Abstract
Medulloblastoma, a central nervous system tumor that predominantly affects children, always requires aggressive therapy. Nevertheless, it frequently recurs as resistant disease and is associated with high morbidity and mortality. While recent efforts to subclassify medulloblastoma based on molecular features have advanced our basic understanding of medulloblastoma pathogenesis, optimal targets to increase therapeutic efficacy and reduce side effects remain largely undefined. Noncoding RNAs (ncRNAs) with known regulatory roles, particularly long noncoding RNAs (lncRNAs) and microRNAs (miRNAs), are now known to participate in medulloblastoma biology, although their functional significance remains obscure in many cases. Here we review the literature on regulatory ncRNAs in medulloblastoma. In providing a comprehensive overview of ncRNA studies, we highlight how different lncRNAs and miRNAs have oncogenic or tumor suppressive roles in medulloblastoma. These ncRNAs possess subgroup specificity that can be exploited to personalize therapy by acting as theranostic targets. Several of the already identified ncRNAs appear specific to medulloblastoma stem cells, the most difficult-to-treat component of the tumor that drives metastasis and acquired resistance, thereby providing opportunities for therapy in relapsing, disseminating, and therapy-resistant disease. Delivering ncRNAs to tumors remains challenging, but this limitation is gradually being overcome through the use of advanced technologies such as nanotechnology and rational biomaterial design.
Keywords: circRNAs, diagnostics therapeutics, lncRNAs, Medulloblastoma, microRNAs
Medulloblastoma (MB) is the most common malignant pediatric brain tumor,1–3 representing 9.2% of all pediatric brain tumor cases but only 1% of adult cases.1,4 MBs arise in the cerebellum and are highly malignant; they commonly metastasize to other parts of the brain and spinal cord and, rarely, to extraneural sites.5,6 Transcriptional programs in MBs mimic developmental cerebellar lineages, highlighting their embryonic origin.7 The clinical management of MB depends on several factors including molecular and histopathological tumor subgroup, stage, extent of resection and location, and overall patient health. Treatment strategies are aggressive, consisting of a mixture of surgical resection, radiotherapy, chemotherapy, and stem cell/bone marrow transplantation. Despite advances in diagnosis and treatment, MB remains deadly in 35–40% of cases, and those that do survive often suffer long-term side effects including organ dysfunction, neurocognitive impairment, endocrine disabilities, and secondary tumors.8–11
Therefore, more effective and less toxic therapies are urgently required to improve clinical outcomes and quality of life for MB patients. In an example of the value of the molecular characterization of cancer, relatively recent molecular classification efforts have refined the clinical and pathological classification of MBs into four clinical/molecular subgroups with distinct driver mutations, cells of origin, and prognoses.3 These subgroups provide targets for personalized therapy, some of which are now being tested clinically, but as with many targeted approaches acquired resistance is common, and some subgroups harbor relatively few somatic mutations.12 Therefore, the assessment of parameters beyond coding DNA is likely to prove useful not only in terms of understanding the basic biology of MB, but also in expanding the repertoire of molecular targets for refined subclassification, biomarker development, and precision medicine. With this in mind, here we review recent advances in the biology of noncoding RNAs, the nontranslated but functionally active portion of the genome now known to participate in tumorigenesis, in the context of MB. In doing so, we explore how these molecules show particular potential as therapeutics targets.
The Molecular Classification of Medulloblastoma: A Clinical and Scientific Success Story
MB was traditionally classified histologically into three major types: classic, nodular/desmoplastic (ND), and large cell/anaplastic (LCA), which had prognostic significance but also issues with specificity and reproducibility.11 However, and in a valuable illustration of the benefit of molecular characterization of cancer, whole genome, transcriptome, and epigenome analyses have identified significant, clinically relevant molecular heterogeneity between MBs in patient subsets. Since 2016, and in a major restructuring of the classification to include genetically defined entities, the World Health Organization (WHO) Classification of Central Nervous System Tumors divided MB into following major molecular subgroups:3 wingless-type (WNT)-activated MB (10%; children and adults, associated with very good prognosis); sonic hedgehog (SHH)-activated MB (30%; intermediate prognosis, infants and adults), further characterized as TP53 mutant or TP53 wild-type; and two provisional non-WNT/SHH subgroups: group 3 MB (25%; poor prognosis, infants, and children) and group 4 MB (35%; intermediate prognosis, children, and adults) (reviewed in refs. 12 and 13). Given that these molecular subgroups have unique clinical and demographic characteristics and prognoses12–14 that outperform traditional histopathological classification or clinical staging,4 diagnosis is now optimally made using a modular and integrated approach that combines histological and molecular features.
The molecular diagnosis of MB has made it easier to differentiate tumor subgroups, with very reliable diagnostic markers available for WNT and SHH MBs, but group 3 and group 4 tumors are more difficult to define. WNT and SHH MBs generally contain mutations activating these pathways and, aside from rare TP53-mutant SHH tumors, are less aggressive than group 3 and 4 tumors.15 Biomarkers for WNT MBs include immunohistochemical evidence of YAP1 and nuclear β-catenin, monosomy 6, as well as identification of activating pathway mutations in β-catenin (CCNTB1) by sequencing.16,17 SHH MBs are generally identified by GAB1 and YAP1 co-expression18,19 and germline or somatic mutations in PTCH1 or SUFU in children as well as recurrent somatic mutations in PTCH1, SMO, and the TERT promoter in adults.20,21 While transcriptional or methylation profiling can also be used to distinguish the four subgroups, this is currently not an accepted clinical assay at most institutions.17,19,22–26
The genetic basis of group 3 and group 4 tumors is much less well defined, so their molecular diagnosis has remained challenging, and perhaps unsurprisingly ongoing transcriptomic and genome-wide methylation studies are revealing even greater heterogeneity in these subgroups.27 There are currently no reliable immunohistochemical markers for group 3 tumors, although NPR3 positivity has been suggested as a potential biomarker25 and MYC levels are also higher in the most clinically aggressive subset of this group. Northcott et al.28 recently characterized somatic copy number aberrations (SCNAs) in 1,087 unique medulloblastomas and found tandem duplication of SNCAIP, a gene normally associated with Parkinson’s disease, as focal copy number gain exquisitely restricted to group 4 tumors. Recurrent translocations of PVT1, including PVT1-MYC and PVT1-NDRG1, arising via chromothripsis (large-scale genomic rearrangements occurring in a single event in confined genomic regions in one or a few chromosomes) were restricted to group 3 MBs. The presence of numerous targetable SCNAs, including recurrent events targeting TGF-β signaling in group 3 MBs and NF-κB signaling in group 4 MBs, suggests that diagnostic biomarkers for group 3 and 4 tumors will be forthcoming.
Potential Role of Regulatory Noncoding RNAs in Medulloblastoma Diagnosis and Treatment
While approximately 80% of the human genome is “active,” i.e., transcribed, only about 2% of the genome is protein coding.29,30 The remaining transcribed but not translated portion is considered noncoding RNA (ncRNA), the most abundant proportion of which comprises housekeeping ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs).29,30 Other nonprotein-coding transcripts were initially regarded and discarded as functionless noise, but ncRNAs are now known to be as important as proteins in regulating cellular function and identity. Based on their size, regulatory noncoding transcripts can be divided into two groups: short noncoding (18–200 nucleotides) and long noncoding RNAs (lncRNAs) (>200 nucleotides). Short noncoding RNAs are further classified into piwi-interacting RNAs (piRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs), and microRNAs (miRNAs or miRs). Similarly, long noncoding RNAs can be classified based on location and direction of transcription into long intergenic ncRNAs (lincRNAs), natural antisense transcripts (NATs), enhancer RNAs (eRNAs), and bidirectional transcripts.31 Circular RNAs (circRNAs) are a further interesting class of recently discovered ncRNA (reviewed in ref. 32; see below) generated from alternative backsplicing of pre-mRNA, with the 3′ and 5′ prime ends of the resulting transcripts covalently binding to produce a circular transcript.
Functionally, regulatory ncRNAs (hereafter referred to as ncRNAs only) regulate transcriptional and post-transcriptional gene expression during development and in disease states. miRNAs regulate gene expression predominantly via post-transcriptional gene silencing through modulating transcript stability.33 LncRNAs are functionally diverse and participate in transcriptional silencing (e.g., XIST34); function as enhancers by regulating three-dimensional chromosomal structure to strengthen interactions between enhancers and promoters (e.g., LUNAR135); and sequester miRNAs from their target sites (e.g., TUG136). LncRNAs can also act as hubs for protein–protein and protein–nucleic acid interactions.37 CircRNAs are still in their nascent phase, but due their exceptional stability and presence of miRNA binding sites are thought to act as miRNA sponges; i.e., they bind to miRNAs and sequester them.32,38 There is now a considerable body of evidence implicating ncRNAs in both health and disease,39–44 including in MB. Here we summarize the latest data on ncRNAs in MB and the implications for therapy.
Mechanistic and Functional Importance of lncRNAs in Medulloblastoma
LncRNAs are defined according to length (>200 nt), are transcribed by RNA polymerase II, and commonly originate from intergenic regions. Although the precise roles of the vast majority of the approximately 40,000 known lncRNAs are still uncertain,45 at least some of these transcripts are now known to be key regulators of cellular differentiation and proliferation. They are also dysregulated in many types of cancer46,47 through their diverse participation in mRNA stability, RNA splicing, chromatin structure, and miRNA‐mediated gene regulation by acting as miRNA sponges48 (Figure 1). However, their role in MB is relatively poorly understood, and there are only a few articles on the topic. Here we discuss recently reported mechanistic insights into how lncRNAs regulate gene expression and contribute to MB formation. Table 1 summarizes the lncRNAs implicated in MB together with their molecular partners or genomic targets that mediate MB phenotypes of proliferation, growth suppression, migration, invasion, and metastasis.
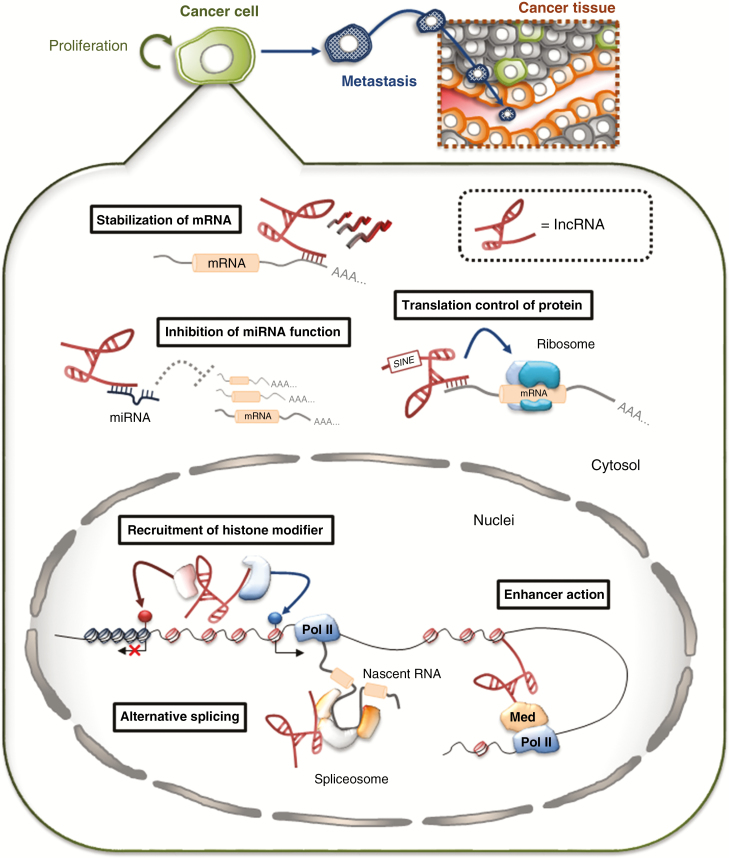
Figure 1.
Schematic showing lncRNA functions. lncRNAs are involved in gene regulation through a variety of mechanisms that rely on interactions with multiple molecules. In the cytoplasm, lncRNAs interact with other types of RNA and affect functions including mRNA stability, mRNA translation, or microRNA (miRNA) sponging. In the nucleus, lncRNAs can regulate transcription by recruiting chromatin-modifying complexes by acting as enhancer RNAs (eRNAs). Moreover, they can regulate gene expression by influencing pre-mRNA splicing. Pol II, RNA polymerase II; Med, Mediator complex.
Table 1.
Summary of functions of the main lncRNAs implicated in medulloblastoma
| lncRNA | Biological roles in medulloblastoma cell | Molecular functions | Target pathway | Reference |
|---|---|---|---|---|
| CCAT1 | Promotion of cell proliferation and metastasis | Unknown | MAPK pathway | 49 |
| CRNDE | Promotion of cell cycle progression | Unknown | Unknown | 50 |
| linc-NeD125 | Promotion of cell proliferation, migration and invasion | miRNA sponge (miR-19a-3p, miR-19b-3p and miR-106a-5p) |
Unknown | 51 |
| LOXL1-AS1 | Promotion of cell proliferation and metastasis | Unknown | PI3K/AKT pathway | 52 |
| NKX2-2AS | Suppression of cell proliferation, migration and invasion | miRNA sponge (miR-103, miR-107 and miR-548m) |
SHH pathway | 53 |
| PVT1 | Promotion of cell proliferation | Host gene for miRNA (miR-1204, miR-1205, miR-1206, and miR-1207) |
Unknown | 28 |
| SPRY4-IT1 | Promotion of cell proliferation, migration and invasion | Unknown | Unknown | 54 |
| UCA1 | Promotion of cell proliferation and migration | Unknown | Unknown | 55 |
Dysregulated lncRNAs in Medulloblastoma
linc-NeD125
linc-NeD125, also known as MIR100HG, is significantly overexpressed in group 4 MBs, the largest and least well characterized molecular medulloblastoma subgroup. Mechanistically, linc-NeD125 recruits the miRNA-induced silencing complex (miRISC) and directly binds miR-19a-3p, miR-19b-3p, and miR-106a-5p. Functionally, linc-NeD125 acts as an miRNA sponge that sequesters these three miRNAs and de-represses their targets CDK6, MYCN, SNCAIP, and KDM6A, which are major driver genes of group 4 MBs. Consistent with the role of linc-NeD125 as an oncogene, ectopic expression of linc-NeD125 promotes medulloblastoma cell proliferation, migration, and invasion in vitro.51
NKX2-2-AS1
NKX2-2-AS1 is involved in SHH-driven MB development. In addition, the SHH pathway transcription factor GLI2 switches on FOXD1 expression, which subsequently represses transcription of NKX2-2-AS1. Specifically, NKX2-2-AS1 functions as an miRNA sponge to sequester miR-103, miR-107, and miR-548m, thereby maintaining expression of their tumor-suppressive targets BTG2, LATS1, and LATS2. Thus, GLI2/FOXD1-mediated NKX2-2-AS1 downregulation contributes to the pathogenesis of SHH-subgroup MB.53
PVT1
PVT1 is a noncoding host gene for four miRNAs (miR-1204, miR-1205, miR-1206, and miR-1207) and is amplified together with MYC in group 3 MBs. PVT1 fusion genes are highly recurrent, restricted to group 3 MB, arise through a chromothripsis-like process, and were the first recurrent translocation reported in MB.28 The PVT1 locus is thought to be genomically fragile, as the majority of MYC-amplified group 3 MBs harbor PVT1 fusions. The identified PVT1 fusions involve only PVT1 exon 1 and miR-1204, and only miR-1204 and not the adjacent miR-1205, miR-1206, and miR-1207 are expressed at higher levels in PVT1-MYC fusion group 3 MBs compared with nonfusion cases. Inhibition of miR-1204 reduced proliferation of group 3 MB cells at a level comparable to MYC knockdown, while an MB cell line with neither MYC amplification nor detectable PVT1-MYC fusion gene was unaffected by miR-1204 knockdown.28 PVT1 stabilizes MYC to promote tumorigenesis, and the PVT1 locus is often amplified in breast cancer.56 However, PVT1 is also frequently disrupted by recurrent translocations or deletions in many cancer types, including MB, suggesting that it may have additional regulatory functions. Of note, a recent study showed that PVT1 lncRNA expression was not required to inhibit MYC transcription; instead, the PVT1 promoter competed with MYC for enhancer binding at the PVT1 locus, preventing MYC promoter firing and suppressing transcriptional elongation of the MYC oncogene and reduced cancer cell growth.57 This might indicate a lncRNA-independent tumor suppressive role for the PVT1 promoter in MB and suggest that regulatory sequences in lncRNA genes may contribute to tumorigenesis.
MicroRNAs as Theranostic Agents in Human Medulloblastoma
MicroRNAs are a family of endogenous small noncoding RNAs, ~18–25 nucleotide in size, with an evolutionarily conserved structure and a predominant role in posttranscriptional mRNA modifications. Since miRNAs were first discovered in Caenorhabditis elegans,58 many miRNAs are now known to be crucial cellular regulators in both health and disease.33
miRNAs are synthesized in the nucleus as parent primary-microRNA (pri-miRNA) transcripts, usually by RNA polymerase II59 and in certain instances by RNA polymerase III,60 with 5′ and 3′ modifications of a normal mRNA61 (Figure 2). Due to the characteristic stem-loop structure of the pri-miRNAs, they are recognized by the miRNA processing machinery composed of DGCR8 and the type III RNase DROSHA to be converted into an approximately 85 nucleotide stem-loop structure called precursor miRNA (pre-miRNA).62 The pre-miRNA is then transported from the nucleus to the cytoplasm where the final processing step by the RNase III enzyme DICER1 produces mature miRNA in duplex form (miRNA/miRNA*, where * indicates the passenger strand).62 The mature single-stranded miRNA is then released from the duplex and incorporated into functional AGO-containing RNA-induced silencing complex (RISC), which guides the complex to target mRNA(s).
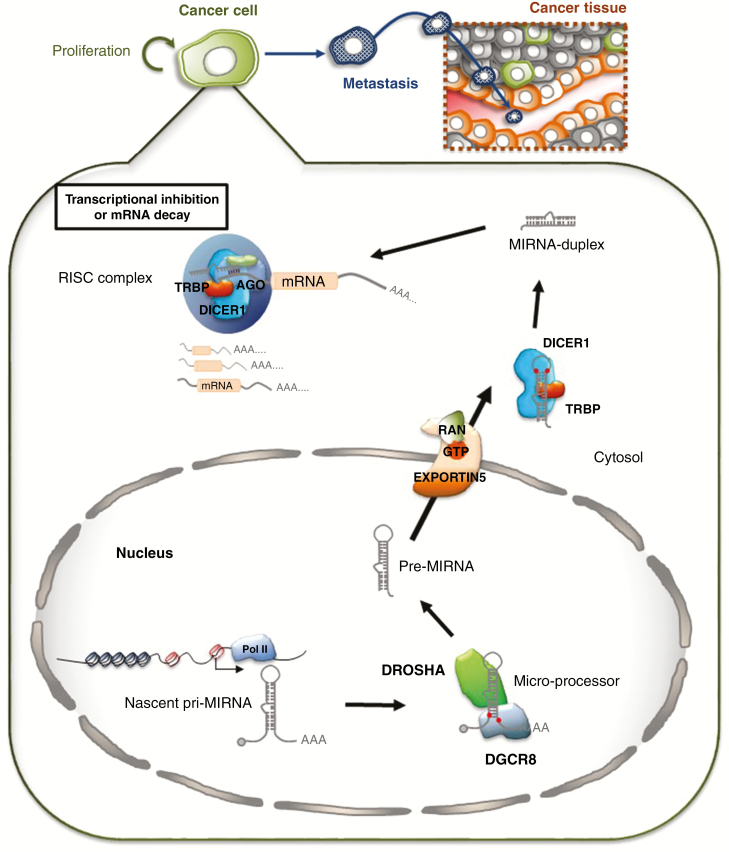
Figure 2.
Schematic showing microRNA biogenesis and function. MicroRNAs are synthesized in the nucleus from microRNA encoding genes as longer primary microRNA (pri-miRNA) precursors that are processed by the DROSHA-DGCR8 complex into pre-microRNA (pre-miRNA) molecules. The pre-miRNA transcript is exported out of the nucleus to the cytoplasm where it undergoes a second round of cleavage by the DICER1 complex to form miRNA-duplexes containing the mature miRNA strand. The mature miRNA complexes with the AGO-containing RISC complex and targets mRNA transcripts to inhibit protein production by targeted mRNA decay or posttranscriptional translational inhibition to regulate physiological processes.
Dysregulated miRNA expression is common in human cancer and is crucial for its progression. Common causes of altered miRNA expression in tumors include copy number changes due to amplification, duplication, or deletion, often involving an entire miRNA cluster. In other cases, miRNA dysregulation can be attributed to changes in expression of upstream transcriptional regulators or signaling pathways regulating their expression. For example, frequently mutated cancer genes including TP5363 or MYC64 affect a number of miRNAs, resulting in their dysregulation. Functionally, miRNAs are predominantly known to regulate posttranscriptional gene regulation by suppressing gene expression via targeted degradation of RISC-bound transcripts and in some cases via posttranscriptional modifications.33,62 miRNAs can act as tumor suppressors or oncogenes depending upon their downstream targets and the cellular context.
MicroRNAs in Medulloblastoma
As in other tumors, miRNAs are the most studied ncRNA in MB. An early study examining the role of miRNAs in MB revealed overexpression of miR18A, 19A, 21, and 25 in 34 MBs.65 Since then, numerous transcriptome-wide comparisons of MBs and control cerebellum tissue have widened the number of candidate miRNAs that might play important roles in MB development.66–70 Some of these profiling studies have also associated specific miRNAs with MB subgroups, suggesting a group-specific role in some cases. Several miRNAs including miR-10B, miR-193A, miR-224–452 cluster, miR-182-183-96 cluster, miR-148A, miR-23B, and miR-365 were specifically enriched in the WNT-MB subgroup.67,69,71 Similarly, overexpression of miRNAs in the miR-17–92 cluster was frequently associated with SHH-MBs and was shown to be oncogenic.72,73 Gershanov et al. found that 12 miRNAs including miR-181A, 135B, and 660 were overexpressed in group 4 MBs compared with other MBs.74 Furthermore, a recent transcriptomic study of the miRNAome of SHH MB cancer stem cells (CSCs) revealed differential up- and downregulation of a number of miRNAs compared with normal neural stem cells including higher expression of miR-20a-5p and miR-193a-5p and lower expression of miR-222-5p, miR-34a-5p, miR-345-5p, miR-210-5p, and miR-200a-3p; some differentially expressed miRNAs in MB CSCs were the same as those dysregulated in primary SHH MB (let-7a, miR-100, miR-132, miR-135a, miR-135b, miR-150, and miR-203).75
These studies suggest that miRNAs might have subgroup-specific functions in MBs. We briefly review some of the miRNAs implicated in MB progression, whether they act as oncogenes or tumor suppressor genes, and the associated in vitro/in vivo evidence (Table 2).
Table 2.
MicroRNAs in medulloblastoma
| miRNA | Expression in MBs | Interactions/Functions | Group specificity | Reference | |
|---|---|---|---|---|---|
| Oncogenic | miR-21 | Upregulated | PDCD4; promotes metastasis | 76 | |
| miR-17~92 cluster | SHH, MYCN; promotes proliferation | SHH | 73,77 | ||
| miR-183~96~182 cluster | SHH, AKT1/2; promotes proliferation and metastasis | 78 | |||
| miR-30b/d | NA | Group 3 | 79 | ||
| miR-10b | BCL2; inhibits apoptosis | 80 | |||
| miR-367 | RYR3, ITGAV, RAB23; promotes proliferation of cancer stem cell | 81 | |||
| miR-106b | PTEN; proliferation | 82 | |||
| Tumor Suppressor | miR-193 | WNT signaling; inhibits cell proliferation | WNT | 67 | |
| miR-224 | 67 | ||||
| miR-124 | Downregulated | CDK6, SCL16A1; inhibits cell proliferation | 83,84 | ||
| miR-199b | HES1, CD15; inhibits cell proliferation | 85,86 | |||
| miR-125b | SHH signaling, LIFRα; inhibits cell proliferation | 72,85,87 | |||
| miR-324 | SMO/GLI1/SHH signaling; inhibits cell proliferation | 72 | |||
| miR-326 | SMO/GLI1/SHH signaling; inhibits cell proliferation and self-renewal | 72,88 | |||
| miR-9 | t-TrkC; promotes cell cycle arrest and apoptosis | 65 | |||
| miR-125a | 65 | ||||
| miR-128a | BMI-1; promotes senescence | 89 | |||
| miR-218 | CDK6, RICTOR, CTSB; promotes cell differentiation | 90,91 | |||
| miR-34a | NOTCH signaling, MYCN, SIRT1, MAGE-A; impairs self-renewal and cell proliferation, promotes apoptosis, sensitizes MB cells to chemotherapy | 92–94 | |||
| miR-31 | MCM2 regulation; reduces proliferation | 95 | |||
| miR-192 | DHFR, CD47; inhibits metastasis | 96 | |||
| miR-135a | ARHGEF6; reduces proliferation | 97 | |||
| miR-494 | MYC/p38 MAPK; reduces proliferation, migration, invasion and increases apoptosis | 98 | |||
| miR-466f-3p | Vegf/Nrp2; epithelial to mesenchymal transition | 99 | |||
| miR-221-3p | EIF5A2; cell proliferation, cell cycle and apoptosis | 100 |
Oncogenic miRNAs
Cell cycle and apoptosis pathways are one of the main targets of oncogenic miRNAs in MB. The miRNAs belonging to the miR-17–92 cluster, namely, miR-17, miR-18A, miR-19A/B, miR-20A, and miR-92A, are frequently upregulated in MB samples.22,73 The encoding locus was found to be amplified in 6% of MB samples, predominantly in SHH MBs.73 In addition, miR17-92 cluster is proposed to be regulated by MYCN, a gene frequently amplified in SHH MBs, and overexpression of this cluster promoted cell proliferation even in the absence of SHH signaling.73 Further, loss of function studies using locked nucleic acid-mediated knockdown reduced cell growth,77 and conditional deletion prevented tumor development in an SHH MB mouse model,101 highlighting the oncogenic role of miRNA-17-92 cluster in SHH-dependent tumor growth. Another pro-proliferative miRNA, miR-10b, was found to be upregulated in ERRB2 (HER2)-overexpressing MBs and in SHH MB and group 3 MB cell lines.65,80 miR-10b expression was found to be positively correlated with that of the anti-apoptotic gene BCL2, possibly in a positive feedback loop as knockdown of one downregulated the expression of the other.80 The authors suggested that miR-10b is important for upregulation/maintenance of BCL2 expression, thereby promoting proliferation and inhibiting apoptosis.
Metastasis is a poor prognostic event in MBs. miR-21, upregulated in MB compared with normal cerebellum, was shown to promote migration and metastasis22,76 by targeting the metastasis suppressor gene PDCD4. miR-21 knockdown upregulated PDCD4, E-cadherin, and TIMP2 and consequently downregulated MAP4K1 and JNK. Another metastasis-associated miRNA cluster, miR183-96-182, was also found to frequently co-occur with MYC amplifications.22,102 Knockdown of these miRNAs individually or together upregulated an apoptosis-associated gene signature and reduced cell viability by impairing DNA repair.78 In addition, the cluster genes were shown to have a pro-metastatic role, possibly by regulating divergent AKT1/2 signaling.78,102 In a mouse SHH-MB model,103 cluster upregulation was frequently associated with Pten loss and the cluster genes acted downstream of SHH signaling to promote proliferation.
Tumor Suppressor miRNAs
Tumor suppressor miRNAs in MB are generally downregulated in response to oncogenic transformations. Some of these miRNAs are involved in promoting neuronal differentiation during normal development. For example, miR-124, a brain-enriched miRNA that usually regulates neurogenesis and neuronal differentiation, is often downregulated in MB.83,104,105 In normal brain, miR-124 regulates cell cycle progression by modulating CDK6, a well-known marker of high-risk MB,84,106 and miR-124 inhibited proliferation in vitro and in vivo upon overexpression in MB cells.105 miR-124 has also been suggested to regulate energy metabolism (glycolysis) by targeting the solute carrier SCL16A1.83 Similarly, miR-218, another differentiated neuron-enriched miRNA,107 was found to be downregulated in MB samples,90 particularly in SHH and group 3 patients. In vitro experiments showed that miR-218 overexpression reduced expression of neural stem cell marker NANOG and increased expression of neuronal differentiation marker MAP2, thereby inhibiting proliferation, clonogenicity, and invasion of SHH MB cells. The study also identified RICTOR, CTSB, and CDK6 as miR-218 targets, further implicating the CDK6-miR-218 network in MB proliferation. Furthermore, miR-125b, a member of the let-7 family and a negative regulator of SMO (smoothened),72 has been shown to promote neuronal differentiation85 and was inversely correlated with MYC expression in SHH-MB cells. miR-125b was shown to target leukemia inhibitory factor repressor alpha (LIFRα), which promotes proliferation in vitro and increases tumor volume in vivo.87
Tumor suppressor miRNAs are often involved in negatively regulating cell cycle progression and cancer cell stemness. miR-326, a negative regulator of SMO, was found to be downregulated in SHH-MBs, with lower expression associated with high-risk patients.65,88 The SHH/GLI-miR-326 network was in particular active in CSCs derived from SHH MBs. CSCs exhibited lower expression of miR-326 and its host gene Arrb1. Upon re-induction in SHH-MB CSCs, miR-326 inhibited SHH/GLI signaling at the receptor and transcription factor levels and impaired the self-renewal and proliferative capacity of the cells.88 miR-192 was found also found to be downregulated in MB patients, with its overexpression inversely correlated with tumor seeding in cerebrospinal fluid, reduced proliferation and anchoring capability of MB cell lines, and it also directly downregulated the expression of target genes DHFR, ITGAV, ITGB1/3, and CD47.96 Furthermore, xenografted MB cells overexpressing miR-192 in nude mice exhibited reduced leptomeningeal seeding, suggesting that miR-192 is a potential metastasis suppressor.96
Notch signaling plays an important role in cell fate decisions in various contexts including neurogenesis and is a frequent direct and indirect target of tumor suppressor miRNAs in MBs. miR-199b, a good prognostic marker, was inversely related to metastases in patients.85 In vitro, miR-199b regulated Notch signaling by targeting HES1, which in turn inhibited miR-199b expression.85,86 miR-199b overexpression in cell lines also decreased the CSC population as judged by reduced expression of CD15 (a direct miR-199B target) and CD133 and inhibited tumor growth in vivo. Another miRNA induced by p53 signaling, miR-34a, targeted Notch ligand Delta-Like 1 (DLL1) in vitro and inhibited CD15+/CD133+ CSC proliferation in vitro and in vivo while promoting neural differentiation.92 miR-34a overexpression also inhibited AKT and STAT3 signaling and was suggested to interact with additional MB pathways such as MYCN, BCL2, and SIRT1.93 miR-34A induced apoptosis and cell cycle arrest by directly targeting the oncogene MAGE,94 the downregulation of the latter inducing p53 signaling to further induce miR-34a to provide positive feedback. In a study specifically examining the CSC compartment in MB, miR-466f-3p was upregulated in CSCs isolated from SHH-MBs derived from Ptch1 heterozygous mice. In this case, miR-466f-3p suppressed a mesenchymal phenotype via downregulation of Vegfa and Nrp2.99 This result might be of particular significance, since CSCs represent a subset of MB cells that not only promote cancer phenotypes but also participate in chemoresistance.108
Therapeutic Potential of miRNAs in Medulloblastoma
Of all the regulatory ncRNAs, having been the first to be discovered, miRNAs are the closest to reaching clinical applicability. miRNAs are, therefore, important therapeutic targets in many different types of cancer, with several candidates being tested in clinical trials.109–112 miRNAs have yet to take a lead in MB therapeutics, but several of the above described miRNAs represent potential therapeutic targets due to their oncogenic or tumor suppressive roles. Here we focus on the therapeutic potential of miRNAs that have shown some promise in in vivo functional validation studies.
miR-34a-dependent downregulation of MAGE-A was shown to sensitize cells to chemotherapeutics such as mitomycin and cisplatin.94 Re-introduction of miR-34a in MB cell lines via adenovirus particles inhibited tumor growth in vivo without exhibiting any toxicity.92 The tumor suppressor effect of miR-34a was shown in part to be due to regulating Notch signaling crucial for CSC proliferation. miR-199b also targets Notch signaling in CSCs by targeting HES1, with similar tumor suppressive results in vivo.85 The negative correlation between miR-199b and metastasis and prognosis suggests that miR-199b-based therapy could be used to improve survival in high-risk patients. The pro-proliferation gene CDK6 is often overexpressed in MB patients and is associated with an unfavorable prognosis, and CDK6 upregulation could be in part due to downregulation of miRNAs targeting CDK6 such as miR-124. Sibler et al. showed that reintroduction of miR-124 reduced tumor growth in vivo, suggesting therapeutic potential in a subset of MBs overexpressing CDK6.105 MCM2, as a component of the multiprotein MCM2-7 complex, is crucial for DNA replication, transcription, and RNA splicing and is a frequently described oncogene.113 miR-31 targeted and downregulated MCM2 in MB cell lines and reduced tumor growth in vivo,95 and miR-192 was found to be upregulated in metastatic MB compared with nonmetastatic MB,96 inhibiting cellular proliferation and tumor dissemination by targeting integrins.
In the case of oncogenic miRNAs, the miR-17-92 cluster was induced by SHH signaling and MYCN and promoted tumor development in vitro and in vivo. Complete knockout of the cluster in SHH-MB mice reduced tumor formation. In addition, locked nucleic acid (LNA)-mediated knockdown of these miRNAs individually or together reduced cell proliferation in vitro and in vivo. Furthermore, intravenous injection of LNAs targeting the miR-17-92 cluster inhibited tumor growth and promoted survival in SHH-MB mice, so may represent a potential therapeutic strategy.77
One notable feature of miRNAs in medulloblastoma is their CSC specificity. CSCs show unlimited self-renewal and differentiation capacity, driving cancer progression not only through the generation of functionally diverse progeny but also by being intimately linked to the processes driving dissemination and metastasis, especially epithelial to mesenchymal transition (EMT).108 Furthermore, as shown in breast cancer and other solid organ tumors, CSCs are often intrinsically resistant to chemoradiotherapy so contribute to posttreatment relapse: this is also true in MB, and there is recent evidence that the frequent relapses and leptomeningeal dissemination seen in MB patients are caused by therapy-resistant CSCs.108 Targeting the CSC compartment is therefore of the utmost importance to fully eradicate the disease. A recent transcriptomic study of the SHH-MB CSC miRNome (compared to background neural stem cells) revealed dysregulation of several KEGG pathways including pathways in cancer, the PI3K-AKT pathway, and the protein processing in endoplasmic reticulum pathway.75 In line with these molecular data, most existing studies in MB focusing on eradicating CSCs have focused on specific signaling pathways active in CSCs such as Notch (γ-secretase inhibitors), PI3K/AKT, and STAT3 (celecoxib), with strong preclinical data in mice. For instance, the highly specific PI3K inhibitor GDC-0941 reduced CD133+ stem-like MB cell numbers and their clonogenicity and also delayed the growth of highly aggressive group 3 MBs in a xenograft model,114 while celecoxib improved the chemoradiosensitivity of CD133+ MB xenografts.115 Given that miRNAs act upstream of these pathways, they represent attractive candidate targets against the most difficult to eradicate component of the disease and might have particular value in recurrent disease that has failed conventional therapy.
Other Regulatory Noncoding RNAs in Medulloblastoma
Enhancer RNAs
Enhancer RNA (eRNA) is a newly identified RNA class that functions as a transcriptional regulator by facilitating high-dimensional DNA structures such that both cis and trans gene interactions link enhancer and super-enhancer DNA sites to transcriptional start sites.116–118 eRNA is most likely involved in target gene regulation and chromosome looping.118–121 Although eRNA does not appear to be essential for all enhancers to function, eRNA appears to affect the regulation of active enhancer transcription, thereby promoting gene expression and affecting cell-specific transcriptional regulation.118,122,123 eRNA is thought to function by directly interacting complexes with RNA pol II as well as DNA- and RNA-binding transcription factors. This interaction induces the formation of genomic looping structures such that enhancers interact with promoters, eRNA, mediator, and cohesin to form 3D tertiary DNA structures and efficient RNA transcription.124 Although the role of eRNAs in cancer generally and medulloblastoma specifically is not yet well defined, eRNA levels correlate with enhancer activity and eRNA can be used as a genome-wide marker of active enhancer elements.125 In addition, recent studies have demonstrated that aberrant regulation of eRNAs transcribed from super-enhancers is often linked to cancer development,126–128 suggesting that eRNAs may be important cancer targets. Using H3K27ac and BRD4 chromatin immunoprecipitation followed by sequencing coupled with tissue-matched DNA methylation and transcriptome data, Liu et al. described the active cis-regulatory landscape across 28 primary MBs.129 Analysis of differentially regulated enhancers and super-enhancers reinforced intersubgroup heterogeneity and revealed novel, clinically relevant insights into medulloblastoma biology.129 eRNA may play an important role in MB phenotypic changes and responses to tumor microenvironmental changes.
Circular RNA
Recent advances in deep-sequencing technology and computational biology have further expanded the repertoire of regulatory ncRNAs, with circular RNAs (circRNAs) one of the latest additions. Besides their unique configuration, circRNAs are distinct from their canonical linear siblings in that they harbor frequent exon scrambling events. circRNAs are products of rare “head-to-tail” back-splicing events catalyzed by the splicing machinery. This back-splicing reaction is favored when pre-mRNAs adopt a noncanonical configuration by juxtaposing the 3′ end of a downstream exon to the 5′ end of an upstream exon. Back-splicing is subject to regulation by cis-regulatory elements and/or trans-acting factors. For example, complementary base-pairing by transposon-derived inverted repeats flanking circRNA-generating exons or RNA elements that contain recognition motifs for select RNA-binding proteins (RBPs) can promote the aforementioned noncanonical pre-RNA configurations, thereby facilitating circRNA biogenesis.130,131 In addition, limited splicing machinery (upon depletion of select spliceosome components) or an increase in the occurrence of transcription read-through events (upon a reduction in levels of components of the pre-mRNA 3′ end processing machinery) also promote back-splicing.132
Originally viewed as incidental by-products of rare “head-to-tail” back-splicing events, circRNAs are now known to be an abundant class of RNAs present in a wide variety of eukaryotes, including nematodes, flies, mice, and humans.131,133–141 An increasing number of circRNAs are being functionally characterized. For example, the mouse circRNA CDR1as/CiRS-7 sequesters miR-7 and affects brain development.135,138,142 The circRNA SRY plays a key role in male sex determination.135,138,142 Furthermore, select intron-containing circRNAs can interact with U1 snRNP and promote host gene transcription.143 In addition, circRNAs can function in gene regulation by competing with linear splicing.144 Select circRNAs can also give rise to functional polypeptides, thereby expanding proteome complexity.145,146 Given that circRNAs are highly enriched in the brain and that select circRNAs have been implicated in cancer,140,147 it is possible that circRNAs participate in the physiopathology of MB. In a recent study by Lv et al.,148 expression profiling of circRNAs in a small sample of MBs found 33 differentially expressed circRNAs compared with control cerebellum. The study identified three of these 33 circRNAs to be overexpressed in the MB tissue. Specifically, MB-enriched circRNAs circ-SKA3 and circ-DTL promoted proliferation and migration in vitro, possibly by regulating host gene expression. As the field of circRNA is in its nascent phase, future studies will shed light on putative candidates for biomarker and therapeutic end goals.
Therapeutic Targeting of ncRNAs in Medulloblastoma: Technical Considerations and Limitations
We have described several lncRNAs and miRNAs that show MB subgroup-specific expression and regulating oncogenes and/or tumor suppressor genes,149 making them putative diagnostic biomarkers and therapeutic targets in MB. While the evidence level for majority of candidates is still in nascent phase lacking in vivo mouse studies, nevertheless in vitro studies highlight the potential of candidate factors.
Several approaches could be used to target lncRNAs in cancer including MB: posttranscriptional degradation through argonaute- and dicer-dependent cleavage with siRNAs or RNase H-dependent degradation with modified antisense oligonucleotides; regulation of lncRNA expression with CRISPR-Cas9 gene editing or promoter blockade; or inhibition of RNA-protein interactions with small molecule inhibitors or antisense oligonucleotides.149 Likewise, similar strategies have been used to target miRNAs: antisense oligonucleotides that anneal to the mature miRNA, often modified (especially with 2’-O- methyl groups) to improve stability, affinity, and specificity; LNA anti-miRNA constructs, which have extremely high affinity to single-stranded RNA; miRNA sponges and miR-masks, transcripts that contain multiple miRNA tandem binding sites (sponges) or single-stranded 2′-O-methyl-modified antisense oligonucleotides complementary to predicted miRNA binding sites to compete with cellular miRNA target sites; or upstream small molecule inhibitors to interfere with their synthesis or processing.150 Conversely, miRNAs or lncRNAs can be reintroduced into tumors as mimicking synthetic oligonucleotides that re-introduce tumor suppressor function.151
There has been some advances in targeting lncRNAs and miRNAs with nucleic acid-based therapies both in vitro and in vivo; 152 however, several challenges stills remains to be addressed. First of all, effective delivery remains an important barrier, particularly in vivo, since (i) nucleic acids require active and protected transport to cross plasma membranes; (ii) cellular nucleases and the innate immune response degrade nucleic acids; and (iii) nucleic acids can become sequestered or degraded in the endosomal compartment.36 Additionally, given the brain location of MBs, the blood-brain-barrier (BBB) presents an MB-specific challenge to delivery, not only for small molecules (most of which cannot cross the BBB) but also for nucleic acids, whose high molecular weight, anionic charge, and instability pose additional challenges.11,153 Several strategies exist to overcome the BBB including viral carriers (e.g., with adeno-associated virus) or nonvirus methods such as hijacking the BBB transcytosis machinery via transferrin receptors and clathrin and caveolae-mediated endocytosis; direct intrathecal injection; or temporarily disrupting the BBB.154
One of the side effects of currently used treatment options is the deterioration/delayed development of cognitive and behavioral potential of a treated child; 8,9 particularly with surgical, chemo, and radiation approaches. RNA-based therapeutic approaches has been suggested to confer reduced toxicity due to reduced systematic exposure and incorporation of modifications for that specific purpose.155–157 For example, while overexpression of siRNA can saturate normal miRNA machinery that could indeed result in severe toxicity,158 designing siRNA as asymmetric DICER substrate could potentially address off-target effects.159 Another source of toxicity in RNA-based therapeutic approaches could be the choice of delivery system, as seen in the case of nanoparticles, in that case, conjugate-based approach offers better alternative.160
Altogether, as in the case of other cancer, ncRNA offers new therapeutic options for MB treatment. While significant challenges remain to identify specific RNA targets for their efficacy and amenability, we must additionally address the cost-to-benefit balance between treatment and overall life quality posttreatment in selecting new therapeutic approaches.
Conclusions
Various ncRNAs are now known to participate in MB biology, although the functional significance of many remains uncertain. Nevertheless, different lncRNAs and miRNAs have oncogenic or tumor suppressive roles in MB. By contributing to the observed heterogeneity between subgroups, these ncRNAs possess subgroup specificity that can be exploited to personalize therapy both by acting as biomarkers and as therapeutic targets. Of note, several of the already identified ncRNAs are specific to MB CSCs, the most difficult-to-treat component of the tumor that drives metastasis and acquired resistance, thereby providing opportunities for therapy in relapsing, disseminating, and therapy-resistant disease. Delivering nucleic acids to tumors—and especially central nervous system tumors behind the blood-brain-barrier—remain a technical challenge but one that is gradually being overcome through the use of advanced technologies such as nanotechnology and rational biomaterial design.
Acknowledgments
This work was supported by National Institutes of Health grants, NCI 5P30CA030199 (SBP), P30 CA006973 (JHU SKCCC) and Florida Department of Health, Bankhead-Coley Cancer Research Program 5BC08 to R.J.P.
Conflict of Interest. The authors declare that they have no competing interests.
Authorship statement. All authors participated in writing, reviewing, and editing the manuscript.
References
- 1. Ostrom QT, Gittleman H, Liao P, et al. . CBTRUS Statistical Report: primary brain and other central nervous system tumors diagnosed in the United States in 2010-2014. Neuro-Oncology. 2017;19(suppl_5):v1–v88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Louis DN, Ohgaki H, Wiestler OD, et al. . The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Diamandis P, Aldape K. World Health Organization 2016 Classification of Central Nervous System Tumors. Neurologic Clinics. 2018;36(3):439–447. [DOI] [PubMed] [Google Scholar]
- 4. Millard NE, De Braganca KC. Medulloblastoma. J child Neurol. 2016;31(12):1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kondoff SI, Milev MD, Laleva LN, et al. . A case of early extraneural medulloblastoma metastases in a young adult. Asian J Neurosurg. 2015;10(4):331–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dufour C, Beaugrand A, Pizer B, et al. . Metastatic medulloblastoma in childhood: chang’s classification revisited. Int J Surg Oncol. 2012;2012:245385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vladoiu MC, El-Hamamy I, Donovan LK, et al. . Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature. 2019;572(7767):67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Braganca KC, Packer RJ. Treatment Options for Medulloblastoma and CNS Primitive Neuroectodermal Tumor (PNET). Curr Treat Options Neurol. 2013;15(5):593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Palmer SL, Reddick WE, Gajjar A. Understanding the cognitive impact on children who are treated for medulloblastoma. J Pediatr Psychol. 2007;32(9):1040–1049. [DOI] [PubMed] [Google Scholar]
- 10. Martin AM, Raabe E, Eberhart C, Cohen KJ. Management of pediatric and adult patients with medulloblastoma. Curr Treat Options Oncol. 2014;15(4):581–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang J, Garancher A, Ramaswamy V, Wechsler-Reya RJ. Medulloblastoma: from molecular subgroups to molecular targeted therapies. Annu Rev Neurosci. 2018;41:207–232. [DOI] [PubMed] [Google Scholar]
- 12. Taylor MD, Northcott PA, Korshunov A, et al. . Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol. 2012;123(4):465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Northcott PA, Dubuc AM, Pfister S, Taylor MD. Molecular subgroups of medulloblastoma. Expert Rev Neurother. 2012;12(7):871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perreault S, Ramaswamy V, Achrol AS, et al. . MRI surrogates for molecular subgroups of medulloblastoma. AJNR Am J Neuroradiol. 2014;35(7):1263–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Louis DN, Perry A, Reifenberger G, et al. . The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 16. Schwalbe EC, Lindsey JC, Straughton D, et al. . Rapid diagnosis of medulloblastoma molecular subgroups. Clin Cancer Res. 2011;17(7):1883–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thompson MC, Fuller C, Hogg TL, et al. . Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24(12):1924–1931. [DOI] [PubMed] [Google Scholar]
- 18. Fernandez-L A, Northcott PA, Dalton J, et al. . YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23(23):2729–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ellison DW, Dalton J, Kocak M, et al. . Medulloblastoma: clinicopathological correlates of SHH, WNT, and non-SHH/WNT molecular subgroups. Acta Neuropathol. 2011;121(3):381–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramaswamy V, Taylor MD. Medulloblastoma: from myth to molecular. J Clin Oncol. 2017;35(21):2355–2363. [DOI] [PubMed] [Google Scholar]
- 21. Northcott PA, Buchhalter I, Morrissy AS, et al. . The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547(7663):311–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cho YJ, Tsherniak A, Tamayo P, et al. . Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J Clin Oncol. 2011;29(11):1424–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kool M, Koster J, Bunt J, et al. . Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. Plos One. 2008;3(8):e3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kool M, Jones DT, Jäger N, et al. ; ICGC PedBrain Tumor Project Genome sequencing of SHH medulloblastoma predicts genotype-related response to smoothened inhibition. Cancer Cell. 2014;25(3):393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Northcott PA, Korshunov A, Witt H, et al. . Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29(11):1408–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Louis DN, Perry A, Burger P, et al. ; International Society Of Neuropathology–Haarlem International Society Of Neuropathology–Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol. 2014;24(5):429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cavalli FMG, Remke M, Rampasek L, et al. . Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell. 2017;31(6):737–754.e736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Northcott PA, Shih DJ, Peacock J, et al. . Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488(7409):49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pennisi E. Genomics. ENCODE project writes eulogy for junk DNA. Science (New York, N.Y.). 2012;337(6099):1159, 1161. [DOI] [PubMed] [Google Scholar]
- 31. Nie L, Wu HJ, Hsu JM, et al. . Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. Am J Transl Res. 2012;4(2):127–150. [PMC free article] [PubMed] [Google Scholar]
- 32. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143(11):1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sahakyan A, Yang Y, Plath K. The Role of Xist in X-Chromosome Dosage Compensation. Trends Cell Biol. 2018;28(12):999–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trimarchi T, Bilal E, Ntziachristos P, et al. . Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158(3):593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Katsushima K, Natsume A, Ohka F, et al. . Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat Commun. 2016;7:13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Long Y, Wang X, Youmans DT, Cech TR. How do lncRNAs regulate transcription? Science Advances. 2017;3(9):eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. [DOI] [PubMed] [Google Scholar]
- 40. Romano G, Veneziano D, Acunzo M, Croce CM. Small non-coding RNA and cancer. Carcinogenesis. 2017;38(5):485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell biol. 2011;21(6):354–361. [DOI] [PubMed] [Google Scholar]
- 42. Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19(7-8):454–492. [DOI] [PubMed] [Google Scholar]
- 43. Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73(13):2491–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stefani G, Slack FJ. Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol. 2008;9(3):219–230. [DOI] [PubMed] [Google Scholar]
- 45. Iyer MK, Niknafs YS, Malik R, et al. . The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12(6):433–446. [DOI] [PubMed] [Google Scholar]
- 47. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. [DOI] [PubMed] [Google Scholar]
- 49. Gao R, Zhang R, Zhang C, Zhao L, Zhang Y. Long noncoding RNA CCAT1 promotes cell proliferation and metastasis in human medulloblastoma via MAPK pathway. Tumori. 2018;104(1):43–50. [DOI] [PubMed] [Google Scholar]
- 50. Song H, Han LM, Gao Q, Sun Y. Long non-coding RNA CRNDE promotes tumor growth in medulloblastoma. Eur Rev Med Pharmacol Sci. 2016;20(12):2588–2597. [PubMed] [Google Scholar]
- 51. Laneve P, Po A, Favia A, et al. . The long noncoding RNA linc-NeD125 controls the expression of medulloblastoma driver genes by microRNA sponge activity. Oncotarget. 2017;8(19):31003–31015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gao R, Zhang R, Zhang C, Liang Y, Tang W. LncRNA LOXL1-AS1 promotes the proliferation and metastasis of medulloblastoma by activating the PI3K/AKT Pathway. Anal Cell Pathol (AMST). 2018;2018:9275685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang Y, Wang T, Wang S, et al. . Nkx2-2as Suppression Contributes to the Pathogenesis of Sonic Hedgehog Medulloblastoma. Cancer Res. 2018;78(4):962–973. [DOI] [PubMed] [Google Scholar]
- 54. Shi PF, Ji HL, Luo YK, Mao TM, Chen X, Zhou KY. [Effect of long noncoding RNA SPRY4-IT1 on proliferation and metastasis of medulloblastoma]. Zhongguo Ying Yong Sheng Li Xue Za Zhi. 2017;33(1):78–82. [DOI] [PubMed] [Google Scholar]
- 55. Zhengyuan X, Hu X, Qiang W, Nanxiang L, Junbin C, Wangming Z. Silencing of urothelial carcinoma associated 1 inhibits the proliferation and migration of medulloblastoma cells. Med Sci Monit. 2017;23:4454–4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tseng YY, Moriarity BS, Gong W, et al. . PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512(7512):82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cho SW, Xu J, Sun R, et al. . Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element. Cell. 2018;173(6):1398–1412.e1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. [DOI] [PubMed] [Google Scholar]
- 59. Lee Y, Kim M, Han J, et al. . MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23(20):4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13(12):1097–1101. [DOI] [PubMed] [Google Scholar]
- 61. Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11(7):537–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Feng Z, Zhang C, Wu R, Hu W. Tumor suppressor p53 meets microRNAs. J Mol Cell Biol. 2011;3(1):44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Psathas JN, Thomas-Tikhonenko A. MYC and the art of microRNA maintenance. Cold Spring Harb Perspect Med. 2014; 4(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ferretti E, De Smaele E, Po A, et al. . MicroRNA profiling in human medulloblastoma. Int J Cancer. 2009;124(3):568–577. [DOI] [PubMed] [Google Scholar]
- 66. Dai J, Li Q, Bing Z, et al. . Comprehensive analysis of a microRNA expression profile in pediatric medulloblastoma. Mol Med Rep. 2017;15(6):4109–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gokhale A, Kunder R, Goel A, et al. . Distinctive microRNA signature of medulloblastomas associated with the WNT signaling pathway. J cancer Res Ther. 2010;6(4):521–529. [DOI] [PubMed] [Google Scholar]
- 68. Genovesi LA, Carter KW, Gottardo NG, Giles KM, Dallas PB. Integrated analysis of miRNA and mRNA expression in childhood medulloblastoma compared with neural stem cells. Plos One. 2011;6(9):e23935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kunder R, Jalali R, Sridhar E, et al. . Real-time PCR assay based on the differential expression of microRNAs and protein-coding genes for molecular classification of formalin-fixed paraffin embedded medulloblastomas. Neuro Oncol. 2013;15(12):1644–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kumar V, Kumar V, Chaudhary AK, Coulter DW, McGuire T, Mahato RI. Impact of miRNA-mRNA profiling and their correlation on medulloblastoma tumorigenesis. Mol Ther Nucleic Acids. 2018;12:490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yogi K, Sridhar E, Goel N, et al. . MiR-148a, a microRNA upregulated in the WNT subgroup tumors, inhibits invasion and tumorigenic potential of medulloblastoma cells by targeting Neuropilin 1. Oncoscience. 2015;2(4):334–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ferretti E, De Smaele E, Miele E, et al. . Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J. 2008;27(19):2616–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Northcott PA, Fernandez-L A, Hagan JP, et al. . The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69(8):3249–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gershanov S, Toledano H, Michowiz S, et al. . MicroRNA-mRNA expression profiles associated with medulloblastoma subgroup 4. Cancer Manag Res. 2018;10:339–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Po A, Abballe L, Sabato C, et al. . Sonic Hedgehog Medulloblastoma Cancer Stem Cells Mirnome and Transcriptome Highlight Novel Functional Networks. Int J Mol Sci. 2018;19(8). Article 2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Grunder E, D’Ambrosio R, Fiaschetti G, et al. . MicroRNA-21 suppression impedes medulloblastoma cell migration. European Journal of Cancer (Oxford, England: 1990). 2011;47(16):2479–2490. [DOI] [PubMed] [Google Scholar]
- 77. Murphy BL, Obad S, Bihannic L, et al. . Silencing of the miR-17~92 cluster family inhibits medulloblastoma progression. Cancer Res. 2013;73(23):7068–7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Weeraratne SD, Amani V, Teider N, et al. . Pleiotropic effects of miR-183~96~182 converge to regulate cell survival, proliferation and migration in medulloblastoma. Acta Neuropathol. 2012;123(4):539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lu Y, Ryan SL, Elliott DJ, et al. . Amplification and overexpression of Hsa-miR-30b, Hsa-miR-30d and KHDRBS3 at 8q24.22-q24.23 in medulloblastoma. Plos One. 2009;4(7):e6159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Pal R, Greene S. microRNA-10b Is Overexpressed and Critical for Cell Survival and Proliferation in Medulloblastoma. Plos one. 2015;10(9):e0137845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kaid C, Silva PB, Cortez BA, Rodini CO, Semedo-Kuriki P, Okamoto OK. miR-367 promotes proliferation and stem-like traits in medulloblastoma cells. Cancer Sci. 2015;106(9):1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Li KK, Xia T, Ma FM, et al. . miR-106b is overexpressed in medulloblastomas and interacts directly with PTEN. Neuropathol Appl Neurobiol. 2015;41(2):145–164. [DOI] [PubMed] [Google Scholar]
- 83. Li KK, Pang JC, Ching AK, et al. . miR-124 is frequently down-regulated in medulloblastoma and is a negative regulator of SLC16A1. Hum Pathol. 2009;40(9):1234–1243. [DOI] [PubMed] [Google Scholar]
- 84. Mendrzyk F, Radlwimmer B, Joos S, et al. . Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. J Clin Oncol. 2005;23(34):8853–8862. [DOI] [PubMed] [Google Scholar]
- 85. Garzia L, Andolfo I, Cusanelli E, et al. . MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. Plos One. 2009;4(3):e4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Andolfo I, Liguori L, De Antonellis P, et al. . The micro-RNA 199b-5p regulatory circuit involves Hes1, CD15, and epigenetic modifications in medulloblastoma. Neuro Oncol. 2012;14(5):596–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Salm F, Dimitrova V, von Bueren AO, et al. . The phosphoinositide 3-kinase p110α isoform regulates leukemia inhibitory factor receptor expression via c-Myc and miR-125b to promote cell proliferation in medulloblastoma. Plos One. 2015;10(4):e0123958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Miele E, Po A, Begalli F, et al. . β-arrestin1-mediated acetylation of Gli1 regulates Hedgehog/Gli signaling and modulates self-renewal of SHH medulloblastoma cancer stem cells. BMC Cancer. 2017;17(1):488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Venkataraman S, Alimova I, Fan R, Harris P, Foreman N, Vibhakar R. MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. Plos One. 2010;5(6):e10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Venkataraman S, Birks DK, Balakrishnan I, et al. . MicroRNA 218 acts as a tumor suppressor by targeting multiple cancer phenotype-associated genes in medulloblastoma. J Biol Chem. 2013;288(3):1918–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shi J, Yang L, Wang T, et al. . miR-218 is downregulated and directly targets SH3GL1 in childhood medulloblastoma. Mol Med Rep. 2013;8(4):1111–1117. [DOI] [PubMed] [Google Scholar]
- 92. de Antonellis P, Medaglia C, Cusanelli E, et al. . MiR-34a targeting of Notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. Plos One. 2011;6(9):e24584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Thor T, Künkele A, Pajtler KW, et al. . MiR-34a deficiency accelerates medulloblastoma formation in vivo. Int J Cancer. 2015;136(10):2293–2303. [DOI] [PubMed] [Google Scholar]
- 94. Weeraratne SD, Amani V, Neiss A, et al. . miR-34a confers chemosensitivity through modulation of MAGE-A and p53 in medulloblastoma. Neuro Oncol. 2011;13(2):165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Jin Y, Xiong A, Zhang Z, et al. . MicroRNA-31 suppresses medulloblastoma cell growth by inhibiting DNA replication through minichromosome maintenance 2. Oncotarget. 2014;5(13):4821–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yang SY, Choi SA, Lee JY, et al. . miR-192 suppresses leptomeningeal dissemination of medulloblastoma by modulating cell proliferation and anchoring through the regulation of DHFR, integrins, and CD47. Oncotarget. 2015;6(41):43712–43730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Hemmesi K, Squadrito ML, Mestdagh P, et al. . miR-135a inhibits cancer stem cell-driven medulloblastoma development by directly repressing Arhgef6 Expression. Stem Cells. 2015;33(5):1377–1389. [DOI] [PubMed] [Google Scholar]
- 98. Xu XH, Zhang SJ, Hu QB, Song XY, Pan W. Effects of microRNA-494 on proliferation, migration, invasion, and apoptosis of medulloblastoma cells by mediating c-myc through the p38 MAPK signaling pathway. Journal of Cellular Biochemistry. 2018;120(2):2594–2606. [DOI] [PubMed] [Google Scholar]
- 99. Besharat ZM, Sabato C, Po A, et al. . Low expression of miR-466f-3p sustains epithelial to mesenchymal transition in sonic hedgehog medulloblastoma stem cells through Vegfa-Nrp2 signaling pathway. Front Pharmacol. 2018;9:1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Yang Y, Cui H, Wang X. Downregulation of EIF5A2 by miR-221-3p inhibits cell proliferation, promotes cell cycle arrest and apoptosis in medulloblastoma cells. Biosci Biotechnol Biochem. 2019;83(3):400–408. [DOI] [PubMed] [Google Scholar]
- 101. Zindy F, Kawauchi D, Lee Y, et al. . Role of the miR-17∼92 cluster family in cerebellar and medulloblastoma development. Biol Open. 2014;3(7):597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bai AH, Milde T, Remke M, et al. . MicroRNA-182 promotes leptomeningeal spread of non-sonic hedgehog-medulloblastoma. Acta Neuropathol. 2012;123(4):529–538. [DOI] [PubMed] [Google Scholar]
- 103. Zhang Z, Li S, Cheng SY. The miR-183∼96∼182 cluster promotes tumorigenesis in a mouse model of medulloblastoma. J Biomed Res. 2013;27(6):486–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci. 2009;12(4):399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Silber J, Hashizume R, Felix T, et al. . Expression of miR-124 inhibits growth of medulloblastoma cells. Neuro Oncol. 2013;15(1):83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Pierson J, Hostager B, Fan R, Vibhakar R. Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol. 2008;90(1):1–7. [DOI] [PubMed] [Google Scholar]
- 107. Thiebes KP, Nam H, Cambronne XA, et al. . miR-218 is essential to establish motor neuron fate as a downstream effector of Isl1-Lhx3. Nat Commun. 2015;6:7718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Huang GH, Xu QF, Cui YH, Li N, Bian XW, Lv SQ. Medulloblastoma stem cells: promising targets in medulloblastoma therapy. Cancer Sci. 2016;107(5):583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yu L, Zhao J, Gao L. Predicting potential drugs for breast cancer based on miRNA and Tissue Specificity. Int J Biol Sci. 2018;14(8):971–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee SS. Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol Ther Nucleic Acids. 2017;8:132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Smith B, Agarwal P, Bhowmick NA. MicroRNA applications for prostate, ovarian and breast cancer in the era of precision medicine. Endocr Relat Cancer. 2017;24(5):R157–R172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Querfeld C, Pacheco T, Foss FM, et al. . Preliminary results of a Phase 1 Trial evaluating MRG-106, a synthetic microRNA antagonist (LNA antimiR) of microRNA-155, in patients with CTCL. Blood. 2016;128(22):1829–1829.27543436 [Google Scholar]
- 113. Fei L, Xu H. Role of MCM2-7 protein phosphorylation in human cancer cells. Cell Biosci. 2018;8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Ehrhardt M, Craveiro RB, Holst MI, Pietsch T, Dilloo D. The PI3K inhibitor GDC-0941 displays promising in vitro and in vivo efficacy for targeted medulloblastoma therapy. Oncotarget. 2015;6(2):802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Yang MY, Lee HT, Chen CM, Shen CC, Ma HI. Celecoxib suppresses the phosphorylation of STAT3 protein and can enhance the radiosensitivity of medulloblastoma-derived cancer stem-like cells. Int J Mol Sci. 2014;15(6):11013–11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Andersson R, Gebhard C, Miguel-Escalada I, et al. . An atlas of active enhancers across human cell types and tissues. Nature. 2014;507(7493):455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kim TK, Hemberg M, Gray JM, et al. . Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Murakawa Y, Yoshihara M, Kawaji H, et al. . Enhanced identification of transcriptional enhancers provides mechanistic insights into diseases. Trends Genet. 2016;32(2):76–88. [DOI] [PubMed] [Google Scholar]
- 119. Kaikkonen MU, Spann NJ, Heinz S, et al. . Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51(3):310–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Li W, Notani D, Ma Q, et al. . Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature. 2013;498(7455):516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell. 2014;56(1):29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Wu H, Nord AS, Akiyama JA, et al. . Tissue-specific RNA expression marks distant-acting developmental enhancers. Plos Genet. 2014;10(9):e1004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Mousavi K, Zare H, Dell’orso S, et al. . eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell. 2013;51(5):606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Shibayama Y, Fanucchi S, Magagula L, Mhlanga MM. lncRNA and gene looping: what’s the connection? Transcription. 2014;5(3):e28658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Cheng JH, Pan DZ, Tsai ZT, Tsai HK. Genome-wide analysis of enhancer RNA in gene regulation across 12 mouse tissues. Sci Rep. 2015;5:12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Jiao W, Chen Y, Song H, et al. . HPSE enhancer RNA promotes cancer progression through driving chromatin looping and regulating hnRNPU/p300/EGR1/HPSE axis. Oncogene. 2018;37(20):2728–2745. [DOI] [PubMed] [Google Scholar]
- 127. Ko JY, Oh S, Yoo KH. Functional enhancers as master regulators of tissue-specific gene regulation and cancer development. Mol Cells. 2017;40(3):169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Liang J, Zhou H, Gerdt C, et al. . Epstein-Barr virus super-enhancer eRNAs are essential for MYC oncogene expression and lymphoblast proliferation. Proc Natl Acad Sci U S A. 2016;113(49):14121–14126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lin CY, Erkek S, Tong Y, et al. . Active medulloblastoma enhancers reveal subgroup-specific cellular origins. Nature. 2016;530(7588):57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Conn SJ, Pillman KA, Toubia J, et al. . The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. [DOI] [PubMed] [Google Scholar]
- 131. Jeck WR, Sorrentino JA, Wang K, et al. . Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Liang D, Tatomer DC, Luo Z, et al. . The output of protein-coding genes shifts to circular RNAs When the Pre-mRNA processing machinery is limiting. Molecular Cell. 2017; 68(5):940–954 e943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. Plos Genet. 2013;9(9):e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. Plos One. 2012;7(2):e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Memczak S, Jens M, Elefsinioti A, et al. . Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. [DOI] [PubMed] [Google Scholar]
- 136. Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Wang PL, Bao Y, Yee MC, et al. . Circular RNA is expressed across the eukaryotic tree of life. Plos One. 2014;9(6):e90859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Hansen TB, Jensen TI, Clausen BH, et al. . Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. [DOI] [PubMed] [Google Scholar]
- 139. Houseley JM, Garcia-Casado Z, Pascual M, et al. . Noncanonical RNAs from transcripts of the Drosophila muscleblind gene. J Hered. 2006;97(3):253–260. [DOI] [PubMed] [Google Scholar]
- 140. Westholm JO, Miura P, Olson S, et al. . Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9(5):1966–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. Plos Genet. 2010;6(12):e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Piwecka M, Glazar P, Hernandez-Miranda LR, et al. . Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science (New York, N.Y.). 2017; 357(6357). [DOI] [PubMed] [Google Scholar]
- 143. Li Z, Huang C, Bao C, et al. . Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. [DOI] [PubMed] [Google Scholar]
- 144. Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. . circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. [DOI] [PubMed] [Google Scholar]
- 145. Legnini I, Di Timoteo G, Rossi F, et al. . Circ-ZNF609 Is a Circular RNA that Can Be translated and functions in myogenesis. Molecular Cell. 2017;66(1):22–37 e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Pamudurti NR, Bartok O, Jens M, et al. . Translation of CircRNAs. Molecular Cell. 2017;66(1):9–21 e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Rybak-Wolf A, Stottmeister C, Glažar P, et al. . Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell. 2015;58(5):870–885. [DOI] [PubMed] [Google Scholar]
- 148. Lv T, Miao YF, Jin K, et al. . Dysregulated circular RNAs in medulloblastoma regulate proliferation and growth of tumor cells via host genes. Cancer Med. 2018;7(12):6147–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Arun G, Diermeier SD, Spector DL. Therapeutic targeting of long non-coding RNAs in Cancer. Trends Mol Med. 2018;24(3):257–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9(10):775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Bader AG, Brown D, Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70(18):7027–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Ji W, Sun B, Su C. Targeting MicroRNAs in Cancer Gene Therapy. Genes (Basel). 2017;8(1). Article 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Wang D, Wu LP. Nanomaterials for delivery of nucleic acid to the central nervous system (CNS). Mater Sci Eng C Mater Biol Appl. 2017;70(Pt 2):1039–1046. [DOI] [PubMed] [Google Scholar]
- 154. Tan JY, Sellers DL, Pham B, Pun SH, Horner PJ. Non-viral nucleic acid delivery strategies to the central nervous system. Front Mol Neurosci. 2016;9:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Rinaldi C, Wood MJA. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat Rev Neurol. 2018;14(1):9–21. [DOI] [PubMed] [Google Scholar]
- 156. Burnett JC, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chem Biol. 2012;19(1):60–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Bramsen JB, Laursen MB, Nielsen AF, et al. . A large-scale chemical modification screen identifies design rules to generate siRNAs with high activity, high stability and low toxicity. Nucleic Acids Res. 2009;37(9):2867–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Grimm D, Wang L, Lee JS, et al. . Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J Clin Invest. 2010;120(9):3106–3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23(2):222–226. [DOI] [PubMed] [Google Scholar]
- 160. Kaczmarek JC, Kowalski PS, Anderson DG. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med. 2017;9(1):60. [DOI] [PMC free article] [PubMed] [Google Scholar]