Abstract
Both aging and Alzheimer’s disease (AD) are associated with widespread epigenetic changes, with most evidence suggesting global hypomethylation in AD. It is, however, unclear how these age-related epigenetic changes are linked to molecular aberrations as expressed in animal models of AD. Here, we investigated age-related changes of epigenetic markers of DNA methylation and hydroxymethylation in a range of animal models of AD, and their correlations with amyloid plaque load. Three transgenic mouse models, including the J20, APP/PS1dE9 and 3xTg-AD models, as well as Caribbean vervets (a non-transgenic non-human primate model of AD) were investigated. In the J20 mouse model, an age-related decrease in DNA methylation was found in the dentate gyrus (DG) and a decrease in the ratio between DNA methylation and hydroxymethylation was found in the DG and cornu ammonis (CA) 3. In the 3xTg-AD mice, an age-related increase in DNA methylation was found in the DG and CA1–2. No significant age-related alterations were found in the APP/PS1dE9 mice and non-human primate model. In the J20 model, hippocampal plaque load showed a significant negative correlation with DNA methylation in the DG, and with the ratio a negative correlation in the DG and CA3. For the APP/PS1dE9 model a negative correlation between the ratio and plaque load was observed in the CA3, as well as a negative correlation between DNMT3A levels and plaque load in the DG and CA3. Thus, only the J20 model showed an age-related reduction in global DNA methylation, while DNA hypermethylation was observed in the 3xTg-AD model. Given these differences between animal models, future studies are needed to further elucidate the contribution of different AD-related genetic variation to age-related epigenetic changes.
Keywords: Alzheimer’s disease, aging, hippocampus, DNA methylation, DNA hydroxymethylation, DNA methyltransferase, animal models
Introduction
Alzheimer’s disease (AD) is a complex age-related neurodegenerative disorder and the most common form of dementia (Sosa-Ortiz et al., 2012), for which presently no effective treatment exists (Lansdall, 2014; Raina et al., 2008). Although recent studies indicate that the widespread neurodegeneration in the AD brain may be initiated in brainstem regions (Stratmann et al., 2015), the development of cognitive impairment is associated with degeneration of the entorhinal cortex and hippocampus (Bartsch and Wulff, 2015). The exact molecular mechanisms underlying the neurodegeneration in AD remain unclear. Nevertheless, there are two pathological hallmarks; extracellular neuritic plaques and intracellular neurofibrillary tangles, which are thought to play a pivotal role in the progression of AD and that are currently the basis of a definitive postmortem AD diagnosis (Defina et al., 2013; Kurz and Perneczky, 2011). These protein aggregates mainly consist of amyloid-β (Aβ) and hyperphosphorylated tau, respectively. Genetic studies have offered important insights and have confirmed the importance of Aβ, especially in the development of familial forms of AD, by identifying mutations in the amyloid precursor protein (APP) and presenilin (PS) genes that are associated with familial AD (Price and Sisodia, 1998).
A growing body of evidence indicates that epigenetics may play a crucial role in complex age-related neurodegenerative diseases such as AD (Iatrou et al., 2016; Lardenoije et al., 2015a; Van den Hove et al., 2014). Epigenetic processes dynamically regulate gene expression at both the transcriptional and translational level (Choudhuri, 2011). They are thought to be able to translate environmental exposures into alterations in gene expression (Liu et al., 2008). In particular, DNA methylation has received attention in the context of AD, and DNA hydroxymethylation has more recently also been increasingly studied (Chouliaras et al., 2010; Lardenoije et al., 2015a; van den Hove et al., 2012). Our group and others have found that with normal aging, region-specific DNA methylation and hydroxymethylation, as well as DNA methyltransferase 3A (DNMT3A) levels rise (Chouliaras et al., 2012a, 2012b, 2011; Hernandez et al., 2011; Lardenoije et al., 2015b; Münzel et al., 2010; Song et al., 2011). In AD, however, overall DNA methylation and hydroxymethylation levels appear to be lowered (Chouliaras et al., 2013; Condliffe et al., 2014; Mastroeni et al., 2010). Depending on the brain region, and likely also methodological differences (e.g. concerning tissue processing), there are, however, also conflicting reports, showing no changes in DNA methylation levels between AD patients and controls (Condliffe et al., 2014), or increased DNA methylation and hydroxymethylation levels (Coppieters et al., 2014). Recent studies employing techniques such as Illumina’s HumanMethylation450 BeadChip assay have provided further insights beyond global changes in epigenetic markers (De Jager et al., 2014; Lunnon et al., 2014). These epigenome-wide association studies on homogenates of brain samples can help to identify important new candidate genes that, through altered epigenetic regulation, may play a role in the pathogenesis of AD.
Many studies investigating epigenetic changes related to AD have studied differences between postmortem brain tissue from diseased and control cases. To go beyond associations and elucidate the exact functional and potentially causal role of epigenetic dysregulation in the course of AD, live model systems are required. To this end, a plethora of AD animal models have been established, including many transgenic rodent models that overexpress mutated human genes that have been associated with rare forms of familial AD (van Goethem et al., 2014), but also non-human primate models that naturally develop Aβ plaque pathology have been used (Frost et al., 2013; Woodruff-Pak, 2008). While these animal models capture some of the molecular, physiological, or behavioral aspects of AD, none of the animal models display the full complexity of AD (Cavanaugh et al., 2014). Most animal models have been characterized based on classical hallmarks of AD, such as plaque development and cognitive impairment, but there are currently no reports comparing different animal models of AD on an epigenetic level. The aim of the present study was therefore to investigate age-related changes in epigenetic markers related to DNA methylation (5-methylcytosine [5mC] and DNMT3A) and DNA hydroxymethylation (5-hydroxymethylcytosine [5hmC]), in three genetically different transgenic mouse models of AD and a non-human primate model that naturally develops Aβ plaque pathology (Frost et al., 2013; Lemere et al., 2004). In addition, correlations between 5mC, 5hmC, and DNMT3A immunoreactivity (IR) and amyloid plaque load were assessed, and compared with findings in humans and other studies related to epigenetic dysregulation in AD.
Materials and methods
Animal models
For this study, 3 transgenic mouse models of AD were used, including J20 mice on a C57BL6 background (Mucke et al., 2000), APP/PS1dE9 mice on a C57BL6J background (Garcia-Alloza et al., 2006; Jankowsky et al., 2001), and 3xTg-AD mice on a C57BL6 background (Oddo et al., 2003). J20 mice harbor the mutated human APP gene (APPK670N/M671L, V717F), APP/PS1dE9 mice express both mutated humanized APP and human PS1 (APPK595N/M596LPS1 deletion of exon 9), and 3xTg-AD mice express 3 mutated human genes, APP, PS1, and microtubule-associated protein tau (MAPT) (APPK670N/M671L, PS1M146V, TauP301L). In addition to these transgenic mouse models, archived fixed brain tissue from 12 Caribbean vervets (Chlorocebus sabaeuss; 12.2 – 32 years of age) was used (Behavioral Science Foundation, St. Kitts) (Lemere et al., 2004). See Table 1 for additional information about the used animal models. The Harvard Medical Area Standing Committee approved of the use of mice at Brigham and Women’s Hospital, which is in line with all state and federal regulations. Vervet brain tissue was retrieved following protocols approved by the Behavioral Science Foundation Animal Care Committee acting under the auspices of the Canadian Council on Animal Care.
Table 1.
Overview of investigated animal models.
| Animal model | Mutations | Promotor | Start plaque deposition (region) | Start cognitive deficits | References | Age |
|---|---|---|---|---|---|---|
| J20 | APPK670N/M671L, V717F | PDGF | 5–7 months (hippocampus, neocortex) | 1–2 months | Mucke et al., 2000; Webster et al., 2014 | 4 months (n = 4) 8 months (n = 4) 16 months (n = 3) 24 months (n = 2) |
| APP/PS1dE9 | APPK595N/M596LPS1 deletion of exon 9 | mPrP | 6 months (hippocampus, cortex) | 4 months | Jankowsky et al., 2001; Jankowsky et al., 2004; Park et al., 2006 | 6 months (n = 4) 16 months (n = 1) 17 months (n = 1) 18 months (n = 1) 23 months (n = 1) 27 months (n = 1) |
| 3xTg-AD | APPK670N/M671L, PS1M146V, TauP301L | mThy-1 | 6 months (frontal cortex)* | 4 months* | Oddo et al., 2003; Billings et al., 2005 | 5 months (n = 4) 14 months (n = 2) 17 months (n = 1) 27 months (n = 3) |
| Caribbean vervet | NA | NA | 15 years (hippocampus) † | 15 years† | This article (Supplementary Figure 9) | 12.2 years (n = 1) 14 years (n = 1) 14.9 years (n = 1) 15 years (n = 2) 16.4 years (n = 1) 17 years (n = 2) 19 years (n = 1) 24 years (n = 1) 27.4 years (n = 1) 32 years (n = 1) |
The 3xTg-AD mice used in this paper appeared to have a 2–3 month delay in Alzheimer’s disease pathology compared to previously published reports, possibly due to a loss of transgene copies with successive breeding (see https://www.jax.org/strain/004807).
This was the youngest vervet with plaques and cognitive impairment, but note that while generally plaque deposition increases with age, there is no clear relationship between age and, onset of plaque pathology, and cognitive decline (see Supplementary Figure 9). NA, not applicable.
Tissue preparation
After anesthetization via CO2 inhalation, mice were perfused with 20 mL ice-cold saline. The brains were then removed and hemisected, fixed in 10% formalin or 4% paraformaldehyde for 2 to 24 hours, paraffin-embedded, and further sectioned into 10 μm-thick slices. The archived nonhuman primate brain tissue was formalin-fixed for months to several years and was divided into nine rostrocaudal regions, paraffin-embedded and further cut into 10 μm-thick coronal sections.
Immunohistochemistry
Ten micron-thick serial sagittal mouse brain sections or coronal vervet brain sections were used for immunohistochemistry. All steps were performed at room temperature unless specified otherwise. Sections were first deparaffinized in Histo-Clear (National Diagnostics, Atlanta, GA) and rehydrated in a series of decreasing ethanol solutions, ending with deionized water. Hydrogen peroxide (0.3%) diluted in methanol was used to quench endogenous peroxidase for 10 minutes. To unmask antigen-binding sites, antigen retrieval was performed with BioGenex citrate buffer (BioGenex, San Ramon, CA), keeping the solution around boiling temperature for 5 minutes in the microwave. Aβ42 staining on the vervet tissue required incubating the sections in 88% formic acid for 10 minutes. The sections were washed with deionized water for 10 minutes and incubated in blocking solution for 20 minutes. The blocking solution consisted of 10% serum dissolved in Tris-buffered saline, with serum from the same species as the secondary antibody host. The sections were subsequently incubated overnight with primary antibody, at 4°C. The following antibodies were used: a mouse monoclonal anti-5mC antibody (1:1000 dilution for mouse sections and 1:500 for vervet sections; GenWay Biotech Inc., San Diego, CA), a rabbit polyclonal anti-5hmC antiserum (1:10,000 dilution; Active Motif, Carlsbad, CA), a rabbit polyclonal anti-DNMT3A antibody (1:200 dilution; Santa Cruz Biotechnology, Dallas, TX), a general monoclonal IgG1 anti-Aβ antibody for staining mouse sections (3A1; 1:1000 dilution, kindly provided by Dr. Brian ÒNaullain at the Ann Romney Center for Neurologic Diseases, Boston, MA), and a mouse monoclonal IgG1 anti-Aβ42 antibody for staining vervet sections (1:500 dilution; BioLegend, San Diego, CA). The 3A1 antibody binds an epitope in the Aβ N-terminus and has previously been fully characterized (Frost et al., 2015). After another wash with deionized water, the slides were incubated with biotinylated secondary antibodies for 30 minutes. Secondary antibodies included a horse anti-mouse secondary antibody (Vector Laboratories, Burlingame, CA) for the 5mC antibody, a goat anti-rabbit secondary antibody (Vector Laboratories) for the 5hmC and DNMT3A antibodies, a goat anti-mouse secondary antibody (Vector Laboratories) for 3A1, and a goat anti-mouse antibody (SouthernBiotech, Birmingham, AL) for Aβ42. The VectorElite horseradish peroxidase ABC kit (Vector Laboratories), with 3,3’-diaminobenzidine tetrahydrochloride (DAB; Sigma-Aldrich, St. Louis, MO) as chromogen, was used to visualize IR. For each staining run, omission of the primary antibody was included as a negative control, which consistently showed no staining (data not shown).
Analysis of 5mC, 5hmC and DNMT3A immunoreactivity and plaque load
For each staining, 3 sagittal hippocampal sections per mouse and 2 coronal hippocampal sections per vervet were examined at approximately equidistant planes. For the IR analysis of 5mC, 5hmC, and DNMT3A stainings, images were captured from hippocampal subregions, including 4 images of the dentate gyrus (DG), 2 of the cornu ammonis (CA) 3, and 2 of the CA1–2 (Supplementary Figure 1), using the 20X objective of a BX50 brightfield microscope (Olympus, Tokyo, Japan) in conjunction with a QIcam digital camera (QImaging, Surrey, BC, Canada). The IR in the regions of interest (DG, CA3, and CA1–2) was analyzed in the images of the hippocampal subregions using ImageJ (version 1.48v, Wayne Rasband, National Institutes of Health, Bethesda, Maryland, USA). For each image, the mean grey value of the region of interest (ROI) was measured after delineating the ROI in the image and setting a fixed threshold for background correction. Additionally, the total ROI area and ROI area above the background threshold was determined. The grey value and area measures were then combined by multiplying the background-corrected mean grey values of the ROI with the fraction of the ROI area with values above the background threshold (i.e. the specifically stained area of the ROI), to get the integrated density. This combined measurement is a more robust representation of protein levels than intensity or area alone, as for instance a decrease in area may lead to the detection of a higher mean intensity while the actual protein levels remained unchanged.
The fraction of the hippocampal area containing plaques was determined in the sections stained for 3A1 with a BIOQUANT image analysis setup (Nashville, TN, USA), and using a fixed threshold of detection. For this analysis, the hippocampus was manually delineated using the 4X objective, after which plaques where automatically detected based on the fixed threshold. Before performing the final measurements, artifacts were manually removed. The sections of the vervet brains varied in plane-cut and often only a part of the hippocampus could be assessed on a single section, which made them unsuited for a BIOQUANT analysis. The plaque load in the vervets was therefore semi-quantitatively scored, with 0 for no plaques, 1 for plaques in the temporal cortex but not inside the hippocampus, 2 for 1 to 5 plaques in the hippocampus, 3 for 6 to 10 plaques in hippocampus, 4 for 11 to 100 plaques in hippocampus, and 5 for more than 100 plaques in hippocampus. All slides and images were processed blinded and in a randomized order.
Statistical analysis
To compare the relative degree of DNA methylation and hydroxymethylation between ages, the ratio of the integrated density of 5mC and 5hmC was calculated (i.e. 5mC IR / by 5hmC IR). Before performing the analyses and generating plots, the data was scaled through division by the root mean square of the data per region, to allow for a better comparison between the different stainings. For each animal model, hippocampal subregion, and epigenetic marker, including the 5mC:5hmC ratio, a linear regression model was fitted with integrated density as the outcome and age as the predictor, coded both as continuous variables. The data was visually inspected for abnormalities and quantile-quantile plots of the regression model residuals were analyzed to check for severe deviations from normality. The correlation between the epigenetic markers and plaque load was determined by calculating Kendall’s tau. For all significance tests, the alpha was set at 0.05. The ImageJ measurements were collected in Microsoft Excel 2013 (Microsoft, Redmond, WA) and processed, normalized, and analyzed in R (version 3.2.2; The R Foundation, Vienna, Austria) and RStudio (version 0.99.486; The Foundation for Open Access Statistics, Boston, MA). In addition to standard R functions, the dplyr package was used for data handling (Wickham and Francois, 2015), the Kendall package was used for the Kendall correlation analysis (McLeod, 2011), and the ggplot2 package was used for generating graphs (Wickham, 2009).
Results
Qualitative analysis of 5mC, 5hmC, and DNMT3A IR
Three main hippocampal subregions (dentate gyrus, DG; cornu ammonis 3, CA3; and CA1–2) were examined by immunohistochemical analysis as shown in Supplementary Figure 1. A few (7%) images were excluded from the analysis due to artifacts. Visual inspection of the analyzed images indicated most cells show nuclear 5mC, 5hmC, and DNMT3A IR (Supplementary Figures 2–5). DNMT3A, however, also appeared to be expressed outside the nucleus, especially in the CA3. The vervet tissue also exhibited some extranuclear 5mC and 5hmC IR. Upon closer observation, the 5hmC and DNMT3A signals appeared diffusely throughout the nucleus, whereas the 5mC signal was limited to a small number of distinct punctuae. Again, the staining pattern in the vervet tissue deviated from this general pattern and appeared diffusely throughout the nucleus for 5mC, 5hmC, and DNMT3A.
A conservative visual comparison of the stainings at the different ages did not reveal obvious age-related changes in IR of the epigenetic markers in the J20 (Supplementary Figure 2) and APP/PS1dE9 models (Supplementary Figure 3). In the 3xTg-AD model an age-related increase of 5mC signal can be observed mainly in the DG, but also the CA3 and CA1–2 (Supplementary Figure 4). The vervet images did not show any consistent age-related alterations in IR (Supplementary Figure 5). The hippocampal DNMT3A signal was generally too variable to draw any conclusions based on visual inspection alone. Some of these observations were confirmed through a semiquantitative analysis.
Semiquantitative analysis of 5mC, 5hmC, and DNMT3A IR
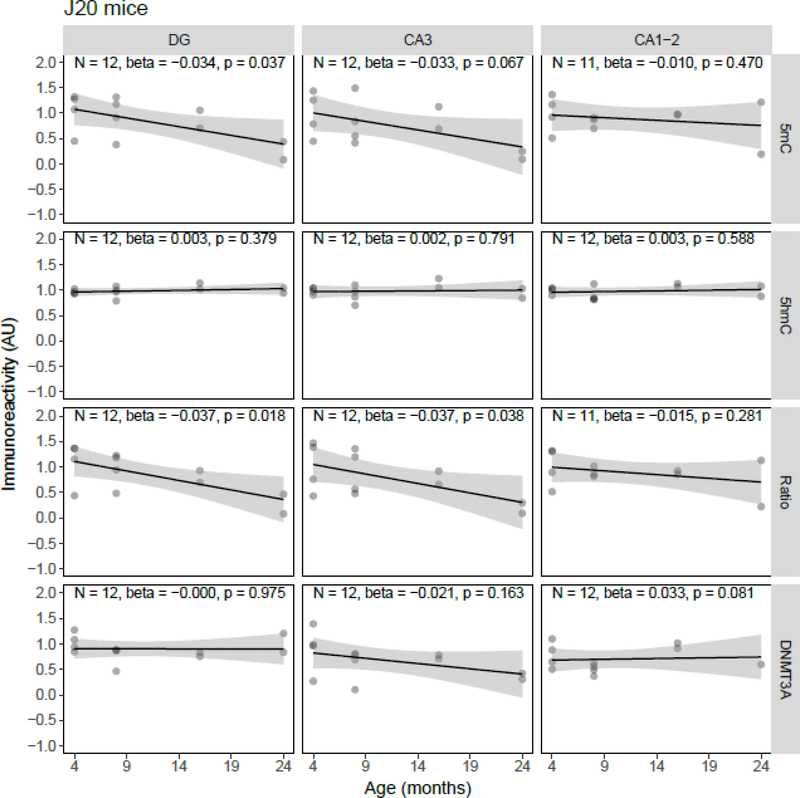
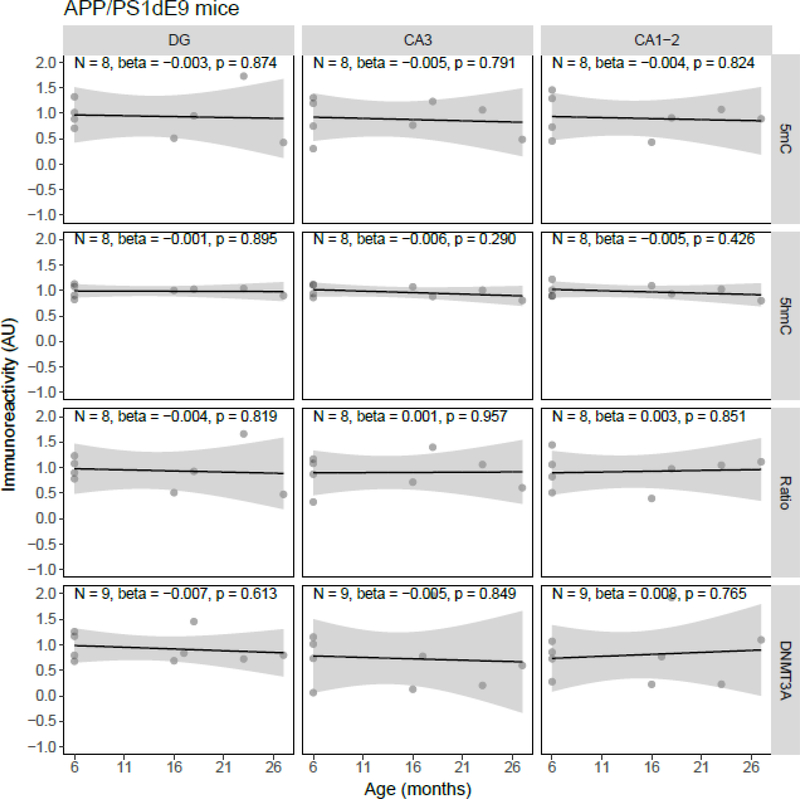
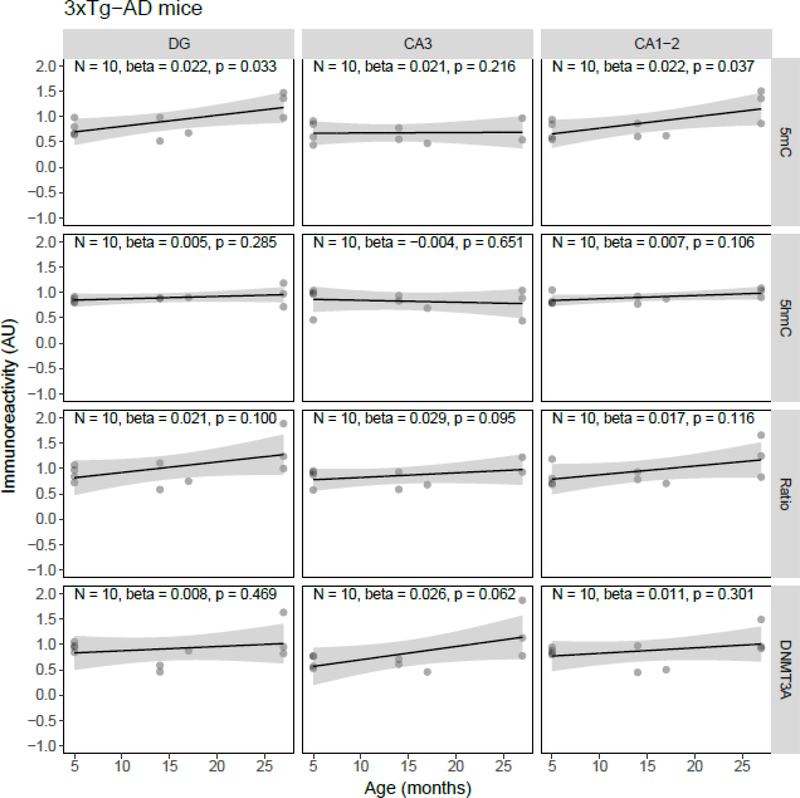
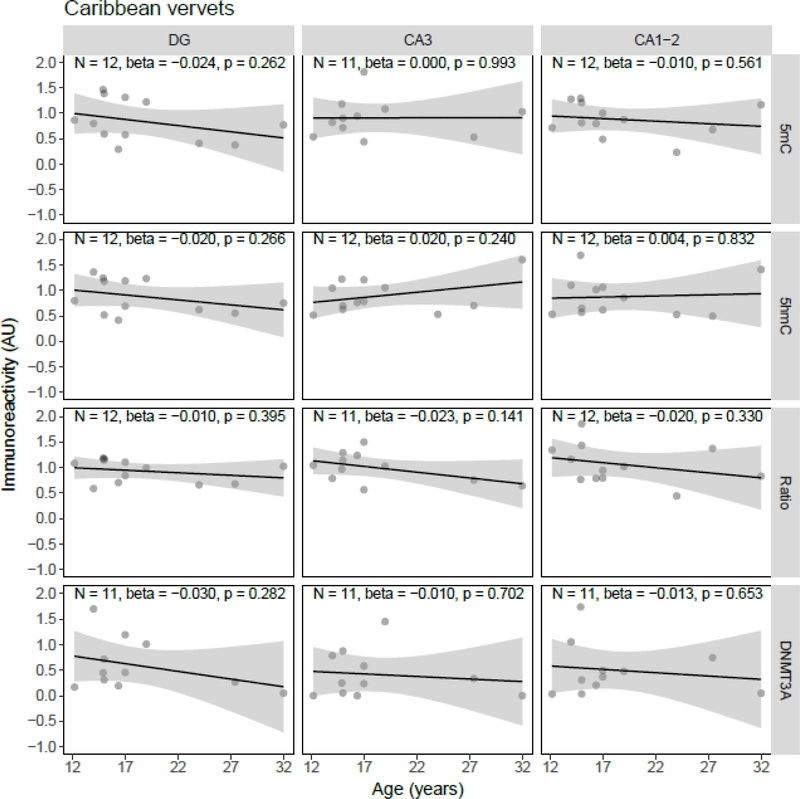
Quantile-quantile plots of the residuals of the regression models were inspected, and although the residuals of some individual models showed deviations from normality, there were no overall indications for either right- or left-skewness of the residuals. Linear regression showed a statistically significant age-related decrease of 5mC IR (β = −0.034, p = 0.037) and the 5mC:5hmC ratio (β = −0.037, p = 0.018) in the DG, and of the 5mC:5hmC ratio in the CA3 (β = −0.037, p = 0.038), in the J20 transgenic mouse model (Figure 1). No statistically significant age-related changes of 5mC IR were observed in APP/PS1dE9 mice (Figure 2). In contrast to the J20 mice, a statistically significant age-related increase of 5mC IR was found in the DG (β = 0.022, p = 0.033) and CA1–2 (β = 0.022, p = 0.037) of the 3xTg-AD model (Figure 3). Also, no statistically significant age-related changes of 5mC IR were observed in vervets (Figure 4), and no changes in 5hmC or DNMT3A were detected in any of the tested animal models (Figures 1–4).
Figure 1.
Semi-quantitative analysis results of age-related alterations in 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC) and DNA methyltransferase 3A (DNMT3A) immunoreactivity (IR), and the 5mC:5hmC ratio in J20 mice. Shown are the background-corrected and scaled integrated density data plotted against the age of the animals, the fitted linear regression lines and the standard error (SE) of the regression lines, for the dentate gyrus (DG), cornu ammonis (CA) 3, and CA1–2 subregions of the hippocampus. A statistically significant effect of age on 5mC IR was found in the DG (p = 0.037), and on the 5mC:5hmC ratio in the DG (p = 0.018) and CA3 (p = 0.038). AU, arbitrary units.
Figure 2.
Semi-quantitative analysis results of age-related alterations in 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC) and DNA methyltransferase 3A (DNMT3A) immunoreactivity (IR), and the 5mC:5hmC ratio in APP/PS1dE9 mice. Shown are the background-corrected and scaled integrated density data plotted against the age of the animals, the fitted linear regression lines and the standard error (SE) of the regression lines, for the dentate gyrus (DG), cornu ammonis (CA) 3, and CA1–2 subregions of the hippocampus. No statistically significant effect of age on any of the investigated epigenetic markers was found. AU, arbitrary units.
Figure 3.
Semi-quantitative analysis results of age-related alterations in 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC) and DNA methyltransferase 3A (DNMT3A) immunoreactivity (IR), and the 5mC:5hmC ratio in 3xTg-AD mice. Shown are the background-corrected and scaled integrated density data plotted against the age of the animals, the fitted linear regression lines and the standard error (SE) of the regression lines, for the dentate gyrus (DG), cornu ammonis (CA) 3, and CA1–2 subregions of the hippocampus. A statistically significant effect of age on 5mC IR was found in the DG (p = 0.022) and CA1–2 (p = 0.037). AU, arbitrary units.
Figure 4.
Semi-quantitative analysis results of age-related alterations in 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC) and DNA methyltransferase 3A (DNMT3A) immunoreactivity (IR), and the 5mC:5hmC ratio in Caribbean vervets. Shown are the background-corrected and scaled integrated density data plotted against the age of the animals, the fitted linear regression lines and the standard error (SE) of the regression lines, for the dentate gyrus (DG), cornu ammonis (CA) 3, and CA1–2 subregions of the hippocampus. No statistically significant effect of age on any of the investigated epigenetic markers was found. AU, arbitrary units.
Plaque load correlates with age-related changes in 5mC IR
As expected, all of the oldest animals of the transgenic mouse models exhibited Aβ plaques (Supplementary Figures 6–8), varying between 17% and 63% of the hippocampus being covered in plaques. The regional distribution of the plaques in the different hippocampal subregions was, however, highly variable between the different mouse models. Hippocampal plaque density was the highest in the J20 model and plaques were observed within (and in vicinity of) the investigated subregions (Supplementary Figure 6). In the APP/PS1dE9 model, Aβ plaques were present throughout the hippocampus, but the load was lower than that seen in the J20 model (Supplementary Figure 7). In the 3xTg-AD model, however, plaques were mainly located in the dorsal subiculum and deeper layers (oriens and alveus) of the hippocampus, with a generally low plaque load close to the investigated regions (Supplementary Figure 8). In the vervets, the oldest animals did not necessarily have the most severe pathology (Supplementary Figure 9). In general, however, the occurrence of plaque pathology increases with age, as the six animals (of 15, 16.4, 19, 24, 27.4 and 32 years old) that had plaque pathology in temporal cortex and/or hippocampus were on average older than the other vervets (of 12.2, 14, 14.9, 15, 17 and 17 years old).
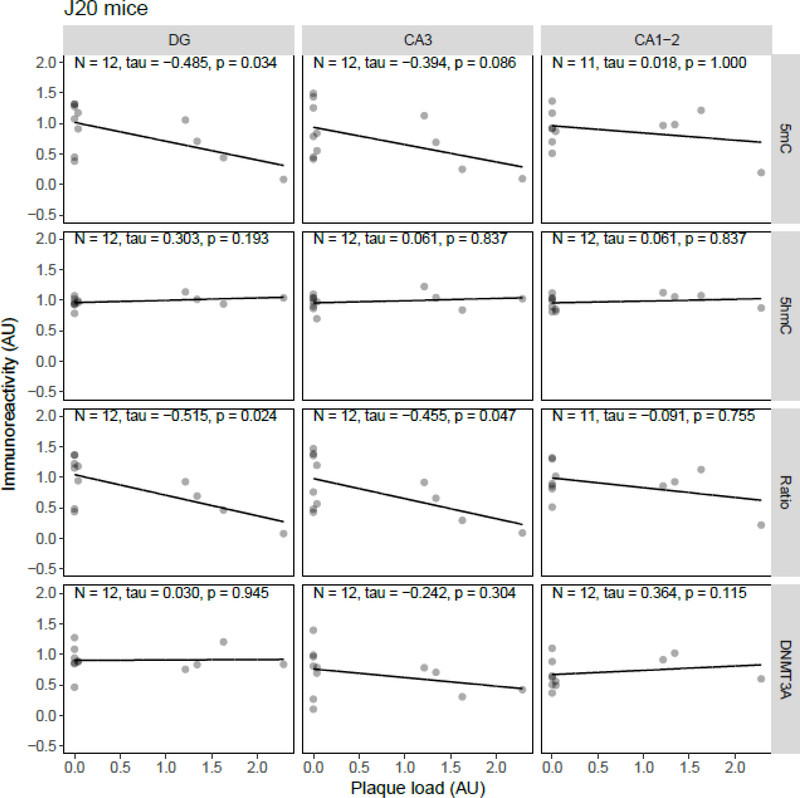
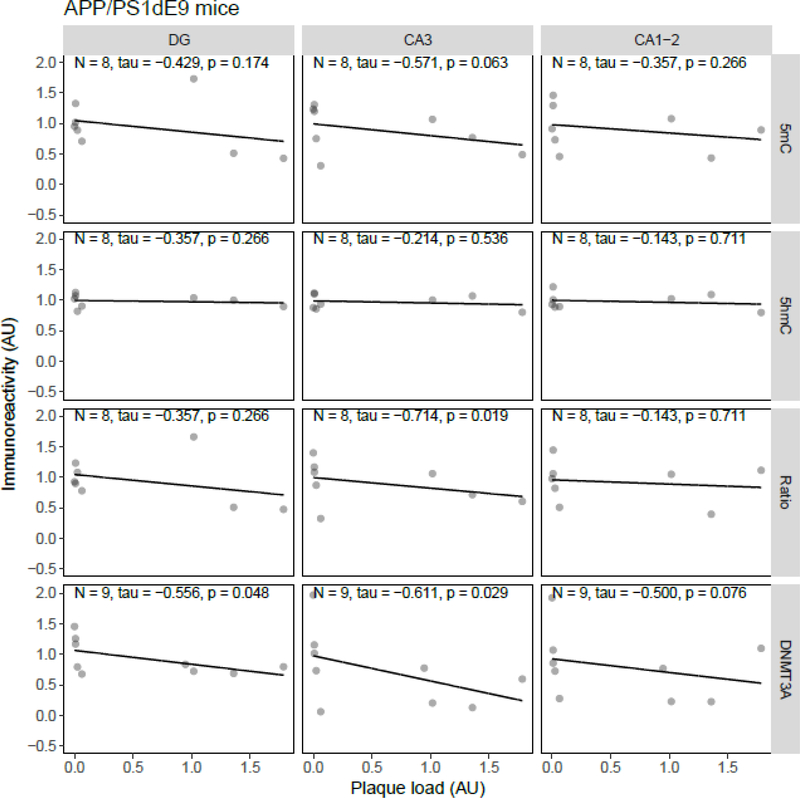
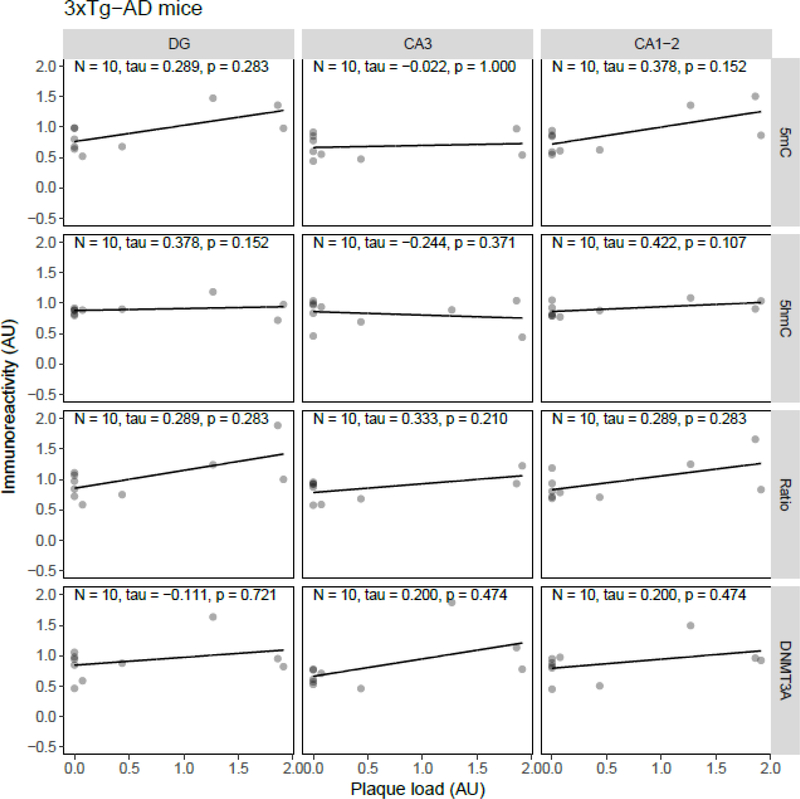
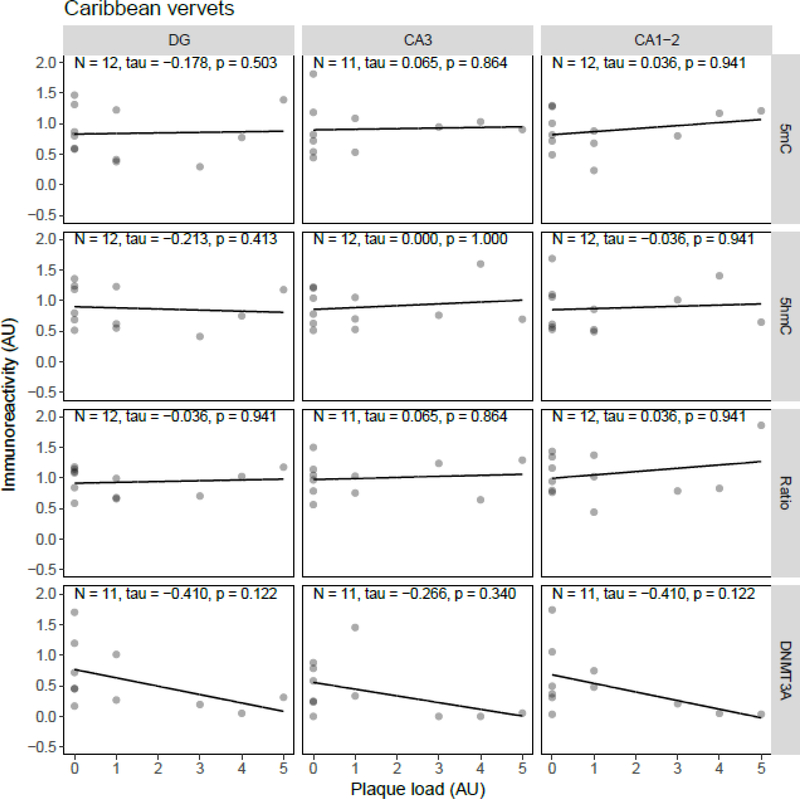
Correlation analysis between plaque load and 5mC, 5hmC, or DNMT3A IR revealed statistically significant inverse correlations in the J20 model between plaque load and 5mC IR in the DG (tau = −0.49, p = 0.034) and between plaque load and the 5mC:5hmC ratio in the DG (tau = −0.52, p = 0.024) and CA3 (tau = −0.46, p = 0.047) (Figure 5). In the APP/PS1dE9 model a significant negative correlation between the 5mC:5hmC ratio and plaque load was observed in the CA3 (tau = −0.71, p = 0.019), and a negative correlation between DNMT3A and plaque levels in the DG (tau = −0.56, p = 0.048) and CA3 (tau = −0.61, p = 0.029) (Figure 6). In the 3xTg-AD model and vervets no statistically significant correlations between plaque load and epigenetic markers were observed (Figure 7 and Figure 8, respectively).
Figure 5.
Correlation analysis results between epigenetic markers 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC) and DNA methyltransferase 3A (DNMT3A) immunoreactivity (IR), and the 5mC:5hmC ratio, and plaque load in J20 mice. Shown are the background-corrected and scaled integrated density data plotted against the scaled fraction of hippocampal area covered by plaques, for the dentate gyrus (DG), cornu ammonis (CA) 3, and CA1–2 subregions of the hippocampus. Fitted linear regression lines are shown for clarity. A statistically significant correlation with plaque load was found for 5mC IR in the DG (tau = −0.49, p = 0.034), and the 5mC:5hmC ratio in the DG (tau = −0.52, p = 0.024) and CA3 (tau = −0.46, p = 0.047). AU, arbitrary units.
Figure 6.
Correlation analysis results between epigenetic markers 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC) and DNA methyltransferase 3A (DNMT3A) immunoreactivity (IR), and the 5mC:5hmC ratio, and plaque load in APP/PS1dE9 mice. Shown are the background-corrected and scaled integrated density data plotted against the scaled fraction of hippocampal area covered by plaques, for the dentate gyrus (DG), cornu ammonis (CA) 3, and CA1–2 subregions of the hippocampus. Fitted linear regression lines are shown for clarity. A statistically significant correlation with plaque load was found for the 5mC:5hmC ratio in the CA3 (tau = −0.71, p = 0.019), and DNMT3A IR in the DG (tau = −0.56, p = 0.048) and CA3 (tau = −0.61, p = 0.029). AU, arbitrary units.
Figure 7.
Correlation analysis results between epigenetic markers 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC) and DNA methyltransferase 3A (DNMT3A) immunoreactivity (IR), and the 5mC:5hmC ratio, and plaque load in 3xTg-AD mice. Shown are the background-corrected and scaled integrated density data plotted against the scaled fraction of hippocampal area covered by plaques, for the dentate gyrus (DG), cornu ammonis (CA) 3, and CA1–2 subregions of the hippocampus. Fitted linear regression lines are shown for clarity. No statistically significant correlation was found between plaque load and any of the investigated epigenetic markers. AU, arbitrary units.
Figure 8.
Correlation analysis results between epigenetic markers 5-methylcytosine (5mC), 5-hydroxymethylcytosine (5hmC) and DNA methyltransferase 3A (DNMT3A) immunoreactivity (IR), and the 5mC:5hmC ratio, and plaque load in Caribbean vervets. Shown are the backgroundcorrected and scaled integrated density data plotted against the scores of hippocampal plaque load, for the dentate gyrus (DG), cornu ammonis (CA) 3, and CA1–2 subregions of the hippocampus. Fitted linear regression lines are shown for clarity. No statistically significant correlation was found between plaque load and any of the investigated epigenetic markers. AU, arbitrary units.
Discussion
Age-related alterations of 5mC, 5hmC, and DNMT3A, three epigenetic markers previously associated with aging, age-related cognitive decline, and/or AD, were investigated in 3 transgenic mouse models of AD and a non-human primate model that is known to develop Aβ plaque pathology with age. Semi-quantitative analysis of 5mC, 5hmC, and DNMT3A IR indicated striking differences in age-related DNA methylation patterns between the different models, while none of the models showed age-related differences in levels of DNA hydroxymethylation and DNMT3A. Plaque load correlated with DNA methylation and 5mC:5hmC ratio in the J20 model, and with the 5mC:5hmC ratio and DNMT3A in the APP/PS1 model, the mouse model with the most severe plaque pathology in the hippocampus.
Age-related decrease of DNA methylation levels in the DG and CA3 of J20 mice
In the present study, aging was associated with a decrease in 5mC IR in the DG and a decrease of the 5mC:5hmC ratio in the DG and CA3. Since 5hmC seems to remain stable with age, this decrease in the 5mC:5hmC ratio is likely due to a decrease in 5mC, even though 5mC IR alone was not decreased in the CA3. Furthermore, correlation analysis between hippocampal plaque load and the investigated epigenetic markers only showed a statistically significant correlation between plaque load and 5mC in the DG and the 5mC:5hmC ratio in the DG and CA3 region; the same areas that showed age-related alterations.
Transgenic J20 mice express human APP with both the Swedish and Indiana mutations associated with the development of familial AD (Mucke et al., 2000). It has previously been reported that these mice already show cognitive deficits starting at 1–2 months of age, but that these impairments do not seem to progress with age up to the development of plaque pathology, which starts around 6 months of age, but ramps up around 10 months of age (Mucke et al., 2000; Webster et al., 2014).
It is most likely that in these models based on mutations seen in familial AD, epigenetic alterations play a role in the progression of the disease and are instigated through other pathological processes directly related to these mutated genes. For instance, Aβ has been reported to influence DNA methylation, inducing global hypomethylation (Chen et al., 2009), which is in line with the observations in the J20 model in the present study. Interestingly, the observation that the promoter of the MAPT gene, which has been implicated in AD, was hypomethylated in J20 mice already at 5 months of age indicates that epigenetic processes may be involved in the development of AD before the occurrence of senile plaques (Coupland et al., 2014). Note, however, that the global changes in DNA methylation observed in the present study occur after the development of extracellular plaque formation.
Another study reports increased levels of histone deacetylase (HDAC) activity in J20 mice 19–20 months of age (Kuo et al., 2012). Although an increase in HDAC activity is generally linked to an increase in DNA methylation (Cedar and Bergman, 2009), this is not consistently observed in AD (Lardenoije et al., 2015a). This inconsistency is also reflected in the present study and may implicate possible methodological differences or the existence of different pathological mechanisms with alternative effects on epigenetic profiles.
Epigenetic markers remain stable with age in APP/PS1dE9 mice
In the present study, extensive plaque deposition was observed in the hippocampus of APP/PS1dE9, but no age-related epigenetic changes were detected. Interestingly, the 5mC:5hmC ratio and DNMT3A levels showed a negative correlation with plaque load. The APP/PS1dE9 transgenic mouse model expresses human PS1 with the deletion of exon 9 and humanized APP with the Swedish mutation (Jankowsky et al., 2003, 2001). This mouse model is widely used and cognitive impairments have been reported for spatial working memory as early as 4 months of age, with additional impairments in reference memory, associative learning, and passive avoidance with increasing age (Frost et al., 2015; Park et al., 2006; Webster et al., 2014).
Recently it was found that in APP/PS1dE9 mice SUMOylation by SUMO1 of HDAC1, which can be induced by Aβ exposure, reduced cognitive impairments and neuropathology, and may thus serve as a defense mechanism against Aβ toxicity (Tao et al., 2017). Another study in the APP/PS1dE9 mice implicated histone deacetylation of Bdnf by HDAC2 in memory performance (Hsiao et al., 2017), and others revealed that treatment with HDAC inhibitors can ameliorate cognitive impairments in these mice (Kilgore et al., 2010). No global differences in hippocampal histone (H) 3 and H4 acetylation levels were detected. Interestingly, studies in similar models with the same AD-related genes, but different mutations, have reported different results. A study using an APP/PS1 model expressing human PS1 with the M146V mutation, found hippocampal decreases in H4 acetylation after fear conditioning, compared to wild-type mice (Francis et al., 2009). Treatment with an HDAC inhibitor was able to both rescue the acetylation levels and behavioral responses in the APP/PS1 mice. Similar findings were obtained in APP/PS1 mice with the APP KM670/671NL and PS1 L166P mutations (Govindarajan et al., 2011), and in APP/PS1 mice with chimeric APP with the K670N/M671L mutation and the PS1 A246E mutation (Wang et al., 2014).
Sanchez-Mut et al. (2013) performed a DNA methylation microarray study in the frontal cortex of similar APP/PS1 mice as used by Wang et al. (2015) and observed F2rl2, Sorbs3, Spnb4, and Tbxa2r to be hypermethylated. A genome-wide analysis of DNA methylation has also been performed in cortex of the APP/PS1dE9 model (Cong et al., 2014). In this study, sites that were observed to be differentially methylated in the APP/PS1dE9 model, as compared to wild-type controls, were mainly hypermethylated. Additionally, they identified transforming growth factor β1 and its associated signaling pathway to be mainly dysregulated. Due to the differences in approach and brain area, it is, however, difficult to compare these results with the present findings.
DNA methylation levels increase in the DG and CA1–2 of 3xTg-AD mice
In contrast to the J20 and APP/PS1 models, the 3xTg-AD model, expressing mutated APP, PS1, and MAPT, not only develops plaques, but also neurofibrillary tangles (Oddo et al., 2003). Contrary to the J20 model, the 3xTg-AD model shows an increase in DNA methylation in the DG and CA1–2 subregions. A possible explanation for this disparity between the models would be the addition of the mutated MAPT transgene, although there is evidence that tau does not cause global hypermethylation, but hypomethylation through oxidative stress and DNA damage (Frost et al., 2014). The 3xTg-AD mice are reported to develop extracellular amyloid deposits in frontal cortex by 6 months, which progressively spread throughout the brain by 12 months of age (Oddo et al., 2003), and tau pathology which appears later, after about 12 to 15 months (Billings et al., 2005). We observed a 2–3 month delay in AD pathology within our 3xTg-AD colony, possibly due to reduced transgene copies with successive breeding (see https://www.jax.org/strain/004807).
However, our findings confirm that hippocampal plaque formation occurs mainly in the deeper layers of the hippocampus and subiculum, and fewer plaques were seen in the DG, CA3, and CA1–2 subregions when compared to the J20 and APP/PS1 models. Importantly, cognitive impairments occur before the development of plaques and tangles, starting around 4 months, and correlate with intraneuronal Aβ (Billings et al., 2005).
The direction of the DNA methylation changes may appear contra intuitive, but it appears that other studies investigating epigenetic changes in this triple transgenic model have made similar observations. For instance, Sanchez-Mut et al. (2013) found an increase in Tbxa2r, F2rl2, Spnb4, and Sorbs3 methylation, with a decrease in the corresponding mRNA levels in the cortex of 3xTg-AD mice. Walker et al. (2013) investigated age-related histone modification changes in neurons from wild-type and 3xTg-AD mice. They found that the repressive H3 lysine (K) 9 methylation marker increased with age in 3xTg-AD neurons, more so than in wild-type neurons. This finding of increased epigenetic repression of gene expression is confirmed by the detection of lower Bdnf gene expression. Importantly, increases in H3K9 methylation were already observed at 4 months, the same age at which cognitive deficits start to emerge (Walker et al., 2013). Another study in the hippocampus of 3xTg-AD mice reported an age-related loss of H3K4 trimethylation, an epigenetic marker that is associated with gene expression (Mastroeni et al., 2015). These observations of increased epigenetic repression are in line with the present study, as DNA methylation is also generally associated with suppression of gene expression (Bird and Wolffe, 1999).
Interestingly, however, in neurons isolated from 3xTg-AD mice, Walker et al. (2013) also observed increases in H3 and H4 acetylation levels with age; epigenetic markers which are associated with enhanced gene expression. As stated previously, these markers decreased in the J20 and APP/PS1 models. A gene-specific analysis showed that the promoter region of the AD-associated Bace1 gene exhibited increased H3 acetylation, concomitant with increased mRNA levels, in the cortex of 3xTg-AD mice (Marques et al., 2012). H4K12 acetylation has also been investigated as a potential biomarker in blood monocytes of 3xTg-AD mice and it was found that this marker was elevated at 10 months of age, during the development of plaque pathology, but not anymore at 20 months of age when plaque pathology is already widespread (Plagg et al., 2015).
Nevertheless, treatment with HDAC inhibitors is also able to improve cognition in the 3xTg-AD mice (Green et al., 2008; Sung et al., 2013). It is therefore necessary for future studies to elucidate which genes are affected by these global changes in histone acetylation, as treatment with HDAC inhibitors may have a beneficial effect independent of AD-related alterations in histone acetylation. This is exemplified by studies showing beneficial effects of HDAC inhibitors during normal aging (Peleg et al., 2010).
In the present study, we do not detect any significant age-related changes in 5hmC IR. Another immunohistochemical study, however, comparing wild-type and 3xTg-AD mice at 17 months detected an increase in cortical 5hmC levels (Cadena-del-Castillo et al., 2014). These differences may be due to differences in methodology, brain area investigated, and age of the studied animals, and clearly, stress the sensitive and complex nature of epigenetic processes and investigations thereof.
No age-related alterations of global epigenetic marks are detected in the hippocampus of Caribbean vervets
We observed no age-related changes in the level of epigenetic marks in the hippocampus of vervets. As opposed to the transgenic mouse models, the vervets serve as a more natural model of AD as some of these non-human primates develop AD-like pathology as they age, without the introduction of mutated transgenes. Interestingly, as AD in humans generally develops at advanced ages, vervets exhibit plaque pathology as early as 15 years, while their lifespan is only 15 to 20 years in the wild (as opposed to 20 to 30 years in captivity) (Lemere et al., 2004). In general, plaque deposition in vervets starts in the frontal cortex and spreads with age (C. Lemere, personal communication), sometimes approaching, by 30 years of age, Aβ pathology as seen in human AD. In the J20 and 3xTg-AD models, the most drastic changes in the levels of the studied epigenetic marks were seen in the extremely old age groups. Thus, it may be the case that AD-pathology in the vervets was not advanced enough, especially in the hippocampus, to induce detectable changes. As the pathology in the vervets was much more variable than in the transgenic mice, the sample size may have been too small to detect significant correlations between age, Aβ pathology, and age. Alternatively, the long-term fixation of the archived vervet brain tissue may have limited the accessibility of antigens for accurate immunohistochemical detection.
Other studies focusing on age-related epigenetic alterations in vervets are scarce, although a study investigating blood DNA methylation in relation to a high fat diet also found no significant association between age and DNA methylation levels (ages between 9.7 and 23.7 years) (Pheiffer et al., 2014).
Translational validity
Given the discrepancy in observations regarding DNA methylation across the different animal models, it is essential to compare the results from the animal models with observations from human and related studies in order to elucidate how the genetically different models may be able to reflect the epigenetic alterations associated with AD. However, as noted previously, differences in brain regions and methodology already hamper comparisons between studies performed on human tissue, let alone comparisons with other species. Additionally, whereas animal models have the advantage to facilitate the study of the temporal sequence of events, human studies focusing on brain markers generally need to rely on post-mortem tissue. Therefore, instead of comparing changes over time, they compare diseased with control brains. It is also important to consider that non-human primates and humans have greater genetic heterozygosity and environmental diversity than homogenous transgenic mouse models.
Targeted approaches aside, there are several studies that investigated AD-related global changes in DNA methylation and hydroxymethylation markers using human brain material. Initial studies, also by our group, have shown an AD-associated global DNA hypomethylation and hypohydroxymethylation in the entorhinal cortex and hippocampus (Chouliaras et al., 2013; Mastroeni et al., 2010). These studies also found an AD-associated decrease in DNA methylation and hydroxymethylation in a monozygotic twin pair of which only one developed AD. Other groups, however, either did not find significant changes in global DNA methylation in the entorhinal cortex (Lashley et al., 2015), or even increases in DNA methylation and hydroxymethylation in the hippocampus, middle frontal gyrus and middle temporal gyrus (Bradley-Whitman and Lovell, 2013; Coppieters et al., 2014). Although these studies do not all show the same direction of change, they observe similar alterations in DNA methylation and hydroxymethylation, whereas Condliffe et al. (Condliffe et al., 2014) found a decrease of only DNA hydroxymethylation in the entorhinal cortex and cerebellum. Rao et al. (2012) only investigated DNA methylation and found an increase in the frontal cortex of AD patients. Using the HumanMethylation450 BeadChip assay on tissue from the dorsolateral prefrontal cortex De Jager et al. (De Jager et al., 2014) reported a modest increase of the methylation value of differentially methylated loci in AD.
Since findings in the same brain region also disagree, it is unlikely that the discrepancies can be explained solely by differences in brain areas. As has been noted previously (Coppieters et al., 2014; Lardenoije et al., 2015a; Lashley et al., 2015), the most likely explanation would be differences in methodology, such as tissue processing and quantification methods. To elucidate how differences in methodology can influence detected DNA methylation and hydroxymethylation levels, different procedures should be systemically tested on the same tissue, and vice versa, the same approach should be used on various tissues. For the present study, however, similar methodology was used to process all of the mouse tissues and to quantify the epigenetic markers. The different observations for the various mouse models would thus point to a genotype effect, possibly due to the expression of different transgenes and/or the use of different promoters. A genotype effect, in turn, is unlikely to explain the differences between the human studies as they generally study mixed samples of, based on the average age, mainly late-onset AD, although due to the large age-range and lack of (reported) genetic tests also familial cases may be included (Coppieters et al., 2014).
To get a better idea of the direction of the epigenetic changes in AD it may help to look at related studies. Observations from in vitro work related to the effect of Aβ and APP mutations on DNA methylation, are more consistent and indicate there is global hypomethylation (Chen et al., 2009; Hodgson et al., 2013; Sung et al., 2011). Additionally, work on tau indicates that it induces global heterochromatin loss, which may lead to aberrant gene expression patterns in AD (Frost et al., 2014). A loss of heterochromatin also points towards a hypomethylated state of the DNA.
DNMTs depend on S-adenosylmethionine (SAM) as methyl donor and some studies have found striking deficiencies of SAM and S-adenosylhomocysteine (SAH), the demethylated metabolite of SAM, throughout the AD-afflicted brain and cerebrospinal fluid (Bottiglieri et al., 1990; Eto et al., 2002; Morrison et al., 1996). In vitro work investigating the relationship between folate, SAM, and DNA methylation indicates a SAM deficiency leads to global hypomethylation (Fuso et al., 2005). Another study found increased levels of brain SAH in AD patients and showed that SAH inhibits methyltransferases, suggesting that increased levels of SAH would also lead to DNA hypomethylation (Kennedy et al., 2004). Others have indeed shown decreased methyltransferase activity in the brain of AD patients (Goggins et al., 1999). Inhibition of DNMTs could also explain alterations in DNA methylation, without changes in the levels of DNMT3A, as seen in the present study.
The direct and indirect evidence mainly points towards a hypomethylated state in AD. Of the investigated models, only the J20 mice exhibit an age-related global DNA hypomethylation in the hippocampus and therefore seems to best capture this view. Of note, in light of previous observations that DNA methylation increases with age (Chouliaras et al., 2012a; Hernandez et al., 2011), the lack of an increase in global DNA methylation in the old mice of the APP/PS1dE9 model could be the result of hypomethylation when compared to normal aging. Similarly, DNA hydroxymethylation and DNMT3A levels have been observed to increase with age (Chouliaras et al., 2012b, 2011; Münzel et al., 2010; Song et al., 2011), therefore, the lack of an age-related change in these markers could be the result of a decrease in comparison to wild-type aged animals. This, however, is beyond the scope of the current study and should be investigated through a direct comparison of transgenic and non-transgenic litter-mates.
Strengths, limitations, and future perspectives
A great strength of the present study is the inclusion of multiple animal models, which has provided crucial insights in how the different mouse models capture AD on an epigenetic level. The inclusion of a non-transgenic non-human primate model provides an additional angle more related to sporadic AD, in contrast to the transgenic mouse models, which are limited to familial forms of AD. Additionally, the use of established immunohistochemistry-based techniques allows the subregion-specific qualitative and semiquantitative analysis of epigenetic markers. Importantly, by using highly specific antibodies 5mC and 5hmC can be reliably distinguished (Chouliaras et al., 2012b; Ficz et al., 2011), which is not possible with other commonly used techniques for 5mC detection (van den Hove et al., 2012).
The study also has its limitations, which thus should be taken into account when interpreting the results. First, although the staining and image-analysis procedures were identical for all the models and performed at the same time, the breeding, sacrificing and tissue processing were not done at the same time, which may have resulted in slight differences between the mouse models. The vervet tissue was differently processed and fixed for a longer time, which may affect immunoreactivity. Additionally, the vervets did not live under strict experimentally controlled conditions such as the mouse models, which may result in more variation. The exclusion of an aging wild-type mouse group may be seen as a limitation, but our group has previously reported extensive epigenetic investigations in normally aging mice (Chouliaras et al., 2012a, 2012b, 2011; Lardenoije et al., 2015b), which were done in a similar manner and can therefore be used to compare the current findings in relation to AD with. Although the used techniques allowed for a subregion-specific analysis of epigenetic markers, recent work has shown that distinguishing between cell-types may also be relevant in the context of neurodegeneration (Mastroeni et al., 2017; Sanchez-Mut et al., 2017).
The borderline significance often observed in the present study suggests that it could have benefitted from larger sample sizes to increase power. However, since this explorative study depended on the availability of animals from other studies, it was not possible to increase the samples sizes. It remains an important point, which has been previously raised (Lunnon and Mill, 2013; Sanchez-Mut and Gräff, 2015), that most current epigenetic studies are relatively small, which may result in the large differences in results. Therefore, there is a need for large, high-powered studies that may provide more conclusive results. Additionally, gene-specific epigenetic investigations would benefit from the integration of different levels of epigenetic regulation, as well as gene expression measurements (e.g. see (Budden et al., 2015)). Nevertheless, the present study serves as an important foundation to guide future AD-related epigenetics research in animal models.
Conclusion
This study set out to determine age-related changes in immunohistochemically detectable markers related to DNA methylation in several widely used and genetically different animal models of AD, and to determine how these models reflect the epigenetic changes observed in AD. In the J20 model global DNA hypomethylation was observed in the hippocampus, while in the 3xTg-AD model global hypermethylation was observed. No alterations in DNA hydroxymethylation or DNMT3A levels were detected in these models. In APP/PS1dE9 and Caribbean vervets, no age-related global epigenetic changes were observed. Although plaque load generally increases with age, a slightly different pattern was seen in the correlation analysis between plaque load and the epigenetic markers. A negative correlation between 5mC and the 5mC:5hmC ratio was found in the J20 model, and in the APP/PS1dE9 model the 5mC:5hmC ratio and DNMT3A showed a negative correlation with hippocampal plaque load. For 3xTg-AD mice and vervets no significant correlations were detected. The main differences between the investigated models were thus in the age-related changes in DNA methylation. Other studies looking at DNA methylation or the effects of Aβ, tau, and SAM/SAH on DNA methylation report mixed results, but appear to lean towards global hypomethylation in AD. Although this would suggest the J20 model best captures the global epigenetic changes related to AD, it appears the different genotypes of the models differentially affect the course of age-related epigenetic changes. Exactly how mutations in the APP, PS1, and MAPT genes interact with the epigenome remains to be elucidated in future studies.
Supplementary Material
Highlights.
The J20 mouse model shows an age-related hippocampal decrease in DNA methylation.
3xTg-AD mice exhibit a hippocampal increase in DNA methylation with age.
No epigenetic changes were detected in aging APP/PS1dE9 mice and Caribbean vervets.
Differences between models suggests interaction genotype and epigenetic state.
Acknowledgements
We kindly thank H Crehan, Q Shi, and S Chowdhury for technical assistance. Funds have been provided by the Internationale Stichting Alzheimer Onderzoek (ISAO) grants 07551 and 11532 (D.L.A.vdH.), by the ISAO grants 09552 and 13515, and the Netherlands Organization for Scientific Research (NWO), grant 916.11.086 (Veni Award) (B.P.F.R.), by an ISAO fellowship and a fellowship as part of NWO grant 022.005.019, (R.L.), and by an Anonymous Foundation and NIH/NIA R01 AG040092 (C.A.L.). Additional funds have been provided by the Joint Programme—Neurodegenerative Disease Research (JPND) for the EPI-AD consortium (http://www.neurodegenerationresearch.eu/wp-content/uploads/2015/10/Factsheet_EPI-AD.pdf). The funding agencies were not involved in the study design, data collection, analysis and interpretation, writing of the report, and the decision to submit the article for publication. The authors declare no conflicts of interest.
Abbreviations
- 5hmC
5-hydroxymethylcytosine
- 5mC
5-methylcytosine
- Aβ
amyloid-β
- AD
Alzheimer’s disease
- APP
amyloid precursor protein
- CA
cornu ammonis
- DG
dentate gyrus
- DNMT3A
DNA methyltransferase 3A
- H
histone
- HDAC
histone deacetylase
- IR
immunoreactivity
- K
lysine
- MAPT
microtubule-associated protein tau
- PS
presenilin
- SAH
S-adenosylhomocysteine
- SAM
S-adenosylmethionine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartsch T, Wulff P, 2015. The hippocampus in aging and disease: From plasticity to vulnerability. Neuroscience. doi: 10.1016/j.neuroscience.2015.07.084 [DOI] [PubMed] [Google Scholar]
- Billings LM, Oddo S, Green KN, McGaugh JL, LaFerla FM, 2005. Intraneuronal Abeta causes the onset of early Alzheimer’s disease-related cognitive deficits in transgenic mice. Neuron 45, 675–88. doi: 10.1016/j.neuron.2005.01.040 [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP, 1999. Methylation-Induced Repression— Belts, Braces, and Chromatin. Cell 99, 451–454. doi: 10.1016/S0092-8674(00)81532-9 [DOI] [PubMed] [Google Scholar]
- Bottiglieri T, Godfrey P, Flynn T, Carney MW, Toone BK, Reynolds EH, 1990. Cerebrospinal fluid S-adenosylmethionine in depression and dementia: effects of treatment with parenteral and oral S-adenosylmethionine. J. Neurol. Neurosurg. Psychiatry 53, 1096–1098. doi: 10.1136/jnnp.53.12.1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley-Whitman MA, Lovell MA, 2013. Epigenetic changes in the progression of Alzheimer’s disease. Mech. Ageing Dev 134, 486–95. doi: 10.1016/j.mad.2013.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budden DM, Hurley DG, Crampin EJ, 2015. Modelling the conditional regulatory activity of methylated and bivalent promoters. Epigenetics Chromatin 8, 21. doi: 10.1186/s13072-015-0013-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadena-del-Castillo C, Valdes-Quezada C, Carmona-Aldana F, Arias C, Bermúdez-Rattoni F, Recillas-Targa F, 2014. Age-dependent increment of hydroxymethylation in the brain cortex in the triple-transgenic mouse model of Alzheimer’s disease. J. Alzheimers. Dis 41, 845–54. doi: 10.3233/JAD-132285 [DOI] [PubMed] [Google Scholar]
- Cavanaugh SE, Pippin JJ, Barnard ND, 2014. Animal models of Alzheimer disease: historical pitfalls and a path forward. ALTEX 31, 279–302. doi: 10.14573/altex.1310071 [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y, 2009. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet 10, 295–304. doi: 10.1038/nrg2540 [DOI] [PubMed] [Google Scholar]
- Chen K-L, Wang SS-S, Yang Y-Y, Yuan R-Y, Chen R-M, Hu C-J, 2009. The epigenetic effects of amyloid-beta(1–40) on global DNA and neprilysin genes in murine cerebral endothelial cells. Biochem. Biophys. Res. Commun 378, 57–61. doi: 10.1016/j.bbrc.2008.10.173 [DOI] [PubMed] [Google Scholar]
- Choudhuri S, 2011. From Waddington’s epigenetic landscape to small noncoding RNA: some important milestones in the history of epigenetics research. Toxicol Mech Methods 21, 252–274. doi: 10.3109/15376516.2011.559695 [DOI] [PubMed] [Google Scholar]
- Chouliaras L, Mastroeni D, Delvaux E, Grover A, Kenis G, Hof PR, Steinbusch HWM, Coleman PD, Rutten BPF, van den Hove DLA, 2013. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer’s disease patients. Neurobiol. Aging 34, 2091–2099. doi: 10.1016/j.neurobiolaging.2013.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouliaras L, Rutten BPF, Kenis G, Peerbooms O, Visser PJ, Verhey F, van Os J, Steinbusch HWM, van den Hove DLA, 2010. Epigenetic regulation in the pathophysiology of Alzheimer’s disease. Prog. Neurobiol 90, 498–510. doi: 10.1016/j.pneurobio.2010.01.002 [DOI] [PubMed] [Google Scholar]
- Chouliaras L, van den Hove DLA, Kenis G, Dela Cruz J, Lemmens MAM, van Os J, Steinbusch HWM, Schmitz C, Rutten BPF, 2011. Caloric restriction attenuates age-related changes of DNA methyltransferase 3a in mouse hippocampus. Brain. Behav. Immun 25, 616–623. doi: 10.1016/j.bbi.2010.11.016 [DOI] [PubMed] [Google Scholar]
- Chouliaras L, van den Hove DLA, Kenis G, Keitel S, Hof PR, van Os J, Steinbusch HWM, Schmitz C, Rutten BPF, 2012a. Prevention of age-related changes in hippocampal levels of 5-methylcytidine by caloric restriction. Neurobiol. Aging 33, 1672–81. doi: 10.1016/j.neurobiolaging.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouliaras L, van den Hove DLA, Kenis G, Keitel S, Hof PR, van Os J, Steinbusch HWM, Schmitz C, Rutten BPF, 2012b. Age-related increase in levels of 5-hydroxymethylcytosine in mouse hippocampus is prevented by caloric restriction. Curr. Alzheimer Res 9, 536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condliffe D, Wong A, Troakes C, Proitsi P, Patel Y, Chouliaras L, Fernandes C, Cooper J, Lovestone S, Schalkwyk L, Mill J, Lunnon K, 2014. Cross-region reduction in 5-hydroxymethylcytosine in Alzheimer’s disease brain. Neurobiol. Aging 35, 1850–1854. doi: 10.1016/j.neurobiolaging.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Jia J, Qin W, Ren Y, Sun Y, 2014. Genome-wide analysis of DNA methylation in an APP/PS1 mouse model of Alzheimer’s disease. Acta Neurol. Belg 114, 195–206. doi: 10.1007/s13760-013-0267-6 [DOI] [PubMed] [Google Scholar]
- Coppieters N, Dieriks BV, Lill C, Faull RL, Curtis MA, Dragunow M, 2014. Global changes in DNA methylation and hydroxymethylation in Alzheimer’s disease human brain. Neurobiol Aging 35, 1334–1344. doi: 10.1016/j.neurobiolaging.2013.11.031 [DOI] [PubMed] [Google Scholar]
- Coupland K, Kim WS, Halliday G, Dobson-Stone C, Kwok JBJ, 2014. MAPT methylation in Alzheimer’s disease. Alzheimer’s Dement. 10, P317–P318. doi: 10.1016/j.jalz.2014.05.274 [DOI] [PubMed] [Google Scholar]
- De Jager PL, Srivastava G, Lunnon K, Burgess J, Schalkwyk LC, Yu L, Eaton ML, Keenan BT, Ernst J, McCabe C, Tang A, Raj T, Replogle J, Brodeur W, Gabriel S, Chai HS, Younkin C, Younkin SG, Zou F, Szyf M, Epstein CB, Schneider JA, Bernstein BE, Meissner A, Ertekin-Taner N, Chibnik LB, Kellis M, Mill J, Bennett DA, 2014. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat. Neurosci 17, 1156–1163. doi: 10.1038/nn.3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defina PA, Moser RS, Glenn M, Lichtenstein JD, Fellus J, 2013. Alzheimer’s disease clinical and research update for health care practitioners. J Aging Res 2013, 207178. doi: 10.1155/2013/207178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto K, Asada T, Arima K, Makifuchi T, Kimura H, 2002. Brain hydrogen sulfide is severely decreased in Alzheimer’s disease. Biochem. Biophys. Res. Commun 293, 1485–8. doi: 10.1016/S0006-291X(02)00422-9 [DOI] [PubMed] [Google Scholar]
- Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W, 2011. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature 473, 398–402. doi: 10.1038/nature10008 [DOI] [PubMed] [Google Scholar]
- Francis YI, Fà M, Ashraf H, Zhang H, Staniszewski A, Latchman DS, Arancio O, 2009. Dysregulation of histone acetylation in the APP/PS1 mouse model of Alzheimer’s disease. J. Alzheimers. Dis 18, 131–9. doi: 10.3233/JAD-2009-1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost B, Hemberg M, Lewis J, Feany MB, 2014. Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci 17, 357–66. doi: 10.1038/nn.3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost JL, Le KX, Cynis H, Ekpo E, Kleinschmidt M, Palmour RM, Ervin FR, Snigdha S, Cotman CW, Saido TC, Vassar RJ, St George-Hyslop P, Ikezu T, Schilling S, Demuth HU, Lemere CA, 2013. Pyroglutamate-3 amyloid-beta deposition in the brains of humans, non-human primates, canines, and Alzheimer disease-like transgenic mouse models. Am J Pathol 183, 369–381. doi: 10.1016/j.ajpath.2013.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost JL, Liu B, Rahfeld J-U, Kleinschmidt M, O’Nuallain B, Le KX, Lues I, Caldarone BJ, Schilling S, Demuth H-U, Lemere CA, 2015. An anti-pyroglutamate-3 Aβ vaccine reduces plaques and improves cognition in APPswe/PS1ΔE9 mice. Neurobiol. Aging 36, 3187–99. doi: 10.1016/j.neurobiolaging.2015.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuso A, Seminara L, Cavallaro RA, D’Anselmi F, Scarpa S, 2005. Sadenosylmethionine/homocysteine cycle alterations modify DNA methylation status with consequent deregulation of PS1 and BACE and beta-amyloid production. Mol. Cell. Neurosci 28, 195–204. doi: 10.1016/j.mcn.2004.09.007 [DOI] [PubMed] [Google Scholar]
- Garcia-Alloza M, Robbins EM, Zhang-Nunes SX, Purcell SM, Betensky RA, Raju S, Prada C, Greenberg SM, Bacskai BJ, Frosch MP, 2006. Characterization of amyloid deposition in the APPswe/PS1dE9 mouse model of Alzheimer disease. Neurobiol Dis 24, 516–524. doi: 10.1016/j.nbd.2006.08.017 [DOI] [PubMed] [Google Scholar]
- Goggins M, Scott JM, Weir DG, 1999. Methylation of cortical brain proteins from patients with HIV infection. Acta Neurol. Scand 100, 326–31. [DOI] [PubMed] [Google Scholar]
- Govindarajan N, Agis-Balboa RC, Walter J, Sananbenesi F, Fischer A, 2011. Sodium butyrate improves memory function in an Alzheimer’s disease mouse model when administered at an advanced stage of disease progression. J. Alzheimers. Dis 26, 187–97. doi: 10.3233/JAD-2011-110080 [DOI] [PubMed] [Google Scholar]
- Green KN, Steffan JS, Martinez-Coria H, Sun X, Schreiber SS, Thompson LM, LaFerla FM, 2008. Nicotinamide restores cognition in Alzheimer’s disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231phosphotau. J. Neurosci 28, 11500–10. doi: 10.1523/JNEUROSCI.3203-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez DG, Nalls MA, Gibbs JR, Arepalli S, van der Brug M, Chong S, Moore M, Longo DL, Cookson MR, Traynor BJ, Singleton AB, 2011. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet 20, 1164–1172. doi: 10.1093/hmg/ddq561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson N, Trivedi M, Muratore C, Li S, Deth R, 2013. Soluble oligomers of amyloid-β cause changes in redox state, DNA methylation, and gene transcription by inhibiting EAAT3 mediated cysteine uptake. J. Alzheimers. Dis. 36, 197–209. doi: 10.3233/JAD-130101 [DOI] [PubMed] [Google Scholar]
- Hsiao Y-H, Hung H-C, Yu Y-J, Su C-L, Chen S-H, Gean P-W, 2017. Co-housing reverses memory decline by epigenetic regulation of brain-derived neurotrophic factor expression in an animal model of Alzheimer’s disease. Neurobiol. Learn. Mem 141, 1–8. doi: 10.1016/j.nlm.2017.02.020 [DOI] [PubMed] [Google Scholar]
- Iatrou A, Kenis G, Rutten BPF, Lunnon K, van Den Hove DLA, 2016. Epigenetic dysregulation of brainstem nuclei in the pathogenesis of Alzheimer’s disease: looking in the correct place at the right time? Cell. Mol. Life Sci 1–15. doi: 10.1007/s00018-016-2361-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky JL, Fadale DJ, Anderson J, Xu GM, Gonzales V, Jenkins NA, Copeland NG, Lee MK, Younkin LH, Wagner SL, Younkin SG, Borchelt DR, 2003. Mutant presenilins specifically elevate the levels of the 42 residue -amyloid peptide in vivo: evidence for augmentation of a 42-specific secretase. Hum. Mol. Genet 13, 159–170. doi: 10.1093/hmg/ddh019 [DOI] [PubMed] [Google Scholar]
- Jankowsky JL, Slunt HH, Ratovitski T, Jenkins NA, Copeland NG, Borchelt DR, 2001. Co-expression of multiple transgenes in mouse CNS: a comparison of strategies. Biomol. Eng 17, 157–165. doi: 10.1016/S1389-0344(01)00067-3 [DOI] [PubMed] [Google Scholar]
- Kennedy BP, Bottiglieri T, Arning E, Ziegler MG, Hansen LA, Masliah E, 2004. Elevated S-adenosylhomocysteine in Alzheimer brain: influence on methyltransferases and cognitive function. J. Neural Transm 111, 547–567. doi: 10.1007/s00702-003-0096-5 [DOI] [PubMed] [Google Scholar]
- Kilgore M, Miller CA, Fass DM, Hennig KM, Haggarty SJ, Sweatt JD, Rumbaugh G, 2010. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology 35, 870–80. doi: 10.1038/npp.2009.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo J-W, Su K-H, Wu C-Y, Chang C-W, Cheng IH-J, Wang H-E, Chang AC, Liu RS, 2012. 18F-FAHA PET signatures of histone deacetylase activity in the transgenic mouse model of Alzheimer’s disease. J. Nucl. Med 53, 1902-. [Google Scholar]
- Kurz A, Perneczky R, 2011. Novel insights for the treatment of Alzheimer’s disease. Prog Neuropsychopharmacol Biol Psychiatry 35, 373–379. doi: 10.1016/j.pnpbp.2010.07.018 [DOI] [PubMed] [Google Scholar]
- Lansdall CJ, 2014. An effective treatment for Alzheimer’s disease must consider both amyloid and tau. Biosci. Horizons 7, hzu002. [Google Scholar]
- Lardenoije R, Iatrou A, Kenis G, Kompotis K, Steinbusch HW, Mastroeni D, Coleman P, Lemere CA, Hof PR, van den Hove DL, Rutten BP, 2015a. The epigenetics of aging and neurodegeneration. Prog Neurobiol 131, 21–64. doi: 10.1016/j.pneurobio.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardenoije R, van den Hove DA, Vaessen TSJ, Iatrou A, Meuwissen KPV, van Hagen BTJ, Kenis G, Steinbusch HWM, Schmitz C, Rutten BPF, 2015b. Epigenetic modifications in mouse cerebellar Purkinje cells: effects of aging, caloric restriction, and overexpression of superoxide dismutase 1 on 5-methylcytosine and 5-hydroxymethylcytosine. Neurobiol. Aging 36, 3079–3089. doi: 10.1016/j.neurobiolaging.2015.08.001 [DOI] [PubMed] [Google Scholar]
- Lashley T, Gami P, Valizadeh N, Li A, Revesz T, Balazs R, 2015. Alterations in global DNA methylation and hydroxymethylation are not detected in Alzheimer’s disease. Neuropathol. Appl. Neurobiol 41, 497–506. doi: 10.1111/nan.12183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemere CA, Beierschmitt A, Iglesias M, Spooner ET, Bloom JK, Leverone JF, Zheng JB, Seabrook TJ, Louard D, Li D, Selkoe DJ, Palmour RM, Ervin FR, 2004. Alzheimer’s disease abeta vaccine reduces central nervous system abeta levels in a non-human primate, the Caribbean vervet. Am J Pathol 165, 283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li Y, Tollefsbol TO, 2008. Gene-environment interactions and epigenetic basis of human diseases. Curr Issues Mol Biol 10, 25–36. [PMC free article] [PubMed] [Google Scholar]
- Lunnon K, Mill J, 2013. Epigenetic studies in Alzheimer’s disease: current findings, caveats, and considerations for future studies. Am. J. Med. Genet. B. Neuropsychiatr. Genet 162B, 789–99. doi: 10.1002/ajmg.b.32201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunnon K, Smith R, Hannon E, De Jager PL, Srivastava G, Volta M, Troakes C, Al-Sarraj S, Burrage J, Macdonald R, Condliffe D, Harries LW, Katsel P, Haroutunian V, Kaminsky Z, Joachim C, Powell J, Lovestone S, Bennett DA, Schalkwyk LC, Mill J, 2014. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat. Neurosci 17, 1164–1170. doi: 10.1038/nn.3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques SCF, Lemos R, Ferreiro E, Martins M, de Mendonça A, Santana I, Outeiro TF, Pereira CMF, 2012. Epigenetic regulation of BACE1 in Alzheimer’s disease patients and in transgenic mice. Neuroscience 220, 256–66. doi: 10.1016/j.neuroscience.2012.06.029 [DOI] [PubMed] [Google Scholar]
- Mastroeni D, Delvaux E, Nolz J, Tan Y, Grover A, Oddo S, Coleman PD, 2015. Aberrant intracellular localization of H3k4me3 demonstrates an early epigenetic phenomenon in Alzheimer’s disease. Neurobiol. Aging 36, 3121–9. doi: 10.1016/j.neurobiolaging.2015.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J, 2010. Epigenetic changes in Alzheimer’s disease: Decrements in DNA methylation. Neurobiol. Aging 31, 2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroeni D, Sekar S, Nolz J, Delvaux E, Lunnon K, Mill J, Liang WS, Coleman PD, 2017. ANK1 is up-regulated in laser captured microglia in Alzheimer’s brain; the importance of addressing cellular heterogeneity. PLoS One 12, e0177814. doi: 10.1371/journal.pone.0177814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLeod AI, 2011. Kendall: Kendall rank correlation and Mann-Kendall trend test. R package version 2.2. [Google Scholar]
- Morrison LD, Smith DD, Kish SJ, 1996. Brain S-adenosylmethionine levels are severely decreased in Alzheimer’s disease. J. Neurochem 67, 1328–31. [DOI] [PubMed] [Google Scholar]
- Mucke L, Masliah E, Yu G-Q, Mallory M, Rockenstein EM, Tatsuno G, Hu K, Kholodenko D, Johnson-Wood K, McConlogue L, 2000. High-level neuronal expression of abeta 1–42 in wild-type human amyloid protein precursor transgenic mice: synaptotoxicity without plaque formation. J Neurosci 20, 4050–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münzel M, Globisch D, Brückl T, Wagner M, Welzmiller V, Michalakis S, Müller M, Biel M, Carell T, 2010. Quantification of the Sixth DNA Base Hydroxymethylcytosine in the Brain. Angew. Chemie Int. Ed 49, 5375–5377. doi: 10.1002/anie.201002033 [DOI] [PubMed] [Google Scholar]
- Oddo S, Caccamo A, Shepherd JD, Murphy MP, Golde TE, Kayed R, Metherate R, Mattson MP, Akbari Y, LaFerla FM, 2003. Triple-transgenic model of Alzheimer’s disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron 39, 409–421. [DOI] [PubMed] [Google Scholar]
- Park JH, Widi GA, Gimbel DA, Harel NY, Lee DHS, Strittmatter SM, 2006. Subcutaneous Nogo receptor removes brain amyloid-beta and improves spatial memory in Alzheimer’s transgenic mice. J. Neurosci 26, 13279–86. doi: 10.1523/JNEUROSCI.4504-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, Salinas-Riester G, Dettenhofer M, Kang H, Farinelli L, Chen W, Fischer A, 2010. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328, 753–6. doi: 10.1126/science.1186088 [DOI] [PubMed] [Google Scholar]
- Pheiffer C, Dias S, Muller C, Louw J, 2014. Decreased global DNA methylation in the white blood cells of high fat diet fed vervet monkeys (Chlorocebus aethiops). J. Physiol. Biochem 70, 725–733. doi: 10.1007/s13105-014-0341-4 [DOI] [PubMed] [Google Scholar]
- Plagg B, Ehrlich D, Kniewallner KM, Marksteiner J, Humpel C, 2015. Increased Acetylation of Histone H4 at Lysine 12 (H4K12) in Monocytes of Transgenic Alzheimer’s Mice and in Human Patients. Curr. Alzheimer Res 12, 752–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DL, Sisodia SS, 1998. Mutant genes in familial Alzheimer’s disease and transgenic models. Annu Rev Neurosci 21, 479–505. doi: 10.1146/annurev.neuro.21.1.479 [DOI] [PubMed] [Google Scholar]
- Raina P, Santaguida P, Ismaila A, Patterson C, Cowan D, Levine M, Booker L, Oremus M, 2008. Effectiveness of cholinesterase inhibitors and memantine for treating dementia: evidence review for a clinical practice guideline. Ann Intern Med 148, 379–397. [DOI] [PubMed] [Google Scholar]
- Rao JS, Keleshian VL, Klein S, Rapoport SI, 2012. Epigenetic modifications in frontal cortex from Alzheimer’s disease and bipolar disorder patients. Transl. Psychiatry 2, e132. doi: 10.1038/tp.2012.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Mut JV, Heyn H, Vidal E, Delgado-Morales R, Moran S, Sayols S, Sandoval J, Ferrer I, Esteller M, Gräff J, 2017. Whole genome grey and white matter DNA methylation profiles in dorsolateral prefrontal cortex. Synapse 71, e21959. doi: 10.1002/syn.21959 [DOI] [PubMed] [Google Scholar]
- Sanchez-Mut JV, Aso E, Panayotis N, Lott I, Dierssen M, Rabano A, Urdinguio RG, Fernandez AF, Astudillo A, Martin-Subero JI, Balint B, Fraga MF, Gomez A, Gurnot C, Roux J-C, Avila J, Hensch TK, Ferrer I, Esteller M, 2013. DNA methylation map of mouse and human brain identifies target genes in Alzheimer’s disease. Brain 136, 3018–3027. doi: 10.1093/brain/awt237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Mut JV, Gräff J, 2015. Epigenetic Alterations in Alzheimer’s Disease. Front. Behav. Neurosci 9, 347. doi: 10.3389/fnbeh.2015.00347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C-X, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen C-H, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C, 2011. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol 29, 68–72. doi: 10.1038/nbt.1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa-Ortiz AL, Acosta-Castillo I, Prince MJ, 2012. Epidemiology of dementias and Alzheimer’s disease. Arch Med Res 43, 600–608. doi: 10.1016/j.arcmed.2012.11.003 [DOI] [PubMed] [Google Scholar]
- Stratmann K, Heinsen H, Korf HW, Del Turco D, Ghebremedhin E, Seidel K, Bouzrou M, Grinberg LT, Bohl J, Wharton SB, den Dunnen W, Rub U, 2015. Precortical phase of Alzheimer’s disease (AD)-related tau cytoskeletal pathology. Brain Pathol. doi: 10.1111/bpa.12289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung HY, Choi EN, Ahn Jo S, Oh S, Ahn J-H, 2011. Amyloid protein-mediated differential DNA methylation status regulates gene expression in Alzheimer’s disease model cell line. Biochem. Biophys. Res. Commun 414, 700–5. doi: 10.1016/j.bbrc.2011.09.136 [DOI] [PubMed] [Google Scholar]
- Sung YM, Lee T, Yoon H, DiBattista AM, Song JM, Sohn Y, Moffat EI, Turner RS, Jung M, Kim J, Hoe H-S, 2013. Mercaptoacetamide-based class II HDAC inhibitor lowers Aβ levels and improves learning and memory in a mouse model of Alzheimer’s disease. Exp. Neurol 239, 192–201. doi: 10.1016/j.expneurol.2012.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao CC, Hsu WL, Ma YL, Cheng SJ, Lee EH, 2017. Epigenetic regulation of HDAC1 SUMOylation as an endogenous neuroprotection against Aβ toxicity in a mouse model of Alzheimer’s disease. Cell Death Differ. 24, 597–614. doi: 10.1038/cdd.2016.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hove DLA, Chouliaras L, Rutten BPF, 2012. The role of 5-hydroxymethylcytosine in aging and Alzheimer’s disease: current status and prospects for future studies. Curr. Alzheimer Res 9, 545–9. [DOI] [PubMed] [Google Scholar]
- Van den Hove DL, Kompotis K, Lardenoije R, Kenis G, Mill J, Steinbusch HW, Lesch KP, Fitzsimons CP, De Strooper B, Rutten BPF, 2014. Epigenetically regulated microRNAs in Alzheimer’s disease. Neurobiol. Aging 35, 731–745. doi: 10.1016/j.neurobiolaging.2013.10.082 [DOI] [PubMed] [Google Scholar]
- van Goethem NP, Lardenoije R, Kompotis K, Rutten BPF, Prickaerts J, Steinbusch HWM, 2014. Cognitive Disorders: Impairment, Aging, and Dementia, in: In Vivo Models for Drug Discovery. Wiley-VCH Verlag GmbH & Co. KGaA, pp. 349–366. doi: 10.1002/9783527679348.ch16 [DOI] [Google Scholar]
- Walker MP, LaFerla FM, Oddo SS, Brewer GJ, 2013. Reversible epigenetic histone modifications and Bdnf expression in neurons with aging and from a mouse model of Alzheimer’s disease. Age (Dordr). 35, 519–31. doi: 10.1007/s11357-011-9375-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J-Y, Gong M-Y, Ye Y-L, Ye J-M, Lin G-L, Zhuang Q-Q, Zhang X, Zhu J-H, 2015. The RIT2 and STX1B polymorphisms are associated with Parkinson’s disease. Parkinsonism Relat. Disord 21, 300–302. doi: 10.1016/j.parkreldis.2014.12.006 [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang X-J, Li T, Li J, Tang Y, Le W, 2014. Valproic acid reduces neuritic plaque formation and improves learning deficits in APP(Swe) /PS1(A246E) transgenic mice via preventing the prenatal hypoxia-induced down-regulation of neprilysin. CNS Neurosci. Ther 20, 209–17. doi: 10.1111/cns.12186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster SJ, Bachstetter AD, Nelson PT, Schmitt FA, Van Eldik LJ, 2014. Using mice to model Alzheimer’s dementia: an overview of the clinical disease and the preclinical behavioral changes in 10 mouse models. Front. Genet 5, 88. doi: 10.3389/fgene.2014.00088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H, 2009. ggplot2 - Elegant Graphics for Data Analysis | Hadley Wickham | Springer. Springer-Verlag, New York. doi: 10.1007/978-0-387-98141-3 [DOI] [Google Scholar]
- Wickham H, Francois R, 2015. dplyr: A Grammar of Data Manipulation. R package version 0.4.3. http://CRAN.R-project.org/package=dplyr. [Google Scholar]
- Woodruff-Pak DS, 2008. Animal models of Alzheimer’s disease: therapeutic implications. J Alzheimers Dis 15, 507–521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.