Abstract
The low therapeutic index of conventional chemotherapy and poor prognosis of patients diagnosed with metastatic cancers are prompting clinicians to adopt newer strategies to simultaneously detect cancer lesions at an early stage and to precisely deliver anticancer drugs to tumor sites. In this study, we employed a novel strategy to engineer a polyvalent theranostic nanocarrier consisting of superparamagnetic iron oxide nanoparticle core (SPIONs) decorated with folic acid-polyamidoamine dendrimers surface (FA-PAMAM). In addition, a highly potent hydrophobic anticancer agent 3,4-difluorobenzylidene-curcumin (CDF) was coloaded in the FA-PAMAM dendrimer to increase its solubility and assess its therapeutic potentials. The resulting targeted nanoparticles (SPIONs@FA-PAMAM-CDF) exhibited high MR contrast. When tested on folate receptor overexpressing ovarian (SKOV3) and cervical (HeLa) cancer cells, the CDF loaded targeted nanoformulations showed higher accumulation with a better anticancer activity as compared to the nontargeted counterparts, possibly due to multivalent folate receptor binding interaction with cells overexpressing the target. The results were corroborated by observation of a larger population of cells undergoing apoptosis due to upregulation of tumor suppressor phosphatase and tensis homologue (PTEN), caspase 3, and inhibition of NF-κB in groups treated with the targeted formulations, which further confirmed the ability of the multivalent theranostic nanoparticles for simultaneous imaging and therapy of cancers.
Graphical Abstract
INTRODUCTION
Worldwide, cancer is accountable for millions of deaths annually. According to the American Cancer Society, cancer is the second most common cause of death in the United States. Approximately 1.7 million new cancer cases are expected and about 600 000 Americans are expected to die of cancer in 2016.1 Chemotherapy, radiotherapy, and photothermal therapy are the most common treatments for cancer. Among them, chemotherapy is the most common strategy in cancer treatment because of its high efficacy as compared to other types of treatments.2–5 In most cases, a cancer diagnosis in the early stage is difficult. Most of the patients are diagnosed at the late stage of cancer with a poor prognosis. In the advanced stage of cancer, chemotherapy and radiotherapy are the only options. However, the development of chemotherapeutic drug resistance is the most common reason that leads to the failure of cancer treatment. The conventional treatment with the systemic distribution of chemotherapeutics is problematic and shows a significant flaw that can make the difference between success and failure.6 High doses are often required to accumulate adequate amounts of chemotherapeutic agents at the tumor site to achieve desirable therapeutic efficacy. However, larger doses can pose higher risk for adverse side-effects with the increase of toxicity in nontargeted sites or normal tissues.
Magnetic resonance imaging is one of the most common clinical diagnostic tools due to its noninvasive, tomographic properties that offer superb spatial resolution without the dangers of ionizing radiation. In recent years, magnetic nanoparticles are getting more attention due to their applications in biology and medicine such as enzyme and protein immobilization, magnetic resonance imaging, tissue engineering, magnetic cell tracking and separation, hyperthermia, and targeted drug and gene delivery. Drug delivery systems based on magnetic nanoparticle carriers possess the ability of magnetic resonance imaging contrast agents as well as the advanced properties of nanocarriers such as the enhanced aqueous solubility, increased systemic circulation time, and targeting delivery of chemotherapeutic drugs with reduced toxicity in normal tissues.7,8
Magnetic iron oxide (Fe3O4) nanoparticles, especially superparamagnetic iron oxide nanoparticles (SPIONs), have been widely studied and have shown great potentials in biotechnology because of their biocompatible, inert, and excellent magnetic properties.9,10 The surface of these magnetic nanoparticles can be modified with polymeric shells such as dextran, polyethylene glycol (PEG), chitosan, and dendrimers for arriving at organometallic hybrids useful for in vivo applications.11–13 The polymeric shells provide not only the biocompatibility for the magnetic nanoparticles, but also the ability for conjugation with biomolecules such as proteins, nucleic acids, enzymes, and targeting ligands as well as loading of drugs.14–17
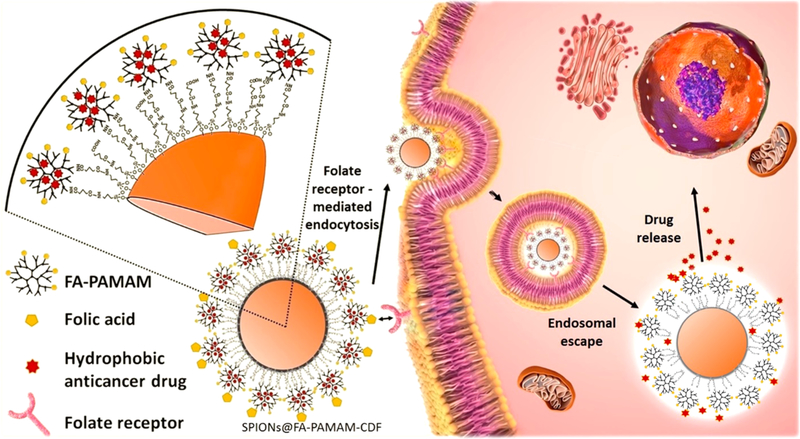
Polyamidoamine (PAMAM) dendrimers are a relatively novel class of polymers with a well-defined, nanosized, highly branched monodispersed structures, having numerous hydrophilic reactive amine groups on the periphery and lipophilic internal cavities. PAMAM dendrimers are known for their ability to encapsulate hydrophobic drugs in their internal cavities to enhance the aqueous solubility of these hydrophobic compounds.18–20 The larger numbers of reactive amine groups on the periphery make PAMAM dendrimers suitable for many biomedical applications such as drug conjugation/loading, siRNA and gene complexation, and conjugation to biorecognition molecules to achieve active targeting ability.21–22 It is reported that PAMAM dendrimers are localized in tumor by enhanced permeation and retention (EPR) effect.23–25 In addition, it is demonstrated that by attaching targeting ligands on the surface, PAMAM dendrimers can achieve active receptor targeting (Figure 1). In this regard, one of the most commonly used and established targeting ligands is folic acid (FA). Many types of cancer cells (such as ovarian, colon, lung, breast, and cervical cancer cells) are known to have a high expression of folate receptors on their membranes.26–29 Drug carriers decorated with FA can thus achieve high binding and internalization into cancer cells overexpressing the folate receptors. Specifically, the cellular internalization is facilitated via folate-receptor mediated endocytosis followed by the release of anticancer drug, thus resulting in a better cancer cell killing with minimized toxicity to normal cells.30,31 In terms of receptor mediated targeting, folate receptor targeting by using FA seems to be a promising strategy for cancer therapy with numerous of drugs going under clinical trials at this moment.32
Figure 1.
Pictorial representation of the folate receptor mediated endocytosis followed by drug release of the targeted theranostic formulation SPIONs@FA-PAMAM-CDF in cancer cells overexpressing folate receptors.
In this study, SPIONs were synthesized by coprecipitation method, followed by a novel method of synthesis and engineering of surface coating with PAMAM-decorated FA. The magnetic nanocarriers were used to encapsulate a poorly water-soluble but highly potent anticancer drug 3,4-difluor-obenzylidene diferuloylmethane (CDF), a synthetic analog of a potent flavonoid anticancer compound diferuloylmethane. In our previous studies, CDF has shown a high anticancer activity when tested on pancreatic, cervical, ovarian, and lung cancer cells.33–36 The improvement in anticancer properties of CDF was attributed to a 16-fold increased half-life, a higher stability, and bioavailability as compared to its natural counterpart, curcumin. It is reported that CDF can inhibit the growth of cancer cells through down-regulation of multiple miRNAs, up-regulation of phosphatase and tensin homologue (PTEN), and attenuation of histone methyltransferase EZH2.37–39 The theranostic capability for cancer imaging and therapy of the magnetic nanocarriers encapsulated CDF was examined by in vitro biological studies, cellular uptake, and T2 relaxation studies. The results demonstrated that the synthesized magnetic nanocarriers could be a promising carrier in active targeting for simultaneous imaging and therapy of premalignant and malignant lesions (Figure. 1).
METHODS
Materials and Cell Lines.
CDF was synthesized as described earlier.33,35,40 Fourth generation (4.0G) PAMAM dendrimer, ferrous chloride (FeCl2·4H2O), ferric chloride (FeCl3·6H2O), N-(3-(dimethylamino)propyl)-N-ethylcarbodiimide hydrochloride (EDC), and 3-[4,5 dimethylthiazol-2-yl]-2,5diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich (St. Louis, MO). FA was purchased from Fisher Scientific. Guava Nexin Reagent for cell apoptosis kit was purchased from EMD Millipore. PTEN, NF-kB antibodies were purchase from Cell Signaling Technology, USA. All other chemicals were of reagent grade and used without any modification.
Human cervical cancer cells (HeLa cells) and human ovarian carcinoma cells (SKOV3 cells) were used in this study due to their high expression of folate receptors.41–44 HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Fisher Scientific, Waltham MA) with 10% fetal bovine serum (FBS) and streptomycin sulfate (10 mg/L). SKOV3 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Thermo Fisher Scientific, USA) with 10% FBS and streptomycin sulfate (10 mg/L). All cell lines were incubated at 37 °C in a 5% CO2 air humidified atmosphere.
Superparamagnetic Iron Oxide Nanoparticles (SPIONs) Synthesis.
Superparamagnetic iron oxide nanoparticles (SPIONs) were synthesized using coprecipitation method.45 Prior to the synthesis, 0.5 M NaOH solution in deionized water (DIW) was prepared in a three-neck 500 mL round-bottom flask (RBF) and degassed by bubbling N2 under stirring at room temperature (RT) for 30 min, followed by degas under vacuum under stirring at RT for another 30 min, and then it was heated to 40 °C. Then ferric chloride FeCl3·H2O (6.56 g, 0.024 mol) and ferrous chloride FeCl2·4H2O (2.48 g, 0.012 mol) were dissolved in 25 mL of degassed 0.4 M HCl solution in DIW and then added to the RBF through a septum. The RBF was heated at 80 °C under strong stirring for 1 h. SPIONs were precipitated using a strong neodymium N52 magnet, and then the reaction mixture was decanted. The SPIONs were washed five times by dispersing them back in EtOH (300 mL) with probe sonication for 10 min, followed by magnet precipitation and decantation of the liquid. A dry powder of SPIONs was obtained by drying under vacuum on a rotary evaporator. The product was characterized by Fourier transform infrared (FTIR) spectroscopy. Hydrodynamic size and zeta potential were characterized by dynamic light scattering using Beckman Coulter Delsa Nano.
FA-PAMAM Decorated SPIONs Fabrication.
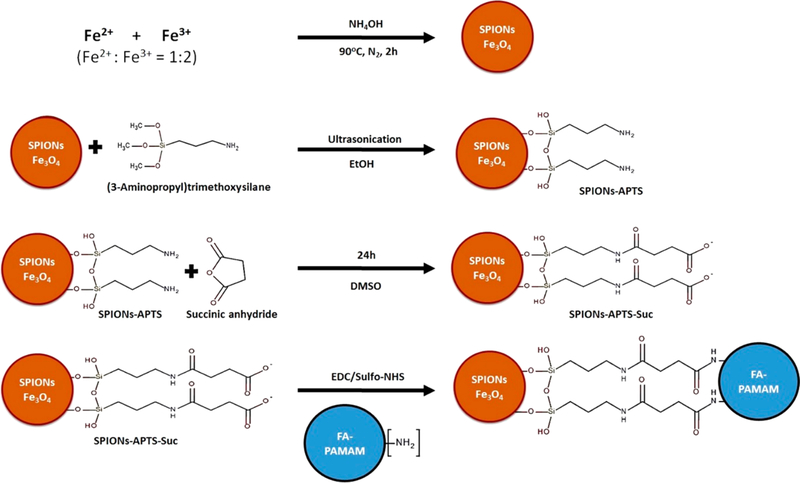
FA was conjugated to fourth generation PAMAM dendrimers through carbodiimide coupling chemistry according to a previously reported method.36 Prior to the FA-PAMAM conjugation, the surface of SPIONs was modified to create the activated carboxyl groups (Figure 2). The fabrication process includes three main steps: (i) amine functionalization, (ii) carboxylation, and (iii) FA-PAMAM conjugation (Figure 2).
Figure 2.
SPIONs decorated FA-PAMAM (SPIONs@FA-PAMAM) synthesis and fabrication process.
Amine Functionalized SPIONs (SPIONs@APTS).
Synthesized SPIONs were functionalized by (3-aminopropyl) trimethoxysilane (APTS) to have peripheral amino groups.46–48 Briefly, 1 g of SPIONs was dispersed in 300 mL of EtOH with probe sonication for 1 h. Then 6 mL of APTS was added to the EtOH solution. The solution was sonicated for another 1 h. The resulting product SPIONs@APTS was precipitated using a strong neodymium N52 magnet, and then the reaction mixture was decanted. The product was washed three times by dispersing in 300 mL of EtOH with probe sonication, followed by magnet decantation. SPIONs@APTS were dried under vacuum on a rotary evaporator. Energy dispersive X-ray spectroscopy (EDS) and FTIR spectroscopy were used to characterize the product. Size and zeta potential were measured by Beckman Coulter Delsa Nano instrument.
Carboxylation of Amine Functionalized SPIONs (SPIONs@COOH).
Succinic anhydride was used to carboxylate the amine groups of SPIONs@APTS according to a previously reported study.49 Briefly, 100 mg of SPIONs@APTS was dispersed in 150 mL of EtOH using probe sonication. Succinic anhydride (500 mg) was added to 150 mL of DMSO under vigorous stirring and then added to the SPIONs@ APTS in EtOH solution. The reaction was stirred at RT for 24 h. The resulting product SPIONs@COOH was precipitated using a strong neodymium N52 magnet, followed by the decantation of the supernatant. The product SPIONs@COOH was washed three times by dispersing in 300 mL of EtOH using probe sonication, followed by decantation, and was dried under vacuum on a rotary evaporator. The product was confirmed by FTIR spectroscopy, size, and zeta potential measurements.
Activation of SPIONs@COOH and Fabrication of FA-PAMAM and SPIONs@COOH Conjugate.
Carboxyl groups of SPIONs@COOH were activated using carbodiimide reaction. In brief, 50 mg of SPIONs@COOH was dispersed in 100 mL of DMSO using probe sonication for 1 h. Then 490 mg of EDC and 735 mg of NHS were added to the solution. The reaction was left for 24 h under vigorous stirring. Activated SPIONs@COOH was conjugated with 100 mg of PAMAM or 100 mg of FA-PAMAM (dissolved in DMSO) to have SPIONs@PAMAM or SPIONs@FA-PAMAM. The final products SPIONs@PAMAM and SPIONs@FA-PAMAM were purified by precipitation by a strong neodymium N52 magnet, followed by decantation of the supernatant. The products were washed three times by dispersing in 300 mL of EtOH and followed by magnet decantation. The products were characterized by size and zeta potential measurements.
To quantify the amount of dendrimers conjugated to SPIONs, PAMAM dendrimers and FA-PAMAM conjugates were labeled with Rhodamine B isothiocyanate according to a previously reported method.50 In short, PAMAM or FA-PAMAM conjugates were dispersed in 50 mL of EtOH. Rhodamine B isothiocyanate was added to the EtOH dispersion. The amount of Rhodamine B isothiocyanate was calculated to have the ratio of Rhodamine B isothiocyanate to PAMAM or FA-PAMAM of 3:1. The reaction was stirred at RT for 1 h, followed by dialysis in 5 L of DIW for three times with the molecular weight cutoff 3.5 kDa. Lyophilization was performed at the end of the dialysis to have the dry powder of Rhodamine B label PAMAM (RhoB-PAMAM) and Rhodamine B labeled FA-PAMAM (RhoB-FA-PAMAM). Fluorescence spectroscopy was employed to measure the fluorescence of the Rhodamine B labeled formulations (SPIONs@RhoB-PAMAM and SPIONs@RhoB-FA-PAMAM). The amounts of PAMAM and FA-PAMAM conjugated to SPIONs were calculated based on the linear equation ofRhodamine B fluorescence and Rho B-PAMAM or Rho B-FA-PAMAM concentrations.
Transmission Electron Microscopic Analysis.
The sizes of the synthesized SPIONs and the fabricated product SPIONs@PAMAM and SPIONs@FA-PAMAM were studied using transmission electron microscopy (TEM). Samples were prepared according to a previous method.36 Briefly, 4 μL of each sample (dispersion of 2 mg powder of the sample in 5 mL of DIW) was applied to a Formvar-coated, carbon-stabilized copper grid (400 mesh). The copper grid was air-dried, negatively stained with 5% aqueous uranyl acetate, and allowed to dry. Samples were analyzed by JEOL transmission electron microscope equipped with LaB6 filament gun (JEM 2010, Tokyo, Japan) at an accelerating voltage of 200 kV.
CDF Encapsulation.
Anticancer drug CDF was encapsulated in SPIONs@PAMAM and SPIONs@FA-PAMAM separately using equilibrium dialysis method as described earlier.5 Briefly, CDF and SPIONs formulations were calculated to have CDF and dendrimer (PAMAM or FA-PAMAM) at the molar ratio of 50:1. Both CDF and SPIONs@PAMAM or SPIONs@FA-PAMAM were dissolved in the mixture of DMSO and phosphate buffered saline (PBS) pH 7.4 (ratio 4:6). The mixed solution was stirred in the dark at a low speed of 50 rpm for 72 h at RT. The CDF encapsulated SPIONs@PAMAM-CDF and SPIONs@FA-PAMAM-CDF were precipitated using a strong neodymium N52 magnet. The supernatant was used for indirect drug loading method. After decantation, the formulations were washed by dispersing in 100 mL of EtOH by vortexing, followed by magnet decantation. Dry products were obtained by drying under vacuum on a rotary evaporator.
The remaining CDF in the supernatant after drug loading was determined by high-performance liquid chromatography (HPLC) method using a C18 column with photodiode array detector (PDA) at 447 nm. For the remaining CDF concentration determination, a standard curve of CDF was made by dissolving known amounts of CDF in DMSO and its successive dilutions in the mobile phase, followed by HPLC analysis at the absorbance of 447 nm. A known amount of the supernatant containing unloaded CDF was diluted in DMSO, followed by further dilution in the mobile phase and HPLC analysis at the absorbance of 447 nm. The amount of remaining CDF was calculated based on the CDF standard curve, and the percentages of CDF loaded in the formulations were calculated based on the subtraction of the initial amount of CDF and the remaining amount of CDF.
Fluorescence Microscopic Study.
Fluorescence microscopic study was performed in SKOV3 cells to compare the targeting ability of SPIONs@FA-PAMAM and SPIONs@PAMAM. In brief, SKOV3 cells were seeded in a four-well chamber slide (5 × 104 cells in each well) and incubated at 37 °C in a 5% CO2 air humidified atmosphere for 24 h. The medium was removed, and Rhodamine B labeled formulations (SPIONs@PAMAM-Rho and SPIONs@FA-PAMAM-Rho) were added and incubated for 6 h. The formulation containing medium was removed, and cells were washed for three times with cold PBS (pH 7.4) and fixed with 3% formaldehyde in PBS pH 7.4 at RT for 10 min. Samples were analyzed qualitatively using a fluorescent microscope (Leica, Germany)51.
T2 Relaxivity and in Vitro Relaxometry and Imaging Studies.
T2 Relaxivity Studies of SPIONs, SPIONs@PAMAM, and SPIONs@FA-PAMAM.
T2 relaxometry was performed using a 7.0 T Bruker ClinScan system. The instrumental parameters were set as follows: a 7.0 T magnetic field strength, pixel spacing at 0.297/0.297, repetition time 2000 ms, echo time 11 ms, and slice thickness of 2 mm. Synthesized SPIONs and their modification SPIONs@PAMAM and SPIONs@FA-PAMAM were analyzed at different iron concentrations 10, 20, 40, 60, 80, and 100 μg/mL. The T2 relaxivity was calculated from the linear slope of the inverse T2 (1/T2) relaxation time according to the iron concentration.
In Vitro MR Relaxometry and Imaging.
HeLa and SKOV3 cells (5 ×105) were incubated with both the nontargeted formulation SPIONs@PAMAM and the targeted formulation SPIONs@FA-PAMAM at iron concentrations of 10, 20, 40, and 80 μg/mL for 30 min at 4 °C according to the previously reported method.17 In short, 5 × 105 HeLa and SKOV3 cells were trypsinized and suspended in cold PBS (in an ice bath) and incubated with the formulations. After 30 min incubation, cells were centrifuged down at 800 rpm for 3 min to form a pellet. Cells were washed three times with cold PBS to remove free particles. Final pellets were resuspended in cold PBS and used for MR imaging. A phantom was constructed consisting of all of the sample vials. The instrumental parameters were set at 7.0 T magnetic field strength, pixel spacing at 0.297/0.297, repetition time 2000 ms, echo time 11 ms, and slice thickness of 2 mm.
in Vitro Cytotoxicity Study.
The in vitro cytotoxicity values of free CDF, SPIONs@PAMAM-CDF, and SPIONs@FA-PAMAM-CDF formulations were evaluated by MTT assay on HeLa and SKOV3 cell lines. In brief, HeLa and SKOV3 cells were seeded in 96-well plates with an average of 3000 cells in each well. After 24 h incubation at 37 °C in a 5% CO2 air humidified atmosphere, cells were treated with various formulations with a concentration range from 0.25–5 μM. Treated cells were incubated for 72 h at 37 °C followed by addition of MTT solution (1 mg/mL) and further incubation at 37 °C for 3 h. Then the media were replaced by DMSO (100 μL in each well). The absorbance was measured at 590 nm using a high-performance multimode plate reader Synergy 2 (BioTek). The percentage of viable cells was determined by comparing the absorbance with appropriate controls.33,40
Folate Receptor Blocking Assay.
The folate receptor blocking assay was performed to understand the mechanism by which the targeting SPIONs@FA-PAMAM-CDF internalize HeLa and SKOV3 cells via folate receptor mediated endocytosis. This assay is based on the principle of the initial blockade of folate receptors of HeLa and SKOV3 by adding an excess amount of free FA (1 mM)36 followed by treatment with formulations (CDF, SPIONs@PAMAM-CDF, and SPIONs@FA-PAMAM-CDF). The cell viability of HeLa and SKOV3 was determined by MTT assay after 72 h incubation at 37 °C. This assay was performed according to the previously reported protocol.52 In short, HeLa and SKOV3 cells were seeded in 96-well plates for 24 h, followed by addition of 100 μL of 1 mM FA in each well and incubation at 37 °C for 3 h. Then cells were washed twice with PBS (pH 7.4), followed by media and addition of formulations. After 72 h incubation at 37 °C, MTT assay was performed to determine the cell viability as stated previously.
Apoptosis Assay by Flow Cytometry.
Apoptosis assay was performed on HeLa cell line according to our previous study.36 In brief, HeLa cells were cultured in six-well plates at 5 × 104 cells in each well and incubated for 24 h at 37 °C under 5% CO2, followed the treatment of plain CDF, SPIONs@PAMAM-CDF, and SPIONs@FA-PAMAM-CDF to induce apoptosis. The concentrations of CDF, SPIONs@PAMAM-CDF, and SPIONs@FA-PAMAM-CDF were chosen based on the IC50 value of CDF on HeLa cells from the in vitro cytotoxicity assay. After 72 h incubation, cells were collected, and sample was prepared according to the protocol for Guava Nexin Annexin V assay (EMD Millipore, USA). In short, media and trypsinized treated cells were collected in 15 mL tubes and centrifuged at 300g for 7 min. Cell pellets were dispersed in PBS pH 7.4 with 1% FBS to have the number of cells in the range of 2 × 105 to 1 × 106 cells/mL. Then 100 μL of cell dispersion of each sample was added 100 μL of the Guava Nexin Reagent and was incubated for 20 min at RT in the dark. The samples were analyzed by Guava Easycyte flow cytometer (EMD Millipore, USA).
Western Blot.
Western blot analysis was performed to determine the level expression of phosphatase and tensin homologue PTEN and nuclear factor kappa B (NF-κB) in HeLa cell line using reported method.53 Briefly, HeLa cells were treated with different formulations as well as free CDF for 24 h and lysed. The protein concentration was determined by the Bio-Rad Protein Assay (Bio-Rad kit). Lysates were electrophoresed by SDS-PAGE, and the proteins were transferred onto the nitrocellulose blotting membrane, followed by blocking with 5% BSA in TBST buffer at RT for 1 h. Primary PTEN, NF-κB, and Caspase 3 antibodies were added and incubated overnight at 4 °C, subsequently washed, and incubated with compatible secondary antibodies. The protein bands were visualized by incubation with chemiluminescent substrate (Thermo scientific) at RT for 2 min, followed by chemiluminescent detection using a digital imaging system ImageQuant LAS 4000 (GE Healthcare Bio-Sciences AB, Sweden).
RESULTS
A vast majority of cancer cells are known to have a high expression of folate receptors, while normal tissues and organs have a very limited expression of folate receptors.54 Many studies have shown an enhancement in anticancer activity employing FA-decorated nanocarriers in different cancer types such as ovarian, lung, cervical, breast, kidney, colorectal, epithelial, and brain cancers.55,56,26,57,58 CDF has been shown to possess a high anticancer activity against various types of cancers as well as the ability to overcome drug resistance.38,53 However, the extremely low aqueous solubility of CDF makes its systemic administration problematic. Our previous study suggested that PAMAM dendrimer conjugated with FA improved the aqueous solubility of CDF dramatically and endowed the active targeting molecule with an enhanced anticancer activity due to the folate receptor-mediated endocytosis.36 In addition, many studies reported the potential usage in biomedical imaging of PAMAM dendrimers when fabricated with magnetic iron oxide nanoparticles.17,59 On the basis of this information, the goal of this present work was to design a theranostic nanoparticle consisting of FA-PAMAM conjugate as the outer shell and iron oxide nanoparticles as the inner core loaded with CDF, which could be used in both cancer imaging and therapy for multiple cancers.
SPIONs Synthesis and Characterization.
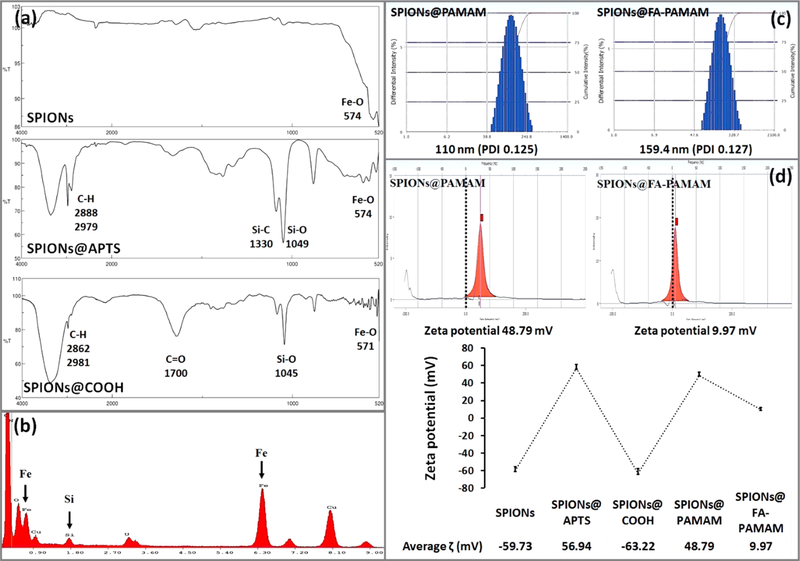
The magnetic iron oxide nanoparticles SPIONs were synthesized by controlled coprecipitation of Fe2+ and Fe3+ ions according to previously reported method.45,60 The synthesized SPIONs were confirmed by FTIR spectroscopy. The presence of Fe3O4 core was identified by the strong stretching absorption band between 408 and 673 cm−1 corresponding to the Fe-O bond of the particles (Figure 3a).46 Dynamic light scattering technique measurement showed a hydrodynamic size of 78.8 nm (PDI 0.177) and a zeta potential at −59.73 mV (Figure 3d).
Figure 3.
(a) FTIR spectra of SPIONs, SPIONs@APTS, and SPIONs@COOH. (b) EDS analysis of SPIONs@APTS. (c) Hydrodynamic size of the fabricated nanoparticles SPIONs@PAMAM and SPIONs@FA-PAMAM. (d) Zeta potential measurement of each step of the fabrication process.
FA-PAMAM Decorated SPIONs Fabrication and Characterization.
Amine Functionalized SPIONs (SPIONs@APTS).
The sonication time was optimized according to previously reported method.46 Sonication of the synthesized SPIONs before modification with APTS helped to decrease the aggregation of SPIONs and improved the magnetic properties and size distribution of the particles. After aminosilane modification of the SPIONs, the achieved product SPIONs@ APTS could be dispersed back in DIW to form a stable dispersion with a hydrodynamic size of 95.9 nm (PDI 0.113). SPIONs@APTS showed a zeta potential of 56.94 mV, which confirmed the presence of the positively charged amine groups of APTS (Figure 3d). EDS spectrum showed the unique peak of Si, which further confirmed the successful coating of the aminosilane APTS on the surface of SPIONs (Figure 3b). FTIR spectrum confirmed the presence of APTS on the surface of SPIONs with the characteristic peaks of C—H at 2888, 2979 cm−1, Si—C at 1330 cm−1, Si—O at 1049 cm−1, and Fe—O at 574 cm−1 (Figure 3a).
Carboxylation of Amine Functionalized SPIONs (SPIONs@COOH).
The carboxylation of primary amine groups on the surface of SPIONs@APTS was confirmed by the change in zeta potential from a positive charge of 56.94 mV (of the amine groups) to a negative charge of −63.22 mV (of the carboxyl groups) (Figure 3d). FTIR spectrum of SPIONs@COOH showed the characteristic peaks of C-H bond at 2981 and 2862 cm−1, Si-O bond at 1045 cm−1, Fe-O bond at 571 cm−1, and the C=O stretching at 1700 cm−1 (Figure 3a). SPIONs@COOH had a hydrodynamic size of 96.2 nm (PDI 0.246).
Activation of SPIONs@COOH and Fabrication of FA-PAMAM and SPIONs@COOH Conjugate.
Zeta potential was used to confirm the conjugation of PAMAM and FA-PAMAM to the activated SPIONs@COOH. Before conjugation, SPIONs@COOH had the zeta potential of −63.22 mV. After the conjugation, the zeta potential values were changed to 48.79 mV and 9.97 mV in case of SPIONs@PAMAM and SPIONs@FA-PAMAM, respectively (Figure 3d). Dynamic light scattering showed an average size of 110.1 nm (PDI 0.125) and 159.4 nm (PDI 0.127) of SPIONs@PAMAM and SPIONs@FA-PAMAM, respectively (Figure 3c). Fluorescence spectroscopy measurement showed an average of 20.37% (w/w) of PAMAM in SPIONs@PAMAM, and 27.61% (w/w) of FA-PAMAM conjugates in SPIONs@FA-PAMAM structure.
Transmission Electron Microscopic Analysis.
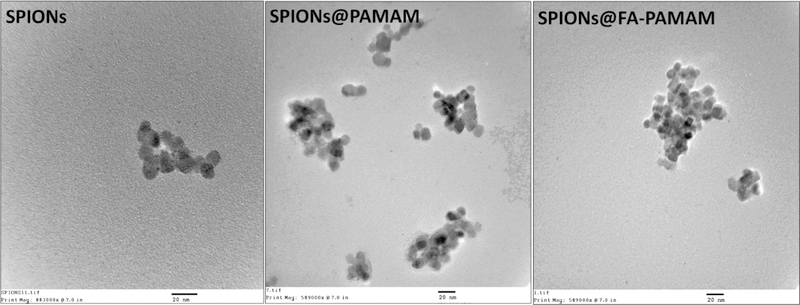
To further determine the size of the nanoformulations, electron microscopic analysis of the synthesized SPIONs and the carrier SPIONs@PAMAM and SPIONs@FA-PAMAM was performed. TEM images showed that the morphology of the fabricated SPIONs remained the same as the unmodified SPIONs. TEM data showed that the inner SPIONs core had the average size of 11 nm, which confirmed the nanometric size of the formulations (Figure 4). The results are in accordance with previously reported methods.46,60,61
Figure 4.
Transmission electron microscopic images of SPIONs, SPIONs@PAMAM, and SPIONs@FA-PAMAM show the morphology of the fabricated nanoparticles.
CDF Drug Loading in Nanoparticles.
The CDF drug loading was studied based on the indirect method. The remaining of CDF in the supernatant after drug loading was measured by HPLC method. A calibration curve of CDF was developed from 10 μg/mL to 250 μg/mL with the R2 value of 0.99. The HPLC method was validated for its accuracy and precision and was used to determine the CDF concentration. The loading of CDF in SPIONs@PAMAM and SPIONs@FA-PAMAM was 12.37% (w/w) and 9.81% (w/w), respectively.
Fluorescence Microscopy Study.
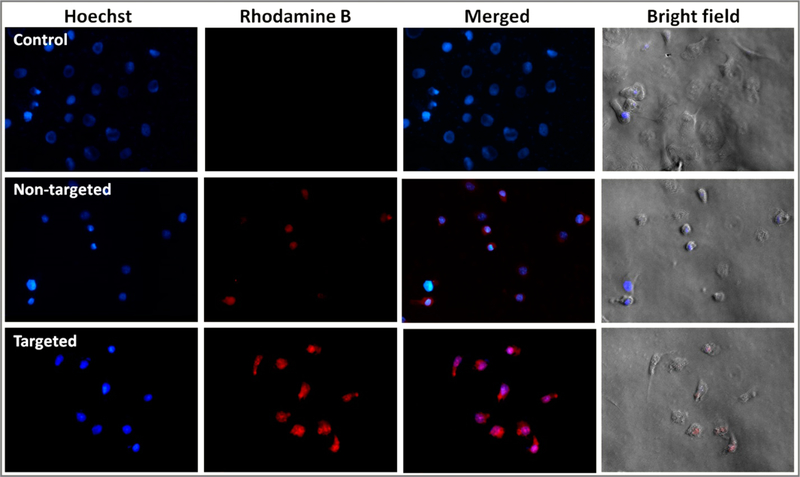
SKOV3 cells were selected for in vitro fluorescence microscopic study based on the results of in vitro cytotoxicity assay and receptor blocking assay to compare the level of cellular internalization of the nontargeted formulation SPIONs@PAMAM-CDF and the targeted formulation SPIONs@FA-PAMAM-CDF. In this cell uptake study, SKOV3 cells were incubated with Rhodamine B (having red fluorescence) labeled nanoformulations and analyzed after 6 h of incubation at 37 °C in the dark. As shown in Figure 5, SKOV3 cells treated with both of the nontargeted and the targeted formulations showed apparent fluorescence. As compared to the nontargeted formulation, there was a significantly higher fluorescence in cells treated with targeted formulations.
Figure 5.
Fluorescence microscopic images (40×) of SKOV3 cells incubated with nuclear stain Hoechst (blue fluorescence) and Rhodamine B (red fluorescence) labeled nontargeted SPIONs@PAMAM and targeted formulations SPIONs@FA-PAMAM at 6 h are shown.
T2 Relaxivity and in Vitro Relaxometry and Imaging Studies.
T2 Relaxivity Studies of SPIONs, SPIONs@PAMAM, and SPIONs@FA-PAMAM.
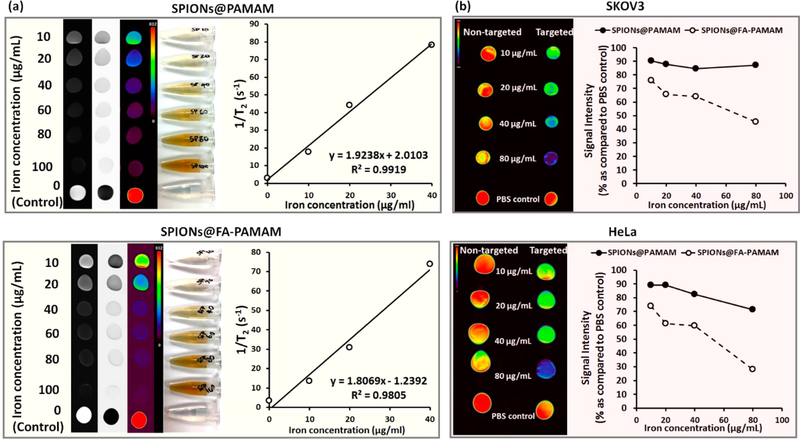
T2 relaxivity studies were performed to examine the magnetic behavior of the synthesized SPIONs and the nanocarrier SPIONs@PAMAM and SPIONs@FA-PAMAM in their biomedical application in MR imaging. The potential of the fabricated magnetic nanoparticles SPIONs@PAMAM and SPIONs@FA-PAMAM to be used as T2-based contrast agent for MR imaging was evaluated using the measured transverse relaxation time (T2) of SPIONs@PAMAM and SPIONs@FA-PAMAM as compared to SPIONs. The T2 values were used to calculate the transverse relaxivity rate (r2) per μg/mL of iron, which showed the efficiency of the fabricated nanoparticles as an MR contrast agent. From Figure 6a, there was a significant decrease in the signal intensity of the T2-weighted MR images with the increase of iron concentration in both of the nontargeted SPIONs@PAMAM and the targeted SPIONs@FA-PAMAM nanoparticles as compared to the control PBS. Pseudocolor MR images showed a decrease in signal intensity for the fabricated nanoparticles from red (high intensity) to purple (low intensity). The T2 relaxation rate (1/T2) increased linearly with the iron concentration (μg/mL) in both cases of the nontargeted SPIONs@PAMAM and the targeted SPIONs@FA-PAMAM nanoparticles. The slope values (r2) were calculated to be 1.92 (μg/mL)−1 s−1 and 1.81(μg/mL)−1 s−1 in the case of SPIONs@PAMAM and SPIONs@FA-PAMAM, respectively. Unmodified SPIONs had the slope value (r2) of 2.01 (μg/mL)−1 s−1. The results suggested that both of the nontargeted nanoparticle SPIONs@ PAMAM and the targeted nanoparticle SPIONs@FA-PAMAM could be used as T2-shortening agents.
Figure 6.
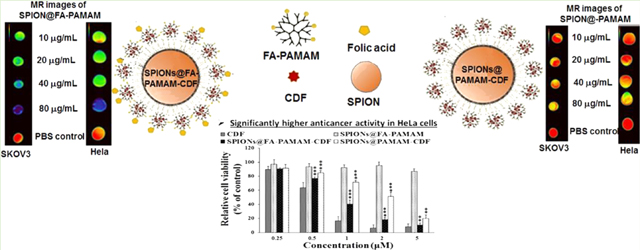
(a) T2-weighted MR images of the aqueous dispersion of SPIONs@PAMAM and SPIONs@FA-PAMAM with the T2 relaxation rate (1/T2) as a function of iron concentration indicating the ability of the fabricated nanoparticles to enhance the contrast in MR images; (b) MR images of SKOV3 and HeLa cell pellets after 30 min incubation with the nontargeted SPIONs@PAMAM and the targeted SPIONs@FA-PAMAM nanoparticles (with the color change from red to purple indicating the gradual decrease of MR signal intensity). The percentage of signal intensity compared to cells in PBS was plotted as a function of iron concentration indicating the faster internalization with higher decrease in MR signal intensity of the targeted SPIONs@FA-PAMAM.
In Vitro MR Relaxometry and Imaging Studies.
In vitro MR relaxometry and imaging studies were performed to examine the effect of the targeting ability of folate-based nanoparticle SPIONs@FA-PAMAM in MR imaging. To study the effect of SPIONs@PAMAM and SPIONs@FA-PAMAM on SKOV3 and HeLa cells, we measured the T2 of SKOV3 and HeLa cells after incubation with different concentration of SPIONs@ PAMAM and SPIONs@FA-PAMAM for 30 min. There was a significant decrease in signal intensity in SKOV3 and HeLa cells when incubated with the targeted nanoparticle SPIONs@FA-PAMAM. In contrast, nontargeted nanoparticle SPIONs@PAMAM showed a very little decrease in the signal intensity in both SKOV3 and HeLa cells. In the T2-weighted MR images, targeted formulation SPIONs@FA-PAMAM at the iron concentration of 80 μg/mL decreased the MR signal intensity to 45.6% in SKOV3 and 28% in HeLa, as compared to 100% of PBS control. However, nontargeted nanoparticle SPIONs@PAMAM at the iron concentration of 80 μg/mL showed a lower decrease in MR signal intensity with 87% in SKOV3 and in 71.4% HeLa, as compared to 100% of control PBS (Figure 6b).
In Vitro Cytotoxicity Study.
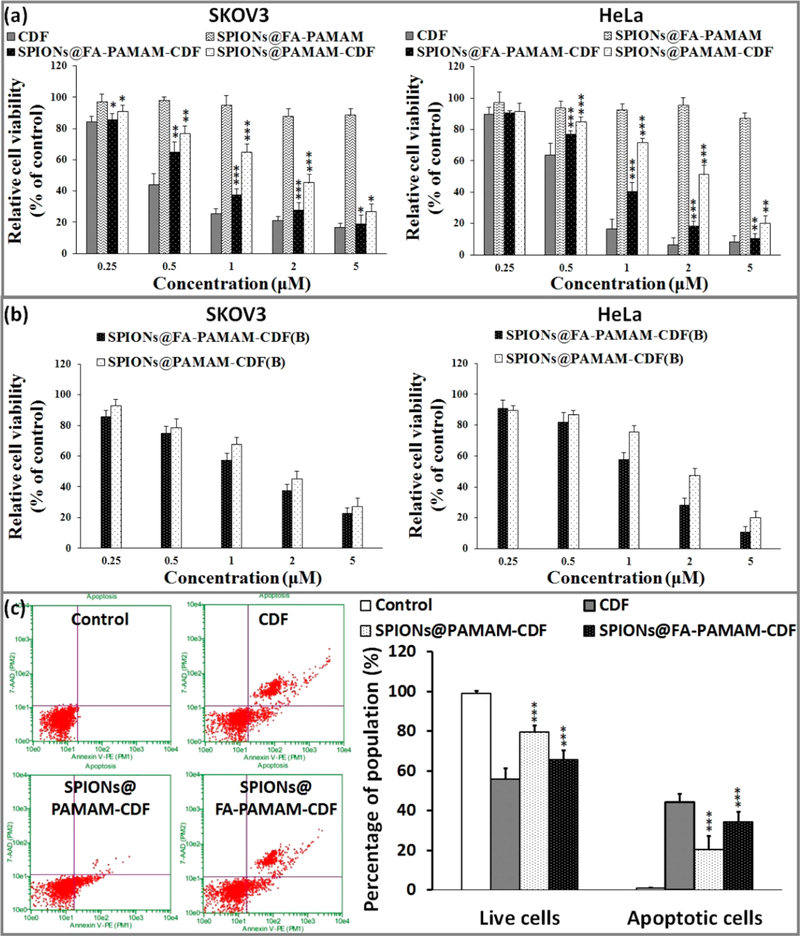
In vitro cytotoxicity of the CDF loaded nanoformulations was examined in two cell lines (SKOV3 (human ovarian carcinoma cell line), HeLa cells (human cervical cancer cells)) with a broad range of CDF concentrations (0.25–5 μM). Plain targeted carrier (SPIONs@FA-PAMAM) showed negligible cytotoxicity with cell viability more than 90%, which confirmed the safety of the targeted carrier. The anticancer activity of the nanoformulations (nontargeted formulation SPIONs@PAMAM-CDF and targeted formulation SPIONs@FA-PAMAM-CDF) was studied and compared with free drug CDF. The results showed a dose-dependent cell killing for both SPIONs@PAMAM-CDF and SPIONs@FA-PAMAM-CDF. The outcome of the study revealed an IC50 (half maximal inhibitory concentration) of 0.45 μM, 0.78 μM, and 1.79 μM for free CDF, SPIONs@FA-PAMAM-CDF, and SPIONs@PAMAM-CDF, respectively, in SKOV3 cells. The noticeably lower IC50 of the targeted SPIONs@FA-PAMAM-CDF as compared to the nontargeted SPIONs@PAMAM-CDF was probably due to the folate receptor specific targeting of SPIONs@FA-PAMAM-CDF. The same pattern was also observed in HeLa cells with the IC50 of 0.66 μM, 0.87 μM, and 1.98 μM for free CDF, SPIONs@FA-PAMAM-CDF, and SPIONs@PAMAM-CDF, respectively (Figure 7a).
Figure 7.
(a) In vitro cytotoxicity assay showing percentage of cell viability observed at 72 h after treating SKOV3 and HeLa cells with various formulations are shown (n = 8). (b) MTT assay observed at 72 h after folate receptor blocking and treating of SKOV3 and HeLa cells with SPIONs@PAMAM-CDF and SPIONs@FA-PAMAM-CDF are shown (n = 8). (c) Induction of apoptosis in HeLa cells when treated with CDF, SPIONs@PAMAM-CDF, and SPIONs@FA-PAMAM-CDF as evaluated by Annexin V/7-AAD dual staining. An increased percentage of the apoptotic cell population was noted when cells were treated with targeted formulation (SPIONs@FA-PAMAM-CDF) as compared to the nontargeted formulation (SPIONs@PAMAM-CDF), which suggested the better killing activity of the targeted formulation SPIONs@FA-PAMAM-CDF. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Folate Receptor Blocking Assay.
Folate receptor blocking assay was performed to examine the fate of the folate-based targeting formulation. SKOV3 and HeLa cells with high expression of folate receptors were treated with an excess amount of FA (1 mM) to overwhelm the folate receptor binding domains on the cell membrane, followed by the treatment with the nanoformulations.41,42 In vitro cytotoxicity MTT assay was used to determine the change in the cell viability in SKOV3 and HeLa treated with the formulations after blocking the folate receptors. It was observed that before blockade of folate receptors, the IC50 values on SKOV3 were 0.80 μM and 1.81 μM for the targeted SPIONs@FA-PAMAM-CDF and the nontargeted SPIONs@PAMAM-CDF, respectively. After the blockade of folate receptors, the IC50 value of the targeted SPIONs@FA-PAMAM-CDF was increased to 1.36 μM. However, there was not a significant change in the IC50 of the nontargeted SPIONs@PAMAM-CDF (1.82 μM). The same outcome was observed in HeLa cells with the IC50 of 0.85 μM and 2 μM for SPIONs@FA-PAMAM-CDF and SPIONs@PAMAM-CDF, respectively, before the folate receptor blockade. After blocking of the folate receptors, the IC50 values were found to be 1.26 μM and 1.94 μM for SPIONs@FA-PAMAM-CDF and SPIONs@PAMAM-CDF, respectively. The results suggested a decrease in anticancer activity of the targeted formulation SPIONs@FA-PAMAM-CDF in SKOV3 cells when folate receptors are blocked (Figure 7b).
Apoptosis Assay.
HeLa cells were selected for this apoptosis assay. Apoptosis induction in HeLa cells of free CDF and the CDF loaded formulations was determined by flow cytometry with Annexin V/7-AAD dual staining. The percentages of Annexin V−/7-AAD− (R5), Annexin V+/7-AAD− (R6), Annexin V−/7-AAD + (R4), and Annexin V−/7-AAD+ (R3) were used to determine the number of live cells, early apoptotic, late apoptotic, and necrotic cells. Apoptosis assay revealed a higher percentage of apoptotic and necrotic HeLa cells (34.2 ± 3.2%) after treatment with the targeted formulation SPIONs@FA-PAMAM-CDF. Nontargeted formulation showed a lower number of apoptotic and necrotic cells at 20.5 ± 2.7% of the cell population. The results suggested a higher apoptosis induction ability of the targeted formulation SPIONs@FA-PAMAM-CDF as compared to the nontargeted formulation SPIONs@PAMAM-CDF (Figure 7c). The results were consistent with the higher anticancer activity of the targeted formulation SPIONs@FA-PAMAM-CDF in in vitro cytotoxicity assay using MTT with higher cellular uptake in fluorescence microscopic studies and in vitro relaxometry and imaging studies.
Western Blot.
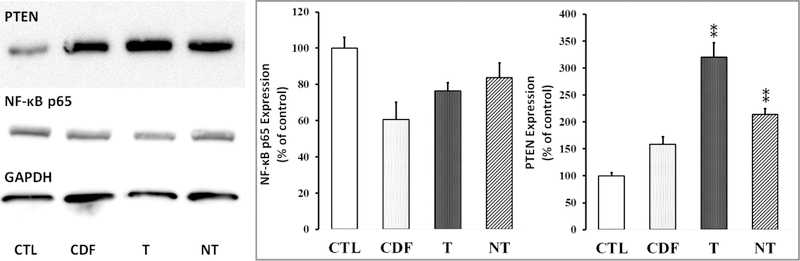
Western blot was performed to examine the level expression of PTEN, NF-κB, Caspase 3 in HeLa cells after treatment with the nanoformulations. In the case of NF-κB, control HeLa cells without treatment showed a higher expression of NF-κB as compared to HeLa cells treated with plain CDF and the nanoformulations. Targeted formulation SPIONs@FA-PAMAM-CDF showed a slightly better down-regulation of NF-κB (76.4 ± 4.6% as of control) as compared to the nontargeted formulation SPIONs@PAMAM-CDF (83.6 ± 8.2% as of control). In the case of PTEN expression, the control HeLa cells without treatment showed a significantly low expression of PTEN. However, after treatment with plain CDF and nanoformulations, PTEN expression was elevated. Noticeably, in comparison to control, HeLa cells treated with the targeted formulation SPIONs@FA-PAMAM-CDF showed the highest upregulation of PTEN expression (319.9 ± 27.1%) as compared to free CDF (158.9 ± 13.5%) and the nontargeted formulation SPIONs@PAMAM-CDF (214.2 ± 10.9%, p < 0.01) (Figure 8). Similarly, pro-apoptotic Caspase 3 protein was up-regulated in case of SPIONs@FA-PAMAM-CDF (T) treated Hela cells than that of other treatment and control groups, as quantified with respect to GAPDH (Figure S1).
Figure 8.
Western blot analysis showing PTEN and NF-κB expression in HeLa cells in control without treatment (CTL) and cells treated with CDF, targeted formulation SPIONs@FA-PAMAM-CDF (T), and nontargeted formulation SPIONs@PAMAM-CDF (NT), (GAPDH expression was used as the protein loading control). **, p < 0.01.
DISCUSSION
The poor outcome for patients diagnosed with metastatic cancer along with the complexity of treating cancers in its advanced stages is driving research in the direction of arriving at newer materials and methods for early cancer detection and prevention. With the advantages of noninvasive MR imaging with superb spatial resolution, targeting MRI contrast agents to cancers are believed to be critical tools that can be exploited for developing cancer theranostics.62,63 The targeting ability of MRI contrast agents depends strongly on their ability to carry specific molecular markers for tumor recognition. As a result, the tumor can be visualized and diagnosed earlier accurately leading to an effective treatment, and ultimately improving patient survival.62,64 In this regard, PAMAM dendrimers with the ability to conjugate targeting ligands through numerous peripheral reactive amine groups have been studied recently for surface modification of iron oxide nanoparticles for targeting MRI.65,66 In terms of targeting ligands, FA has received considerable attention as a targeting ligand to target folate receptors overexpressed on numerous types of tumors.27,28,56 PAMAM dendrimers conjugated FA can take advantage of the favorable binding of targeting ligand FA to folate receptors on tumor cells to enhance the cellular uptake via folate receptor mediated endocytosis. Many studies have shown that iron oxide nanoparticles decorated with folate-based PAMAM dendrimer could achieve MRI targeting with higher cellular internalization in cancer cells with a higher contrast in T2-weighted MR images.17,46,67 However, in most of the studies, the ability of PAMAM dendrimers in drug delivery was not fully examined.
The poor aqueous solubility of several potent anticancer compounds could be a major issue and deciding factor in realizing its translation potential from the bench to the bedside. The nonspecific distribution of chemotherapeutic agents in the systemic circulation gives the conventional chemotherapy a low therapeutic index and severe side effects, which cause the treatment to be ineffective and lead to the recurrence of tumor after initial treatment. Recently, CDF has been shown to be a very potent anticancer compound with the ability to treat different types of cancers.36,39,40 However, low solubility profile of CDF makes its systemic administration problematic. In this regard, PAMAM dendrimers have revealed promising potentials. Our earlier reported study suggested that folate-decorated PAMAM dendrimers could encapsulate CDF in their hydrophobic cavities for aqueous solubility enhancement and deliver CDF specifically to the tumor site with minimum adverse side effects.36 The aim of this study was to design a theranostic carrier consisting of iron oxide nanoparticles with folate-based PAMAM dendrimers for active targeting MR imaging and anticancer drug delivery. As a result, the targeted nanoparticles SPIONs@FA-PAMAM can enhance the aqueous solubility of CDF and deliver it specifically to cancer cells with a higher contrast in T2-weight MR images.
The synthesis of SPIONs was confirmed by FTIR spectrum with the characteristic peak of Fe-O at 574 cm−1 (Figure 3). The synthesized SPIONs could be dispersed back in DIW or EtOH to form a stable dispersion with a hydrodynamic size of 78.8 nm (PDI 0.177) and a zeta potential of −59.73 mV. TEM images of SPIONs showing an average size at 10.5 ± 1 nm further reconfirmed the success of the synthesis. Surface modification of SPIONs was confirmed by zeta potential with a change from an initial zeta potential of −59.73 mV of SPIONs to 56.94 mV of SPIONs@APTS, and −63.22 mV of SPIONs@ COOH (Figure 3d). In EDX analysis of SPIONs@APTS, the atomic ratio of Fe/Si was shown to be 17.55:1 (Figure 3b). The mathematic calculation was used to estimate the number of silanes per gram of particles according to the reported method.45 From TEM images, the synthesized SPIONs were shown to have an average diameter (rSPIONs) of 5.5 nm; therefore, the surface area (SASPIONs) of one particle was 380 nm2 (SASPIONs = 4πrSPIONs2). One silane is known to cover approximately 0.4 nm2 of the particle’s surface area, so we have an average of 950 silanes covering the surface of one SPION.45 With the atomic ratio 17.55:1 of Fe/Si in SPIONs@APTS from EDX analysis, it is estimated that 950 silanes would cover one particle with an average of 16 672.5 iron atoms. Therefore, 1 g of SPIONs would theoretically have an average of 0.099 g of APTS coating.
PAMAM and FA-PAMAM conjugation were confirmed by a change in zeta potential (Figure 3). After conjugation, SPIONs@PAMAM and SPIONs@FA-PAMAM had a zeta potential values of 48.79 mV and 9.97 mV, respectively (Figure 3d). FA conjugation helped the targeted carrier SPIONs@FA-PAMAM reduce the highly positive charge, which is known to be associated with the toxicity of PAMAM dendrimers.20 The morphology of nanoparticles could be affected by several factors including the fabrication and reaction involved. The shape and size of nanoparticles could have a high impact on their biodistribution, clearance, and biocompatibility.68 TEM images suggested that the morphology of the particles remained unchanged after the fabrication. Dynamic light scattering measurement showed that both of the nontargeted and targeted formulations were in the nanosized range with an average hydrodynamic size of 110.1 nm (PDI 0.125) and 159.4 nm (PDI 0.127) of SPIONs@PAMAM and SPIONs@FA-PAMAM, respectively (Figure 3c). A higher value in hydrodynamic size of SPIONs@FA-PAMAM could be attributed to the presence of FA resulting in a low positive charge of the particles. Drug loading studies showed a little higher in CDF encapsulation in SPIONs@FA-PAMAM-CDF as compared to SPIONs@PAMAM-CDF. The presence of FA might be responsible for a higher drug loading with a shielding effect outside the hydrophobic cavities; thus, a higher amount of CDF was encapsulated inside the hydrophobic cavities.
Targeting ability of folate-decorated nanoformulations was examined in fluorescence microscopic study on SKOV3 cell line. SKOV3 cells treated with Rhodamine B labeled targeted formulation SPIONs@FA-PAMAM-CDF showed a significantly higher red fluorescence intensity of Rhodamine B dye as compared to the Rhodamine B labeled nontargeted formulation SPIONs@PAMAM. The fluorescence microscopic images were taken after only 6 h incubation of SKOV3 cells with the Rhodamine B labeled formulations, which proved the ability of the targeted formulation to internalize the cells within such a short time frame (Figure 5).
The magnetic behavior of the nontargeted SPIONs@PAMAM and the targeted SPIONs@FA-PAMAM was examined in the T2 relaxivity studies to test their ability to enhance the contrast in MR images (Figure 6). The T2 relaxation rate (1/T2) as a function of the iron concentration (from 10–40 μg/mL) of both of the carriers showed that the relaxation rate increases linearly with the iron concentration with a slope (r2) of 1.92 (μg/mL)−1 s−1 and 1.81(μg/mL)−1 s−1 for SPIONs@PAMAM and SPION@FA-PAMAM, respectively. The slightly lower r2 of SPIONs@FA-PAMAM may be due to the presence of FA on the periphery of the particles, which shields water molecules from accessing the surface of SPIONs. The results suggested that SPIONs@PAMAM and SPIONs@FA-PAMAM could be used as T2-shortening agents, which helped enhance the contrast in the MR images. To study the effect of the nontargeted SPIONs@PAMAM and the targeted SPIONs@FA-PAMAM on the MR images of SKOV3 and HeLa cells, T2 of SKOV3 and HeLa cells was measured after 30 min incubation with various concentrations of SPIONs@PAMAM and SPIONs@FA-PAMAM. The T2 values of SKOV3 and HeLa cell pellets treated with SPIONs@FA-PAMAM particles decreased dramatically as a function of iron concentration. In contrast, there was a small decrease in the T2 values of SKOV3 and HeLa cell pellets treated with the nontargeted SPIONs@PAMAM particles. In the T2-weighted MR images (the color change from red to purple indicates a decrease in MR signal intensity) observed in SKOV3 and HeLa cells, targeted SPIONs@FA-PAMAM nanoparticles at 80 μg/mL reduced the signal intensity to 45.6% and 28% of the initial value (PBS control) in SKOV3 and HeLa cells, respectively, whereas nontargeted SPIONs@PAMAM nanoparticles at 80 μg/mL reduced the signal intensity to 87% and 71.4% of the initial value in SKOV3 and HeLa cells, respectively. The results suggested the targeted nanoparticles SPIONs@FA-PAMAM can specifically hamper the MR signal intensity through folate receptor mediated endocytosis. The results are in accordance with the higher cellular internalization of the targeted nanoparticles in the fluorescence microscopic studies.
The anticancer activity of the CDF loaded nanoformulations was examined using the in vitro cytotoxicity assay on SKOV3 and Hela cells. The MTT assay results showed a high anticancer activity on SKOV3 and HeLa cells with both the formulations with relatively low IC50. It should be noted that the targeted formulation SPIONs@FA-PAMAM-CDF showed a higher anticancer activity with lower IC50 as compared to the nontargeted formulation SPIONs@PAMAM-CDF when tested at a concentration range from 0.25–5 μM. IC50 values of SPIONs@FA-PAMAM-CDF were found to be 0.78 μM and 0.87 μM in SKOV3 and HeLa, which were lower by 2.29-fold and 2.27-fold as compared to the nontargeted SPIONs@ PAMAM-CDF in SKOV3 and HeLa, respectively. The better anticancer activity of the targeted formulation could be attributed to the targeting effect of FA, which helped the formulations have a higher degree of cellular internalization via folate receptor mediated endocytosis. Fourth generation PAMAM dendrimers are known to be cytotoxic due to the highly positive charge of 64 amino groups on the periphery. FA conjugation gave the targeted formulation an ability to target folate receptors overexpressed on cancer cells and also helped reduce the positive charge of the nanoconstruct. As a result, SPIONs@FA-PAMAM carrier showed a relatively safe profile on both SKOV3 and HeLa cells with more than 90% viable cells at all tested concentrations. The results proved that SPIONs@FA-PAMAM could be a promising carrier for anticancer drug delivery (Figure 7a). The folate receptor assay was performed to evaluate the fate of the nanoformulations under blocking the folate receptors. An increase in IC50 values of the targeted formulation SPIONs@FA-PAMAM-CDF in SKOV3 and HeLa cells after blocking folate receptors was observed. In contrast, IC50 values of the nontargeted formulation SPIONs@PAMAM-CDF did not show a significant change after the blocking of folate receptors. The results suggested that the higher activity of the targeted formulation SPIONs@FA-PAMAM-CDF was due to the active targeting ability of the folate-decorated nanoformulations. However, it should be noted that the IC50 value of SPIONs@PAMAM-CDF was still higher than SPIONs@FA-PAMAM-CDF in both SKOV3 and HeLa after the blocking the folate receptors (Figure 7b). The results suggested that the blocking of folate receptors on SKOV3 and HeLa cells could decrease the cellular uptake of the targeted formulation to a certain degree, and when the folate receptors are recycled, the folate receptor mediate endocytosis pathway was recovered and the targeting ability of SPIONs@FA-PAMAM was regained. Some reports have stated that amine functionalized polymers are thought to mediate endosomal escape through “proton sponge” effect at acidic tumor endosomal pH.69 Thus, polyamine functionalized SPIONs@FA-PAMAM-CDF could be able to escape the endosome and deliver the drug in the cytosol.
As a next step, apoptosis assay and Western blot studies were performed to further examine the targeting ability of the targeted formulation SPIONs@FA-PAMAM-CDF in anti-cancer activity against HeLa cells. It should be noted that free CDF showed the highest percentage of apoptotic and necrotic cells. This could be explained by the small molecular weight and the highly lipophilic profile of CDF, which gives CDF a high rate of internalization of passive diffusion. Apoptosis assay showed a higher percentage of apoptotic and necrotic cells in HeLa after treatment with the targeted formulation SPIONs@FA-PAMAM-CDF as compared to the nontargeted formulation SPIONs@PAMAM-CDF. A better apoptosis induction ability of the targeted formulation SPIONs@FA-PAMAM-CDF could be explained by a higher cellular uptake of the formulation due to the targeting ability of FA to the folate receptors overexpressed on HeLa cells (Figure 7c). The results from apoptosis assay were in accordance with higher anticancer activity of the targeted formulation with higher cellular uptake in fluorescence microscopic studies and in vitro T2 relaxation studies.
Western blot studies were performed on HeLa cells to determine the expression levels of PTEN, NF-κB and Caspase 3 after treatment with the formulations. CDF is known to upregulate PTEN, which is a tumor suppressor gene with a key role in stem cell self-renewal.53 PTEN can dephosphorylate phosphatidylinositol 3,4,5-triphosphate (PIP3) and antagonize the PI3K/Akt pathway. It is believed that PTEN down-regulation is a key factor contributing to the development of chemotherapy resistance and recurrence of various human tumors.70 PTEN is known to regulate many cellular processes including growth, adhesion, migration, invasion, and apoptosis.38,53 Nuclear factor kappa B (NF-κB) is known to upregulate antiapoptotic genes such as bcl-xL and X-linked inhibitor of apoptosis (XIAP) leading to induce cell survival. Activation of NF-κB was observed in many types of cancer and was shown to contribute to apoptosis resistance in cancer cells.71 In the case of NF-κB expression, control HeLa cells without treatment showed a high expression of NF-κB. After treatment with CDF and the formulations, NF-κB levels in HeLa were decreased. Higher downregulation of NF-κB in HeLa cells was found when cells treated with the targeted formulation SPIONs@FA-PAMAM-CDF (decreased by 1.3-fold as compared to control) as compared to the nontargeted formulation SPIONs@PAMAM-CDF (decreased by 1.19-fold as compared to control), which indicated a better anticancer activity of the targeted formulation. In the case of PTEN expression, Western blot results showed a significantly low expression of PTEN in control cells. However, there was an increase in PTEN expression in cells treated with CDF and both of the formulations. Noticeably, PTEN level in cells treated with the targeted formulation SPIONs@FA-PAMAM-CDF was higher than CDF and the nontargeted formulation SPIONs@PAMAM-CDF. As compared to control, PTEN expression was upregulated by 3.2-fold in HeLa treated with the targeted formulation, whereas in HeLa treated with free CDF and the nontargeted formulation, PTEN expression was increased by 1.58- and 2.14-fold, respectively (Figure 8). Another crucial apoptotic factor, Caspase 3, was upregulated in treatment with targeted formulation SPIONs@FA-PAMAM-CDF (T). Thus, the results suggested the ability of the targeted formulation SPIONs@FA-PAMAM-CDF to upregulate PTEN, Caspase 3 and downregulate NF-κB, which is known to play an important role in tumor suppressor activity and cancer cell death.
CONCLUSIONS
In this study, theranostic nanoparticles for simultaneous cancer imaging and therapy were successfully performed with SPIONs as the core and FA-PAMAM conjugates on the periphery. To our knowledge, this is the first study that employs a simple and efficient chemistry to fabricate SPIONs decorated PAMAM dendrimers loaded with an active drug for MR imaging and targeted anticancer drug delivery. This method could be employed to encapsulate other hydrophobic anticancer drugs for targeted MRI and therapy in various type of cancers by using appropriate targeting ligands for specific recognition of unique biomarker overexpressed on cancer cells. In this study, the engineered magnetic nanoparticles SPIONs@FA-PAMAM showed great potential to be a promising MR contrast agent as well as an anticancer drug delivery system for CDF, a potent anticancer but highly lipophilic compound. The targeted nanoparticles SPIONs@FA-PAMAM possess numerous favorable characteristics such as improved aqueous solubility for delivering a high dose of CDF with high specificity to target cancer cells expressing folate receptors. As compared to SPIONs@PAMAM, SPIONs@FA-PAMAM showed a better ability to enhance MR contrast with a faster cellular uptake in in vitro T2-weighted images and fluorescence microscopic studies. In addition, targeted formulation SPIONs@FA-PAMAM-CDF showed a higher anticancer activity, a higher percentage of apoptotic and necrotic cells with the ability to upregulate PTEN, Caspase 3, and downregulate NF-κB, which could help overcome anticancer drug resistance and the recurrence of cancer after initial treatment. The results demonstrate promising potential of SPIONs@FA-PAMAM-CDF for simultaneous targeted MRI and therapy of folate receptor expressing cancers, warranting further in vivo investigations are underway in our laboratory.
Supplementary Material
ACKNOWLEDGMENTS
The authors wish to acknowledge the funding support from National Cancer Institute’s (NCI’s) Grant No. 1R21CA179652–01A1. We acknowledge partial support for this work by Wayne State University Start-up funding to A.K.I. We also acknowledge Dr. Fei Chen’s group at the Department of Pharmaceutical Sciences, Eugene Applebaum College of Pharmacy and Health Sciences, Wayne State University, for providing Caspase-3 antibody.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-mac.6b01885.
Western blot analysis and quantification of Caspase 3 (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Cancer Facts and Figures; American Cancer Society, 2015. [Google Scholar]
- (2).Mura S; Nicolas J; Couvreur P. Nat. Mater 2013, 12, 991. [DOI] [PubMed] [Google Scholar]
- (3).Peer D; Karp JM; Hong S; Farokhzad OC; Margalit R; Langer R. Nat. Nanotechnol 2007, 2, 751. [DOI] [PubMed] [Google Scholar]
- (4).Maeda H; Wu J; Sawa T; Matsumura Y; Hori KJ Controlled Release 2000, 65, 271. [DOI] [PubMed] [Google Scholar]
- (5).Kesharwani P; Xie L; Banerjee S; Mao G; Padhye S; Sarkar FH; Iyer AK Colloids Surf., B 2015, 136, 413. [DOI] [PubMed] [Google Scholar]
- (6).Liebowitz MR; Keller MB J. Clin. Psychiatry 1993, 54, 10. [Google Scholar]
- (7).Barick KC; Singh S; Bahadur D; Lawande MA; Patkar DP; Hassan PA J. Colloid Interface Sci 2014, 418, 120. [DOI] [PubMed] [Google Scholar]
- (8).Sharma P; Rana S; Barick KC; Kumar C; Salunke HG; Hassan PA New J. Chem 2014, 38, 5500. [Google Scholar]
- (9).Song EQ; Hu J; Wen CY; Tian ZQ; Yu X; Zhang ZL; Shi YB; Pang DW ACS Nano 2011, 5, 761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ren Y; Rivera JG; He L; Kulkarni H; Lee D-K; Messersmith PB BMC Biotechnol. 2011, 11, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Veiseh O; Gunn JW; Zhang M. Adv. Drug Delivery Rev 2010, 62, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Anirudhan TS; Dilu D; Sandeep SJ Magn. Magn. Mater 2013, 343, 149. [Google Scholar]
- (13).Laurent S; Forge D; Port M; Roch A; Robic C; Vander Elst L; Muller RN Chem. Rev 2008, 108, 2064. [DOI] [PubMed] [Google Scholar]
- (14).Arias JL; Gallardo V; Gomez-Lopera S. a; Plaza RC; Delgado a V J. Controlled Release 2001, 77, 309. [DOI] [PubMed] [Google Scholar]
- (15).Shimomura M; Abe T; Sato Y; Oshima K; Yamauchi T; Miyauchi S. Polymer 2003, 44, 3877. [Google Scholar]
- (16).Tanyolaç D; Özdural AR React. Funct. Polym 2000, 45, 235. [Google Scholar]
- (17).Wang SH; Shi X; Van Antwerp M; Cao Z; Swanson SD; Bi X; Baker JR Adv. Funct. Mater 2007, 17, 3043. [Google Scholar]
- (18).Kukowska-Latallo JF; Candido KA; Cao Z; Nigavekar SS; Majoros IJ; Thomas TP; Balogh LP; Khan MK; Baker JR Cancer Res. 2005, 65, 5317. [DOI] [PubMed] [Google Scholar]
- (19).Malik N; Evagorou EG; Duncan R. Anti-Cancer Drugs 1999, 10, 767. [PubMed] [Google Scholar]
- (20).Luong D; Kesharwani P; Deshmukh R; Mohd Amin MCI; Gupta U; Greish K; Iyer AK Acta Biomater. 2016, 43, 14. [DOI] [PubMed] [Google Scholar]
- (21).Ganta S; Devalapally H; Shahiwala A; Amiji MJ Controlled Release 2008, 126, 187. [DOI] [PubMed] [Google Scholar]
- (22).El-Sayed M; Ginski M; Rhodes C; Ghandehari HJ Controlled Release 2002, 81, 355. [DOI] [PubMed] [Google Scholar]
- (23).Matsumura Y; Maeda H. Cancer Res. 1986, 46, 6387. [PubMed] [Google Scholar]
- (24).Malik N; Wiwattanapatapee R; Klopsch R; Lorenz K; Frey H; Weener JW; Meijer EW; Paulus W; Duncan RJ Controlled Release 2000, 65, 133. [DOI] [PubMed] [Google Scholar]
- (25).Sato N; Kobayashi H; Hiraga A; Saga T; Togashi K; Konishi J; Brechbiel MW Magn. Reson. Med 2001, 46, 1169. [DOI] [PubMed] [Google Scholar]
- (26).Lu Y; Low PS Adv. Drug Delivery Rev 2012, 64, 342. [Google Scholar]
- (27).Holm J; Hansen SI; HoierMadsen M; Helkjaer PE; Nichols CW Biosci. Rep 1997, 17, 415. [DOI] [PubMed] [Google Scholar]
- (28).Holm J; Hansen SI; Hoier-Madsen M; Helkjaer PE; Bzorek M. APMIS 1995, 103, 663. [DOI] [PubMed] [Google Scholar]
- (29).Holm J; Hansen SI; Hoier-madsen M; Helkjaer PE; Bzorek M. Apmis 1995, 103, 663. [DOI] [PubMed] [Google Scholar]
- (30).Pan J; Feng SS Biomaterials 2008, 29, 2663. [DOI] [PubMed] [Google Scholar]
- (31).Mi Y; Liu Y; Feng SS Biomaterials 2011, 32, 4058. [DOI] [PubMed] [Google Scholar]
- (32).Srinivasarao M; Galliford CV; Low PS Nat. Rev. Drug Discovery 2015, 14, 203. [DOI] [PubMed] [Google Scholar]
- (33).Kesharwani P; Banerjee S; Padhye S; Sarkar FH; Iyer AK Colloids Surf., B 2015, 132, 138. [DOI] [PubMed] [Google Scholar]
- (34).Padhye S; Banerjee S; Chavan D; Pandye S; Swamy KV; Ali S; Li J; Dou QP; Sarkar FH Pharm. Res 2009, 26, 2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Padhye S; Yang H; Jamadar A; Cui QC; Chavan D; Dominiak K; McKinney J; Banerjee S; Dou QP; Sarkar FH Pharm. Res 2009, 26, 1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Luong D; Kesharwani P; Killinger BA; Moszczynska A; Sarkar FH; Padhye S; Rishi AK; Iyer AK J. Colloid Interface Sci. 2016, 484, 33. [DOI] [PubMed] [Google Scholar]
- (37).Li L; Braiteh FS; Kurzrock R. Cancer 2005, 104, 1322. [DOI] [PubMed] [Google Scholar]
- (38).Bao B; Ali S; Kong D; Sarkar SH; Wang Z; Banerjee S; Aboukameel A; Padhye S; Philip PA; Sarkar FH PLoS One 2011, 6, e17850. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- (39).Bao B; Ali S; Banerjee S; Wang Z; Logna F; Azmi AS; Kong D; Ahmad A; Li Y; Padhye S; Sarkar FH Cancer Res. 2012, 72, 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Kesharwani P; Banerjee S; Padhye S; Sarkar FH; Iyer AK Biomacromolecules 2015, 16, 3042. [DOI] [PubMed] [Google Scholar]
- (41).García-Díaz M; Nonell S; Villanueva Á; Stockert JC; Cañete M; Casadó A; Mora M; Sagristá ML Biochim. Biophys. Acta, Biomembr 2011, 1808, 1063. [DOI] [PubMed] [Google Scholar]
- (42).Mornet E; Carmoy N; Lainé C; Lemiègre L; Le Gall T; Laurent I; Marianowski R; Férec C; Lehn P; Benvegnu T; Montier T. Int. J. Mol Sci 2013, 14, 1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Leamon CP; Low PS Biochem. J 1993, 291 (3), 855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Zhao Y; Liu S; Li Y; Jiang W; Chang Y; Pan S; Fang X; Wang YA; Wang JJ Colloid Interface Sci. 2010, 350, 44. [DOI] [PubMed] [Google Scholar]
- (45).McCarthy S. a; Davies G-L; Gun’ko YK Nat. Protoc 2012, 7, 1677. [DOI] [PubMed] [Google Scholar]
- (46).Khodadust R; Unsoy G; Yalcin S; Gunduz G; Gunduz UJ Nanopart. Res 2013, 15, 1488. [Google Scholar]
- (47).Gao F; Pan BF; Zheng WM; Ao LM; Gu HC J. Magn. Magn. Mater 2005, 293, 48. [Google Scholar]
- (48).Mashhadizadeh MH J. Nanomed. Nanotechnol 2012, 03, 3. [Google Scholar]
- (49).Shen M; Cai H; Wang X; Cao X; Li K; Wang SH; Guo R; Zheng L; Zhang G; Shi X. Nanotechnology 2012, 23, 105601. [DOI] [PubMed] [Google Scholar]
- (50).Ignacio-de Leon PAA; Zharov I. Chem. Commun. (Cambridge, U. K.) 2011, 47, 553. [DOI] [PubMed] [Google Scholar]
- (51).Iyer AK; Greish K; Fang J; Murakami R; Maeda H. Biomaterials 2007, 28, 1871. [DOI] [PubMed] [Google Scholar]
- (52).Ganesh S; Iyer AK; Weiler J; Morrissey DV; Amiji MM Mol. Ther.-Nucleic Acids 2013, 2, e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Roy S; Yu Y; Padhye SB; Sarkar FH; Majumdar AP N. PLoS One 2013, 8, e68543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Yoo HS; Park TG J. Controlled Release 2004, 100, 247. [DOI] [PubMed] [Google Scholar]
- (55).Guo M; Que C; Wang C; Liu X; Yan H; Liu K. Biomaterials 2011, 32, 185. [DOI] [PubMed] [Google Scholar]
- (56).Kalli KR; Oberg AL; Keeney GL; Christianson TJH; Low PS; Knutson KL; Hartmann LC Gynecol. Oncol 2008, 108, 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Asadishad B; Vossoughi M; Alemzadeh I. Ind. Eng. Chem. Res 2010, 49, 1958. [Google Scholar]
- (58).Torchilin VP Multifunctional nanocarriers. Adv. Drug Delivery Rev 2012, 64, 302–315. [DOI] [PubMed] [Google Scholar]
- (59).Sun W; Mignani S; Shen M; Shi X. Drug Discovery Today 2016, 21, 1873. [DOI] [PubMed] [Google Scholar]
- (60).Wei Y; Han B; Hu X; Lin Y; Wang X; Deng X. Procedia Eng. 2012, 27, 632. [Google Scholar]
- (61).Liu W-M; Xue Y-N; Peng N; He W-T; Zhuo R-X; Huang S-WJ Mater. Chem 2011, 21, 13306. [Google Scholar]
- (62).Berry CC J. Mater. Chem 2005, 15, 543. [Google Scholar]
- (63).Sosnovik DE; Weissleder R. Curr. Opin. Biotechnol 2007, 18, 4. [DOI] [PubMed] [Google Scholar]
- (64).Konda SD; Aref M; Wang S; Brechbiel M; Wiener EC Magma 2001, 12, 104. [DOI] [PubMed] [Google Scholar]
- (65).Basly B; Felder-Flesch D; Perriat P; Billotey C; Taleb J; Pourroy G; Begin-Colin S. Chem. Commun 2010, 46, 985. [DOI] [PubMed] [Google Scholar]
- (66).Lee H; Shao H; Huang Y; Kwak B. IEEE Trans. Magn 2005, 41, 4102–4104. [Google Scholar]
- (67).Shi X; Wang SH; Swanson SD; Ge S; Cao Z; Van Antwerp ME; Landmark KJ; Baker JR Adv. Mater 2008, 20, 1671. [Google Scholar]
- (68).Huang X; Li L; Liu T; Hao N; Liu H; Chen D; Tang F. ACS Nano 2011, 5, 5390. [DOI] [PubMed] [Google Scholar]
- (69).Waite CL; Sparks SM; Uhrich KE; Roth CM BMC Biotechnol. 2009, 9, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Nagata Y; Lan KH; Zhou X; Tan M; Esteva FJ; Sahin AA; Klos KS; Li P; Monia BP; Nguyen NT; Hortobagyi GN; Hung MC; Yu D. Cancer Cell 2004, 6, 117. [DOI] [PubMed] [Google Scholar]
- (71).Collett GP; Campbell FC Carcinogenesis 2006, 27, 1285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.