Abstract
Podophyllotoxin is the natural product precursor of the chemotherapeutic etoposide, yet only part of its biosynthetic pathway is known. We used transcriptome mining in Podophyllum hexandrum (mayapple) to identify biosynthetic genes in the podophyllotoxin pathway. We selected 29 candidate genes to combinatorially express in Nicotiana benthamiana (tobacco) and identified six pathway enzymes, including an oxoglutarate-dependent dioxygenase that closes the core cyclohexane ring of the aryltetralin scaffold. By coexpressing 10 genes in tobacco—these 6 plus 4 previously discovered—we reconstitute the pathway to (−)-4′-desmethylepipodophyllotoxin (the etoposide aglycone), a naturally occurring lignan that is the immediate precursor of etoposide and, unlike podophyllotoxin, a potent topoisomerase inhibitor. Our results enable production of the etoposide aglycone in tobacco and circumvent the need for cultivation of mayapple and semisynthetic epimerization and demethylation of podophyllotoxin.
Although numerous clinically used drugs derive from plant natural products, little is known about their biosynthetic genes, which prevents access to engineered hosts for their production (1). Very few complete pathways exist, and only three—artemisinic acid (2), the benzylisoquinoline alkaloids (3, 4), and the monoterpenoid indole alkaloids (5, 6)—have been transferred to a heterologous host for current or future industrial production. Knowledge of plant pathways is especially stark in comparison with the >700 bacterial and fungal biosynthetic pathways that have been characterized (7).
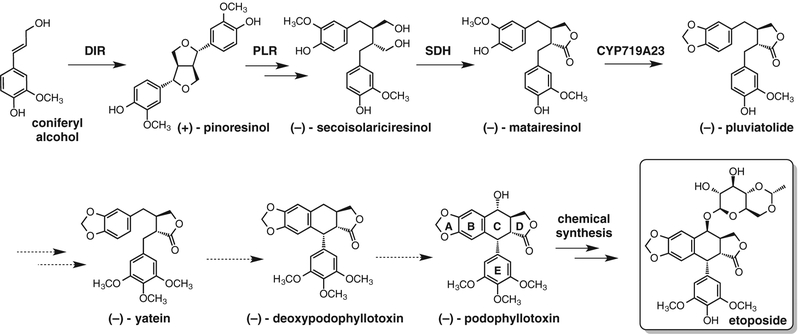
Podophyllotoxin, a lignan from mayapple, is the natural product precursor to the topoisomerase inhibitor etoposide (8–10), which is used in dozens of chemotherapy regimens for a variety of malignancies. Although etoposide is on the World Health Organization’s list of essential medicines, production requires isolation of (−)-podophyllotoxin from the medicinal plant Podophyllum (11). Subsequent semisynthetic steps to produce etoposide are required for topoisomerase inhibitory activity not present in podophyllotoxin. A complete biosynthetic route would enable more facile access to etoposide and natural and unnatural derivatives that are difficult to produce synthetically (12). Early steps of podophyllotoxin biosynthesis (13–16) involve the unusual enantio- and site-selective dimerization of coniferyl alcohol to form (+)-pinoresinol and provide a starting point for identifying additional biosynthetic genes (Fig. 1). However, biosynthetic gene discovery in Podophyllum is a challenge, because the plant grows slowly, the genome is large [~16 Gb (17)] and unsequenced, and methods for constructing mutants are laborious (18).
Fig. 1. Biosynthetic pathway of (−)-podophyllotoxin in P. hexandrum.
Uncharacterized steps are indicated by dashed lines.
We used Agrobacterium-mediated transient expression in N. benthamiana to test candidate genes for the podophyllotoxin pathway for two reasons. First, this versatile plant host would likely produce correctly folded, active proteins from a variety of enzyme superfamilies without optimization. Second, we wanted to rapidly and combinatorially express candidate enzymes without knowing the order of steps or identities of metabolic intermediates and without additional cloning. Combinatorial expression can be accomplished by coinfiltrating multiple Agrobacterium strains—each harboring a different expression construct—and analyzing the resulting plant tissue extracts by using untargeted metabolomics to identify products.
In our initial approach to produce the pathway intermediate (−)-pluviatolide in N. benthamiana leaves, we coexpressed three of the four known podophyllotoxin biosynthetic enzymes: pinoresinollariciresinol reductase (PLR), secoisolariciresinol dehydrogenase (SDH), and CYP719A23 [dirigent protein (DIR) was not required]. Although we observed low levels of (−)-pluviatolide in the resulting leaf extracts, the amount was insufficient for detecting downstream intermediates produced when coexpressing candidate enzymes (fig. S1). No pluviatolide was detected in control experiments when only green fluorescent protein (GFP) is expressed. To enhance (−)-pluviatolide production in planta, we infiltrated leaves expressing CYP719A23 with (−)-matairesinol (isolated from Forsythia × intermedia) 5 days after Agrobacterium infiltration. After 1 day, (−)-pluviatolide concentrations were ~75 times those in leaves expressing PLR, SDH, and CYP719A23, without substrate infiltration (fig. S2), which provided sufficient (−)-pluviatolide to enable candidate enzyme screening.
To select candidate enzymes for conversion of (−)-pluviatolide to the next pathway intermediate, we mined the publicly available P. hexandrum RNA-sequencing (RNA-Seq) data set from the Medicinal Plants Consortium. We noted that all known podophyllotoxin genes were highly expressed in rhizome, stem, and leaf tissues, and we selected candidate genes with similar expression profiles (fig. S3). As the order of steps in the pathway was not known, we chose four putative O-methyltransferases (OMT1–4), 12 cytochromes P450 (CYP), and a 2-oxoglutarate/Fe(II)-dependent dioxygenase (2-ODD). We infiltrated (−)-matairesinol into leaves each coexpressing CYP719A23 and a single candidate enzyme. Liquid chromatography–mass spectrometry (LC-MS) analysis revealed the consumption of (−)-pluviatolide in tobacco leaves coexpressing just one of the candidates, OMT3 (fig. S4). By computationally comparing untargeted metabolomics data from tissue extracts, we identified two compound mass signals unique to CYP719A23 + OMT3 samples relative to CYP719A23 alone: One corresponds to (−)-5′-desmethoxy-yatein (fig. S5); the other, with much lower ion abundance, likely derives from the double methylation of (−)-matairesinol (fig. S6). Expression of OMT3 alone, followed by infiltration of (−)-matairesinol, results in greater amounts of the doubly methylated product, which suggests that this enzyme can accept multiple substrates. We recombinantly expressed OMT3 in Escherichia coli and measured its kinetic parameters for (−)-pluviatolide methylation [apparent Michaelis constant (Km) = 1.4 μM and enzymatic rate (kcat) = 0.72 s−1] (fig. S7). OMT3 accepts (−)-matairesinol and (−)-arctigenin with much lower efficiency and cannot turn over (+)-pinoresinol; these data suggest that OMT3 catalyzes methylation of pluviatolide to generate (−)-5′-desmethoxy-yatein as the next step in the pathway.
We next coexpressed individual candidate CYP and 2-ODD enzymes with CYP719A23 and OMT3; however, we did not observe consumption of (−)-5′-desmethoxy-yatein in leaf extracts, which suggested that our set of candidate genes was incomplete. We reasoned that additional transcriptome data from P. hexandrum tissue samples with differential expression of pathway genes could aid candidate selection.
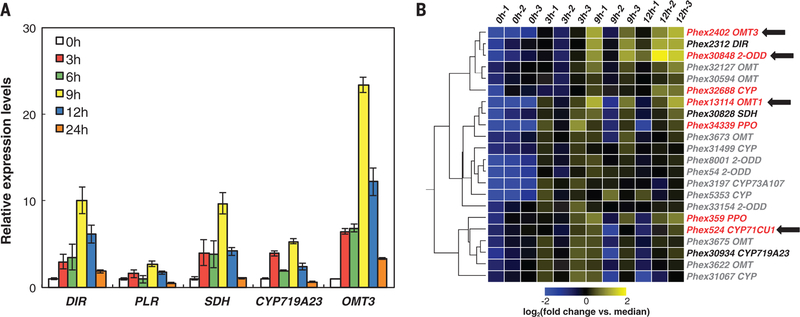
The expression of known (−)-podophyllotoxin biosynthetic genes is up-regulated in P. hexandrum leaves after wounding (19) (Fig. 2A and fig. S8). LC-MS analysis of metabolites in wounded leaves (removed from the stem to eliminate the possibility of metabolite transport) revealed that both (−)-yatein and (−)-deoxypodophyllotoxin [proposed precursors to (−)-podophyllotoxin (20, 21)] accumulate and reach a maximum level 12 to 24 hours after wounding (fig. S9). Consistent with previous reports (16), we did not detect (−)-podophyllotoxin or its glucoside in leaf tissues.
Fig. 2. Expression analysis to identify candidate genes.
(A) qRT-PCR analysis of podophyllotoxin biosynthetic genes after P. hexandrum leaf wounding (at t = 0 hour). Relative expression levels were normalized to t = 0 hour. Data are average values (three technical replicates) ± one SD. (B) Hierarchical clustering of RNA-Seq expression data after filtering by enzyme family and expression level. Heat map depicts the expression levels from a single node from the resulting cluster. Color key: Known biosynthetic genes (black), candidate genes (red), genes identified in this report (red with black arrows).
We took advantage of the pathway’s inducibility and performed RNA-Seq on triplicate P. hexandrum leaf samples, 0, 3, 9, and 12 hours after wounding, from a single plant with the strongest metabolite response. We assembled a leaf transcriptome, determined expression levels, and used predicted enzyme activities required for the missing pathway steps to mine the data for gene sequences encoding OMTs, CYPs, 2-ODDs, and polyphenol oxidases (PPOs). A computational analysis based on expression profile similarity with known pathway genes DIR and CYP719A23 and overall expression level yielded seven candidate pathway genes: Phex30848 (2-ODD); Phex32688 (CYP); Phex13114 (OMT1, previously tested); Phex359 (PPO); Phex34339 (PPO); Phex524 (CYP71CU1); and Phex15199 (CYP) (fig. S10 to S12). Hierarchical clustering analysis of 336 expressed genes, selected by filtering all data (34,384 total genes; see table S1) by enzyme family, revealed a single clade of 91 genes; further filtering by expression level condensed this clade to 22 genes containing six of these seven candidates, three of four known pathway genes, and OMT3 (Fig. 2B).
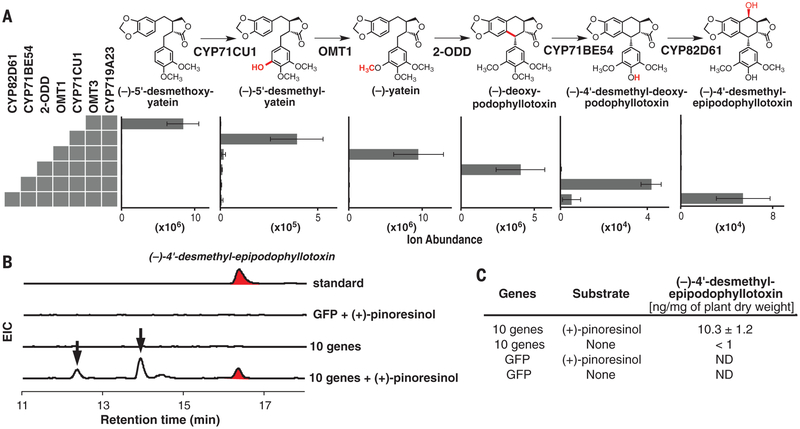
We individually coexpressed six of these seven candidate enzymes (the putative hydroxylases) with CYP719A23 and OMT3 in tobacco leaves to test for a (−)-5′-desmethoxy-yatein hydroxylase. We infiltrated leaves with (−)-matairesinol 4 days after infiltration and harvested a day later for LC-MS analysis. In samples coexpressing Phex524 (CYP71CU1), we observed turnover of (−)-5′-desmethoxy-yatein (Fig. 3). A comparison of the leaf metabolomes revealed two CYP71CU1-dependent compound mass signals that correspond to the calculated m/z of (−)-5′-desmethyl-yatein [assignment supported by tandem mass spectrometry (MS/MS)] (fig. S13). The earlier eluting mass signal is likely an in-source fragmentation ion originating from a glycosylated derivative of (−)-5′-desmethyl-yatein produced by endogenous tobacco enzymes. Thus, CYP71CU1 likely catalyzes the next pathway step as part of E-ring functionalization.
Fig. 3. Six genes identified for the biosynthesis of the etoposide aglycone.
(A) Average LC-MS ion abundance ± one SD (three biological replicates) are shown for (−)-podophyllotoxin intermediates and derivatives produced in tobacco after expression of indicated enzymes and (−)-matairesinol infiltration. (B) Extracted ion chromatograms (EIC) for the etoposide aglycone, (−)-4′-desmethylepipodophyllotoxin (m/z = 401) in tobacco leaves expressing GFP or DIR, PLR, SDH, CYP719A23, and the six genes identified in this report with and without the infiltration of (+)-pinoresinol. Arrows indicate glycosylated derivatives of (−)-4′-desmethylepipodophyllotoxin. (C) Average amounts of (−)-4′-desmethylepipodophyllotoxin detected in tobacco ± one SD (three biological replicates).
To complete the biosynthesis of (−)-yatein, a proposed intermediate in the podophyllotoxin pathway (21), we tested Phex13114 (OMT1) for the ability to methylate (−)-5′-desmethyl-yatein. We infiltrated (−)-matairesinol into tobacco leaves expressing OMT1 in combination with CYP719A23, OMT3, and CYP71CU1. (−)-5′-Desmethyl-yatein could not be detected in leaf extracts in which OMT1 had been coexpressed (Fig. 3 and fig. S14); instead, we detected the accumulation of (−)-yatein. Thus, OMT1 likely converts (−)-5′-desmethylyatein to (−)-yatein as the seventh step in the pathway.
The remainder of the pathway involves closing the central six-membered ring in the aryltetralin scaffold and oxidative tailoring. In our initial screen (coexpression with CYP719A23 and OMT3), we observed substantial consumption of (−)-5′-desmethoxy-yatein in samples coexpressing Phex30848 (2-ODD). Computational comparison of leaf metabolomes revealed a new 2-ODD–dependent compound mass signal that corresponds to 5′-desmethoxy-deoxypodophyllotoxin bearing the required aryltetralin scaffold (assignment supported by MS/MS analysis) (fig. S14). We hypothesize that the reaction mechanism involves activation of the 7′ carbon by hydroxylation, followed by dehydration and carbon-carbon bond formation via a quinone methide intermediate (fig. S16).
Prior feeding studies (21) and our P. hexandrum wounding metabolomics data suggest that (−)-yatein is the native substrate for ring closure. Therefore, we tested whether 2-ODD could also catalyze the conversion of (−)-yatein to (−)-deoxypodophyllotoxin in planta. We expressed 2-ODD in tobacco leaves along with CYP719A23, OMT3, CYP71CU1, and OMT1. Four days after Agrobacterium infiltration, we infiltrated leaves with (−)-matairesinol and, a day later, harvested them for LC-MS analysis. We observed that (−)-yatein was consumed in a 2-ODD–dependent fashion, and a computational comparison of metabolite extracts confirmed the accumulation of (−)-deoxypodophyllotoxin in tobacco leaves coexpressing 2-ODD (Fig. 3 and fig. S17). Thus 2-ODD catalyzes oxidative ring closure to establish the core of the aryltetralin scaffold.
We sought to confirm the activities of these enzymes by biochemical analysis. We isolated microsomes enriched with Phex524 (CYP71CU1) after expression in Saccharomyces cerevisiae WAT11, and purified Phex13114 (OMT1) and Phex30848 (2-ODD) with C-terminal hexahistidine tags after expression in E. coli. As expected, incubation of (−)-5′-desmethoxy-yatein with CYP71CU1 and the reduced form of nicotinamide adenine dinucleotide phosphate (NADPH) gave the hydroxylated product, (−)-5′-desmethyl-yatein; incubation with CYP71CU1 and OMT1, and with the cofactors, NADPH and S-adenosylmethionine, gave (−)-yatein (fig. S18). Incubation of 2-ODD with (−)-yatein as the substrate in the presence of 2-oxoglutarate and Fe2+ yielded (−)-deoxypodophyllotoxin. All enzymes showed little to no activity on similar substrates under identical assay conditions. These data confirm the enzyme activities and order of reactions for the pathway through (−)-deoxypodophyllotoxin (Fig. 3 and fig. S19).
To identity the enzyme involved in what we hypothesized to be the final step of (−)-podophyllotoxin biosynthesis, the hydroxylation of (−)-deoxypodophyllotoxin, we returned to the publicly available transcriptome data to identify CYPs predominantly and highly expressed in P. hexandrum rhizomes, the tissue in which (−)-podophyllotoxin is primarily produced. We identified six CYP candidates that matched our criterion (fig. S20). We screened the candidates in tobacco by individual coexpression with the five (−)-deoxypodophyllotoxin biosynthetic genes, starting from CYP719A23 and infiltration of (−)-matairesinol. By a comparative metabolomic analysis, we observed consumption of (−)-deoxypodophyllotoxin in leaves coexpressing the candidate enzyme, Ph14372 (CYP71BE54), but—contrary to our expectation—no (−)-podophyllotoxin was detected (Fig. 3). Instead, we observed CYP71BE54-dependent accumulation of two compound mass signals with predicted molecular formulas and MS/MS data that correlate to compounds derived from the demethylation of (−)-deoxypodophyllotoxin, formally (−)-4′-desmethyldeoxypodophyllotoxin (fig. S21). The earlier eluting mass signal is likely derived from a glycosylated derivative. The observed activity of CYP71BE54 implies that the demethylated lignans found in P. hexandrum (22–24) are a result of enzymatic demethylation rather than the failure of OMT3 to methylate a portion of the lignan flux. Consistent with this view, CYP71CU1-enriched microsomes cannot accept (−)-pluviatolide as a substrate, which indicates a need for fully methylated substrate earlier in the pathway. Despite poor expression in yeast, isolated CYP71BE54 microsomes accepted (−)-deoxypodophyllotoxin as a substrate but not other similar molecules (fig. S22).
Upon screening an additional candidate P450, Ph35407 (CYP82D61), we also observed consumption of (−)-deoxypodophyllotoxin. However, we did not detect formation of (−)-podophyllotoxin; instead, we observed accumulation of its epimer, (−)-epipodophyllotoxin (fig. S23). To confirm the activity of CYP82D61 in the context of the late pathway enzymes, we infiltrated (−)-matairesinol into tobacco leaves expressing CYP71BE54, CYP82D61, and the five (−)-deoxypodophyllotoxin biosynthetic genes starting from CYP719A23. Comparative metabolomics demonstrated the accumulation of (−)-4′-desmethylepipodophyllotoxin, along with two other earlier eluting compound mass signals that are likely derived from glycosylated (−)-4′-desmethylepipodophyllotoxin derivatives (Fig. 3 and fig. S24). (−)-4′-Desmethylepipodophyllotoxin is the direct precursor to etoposide, which currently is made by chemical modification of podophyllotoxin. Potent topoisomerase activity of etoposide was discovered by serendipitous derivatization of trace amounts of (−)-4′-desmethylepipodophyllotoxin glucoside, present in P. hexandrum rhizome extracts (8).
Having discovered six enzymes that complete the pathway to (−)-4′-desmethylepipodophyllotoxin, we then sought to reconstitute the pathway in N. benthamiana from (+)-pinoresinol. We expressed DIR, PLR, SDH, CYP719A23, and the six enzymes that we identified in tobacco leaves and subsequently infiltrated 100 μM (+)-pinoresinol, yielding 10.3 ng of (−)-4′-desmethylepipodophyllotoxin per mg of plant dry weight. The total amount produced is likely even higher, as some of the product is derivatized by tobacco enzymes and could not be quantified. Less than 1 ng of product per mg of plant dry weight was obtained without infiltration of (+)-pinoresinol, which suggested that native production of this intermediate in tobacco is limiting (Fig. 3, B and C). We also produced (−)-deoxypodophyllotoxin and (−)-epipodophyllotoxin starting from (+)-pinoresinol in N. benthamiana by omitting CYP71BE54 and CYP82D61, and CYP71BE54, respectively (figs. S25 and S26); pathway intermediates do not accumulate in either case (fig. S27). The yield of (−)-deoxypodophyllotoxin in tobacco (~90 ng/mg dry weight) is more than one-third of the yield from wound-induced leaves of Podophyllum.
Thus, the etoposide aglycone, (−)-4′-desmethylepipodophyllotoxin, can be produced in N. benthamiana, which circumvents the current need for mayapple cultivation and subsequent semisynthetic epimerization and demethylation (fig. S28). By coupling transcriptome mining with combinatorial expression of candidate enzymes in tobacco, we identified six biosynthetic enzymes, including a 2-ODD that catalyzes the novel C-C bond-forming step for stereoselective cyclization to close the aryltetralin scaffold and a late-stage P450 to unmask the E-ring phenol. A similar approach could be used to engineer synthetic pathways that produce podophyllotoxin derivatives with improved bioactive properties.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the NIH grants R00 GM089985and DP2 AT008321 (E.S.S.). We thank J. Rajniak, A. P. Klein, and M. Voges (Stanford) for critical reading of the manuscript, and members of the Sattely lab for helpful discussions. Wethank J.-G. Kim (Stanford) for assistance with N. benthamiana transient expression, quantitative reverse transcription polymerase chain reactions (qRT-PCR) and RNA-Seq library preparation, G. Lomonossoff (John Innes Centre) for plasmid pEAQ, and D. Nelson for CYP nomenclature assignment. We thank the Stanford Center for Genomics and Personalized Medicine for RNA sequencing services and the Stanford Genetics Bioinformatics Service Center for computational resources. Gene sequences have been deposited into GenBank (accession numbers: KT390155 to KT390182). RNA Sequencing data have been deposited into the National Center for Biotechnology Information, NIH, Sequence Read Archive (accession number: SRP061783). E.S.S., W.L., and Stanford University have filed a provisional patent application. Supplementary materials contain additional data.
Footnotes
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.De Luca V, Salim V, Atsumi SM, Yu F, Science 336, 1658–1661 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Paddon CJ et al. , Nature 496, 528–532 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Thodey K, Galanie S, Smolke CD, Nat. Chem. Biol 10, 837–844 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeLoache WC et al. , Nat. Chem. Biol 11, 465–471 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Brown S, Clastre M, Courdavault V, O’Connor SE, Proc. Natl. Acad. Sci. U.S.A 112, 3205–3210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu Y et al. , Proc. Natl. Acad. Sci. U.S.A 112, 6224–6229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cimermancic P et al. , Cell 158, 412–421 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stähelin HF, von Wartburg A, Cancer Res 51, 5–15 (1991). [PubMed] [Google Scholar]
- 9.Canel C, Moraes RM, Dayan FE, Ferreira D, Phytochemistry 54, 115–120 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Gordaliza M, García PA, Miguel del Corral JM, Castro MA, Gómez-Zurita MA, Toxicon 44, 441–459 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Lata H, Mizuno CS, Moraes RM, in Protocols for in Vitro Cultures and Secondary Metabolite Analysis of Aromatic and Medicinal Plants (Springer, 2009), pp. 387–402. [Google Scholar]
- 12.Kamal A, Ali Hussaini SM, Rahim A, Riyaz S, Podophyllotoxin derivatives: A patent review (2012–2014). Expert Opin. Ther. Patents 10.1517/13543776.2015.1051727 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Dinkova-Kostova AT et al. , J. Biol. Chem 271, 29473–29482 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Davin LB et al. , Science 275, 362–367 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Xia Z-Q, Costa MA, Pélissier HC, Davin LB, Lewis NG, J. Biol. Chem 276, 12614–12623 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Marques JV et al. , J. Biol. Chem 288, 466–479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nag A, Chanda S, Subramani R, J. Cell Plant Sci 2, 19–23 (2011). [Google Scholar]
- 18.Rajesh M et al. , Plant Cell Tissue Organ Cult 114, 71–82 (2013). [Google Scholar]
- 19.Wankhede DP, Biswas DK, Rajkumar S, Sinha AK, Protoplasma 250, 1239–1249 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Kamil WM, Dewick PM, Phytochemistry 25, 2089–2092 (1986). [Google Scholar]
- 21.Kamil WM, Dewick PM, Phytochemistry 25, 2093–2102 (1986). [Google Scholar]
- 22.Broomhead AJ, Rahman MMA, Dewick PM, Jackson DE, Lucas JA, Phytochemistry 30, 1489–1492 (1991). [Google Scholar]
- 23.Jackson DE, Dewick PM, Phytochemistry 23, 1147–1152 (1984). [Google Scholar]
- 24.Jackson DE, Dewick PM, Phytochemistry 23, 1037–1042 (1984). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.