Abstract
Multiple myeloma (MM), the disorder of plasma cells, is the second most common type of hematological cancer and is responsible for approximately 20% of deaths from hematological malignancies. The current gold standard for MM diagnosis includes invasive bone marrow aspiration. However, it lacks the sensitivity to detect minimal residual disease, and the nonuniform distribution of clonal plasma cells (CPCs) within bone marrow also often results in inaccurate reporting. Serum and urine assessment of monoclonal proteins, such as Kappa light chains, is another commonly used approach for MM diagnosis. Although it is noninvasive, the level of paraprotein elevation is still too low for detecting minimal residual disease and nonsecretive MM. Circulating CPCs (cCPCs) have been reported to be present in the peripheral blood of MM patients, and high levels of cCPCs were shown to correlate with poor survival. This suggests a potential noninvasive approach for MM disease progress monitoring and prognosis. In this study, we developed a mechanical property-based microfluidic platform to capture cCPCs. Using human myeloma cancer cell lines spiked in healthy donor blood, the microfluidic platform demonstrates high enrichment ratio (>500) and sufficient capture efficiency (40%–55%). Patient samples were also assessed to investigate the diagnostic potential of cCPCs for MM by correlating with the levels of Kappa light chains in patients.
INTRODUCTION
Multiple myeloma (MM) is a malignant of plasma cells that accumulate in the bone marrow, leading to bone destruction and marrow failure. Currently, it remains incurable, with a treatment-refractory state eventually developing in all patients. MM accounts for about 1.8% of all cancer and slightly over 17% of hematologic malignancies in the United States.1 The American Cancer Society has estimated that there were 26 850 new cases of MM and 11 240 MM-associated deaths in the United States in 2014.1
Recent studies showed that some myeloma cells leave the bone marrow and enter the peripheral blood.2 These clonal circulating plasma cells (cCPCs) can be identified in the blood collected from MM patients, and the presence of cCPCs in newly diagnosed MM patients correlates with shorter survival.2,3 Studies have shown the occurrence of cCPCs in MM patients to be around 1–10 per 50 000 cells in whole blood and dependent on the stage of disease.4 Slide-based immunofluorescence assay is the most commonly used method to detect cCPCs, but it is complex and labor-intensive.4 Multiparametric flow cytometry (MFC) is a more sensitive tool to detect and quantify the concentration of cCPCs in whole blood. Quantification of cCPCs using multiparametric flow cytometry (MFC) is capable of identifying patients with smoldering MM that are likely to progress to active MM within 2 years.5
Although MFC is, currently, the most widely accepted method for cCPCs detection, there are several limitations associated with it. First, the sensitivity of MFC at the magnitude of 105 (1 cell per 105 cells) is not high enough to detect cCPCs in all MM patients or to quantify cCPCs accurately.4 Second, the interpretation of MFC results lacks standardization and is subjective. Third, MFC indicates the presence of cCPCs, but provides limited cellular and molecular information of cCPCs,6 which is crucial for monitoring clonal evolution and predicting prognosis. Moreover, it requires a large volume of samples (7.5 ml blood) to produce accurate results.4 Furthermore, it requires the RBCs in the samples to be lysed, inevitably causing CPC loss in the samples.
For nearly all hematologic malignancies, a direct correlation exists between the depth of response to the treatment and prolonging the survival.7,8 Thus, the ability to detect and monitor minimal residual disease for patients, particularly ones in the complete remission stage, can greatly benefit the evaluation of treatment effect and the prognosis of disease. The in-depth cellular and molecular analysis of isolated cCPCs will be critical in understanding the mechanisms of relapse, metastasis, drug resistance, and clonal evolution of MM.4
Microfluidics have shown great potential in cell sorting in general. Affinity-based sorting is one of the most classic and common cell sorting technologies in microfluidic chips.8–10 Indeed, a CD138 antibody-based microfluidic device had been reported in isolating circulating plasma cells from the peripheral blood of MM patients.9 With this device, the number of circulating plasma cells captured from relapsing patients ranged from 45 to 184 cells/ml with a purity of 1%–5%. Unfortunately, this device is not able to differentiate normal circulating plasma cells from cCPCs. Although the authors of this study suggest that the number of circulating plasma cells captured by their device can be used to diagnose MM progression, the direct correlation to disease stages was not demonstrated in the study. In contrast, previous clinical studies have demonstrated that cCPCs are highly relevant to the progress of MM.10 The presence of cCPCs is also a marker of high risk in newly diagnosed MM patients.4 Thus, in this study, we aim to capture cCPCs in patients' blood samples.
One of the limitations of affinity-based sorting is the lack of the ability to retrieve captured cells without compromised cell integrity, which is critical for a later disease mechanism study. To address this limitation, we designed a label-free microfluidic system for capturing cCPCs from the peripheral blood. Although there are existing label-free microfluidic technologies (Dean flow-based platforms, sieve or filtration-based platforms, electrophoresis, and acoustophoresis) in the application of isolation circulating tumor cells (CTCs),11–13 to the best of our knowledge, we are the first group to design a label-free microfluidic device specifically for cCPCs based on their unique physical and mechanical properties.
METHODS
Cancer cell line and clinical samples
U266 human myeloma cells (American Type Culture Collection) were cultured in suspension in RPMI-1640 (Gibco™) supplemented with 10% fetal bovine serum (FBS, Gibco™), incubated at 37 °C with 5% carbon dioxide. Cells were passaged from 8 to 14 generations. The diameters of U266 cells were estimated with the image-based assay (Axio Vision software) integrated with an inverted microscope (ZEISS Axio Vert.A1).
Blood samples from 2 MM patients and 4 healthy donors were analyzed. All the blood samples were preserved in Vacutainer collection tubes (BD—Becton Dickinson) filled with ethylenediaminetetraacetic acid (EDTA) solution. Fresh blood samples were placed on a nutator in room temperature (within 1 h of sample collection) prior to experiments. The blood sample was diluted 1:1 with PBS (Phosphate-buffered saline). This study was approved by the institutional review board of the participating center, and written informed consent was obtained from all patients and healthy donors.
Microfluidic device fabrication
Soft lithography was used to create the array structure. Channel and pillar features of the device are designed in the photomask and then transferred onto the SU8-2000 (Mirochem, USA) photoresist. After that, the photoresist layer was exposed to ultraviolet (UV) light to create the designated pattern and then the silicon master was fabricated to produce the microfluidic chips. Polydimethylsiloxane (PDMS) was selected as the material for prototyping the microfluidic devices because it is a moderately stiff elastomer and commercially available at low cost. Moreover, it is nontoxic and optically transparent, which is essential for imaging.
Microfluidic system setup
The system is composed of syringes, Cole-Parmer Stopcock with Luer connections, tubes, a microfluidic chip, and a TJ-3A longer syringe pump. Stopcocks and precision tips were used to connect the tubes on the chip with the syringes on the pump. The syringe pump was then set to withdraw flow from the inlet to the outlet, and the chip was placed on the microscope stage for real-time monitoring of the cCPC-isolation process.
Computational simulation
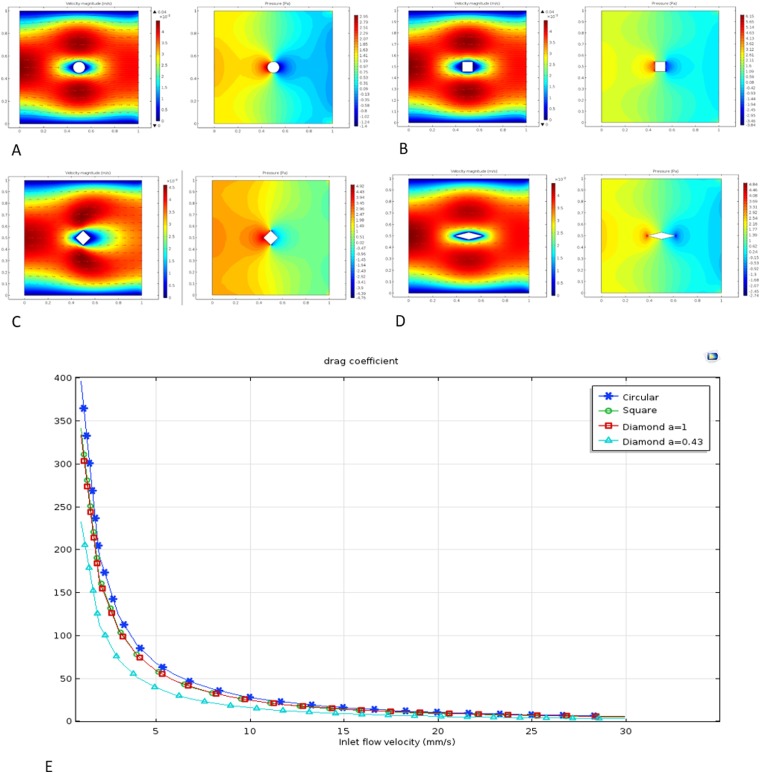
Fluid computational analysis was used to assist the microfluidic chip design. Since blood normally coagulates upon exposure to a foreign interface,14 pillars in our microfluidic channel would increase blood coagulation by hindering the flow and increasing the adhesion of cells, such as platelets. Therefore, it is important to design appropriate pillar geometry to minimize resistance. Circular, square, and diamond pillar shapes in the flow field were analyzed using COMSOL Multiphysics. A 2-dimensional simplified model and fluid properties of blood (density of 1050 kg/m3 and viscosity of 0.005 Pa s at 25 °C15) were used in simulation. Furthermore, since a previously published cell-capturing microfluidic device reported flow velocity at 0.5–10 mm/s,16 and Reynold's numbers for this range of velocity are less than 1, a Stokes flow model was used to construct the simulation. With these parameters, when the inlet flow speed was set at 1 mm/s, the system reached a steady state within 1 s. Thus, the steady state and the no-slip boundary condition were used for further analysis. We then simulated the flow at 0.1–30 mm/s to generate velocity and fluid stress profiles for each pillar geometry [Figs. 1(a)–1(d)]. Based on the simulation results, the drag coefficient Cd of each pillar shape was then calculated using Eq. (1) and plotted against the flow rate [Fig. 1(e)]. This plot showed that the circular shape creates the biggest drag coefficient, whereas the diamond shape yields the smallest resistance. The drag coefficient could be further reduced by adjusting the diagonal ratio of the diamond shape. However, on the other hand, we also need the pillar to have enough longitudinal width to intercept the cells. Therefore, instead of further adjusting the diagonal ratio, we directly selected the diamond shape with a diagonal ratio of 0.43 as the basic geometry for the micropillar design,
| (1) |
where is the drag force, ρ is the mass density of the fluid, v is the flow speed of the object relative to the fluid, and A is the area of the geometry.
FIG. 1.
Velocity and pressure profile of circular (a), square (b), and diamond shape with diagonal ratio (c) a = 1 and (d) a = 0.43 under the microfluidic flow field. (e) Drag coefficient varies with the liquid flow velocity.
Simulation of cell-and-pillar contact in channel
Studies showed that MM cells have unique physical and mechanical properties that distinguish them from ordinary blood cells. At a size of 30–50 μm,17 they are bigger than red blood cells (RBCs) and white blood cells (WBCs), the main components of peripheral blood. Human RBCs are approximately 6–8 μm in diameter with a biconcave disks shape,18 while WBCs' diameter ranges from 7 to 30 μm.19 Some of the WBCs, such as macrophages, can be as large as the MM cells in diameter. However, MM cells are much stiffer in terms of elastic's modulus.20 To isolate the MM cells based on size and stiffness, the microfluidic channel is composed of numerous arrays of micropillars. Three micropillars are patterned together to make up one capture unit. The capture unit is designed to filter out most of the normal blood cells by controlling the gap between the micropillars. RBCs can easily pass through the capture unit due to its small size, while other large WBCs can deform and squeeze through due to their lower elastic modulus (20–200 Pa).21 On the other hand, MM cells will be trapped within the capture unit because of its larger size and high cell membrane elastic modulus (around 540 Pa).20
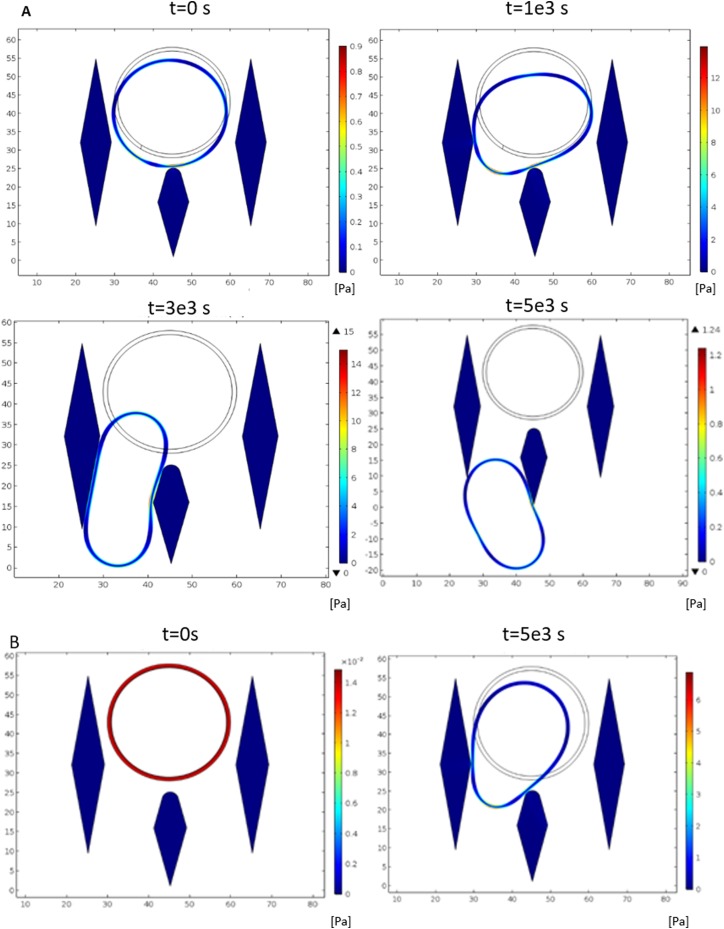
To determine the appropriate gap space for capturing MM cells, we first employed computational fluid dynamics simulation to guide our design. Blood exhibits a substantial non-Newtonian behavior due to a mixture of cells and protein components.22 However, in the simulation performed here, it focusses on how the MM cells and other hematopoietic cells squeeze through a microfluidic filtering channel, and thus whole blood is considered a Newtonian fluid (plasma) with particle suspension (hematopoietic cells) in it. In order to ensure that all WBCs pass through the capture unit, we simulated the most difficult situation, which used the upper limit (30 μm and 200 Pa) of the WBCs' size and elastic modulus. In the simulation, a particle with such mechanical properties was traced in the flow field with velocity ranging from 1 to 30 mm/s [Fig. 2(a)]. Figure 2(a) shows that the particle can comfortably squeeze through the micropillars when the gap is wider than 11 μm, and the flow velocity is greater than 6 mm/s. Furthermore, we also simulated the MM cells passing scenario under the same flow velocity. The MM cell's diameter is set at 30 μm with a cell membrane elastic modulus of 540 Pa, to check whether it would pass through the capture unit [Fig. 2(b)]. The simulation showed that within such flow velocity (6 mm/s), the MM cells were not able to squeeze through the micropillars due to their higher stiffness. Therefore, we can ensure that micropillars with such design setup and flow condition would be able to trap MM cells and filter out the other ordinary blood cells. Particle tracing, Fluid-Structure Interaction, and Solid mechanics are the packages used to complete this simulation.
FIG. 2.
Plots showing WBCs and CPCs passing through micropillars in simulations under the same capture unit configurations. (a) WBCs passing through. (b) CPCs being captured with the free stream velocity at 6 mm/s, simulation time frame from 0 to 5 × 10−3 s.
Design overview
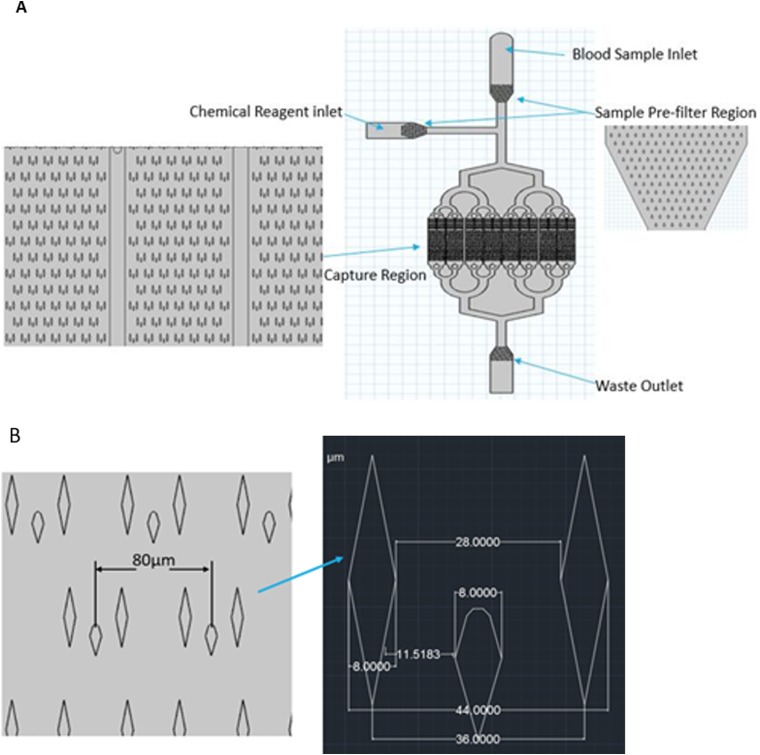
Figure 3(a) shows the configuration of the cCPC-capturing device. Blood, liquid, and chemical reagents enter the channel separately to avoid any contamination. However, in the preliminary test of the porotype, it was noticed that the debris or particles larger than the average cell size entered the capture region and got stuck in the flow chamber, adversely affecting the operation of the device. This may be inevitable since the chip was already operating in a dust-free environment. Therefore, a prefilter region was added that consists of multiple layers of micropillars with gap spaces decreasing stepwise from 60 to 45 μm. Next, the capture region is composed of 8 separate channels; using such multichamber configuration was to increase throughput and provide uniform flow compared to one big chamber. In addition, there are over 5000 capture units located in parallel and repeated in rows to ensure a large capacity. Detailed geometrical information is demonstrated in Fig. 3(b).
FIG. 3.
(a) Overview layout of the microfluidic chips. (b) Detailed dimensions of micropillars.
Experimental design
The cCPC-capturing microfluidic device was designed based on the size and stiffness of CPCs. We first tested the device's ability to capture MM cells and quantitatively measured its capture efficiency under different volumetric flow rates. To do this, we ran U266 cell suspension (1000 cells/ml in PBS) through the microfluidic device at volumetric flow rates of 0.5, 1, and 1.5 ml/h. Next, we mixed U266 cells into 1 ml of the peripheral blood from healthy donors to test the devices' ability to isolate MM cells. The peripheral blood sample without U266 cells was also tested as a control. Finally, blood samples from MM patients at releasing and remission stages were used to test the device's ability to differentiate patients at different stages.
Immunostaining and immunophenotyping of CPCs
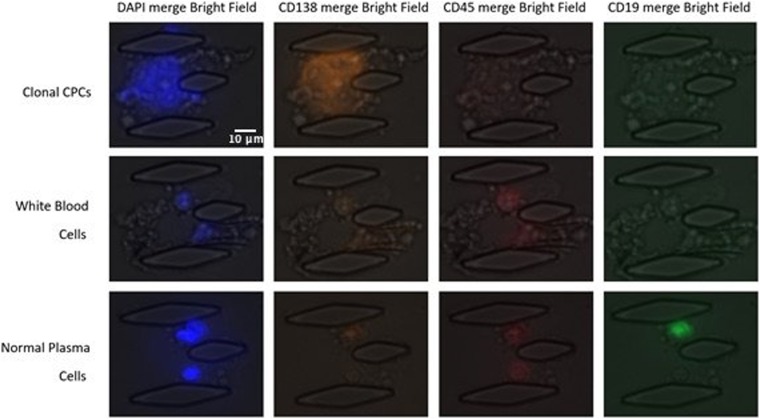
Immunostaining was performed to identify CPCs from normal plasma cells. Cells captured by the microfluidic device were fixed with 5% paraformaldehyde and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) in PBS. After blocking for 20 min with 5% FBS (Wisent), CPCs were identified using a combination of stains: 4′,6-diamidino-2-phenylindole (DAPI), phycoerythrin-conjugated CD138 mouse monoclonal antibody, Alexa Fluor 647-labeled CD45 mouse monoclonal antigen, and Alexa Fluor 488-labeled CD19 mouse monoclonal antibodies (DAPI+, CD138+, CD45−, CD19−).10 All those reagents entered through the chemical inlet of the microfluidic chip. A Zeiss Axio Observer inverted fluorescence microscope was used for imaging. DAPI and CD138 were positive for the identification of circulating plasma cells. It has been shown that normal plasma cells are generally positive for these antigens, whereas abnormal plasma cells characteristically lack CD19 and variably express of CD45.23
RESULT AND DISCUSSION
Capture efficiency
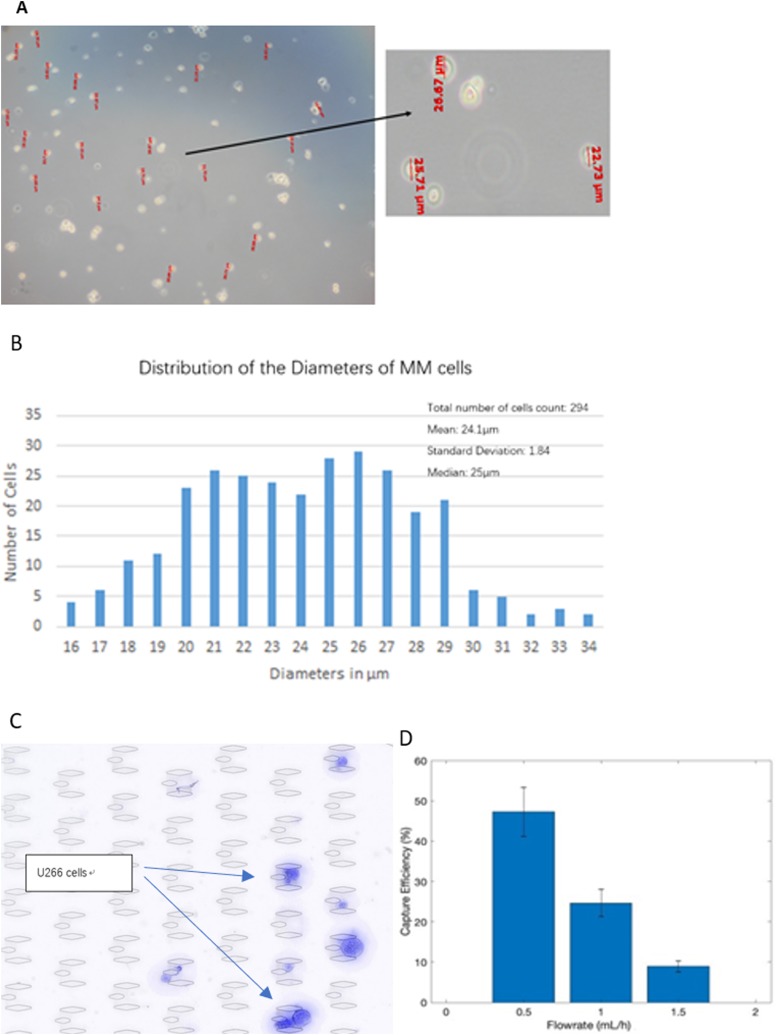
The concentration of U266 cell suspensions was determined by the BIO-RAD TC20TM Automated CellCounter and then diluted to 1000 cells/ml in PBS, mimicking the low level of cCPCs in the peripheral blood. The sizes of U266 cells were then measured (Fig. 4) to confirm that the sizes of U266 cells fall within the range of cCPCs and that results using U266 cells are a good reference. To determine the effect of flow velocity on the capture efficiency, we pipetted 1 ml of U266 cell suspension to the microfluidic chip at velocities 1.5, 1.0, and 0.5 ml/h. DAPI was used to identify the captured cells, and the capture efficiency was calculated by Eq. (2). A representative image of successfully captured U266 cells is shown in Fig. 4(c). Enumeration of U266 cells was done by screening and counting each of the capture units. The results are summarized in Fig. 4(d), which shows that capture efficiency significantly increased as the volumetric flow rate decreased. At 0.5 ml/h, the device's capture efficiency was around 50%. However, the volumetric flow rate could not be further reduced since it would be lower than the critical velocity (6 mm/s) required for the filtering function. The capture efficiency measured from the MM cell line experiments using our device is lower than other existing devices for capturing solid tumor cells.24 One reason for this is that the myeloma cells originated from hematopoietic cells result in similar characteristics compared to peripheral blood cells than solid tumor cells. For example, the Elastic's modulus for ovarian cancer cells is at the magnitude of 1–2 kPa, whereas it is only 500 Pa for MM cells.20,25 Thus, it is more difficult to capture MM cells due to its lower rigidity,
| (2) |
FIG. 4.
Diameters of U266 cells. (a) Diameters measured by a compound microscope. (b) Distribution of cell diameters. (c) A snapshot of U266 cells being trapped. (d) Capture efficiency of U266 cell line testing through various flow rates. Error bars indicate standard deviation.
cCPCs enrichment and purity
To evaluate the devices' capability of capturing clonal plasma cells in the blood, we spiked U266 cells in 1 ml of peripheral blood from a healthy donor at varying concentrations to mimic samples from MM patients. Three samples were individually processed in 3 microfluidic chips. 1500 and 500 U266 cells were resuspended in samples 1 and 2, respectively. Sample 3 did not contain any U266 cells and was used as a control. To ensure consistency, the blood used in these 3 samples was taken from the same donor. A buffer solution was mixed with the blood samples at a volume ratio of 1:1 to prevent blood coagulation and reduce nonspecific cell adhesion. This is important as it takes more than 4 h to process the sample at a flow rate of 0.5 ml/h. EDTA, heparin, and citrate are the most commonly used anticoagulants, with EDTA being the best for long-time blood processing.26 Studies showed that, for PDMS elastomers, the level of protein adsorption is positively correlated with cell attachment and can be greatly reduced with Pluronic F68 surfactant treatment.27 Therefore, the buffer solution is made up of 0.1M Tris-EDTA (Sigma-Aldrich) and 1% w/v Pluronic F68 (Gibco™) in PBS.
Due to the complicated nature of the blood's component, it is expected that there is a decrease in capture efficiency for MM cell lines once it gets mixed with blood, which is shown in Table I. While 510 and 107 U266 cells were captured and identified in samples 1 and 2, respectively, only 1 captured cell in the negative control (sample 3) was identified to have MM phenotypes based on immunostaining. Furthermore, only around 2000 WBCs were retained by the chip, filtering out 99.99% of WBCs as there are approximately 5 million WBCs/ml of peripheral blood.28 After the enrichment process, the ratio between U266 cancer cells to the normal blood cells increased by up to 1:102–103. It enables the functional characterization of cCPCs that are critical for disease interrogation and target therapy.29 Additionally, through immunostaining, we confirmed the identity of cCPCs from the background cells as illustrated in Fig. 5, demonstrating that this device can separate myeloma cells from the whole blood.
TABLE I.
Enrichment performance of the microfluidic device to enrich the concentration of clonal plasma cells from the peripheral blood.
| Trial No. | Clonal CPCs captured enumeration | Enrichment ratio | Capture efficiency (%) |
|---|---|---|---|
| T1 (mix around 1500 U266 cells) | 510 | 1133 | 34 |
| T2 (mix around 500 U266 cells) | 107 | 713 | 21 |
| T3 (negative control, not mix U266 cells) | 1 | N/A | N/A |
FIG. 5.
Immunostaining of cCPCs. After capture, cells were stained for DAPI, CD138, CD45, and CD19. CPCs were identified as DAPI+/CD138+/CD45−/CD19−, WBCs were identified as DAPI+/CD138+/CD45+, and ordinary plasma cells can be identified as DAPI+/CD138−/CD19+.
Capturing cCPCs from MM patients
We then proceeded to test the device using blood samples from MM patients. To further challenge the limit of the microfluidic chip, we only used 0.5 ml of peripheral blood for MM patients. The cCPCs captured from patients with high tumor burden were drastically different with that from remission patients (low tumor burden) or healthy donors, as shown in Table II. 117 cCPCs were detected in the peripheral blood sample from the relapsing MM patient, 2 cCPCs from the remission MM patient, and 0–1 cCPC from 2 healthy donors. Prior to our study, Qasaimeh et al. demonstrated that the concentration of CPCs is low in the peripheral blood of a healthy human, at 2–5 cells/ml.10 This is consistent with our results showing the low number of CPCs detected in healthy donors and the remission patient. Based on hospital reports, the 2 patients included in our study were clinically diagnosed as Kappa-type MM, with Kappa light chain concentrations included in Table II. With this information, we observed that a larger number of captured cCPCs may correspond to an elevation of the Kappa paraprotein level. It is understandable that our device is not able to differentiate MM remission patients from healthy donors. For those remission patients, particularly in the complete remission stage with very low tumor burden, the cPC levels are outside the detectable ranges of microfluidic devices and other available tests such as flow cytometry.4 Healthy donors are used as the blank control group in our study. If the number of cCPCs captured from patients is not significantly different from healthy donors, it would suggest very low tumor burden in these patients. We believe our devices are more suitable for monitoring disease status (the tumor burden) in myeloma patients rather than early diagnosis to screen healthy individuals.
TABLE II.
Number of captured cCPCs from the blood of MM patients and healthy donors, with Kappa light chain concentrations (normal range of Kappa light chains: 3.3–19.4 mg/l30).
| Sample sources | Total blood volume processed (mL) | Volumetric flow rate (ml/h) | Total number of cCPCs captured | Kappa (mg/l) |
|---|---|---|---|---|
| MM relapsing patient | 0.5 | 0.5 | 117 | 21 792 |
| MM remission patient | 0.5 | 0.5 | 2 | 11.5 |
| Healthy donor 1 | 0.5 | 0.5 | 0 | N/A |
| Healthy donor 2 | 0.5 | 0.5 | 1 | N/A |
Using the physical sorting technique to capture CTCs is one of the common approaches in this field. Based on the difference between CTCs and blood cell physical properties (such as deformability, size, hydrodynamic properties, etc.), by patterning some microstructural units in the chip, CTCs can be separated from the blood. Commonly used microstructure includes micropores, microfilters, and micropillars.31–33 The tumor cells originated from solid tissues and are generally larger than blood cells. Most epithelial-derived CTCs range in diameter from 14 to 26 μm, while white blood cell diameters range from 8 to 20 μm.13 Using different micropores, microfilters, micropillars, etc., which in terms of size are smaller than the diameter of CTCs, the CTCs get stuck due to the large diameter while the blood cells flow out together with the buffer. Lim et al. designed a microfluidic chip that consisted of hundreds of microarrays.24 The space between each micropillar ranged from 8 to 9 μm. The syringe pump pumped blood samples from the inlet to the outlet of the chip. The system captures more than 80% of MCF-7 and HepG2 cell lines in healthy human blood. The author further validated the reliability of the system with blood samples from 8 cancer patients and successfully isolated CTCs from the patient's blood. The entire process took only 1.5 h. However, there is no such universal device that can be used to capture all types of CTCs, particularly for cCPCs, a hematological cancer not originating from solid tissues, having their own unique physical properties.
In contrast, our micropillar-integrated microfluidic chip can accurately detect and enumerate cCPCs using much less sample (0.5 ml) and without preprocessing. Furthermore, this isolation is based purely on mechanical properties and, therefore, allows cCPCs to be captured without compromised cell integrity. It can potentially recover cCPCs for further analysis by reversing the direction of flow.
It is worth to note that the timing for clonal plasma cells released from the bone marrow is rather random. It is very difficult to ensure that the cCPC level in the peripheral blood is consistent at any time and any point. Therefore, it is difficult to determine a threshold value to differentiate a patient’s disease stages based on the number of cCPCs captured. One potential solution is to directly connect the microfluidic chip to the arm vein and allow the blood to pass through the chip for about 1 h, similar to the hemodialysis process used for uremic patients. However, this would require the material to be perfectly biocompatible and not induce any coagulation.
To summarize, the microfluidic device presented here is a highly sensitive method for quantifying cCPCs that also allows for cytogenetic and molecular characterization of cCPCs. More comprehensive characterization of cCPCs in MM may identify molecular mechanisms of drug resistance. cCPC evaluation including chromosome transplantation, gene mutation, and drug resistance could be done to guide individualized therapeutic options.
CONCLUSION
cCPC is a promising biomarker for MM, where the elevation of cCPCs in the peripheral blood is associated with faster progression to active MM and shorter survival. Our device successfully demonstrated the great potential of cCPCs to be used in early and noninvasive diagnosis for MM. Further investigation with a larger sample size is still needed to validate the results and evaluate the relationships between the number of cCPCs and the types, stages, and recurrence of MM. However, the micropillar device described here should be considered a clinically relevant method for capturing cCPCs.
ACKNOWLEDGMENTS
This work was supported by NSERC DG (Grant No. 06465-14), the Southern University of Science and Technology Startup Grant (No. Y01231004), a Basic Research Grant (No. JCYJ20170817104516358) from the Shenzhen Municipal Science Foundation, the Shenzhen Basic Research Grant (No. JCYJ20160331115633182), the Guangzhou Science and Technology Project (Project No. 201804010284), and the Natural Science Foundation of Guangdong Province (Project No. 2018A0303130042).
Contributor Information
Jaewon Park, Email: .
Lidan You, Email: .
REFERENCES
- 1.Siegel R., Miller K. D., and Ahmedin J., “Cancer statistics,” CA Cancer J. Clin. 67(1), 7–30 (2017). 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Nowakowski G. S. et al. , “Circulating plasma cells detected by flow cytometry as a predictor of survival in 302 patients with newly diagnosed multiple myeloma,” Blood 106(7), 2276–2279 (2005). 10.1182/blood-2005-05-1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bezděková R., Penka M., Hájek R., and Říhová L., “Circulating plasma cells in monoclonal gammopathies,” Klin. Onkol. 30(Suppl 2), 2S29–2S34 (2017). 10.14735/amko20172S29 [DOI] [PubMed] [Google Scholar]
- 4.Gonsalves W. I. et al. , “Quantification of clonal circulating plasma cells in relapsed multiple myeloma,” Br. J. Haematol. 167(4), 500–505 (2014). 10.1111/bjh.13067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonsalves W. I. et al. , “Quantification of circulating clonal plasma cells via multiparametric flow cytometry identifies patients with smoldering multiple myeloma at high risk of progression,” Leukemia 31(1), 130–135 (2017). 10.1038/leu.2016.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mailankody S. et al. , “Minimal residual disease in multiple myeloma: Bringing the bench to the bedside,” Nat. Rev. Clin. Oncol. 12(5), 286–295 (2015). 10.1038/nrclinonc.2014.239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger N., Kim-Schulze S., and Parekh S., “Minimal residual disease in multiple myeloma: Impact on response assessment, prognosis and tumor heterogeneity,” Adv. Exp. Med. Biol. 1100, 141–159 (2018). 10.1007/978-3-319-97746-1_9 [DOI] [PubMed] [Google Scholar]
- 8.Paiva B., Van Dongen J. J. M., and Orfao A., “New criteria for response assessment: Role of minimal residual disease in multiple myeloma,” Blood 125(20), 3059–3069 (2016). 10.1182/blood-2014-11-568907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qasaimeh M. A. et al. , “Isolation of circulating plasma cells in multiple myeloma using CD138 antibody-based capture in a microfluidic device,” Sci. Rep. 7, 45681 (2017). 10.1038/srep45681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foulk B. et al. , “Enumeration and characterization of circulating multiple myeloma cells in patients with plasma cell disorders,” Br. J. Haematol. 180(1), 71–81 (2018). 10.1111/bjh.15003 [DOI] [PubMed] [Google Scholar]
- 11.Hou H. W. et al. , “Isolation and retrieval of circulating tumor cells using centrifugal forces,” Sci. Rep. 3(1), 1259 (2013). 10.1038/srep01259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayashi M. et al. , “Size-based detection of sarcoma circulating tumor cells and cell clusters,” Oncotarget 8(45), 78965–78977 (2017). 10.18632/oncotarget.20697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huebner H. et al. , “Filtration based assessment of CTCs and CellSearch® based assessment are both powerful predictors of prognosis for metastatic breast cancer patients,” BMC Cancer 18(1), 204 (2018). 10.1186/s12885-018-4115-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregory K. and Basmadjian D., “An analysis of the contact phase of blood coagulation: Effects of shear rate and surface are intertwined,” Ann. Biomed. Eng. 22(2), 184–193 (1994). 10.1007/BF02390376 [DOI] [PubMed] [Google Scholar]
- 15.Cinar Y., Senyol A. M., and Duman K., “Blood viscosity and blood pressure: role of temperature and hyperglycemia,” Am. J. Hypertens. 14, 433 (2001). 10.1016/s0895-7061(00)01260-7 [DOI] [PubMed] [Google Scholar]
- 16.Wan Y., Tan J., Asghar W., Kim Y. T., Liu Y., and Iqbal S. M., “Velocity effect on aptamer-based circulating tumor cell isolation in microfluidic devices,” J. Phys. Chem. B 115(47), 13891–13896 (2011). 10.1021/jp205511m [DOI] [PubMed] [Google Scholar]
- 17.Chen H. and Sun X., “Laboratory characteristics of multiple myeloma and analysis of misdiagnosis,” Chin. J. Clin. Pathol. 4(4), 1 (2012). 10.3969/j.issn.1674-7151.2012.04.014 [DOI] [Google Scholar]
- 18.Turgeon M. L., Clinical Hematology: Theory and Procedures (Lippincott Williams & Wilkins, 2004). [Google Scholar]
- 19.Daniels H., Wheater V. G., and Burkitt P. R., Functional Histology: A Text and Colour Atlas (Churchill Livingstone, Edinburgh, 1979). [Google Scholar]
- 20.Feng Y. et al., “Unique biomechanical interactions between myeloma cells and bone marrow stroma cells,” Prog. Biophys. Mol. Biol. 103, 148 (2010). 10.1016/j.pbiomolbio.2009.10.004 [DOI] [PubMed] [Google Scholar]
- 21.Dong C. and Skalak R., “Leukocyte deformability: Finite element modeling of large viscoelastic deformation,” J. Theor. Biol. 158(2), 173–193 (1992). 10.1016/S0022-5193(05)80716-7 [DOI] [PubMed] [Google Scholar]
- 22.Zeng H. and Zhao Y., “Rheological analysis of non-Newtonian blood flow using a microfluidic device,” Sens. Actuators A 166, 207–213 (2010). 10.1016/j.sna.2010.01.031 [DOI] [Google Scholar]
- 23.Li J. L., Liu Y. R., Chang Y., Fu J. Y., and Chen S. S., “[Immunophenotypic characteristics of multiple myeloma cells],” Zhongguo Shi Yan Xue Ye Xue Za Zhi 10(3), 226–228 (2002). 10.3969/j.issn.1009-2137.2002.03.012 [DOI] [PubMed] [Google Scholar]
- 24.Tan S. J., Yobas L., Lee G. Y. H., Ong C. N., and Lim C. T., “Microdevice for the isolation and enumeration of cancer cells from blood,” Biomed. Microdevices 11(4), 883–892 (2009). 10.1007/s10544-009-9305-9 [DOI] [PubMed] [Google Scholar]
- 25.Xu W., Mezencev R., Kim B., Wang L., and Mcdonald J., “Cell stiffness is a biomarker of the metastatic potential of ovarian cancer cells,” PLoS One 7(10), 46609 (2012). 10.1371/journal.pone.0046609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam N. Y. L., Rainer T. H., Chiu R. W. K., Lo Y. M. D., Lam C. W. K., and Lo Y. M. D., “EDTA is a better anticoagulant than heparin or citrate for delayed blood processing for plasma DNA analysis,” Clin. Chem. 50(1), 256–257 (2004). 10.1373/clinchem.2003.026013 [DOI] [PubMed] [Google Scholar]
- 27.Boxshall K., Wu M.-H., Cui Z., Cui Z., Watts J. F., and Baker M. A., “Simple surface treatments to modify protein adsorption and cell attachment properties within a poly(dimethylsiloxane) micro-bioreactor,” Surf. Interface Anal. 38(4), 198–201 (2006). 10.1002/sia.2274 [DOI] [Google Scholar]
- 28.Hollowell J. G., van Assendelft O. W., Gunter E. W., Lewis B. G., Najjar M., and Pfeiffer C., “Hematological and iron-related analytes–reference data for persons aged 1 year and over: United States, 1988-94,” Vital Health Stat. 11(247), 1–156 (2005). [PubMed] [Google Scholar]
- 29.Lê G. N. et al. , “Current and future biomarkers for risk-stratification and treatment personalisation in multiple myeloma,” Mol. Omics 15(1), 7–20 (2019). 10.1039/C8MO00193F [DOI] [PubMed] [Google Scholar]
- 30.Cotten S. W., Shajani-Yi Z., Cervinski M. A., Voorhees T., Tuchman S. A., and Korpi-Steiner N., “Reference intervals and diagnostic ranges for serum free κ and free λ immunoglobulin light chains vary by instrument platform: Implications for classification of patient results in a multi-center study,” Clin. Biochem. 58, 100–107 (2018). 10.1016/j.clinbiochem.2018.06.003 [DOI] [PubMed] [Google Scholar]
- 31.Sarioglu A. F. et al. , “A microfluidic device for label-free, physical capture of circulating tumor cell clusters,” Nat. Methods 12(7), 685–691 (2015). 10.1038/nmeth.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang G., Crawford K., Turbyfield C., Lam W., Alexeev A., and Sulchek T., “Microfluidic cellular enrichment and separation through differences in viscoelastic deformation,” Lab Chip 15(2), 532–540 (2015). 10.1039/C4LC01150C [DOI] [PubMed] [Google Scholar]
- 33.Gossett D. R. et al. , “Label-free cell separation and sorting in microfluidic systems,” Anal. Bioanal. Chem. 397(8), 3249 (2010). 10.1007/s00216-010-3721-9 [DOI] [PMC free article] [PubMed] [Google Scholar]