Abstract
Circulating tumor DNA (ctDNA) is a promising prognostic biomarker in various cancers. Due to the high recurrence rate of resectable pancreatic ductal adenocarcinoma (PDAC), effective strategies for prognostic stratification are necessary. Yet, for resectable PDAC, prognostic impact of ctDNA lacks systemic evidence. We sought to investigate the prognostic significance of baseline ctDNA and postoperative ctDNA in patients with resectable PDAC. PubMed, EMBASE, and the Cochrane library were searched up to March 2019. Five studies met the inclusion criteria, and 375 patients were pooled for the meta-analysis. Positive ctDNA significantly indicated poor overall survival (at baseline, hazard ratio [HR] 2.27, 95% confidence interval [CI] 1.13–4.56; postoperative, HR 3.66, 95% CI 1.45–9.28). Patients with detectable ctDNA showed the trend to have higher risk for disease recurrence than those without detectable ctDNA (at baseline, HR 1.96, 95% CI 0.65–5.87; postoperative, HR 2.20, 95% CI 0.99–4.87). The results were consistent regardless of pre- or post-operative ctDNA. There was no significant heterogeneity among the included studies. In conclusion, our meta-analysis revealed that ctDNA, either at baseline or postoperative, might be a useful prognostic biomarker for stratifying risk of death and recurrence in resectable PDAC.
Subject terms: Pancreatic cancer, Prognostic markers, Cancer genomics, Cancer genetics, Pancreatic cancer
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most lethal cancers, where the five-year survival rate for all stages of PDAC as low as 6–8%1. Surgical resection remains the only chance for cure, increasing the five-year survival rate to 15–25%2. However, tumors recur in 85% of resected cases; therefore, identifying patients with a high risk of recurrence is a major challenge3. Hence, it is important to find out effective strategies for evaluating the risk of recurrence and mortality in resectable PDAC.
Circulating tumor DNA (ctDNA) is a promising blood-based biomarker in cancer management4,5. The majority of previous reports have summarized the benefit of ctDNA as a non-invasive marker for treatment selection, real-time disease monitoring, detection of residual disease, and estimation of prognosis6–8. Recently, there has been increasing attention towards the emerging role of ctDNA in early-stage cancers; pre- and post-operative ctDNA in various cancers has been introduced as a useful prognostic biomarker to indicate recurrence after resection9. Regarding PDAC, a few studies have focused on the prognostic value of ctDNA in patients undergoing curative resection7,10. Yet, the results have been controversial, and there is lack of evidence that systematically demonstrates the prognostic value of ctDNA in resectable PDAC.
Therefore, we performed a meta-analysis to evaluate the prognostic significance of ctDNA in patients with resectable PDAC in terms of overall survival (OS) and disease-free survival (DFS).
Results
Search results
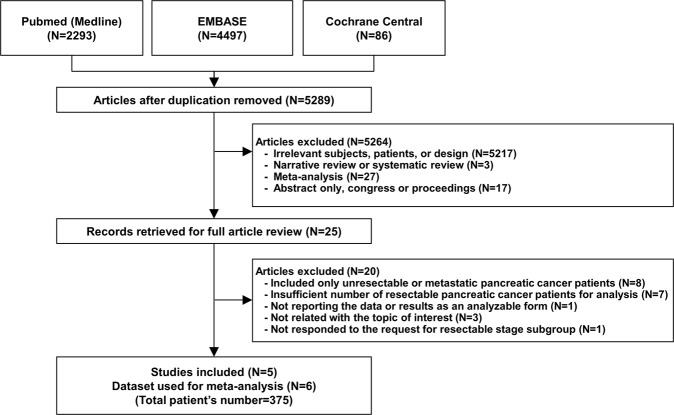
A total of 5,289 citations were identified after removing duplicates. Among these, 25 articles were retrieved for a full review, and five were selected for the analysis (Fig. 1). The characteristics of the 20 studies excluded after the full review are summarized in the Supplementary Material. The final five studies with six results encompassed 375 patients with resectable PDAC grouped based on the time point of measuring ctDNA (patients measured with baseline sample, n = 299; patients measured with postoperative sample, n = 76). Among the six results, four results provided HRs and 95% CIs in patients with detectable ctDNA at baseline, and the other two results provided HRs and 95% CIs in patients with detectable ctDNA after surgery.
Figure 1.
Flow diagram of study selection. The flow diagram is presented according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.
Study characteristics and risk of bias within studies
The main characteristics of the individual studies are summarized in Table 1 and in Supplementary Table 1. All studies included a significant proportion of resectable patients, and per our request, all authors provided survival data of patients in the resectable stage. The positive ctDNA detection rate varied among studies from 8.33% to 68.57%. Three studies confirmed KRAS mutation status with matched tissue where the prevalence of KRAS mutations ranged from 81.9% to 94.3%. Except one study that performed Peptide Nucleic Acid-directed PCR clamping analysis using serum samples, all studies performed either dPCR or NGS using plasma samples. Mean or median age ranged from 66 to 70 years, and the proportion of males from 52.4 to 64.4%. Four studies analyzed Asian populations. All studies were retrospective investigations, while four provided adjusted HRs and 95% CIs using multivariable regression.
Table 1.
Characteristics of studies selected for analysis.
| Source (Year) | Study period | Patient No. | Time point measuring ctDNA | ctDNA (+) rate | Follow-up duration | Median survival | Sample type | Detection method | Platform | Target mutation | Age (Y) | Male (%) | Ethnicity | Tissue mutation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Takai et al.23 | 2011–2014 | 108 | Baseline | 8.33% | Up to 40mo | NR | Plasma | Digital PCR | RainDrop |
KRAS - G12D - G12R - G12V - G13D |
66 | 61.8 | Asian | NA |
| Hadano et al.7 | 2007–2013 | 105 | Baseline | 31.43% |
14–96mo (mean 54mo) |
13.6mo/27.6mo | Plasma | ddPCR | Bio-Rad QX100 |
KRAS - G12D - G12V - G12R |
69 | 52.4 | Asian | 81.9% (86/105) |
| Pietrasz et al.8 | 2011–2015 | 31 | Postoperative | 19.35% | Median 33.3mo | 19.3mo/32.2mo | Plasma | Amplicon-based NGS* | Ion Proton | 22 genes** | 68 | 54.8 | Non-Asian | NA |
| Kim et al.24 | 2015–2017 | 41 | Baseline | 68.57% | Median 10.03mo | NR | Plasma | ddPCR | Bio-Rad QX200 |
KRAS - G12A - G12C - G12D - G12R - G12S - G12V - G13D |
66 | 63.2 | Asian | 94.3% (33/35)** |
| Nakano et al.10 | 2013–2016 | 45 | Both baseline and postoperative | 24.44% (baseline)/44.44% (post) | Up to 42mo | NR | Serum | PNA-directed PCR clamping |
KRAS codon 12/13 |
70 | 64.4 | Asian | 83.3% (35/42) |
Abbreviations: ctDNA, circulating-tumor DNA; mo, month; NR, not reached; ddPCR, droplet digital PCR; NGS, next-generation sequencing; PNA, peptide nucleic acid.
*Sequencing library was prepared using Ion AmpliSeq Colon and Lung Cancer Research Panel v2 (Thermo Fisher). The libraries were processed on Ion Chef system and sequenced on the Ion Proton system.
**KRAS, EGFR, BRAF, PIK3CA, AKT1, ERBB2, PTEN, NRAS, STK11, MAP2K1, ALK, DDR2, CTNNB1, MET, TP53, SMAD4, FBX7, FGFR3, NOTCH1, ERBB4, FGFR1 and FGFR2.
The quality of studies included was assessed via NOS for non-randomized studies and the results are presented in Supplementary Table 2. All five studies received at least seven stars, and thus fulfilled the adequacy criteria for non-randomized studies.
Impact of ctDNA detection on the prognosis of resectable PDAC
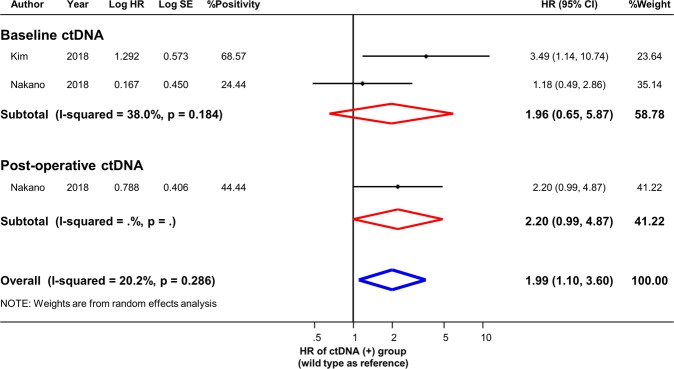
Six study results (four using preoperative sample, and two using post-operative sample) indicated the association of ctDNA detection with OS, three (two using preoperative sample, and one using post-operative sample) of which evaluated the association of ctDNA with DFS. Pooled results from the random-effects models of OS and DFS are presented in Figs 2 and 3, respectively. The risk was significantly higher in the ctDNA-positive group than in the ctDNA-negative group in terms of mortality, regardless of the time point (at baseline or postoperative) of sample collection (detectable baseline ctDNA, pooled HR 2.27, 95% CI 1.13–4.56; detectable postoperative ctDNA, pooled HR 3.66, 95% CI 1.45–9.28). The risk for disease recurrence showed the trend of increase in the ctDNA-positive group than in the ctDNA-negative group for both preoperative and postoperative measurement (detectable baseline ctDNA, pooled HR 1.96, 95% CI 0.65–5.87; detectable postoperative ctDNA, pooled HR 2.20, 95% CI 0.99–4.87). The I-squared statistical heterogeneity was not significant for either outcome (OS, baseline ctDNA I2 = 38.0%, p = 0.184, postoperative ctDNA I2 = 0.0%, p = 0.722; DFS, baseline ctDNA I2 = 38.0%, p = 0.184). A funnel plot and results of Egger’s and Begg’s tests demonstrated that there was no significant publication bias in qualitative or quantitative terms (Supplementary Fig. 1).
Figure 2.
Prognostic effect of baseline or post-operative ctDNA in resectable PDAC. Hazard ratios with 95% confidence intervals are displayed by individual studies, describing pooled overall effects for baseline ctDNA and for post-operative ctDNA, respectively. Abbreviations: HR, hazard ratio; SE, standard error; CI, confidence interval.
Figure 3.
Effect of ctDNA on disease-free survival in resectable PDAC. Hazard ratios with 95% confidence intervals are displayed by individual studies, describing pooled overall effects for baseline ctDNA and for post-operative ctDNA, respectively. Abbreviations: HR, hazard ratio; SE, standard error; CI, confidence interval.
Subgroup analysis and meta-regression analysis
The results of subgroup analysis stratified based on several variables are shown in Fig. 4. The pooled results were consistent in both random- and fixed-effects model. Stratification based on ethnicity (Asian or non-Asian population) showed that the prognostic impact of ctDNA on OS was significant both in Asian and non-Asian patients. The source of heterogeneity was identified in the subgroups of specimen type and statistical method. Excluding one study with two results that used serum samples and univariable regression analysis, the pooled results were significant with heterogeneity of I2 = 0.0%. In meta-regression analysis, no significant effects were observed based on demographic characteristics including the number of patients, ctDNA positivity rate, age, or proportion of males (Table 2).
Figure 4.
Subgroup analysis. Effect of ctDNA on overall survival according to the various subgroups is presented. Abbreviations: HR, hazard ratio; CI, confidence interval.
Table 2.
Results of Meta-Regression Analysis.
| Meta-regression OR | 95% CI | P value | |
|---|---|---|---|
| By patient number | 1.006 | 0.974–1.039 | 0.594 |
| By positivity rate | 0.996 | 0.935–1.062 | 0.866 |
| By mean/median age | 0.822 | 0.406–1.664 | 0.442 |
| By proportion of men | 0.961 | 0.812–1.137 | 0.505 |
Abbreviations: OR, odds ratio; CI, confidence interval.
Discussion
In the present meta-analysis, the prognostic significance of ctDNA in baseline and postoperative samples from the patients with resectable PDAC was evaluated, respectively. The results can be summarized as following. (1) Detectable ctDNA in resectable PDAC had a significant negative effect on prognosis in terms of both OS and DFS; (2) These results were consistently observed using either pre- or post-operative ctDNA; (3) No significant heterogeneity was observed among the included studies, and heterogeneity decreased to 0.0% after excluding one study that used serum samples and univariable analysis; (4) There were no differences between the results according to ethnicity, age, sex, rate of positive ctDNA detection, or sample size in subgroup analysis and meta-regression. To the best of our knowledge, this is the first meta-analysis to identify the prognostic role of ctDNA in patients with resectable PDAC.
PDAC is well-known as a digestive system cancer with poor prognosis, and even resectable stage PDAC shows a five-year survival rate of only 15–25%2. Patients with early stage PDAC are usually recommended to undergo extensive surgery as well as adjunctive chemotherapy before and/or after surgery. Nevertheless, up to 85% of resectable PDAC results in recurrence3 and therefore, the use of biomarkers to predict the prognosis and recurrence of PDAC is essential. Recently, ctDNA has been suggested as a promising prognostic biomarker in various cancers including PDAC. Significant associations between ctDNA and OS/DFS were consistently shown in various studies and meta-analyses11–13. In a recent review, Lee et al. have documented clinical application of ctDNA in PDAC in various aspects including detection of ctDNA in patients with premalignant lesion (i.e., intraductal papillary mucinous neoplasm), and use of ctDNA as prognostic and predictive markers without distinguishing between resectable and advanced stages14. However, most studies to date have focused on patients with advanced or metastatic stage PDAC15–20, or heterogeneous populations including a small proportion of early stage PDAC21–25. There are only a few studies demonstrating the clinical significance of ctDNA in resectable PDAC, and it is necessary to establish comprehensive evidence in this specific population. Therefore, in this study, we exclusively analyzed patients undergoing curative resection, which provided novel insights into the prognostic significance of ctDNA in early stage PDAC. To incorporate relevant study results to the extent possible, we also collected subgroup data limited to resectable PDAC not reported in the original articles by individually contacting the authors of each study. Thus, incorporating unique data that have not been reported elsewhere, the present study succeeded to show, for the first time, the pooled prognostic effect of detectable ctDNA in resectable PDAC.
The present study highlights the fact that either baseline or postoperative ctDNA can be used as a prognostic indicator in resectable PDAC in terms of OS. We found similar trend for the disease recurrence, although not reaching statistical significance due to the limited number of studies that reported DFS in patients with resectable PDAC. Further studies focusing on the effect of ctDNA on the disease recurrence of early stage PDAC are required to solidify our findings. There is increasing attention towards the emerging role of ctDNA in risk stratification and residual tumor monitoring after curative resection in early stage PDAC patients26. It is important to note that if we could identify patients who are at high risk of recurrence at baseline, we can then focus on active surveillance allowing early treatment to prevent or delay recurrence. Furthermore, monitoring ctDNA serially, at multiple time-points after surgery, could be valuable in prediction of clinical outcome and adjusting further treatment strategies. In summary, our results demonstrate that both baseline and post-operative ctDNA can be an important prognostic biomarker if used in parallel or in a complementary. Nakano et al. reported contrasting results regarding the association of ctDNA in preoperative serum with OS10. We speculate that the different choice of matrix might have contributed to this outcome, because the above study is the only one to use serum. Therefore, we performed a subgroup analysis stratifying the studies based on the specimen type. A considerable difference was found between plasma ctDNA and serum ctDNA; plasma ctDNA was predictive of poor OS without any heterogeneity, but not serum ctDNA. This finding suggests that plasma is an optimal ctDNA source. Notably, during clotting process, leukocyte lysis more frequently occurs in serum than in plasma, possibly leading to dilution of ctDNA with genomic DNA from leukocytes26,27. Thus, the mutant allelic fraction of cfDNA in serum may have been underestimated in terms of sensitivity.
Several technologies (e.g., real-time or digital PCR, NGS, and BEAMing [beads, emulsion, amplification, and magnetics]) have been developed for the detection of ctDNA, and each of these technologies has different assay performance characteristics26. We attempted to determine if the different assay platforms lead to different prognostic values of ctDNA in resectable PDAC. Our subgroup analysis included three studies that used dPCR (droplet digital PCR, BioRad, n = 2; microfluidic digital PCR, RainDrop®, n = 1) and comparison of the pooled result using NGS (Ion ProtonTM). Although dPCR methods more frequently detected ctDNA (pooled positivity, 27.6%) than the NGS method (pooled positivity, 19.4%), significant prognostic value was achieved with both methods. While dPCR focuses on the detection of rare mutations at specific loci and attains high sensitivity ranging from 0.001% to 0.1%, NGS-based approaches have the potential to detect a broad range of molecular targets (e.g., TP53) in PDAC other than KRAS28. Moreover, advanced NGS strategies that minimize artifacts by using molecular barcodes or digital error suppression enable highly sensitive detection of ctDNA with allele frequencies theoretically ranging from 0.00025% to 0.1%29,30. We speculate that these technologies may provide benefits in early stages of cancer (e.g., MRD monitoring and early diagnosis of relapse)31. Further studies may be needed to verify the prognostic significance of ctDNA in resectable PDAC, using advanced NGS strategies.
The present meta-analysis has several limitations. First, the selected studies varied in their clinical and methodological characteristics. Second, the number of included studies and pooled patients were relatively small. However, despite using a limited number of studies on ctDNA in resectable PDAC, our results were consistent without any significant heterogeneity. Third, although four out of five studies provided multivariable-adjusted results, the effect of unmeasured confounders could exist due to non-randomized baseline characteristics.
In conclusion, our meta-analysis demonstrated the significant prognostic impact of detectable ctDNA (either at baseline or postoperative) in resectable PDAC. Our findings suggest that ctDNA might be a useful predictive biomarker of both mortality and recurrence in resectable PDAC.
Methods
Data sources and searches
We conducted systematic searches of the databases PubMed, EMBASE, and the Cochrane Central Register of Controlled Trials for published or unpublished studies. A manual search of references cited in articles, recent reviews, editorials, and meta-analyses was performed. We did not apply any restrictions on the language, study period, or sample size during the search process.
Study selection and outcome definition
The eligibility criteria for study selection were 1) execution until March 2019, 2) inclusion of patients with resectable PDAC, 3) ctDNA assessment using plasma or serum, and 4) hazard ratios (HR) and 95% confidence intervals (CI) of survival outcomes in resectable groups being directly accessible, statistically estimable, or available from the authors on request. We excluded studies that included only patients with unresectable or metastatic cancer, had a small number of patients (n < 30) insufficient for statistical analysis, or did not report data or results in an analyzable form. Two investigators independently screened titles and abstracts, identified duplicates, reviewed full articles, and determined their eligibility. Disagreements were resolved via discussion. The last search was performed in March 2019. The primary outcome was OS defined as the interval from the defined initiation in each study to death, and the secondary outcome was DFS calculated from the initiation to recurrence.
Data extraction and quality assessment
We extracted the following characteristics from each eligible study; study design, number of patients who underwent curative resection, ctDNA positivity rate, duration of follow-up, specimen type, method/platform of ctDNA detection, target genes/variants, patient demographics, and HR with 95% CI in the ctDNA-positive group (with the ctDNA-negative group as reference). Of note, one study assessed ctDNA at two different time points (at baseline and after surgery) and we incorporated both results in the analysis (five studies with six results). For the studies that reported survival outcomes irrespective of clinical stage, we contacted each corresponding author individually and requested the adjusted HRs and 95% CIs in the subgroup limited to the resectable stage. The quality of eligible studies was assessed using the Newcastle–Ottawa Scale (NOS) checklist for non-randomized studies.
Data synthesis and analysis
Random-effect models were applied for all analyses, and pooled HRs with 95% CIs were represented as statistical summaries. The pooled HRs and 95% CIs of the random- and fixed-effects models were calculated using the restricted maximum likelihood (REML) and Mantel–Haenszel methods.
We quantified the statistical heterogeneity using I2 statistics. Publication bias was assessed through funnel plot asymmetry using Egger’s and Begg’s tests. Meta-regression analysis was performed to assess the relationship between effect size (log-ORs) and patient number, ctDNA positivity rate, mean or median age, and proportion of males.
To gain insights into prognostic significance of sampling time points, we assessed the HRs of detectable baseline ctDNA and of detectable postoperative ctDNA, respectively. Subgroup analyses were performed as follows to explore the source of heterogeneity; (1) meta-analysis model (random vs. fixed effects); (2) ethnicity of the study population (Asian vs. Non-Asian); (3) specimen type used in the study (plasma vs. serum); (4) detection method (digital PCR [dPCR] vs. next-generation sequencing [NGS]); and (5) statistical method used in the study (multivariable vs. univariable regression analysis). Two-sided p-values < 0.05 were considered to be statistically significant. Statistical computations used the standard software STATA/SE v12.0 (StataCorp LP, College Station, Texas, USA). The present study followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Supplementary Table 3) and the Meta-analysis of Observational Studies in Epidemiology (MOOSE) guidelines.
Supplementary information
Acknowledgements
This work was supported by Hallym University Research Fund (HURF-2017-07).
Author contributions
Design and direction of the study: Y.K. Lee and H.J. Kang. Acquisition of data: J.S. Lee and T.M. Rhee. Analysis and interpretation of data: K. Jeon and J. Lee. Writing the manuscript: J.S. Lee and T.M. Rhee. Administrative, technical, or material supports: D. Pietrasz, J.B. Bachet, P. Laurent-Puig, E. Takai, S. Yachida, T. Shibata, S.Y. Kong, J.W. Lee, H.C. Park, D.Y. Zang, M. Kim and H.S. Kim. Study supervision: Y.K. Lee.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-53271-6.
References
- 1.Miller KD, et al. Cancer Statistics for Hispanics/Latinos, 2018. CA Cancer J Clin. 2018;68:425–445. doi: 10.3322/caac.21494. [DOI] [PubMed] [Google Scholar]
- 2.Schlitter AM, et al. Molecular, morphological and survival analysis of 177 resected pancreatic ductal adenocarcinomas (PDACs): Identification of prognostic subtypes. Sci Rep. 2017;7:41064. doi: 10.1038/srep41064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JC, et al. Management of recurrent pancreatic cancer after surgical resection: a protocol for systematic review, evidence mapping and meta-analysis. BMJ Open. 2018;8:e017249. doi: 10.1136/bmjopen-2017-017249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem. 2015;61:112–123. doi: 10.1373/clinchem.2014.222679. [DOI] [PubMed] [Google Scholar]
- 5.Ignatiadis M, Lee M, Jeffrey SS. Circulating Tumor Cells and Circulating Tumor DNA: Challenges and Opportunities on the Path to Clinical Utility. Clin Cancer Res. 2015;21:4786–4800. doi: 10.1158/1078-0432.CCR-14-1190. [DOI] [PubMed] [Google Scholar]
- 6.Cohen JD, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359:926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hadano N, et al. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br J Cancer. 2016;115:59–65. doi: 10.1038/bjc.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pietrasz D, et al. Plasma Circulating Tumor DNA in Pancreatic Cancer Patients Is a Prognostic Marker. Clin Cancer Res. 2017;23:116–123. doi: 10.1158/1078-0432.CCR-16-0806. [DOI] [PubMed] [Google Scholar]
- 9.Isbell JM, Jones DR, Li BT. Circulating tumor DNA: A promising biomarker to guide postoperative treatment and surveillance of non-small cell lung cancer. J Thorac Cardiovasc Surg. 2018;155:2628–2631. doi: 10.1016/j.jtcvs.2017.12.157. [DOI] [PubMed] [Google Scholar]
- 10.Nakano Y, et al. KRAS mutations in cell-free DNA from preoperative and postoperative sera as a pancreatic cancer marker: a retrospective study. Br J Cancer. 2018;118:662–669. doi: 10.1038/bjc.2017.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li T, et al. K-Ras mutation detection in liquid biopsy and tumor tissue as prognostic biomarker in patients with pancreatic cancer: a systematic review with meta-analysis. Med Oncol. 2016;33:61. doi: 10.1007/s12032-016-0777-1. [DOI] [PubMed] [Google Scholar]
- 12.Chen L, et al. Prognostic value of circulating cell-free DNA in patients with pancreatic cancer: A systemic review and meta-analysis. Gene. 2018;679:328–334. doi: 10.1016/j.gene.2018.09.029. [DOI] [PubMed] [Google Scholar]
- 13.Zhuang R, et al. The prognostic value of KRAS mutation by cell-free DNA in cancer patients: A systematic review and meta-analysis. PLoS ONE. 2017;12:e0182562. doi: 10.1371/journal.pone.0182562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JS, Park SS, Lee YK, Norton JA, Jeffrey SS. Liquid biopsy in pancreatic ductal adenocarcinoma: current status of circulating tumor cells and circulating tumor DNA. Mol Oncol. 2019;13:1623–1650. doi: 10.1002/1878-0261.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perets R, et al. Mutant KRAS Circulating Tumor DNA Is an Accurate Tool for Pancreatic Cancer Monitoring. Oncologist. 2018;23:566–572. doi: 10.1634/theoncologist.2017-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Laethem JL, et al. Phase I/II Study of Refametinib (BAY 86-9766) in Combination with Gemcitabine in Advanced Pancreatic cancer. Target Oncol. 2017;12:97–109. doi: 10.1007/s11523-016-0469-y. [DOI] [PubMed] [Google Scholar]
- 17.Tjensvoll K, et al. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol Oncol. 2016;10:635–643. doi: 10.1016/j.molonc.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinugasa H, et al. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer. 2015;121:2271–2280. doi: 10.1002/cncr.29364. [DOI] [PubMed] [Google Scholar]
- 19.Semrad T, et al. Pharmacodynamic separation of gemcitabine and erlotinib in locally advanced or metastatic pancreatic cancer: therapeutic and biomarker results. Int J Clin Oncol. 2015;20:518–524. doi: 10.1007/s10147-014-0730-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, et al. K-ras mutational status predicts poor prognosis in unresectable pancreatic cancer. Eur J Surg Oncol. 2010;36:657–662. doi: 10.1016/j.ejso.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 21.Singh N, Gupta S, Pandey RM, Chauhan SS, Saraya A. High levels of cell-free circulating nucleic acids in pancreatic cancer are associated with vascular encasement, metastasis and poor survival. Cancer Invest. 2015;33:78–85. doi: 10.3109/07357907.2014.1001894. [DOI] [PubMed] [Google Scholar]
- 22.Earl J, et al. Circulating tumor cells (Ctc) and kras mutant circulating free Dna (cfdna) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer. 2015;15:797. doi: 10.1186/s12885-015-1779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takai E, et al. Clinical utility of circulating tumor DNA for molecular assessment in pancreatic cancer. Sci Rep. 2015;5:18425. doi: 10.1038/srep18425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim MK, et al. Prognostic Implications of Multiplex Detection of KRAS Mutations in Cell-Free DNA from Patients with Pancreatic Ductal Adenocarcinoma. Clin Chem. 2018;64:726–734. doi: 10.1373/clinchem.2017.283721. [DOI] [PubMed] [Google Scholar]
- 25.Lin M, et al. Circulating Tumor DNA as a Sensitive Marker in Patients Undergoing Irreversible Electroporation for Pancreatic Cancer. Cell Physiol Biochem. 2018;47:1556–1564. doi: 10.1159/000490874. [DOI] [PubMed] [Google Scholar]
- 26.Merker JD, et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol. 2018;36:1631–1641. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 27.Meddeb, R., Pisareva, E. & Thierry, A. R. Guidelines for the Preanalytical Conditions for Analyzing Circulating Cell-Free DNA. Clin Chem (2019). [DOI] [PubMed]
- 28.Gorgannezhad L, Umer M, Islam MN, Nguyen NT, Shiddiky MJA. Circulating tumor DNA and liquid biopsy: opportunities, challenges, and recent advances in detection technologies. Lab Chip. 2018;18:1174–1196. doi: 10.1039/C8LC00100F. [DOI] [PubMed] [Google Scholar]
- 29.Elazezy M, Joosse SA. Techniques of using circulating tumor DNA as a liquid biopsy component in cancer management. Comput Struct Biotechnol J. 2018;16:370–378. doi: 10.1016/j.csbj.2018.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zill OA, et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin Cancer Res. 2018;24:3528–3538. doi: 10.1158/1078-0432.CCR-17-3837. [DOI] [PubMed] [Google Scholar]
- 31.Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14:531–548. doi: 10.1038/nrclinonc.2017.14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.