Abstract
Autism spectrum disorder (ASD) is a neurodevelopmental disorder with considerable neuroanatomical heterogeneity. Thus, how and to what extent the brains of individuals with ASD differ from each other is still unclear. In this study, brain structural MRI data from 356 right‐handed, male subjects with ASD and 403 right‐handed male healthy controls were selected from the Autism Brain Image Data Exchange database (age range 5–35 years old). Voxel‐based morphometry preprocessing steps were conducted to compute the gray matter volume maps for each subject. Individual neuroanatomical difference patterns for each ASD individual were calculated. A data‐driven clustering method was next utilized to stratify individuals with ASD into several subtypes. Whole‐brain functional connectivity and clinical severity were compared among individuals within the ASD subtypes identified. A searchlight analysis was applied to determine whether subtyping ASD could improve the classification accuracy between ASD and healthy controls. Three ASD subtypes with distinct neuroanatomical difference patterns were revealed. Different degrees of clinical severity and atypical brain functional connectivity patterns were observed among these three subtypes. By dividing ASD into three subtypes, the classification accuracy between subjects of two out of the three subtypes and healthy controls was improved. The current study confirms that ASD is not a disorder with a uniform neuroanatomical signature. Understanding neuroanatomical heterogeneity in ASD could help to explain divergent patterns of clinical severity and outcomes.
Keywords: autism spectrum disorder, data‐driven, neuroanatomical heterogeneity, searchlight classification
1. INTRODUCTION
Autism spectrum disorder (ASD) is a condition characterized by deficits in social communication skills and restricted patterns of behavior, interests, or activities. Although a clear definition and reliable diagnostic criteria for ASD are available in the current version of Diagnostic and Statistical Manual of Mental Disorders (DSM), additional features are also associated with ASD including atypical language development and abilities, motor abnormalities and increased attention to detail (Lai, Lombardo, & Baron‐Cohen, 2014). Clinical manifestation in individuals with ASD is highly variable (Huerta & Lord, 2012). These individual differences within ASD have hindered attempts at understanding the neurobiological mechanisms of the disorder (Amaral, 2011).
It is generally recognized that ASD is a neurodevelopmental disorder, and atypical neuroanatomy has been widely documented in individuals with ASD (Radua, Via, Catani, & Mataix‐Cols, 2011). However, inconsistencies exist in the regional specificity of these neuroanatomical findings. Carper and Courchesne reported increased gray matter volume in dorsolateral prefrontal regions (Carper & Courchesne, 2005), while Girgis found decreased gray matter volume in the right lateral orbitofrontal cortex (Girgis et al., 2007), and another study exploring the size of orbitofrontal cortex in ASD reported no difference at the significance level of p < .05 (Hardan, et al., 2006). Factors such as age, gender, and IQ are thought to explain these inconsistent results (Amaral, Schumann, & Nordahl, 2008). Zhang et al.’s recent work based on a large‐scale multi‐site dataset reported a significant three‐way interaction of diagnosis by age by sex on hippocampal volumes. However, another multisite meta‐analysis reported no significant three‐way interaction of diagnosis by age by sex on subcortical volumes (Van Rooij, et al., 2017). Therefore at present, the effect of age and gender on brain volume in ASD is still unclear.
We hypothesized that individual differences among high‐functioning individuals with ASD might be another factor contributing to the highly variable neuroanatomical deficits reported in ASD. At the genetic level, ASD is thought to be linked with more than 400 kinds of genetic mutations (Bourgeron, 2015). A recent study using 26 different autism mouse models showed that different autism‐related genes caused different neuroanatomical abnormalities (Ellegood et al., 2015), indicating the genetic bases of structural heterogeneity in ASD. Subjects with ASD showing different clinical symptoms exhibited different neuroanatomical abnormalities (Katuwal, Baum, Cahill, & Michael, 2016). However, the extent of brain structural abnormality differences among individuals with ASD is unclear.
We hypothesized that high‐functioning individuals with ASD would exhibit different atypical patterns of neuroanatomy. Based on the different individual neuroanatomical patterns, we predicted that individuals with ASD could be subdivided into several subtypes. We further tested whether different neuroanatomical subtypes of ASD could be a source of the variance in the clinical severity and functional connectivity patterns observed within ASD. Finally, we tested whether this subtyping of ASD could improve the ability to identify structural imaging‐based biomarkers of ASD.
2. MATERIALS AND METHODS
2.1. Participant selection
Structural MRI data, resting‐state fMRI data and phenotype data were downloaded from the ABIDE‐I and ABIDE‐II databases (http://fcon_1000.projects.nitrc.org/indi/abide/) (Di Martino et al., 2017; Di Martino et al., 2014). The exclusion criteria were as follows: (i) female subjects; (ii) left‐ or mixed‐handed subjects and subjects with no handedness information; (iii) subjects older than 35 years of age; (iv) subjects missing full scale IQ (FIQ) data; (v) subjects with low quality structural images, as determined by an initial manual check on all structural images and a second manual check on voxel‐based morphometry (VBM) maps exhibiting low homogeneity; (vi) data collection sites with less than 25 Healthy Controls (HCs). After applying these exclusion criteria, data from 781 subjects (ASD = 356; HC = 425) from eight sites remained. Detailed information regarding participant demographics can be found in Table 1 and subject IDs are listed in anat_ID.txt.
Table 1.
Participant demographics
| Number of participants | ASD | HCa | p value | ||||
|---|---|---|---|---|---|---|---|
| 356 | 425 | – | |||||
| Mean | SD | Range | Mean | SD | Range | ||
| Age | 14.2 | 5.52 | 5–35 | 14.72 | 5.73 | 6–34 | 0.23 |
| FIQ | 105.1 | 16.1 | 69–148 | 112.5 | 12.0 | 79–144 | <0.001 |
| ADOSb: | – | ||||||
| Social | 7.70 | 2.73 | 2–14 | – | |||
| Communication | 3.58 | 1.59 | 0–9 | – | |||
| RRB | 1.95 | 1.68 | 0–8 | – | |||
| Total | 11.33 | 3.78 | 2–23 | ||||
| Severity | 7.18 | 2.08 | 1–10 | ||||
The p values were calculated using a two‐sample t‐test.
ADOS, autism diagnostic observation schedule; FIQ, full‐scale IQ; RRB, restricted repetitive behavior.
HC: an individual‐level match strategy was used in our study. Here HC means the candidate HC for ASD individuals.
ADOS total scores were available for 247 subjects; ADOS social scores were available for 223 subjects; ADOS communication scores were available for 223 subjects; ADOS RRB scores were available for 216 subjects. ADOS Severity scores were available for 211 subjects.
2.2. VBM map construction
VBM maps were constructed using structural MRI data from each subject. The Computational Anatomy Toolbox 12 (CAT12 http://dbm.neuro.uni-jena.de/cat/) was utilized to calculate the gray matter volumes of voxels in individual brains. The steps were: (1) manual coregistration to set the anterior commissure of each T1 image at the origin of the Montreal Neurological Institute (MNI) coordinate system; (2) segmentation of the coregistered images into gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF); (3) normalization of the GM and WM maps into MNI space; (4) modulation to convert the voxel values of tissue concentration (density) to volume; (5) smoothing with an isotropic Gaussian kernel (full width at half maximum = 8 mm).
We calculated the homogeneity of the segmented GM maps between subjects using the CAT12 toolbox. Maps with low mean homogeneity (lower than mean – 2 * SD of the maps from same site) with others were rechecked manually. Homogeneity is defined as the Pearson correlation coefficients between the normalized gray matter maps of each pair of subjects.
2.3. Structural difference map
As previous studies have revealed significant relationships between age, FIQ, gender, handedness, and brain structure (Gong, Sluming, Cezayirli, Mayes, & Roberts, 2001; Smith, Chebrolu, Wekstein, Schmitt, & Markesbery, 2007; Srinivasan, 1993), we first constructed the individual structural difference map for each subject with ASD (Figure 1). For each subject with ASD, we first selected n HCs from the same site with the most similar age and FIQ to the corresponding ASD subject. Gender and handedness were not considered, as we only included male, right‐handed subjects. Here, n is set to 20, as this number produced the most stable performance (details of the selection of n can be found in Supporting Information: Selection of the number of the corresponding HCs for each ASD subject).
Figure 1.
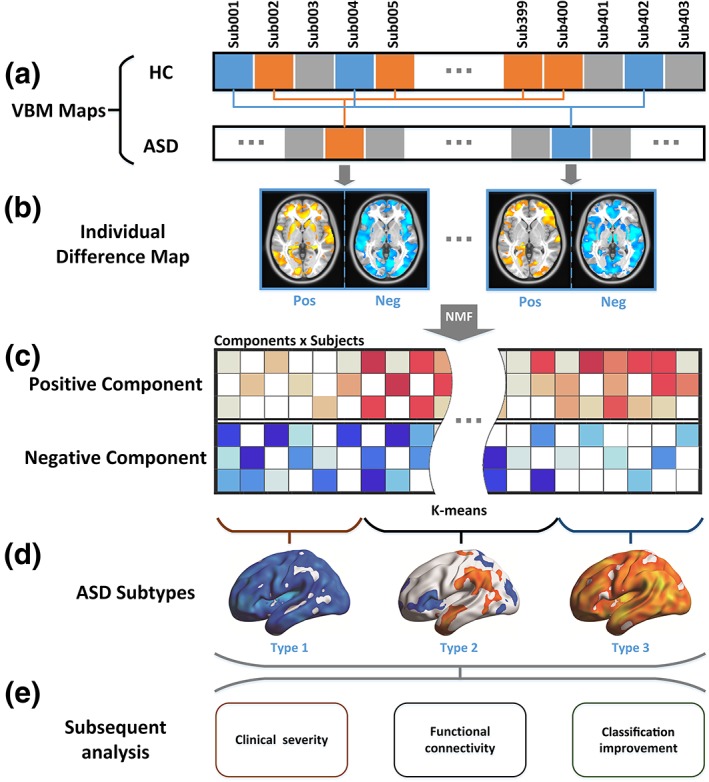
Data analysis flowchart analysis flowchart. (a) For each subject, a VBM map was calculated. For each ASD subject, 20 HC subjects who were most matched with the corresponding ASD subject were selected (see Methods: Structural difference map). In panel a, HC subjects with the same color with ASD subjects were selected as the most matched HC subjects. (b) For each ASD subject, a structural difference map was calculated. Then the structural difference maps were divided into positive and negative (see Methods: Structural difference map). (c) A non‐negative matrix factorization (NMF) method was used to reduce the feature dimensions. Red cells represent the coefficients of positive components and blue cells represent the coefficients of negative components. (d) The coefficients corresponding to the NMF components of each subject were used as features in cluster analyses (see Methods: Cluster analysis). (e) Subsequent analyses compared the behavior and brain functional traits of ASD subtypes (see Methods: Clinical severity and neuroanatomical subtypes of ASD and methods: Functional connectivity and neuroanatomical subtypes of ASD), and explored the advantages of subtyping ASD to classify ASD and HC based on the neuroanatomical images (see Method: Classification between subtypes of ASD and HC) [Color figure can be viewed at http://wileyonlinelibrary.com]
For each voxel of the gray matter map of each subject, we defined the structural difference value as:
where, VASD denotes the value of volume of the subject with ASD, represents the mean value of volumes of the HCs corresponding to the subject with ASD, std(VHC) represents the standard deviation of the values of volume of the HCs corresponding to the subject with ASD. The structural difference values Vd were then calculated to measure the deviation level of the volume of a given subject with ASD from matched HCs. To exclude the effects of covariates such as age and FIQ, a GLM method was used to regress out age, FIQ, site (using a dummy coding scheme such that the site of interest was assigned a value of 1 while all other sites were assigned 0) (Cohen, Cohen, West, & Aiken, 2003), the difference of age, FIQ between the ASD subjects and the corresponding HCs, the standard deviation of age, FIQ of the corresponding HCs for each ASD subject and a vector of ones representing the mean value for each voxel. As early overgrowth of brain volume is one feature of ASD (Courchesne, 2004), the total brain volume was not regressed here. After regression, the mean values were added back from the beta value of the vector of ones from the regression model. These steps were constrained in the gray matter mask obtained from the tissue probability map of SPM12 (probability threshold = 0.25). The cerebellum was excluded from analyses.
2.4. Non‐negative matrix factorization decomposition
We utilized a non‐negative matrix factorization (NMF) method to extract the major patterns of the structural difference patterns of ASD compared with HC (Lee & Seung, 1999). Compared with other decomposition methods like the Principal Components Analysis (PCA) and Independent Components Analysis (ICA), NMF is able to capture the non‐negative regional structural difference patterns (Lee & Seung, 1999). As sparsity is introduced by the non‐negative property, only the structural difference patterns that were commonly distributed in ASD subjects remained.
The NMF is defined as:
where, M is the original non‐negative matrix (here, the stacked difference map across subjects, where the matrix dimensions are subject number × voxel number), and W and H are the non‐negative factors which need to be calculated. Here, W represents the factorized component and H is the coefficient for each column of W for each subject.
We first divided the structural difference maps into positive and negative parts for each subject and then applied the NMF on the positive and negative structural difference maps separately to extract the major structural difference patterns across ASD subjects compared with HCs. As the optimal component number is unknown, we tried 20, 30, and 40 in accordance with previous comparable component factorization studies (Smith et al., 2009). We next manually checked the factorized patterns and chose a value of 30, resulting in 60 components of the structural difference map (30 positive and 30 negative) and the corresponding coefficients that represent the level of structural difference components. The cluster analysis was also conducted based on 20 and 40 components.
2.5. Cluster analysis
For each subject, we extracted 60 coefficients that measured the degree of the corresponding local structural difference patterns. We then used these 60 coefficients as features to explore the difference patterns across subjects using a cluster analysis.
A k‐means clustering method was utilized based on the 356 × 60 matrix (356 represents the subject number, 60 represents 60 features). To determine k, we calculated the Silhouette values using k from 2 to 20. According to the Silhouette criteria we found the optimal value of k to be 3 (Silhouette values are plotted in Supporting Information Figure S6). To avoid local minima, we repeated the clustering procedure 1,000 times and chose the best one. To validate the stability of the clustering solutions, we randomly picked 80% of the subjects and repeated the k‐means clustering on this randomly chosen subset. We then compared the resulting cluster index with the original cluster index and repeated this procedure 10 times. Results based on 40 (NMF components = 20) and 80 (NMF components = 40) features are shown in Supporting Information Figures S2 and S3.
To evaluate whether the cluster solution is affected by factors such as age and FIQ, we used a one‐way ANOVA to test the difference in age, FIQ, the difference of age, FIQ between ASD subjects and corresponding HCs, the standard deviation of age, and FIQ of the corresponding HCs for each ASD subject between the ASD subtypes obtained from the cluster analysis. We applied a chi‐square test to determine the difference of the ratio of sites between ASD subtypes.
2.6. Clinical severity and neuroanatomical subtypes of ASD
To assess whether symptom severity differed between subtypes of ASD, a one‐way ANOVA was utilized on the ADOS total, social, communication, RRB and severity (Gotham, Pickles, & Lord, 2009) scores, respectively. During this step, subjects with missing ADOS scores were excluded.
We then subdivided the ASD subjects into three sub‐groups according to the ADOS total score [44 subjects with the lowest ADOS total scores (below 8), 132 subjects with moderate ADOS total scores (8–13), and 71 subjects with the highest ADOS total scores (above 13); the number of subjects in each severity‐based subtype is the same as that of subjects in corresponding neuroanatomical subtypes who had ADOS information] and explored whether the different neuroanatomical difference patterns of the three ASD subtypes were fully driven by differences in clinical symptom severity. Details are shown in Supporting Information: Comparison of ASD subtypes based on clinical severity and neuroanatomic pattern.
2.7. Functional connectivity and neuroanatomical subtypes of ASD
We analyzed resting‐state fMRI data from the same subjects included in the structural MRI dataset. All resting‐state fMRI data were preprocessed using the DPARSF advanced edition (http://www.rfmri.org/DPARSF). The preprocessing, subject exclusion and functional network construction steps are shown in Supporting Information: Resting‐state fMRI data processing.
Next we tested whether individuals within the three different ASD subtypes would show different atypical patterns of FC networks. Here we utilized network based statistics (NBS) to assess atypical patterns at the connection level and graph theory to evaluate atypical patterns at the network level.
NBS offers substantially greater power than generic procedures for controlling family‐wise error rates (Zalesky, Fornito, & Bullmore, 2010). A four‐level one‐way ANOVA was applied on the HC subjects and subjects with the three ASD subtypes with age, FIQ, meanFD, site (using a dummy coding scheme) (Cohen, Cohen, West, & Aiken, 2003) as covariates (Li et al., 2018). Clusters with connection level p < .05 and cluster level p < .05 were considered as statistically significant. Simple effects analyses were performed to explore the differences between ASD subtypes and HC separately within the mask obtained from the one‐way ANOVA. For the simple effects analysis, clusters with connection level p < .05 and cluster level p < .05/3 (three simple effects were tested: HC vs. ASD subtype1, HC vs. ASD subtype2 and HC vs. ASD subtype3) were considered as statistically significant.
We next investigated FC network efficiency between HC and the three ASD subtypes. Here we calculated the global efficiency and local efficiency for each subject. First, the FC network was converted to a binary network according to a predefined cost value (range from 0.1 to 0.5). For each cost, a two‐sample t‐test was applied to assess the difference between HC and corresponding ASD subtypes (p < .05/3 was considered statistically significant).
2.8. Classification between subtypes of ASD and HC
A searchlight algorithm was applied to classify ASD and HC (Uddin et al., 2011) based on the VBM maps (details of the searchlight method can be found in Supporting Information). Here we performed four procedures: all ASD individuals versus HCs; individuals with ASD type1 versus HCs; individuals with ASD type2 versus HCs; individuals with ASD type3 versus HCs and compared the classification performance across these four trials.
We used a support‐vector machine (SVM) implemented in the LIBSVM software (http://www.csie.ntu.edu.tw/~cjlin/libsvm) with default parameters (Liao et al., 2018). Accuracy was assessed using a five‐fold cross‐validation. As the number of HCs is larger than the numbers of ASD and subtypes of ASD, we randomly chose HCs of the same number of ASD and performed the searchlight procedure. To exclude the bias caused by this random selection, this step was repeated 10 times and the final accuracy was defined as the mean accuracy of these 10 iterations. The values of the accuracy map were then converted to p values based on the Binomial distribution (Uddin et al., 2011). A Bonferroni FWE correction was applied to the converted p maps. p < .05 (FWE corrected) was considered statistically significant.
3. RESULTS
3.1. Clustering analysis based on individual structural difference patterns
We divided subjects with ASD into three subtypes using k‐means clustering applied to the individual structural difference patterns. The one‐sample T map of the structural difference maps for the three subtypes are shown in Figure 2b. Individuals with ASD subtype1 (63 subjects) showed decreased gray matter volume (GMV) particularly in the prefrontal lobes. Individuals with ASD subtype2 (190 subjects) showed both increased and decreased GMV; increases were observed in the temporal lobes while decreases were observed in occipital and prefrontal lobes. Individuals with ASD subtype3 (103 subjects) showed increased GMV compared with HCs in the temporal lobe. More detailed atypical neuroanatomical pattern descriptions can be found in Supporting Information. When repeating clustering on a subset of 80% of the whole data, we found that during all 10 iterations, at least 95% of subjects were clustered to the same subtype as when clustering was applied to 100% of the subjects.
Figure 2.
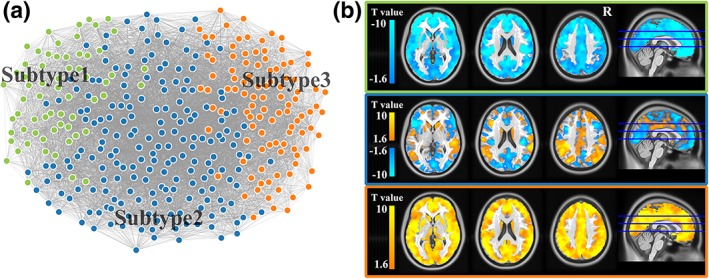
Distinct neuroanatomical difference patterns across three ASD subtypes. (a) Force‐directed graph of all ASD subjects created using d3.Js (https://d3js.org/). Green nodes represent ASD subjects belonging to subtype1; blue nodes represent ASD subjects belonging to subtype2; red nodes represent ASD subjects belonging to subtype3. Details of the force‐directed graph construction are in Supporting Information. (b) One‐sample t‐test of the neuroanatomical difference maps of each ASD subtype. Green box represents subtype1; blue box represents subtype2; red box represent subtype3 [Color figure can be viewed at http://wileyonlinelibrary.com]
We did not find significant differences in age, FIQ, site, the difference of age, FIQ between the ASD subjects and the mean of the corresponding HCs, the standard deviation of age, FIQ of the corresponding HCs for each ASD subject among the three ASD subtypes. Demographic information for subjects within the three subtypes is show in Table 2.
Table 2.
Participant demographics of three ASD subtypes
| ASD subtype1 | ASD subtype2 | ASD subtype3 | Group comparison (p value) | |
|---|---|---|---|---|
| Count | 63 | 190 | 103 | |
| Age | 13.96 ± 5.45 | 14.23 ± 5.56 | 14.40 ± 5.54 | 0.88a |
| FIQ | 104.60 ± 15.20 | 104.90 ± 16.70 | 105.77 ± 15.61 | 0.87a |
| Site | 2/8/3/8/7/10/10/6 | 4/20/9/46/16/26/25/15 | 3/9/2/23/9/8/18/15 | 0.73b |
| ADOS | ||||
| Social | 6.88 ± 2.85 | 7.52 ± 2.55 | 8.55 ± 2.79 | 0.005a |
| Communication | 3.20 ± 1.40 | 3.41 ± 1.44 | 4.14 ± 1.82 | 0.003a |
| RRB | 1.80 ± 1.83 | 1.72 ± 1.46 | 2.45 ± 1.85 | 0.017a |
| Total | 10.16 ± 3.53 | 11.03 ± 3.53 | 12.62 ± 4.98 | 0.001a |
| Severity | 6.94 ± 2.20 | 6.89 ± 2.14 | 7.84 ± 1.77 | 0.011a |
All numbers were presented as mean ± SD.
One‐way ANOVA test.
Chi‐square test.
3.2. Clinical symptom severity across three ASD subtypes
Using an ANOVA, we found significant differences in the ADOS total score (F = 6.97, p = .001), social score (F = 5.41, p = .005), communication score (F = 6.16, p = .003), RRB score (F = 4.18, p = .017), and severity score (F = 4.62, p = .011) among the three ASD subtypes (Figure 3). Using post‐hoc analyses, we found that the severity level of subtype3 was significantly higher than those of subtype1 and subtype2 (Figure 3 and Table 2).
Figure 3.
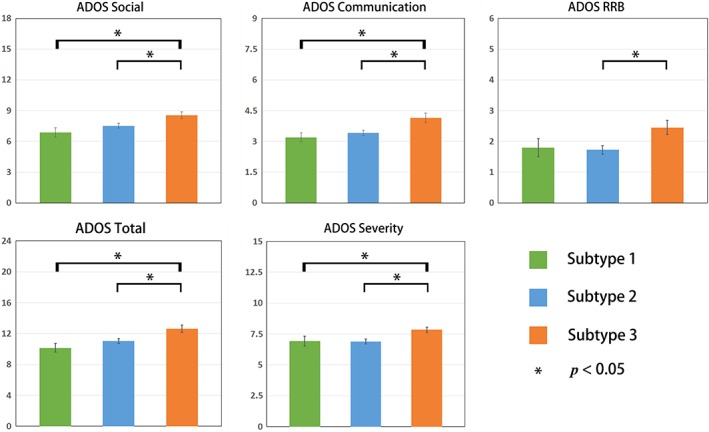
Different clinical symptom severity across three ASD subtypes. Mean ADOS scores for each ASD subtype. Error bars represent the standard error of the ADOS scores for each ASD subtype. * represents significant differences (two‐sample t‐test, p < .05) [Color figure can be viewed at http://wileyonlinelibrary.com]
We found that the neuroanatomical patterns across three severity–based subtypes were more similar (average correlation coefficient = 0.79, r 2 = 0.62) than the ones across the three neuroanatomical subtypes (average correlation coefficient = 0.49, r 2 = 0.24). The explanation rate (r 2) of neuroanatomy between severity‐based subtypes is much higher than the neuroanatomical subtypes, showing that the neuroanatomy between the neuroanatomical subtypes is more distinct than that observed in severity‐based subtypes. These results might imply that severity cannot fully explain the distinct neuroanatomy patterns observed between subjects within the three neuroanatomical subtypes identified.
3.3. Atypical functional connectivity patterns of three ASD subtypes
Using an NBS method, we found clusters of significant atypical functional connections in ASD subtype1 and subtype3 compared with HCs (connection level p < .05, cluster level p < .05/3, Figure 4a). Different atypical fronto‐parietal network connectivity patterns were found between subtype1 and subtype3 (Figure 4a). We also found atypical local efficiency in individuals with ASD subtype2 and subtype3 (Figure 4b) compared with HCs.
Figure 4.
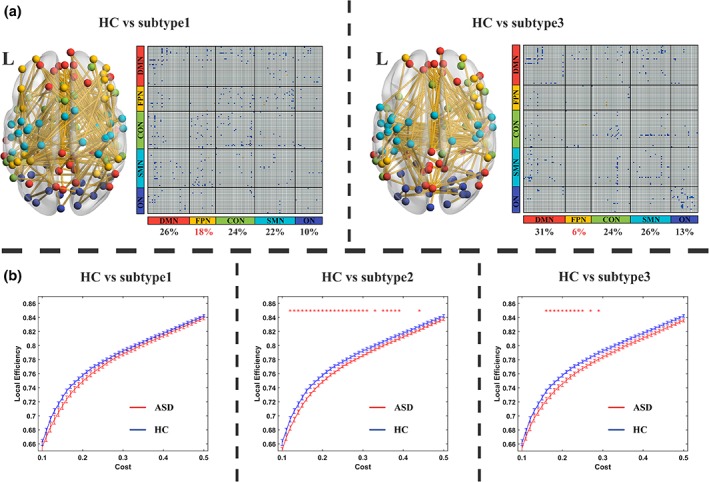
Different atypical functional connectivity patterns across three ASD subtypes. (a) Different atypical functional connectivity patterns between HC and ASD subtypes. Blue cells in the functional connectivity matrix represent the connectivity of ASD subtype, which is lower than HCs. The nodes in the brain maps represent the ROIs located in different sub‐networks: Red nodes represent ROIs in default mode network; yellow nodes represent ROIs in fronto‐parietal network; green nodes represent ROIs in cingulo‐opercular network; blue nodes represent ROIs in sensori‐motor network and purple nodes represent ROIs in occipital network. We found no FC clusters showing significant differences between ASD subtype2 and HC. (b) Local efficiency among three ASD subtypes. Red stars represent significant differences found at corresponding cost. Error bars represent the standard error [Color figure can be viewed at http://wileyonlinelibrary.com]
3.4. Improvement of classification between ASD and HC
Using a searchlight method, we achieved an accuracy of 58.8% when classifying the entire group of HC and ASD subjects. This accuracy indicates insignificant capacity to distinguish HC and ASD (no significant clusters were found at the threshold of p < .05, FWE corrected). After dividing the ASD subjects into three subtypes, the classification accuracies were improved (HC vs. ASD subtype1: 79.0%; HC vs. ASD subtype2: 60.2%; HC vs. ASD subtype3: 73.8%). Significant clusters at the threshold of p < .05 (FWE corrected) were found for HC versus ASD subtype1 and HC versus ASD subtype3. Brain regions are listed in Supporting Information Table S2, and Supporting Information Figures S4 and S5.
4. DISCUSSION
4.1. Analytic overview
In the present study, we clustered male, high‐functioning subjects with ASD into three subtypes based on individual neuroanatomical difference patterns. We found different profiles of clinical symptom severity and brain functional connectivity among these three ASD subtypes. After dividing the ASD group into three subtypes, we found that accuracy of classification between two out of the three ASD subtypes and HC was improved. These results provide evidence of ASD subtypes at the neuroanatomical level, and demonstrate different clinical presentation and brain function associated with different neuroanatomical subtypes of ASD. Moreover, we illustrate the potential benefit of subdividing ASD for the development of brain‐based biomarkers of the disorder.
4.2. Neuroanatomical subtypes of ASD
Accumulating studies have suggested that brain structural heterogeneity might be one characteristic of ASD (Amaral, 2011; Ellegood et al., 2015). Our results support this view, revealing neuroanatomical difference patterns across a large sample of ASD subjects. Moreover, we showed that the neuroanatomical difference patterns across ASD subjects can be clustered into three major distinct patterns.
From the onset of ASD, development heterogeneity exists. Some children who develop ASD later showed signs of developmental delays within the first 18 months of life. However, 25–40% of children with ASD showed near‐normal development at early ages and regress into ASD after 18 months (Werner & Dawson, 2005). A head circumference study reported the existence of three different ASD subtypes with different head circumference at early ages: increased (most children with ASD), decreased (only a small number of ASD children) and moderate head circumference (Muratori et al., 2009). A recent study using cortical surface information also supported the existence of distinct subtypes of ASD. Based on a dataset of 107 ASD subjects, they found three ASD subtypes with different cortical surface measures (Hong, Valk, Di Martino, Milham, & Bernhardt, 2017). For children with ASD who showed enlarged brain volume, inconsistent results were reported regarding whether this enlargement persists into later childhood and adolescence (Amaral et al., 2008). This inconsistency might be another source of the different neuroanatomical difference patterns we observe in ASD in this study. Studies attributed this variance to age (Aylward, Minshew, Field, Sparks, & Singh, 2002), intelligence level (Salmond, Vargha‐Khadem, Gadian, de Haan, & Baldeweg, 2007), gender (Lai et al., 2013), and other factors. Previous study showed that the development is an important source of neuroanatomical heterogeneity of ASD (Lin, Ni, Lai, Tseng, & Gau, 2015). Our results indicate that after controlling for these factors by only including male subjects, regression of age, fIQ and no difference of age, fIQ between clustered subtypes, the ASD group still showed inconsistent neuroanatomical difference patterns. Another ASD subtyping study based on surface morphometry which reported no significant difference of age, IQ, and site between ASD subtypes (Hong et al., 2017). These results might suggested that although age plays an important role on the neuroanatomical heterogeneity of ASD, the inter‐subjects difference exists during whole age range and covered the age‐heterogeneity in our study. Meanwhile more than 70% of individuals with ASD have concurrent medical, developmental, or psychiatric conditions (Lai et al., 2014), such as intellectual disability, attention‐deficit hyperactivity, epilepsy, anxiety, and so on. Complex comorbidity might be another source of the neuroanatomical subtypes of ASD observed here.
4.3. Clinical symptom severity and brain functional connectivity across ASD subtypes
Neuroanatomy is related to human cognition (Loh & Kanai, 2014; Omura, Todd Constable, & Canli, 2005) and ASD clinical symptomatology (Amaral et al., 2008). It is reasonable to assume that subjects within the three subtypes with different neuroanatomical patterns would exhibit different clinical patterns. By comparing mean ADOS scores, we found that subjects within different ASD subtypes did exhibit different clinical symptom severity levels. Although characteristics and diagnosis of ASD are clearly defined in the current version of the DSM (American Psychiatric Association, 2013), several studies report wide clinical heterogeneity among subjects with ASD, and this variance hinders the diagnosis and treatment of ASD (Cholemkery, Medda, Lempp, & Freitag, 2016; Georgiades et al., 2013; Kim, Macari, Koller, & Chawarska, 2015). Our results demonstrate that the different neuroanatomical difference patterns across ASD subjects may contribute to the observed heterogeneity in clinical presentation.
Brain structure constrains brain function. In the human brain, completing a specific task is associated with the functional integration of separate brain regions. We examined brain functional connectivity and found different atypical brain functional connectivity patterns across the three subtypes. For ASD, the inconsistent findings of brain functional connectivity have become a major obstacle to understanding the brain functional mechanisms of ASD (Hull, Jacokes, Torgerson, Irimia, & Van Horn, 2016). A recent study reported higher inter‐subject variance of brain functional connectivity in ASD and suggested that the higher inter‐subject variance might account for the inconsistent findings (Hahamy, Behrmann, & Malach, 2015). However, how inter‐subject variance influences brain functional connectivity in subjects with ASD is still unknown. Our results suggest that the variance of atypical brain function is related to the different atypical neuroanatomical patterns across subjects with ASD.
Our results showed different clinical characteristics and brain functional connectivity patterns across three neuroanatomical ASD subtypes. These findings might help us to understand inter‐subject clinical and brain functional variance in ASD.
4.4. Development of brain‐based biomarkers
To explore the potential application of subdividing ASD for aiding in ASD diagnosis, we showed that the classification accuracy between ASD and HC could be significantly improved after subdividing ASD group into subtypes. The diagnosis of ASD is currently based on the observation of behavioral features, which are scored by subjective criteria (American Psychiatric Association, 2013). It is inevitable that subjective bias can contribute to the outcome of ASD diagnosis, despite the careful application of established criteria (Uddin et al., 2017). Neuroanatomical images can provide potential objective biomarkers for ASD, and early attempts on smaller datasets have achieved high accuracy in distinguishing ASD and HC (Uddin et al., 2011). However, when applied to large multisite datasets, the reported accuracy decreases to a lower level (about 60%) (Haar, Berman, Behrmann, & Dinstein, 2016; Katuwal, Cahill, Baum, & Michael, 2015), close to the classification accuracy (58.8%) based on the entire ASD dataset in the current study. The improved accuracy after subdividing ASD into three subtypes suggests that neuroanatomical heterogeneity contributes to the low classification accuracy levels reported in earlier studies. The current results demonstrate the utility of subdividing ASD for the development of neuroanatomical ASD biomarkers.
In the present study, the subjects' ages ranged from 5 to 35 years old, and the mean age was much older than the age of ASD diagnosis (usually at the age of 4; Lai et al., 2014). Our results show that subtyping of ASD is not age‐related, and that accounting for individual neuroanatomical differences could potentially improve classification accuracy between ASD and HC. We hypothesized that subtyping could also improve classification between ASD and HCs. However, as there is a current lack of large datasets from young children with ASD, future studies based on children with ASD are needed.
4.5. Methodological considerations
Previous studies clustering ASD based on cortical features of ASD subjects have been conducted; one included 64 subjects with ASD (Hrdlicka et al., 2005) while another one included 107 subjects with ASD (Hong et al., 2017). Compared with these studies which used the original or normalized cortical features of ASD subjects, in the current work we characterized individual neuroanatomical atypical patterns of ASD individuals. Based on these individual neuroanatomical atypical patterns compared with HC and utilizing a purely data‐driven method without a priori knowledge, we obtained ASD subtypes with distinct neuroanatomical atypical patterns. The relatively large dataset compromising 356 individuals with ASD helped produce more stable results when exploring the heterogeneity of ASD. Moreover, we demonstrate improvements on previous work aiming to identify potential neuroanatomical ASD biomarkers by subtyping ASD. Additionally, the results of reproducibility analysis, such as the replication of different NMF components and clustering based on sub‐datasets, demonstrate the generalizability of our method.
4.6. Limitations
To our knowledge, the present study is the largest dataset to date that has been used to study neuroanatomical heterogeneity in ASD. However, the biased gender and handedness distribution made it hard to find female non‐right handed subjects within each site in the ABIDE database. Only including male right‐handed subjects limits our ability to investigate these factors that might further contribute to the heterogeneity of ASD. To overcome this problem, larger datasets which contain female and left‐handed subjects are needed.
5. CONCLUSION
In the present study, we explored the neuroanatomical heterogeneity of ASD and revealed three subtypes with distinct neuroanatomical difference patterns. Moreover, ASD subjects belonging to different subtypes also showed different clinical symptom severity and brain functional connectivity. By taking these ASD subtypes into account, ASD‐HC classification could be significantly improved. This work suggests the existence of ASD subtypes at the neuroanatomical level, and reveals the potential for such findings to not only improve our understanding of the neurobiology underlying ASD, but also to lead to the development of potential brain‐based biomarkers of the disorder.
Supporting information
Table S1. Participant demographics: resting state fMRI
Table.S2 Results of searchlight classification analysis
Fig.S1 Lines with colors represents the neuroanatomical difference value of each ASD subject. Black lines represent the mean first‐order difference of neuroanatomical difference values across ASD subjects.
Fig.S2 Distinct neuroanatomical difference patterns across three ASD subtypes based on 40 features
Fig.S3 Distinct neuroanatomical difference patterns across three ASD subtypes based on 80 features
Fig.S3 Searchlight results: HC vs. ASD subtype1
Fig.S4 Searchlight results: HC vs. ASD subtype3
Fig.S2 Silhouette plot of the kmeans cluster
Fig.S7 NMF components
Fig.S8 Detailed neuroanatomical difference patterns of the three ASD subtypes
ACKNOWLEDGMENT
This work was supported by the 863 project (2015AA020505), the National Natural Science Foundation of China (61533006, 61673089 and 81871432) and the Fundamental Research Funds for the Central Universities (Nos. ZYGX2016J187 and 2672018ZYGX2018J079), and the National Institute of Mental Health (NIMH R01MH107549 to LQU). Funding sources for the datasets comprising the 1000 Functional Connectome Project are listed at fcon_1000.projects.http://nitrc.org/fcpClassic/FcpTable.html. Funding sources for the ABIDE dataset are listed at fcon_1000.projects.http://nitrc.org/indi/abide. All authors declared no interest of conflicts.
Chen H, Uddin LQ, Guo X, et al. Parsing brain structural heterogeneity in males with autism spectrum disorder reveals distinct clinical subtypes. Hum Brain Mapp. 2019;40:628–637. 10.1002/hbm.24400
Contributor Information
Xujun Duan, Email: duanxujun@uestc.edu.cn.
Huafu Chen, Email: chenhf@uestc.edu.cn.
REFERENCES
- Amaral, D. G. (2011). The promise and the pitfalls of autism research: An introductory note for new autism researchers. Brain Research, 1380, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral, D. G. , Schumann, C. M. , & Nordahl, C. W. (2008). Neuroanatomy of autism. Trends in Neurosciences, 31, 137–145. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association . (2013). The Diagnostic and Statistical Manual of Mental Disorders: DSM 5. bookpointUS (p. 991). Washington, DC: Author. [Google Scholar]
- Aylward, E. H. , Minshew, N. J. , Field, K. , Sparks, B. F. , & Singh, N. (2002). Effects of age on brain volume and head circumference in autism. Neurology, 59, 175–183. [DOI] [PubMed] [Google Scholar]
- Bourgeron, T. (2015). From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nature Reviews. Neuroscience, 16, 551–563. [DOI] [PubMed] [Google Scholar]
- Carper, R. A. , & Courchesne, E. (2005). Localized enlargement of the frontal cortex in early autism. Biological Psychiatry, 57, 126–133. [DOI] [PubMed] [Google Scholar]
- Cholemkery, H. , Medda, J. , Lempp, T. , & Freitag, C. M. (2016). Classifying autism Spectrum disorders by ADI‐R: Subtypes or severity gradient? Journal of Autism and Developmental Disorders, 46, 2327–2339. [DOI] [PubMed] [Google Scholar]
- Cohen, J. , Cohen, P. , West, S. G. , & Aiken, L. S. (2003). Applied multiple regression/correlation analysis for the behavioral sciences (3rd ed., pp. 227–229). Mahwah, NJ: L. Erlbaum Associates. [Google Scholar]
- Courchesne, E. (2004). Brain development in autism: Early overgrowth followed by premature arrest of growth. Mental Retardation and Developmental Disabilities Research Reviews, 10, 106–111. [DOI] [PubMed] [Google Scholar]
- Di Martino, A. , O'Connor, D. , Chen, B. , Alaerts, K. , Anderson, J. S. , Assaf, M. , … Milham, M. P. (2017). Enhancing studies of the connectome in autism using the autism brain imaging data exchange II. Scientific Data, 4, 170010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino, A. , Yan, C. G. , Li, Q. , Denio, E. , Castellanos, F. X. , Alaerts, K. , … Milham, M. P. (2014). The autism brain imaging data exchange: Towards a large‐scale evaluation of the intrinsic brain architecture in autism. Molecular Psychiatry, 19, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegood, J. , Anagnostou, E. , Babineau, B. A. , Crawley, J. N. , Lin, L. , Genestine, M. , … Lerch, J. P. (2015). Clustering autism: Using neuroanatomical differences in 26 mouse models to gain insight into the heterogeneity. Molecular Psychiatry, 20, 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades, S. , Szatmari, P. , Boyle, M. , Hanna, S. , Duku, E. , Zwaigenbaum, L. , … Pathways A.S.D.S.T . (2013). Investigating phenotypic heterogeneity in children with autism spectrum disorder: A factor mixture modeling approach. Journal of Child Psychology and Psychiatry, and Allied Disciplines, 54, 206–215. [DOI] [PubMed] [Google Scholar]
- Girgis, R. R. , Minshew, N. J. , Melhem, N. M. , Nutche, J. J. , Keshavan, M. S. , & Hardan, A. Y. (2007). Volumetric alterations of the orbitofrontal cortex in autism. Progress in Neuro‐Psychopharmacology & Biological Psychiatry, 31, 41–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Q. , Sluming, V. , Cezayirli, E. , Mayes, A. R. , & Roberts, N. (2001). Voxel based morphometry study of relationship between brain structure and IQ. NeuroImage, 13, 411. [Google Scholar]
- Gotham, K. , Pickles, A. , & Lord, C. (2009). Standardizing ADOS scores for a measure of severity in autism spectrum disorders. Journal of Autism and Developmental Disorders, 39, 693–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haar, S. , Berman, S. , Behrmann, M. , & Dinstein, I. (2016). Anatomical abnormalities in autism? Cerebral Cortex, 26, 1440–1452. [DOI] [PubMed] [Google Scholar]
- Hahamy, A. , Behrmann, M. , & Malach, R. (2015). The idiosyncratic brain: Distortion of spontaneous connectivity patterns in autism spectrum disorder. Nature Neuroscience, 18, 302–309. [DOI] [PubMed] [Google Scholar]
- Hardan, A. Y. , Girgis, R. R. , Lacerda, A. L. , Yorbik, O. , Kilpatrick, M. , Keshavan, M. S. , & Minshew, N. J. (2006). Magnetic resonance imaging study of the orbitofrontal cortex in autism. Journal of Child Neurology, 21, 866–871. [DOI] [PubMed] [Google Scholar]
- Hong, S. J. , Valk, S. L. , Di Martino, A. , Milham, M. P. , & Bernhardt, B. C. (2017). Multidimensional neuroanatomical subtyping of autism Spectrum disorder. Cerebral Cortex, 14, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdlicka, M. , Dudova, I. , Beranova, I. , Lisy, J. , Belsan, T. , Neuwirth, J. , … Urbanek, T. (2005). Subtypes of autism by cluster analysis based on structural MRI data. European Child & Adolescent Psychiatry, 14, 138–144. [DOI] [PubMed] [Google Scholar]
- Huerta, M. , Lord, C. (2012) Diagnostic evaluation of autism spectrum disorders. Pediatric Clinics of North America, 59:103–11, xi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull, J. V. , Jacokes, Z. J. , Torgerson, C. M. , Irimia, A. , & Van Horn, J. D. (2016). Resting‐state functional connectivity in autism Spectrum disorders: A review. Frontiers in Psychiatry, 7, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katuwal, G. J. , Baum, S. A. , Cahill, N. D. , & Michael, A. M. (2016). Divide and conquer: Sub‐grouping of ASD improves ASD detection based on brain Morphometry. PLoS One, 11, e0153331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katuwal, G. J. , Cahill, N. D. , Baum, S. A. , & Michael, A. M. (2015) The predictive power of structural MRI in Autism diagnosis. Conference proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society (pp. 4270–4233). 2015.. [DOI] [PubMed]
- Kim, S. H. , Macari, S. , Koller, J. , & Chawarska, K. (2015). Examining the phenotypic heterogeneity of early autism Spectrum disorder: Subtypes and short‐term outcomes. Journal of Child Psychology and Psychiatry, and Allied Disciplines, 57, 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, M. C. , Lombardo, M. V. , & Baron‐Cohen, S. (2014). Autism. Lancet, 383, 896–910. [DOI] [PubMed] [Google Scholar]
- Lai, M. C. , Lombardo, M. V. , Suckling, J. , Ruigrok, A. N. , Chakrabarti, B. , Ecker, C. , … Baron‐Cohen, S. (2013). Biological sex affects the neurobiology of autism. Brain: A Journal of Neurology, 136, 2799–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D. D. , & Seung, H. S. (1999). Learning the parts of objects by non‐negative matrix factorization. Nature, 401, 788–791. [DOI] [PubMed] [Google Scholar]
- Li, R. , Liao, W. , Yu, Y. , Chen, H. , Guo, X. , Tang, Y. L. , & Chen, H. (2018). Differential patterns of dynamic functional connectivity variability of striato‐cortical circuitry in children with benign epilepsy with centrotemporal spikes. Human Brain Mapping, 39, 1207–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, W. , Li, J. , Duan, X. , Cui, Q. , Chen, H. , & Chen, H. (2018). Static and dynamic connectomics differentiate between depressed patients with and without suicidal ideation. Human Brain Mapping, 39, 300–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H. Y. , Ni, H. C. , Lai, M. C. , Tseng, W. I. , & Gau, S. S. (2015). Regional brain volume differences between males with and without autism spectrum disorder are highly age‐dependent. Molecular Autism, 6, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh, K. K. , & Kanai, R. (2014). Higher media multi‐tasking activity is associated with smaller gray‐matter density in the anterior cingulate cortex. PLoS One, 9, e106698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratori, F. , Telleschi, M. , Santocchi, E. , Tancredi, R. , Igliozzi, R. , Parrini, B. , … Calderoni, S. . (2009). Early development of head circumference in autistic children: Searching clinical subtypes. Conference: International Meeting for Autism Research; Filippo Muratori at Università di Pisa. Filippo Muratori. 40.51.
- Omura, K. , Todd Constable, R. , & Canli, T. (2005). Amygdala gray matter concentration is associated with extraversion and neuroticism. Neuroreport, 16, 1905–1908. [DOI] [PubMed] [Google Scholar]
- Radua, J. , Via, E. , Catani, M. , & Mataix‐Cols, D. (2011). Voxel‐based meta‐analysis of regional white‐matter volume differences in autism spectrum disorder versus healthy controls. Psychological Medicine, 41, 1539–1550. [DOI] [PubMed] [Google Scholar]
- Salmond, C. H. , Vargha‐Khadem, F. , Gadian, D. G. , de Haan, M. , & Baldeweg, T. (2007). Heterogeneity in the patterns of neural abnormality in autistic spectrum disorders: Evidence from ERP and MRI. Cortex, 43, 686–699. [DOI] [PubMed] [Google Scholar]
- Smith, C. D. , Chebrolu, H. , Wekstein, D. R. , Schmitt, F. A. , & Markesbery, W. R. (2007). Age and gender effects on human brain anatomy: A voxel‐based morphometric study in healthy elderly. Neurobiology of Aging, 28, 1075–1087. [DOI] [PubMed] [Google Scholar]
- Smith, S. M. , Fox, P. T. , Miller, K. L. , Glahn, D. C. , Fox, P. M. , Mackay, C. E. , … Beckmann, C. F. (2009). Correspondence of the brain's functional architecture during activation and rest. Proceedings of the National Academy of Sciences of the United States of America, 106, 13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan, K. (1993). Lateralisation of speech Centre in left‐handedness due to cerebral and extracerebral lesions. Acta Neurochirurgica. Supplementum, 56, 83–84. [DOI] [PubMed] [Google Scholar]
- Uddin, L. Q. , Dajani, D. R. , Voorhies, W. , Bednarz, H. , & Kana, R. K. (2017). Progress and roadblocks in the search for brain‐based biomarkers of autism and attention‐deficit/hyperactivity disorder. Translational Psychiatry, 7, e1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin, L. Q. , Menon, V. , Young, C. B. , Ryali, S. , Chen, T. , Khouzam, A. , … Hardan, A. Y. (2011). Multivariate searchlight classification of structural magnetic resonance imaging in children and adolescents with autism. Biological Psychiatry, 70, 833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rooij, D. , Anagnostou, E. , Arango, C. , Auzias, G. , Behrmann, M. , Busatto, G. F. , … Buitelaar, J. K. (2017). Cortical and subcortical brain morphometry differences between patients with autism Spectrum disorder and healthy individuals across the lifespan: Results from the ENIGMA ASD working group. The American Journal of Psychiatry, 175, 359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, E. , & Dawson, G. (2005). Validation of the phenomenon of autistic regression using home videotapes. Archives of General Psychiatry, 62, 889–895. [DOI] [PubMed] [Google Scholar]
- Zalesky, A. , Fornito, A. , & Bullmore, E. T. (2010). Network‐based statistic: Identifying differences in brain networks. NeuroImage, 53, 1197–1207. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Participant demographics: resting state fMRI
Table.S2 Results of searchlight classification analysis
Fig.S1 Lines with colors represents the neuroanatomical difference value of each ASD subject. Black lines represent the mean first‐order difference of neuroanatomical difference values across ASD subjects.
Fig.S2 Distinct neuroanatomical difference patterns across three ASD subtypes based on 40 features
Fig.S3 Distinct neuroanatomical difference patterns across three ASD subtypes based on 80 features
Fig.S3 Searchlight results: HC vs. ASD subtype1
Fig.S4 Searchlight results: HC vs. ASD subtype3
Fig.S2 Silhouette plot of the kmeans cluster
Fig.S7 NMF components
Fig.S8 Detailed neuroanatomical difference patterns of the three ASD subtypes


