Abstract
The present study aimed to identify the core genes and pathways involved in depression in patients with ovarian cancer (OC) who suffer from high or low-grade depression. The dataset GSE9116 from Gene Expression Omnibus database was analyzed to identify differentially expressed genes (DEGs) in these patients. To elucidate how certain genes could promote depression in patients with OC, pathway crosstalk, protein-protein interaction (PPI) and comprehensive gene-pathway analyses were determined using WebGestalt, ToppGene and Search Tool for the Retrieval of Interacting Genes and gene ontology analysis. Key genes and pathways were extracted from the gene-pathway network, and gene expression and survival analysis were evaluated. A total of 93 DEGs were identified from GSE9116 dataset, including 84 upregulated genes and nine downregulated genes. The PPI, pathway crosstalk and comprehensive gene-pathway analyses highlighted C-C motif chemokine ligand 2 (CCL2), Fos proto-oncogene, AP-1 transcription factor subunit (FOS), serpin family E member 1 (SERPINE1) and serpin family G member 1 (SERPING1) as core genes involved in the promotion of depression in patients with OC. These core genes were involved in the following four pathways ‘Ensemble of genes encoding ECM-associated proteins including ECM-affiliated proteins’, ‘ECM regulators and secreted factors’, ‘Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins’ and ‘MAPK signaling pathway and IL-17 signaling pathway’. The results from gene expression and survival analysis demonstrated that these four key genes were upregulated in patients with OC and high-grade depression and could worsen patients' survival. These results suggested that CCL2, FOS, SERPINE1 and SERPING1 may serve a crucial role in the promotion of depression in patients with OC. This finding may provide novel markers for predicting and treating depression in patients with OC; however, the underlying mechanisms remain unknown and require further investigation.
Keywords: oncogenes, depression, ovarian cancer, bioinformatics, molecular mechanism
Introduction
Ovarian cancer (OC) is a fatal malignancy of the female reproductive system (1). In 2018, there were 22,240 new cases of ovarian malignancies, and 14,070 OC-associated mortality cases (2). Furthermore, the incidence and mortality rates of malignant ovarian tumors in China reached 52.1/100,000 and 22.5/100,000, respectively, in 2015 (3). Due to the non-specific symptoms of OC, 70% of Chinese patients are diagnosed with a later stage of the disease at first diagnosis (4).
Previous studies demonstrated that ~38% of patients with cancer display major emotional distress, including anxiety and depression (5–7). In patients with OC, the incidence of depression and anxiety is of 82 and 92%, respectively (8). In addition, the degree of depression or anxiety is highly variable in patients with OC (9).
At present, the majority of studies have focused on how depression affects the prognosis of OC (10–12); however, the pathogenesis of depression in OC remains poorly investigated, particularly at a molecular level. By using a miRNA-mRNA regulation network, Wu et al (13) identified 12 miRNA-mRNAs pairs (miR-629-5p-FGF1, miR-629-5p-AKT3, miR-629-5p-MAGI2, miR-933-BDNF, miR-933-MEF2A, miR-23b-3p-TJP1, miR-23b-3p-JMJD1, miR-23b-3p-APAF1, miR-23b-3p-CAB39, miR-1265-CDKN1B, miR-33b-3p-CDKN1B and miR-33b-3p-F2R) that could be associated with the development of major depressive disorder (MDD) in patients with OC. Furthermore, Rahman et al (14) identified 34 differentially expressed genes (DEGs) associated with depression in patients with OC and demonstrated that CXCL12, ARL4C, NQO2 were associated with worse survival in patients with OC. As depressed patients display higher mortality rates (15) and since a different mental status can lead to different clinical outcomes (16) and prognosis (17), it is crucial to further understand the molecular mechanisms underpinning the onset of depression in patients with OC.
The present study aimed to identify DEGs in patients with OC and high or low-grade depression using bioinformatics analyses, and to determine the potential hub genes and pathways that may serve critical roles in the onset of depression in patients with OC.
Materials and methods
Study design
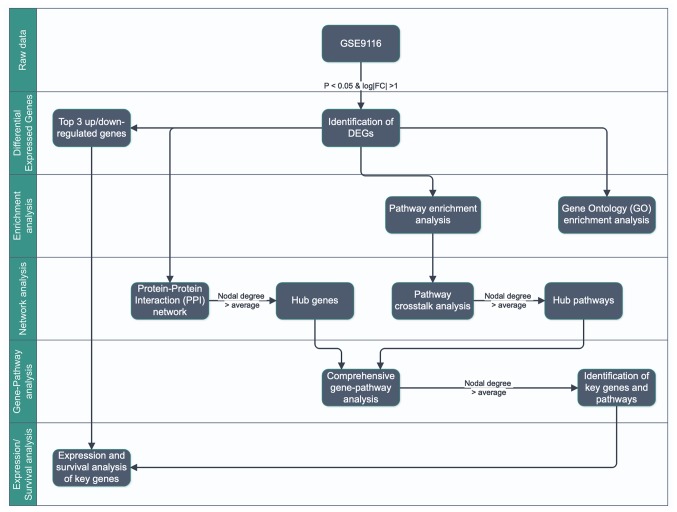
The flowchart for this study is presented in Fig. 1. DEGs were first identified in GSE9116 dataset (18,19) according to the following criteria: P<0.05 and absolute log2 value of fold change >1 (|log2(FC)|>1). The top three up- and downregulated DEGs were collected as potential key genes for further analysis. Subsequently, Gene Ontology (GO) (https://www.webgestalt.org/) and pathways enrichment analyses (https://toppgene.cchmc.org/) were performed using WebGestalt (20) and ToppGene (21) tools separately, and the protein-protein interaction (PPI) and pathway crosstalk networks were constructed. The gene-pathway network was constructed by mapping the hub genes into hub pathways extracted from PPI and pathway crosstalk network according to the criterion of nodal degree > average. Subsequently, the core genes with pathways were extracted from the gene-pathway network according to the criterion node degree > average. Eventually, the core genes and the top three up- and downregulated DEGs, which were defined as key genes, were studied by expression and survival analysis.
Figure 1.
Workflow for the identification of core genes promoting depression in patients with ovarian cancer. FC, fold change; DEGs, differentially expressed genes.
Microarray dataset
The GSE9116 dataset describing the gene expression profile of OC (18,19) and established on the platform of GPL96 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL96), was downloaded from Gene Expression Omnibus (GEO) database (22). To the best of our knowledge, this dataset is the only one that studied the impact of depression on gene expression in patients with OC. This dataset contains ten samples of primary OC (grade 3 serous cancer or serous adenocarcinoma), including five samples from patients with low-grade depression (LD) and five samples from patients with high-grade depression (HD). The samples and psychosocial data were collected from patients who were diagnosed with ovarian epithelial cancer, peritoneal cancer, or cancer of the fallopian tube, and who underwent primary surgical resection of ovarian carcinoma (14). A patient with a Center for Epidemiologic Studies Depression Scale score (23) ≥16 and a Social Provisions Scale-Attachment score (24) ≤15 was defined as having high-grade depression. Otherwise, patients were defined as having low-grade depression (18).
Identification of DEGs
The GSE9116 dataset was divided into two groups: HD and LD groups. Genes were considered differentially expressed between HD and LD groups if they exhibited a |log2(FC)|>1 and P<0.05 calculated using GEO2R (16) with the limma package (25). The top three up- and downregulated DEGs were considered as potential key genes for further analysis.
GO enrichment analysis
GO analysis was performed using WebGestalt tool (20) on the DEGs. Enriched biological terms for cellular components (CC), biological process (BP) and molecular functions (MF) with a P<0.05 were identified.
PPI network and identification of hub genes
The Search Tool for the Retrieval of Interacting Genes (STRING) (26) database and Cytoscape (version 3.5.1) (27) were used to identify hub genes. The PPI network was searched by gene symbols and the minimum required interaction score was set at 0.7 to ensure high confidence in the results. Nodes that were not connected to the major network were removed to reduce the error detection rate. The CentiScaPe plug-in (28) was used to calculate the degree of each node. Nodes were considered as hub if their degree was larger than the average.
Pathway enrichment and crosstalk analysis
For pathway enrichment, DEGs were mapped using the Kyoto Gene and Genome Encyclopedia (KEGG) database (29) and BioCarta (30) using the online analysis tool (21). Pathways with a P<0.05 were considered significant.
Pathway crosstalk analysis was conducted on the pathways defined above based on the assumption that two pathways are considered crosstalking if they share a proportion of genes (31,32). Briefly, pathways containing <3 genes were excluded as pathways with too few genes may contain insufficient biological information. Two indicators named Jaccard Coefficient (33) and Overlapping Coefficient (34), were used to measure the overlap between two pathways and were calculated as follows: Jaccard Coefficient = |(A∩B)/(A∪B) and Overlapping Coefficient = [(|A∩B|)/(min(|A|,|B|)], where A and B represent the list of genes in the two pathways, and min indicated the minimum number of genes between |A| and |B|. Subsequently, pairs of pathways with more than one overlapping gene were retained. Once the pathway crosstalking network was obtained, a subnetwork representing the hub pathways was identified according to the criterion nodal degree > average.
Comprehensive gene-pathway analysis
By mapping hub genes into the subnetwork of pathway crosstalk using KEGG (29) and BioCarta (30), a comprehensive gene-pathway network was obtained to further investigate the association between genes and pathways. The core genes and pathways in the gene-pathway network were identified according to the criterion nodal degree > average.
Expression and survival analysis of key genes
The expression of core genes in the sub gene-pathway network and of the top three up and downregulated DEGs were extracted from the GSE9116 dataset and evaluated for regulations. Furthermore, to analyze the effect of the key genes on patients' overall survival (OS), progression-free survival (PFS) and post-progression survival (PPS), patients were stratified according to the expression of the key genes and evaluated using Kaplan-Meier analysis (35). Kaplan-Meier analysis provides a survival assessment of prognosis-related genes in OC patients whose data obtained from The Cancer Genome Atlas (36) and 14 GSE profiles [GSE14764 (37), GSE15622 (38), GSE18520 (39), GSE19829 (40), GSE23554 (41), GSE26193 (42,43), GSE26712 (44,45), GSE27651 (46), GSE30161 (47), GSE3149 (48), GSE51373 (49), GSE63885 (50), GSE65986 (51) and GSE9891 (52)]. Patients were grouped by median gene expression and hazard ratio (HR) with 95% confidence intervals (CI) and log-rank P-values were calculated. P<0.05 was considered to indicate a significant difference.
Results
Identification of DEGs
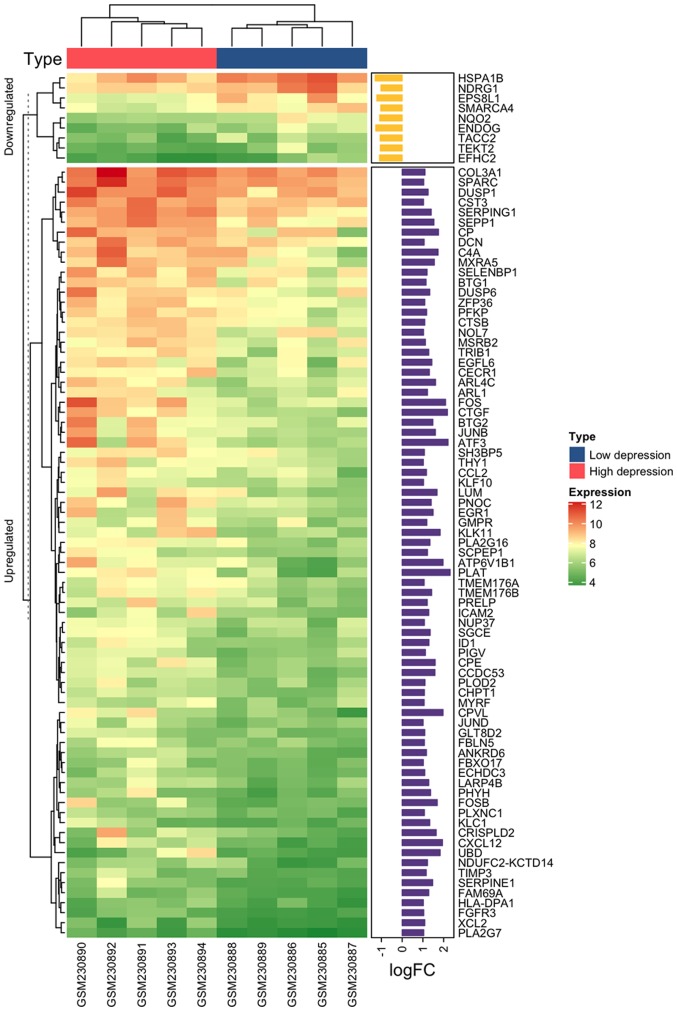
According to the criteria P<0.05 and |Log2(FC)|>1, 93 DEGs were identified when comparing the HD and LD groups. A total of 84 DEGs were upregulated and nine were downregulated. Plasminogen activator, tissue type (PLAT), activating transcription factor 3 (ATF3) and cellular communication network factor 2 (CTGF) were the top three upregulated genes with the highest change in expression. Heat shock 70 kDa protein 1B (HSPA1B), endonuclease G (ENDOG) and EPS8 like 1 (EPS8L1) were the top three downregulated genes. A heatmap with logFC values of the 93 DEGs is presented in Fig. 2.
Figure 2.
Heatmap of all differentially expressed genes from the GSE9116 dataset. The groups indicated at the top of the figure in blue and red represent groups with low-grade depression and high-grade depression, respectively. The numbers in the expression key represent the log2 value of reads for each gene.
GO enrichment analysis
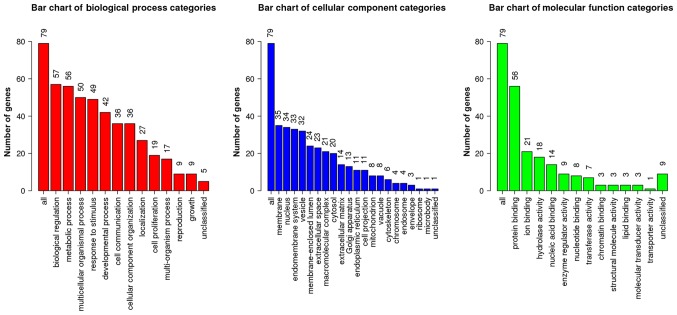
GO enrichment analysis was performed on the 93 DEGs using WebGestalt (Fig. 3). In terms of BP, DEGs were primarily involved in ‘biological regulation’, ‘metabolic process’, ‘multicellular organismal process’ and ‘response to stimulus’. In terms of CC, DEGs were enriched in ‘membrane’, ‘nucleus’, ‘endomembrane system’ and ‘vesicle’. In terms of MF, DEGs mainly participated in ‘protein binding’, ‘ion binding’ and ‘hydrolase activity’.
Figure 3.
Results of GO enrichment analysis for ‘Biological Process’, ‘Cellular Component’ and ‘Molecular Function’ categories. The number on each bar indicates the number of enriched genes annotated with the corresponding GO term. Go, Gene Ontology.
Construction of the PPI network and identification of hub genes
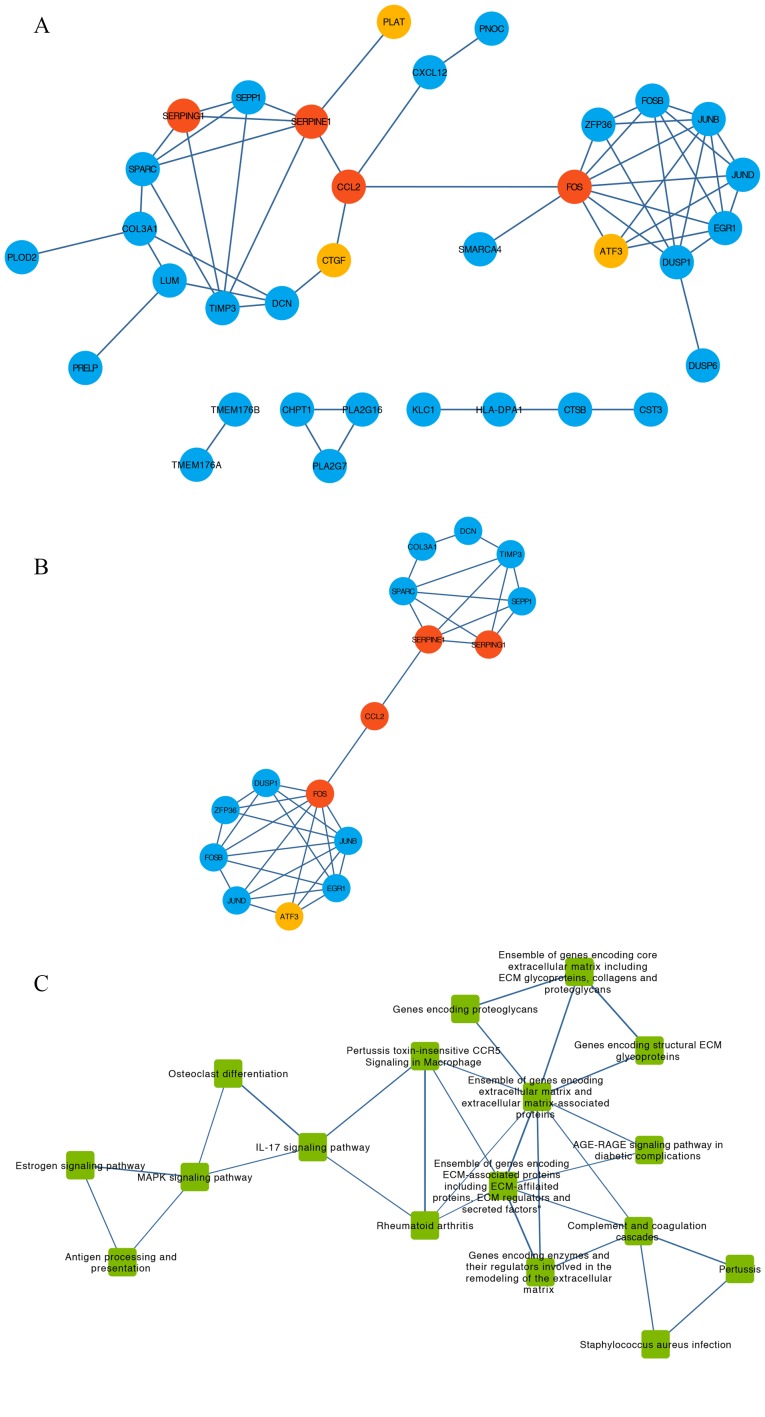
Using DEGs and the STRING database, a PPI network containing 34 nodes and 55 edges was obtained (Fig. 4A). The degree of each node was calculated using CentiScaPe (Table I). A total of 16 hub genes of which degree > average were extracted from the PPI network (Fig. 4B). This subnetwork was divided into two clusters that contained eight and seven genes, respectively. These two clusters were connected by C-C motif chemokine ligand 2 (CCL2) (Fig 4B).
Figure 4.
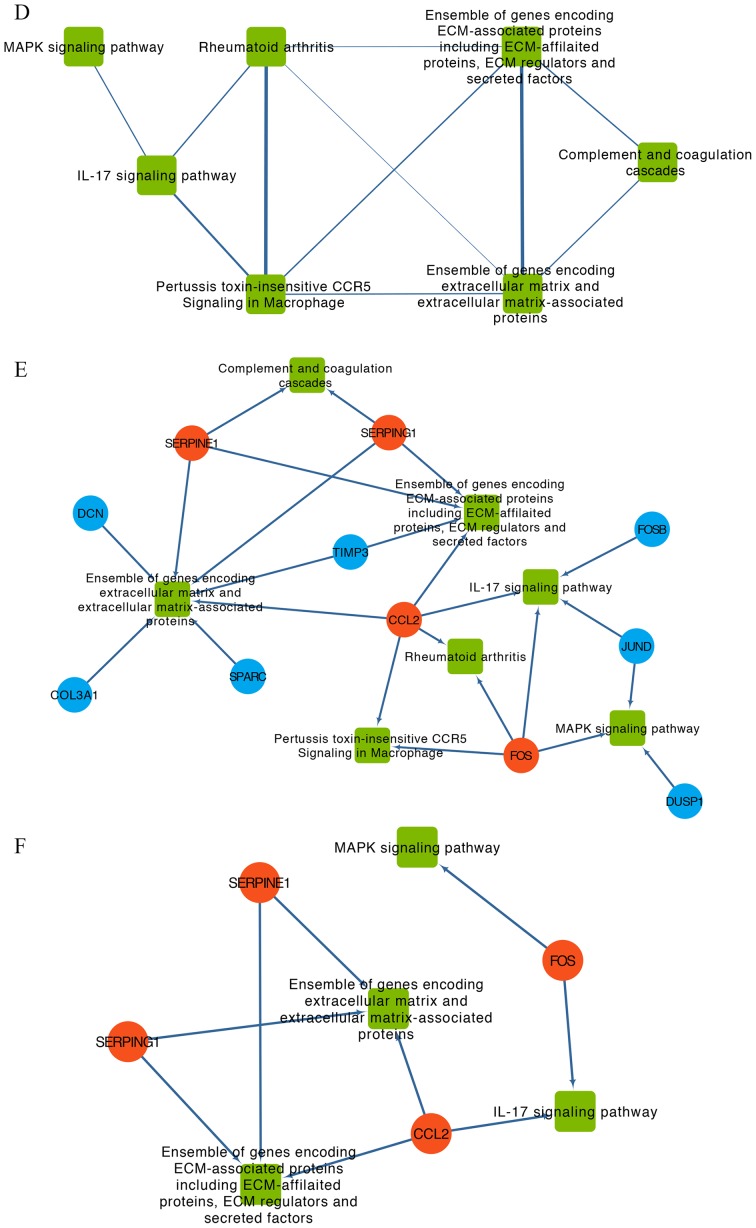
PPI network, pathway crosstalk and gene-pathway analyses of the DEGs from the GSE9116 dataset. (A) PPI network of candidate genes. Red, core genes. Orange, top three up-regulated genes. Blue, other DEGs. (B) Subnetwork of PPI network for hub genes of which nodal degree > average. Red, core genes. Orange, top three up-regulated genes. Blue, other DEGs. (C) Pathway crosstalk analysis of DEGs. Edge thickness represents the average value of Jaccard and Overlapping Coefficient. (D) Subnetwork of pathway crosstalk extracted by the criterion nodal degree > average. (E) Comprehensive gene-pathway network established by mapping the hub genes to the subnetwork. The arrow direction between gene and pathway was determined from Kyoto Gene and Genome Encyclopedia database and BioCarta. Red circle, core genes. Blue circle, other DEGs. Green square, pathways. (F) Subnetwork of gene-pathway collected according to the criterion nodal degree > average; Red circle, core genes. Green square, pathways. ATF3, activating transcription factor 3; CCL2, C-C motif chemokine ligand 2; CTGF, cellular communication network factor 2; DEGs, differentially expressed genes; FOS, Fos proto-oncogene, AP-1 transcription factor subunit; PLAT, plasminogen activator, tissue type; PPI, protein-protein interaction; SERPINE1, serpin family E member 1; SERPING1, serpin family G member 1.
Table I.
Characteristics of genes identified from protein-protein interaction network.
| Rank | Gene | Degree of each node |
|---|---|---|
| Overall average value | 3.2352 | |
| 1 | FOS | 9 |
| 2 | JUNB | 7 |
| 3 | DUSP1 | 6 |
| 4 | EGR1 | 6 |
| 5 | FOSB | 6 |
| 6 | SERPINE1 | 6 |
| 7 | SPARC | 5 |
| 8 | TIMP3 | 5 |
| 9 | JUND | 5 |
| 10 | SEPP1 | 4 |
| 11 | SERPING1 | 4 |
| 12 | ZFP36 | 4 |
| 13 | ATF3 | 4 |
| 14 | COL3A1 | 4 |
| 15 | DCN | 4 |
| 16 | CCL2 | 4 |
| 17 | LUM | 3 |
| 18 | CTGF | 2 |
| 19 | CXCL12 | 2 |
| 20 | CTSB | 2 |
| 21 | HLA-DPA1 | 2 |
| 22 | CHPT1 | 2 |
| 23 | PLA2G7 | 2 |
| 24 | PLA2G16 | 2 |
| 25 | SMARCA4 | 1 |
| 26 | CST3 | 1 |
| 27 | KLC1 | 1 |
| 28 | PNOC | 1 |
| 29 | TMEM176A | 1 |
| 30 | TMEM176B | 1 |
| 31 | PRELP | 1 |
| 32 | DUSP6 | 1 |
| 33 | PLOD2 | 1 |
| 34 | PLAT | 1 |
Pathway enrichment and crosstalk analysis
Pathway enrichment analysis was performed using ToppGene database. The results indicated that the top five significantly enriched pathways were ‘Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins’, ‘Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans’, ‘Complement and coagulation cascades’, ‘Pertussis toxin-insensitive CCR5 Signaling in Macrophage’ and ‘Rheumatoid arthritis’ (Table II).
Table II.
Results from pathway enrichment analysis.
| Pathways | Source | P-value | Genes in the pathway |
|---|---|---|---|
| Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins | BIOCARTA | 1.19×10−08 | COL3A1, CRISPLD2, FBLN5, SPARC, MXRA5, DCN, EGFL6, TIMP3, PLXNC1, PRELP, SERPINE1, XCL2, CST3, SERPING1, CCL2, PLAT, CTGF, LUM, CTSB, XCL1, PLOD2, CXCL12 |
| Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans | BIOCARTA | 1.73×10−05 | COL3A1, CRISPLD2, FBLN5, SPARC, MXRA5, DCN, PRELP, CTGF, LUM |
| Complement and coagulation cascades | KEGG | 6.90×10−05 | SERPINE1, SERPING1, PLAT, C4A, C4B |
| Pertussis toxin-insensitive CCR5 signaling in Macrophage | BIOCARTA | 1.20×10−04 | FOS, CCL2, CXCL12 |
| Rheumatoid arthritis | KEGG | 1.29×10−04 | ATP6V1B1, HLA-DPA1, FOS, CCL2, CXCL12 |
| Ensemble of genes encoding ECM-associated proteins including ECM-affiliated proteins, ECM regulators and secreted factors | BIOCARTA | 1.85×10−04 | EGFL6, TIMP3, PLXNC1, SERPINE1, XCL2, CST3, SERPING1, CCL2, PLAT, CTSB, XCL1, PLOD2, CXCL12 |
| Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix | BIOCARTA | 3.07×10−04 | TIMP3, SERPINE1, CST3, SERPING1, PLAT, CTSB, PLOD2 |
| MAPK signaling pathway | KEGG | 4.66×10−04 | FOS, DUSP1, DUSP6, JUND, FGFR3, HSPA1A, HSPA1B |
| Pertussis | KEGG | 7.77×10−04 | FOS, SERPING1, C4A, C4B |
| Antigen processing and presentation | KEGG | 8.16×10−04 | HLA-DPA1, CTSB, HSPA1A, HSPA1B |
| Genes encoding proteoglycans | BIOCARTA | 9.00×10−04 | DCN, PRELP, LUM |
| IL-17 signaling pathway | KEGG | 1.65×10−03 | FOS, FOSB, JUND, CCL2 |
| Lectin Induced Complement Pathway | BIOCARTA | 1.87×10−03 | C4A, C4B |
| Fibrinolysis Pathway | BIOCARTA | 1.87×10−03 | SERPINE1, PLAT |
| Ether lipid metabolism | KEGG | 1.88×10−03 | PLA2G7, PLA2G16, CHPT1 |
| Estrogen signaling pathway | KEGG | 2.00×10−03 | FOS, HSPA1A, HSPA1B, GABBR1 |
| AGE-RAGE signaling pathway in diabetic complications | KEGG | 2.08×10−03 | COL3A1, EGR1, SERPINE1, CCL2 |
| Classical Complement Pathway | BIOCARTA | 2.56×10−03 | C4A, C4B |
| Platelet Amyloid Precursor Protein Pathway | BIOCARTA | 2.56×10−03 | SERPINE1, PLAT |
| B Cell Survival Pathway | BIOCARTA | 3.36×10−03 | FOS, JUND |
| Staphylococcus aureus infection | KEGG | 3.51×10−03 | HLA-DPA1, C4A, C4B |
| Genes encoding structural ECM glycoproteins | BIOCARTA | 4.29×10−03 | CRISPLD2, FBLN5, SPARC, MXRA5, CTGF |
| Complement Pathway | BIOCARTA | 4.73×10−03 | C4A, C4B |
| Osteoclast differentiation | KEGG | 5.52×10−03 | FOS, FOSB, JUNB, JUND |
For the pathway crosstalk analysis, 17 out of 24 pathways that contained ≥2 genes met the crosstalk analysis criteria and were selected to construct the network (Fig. 4C). The thickness of the edges indicated the average values of Jaccard and the Overlapping Coefficient that represented the overlapping level of genes between two pathways. By selecting the nodes with degree > average, a subnetwork of pathway crosstalk with 7 nodes and 11 edges was constructed (Fig. 4D).
Comprehensive gene-pathway analysis
After mapping the hub genes onto the subnetwork of pathways provided by KEGG and BioCarta, a potential gene-pathway network including 7 essential pathways and 11 hub genes was constructed (Fig. 4E). The results from this network demonstrated that CCL2 and Fos proto-oncogene, AP-1 transcription factor subunit (FOS) participated in most of the pathways. Based on the number of genes involved, the top three pathways were ‘Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins’, ‘Ensemble of genes encoding ECM-associated proteins including ECM-affiliated proteins, ECM regulators and secreted factors’ and ‘IL-17 signaling pathway’ (Fig. 4E).
To identify the main nodes (including genes and pathways) in the gene-pathway network, nodes with degree > average were selected (Fig. 4F). The results demonstrated that CCL2, FOS, serpin family G member 1 (SERPING1) and serpin family E member 1 (SERPINE1) and the four pathways ‘Ensemble of genes encoding ECM-associated proteins including ECM-affiliated proteins, ECM regulators and secreted factors’, ‘Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins’, ‘MAPK signaling pathway’ and ‘IL-17 signaling pathway’ were identified and may serve a crucial role in the development of depression in patients with OC.
Association between key genes and the six top DEGs
The evaluation of the interaction between the four key genes (CCL2, FOS, SERPING1 and SERPINE1) and the six top DEGs (upregulated; PLAT, ATF3 and CTGF and downregulated; HSPA1B, ENDOG and EPS8L1) in the PPI (Fig 4A) demonstrated that HSPA1B, ENDOG, EPS8L1 were not found since they were not connected to the major network. Furthermore, PLAT was interplayed with SERPINE1, CTGF interacted with CCL2 and ATF3 was connected to FOS.
Expression and survival analysis of key genes
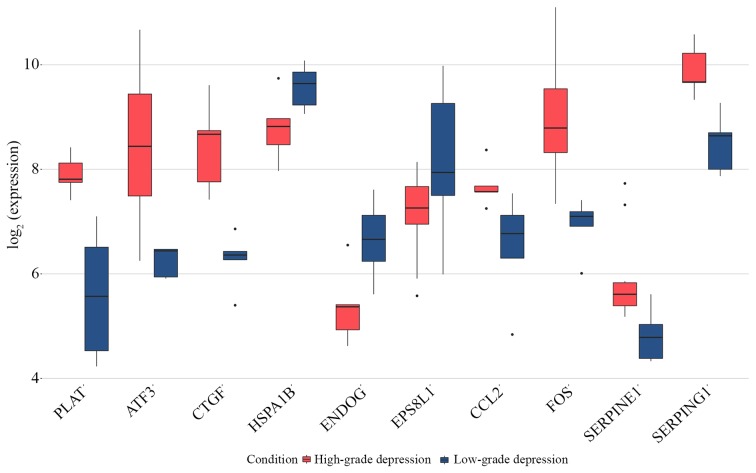
As presented in Fig. 5, all core genes and top three upregulated DEGs were significantly upregulated in the HD group, which was not the case for the top three downregulated DEGs compared with the LD group.
Figure 5.
Expression of the core genes CCL2, FOS, SERPINE1 and SERPING1 and the top three up and downregulated genes PLAT, ATF3, CTGF, HSPA1B, ENDOG and EPS8L1 obtained from GSE9116. All core genes were upregulated in the high-grade depression group. ATF3, activating transcription factor 3; CCL2, C-C motif chemokine ligand 2; CTGF, cellular communication network factor 2; DEGs, differentially expressed genes; ENDOG, endonuclease G; EPS8L1, EPS8 like 1; FOS, Fos proto-oncogene, AP-1 transcription factor subunit; HSPA1B, heat shock protein family A (Hsp70) member 1B; PLAT, plasminogen activator, tissue type; PPI, protein-protein interaction; SERPINE1, serpin family E member 1; SERPING1, serpin family G member 1.
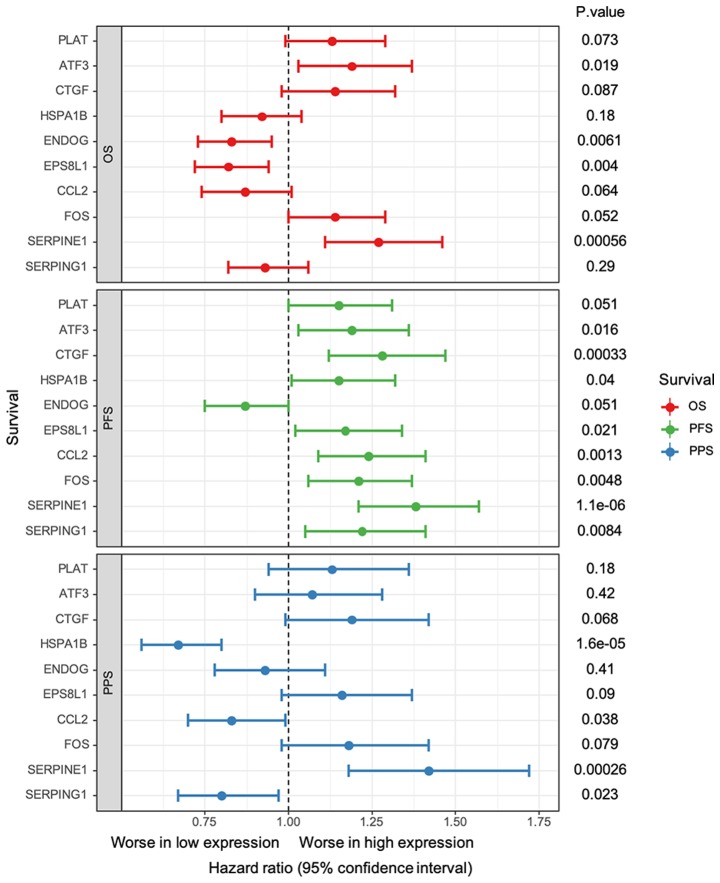
The results from survival analysis indicated that the top 3 upregulated DEGs had similar effects as the core genes, which was not the case for the top 3 downregulated DEGs (Fig. 6). In particular, higher expression of ATF3 (HR, 1.19; 95% CI, 1.03–1.37; P=0.019) and SERPINE1 (HR, 1.27; 95% CI, 1.11–1.46; P=0.00056) and lower expression of ENDOG (HR, 0.83; 95% CI, 0.73–0.95; P=0.0061) and EPS8L1 (HR, 0.82; 95% CI, 0.72–0.94; P=0.004) were significantly associated with worse OS. Furthermore, ATF3 (HR, 1.19; 95% CI, 1.03–1.36; P=0.016), CTGF (HR, 1.28; 95% CI, 1.12–1.47; P=0.00033), HSPA1B (HR, 1.15; 95% CI, 1.01–1.52; P=0.04), EPS8L1 (HR, 1.17; 95% CI, 1.12–1.34; P=0.0021) and all core genes, including CCL2 (HR, 1.24; 95% CI, 1.09–1.41; P=0.0013), FOS (HR: 1.21, 95%CI: 1.06–1.37; P=0.0048), SERPINE1 (HR, 1.38, 95% CI, 1.21–1.57; P=1.1e-06) and SERPING1 (HR, 1.22; 95% CI, 1.05–1.41; P=0.0084), were significantly associated with a lower PFS. However, lower expression of HSPA1B (HR, 0.67; 95% CI, 0.56–0.8; P=0.000016), CCL2 (HR, 0.83; 95% CI, 0.7–0.99; P=0.038) and SERPING1 (HR, 0.8; 95% CI, 0.67–0.97; P=0.023) and high expression of SERPINE1 (HR, 1.42; 95% CI, 1.18–1.72; P=0.00026) were significantly associated with worse PPS. The other genes had no significant effect on OS, PFS or PPS.
Figure 6.
Survival analysis of the core genes; CCL2, FOS, SERPINE1 and SERPING1, and the top three up/downregulated genes; PLAT, ATF3, CTGF, HSPA1B, ENDOG and EPS8L1. Vertical dotted line represents no effect for the pooled relative risk estimation. If the confidence intervals for individual gene overlap with this line, it means that the survival between low and high expression is not significant. ATF3, activating transcription factor 3; CCL2, C-C motif chemokine ligand 2; CTGF, cellular communication network factor 2; DEGs, differentially expressed genes; ENDOG, endonuclease G; EPS8L1, EPS8 like 1; FOS, Fos proto-oncogene, AP-1 transcription factor subunit; HSPA1B, heat shock protein family A (Hsp70) member 1B; OS, overall survival; PFS, progression-free survival; PLAT, plasminogen activator, tissue type; PPI, protein-protein interaction; PPS, post-progression survival; SERPINE1, serpin family E member 1; SERPING1, serpin family G member 1.
Discussion
The incidence of cancer continues to rise annually (53). With the advancement of psychological research, increasing attention is being paid to the mental health of patients with malignant tumors (54). Depression is a common psychological issue observed in cancer patients. It can reduce the efficacy of treatments, delay the recovery time and reduce the quality of life of patients (55). At similar stages of cancer cell differentiation or treatments, patients with depression may have a worse prognosis compared with non-depressed patients (56). It has been reported that patients with OC whom exhibit a positive attitude have an improved quality of life and prognosis compared with patients with negative emotions (57). It is therefore crucial to determine the underlying molecular mechanisms linking depression with poorer prognosis for patients with ovarian cancer.
Whereas the original study of GSE9116 used a promoter-based bioinformatics strategy to investigate the effect of β-adrenergic signaling (18) and focused on the genetic interaction locus in the human interleukin (IL)-6 promoter (single nucleotide polymorphisms rs1800795) (19), the present study aimed to identify key genes and pathways associated with depression in patients with OC using PPI networks, pathway crosstalk and gene-pathway analyses. The results from this study may provide an improved understanding underlying the link between depression in and a poorer prognosis in patients with OC.
The results from the present study demonstrated that CCL2 may induce depression in patients with OC through ECM-related and IL-17 signaling pathways. Previous studies indicated that high expression of CCL2 is associated with depression. Leighton et al (58) performed a meta-analysis, which included 4688 subjects (765 depressed healthy patients vs. 1528 not depressed healthy patients, and 742 depressed patients with inflammatory illness vs. 1653 not depressed patients with inflammatory illness) and reported that CCL2 blood levels were significantly higher in depressed patients with or without inflammatory illness compared with the control. Another meta-analysis reported similar results for CCL2 (59). In addition, CCL2 was upregulated in cancerous cells, and this required the participation of IL-17 (60), and CCL2 induced tumor cell proliferation and stimulated tumor cell migration and invasion into the surrounding extracellular matrix in order to promote tumor cell intravasation into the circulation (61).
The present study demonstrated an association between FOS expression and depression via MAPK and IL-17 signaling pathways in patients with OC. Yu et al (62) reported that human depression is associated with cognitive deficits, and that FOS is upregulated in cognitively impaired mice. Kung et al (63) demonstrated that FOS expression is higher in preproenkephalin-knockout mice, and induced anxiety and depression-like symptoms of post-traumatic stress disorder compared with wild-type mice. In addition, it was reported that FOS expression is regulated by the MAPK pathway (64,65). Furthermore, IL-17 can mediate inflammatory responses via FOS activation (66) and enhance the recruitment of activated FOS in synergy with IL-6 (67).
The results from the present study indicated that SERPINE1 and SERPING1 could induce depression in patients with OC through ECM-related pathways. Both SERPINE1 and SERPING1 belong to the SERPINs family of serine protease inhibitors that regulate proteases involved in fibrinolysis, coagulation, inflammation, cell mobility, cellular differentiation and apoptosis (68). SERPINE1, which is known as plasminogen activator inhibitor-1 (PAI-1), is described as a major physiological inhibitor of endogenous plasminogen activator which inhibits fibrin degradation, promotes fibrin deposition on blood vessel walls and stimulates smooth muscle cell proliferation (69). Furthermore, SERPINE1 may be involved in the pathogenic and therapeutic mechanisms of MDD. Yamamoto et al (70) reported that stress induced SERPINE1 upregulation in a tissue- and cell type-specific manner. In addition, previous studies demonstrated that both women (71) and men (72) with MDD present higher blood PAI-1 levels compared with healthy subjects. Although these findings were not described in patients with OC, they are similar to the results from the present study. Previous studies indicated that SERPINE1 is a stress-associated gene and that its genetic variants may contribute to the causes of depression and the acute therapeutic response to selective serotonin reuptake inhibitors in MDD (73). However, the promoter polymorphisms of SERPINE1 gene were not associated with Alzheimer's disease-related depression, but they may be associated with the response to antidepressant treatments (74). Therefore, the mechanism by which SERPINE1 induces depression in patients with OC requires further investigation. SERPING1 is also referred to as a complement system C1 inhibitory gene [complement component 1 inhibitor (C1INH)], which is located on chromosome 11q.11-q13.1 on GeneMap (75). SERPING1 has been reported to be associated with plasma protein supplementation and to be a member of the serine protease inhibitor gene family (76). Numerous studies have reported that SERPING1 is downregulated in patients with hereditary angioedema and depression (77–79). Furthermore, Ditzen et al (80) demonstrated that SERPING1 expression levels were decreased in the cerebrospinal fluid of depressed patients compared with healthy controls. Since the results from the present study demonstrated that patients with OC and depression exhibited higher SERPING1 expression, SERPING1 may cause depression through other pathways in cancer microenvironment. In addition, SERPINE1 and SERPING1 are associated with extracellular matrix regulation (68). The plasminogen activator/plasmin system serves a crucial role in ECM degradation, and SERPINE1 is a physiological inhibitor of plasminogen activators (81). Furthermore, SERPINE1 upregulation promotes the deposition of ECM components (82), and SERPING1 interacts with extracellular matrix components to inhibit protease activity (83). In addition, interactions between SERPING1 and extracellular matrix components may result in an increase in C1INH concentration at inflammation sites (84). However, as SERPINE1/SERPING1 interaction with ECM remains unclear, further investigation is required.
ATF3 and CTGF upregulation have been demonstrated to be associated with depression. Green et al (85) reported that ATF3 upregulation in nucleus accumbens decreases emotional reactivity and increases depression-like behavior. Similarly, Turner et al (86) demonstrated that CTGF expression in human amygdala is significantly increased in patients with major depressive disorder compared with healthy subjects. In addition, CTGF administration increases depression-like behavior in outbred rats (86). Only few studies have focused on PLAT association with depression. The results from the present study demonstrated that PLAT, ATF3 and CTGF were co-expressed with SERPINE1, FOS and CCL2. PLAT, ATF3 and CTGF may therefore induce depression through SERPINE1, FOS and CCL2 in patients with OC. However, the underlying mechanisms remain unknown and require further investigation.
Previous studies reported that depression is detrimental to cancer patients' survival (87,88). The results from the present study revealed that upregulation of CCL2, FOS, SERPINE1 and SERPING1 was associated with worse survival. However, Wojnarowicz et al (89) demonstrated that CCL2 protein expression is not correlated with overall or disease-free survival. Conversely, Mahner et al (90) reported that reduced FOS expression is associated with unfavorable PFS and OS in patients with epithelial ovarian carcinoma. However, it has been found that elevated tumor SERPINE1 levels are associated with a poor prognosis and reduced disease-free survival in patients with ovarian carcinoma (91,92). To the best of our knowledge, no study has investigated the association between SERPING1 and survival in patients with OC. However, Peng et al (93) demonstrated that lower SERPING1 mRNA levels predicted worse OS and disease-free survival in patients with prostate cancer compared with healthy controls; however, Mejia et al (94) reported that OS is significantly improved in a mice model of malaria treated with SERPING1 compared with controls. Since it is not clear whether these studies included patients with depression or whether the core genes serve similar functions in different types of cancer, determining how CCL2, FOS, SERPINE1 and SERPING1 could influence patients' survival requires further investigation.
In conclusion, the present study demonstrated that the four core genes CCL2, FOS, SERPINE1 and SERPING1 were upregulated in the HD group, which suggested that they may promote depression and worsen survival in patients with OC through four pathways (‘Ensemble of genes encoding ECM-associated proteins including ECM-affiliated proteins’, ‘ECM regulators and secreted factors’, ‘Ensemble of genes encoding extracellular matrix and extracellular matrix-associated proteins’ and ‘MAPK signaling pathway and IL-17 signaling pathway’). These findings may provide novel markers and methods for predicting and treating depression in patients with OC; however, the determination of specific mechanisms requires further investigated.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in the published article.
Authors' contributions
YY, YL and WZ designed the study and drafted the manuscript. WW and KW performed the literature search, data extraction and statistical analysis. All authors contributed to the interpretation of the data and the redaction of the manuscript. The final version of the manuscript was reviewed and approved by all authors.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Reid BM, Permuth JB, Sellers TA. Epidemiology of ovarian cancer: A review. Cancer Biol Med. 2017;14:9–32. doi: 10.20892/j.issn.2095-3941.2016.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in china. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L, Luo M, Yang H, Zhu S, Cheng X, Qing C. Next-generation sequencing-based genomic profiling analysis reveals novel mutations for clinical diagnosis in chinese primary epithelial ovarian cancer patients. J Ovarian Res. 2019;12:19. doi: 10.1186/s13048-019-0494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson LE, Angen M, Cullum J, Goodey E, Koopmans J, Lamont L, MacRae JH, Martin M, Pelletier G, Robinson J. High levels of untreated distress and fatigue in cancer patients. Br J Cancer. 2004;90:2297–2304. doi: 10.1038/sj.bjc.6601887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sellick SM, Crooks DL. Depression and cancer: An appraisal of the literature for prevalence, detection, and practice guideline development for psychological interventions. Psychooncology. 1999;8:315–333. doi: 10.1002/(SICI)1099-1611(199907/08)8:4<315::AID-PON391>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 7.Zabora J, BrintzenhofeSzoc K, Curbow B, Hooker C, Piantadosi S. The prevalence of psychological distress by cancer site. Psychooncology. 2001;10:19–28. doi: 10.1002/1099-1611(200101/02)10:1<19::AID-PON501>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 8.Greimel E, Thiel I, Peintinger F, Cegnar I, Pongratz E. Prospective assessment of quality of life of female cancer patients. Gynecol Oncol. 2002;85:140–147. doi: 10.1006/gyno.2002.6586. [DOI] [PubMed] [Google Scholar]
- 9.Lutgendorf SK, Slavich GM, Degeest K, Goodheart M, Bender D, Thaker PH, Penedo F, Zimmerman B, Lucci J, III, Mendez L, et al. Non-cancer life stressors contribute to impaired quality of life in ovarian cancer patients. Gynecol Oncol. 2013;131:667–673. doi: 10.1016/j.ygyno.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watts S, Prescott P, Mason J, McLeod N, Lewith G. Depression and anxiety in ovarian cancer: A systematic review and meta-analysis of prevalence rates. BMJ Open. 2015;5:e007618. doi: 10.1136/bmjopen-2015-007618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang T, Poole EM, Okereke OI, Kubzansky LD, Eliassen AH, Sood AK, Wang M, Tworoger SS. Depression and risk of epithelial ovarian cancer: Results from two large prospective cohort studies. Gynecol Oncol. 2015;139:481–486. doi: 10.1016/j.ygyno.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mielcarek P, Nowicka-Sauer K, Kozaka J. Anxiety and depression in patients with advanced ovarian cancer: A prospective study. J Psychosom Obstet Gynaecol. 2016;37:57–67. doi: 10.3109/0167482X.2016.1141891. [DOI] [PubMed] [Google Scholar]
- 13.Wu C, Zhao Y, Liu Y, Yang X, Yan M, Min Y, Pan Z, Qiu S, Xia S, Yu J, et al. Identifying miRNA-mRNA regulation network of major depressive disorder in ovarian cancer patients. Oncol Lett. 2018;16:5375–5382. doi: 10.3892/ol.2018.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahman MR, Islam T, Al-Mamun MA, Zaman T, Karim MR, Moni MA. The influence of depression on ovarian cancer: Discovering molecular pathways that identify novel biomarkers and therapeutic targets. Informatics Med Unlocked. 2019;16:100207. doi: 10.1016/j.imu.2019.100207. [DOI] [Google Scholar]
- 15.Watson M, Haviland JS, Greer S, Davidson J, Bliss JM. Influence of psychological response on survival in breast cancer: A population-based cohort study. Lancet. 1999;354:1331–1336. doi: 10.1016/S0140-6736(98)11392-2. [DOI] [PubMed] [Google Scholar]
- 16.DiMatteo MR, Lepper HS, Croghan TW. Depression is a risk factor for noncompliance with medical treatment: Meta-analysis of the effects of anxiety and depression on patient adherence. Arch Intern Med. 2000;160:2101–2107. doi: 10.1001/archinte.160.14.2101. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto T, Yanagimoto H, Satoi S, Toyokawa H, Hirooka S, Yamaki S, Yui R, Yamao J, Kim S, Kwon AH. Circulating CD4+CD25+ regulatory T cells in patients with pancreatic cancer. Pancreas. 2012;41:409–415. doi: 10.1097/MPA.0b013e3182373a66. [DOI] [PubMed] [Google Scholar]
- 18.Lutgendorf SK, DeGeest K, Sung CY, Arevalo JM, Penedo F, Lucci J, III, Goodheart M, Lubaroff D, Farley DM, Sood AK, Cole SW. Depression, social support, and beta-adrenergic transcription control in human ovarian cancer. Brain Behav Immun. 2009;23:176–183. doi: 10.1016/j.bbi.2008.04.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole SW, Arevalo JMG, Takahashi R, Sloan EK, Lutgendorf SK, Sood AK, Sheridan JF, Seeman TE. Computational identification of gene-social environment interaction at the human IL6 locus. Proc Natl Acad Sci. 2010;107:5681–5686. doi: 10.1073/pnas.0911515107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Vasaikar S, Shi Z, Greer M, Zhang B. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017;45:W130–W137. doi: 10.1093/nar/gkx356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radloff LS. The CES-D scale: A self-report depression scale for research in the general population. Appl Psychol Meas. 1977;1:385–401. doi: 10.1177/014662167700100306. [DOI] [Google Scholar]
- 24.Cutroa CE, Rusell DW. The provisions of social relationships and adaptation to stress. Adv Pers Relationships. 1987;1:37–67. [Google Scholar]
- 25.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shannon P, Markiel A, Owen O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scardoni G, Petterlini M, Laudanna C. Analyzing biological network parameters with centiScaPe. Bioinformatics. 2009;25:2857–2859. doi: 10.1093/bioinformatics/btp517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fox R. A view from the web. J R Soc Med. 1997;90:473. doi: 10.1177/014107689709000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jia P, Kao CF, Kuo PH, Zhao Z. A comprehensive network and pathway analysis of candidate genes in major depressive disorder. BMC Syst Biol. 2011;5(Suppl 3):S12. doi: 10.1186/1752-0509-5-S3-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Y, Pan Z, Hu Y, Zhang L, Wang J. Network and pathway-based analyses of genes associated with parkinson's disease. Mol Neurobiol. 2017;54:4452–4465. doi: 10.1007/s12035-016-9998-8. [DOI] [PubMed] [Google Scholar]
- 33.Niwattanakul S, Singthongchai J, Naenudorn E, Wanapu S. Using of jaccard coefficient for keywords similarity. Proc Int Multiconference Eng Comput Sci. 2013;1:13–15. [Google Scholar]
- 34.Mizuno S, Yamaguchi T, Fukushima A, Matsuyama Y, Ohashi Y. Overlap coefficient for assessing the similarity of pharmacokinetic data between ethnically different populations. Clin Trials. 2005;2:174–181. doi: 10.1191/1740774505cn077oa. [DOI] [PubMed] [Google Scholar]
- 35.Gyorffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Denkert C, Budczies J, Darb-Esfahani S, Györffy B, Sehouli J, Könsgen D, Zeillinger R, Weichert W, Noske A, Buckendahl AC, et al. A prognostic gene expression index in ovarian cancer-validation across different independent data sets. J Pathol. 2009;218:273–280. doi: 10.1002/path.2547. [DOI] [PubMed] [Google Scholar]
- 38.Ahmed AA, Mills AD, Ibrahim AE, Temple J, Blenkiron C, Vias M, Massie CE, Iyer NG, McGeoch A, Crawford R, et al. The extracellular matrix protein TGFBI induces microtubule stabilization and sensitizes ovarian cancers to paclitaxel. Cancer Cell. 2007;12:514–527. doi: 10.1016/j.ccr.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mok SC, Bonome T, Vathipadiekal V, Bell A, Johnson ME, Wong KK, Park DC, Hao K, Yip DK, Donninger H, et al. A gene signature predictive for outcome in advanced ovarian cancer identifies a survival factor: Microfibril-associated glycoprotein 2. Cancer Cell. 2009;16:521–532. doi: 10.1016/j.ccr.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konstantinopoulos PA, Spentzos D, Karlan BY, Taniguchi T, Fountzilas E, Francoeur N, Levine DA, Cannistra SA. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J Clin Oncol. 2010;28:3555–3561. doi: 10.1200/JCO.2009.27.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchion DC, Cottrill HM, Xiong Y, Chen N, Bicaku E, Fulp WJ, Bansal N, Chon HS, Stickles XB, Kamath SG, et al. BAD phosphorylation determines ovarian cancer chemosensitivity and patient survival. Clin Cancer Res. 2011;17:6356–6366. doi: 10.1158/1078-0432.CCR-11-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gentric G, Kieffer Y, Mieulet V, Goundiam O, Bonneau C, Nemati F, Hurbain I, Raposo G, Popova T, Stern MH, et al. PML-regulated mitochondrial metabolism enhances chemosensitivity in human ovarian cancers. Cell Metab. 2019;29:156–173. doi: 10.1016/j.cmet.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mateescu B, Batista L, Cardon M, Gruosso T, de Feraudy Y, Mariani O, Nicolas A, Meyniel JP, Cottu P, Sastre-Garau X, Mechta-Grigoriou F. MiR-141 and miR-200a act on ovarian tumorigenesis by controlling oxidative stress response. Nat Med. 2011;17:1627–1635. doi: 10.1038/nm.2512. [DOI] [PubMed] [Google Scholar]
- 44.Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, Ozbun L, Brady J, Barrett JC, Boyd J, Birrer MJ. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–5486. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vathipadiekal V, Wang V, Wei W, Waldron L, Drapkin R, Gillette M, Skates S, Birrer M. Creation of a human secretome: A novel composite library of human secreted proteins: Validation using ovarian cancer gene expression data and a virtual secretome array. Clin Cancer Res. 2015;21:4960–4969. doi: 10.1158/1078-0432.CCR-14-3173. [DOI] [PubMed] [Google Scholar]
- 46.King ER, Tung CS, Tsang YT, Zu Z, Lok GT, Deavers MT, Malpica A, Wolf JK, Lu KH, Birrer MJ, et al. The anterior gradient homolog 3 (AGR3) gene is associated with differentiation and survival in ovarian cancer. Am J Surg Pathol. 2011;35:904–912. doi: 10.1097/PAS.0b013e318212ae22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferriss JS, Kim Y, Duska L, Birrer M, Levine DA, Moskaluk C, Theodorescu D, Lee JK. Multi-gene expression predictors of single drug responses to adjuvant chemotherapy in ovarian carcinoma: Predicting platinum resistance. PLoS One. 2012;7:e30550. doi: 10.1371/journal.pone.0030550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 49.Koti M, Gooding RJ, Nuin P, Haslehurst A, Crane C, Weberpals J, Childs T, Bryson P, Dharsee M, Evans K. Identification of the IGF1/PI3K/NF κB/ERK gene signalling networks associated with chemotherapy resistance and treatment response in high-grade serous epithelial ovarian cancer. BMC Cancer. 2013;13:549. doi: 10.1186/1471-2407-13-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lisowska KM, Olbryt M, Dudaladava V, Pamuła-Piłat J, Kujawa K, Grzybowska E, Jarząb M, Student S, Rzepecka IK, Jarząb B, Kupryjańczyk J. Gene expression analysis in ovarian cancer-faults and hints from DNA microarray study. Front Oncol. 2014;4:6. doi: 10.3389/fonc.2014.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uehara Y, Oda K, Ikeda Y, Koso T, Tsuji S, Yamamoto S, Asada K, Sone K, Kurikawa R, Makii C, et al. Integrated copy number and expression analysis identifies profiles of whole-arm chromosomal alterations and subgroups with favorable outcome in ovarian clear cell carcinomas. PLoS One. 2015;10:e0128066. doi: 10.1371/journal.pone.0128066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 53.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. 2018:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 54.Chaturvedi SK. Psychiatric oncology: Cancer in mind. Indian J Psychiatry. 2012;54:111–118. doi: 10.4103/0019-5545.99529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pasquini M, Biondi M. Depression in cancer patients: A critical review. Clin Pract Epidemiol Ment Health. 2007;3:2. doi: 10.1186/1745-0179-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prasad SM, Eggener SE, Lipsitz SR, Irwin MR, Ganz PA, Hu JC. Effect of depression on diagnosis, treatment, and mortality of men with clinically localized prostate cancer. J Clin Oncol. 2014;32:2471–2478. doi: 10.1200/JCO.2013.51.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Colombo N, Lorusso D, Scollo P. Impact of recurrence of ovarian cancer on quality of life and outlook for the future. Int J Gynecol Cancer. 2017;27:1134–1140. doi: 10.1097/IGC.0000000000001023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leighton SP, Nerurkar L, Krishnadas R, Johnman C, Graham GJ, Cavanagh J. Chemokines in depression in health and in inflammatory illness: A systematic review and meta-analysis. Mol Psychiatry. 2018;23:48–58. doi: 10.1038/mp.2017.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eyre HA, Air T, Pradhan A, Johnston J, Lavretsky H, Stuart MJ, Baune BT. A meta-analysis of chemokines in major depression. Prog Neuropsychopharmacol Biol Psychiatry. 2016;68:1–8. doi: 10.1016/j.pnpbp.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen K, Pociask DA, McAleer JP, Chan YR, Alcorn JF, Kreindler JL, Keyser MR, Shapiro SD, Houghton AM, Kolls JK, Zheng M. IL-17RA is required for CCL2 expression, macrophage recruitment, and emphysema in response to cigarette smoke. PLoS One. 2011;6:e20333. doi: 10.1371/journal.pone.0020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lim SY, Yuzhalin AE, Gordon-Weeks AN, Muschel RJ. Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget. 2016;7:28697–28710. doi: 10.18632/oncotarget.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu T, Guo M, Garza J, Rendon S, Sun X, Zhang W, Lu XY. Cognitive and neural correlates of depression-like behaviour in socially defeated mice: An animal model of depression with cognitive dysfunction. Int J Neuropsychopharmacol. 2011;14:303–317. doi: 10.1017/S1461145710000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kung JC, Chen TC, Shyu BC, Hsiao S, Huang AC. Anxiety- and depressive-like responses and c-fos activity in preproenkephalin knockout mice: Oversensitivity hypothesis of enkephalin deficit-induced posttraumatic stress disorder. J Biomed Sci. 2010;17:29. doi: 10.1186/1423-0127-17-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yokoyama K, Hiyama A, Arai F, Nukaga T, Sakai D, Mochida J. C-Fos regulation by the MAPK and PKC pathways in intervertebral disc cells. PLoS One. 2013;8:1–14. doi: 10.1371/journal.pone.0073210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitmarsh AJ. Regulation of gene transcription by mitogen-activated protein kinase signaling pathways. Biochim Biophys Acta. 2007;1773:1285–1298. doi: 10.1016/j.bbamcr.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 66.Li JK, Nie L, Zhao YP, Zhang YQ, Wang X, Wang SS, Liu Y, Zhao H, Cheng L. IL-17 mediates inflammatory reactions via p38/c-Fos and JNK/c-Jun activation in an AP-1-dependent manner in human nucleus pulposus cells. J Transl Med. 2016;14:77. doi: 10.1186/s12967-016-0833-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ma X, Reynolds SL, Baker BJ, Li X, Benveniste EN, Qin H. IL-17 enhancement of the IL-6 signaling cascade in astrocytes. J Immunol. 2010;184:4898–4906. doi: 10.4049/jimmunol.1000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hayashi KG, Ushizawa K, Hosoe M, Takahashi T. Differential gene expression of serine protease inhibitors in bovine ovarian follicle: Possible involvement in follicular growth and atresia. Reprod Biol Endocrinol. 2011;9:72. doi: 10.1186/1477-7827-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lassila M, Fukami K, Jandeleit-Dahm K, Semple T, Carmeliet P, Cooper ME, Kitching AR. Plasminogen activator inhibitor-1 production is pathogenetic in experimental murine diabetic renal disease. Diabetologia. 2007;50:1315–1326. doi: 10.1007/s00125-007-0652-x. [DOI] [PubMed] [Google Scholar]
- 70.Yamamoto K, Takeshita K, Shimokawa T, Yi H, Isobe K, Loskutoff DJ, Saito H. Plasminogen activator inhibitor-1 is a major stress-regulated gene: Implications for stress-induced thrombosis in aged individuals. Proc Natl Acad Sci. 2002;99:890–895. doi: 10.1073/pnas.022608799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eskandari F, Mistry S, Martinez PE, Torvik S, Kotila C, Sebring N, Drinkard BE, Levy C, Reynolds JC, Csako G, et al. Younger, premenopausal women with major depressive disorder have more abdominal fat and increased serum levels of prothrombotic factors: Implications for greater cardiovascular risk. Metabolism. 2005;54:918–924. doi: 10.1016/j.metabol.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 72.Lahlou-Laforet K, Alhenc-Gelas M, Pornin M, Bydlowski S, Seigneur E, Benetos A, Kierzin JM, Scarabin PY, Ducimetiere P, Aiach M, et al. Relation of depressive mood to plasminogen activator inhibitor, tissue plasminogen activator, and fibrinogen levels in patients with versus without coronary heart disease. Am J Cardiol. 2006;97:1287–1291. doi: 10.1016/j.amjcard.2005.11.062. [DOI] [PubMed] [Google Scholar]
- 73.Tsai SJ, Hong CJ, Liou YJ, Yu YW, Chen TJ. Plasminogen activator inhibitor-1 gene is associated with major depression and antidepressant treatment response. Pharmacogenet Genomics. 2008;18:869–875. doi: 10.1097/FPC.0b013e328308bbc0. [DOI] [PubMed] [Google Scholar]
- 74.Fang Y, Zhang L, Zeng Z, Lian Y, Jia Y, Zhu H, Xu Y. Promoter polymorphisms of SERPINE1 are associated with the antidepressant response to depression in alzheimer's disease. Neurosci Lett. 2012;516:217–220. doi: 10.1016/j.neulet.2012.03.090. [DOI] [PubMed] [Google Scholar]
- 75.Haslund D, Ryø LB, Seidelin Majidi S, Rose I, Skipper KA, Fryland T, Bohn AB, Koch C, Thomsen MK, Palarasah Y, et al. Dominant-negative SERPING1 variants cause intracellular retention of C1 inhibitor in hereditary angioedema. J Clin Invest. 2019;129:388–405. doi: 10.1172/JCI98869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Madsen DE, Hansen S, Gram J, Bygum A, Drouet C, Sidelmann JJ. Presence of C1-inhibitor polymers in a subset of patients suffering from hereditary angioedema. PLoS One. 2014;9:e112051. doi: 10.1371/journal.pone.0112051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lumry WR, Craig T, Zuraw B, Longhurs H, Baker J, Li HH, Bernstein JA, Anderson J, Riedl MA, Manning ME, et al. Health-related quality of life with subcutaneous C1-inhibitor for prevention of attacks of hereditary angioedema. J Allergy Clin Immunol Pract. 2018;112:1733–1741. doi: 10.1016/j.jaip.2017.12.039. [DOI] [PubMed] [Google Scholar]
- 78.Fouche AS, Saunders EF, Craig T. Depression and anxiety in patients with hereditary angioedema. Ann Allergy Asthma Immunol. 2014;112:371–375. doi: 10.1016/j.anai.2013.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Varga L, Széplaki G, Laki J, Kocsis A, Kristóf K, Gál P, Bajtay Z, Wieslander J, Daha MR, Garred P, et al. Depressed activation of the lectin pathway of complement in hereditary angioedema. Clin Exp Immunol. 2008;153:68–74. doi: 10.1111/j.1365-2249.2008.03671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ditzen C, Tang N, Jastorff AM, Teplytska L, Yassouridis A, Maccarrone G, Uhr M, Bronisch T, Miller CA, Holsboer F, Turck CW. Cerebrospinal fluid biomarkers for major depression confirm relevance of associated pathophysiology. Neuropsychopharmacology. 2012;37:1013–1025. doi: 10.1038/npp.2011.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu RM. Oxidative stress, plasminogen activator inhibitor 1, and lung fibrosis. Antioxid Redox Signal. 2008;10:303–319. doi: 10.1089/ars.2007.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oh CK, Ariue B, Alban RF, Shaw B, Cho SH. PAI-1 promotes extracellular matrix deposition in the airways of a murine Asthma model. Biochem Biophys Res Commun. 2002;294:1155–1160. doi: 10.1016/S0006-291X(02)00577-6. [DOI] [PubMed] [Google Scholar]
- 83.Singer M, Jones AM. Bench-to-bedside review: The role of C1-esterase inhibitor in sepsis and other critical illnesses. Crit Care. 2011;15:203. doi: 10.1186/cc9304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davis AE, III, Lu F, Mejia P. C1 inhibitor, a multi-functional serine protease inhibitor. Thromb Haemost. 2010;104:886–893. doi: 10.1160/TH10-01-0073. [DOI] [PubMed] [Google Scholar]
- 85.Green TA, Alibhai IN, Unterberg S, Neve RL, Ghose S, Tamminga CA, Nestler EJ. Induction of activating transcription factors (ATFs) ATF2, ATF3, and ATF4 in the nucleus accumbens and their regulation of emotional behavior. J Neurosci. 2008;28:2025–2032. doi: 10.1523/JNEUROSCI.5273-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Turner CA, Sharma V, Hagenauer MH, Chaudhury S, O'Connor AM, Hebda-Bauer EK, Thompson RC, Myers RM, Bunney WE, Barchas JD. Connective tissue growth factor is a novel prodepressant. Biol Psychiatry. 2018;84:555–562. doi: 10.1016/j.biopsych.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Giese-Davis J, Collie K, Rancourt KM, Neri E, Kraemer HC, Spiegel D. Decrease in depression symptoms is associated with longer survival in patients with metastatic breast cancer: A secondary analysis. J Clin Oncol. 2011;29:413–420. doi: 10.1200/JCO.2010.28.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Raison CL, Miller AH. The evolutionary significance of depression in pathogen host defense (PATHOS-D) Mol Psychiatry. 2013;18:15–37. doi: 10.1038/mp.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wojnarowicz P, Gambaro K, de Ladurantaye M, Quinn MC, Provencher D, Mes-Masson AM, Tonin PN. Overexpressing the CCL2 chemokine in an epithelial ovarian cancer cell line results in latency of in vivo tumourigenicity. Oncogenesis. 2012;1:e27. doi: 10.1038/oncsis.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mahner S, Baasch C, Schwarz J, Hein S, Wölber L, Jänicke F, Milde-Langosch K. C-Fos expression is a molecular predictor of progression and survival in epithelial ovarian carcinoma. Br J Cancer. 2008;99:1269–1275. doi: 10.1038/sj.bjc.6604650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Andreasen PA, Kjøller L, Christensen L, Duffy MJ. The urokinase-type plasminogen activator system in cancer metastasis: A review. Int J cancer. 1997;72:1–22. doi: 10.1002/(SICI)1097-0215(19970703)72:1<1::AID-IJC1>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 92.Klein RM, Bernstein D, Higgins SP, Higgins CE, Higgins PJ. SERPINE1 expression discriminates site-specific metastasis in human melanoma. Exp Dermatol. 2012;21:551–554. doi: 10.1111/j.1600-0625.2012.01523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Peng S, Du T, Wu W, Chen X, Lai Y, Zhu D, Wang Q, Ma X, Lin C, Li Z, et al. Decreased expression of serine protease inhibitor family G1 (SERPING1) in prostate cancer can help distinguish high-risk prostate cancer and predicts malignant progression. Urol Oncol. 2018;36:366.e1–366.e9. doi: 10.1016/j.urolonc.2018.05.021. [DOI] [PubMed] [Google Scholar]
- 94.Mejia P, Diez-Silva M, Kamena F, Lu F, Fernandes SM, Seeberger PH, Davis AE, III, Mitchell JR. Human C1-inhibitor suppresses malaria parasite invasion and cytoadhesion via binding to parasite glycosylphosphatidylinositol and host cell receptors. J Infect Dis. 2016;213:80–89. doi: 10.1093/infdis/jiv439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in the published article.