Abstract
Legionella spp. is an important pulmonary pathogen but rarely causes extra-pulmonary infections. We report a case of joint infection caused by Legionella micdadei and Staphylococcus aureus in a 54-year-old male with medication history of oral steroid for systemic lupus erythematosus (SLE). He developed arthritis in his right metacarpophalangeal (MCP) joints without precursor pneumonia. In the joint aspirate, S. aureus was detected through culture. The existence of L. micdadei and S. aureus were indicated by metagenomic next-generation sequencing (mNGS) and confirmed by 16S rRNA sequence analysis. After oral levofloxacin treatment for 54 days, the patient’s symptoms ameliorated and blood test results improved, which were consistent with the dynamic trend of reads numbers in mNGS data. Our case included, arthritis caused by Legionella spp. have been reported in 11 patients. However, our case is the first to report septic arthritis caused by L. micdadei in native joints and monitored by mNGS. This case demonstrated an application of mNGS for etiological diagnosis and semi-quantification in joint aspirate. mNGS may serve as a promising tool for rapid and accurate etiological diagnosis and surveillance, contributing to appropriate antimicrobial drug applications and timely medication adjustments when necessary.
Keywords: Arthritis, legionella, high-throughput nucleotide sequencing
Introduction
Legionella spp. is widely known as the causative pathogen of community-acquired pneumonia, the common symptoms of which include fever, cough, expectoration, dyspnea and radiological signs. Extra-pulmonary infections due to Legionella spp. are rare. The difficulties in culture remains to be a challenge in clinical diagnosis.
Metagenomics Next-generation sequencing (mNGS) is a new technique which is increasingly used for infectious diagnosis. It allows for culture-independent identification of various pathogens in complex microbial samples and semi-quantification of pathogen loads. Now we report a case of septic arthritis caused by L. micdadei and Staphylococcus aureus unexpectedly diagnosed by mNGS.
Case presentation
A 54-year-old male visit the clinic of the Infectious Diseases of Zhongshan Hospital because of new-onset swelling with progressive pain in the 1st and 3rd metacarpophalangeal (MCP) joints of his right hand for 16 days. He denied any symptom of local trauma, fever, respiratory disorder or febrile illness before the onset. He had medication history of oral corticosteroid for systemic lupus erythematosus (SLE). After the onset, he increased the steroid doses without doctor’s advice. In fact, he developed progressive pain and swelling in the right elbow joint one month earlier, which was ended up with spontaneous rupture of the joint mass and discharge of dark red pus.
He had a visit to the rheumatologic clinic on the 9th day of the onset. The initial analysis detected a leukocytosis count of 13.39×109/L, neutrophil proportion of 93.1%, a slight increase in erythrocyte sedimentation rate (ESR, 52 mm/h), significant increase in C-reactive protein (CRP, 117.4 mg/L), undetected pro-calcitonin (PCT) and a similar level of autoantibodies as before (ANA 1:100, dsDNA 100 IU/mL). CT of hands showed some swelling of the 1st and 3rd MCP joints with subluxation (Figure 1). His SLE was evaluated as stable, with a possibility of infection. He was not admitted to the rheumatologic ward. The rheumatologic clinician ordered a decreased dose of corticosteroid and a normal dose of oral levofloxacin, which slightly relieved the symptoms in the joints.
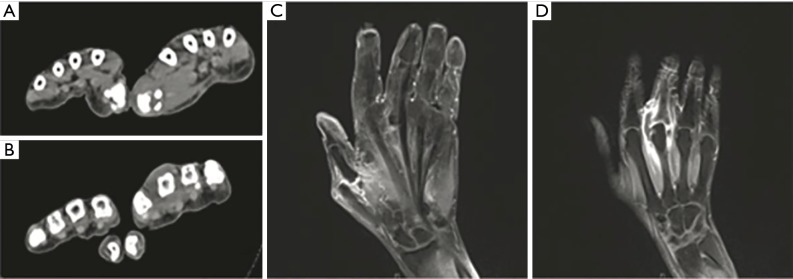
Figure 1.
CT and MRI of the patient’s 1st and 3rd MCP joints on admission. (A,C) Swelling and subluxation of the patient’s 1st MCP joints shown in CT on day 0 and MRI-T2 on day 6, respectively. (B,D) Swelling and subluxation of the patient’s 3rd MCP joints shown in CT on day 0 and MRI-T2 on day 6, respectively. MCP, metacarpophalangeal.
The patient was admitted into the ward of Infectious Diseases on the day of the clinic. He was afebrile and normotensive. His MCP joints were swollen, tender and warmer than surrounding tissues, especially in the 3rd MCP joint. Blood tests indicated increased levels (leukocytosis count 19.79×109/L, neutrophil proportion 90.9%, ESR 78 mm/h, CRP 242.1 mg/L, PCT 0.33 ng/mL). A purulent aspirate (about 3ml in all) obtained by arthrocentesis on admission was sent to microbiological lab for conventional microbial culture and to a tertiary sequencing lab for mNGS test simultaneously (Figure 2).
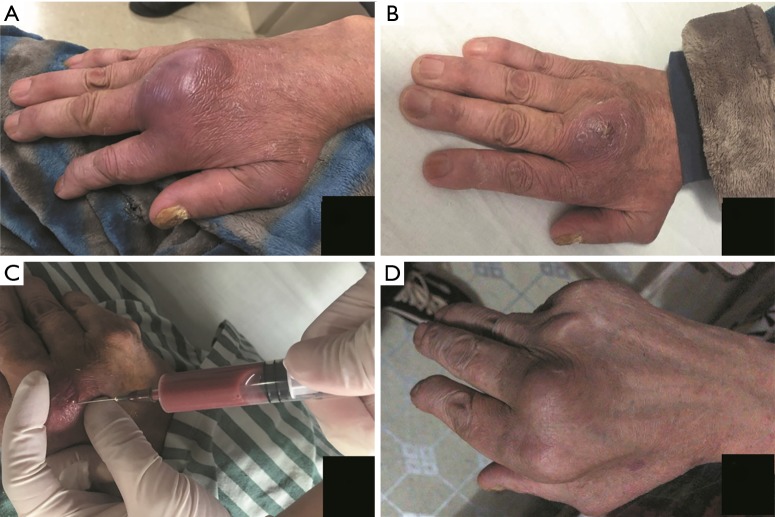
Figure 2.
The patient’s 3rd MCP joint before, during and after treatment. (A) Day 0. The obviously swollen joint on admission. (B) Day 6. The swelling alleviated significantly after first arthrocentesis and treatment of oral levofloxacin. (C) Day 6. About 10 mL dark red aspirate obtained in the 2nd arthrocentesis. (D) Day 56. The joint was almost recovered. MCP, metacarpophalangeal.
Brief methods of mNGS were according to the standard protocol of the BGISEQ-100 sequencing platform (BGI, China). (I) DNA Extraction. (II) Library Construction and Sequencing. The DNA extracted was sonicated to generate 200–300 bp fragments. The fragments were then end-repaired, added specific barcodes, amplified by PCR and purified. Quality control was performed with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). Reads of short (<35 bp) and low-quality were filtered out. Human host sequences (mapped to the human reference genomes hg19) were eliminated. (III) Data processing and analysis. High-quality sequencing data were aligned to the Microbial Genome Database. The mapping sequences were then advanced analyzed. The reference genomes were downloaded from the National Center for Biotechnology Information (ftp://ftp.ncbi.nlm.nih.gov/genomes/) including 3,297 bacteria, 4,152 viruses, 206 fungi, 140 parasites, 104 mycobacteria and 45 mycoplasma/chlamydia, all associated to human diseases (1).
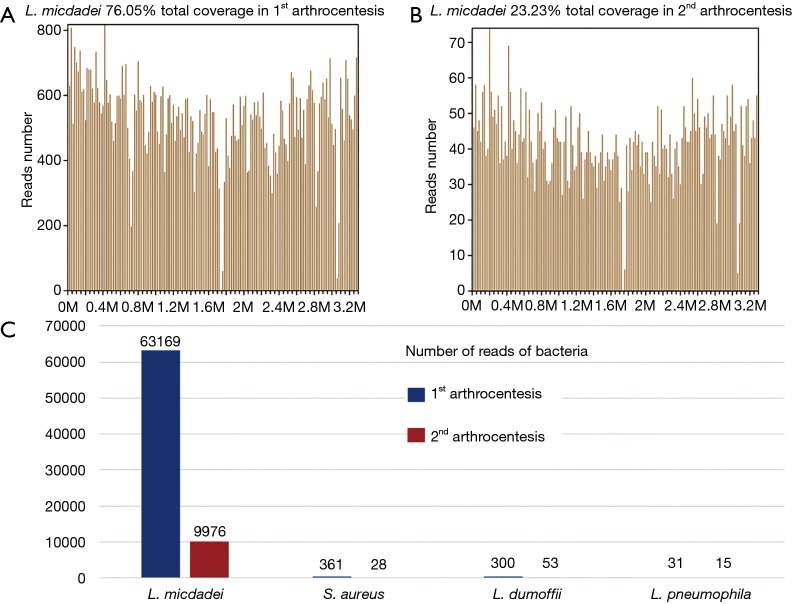
Twenty-four hours later, we received the mNGS report. A large number of standardized strict mapping reads (SDSMR) of L. micdadei and small numbers of SDSMR of S. aureus, L. dumoffii and L. pneumophila were detected. The SDSMR numbers of L. micdadei, S. aureus, L. dumoffii and L. pneumophila were 63,169, 361, 300 and 31; with genomic coverage rate of 76.05%, 2%, 0.4143% and 0.1958%, respectively (Figure 3). In consideration of microorganism genome’s rupture into short fragments and the sequence homology among different Legionella species, the non-micdadei Legionella reads were assumed as micdadei reads that have mapped to non-micdadei reference sequences. In addition, several skin colonizers such as Propionibacterium acnes and coagulase negative staphylococcus were also present in the results (4 and 1, respectively), yet were considered as contamination due to the small numbers of reads and mis-mapping due to the sequence homology among different staphylococcus. The following 16s rRNA sequence analysis confirmed the presence of L. micdadei. Oral levofloxacin treatment was continued after mNGS test.
Figure 3.
mNGS results in 1st arthrocentesis and 2nd arthrocentesis. (A) In 1st arthrocentesis, 63,169 of bacterial reads corresponded to L. micdadei, with a coverage of 76.05%. (B) In 2nd arthrocentesis, 9,976 of bacterial reads corresponded to L. micdadei, with a coverage of 76.05%. (C) The most abundant four bacteria in the two arthrocentesis remained the same. The number of reads in 2nd arthrocentesis decreased significantly, which was consistent with the alleviation of patients’ symptoms and improvement of laboratory test results. mNGS, metagenomics next-generation sequencing.
Sixty hours later, a growth of methicillin-sensitive S. aureus (MSSA) in standard culture medium was reported. The S. aureus was sensitive to levofloxacin. In consideration of the patient’s economic situation, we did not add new antibiotics although levofloxacin is not a first-choice for S. aureus. Oral levofloxacin treatment was continued.
On day 6, the patient’s pain and swelling in the joint were alleviated significantly. Blood tests showed decreases in routine blood test indexes and inflammatory biomarkers (leukocytosis count 14.14×109/L, neutrophil proportion 89%, ESR 62 mm/h, CRP 43.8 mg/L, PCT 0.09 ng/mL). MRI still showed swelling and subluxation of the 1st and 3rd MCP joints (Figure 1). Purulent aspirate (about 10 mL) obtained by a complete arthrocentesis was sent to the two labs again (Figure 2).
Twenty-four hours later, mNGS reported Legionella species and S. aureus again. Compared to the previous report, most of the bacteria had decreased significantly in sequencing reads (Figure 3), which were consistent with the alleviation of symptoms and improvement of laboratory test results. Propionibacterium acnes and coagulase negative staphylococcus were negative this time. 54 h later, the conventional culture reported a single MSSA again. The patient was prescribed with another 60 days’ oral levofloxacin. Eight weeks later, the patient revisited the outpatient clinic and was found almost fully recovered.
Discussion
This report describes a case of septic arthritis caused by L. micdadei and S. aureus in an immunocompromised patient. The incidence of legionellosis has been increasing in the United States and in Europe (2-4), with a nearly 3.5-fold increase between 2000 and 2011 in the United States (3). The typical mode of transmission for Legionella is inhalation of aerosols which leads to pneumonia. Although rare in frequency, extra-pulmonary infections caused by Legionella species includes prosthetic valve endocarditis (5), pyelonephritis (6), sinusitis (7) and cellulitis (8). Legionella species may also cause wound infections due to direct inoculation and skin contamination (9,10). The majority of extra-pulmonary infections caused by Legionella species occur in immunocompromised individuals (11).
To date, ten cases of Legionella arthritis have been reported, including eight septic (12-19) and two reactive (20,21) arthritis (Table 1). However, we present the first case of septic arthritis in native joint caused by L. micdadei, accompanied with S. aureus. Our case included, 8/11 patients were immunocompromised. Of the other three cases, two were diagnosed as reactive arthritis and had pneumonia history about 2 weeks before the onset. L. pneumophila is the leading pathogen (5/11). The others are L. longbeachae (one case) (14), L. Dumoffii (one case) (15), L. micdadei (two cases, one in prosthetic joint (19) and our case of septic arthritis in native joint), L. bozemanae (one case) (16) and L. cincinnatiensis (one case) (18). Both small and large joints could be affected. Except for one 32-year-old patient, who had dived using a compressed air breathing apparatus 2 weeks before a CT detected atypical pneumonia and clinical examination detected arthritis (20), all patients were more than 50 years old. Intra-articular injection and aerosolized water therapy may be the potential risk factors. Besides SLE, we did not find other risk factors for Legionella micdadei infection.
Table 1. Case reports of arthritis caused by Legionella species including the present case.
| No. | Age | Gender | Medical history and possible risk factors | Species | Diagnostic methods |
|---|---|---|---|---|---|
| 1 (12) | 51 | Male | Late recurrence of thymoma | L. pneumophila serogroup 1 | Serology (IgG, IgM)+; UAT+ |
| Chemotherapy | Culture (aspirate and blood, medium specific for mycobacteria +BCYE. medium)+; Culture (sputum)− | ||||
| 2 (13) | 80 | Female | Osteoarthritis | L. pneumophila serogroup 4 | Serology (IgG)+; Culture (Joint tissue, chocolate agar)+ |
| Immunofluorescence revealed L. pneumophila serogroup 4 (insufficient material for culture in BCYE medium) | |||||
| Culture (blood)-; UAT- | |||||
| 3 (20) | 32 | Male | None | L. pneumophila | Serology (L. pneumophila antibody serogroup 1)+ |
| Diving using a compressed air breathing apparatus 2w before a CT detected atypical pneumonia and clinical examination detected arthritis | UAT+ | ||||
| Culture (aspirate,medium not mentioned)− | |||||
| 4 (21) | 80 | Female | Chronic renal failure | L. pneumophila serogroup 1 | UAT+; Gram staining (aspirate)− |
| Pneumonia 16d before onset | Culture (aspirate, standard and BCYE medium)− | ||||
| 16S rRNA PCR+, sequence analysis revealed L. pneumophila serogroup 1 | |||||
| 5 (14) | 56 | Male | RA + type 2 diabetes mellitus | L. longbeachae | Gram staining(aspirate)+ |
| Methotrexate + prednisolone | Culture (aspirate, BCYE medium)+, 16S rRNA PCR+, sequence analysis revealed L. longbeachae | ||||
| No environmental risk factors or precedent pneumonia | Culture (blood)− | ||||
| 6 (15) | 58 | Female | SLE-like disease | L. dumoffii | Culture (aspirate, BCYE medium)+ |
| Methotrexate + prednisolone | Culture (blood)- | ||||
| Intra-articular injection of triamcinolone acetonide in the week prior to admission | 16S rRNA PCR+, sequence analysis revealed L. dumoffii | ||||
| 7 (19) | 83 | Female | RA | L. micdadei | Gram staining (aspirate)− |
| Methotrexate + deflazacort | Culture (aspirate, standard medium)− | ||||
| Replacement of the old prosthesis 32m before, progressive pain followed by swelling in her right knee 20m before, pneumonia 8m before admission; aerosolized water therapy at a spa in the year of onset and 2 arthrocentesis at the onset; no large air-conditioning units near the residence | Culture (aspirate, BCYE medium)+ | ||||
| 16S rRNA PCR+, sequence analysis revealed L. micdadei | |||||
| Culture (sputum)− | |||||
| UAT− | |||||
| 8 (16) | 71 | Female | Amyopathic dermatomyositis | L. bozemanae | Serology (IgG, IgM)+ |
| Methotrexate+ prednisolone | 16S rRNA PCR (aspirate)+, sequence analysis revealed L. bozemanae | ||||
| A rehabilitation stay at a rheumatologic hospital. L. bozemanae not detected from water samples from the patient’s home, the rehabilitation hospital or the department | Culture (aspirate, MWY medium)+, MALDI-TOF MS revealed L. bozemanae by | ||||
| Culture (blood + urine, standard medium)- | |||||
| 9 (17) | 70 | Female | Thymoma, lymphocytes, low IgG and IgA | L. pneumophila serogroup 1 | Culture (blood + aspirate, standard medium)- |
| No detectable CD19 and CD20 | Culture (aspirate, BCYE medium)+ | ||||
| Pneumonia 2y before the first onset, native valve endocarditis at the recurrence | 16S rRNA PCR (aspirate)+, sequence analysis revealed L. pneumophila serogroup 1 | ||||
| UAT- | |||||
| 10 (18) | 90 | Female | Chronic kidney disease + presumed chondrocalcinosis | L. cincinnatiensis | Gram staining (intraoperative pus specimen)+ |
| Prednisone + etoricoxib | Culture (intraoperative pus specimen, standard medium)- | ||||
| Intra-articular corticosteroid before onset. No contact with soil, potting mix or fountains, no respiratory symptoms or febrile illness before onset | 16S rRNA PCR (intraoperative pus specimen)+, sequence analysis revealed L. cincinnatiensis/longbeachae | ||||
| Culture (intraoperative pus specimen, BCYE medium)+, MALDI-TOF MS and MIP gene sequencing revealed L. cincinnatiensis | |||||
| 11 (current case) | 54 | Male | SLE | L. micdadei | Culture (aspirate, standard medium) revealed S. aureus |
| Corticosteroid | mNGS revealed L. micdadei and aureus, L. micdadei was confirmed by 16S rRNA sequence analysis |
L., Legionella; UAT, urinary antigen test; BCYE medium, buffered charcoal yeast extract medium; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; MWY medium, modified Wadowsky Yee medium; MALDI-TOF MS, matrix-assisted laser desorption/ionization time of flight mass spectrometry; MIP gene, macrophage infectivity potentiator gene.
The urinary antigen test (UAT) and serum antibody examination are common methods for Legionella detection. UAT is designed for L. pneumophila serogroup 1. To a great extent, it is the availability of this test that L. pneumophila serogroup 1 had most confirmed cases. While other species do not have their specific tests and could not achieve such accurate etiological diagnosis. Serum antibody examination requires several weeks for seroconversion. Most microbiological diagnosis was established upon joint specimen culture and 16s rRNA sequence analysis. Legionella species grow on special culture media (buffered charcoal yeast extract, BCYE) instead of standard ones, impeding the detection of this pathogen in routine culture. Hence the true frequency of infection may be significantly underestimated. Interestingly, Legionella species also grew unexpectedly on other culture media, such as chocolate agar (13) and mycobacteria growth medium (12). For culture-negative infections, 16s rRNA sequence analysis may help. Since cases of Legionella infections are still few, it is hard to tell the differences on epidemiology, toxicity, pathogenicity or other biological characteristics between Legionella micdadei and other Legionella species. As mNGS gaining its popularity, it would be possible to summarize the characteristics of different Legionella species in future.
Given the single positive result in conventional culture, Legionella infections would not be taken into consideration for the patient. mNGS detected the mixed infection and identified the species, making a great supplement to conventional culture. Besides, mNGS is time-saving because it’s culture-independent. In immunocompromised patients, complex mixed infections are very common, where conventional culture-dependent methods might not be enough for etiological diagnosis. mNGS is a new technique that is increasingly used for the clinical diagnosis of infectious diseases. Theoretically, with sufficiently long reads, multiple hits in the microbial genome, and a complete reference database, most microorganisms can be uniquely identified through mNGS (22). Althrough PCR based testing other than next generation, full-genome sequencing can be used to seek viral and fungal pathogens, mNGS has its superiority in diagnosis: (I) that a single test is able to identify bacterial, viral, fungal and parasitic pathogens, (II) that it is able to identify the presence of multiple organisms, (III) that it is able to provide a relative quantification of the multiple organisms present in a single sample and (IV) that it greatly saves time, especially for the mycobacteria infection. Several case reports and clinical studies have demonstrated the use of mNGS as a promising diagnostic tool for infectious diseases (22-24). Our case is the first one in which septic arthritis was diagnosed through mNGS of the aspirate sample.
Currently, clinical surveillance of infection progression is mainly guided by clinical manifestations, routine laboratory tests and culture. However, we still need a tool to directly monitor the dynamics of the pathogen. Apart from etiological diagnosis, mNGS offers a quick way to semi-quantitively monitor the changes in pathogen loads. Thus we consider mNGS a rapid method for disease surveillance (25). Our case implies the promising potential of mNGS as an effective tool for rapid and accurate infection diagnosis and surveillance.
Sample contamination is a common problem in mNGS. There were several possible contaminating microorganisms in this study. In consideration of the high sensitivity of mNGS, the relatively small reads numbers and the sequence homology among different staphylococcus, P. acnes and coagulase negative staphylococcus were considered as contamination. This contamination mainly caused by commensal microbes on the skin surface and are also very common in culture (especially coagulase negative staphylococcus). Similarly, in sputum sample, commensal oral flora such as Streptococci and anaerobe are frequently reported. Clinicians should be familiar with the background microorganisms in various parts of human body where specimens are collected. However, sometimes commensal microorganisms and pathogens can perform mutual transformations.
In conclusion, we present the first case of septic arthritis in native joint caused by L. micdadei, accompanied with S. aureus, diagnosed through mNGS. Conventional culture may not be able to detect all the pathogens. mNGS is a good supplement for rapid and accurate etiological diagnosis and surveillance. mNGS may play an important role in complicated infections, especially in immune-compromised patients.
Acknowledgments
Funding: This work was supported by the 4th Three-year Action Plan for Public Health of Shanghai (grant number 15GWZK0101); and the Zhongshan Hospital Outstanding Youth Program (grant number 2017ZSYXQN03).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Written informed consent was obtained from the patient for publication of this manuscript and any accompanying images.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Miao Q, Ma YY, Wang QQ, et al. Microbiological Diagnostic Performance of Metagenomic Next-generation Sequencing When Applied to Clinical Practice. Clin Infect Dis 2018;67:S231-40. 10.1093/cid/ciy693 [DOI] [PubMed] [Google Scholar]
- 2.Mercante JW, Winchell JM. Current and emerging Legionella diagnostics for laboratory and outbreak investigations. Clin Microbiol Rev 2015;28:95-133. 10.1128/CMR.00029-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dooling KL, Toews KA, Hicks LA, et al. Active Bacterial Core Surveillance for Legionellosis - United States, 2011-2013. MMWR Morb Mortal Wkly Rep 2015;64:1190-3. 10.15585/mmwr.mm6442a2 [DOI] [PubMed] [Google Scholar]
- 4.Beer KD, Gargano JW, Roberts VA, et al. Surveillance for Waterborne Disease Outbreaks Associated with Drinking Water - United States, 2011-2012. MMWR Morb Mortal Wkly Rep 2015;64:842-8. 10.15585/mmwr.mm6431a2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen TT, Schapiro JM, Loutit J. Prosthetic valve endocarditis due to Legionella pneumophila. The Journal of cardiovascular surgery 1996;37:631-3. [PubMed] [Google Scholar]
- 6.Dorman SA, Hardin NJ, Winn WC., Jr Pyelonephritis associated with Legionella pneumophila, serogroup 4. Ann Intern Med 1980;93:835-7. 10.7326/0003-4819-93-6-835 [DOI] [PubMed] [Google Scholar]
- 7.Schlanger G, Lutwick LI, Kurzman M, et al. Sinusitis caused by Legionella pneumophila in a patient with the acquired immune deficiency syndrome. Am J Med 1984;77:957-60. 10.1016/0002-9343(84)90551-5 [DOI] [PubMed] [Google Scholar]
- 8.Waldor MK, Wilson B, Swartz M. Cellulitis caused by Legionella pneumophila. Clin Infect Dis 1993;16:51-3. 10.1093/clinids/16.1.51 [DOI] [PubMed] [Google Scholar]
- 9.Brabender W, Hinthorn DR, Asher M, et al. Legionella pneumophila wound infection. JAMA 1983;250:3091-2. 10.1001/jama.1983.03340220059036 [DOI] [PubMed] [Google Scholar]
- 10.Lowry PW, Blankenship RJ, Gridley W, et al. A cluster of legionella sternal-wound infections due to postoperative topical exposure to contaminated tap water. N Engl J Med 1991;324:109-13. 10.1056/NEJM199101103240207 [DOI] [PubMed] [Google Scholar]
- 11.Sivagnanam S, Podczervinski S, Butler-Wu SM, et al. Legionnaires' disease in transplant recipients: A 15-year retrospective study in a tertiary referral center. Transpl Infect Dis 2017;19. doi: . 10.1111/tid.12745 [DOI] [PubMed] [Google Scholar]
- 12.Bemer P, Leautez S, Ninin E, et al. Legionella pneumophila Arthritis: use of medium specific for Mycobacteria for isolation of L. pneumophila in culture of articular fluid specimens. Clin Infect Dis 2002;35:E6-7. 10.1086/340713 [DOI] [PubMed] [Google Scholar]
- 13.Linscott AJ, Poulter MD, Ward K, et al. Legionella pneumophila serogroup 4 isolated from joint tissue. J Clin Microbiol 2004;42:1365-6. 10.1128/JCM.42.3.1365-1366.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dugar M, Rankin WA, Rowe E, et al. "My foot hurts": a flare of rheumatoid arthritis? Med J Aust 2009;190:392-3. 10.5694/j.1326-5377.2009.tb02459.x [DOI] [PubMed] [Google Scholar]
- 15.Flendrie M, Jeurissen M, Franssen M, et al. Septic arthritis caused by Legionella dumoffii in a patient with systemic lupus erythematosus-like disease. J Clin Microbiol 2011;49:746-9. 10.1128/JCM.00606-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Just SA, Knudsen JB, Uldum SA, et al. Detection of Legionella bozemanae, a new cause of septic arthritis, by PCR followed by specific culture. J Clin Microbiol 2012;50:4180-2. 10.1128/JCM.01899-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thurneysen C, Boggian K. Legionella pneumophila serogroup 1 septic arthritis with probable endocarditis in an immunodeficient patient. J Clin Rheumatol 2014;20:297-8. 10.1097/RHU.0000000000000128 [DOI] [PubMed] [Google Scholar]
- 18.Banderet F, Blaich A, Soleman E, et al. Septic arthritis due to Legionella cincinnatiensis: case report and review of the literature. Infection 2017;45:551-5. 10.1007/s15010-016-0964-1 [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Cruz A, Marin M, Castelo L, et al. Legionella micdadei, a new cause of prosthetic joint infection. J Clin Microbiol 2011;49:3409-10. 10.1128/JCM.00770-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andereya S, Schneider U, Siebert CH, et al. Reactive knee and ankle joint arthritis: abnormal manifestation of Legionella pneumophila. Rheumatol Int 2004;24:182-4. 10.1007/s00296-003-0382-4 [DOI] [PubMed] [Google Scholar]
- 21.Naito T, Suda T, Saga K, et al. Reactive Legionella pneumophila arthritis diagnosed by polymerase chain reaction. Rheumatol Int 2007;27:415-6. 10.1007/s00296-006-0223-3 [DOI] [PubMed] [Google Scholar]
- 22.Fan S, Ren H, Wei Y, et al. Next-generation sequencing of the cerebrospinal fluid in the diagnosis of neurobrucellosis. Int J Infect Dis 2018;67:20-4. 10.1016/j.ijid.2017.11.028 [DOI] [PubMed] [Google Scholar]
- 23.Yao M, Zhou J, Zhu Y, et al. Detection of Listeria monocytogenes in CSF from Three Patients with Meningoencephalitis by Next-Generation Sequencing. J Clin Neurol 2016;12:446-51. 10.3988/jcn.2016.12.4.446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiu CY, Coffey LL, Murkey J, et al. Diagnosis of Fatal Human Case of St. Louis Encephalitis Virus Infection by Metagenomic Sequencing, California, 2016. Emerg Infect Dis 2017;23:1964-8. 10.3201/eid2310.161986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ai JW, Zhang HC, Cui P, et al. Dynamic and direct pathogen load surveillance to monitor disease progression and therapeutic efficacy in central nervous system infection using a novel semi-quantitive sequencing platform. J Infect 2018;76:307-10. 10.1016/j.jinf.2017.11.002 [DOI] [PubMed] [Google Scholar]