Abstract
l-arabinofuranose is a ubiquitous component of the cell wall and various natural products in plants, where it is synthesized from cytosolic UDP-arabinopyranose (UDP-Arap). The biosynthetic machinery long remained enigmatic in terms of responsible enzymes and subcellular localization. With the discovery of UDP-Arap mutase in plant cytosol, the demonstration of its role in cell-wall arabinose incorporation and the identification of UDP-arabinofuranose transporters in the Golgi membrane, it is clear that the cytosolic UDP-Arap mutases are the key enzymes converting UDP-Arap to UDP-arabinofuranose for cell wall and natural product biosynthesis. This has recently been confirmed by several genotype/phenotype studies. In contrast to the solid evidence pertaining to UDP-Arap mutase function in vivo, the molecular features, including enzymatic mechanism and oligomeric state, remain unknown. However, these enzymes belong to the small family of proteins originally identified as reversibly glycosylated polypeptides (RGPs), which has been studied for >20 years. Here, we review the UDP-Arap mutase and RGP literature together, to summarize and systemize reported molecular characteristics and relations to other proteins.
Keywords: arabinofuranose, arabinose, reversibly glycosylated polypeptide, UDP-arabinopyranose mutase, UDP-arabinose
Introduction
The polysaccharides of the plant cell wall have been the subject of intense scientific study through several decades, due to their role in interesting plant properties on the cellular and organismal level, such as mechanical strength, chemical resilience, plasticity and flexibility and pathogen defense. These molecules also represent one of the most abundant resources on earth for food, feed and biofuels. A major carbohydrate component of the plant cell wall polysaccharides is l-arabinose, which is found in pectins and hemicelluloses in cell walls across the plant kingdom but has not been identified in animals (Kotake et al. 2016). Like other pentoses, l-arabinose is not readily fermented and thus represents a challenge for biomass utilization (Rennie and Scheller 2014). l-Arabinose moieties are derived from UDP-arabinopyranose (UDP-Arap) but are found predominantly in the furanose form (Araf) in the cell wall (Kotake et al. 2016). The route from pyranose to furanose has long been elusive to researchers. The discovery of UDP-Araf transporters (UAfTs) in the Golgi membrane (Rautengarten et al. 2017), together with recent genotype/phenotype studies (Dugard et al. 2016; Willis et al. 2016; Honta et al. 2018), has contributed to a clear picture of the cytosolic UDP-Arap mutases (UAMs) as the key enzymes converting UDP-Arap to UDP-Araf for cell wall and glycoprotein biosynthesis.
UAM activity was first demonstrated in rice cytosol in a seminal paper by Konishi et al. (2007). They isolated a 41 kDa protein from rice seedlings and showed that it was capable of in vitro interconversion of UDP-Arap and UDP-Araf. Similar proteins (Table I) have since been characterized in Arabidopsis thaliana (Rautengarten et al. 2017) Chlamydomonas reinhardtii (Kotani et al. 2013) and Hordeum vulgare (Hsieh et al. 2016), and sequence analysis suggests a kingdom-wide distribution (Fedosejevs et al. 2017).
Table I.
Summary of characterized proteins with UAM or RGP activity
| Organism, name | UAM activity | RGP activity | Co-factors | References |
|---|---|---|---|---|
| Daucus carota | (Ingold and Seitz 1985; Quentmeier et al. 1987) | |||
| GT IsU | UDP-Glc, UDP-Gal and UDP-Xyl | Mg2+, Mn2+, Zn2+ | ||
| Zea mays | (Singh et al. 1995) | |||
| Amylogenin | UDP-Glc | |||
| Pisum sativum | (Dhugga et al. 1991, 1997) | |||
| PsRGP1 | UDP-Glc, UDP-Gal and UDP-Xyl | Mg2+, Mn2+ | ||
| Arabidopsis thaliana | (Delgado et al. 1998; Kuttiyatveetil and Sanders 2017; Rautengarten et al. 2011) | |||
| AtRGP1 | UDP-Araf, UDP-Arap | UDP-Glc, UDP-Gal, and UDP-Xyl | Mn2+ | |
| AtRGP2 | UDP-Araf, UDP-Arap | Mn2+ | ||
| AtRGP3 | UDP-Araf, UDP-Arap | Mn2+ | ||
| AtRGP4 | ||||
| AtRGP5 | ||||
| Triticum aestivum | (Langeveld et al. 2002; Zeng et al. 2010) | |||
| TaRGP1 | UDP-Glc, UDP-Gal and UDP-Xyl | |||
| TaRGP2 | ||||
| Oryzae sativa | (Konishi et al. 2007; Konishi, Miyazaki et al. 2010; Kuttiyatveetil and Sanders 2017; Langeveld et al. 2002) | |||
| OsUAM1 | UDP-Araf, UDP-Arap and UDP-Galf | UDP-Glc, UDP-Gal, UDP-Xyl, UDP-Araf and UDP-Arap | Mn2+ | |
| OsUAM2 | ||||
| OsUAM3 | UDP-Araf and UDP-Arap | UDP-Glc, UDP-Gal, UDP-Xyl, UDP-Araf, and UDP-Arap | Mn2+ | |
| Solanum tuberosum | (De Pino et al. 2007) | |||
| StRGP | UDP-Glc, UDP-Gal and UDP-Xyl | |||
| Solanum lycopersicum | (Selth et al. 2006) | |||
| SlUPTG1 | UDP-Glc | |||
| Chlamydomonas reinhardtii | (Kotani et al. 2013) | |||
| CrUAM | UDP-Araf and UDP-Arap | UDP-Glc | ||
| Hordeum vulgare | (Hsieh et al. 2016) | |||
| HvUAM1 | UDP-Araf and UDP-Arap | UDP-Glc | Mn2+, Mg2+, | |
| HvUAM2 | UDP-Araf and UDP-Arap | |||
| HvUAM3 | UDP-Araf and UDP-Arap | |||
| HvUAM4 |
The enzymatic mechanism and other features such as protein structure, oligomeric state and regulation are unknown. However, UAMs were originally identified as a small family of reversibly glycosylated polypeptides (RGPs), which have been studied for >20 years. Here, we put the spotlight on these enigmatic enzymes that are emerging as key players in cell wall and glycoprotein biosynthesis. We seek to stimulate interest and further research to address the many open questions that remain.
Biological importance of UAMs
UAM activity represents the only known route to Araf in plants. Araf is a constituent of a wide variety of cell wall components, including glycans (e.g. xylans, pectic arabinans and rhamnogalacturonans) and glycoproteins (e.g. arabinogalactan proteins (AGPs), and extensin-like proteins) (Carpita and Gibeaut 1993). In addition, it is found in natural products such as triterpenes (Jacobsen et al. 1996). Thus, it is not surprising that UAM activity is essential; Arabidopsis double loss-of-function mutants in AtRGP1 and AtRGP2 are not viable (Drakakaki et al. 2006), and RNAi targeting the two genes leads to severely restricted growth and low arabinose levels in the cell walls (Rautengarten et al. 2011). Further, UAMs have been implicated in various important biological processes, including development (Zhao et al. 2001; Gallardo et al. 2003; Dai et al. 2006; Wu et al. 2006; Zavaliev et al. 2009; Sumiyoshi et al. 2014), viral defense (Selth et al. 2006; Zavaliev et al. 2009; Burch-Smith et al. 2012), abiotic stress response (Wu et al. 2006; Zeng et al. 2014) and gravitropic bending (Hu et al. 2009).
Much of the plant cell wall Araf is contained in pectic rhamnogalacturonan I (RGI) (Mohnen 2008). It is not obvious, however, that this feature of RGI is indispensable, since mutants that are specifically deficient in RGI arabinans show no obvious phenotype (Harholt et al. 2006, 2012). Araf is also found in the complex glycan RGII, which typically includes four Araf residues (Ndeh et al. 2017). In general, there is little flexibility in RGII structure, and mutants in RGII biosynthesis tend to be lethal. However, the Araf residues are all terminal (Ndeh et al. 2017), and the specific arabinosyltransferases responsible for adding them are not known. Hence, it is possible but not documented that some of the Araf residues in RGII are essential.
Extensins are cell wall glycoproteins that are decorated with short arabinose-containing sidechains (Møller et al. 2017). Similar structures are found in extracellular peptides such as CLAVATA3. Arabinosylation of these proteins and peptides is clearly essential, especially for pollen tube growth, as evidenced by analysis of Arabidopsis mutants in the HPAT arabinosyltransferases that add the first Araf residue to hydroxyproline in the protein backbone (MacAlister et al. 2016). Accordingly, UAM is associated with pollen development in rice (Dai et al. 2006; Sumiyoshi et al. 2014). Arabinosylation of CLAVATA3 is also required for its function in meristem development (Ohyama et al. 2009). Finally, Araf is present in AGPs, but the biosynthesis of their glycans is poorly understood. The Arabidopsis ray1 mutant is deficient in AGP Araf, and the RAY1 protein has arabinofuranosyltransferase activity (Gille et al. 2013). Growth of the ray1 mutant plants is only slightly affected. Nevertheless, given the role of AGPs in many biological functions it is likely that other and more specific functions of AGP arabinofuranosylation will be found.
The UAM reaction
Intriguingly, the biologically essential UDP-Araf is generated by UAM activity on UDP-Arap (Figure 1) through a highly unfavorable equilibrium of 1:9 furanose:pyranose (Konishi, Ohnishi-Kameyama et al. 2010; Hsieh et al. 2016). UAM activity has been detected solely in the plant cytosol and associated with the cytosolic side of the Golgi apparatus (Konishi et al. 2007; Rautengarten et al. 2011; Kotake et al. 2016). This is counterintuitive, since most of the UDP-Arap substrate is synthesized in the Golgi lumen by UDP-xylose-4-epimerase (Burget et al. 2003). Hence, plants must transport UDP-Arap out of the Golgi apparatus by yet unknown transporters. Once in the cytosol, the UAM-generated UDP-Araf can be used in natural product biosynthesis. However, this is the exception, since all the other biosynthetic reactions that use UDP-Araf take place in the Golgi lumen. To enable these reactions, plants have specific UAfTs in the Golgi membrane that transports the UDP-Araf generated by UAMs back into the Golgi lumen (Rautengarten et al. 2017). It is possible that the action of the UAfTs drives the UAM reaction by continuously removing UDP-Araf from the equilibrium.
Fig. 1.
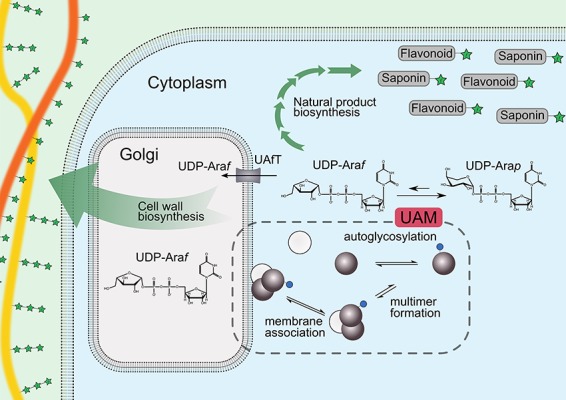
Schematic of UAM in the plant cell. The catalyzed reaction is shown above a hypothetical model (in stipled enclosure) of UAM autoglycosylation, oligomerization and membrane association. UAfT represents UDP-araf transporter; gray spheres, class 1 UAM; white spheres, class 2 UAM; small blue sphere, glucose; green star, arabinofuranose; orange line, xylan; yellow line, pectin.
While some nucleotide sugar-active enzymes can catalyze reactions with different nucleotide sugars, the UAM reaction seems to have a quite limited substrate scope; UDP-Ara has been repeatedly demonstrated to be tautomerized by different UAMs (Konishi et al. 2007; Rautengarten et al. 2011; Kotani et al. 2013; Hsieh et al. 2016), but no mutase activity on UDP-Gal, UDP-Glc, UDP-Xyl, GDP-Fuc or GDP-Man was found in Arabidopsis UAMs (Rautengarten et al. 2011). To the best of our knowledge, the only other described substrate for the UAM reaction is UDP-Gal, which has been demonstrated to be inefficiently tautomerized by OsUAM1 (Kuttiyatveetil and Sanders 2017). The low activity with UDP-Gal may not be relevant in vivo since Galf has not been reported in plants.
To the best of our knowledge, kinetically characterized UAMs are limited to rice UAM1 (Konishi et al. 2007; Kuttiyatveetil and Sanders 2017), three barley UAMs (HvUAM1-3) (Hsieh et al. 2016), and a green algae UAM (CrUAM) (Kotani et al. 2013). These studies indicate that UAM is an enzyme class of average to low catalytic efficiency. Thus, the reaction that provides Araf for the plant cell wall—conversion of UDP-Arap to UDP-Araf—has been reported to occur at rates in the 0.1–4 s-1 range, while the reverse reaction seems to be around an order of magnitude faster (1.5–67 s-1) (Kotani et al. 2013; Hsieh et al. 2016; Kuttiyatveetil and Sanders 2017). Furthermore, KM values for the two substrates appear to be similar and in the micromolar range, albeit lower for UDP-Araf (UDP-Araf: 4–390 μM; UDP-Arap: 127–1290 μM) (Konishi et al. 2007; Kotani et al. 2013; Hsieh et al. 2016; Kuttiyatveetil and Sanders 2017). These four studies also reported pH optima varying between 5 and 7.7 and temperature optima between 55°C and 65°C, with no obvious correlation between the temperature and pH tolerance and source organism.
Reversible autoglycosylation
The mechanism of the UAM reaction is unknown, and it can be speculated whether it involves transient separation from the UDP moiety, as discussed later. In line with this idea, rice UAMs have been shown to be reversibly auto-arabinosylated upon reaction with UDP-Ara, providing perhaps a glimpse of an enzyme-substrate reaction intermediary (Konishi et al. 2007). However, auto-arabinosylation is just one example of a widely observed phenomenon; UAM activity seems to go hand-in-hand with reversible autoglycosylation with a variety of sugars (Figure 1) of a conserved arginine residue (Konishi et al. 2007; Rautengarten et al. 2011; Kotani et al. 2013; Hsieh et al. 2016). Indeed, UAMs were initially identified as RGPs, an activity which was subject of many studies prior to the realization of the UAM activity. The RGPs have been characterized as wide-spread plant-specific enzymes that reversibly autoglucosylate a conserved arginine residue (Singh et al. 1995; Dhugga et al. 1997; Delgado et al. 1998), the biological significance of which is unknown.
RGP activity has a broader substrate scope than UAM activity, with UDP-Glc, UDP-Gal, UDP-Xyl, UDP-Arap and UDP-Araf all being able to chase out labeling of OsUAM1 with radioactive UDP-Glc (Konishi et al. 2007). This is in line with previous RGP-focused studies showing reversible autoglycosylation activity with UDP-Glc, UDP-Gal and UDP-Xyl of RGPs from various plants (Dhugga et al. 1991; Singh et al. 1995; Delgado et al. 1998; Langeveld et al. 2002; Testasecca et al. 2004; De Pino et al. 2007). In addition, these studies indicate that uridine is the only accepted nucleoside, and that nucleotide mannosides are not RGP substrates (Dhugga et al. 1991; Singh et al. 1995; Dhugga et al. 1997; Delgado et al. 1998; Langeveld et al. 2002).
The biological role of the RGP activity of UAMs is unclear. In the current context, we can hypothesize that this autoglycosylation is an intermediary or by-product of the primary mutase reaction. For example, the reaction could be speculated to proceed via three steps (transfer of sugar moiety to enzyme, tautomerization and release), where step 2 and/or 3 has stricter stereochemical requirements for catalysis than step 1. However, since no proteins with only one of the two activities have been identified or generated it has so far not been possible to address this experimentally. It is also possible that RGP activity has a separate role, for example regulatory or structural, or the enzymes themselves have separate biological functions mediated by the two activities.
Class 2 UAMs
Intriguingly, plants have versions of UAMs with no UAM or RGP activity. For example, rice contains three UAMs, where only OsUAM1 and OsUAM3 have been demonstrated to be active (Konishi et al. 2007). Likewise, two of Arabidopsis’ five UAMs (AtRGP4 and AtRGP5) and one of barleys four UAMs (HvUAM4) seem to be noncatalytic (Rautengarten et al. 2011; Hsieh et al. 2016). Langeveld et al. (2002) observed a similar pattern for wheat RGPs, where one was apparently catalytically inactive. Based on sequence analysis, the authors proposed to divide RGP sequences into class 1 and 2, with the two classes having ~40% sequence identity. Sequences reported to be devoid of UAM and RGP activities exclusively belong to class 2 (OsUAM2, TaUAM2, AtUAM5 and HvUAM4) (Zhao et al. 2001; Langeveld et al. 2002; Sagi et al. 2005; Drakakaki et al. 2006; De Pino et al. 2007; Konishi et al. 2007; Rautengarten et al. 2011; Kotani et al. 2013; Hsieh et al. 2016) with the notable exception of class 1 AtRGP4 (77% sequence identity to OsUAM1 and only 46% to OsUAM2), which seems devoid of activity (Rautengarten et al. 2011). The biological role of these seemingly non-enzymatic proteins remains unknown and can be speculated to be regulatory or related to scaffolding (Figure 1), since they have been shown to participate in large multimeric complexes with active UAMs, as described below.
Similarities to other enzymes
Owing to their RGP activity, UAMs can be classified as Leloir glycosyltransferases (Lairson et al. 2008), in that they catalyze glycosidic bond formation using an activated sugar donor (a UDP-sugar in this case) to transfer the sugar moiety to an acceptor molecule (the UAM itself in this case). Based on sequence homology, UAMs are classified into glycosyltransferase family 75 (GT75) in the Carbohydrate-Active Enzymes database (www.cazy.com) (Lombard et al. 2013). GT75 currently contains 161 entries, including 109 from the plant kingdom, 9 bacterial and 43 archaeal sequences. UAMs contain three DxD glycosyltransferase signature motifs at positions 52, 110 and 182 (OsUAM1 numbering; Figure 2). While the first motif—which is DGD in all characterized class 1 UAMs and DPE/D in class 2 (Figure 2)—is not completely conserved across GT75 (Supplementary data Figure S1). The next motif—DDD/DDN in all class 1/2 GT75 sequences, respectively—is highly conserved and sits in a highly conserved sequence area. This motif was shown to be essential for UAM activity in OsUAM1 (Kuttiyatveetil and Sanders 2017) but not sufficient to install UAM activity in OsUAM2 (Konishi, Ohnishi-Kameyama et al. 2010). The last motif is DYD in all plant sequences, DLD in all bacterial sequences and DVD in all archaeal sequences (Supplementary data Figure S1). In glycosyltransferases, the DxD motif interacts with the nucleotide sugar substrate through a coordinated divalent cation (Lairson et al. 2008). Accordingly, both UAM and RGP activities are dependent on divalent cations, specifically Mn2+ and Mg2+ (Ingold and Seitz 1986; Dhugga et al. 1991; Konishi et al. 2007; Hsieh et al. 2016; Kuttiyatveetil and Sanders 2017). . In addition, UAMs from rice (Konishi et al. 2007) and barley (Hsieh et al. 2016) have been shown to be inhibited by ethylenediaminetetraacetic acid treatment. Hsieh et al. (2016) showed that addition of Mn2+ decreased KM of barley UAMs toward UDP-Araf between 3.5 and 41 times. In glycosyltransferases, this interaction is thought to electrostatically stabilize the negative charge developing on the phosphates after glycosidic cleavage and to enable leaving group departure (Lairson et al. 2008). On this basis, one can speculate that the UAM mechanism likewise involves a glycosidic cleavage and subsequent UDP departure, followed by UDP re-entry after ring-chain tautomerization. Another interpretation could be that a DxD:metal ion complex simply interacts with the diphosphate group to orient UDP-Ara for catalysis. However, no direct evidence exists for an interaction between the DxD motif and a metal ion in UAMs.
Fig. 2.
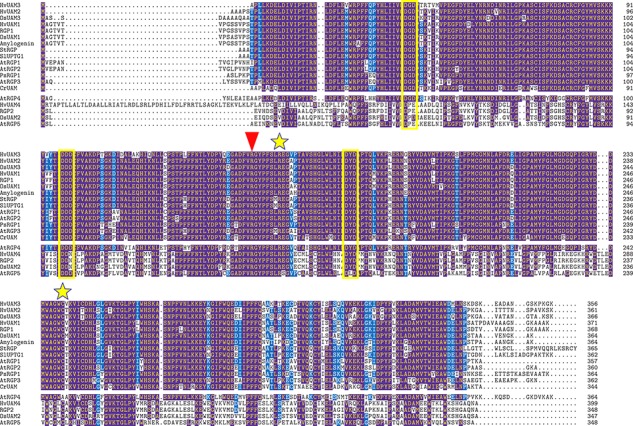
Sequence alignment of characterized UAMs. The top 14 sequences are class 1, and the bottom 5 sequences are class 2. Residues with blue background are >50% conserved, while those with purple background are >70% conserved. DxD motifs are marked with a black bar, the autoglycosylation site is marked with a red triangle, and two residues shown to be important for function are marked with yellow stars.
The interconversion of UDP-Arap and UDP-Araf is analogous to the interconversion of UDP-Galp and UDP-Galf catalyzed by UDP-galactopyranose mutases (UGMs) (Tanner et al. 2014). In fact, UGMs can inefficiently interconvert arabinose (Konishi et al. 2007), and at least one UAM can inefficiently interconvert galactose (Kuttiyatveetil and Sanders 2017). UGMs are flavoenzymes found in bacteria, fungi and unicellular protozoan parasites. They possess a unique and conserved 3D fold and display a variety of oligomeric states (Tanner et al. 2014), not unlike UAMs, as described later. The UGM-catalyzed reaction proceeds through a nucleophilic attack on the anomeric carbon mediated by an enzyme-bound FAD cofactor. However, FAD is not present in UAMs from barley or rice, and its addition had no effect on UAM activity (Konishi et al. 2007; Hsieh et al. 2016). On the amino acid sequence level, UGMs are unrelated to UAMs, making it hard to infer mechanistic detail, and it is likely that the mechanisms differ entirely between these two sets of enzymes.
Structure–function relationships
To elucidate the enzymatic and regulatory mechanisms of UAMs, structural information would be very helpful. Although we recently reported preliminary crystallization conditions for the catalytically inactive OsUAM2 (Welner et al. 2017b), no crystal structure has been solved for any UAM. In the following, we will summarize insights from in silico and mutational analyses.
Analyses of the primary structures of UAMs (Figure 2) assign the amino acid sequences to the RGP family (pfam 03214) in their entire length. Disorder predictions reveal no obvious linker regions, indicating a one-domain structure. Likewise, there are generally no recognizable transmembrane regions or signal peptides. In addition to the mentioned conserved DxD motifs (Figure 2), UAMs have a conserved arginine residue (Arg158 in OsUAM1), which has been shown to be the site of autoglycosylation (Singh et al. 1995; Konishi, Ohnishi-Kameyama 2010; Hsieh et al. 2016). Mutation of this residue resulted in complete loss of mutase activity for HvUAM1 (Hsieh et al. 2016), and both autoglycosylation and mutase activities for OsUAM1 and OsUAM3 (Konishi, Ohnishi-Kameyama et al. 2010). Other noteworthy amino acids are Cys257 of AtRGP2, which has been found to be mutated to a tyrosine in the cell wall arabinose-deficient Arabidopsis mutant murus5, most likely impacting AtRGP2 folding or oligomerization (Dugard et al. 2016), and a conserved arginine adjacent to the autoglycosylated residue (Arg165 in OsUAM1), which was shown to be essential for mutase activity (Konishi, Ohnishi-Kameyama et al. 2010; Hsieh et al. 2016) and hypothesized to play a role in phosphoryl coordination of the nucleotide sugar, analogous to what has been suggested for Klebsiella pneumonia UGM (Chad et al. 2007).
Secondary structure predictions and hydrophobic cluster analyses indicate that UAMs contain both α-helices and β-sheets, and that at least class 1 UAMs could adopt a Rossmann fold commonly found in nucleotide binding proteins (De Pino et al. 2007; Dugard et al. 2016; Kuttiyatveetil and Sanders 2017). Circular dichroism further corroborated the existence of significant amounts of both α- and β-structural elements (17 and 29%, respectively) in OsUAM1 (Kuttiyatveetil and Sanders 2017). The presence of a Rossmann fold is supported by 3D modeling, since both StRGP and AtRGP2 have been threaded on glycosyltransferase templates containing such a nucleotide-binding domain (De Pino et al. 2007; Dugard et al. 2016). In both cases, the sequence identity between the template and the query was quite low (12 and 14%, respectively), limiting the reliability of the resulting models. Nonetheless, De Pino et al. (2007) note that their model supports the glycosylation of Arg158, in that there is no steric hindrance and it is in the vicinity of a DxD motif. Ab initio modeling of OsUAM1 yields a β/α-containing structure, although the topology does not seem to correspond to a classical Rossmann fold (Kuttiyatveetil and Sanders 2017). A search for functional homologs using the ab initio model did, however, yield glycosyltransferase hits with Rossmann folds (Kuttiyatveetil and Sanders 2017). Given the similar reaction catalyzed by UAMs and UGMs, Dugard et al. (2016) also did an attempt to model AtRGP2 on an Escherichia coli UGM template, despite the low sequence identity (21%). However, this approach placed the DxD motif far from the active site and the resulting model seems unlikely.
Oligomerization and protein–protein interactions
UAM structure–function is perhaps most well studied on the quarternary level. This stems from the fact that oligomerization seems to be a prominent feature of most UAMs, including those from rice, wheat, potato, Arabidopsis and green algae (Bocca et al. 1999; Langeveld et al. 2002; Drakakaki et al. 2006; De Pino et al. 2007; Konishi et al. 2007; Rautengarten et al. 2011; Kotani et al. 2013). In fact, the only UAMs reported not to form multimeric complexes are the four from barley (Hsieh et al. 2016). The first indication came already in 1987, when Quentmeier et al. (1987) identified ~40 kDa autocatalytic glycosyltransferase in soluble and membrane-associated fractions of carrot cell suspensions, which formed large molecular complexes. While reversible glycosylation was not investigated in this study, it is likely that they were in fact observing glycosylation of RGP (Ingold and Seitz 1986). Following this observation, the RGP gene family was defined and RGPs reported to migrate in native page or size-exclusion chromatography as 230–260 kDa complexes corresponding to penta- or hexamers (Bocca et al. 1999; Langeveld et al. 2002; De Pino et al. 2007). Studies of StRGP pointed to a role for disulfide bridges in oligomerization and hypothesized that the aforementioned C257 (C251 in StRGP) might play a role (De Pino et al. 2007). RGP complex formation was further substantiated by the report of in vivo co-localization and co-immunoprecipitation of AtRGP1 and AtRGP2 (Drakakaki et al. 2006). When UAM activity was discovered, this too was demonstrated to localize to high molecular weight complexes in rice (Konishi et al. 2007; Konishi, Miyazaki et al. 2010), Arabidopsis (Rautengarten et al. 2017) and green algae (Kotani et al. 2013).
The functional implications of oligomerization are unknown. The first study indicating a functional role showed that monomeric StRGP was more active than the multimeric version, that glycosylation promoted oligomerization, and that Golgi-associated StRGP was predominantly multimeric (De Pino et al. 2007). Based on their observations, the authors hypothesized that glycosylation of StRGP leads to the formation of oligomers that target soluble StRGP to the Golgi membrane (Figure 1). Conversely, Langeveld et al. (2002) previously reported that TaRGP2 expressed in tobacco leaves is active only when in high molecular weight complexes. With today’s knowledge, their observation can be explained by a potential interaction between endogenous, active tobacco RGP and inactive recombinant TaRGP2, which belongs to UAM class 2. In 2010, Konishi, Miyazaki et al. (2010) reported that rice UAMs work synergistically in complexes, since in vitro mixing of insect-cell expressed OsUAM1, 2 and 3 resulted in a nonlinear increase in mutase activity. This contrasts with the authors’ own observation from 2007 that mixing of E.coli-expressed OsUAM1, 2 and 3 had no synergistic effect (Konishi et al. 2007) and with the work of Rautengarten et al. (2011), who detected no synergy when mixing Arabidopsis UAMs.
The biological consequences of UAM oligomerization is likewise an enigma. It could play a regulatory role, leveraging the spatially and temporally differentiated expression of UAM isozymes (Wu et al. 2006; Rautengarten et al. 2011; Sumiyoshi et al. 2014; Gupta et al. 2018) as a way to modulate UAM complex composition and thereby fine-tune regulation. This is in line with the observation that a ~116-kDa rice UAM-containing protein complex emerges at day 4 after rice seed germination (De Pino et al. 2007), and, thus, oligomerization seems to be correlated with rice plant development. The occurrence of the apparently non-enzymatic class 2 UAMs could then be explained by a scaffolding role in these regulatory complexes (Figure 1).
Finally, it is worth mentioning that even though Langeveld et al. (2002) found no other protein to be part of RGP containing complexes from wheat, UAM has been found in co-immunoprecipitations of sucrose synthase from various seeds and flowers (Fedosejevs et al. 2017) and in a xylan synthase complex from wheat (Zeng et al. 2010). Finally, UAMs have repeatedly been shown to associate with the Golgi surface (Dhugga et al. 1991; Epel et al. 1996; Dhugga et al. 1997; Sagi et al. 2005; Drakakaki et al. 2006; Rautengarten et al. 2011), and it is tempting to speculate that this is mediated by interactions with the UAfTs, representing the next step in arabinofuranosylation pathways, through UAM surface patches with protein affinity. No experimental validation of such an interaction has been reported.
Biotechnological implications
UDP-Araf is the precursor for the biosynthesis of many polysaccharides, proteoglycans and glycoproteins (Konishi et al., 2006). Even though UDP-Araf can be chemically synthesized in low yields, the procedure is complex, and the compound is very expensive (~1600 USD per mg). Therefore, a simple and efficient route to UDP-Araf would be useful. The unique FAD-independent mutase activity presented by UAMs might be harnessed in biocatalyst applications for such furanose synthesis and production. In addition, UAMs have potential applications in biofuels production, by offering a tool to control cell wall composition. It was recently shown that downregulating UAM1 alters the amount of arabinoxylan in switchgrass cell walls and thereby potentially decreases recalcitrance in this important biofuel feedstock (Willis et al., 2016).
Outlook
With the biotechnological potential and importance in plant biology, there is an immediate need to understand the molecular aspects of UAMs. Open questions include the enzymatic mechanism, regulatory aspects and the roles of autoglycosylation and oligomerization. So far, structural and functional characterization has been impaired by difficulties in in vitro handling of these enzymes, with only limited yields from recombinant expression (Welner et al., 2017a). However, modern high-throughput and in vivo methods are likely to overcome this, and it will be exciting to see what future UAM research brings.
Supplementary Material
Abbreviations
- AGP
arabinogalactan protein
- Araf
arabinofuranose
- GT75
glycosyltransferase family 75
- RGI
rhamnogalacturonan I
- RGP
reversibly glycosylated polypeptide
- UAfT
UDP-Araf transporter
- UAM
UDP-arabinopyranose mutase
- UDP-Arap
UDP-arabinopyranose
- UGM
UDP-galactopyranose mutase
Funding
Novo Nordisk Foundation (NNF10CC1016517) To the Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark; Joint BioEnergy Institute supported by the US Department of Energy, Office of Science, Office of Biological and Environmental Research through contract (DE-AC02-05CH11231) between Lawrence Berkeley National Laboratory and the US Department of Energy.
Conflict of interest statement
None declared
References
- Beitz E. 2000. TeXshade: Shading and labeling of multiple sequence alignments using LaTeX2. Bioinformatics. 16:135–139. [DOI] [PubMed] [Google Scholar]
- Bocca SN, Kissen R, Rojas-Beltrán JA, Noel F, Gebhardt C, Moreno S, Du Jardin P, Tandecarz JS. 1999. Molecular cloning and characterization of the enzyme UDP-glucose: Protein transglucosylase from potato. Plant Physiol Biochem. 37:809–819. [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Cui Y, Zambryski PC. 2012. Reduced levels of class 1 reversibly glycosylated polypeptide increase intercellular transport via plasmodesmata. Plant Signal Behav. 7:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burget EG, Verma R, Mølhøj M, Reiter W-D. 2003. The biosynthesis of L-arabinose in plants: Molecular cloning and characterization of a Golgi-localized UDP-D-xylose 4-epimerase encoded by the MUR4 gene of Arabidopsis. Plant Cell. 15:523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. 1993. Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3:1–30. [DOI] [PubMed] [Google Scholar]
- Chad JM, Sarathy KP, Gruber TD, Addala E, Kiessling LL, Sanders DA. 2007. Site-directed mutagenesis of UDP-galactopyranose mutase reveals a critical role for the active-site, conserved arginine residues. Biochem. 46:6723–6732. [DOI] [PubMed] [Google Scholar]
- Dai S, Li L, Chen T, Chong K, Xue Y, Wang T. 2006. Proteomic analyses of Oryza sativa mature pollen reveal novel proteins associated with pollen germination and tube growth. Proteomics. 6:2504–2529. [DOI] [PubMed] [Google Scholar]
- De Pino V, Borán M, Norambuena L, González M, Reyes F, Orellana A, Moreno S. 2007. Complex formation regulates the glycosylation of the reversibly glycosylated polypeptide. Planta. 226:335–345. [DOI] [PubMed] [Google Scholar]
- Delgado IJ, Wang Z, de Rocher A, Keegstra K, Raikhel NV. 1998. Cloning and characterization of AtRGP1: A reversibly autoglycosylated Arabidopsis protein implicated in cell wall biosynthesis. Plant Physiol. 116:1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhugga K, Ulvskov P, Gallagher S, Ray P. 1991. Plant polypeptides reversibly glycosylated by UDP-glucose. Possible components of Golgi beta-glucan synthase in pea cells. J Biol Chem. 266:21977–21984. [PubMed] [Google Scholar]
- Dhugga KS, Tiwari SC, Ray PM. 1997. A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: Purification, gene cloning, and trans-Golgi localization. Proc Natl Acad Sci USA. 94:7679–7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G, Zabotina O, Delgado I, Robert S, Keegstra K, Raikhel N. 2006. Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiol. 142:1480–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugard CK, Mertz RA, Rayon C, Mercadante D, Hart C, Benatti MR, Olek AT, SanMiguel PJ, Cooper BR, Reiter W-D. 2016. The cell wall arabinose-deficient Arabidopsis thaliana mutant murus5 encodes a defective allele of REVERSIBLY GLYCOSYLATED POLYPEPTIDE2. Plant Physiol. 171:1905–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epel BL, van Lent JW, Cohen L, Kotlizky G, Katz A, Yahalom A. 1996. A 41 kDa protein isolated from maize mesocotyl cell walls immunolocalizes to plasmodesmata. Protoplasma. 191:70–78. [Google Scholar]
- Fedosejevs ET, Liu LN, Abergel M, She YM, Plaxton WC. 2017. Coimmunoprecipitation of reversibly glycosylated polypeptide with sucrose synthase from developing castor oilseeds. FEBS Lett. 591:3872–3880. [DOI] [PubMed] [Google Scholar]
- Gallardo K, Le Signor C, Vandekerckhove J, Thompson RD, Burstin J. 2003. Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiol. 133:664–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gille S, Sharma V, Baidoo EE, Keasling JD, Scheller HV, Pauly M. 2013. Arabinosylation of a Yariv-precipitable cell wall polymer impacts plant growth as exemplified by the Arabidopsis glycosyltransferase mutant ray1. Mol Plant. 6:1369–1372. [DOI] [PubMed] [Google Scholar]
- Gupta M, Kaul S, Dhar MK. 2018. Identification and characterization of some putative genes involved in arabinoxylan biosynthesis in Plantago ovata. 3 Biotech. 8:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Sørensen SO, Orfila C, Pauly M, Scheller HV. 2006. ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol. 140:49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Verhertbruggen Y, Søgaard C, Bernard S, Nafisi M, Poulsen CP, Geshi N, Sakuragi Y, Driouich A. 2012. ARAD proteins associated with pectic Arabinan biosynthesis form complexes when transiently overexpressed in planta. Planta. 236:115–128. [DOI] [PubMed] [Google Scholar]
- Honta H, Inamura T, Konishi T, Satoh S, Iwai H. 2018. UDP-arabinopyranose mutase gene expressions are required for the biosynthesis of the arabinose side chain of both pectin and arabinoxyloglucan, and normal leaf expansion in Nicotiana tabacum. J Plant Res. 131:307–317. [DOI] [PubMed] [Google Scholar]
- Hsieh YS, Zhang Q, Yap K, Shirley NJ, Lahnstein J, Nelson CJ, Burton RA, Millar AH, Bulone V, Fincher GB. 2016. Genetics, transcriptional profiles, and catalytic properties of the UDP-arabinose mutase family from barley. Biochemistry. 55:322–334. [DOI] [PubMed] [Google Scholar]
- Hu L, Cui D, Zang A, Neill S, Cai W. 2009. Auxin-regulated OsRGP1 and OsSuS are involved in gravitropic bending of rice shoot bases. Mol Cell Biol. 42:27–34. [PubMed] [Google Scholar]
- Ingold E, Seitz HU. 1986. Characterization and properties of different glucosyltransferases isolated from suspension-cultured cells of Daucus carota. Z Naturforsch C. 41:409–420. [Google Scholar]
- Jacobsen NE, Fairbrother WJ, Kensil R, Lim A, Wheeler DA, Powell MF. 1996. Structure of the saponin adjuvant QS-21 and its base-catalyzed isomerization product by 1H and natural abundance 13C NMR spectroscopy. Carbohydr Res. 280:1–14. [DOI] [PubMed] [Google Scholar]
- Konishi T, Miyazaki Y, Yamakawa S, Iwai H, Satoh S, Ishii T. 2010a. Purification and biochemical characterization of recombinant rice UDP-arabinopyranose mutase generated in insect cells. Biosci Biotechnol Biochem. 74:191–194. [DOI] [PubMed] [Google Scholar]
- Konishi T, Ohnishi-Kameyama M, Funane K, Miyazaki Y, Konishi T, Ishii T. 2010b. An arginyl residue in rice UDP-arabinopyranose mutase is required for catalytic activity and autoglycosylation. Carbohydr Res. 345:787–791. [DOI] [PubMed] [Google Scholar]
- Konishi T, Ono H, Ohnishi-Kameyama M, Kaneko S, Ishii T. 2006. Identification of a mung bean arabinofuranosyltransferase that transfers arabinofuranosyl residues onto (1, 5)-linked α-L-arabino-oligosaccharides. Plant Physiol. 141:1098–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Takeda T, Miyazaki Y, Ohnishi-Kameyama M, Hayashi T, O’neill MA, Ishii T. 2007. A plant mutase that interconverts UDP-arabinofuranose and UDP-arabinopyranose. Glycobiology. 17:345–354. [DOI] [PubMed] [Google Scholar]
- Kotake T, Yamanashi Y, Imaizumi C, Tsumuraya Y. 2016. Metabolism of L-arabinose in plants. J Plant Res. 129:781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani A, Tsuji M, Azama Y, Ishii T, Takeda T, Yamashita T, Shimojima M, Konishi T. 2013. Purification and characterization of UDP-arabinopyranose mutase from Chlamydomonas reinhardtii. Biosci Biotechnol Biochem. 77:1874–1878. [DOI] [PubMed] [Google Scholar]
- Kuttiyatveetil JR, Sanders DA. 2017. Analysis of plant UDP-arabinopyranose mutase (UAM): Role of divalent metals and structure prediction. Biochim Biophys Acta. 1865:510–519. [DOI] [PubMed] [Google Scholar]
- Lairson L, Henrissat B, Davies G, Withers S. 2008. Glycosyltransferases: Structures, functions, and mechanisms. Annu Rev Biochem. 77:521–555. [DOI] [PubMed] [Google Scholar]
- Langeveld SM, Vennik M, Kottenhagen M, van Wijk R, Buijk A, Kijne JW, de Pater S. 2002. Glucosylation activity and complex formation of two classes of reversibly glycosylated polypeptides. Plant Pathol. 129:278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2013. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42:D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister CA, Ortiz-Ramírez C, Becker JD, Feijó JA, Lippman ZB. 2016. Hydroxyproline O-arabinosyltransferase mutants oppositely alter tip growth in Arabidopsis thaliana and Physcomitrella patens. Plant J. 85:193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohnen D. 2008. Pectin structure and biosynthesis. Curr Opin Plant Biol. 11:266–277. [DOI] [PubMed] [Google Scholar]
- Møller SR, Yi X, Velásquez SM, Gille S, Hansen PLM, Poulsen CP, Olsen CE, Rejzek M, Parsons H, Yang Z. 2017. Identification and evolution of a plant cell wall specific glycoprotein glycosyl transferase, ExAD. Sci Rep. 7: 45341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndeh D, Rogowski A, Cartmell A, Luis AS, Baslé A, Gray J, Venditto I, Briggs J, Zhang X, Labourel A. 2017. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature. 544:65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. 2009. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol. 5:578–580. [DOI] [PubMed] [Google Scholar]
- Quentmeier H, Ingold E, Seitz HU. 1987. Purification of an autocatalytic protein-glycosylating enzyme from cell suspensions of Daucus carota L. Planta. 171:483–488. [DOI] [PubMed] [Google Scholar]
- Rautengarten C, Birdseye D, Pattathil S, McFarlane HE, Saez-Aguayo S, Orellana A, Persson S, Hahn MG, Scheller HV, Heazlewood JL. 2017. The elaborate route for UDP-arabinose delivery into the Golgi of plants. Proc Natl Acad Sci USA. 114:4261–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Ebert B, Herter T, Petzold CJ, Ishii T, Mukhopadhyay A, Usadel B, Scheller HV. 2011. The interconversion of UDP-arabinopyranose and UDP-arabinofuranose is indispensable for plant development in Arabidopsis. Plant Cell. 23:1373–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennie EA, Scheller HV. 2014. Xylan biosynthesis. Curr Opin Biotechnol. 26:100–107. [DOI] [PubMed] [Google Scholar]
- Sagi G, Katz A, Guenoune-Gelbart D, Epel BL. 2005. Class 1 reversibly glycosylated polypeptides are plasmodesmal-associated proteins delivered to plasmodesmata via the Golgi apparatus. Plant Cell. 17:1788–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selth LA, Dogra SC, Rasheed MS, Randles JW, Rezaian MA. 2006. Identification and characterization of a host reversibly glycosylated peptide that interacts with the tomato leaf curl virus V1 protein. Plant Mol Biol. 61:297–310. [DOI] [PubMed] [Google Scholar]
- Singh DG, Lomako J, Lomako WM, Whelan WJ, Meyer HE, Serwe M, Metzger JW. 1995. β-Glucosylarginine: A new glucose–protein bond in a self-glucosylating protein from sweet corn. FEBS Lett. 376:61–64. [DOI] [PubMed] [Google Scholar]
- Sumiyoshi M, Inamura T, Nakamura A, Aohara T, Ishii T, Satoh S, Iwai H. 2014. UDP-arabinopyranose mutase 3 is required for pollen wall morphogenesis in rice (Oryza sativa). Plant Cell Physiol. 56:232–241. [DOI] [PubMed] [Google Scholar]
- Tanner JJ, Boechi L, McCammon JA, Sobrado P. 2014. Structure, mechanism, and dynamics of UDP-galactopyranose mutase. Arch Biochem Biophys. 544:128–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testasecca P, Wald FA, Cozzarín ME, Moreno S. 2004. Regulation of self-glycosylation of reversibly glycosylated polypeptides from Solanum tuberosum. Plant Physiol. 121:27–34. [DOI] [PubMed] [Google Scholar]
- Welner DH, Shin D, Tomaleri GP, DeGiovanni AM, Tsai AY-L, Tran HM, Hansen SF, Green DT, Scheller HV, Adams PD. 2017a. Plant cell wall glycosyltransferases: High-throughput recombinant expression screening and general requirements for these challenging enzymes. PLoS One. 12:e0177591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welner DH, Tsai AY-L, DeGiovanni AM, Scheller HV, Adams PD. 2017b. X-ray diffraction analysis and in vitro characterization of the UAM2 protein from Oryza sativa. Acta Crystallogr F Struct Biol Commun. 73:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JD, Smith J, Mazarei M, Zhang J, Turner G, Decker SR, Sykes R, Poovaiah C, Baxter H, Mann D. 2016. Downregulation of the UDP-arabinomutase gene in switchgrass (Panicum virgatum L.) results in increased cell wall lignin while reducing arabinose-glycans. Front Plant Sci. 7:1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A-M, Ling C, Liu J-Y. 2006. Isolation of a cotton reversibly glycosylated polypeptide (GhRGP1) promoter and its expression activity in transgenic tobacco. J Plant Physiol. 163:426–435. [DOI] [PubMed] [Google Scholar]
- Zavaliev R, Sagi G, Gera A, Epel BL. 2009. The constitutive expression of Arabidopsis plasmodesmal-associated class 1 reversibly glycosylated polypeptide impairs plant development and virus spread. J Exp Bot. 61:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Wu X, Qiu B, Wu F, Jiang L, Zhang G. 2014. Physiological and proteomic alterations in rice (Oryza sativa L.) seedlings under hexavalent chromium stress. Planta. 240:291–308. [DOI] [PubMed] [Google Scholar]
- Zeng W, Jiang N, Nadella R, Killen TL, Nadella V, Faik A. 2010. A glucurono (arabino) xylan synthase complex from wheat contains members of the GT43, GT47, and GT75 families and functions cooperatively. Plant Pathol. 154:78–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G-R, Liu J-Y, Du X-M. 2001. Molecular cloning and characterization of cotton cDNAs expressed in developing fiber cells. Biosci Biotechnol Biochem. 65:2789–2793. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


