Abstract
The landmark 1969 discovery of nuclear RNA polymerases I, II and III in diverse eukaryotes represented a major turning point in the field that, with subsequent elucidation of the distinct structures and functions of these enzymes, catalyzed an avalanche of further studies. In this Review, written from a personal and historical perspective, I highlight foundational biochemical studies that led to the discovery of an expanding universe of the components of the transcriptional and regulatory machineries, and a parallel complexity in gene-specific mechanisms that continue to be explored to the present day.
The cell-specific expression of protein-coding genes (~20,000 in humans) is central to many biological processes and is regulated primarily at the level of transcription. The earliest insights into the mechanism and regulation of transcription came from studies of bacteria and bacteriophages, and most of these had their origins in genetic studies, exemplified by the landmark Jacob and Monod publication on the lac operon in 19611. Complementary genetic and biochemical studies then led to the biochemical identification and characterization of both negative and positive gene-specific regulatory factors that included the classic lac repressor, lambda repressor, araC and CAP proteins. Given the rapid pace of bacterial transcription studies relative to eukaryotic transcription in the 1960s, it is relevant to note that the first true RNA polymerase activity, showing incorporation of all four nucleotides from nucleoside triphosphates, was discovered by Sam Weiss in rat liver nuclei in 1959 (ref.2) and later characterized as a derived aggregate (likely chromatin)3. However, most of the immediately following studies were focused on newly reported bacterial RNA polymerase preparations that, unlike the liver enzyme, were soluble, far more abundant and showed a clear DNA dependence4. Remarkably, despite the importance and relative abundance of the single bacterial RNA polymerase, it was not until 1969 that it was finally purified to homogeneity by Richard Burgess and then shown to execute specific transcription initiation dependent on the dissociable sigma subunit5.
In contrast to the progress in understanding bacterial transcription, we had only a primitive understanding of eukaryotic transcription by the late 1960s, with a basic appreciation of three general classes of RNA (rRNA, tRNA and mRNA), as in bacteria, but little else. Having gotten interested in transcription in animal cells as a graduate student with Bill Rutter, whose lab was focused on problems other than transcription, the big question to me was whether eukaryotic transcription would be fundamentally different from prokaryotic transcription with respect to the enzymology and regulatory factors. An initial and surprising answer came in 1969, with my discovery of three chromatographically distinct nuclear RNA polymerases that later proved to have distinct subunit structures and functions in the synthesis of the major classes of RNA6. This was, at least to me, the ‘big bang’ that immediately generated a broad surge of interest in eukaryotic transcription mechanisms. In the following sections, I provide a historical perspective on subsequent transcription studies that, at least in my laboratory, emphasized a reductionist biochemical approach based on the Feynman philosophy of “what I cannot create [build], I cannot understand,” and revealed an unexpected, ever-increasing complexity of the basic transcriptional machinery and of the associated regulatory proteins and mechanisms. These biochemical approaches were instrumental in setting the stage for the large number of later (for example, genomic, genetic, biophysical/structural and imaging) approaches by identifying the major components of the transcriptional and regulatory machineries used as key reagents in these analyses and by providing initial insights and concepts regarding their mechanisms.
RNA polymerases: diversity, structure and general functions
After the seminal Weiss discovery of the rat liver RNA polymerase activity, and despite several reports of a soluble DNA-dependent mammalian polymerase activity, most studies of eukaryotic RNA polymerase focused on RNA synthesis from endogenous templates in isolated nuclei (nuclear ‘run-on’ assays). In this regard, Widnell and Tata7 reported two enzyme ‘activities’ based on the levels and base compositions of the RNAs synthesized by endogenous RNA polymerase in isolated nuclei at drastically different salt and metal ion conditions. However, these analyses did not distinguish the function of distinct enzymes on different chromosomal templates from the function of a common enzyme on distinct chromosomal templates that were differentially sensitive to salt and metal ion affects. In my early graduate studies, I too had been using isolated nuclei to study levels of RNA synthesis during hormonal responses in rat liver and during sea urchin development. However, an inability in that pre-cloning era to adequately study the synthesis of specific RNAs, in particular mRNAs, led me to think about getting to the heart of the matter—the RNA polymerase. Careful consideration of the 1960 Weiss paper, as well as other mammalian RNA polymerase studies that reported only trace amounts of soluble enzyme following the standard low-salt extraction procedures used for bacteria, led me to suspect (correctly) that most of the mammalian polymerase was chromatin-bound. After the systematic development of methods for the complete high-salt solubilization of the nuclear RNA polymerase activity, and separation from DNA and histones, I was able to identify three chromatographically distinct RNA polymerases (in sea urchin embryos, rat liver and yeast8) that exhibited different metal ion and DNA template preferences. This was a truly ‘eureka’ moment for me—both assuring me a respectable thesis and swelling my mind with many future experiments. We reported this exciting discovery at an international meeting in April 1969 (ref.9) and later in a 1969 Nature article6 that, remarkably and memorably, was originally rejected on the grounds that it was not of general interest! In subsequent studies further indicative of distinct enzymes, the laboratory of Pierre Chambon reported two forms of RNA polymerase that, respectively, were insensitive (Pol A) and sensitive (Pol B) to a low concentration of the mushroom toxin α-amanitin10·, and the Rutter lab similarly showed that Pols I and III were resistant, while Pol II was sensitive, to low concentrations of α-amanitin11. The discovery of three nuclear RNA polymerases was the first of many clues that the logistics of transcriptional regulation might be fundamentally different in prokaryotes and eukaryotes.
The identification of the three nuclear RNA polymerases clearly set the stage for further studies of structure and function, and the Rutter12 and Chambon13 labs provided initial evidence for distinct (but grossly incomplete) multisubunit structures for mammalian Pol I and Pol II. Although I, too, appreciated the importance of understanding the structures of the enzymes, I nonetheless felt that an exciting next step would be to show that a specific polymerase could selectively transcribe a specific gene in vitro—a seemingly reasonable possibility in light of the recent success with the bacterial RNA polymerase5. Having established that Pol I was localized to the nucleolus and thus likely responsible for ribosomal RNA synthesis14, I proceeded to the laboratory of Don Brown for postdoctoral studies, anticipating that I would soon have purified Pol I accurately transcribing the large ribosomal RNA genes (the exclusive Pol I targets) that had been purified in his laboratory. Surprisingly—and disappointingly—these analyses revealed only a low level of random symmetric transcription by both Pol I and the control Pol II15, leading me to suspect requirements for still other factors and providing an early foreboding of the difficult task that lay ahead. Consequently, I thought it imperative to better define both the structures and, especially, the functions of the three enzymes; and subsequent studies in my own lab at Washington University were directed toward these objectives. By 1974 we had purified the complete trio of Pols I, II and III to homogeneity for the first time and had shown that they contain distinct subunit structures, but with some apparently common subunits based on size16. In parallel studies—and with our discovery that mammalian Pol III showed an α-amanitin sensitivity between that of Pol II (highly sensitive) and Pol I (completely insensitive)—we rigorously (with titration curves) compared sensitivities of the synthesis of specific classes of RNA in isolated nuclei with sensitivities of the purified enzymes, and established distinct functions for Pol I, Pol II, and Pol III in the synthesis, respectively, of large ribosomal RNAs, pre-mRNA and 5S and tRNAs17,18. These critical results immediately confirmed my earlier rationale for the evolution of three structurally and functionally distinct RNA polymerases, namely to provide a mechanism to independently control synthesis levels of the major classes of RNA (for example, in growth-state changes). Of more immediate importance, these results provided clear direction for further attempts, in my own and other labs, to establish specific transcription initiation by focusing on specific polymerase-gene combinations.
With the advent of advances in biochemistry, molecular biology, cloning and genome sequencing, the polypeptide sequences of the Pol I, II and III subunits were revealed—initially for yeast in the early 1990s in the laboratories of Rick Young and Andre Sentenec, and later for higher organisms (including mammals)19–21. Notably, these studies revealed a strong evolutionary conservation of the subunits of the individual enzymes, including both common subunits and enzyme-specific subunits that account for the diverse functions in transcription. Moreover, some of the enzyme-specific subunits were found to be related to each other and also to be counterparts of bacterial polymerase subunits, reflecting conserved catalytic mechanisms. A particularly pleasing development, especially with my longstanding commitment to the eukaryotic RNA polymerases, was the solution of high-resolution structures of the enzymes—beginning with the groundbreaking studies of Roger Kornberg and colleagues on Pol II22,23, and continuing with related studies of Pol I24,25 and Pol III26. Apart from revealing fundamental principles of structure and catalysis similar to those established earlier for the bacterial RNA polymerase27, these studies have provided important insights into similarities and differences between the three enzymes.
RNA polymerase accessory factors: diversity, structure and function
Given the enormous complexity of Pols I, II and III (14, 12 and 17 subunits, respectively) relative to the initiation-competent five-subunit bacterial RNA polymerase, an early surprise to me was the inability of these enzymes to accurately initiate transcription on defined core promoter elements, especially in the case of Pol I with only one target gene. However, a major breakthrough was our 1976 demonstration that the highly reiterated oocyte-specific 5S RNA genes (20,000 genomic copies) in purified Xenopus oocyte chromatin (but not in the histone-free DNA) could be accurately and selectively transcribed by purified Pol III (but not by Pols I and II)—clearly indicating the existence of essential accessory factors stably associated with active chromatin and the feasibility of successful in vitro transcription assays with purified polymerases28,29. These gratifying and highly stimulating studies were soon followed by our demonstration that purified or cloned DNA templates could be accurately transcribed by purified Pol III30 and Pol II31 in conjunction with RNA-polymerase-free subcellular extracts, again indicating the involvement of essential undefined factors that I was convinced could be identified through further biochemistry. Contemporaneous studies by others also reported accurate transcription of purified or cloned genes in crude cellular extracts, although they provided no indication that any factors other than the endogenous Pol III32,33 or Pol II34 were necessary for the observed transcription. Slightly later studies by Ingrid Grummt, after a sojourn in my laboratory, also reported the ability of our purified Pol I to specifically transcribe large ribosomal RNA genes (rDNA) in the presence of a cellular extract35.
These studies provided systems for many labs to begin mapping corresponding core promoter elements in vitro, in parallel to other promoter mapping studies using cell-based assays with transfected or microinjected genes. However, they immediately raised the question of the nature of the essential factors, and I wondered whether those factors might be similar or different for the three polymerases and possibly something so simple as a missing sigma-like factor as in the case of the bacterial enzyme. This was the next most interesting question for me, and immediate studies were focused on Pol III and Pol II. In further seminal studies, the biochemical fractionation of mammalian cell extracts in my laboratory led to the first identification of general initiation factors specific for individual RNA polymerases, including TFIIIB and TFIIIC for Pol III36, and several factors (including TFIID) for Pol II37. And confirming predictions from my 1970 studies15, but still somewhat surprising in view of the dedication of Pol I to the large rRNA genes, a later study from the lab of Masami Muramatsu also reported multiple accessory factors for Pol I transcription38. Notably, these pioneering studies, with their foundations in my laboratory, opened the floodgates for further studies of the Pol I, II and III factors in organisms from yeast to man, and contributions from many laboratories led to their complete resolution, purification and (in yeast and human) cognate cDNA cloning. It deserves emphasis that, with few exceptions, the identification of these factors was based on functional in vitro complementation assays with purified polymerases and chromatographic fractions and that the ultimate (and crucial) validation of the functional relevance of the identified polypeptides involved cognate cDNA cloning, expression and functional analyses.
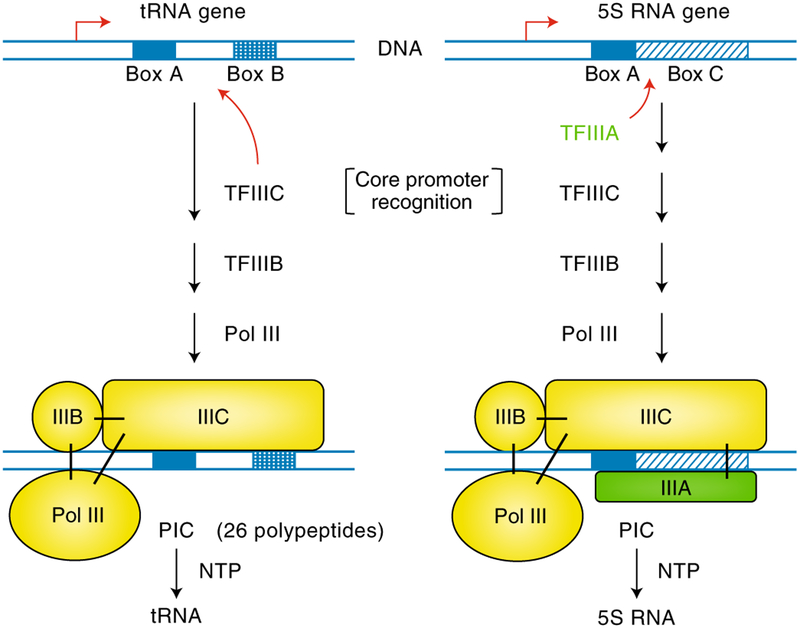
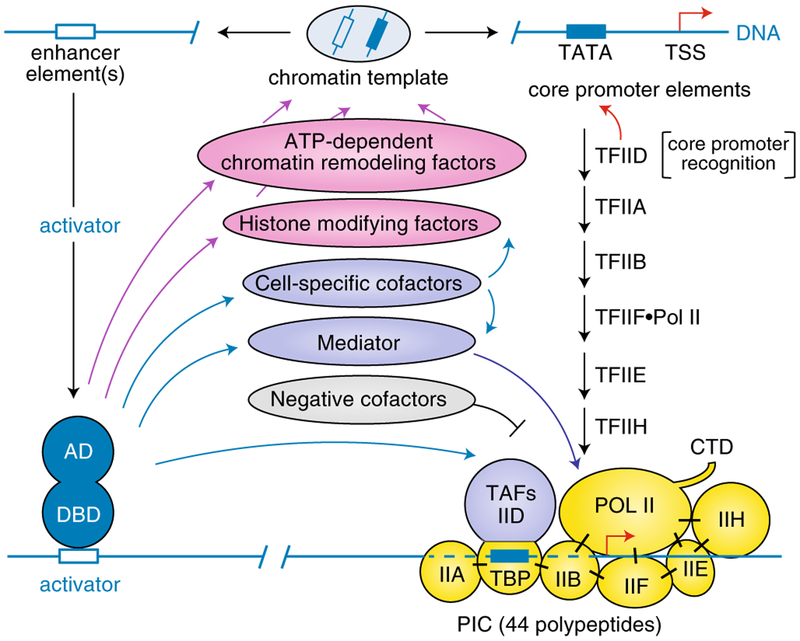
A surprising finding from these studies, again emphasizing divergence from the bacterial paradigm, was the large number and subunit complexity of the general initiation factors: TFIIIC and TFIIIB (9 polypeptides) for Pol III39–41, TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH (32 polypeptides) for Pol II42–44, and SL1/TIF1B/core factor, TIF1A/rRn3 and UBF/UAF (~6 polypeptides) for Pol I45,46. The availability of these factors, even before complete purification, led to elucidation of pathways for the assembly of functional preinitiation complexes (PICs), initially for Pol III in my lab47,48 (reviewed in refs.39–41) and later for Pol II49–51 (reviewed in refs.42–44) and Pol I (reviewed in refs.45,46). Pathways for PIC formation on a Pol III-transcribed tRNA gene promoter and on a Pol II-transcribed TATA-containing promoter are shown, respectively, in Fig. 1 (left) and Fig. 2 (right). Consistent with the elucidation of critical core promoter elements, key factors within each group were found to bind independently to these elements to nucleate PIC formation: TFIIIC for Pol III47, TFIID for Pol II52,53 and SL1/TIF-IB for Pol I54,55. Noteworthy is the fact that for both Pol III and Pol II (Figs. 1 and 2), and also for Pol I46, recruitment of the RNA polymerase to the core promoter is mediated through interactions with specific initiation factors. This reflects another major divergence from the simple bacterial paradigm of direct RNA polymerase-promoter interactions. Beyond their necessity for elucidating the PIC assembly pathways, biochemical studies also provided key insights into the fate of the initiation factors during the transcription cycle. This is exemplified by a seminal paper, from the lab of Danny Reinberg, that demonstrated the sequential release of initiation factors from the PIC during the initiation to elongation transition, as well as recycling to a stable TFIID-promoter complex56. Biochemical studies have also provided key insights into the complete Pol III transcription cycle of initiation, elongation and termination, which is readily observed in vitro, with indications that the Pol III PIC is much more stable than the Pol II PIC and may thus facilitate robust transcription and Pol III recycling41. As discussed below, these fundamental biochemical studies, providing well-characterized initiation factors and initiation mechanisms, set the stage for high resolution structural studies.
Fig. 1 |. General initiation factors and PIC assembly pathways for Pol III-transcribed genes with internal promoter elements, and activation by a gene-specific activator.
On the tRNA gene (left), assembly of a functional PIC containing Pol III and general initiation factors (yellow) is nucleated by stable binding of TFIIIC to the Box A and Box B elements. In contrast, assembly of a PIC on the divergent promoter of the 5S RNA gene is more highly regulated and requires prior binding to Box A and Box C of the 5S RNA gene-specific transcriptional activator TFIIIA, which interacts with and stabilizes TFIIIC binding. For both genes, TFIIIB is recruited by TFIIIC and Pol III is recruited by TFIIIB and TFIIIC. Solid black bars indicate interactions between Pol III and the various factors, NTP, ribonucleoside triphosphates. Adapted from ref.155.
Fig. 2 |. General initiation factors and PIC assembly pathway for a Pol II-transcribed gene with a strong TATA-containing core promoter, and regulation by gene-specific factors and interacting cofactors.
Assembly of a PIC containing Pol II and general initiation factors (yellow) is nucleated by binding of TFIID to the TATA element of the core promoter. A model for the regulation of PIC assembly and function involves, sequentially: (i) binding of regulatory factors to distal control elements; (ii) regulatory factor interactions with cofactors that modify chromatin structure to facilitate additional factor interactions; and (iii) regulatory factor interactions with cofactors that act after chromatin remodeling to facilitate, through direct interactions, recruitment or function of components the general transcription machinery. Solid black bars indicate direct interactions between the indicated factors. TAFs, TBP-associated factors. AD, activation domain. DBD, DNA binding domain. TSS, transcription start site. Adapted from ref.155.
Following establishment of these basic principles, several other features of transcription by the three polymerases were discovered, largely through biochemical studies, and deserve mention. First, in yet another surprise, given its preeminent and early defined role in TFIID binding to TATA elements in Pol II-transcribed genes, TATA-binding polypeptide (TBP) was found to reside not only in TFIID (with ~14 TBP-associated factors; TAFs), but also in SL1 (with 3 TAFs) and in TFIIIB (with BDP1, and either of the two TFIIB-related factors BRF1 and BRF2). Notably, like the TFIID TAFs implicated in TFIID binding to variant core promoters in Pol II-transcribed genes (below), the SL1 TAFs were found to bind promoter sequences and also to constrain TBP-TATA interactions to facilitate promoter selectivity57,58. Second, variations in core promoter elements, necessitating variations in essential initiation factors, were described for both Pol III-transcribed39,40 and Pol II-transcribed44,59 genes. Particularly noteworthy is the absence of consensus TATA elements, directly recognized by the TBP component of TFIID, in the majority of mammalian and Drosophila Pol II-transcribed genes; this led to the discovery of modified pathways involving different core promoter elements (for example, initiator, DPE and MTE) and novel factor interactions for TFIID recruitment—including direct TAF interactions not only with core promoter elements44,59,60 but also with acetylated61 and methylated62,63 nucleosomes. The alternate forms of TFIIIB likewise function on different classes of Pol III-transcribed genes, namely tRNA and 5S RNA genes versus 7SK and U6 RNA genes, and provide an example of the evolutionary repurposing of a factor (TFIIB) for different transcription specificities39,40. Third, an unanticipated finding that helps explain the surprising subunit complexity of Pol I and Pol III relative to that of Pol II, with its far greater diversity of target genes, is that counterparts to some of the Pol II initiation factors are found as stably integrated subunits of Pol I and Pol III, where they may potentially perform similar functions21. Fourth, in our early Pol II transcription studies, a surprise was our discovery that cell-specific genes, including the adenovirus ML31 and β-globin64 genes, could be accurately and robustly transcribed (as histone-free DNA templates) by the basal (ubiquitous) transcription machinery. But on careful reflection, it appeared to me that these genes simply had common, widely used core promoter elements and, most importantly, that there must be both a general repression mechanism to suppress the promiscuous transcription machinery and gene- and cell-specific transcription factors to reverse this repression—both of which were ultimately demonstrated (see below). Fifth, relative to my earlier rationale for the evolution of structurally and functionally distinct RNA polymerases, biochemical and genetic analyses have revealed general regulatory factors and modifications, as well as underlying mechanisms, for the different transcription systems. Examples include: (i) for Pol I, the growth state-related regulation of SL1/TIF-1B and the tumor-suppressor-mediated repression of UBF65,66; for Pol III, the nutrient-regulated regulation of the MAF1 repressor that acts through inhibitory interactions with TFIIIB and Pol III67, as well as tumor-suppressor-mediated repression through interactions with TFIIIB66; for Pol II, the Pol II-binding repressor Gdown1 that we demonstrated to act by occlusion of TFIIB and TFIIF binding sites on Pol II and to elicit a strong Mediator requirement for transcription68,69 and factors (MOT1, NC2) that show context-dependent repression (or activation) through interactions with basal initiation factors (TBP, TFIIA) that alter PIC assembly44,70 (Fig. 2).
Recent years have seen an explosion of structural studies revealing remarkable structural and mechanistic detail related to the function of macromolecular assemblies, and nowhere has this been more apparent than in the transcription field. With respect to Pol II transcription, early (1992–1996) crystallographic studies in the labs of Stephen Burley (in collaboration with my lab) and Paul Sigler revealed intriguing structures for the TATA-binding TFIID subunit TBP, the TBP-TATA DNA complex (with TBP binding in the minor groove and inducing a 90° bend in the DNA), the TBP-TATA-TFIIB complex and the TBP-TATA-TFIIA complex (reviewed in ref.71), all of which confirmed interactions established in the biochemical analyses and revealed further insights into the initial steps of PIC assembly. Building on these initial studies, various crosslinking studies to map positions of PIC components72 and the RNA Pol II structural studies mentioned above, several groups have provided high resolution cryo-EM structures for the complete TBP-containing yeast73–75 and human76,77 Pol II PICs. While confirming the biochemically established interactions and PIC assembly pathways, these analyses have provided additional insights into core promoter recognition, transcription initiation and interactions (notably those of TFIIB) involved in the initiation-to-elongation transition. Highly informative cryo-EM structures for the yeast Pol I78,79 and Pol III80 PICs have also been reported. More recently, single-molecule approaches are being applied to the study of transcription factors both in vivo and in vitro, of special importance for an understanding of the dynamics of transcription factor interactions and functions. With respect to Pol II-related PICs, one interesting in vitro analysis confirmed the stability of promoter-TFIID-TFIIA complexes but showed a surprising and highly dynamic interaction of TFIIB, a key regulator of Pol II functions whose stable association with the early stage complex was found to depend on interactions with the Pol II-TFIIF complex81. Future emphasis on such studies, coupled with structural information, should provide profound new insights into the dynamics of PIC formation and function in conjunction with transcriptional activators and coactivators.
Gene- and cell-specific transcriptional regulatory factors: key determinants of gene control
Identification and characterization of site-specific DNA-binding regulatory factors in bacteria and bacteriophages in the late 1960s and 1970s raised the obvious possibility that eukaryotic genes might be similarly regulated. However, the presence and rapid turnover of heterogeneous nuclear RNA82 and the discovery of RNA splicing raised the possibility that cell-specific RNAs may be derived largely from global transcription followed by selective splicing and stabilization of specific RNAs rather than gene-specific transcription. Although this possibility seemed unlikely82 and indeed was unappealing to me, it was something I did think about in the absence of any identified gene- or cell-specific factors. But fortunately, biochemical studies in 1979 led to our discovery of the 5S RNA gene-specific TFIIIA, the first-established gene-specific transcriptional activator in eukaryotes83. Mechanistically, TFIIIA was shown to bind specifically to the gene-internal promoter83 that had just been defined84 and to facilitate recruitment of general initiation factors (TFIIIC and TFIIIB) that in turn recruit Pol III47,48 (Fig. 1, right). This again indicated a paradigm shift from the bacterial mechanism in which transcriptional activators interact directly with the polymerase. Notably, these studies were the first to establish the fundamentally important paradigm of gene-specific transcription through a gene-specific activator in eukaryotes, and also the first to establish the mechanism of action of a eukaryotic transcriptional regulatory factor (enhanced recruitment of an initiation factor that in turn recruits RNA polymerase). Our cognate cDNA cloning of TFIIIA in 1984 (ref.85) also represents the first cloning and sequence identification of an established eukaryotic transcription factor and led to the deduction by Aaron Klug of the zinc-finger motif86, the most common DNA-binding motif in eukaryotic transcription factors87.
These seminal observations on TFIIIA gave me confidence that there must also be gene- and cell-specific factors for Pol II-transcribed genes and prompted subsequent studies in many laboratories that led to identification of gene-specific promoter/enhancer-binding factors for the more numerous Pol Ii-transcribed genes. The earliest-described Pol II activators that were shown to bind and/or act through specific promoter sites include glucocorticoid receptor88, Sp1 (ref.89), HSF90, USF/MLTF53,91 and Gal4 (ref.92); and all were eventually cloned and shown to function as recombinant proteins. Our studies of USF, showing cooperative interactions with promoter-bound TFIID53, also provided the earliest example of a target and mechanism of action of an activator for Pol II target genes, again reinforcing the paradigm of activator function through a general transcription factor rather than through the RNA polymerase. Our later studies of other activators extended these results implicating TFIID as an activator target, and indicated a mechanism (validated later by other groups) involving an activator-induced TFIID isomerization on the promoter93. Although more recent studies have focused on coactivators as primary targets of transcriptional activators (discussed below), it seemed to me perfectly logical, in view of the complexity of the Pol II initiation factors and the precedent established for TFIIIA interactions with Pol III initiation factors, that some activators would directly target Pol II initiation factors. Indeed, there are many early reports of activator-initiation factor interactions that in some cases (for example, refs.94,95) have been strongly implicated in activator function. My expectation is that many more examples will be uncovered as the actual mechanisms of action of the many Pol II transcription factors are established. Although not emphasized here, site-specific DNA-binding repressors are also an integral part of eukaryotic transcriptional regulation, and an early biochemical analysis of the viral SV40 T antigen in the laboratory of Bob Tjian established a direct repression of active SV40 promoters that was dependent on site-specific binding96.
Notably, beyond the early identification of transcription factors based largely on in vitro binding/transcription assays and subsequent biochemical purification and cloning, many approaches have been used to identify the large number (~1,600 in human) of established or predicted transcription factors (activators and repressors)87. It remains a major challenge to understand both the biological functions of many of these factors as well as the mechanistic details of how they function on their respective target genes. In this regard, some of the exciting developments since the original discovery of eukaryotic transcription factors include: (i) the discovery that many of these factors are master regulators of development and cell differentiation97,98, emphasizing their profound powers as well as their physiological importance; (ii) the early discovery of conventional enhancers acting at both short and long distances99, and the more recent recognition of ‘super-enhancers’ with very high densities of transcription factors and associated coactivators100; (iii) the discovery of enhanceosomes as clusters of interacting enhancer-bound transcription factors101, allowing not only cooperative binding but potential cooperativity through recognition of different coactivator and general initiation factor targets; (iv) the discovery of topological-associated domains (TADs) that constrain the target gene functions of contained enhancers102,103.
Transcriptional cofactors: bridges between enhancer-bound regulatory factors and the basic transcription machinery
Given their function in diverse gene regulatory pathways, transcription factors that activate genes transcribed by Pol II have been of broad interest over the last three to four decades, and a key question has been their mechanism of action. Studies over the last three decades have shown that despite the complexity of the general transcription machinery and several documented interactions of activators with this machinery (their ultimate targets), activator function requires diverse coactivators that include chromatin-modifying factors as well as factors that interact more directly with the general transcription machinery. In relation to the latter, early DNA-templated in vitro transcription assays with highly purified basal transcription factors revealed requirements for additional factors that included the 20–30-subunit Mediator complex104,105 and the TAFs within the 15-subunit TFIID complex44,106.
The Mediator was first identified in yeast through genetic and biochemical approaches in the Kornberg and Young labs107,108, and ultimately purified and shown to interact directly with Pol II109. The mammalian counterpart was first evidenced in my lab as the human USA coactivator activity110, the principal component (PC2) of which proved to be the Mediator111. The human Mediator was first purified in my lab as the thyroid hormone receptor-associated protein (TRAP) complex112 and subsequently shown to interact with several nuclear hormone receptors through the MED1/TRAP220 subunit113. The physiological relevance and gene/activator specificity of MED1 was established by genetic analyses that demonstrated its necessity for PPARγ-mediated adipogenesis but not for MyoD-mediated myogenesis or precursor cell (fibroblast) viability114. Collectively, the independent discoveries of yeast Mediator-Pol II interactions by the Kornberg and Young labs, and of physical and functional mammalian Mediator-activator interactions by my lab, clearly indicated a mechanism in which the Mediator serves as a bridge to facilitate physical and functional interactions between enhancer-bound activators and the general transcription machinery (principally Pol II) at the core promoter (Fig. 2). More recent studies have confirmed the critical role for the Mediator in facilitating enhancer-promoter interactions115. Moreover, the Mediator, and especially the MED1 subunit, has proved to be an important diagnostic in chromatin immunoprecipitation and sequencing (ChIP-seq) analyses for the identification of candidate enhancers100. Structural studies have identified head, middle, tail and kinase modules116, with the tail module serving as the principal site of activator interactions104,105. Supporting the view that the tail module serves primarily to anchor the Mediator to enhancer-bound activators and establishing the basic functional components of the Mediator, our biochemical studies with reconstituted Mediator complexes identified a minimal 15-subunit mammalian core Mediator complex, comprising subsets of head and middle subunits, that is necessary and sufficient for carrying out the effector function in stimulating the basal transcription machinery117. Ongoing biochemical and genetic analyses in many labs continue to elucidate the multiple cellular functions and mechanisms of the Mediator, whereas cryo-EM studies have provided high-resolution structures of the Mediator alone116 and in association with Pol II118 and the basal PIC74,119,120. These remarkable studies have provided key insights into Mediator-PIC interactions important for assembly and function.
TFIID, now known to be comprised of TBP and ~14 TAFs44,106, was first identified as an essential initiation factor in our original discovery of Pol II accessory factors37 and was always of special interest to me in view of its direct interactions with TATA-containing core promoters53. Ultimately, its complete purification and characterization was greatly facilitated by (i) the identification in yeast of a single polypeptide (TBP) that could substitute for human TFIID in the basal (activator-independent) transcription assays that we had developed121,122, and (ii) the subsequent cloning of yeast, fly and human TBPs and the exploitation of affinity methods for TFIID purification106. In initial comparative studies of TBP and native TFIID with Drosophila factors in the Tjian lab and human factors in my lab, TFIID displayed both basal and activator-enhanced activities, whereas TBP manifested only basal activity123,124. These seminal studies indicated the presence of coactivators in the natural TFIID preparations that were later identified as the tightly associated TAFs in homogeneous (affinity-purified) TFIID complexes (Fig. 2). Following the cloning and characterization of the Drosophila and human TAFs (primarily in the Tjian, Roeder and Yoshihiro Nakatani labs), and as an extension of our earlier demonstration of promoter-associated activator-TFIID interactions53,93, several studies reported specific activator-TAF interactions that were implicated in activator function44,106. As a clear example, a more recent study from my laboratory provided compelling evidence, from both biochemical and cell-based assays, for the role of TAF4 in mediating, through direct interactions, the activator function of E2A and provided the first clear evidence for a specific TAF coactivator mechanism that, in this case, involved direct E2A-enhanced TFIID binding as well as a reciprocal stabilizing effect of TFIID on E2A binding125. To date, and as expected from their complexity, in vitro and in vivo studies have clearly implicated TAFs in many gene activation pathways, not only as conventional transcriptional coactivators but also (as mentioned above) as core promoter recognition factors that may be especially important on TATA-less promoters59,61. Of special significance toward a mechanistic understanding of the multiple functions of TFIID, recent cryo-EM studies of human TFIID have provided important structural details regarding overall organization of component subunits within distinct lobes, specific DNA interactions and dynamic conformational changes associated with core promoter recognition and stable TFIID binding60,126.
Although some individual subunits of the Mediator and TFIID clearly function as activator/gene-specific cofactors, these large multisubunit complexes are generally ubiquitous—the main exception being some cell-specific TAFs and TBP-related factors that allow for the assembly of variant forms of TFIID44,61. In contrast, there also exist more dedicated cell-specific coactivators. The prototype, OCA-B, was discovered in my lab as a B-cell-specific, OCT1/2-interacting factor that mediated activation of B-cell-specific genes by promoter/enhancer-bound OCT1/2 in biochemical assays127, and then was shown by genetic analyses to be critical for germinal-center formation and immune responses128. Another prime example is the nuclear receptor-interacting PGC-1 that was discovered by Bruce Spiegelman and shown to have a key role in many metabolic processes that include brown fat thermogenesis129. These types of cofactors, which do not directly bind DNA regulatory sequences, offer additional mechanisms for gene- or cell-specific regulation. Mechanistically, PGC-1 (ref.130) and OCA-B (unpublished observations) have been shown to act through interactions with Mediator and other general cofactors (Fig. 2). Our biochemical analyses also identified other general coactivators such as PC4 (ref.131) and p75/PSIP/LEDGF132, the latter of particluar importance because of its role in MLL1 leukemic fusion protein function and in directing HIV integrase to transcribed regions. In considering the discovery, importance and mechanisms of these cellular cofactors, it may be noted that general precedent was established from studies of the viral E1A and VP16 proteins that act as potent transcriptional (co) activators through interactions with cellular proteins133.
Extensions of biochemical transcription studies from DNA to chromatin templates
Although much has been learned from biochemical studies of transcription with DNA templates, it was logical to extend our studies to more physiological chromatin templates, especially in light of the discoveries indicating transcriptional regulation through chromatin structure134. As this topic is covered in an accompanying article by Jim Kadonaga, I highlight primarily key, and unique, biochemical experiments from my laboratory. The in vitro biochemical studies were facilitated by the development of methods for chromatin assembly, first with crude extracts and subsequently with defined systems established by Kadonaga135. An initial and seminal group of experiments in the Luse, Kornberg and Roeder labs demonstrated chromatin-structure-mediated repression of the otherwise promiscuous activity of the general transcription machinery136–138. These results confirmed my earlier prediction, based on the promiscuity of the general transcription machinery on DNA templates31,64, of the presence of a general repression mechanism, and were later confirmed by genetic analyses in vivo139. Our own in vitro studies further indicated the ability of pre-bound initiation factors (notably TFIID) to preclude chromatin-assembly-mediated repression138, as well as the ability of activators to facilitate PIC assembly to escape this repression in competitive chromatin assembly transcription assays140, thereby indicating mechanisms to induce/maintain active promoters.
Given many reports of diverse histone-modifying factors as transcriptional cofactors and correlations of histone modifications (acetylation/methylation) with changes in transcription in cells, and following biochemical studies by Kadonaga141 and Jerry Workman142 demonstrating histone acetyl transferase (HAT)-dependent transcription of chromatin templates in response to various activators, we used recombinant chromatin-templated assays to assess fundamental questions regarding underlying mechanisms. First, through the use of templates assembled with mutant or pre-modified histones, we demonstrated both a core nucleosome block to transcription that is independent of the histone tails143 and an absolute requirement of the tails for transcriptional activation through histone acetylation143,144 and methylation63,145. Notably, these results provided the first formal proof that histone modifications (acetylation and methylation) can indeed be causal for transcription. This was important in light of our seminal 1997 discovery146, and many other reports since147, that transcription factors can be functionally modified by histone-modifying enzymes, such that the loss of transcription with the loss of catalytic activity of an acetyl- or methyl-transferase (in vitro or in vivo) does not necessarily prove a mechanism involving histone modifications. Second, through rigorous biochemical analyses, including the elucidation of key protein-protein interactions, we established mechanisms underlying the cooperative functions of different histone-modifying cofactors in transcription145,148. This included elucidation of the mechanism underlying a previously reported catalytic-activity-independent function of an important MLL4-associated histone demethylase (UTX) in gene activation in embryonic stem cells, and provides a prime example of how biochemical analyses can establish mechanisms not readily elucidated in cellular assays148. Third, following the recent identification of many novel histone acylation modifications149, our cell-free systems provided clear proof of principle of a direct effect of activator-dependent, p300-mediated novel acylation events on transcription150. Fourth, using totally defined transcription initiation-dependent assays, we demonstrated direct functions of TFIIS and the PAF1 complex, acting either independently or synergistically through cooperative Pol II interactions, in Pol II transcription elongation through long nucleosomal arrays151,152. Fifth, in a recent extension to higher-order, linker histone H1-compacted/repressed chromatin, and complementing an earlier study implicating SWI/SNF and p300 in activation of histone H1-chromatin153, our biochemical analyses established a novel gene-specific mechanism for reactivation involving an activator→p300→NAP1→H1 pathway154. The use of these in vitro systems by us and others has provided important mechanistic insights into chromatin-related regulatory mechanisms that cannot easily be established from cell-based studies. Notably, they provide new opportunities for continued biochemical/mechanistic studies, especially with the application of chemical biology approaches for synthesis of specifically modified (‘designer’) histones, and assembly and functional analysis of derived chromatin templates.
Next frontiers: new macromolecules, pathways, concepts and experimental approaches in transcriptional regulation
It is apparent that the 1969 discovery of three nuclear RNA polymerases represented only the tip of the iceberg, with ensuing decades of studies revealing an expanding universe of factors and mechanisms that are important for the execution and regulation of transcription and that were completely unanticipated five decades ago155. Beginning with foundational biochemical studies, we have proceeded from multiple enzymes, to cognate initiation factors, to diverse cofactors for effecting the functions of the hundreds of documented transcriptional regulatory factors. But beyond the diverse proteins and protein complexes discussed here, and based in large part on more recent genomic technologies102, we also have witnessed the emergence of other demonstrated or candidate transcriptional regulatory macromolecules that include enhancer RNAs and long noncoding RNAs156, whose no doubt varied functions and mechanisms remain to be fully elucidated both genetically and biochemically. Genomic analyses based on earlier biochemically defined transcription factors and cofactors have also given us insights into increasingly complex genetic regulatory elements, in particular the locus control regions/super-enhancers that may be located kilobases to megabases from target promoters99,100, as well as the three-dimensional organization of the genome into active and inactive compartments and associated TADs, and various proteins and processes important for their formation and maintenance103,157. Key related questions are cause versus effect relationships between these domains and transcription factors/processes, and especially the factors and the processes that are most critical for productive enhancer-promoter interactions leading to transcription. In this regard, it clearly will be important not only to continue to apply advanced genomic techniques102 but also, and perhaps most importantly, to deploy advanced single-molecule imaging technologies158 to follow the interactions of enhancers and promoters (and associated factors) for individual genes in time and space. The complementary use of CRISPR-Cas9-based technologies159 to modify specific proteins and DNA regulatory elements will likewise contribute to an understanding of the relevant macromolecular interactions. Another new frontier relates to the possible role and mechanism of action of intrinsically disordered regions, common to many transcription factors, in gene activation and repression through the formation of biomolecular condensates (phase separation)160,161.
Although it is unlikely that some of the above-mentioned higher-order phenomena would have been revealed by the standard biochemical studies that did set the stage for the study of transcription at many levels, I remain confident that biochemistry will continue to play a key role in unraveling underlying mechanisms. Superficially, it may appear that the ‘emergent properties’ of some of these structures may simply not be amenable to the reductionist approaches that are typical of biochemistry. However, experience over the last several decades that biochemical and structural approaches continue to be refined to meet the most daunting of challenges gives me hope that some of the most complex phenomena, such as super-enhancer-mediated transcription regulation, will soon be reconstituted in vitro and thereby clearly understood with respect to the essential factors and their mechanisms of action.
Acknowledgements
I thank S. Malik for a critical reading and suggestions on the manuscript. I am indebted to my mentors W. J. Rutter and D. D. Brown for their support and encouragement in the earliest phase of my scientific career, and to the numerous students and fellows who contributed so profoundly to the studies from my laboratory over the last five decades. I apologize to the many colleagues in the transcription field whose work could not be cited because of space limitations and the focus of the manuscript. Work in my laboratory over the past decades has been generously supported by the National Institutes of Health, the American Cancer Society, the Leukemia and Lymphoma Society, the Starr Cancer Consortuim, many other private foundations and The Rockefeller University.
Footnotes
Competing interests
The author declares no competing interests.
Peer review information: Beth Moorefield was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jacob F & Monod J Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol 3, 318–356 (1961). [DOI] [PubMed] [Google Scholar]
- 2.Weiss SB & Gladstone L A mammalian system for the incorporation of cytidine triphosphate into ribonucleic acid. J. Am. Chem. Soc 81, 4118–4119 (1959). [Google Scholar]
- 3.Weiss SB Enzymatic incorporation of ribonucleoside triphosphates into the interpolynucleotide linkages of ribonucleic acid. Proc. Natl Acad. Sci. USA 46, 1020–1030 (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hurwitz J The discovery of RNA polymerase. J. Biol. Chem 280, 42477–42485 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Burgess RR, Travers AA, Dunn JJ & Bautz EK Factor stimulating transcription by RNA polymerase. Nature 221, 43–46 (1969). [DOI] [PubMed] [Google Scholar]
- 6.Roeder RG & Rutter WJ Multiple forms of DNA-dependent RNA polymerase in eukaryotic organisms. Nature 224, 234–237 (1969). [DOI] [PubMed] [Google Scholar]
- 7.Widnell CC & Tata JR Evidence for two DNA-dependent RNA polymerase activities in isolated rat liver nuclei. Biochim. Biophys. Acta 87, 531–533 (1964). [DOI] [PubMed] [Google Scholar]
- 8.Roeder RG Multiple RNA Polymerases and RNA Synthesis in Eukaryotic Systems Ph.D. Thesis. University of Washington; (1969). [Google Scholar]
- 9.Roeder RG & Rutter WJ DNA dependent RNA polymerase in sea urchin development. Fed. Proc 28, 599 (1969). [Google Scholar]
- 10.Kedinger C, Gniazdowski M, Mandel JL Jr., Gissinger F & Chambon P Alpha-amanitin: a specific inhibitor of one of two DNA-dependent RNA polymerase activities from calf thymus. Biochem. Biophys. Res. Commun 38, 165–171 (1970). [DOI] [PubMed] [Google Scholar]
- 11.Lindell TJ, Weinberg F, Morris PW, Roeder RG & Rutter WJ Specific inhibition of nuclear RNA polymerase II by alpha-amanitin. Science 170, 447–449 (1970). [DOI] [PubMed] [Google Scholar]
- 12.Blatti SP et al. Structure and Regulatory Properties of Eucaryotic RNA Polymerase. Cold Spring Harb. Symp. Quant. Biol 35, 649–657 (1970). [Google Scholar]
- 13.Chambon P et al. Purification and Properties of Calf Thymus DNA-Dependent RNA Polymerases A and B. Cold Spring Harb. Symp. Quant. Biol 35, 693–707 (1970). [Google Scholar]
- 14.Roeder RG & Rutter WJ Specific nucleolar and nucleoplasmic RNA polymerases. Proc. Natl Acad. Sci. USA 65, 675–682 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roeder RG, Reeder RH & Brown DD Multiple forms of RNA polymerase in Xenopus laevis: Their relationship to RNA synthesis in vivo and their fidelity of transcription in vitro. Cold Spring Harb. Symp. Quant. Biol 35, 727–735 (1970). [Google Scholar]
- 16.Sklar VE, Schwartz LB & Roeder RG Distinct molecular structures of nuclear class I, II, and III DNA-dependent RNA polymerases. Proc. Natl Acad. Sci. USA 72, 348–352 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinmann R, Raskas HJ & Roeder RG Role of DNA-dependent RNA polymerases II and III in transcription of the adenovirus genome late in productive infection. Proc. Natl Acad. Sci. USA 71, 3426–3439 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinmann R & Roeder RG Role of DNA-dependent RNA polymerase 3 in the transcription of the tRNA and 5S RNA genes. Proc. Natl Acad. Sci. USA 71, 1790–1794 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sentenac A et al. Yeast RNA polymerase subunits and genes, 27–54 (Cold Spring Harbor Press, Cold Spring Harbor Laboratory, N.Y, 1992). [Google Scholar]
- 20.Young RA RNA polymerase II. Annu. Rev. Biochem 60, 689–715 (1991). [DOI] [PubMed] [Google Scholar]
- 21.Vannini A & Cramer P Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell 45, 439–446 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Cramer P et al. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288, 640–649 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Cramer P, Bushnell DA & Kornberg RD Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292, 1863–1876 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Kuhn CD et al. Functional architecture of RNA polymerase I. Cell 131, 1260–1272 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Pilsl M et al. Structure of the initiation-competent RNA polymerase I and its implication for transcription. Nat. Commun 7, 12126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jasiak AJ, Armache KJ, Martens B, Jansen RP & Cramer P Structural biology of RNA polymerase III: subcomplex C17/25 X-ray structure and 11 subunit enzyme model. Mol. Cell 23, 71–81 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Zhang G et al. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell 98, 811–824 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Parker CS, Ng SY & Roeder RG Selective transcription of the 5S RNA genes in isolated chromatin by RNA polymerase III in Molecular Mechanisms in the Control of Gene Expression (eds. Nierlich DP, Rutter WJ & Fox CF) 223–42 (Academic Press, New York, 1976). [Google Scholar]
- 29.Parker CS & Roeder RG Selective and accurate transcription of the Xenopus laevis 5S RNA genes in isolated chromatin by purified RNA polymerase III. Proc. Natl Acad. Sci. USA 74, 44–48 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng SY, Parker CS & Roeder RG Transcription of cloned Xenopus 5S RNA genes by X. laevis RNA polymerase III in reconstituted systems. Proc. Natl Acad. Sci. USA 76, 136–140 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weil PA, Luse DS, Segall J & Roeder RG Selective and accurate initiation of transcription at the Ad2 major late promotor in a soluble system dependent on purified RNA polymerase II and DNA. Cell 18, 469–484 (1979). [DOI] [PubMed] [Google Scholar]
- 32.Birkenmeier EH, Brown DD & Jordan E A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell 15, 1077–1086 (1978). [DOI] [PubMed] [Google Scholar]
- 33.Wu GJ Adenovirus DNA-directed transcription of 5.5S RNA in vitro. Proc. Natl Acad. Sci. USA 75, 2175–2179 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manley JL, Fire A, Cano A, Sharp PA & Gefter ML DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc. Natl Acad. Sci. USA 77, 3855–3859 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grummt I Specific transcription of mouse ribosomal DNA in a cell-free system that mimics control in vivo. Proc. Natl Acad. Sci. USA 78, 727–731 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segall J, Matsui T & Roeder RG Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J. Biol. Chem 255, 11986–11991 (1980). [PubMed] [Google Scholar]
- 37.Matsui T, Segall J, Weil PA & Roeder RG Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J. Biol. Chem 255, 11992–11996 (1980). [PubMed] [Google Scholar]
- 38.Mishima Y, Financsek I, Kominami R & Muramatsu M Fractionation and reconstitution of factors required for accurate transcription of mammalian ribosomal RNA genes: identification of a species-dependent initiation factor. Nucleic Acids Res 10, 6659–6670 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dumay-Odelot H, Durrieu-Gaillard S, El Ayoubi L, Parrot C & Teichmann M Contributions of in vitro transcription to the understanding of human RNA polymerase III transcription. Transcription 5, e27526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schramm L & Hernandez N Recruitment of RNA polymerase III to its target promoters. Genes Dev 16, 2593–2620 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Geiduschek EP & Kassavetis GA The RNA polymerase III transcription apparatus. J. Mol. Biol 310, 1–26 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Orphanides G, Lagrange T & Reinberg D The general transcription factors of RNA polymerase II. Genes Dev 10, 2657–2683 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Roeder RG The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci 21, 327–335 (1996). [PubMed] [Google Scholar]
- 44.Thomas MC & Chiang CM The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol 41, 105–178 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Drygin D, Rice WG & Grummt I The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol 50, 131–156 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Goodfellow SJ & Zomerdijk JC Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA. genes. Subcell. Biochem 61, 211–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lassar AB, Martin PL & Roeder RG Transcription of class III genes: formation of preinitiation complexes. Science 222, 740–748 (1983). [DOI] [PubMed] [Google Scholar]
- 48.Bieker JJ, Martin PL & Roeder RG Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell 40, 119–127 (1985). [DOI] [PubMed] [Google Scholar]
- 49.Van Dyke MW, Roeder RG & Sawadogo M Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science 241, 1335–1338 (1988). [DOI] [PubMed] [Google Scholar]
- 50.Buratowski S, Hahn S, Guarente L & Sharp PA Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56, 549–561 (1989). [DOI] [PubMed] [Google Scholar]
- 51.Flores O, Lu H & Reinberg D Factors involved in specific transcription by mammalian RNA polymerase II. Identification and characterization of factor IIH. J. Biol. Chem 267, 2786–2793 (1992). [PubMed] [Google Scholar]
- 52.Parker CS & Topol J A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell 36, 357–369 (1984). [DOI] [PubMed] [Google Scholar]
- 53.Sawadogo M & Roeder RG Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell 43, 165–175 (1985). [DOI] [PubMed] [Google Scholar]
- 54.Learned RM, Cordes S & Tjian R Purification and characterization of a transcription factor that confers promoter specificity to human RNA polymerase I. Mol. Cell. Biol 5, 1358–1369 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clos J, Buttgereit D & Grummt I A purified transcription factor (TIF-IB) binds to essential sequences of the mouse rDNA promoter. Proc. Natl Acad. Sci. USA 83, 604–608 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zawel L, Kumar KP & Reinberg D Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev 9, 1479–1490 (1995). [DOI] [PubMed] [Google Scholar]
- 57.Rudloff U, Eberhard D, Tora L, Stunnenberg H & Grummt I TBP-associated factors interact with DNA and govern species specificity of RNA polymerase I transcription. EMBO J 13, 2611–2616 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beckmann H, Chen JL, O’Brien T & Tjian R Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science 270, 1506–1509 (1995). [DOI] [PubMed] [Google Scholar]
- 59.Kadonaga JT Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip. Rev. Dev. Biol 1, 40–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Louder RK et al. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature 531, 604–609 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goodrich JA & Tjian R Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat. Rev. Genet 11, 549–558 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vermeulen M et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Lauberth SM et al. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152, 1021–1036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luse DS & Roeder RG Accurate transcription initiation on a purified mouse beta-globin DNA fragment in a cell-free system. Cell 20, 691–699 (1980). [DOI] [PubMed] [Google Scholar]
- 65.Grummt I Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev 17, 1691–1702 (2003). [DOI] [PubMed] [Google Scholar]
- 66.White RJ RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol 6, 69–78 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Willis IM & Moir RD Signaling to and from the RNA polymerase III transcription and processing machinery. Annu. Rev. Biochem 87, 75–100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu X et al. A Mediator-responsive form of metazoan RNA polymerase II. Proc. Natl Acad. Sci. USA 103, 9506–9511 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jishage M et al. Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1. Nat. Struct. Mol. Biol 25, 859–867 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sikorski TW & Buratowski S The basal initiation machinery: beyond the general transcription factors. Curr. Opin. Cell Biol 21, 344–351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nikolov DB & Burley SK RNA polymerase II transcription initiation: a structural view. Proc. Natl Acad. Sci. USA 94, 15–22 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cramer P et al. Structure of eukaryotic RNA polymerases. Annu. Rev. Biophys 37, 337–352 (2008). [DOI] [PubMed] [Google Scholar]
- 73.Murakami K et al. Structure of an RNA polymerase II preinitiation complex. Proc. Natl Acad. Sci. USA 112, 13543–13548 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robinson PJ et al. Structure of a complete mediator-RNA polymerase II pre-initiation complex. Cell 166, 1411–1422.e16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Plaschka C et al. Transcription initiation complex structures elucidate DNA opening. Nature 533, 353–358 (2016). [DOI] [PubMed] [Google Scholar]
- 76.He Y, Fang J, Taatjes DJ & Nogales E Structural visualization of key steps in human transcription initiation. Nature 495, 481–486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.He Y et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533, 359–365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Engel C et al. Structural basis of RNA polymerase I transcription initiation. Cell 169, 120–131.e22 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Sadian Y et al. Structural insights into transcription initiation by yeast RNA polymerase I. EMBO J 36, 2698–2709 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abascal-Palacios G, Ramsay EP, Beuron F, Morris E & Vannini A Structural basis of RNA polymerase III transcription initiation. Nature 553, 301–306 (2018). [DOI] [PubMed] [Google Scholar]
- 81.Zhang Z et al. Rapid dynamics of general transcription factor TFIIB binding during preinitiation complex assembly revealed by single-molecule analysis. Genes Dev 30, 2106–2118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Darnell JE, Jelinek WR & Molloy GR Biogenesis of mRNA: genetic regulation in mammalian cells. Science 181, 1215–1221 (1973). [DOI] [PubMed] [Google Scholar]
- 83.Engelke DR, Ng SY, Shastry BS & Roeder RG Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell 19, 717–728 (1980). [DOI] [PubMed] [Google Scholar]
- 84.Sakonju S, Bogenhagen DF & Brown DD A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5′ border of the region. Cell 19, 13–25 (1980). [DOI] [PubMed] [Google Scholar]
- 85.Ginsberg AM, King BO & Roeder RG Xenopus 5S gene transcription factor, TFIIIA: characterization of a cDNA clone and measurement of RNA levels throughout development. Cell 39, 479–489 (1984). [DOI] [PubMed] [Google Scholar]
- 86.Miller J, McLachlan AD & Klug A Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J 4, 1609–1614 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lambert SA et al. The human transcription factors. Cell 175, 598–599 (2018). [DOI] [PubMed] [Google Scholar]
- 88.Payvar F et al. Purified glucocorticoid receptors bind selectively in vitro to a cloned DNA fragment whose transcription is regulated by glucocorticoids in vivo. Proc. Natl Acad. Sci. USA 78, 6628–6632 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dynan WS & Tjian R Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell 32, 669–680 (1983). [DOI] [PubMed] [Google Scholar]
- 90.Parker CS & Topol J A Drosophila RNA polymerase II transcription factor binds to the regulatory site of an hsp 70 gene. Cell 37, 273–283 (1984). [DOI] [PubMed] [Google Scholar]
- 91.Carthew RW, Chodosh LA & Sharp PA An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell 43, 439–448 (1985). [DOI] [PubMed] [Google Scholar]
- 92.Bram RJ & Kornberg RD Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc. Natl Acad. Sci. USA 82, 43–47 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Horikoshi M, Hai T, Lin YS, Green MR & Roeder RG Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell 54, 1033–1042 (1988). [DOI] [PubMed] [Google Scholar]
- 94.Roberts SG, Ha I, Maldonado E, Reinberg D & Green MR Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature 363, 741–744 (1993). [DOI] [PubMed] [Google Scholar]
- 95.Rochette-Egly C, Adam S, Rossignol M, Egly JM & Chambon P Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell 90, 97–107 (1997). [DOI] [PubMed] [Google Scholar]
- 96.Rio D, Robbins A, Myers R & Tjian R Regulation of simian virus 40 early transcription in vitro by a purified tumor antigen. Proc. Natl Acad. Sci. USA 77, 5706–5710 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Davis RL, Weintraub H & Lassar AB Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000 (1987). [DOI] [PubMed] [Google Scholar]
- 98.Takahashi K & Yamanaka S Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006). [DOI] [PubMed] [Google Scholar]
- 99.Schaffner W Enhancers, enhancers - from their discovery to today’s universe of transcription enhancers. Biol. Chem 396, 311–327 (2015). [DOI] [PubMed] [Google Scholar]
- 100.Hnisz D et al. Super-enhancers in the control of cell identity and disease. Cell 155, 934–947 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Maniatis T et al. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb. Symp. Quant. Biol 63, 609–620 (1998). [DOI] [PubMed] [Google Scholar]
- 102.Yu M & Ren B The three-dimensional organization of mammalian genomes. Annu. Rev. Cell Dev. Biol 33, 265–289 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.van Steensel B & Furlong EEM The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol 20, 327–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Malik S & Roeder RG The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet 11, 761–772 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Allen BL & Taatjes DJ The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol 16, 155–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Albright SR & Tjian R TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242, 1–13 (2000). [DOI] [PubMed] [Google Scholar]
- 107.Flanagan PM, Kelleher RJ III, Sayre MH, Tschochner H & Kornberg RD A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350, 436–438 (1991). [DOI] [PubMed] [Google Scholar]
- 108.Thompson CM, Koleske AJ, Chao DM & Young RA A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73, 1361–1375 (1993). [DOI] [PubMed] [Google Scholar]
- 109.Kim YJ, Björklund S, Li Y, Sayre MH & Kornberg RD A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599–608 (1994). [DOI] [PubMed] [Google Scholar]
- 110.Meisterernst M, Roy AL, Lieu HM & Roeder RG Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell 66, 981–993 (1991). [DOI] [PubMed] [Google Scholar]
- 111.Malik S, Gu W, Wu W, Qin J & Roeder RG The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol. Cell 5, 753–760 (2000). [DOI] [PubMed] [Google Scholar]
- 112.Fondell JD, Ge H & Roeder RG Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl Acad. Sci. USA 93, 8329–8333 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen W & Roeder RG Mediator-dependent nuclear receptor function. Semin. Cell Dev. Biol 22, 749–758 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ge K et al. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature 417, 563–567 (2002). [DOI] [PubMed] [Google Scholar]
- 115.Malik S & Roeder RG Mediator: a drawbridge across the enhancer-promoter divide. Mol. Cell 64, 433–434 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tsai KL et al. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell 157, 1430–1444 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cevher MA et al. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nat. Struct. Mol. Biol 21, 1028–1034 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tsai KL et al. Mediator structure and rearrangements required for holoenzyme formation. Nature 544, 196–201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Plaschka C et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature 518, 376–380 (2015). [DOI] [PubMed] [Google Scholar]
- 120.Nozawa K, Schneider TR & Cramer P Core Mediator structure at 3.4 Å extends model of transcription initiation complex. Nature 545, 248–251 (2017). [DOI] [PubMed] [Google Scholar]
- 121.Buratowski S, Hahn S, Sharp PA & Guarente L Function of a yeast TATA element-binding protein in a mammalian transcription system. Nature 334, 37–42 (1988). [DOI] [PubMed] [Google Scholar]
- 122.Cavallini B et al. A yeast activity can substitute for the HeLa cell TATA box factor. Nature 334, 77–80 (1988). [DOI] [PubMed] [Google Scholar]
- 123.Hoffman A et al. Highly conserved core domain and unique N terminus with presumptive regulatory motifs in a human TATA factor (TFIID). Nature 346, 387–390 (1990). [DOI] [PubMed] [Google Scholar]
- 124.Pugh BF & Tjian R Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell 61, 1187–1197 (1990). [DOI] [PubMed] [Google Scholar]
- 125.Chen WY et al. A TAF4 coactivator function for E proteins that involves enhanced TFIID binding. Genes Dev 27, 1596–1609 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Patel AB et al. Structure of human TFIID and mechanism of TBP loading onto promoter DNA. Science 362, eaau8872 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Luo Y, Fujii H, Gerster T & Roeder RG A novel B cell-derived coactivator potentiates the activation of immunoglobulin promoters by octamer-binding transcription factors. Cell 71, 231–241 (1992). [DOI] [PubMed] [Google Scholar]
- 128.Kim U et al. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature 383, 542–547 (1996). [DOI] [PubMed] [Google Scholar]
- 129.Lin J, Handschin C & Spiegelman BM Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 1, 361–370 (2005). [DOI] [PubMed] [Google Scholar]
- 130.Wallberg AE, Yamamura S, Malik S, Spiegelman BM & Roeder RG Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol. Cell 12, 1137–1149 (2003). [DOI] [PubMed] [Google Scholar]
- 131.Ge H & Roeder RG Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell 78, 513–523 (1994). [DOI] [PubMed] [Google Scholar]
- 132.Ge H, Si Y & Roeder RG Isolation of cDNAs encoding novel transcription coactivators p52 and p75 reveals an alternate regulatory mechanism of transcriptional activation. EMBO J 17, 6723–6729 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wysocka J & Herr W The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem. Sci 28, 294–304 (2003). [DOI] [PubMed] [Google Scholar]
- 134.Li B, Carey M & Workman JL The role of chromatin during transcription. Cell 128, 707–719 (2007). [DOI] [PubMed] [Google Scholar]
- 135.Ito T, Bulger M, Pazin MJ, Kobayashi R & Kadonaga JT ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90, 145–155 (1997). [DOI] [PubMed] [Google Scholar]
- 136.Knezetic JA & Luse DS The presence of nucleosomes on a DNA template prevents initiation by RNA polymerase II in vitro. Cell 45, 95–104 (1986). [DOI] [PubMed] [Google Scholar]
- 137.Lorch Y, LaPointe JW & Kornberg RD Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell 49, 203–210 (1987). [DOI] [PubMed] [Google Scholar]
- 138.Workman JL & Roeder RG Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell 51, 613–622 (1987). [DOI] [PubMed] [Google Scholar]
- 139.Han M & Grunstein M Nucleosome loss activates yeast downstream promoters in vivo. Cell 55, 1137–1145 (1988). [DOI] [PubMed] [Google Scholar]
- 140.Workman JL, Abmayr SM, Cromlish WA & Roeder RG Transcriptional regulation by the immediate early protein of pseudorabies virus during in vitro nucleosome assembly. Cell 55, 211–219 (1988). [DOI] [PubMed] [Google Scholar]
- 141.Kraus WL & Kadonaga JT p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev 12, 331–342 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Utley RT et al. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394, 498–502 (1998). [DOI] [PubMed] [Google Scholar]
- 143.An W, Palhan VB, Karymov MA, Leuba SH & Roeder RG Selective requirements for histone H3 and H4 N termini in p300-dependent transcriptional activation from chromatin. Mol. Cell 9, 811–821 (2002). [DOI] [PubMed] [Google Scholar]
- 144.An W, Kim J & Roeder RG Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117, 735–748 (2004). [DOI] [PubMed] [Google Scholar]
- 145.Tang Z et al. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell 154, 297–310 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gu W & Roeder RG Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 (1997). [DOI] [PubMed] [Google Scholar]
- 147.Sheikh BN & Akhtar A The many lives of KATs - detectors, integrators and modulators of the cellular environment. Nat. Rev. Genet 20, 7–23 (2019). [DOI] [PubMed] [Google Scholar]
- 148.Wang SP et al. A UTX-MLL4-p300 Transcriptional Regulatory Network Coordinately Shapes Active Enhancer Landscapes for Eliciting Transcription. Mol. Cell 67, 308–321.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tan M et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sabari BR et al. Intracellular crotonyl-CoA stimulates transcription through p300-catalyzed histone crotonylation. Mol. Cell 58, 203–215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Guermah M, Palhan VB, Tackett AJ, Chait BT & Roeder RG Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell 125, 275–286 (2006). [DOI] [PubMed] [Google Scholar]
- 152.Kim J, Guermah M & Roeder RG The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140, 491–503 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Li G et al. Highly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivo. Mol. Cell 38, 41–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shimada M et al. Gene-Specific H1 Eviction through a Transcriptional activator→p300→NAP1→H1 Pathway. Mol. Cell 74, 268–283.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Roeder RG Lasker Basic Medical Research Award. The eukaryotic transcriptional machinery: complexities and mechanisms unforeseen. Nat. Med 9, 1239–1244 (2003). [DOI] [PubMed] [Google Scholar]
- 156.Kaikkonen MU & Adelman K Emerging roles of non-coding RNA transcription. Trends Biochem. Sci 43, 654–667 (2018). [DOI] [PubMed] [Google Scholar]
- 157.Rowley MJ & Corces VG Organizational principles of 3D genome architecture. Nat. Rev. Genet 19, 789–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Coleman RA et al. Imaging transcription: past, present, and future. Cold Spring Harb. Symp. Quant. Biol 80, 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang H, La Russa M & Qi LS CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem 85, 227–264 (2016). [DOI] [PubMed] [Google Scholar]
- 160.Hnisz D, Shrinivas K, Young RA, Chakraborty AK & Sharp PA A phase separation model for transcriptional control. Cell 169, 13–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hahn S Phase separation, protein disorder, and enhancer function. Cell 175, 1723–1725 (2018). [DOI] [PubMed] [Google Scholar]