ABSTRACT
Dark septate endophytes (DSEs) present a group of widespread root-colonizing fungi. The role of these endophytes in ecosystems and their interactions with plant pathogens are not well understood. In the current study, we assessed the antagonistic potential of the model DSE Cadophora sp. against the tomato soilborne pathogens Rhizoctonia solani, Pythium aphanidermatum and Verticillium dahliae. To investigate their interactions, we conducted in vitro assays followed by a greenhouse experiments in which tomato plants were inoculated with different combinations of the DSE and pathogens. RNA accumulation of selected tomato pathogenesis-related genes and of Cadophora sp. genes with putative antifungal function was analyzed. Cadophora sp. inhibited the growth of the fungal pathogens in vitro and vice versa; a negative impact of the pathogens on the growth of the DSE was also detected. In roots, however, this mutual negative interaction could not be observed. Expression analyses of plant genes could not explain this differential effect, but among the Cadophora sp. genes analyzed, a gene coding for a chalcone synthase was downregulated in planta. The data indicate that plants can change the interaction between fungi and, therefore, in vitro detected antagonism does not necessarily reflect the situation inside the plant.
Keywords: dark septate endophytes, fungi, plant pathogens, Cadophora, biocontrol
In vitro detected antagonism does not necessarily reflect the situation in planta.
INTRODUCTION
Fungal plant pathogens are detrimental factors in agriculture. They reduce crop production and consequently cause considerable economic losses (Fradin and Thomma 2006). Fungal pathogens are mainly controlled by the application of fungicides, about which concerns are growing for not only their environmental damage but also their effects on human health (Soares and Porto 2009), besides the increasing emergence of fungicide-resistant pathogens.
Microbes can protect plants from pathogens by enhancing plant resistance or tolerance, or by their direct antagonism toward plant pathogens. The enhancement of plant resistance or tolerance can be achieved when microbes alter plant biochemistry (Thomma et al. 2001; Caplan, Padmanabhan and Dinesh-Kumar 2008). Plant recognition of non-pathogenic microbes activates signaling pathways (cascades), which finally lead to enhanced plant resistance. Plant tolerance is often increased by the activation of antioxidative activities during colonization by non-pathogenic microbes (White and Torres 2010). This does not lead to lower pathogen infection but reduces symptoms development. Direct antagonism involves different mechanisms including competition for iron and other nutrients, parasitism by the secretion of proteins (e.g. enzymes) and the secretion of secondary metabolites with antibiotic activity (Heydari and Pessarakli 2010).
Numerous studies have spotted the use of microbes to control soilborne diseases but the number of available products does not meet the market's need (Alabouvette et al. 2009). In many cases, the effectiveness of microbes in protecting plants cannot be confirmed in practice (Ojiambo and Scherm 2006). Therefore, understanding the interactions between plant roots, beneficial microorganisms and soilborne pathogens, and how these interactions are influenced by environmental factors are of high importance for the future selection of effective biocontrol agents.
Endophytic fungi colonize plant tissues without causing symptoms, and exhibit a wide spectrum of interactions with their host plants (Saikkonen et al. 1998). Generally, fungal endophytes benefit plant performance in different ways. This includes the enhancement of plant growth through nutrient acquisition (Jumpponen 2001; Mandyam and Jumpponen 2005; Mahmoud and Narisawa 2013), and through the production of phytohormones (Schulz et al. 1998; Schulz et al. 2002; Schulz and Boyle 2005). Besides increasing tolerance and resistance of plants to abiotic and biotic stress, fungal endophytes can produce antimicrobial secondary metabolites inhibiting the growth of pathogens (Yu et al. 2010).
Dark septate endophytes (DSEs) are an abundant and widely distributed group of fungal root colonizers, belonging to the Class 4 of non-clavicipitaceous fungal endophytes (Rodriguez et al. 2009). They are known for their septated and melanized hyphae, and their ability to form microsclerotia in plant roots. The role of DSEs in ecosystem functioning is still ambiguous (Mandyam and Jumpponen 2005) and knowledge about their effects on plants is scarce. Inoculation experiments were carried out with several DSE species to improve the understanding of their interaction with their host plant; some have shown to positively affect tomato roots and fruit quality (Andrade-Linares et al. 2011), and to improve tomato growth under organic N conditions (Mahmoud and Narisawa 2013). Generally, an overall positive effect of DSEs on plants growing on sterile substrates was revealed by a meta-analysis (Newsham 2011).
Some studies have shown a potential for DSEs to control fungal plant diseases. The dark septate endophyte Harpophora oryzae protects rice plants from Magnaporthe oryzae by accumulating H2O2 and inducing plant systematic resistance (Su et al. 2013). Phialocephala fortinii reduced the symptoms caused by Verticillium longisporum in cabbage grown in vitro (Narisawa, Usuki and Hashiba 2004), Leptodontidium orchidicola decreased the wilting disease in tomato plants infected with Verticilliumdahliae (Andrade-Linares et al. 2011), and a Cadophora sp. (different than the one used in the present study) protected melon seedlings against Fusarium oxysporum f. sp. melonis in media supplied with an organic nitrogen source (Khastini et al. 2014). Furthermore, DSE fungi in the PAC group were shown to reduce mortality and to prevent the infection of Picea abies seedlings challenged with different pathogens in vitro (Tellenbach and Sieber 2012; Terhonen, Sipari and Asiegbu 2016).
Tomato is one of the most consumed vegetables worldwide, whereas its global production has reached almost 165 million tons in 2013 (http://faostat.fao.org). With the availability of genomic data and many mutant lines (Sato et al. 2012), tomato is now considered a model plant (Kimura and Sinha 2008). Root diseases can have a huge impact on the production of tomato. Rhizoctonia solani, Pythium aphanidermatum and Verticillium dahliae are good examples of soilborne pathogens, which cause major yield losses to tomatoes and are difficult to control (Barbara 2003; Klosterman et al. 2009; Warner 2012).
In this study, we investigated the 3-fold interaction between the DSE Cadophora sp., tomato plants and the three distinct pathogens: the oomycete P. aphanidermatum (phylum: Heterokontophyta), the true fungi R. solani (phylum: Basidiomycota) and V. dahliae (phylum: Ascomycota). The DSE Cadophora sp. was chosen because its genome sequence is available (Knapp et al. 2018). Together with the genome data of at least two of the three pathogens (Klosterman et al. 2011; Wibberg et al. 2015) and of the model tomato (Sato et al. 2012), it will be possible to study the molecular mechanisms underlying the interactions in the future. In vitro assays as well as greenhouse inoculation experiments were conducted to assess the biocontrol potential of the DSEs model Cadophora sp. Growth of the interacting fungi was monitored by morphological parameters (in vitro) or by quantitative PCR (in planta). We hypothesize that the presence of Cadophora sp. in natural ecosystem is accompanied by its ability to compete with other fungi, some of which are phytopathogenic. A second hypothesis is that the restriction of pathogens’ growth can also be observed during the colonization of the host plant. The third hypothesis states that differential interaction of DSEs with pathogenic fungi inside and outside the plant is based on differences in plant and/or fungal gene expression.
MATERIALS AND METHODS
Fungal strains
Cadophora sp. DSE1049 (Knapp, Pintye and Kovacs 2012) was maintained on a complete medium (Pontecorvo et al. 1953). The phytopathogenic fungus V. dahliae TomIGZ (accession GU060637, Race 1) R. solani AG3PT isolate Ben3 and P. aphanidermatum (BBA 70417) were obtained from the strain collection of Leibniz Institute for Vegetables and Ornamental Crops, Großbeeren, Germany. The pathogens were maintained on PDA medium and used in the experiments.
In vitro antagonism test between DSEs and fungal pathogens
Cadophora sp. was tested for its antifungal activity against the pathogens R. solani, P. aphanidermatum and V. dahliae on a solid medium. Twenty-five percent PDA plates were inoculated with 5 mm diameter plugs of Cadophora sp. at one side, and a pathogen plug on the other side allowing a distance of ∼6 cm between the two plugs. The pathogens and Cadophora sp. were grown in axenic cultures as a control, and each combination was repeated four times. Plates were inoculated with Cadophora sp. and V. dahliae at the same time, whereas in the case of R. solani and P. aphanidermatum, due to their faster growth, Cadophora sp. was transferred to the media first, incubated at 26°C for 5 days to allow the colonies to get established and thereafter the pathogens plugs were introduced. The colonies were cultivated until the pathogens have covered all the control plates (in the case of R. solani and P. aphanidermatum) or when no further growth alterations were observed in the dual culture (in the case of V. dahliae). The radii of fungal colonies were measured toward the center of the opposing colony and compared to the radii of the same species in the axenic culture. The viability of pathogens in the Cadophora sp. confronted mycelium was tested by transferring a 2 mm fungal plug from the ‘contact zone’ and plating it on new agar plates.
The effect of secreted metabolites on the growth of the phytopathogenic fungi
Cadophora sp. was inoculated into flasks containing 250 mL complete medium, and was shaken at 120 rpm for 3 weeks at 27°C. Fungal mycelia were removed by simple filtration and 100 mL of the filtrate was extracted with 2× the amount of ethyl acetate and concentrated to a volume of 5 mL using a rotary evaporator (KNF, Trenton, New Jersey). Five millimeter filter paper disks were immersed in the extract or in pure ethyl acetate as a control. Ethyl acetate was left to evaporate from the filter papers under the clean bench, and one extract-immersed filter paper was put on the side of a 25% PDA Petri dish, with a control filter paper on the opposing side allowing 6 cm in between. Five millimeter plug of each pathogen was transferred to the center of the plate allowing a distance of 3 cm from each of the filter papers. Colonies were let to grow for 2 days in the case of P. aphanidermatum, 3 days in the case R. solani and 14 days in the case of V. dahliae. The growth of the fungal colonies toward the extract-immersed filter disk was measured and compared with the growth toward the control filter disk. The test was carried out in three replicates.
Greenhouse pot experiments
The experiments were carried out in Großbeeren (52°N, 13°E) in greenhouse conditions (see below). The first experiment had 10–12 repetition per treatment and two factors ‘Pathogen’ (R. solani, P. aphanidermatum, V. dahliae or control) and ‘DSE’ (Cadophora sp. or control). Tomato seeds (Solanum lycopersicum cv. Hildares F1, Hild Samen GmbH, Marbach, Germany) were surface sterilized by soaking for 4 min in 2.5% NaClO, followed by washing with sterilized water for five times. Seeds were germinated on MS medium for 2 weeks in a growth chamber (Adaptis A1000, Conviron Germany GmbH, Berlin, Germany) set on 16 h day/8 h night at 23/21 °C, respectively, and 110 μmol m−2 s−1 light intensity, and were transplanted into pots containing 700 g per pot of quartz sand (RIGK GmbH, Wiesbaden, Germany) with 35% big particles (2 to 3 mm) and 65% small particles (0.5 to 1 mm). The seedlings were inoculated by drenching (see below) with DSEs or with a mock solution. The plants were challenged with the pathogens 2 weeks after DSE inoculation. In the first 2 weeks of the experiment, pots were irrigated daily with 50 mL of Hoagland's solution (De Kreij et al. 1997). Starting from the 3rd week, the daily supply of Hoagland's solution was increased by 50 mL every week reaching 300 mL on the 7th week. Pot saucers were kept full of osmotic water to ensure plant water supply. Plants were cultivated in the greenhouse for 6 weeks after inoculation with Cadophora sp. (4 weeks after inoculation with pathogens) before harvest. Fresh and dry biomass were measured, and the colonization rates of the pathogens as well as of Cadophora sp. were assessed using qPCR assays.
A second experiment was carried out where plants were inoculated with Cadophora sp. or a mock solution, and challenged with the pathogen V. dahliae or with a mock solution of autoclaved inoculum. Seeds were sterilized and germinated as previously described. The seedlings were transplanted on sand (particle size: 0.5 to 1 mm)/vermiculite (50/50) (RIGK GmbH, Wiesbaden, Germany) and inoculated with Cadophora sp. by drenching. Ten days after DSE inoculation, the plants were challenged with V. dahliae and were cultivated for 14 days (24 days in total). Fresh and dry shoot biomass were assessed. DNA was extracted from the roots to assess the colonization rate of V. dahliae as well as of the two DSEs using qRT-PCR. RNA was extracted from roots to assess the expression of tomato pathogenesis-related (PR) genes.
In the first greenhouse experiment, greenhouse day/night temperatures were 19.8/16.15°C with a relative humidity of 57/55% and a mean daily radiation of 33 mol m−2 day−1. In the second greenhouse experiment, temperatures were 25/20°C, relative humidity was 70/78% and a mean daily radiation was 45.7 mol m−2 day−1.
Inoculation and inoculum preparation
Cadophora sp. inoculum was prepared by introducing three fungal plugs to 200 mL liquid complete medium (Pontecorvo et al. 1953) in a flask. Plugs were taken from the freshly grown mycelia on complete medium agar plates. Mycelia were grown for 3 to 4 weeks on a shaker (120 rpm) at 27°C and were collected through filtration, washed, suspended in distilled water and mixed for 45 seconds using a blender at minimal speed (Model D72, Moulinex, Leipzig, Germany). The number of propagules (spores and/or hyphal fragments) was calculated by spreading 1:100–1:10 000 dilutions of each suspension on complete medium agar plates supplied with ampicillin. The concentrations used were 0.9 × 105 cfu/mL for the first greenhouse experiment and 3 × 105 cfu/mL for the second. In the first experiment, plants were inoculated by drenching 10 mL of propagules suspension on the roots, while in the second experiment, 25 mL were drenched. In both experiments, an autoclaved suspension served as mock control.
Verticillium dahliae inoculation was prepared by introducing three fungal plugs into sucrose sodium nitrate (SSN) liquid medium (Sinha and Wood 1968) and was cultivated for 4 weeks. Pythium aphanidermatum inoculum was prepared by growing the fungus in carrot broth medium (50 g/L). The inoculum and inoculation density for both fungi were calculated as previously mentioned. Verticillium dahliae inoculation in the first greenhouse experiment was done by drenching 16 mL of 5 × 105 cfu/mL around the roots, and in the second experiment by drenching 30 mL of 7.4 × 105 cfu/mL. Plants were infected with P. aphanidermatum by drenching 20 mL of 6 × 104 cfu/mL into each pot.
In case of R. solani, the inoculum was prepared by growing the R. solani strain on autoclaved barley kernels for at least 2 weeks, and inoculation was done by introducing six infected kernels to each pot at a depth of ∼5 cm.
Quantification of fungal DNA in the roots
qRT-PCR-based assay was used to quantify fungal colonization of the roots. Fungal DNA of each of the studied species was quantified using species-specific primers (Table S1, Supporting Information) in a relative quantification method, in which the relative abundance was assessed with respect to tomato ubiquitin gene.
Total genomic DNA was extracted from the randomly sampled roots of three biological replicates per treatment in both greenhouse experiments, using DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. The reactions were performed on a real-time PCR system (model 7500; ABI, Warrington, UK) in three technical replicates in 10 µL volume and composed of 5 µL 2× SensiMix SYBR® Low-ROX kit (Bioline, Luckenwalde, Germany), 600 nM each primer and 50−200 ng template DNA. The PCR programs comprised a 3, 5 or 10 min denaturation at 95°C followed by 40 cycles of 95°C/15 s, 64.5°C/30 s, 72°C/30 s for Cadophora sp., 95°C/15 s, 58.5°C/30 s, 72°C/45 s for P. aphanidermatum, 95°C/30 s, 57°C/30 s, 72°C/1 min for R. solani, 95°C/30 s, 66°C/30 s, 72°C/30 s for V. dahliae and 95°C/15 s, 60°C/1 min for the tomato ubiquitin gene. All programs were followed by a melting curve analysis (60–95°C) to insure the amplification of a single product.
Normalized fungal abundance was assessed using a ∆Ct method modified from De Coninck et al. (2012); ∆Ct = Ct tomato ubiquitin − Ct fungal primer. 2∆Ct values were further used as indicators of fungal abundance and compared between treatments (Su et al. 2013).
Expression of tomato pathogenesis-related genes
In order to test whether the colonization by Cadophora sp. and V. dahliae influence the RNA accumulation of selected plant defense genes, total RNA was extracted from root tissues of the second greenhouse experiment using the innuPREP Plant RNA Kit (Analytik Jena AG, Jena, Germany) following the manufacturer's protocol. DNA was removed by on-column digestion using the innuPREP DNase I Kit produced by the same manufacturer. The extracted RNA was quantified using NanoDrop1000 Spectrophotometer (Thermo Fischer, Dreieich, Germany), 1–2 µg were reverse transcribed into cDNA using M-MLV Reverse Transcriptase kit (Promega GmbH, Mannheim, Germany) in a 25 µL reaction following the supplier's instructions. One microliter aliquot of 50 times diluted cDNA served as template for every qRT-PCR reaction, which also contained 5 µL 2× SensiMix SYBR® Low-ROX kit (Bioline, Luckenwalde, Germany), and 600 nM of each primer. Six tomato pathogenesis-related genes were assayed (Table S2, Supporting Information) encoding for proteinase inhibitor II (Pin2), endochitinase (P3), pathogenesis-related protein 6 (P6), ß-1,3-glucanase (Glu), chitinase (Chi) and PR-5 protein (NP24). Actin and GADPH were used after showing high expression stability (see below). The reactions were performed in a Real–Time PCR System (Applied Biosystems, Warrington, UK) and performed in three technical replicates. The PCR programs comprised a 7 min denaturation at 95°C followed by 40 cycles of 95°C/15 s then 1 min at annealing temperature (Tm) according to the primers as shown in Table S2 (Supporting Information). Reference genes expression stability was assessed by calculating the expression stability (M) value and coefficient of variation (CV) using Biogazelle qBase+ version 3.0 (Biogazelle, Zwijnaarde, Belgium—www.qbaseplus.com), with the chosen thresholds M < 0.5 and CV < 0.25. Using the same software, calibrated normalized relative quantity (CNRQ) values of the six target genes were obtained to be compared between treatments.
Expression of Cadophora sp. genes in vitro vs. in planta
RNA accumulation of nine selected genes of Cadophora sp. was analyzed in an in vitro system and compared between fungal growth on a medium versus growth in plant roots.
The annotated Cadophora sp. genome (Knapp et al. 2018) was screened for genes of putative antifungal function. Genes were collected using the search terms ‘thaumatin’, ‘chitinase’ and ‘chalcone synthase’ in the MycoCosm resource (Grigoriev et al. 2014). The protein sequences of selected genes were screened for the presence of signal peptides using SignalP 4.1 server (Petersen et al. 2011) using standard eukaryotic settings. Three thaumatin-like proteins (TLPs), three chitinases (chi) and two chalcone synthase genes (CHS) were further analyzed (Table S3, Supporting Information).
Cadophora sp. was grown on solid MS medium supplied with 10% sucrose, or with tomato seedlings grown in vitro on carbon-free MS medium. Fifty milliliter MS media was poured in 580 mL round glass jars (WECK, Wehr, Germany) and autoclaved cellophane membranes were placed on the top of the media to allow the harvest of fungus and roots. Inoculation with Cadophora sp. was done by adding and spreading 100 µL of a propagule solution (4 × 105 cfu/mL) on cellophane's surface. Four 12 days old tomato seedlings, germinated in vitro as described before, were transferred to the carbon-free MS medium to allow the growth of Cadophora sp. in planta. Cadophora sp. was grown in a growth chamber (12 h day/12 h night 20 °C and 317 µmol m−2 s−1 light intensity) for 12 days.
The propagules solution was prepared by adding distilled autoclaved water on a PDA plate fully covered with mycelium of Cadophora sp. A sterilized artist's brush according to Silman, Nelse and Bothast (1991) was used to scrub the mycelium and free the propagules/ spores. The solution was then filtered using two layers of cheese cloth to remove big hyphal fragments. The concentration was estimated by plating a series of dilutions as previously mentioned.
The growth of Cadophora sp. into the roots was confirmed microscopically after staining root fragments with wheat germ agglutinin-Alexa Fluor 488 (WGA-AF 488) (Molecular Probes, Karlsruhe, Germany) in root tissues stained with Congo red according to Deshmukh et al. (2006). RNA was extracted from medium grown and roots-colonizing Cadophora sp. using innuPREP Bacteria/Fungi RNA Kit (Analytik Jena AG, Jena, Germany) following the manufacturer's instruction in addition to DNA removal using innuPREP DNase I Kit. cDNA was synthesized as previously described. A dilution of 1/10 of roots cDNA and 1/100 of medium-grown Cadophora sp. served as templates for qRT-PCR reactions. The qRT-PCR programs comprised a 7 min denaturation at 95°C followed by 40 cycles of 95°C/15 s then 1 min at 60°C; primer sequences are shown in Table S3 (Supporting Information). Genes coding for fungal actin and β-tubulin were chosen as reference genes as they showed high stability (M < 0.5 and CV < 0.25). CNRQ values were obtained using Biogazelle qBase+ version 3.0 (Biogazelle, Zwijnaarde, Belgium—www.qbaseplus.com) as mentioned above.
Primer design and validation
Cadophora sp. ITS-based primer pair was used for the assessment of its abundance; sequences of all fungal genes were obtained from the MycoCosm database (Grigoriev et al. 2014). Sequences for the tomato gene encoding an endochitinase (PR3; accession number: XM_004237785.3) and for the Cadophora sp. ITS (accession number: JN859258) were obtained from the NCBI database (Coordinators 2016). All primers were designed using the NCBI primer design tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/).
The specificity validation of the newly designed primers was done by running PCR reactions using pure DNA templates of the other fungi included in this study, as well as non-inoculated tomato roots obtained from the greenhouse as negative controls. The products of all newly designed primers were cloned into the pGEM®-T Vector (Promega, Mannheim, Germany) and sequenced (Eurofins Genomics, Ebersberg, Germany) to confirm the target sequences. Primers efficiency was determined for all qRT-PCR reactions using LinReg software, which uses the amplification data recorded during the PCR reaction (Ramakers et al. 2003).
Statistical analyses
All statistical analyses were performed using STATISTICA program version 11 (StatSoft Inc., Tulsa, Oklahoma). The comparison between the growth of colonies in the in vitro experiments, as well as comparing the colonization rates were done using one way analysis of variance (ANOVA), followed by Tukey's test at P = 0.05. In the greenhouse experiments, two ways ANOVA (P = 0.05) was used for plant growth parameters, and for plant gene expression data to assess the impact of factors and their interaction. T-test at P = 0.05 was performed to compare the expression of Cadophora sp. genes in planta vs. in vitro. All data sets were subjected to Kolmogorov-Smirnov test for normality and Levene's test for the homogeneity of variance. Kruskal–Wallis test (P = 0.05) was used on parameters lacking homogeneity of variance based on Levene's test.
RESULTS
In vitro antagonism between Cadophora sp. and fungal pathogens, and the effect of crude extracts of the secreted metabolites on the growth of pathogenic fungi
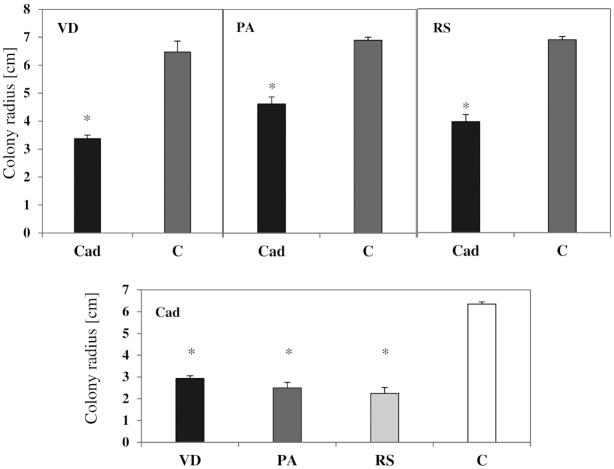
In all combinations when the antagonism of Cadophora sp. with R. solani, P. aphanidermatum and V. dahliae was tested, an inhibition of fungal colonies was observed (Fig. 1A, Fig. 2). Cadophora sp. was able to diminish the growth of the three pathogens tested. On the other hand, the three pathogens significantly reduced the growth of Cadophora sp. colonies. When media plugs obtained from the confrontation line between the two colonies were transferred to a new PDA plate, all fungi emerged and were able to establish new colonies.
Figure 1.
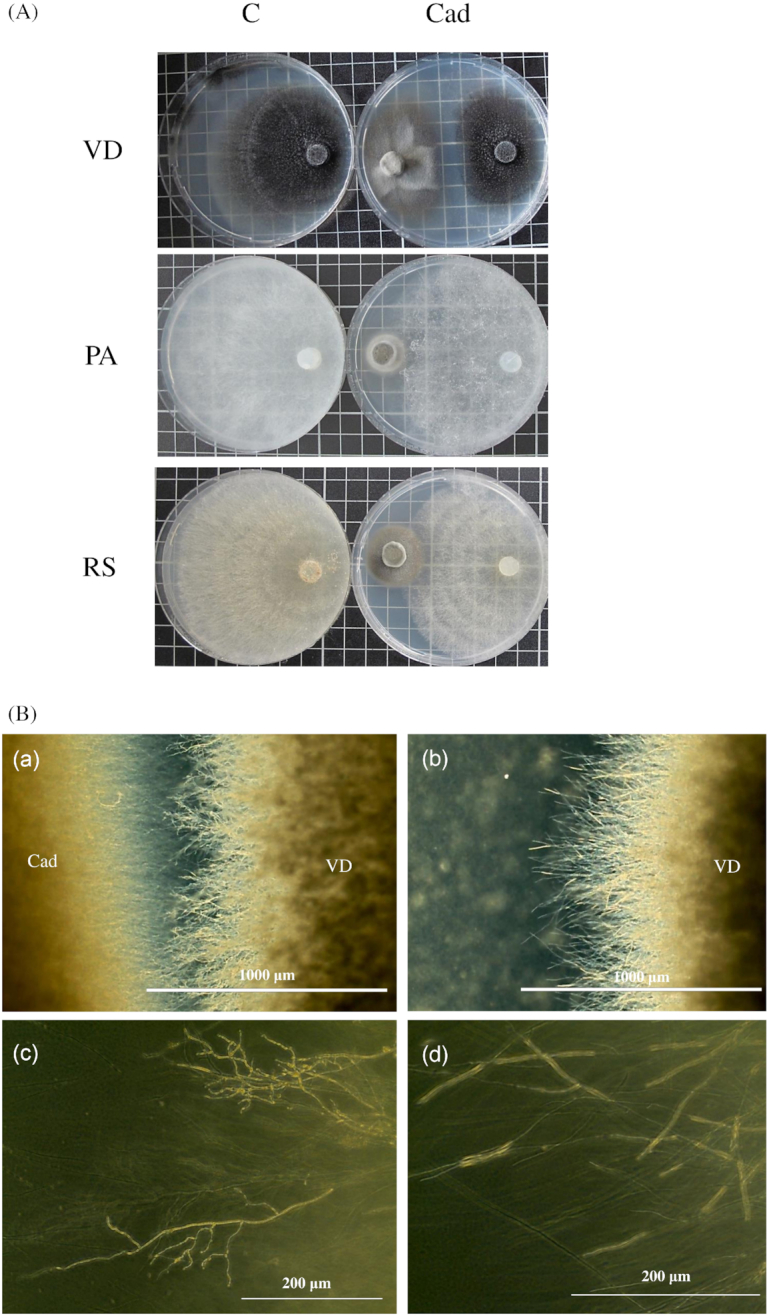
(A) The growth of fungal pathogens in the presence and absence of Cadophora sp. The three pathogens V. dahliae (VD), P. aphanidermatum (PA) and R. solani (RS) were grown in axenic cultures (C) or in dual cultures with Cadophora sp. (Cad). (B) The growth characteristics of V. dahliae’s hyphae. Verticillium dahliae growing on 25% PDA in the presence of Cadophora sp. (Cad) (a, c) or when growing alone (b, d).
Figure 2.
The growth inhibitory effects of Cadophora sp. and pathogenic fungi. The DSE Cadophora sp. (Cad) was grown in dual cultures with the three pathogens V. dahliae (VD), P. aphanidermatum (PA) and R. solani (RS). Each of the pathogens was grown in the presence and absence of Cadophora sp. (Cad). Shown are the means and standard deviations of the radii of fungal colonies measured toward the direction of the opposing colony in the co-culture, and compared to the growth of the same fungus in axenic culture (C). T-test (P = 0.05, n = 4) revealed an inhibitory effect of Cadophora sp. (Cad) on the growth of all pathogenic fungi. An inhibitory effect of pathogens on the radius of Cadophora sp. (Cad) was also observed. Significant differences between fungal growth in the axenic and the co-culture are indicated by asterisks.
In addition to reduction in growth, V. dahliae exhibited alteration in hyphal morphology in the co-culture (Fig. 1B). Fungal hyphae tended to be more branched, curled and swollen in comparison to the control colonies. These hyphal growth patterns were observed in all species upon encountering an opposing colony in the co-culture but were less notable in comparison to V. dahliae.
The crude extract of Cadophora sp. altered the growth of P. aphanidermatum and V. dahliae colonies (Figure S1, Supporting Information), but this effect was not statistically significant.
Greenhouse experiments
Some significant influences of the pathogens were detected on fresh and dry weights of shoots and roots in the greenhouse experiments. Plants growth was generally inhibited by the pathogens P. aphanidermatum and R. solani as they both significantly reduced shoot fresh and dry weight (Figure S2, Supporting Information), while V. dahliae only reduced shoot fresh weight (fresh weight is not shown) but not dry weight (Figure S2, Supporting Information). Concerning the roots, no effect on fresh and dry weights was observed, but a significant interaction was found between the factors ‘Cad’ and ‘pathogen’ for both parameters.
In the second greenhouse experiment, Cadophora sp. and V. dahliae exhibited no effect on plant growth (data not shown).
Quantification of fungal DNA in the roots
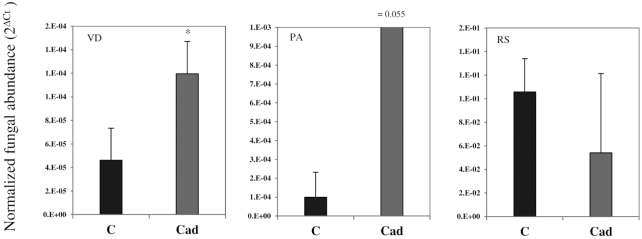
The pathogens were detected in all sampled inoculated plants. The colonization rates of P. aphanidermatum and R. solani did not significantly differ between Cadophora sp.-inoculated plants and controls. Colonization of tomato roots by V. dahliae was, however, significantly higher in Cadophora sp.-inoculated roots (Fig. 3). Cadophora sp. was quantified also to compare its colonization between the pathogen-infected roots and the controls and was found to be influenced by the pathogens (Figure S3, Supporting Information).
Figure 3.
The colonization of tomato by pathogenic fungi. DNA was extracted from non-inoculated roots (C), or roots inoculated with Cadophora sp. (Cad), which were challenged with V. dahliae (VD), P. aphanidermatum (PA) or R. solani (RS). Pathogens were quantified using qRT-PCR assays and compared between treatments. T-test (P = 0.05, n = 3) was carried out and showed higher V. dahliae colonization in Cadophora sp.-inoculated roots. Significant differences in pathogens colonization rate between Cadophora sp.-inoculated and control plants are indicated by asterisks.
In the second greenhouse experiment, no significant effect on V. dahliae colonization in Cadophora sp.-inoculated roots was detected. In addition, the quantification of Cadophora sp. did not reveal any significant differences in colonization rates between V. dahliae-inoculated roots and the control treatment.
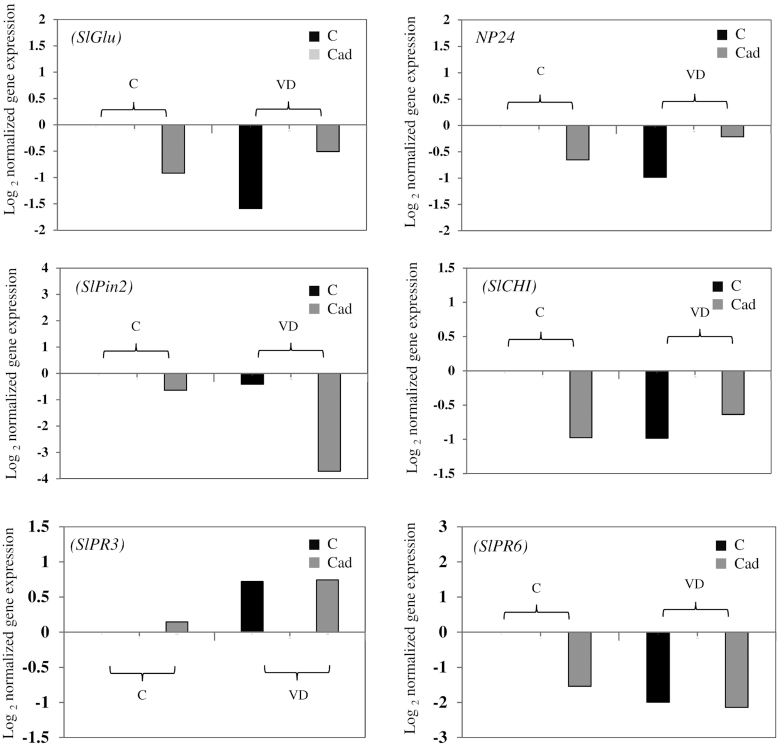
RNA accumulation of tomato PR genes
Our quantification showed that inoculation with Cadophora sp. did not influence the expression of the six genes analyzed, while colonization by V. dahliae resulted in the upregulation of the endochitinase gene SlPR3 (Fig. 4). No interaction between factors was observed.
Figure 4.
Normalized expression levels of defense-related genes in tomato. Tomato seedlings were not inoculated (C), or inoculated with Cadophora sp. (Cad) and challenged after 10 days with V. dahliae (VD) or were mock inoculated (C) (second greenhouse experiment) and grown for 2 weeks. RNA was extracted from roots and cDNA was synthesized. Shown are the Log2 CNRQ means of all treatments divided by that of control (C). Two ways ANOVA (P = 0.05, n = 3) on CNRQ values showed an impact of V. dahliae on the RNA accumulation of the endochitinase (SlPR3) gene. No other significant impacts were observed.
Expression of Cadophora sp. genes in vitrovs.in planta
One hundred forty-three chitinase-encoding genes were found in the Cadophora sp. genome; 58 are putative secreted chitinases and contained a signal peptide. Three genes that contained carbohydrate-binding module 18 and a signal peptide were assessed. Three genes coding for thaumatin proteins were found in the Cadophora sp. genome and contained signal peptides. In addition, two chalcone/ stilbene synthase-encoding genes were found. These genes were also involved in the analysis.
RNA accumulation levels of genes encoding for thaumatin-like proteins (CdTLPs) and chitinases (CdChi) and one chalcone synthase gene (CdCHS2) were not differentially expressed between the two treatments. The chalcone synthase gene (CdCHS1) was significantly downregulated in planta in comparison to in vitro.
DISCUSSION
Cadophora sp.—pathogen interactions in vitro
Cadophora sp. was shown to suppress the growth of the tomato pathogens P. aphanidermatum, V. dahliae and R. solani belonging to distant evolutionary lineages (Fig. 1, Fig. 2). The in vitro suppression was accompanied with a clear inhibition zones between the colonies of Cadophora sp. and V. dahliae, which could be explained by the production of antibiotics or toxic metabolites (Swadling and Jeffries 1996). The suppression of fungal growth was not accompanied with killing the fungus during the experiment, as further growth was observed when plugs taken from the contact zones of the two colonies were transferred to a new plate. Fungi belonging to the genus Cadophora were scarcely investigated for the production of secondary metabolites (Almeida et al. 2010). A Cadophora fungus has been shown to produce isochromanones with antimicrobial activity (Rusman et al. 2015, 2018), and the isolate Cadophora sp. DSE1049 might be a good candidate for the production of antifungal compounds as in the dual cultures alterations in hyphal growth (e.g. intensive lateral branching) in all species were observed. Induced lateral branching and curling of hyphae are known fungal response to antibiotics (Baráthová, Betina and Nemec 1969; Gunji, Arima and Beppu 1983) and metabolites derived from dark septate endophytes were shown to induce such a morphogenesis (Terhonen, Sipari and Asiegbu 2016). Lateral branching has been found to be an avoidance strategy by the human pathogen Aspergillus fumigatus to prevent a contact with neutrophils (Ellett et al. 2017). In the case of V. dahliae, intensive branching and the observed alteration might be a strategy to reduce exposure to compounds produced by Cadophora sp. that are unfavorable for growth.
The crude extract of Cadophora sp. was shown to slightly alter fungal growth (Figure S1, Supporting Information), which indicated that bioactive secondary metabolites might have been produced in the liquid culture. An optimization of fungal growth conditions and crude extraction method may lead to the identification of antifungal secondary metabolites produced by Cadophora sp.
DSE–pathogen interactions in planta, and impact on plant growth
The antagonism observed in vitro gave rise to the hypothesis that Cadophora sp. would be able to suppress the growth of pathogens in planta. Our first experiment showed a negative impact of the pathogens on plant growth and no impact of Cadophora sp. Positive effects of DSEs on plant growth have been mainly linked to improved plant nutrition (Haselwandter and Read 1982; Jumpponen, Mattson and Trappe 1998; Newsham 2000; Yakti et al. 2018; Yakti et al. 2019), for example, DSEs had significant effects on plant biomass when N was supplied in organic form (Newsham 2011) pointing out to their ability to solubilize nutrients. This could not be expected in our experimental system where all nutrients were supplied in their plant-available forms. Nevertheless, it is assumed that several factors, including environmental conditions and plant physiology, produce different host responses to endophytic colonization (Redman, Dunigan and Rodriguez 2001; Mandyam and Jumpponen 2015). Other studies have shown that DSEs can have negative impacts on different parameters of their host (Wilcox and Wang 1987; Tellenbach, Grünig and Sieber 2011; Reininger, Grünig and Sieber 2012; Terhonen, Sipari and Asiegbu 2016). In our experiments, Cadophora sp. had a negative effect on root biomass in the presence of V. dahliae which might be even an advantageous effect in the presence of a soilborne pathogen (Figure S2, Supporting Information).
In contrast to the in vitro observations, Cadophora sp. and pathogens did not show antagonistic effects on each other in planta. Moreover, plants inoculated with Cadophora sp. tended to have a higher level of V. dahliae colonization in the first greenhouse experiment (Fig. 3 ), combined with growth reduction in roots colonized by both fungi. In the case of P. aphanidermatum, a trend for more colonization was observed, but was not significant with a notably high standard deviation. Obtaining a representative root sample for the quantification of P. aphanidermatum is challenging (Kyuchukova et al. 2006), which might be an issue in our experiment, too.
To the best of our knowledge, the current study is the first to implement a qPCR assay to directly assess the interaction of DSE and pathogenic fungi in the roots. Tellenbach and Sieber (2012) have used quantitative PCR to assess only the abundance of the DSE Phialocephala subalpina with and without the presence of pathogenic oomycetes and found no effect of pathogens’ occurrence on the DSE's abundance, which is in accordance with our study. Whitaker and Bakker (2018) have also shown that in vitro antagonism of bacterial endophytes could not be observed in live plant tissues and pointed out that the commonly used in vitro assays cannot always predictin planta scenarios.
The effect of co-inoculations on the expression of plant PR genes
The majority of studies that dealt with plant defense responses have been carried out with an infection of a sole pathogen or with co-inoculation with beneficial symbionts and fungal pathogens (Van Wees, Van der Ent and Pieterse 2008; Jiang, Zheng and Chen 2009; Jung et al. 2012; Muthamilarasan and Prasad 2013; Vos et al. 2013; Pusztahelyi, Holb and Pócsi 2015). Only limited knowledge is, however, available about the situation when more than one fungus is present showing negative effects in the plant (Tsror and Hazanovsky 2001; Abdullah et al. 2017). It has been observed that a plant-colonizing microbe could facilitate the invasion of a pathogen (Spoel, Johnson and Dong 2007). In one of these cases, it has been shown that a biotrophic fungus, Albugo candida, repressed the expression of defense-related genes allowing more microbial invasions (Cooper et al. 2008). We, therefore, hypothesized that the less negative or even positive interactions between the pathogens and the DSE could be due to a defense repression caused by the fungi. This hypothesis was tested for plants of the second greenhouse experiment, where control plants or plants inoculated with Cadophora sp. were challenged or non-challenged with V. dahliae. Genes for an endochitinase (SlPR3) and for the PR protein 6 (SlPR6) were selected as they have been upregulated in stems of V. dahliae infected plants (Robb et al. 2009). In the current study, only the expression of SlPR3 was induced in roots infected with V. dahliae. Nevertheless, the hypotheses that DSEs could repress the expression of the selected genes had to be rejected.
Cadophorasp. gene expression in plantavs.in vitro
Besides the effects of co-inoculation on plant defense, the lack of mutual suppression in planta compared to in vitro could be also based on differences in the direct interactions between the DSE and the pathogens. Even though the establishment of DSE colonization of the roots was allowed in the greenhouse experiments before plants were infected with the pathogen, no suppression of pathogenic growth was observed (Fig. 3). This gave rise to the hypothesis that the production of antifungal compounds by the DSE is downregulated in planta. To test this hypothesis, we grew Cadophora sp. in vitro and in planta and assessed the expression of genes putatively involved in the antagonistic activity of the fungus (Table S4, Supporting Information). Three secreted chitinase-encoding genes, three genes for thaumatin-like proteins (TLC) and two chalcone synthase-encoding genes (CHS) were selected to test the hypothesis by expression analyses. Chitinases hydrolyze chitin, a main component of fungal cell wall. In plants, chitinases play crucial role in defense against fungal pathogens (Punja and Zhang 1993), while in fungi they could have many functions including nutrition and morphogenesis (Adams 2004). TLCs are low molecular weight proteins that have been shown to be upregulated in plants in response to biotic stress (Singh et al. 2013). TLCs possess antibiotic activity against many fungal pathogens (Garcia‐Casado et al. 2000; Chu and Ng 2003; Ho, Wong and Ng 2007). CHS belong to the type III polyketide synthases which are involved in the production of a wide variety of bioactive secondary metabolites in microbes (Yu et al. 2012). In plants, it is a key enzyme in the biosynthesis of flavonoids (Knogge, Schmelzer and Weissenböck 1986; Dao, Linthorst and Verpoorte 2011) that has been shown to possess antimicrobial activity (Orhan et al. 2010; Mousa and Raizada 2013), playing a role in plant defense against pathogens. Nevertheless, no differences were found in the RNA accumulation levels of chitinases and TLPs, but one CHS was significantly downregulated in planta (Fig. 5). The results indicate a shift in the production of bioactive secondary metabolites by Cadophora sp. during its endophytic lifestyle. The products of the corresponding pathway in Cadophora sp. and their role in its antagonistic activity have to be tested.
Figure 5.
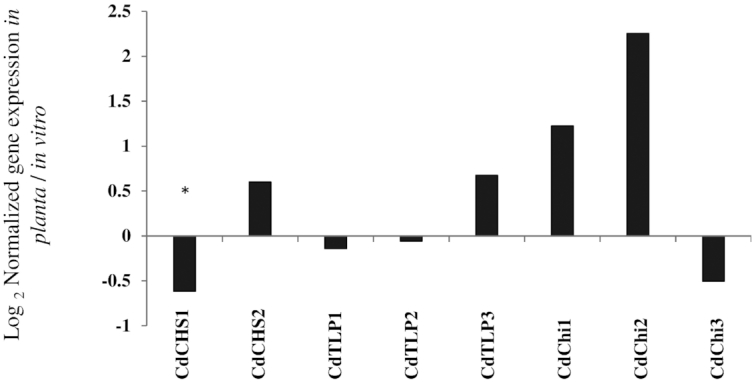
The expression of Cadophora sp. genes in planta vs. in vitro. Cadophora sp. was grown for 12 days on MS medium supplied with 10% sucrose (in vitro treatment), or in tomato seedlings grown on MS medium (in planta treatment). RNA was extracted and cDNA was synthesized. Three genes coding for thaumatin-like proteins (CdTLP1–3), three coding for lectins (CdChi1–3) and two genes coding for chalcone synthase (CdCHS1,2) were analyzed. Shown are the CNRQ values of in planta grown Cadophora sp. divided by thein vitro CNRQ values. T-test (P = 0.05, n = 3) was performed on CNRQ values and compared between treatments. Significant differences are indicated by asterisks.
CONCLUSION
Compared to the other well-explored groups of root-associated fungi, the agricultural application potential of DSEs seems to be still enigmatic. DSEs possess high genetic and functional diversity, and their effects could vary based on the DSE–plant species combination and plant nutritional status (Mandyam and Jumpponen 2005; Newsham 2011; Mayerhofer, Kernaghan and Harper 2013). Therefore, more research is needed to build an overall idea about their function and potential application. It could be, however, shown in the current study that DSEs could have antagonistic activities, which is in clear contrast to arbuscular mycorrhizal fungi or other root-colonizing fungi where such activities could never be detected.
Our data reveal that the antagonism between the model DSE Cadophora sp. and pathogens observed in vitro could not be observed in greenhouse pot cultures. Nevertheless, different outcomes could be expected with different host plants, cultivation conditions or different pathogens. Expression data of plant defense-related genes did not provide an explanation because downregulation during co-inoculation was not observed. In contrast, the expression of a Cadophora sp. chalcone synthase encoding gene, a key enzyme in flavonoid biosynthesis in plants, showed a decreased expression in planta. Products of Cadophora sp.’s chalcone synthase, most probably fungal flavonoids as chalcone derivatives, might play a role in the observed antagonistic activity in vitro and the decreased mRNA accumulation of this gene in planta might have led to the loss of antifungal activity. Nevertheless, the antagonistic activity of Cadophora sp. may still be beneficial in terms of plant protection in a soil environment. While occupying soil niches and with their wide enzymatic arsenal (Mandyam, Loughin and Jumpponen 2010; Knapp and Kovács 2016; Knapp et al. 2018), DSEs might be able to limit the growth and persistence of plant-damaging fungi and prevent severe infestations. The ability of Cadophora sp. and other DSEs to produce antimicrobial metabolites in their saprophytic lifestyle in the soil is a topic for future research.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
Supplementary Material
FUNDING
This research was partially funded by the Ministries of Consumer Protection, Food and Agriculture of the Federal Republic of Germany, the Ministry of Science, Research and Culture of the State of Brandenburg, the Thuringian Ministry of Infrastructure and Agriculture, Germany, the Hungarian National Research, Development and Innovation Office (NKFIH/OTKA/K109102) and the ELTE Institutional Excellence Program (1783–3/2018/FEKUTSRAT) of the Hungarian Ministry of Human Capacities. The first author was supported by ERASMUS MUNDUS grant and a short trip scientific mission in the frame of COST Action (FA1103).
Conflict of interest. None declared.
REFERENCES
- Abdullah AS, Moffat CS, Lopez-Ruiz FJ et al.. Host–multi-pathogen warfare: pathogen interactions in co-infected plants. Front Plant Sci. 2017;8:1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams DJ. Fungal cell wall chitinases and glucanases. Microbiology. 2004;150:2029–35. [DOI] [PubMed] [Google Scholar]
- Alabouvette C, Olivain C, Migheli Q et al.. Microbiological control of soilborne phytopathogenic fungi with special emphasis on wilt‐inducing Fusarium oxysporum. New Phytol. 2009;184:529–44. [DOI] [PubMed] [Google Scholar]
- Almeida C, Eguereva E, Kehraus S et al.. Hydroxylated sclerosporin derivatives from the marine-derived fungus Cadophora malorum. J Nat Prod. 2010;73:476–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade-Linares DR, Grosch R, Restrepo S et al.. Effects of dark septate endophytes on tomato plant performance. Mycorrhiza. 2011;21:413–22. [DOI] [PubMed] [Google Scholar]
- Baráthová H, Betina V, Nemec P. Morphological changes induced in fungi by antibiotics. Folia Microbiol. 1969;14:475. [DOI] [PubMed] [Google Scholar]
- Barbara D. Verticillium Wilts-GF Pegg and BL Brady; CABI Publishing, CAB International, Wallingford, Oxon OX10 8DE, UK & 10 East 40th Street, Suite 3203, New York, NY 10016, USA, 552 pages. ISBN 0 85199 529 2 Physiol Mol Plant Pathol. 2003;62:51–2. [Google Scholar]
- Caplan J, Padmanabhan M, Dinesh-Kumar SP. Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe. 2008;13:126–35. [DOI] [PubMed] [Google Scholar]
- Chu K, Ng T. Isolation of a large thaumatin-like antifungal protein from seeds of the Kweilin chestnut Castanopsis chinensis. Biochem Biophys Res Commun. 2003;301:364–70. [DOI] [PubMed] [Google Scholar]
- Cooper AJ, Latunde-Dada AO, Woods-Tor A et al.. Basic compatibility of Albugo candida in Arabidopsis thaliana and Brassica juncea causes broad-spectrum suppression of innate immunity. Mol Plant Microbe Interact. 2008;21:745–56. [DOI] [PubMed] [Google Scholar]
- Coordinators NR. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44:D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao T, Linthorst H, Verpoorte R. Chalcone synthase and its functions in plant resistance. Phytochem Rev. 2011;10:397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coninck B, Amand O, Delauré S et al.. The use of digital image analysis and real‐time PCR fine‐tunes bioassays for quantification of Cercospora leaf spot disease in sugar beet breeding. Plant Pathol. 2012;61:76–84. [Google Scholar]
- De Kreij C, Voogt W, van den Bos AL et al.. Voedingsoplossingen Voor de Teelt van Tomaat in Gesloten Teeltsystemen. The Netherlands: Brochure VG Tomaat, PBG Naaldwijk, 1997. [Google Scholar]
- Deshmukh S, Hueckelhoven R, Schaefer P et al.. The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. Proc Natl Acad Sci USA. 2006;103:18450–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellett F, Jorgensen J, Frydman GH et al.. Neutrophil interactions stimulate evasive hyphal branching by Aspergillus fumigatus. PLoS Path. 2017;13:e1006154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin EF, Thomma BPHJ. Physiology and molecular aspects of Verticillium wilt diseases caused by Verticillium dahliae and Verticillium albo-atrum. Mol Plant Pathol. 2006;7:71–86. [DOI] [PubMed] [Google Scholar]
- Garcia‐Casado G, Collada C, Allona I et al.. Characterization of an apoplastic basic thaumatin‐like protein from recalcitrant chestnut seeds. Physiol Plant. 2000;110:172–80. [Google Scholar]
- Grigoriev IV, Nikitin R, Haridas S et al.. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42:D699–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunji S, Arima K, Beppu T. Screening of antifungal antibiotics according to activities inducing morphological abnormalities. Agric Biol Chem. 1983;47:2061–9. [Google Scholar]
- Haselwandter K, Read DJ. The significance of a root–fungus association in two Carex species of high-alpine plant communities. Oecologia. 1982;53:352–4. [DOI] [PubMed] [Google Scholar]
- Heydari A, Pessarakli M. A review on biological control of fungal plant pathogens using microbial antagonists. J Biol Sci. 2010;10:273–90. [Google Scholar]
- Ho VS, Wong JH, Ng T. A thaumatin-like antifungal protein from the emperor banana. Peptides. 2007;28:760–6. [DOI] [PubMed] [Google Scholar]
- Jiang F, Zheng XD, Chen JS. Microarray analysis of gene expression profile induced by the biocontrol yeast Cryptococcus laurentii in cherry tomato fruit. Gene. 2009;430:12–6. [DOI] [PubMed] [Google Scholar]
- Jumpponen A. Dark septate endophytes—are they mycorrhizal? Mycorrhiza. 2001;11:207–11. [Google Scholar]
- Jumpponen A, Mattson KG, Trappe JM. Mycorrhizal functioning of Phialocephala fortinii with Pinus contorta on glacier forefront soil: interactions with soil nitrogen and organic matter. Mycorrhiza. 1998;7:261–5. [DOI] [PubMed] [Google Scholar]
- Jung SC, Martinez-Medina A, Lopez-Raez JA et al.. Mycorrhiza-induced resistance and priming of plant defenses. J Chem Ecol. 2012;38:651–64. [DOI] [PubMed] [Google Scholar]
- Khastini RO, Ogawara T, Sato Y et al.. Control of Fusarium wilt in melon by the fungal endophyte, Cadophora sp. Eur J Plant Pathol. 2014;139:339–48. [Google Scholar]
- Kimura S, Sinha NT. Tomato (Solanum lycopersicum): a model fruit-bearing crop. CSH Protoc. 2008;2008:emo105. [DOI] [PubMed] [Google Scholar]
- Klosterman SJ, Atallah ZK, Vallad GE et al.. Diversity, pathogenicity; and management of Verticillium species. Annu Rev Phytopathol. 2009;47:39–62. [DOI] [PubMed] [Google Scholar]
- Klosterman SJ, Subbarao KV, Kang S et al.. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Path. 2011;7:e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp DG, Kovács GM. Interspecific metabolic diversity of root-colonizing endophytic fungi revealed by enzyme activity tests. FEMS Microbiol Ecol. 2016;92:12. [DOI] [PubMed] [Google Scholar]
- Knapp DG, Németh JB, Barry K et al.. Comparative genomics provides insights into the lifestyle and reveals functional heterogeneity of dark septate endophytic fungi. Sci Rep. 2018;8:6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp DG, Pintye A, Kovacs GM. The dark side is not fastidious—dark septate endophytic fungi of native and invasive plants of semiarid sandy areas. PLoS One. 2012;7:e32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knogge W, Schmelzer E, Weissenböck G. The role of chalcone synthase in the regulation of flavonoid biosynthesis in developing oat primary leaves. Arch Biochem Biophys. 1986;250:364–72. [DOI] [PubMed] [Google Scholar]
- Kyuchukova MA, Büttner C, Gabler J et al.. Evaluation of a method for quantification of Pythium aphanidermatum in cucumber roots at different temperatures and inoculum densities. J Plant Dis Protect. 2006;113:113–9. [Google Scholar]
- Mahmoud RS, Narisawa K. A new fungal endophyte, Scolecobasidium humicola, promotes tomato growth under organic nitrogen conditions. Plos One. 2013;8:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandyam K, Jumpponen A. Seeking the elusive function of the root-colonising dark septate endophytic fungi. Stud Mycol. 2005;53:173–89. [Google Scholar]
- Mandyam K, Loughin T, Jumpponen A. Isolation and morphological and metabolic characterization of common endophytes in annually burned tallgrass prairie. Mycologia. 2010;102:813–21. [DOI] [PubMed] [Google Scholar]
- Mandyam KG, Jumpponen A. Mutualism–parasitism paradigm synthesized from results of root-endophyte models. Front Microbiol. 2015;5:776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayerhofer MS, Kernaghan G, Harper KA. The effects of fungal root endophytes on plant growth: a meta-analysis. Mycorrhiza. 2013;23:119–28. [DOI] [PubMed] [Google Scholar]
- Mousa W, Raizada M. The diversity of anti-microbial secondary metabolites produced by fungal endophytes: an interdisciplinary perspective. Front Microbiol. 2013;4:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthamilarasan M, Prasad M. Plant innate immunity: an updated insight into defense mechanism. J Biosci. 2013;38:433–49. [DOI] [PubMed] [Google Scholar]
- Narisawa K, Usuki F, Hashiba T. Control of Verticillium yellows in Chinese cabbage by the dark septate endophytic fungus LtVB3. Phytopathology. 2004;94:412–8. [DOI] [PubMed] [Google Scholar]
- Newsham KK. Phialophora graminicola, a dark septate fungus, is a beneficial associate of the grass Vulpia ciliata ssp. ambigua. New Phytol. 2000;144:517–24. [DOI] [PubMed] [Google Scholar]
- Newsham KK. A meta-analysis of plant responses to dark septate root endophytes. New Phytol. 2011;190:783–93. [DOI] [PubMed] [Google Scholar]
- Ojiambo PS, Scherm H. Biological and application-oriented factors influencing plant disease suppression by biological control: a meta-analytical review. Phytopathology. 2006;96:1168–74. [DOI] [PubMed] [Google Scholar]
- Orhan DD, Özçelik B, Özgen S et al.. Antibacterial, antifungal, and antiviral activities of some flavonoids. Microbiol Res. 2010;165:496–504. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G et al.. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G, Roper JA, Hemmons LM et al.. The genetics of Aspergillus nidulans. Adv Genet. 1953;5:141–238. [DOI] [PubMed] [Google Scholar]
- Punja ZK, Zhang Y-Y. Plant chitinases and their roles in resistance to fungal diseases. J Nematol. 1993;25:526. [PMC free article] [PubMed] [Google Scholar]
- Pusztahelyi T, Holb IJ, Pócsi I. Secondary metabolites in fungus–plant interactions. Front Plant Sci. 2015;6:573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL et al.. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–6. [DOI] [PubMed] [Google Scholar]
- Redman RS, Dunigan DD, Rodriguez RJ. Fungal symbiosis from mutualism to parasitism: who controls the outcome, host or invader?. New Phytol. 2001;151:705–16. [DOI] [PubMed] [Google Scholar]
- Reininger V, Grünig CR, Sieber TN. Host species and strain combination determine growth reduction of spruce and birch seedlings colonized by root-associated dark septate endophytes. Environ Microbiol. 2012;14:1064–76. [DOI] [PubMed] [Google Scholar]
- Robb J, Castroverde CD, Shittu HO et al.. Patterns of defence gene expression in the tomato–Verticillium interaction. Botany. 2009;87:993–1006. [Google Scholar]
- Rodriguez RJ, White JF Jr., Arnold AE et al.. Fungal endophytes: diversity and functional roles. New Phytol. 2009;182:314–30. [DOI] [PubMed] [Google Scholar]
- Rusman Y, Held BW, Blanchette RA et al.. Cadopherone and colomitide polyketides from Cadophora wood-rot fungi associated with historic expedition huts in Antarctica. Phytochemistry. 2018;148:1–10. [DOI] [PubMed] [Google Scholar]
- Rusman Y, Held BW, Blanchette RA et al.. Soudanones A–G: antifungal isochromanones from the ascomycetous fungus Cadophora sp. isolated from an iron mine. J Nat Prod. 2015;78:1456–60. [DOI] [PubMed] [Google Scholar]
- Saikkonen K, Faeth SH, Helander M et al.. Fungal endophytes: a continuum of interactions with host plants. Annu Rev Ecol Syst. 1998;29:319–43. [Google Scholar]
- Sato S, Tabata S, Hirakawa H et al.. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz B, Boyle C. The endophytic continuum. Mycol Res. 2005;109:661–86. [DOI] [PubMed] [Google Scholar]
- Schulz B, Boyle C, Draeger S et al.. Endophytic fungi: a source of novel biologically active secondary metabolites. Mycol Res. 2002;106:996–1004. [Google Scholar]
- Schulz B, Guske S, Dammann U et al.. Endophyte–host interactions. II. Defining symbiosis of the endophyte–host interaction. Symbiosis. 1998;25:213–27. [Google Scholar]
- Silman RW, Nelsen T, Bothast R. Comparison of culture methods for production of Colletotrichum truncatum spores for use as a mycoherbicide. FEMS Microbiol Lett. 1991;79:69–74. [Google Scholar]
- Singh NK, Kumar KRR, Kumar D et al.. Characterization of a pathogen induced thaumatin-like protein gene AdTLP from Arachis diogoi, a wild peanut. Plos One. 2013;8:e83963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha AK, Wood RKS. Studies on nature of resistance in tomato plants to Verticillium albo-artrum. Ann Appl Biol. 1968;62:319–27. [Google Scholar]
- Soares WL, Porto MFD. Estimating the social cost of pesticide use: an assessment from acute poisoning in Brazil. Ecol Econ. 2009;15:2721–8. [Google Scholar]
- Spoel SH, Johnson JS, Dong X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci USA. 2007;104:18842–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z-Z, Mao L-J, Li N et al.. Evidence for biotrophic lifestyle and biocontrol potential of dark septate endophyte Harpophora oryzae to rice blast disease. Plos One. 2013;8:e61332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swadling IR, Jeffries P. Isolation of microbial antagonists for biocontrol of grey mould disease of strawberries. Biocontrol Sci Technol. 1996;6:125–36. [Google Scholar]
- Tellenbach C, Grünig CR, Sieber TN. Negative effects on survival and performance of Norway spruce seedlings colonized by dark septate root endophytes are primarily isolate-dependent. Environ Microbiol. 2011;13:2508–17. [DOI] [PubMed] [Google Scholar]
- Tellenbach C, Sieber TN. Do colonization by dark septate endophytes and elevated temperature affect pathogenicity of oomycetes?. FEMS Microbiol Ecol. 2012;82:157–68. [DOI] [PubMed] [Google Scholar]
- Terhonen E, Sipari N, Asiegbu FO. Inhibition of phytopathogens by fungal root endophytes of Norway spruce. Biol Control. 2016;99:53–63. [Google Scholar]
- Thomma BP, Penninckx IA, Cammue BP et al.. The complexity of disease signaling in Arabidopsis. Curr Opin Immunol. 2001;13:63–8. [DOI] [PubMed] [Google Scholar]
- Tsror L, Hazanovsky M. Effect of coinoculation by Verticillium dahliae and Colletotrichum coccodes on disease symptoms and fungal colonization in four potato cultivars. Plant Pathol. 2001;50:483–8. [Google Scholar]
- Van Wees SCM, Van der Ent S, Pieterse CMJ. Plant immune responses triggered by beneficial microbes. Curr Opin Plant Biol. 2008;11:443–8. [DOI] [PubMed] [Google Scholar]
- Vos C, Schouteden N, Van Tuinen D et al.. Mycorrhiza-induced resistance against the root-knot nematode Meloidogyne incognita involves priming of defense gene responses in tomato. Soil Biol Biochem. 2013;60:45–54. [Google Scholar]
- Warner JS. Principal characteristics of pathogenic agents and methods of control. In: Blancard D.(ed). Tomato Diseases, 2nd edn. San Diego: Academic Press, 2012, 413–650. [Google Scholar]
- Whitaker BK, Bakker MG. Bacterial endophyte antagonism toward a fungal pathogen in vitro does not predict protection in live plant tissue. FEMS Microbiol Ecol. 2018;95:fiy237. [DOI] [PubMed] [Google Scholar]
- White JF, Torres MS. Is plant endophyte‐mediated defensive mutualism the result of oxidative stress protection? Physiol Plant. 2010;138:440–6. [DOI] [PubMed] [Google Scholar]
- Wibberg D, Rupp O, Blom J et al.. Development of a Rhizoctonia solani ag1-ib specific gene model enables comparative genome analyses between phytopathogenic R. solani ag1-ia, ag1-ib, ag3 and ag8 isolates. Plos One. 2015;10:e0144769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox HE, Wang CJK. Mycorrhizal and pathological associations of dematiaceous fungi in roots of 7-month-old tree seedlings. Can J Forest Res. 1987;17:884–99. [Google Scholar]
- Yakti W Andrade-Linares DR, Ngwene B et al.. Phosphate nutrition in root–fungus interactions. In: Hodkinson T, Doohan F, Saunders M, Murphy B(eds). Endophytes for a Growing World. Cambridge: Cambridge university press, 2019, 120–42. [Google Scholar]
- Yakti W, Kovács GM, Vági P et al.. Impact of dark septate endophytes on tomato growth and nutrient uptake. Plant Ecol Divers. 2018;11:637–48. [Google Scholar]
- Yu D, Xu F, Zeng J et al.. Type III polyketide synthases in natural product biosynthesis. IUBMB life. 2012;64:285–95. [DOI] [PubMed] [Google Scholar]
- Yu H, Zhang L, Li L et al.. Recent developments and future prospects of antimicrobial metabolites produced by endophytes. Microbiol Res. 2010;165:437–49. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.