Abstract
Atypical enteropathogenic (serotypes O4:H16, O8:H25, O68:H2, O105:H7, and OR:H25) and Shigatoxigenic (ONT:H46) Escherichia coli were isolated from samples of ground beef and poultry breast purchased in Botucatu, Brazil. Phenotypic and molecular characterization indicated the potential of these isolates to adhere to host epithelial cells and cause damage.
Electronic supplementary material
The online version of this article (10.1007/s42770-019-00101-6) contains supplementary material, which is available to authorized users.
Keywords: EPEC, STEC, Pathogenesis, Genotyping, Ground beef, Poultry breast and diarrhea
On the basis of virulence features, diarrheagenic Escherichia coli (DEC) are divided into six distinct pathotypes: enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), Shiga toxin-producing E. coli (STEC), enteropathogenic E. coli (EPEC), and diffusely adherent E. coli (DAEC) [1]. EPEC is sub-grouped into typical (tEPEC) and atypical (aEPEC) according to the occurrence of the virulence plasmid pEAF (EPEC adherence factor plasmid) only in tEPEC [2, 3]. pEAF harbors the bfp operon, which encodes the bundle-forming pilus (BFP); BFP meditates the localized adherence (LA) pattern, characterized by the presence of compact bacterial microcolonies on the epithelial cell surface [4–6].
STEC is characterized by the production of potent cytotoxins, Stx1 and/or Stx2, which inhibit protein synthesis in eukaryotic cells [7]. In the early 1980s, STEC O157:H7 was implicated as the cause of two different outbreaks of hemorrhagic colitis [8]. However, it has become evident that non-O157 STEC, particularly those belonging to serogroups O26, O45, O103, O111, O121, and O145, can cause similar illnesses as STEC O157:H7 [9]. A large outbreak of gastroenteritis and HUS (hemolytic uremic syndrome), caused by a hybrid EAEC/STEC (serotype O104:H4), occurred in Germany, during 3 months of 2011. In this outbreak, 3816 patients were reported with diarrhea, of which 845 (22%) developed HUS. In addition, 54 deaths were registered, thus indicating the high virulence of this hybrid EAEC/STEC pathogen [10, 11].
EPEC and some STEC isolates, known as enterohemorrhagic E. coli (EHEC), can also produce attaching and effacing (AE) lesions, which are characterized by intimate bacterial attachment, localized destruction of brush border microvilli, and accumulation of F-actin filaments underneath adherent bacteria resulting in pedestal-like structures on the surface of infected epithelial cells [1]. The proteins necessary for AE lesion formation are encoded by genes located in a 35.5-kb chromosomal pathogenicity island (PAI) termed locus of enterocyte effacement (LEE) [12]. The eae (E. coli attaching and effacing) gene, located within the LEE region, encodes an adhesive protein known as intimin, which is required for intimate adhesion and cytoskeletal reorganization in AE lesion formation [13].
In this study, we checked for EPEC and STEC pathotypes in 203 food samples (100 ground beef and 103 poultry breast) purchased in Botucatu, São Paulo State, Brazil. Additionally, the EPEC and STEC isolates were phenotypically and molecularly characterized, mainly emphasizing their ability to interact with host epithelial cells and cause damage.
We analyzed 100 samples of ground beef (bought from May 2013 to April 2014) and 103 samples of poultry breast (33 samples bought in 2013 and 70 samples from January 2017 to May 2017). The samples were bought from 56 different retailers located in Botucatu, São Paulo State, Brazil, and were transported to the laboratory, within 2 h after being acquired, in isothermal refrigerated boxes. In the laboratory, 25 g of the samples were homogenized with 225 ml of buffered peptone water (1%), using a Stomacher Lab Blender 400 for 30 s, and incubated for approximately 18 h at 37 °C. One loopful of the culture was streaked onto Chromocult Coliform Agar (Merck, Darmstadt, Germany) and incubated for 18 h at 37 °C. After incubation, five to ten well-isolated colonies, presumably E. coli due to taking on dark blue color, were selected and confirmed to be E. coli by using standard biochemical techniques [14, 15].
A total of 929 E. coli isolates were obtained from 86.7% of the food samples analyzed (Table 1). On the basis of the detection of the virulence factor-encoding genes used for EPEC (eae and bfpB) and STEC (eae, stx1, and stx2) identification, performed as previously described [16], six isolates were classified as aEPEC and one as STEC (Table 2). Of note, none of the 929 E. coli isolates harbored the aatA and/or aggR genes, thus indicating the absence of the EAEC pathotype among the meat samples analyzed (data not shown). The isolation of STEC and aEPEC from meat products observed in this study corroborates reports of previous studies performed in Brazilian cities, including São Paulo, Rio de Janeiro, Ribeirão Preto, Campinas, and São José do Rio Preto [17–20]. Regarding the classification of the E. coli isolates in the different phylo-groups, using a quadruplex PCR method [21], we found that five aEPEC isolates were assigned to phylo-group A, while the aEPEC serotype O105:H7 and STEC ONT:H46 were assigned to phylo-group B1 (Table 2).
Table 1.
Number of food samples analyzed and diarrheagenic Escherichia coli (DEC) isolates obtained in the present study
| Food sample | No. of food samples positive for: | No. of isolates obtained: | ||
|---|---|---|---|---|
| E. coli (%) | DECa (%) | E. coli | DECa | |
| Ground beef (n = 100) | 76 (76.0) | 3 (3.0) | 471 | 3 |
| Poultry breast (n = 103) | 100 (97.1) | 3 (2.9) | 458 | 4b |
| Total (n = 203) | 176 (86.7) | 6 (3.0) | 929 | 7 |
aDiarrheagenic Escherichia coli
bTwo DEC isolates (aEPEC of serotype O8:H25) were obtained from the same poultry breast sample
Table 2.
Phenotypic and genetic features of atypical enteropathogenic (aEPEC) and Shiga toxin-producing (STEC) Escherichia coli isolates obtained from ground beef and poultry breast purchased in Botucatu, São Paulo, Brazil
| Origin | Food sample | E. coli isolates | DEC pathotypea | Serotypeb | DEC virulence markers | Additional virulence factor-encoding genesd | HeLa cells | Stxg | Phylo-group | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| eae (intimin subtype)c | bfpB | stx1 | stx2 | Adherence patterne | FASf | ||||||||
| Ground beef | 11 | 11–1 | aEPEC | O105:H7 | + (θ) | – | – | – | nleB, nleE | UND | + | NT | B1 |
| 18 | 18–5 | aEPEC | O4:H16 | + (β1) | – | – | – | – | LAL | + | NT | A | |
| 20 | 20–1 | STEC | ONT:H46 | – | – | – | + | iha, saa, ehxA, espP | UND | – | + | B1 | |
| Poultry breast | 28 | 28–3 | aEPEC | O68:H12 | + (β1) | – | – | – | – | UND | + | NT | A |
| 45 | 45–3 | aEPEC | OR:H25 | + (NT) | – | – | – | – | UND | + | NT | A | |
| 47 | 47–1 | aEPEC | O8:H25 | + (NT) | – | – | – | – | UND | + | NT | A | |
| 47–3 | aEPEC | O8:H25 | + (NT) | – | – | – | – | UND | + | NT | A | ||
aDEC: diarrheagenic E. coli; aEPEC: atypical enteropathogenic E. coli; STEC: Shiga toxin-producing E. coli
bONT: somatic antigen O non-typeable with the antisera used (O1-O188); OR: rough isolates
cIntimin subtypes: β1: beta1, θ: theta, NT: non-typeable
dAdditional virulence factor-encoding genes investigated: iha, paa, saa, efa1/lifA, nleB, nleE, ehxA, and espP
eAdherence patterns exhibited in HeLa cells (6 h assay) in the presence of D-mannose (2%): LAL (localized adherence-like) and UND (undefined adherence)
fFAS, fluorescent-actin staining test
gShiga toxin (Stx) production (Vero cells toxicity assay). NT, not tested
O:H serotyping, performed with absorbed somatic (O1-O188) and flagellar (H1-H56) antisera produced at Adolfo Lutz Institute [22], demonstrated that the six aEPEC isolates belonged to five distinct serotypes, namely O4:H16, O68:H12, O105:H7, OR:H25 (1 isolate each), and O8:H25 (2 isolates), whereas the STEC isolate was ONT:H46 (Table 2). According to the literature, aEPEC O105:H7 was previously isolated from a diarrheic child in an epidemiological study conducted in Northeast Brazil [23], while aEPEC O4:H16 was obtained from a non-diarrheic child living in the city of Rio de Janeiro [24].
STEC ONT:H46 was previously isolated from beef and dairy cattle in different Brazilian states [25], as well as, from a patient with diarrheal disease during the year 2015 in Brazil [22]. For comparative purpose, three STEC ONT:H46, one from a diarrheic patient [22] and two from cattle [25], were evaluated in order to investigate if these isolates share virulence markers and clonal relatedness with the STEC ONT:H46 (20-1) obtained from ground beef in this study. The four STEC ONT:H46 isolates sharing an identical virulence-encoding gene profile (stx2, iha, saa, ehxA, and espP) were assigned in the phylo-group B1 and presented more than 80% similarity in pulsed-field gel electrophoresis (PFGE) analyze (Fig. S1). Together, the previously mentioned results may suggest that STEC ONT:H46, isolated from distinct sources in Brazil, are phylogenetically related.
The aEPEC isolates serotypes O4:H16 and O68:H12 harbored intimin subtype β1, while the aEPEC serotype O105:H7 harbored intimin-subtype θ (Table 2). The eae gene from the three other aEPEC isolates (45-3, 47-1, and 47-3) was non-typeable (NT) with the primers and PCR conditions tested [26]. Intimin subtypes β1 and θ appear as the most common types identified in aEPEC isolates obtained worldwide [3, 27, 28].
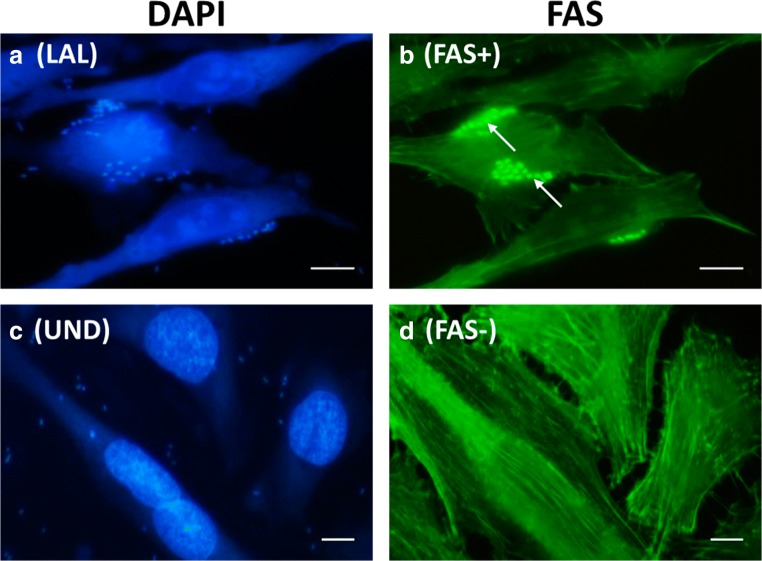
Furthermore, the six aEPEC and one STEC isolates were tested regarding their ability to adhere to HeLa cells (adherence assay), and to induce F-actin polymerization underneath adherent bacteria (fluorescence actin staining-FAS test), as previously described [29, 30]. aEPEC 18-5 (O4:H16) adhered to HeLa cells, forming loose microcolonies on the cell surface and producing an adherence pattern termed localized adherence-like (LAL) (Table 2, Fig. 1). The LAL pattern was first described in aEPEC serogroup O55 [31] and is considered the most common adherence pattern among aEPEC isolates [23, 32]. On other hand, adherence of five aEPEC isolates (11-1, 28-3, 45-3, 47-1, and 47-3) and STEC ONT:H46 (20-1) showed few bacteria interacting with the epithelial cells, thus not resulting in any of the adherence patterns described in the literature, which was referred as undefined (UND) adherence (Table 2, Fig. 1). The report of the number of aEPEC isolates exhibiting a UND adherence to HeLa/HEp2 cells has significantly increased, as observed in studies performed in the last 10 years [23, 33].
Fig. 1.
Fluorescent actin staining (FAS) test performed with two representative isolates: a, b aEPEC 18-5 and c, d STEC 20-1. aEPEC 18-5 adheres to HeLa cells, forming loose microcolonies (a), characteristic of the LAL (localized adherence-like) pattern, while STEC 20–1 exhibits a UND (undefined) adherence (c). Note foci of intense fluorescence in panel B (FAS-positive), indicating F-actin accumulation beneath attached bacteria (arrows), while in panel D (FAS-negative) this feature is not observed. Host cells and adherent bacteria were visualized with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) staining (a and c). Bar scale = 10 μm
The FAS test demonstrated that all six aEPEC isolates were able to induce F-actin polymerization at the site of attachment, independently of the adherence pattern exhibited by these isolates on HeLa cells, indirectly indicating the ability of these isolates to promote AE lesions in infected epithelial cells (Table 2, Fig. 1). The AE lesion is the main virulence attribute of typical and atypical EPEC isolates [1–3]. A study with adult volunteers revealed that the administration of an EPEC isogenic mutant strain, unable to induce AE lesion, caused diarrhea in only 36.6% of the subjects challenged, thus confirming the importance of this virulence property in the establishment of diarrheal disease [34]. We also demonstrated that sterile supernatant, prepared with STEC 20-1, was able to induce a characteristic cytotoxic effect in Vero cells monolayers, confirming the ability of this pathogen to produce and secret Stx2 (Table 2). Vero cells toxicity assay was performed as previously described [35]. Therefore, we clearly verified that the aEPEC and STEC isolates obtained in this study retain the main virulence features associated with these pathogens for causing disease in humans.
With the objective to determine if the aEPEC and STEC isolates obtained in this study harbored genes encoding additional virulence factors associated with the pathogenicity of these DEC pathotypes, we tested for the presence of iha, paa, saa, efa1/lifA, nleB, nleE, ehxA, and espP genes using primers and PCR conditions as previously described [27]. Besides the LEE region, we found that aEPEC 11-1 (O105:H7) harbored the nleB and nleE genes (Table 2), which are located in a PAI known as OI-122, in contrast with the other five aEPEC isolates, which lacked all the additional virulence factor-encoding genes investigated here (Table 2). PAI OI-122 has been identified more frequently in aEPEC isolates obtained from diarrheic patients than from healthy subjects [36, 37], and more recently, this PAI was statistically associated with diarrheal episodes of higher severity [38].
STEC 20-1 (ONT:H46), which lacked the LEE region, harbored the iha and saa genes, encoding proteins associated with the adherence of STEC isolates to epithelial cells and the ehxA and espP genes from the O157-EHEC plasmid (pO157), encoding for a EHEC-hemolysin and a serine protease with adhesive capability, respectively (Table 2).
Despite the low rate of isolation, we observed that the aEPEC and STEC isolates obtained in this study harbored virulence markers associated with the ability of these pathogens to cause disease in human hosts. Moreover, some of these isolates belonged to serotypes previously obtained from human infections in Brazil, thus indicating the circulation of these pathogens in Brazilian settings. These findings should serve as an alert for food risk assessment, so that appropriate strategies can be adopted to guarantee the food quality before consumption.
Electronic supplementary material
(PDF 117 kb)
Acknowledgments
This study was supported by a grant from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, grant no. 2012/19077-0). M.A.V. and N.A.B.M. received a fellowship from FAPESP, grant no. 2013/23414-5 and 2013/03460-2, respectively. We thank Tânia A. T. Gomes for the helpful suggestions. Dr. A. Leyva (USA) helped with English editing of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/CMR.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trabulsi LR, Keller R, Gomes TA. Typical and atypical enteropathogenic Escherichia coli. Emerg Infect Dis. 2002;8:508–513. doi: 10.3201/eid0805.010385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hernandes RT, Elias WP, Vieira MA, Gomes TA. An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiol Lett. 2009;297:137–149. doi: 10.1111/j.1574-6968.2009.01664.x. [DOI] [PubMed] [Google Scholar]
- 4.Scaletsky IC, Silva ML, Trabulsi LR. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect Immun. 1984;45:534–536. doi: 10.1128/IAI.45.2.534-536.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Girón JA, Ho AS, Schoolnik GK. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254(5032):710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 6.Donnenberg MS, Girón JA, Nataro JP, Kaper JB. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol Microbiol. 1992;6:3427–3437. doi: 10.1111/j.1365-2958.1992.tb02210.x. [DOI] [PubMed] [Google Scholar]
- 7.Karch H, Tarr PI, Bielaszewska M. Enterohaemorrhagic Escherichia coli in human medicine. Int J Med Microbiol. 2005;295:405–418. doi: 10.1016/j.ijmm.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, Hebert RJ, Olcott ES, Johnson LM, Hargrett NT, Blake PA, Cohen ML. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. 1983;308:681–685. doi: 10.1056/NEJM198303243081203. [DOI] [PubMed] [Google Scholar]
- 9.Gould LH, Mody RK, Ong KL, Clogher P, Cronquist AB, Garman KN, Lathrop S, Medus C, Spina NL, Webb TH, White PL, Wymore K, Gierke RE, Mahon BE, Griffin PM. Increased recognition of non-O157 Shiga toxin-producing Escherichia coli infections in the United States during 2000-2010: epidemiologic features and comparison with E. coli O157 infections. Foodborne Pathog Dis. 2013;10:453–460. doi: 10.1089/fpd.2012.1401. [DOI] [PubMed] [Google Scholar]
- 10.Frank C, Werber D, Cramer JP, Askar M, Faber M, an der Heiden M, Bernard H, Fruth A, Prager R, Spode A, Wadl M, Zoufaly A, Jordan S, Kemper MJ, Follin P, Müller L, King LA, Rosner B, Buchholz U, Stark K, Krause G. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med. 2011;365(19):1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 11.Kaper JB, O'Brien AD (2014) Overview and historical perspectives. Microbiol Spectr [DOI] [PMC free article] [PubMed]
- 12.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci U S A. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jerse AE, Yu J, Tall BD, Kaper JB. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci U S A. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toledo MRF, Fontes CF, Trabulsi LR. EPM-modificação do meio de Rugai e Araújo para a realização simultânea dos testes de produção de gás a partir da glicose, H2S, urease e triptofanodesaminase. Rev Microbiol. 1982;13:309–315. [Google Scholar]
- 15.Toledo MRF, Fontes CF, Trabulsi LR. MILi-um meio para a realização dos testes de motilidade, indol e lisina descarboxilase. Rev. Microbiol13. 1982. pp. 230–235. [Google Scholar]
- 16.Dias RC, Dos Santos BC, Dos Santos LF, Vieira MA, Yamatogi RS, Mondelli AL, Sadatsune T, Sforcin JM, Gomes TA, Hernandes RT. Diarrheagenic Escherichia coli pathotypes investigation revealed atypical enteropathogenic E. coli as putative emerging diarrheal agents in children living in Botucatu, São Paulo State, Brazil. APMIS. 2016;124(4):299–308. doi: 10.1111/apm.12501. [DOI] [PubMed] [Google Scholar]
- 17.Cerqueira AMF, Tibana A, Guth BE. High occurrence of Shiga-like toxin-producing strains among diarrheagenic Escherichia coli isolated from raw beef products in Rio de Janeiro City, Brazil. J Food Prot. 1997;60:177–180. doi: 10.4315/0362-028X-60.2.177. [DOI] [PubMed] [Google Scholar]
- 18.Bergamini AMM, Simões M, Irino K, Gomes TA, Guth BE. Prevalence and characteristics of Shiga toxin-producing Escherichia coli (STEC) strains in ground beef in São Paulo, Brazil. Braz J Microbiol. 2007;38:553–556. doi: 10.1590/S1517-83822007000300032. [DOI] [Google Scholar]
- 19.Peresi JTM, De Almeida IAZC, Vaz TMI, Hernandes RT, Teixeira ISC, Silva SIL, Graciano RAS, Pinheiro SR, Dos Santos LF. Search for diarrheagenic Escherichia coli in raw kibbe samples reveals the presence of Shiga toxin-producing strains. Food Control. 2016;63:165–170. doi: 10.1016/j.foodcont.2015.11.018. [DOI] [Google Scholar]
- 20.Ristori CA, Rowlands REG, Martins CG, Barbosa ML, dos Santos LF, Jakabi M, de Melo Franco BDG. Assessment of consumer exposure to Salmonella spp., Campylobacter spp., and Shiga toxin-producing Escherichia coli in meat products at retail in the city of São Paulo, Brazil. Foodborne Pathog Dis. 2017;14:447–453. doi: 10.1089/fpd.2016.2270. [DOI] [PubMed] [Google Scholar]
- 21.Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5:58–65. doi: 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 22.Ori EL, Takagi EH, Andrade TS, Miguel BT, Cergole-Novella MC, Guth BEC, Hernandes RT, Dias RCB, Pinheiro SRS, Camargo CH, Romero EC, Dos Santos LF (2019) Diarrhoeagenic Escherichia coli and Escherichia albertii in Brazil: pathotypes and serotypes over a 6-year period of surveillance. Epidemiol Infect 147:1–9 [DOI] [PMC free article] [PubMed]
- 23.Abe CM, Trabulsi LR, Blanco J, Blanco M, Dahbi G, Blanco JE, Mora A, Franzolin MR, Taddei CR, Martinez MB, Piazza RM, Elias WP. Virulence features of atypical enteropathogenic Escherichia coli identified by the eae(+) EAF-negative stx(−) genetic profile. Diagn Microbiol Infect Dis. 2009;64:357–365. doi: 10.1016/j.diagmicrobio.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 24.Gomes TA, Irino K, Girão DM, Girão VB, Guth BE, Vaz TM, Moreira FC, Chinarelli SH, Vieira MA. Emerging enteropathogenic Escherichia coli strains? Emerg Infect Dis. 2004;10:1851–1855. doi: 10.3201/eid1010.031093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliveira MG, Brito JR, Gomes TA, Guth BE, Vieira MA, Naves ZV, Vaz TM, Irino K. Diversity of virulence profiles of Shiga toxin-producing Escherichia coli serotypes in food-producing animals in Brazil. Int J Food Microbiol. 2008;127:139–146. doi: 10.1016/j.ijfoodmicro.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 26.Blanco M, Blanco JE, Dahbi G, Mora A, Alonso MP, Varela G, Gadea MP, Schelotto F, González EA, Blanco J. Typing of intimin (eae) genes from enteropathogenic Escherichia coli (EPEC) isolated from children with diarrhoea in Montevideo, Uruguay: identification of two novel intimin variants (muB and xiR/beta2B) J Med Microbiol. 2006;55:1165–1174. doi: 10.1099/jmm.0.46518-0. [DOI] [PubMed] [Google Scholar]
- 27.Vieira MA, Dos Santos LF, Dias RC, Camargo CH, Pinheiro SR, Gomes TA, Hernandes RT. Atypical enteropathogenic Escherichia coli as aetiologic agents of sporadic and outbreak-associated diarrhoea in Brazil. J Med Microbiol. 2016;65(9):998–1006. doi: 10.1099/jmm.0.000313. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y, Bai X, Zhao A, Zhang W, Ba P, Liu K, Jin Y, Wang H, Guo Q, Sun H, Xu J, Xiong Y. Genetic diversity of Intimin gene of atypical enteropathogenic Escherichia coli isolated from human, animals and raw meats in China. PLoS One. 2016;11(3):e0152571. doi: 10.1371/journal.pone.0152571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cravioto A, Gross RJ, Scotland SM, Rowe B. An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr Microbiol. 1979;3:95–99. doi: 10.1007/BF02602439. [DOI] [Google Scholar]
- 30.Knutton S, Baldwin T, Williams PH, McNeish AS. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/IAI.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues J, Scaletsky IC, Campos LC, Gomes TA, Whittam TS, Trabulsi LR. Clonal structure and virulence factors in strains of Escherichia coli of the classic serogroup O55. Infect Immun. 1996;64:2680–2686. doi: 10.1128/IAI.64.7.2680-2686.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vieira MA, Andrade JR, Trabulsi LR, Rosa AC, Dias AM, Ramos SR, Frankel G, Gomes TA. Phenotypic and genotypic characteristics of Escherichia coli strains of non-enteropathogenic E. coli (EPEC) serogroups that carry eae and lack the EPEC adherence factor and Shiga toxin DNA probe sequences. J Infect Dis. 2001;183:762–772. doi: 10.1086/318821. [DOI] [PubMed] [Google Scholar]
- 33.Scaletsky IC, Aranda KR, Souza TB, Silva NP. Adherence factors in atypical enteropathogenic Escherichia coli strains expressing the localized adherence-like pattern in HEp-2 cells. J Clin Microbiol. 2010;48(1):302–306. doi: 10.1128/JCM.01980-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donnenberg MS, Tacket CO, James SP, Losonsky G, Nataro JP, Wasserman SS, Kaper JB, Levine MM. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J Clin Invest. 1993;92(3):1412–1417. doi: 10.1172/JCI116717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Konowalchuk J, Speirs JI, Stavric S. Vero response to a cytotoxin of Escherichia coli. Infect Immun. 1977;18:775–779. doi: 10.1128/IAI.18.3.775-779.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afset JE, Bruant G, Brousseau R, Harel J, Anderssen E, Bevanger L, Bergh K. Identification of virulence genes linked with diarrhea due to atypical enteropathogenic Escherichia coli by DNA microarray analysis and PCR. J Clin Microbiol. 2006;44:3703–3711. doi: 10.1128/JCM.00429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieira MA, Salvador FA, Silva RM, Irino K, Vaz TM, Rockstroh AC, Guth BE, Gomes TA. Prevalence and characteristics of the O122 pathogenicity island in typical and atypical enteropathogenic Escherichia coli strains. JClin Microbiol. 2010;48:1452–1455. doi: 10.1128/JCM.01944-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercado EH, Piscoche C, Contreras C, Durand D, Riveros M, Ruiz J, Ochoa TJ. Pathogenicity Island O-122 in enteropathogenic Escherichia coli strains is associated with diarrhea severity in children from Lima Peru. Int J Med Microbiol. 2016;306:231–236. doi: 10.1016/j.ijmm.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 117 kb)