti VS, Sundell GN, Ivarsson Y, Genovese I, Colotti G. 2015. Structural basis of Sorcin-mediated calcium-dependent signal transduction. Sci Rep 5: 16828 10.1038/srep16828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MN, Fox D, Guo R, Enomoto T, Wang W. 2010. RecQL5 promotes genome stabilization through two parallel mechanisms–interacting with RNA polymerase II and acting as a helicase. Mol Cell Biol 30: 2460–2472. 10.1128/MCB.01583-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jyotsana N, Heuser M. 2018. Exploiting differential RNA splicing patterns: a potential new group of therapeutic targets in cancer. Expert Opin Ther Targets 22: 107–121. 10.1080/14728222.2018.1417390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S. 2013. Function of alternative splicing. Gene 514: 1–30. 10.1016/j.gene.2012.07.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch S, Acebron SP, Herbst J, Hatiboglu G, Niehrs C. 2015. Post-transcriptional Wnt signaling governs epididymal sperm maturation. Cell 163: 1225–1236. 10.1016/j.cell.2015.10.029 [DOI] [PubMed] [Google Scholar]
- Kohn AD, Moon RT. 2005. Wnt and calcium signaling: β-catenin-independent pathways. Cell Calcium 38: 439–446. 10.1016/j.ceca.2005.06.022 [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR. 2006. Chromatin, transcript elongation and alternative splicing. Nat Struct Mol Biol 13: 5–7. 10.1038/nsmb0106-5 [DOI] [PubMed] [Google Scholar]
- Lee SC-W, Abdel-Wahab O. 2016. Therapeutic targeting of splicing in cancer. Nat Med 22: 976–986. 10.1038/nm.4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rio DC. 2015. Mechanisms and regulation of alternative Pre-mRNA splicing. Annu Rev Biochem 84: 291–323. 10.1146/annurev-biochem-060614-034316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen L, Feng B, Liu G. 2014. Reversing effect of sorcin in the drug resistance of human nasopharyngeal carcinoma. Anat Rec (Hoboken) 297: 215–221. 10.1002/ar.22832 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan B, Virshup DM. 2015. Targeting Wnts at the source-new mechanisms, new biomarkers, new drugs. Mol Cancer Ther 14: 1087–1094. 10.1158/1535-7163.MCT-14-1038 [DOI] [PubMed] [Google Scholar]
- Madan B, Ke Z, Harmston N, Ho SY, Frois AO, Alam J, Jeyaraj DA, Pendharkar V, Ghosh K, Virshup IH, et al. 2016. Wnt addiction of genetically defined cancers reversed by PORCN inhibition. Oncogene 35: 2197–2207. 10.1038/onc.2015.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan B, Harmston N, Nallan G, Montoya A, Faull P, Petretto E, Virshup DM. 2018a. Temporal dynamics of Wnt-dependent transcriptome reveal an oncogenic Wnt/MYC/ribosome axis. J Clin Invest 128: 5620–5633. 10.1172/JCI122383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan B, McDonald MJ, Foxa GE, Diegel CR, Williams BO, Virshup DM. 2018b. Bone loss from Wnt inhibition mitigated by concurrent alendronate therapy. Bone Res 6: 17 10.1038/s41413-018-0017-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalena F, Laudiero G, Piscazzi A, Secondo A, Scorziello A, Lombardi V, Matassa DS, Fersini A, Neri V, Esposito F, et al. 2011. Sorcin induces a drug-resistant phenotype in human colorectal cancer by modulating Ca2+ homeostasis. Cancer Res 71: 7659–7669. 10.1158/0008-5472.CAN-11-2172 [DOI] [PubMed] [Google Scholar]
- Mah AT, Yan KS, Kuo CJ. 2016. Wnt pathway regulation of intestinal stem cells. J Physiol (Lond) 594: 4837–4847. 10.1113/JP271754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez NM, Pan Q, Cole BS, Yarosh CA, Babcock GA, Heyd F, Zhu W, Ajith S, Blencowe BJ, Lynch KW. 2012. Alternative splicing networks regulated by signaling in human T cells. RNA 18: 1029–1040. 10.1261/rna.032243.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Itou J, Sato F, Toi M. 2018. SALL -KHDRBS3 network enhances stemness by modulating CD44 splicing in basal-like breast cancer. Cancer Med 7: 454–462. 10.1002/cam4.1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalfe C, Bienz M. 2011. Inhibition of GSK3 by Wnt signalling—two contrasting models. J Cell Sci 124: 3537–3544. 10.1242/jcs.091991 [DOI] [PubMed] [Google Scholar]
- Michels BE, Mosa MH, Grebbin BM, Yepes D, Darvishi T, Hausmann J, Urlaub H, Zeuzem S, Kvasnicka HM, Oellerich T, et al. 2019. Human colon organoids reveal distinct physiologic and oncogenic Wnt responses. J Exp Med 216: 704–720. 10.1084/jem.20180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Choileain S, Hay J, Thomas J, Williams A, Vermeren MM, Benezech C, Gomez-Salazar M, Hugues OR, Vermeren S, Howie SEM, et al. 2017. TCR-stimulated changes in cell surface CD46 expression generate type 1 regulatory T cells. Sci Signal 10: eaah6163 10.1126/scisignal.aah6163 [DOI] [PubMed] [Google Scholar]
- Nusse R, Clevers H. 2017. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169: 985–999. 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- Ortis F, Naamane N, Flamez D, Ladrière L, Moore F, Cunha DA, Colli ML, Thykjaer T, Thorsen K, Orntoft TF, et al. 2010. Cytokines interleukin-1β and tumor necrosis factor-α regulate different transcriptional and alternative splicing networks in primary β-cells. Diabetes 59: 358–374. 10.2337/db09-1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415. 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- Patterson K, Arya L, Bottomley S, Morgan S, Cox A, Catto J, Bryant HE. 2016. Altered RECQL5 expression in urothelial bladder carcinoma increases cellular proliferation and makes RECQL5 helicase activity a novel target for chemotherapy. Oncotarget 7: 76140–76150. 10.18632/oncotarget.12683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popuri V, Tadokoro T, Croteau DL, Bohr VA. 2013. Human RECQL5: guarding the crossroads of DNA replication and transcription and providing backup capability. Crit Rev Biochem Mol Biol 48: 289–299. 10.3109/10409238.2013.792770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan AJ, Lamlum H, Ilyas M, Wheeler J, Straub J, Papadopoulou A, Bicknell D, Bodmer WF, Tomlinson IP. 2000. APC mutations in sporadic colorectal tumors: a mutational “hotspot” and interdependence of the “two hits”. Proc Natl Acad Sci 97: 3352–3357. 10.1073/pnas.97.7.3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan M, Wong WC, Brown R, Akbani R, Su X, Broom B, Melott J, Weinstein J. 2016. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res 44: D1018–D1022. 10.1093/nar/gkv1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma K, Sasaki E, Kimura K, Komori K, Shimizu Y, Yatabe Y, Aoki M. 2018a. HNRNPLL stabilizes mRNA for DNA replication proteins and promotes cell cycle progression in colorectal cancer cells. Cancer Sci 109: 2458–2468. 10.1111/cas.13660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma K, Sasaki E, Kimura K, Komori K, Shimizu Y, Yatabe Y, Aoki M. 2018b. HNRNPLL, a newly identified colorectal cancer metastasis suppressor, modulates alternative splicing of CD44 during epithelial-mesenchymal transition. Gut 67: 1103–1111. 10.1136/gutjnl-2016-312927 [DOI] [PubMed] [Google Scholar]
- Saponaro M, Kantidakis T, Mitter R, Kelly GP, Heron M, Williams H, Söding J, Stewart A, Svejstrup JQ. 2014. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell 157: 1037–1049. 10.1016/j.cell.2014.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer S, Miao K, Benson CC, Heinig M, Cook SA, Hubner N. 2015. Alternative splicing signatures in RNA-seq data: percent spliced in (PSI). Curr Protoc Hum Genet 87: 11.16.1–11.16.14. 10.1002/0471142905.hg1116s87 [DOI] [PubMed] [Google Scholar]
- Schwendener S, Raynard S, Paliwal S, Cheng A, Kanagaraj R, Shevelev I, Stark JM, Sung P, Janscak P. 2010. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J Biol Chem 285: 15739–15745. 10.1074/jbc.M110.110478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebestyén E, Singh B, Miñana B, Pagès A, Mateo F, Pujana MA, Valcárcel J, Eyras E. 2016. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res 26: 732–744. 10.1101/gr.199935.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamoto A, Nishikawa K, Kitao S, Furuichi Y. 2000. Human RecQ5α, a large isomer of RecQ5 DNA helicase, localizes in the nucleoplasm and interacts with topoisomerases 3α and 3β. Nucleic Acids Res 28: 1647–1655. 10.1093/nar/28.7.1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinde MY, Sidoli S, Kulej K, Mallory MJ, Radens CM, Reicherter AL, Myers RL, Barash Y, Lynch KW, Garcia BA, et al. 2017. Phosphoproteomics reveals that glycogen synthase kinase-3 phosphorylates multiple splicing factors and is associated with alternative splicing. J Biol Chem 292: 18240–18255. 10.1074/jbc.M117.813527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks AB, Morin PJ, Vogelstein B, Kinzler KW. 1998. Mutational analysis of the APC/β-catenin/Tcf pathway in colorectal cancer. Cancer Res 58: 1130–1134. [PubMed] [Google Scholar]
- Srebrow A, Kornblihtt AR. 2006. The connection between splicing and cancer. J Cell Sci 119: 2635–2641. 10.1242/jcs.03053 [DOI] [PubMed] [Google Scholar]
- Su Y, Liu Y, Behrens CR, Bidlingmaier S, Lee N-K, Aggarwal R, Sherbenou DW, Burlingame AL, Hann BC, Simko JP, et al. 2018. Targeting CD46 for both adenocarcinoma and neuroendocrine prostate cancer. JCI Insight 3: 1458 10.1172/jci.insight.121497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, Skotheim RI. 2016. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 35: 2413–2427. 10.1038/onc.2015.318 [DOI] [PubMed] [Google Scholar]
- Tadokoro T, Ramamoorthy M, Popuri V, May A, Tian J, Sykora P, Rybanska I, Wilson DM, Croteau DL, Bohr VA. 2012. Human RECQL5 participates in the removal of endogenous DNA damage. Mol Biol Cell 23: 4273–4285. 10.1091/mbc.e12-02-0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taelman VF, Dobrowolski R, Plouhinec J-L, Fuentealba LC, Vorwald PP, Gumper I, Sabatini DD, De Robertis EM. 2010. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell 143: 1136–1148. 10.1016/j.cell.2010.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang SJ, Luo S, Ho JXJ, Ly PT, Goh E, Roca X. 2016. Characterization of the regulation of CD46 RNA alternative splicing. J Biol Chem 291: 14311–14323. 10.1074/jbc.M115.710350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsen K, Mansilla F, Schepeler T, Øster B, Rasmussen MH, Dyrskjøt L, Karni R, Akerman M, Krainer AR, Laurberg S, et al. 2011. Alternative splicing of SLC39A14 in colorectal cancer is regulated by the Wnt pathway. Mol Cell Proteomics 10: M110.002998 10.1074/mcp.M110.002998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Liu Y, Zhu B, Tian Y, Zhong R, Chen W, Lu X, Zou L, Shen N, Qian J, et al. 2015. SF3A1 and pancreatic cancer: new evidence for the association of the spliceosome and cancer. Oncotarget 6: 37750–37757. 10.18632/oncotarget.5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Wang Z, Mei S, Yang N, Yang Y, Ke J, Zhu Y, Gong Y, Zou D, Peng X, et al. 2019. CancerSplicingQTL: a database for genome-wide identification of splicing QTLs in human cancer. Nucleic Acids Res 47: D909–D916. 10.1093/nar/gky954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltri A, Lang C, Lien W-H. 2018. Concise review: Wnt signaling pathways in skin development and epidermal stem cells. Stem Cells 36: 22–35. 10.1002/stem.2723 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielenga VJ, Smits R, Korinek V, Smit L, Kielman M, Fodde R, Clevers H, Pals ST. 1999. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am J Pathol 154: 515–523. 10.1016/S0002-9440(10)65297-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Pan W. 2010. GSK3: a multifaceted kinase in Wnt signaling. Trends Biochem Sci 35: 161–168. 10.1016/j.tibs.2009.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi N, Nakao R, Kondo R, Nishitsuji M, Saito Y, Kuga T, Hatayama T, Nakayama Y. 2014. Increased expression of sorcin is associated with multidrug resistance in leukemia cells via up-regulation of MDR1 expression through cAMP response element-binding protein. Biochem Biophys Res Commun 448: 430–436. 10.1016/j.bbrc.2014.04.125 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Fara AF, Dasgupta P, Kemper C. 2013. CD46: the “multitasker” of complement proteins. Int J Biochem Cell Biol 45: 2808–2820. 10.1016/j.biocel.2013.09.016 [DOI] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y. 2012. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HH, Lipovsky AI, Dibble CC, Sahin M, Manning BD. 2006. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol Cell 24: 185–197. 10.1016/j.molcel.2006.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B-B, Zhang P, Jia W-W, Yu L-G, Guo X-L. 2012. Sorcin, a potential therapeutic target for reversing multidrug resistance in cancer. J Physiol Biochem 68: 281–287. 10.1007/s13105-011-0140-0 [DOI] [PubMed] [Google Scholar]
- Zhou H-L, Luo G, Wise JA, Lou H. 2014. Regulation of alternative splicing by local histone modifications: potential roles for RNA-guided mechanisms. Nucleic Acids Res 42: 701–713. 10.1093/nar/gkt875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Ma N, Jiang H, Rong Y, Deng Y, Feng Y, Zhu H, Kuang T, Lou W, Xie D, et al. 2017. SF3B4 is decreased in pancreatic cancer and inhibits the growth and migration of cancer cells. Tumour Biol 39: 1010428317695913 10.1177/1010428317695913 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Xu G, Yang YT, Xu Z, Chen X, Shi B, Xie D, Lu ZJ, Wang P. 2019. POSTAR2: deciphering the post-transcriptional regulatory logics. Nucleic Acids Res 47: D203–D211. 10.1093/nar/gky830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerli D, Cecconi V, Valenta T, Hausmann G, Cantù C, Restivo G, Hafner J, Basler K, van den Broek M. 2018. WNT ligands control initiation and progression of human papillomavirus-driven squamous cell carcinoma. Oncogene 37: 3753–3762. 10.1038/s41388-018-0244-x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.