Abstract
Arbuscular mycorrhizal (AM) fungi show high promiscuity in terms of host. Effector proteins expressed by AM fungi are found important in establishing interaction with host. However, the mechanistic underlying host-specific interactions of the fungi remain unknown. The present study aimed (i) to identify effectors encoded by Rhizophagus proliferus and (ii) to understand molecular specificity encoded in effectors for interaction with specific plant species. The effectors predicted from the whole genome sequence were annotated by homology search in NCBI non-redundant protein, Interproscan, and pathogen-host interaction (PHI) databases. In total, 416 small secreted peptides (SSPs) were predicted, which were effector peptides with presence of nuclear localization signal, small cysteine-rich, and repeat-containing proteins domains. Similar to the functionally validated SP7 effectors in Rhizophagus irregularis, two proteins (RP8598 and RP23081) were identified in R. proliferus. To understand whether interaction between SP7 and the plant target protein, ERF19, is specific in nature, we examined protein-peptide interaction using in silico molecular docking. Pairwise interaction of RP8598 and RP23081 with the ethylene-responsive factors (ERF19) coded by five different plant species (Lotus japonicus, Solanum lycopersicum, Ocimum tenuiflorum, Medicago truncatula, Diospyros kaki) was investigated. Prediction of high-quality interaction of SP7 effector with ERF19 protein expressed only by specific plant species was observed in in silico molecular docking, which may reiterate the role of effectors in host specificity. The outcomes from our study indicated that sequence precision encoded in the effector peptides of AM fungi and immunomodulatory proteins of host may regulate host specificity in these fungi.
Electronic supplementary material
The online version of this article (10.1007/s42770-019-00099-x) contains supplementary material, which is available to authorized users.
Keywords: Arbuscular mycorrhizal fungi (AM fungi), Effector proteins, AM-plant interaction, SP7, ERF19, Protein-peptide molecular docking
Introduction
Arbuscular mycorrhiza (AM) is a widespread symbiosis between fungi belonging to Glomeromycota and a large diversity of terrestrial plant species. AM fungi play a crucial role in the adaptation of > 80% of land plants to different environmental conditions [1]. Under natural environments, the AM fungi are involved in structuring terrestrial ecosystems, from grasslands to forests and deserts [2]. In agro-ecosystems, these fungi are crucial in plant nutrition (especially in acquiring phosphorus), mitigation of abiotic and biotic stresses, and carbon sequestration [3]. Experimental evidences confirm that each species of AM fungi can be hosted by a large spectrum of plant species [4]. Molecular cross talks originating from fungal and plant partners have been reported to regulate interaction between AM fungi and plants [5]. A high degree of genetic and/or metabolic coordination between both the partners has been suggested in establishment of the mycorrhizal association. Activation of the plant immune responses seems to occur both locally and systemically when AM fungi first come into contact with a host plant root [6]. Production of strigolactone by plant roots has been suggested as the first stimulus from plants towards the initiation of endosymbiotic interaction [7]. Functions of plants expressed signaling molecules during interaction with AM fungi are being widely explored and their roles have been well-elucidated [5–10]. However, the molecular signals relayed by AM fungi to establish association with enormously large variety of plants remains yet to be completely deciphered. In an initial study, a small secreted protein, SP7, coded by Rhizophagus irregularis was found to translocate in plant nucleus and interact with the pathogenesis-related transcription factor (ERF19) in Medicago truncatula [11]. This study suggested the role of AM effector in prohibiting the plant defense mechanism in favor of fungal entry into plant cells. Upregulation of another putative secreted protein, strigolactone-induced putative secreted protein 1 (SIS1) in R. irregularis, was demonstrated to be important in host colonization by knock-out studies [12]. In an investigation for identifying differential responses of host and non-host to AM fungi, receptors with lysin motif domain (LysM) were found to over-express in AM-host roots [13]. RiLySM, lysin motif domain containing effector from R. irregularis, was found to bind chitin oligosaccharides during root colonization. This suggested to have a role in downregulation of plant defense responses [14]. Downregulation of RiCRN1, a crinkler (CRN) effector protein of R. irregularis, has been found responsible for impairment of the symbiosis in M. truncatula [15]. In addition, the recently available genome and secretome studies [16–19] have proposed that the AM fungi express a smaller number of genes for glycosyl hydrolases in comparison with pathogenic fungi. A lower number of genes encoding glycosyl hydrolases have also been reported in other mycorrhizal fungi [20, 21], which could be a strategy for downregulation of DAMPs (damage-associated molecular patterns) expression, in order to remain largely innocuous and thus undetected by plants during mycorrhizal infection. The next-generation genome and transcriptome sequencing studies have also ascertained presence/expression of a large variety of effector proteins in AM fungi. Effectors efficiently regulate plant defense mechanism in favor of intracellular colonization of AM fungi in a broad range of host plants [12, 16, 17]. Effectors play important roles in many other plant-associated organisms, including microbes, nematodes, and insects. This topic has been very well reviewed by Mesarich et al. [22]. Many plant-associated organisms deliver effector proteins into the plant cells to achieve colonization similar to fungal plant pathogens; interaction of AM fungi expressed effector peptides with immunoresponsive molecule of host has been suggested in inhibition/alteration of defense responses against microbial associated molecular patterns (MAMPs) [12, 13, 16], which may facilitate mycorrhization inside host.
Of the several hundred predicted SSPs and effector proteins from the genome sequencing projects of AM fungi, only a few are functionally validated for their role in interaction with host. The mechanism adapted for regulation of plant immune response by majority of the identified effector proteins remains unknown. A comparison between SSPs encoded by R. irregularis and R. clarus showed that a large proportion of the effector proteins were remarkably conserved between the two species and were proposed to play a common fundamental role during establishment of fungus-plant interaction [18]. A subsequent comparison between distantly related species of AM fungi, R. irregularis and Gigaspora rosea, reported that the two species shared only a small set of conserved SSPs, which were commonly upregulated to establish mycorrhization with hosts [23]. Examination for expression of host-specific classes of SSPs among the two fungi revealed a lower fraction of host-specific secreted proteins in R. irregularis in comparison with G. rosea.
Several questions remain unresolved regarding the molecular mechanisms underlying AM fungi-plant interaction. Available studies suggest limited host specificity and varying symbiotic efficiencies during interaction between AM fungi and host. Recently, using RNA-seq datasets [24] validated 338 genes that encoded putative secreted proteins in R. irregularis. An investigation carried out to unravel general and host-dependent expression of the effectors found that a majority of these peptides (n = 254) were commonly expressed among three evolutionary distant plant hosts, Medicago truncatula, Nicotiana benthamiana, and Allium schoenoprasum. However, expression of a set of effectors (n = 42) showed significant differential expression between the different plant species. With these findings, the study proposed that the fungal SSPs may act as effectors to control symbiotic efficiency in a host-dependent manner.
Our study presents prediction for SSPs and effector peptides encoded by Rhizophagus proliferus. Limited information on genetic composition and secretome/effectome expressed by this important species of AM fungi is present in the public domain. Such unavailability of genetic information gave us impetus to carry out investigation of the secretome of R. proliferus. Also, the mechanism underlying effector protein-mediated change in immunomodulatory response in host is largely unknown in arbuscular mycorrhiza. The key goal of the study, therefore, was to carry out functional annotation of the effector peptides encoded by Rhizophagus proliferus and investigate their role in interaction with different host plants, which may provide clue for molecular factors responsible for host range and host specificity in AM fungi.
Methods
Fungal isolate and genomic information
An isolate of Rhizophagus proliferus (AM-1901) from the Centre for Mycorrhizal Culture Collection (CMCC), TERI, India, was used in this study. Spores were produced in monoxenic cultures that were maintained on Agrobacterium rhizogenes–transformed roots of carrot. High molecular weight (HMW) genomic DNA was extracted using cetyltrimethylammonium bromide (CTAB) method. For whole genome sequencing (reported elsewhere), genomic library was constructed and paired-end (2 × 150 bp) sequencing was performed using the services of a commercial service provider (AgriGenome Labs Pvt. Ltd., Kerala, India) on a HiSeq 2500 sequencing platform. Good-quality sequencing reads were used to create de novo assembly by SOAPdenovo (http://soapdenovo2.sourceforge.net/). Repeat masking was performed using repeatmasker (http://www.repeatmasker.org/RepeatModeler/) before structural annotation. Gene prediction and annotation was performed using AUGUSTUS 3.1.0 with the gene model of Saccharomyces cerevisiae [25]. The predicted gene models were annotated by homology search using Diamond (v0.9.3.104) (https://ab.inf.uni-tuebingen.de/software/diamond) with uniprot, NCBI Protein, KEGG, InterPro, Pfam, and Gene Ontology databases.
Prediction of effector proteins
Previously reported methods [18, 19] were employed to identify the putative effector candidates coded by R. proliferus. Briefly, the secreted proteins predicted by SignalP 4.0 [26] were considered for investigation. Proteins with predicted transmembrane domains (TMDs) and Prosite motif “PS00014” (KDEL) were excluded [27], except for those proteins in which predicted TMDs overlapped with sequence of signal peptides (D-score > 0.450). Proteins were then subjected to target prediction by the TargetP 1.1 tool [28]. Candidate proteins of length between 30 and 150 amino acids and starting with methionine amino acid were further probed and grouped into nuclear localization signal (NLS)–containing proteins, small cysteine-rich (SCR) proteins, and tandem repeat–containing proteins (RCP). The predicted effector proteins were annotated by subjecting to homology search in various databases including NCBI non-redundant protein, Interproscan, and pathogen-host interaction (PHI) db.
In silico protein-peptide interaction by molecular docking
Protein modeling was done for plant-coded ERF19 and SP7 effector of R. proliferus by I-Tasser web interface [29]. Protein-peptide docking was performed using the CABS-dock web server [30, 31], which utilized Replica Exchange Monte Carlo dynamics for flexible protein-peptide docking without previous knowledge about the binding site. The CABS-dock simulation method, based on the coarse-grained CABS model, enabled docking search of fully flexible peptides over the entire surface of flexible proteins.
Comparative genomics analysis
A comparative analysis using information from secretomes of R. irregularis and R. clarus was carried out to identify commonly expressed and uniquely coded sets of effector proteins in R. proliferus. Unique effectors of R. proliferus were annotated by homology search in pathogen-host interaction (PHI), NCBI non-redundant protein, and Interproscan (IPR). Two SP7 proteins (RP8598 and RP23081), identified based on significant homology by blastp (1e-5) to previously characterized SP7 proteins encoded by R. irregularis [11], were tested for their role in conferring host specificity to AM fungi. For this, in silico protein structure modeling and protein-peptide docking predictions were carried out. For molecular docking analysis, the ethylene-responsive factors (ERF) expressed by five different plant species (Lotus japonicus, Solanum lycopersicum, Ocimum tenuiflorum, Medicago truncatula, Diospyros kaki) were modeled and used in a pairwise combination to study interaction with the two SP7 proteins encoded by R. proliferus.
Results
A total of 663 proteins with presence of N-terminal signal peptides and with no transmembrane domain were predicted from the genomic assembly of R. proliferus. A total of 416 of 663 proteins were strongly predicted to be located in the secretory pathway (parameter LOC = S and reliability class = 1). We have deposited the sequence of these 416 secretory peptides in GenBank (https://www.ncbi.nlm.nih.gov/genbank/submission ID is 2224587). Of these, 171, 47, and 19 SSPs could be grouped as effector proteins with NLS, RCP, and SCR domains respectively (table S1).
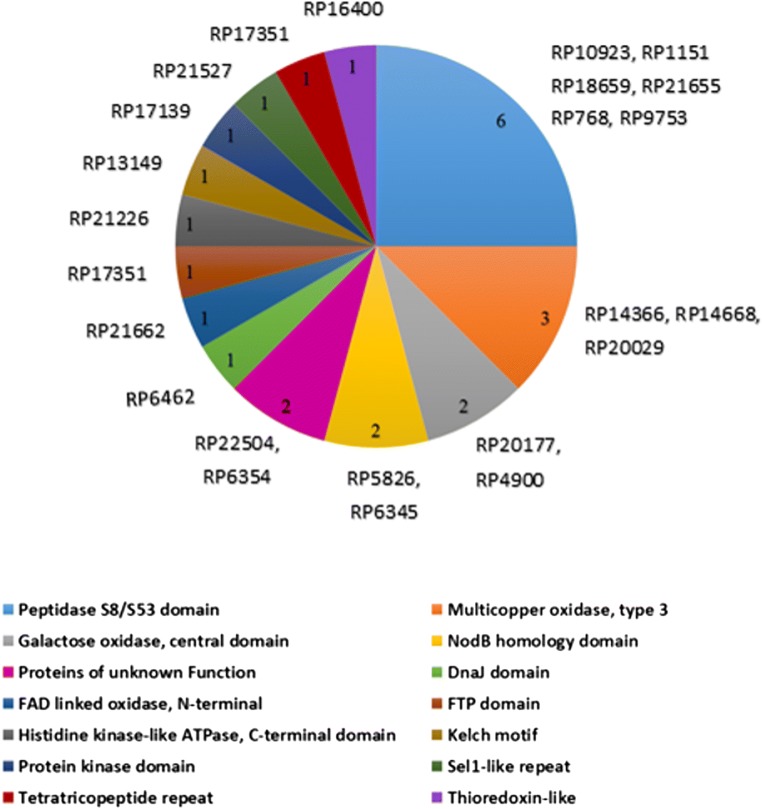
A blastp cut-off of (1e-5) was applied for all sequence-homology searches. A group of 22 SPs were previously identified as specific to AM fungi and found to be present only in R. irregularis but not in pathogens [17]. In our analysis, five of these 22 SPs were also encoded in R. proliferus genome, indicating their conserved functions in AM symbiosis (Fig. 1). A total of n = 92 (23%) and n = 84 (20%) effectors encoded by R. proliferus demonstrated significant sequence homology with the effectors of R. irregularis (n = 338) [24] and R. clarus (n = 218) [18] respectively (table S2). A total of 76 effector proteins (Fig. 2, table S2) were commonly shared between all three species of Rhizophagus included for comparison in our study. Approximately 31% (n = 24/76) of the commonly shared effector candidates between all the three species of Rhizophagus had homologs present in the PHI database (Fig. 3, table S3). Further, information present in the PHI db revealed that mutations in the homologs of n = 18/24 conserved effectors have been experimentally validated to affect virulence of microbes. Functions for the remaining 317 effector proteins that were uniquely coded by R. proliferus (when compared with the secreted protein sets of R. irregularis and R. clarus) were inferred by Interproscan analysis (table S4). Twenty-two out of 24 effectors could be functionally annotated. Of these, 15 effectors (Fig. 3, table S3) presented functions appropriate for microbial-host interactions: five effectors were S8/S53 domain-containing peptidases, three were laccases, one contained protein kinase domain, and another protein had a fungalysin-thermolysin-propeptide (FTP) domain, and the other was a histidine kinase–like ATPase. A solenoid protein with Sel1-like repeat was also found; two effector proteins contained NodB homology domain.
Fig. 1.
R. proliferus effectors sharing homology with five proteins that belong to the set of 22 AMF protein tribe in R. irregularis. RP indicates effectors of R. proliferus and Rir indicates effectors of R. irregularis
Fig. 2.
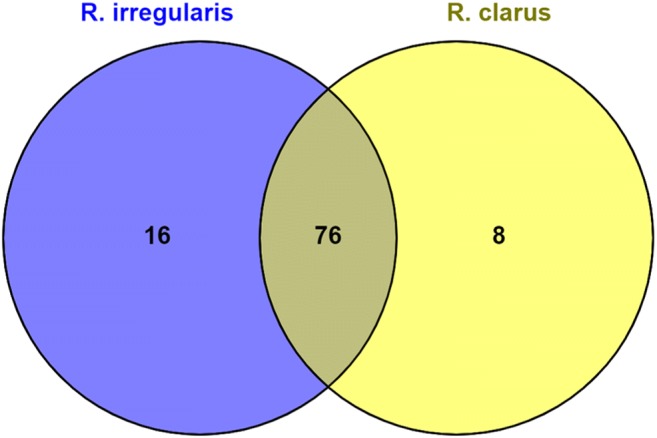
Homologous effector proteins among R. proliferus, R. irregularis, and R. clarus. Venn diagram shows homologs for effectors identified in R. proliferus among R. irregularis (marked in blue) and R. clarus (marked in yellow) and region marked in gray indicates common effectors among all the three species
Fig. 3.
Functions of the 15 functionally conserved effectors
In comparison with the previously reported functionally validated effectors reported in other AM fungi, two LysM (RP7182, RP23885; figure S1), one SIS1 (RP5293; figure S1), and two SP7 (RP23081 and RP8598; Fig. 4) effectors were identified in R. proliferus. Multiple alignment analysis of SP7 coded by R. proliferus (RP8598 and RP23081) and R. irregularis (AEK82120.1, AEK82121.1, and AEK8212.1) revealed conservation in amino acid sequence within the repeat motifs and presence of the conserved Kex2 cleavage site [23, 32]. Due to the presence of Kex2 protease cleavage sites, the two SP7 effectors are expected to be cleaved and secreted as short peptides [23].
Fig. 4.
Multiple sequence alignment of SP7 proteins coded by R. proliferus and R. irregularis genomes. RP23081 and RP8598: R. proliferus-coded SP7 proteins; AEK82120.1, AEK82121.1, and AEK8212.1: R. irregularis-coded SP7 proteins; green color represents a conserved region, red boxes indicate Kex2 cleavage sites, and residues in yellow color indicate the presence of tandem repeats
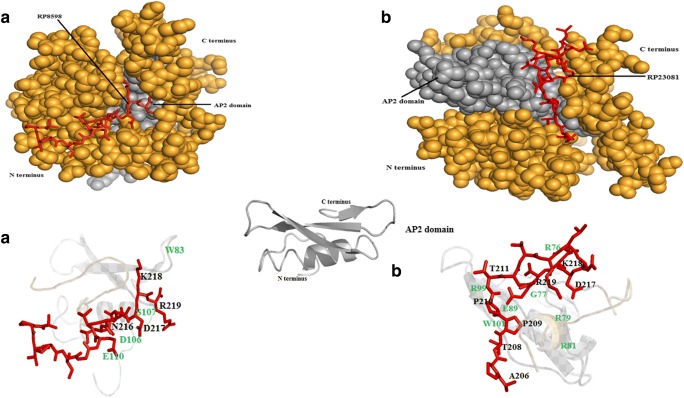
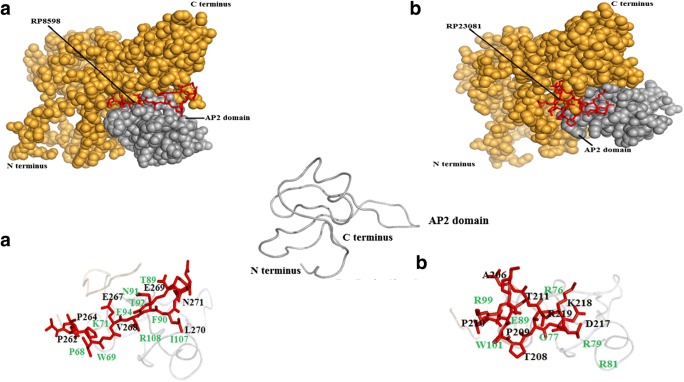
In order to achieve better understanding on host specificity inscribed in the AM effector sequences, SP7 effectors were investigated for their probability to specifically interact with plant-encoded ERF19 proteins. We investigated the possibility of interaction between the NLS domain–containing short peptide (p14), which is produced by cleavage of SP7 effectors at the first Kex2 site from the N-terminus, with the ERF19 proteins using an in silico molecular docking approach. Outcomes of molecular docking analysis are presented in table S5. ERF19 proteins coded by five plant species (Lotus japonicus, Solanum lycopersicum, Ocimum tenuiflorum, Medicago truncatula, Diospyros kaki) were investigated for interaction possibilities with two SP7 effectors coded by R. proliferus. Inference on significant interaction between SP7 effector and ERF19 protein was based on RMSD and cluster density values. High probabilities for specific interactions for SP7-RP8598 (p14) with the AP2 domain of the ERF19 proteins of three plant species, namely, L. japonicus, S. lycopersicum, and D. kaki were predicted. However, RP23081 (p14) showed high-quality prediction with ERF19 protein coded by two plant species only, i.e., L. japonicus and S. lycopersicum (Fig. 5, table S5). Amino acids of SP7 coded by R. proliferus and ERF19 proteins expressed by different hosts that participated in significant interactions are depicted in Fig. 5.
Fig. 5.
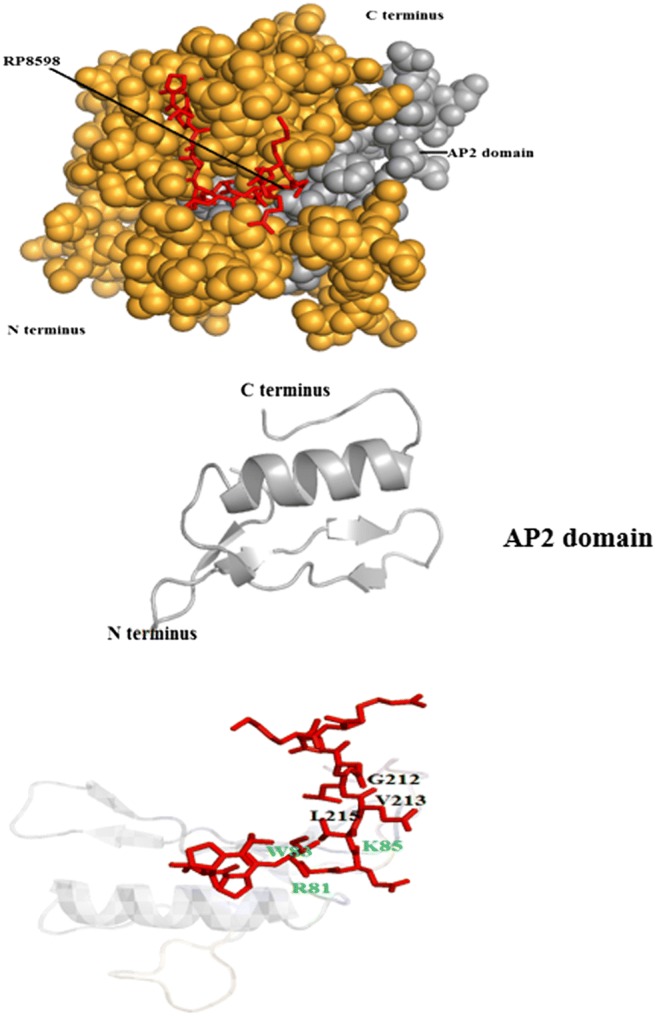
In silico molecular docking analysis. (i) Interaction between SP7 (a RP8598; b RP23081) and ERF19 of L. japonicus; (ii) interaction between SP7 (a RP8598; b RP23081) and ERF19 of S. lycopersicum; (iii) interaction between SP7 (RP8598) and D. kaki. Amino acids of the SP7 (p14) present at the binding site are marked in black color and the interacting amino acids of the AP2 domain of ERF19 are marked in green color
Discussion
AM symbiosis is initiated by signal exchange between plant roots and germinating fungal spores, which triggers coherent differentiation of both partners to enable their interaction. In order to short-circuit the host defense responses or gain stealth entry into host by deceiving its molecular mechanisms, AM fungi express effector proteins [11, 12, 14–19]. AM fungi exhibit an extremely broad host range and expression of combinations of conserved and unique effectors in a species-specific manner is speculated to regulate host range [18, 19, 24].
Our comparative genomic analysis revealed that the effectome of R. proliferus is similar in abundance (n = 416) to G. rosea (n = 441) and R. irregularis (n = 566). Common effector peptides (table S5) identified between all three species of Rhizophagus (R. proliferus, R. irregularis, and R. clarus) could be involved in conserved and basic functions required for establishment of AM-plant interaction. The presence of homologs of 31% of the common effector peptides in the PHI database further underscored importance of these effectors in AM fungi-host interactions (table S3). The PHI db catalogs virulence and effector genes with functional validation from fungi, oomycetes, and bacterial pathogens of fungi, insect, plant, and animal hosts. Homolog identification in PHI db, therefore, suggests a putative role of the effector protein in the interaction between microbe and host. The role of n = 18/24 conserved effectors seemed more crucial due to availability of experimental validation data in PHI db, which suggested that mutations in these proteins may regulate fungi-plant interaction. In addition, 15/24 effectors with Interproscan annotation (Fig. 3, table S3) presented functions appropriate for microbe-host interaction. S8/S53 domain–containing peptidases function as essential hydrolytic enzymes that utilize the catalytic serine residue for cleaving peptide bonds in proteins. These peptidases are identified in both pathogenic and symbiotic fungi-host interactions to help the fungal partner evade host’s immune system by degrading chitinases that destroy fungal cell wall [32]. Laccases are involved in the degradation of lignin and humic acids, and therefore may play important roles in fungi-plant interactions. Protein kinases are crucial in signal transduction; these proteins help fungi detect and respond to a diversity of stimulus such as the presence of a host and host defense molecules, osmotic or oxidative stress, light, and other environmental cues. The FTP, M36 class of proteins is generally synthesized as pre-propeptides with fairly long prodomains (or propeptides). Studies indicate a role of these proteins in inhibition of mature fungal serine proteinases and metalloproteinases [33]. Histidine kinases (HKs) act as primary sensors for various environmental stimuli which upon activation, initiate phosphate transfer events between proteins towards cell-signaling and adaptive response [34]. Solenoid proteins are important in signal transduction pathways [35]. NodB genes have been proposed to be involved in the synthesis of oligosaccharide signal molecules [36, 37]. Interaction of AM fungi-expressed NodB signal molecules with nucleotide-binding oligomerization domain (NOD)-like receptors of plants activates specific signaling pathways that leads to expression of genes that tailor immune responses.
The presence of the previously reported and experimentally validated effectors in AM fungi, LysM (RP7182, RP23885; figure S1), SIS1 (RP5293; figure S1), and SP7 (RP23081 and RP8598; Fig. 4), in R. proliferus underscores genetic conservation of these effectors. LysM effectors have also been found in many species of pathogenic fungi and have been identified crucial in downregulation of chitin-triggered immune responses [38, 39]. A RiLySM in R. irregularis has been demonstrated to bind chitin and plant immune response [14].
The R. irregularis SP7 effector has been suggested to localize in the plant nucleus, interact with the plant-coded ethylene-responsive factor 19 (ERF19), and regulate the plant immune response [11]. ERF19 is a single AP2 domain–containing ERF protein belonging to the EREBP subfamily, which function in the signal transduction pathways of stress responses and cambial tissue development [39, 40]. Molecular docking analysis carried by us to test if host range is governed by AM effectors, particularly through specificity inscribed in their amino acid sequence, was supported by specific interactions of two SP7 proteins, RP8598 and RP23081, with EFR19 proteins of limited number of host species only. Our findings corroborated previous report [11] that SP7 effector interacts specifically with plant-expressed ERF19 protein, which may regulate plant immune responses during colonization of AM fungi in host root. In addition, in silico prediction of high-quality interaction between SP7 and ERF19 proteins expressed only by specific plant species may reiterate role of effectors in governing host specificity. Our results are also in agreement with a recent finding reported by Zeng et al. (2018) [24] wherein interaction stage–dependent differential expression of R. irregularis-encoded secreted proteins in three evolutionarily distant host species, Medicago truncatula, Nicotiana benthamiana, and Allium schoenoprasum [24], was observed.
Findings in AM fungi further draw support from certain other plant-microbe associations, where in order to overcome molecular recognition hurdles, plant-associated organisms undergo adaptive evolution (due to host-imposed selection pressure) and modify their effector peptides [41–43].
Conclusion
Our study provides a comprehensive analysis of the secretome of AM fungal species R. proliferus. Outcome from our survey may support role of AM fungi–coded effector proteins in conferring host specificity. The consequences hold significance for researchers exploring the molecular mechanism underlying arbuscular mycorrhizal association in particular and other plant-microbe interactions in general.
Electronic supplementary material
RiLySM and RiSIS1 proteins coded by R. proliferus and their homologs identified in R. irregularis (PNG 85 kb)
Effector proteins identified in R. proliferus (DOCX 47 kb)
Homologous effector proteins among R. proliferus, R. irregularis and R. clarus (DOCX 46 kb)
Functions of the 15 functionally conserved effectors in all three species of Rhizophagus and had a homolog in PHI db (DOCX 33 kb)
Annotation of effector proteins unique to R. proliferus (DOCX 65 kb)
Molecular docking interaction between SP7 and ERF19 proteins (DOCX 37 kb)
Acknowledgments
The authors thank Ms. Deepti Varshney and Mr. Aditya Gaur, TERI Deakin Nanobiotechnology Centre, Gwal Pahari, Haryana, India, for software support on high-performance computing cluster required in this study. The authors also thank Ms. Sadhana Shukla and Dr. Reena Singh for maintaining and providing monosporal culture of R. proliferus. Sequencing was performed at Agrigenome Labs, Kochi, and Kerala, India.
Authors’ contributions
All authors have read and approved the final manuscript. PPS was involved in conceptualization of the project, study design, data analyses, data compilation, manuscript writing, critical inputs, and finalization of the manuscript. DS and AJ were involved in genomics data analysis, data compilation, and manuscript writing. AA gave critical comments and reviewed the work.
Funding information
This work was financially supported by the grant-in-aid for research by the Department of Science and Technology (DST), India, under the grant number “EMR/2017/000657”.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Smith SE, Read DJ. Mycorrhizal symbiosis. 3. London: Academic; 2008. [Google Scholar]
- 2.Öpik M, Moora M, Liira J, Zobel M. Composition of root-colonizing arbuscular mycorrhizal fungal communities in different ecosystems around the globe. J Ecol. 2006;94:778–790. doi: 10.1111/j.1365-2745.2006.01136.x. [DOI] [Google Scholar]
- 3.Cheng L, Booker FL, Tu C, Burkey KO, Zhou L, Shew HD, Rufty TW, Hu S. Arbuscular mycorrhizal fungi increase organic carbon decomposition under elevated CO2. Science. 2012;337(6098):1084–1087. doi: 10.1126/science.1224304. [DOI] [PubMed] [Google Scholar]
- 4.Öpik M, Vanatoa A, Vanatoa E, Moora M, Davison J, Kalwij JM, Reier Ü, Zobel M. The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota) New Phytol. 2010;188:223–241. doi: 10.1111/j.1469-8137.2010.03334.x. [DOI] [PubMed] [Google Scholar]
- 5.Gutjahr C, Parniske M. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annu Rev Cell Dev Biol. 2013;29:593–617. doi: 10.1146/annurev-cellbio-101512-122413. [DOI] [PubMed] [Google Scholar]
- 6.Jung SC, Martinez-Medina A, Lopez-Raez JA, Pozo MJ. Mycorrhiza-induced resistance and priming of plant defenses. J Chem Ecol. 2012;38(6):651–664. doi: 10.1007/s10886-012-0134-6. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama K, Matsuzaki K, Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435(7043):824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 8.Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais JC, Roux C, Bécard G, Séjalon-Delmas N. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 2006;4(7):e226. doi: 10.1371/journal.pbio.0040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kretzschmar T, Kohlen W, Sasse J, Borghi L, Schlegel M, Bachelier JB, Reinhardt D, Bours R, Bouwmeester HJ, Martinoia E. A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature. 2012;483(7389):341–344. doi: 10.1038/nature10873. [DOI] [PubMed] [Google Scholar]
- 10.Harrison MJ. Cellular programs for arbuscular mycorrhizal symbiosis. Curr Opin Plant Biol. 2012;15(6):691–698. doi: 10.1016/j.pbi.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Kloppholz S, Kuhn H, Requena N. A secreted fungal effector of Glomus intraradices promotes symbiotic biotrophy. Curr Biol. 2011;21(14):1204–1209. doi: 10.1016/j.cub.2011.06.044. [DOI] [PubMed] [Google Scholar]
- 12.Tsuzuki S, Handa Y, Takeda N, Kawaguchi M. Strigolactone-induced putative secreted protein 1 is required for the establishment of symbiosis by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mol Plant-Microbe Interact. 2016;29(4):277–286. doi: 10.1094/MPMI-10-15-0234-R. [DOI] [PubMed] [Google Scholar]
- 13.Fiorilli V, Vallino M, Biselli C, Faccio A, Bagnaresi P, Bonfante P. Host and non-host roots in rice: cellular and molecular approaches reveal differential responses to arbuscular mycorrhizal fungi. Front Plant Sci. 2015;6:636. doi: 10.3389/fpls.2015.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitz AM, Teresa PE, Harrison MJ. A short LysM protein with high molecular diversity from an arbuscular mycorrhizal fungus, Rhizophagus irregularis. Mycoscience. 2019;60(1):63–70. doi: 10.1016/j.myc.2018.09.002. [DOI] [Google Scholar]
- 15.Voß S, Betz R, Heidt S, Corradi N, Requena N. RiCRN1, a crinkler effector from the arbuscular mycorrhizal fungus Rhizophagus irregularis, functions in arbuscule development. Front Microbiol. 2018;9:2068. doi: 10.3389/fmicb.2018.02068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tisserant E, Malbreil M, Kuo A, Kohler A, Symeonidi A, Balestrini R, Charron P, Duensing N, Frei dit Frey N, Gianinazzi-Pearson V, Gilbert LB, Handa Y, Herr JR, Hijri M, Koul R, Kawaguchi M, Krajinski F, Lammers PJ, Masclaux FG, Murat C, Morin E, Ndikumana S, Pagni M, Petitpierre D, Requena N, Rosikiewicz P, Riley R, Saito K, San Clemente H, Shapiro H, van Tuinen D, Bécard G, Bonfante P, Paszkowski U, Shachar-Hill YY, Tuskan GA, Young JP, Sanders IR, Henrissat B, Rensing SA, Grigoriev IV, Corradi N, Roux C, Martin F (2013) Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc Natl Acad Sci U S A 110(50):2011 7–2011 201122
- 17.Lin K, Limpens E, Zhang Z, Ivanov S, Saunders DG, Mu D, Pang E, Cao H, Cha H, Lin T, Zhou Q, Shang Y, Li Y, Sharma T, van Velzen R, de Ruijter N, Aanen DK, Win J, Kamoun S, Bisseling T, Geurts R, Huang S. Single nucleus genome sequencing reveals high similarity among nuclei of an endomycorrhizal fungus. PLoS Genet. 2014;10(1):e1004078. doi: 10.1371/journal.pgen.1004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sędzielewska Toro K, Brachmann A. The effector candidate repertoire of the arbuscular mycorrhizal fungus Rhizophagus clarus. BMC Genomics. 2016;17(101):101. doi: 10.1186/s12864-016-2422-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang N, San Clemente H, Roy S, Bécard G, Zhao B, Roux C. A survey of the gene repertoire of Gigaspora rosea unravels conserved features among Glomeromycota for obligate biotrophy. Front Microbiol. 2016;7:233. doi: 10.3389/fmicb.2016.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin F, Aerts A, Ahrén D, Brun A, Danchin EG, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V, Salamov A, Shapiro HJ, Wuyts J, Blaudez D, Buée M, Brokstein P, Canbäck B, Cohen D, Courty PE, Coutinho PM, Delaruelle C, Detter JC, Deveau A, DiFazio S, Duplessis S, Fraissinet-Tachet L, Lucic E, Frey-Klett P, Fourrey C, Feussner I, Gay G, Grimwood J, Hoegger PJ, Jain P, Kilaru S, Labbé J, Lin YC, Legué V, Le Tacon F, Marmeisse R, Melayah D, Montanini B, Muratet M, Nehls U, Niculita-Hirzel H, Oudot-Le Secq MP, Peter M, Quesneville H, Rajashekar B, Reich M, Rouhier N, Schmutz J, Yin T, Chalot M, Henrissat B, Kües U, Lucas S, Van de Peer Y, Podila GK, Polle A, Pukkila PJ, Richardson PM, Rouzé P, Sanders IR, Stajich JE, Tunlid A, Tuskan G, Grigoriev IV. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 2008;452:88–92. doi: 10.1038/nature06556. [DOI] [PubMed] [Google Scholar]
- 21.Martin F, Kohler A, Murat C, Balestrini R, Coutinho PM, Jaillon O, Montanini B, Morin E, Noel B, Percudani R, Porcel B, Rubini A, Amicucci A, Amselem J, Anthouard V, Arcioni S, Artiguenave F, Aury JM, Ballario P, Bolchi A, Brenna A, Brun A, Buée M, Cantarel B, Chevalier G, Couloux A, Da Silva C, Denoeud F, Duplessis S, Ghignone S, Hilselberger B, Iotti M, Marçais B, Mello A, Miranda M, Pacioni G, Quesneville H, Riccioni C, Ruotolo R, Splivallo R, Stocchi V, Tisserant E, Viscomi AR, Zambonelli A, Zampieri E, Henrissat B, Lebrun MH, Paolocci F, Bonfante P, Ottonello S, Wincker P. Périgord black truffle genome uncovers evolutionary origins and mechanisms of symbiosis. Nature. 2010;464:1033–1038. doi: 10.1038/nature08867. [DOI] [PubMed] [Google Scholar]
- 22.Mesarich CH, Bowen JK, Hamiaux C, Templeton MD (2015) Repeat-containing protein effectors of plant-associated organisms. Front Plant Sci 6(872) [DOI] [PMC free article] [PubMed]
- 23.Kamel L, Tang N, Malbreil M, San Clemente H, Le Marquer M, Roux C, Frei Dit Frey N. The comparison of expressed candidate secreted proteins from two arbuscular mycorrhizal fungi unravels common and specific molecular tools to invade different host plants. Front Plant Sci. 2017;8:124. doi: 10.3389/fpls.2017.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng T, Holmer R, Hontelez J, Te Lintel-Hekkert B, Marufu L, de Zeeuw T, Wu F, Schijlen E, Bisseling T, Limpens E. Host- and stage-dependent secretome of the arbuscular mycorrhizal fungus Rhizophagus irregularis. Plant J. 2018;94(3):411–425. doi: 10.1111/tpj.13908. [DOI] [PubMed] [Google Scholar]
- 25.Hoff KJ, Stanke M. Predicting genes in single genomes with AUGUSTUS. Curr Protoc Bioinformatics. 2018;22:e57. doi: 10.1002/cpbi.57. [DOI] [PubMed] [Google Scholar]
- 26.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 27.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34(Web Server issue):W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300(4):1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinf. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurcinski M, Jamroz M, Blaszczyk M, Kolinski A, Kmiecik S. CABS-dock web server for the flexible docking of peptides to proteins without prior knowledge of the binding site. Nucleic Acids Res. 2015;43:W419–W424. doi: 10.1093/nar/gkv456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mueller O, Kahmann R, Aguilar G, Trejo-Aguilar B, Wu A, de Vries RP. The secretome of the maize pathogen Ustilago maydis. Fungal Genet Biol. 2008;45:S63–S70. doi: 10.1016/j.fgb.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 32.Sánchez-Vallet A, Mesters JR, Thomma BPHJ. The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol Rev. 2014;39:171–183. doi: 10.1093/femsre/fuu003. [DOI] [PubMed] [Google Scholar]
- 33.Kabbara S, Hérivaux A, de Bernonville TD, Courdavault V, Clastre M, Gastebois A, Osman M, Hamze M, Cock JM, Schaap P, Papon N. Diversity and evolution of sensor histidine kinases in eukaryotes. Genome Biol Evol. 2019;11(1):86–108. doi: 10.1093/gbe/evy213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mittl PR, Schneider-Brachert W. Sel1-like repeat proteins in signal transduction. Cell Signal. 2007;19(1):20–31. doi: 10.1016/j.cellsig.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 35.Franchi L, Warner N, Viani K, Nuñez G. Function of Nod-like receptors in microbial recognition and host defense. Immunol Rev. 2009;227(1):106–128. doi: 10.1111/j.1600-065X.2008.00734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt J, Röhrig H, John M, Wieneke U, Stacey G, Koncz C, Schell J. Alteration of plant growth and development by Rhizobium nodA and nodB genes involved in the synthesis of oligosaccharide signal molecules. Plant J. 1993;4(4):651–658. doi: 10.1046/j.1365-313X.1993.04040651.x. [DOI] [Google Scholar]
- 37.Langner T, Göhre V. Fungal chitinases: function, regulation, and potential roles in plant/pathogen interactions. Curr Genet. 2016;62:243–254. doi: 10.1007/s00294-015-0530-x. [DOI] [PubMed] [Google Scholar]
- 38.Markaryan A, Lee JD, Sirakova TD, Kolattukudy PE. Specific inhibition of mature fungal serine proteinases and metalloproteinases by their propeptides. J Bacteriol. 1996;178(8):2211–2215. doi: 10.1128/jb.178.8.2211-2215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819(2):86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Licausi F, Ohme-Takagi M, Perata P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 2013;199:639–649. doi: 10.1111/nph.12291. [DOI] [PubMed] [Google Scholar]
- 41.Stergiopoulos I, De Kock MJ, Lindhout P, De Wit PJ. Allelic variation in the effector genes of the tomato pathogen Cladosporium fulvum reveals different modes of adaptive evolution. Mol Plant-Microbe Interact. 2007;20:1271–1283. doi: 10.1094/MPMI-20-10-1271. [DOI] [PubMed] [Google Scholar]
- 42.Win J, Morgan w BJ, Ksenia VK, Cano LM, Chaparro-Garcia A, Ammar R, Staskawicz JB, Kamoun S. Adaptive evolution has targeted the C-terminal domain of the RXLR effectors of plant pathogenic oomycetes. Plant Cell. 2007;19:2349–2369. doi: 10.1105/tpc.107.051037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong S, Stam R, Cano LM, Song J, Sklenar J, Yoshida K, Bozkurt TO. Oliva R, Liu Z, Tian M, Win J, Banfield MJ, Jones AM, van der Hoorn RA, Kamoun S. Effector specialization in a lineage of the Irish potato famine pathogen. Science. 2014;343:552–555. doi: 10.1126/science.1246300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RiLySM and RiSIS1 proteins coded by R. proliferus and their homologs identified in R. irregularis (PNG 85 kb)
Effector proteins identified in R. proliferus (DOCX 47 kb)
Homologous effector proteins among R. proliferus, R. irregularis and R. clarus (DOCX 46 kb)
Functions of the 15 functionally conserved effectors in all three species of Rhizophagus and had a homolog in PHI db (DOCX 33 kb)
Annotation of effector proteins unique to R. proliferus (DOCX 65 kb)
Molecular docking interaction between SP7 and ERF19 proteins (DOCX 37 kb)