Abstract
With the advancing age of humans and with it, growing numbers of age-related diseases, aging has become a major focus in recent research. The lack of fitting aging models, especially in neurological diseases where access to human brain samples is limited, has highlighted direct conversion into induced neurons (iN) as an important method to overcome this challenge. Contrary to iPSC reprogramming and its corresponding cell rejuvenation, the generation of iNs enables us to retain aging signatures throughout the conversion process and beyond. In this review, we explore different cell reprogramming methods in light of age-associated neurodegenerative diseases and discuss different approaches, advances, and limitations.
Keywords: brain aging, induced neurons (iNs), disease modeling, age-associated neurodegeneration, induced pluripotent stem cells (iPSCs), rejuvenation
1. Introduction
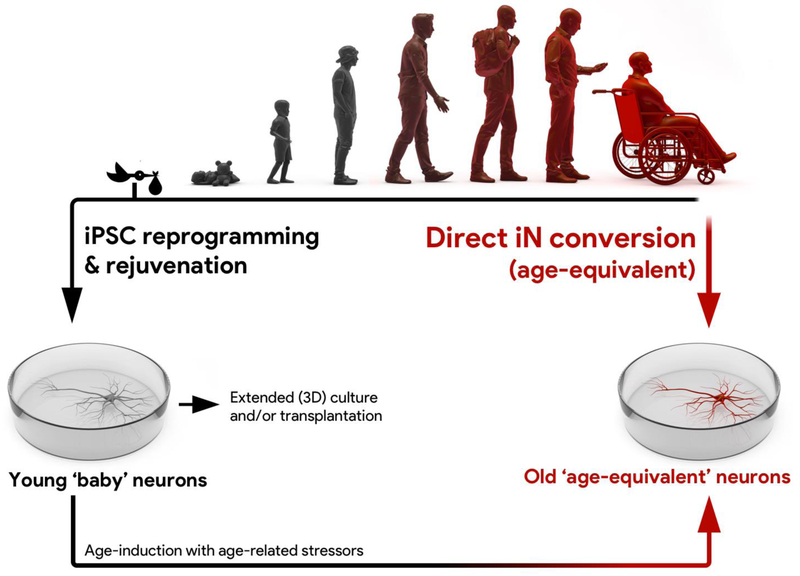
Aging is one of the most fundamental biological processes, and by far the most crucial risk factor for several human age-associated diseases, including cancer, cardiovascular diseases, diabetes and neurodegenerative diseases [1]. As the numbers of elderly increase, so too does the need for appropriate human disease model systems to advance our understanding of the contribution of age to the onset of cellular dysfunction and disease (Fig. 1) [2]. As old age does not cause disease solitarily, a ‘multiple-hit theory’ appears helpful when trying to better describe the interface between aging and age-related diseases. Here, several ‘hits’ on a cell or organism are necessary in order for an age-related disease to unfold, and at least one of these ‘hits’ is an aging phenotype, while other ‘hits’ are of genomic predisposition, environmental factor, or unknown nature [3,4]. Animal models for age-related diseases have yielded important results, but are limited by comparatively short life spans relative to humans. While the age-related mechanisms compromising the physiology of a one-month-old nematode, a two-years-old mouse, and an 80-years-old human might partially overlap, the transferability of results is not trivial, and is further complicated by differences in physiology. The discovery of induced pluripotent stem cells (iPSCs) 11 years ago has sparked the development of numerous human patient-specific disease models. The general strategy is to reprogram patient and control somatic cells, typically skin fibroblasts, into iPSCs, and subsequently differentiate them into the target cell type(s) affected by the actual disease [5]. Improved differentiation protocols have made it possible to generate subtype-specific and mature neuronal cultures from iPSCs [6–8]. This technology has provided a unique opportunity to investigate important aspects of neurodegenerative diseases, and has proven particularly useful for conditions with a strong genetic component [9–11]. However, biological aging is very complex and in addition to genomic mutations, which contribute a small fraction to the cellular aging phenotype, also involves changes in epigenetic control, damage of long-lived cellular structures, impaired energy metabolism, and other cellular mechanisms [12,13]. During cellular reprogramming however, iPSCs transit an embryo-like state, which is a highly selective process associated with complete epigenetic remodeling and immense cell proliferation [14]. Consequently, iPSCs and their derivatives have been found to be largely rejuvenated [15–17] (Figure 1). This fact makes it very challenging to study potentially critical aging-associated phenotypes, which are likely instrumental in sporadic forms of Parkinson’s Disease or Alzheimer’s Disease [18,19]. By contrast, direct transcription factor-based conversion of fibroblasts into induced neurons (iNs), a technology invented six years ago, was shown to maintain the cellular age of donor cells (Figure 1), and therefore appears to be a suitable model to study age-associated diseases of the brain [20–22]. This review will focus on the opportunities that arise from direct conversion as a relatively young technology for studying neurodegeneration. In each section, we highlight one aspect of cellular aging present in iNs, and summarize related work where iNs have been applied.
Figure 1 |. Direct iN conversion and iPSC reprogramming to study neuronal aging and age-related diseases.
Cultures of functional human neurons can be generated via direct conversion of human somatic cells into induced neurons (iNs), as well as via iPSC reprogramming and differentiation. Directly converted iNs from fibroblasts from elderly humans preserve key features of cellular aging and are in many aspects considered an ‘age-equivalent’ reflection of an old person’s brain cells. As a result, iNs represent a new and upcoming model system to study cellular and molecular aspects of brain aging, as well as of age-related diseases in a patient-specific context. By contrast, iPSC reprogramming is associated with a widespread rejuvenation of cellular and molecular aging phenotypes, as iPSC-derived neurons even from very old human donors resemble rejuvenated and prenatal-like ‘baby’ neurons. Extended in vitro or in vivo culture of iPSC derivatives as monolayers or 3D cultures could lead to cellular phenotypes that more closely resemble adult or even aged cells. Further, rejuvenated iPSC-derived neuronal cultures can be ‘made old’ by the application of age-inducing stressors; a strategy that might help to uncover aging-related triggers of diseases.
2. Aspects of cellular aging
2.1. Epigenetic aging
In the course of a neuron’s lifespan, neurons must preserve their cellular identity over a long period of time. With advancing age, a cell’s grip on its epigenetic state appears to loosen; a phenomenon also referred to as epigenetic drift [23,24]. Since the epigenome acts as a unifying platform for intrinsic as well as extrinsic factors, and in turn controls a cell’s transcriptional state, epigenetic changes are a major focus in aging research [15,23–27]. During the conversion of fibroblasts to iNs, extensive epigenomic reorganization has been noted [28,29]. Remarkably however, many epigenetic signatures adopted throughout a person’s life can be transferred from fibroblasts to iNs. A recent study found that the epigenetic age (or more precisely termed ‘DNA methylation (DNAm) age’), a distinctive pattern of DNA methylation marks from which a donor’s age can be predicted, was the same between iNs and their parental fibroblast [30]. Consequently, DNAm age appears to be at least in part unaffected by direct conversion. On the contrary, iPSCs from both young and old donors consistently show prenatal DNAm age, and retain minimal age-related epigenetic memory only during initial passages [16]. Consistently, while iPSCs generated from young and old donors don’t show significant age-related gene expression changes, iNs and fibroblasts show comparable numbers of genes significantly differentially expressed by age [31]. Importantly, iNs displayed a seven-fold increase in overlap with human brain aging genes compared to their parental fibroblasts, and also presented age-dependent regulation of gene categories previously noted in the aging human brain [31,32]. Taken together, direct conversion represents an interesting model to study aging-related epigenetic drift, as it enables the visualization of aging-specific epigenetic changes in different cell types from the same donor Another upcoming technology to generate human neural cell types is the direct conversion of fibroblasts into induced neural stem cells (iNSCs), which represents a hybrid technology of direct cell type conversion and reprogramming [33–36]. Interestingly, while iNSCs have been shown to erase age-associated epigenetic signatures similar to iPSCs, iNSCs appear to be stuck at a DNAm age of around five years, and extensive passaging does not further decrease iNSCs’ DNAm ages [37]. Considering that passaging of immortalized fibroblasts increases their mDNA age several folds faster than in vivo [38,39], these results indicate that DNAm age is not a direct function of replicative aging or dilution of age-related damage through cell division, and provides fascinating insights into how cell identity and age appear to be interdependent. Now, more work needs to be done to fully exploit the potential of iNs to study epigenetic aging in the postmitotic cellular state. The methylation marks that constitute the epigenetic clock only represent a minor fraction of age-related epigenetic marks on the human genome. Further, they do not directly account for changes in histone modifications, nucleosome positioning, and nuclear architecture, all of which impact chromatin accessibility and gene expression. iNs represent an interesting model system to explore aging-related epigenetic changes that are consequential for cellular fitness. Further, single-cell sequencing techniques provided insight into a loss of cellular fitness triggered by accumulated mutations and transcriptional noise [40,41]. The extent of this transcriptional noise, and the transcriptional consequences of erosion of epigenetic marks in age-related diseases in iNs, further remain to be investigated.
2.2. Proteins and aging
Age-related loss of cellular proteostasis mechanisms and accumulation of age-related damage on long-lived proteins have been regarded as major contributors to cellular aging [42], and have both been in the focus of studies using iNs [31]. In contrast to iPSCs, the iN conversion process does not involve any cell divisions, thus preventing the dilution of aged macromolecular structures that have accumulated damage over time. Further, iN conversion is a rather non-selective and therefore highly efficient process, as a fraction of typically >50% of the starting cells actually convert into neurons, which is by far less selection pressure than during iPSC reprogramming, which typically performs at <1% efficiency [43]. iNs thus represent an interesting model to study aging-related signatures that relate to the accumulation of damaged protein structures.
Recently, a model of Huntington’s Disease of directly induced medium spiny neurons (MSNs) showed that mutant huntingtin (mHTT) spontaneously aggregates specifically in neurons from patients, but not in their corresponding fibroblasts or iPSC-derived counterparts. [44]. This is an important finding, as earlier attempts to model Huntington’s Disease with iPSC-derived MSNs did not result in spontaneous mHTT aggregates [45]. Interestingly, the difference between these two models can be attributed to the inherent age-related collapse in proteostasis found in the induced MSNs, but not in their iPSC-derived counterparts [44].
Recently, nuclear pores have moved into the spotlight as potential drivers and/or targets of age-related dysregulation of protein quality. Nuclear pore complexes consist of proteins, the so-called nucleoporins, which are incorporated into the nucleus during mitosis. They regulate nucleo-cytoplasmic compartmentalization of nucleic acids and proteins within cells, and function as an anchor point for gene regulation, transcription, and global nuclear organization [46]. Nucleoporins represent a central target for aging, because they are exceptionally long-lived proteins, and were shown deteriorate and become ‘leaky’ with progressive aging [47–49]. As iNs are induced directly from fibroblasts without cell division, aging features encoded at the nuclear pore level are preserved: In the first study describing age-retention in iNs, we detected an age-related loss of nucleo-cytoplasmic compartmentalization, meaning that proteins from the cytoplasm enter the nucleus and vice versa. Interestingly, both fibroblasts and iNs showed age-related nuclear pore dysfunction similar to the age-dependent loss of the importin-β family transport factor RanBP17, while iPSCs did not [31]. In another study, direct reprogramming of fibroblasts was used to generate induced motor neurons (MNs) as an age-equivalent in vitro model for MN diseases [50]. In contrast to iPSC-derived MNs, the aging hallmarks of old donors were conserved in their corresponding induced MNs, including increased DNA damage, loss of heterochromatin, and changes in nuclear morphology. Nuclear structural protein LaminB1 displayed folded structures and blebbing in the nuclear envelope of induced MNs from old donors, but not in iPSC-derived MNs from the same donors [50]. Further, tau protein has been shown to interact with the nuclear pore complex, with pathological tau leading to dissociation of nucleoporins and impaired nuclear transport [51]. Also, in two independent iN models for amyotrophic lateral sclerosis (ALS), one based on C9orf72 repeat-expansion patients and the other on FUS mutation-carrying patients, nucleocytoplasmic mislocalization of RCC1/RanGEF or FUS were reported [52,53].
2.3. Energy metabolism and aging
Despite its comparatively small size, the brain consumes around 20% of the body’s energy [54]. During the differentiation of a neuron from stem cells, newborn and developing neurons undergo a metabolic switch in order to fulfill this huge energy demand. Neural stem cells and progenitors rely on glycolysis, reflected by high levels of glycolytic genes such as HK2 or LDHA. During differentiation, metabolic regulators like PGC-1α or ERRγ facilitate the expression of genes involved in mitochondrial biomass generation, tricarboxylic acid cycle and the electron transport chain. This metabolic switch allows for high rates of oxidative phosphorylation (OXPHOS) within mitochondria, ultimately generating approximately 18 times as much ATP as glycolysis would provide [55,56]. The downregulation of glycolysis during the differentiation process is crucial for neuronal specification and survival, and forced overexpression of glycolysis genes in neural progenitor cells results in their inability to differentiate [56]. This highlights the dramatic importance of cellular metabolism for neural function, and further, age-related mitochondrial dysfunction has been well documented as a common component of neurodegenerative diseases, like Parkinson’s disease or Alzheimer’s disease [57,58]. Consistently, iNs derived from old donors show marked decreases in the expression of mitochondrial genes and show dysfunctional OXPHOS, accompanied by decreased mitochondrial membrane potential, morphological changes of neuronal mitochondria, and low levels of ATP [59]. These changes were not present in rejuvenated iPSC-derived neurons from the same donors, and were found to be only weakly developed in glycolysis-dependent fibroblasts. In addition to compromised energy supply, mitochondrial aging is further associated with elevated levels of reactive oxygen species, which in turn further damage the mitochondria and also other cellular structures such as long-lived protein complexes and genomic DNA [59]. Neurons are particularly susceptible to DNA damage that can be caused by ROS, as they are post-mitotic and therefore lack division-mediated DNA repair (20), making them dependent on other repair mechanisms, such as the NAD+ dependent DNA repair machinery to maintain genome stability [60]. This connection is also highlighted by diseases that involve inefficient DNA repair, which in turn results in NAD+ shortage and metabolic dysfunctions [61,62]. Increasing DNA mutation rates with age suggests similar mechanisms apply in normal aging, as caloric restriction or supplementation of NAD+ precursors have been shown to increase lifespan of mice or other model animals, further stressing the importance of cellular energy homeostasis in old age [63–66]. Thus far, studies have shown that iNs show age-related accumulation of ROS and elevated DNA damage represented by long comet tail lengths [30,50], together suggesting that age-related neuronal metabolic dysfunctions can be studied in the iN system. Further studies will aim to explore metabolic aging in human neurons to better understand how the age-related increase of ROS and a global cellular energy crisis underlie the neuronal sensitivity for DNA damage in the aging human brain in order to help determine where mechanistic intervention is possible.
3. iPSC-based models to identify age-related disease triggers
iNs are just entering the stage as a model system for age-related neurodegenerative disorders. Although they are rapidly being appreciated as a highly useful model that allows investigators to perform studies in an age-equivalent human neuronal context, the most widely used in vitro models in neurodegeneration research remain iPSC-based.
Most genetic or sporadic neurodegenerative diseases do not affect young people, but are thought to rely on one or more age-related triggers that jump-start or permit the development of a pathogenic cascade. Obviously, identification and interference with those triggers holds a tremendous biomedical potential. In this context, the rejuvenated identity of iPSCs, thus far seen as a hurdle for aging research, might turn into a particular strength. The first generation of iPSC-based disease models for neurodegenerative diseases typically detected no disease-associated phenotypes in normal culture conditions. For example, antioxidant culture supplements had to be omitted from dopaminergic neurons to model phenotypes associated with Parkinson’s disease [67]. Further, glutamate-based overexcitation or oxidative stress have been used to tease out patient-specific neuronal dysfunctions, including aggregate formation in iPSC models for Huntington’s Disease, spinocerebellar ataxia, or Parkinson’s Disease [45,68,69]. Additionally, pharmacological insults were shown to increased proteostasis-related defects in ALS iPSC-derived MNs [70]. These studies have demonstrated the unique value of iPSC-based models for exploring neurodegenerative diseases, as they allow for the detection of patient- and disease-specific cellular phenotypes. However, these studies also emphasized that iPSC-derived neurons represent prenatal neurons that lack both adult neuron-specific identity as well as aging-related signatures, both of which are thought to be essential for modeling age-related diseases, and thus rely on an exogenous trigger to elicit disease-specific phenotypes. Following this concept, laboratories have started to more generally induce ‘age’ rather than one particular ‘stressor’ in their iPSC-based models (Figure 1). Using overexpression of the premature aging-associated protein progerin, which impairs nuclear envelope structures, dopaminergic neurons from Parkinson’s patients developed increased changes in nuclear morphology including folding and blebbing as well as DNA damage, mtROS and breakdown of established neurites, while no comparable phenotypes were observed without progerin-mediated age induction [18]. Similarly, inhibition of telomerase during the stem cell stages was used to generate neurons with short telomeres that manifested aging- and disease-related phenotypes in otherwise rejuvenated iPSC-derived neurons [19]. Further, induction of DNA damage with chemical stressors resulted in dysfunction of DNA damage repair and increased ROS among others phenotypes [71]. It appears conceivable that these ‘age-inducing’ strategies might more broadly simulate aging than, for example, a ‘proteostasis-only’ approach. However, the concept of adding ‘old age’ piece by piece, and in different combinations into rejuvenated neurons until a disease-defining phenotype is detected, appears to warrant a more systematic approach, and such experiments, however, could help elucidate the most upstream and disease-relevant aging triggers that permit and promote the variety of age-related neurodegenerative diseases (Figure 1).
4. Outlook
The possibility of directly converting human fibroblasts into various neuronal cell types represents a fascinating opportunity for studying age-related diseases, as iNs retain a broad spectrum of aging signatures which their iPSC-derived counterparts are lacking. Age-related neurodegenerative disorders often follow certain stages of development where some brain regions are affected earlier than others, and some diseases even exclusively affect certain brain regions. It thus appears conceivable, that different neuronal subtypes show differential susceptibility to aging mechanisms – they might age differently. Midbrain dopaminergic neurons might be specifically vulnerable to mitochondrial aging, while cortical excitatory neurons might have trouble coping with age-related nuclear pore deterioration in particular [48,72]. While yet this notion remains mainly speculative, iNs represent an excellent opportunity to address this concept experientially, as protocols for the conversion of human fibroblasts into several specific neuronal subtypes using co-expression of region-specific transcription factors have been developed [73–75].
One major challenge to overcome in this model, however, is the limitation of cell numbers. Contrary to the inexhaustible cell source for the generation of iPSC derivatives, the cell source for iN experiments are adult human skin fibroblasts. Since fibroblasts are the only proliferating cell type during direct conversion and their expandability is limited to typically under 20 – 30 passages, high conversion efficiencies and optimized downstream protocols are necessary. Indeed, the translation of direct conversion into pharmacological drug discovery faces exactly these obstacles, as large-scale high-throughput drug screenings require multiple hundreds of millions of cells. Where iPSC-derived neurons can be relatively easily generated, generating the same number of iNs currently represents a challenge. Additionally, as proliferating precursor cells that recapitulate human development are lacking during direct conversion, an iN model for an organoid-like 3D system appears technically harder to obtain. Further, in addition to neurons, glial cell types such as astrocytes, oligodendrocytes and microglia play pivotal roles in neurodegenerative diseases. It will thus be important to develop age-retaining human direct conversion strategies for glial cells in order to study aspects of age-related diseases that are not autonomously present in neurons [76]. Next, even though one of the major strengths of iNs is the possibility to generate neurons from large cohorts of patients, targeted genetic studies might also become important in several cases. Here, the lack of a clonal and proliferating intermediate during iN conversion limits the applicability of classical genetic tools such as zinc finger nucleases, adeno-associated viruses, TALENs and CRISPR. However, new optimized tools that have high-efficiencies, low off-target effects, and that can be packaged into viruses are becoming available right now [77–80].
Taken together, new technologies that are advancing towards ‘low-input-big-data’ strategies, complex 3D culture systems, and precise and efficient (epi)genetic tools, will all ultimately also benefit iN studies to better understand age-related diseases of the nervous system and to define new treatment strategies.
Acknowledgements
We thank Drs. S. T. Schäfer, D. Reid, F. Edenhofer, J. C. M. Schlachetzki, K. Günther, and F. H. Gage for fruitful discussions, and V. Mertens for graphical illustrations. This review was supported by the National Institutes of Health Pathway to Independence Award K99-AG056679–01 and the Leopold-Franzens University of Innsbruck to J.M.. L.B. is supported by the AGE_REG Doctoral Program of the University of Innsbruck, L. Traxler is supported by the Austrian Science Fund FWF-funded excellence Doctoral Program SPIN (DK-W1206), and J. R. Herdy by the UCSD neuroscience grad school.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Niccoli T, Partridge L. Ageing as a Risk Factor for Disease. Curr Biol 2012;22:R741–52. doi: 10.1016/j.cub.2012.07.024. [DOI] [PubMed] [Google Scholar]
- [2].López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The Hallmarks of Aging. Cell 2013;153:1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gladyshev TV, Gladyshev VN. A Disease or Not a Disease? Aging As a Pathology. Trends Mol Med 2016;22:995–6. doi: 10.1016/J.MOLMED.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zhu X, Raina AK, Perry G, Smith MA. Alzheimer’s disease: the two-hit hypothesis. Lancet Neurol 2004;3:219–26. doi: 10.1016/S1474-4422(04)00707-0. [DOI] [PubMed] [Google Scholar]
- [5].Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- [6].Kriks S, Shim J-W, Piao J, Ganat YM, Wakeman DR, Xie Z, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 2011;480:547–51. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc 2012;7:1836–46. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- [8].Du Z-W, Chen H, Liu H, Lu J, Qian K, Huang C-L, et al. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat Commun 2015;6:6626. doi: 10.1038/ncomms7626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lim RG, Salazar LL, Wilton DK, King AR, Stocksdale JT, Sharifabad D, et al. Developmental alterations in Huntington’s disease neural cells and pharmacological rescue in cells and mice. Nat Neurosci 2017;20:648–60. doi: 10.1038/nn.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y, et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Aβ and differential drug responsiveness. Cell Stem Cell 2013;12:487–96. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- [11].Ebert AD, Yu J, Rose FF, Mattis VB, Lorson CL, Thomson JA, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature 2009;457:277–80. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gladyshev VN. Aging: progressive decline in fitness due to the rising deleteriome adjusted by genetic, environmental, and stochastic processes. Aging Cell 2016;15:594–602. doi: 10.1111/acel.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yankner BA, Lu T, Loerch P. The Aging Brain. Annu Rev Pathol Mech Dis 2008;3:41–66. doi: 10.1146/annurev.pathmechdis.2.010506.092044. [DOI] [PubMed] [Google Scholar]
- [14].Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature 2013;502:462–71. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Horvath S DNA methylation age of human tissues and cell types. Genome Biol 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Frobel J, Hemeda H, Lenz M, Abagnale G, Joussen S, Denecke B, et al. Epigenetic Rejuvenation of Mesenchymal Stromal Cells Derived from Induced Pluripotent Stem Cells. Stem Cell Reports 2014;3:414–22. doi: 10.1016/J.STEMCR.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Studer L, Vera E, Cornacchia D. Programming and Reprogramming Cellular Age in the Era of Induced Pluripotency. Cell Stem Cell 2015;16:591–600. doi: 10.1016/J.STEM.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell 2013;13:691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Vera E, Bosco N, Studer L. Generating Late-Onset Human iPSC-Based Disease Models by Inducing Neuronal Age-Related Phenotypes through Telomerase Manipulation. Cell Rep 2016;17:1184–92. doi: 10.1016/j.celrep.2016.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ladewig J, Mertens J, Kesavan J, Doerr J, Poppe D, Glaue F, et al. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat Methods 2012;9:575–8. doi: 10.1038/nmeth.1972. [DOI] [PubMed] [Google Scholar]
- [21].Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010;463:1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Abernathy DG, Kim WK, McCoy MJ, Lake AM, Ouwenga R, Lee SW, et al. MicroRNAs Induce a Permissive Chromatin Environment that Enables Neuronal Subtype-Specific Reprogramming of Adult Human Fibroblasts. Cell Stem Cell 2017;21:332–348.e9. doi: 10.1016/j.stem.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang S-C, Oelze B, Schumacher A. Age-Specific Epigenetic Drift in Late-Onset Alzheimer’s Disease. PLoS One 2008;3:e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Teschendorff AE, West J, Beck S. Age-associated epigenetic drift: implications, and a case of epigenetic thrift? Hum Mol Genet 2013;22:R7–15. doi: 10.1093/hmg/ddt375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zheng SC, Widschwendter M, Teschendorff AE. Epigenetic drift, epigenetic clocks and cancer risk. Epigenomics 2016;8:705–19. doi: 10.2217/epi-2015-0017. [DOI] [PubMed] [Google Scholar]
- [26].Ma Z, Wang H, Cai Y, Wang H, Niu K, Wu X, et al. Epigenetic drift of H3K27me3 in aging links glycolysis to healthy longevity in Drosophila. Elife 2018;7. doi: 10.7554/eLife.35368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Veitia RA, Govindaraju DR, Bottani S, Birchler JA. Aging: Somatic Mutations, Epigenetic Drift and Gene Dosage Imbalance. Trends Cell Biol 2017;27:299–310. doi: 10.1016/J.TCB.2016.11.006. [DOI] [PubMed] [Google Scholar]
- [28].Smith DK, Yang J, Liu M-L, Zhang C-L. Small Molecules Modulate Chromatin Accessibility to Promote NEUROG2-Mediated Fibroblast-to-Neuron Reprogramming. Stem Cell Reports 2016;7:955–69. doi: 10.1016/j.stemcr.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, Fuentes DR, et al. Hierarchical Mechanisms for Direct Reprogramming of Fibroblasts to Neurons. Cell 2013;155:621–35. doi: 10.1016/J.CELL.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Huh CJ, Zhang B, Victor MB, Dahiya S, Batista LF, Horvath S, et al. Maintenance of age in human neurons generated by microRNA-based neuronal conversion of fibroblasts. Elife 2016;5. doi: 10.7554/eLife.18648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mertens J, Paquola ACM, Ku M, Hatch E, Böhnke L, Ladjevardi S, et al. Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell 2015;17:705–18. doi: 10.1016/j.stem.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lu T, Pan Y, Kao S-Y, Li C, Kohane I, Chan J, et al. Gene regulation and DNA damage in the ageing human brain. Nature 2004;429:883–91. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- [33].Thier M, Wörsdörfer P, Lakes YB, Gorris R, Herms S, Opitz T, et al. Direct Conversion of Fibroblasts into Stably Expandable Neural Stem Cells. Cell Stem Cell 2012;10:473–9. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- [34].Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G, et al. Direct Reprogramming of Mouse and Human Fibroblasts into Multipotent Neural Stem Cells with a Single Factor. Cell Stem Cell 2012;11:100–9. doi: 10.1016/J.STEM.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Karow M, Sánchez R, Schichor C, Masserdotti G, Ortega F, Heinrich C, et al. Reprogramming of Pericyte-Derived Cells of the Adult Human Brain into Induced Neuronal Cells. Cell Stem Cell 2012;11:471–6. doi: 10.1016/J.STEM.2012.07.007. [DOI] [PubMed] [Google Scholar]
- [36].Han DW, Tapia N, Hermann A, Hemmer K, Höing S, Araúzo-Bravo MJ, et al. Direct Reprogramming of Fibroblasts into Neural Stem Cells by Defined Factors. Cell Stem Cell 2012;10:465–72. doi: 10.1016/J.STEM.2012.02.021. [DOI] [PubMed] [Google Scholar]
- [37].Sheng C, Jungverdorben J, Wiethoff H, Lin Q, Flitsch LJ, Eckert D, et al. A stably self-renewing adult blood-derived induced neural stem cell exhibiting patternability and epigenetic rejuvenation. Nat Commun 2018;9:4047. doi: 10.1038/s41467-018-06398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lowe D, Horvath S, Raj K, Lowe D, Horvath S, Raj K. Epigenetic clock analyses of cellular senescence and ageing. Oncotarget 2016;7:8524–31. doi: 10.18632/oncotarget.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kabacik S, Horvath S, Cohen H, Raj K. Epigenetic ageing is distinct from senescence-mediated ageing and is not prevented by telomerase expression. Aging (Albany NY) 2018;10:2800–15. doi: 10.18632/aging.101588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lodato MA, Rodin RE, Bohrson CL, Coulter ME, Barton AR, Kwon M, et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 2018;359:555–9. doi: 10.1126/science.aao4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Enge M, Arda HE, Mignardi M, Beausang J, Bottino R, Kim SK, et al. Single-Cell Analysis of Human Pancreas Reveals Transcriptional Signatures of Aging and Somatic Mutation Patterns. Cell 2017;171:321–330.e14. doi: 10.1016/J.CELL.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Mattson MP, Magnus T. Ageing and neuronal vulnerability. Nat Rev Neurosci 2006;7:278–94. doi: 10.1038/nrn1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Malik N, Rao MS. A review of the methods for human iPSC derivation. Methods Mol Biol 2013;997:23–33. doi: 10.1007/978-1-62703-348-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Victor MB, Richner M, Olsen HE, Lee SW, Monteys AM, Ma C, et al. Striatal neurons directly converted from Huntington’s disease patient fibroblasts recapitulate age-associated disease phenotypes. Nat Neurosci 2018;21:341–52. doi: 10.1038/s41593-018-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].The HD iPSC Consortium. Induced Pluripotent Stem Cells from Patients with Huntington’s Disease Show CAG-Repeat-Expansion-Associated Phenotypes. Cell Stem Cell 2012;11:264–78. doi: 10.1016/J.STEM.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Capelson M, Hetzer MW. The role of nuclear pores in gene regulation, development and disease. EMBO Rep 2009;10:697–705. doi: 10.1038/embor.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-Dependent Deterioration of Nuclear Pore Complexes Causes a Loss of Nuclear Integrity in Postmitotic Cells. Cell 2009;136:284–95. doi: 10.1016/J.CELL.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Savas JN, Toyama BH, Xu T, Yates JR, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Toyama BH, Savas JN, Park SK, Harris MS, Ingolia NT, Yates JR, et al. Identification of Long-Lived Proteins Reveals Exceptional Stability of Essential Cellular Structures. Cell 2013;154:971–82. doi: 10.1016/J.CELL.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Tang Y, Liu M-L, Zang T, Zhang C-L. Direct Reprogramming Rather than iPSC-Based Reprogramming Maintains Aging Hallmarks in Human Motor Neurons. Front Mol Neurosci 2017;10:359. doi: 10.3389/fnmol.2017.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Eftekharzadeh B, Daigle JG, Kapinos LE, Coyne A, Schiantarelli J, Carlomagno Y, et al. Tau Protein Disrupts Nucleocytoplasmic Transport in Alzheimer’s Disease. Neuron 2018;99:925–940.e7. doi: 10.1016/J.NEURON.2018.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 2015;18:1226–9. doi: 10.1038/nn.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Liu M-L, Zang T, Zhang C-L. Direct Lineage Reprogramming Reveals Disease-Specific Phenotypes of Motor Neurons from Human ALS Patients. Cell Rep 2016;14:115–28. doi: 10.1016/j.celrep.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mergenthaler P, Lindauer U, Dienel GA, Meisel A. Sugar for the brain: the role of glucose in physiological and pathological brain function. Trends Neurosci 2013;36:587–97. doi: 10.1016/j.tins.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Magistretti PJ, Allaman I. A cellular perspective on brain energy metabolism and functional imaging. Neuron 2015;86:883–901. doi: 10.1016/j.neuron.2015.03.035. [DOI] [PubMed] [Google Scholar]
- [56].Zheng X, Boyer L, Jin M, Mertens J, Kim Y, Ma L, et al. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife 2016;5. doi: 10.7554/eLife.13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Area-Gomez E, de Groof A, Bonilla E, Montesinos J, Tanji K, Boldogh I, et al. A key role for MAM in mediating mitochondrial dysfunction in Alzheimer disease. Cell Death Dis 2018;9:335. doi: 10.1038/s41419-017-0215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Reeve AK, Grady JP, Cosgrave EM, Bennison E, Chen C, Hepplewhite PD, et al. Mitochondrial dysfunction within the synapses of substantia nigra neurons in Parkinson’s disease. NPJ Park Dis 2018;4:9. doi: 10.1038/s41531-018-0044-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kim Y, Zheng X, Ansari Z, Bunnell MC, Herdy JR, Traxler L, et al. Mitochondrial Aging Defects Emerge in Directly Reprogrammed Human Neurons due to Their Metabolic Profile. Cell Rep 2018. doi: 10.1016/j.celrep.2018.04.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Madabhushi R, Pan L, Tsai L-H. DNA damage and its links to neurodegeneration. Neuron 2014;83:266–82. doi: 10.1016/j.neuron.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Fang EF, Kassahun H, Croteau DL, Scheibye-Knudsen M, Marosi K, Lu H, et al. NAD+ Replenishment Improves Lifespan and Healthspan in Ataxia Telangiectasia Models via Mitophagy and DNA Repair. Cell Metab 2016;24:566–81. doi: 10.1016/j.cmet.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Scheibye-Knudsen M, Mitchell SJ, Fang EF, Iyama T, Ward T, Wang J, et al. A high-fat diet and NAD(+) activate Sirt1 to rescue premature aging in cockayne syndrome. Cell Metab 2014;20:840–55. doi: 10.1016/j.cmet.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Liang Y, Liu C, Lu M, Dong Q, Wang Z, Wang Z, et al. Calorie restriction is the most reasonable anti-ageing intervention: a meta-analysis of survival curves. Sci Rep 2018;8:5779. doi: 10.1038/s41598-018-24146-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Vermeij WP, Dollé MET, Reiling E, Jaarsma D, Payan-Gomez C, Bombardieri CR, et al. Restricted diet delays accelerated ageing and genomic stress in DNA-repair-deficient mice. Nature 2016;537:427–31. doi: 10.1038/nature19329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Mattison JA, Colman RJ, Beasley TM, Allison DB, Kemnitz JW, Roth GS, et al. Caloric restriction improves health and survival of rhesus monkeys. Nat Commun 2017;8:14063. doi: 10.1038/ncomms14063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Schöndorf DC, Ivanyuk D, Baden P, Sanchez-Martinez A, De Cicco S, Yu C, et al. The NAD+ Precursor Nicotinamide Riboside Rescues Mitochondrial Defects and Neuronal Loss in iPSC and Fly Models of Parkinson’s Disease. Cell Rep 2018;23:2976–88. doi: 10.1016/J.CELREP.2018.05.009. [DOI] [PubMed] [Google Scholar]
- [67].Reinhardt P, Schmid B, Burbulla LF, Schöndorf DC, Wagner L, Glatza M, et al. Genetic Correction of a LRRK2 Mutation in Human iPSCs Links Parkinsonian Neurodegeneration to ERK-Dependent Changes in Gene Expression. Cell Stem Cell 2013;12:354–67. doi: 10.1016/J.STEM.2013.01.008. [DOI] [PubMed] [Google Scholar]
- [68].Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, Gujar P, et al. LRRK2 Mutant iPSC-Derived DA Neurons Demonstrate Increased Susceptibility to Oxidative Stress. Cell Stem Cell 2011;8:267–80. doi: 10.1016/J.STEM.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Koch P, Breuer P, Peitz M, Jungverdorben J, Kesavan J, Poppe D, et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature 2011;480:543–6. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- [70].Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, et al. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell 2014;14:781–95. doi: 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Dong C-M, Wang X-L, Wang G-M, Zhang W-J, Zhu L, Gao S, et al. A stress-induced cellular aging model with postnatal neural stem cells. Cell Death Dis 2017;5:e1116–e1116. doi: 10.1038/cddis.2014.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Rango M, Bresolin N, Rango M, Bresolin N. Brain Mitochondria, Aging, and Parkinson’s Disease. Genes (Basel) 2018;9:250. doi: 10.3390/genes9050250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Tsunemoto R, Lee S, Szucs A, Chubukov P, Sokolova I, Blanchard JW, et al. Diverse reprogramming codes for neuronal identity. Nature 2018:1. doi: 10.1038/s41586-018-0103-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Mertens J, Marchetto MC, Bardy C, Gage FH. Evaluating cell reprogramming, differentiation and conversion technologies in neuroscience. Nat Rev Neurosci 2016;17:424–37. doi: 10.1038/nrn.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Mertens J, Reid D, Lau S, Kim Y, Gage FH. Aging in a Dish: iPSC-Derived and Directly Induced Neurons for Studying Brain Aging and Age-Related Neurodegenerative Diseases. Annu Rev Genet 2018;52:annurev-genet-120417–031534. doi: 10.1146/annurev-genet-120417-031534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Caiazzo M, Giannelli S, Valente P, Lignani G, Carissimo A, Sessa A, et al. Direct conversion of fibroblasts into functional astrocytes by defined transcription factors. Stem Cell Reports 2015;4:25–36. doi: 10.1016/j.stemcr.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018;173:665–676.e14. doi: 10.1016/J.CELL.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Akcakaya P, Bobbin ML, Guo JA, Malagon-Lopez J, Clement K, Garcia SP, et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 2018;561:416–9. doi: 10.1038/s41586-018-0500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Black JB, Adler AF, Wang H-G, D’Ippolito AM, Hutchinson HA, Reddy TE, et al. Targeted Epigenetic Remodeling of Endogenous Loci by CRISPR/Cas9-Based Transcriptional Activators Directly Converts Fibroblasts to Neuronal Cells. Cell Stem Cell 2016;19:406–14. doi: 10.1016/J.STEM.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 2015;12:326–8. doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]