Abstract
We introduce a novel method for delineating context‐dependent functional brain networks whose connectivity dynamics are synchronized with the occurrence of a specific psychophysiological process of interest. In this method of context‐related network dynamics analysis (CRNDA), a continuous psychophysiological index serves as a reference for clustering the whole‐brain into functional networks. We applied CRNDA to fMRI data recorded during the viewing of a sadness‐inducing film clip. The method reliably demarcated networks in which temporal patterns of connectivity related to the time series of reported emotional intensity. Our work successfully replicated the link between network connectivity and emotion rating in an independent sample group for seven of the networks. The demarcated networks have clear common functional denominators. Three of these networks overlap with distinct empathy‐related networks, previously identified in distinct sets of studies. The other networks are related to sensorimotor processing, language, attention, and working memory. The results indicate that CRNDA, a data‐driven method for network clustering that is sensitive to transient connectivity patterns, can productively and reliably demarcate networks that follow psychologically meaningful processes. Hum Brain Mapp 37:4654–4672, 2016. © 2016 Wiley Periodicals, Inc.
Keywords: fMRI, network dynamics, functional connectivity, emotion
Abbreviations
- CRNDA
Context‐related network dynamics analysis
- DCM
Dynamic causal modeling
- ERQ
Emotion regulation questionnaire
- ICA
Independent component analysis
- NCI
Network cohesion index
- PLS
Partial least squares
- PPI
Psychophysiological interaction
- SPGR
Spoiled gradient echo
- TPJ
Temporo‐parietal junction
INTRODUCTION
The research of large‐scale brain networks is a growing trend within cognitive neuroscience, especially in functional magnetic resonance imaging (fMRI) studies. Recent works employing various unsupervised methods suggest replicable parcellation of the brain into functionally meaningful networks including the “executive control”, “sensory‐motor”, “default mode”, and “salience” networks [Allen et al., 2012; Seeley et al., 2007; Shirer et al., 2012].
Although these networks are often regarded as “intrinsic networks”, having a stationary connectivity level over time (at least partially due to an underlying anatomical setup), evidence increasingly points to the time‐varying nature of this connectivity as a key feature of the brain (for a review, see Hutchison et al., 2013]. Thus, computational models of spontaneous neural interactions [Deco et al., 2009; Honey et al., 2007] suggest that functional connectivity fluctuation, as measured by fMRI, is a fundamental characteristic of network functioning. Moreover, using sliding window correlation estimates to analyze the magnitude and spectral characteristics of correlation patterns, reveals a meaningful variance of these factors across brain systems and populations. For example, such analyses indicated the existence of prototypical connectivity patterns [Allen et al., 2012], the dynamic interplay between specific functional networks [Gonzalez‐Castillo et al., 2014; Leonardi et al., 2014; Raz et al., 2014], and the links between abnormalities in network dynamics and pathological states [Sakoğlu et al., 2010; Shen et al., 2014].
A common practice in the increasing use of sliding window correlations is to define the networks in advance. This is done on the basis of connectivity analysis applied over entire epochs of resting state data [Allen et al., 2012; Sakoğlu et al., 2010; Smith et al., 2012; Sripada et al., 2014], rather than on the basis of connectivity dynamics. In this case, intrinsic networks are first delineated by applying independent component analysis (ICA) or correlation‐based clustering to resting‐state data. Their temporal connectivity indices are then compared across different physiological and psychological states or across groups of subjects.
These approaches use separate criteria to form the networks than those applied to estimation of the dependency between their connectivity dynamics and processes of interest. Thus, they are not optimized by design to maximize the synchrony between network connectivity dynamics and reference signals. Therefore, they may be less sensitive to the emergence of transient context‐related neural constellations which may underlie flexible and adaptive behavioral reactions [Durstewitz and Deco, 2008]. It should be noted that ICA was also used for delineating transient connectivity patterns in a subset of time windows (see [Esposito et al., 2003] for a sliding‐window approach). However, such approach does not allow for the probing of connectivity dynamics of the same network, but rather demarcates ad‐hoc networks in different time points.
Psychophysiological interaction (PPI) and dynamic causal modeling (DCM) are apparently the most commonly used methods for analyzing network connectivity dynamics in fMRI. PPI analysis [Friston et al., 1997] investigates the interactions between brain responses and a continuous experimental parameter. DCM [Stephan et al., 2007] performs a Bayesian comparison between connectivity models of hypothesized networks and the influence of experimental conditions on the connectivity; it thereby enables the analysis of the temporal aspects of connectivity within a set of regions.
These methods, at least in their classical formats, do not suggest a whole‐brain data‐driven delineation of distinct networks related to specific functions. PPI analysis traces pairwise correlations between the signals of a seed region of interest and other voxels in the brain. However, it does not account for the connectivity state of a network (i.e., a set of interacting nodes). Moreover, in PPI analysis, the seed regions have to be a‐priori defined, contrary to data‐driven analysis. Similarly, DCM is limited to the analysis of a given network.
Behavioral partial least squares (PLS) analysis [Krishnan et al., 2011] is another data driven approach that finds matched pairs of components, one over brain activity and the other over behavioral conditions (or time points in the case of a continuous behavioral measure). PLS is not typically used with dynamic connectivity data, but in principle it can be adapted to account for connectivity dynamics by replacing the brain activity values with inter‐regional pairwise correlation indices. Such an adapted method would provide matched pairs of components wherein one is over pairs of ROIs (or ROI graph edges) and the other is over behavioral conditions (or time points for a continuous measure). Interpreting such a result would need further processing such as thresholding the component values and comparing the components over time points with relation to the behavioral measure.
The alternative approach presented in this paper produces clusters of ROIs with a more clearly defined meaning that need not be interpreted in relation to a time‐wise component. It uses contextual information already at the network delineation stage, rather than first selecting the networks and then assessing their connectivity in relation to context. This method is especially sensitive to context‐dependent transient connectivity fluctuations. It offers a whole‐brain parcellation into networks whose connectivity level covaries with continuous indices that reflect specific psychological and physiological states. Our approach is in line with the notion that a specific set of inter‐regional interactions in the brain varies depending on the relevant context.
In our method for Context‐Related Network Dynamics Analysis (CRNDA), a set of regions is considered a context‐related network when the functional connectivity between these regions unfolds hand in hand with a continuous measure of a specific concurrent psychological or physiological process related to the context. CRNDA is an extension of the network cohesion analysis approach [Raz et al., 2012, 2014]. In contrast to previous work on dynamic functional connectivity, our network formation procedure actively searches for the relevant networks, supervised by the context index of interest. Our method is geared towards stimulus‐driven experimental paradigms, while most other dynamic connectivity studies to date are based on resting‐state experiments.
CRNDA (Fig. 1) starts with parcellation of the brain into regions that serve as network nodes. We then calculate a pairwise affinity measure between regions so that regions whose temporal connectivity is in sync with an independent time series of interest across subjects have higher pairwise affinity. We cluster the nodes into networks that are then tested for statistical significance on a data set that was kept aside. The process can be repeated in a cross‐validation paradigm to assess the stability of the networks and allow for overlap between resulting networks. Finally, we test the statistical significance of the findings on an independent data set.
Figure 1.
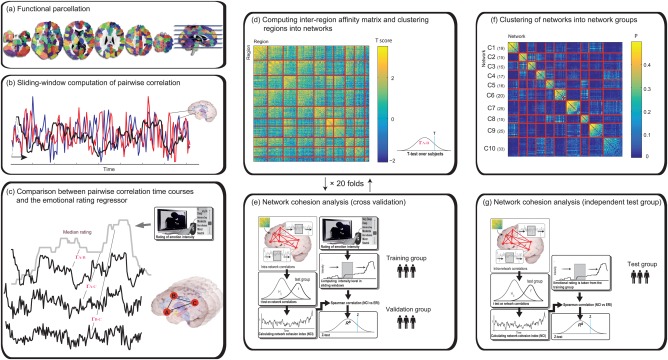
Context‐Dependent Network Dynamics Analysis scheme. (a) Gray matter was parceled into elementary regions of interest using the Iclust algorithm. The different color patches represent distinct ROIs in this partition of 150 ROIs per hemisphere. (b) Correlation between the signals of each pair of ROIs was computed in sliding windows of 30 seconds (c) For each pair of regions and each subject, the correlation time course was correlated with the median rating using Spearman's test. (d) A t‐statistic was used to summarize the latter correlations for each ROI pair. The heatmap represents an affinity matrix based on the resulting t‐scores. Rows and columns are sorted and partitioned according to the clustering results. (e) NCIs of the resulting clusters were computed for the training and validation groups as described in the main text. Actions (d) and (e) were repeated 20 times with random splits of the subjects into training and validation groups. (f) A similarity matrix of Jaccard scores of the spatial overlap between each pair of networks resulted from the 20 cross‐validation folds. Rows and columns are sorted and partitioned according to the network clustering. (g) The link between the NCI of the final networks and the median rating was validated on an independent test group. [Color figure can be viewed at http://wileyonlinelibrary.com]
We applied CRNDA to fMRI data and a moment‐to‐moment self‐reported rating of emotion intensity by subjects who viewed an excerpt from the commercial film Stepmom [Columbus]. This movie was previously shown to induce an experience of sadness [Raz et al., 2012]. The experimental setup followed previous fMRI research [Bartels and Zeki, 2005], which demonstrated the power of a life‐like cinematic experimental paradigm relative to resting state to expose functional segregation and specialization.
To further validate the relevance of networks delineated by CRNDA for emotion‐related processing, we examined whether their cohesion index covaries with an independent index of emotion regulation tendencies; this index is related to the context of the emotional movie. We expected the emotion‐related networks we identified to show lower connectivity among individuals with higher tendency to regulate their emotions, especially during the film's dramatic peaks.
METHODS
Participants
We collected data from two samples of healthy volunteers with no known history of neurological or psychiatric disorder. All volunteers had at least 12 years of education, with Hebrew as their spoken language. The participants signed an informed consent form approved by the ethical committees of the Tel Aviv Sourasky Medical Center. Valid fMRI data were obtained from 53 subjects (age: 26.75 ± 4.86 years, 21 females) and from 31 additional subjects (age: 32.6 ± 5.57 years, 7 females) in Samples 1 and 2, respectively after we excluded 21 subjects' data sets due to technical failures and exaggerated head motions. Parts of the data collected in Sample 1 were used in previous studies [Raz et al., 2012, 2013b].
Task
The participants passively viewed a clip from the movie Stepmom (Columbus, [Link]; 8:21 min) to elicit the experience of sadness. The clip included two scenes in which a terminally‐ill mother talks about her impending death with each of her two children. The display was preceded and followed by a three minute epoch, during which the participants passively watched an all‐black slide.
Continuous Emotion Intensity Ratings
The participants watched the same movie clip again in a post‐scan session outside of the scanner. They were instructed to report, in retrospect, on the intensity of sadness they experienced during the first viewing. Using in‐house software, the participants rated their sadness intensity on a 21‐point scale ranging from “neutral” to “very high” sadness intensity. This protocol was selected to avoid interfering with the emotional experience during the scan, following evidence on high correlations between original and retrospective ratings [Raz et al., 2012].
fMRI Acquisition and Preprocessing
Data were collected using a GE 3T Signa Excite echo speed scanner with an eight‐channel head coil, located at the Wohl Institute for Advanced Imaging at the Tel‐Aviv Sourasky Medical Center. We used a T1‐weighted 3D axial spoiled gradient echo (SPGR) pulse sequence for structural scanning, with the following parameters: TR/TE = 7.92/2.98 ms, slice thickness = 1 mm, flip angle = 15º, pixel size = 1 mm, FOV = 256 × 256 mm. We performed functional whole‐brain scans in interleaved order with a T2*‐weighted gradient echo planar imaging pulse sequence (time repetition [TR]/TE = 3,000/35 ms, flip angle = 90º, pixel size = 1.56 mm, FOV = 200 × 200 mm, slice thickness = 3 mm, 39 slices per volume). The subjects were given active noise cancelling headphones (Optoacoustics).
We pre‐processed the resulting data using Brain Voyager QX version 2.4. Head motions were detected and corrected using trilinear and sinc interpolations, respectively, applying rigid body transformations with three translation and three rotation parameters. The criterion for exclusion due to exaggerated head motions was deviations higher than 1.5 mm and 1.5° from the reference point. Valid data were high pass filtered at 0.008 Hz, and we applied a spatial smoothing with a 6 mm FWHM kernel. To avoid the confounding effect of fluctuations in the whole‐brain BOLD signal, the mean white matter and ventricle signals were regressed out of every data set. We then used an ICBM 452 probability map (http://www.loni.usc.edu/atlases/Atlas_Detail.php?atlas_id=6; Last accessed on 27/07/2016) to generate the white matter and ventricle masks. The probability thresholds were set to 95% and 99% for the ventricles and white matter masks, respectively.
We co‐registered SPGR images with the corresponding functional maps (via Brain Voyager QX automatic algorithm and manual verification) after standardizing them to 1 × 1 × 1 mm and transforming them into Talairach space.
ROI Signal Extraction
We first parceled gray matter voxels into regions of interest (ROIs), which served as candidate nodes in the CRNDA network‐forming procedure (see Supporting Information for details). A simple method of extracting ROI signals is to average the signal over the voxels. To further ensure that each brain region represented a single coherent signal, we also performed an intra‐region voxel weighing step. The top 10% of voxels with the highest mean intra‐region similarity (i.e., average correlations) were selected to be the core voxels. All the voxels were then weighted by their mean correlation to the core voxels (Supporting Information Fig. S1). Regional voxels that were negatively correlated to the core (if any existed) received zero weight, and were thus effectively removed. We used this weighting to linearly combine the voxel activations into a regional signal.
Context‐Related Network Dynamics Analysis
Given an ROI map that clustered voxels into regions, a data set of subjects' fMRI, a regressor of interest (subjects' emotional rating), and the requested number of networks, our network forming procedure involves the following basic steps:
Use the training set to calculate a suitable inter‐region affinity measure that captures the hypothesized function of the networks.
Use the affinity measure and a clustering procedure to cluster the regions into networks.
Assess the correlation between the network's dynamic connectivity (cohesion) index and the continuous psychophysiological measure of interest to produce a fitness score.
Steps 1 and 2 generate candidate networks from training data. Since clustering methods can produce results that are sensitive to data perturbations [Rand, 1971], we repeated steps 1 and 2 twenty times on random subsets of training set subjects and summarized the results to enhance the robustness of our analysis. To summarize the results, we assessed the tendency of each of the ROIs to stay clustered in the same network across repetitions and integrated the multiple clusterings into a single stable set of networks.
Step 3 assesses the strength of the relationship between a network's dynamic connectivity and the psychophysiological measure. We applied this step to a held‐out test set with a completely new set of subjects, to obtain statistical significance.
The abovementioned steps are described in detail below.
Basic step #1, computing inter‐region affinity matrix
This first step uses an affinity measure to derive the networks. It operationalizes the notion that regions belong to the same functional network if their connectivity fluctuates in concert with a relevant variable of interest.
For each subject, s, and each pair of regions, i and j whose time series xi and xj were obtained in the ROI forming procedure, let
| (1) |
Be a pairwise time series of normalized dynamic covariance, where the covariance is taken over a running time window of size ending at time t. The normalizing and are the standard deviations of the full time series xi and xj, respectively. We computed the standard deviations over the entire time series instead of the running time window, because we found this to yield a less noisy signal in preliminary tests (due to higher inter‐subject correlation of the connectivity index time series)1.
We selected the variable of interest to be the median emotion intensity rating, taken at each time point over all the subjects. Let
| (2) |
Be the Spearman correlation between the median rating and each subject's pairwise dynamic covariance of regions i and j for subject s. The i,j entry of the affinity matrix S is
| (3) |
is, therefore, the result of a t test, applied over the Fisher‐z transformed set of correlations calculated for each subject using Eq. (2). Consequently, this measure captures the significance of non‐zero correlation between inter‐region dynamic connectivities and the median emotion rating.
In this analysis, the window size in Eq. (1), , was set to 30 seconds (10 TRs), following evidence on functionally meaningful connectivity fluctuations at this time‐scale [Hutchison et al., 2013]. We calculated the median rating values in corresponding 30 second sliding windows, so the emotional regressor would reflect the rating in the same time window (of size ) as the connectivity.
Basic step #2, clustering regions into networks
We used the Iclust algorithm to form networks of regions with the inter‐region affinity matrix S as the similarity matrix and n c = 10 as the requested number of clusters. (Fig. 1d). We defined the resulting clusters as candidate functional networks. These networks are the result of a search in the huge space of all possible partitions of all the regions into clusters. The score of each cluster is the mean intra‐cluster similarity. The solution is locally optimal in that the total intra‐cluster similarity would decrease if any single region were to be reassigned to a different cluster. To improve the chance of finding a globally optimal solution, Iclust was set to perform its internal optimization 10 times from random initial conditions and to report on the best‐scoring result.
Basic step #3, network cohesion analysis (assessing fitness score of candidate networks)
To assess the statistical significance of the synchrony between candidate networks and the emotional intensity index, we first defined an index of network dynamic connectivity (or Cohesion), similar to the network cohesion index (NCI) of Raz et al. [Raz et al., 2012]. This parallels the pairwise dynamic connectivity , used earlier for clustering. For each network, , subject s and time point t let
| (4) |
where calculated as
| (5) |
With being the sample mean, s the sample standard deviation and n the sample size ( in this case). That is, is the t‐statistic of the set of intra‐network pairwise dynamic covariances [defined in Eq. (1)].
Next, we measured the synchrony of the NCI with the measure of emotional intensity. For every individual subject and network, we compared the NCI with the median rating of sadness intensity using Spearman's correlation.
| (6) |
We defined the fitness of network k for the subject group as the t‐statistic of their Fisher‐z transformed correlations:
| (7) |
Equations (4), (6), and (7) extract a fitness score for the network which is mirrored by the pairwise affinity measure extracted for pairs of regions in Eqs. (1), (2), (3), respectively. The pairwise normalized covariance of Eq. (1) is replaced by a group t‐statistic in Eq. (4). Eqs. (3) and (5) are identical except for the replacement of the pairwise dynamic connectivity of Eq. (1) with that of the network dynamic connectivity from Eq. (4). Equations (3) and (7) are similarly matched, calculating a group statistic over the results of Eqs. (2) and (6), respectively.
As noted in the method outline, we performed this step on training sets to select the best number of ROIs. We repeated it again after the results of 20 folds of network clustering were integrated (see below). This allowed us to test the significance of the correlation between cohesion of the integrated networks and the emotion rating in an independent sample of subjects.
Stable Network Extraction and Significance Testing
The 53 subjects of Sample 1 were randomly subsampled in each of the 20 clustering folds, (selecting approximately half of the subjects). We performed the network forming procedure (Steps 1 and 2) and estimated the fitness score on the same sample (Step 3). This procedure produced 20 folds × 10 networks = 200 networks. While networks from the same fold were necessarily disjoint, 2 folds would often yield very similar, yet always slightly different, networks. To produce a final set of networks containing regions that are robust to the sampling of training subjects for the affinity matrix, we performed the following integration procedure:
- We clustered the networks into 10 groups (see Fig. 1f), using Iclust. We provided Iclust with a similarity matrix comprised of the Jaccard similarity indices, defined as the number of regions in the intersection of the 2 networks divided by the number of regions in their union:
(8) For each group of networks i and each region j, we defined a region membership score to be the proportion of networks containing region j that fell in group i.
We assessed a null distribution of membership scores using a permutation sampling procedure that kept the sizes of the networks, but randomly permuted the assignment of regions to networks (repeating steps I and II). The 95% confidence threshold for membership was found to be 0.3.
We then constructed a final set of 10 integrated networks from the 10 groups, by assigning each region to the groups that contained it with a membership score above the confidence threshold. Some regions were assigned to more than a single network and some were not assigned to any network in this step, since their highest membership score was below the 0.3 threshold.
The significance of the correlation between the rating and the integrated networks NCI was then assessed (as explained in basic step 3) using a completely new test set of 31 subjects (Sample 2) not used during the network forming procedure. We performed the statistical test as specified in Eqs. (6) and (7), and applied FDR correction for multiple (10) comparisons [Benjamini and Hochberg, 1995].
Test of Network Spatial Specificity
The previous steps ensured that the final networks had a statistically significant positive correlation of connectivity dynamics with the measure of interest (sadness rating). It also verified that the regions making up the networks (i.e., the clustering results) were robust to selection of the training set used to learn the clustering. We applied a third statistical test to assess the spatial specificity of the results. This served to examine the extent to which the strength of the rating‐cohesion link was unique to the specific spatial constellation of the networks identified in the previous stage. For each of the final networks that showed significant NCI‐rating association, we created 1000 corresponding pseudo‐networks, each containing ROIs that were randomly selected from the full ROI map. The number of ROIs selected remained constant and equal to that of the original network. We computed the NCI‐rating association in the second sample group for each of the random networks and compared the strength of the original network's association to this background distribution. The spatial specificities of the networks were defined as the proportion of random networks that had a lower NCI‐rating association score.
Functional Profiling of the Networks
We used the Neurosynth database (http://neurosynth.org/, Last accessed on 27/07/2016) [Yarkoni et al., 2011] to obtain evidence for the functional characteristics of the final networks. Neurosynth is a meta‐analytical platform that aggregates findings from over 10000 neuroimaging studies. Our functional profiling procedure tagged the networks with psychologically meaningful labels by summarizing the tags assigned by Neurosynth to each of the network's nodes (ROIs).
For each ROI, a representative voxel was defined as: the voxel with the maximal average correlation coefficient with the other voxels in the ROI. Functional tagging was performed for each of these representative voxels in the network. Using Neurosynth, we computed posterior probability indices that reflect the likelihood of any term in the database being used in a study that reports on activation of this voxel. For each term, we averaged the posterior probability indices over the network's nodes to indicate the extent of relevance of that term to the entire network.
Following the functional tagging results, we performed a post‐hoc analysis to examine the link between the networks and the empathy‐related notions found relevant to some of the networks. There is growing evidence for the existence of two distinct neural circuits: cognitive empathy [also called “Theory of Mind” (ToM)] and sharing other's internal states [Zaki and Ochsner, 2012]. We specifically tested the similarity of our ‘empathy‐related’ networks to available statistical maps obtained from two sources: (i) a recent ToM functional localizer experiment [Spunt and Adolphs, 2014; thresholded and binarized at Q FDR < 0.05) contrasting “how” and “why” questions.; (ii) meta‐analytic conjunction analysis demarcating regions activated both when an individual experiences pain and observes other's pain [Lamm et al., 2011; the map was kindly provided by the author and thresholded and binarized at P < 0.05, FWE‐corrected on cluster‐level).
Recent neuroimaging studies associate interoceptive and affective processes related to empathy for pain with a “salience network,” which is more broadly implicated in the detection of stimuli relevant for bodily homeostasis (see [Menon, 2011]. Therefore, we also tested the overlap between the final networks and a statistical map representing the “salience network” as originally reported by Seeley and colleagues [Seeley et al., 2007]. We expected that a network that overlapped with the ES circuit would also show similarity with the salience networks.
To assess the spatial overlap between the seven final networks that passed the cross‐validation test and these previously delineated networks, we computed the Jaccard index [see Eq. (8)] between each of our networks and the three maps. To assess the statistical significance of the results, we compared these scores with Jaccard indices computed in the same manner for permuted networks. Thus, for each comparison between a final network and one of the two statistical maps, the result was compared with Jaccard scores computed for 1000 corresponding networks generated as described in the previous section (preserving the number of ROIs but otherwise random).
Relations Between Emotion‐Related Network Cohesion and Emotion Regulation
We wanted to further explore the relevance of the networks delineated by CRNDA to emotion‐related processing. To do this, we examined whether there were times during the movie presentations when the networks' cohesion indices covaried across subjects with independently measured emotion‐related trait indices; these indices were obtained from 37 subjects in Sample 1. The subjects filled in the emotion regulation questionnaire (ERQ; [Gross and John, 2003], which assesses the individual's tendency to regulate emotions using two strategies: cognitively reappraising the situation (Reappraisal) and suppressing the expression of emotion once it is elicited (Suppression).
We performed a post‐hoc testing of the covariance between these indices of emotional reaction and the connectivity measures of networks identified by CRNDA [as measured with Eq. (4)]. This was done to account for the use of emotion regulation processes to modulate empathic responsiveness [Decety and Lamm, 2006; Engen and Singer, 2013]. To avoid an excess of redundant hypotheses testing, we limited the analysis to networks that were linked by our functional profiling procedure with the emotion‐related domain of empathy. We excluded other networks related to domain‐general processes. We specifically tested Networks #1, #2, and #9, which were tagged as related to either ToM and mentalizing (Networks #1 and #9), or to empathy for pain (Network #2; see Fig. 4 and Bartels and Zeki, 2005).
Figure 4.
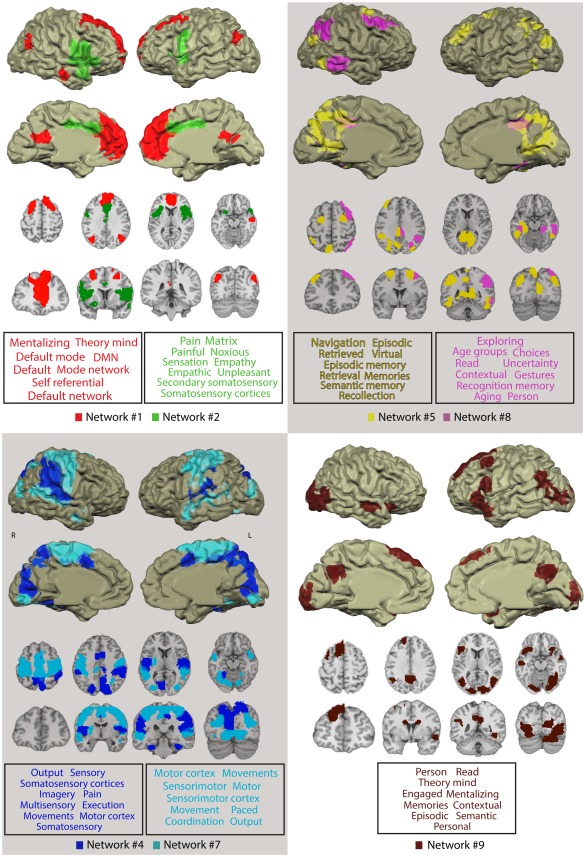
Networks delineated by CRNDA whose NCI showed reliable correlation with the sadness intensity rating and their functional profiling. For clarity, we superimposed no more than two networks on each anatomical image. The font size in the word cloud is proportional to the average probability with which the term was assigned to peak voxels in the network by the Neurosynth database. Only the top 10 functional terms are visualized (for the entire list of 100 terms, see Supporting Information Table SI). [Color figure can be viewed at http://wileyonlinelibrary.com]
Using Spearman's coefficient, we tested the relations between the NCIs of these networks and the reappraisal and suppression scores in each of the time windows across the movie. We did this for each of the two families of hypotheses: Suppression and Reappraisal. An FDR correction was applied to control the number of comparisons (i.e., 151 time‐windows × 3 networks = 453 comparisons).
RESULTS
Efficacy of the Movie Clip as an Emotion Elicitor
The continuous rating of sadness intensity induced by the movie indicates that the experimental stimulus used in the study was effective in triggering a dynamic emotional experience. The median rating over Sample 1 (Fig. 2) had two peaks, corresponding to “moderate” and “moderate to high” levels of sadness intensity. As mentioned in the introduction, this result and its robustness for this stimulus has already been studied [Raz et al., 2013b].
Figure 2.
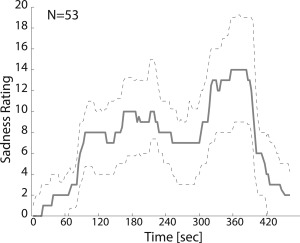
Emotional rating. The solid line indicates the median sadness intensity rating by participants who watched a scene from the movie Stepmom (Columbus, 1998). The interquartile range is represented by the dashed lines.
Delineation of Emotion‐Related Context‐Dependent Networks
Gray matter was parceled into regions with functional clustering using the Iclust algorithm (Fig. 1a). We ran the CRNDA network forming procedure in 20‐folds, using the affinity matrices resulting from an assessment of the correlations between pairwise dynamic connectivity and the median sadness rating across all Sample 1 subjects (Fig. 1b–d).
We repeated the procedure for ROI maps containing 100, 160, 200, 300, and 400 parcels (divided evenly across hemispheres). The networks' fitness scores, measuring the significance of the correlations between network dynamic connectivities (NCIs) and the median sadness rating, were calculated on training set subjects per fold (basic‐step #3). The ROI map with the best average fitness score across folds was that with 300 (with a t‐score of 3.32 compared with 3.23, 3.31, 3.31 and 3.29 for ROI maps with 100, 160, 200, and 400 ROIs, respectively).
We assimilated the networks obtained across the 20 folds into 10 groups (Fig. 1f). This procedure resulted in the demarcation of 10 final networks. We employed a permutation filtering procedure (Fig. 1f) to exclude ROIs that were weakly related to this final set of network groups. We computed the membership scores for each of the regions and thresholded them according to a 95% confidence interval, indicating the probability of assigning ROI to a network in a random null membership distribution. This threshold was empirically found to be 0.3.
We examined the link between the resulting networks and the rating in the independent Sample 2. After FDR correction for multiple comparisons, the NCI of seven out of the ten networks showed significant correlation with the rating (Fig. 3). These networks have widespread and mostly bilaterally symmetric configurations (Fig. 4 and Table 1).
Figure 3.
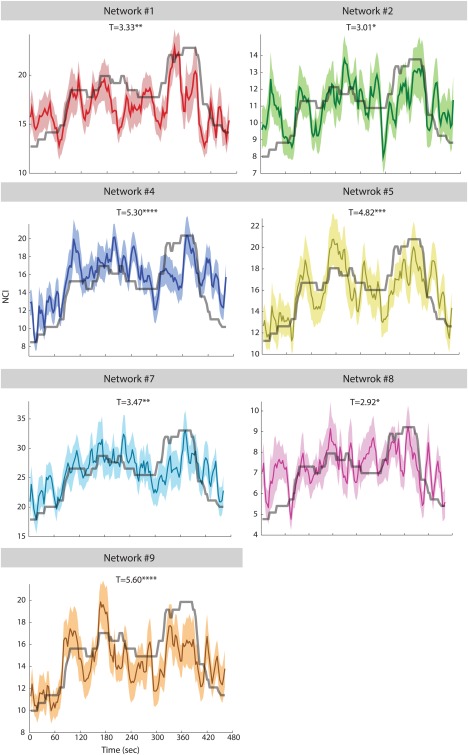
Relations between median sadness rating for a scene from the movie Stepmom (Columbus, 1998) and the NCIs of the networks delineated by CRNDA. The data presented here were acquired from a second independent sample group (N = 31). Colored surfaces indicate the standard errors. *P < 0.02 (Q FDR < 0.05), *P < 0.005 (Q FDR < 0.05), **P < 10 − 5 (Q FDR < 0.05). [Color figure can be viewed at http://wileyonlinelibrary.com]
Table 1.
Details on the components of the final networks delineated by CRNDA
| Region label | Region size (voxels) | Region peak | ||
|---|---|---|---|---|
| X | Y | Z | ||
| Network #1 | ||||
| L Anterior Cingulate (Brodmann area 32) | 2521 | −6 | 44 | 7 |
| L Medial Frontal Gyrus (Brodmann area 9) | 217 | −3 | 53 | 28 |
| L Medial Frontal Gyrus (Brodmann area 8) | 161 | −6 | 35 | 37 |
| L Middle Frontal Gyrus (Brodmann area 8) | 289 | −21 | 26 | 46 |
| R Middle Frontal Gyrus (Brodmann area 6) | 255 | 21 | 17 | 61 |
| R Medial Frontal Gyrus (Brodmann area 10) | 28 | 6 | 50 | 10 |
| R Superior Frontal Gyrus (Brodmann area 10) | 41 | 21 | 59 | 19 |
| R Superior Frontal Gyrus (Brodmann area 10) | 32 | 12 | 35 | 55 |
| R Superior Frontal Gyrus (Brodmann area 8) | 30 | 12 | 53 | 37 |
| R Anterior Cingulate (Brodmann area 32) | 45 | 9 | 41 | 1 |
| L Posterior Cingulate (Brodmann area 23) | 152 | −6 | −55 | 19 |
| R Angular Gyrus (Brodmann area 39) | 153 | 45 | −64 | 28 |
| L Angular Gyrus (Brodmann area 39) | 111 | −39 | −70 | 31 |
| R Anterior Middle Temporal Gyrus (Brodmann area 21) | 79 | 57 | −13 | −17 |
| Network #2 | ||||
| L Middle Cingulate Gyrus (Brodmann area 32) | 348 | −3 | 23 | 31 |
| L Middle Cingulate Gyrus (Brodmann area 24) | 104 | −3 | −13 | 40 |
| R Middle Cingulate Gyrus (Brodmann area 24) | 5 | 3 | −4 | 37 |
| R Inferior Frontal Gyrus (Brodmann area 44) | 1141 | 54 | 17 | 19 |
| R Anterior Insula (Brodmann area 13) | 427 | 36 | 20 | 1 |
| R Precentral Gyrus (Brodmann area 44) | 110 | 48 | 2 | 19 |
| R Middle Frontal Gyrus (Brodmann area 46) | 21 | 39 | 23 | 19 |
| R Claustrum | 20 | 30 | 5 | 16 |
| R middle Insula (Brodmann area 13) | 34 | 42 | −1 | 4 |
| R Anterior Superior Temporal Gyrus (Brodmann area 38) | 77 | 51 | 17 | −17 |
| L Inferior Frontal Gyrus (Brodmann area 44) | 509 | −51 | 5 | 1 |
| L Anterior Insula (Brodmann area 13) | 97 | −36 | 14 | 10 |
| L Lentiform Nucleus (Putamen) | 50 | −24 | 11 | −14 |
| Network #4 | ||||
| R Cuneus (Brodmann area 18) | 1506 | 9 | −73 | 34 |
| R Precuneus (Brodmann area 7) | 202 | 6 | −55 | 40 |
| L Cuneus (Brodmann area 17) | 250 | −6 | −76 | 7 |
| L Precuneus (Brodmann area 19) | 93 | −18 | −76 | 40 |
| R Posterior Cingulate Gyrus (Brodmann area 31) | 61 | 9 | −28 | 46 |
| R Supramarginal Gyrus/Postcentral Gyrus (Brodmann areas 1,4,40) | 1171 | 57 | −37 | 40 |
| R Inferior Parietal Lobule (Brodmann area 40) | 118 | 57 | −28 | 28 |
| R Postcentral Gyrus (Brodmann area 2) | 32 | 51 | −25 | 40 |
| R Posterior Insula (Brodmann area 13) | 32 | 48 | −28 | 19 |
| R Superior Temporal Gyrus (Brodmann area 41) | 195 | 60 | −19 | 10 |
| L Inferior Parietal Lobule/Postcentral Gyrus (Brodmann areas 1,40) | 691 | −54 | −31 | 37 |
| L Cingulate Gyrus (Brodmann area 24) | 195 | −3 | 5 | 43 |
| L Posterior Insula (Brodmann area 13) | 266 | −39 | −10 | 4 |
| L Superior Temporal Gyrus (Brodmann area 41) | 118 | −54 | −22 | 10 |
| L Parahippocampal Gyrus (Brodmann area 19) | 121 | −27 | −55 | −11 |
| R Parahippocampal Gyrus (Brodmann area 36) | 60 | 36 | −34 | −23 |
| Network #5 | ||||
| R Inferior Temporal Gyrus (Brodmann area 20) | 183 | 51 | −55 | −11 |
| R Middle Frontal Gyrus (Brodmann area 6) | 133 | 27 | 2 | 61 |
| R Superior Occipital Gyrus (Brodmann area 19) | 129 | 39 | −76 | 26 |
| R + L Cuneus/Precuneus/Cingulate Gyrus/Parahippocampal Gyrus (Brodmann areas 7, 28,30,31, 36) | 1840 | −7 | −71 | 33 |
| L Middle Frontal Gyrus (Brodmann area 6) | 125 | −24 | −1 | 61 |
| L Middle Frontal Gyrus (Brodmann area 9) | 157 | −36 | 38 | 37 |
| L Inferior Parietal Lobule/Superior Occipital Gyrus (Brodmann areas 19,40) | 555 | −47 | −47 | 38 |
| L Fusiform Gyrus (Brodmann area 37) | 227 | −49 | 49 | −17 |
| Network #7 | ||||
| R Inferior Parietal Lobule (Brodmann area 7) | 110 | 33 | −58 | 47 |
| R Lingual Gyrus, Fusiform Gyrus (Brodmann areas 18,19) | 816 | 12 | −80 | −8 |
| R Superior Temporal Gyrus (Brodmann area 22) | 528 | 50 | −56 | 16 |
| R Middle Occipital Gyrus (Brodmann area 19) | 206 | 30 | −82 | 18 |
| R Superior Temporal Gyrus/Middle Temporal Gyrus (Brodmann areas 21,38) | 185 | 48 | 3 | −20 |
| R + L Precentral Gyrus, Middle Frontal Gyrus, Medial Frontal Gyrus, Superior Frontal Gyrus (Brodmann areas 2,3,4,5,6) | 3756 | −45 | 1 | 46 |
| L Superior Temporal Gyrus, Middle Temporal Gyrus (Brodmann area 13, 21, 22) | 747 | −52 | −49 | 10 |
| L Middle Occipital Gyrus (Brodmann area 19) | 216 | −28 | −82 | 18 |
| L Lentiform Nucleus (Putamen) | 109 | −27 | −10 | 10 |
| Network #8 | ||||
| R Inferior Parietal Lobule (Brodmann area 40) | 617 | 48 | −58 | 43 |
| R Supramarginal Gyrus (Brodmann area 40) | 32 | 51 | −52 | 28 |
| R Superior Parietal Lobule (Brodmann area 7) | 145 | 36 | −73 | 46 |
| R Posterior Cingulate Gyrus (Brodmann area 31) | 108 | 6 | −37 | 37 |
| R Posterior Middle/Inferior Temporal Gyrus (Brodmann area 21) | 313 | 60 | −40 | −11 |
| R Middle Frontal Gyrus (Brodmann area 6) | 295 | 42 | 8 | 52 |
| R Superior Frontal Gyrus (Brodmann area 8) | 83 | 39 | 23 | 49 |
| R Parahippocampal Gyrus (Brodmann area 36) | 111 | 27 | −34 | −14 |
| Network #9 | ||||
| R Middle Temporal Gyrus (Brodmann area 21) | 125 | 57 | −7 | −6 |
| R Inferior Frontal Gyrus (Brodmann area 47) | 148 | 32 | 20 | −10 |
| R Caudate | 46 | 15 | −1 | 21 |
| R+L Middle Temporal Gyrus/Middle Occipital Gyrus/Inferior Occipital Gyrus (Brodmann areas 17, 18, 19, 37) | 2344 | 48 | −65 | 7 |
| R+L Precuneus/Cingulate Gyrus (Brodmann area 7,31) | 268 | −3 | −55 | 31 |
| L Middle Frontal Gyrus (Brodmann area 6) | 119 | −39 | 14 | 49 |
| L Inferior Frontal Gyrus/Superior Temporal Gyrus/(Brodmann areas 6, 38, 44, 45,47) | 503 | −51 | 17 | 17 |
| L Superior Frontal Gyrus/Middle Frontal Gyrus (Brodmann area 6,8,9) | 600 | −18 | 49 | 31 |
| L Middle Temporal Gyrus (Brodmann area 21) | 74 | −54 | −17 | −15 |
| L Caudate | 38 | −12 | −1 | 21 |
Because t‐scores indicate the strength of NCI‐rating association, we computed them for 1000 random sets of ROIs to assess the spatial specificity of the results for each network. We then compared the t‐score obtained for the original networks with the background distributions generated by this spatial permutation. The spatial specificity of all of the networks was higher than 91%, with maximal specificity for Networks #4, #5, and #9; the t‐scores of those networks were higher than all of the corresponding randomly sampled networks (Fig. 5).
Figure 5.
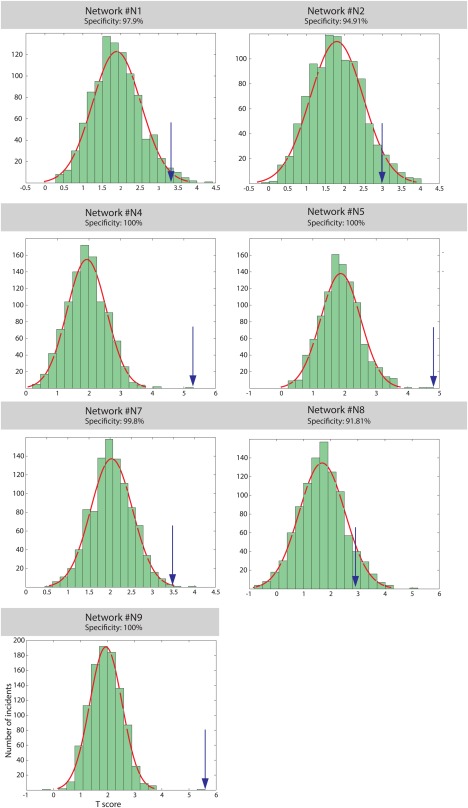
Spatial specificity of the NCI‐rating link as indicated by a permutation test. The blue arrows indicate the t‐scores of the original networks relative to a histogram of t‐scores for randomized networks. [Color figure can be viewed at http://wileyonlinelibrary.com]
Functional Profiling of the Networks
To gain insight into the set of functions in which each of the networks is commonly implicated, we used a meta‐analytical tool available at Neurosynth [Yarkoni et al., 2011]. This tool helped us examine the relevance of more than 3000 terms to specific brain voxels. Examining the posterior probability indices estimated by Neurosynth for each of these terms, we gained evidence for the tendency of voxels in the network's nodes to be preferentially involved in specific functions related to this term relative to the alternatives. We examined how frequently each of the terms was associated with the activation of the peak voxel in each of the network's nodes across the neuroimaging studies filed by Neurosynth. Figure 4 visualizes the probability of the most relevant functional terms (pure anatomical terms were not visualized) after averaging over all of the network's nodes. For an extended report on the tagging results, see Supporting Information Table SI.
The functional tagging results indicate that Networks #1 and #9 on the one hand, and Network #2 on the other, are preferably associated with different types of empathy. It showed a link between cognitive mentalizing and theory of mind for Networks #1 and #9, and empathic sharing of other's bodily state (e.g., pain) for Network #2 (Fig. 4).
To further test the specificity of this functional link, we compared these networks with statistical maps from relevant previous studies on theory of mind [Spunt and Adolphs, 2014], embodied simulation [Lamm et al., 2011], and salience detection [Seeley et al., 2007].
A permutation analysis indicated a highly spatially specific link between Network #1 and the ToM localizer map. The Jaccard score for this pair of maps was higher than any similarity index computed when comparing the ToM localizer map with spatially permuted Network #1 maps. (See Table 2 and Figure 6; while the Network #1‐ToM Jaccard score was 0.21, the highest score for a corresponding permuted network was 0.12). This ToM map also showed significant overlap with Network #9. A highly specific link was also found between Network #2 and the embodied simulation and salience map (P < 0.001). No other final network showed significant overlap with these maps.
Table 2.
Spatial overlap between final networks and statistical maps representing relevant functions and states
| ToM | ES (empathy for pain) | Salience | ||||
|---|---|---|---|---|---|---|
| JS | P | JS | P | JS | P | |
| Network #1 | 0.21 | <0.001 | 0.02 | 0.34 | 0.05 | 0.61 |
| Network #2 | 0.01 | 0.94 | 0.09 | <0.001 | 0.15 | <0.001 |
| Network #4 | 0.01 | 1 | 0.03 | 0.04 | 0.1 | 0.03 |
| Network #5 | 0.01 | 1 | 0 | 0.95 | 0.02 | 0.99 |
| Network #7 | 0.05 | 0.88 | 0 | 0.97 | 0.06 | 0.96 |
| Network #8 | 0.01 | 0.87 | 0 | 1 | 0.01 | 0.97 |
| Network #9 | 0.18 | <0.001 | 0 | 0.81 | 0.03 | 1 |
For references to the sources of the maps, see the main text.
Abbreviations: ToM—theory of mind, DMN—default mode network, ES—embodied simulation, JS—Jaccard score. Results surviving Q FDR < 0.05 are emphasized in bold.
Figure 6.
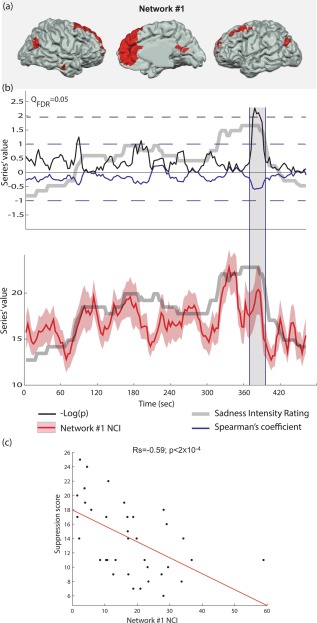
Emotional suppression index correlates with emotion‐related network cohesion during emotional peak. (a) Anatomical location of Network #1 nodes. (b) Spearman's correlation between Network #1 NCI and the suppression scores was calculated for each time‐window (the solid blue line indicates the coefficient value; the blue dashed line indicates the coefficient boundaries [−1 1]). Significant effect is evident when –log(p) (solid black line) crosses the Q FDR = 0.05 significance threshold (dashed black line). This effect occurs at the ending of the emotional rating peak (gray line). Network #1 NCI is indicated in the lower panel (the solid red line indicates the average over the group, while the pink surface indicates the standard error). (c) Scatter plot of the suppression scores and Network #1 NCI during the peak effect time‐window. [Color figure can be viewed at http://wileyonlinelibrary.com]
Association Between Emotion‐Related Network Cohesion and Emotion Regulation Indices
The analysis described above examines the correlation between NCI and a moment‐to‐moment measure of emotion (rating) within the subject. We hypothesized that the NCI of emotion‐related networks will also covary with relevant inter‐subject variability. Specifically, we tested the NCIs of Networks #1, #2, and #9, which are associated with empathy‐related tags and overlap with well‐documented empathy‐related networks (Figs. 4 and 6). This analysis examined whether they correlate with individual emotion regulation indices of reappraisal and suppression tendencies.
We found significant negative correlation between the NCI and suppression scores for Network #1 in five consecutive time‐windows around the movie's dramatic peak ( between −0.55 and −0.59, Q FDR < 0.05; Fig. 7). The first three of these time‐windows corresponded to the last time‐windows that had the maximal level of median rating. The results showed no other significant correlation between any of the networks and the emotion regulation indices. It should be noted that this analysis was limited to the three empathy‐related networks since we hypothesized their relevance to emotion regulation. Extending this test to all seven networks yields no results that survive the threshold of Q FDR < 0.05.
Figure 7.
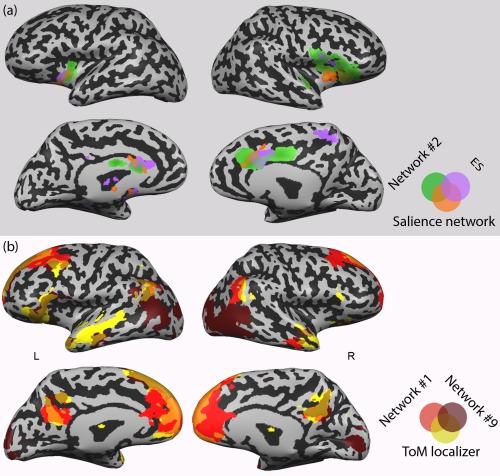
Networks delineated by CRNDA overlap with previously demarcated functional networks. (a) Network #2, the “salience network” [Seeley et al., 2007], and a conjunction map representing embodied simulation (ES) and empathy for pain (Lamm et al., 2011); (b) Network #1, Network #9, and ToM localizer map (Spunt and Adolphs, 2014). [Color figure can be viewed at http://wileyonlinelibrary.com]
DISCUSSION
This article introduces CRNDA, a novel method for identifying networks whose functional connectivity dynamics are related to a concurrent unfolding process. It demonstrates that CRNDA facilitates replicable delineation of brain networks related to a behavioral measure of interest. This method also yields time courses of network connectivity indices that allow the investigation of effects taking place during specific time‐windows.
Our approach is based on the assumption that when segregated brain regions participate in specific psychophysiological processes, their activity becomes synchronized as they form context‐related networks. As the relevant processes intensify, weaken, and are replaced by other processes, so does the connectivity between regions whose functions may be co‐dependent in certain situations (yet separate in others). By searching for a match between inter‐region connectivity and the intensity of a psychophysiological process, our approach makes this assumption explicit.
CRNDA searches across all the provided ROIs for multiple networks. We assume that although all of the resulting networks have dynamic connectivity, which is correlated with the variable of interest, they support separate concurrent processes. The separation between processes would then be reflected by the clustering procedure's affinity matrix as the difference between the intra‐cluster pairwise affinity values and the inter‐cluster affinities.
While our approach deviates from the common methods for investigating “static” connectivity patterns over entire resting‐state blocks, its application in the case reported here, yields findings that are largely congruent with existing literature, along with added valuable functional information. Thus, Network #2, which encompasses the bilateral mid‐anterior insula and the anterior cingulate, considerably overlaps with the “salience network” whose action has been related to a degree of subjective salience during interoception, pain, and emotions [Legrain et al., 2011; Seeley et al., 2007; Taylor et al., 2009]. It also shows considerable similarity with a network that has been associated with a mode of empathy, based on a mirroring‐like effect. In this network, overlapping somato‐visceral neural circuits are activated both in the individual who experiences empathy and her object of empathy [Lamm et al., 2011; Raz et al., 2013b]. The functional tagging analysis supports this link, as it associates Network #2 with the notions of pain, empathy, and somatosensory processing (Fig. 4). Thus, it is likely that the coupling of the increasing connectivity of Network #2, with the intensification of sadness experience, reflects somato‐visceral empathic reaction to the movies.
The neuroscientific literature on empathy distinguishes this mode of empathy from a higher‐level cognitively‐driven empathy [Zaki and Ochsner, 2012]. The latter, often called “mentalization” or “Theory of Mind” (ToM), involves inferences about the mental traits and states of other individuals [Frith and Frith, 2006]. While somato‐visceral empathy is associated with an insulary‐cingulate circuit, cognitive empathy is identified with a network that includes the medial prefrontal cortex, temporo‐parietal junction (TPJ), the posterior cingulate/precuneus, and the anterior temporal lobes. A recent meta‐analysis [Schurz et al., 2014] suggests that the medial prefrontal cortex and the bilateral posterior TPJ comprise a “core ToM” network, which is consistently activated across various mentalization tasks. This set of ToM‐related regions clearly overlaps with Network #1, delineated here by CRNDA (Fig. 6, Table 2). Indeed, network #1 is associated with the terms “theory of mind” and “mentalization”, as well as with the default mode network (Fig. 4), which has been related to these processes [Spreng et al., 2013].
This network is involved in the processing of emotion‐related content, as supported by the finding that its NCI negatively correlates with the emotion regulation index of suppression scores when the cinematic drama peaks and starts to wane. Interestingly, the inter‐subject variability in suppression tendency is exposed when the Network #1 NCI shows a decreasing trend, coinciding with the termination of the emotional cinematic scene (see Figs. 3a and 6). In other words, these results indicate that individuals who tend to suppress their emotional experiences were faster in “disintegrating” Network #1. The evidence that this effect is specific to Network #1, whose core regions have been repeatedly associated with the cognitive mode of empathy, and not to the insulary‐cingulate Network #2, provides an intriguing clue about the involvement of high‐order mechanisms in this mode of emotion regulation.
The functional profiling analyses pointed to the involvement of Network #9 in mentalizing and theory of mind as well. This network also contains a left lateralized set of classical language‐related regions, including Brodmann areas 39, 44, and 45. Two other Network #9 nodes, located in the left superior frontal gyrus and the superior medial frontal cortex, have been repeatedly associated with comprehension and inference processes [Chan et al., 2012; Moss et al., 2011; Yarkoni et al., 2008][Moss et al., 2011; Newman et al., 2013; Shibata et al., 2012]. This network also includes bilateral caudal nodes and visual‐related regions across the calcarine sulcus and the extrastriate cortex.
It is possible that this network mediates context‐dependent comprehension by integrating perceptual and conceptual processes. This is observed in light of evidence on the role of the caudate in modulating distributed language‐related regions in general, and specifically in sequencing temporal and cognitive processes [Chan et al., 2013; Duffau, 2008; Kotz et al., 2009]. This interpretation is supported by the association of Network #9 to the terms Contextual, Semantic, and Read (Fig. 4). Interestingly, CRNDA clustered the ToM‐related regions, including the posterior cingulate and temporal regions, with language‐related areas rather than with other components of the common ToM circuit in Network #1. This is in line with the intriguing evidence on functional segregation within this network, as suggested by Schurz and colleagues [Schurz et al., 2014].
The resemblance of Networks #1, #2, and #9 to known empathy‐related networks demonstrates the efficiency of CRNDA in delineating functionally‐relevant brain networks. Although separate experiments are generally employed to demarcate these different empathy‐related networks, CRNDA identified them as distinct functionally‐relevant networks within a single naturalistic experiment.
Apart from identifying networks related to empathy, CRNDA demarcated sets of regions that have been associated with domain‐general functions. Network #8 includes right interior parietal and dorsolateral loci, which are parts of a “right executive control network” (see Fig. 6c). This network is thought to have a key role in exerting cognitive control on goal‐directed behavior as suggested by Shirer and colleagues, for instance [Krmpotich et al., 2013; Shirer et al., 2012; Spreng et al., 2013]. A third node of Network #8, which is located in the middle/inferior temporal cortex, has been associated with related context‐sensitive attention responses [Mattler et al., 2006; Mitchell, 2007; Mobbs et al., 2006]. A fourth node in Network #8 is located in the posterior cingulate (Fig. 6c). This region, which is part of the executive control network [Krmpotich et al., 2013; Shirer et al., 2012; Spreng et al., 2013] has been specifically implicated in episodic encoding and retrieval [Spaniol et al., 2009]. Similarly, two other Network #8 nodes are located in the left fusiform and the right parahippocampal gyri, which are often activated in studies of memory encoding and retrieval [Burgmans et al., 2010; Hämäläinen et al., 2007; Kensinger and Schacter, 2007; Krug et al., 2010; Sakai et al., 2002; Spaniol et al., 2009].
Taken together, the evidence suggests that Network #8 integrates related cognitive functions of executive control, attention, and memory. This is also reflected by the functional profiling analysis, which associates this network with terms such as Exploring, Choices, Contextual, and Recognition (Fig. 4). This neural constellation, which seems to be unspecific to emotion, is strengthened during moments of high emotional intensity in which the coordination between these processes is particularly important to facilitate adaptive reaction.
While Network #8 is lateralized to the right hemisphere, Network #5 is spread over large left posterior cingulate, inferior parietal, and dorsolateral prefrontal areas (Fig. 4). It also contains the bilateral frontal eye field. This set of regions shows similarity to the “dorsal attention system”, which is implicated in top‐down attention allocation [Corbetta et al., 2008; Spreng et al., 2013; Vossel et al., 2014]. The inclusion of bilateral visual association regions in Network #5 may reflect the context‐dependent top‐down effect of the core dorsal attention network on early visual processing.
Network #4 includes large areas across the primary and associative auditory and visual cortices, the sensorimotor strip, and the supplementary motor area (SMA). It also contains bilateral parahippocampal foci (Fig. 6c). Network #7 includes widespread neighboring bilateral areas across the dorsal precentral and postcentral gyri, supplementary motor complex, and the middle and superior temporal cortex. It also contains visual‐related regions across the lateral and superior occipital gyri, and the lingual and fusiform gyri.
Functional tagging of these networks suggests that while Network #4 is associated more with sensory functions, motor processing is a salient aspect in Network #7. Again, the functions associated with the components of these networks are inter‐related but unspecific to emotion. Relying on early visual, auditory, and tactile processing on the one hand and on motor processing and planning on the other, both networks may function as perceptual‐motor circuits. Network #4 may be more focused on primary audiovisual processing and spatial orientation, with the parahippocampal nodes playing a specific role in scene recognition [Epstein, 2008]. Network #7, which includes no primary visual and auditory regions, but is spread over large association areas across the occipital and temporal lobes, may be related to higher‐order sensorimotor control functions. The finding that the connectivity of these networks increases with emotional intensity is in line with the notion that intense cinematic drama may trigger sensorimotor simulation [Grodal, 2009].
The functional characterization of the networks identified in this study can be further refined using relevant inter‐subject measures (of motor arousal or comprehension, for example) similar to the comparison of the NCI of Network #1 with emotion regulation indices. The option of extracting a continuous network connectivity index and testing its covariance with various inter‐subject variables in a given time‐window is an additional advantage of CRNDA relative to the common “static” functional connectivity approaches. The added value of this analytical feature is emphasized in light of evidence that individual differences—including pathological factors—are linked not only to the strength of network connectivity, but also to specific temporal patterns of connectivity [Hutchison et al., 2013]. Thus, CRNDA, which allows the extraction of temporal connectivity patterns on an individual basis, is particularly valuable in analyzing network connectivity dynamics.
The moment‐to‐moment rating of emotional intensity used here as the basis for CRNDA, is only one example for a possible psychophysiological measure. Future CRNDA studies may be based on other continuous indices of various relevant processes that take place during the scan. Moreover, in a modified CRNDA version, several indices may be combined into a general linear model so the network clustering will be based on the beta fit of these regressors.
Another issue for future inquiry is whether clustering into a smaller or larger number of networks can be reliable. This issue can be examined by varying the number of final networks (10 in the current study) and examining the replicability of the resulting clustering. Atlas‐based anatomical segmentation can be combined with the functional clustering procedure. Such an analysis may show that there is no single correct number, as the amount of separation between networks may be dependent on the number of clusters in a gradual fashion without a single clear cutoff. In general, CRNDA should be considered as a pipeline method, whose components are empirically optimized. Various additional parameters can be targets for optimization, such as preprocessing parameters (e.g., detrending, spatial smoothing, regress‐out of global signals), moving‐window size, and how the measure of interest is calculated. Entire components of the procedure may also be swapped. For example, the Iclust algorithm could be replaced with another clustering engine, such as spectral clustering or simple K‐means. Alternative methods for computing network dynamic connectivity may also be tested to examine the improvement of signal‐to‐noise ratio for such measures [Lindquist et al., 2014].
An additional caveat of the CRNDA application presented here is related to the fact that it was optimized to identify networks with mesh topology, i.e., it equally accounts for all possible edges between the network's nodes. Future CRNDA adaptations may facilitate sensitivity to other network topologies (e.g., small‐world, star). This may be achieved for instance by basing the clustering procedure on a modified version of PLS, so that the network will be defined based on its edges (so it is not necessarily fully connected) rather than on its nodes.
To note, while in the current application of CRNDA we used the median emotion rating as the reference, our method also facilitates network demarcation based on covariance of functional connectivity time series with subject‐specific emotion ratings. This option offers an increased sensitivity to individual differences in emotional responsiveness, but possibly at the expense of reproducibility and robustness to idiosyncratic noise resulting from inaccuracy of self‐assessment of emotional intensity. In our case, the increased robustness of using the group rating is indicated by the fact that the individual ratings in the test group have higher correlation with the median rating of the training group (mean Pearson coefficient ± std: 0.73 ± 0.21) than with the individual ratings of this group (0.57 ± 0.25). While in this proof‐of‐concept study we preferred robustness over sensitivity to individual variability, we repeated CRNDA using the individual ratings as the reference. The resulting networks are showed in Supporting Information Figure S2. While some of these networks considerably overlap with networks delineated using the median rating, other networks have no similar correspondence. A comprehensive discussion in the results and their comparison across protocols is beyond the scope of this paper.
To conclude, this study demonstrated that CRNDA, a method which is sensitive to connectivity dynamics, yields a replicable delineation of brain networks that are functionally relevant to the psychophysiological process used as the basis for the delineation. The composition of the resulting networks is congruent with existing literature, and includes brain areas with an established common denominator—either specific or unspecific to emotion. In addition, CRNDA indicates that the connectivity of these networks is not stationary; it develops in correspondence with emotional intensity, over time windows. Encouraged by these findings, we hope that CRNDA will be adopted as an analytical tool within the growing investigation of brain network dynamics.
Supporting information
Supporting Information
Supporting Information Table 1.
ACKNOWLEDGMENTS
The authors thank Chani Sacharen for her assistance with editing the manuscript. This work would not have been possible without support from the University of Chicago's Arete Initiative and its program for 'The Science of Virtues', given to Talma Hendler, Gal Raz, and Gadi Gilam. Further support was provided by the Dan David Scholarship awarded to Gal Raz.
The first two authors contributed equally to this work.
Footnotes
Note however that is therefore not limited to the range [‐1, 1]
REFERENCES
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD (2012): Tracking whole‐brain connectivity dynamics in the resting state. Cereb Cortex:bhs 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels A, Zeki S (2005): The chronoarchitecture of the cerebral cortex. Philos Trans R Soc B Biol Sci 360:733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995): Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 289–300. [Google Scholar]
- Burgmans S, van Boxtel MPJ, Vuurman EFPM, Evers EAT, Jolles J (2010): Increased neural activation during picture encoding and retrieval in 60‐year‐olds compared to 20‐year‐olds. Neuropsychologia 48:2188–2197. [DOI] [PubMed] [Google Scholar]
- Chan Y‐C, Chou T‐L, Chen H‐C, Liang K‐C (2012): Segregating the comprehension and elaboration processing of verbal jokes: An fMRI study. NeuroImage 61:899–906. [DOI] [PubMed] [Google Scholar]
- Chan S‐H, Ryan L, Bever TG (2013): Role of the striatum in language: Syntactic and conceptual sequencing. Brain Lang 125:283–294. [DOI] [PubMed] [Google Scholar]
- Columbus C Stepmom.
- Corbetta M, Patel G, Shulman GL (2008): The reorienting system of the human brain: From environment to theory of mind. Neuron 58:306–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decety J, Lamm C (2006): Human empathy through the lens of social neuroscience. Sci World J 6:1146–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Jirsa V, McIntosh AR, Sporns O, Kötter R (2009): Key role of coupling, delay, and noise in resting brain fluctuations. Proc Natl Acad Sci U S A 106:10302–10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffau H (2008): The anatomo‐functional connectivity of language revisited. New insights provided by electrostimulation and tractography. Neuropsychologia 46:927–934. [DOI] [PubMed] [Google Scholar]
- Durstewitz D, Deco G (2008): Computational significance of transient dynamics in cortical networks. Eur J Neurosci 27:217–227. [DOI] [PubMed] [Google Scholar]
- Engen HG, Singer T (2013): Empathy circuits. Curr Opin Neurobiol 23 Macrocircuits: 275–282. [DOI] [PubMed] [Google Scholar]
- Epstein RA (2008): Parahippocampal and retrosplenial contributions to human spatial navigation. Trends Cogn Sci 12:388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito F, Seifritz E, Formisano E, Morrone R, Scarabino T, Tedeschi G, Cirillo S, Goebel R, Di Salle F (2003): Real‐time independent component analysis of fMRI time‐series. NeuroImage 20:2209–2224. [DOI] [PubMed] [Google Scholar]
- Friston K, Buechel C, Fink G, Morris J, Rolls E, Dolan R (1997): Psychophysiological and Modulatory Interactions in Neuroimaging. NeuroImage 6:218–229. [DOI] [PubMed] [Google Scholar]
- Frith CD, Frith U (2006): The Neural Basis of Mentalizing. Neuron 50:531–534. [DOI] [PubMed] [Google Scholar]
- Gonzalez‐Castillo J, Handwerker DA, Robinson ME, Hoy CW, Buchanan LC, Saad ZS, Bandettini PA (2014): The spatial structure of resting state connectivity stability on the scale of minutes. Front Neurosci 8:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grodal TK (2009): Embodied Visions: Evolution, Emotion, Culture, and Film. New York: Oxford University Press. [Google Scholar]
- Gross JJ, John OP (2003): Individual differences in two emotion regulation processes: Implications for affect, relationships, and well‐being. J Pers Soc Psychol 85:348–362. [DOI] [PubMed] [Google Scholar]
- Hämäläinen A, Pihlajamäki M, Tanila H, Hänninen T, Niskanen E, Tervo S, Karjalainen PA, Vanninen RL, Soininen H (2007): Increased fMRI responses during encoding in mild cognitive impairment. Neurobiol Aging 28:1889–1903. [DOI] [PubMed] [Google Scholar]
- Honey CJ, Kötter R, Breakspear M, Sporns O (2007): Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proc Natl Acad Sci 104:10240–10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, Della Penna S, Duyn JH, Glover GH, Gonzalez‐Castillo J, Handwerker DA, Keilholz S, Kiviniemi V, Leopold DA, de Pasquale F, Sporns O, Walter M, Chang C (2013): Dynamic functional connectivity: Promise, issues, and interpretations. NeuroImage 80:360–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensinger EA, Schacter DL (2007): Remembering the specific visual details of presented objects: Neuroimaging evidence for effects of emotion. Neuropsychologia 45:2951–2962. [DOI] [PubMed] [Google Scholar]
- Kotz SA, Schwartze M, Schmidt‐Kassow M (2009): Non‐motor basal ganglia functions: A review and proposal for a model of sensory predictability in auditory language perception. Cortex J Devoted Study Nerv Syst Behav 45:982–990. [DOI] [PubMed] [Google Scholar]
- Krishnan A, Williams LJ, McIntosh AR, Abdi H (2011): Partial least squares (PLS) methods for neuroimaging: A tutorial and review. NeuroImage 56:455–475. Multivariate Decoding and Brain Reading: [DOI] [PubMed] [Google Scholar]
- Krmpotich TD, Tregellas JR, Thompson LL, Banich MT, Klenk AM, Tanabe JL (2013): Resting‐state activity in the left executive control network is associated with behavioral approach and is increased in substance dependence. Drug Alcohol Depend 129:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug A, Markov V, Krach S, Jansen A, Zerres K, Eggermann T, Stöcker T, Shah NJ, Nöthen MM, Treutlein J, Rietschel M, Kircher T (2010): The effect of Neuregulin 1 on neural correlates of episodic memory encoding and retrieval. NeuroImage 53 Imaging Genetics: 985–991. [DOI] [PubMed] [Google Scholar]
- Lamm C, Decety J, Singer T (2011): Meta‐analytic evidence for common and distinct neural networks associated with directly experienced pain and empathy for pain. NeuroImage 54:2492–2502. [DOI] [PubMed] [Google Scholar]
- Legrain V, Iannetti GD, Plaghki L, Mouraux A (2011): The pain matrix reloaded: A salience detection system for the body. Prog Neurobiol 93:111–124. [DOI] [PubMed] [Google Scholar]
- Leonardi N, Shirer WR, Greicius MD, Van De Ville D (2014): Disentangling dynamic networks: Separated and joint expressions of functional connectivity patterns in time. Hum Brain Mapp 35:5984–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist MA, Xu Y, Nebel MB, Caffo BS (2014): Evaluating dynamic bivariate correlations in resting‐state fMRI: A comparison study and a new approach. NeuroImage 101:531–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattler U, Wüstenberg T, Heinze H‐J (2006): Common modules for processing invalidly cued events in the human cortex. Brain Res 1109:128–141. [DOI] [PubMed] [Google Scholar]
- Menon V (2011): Large‐scale brain networks and psychopathology: A unifying triple network model. Trends Cogn Sci 15:483–506. [DOI] [PubMed] [Google Scholar]
- Mitchell RLC (2007): fMRI delineation of working memory for emotional prosody in the brain: Commonalities with the lexico‐semantic emotion network. NeuroImage 36:1015–1025. [DOI] [PubMed] [Google Scholar]
- Mobbs D, Weiskopf N, Lau HC, Featherstone E, Dolan RJ, Frith CD (2006): The Kuleshov Effect: The influence of contextual framing on emotional attributions. Soc Cogn Affect Neurosci 1:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss J, Schunn CD, Schneider W, McNamara DS, VanLehn K (2011): The neural correlates of strategic reading comprehension: Cognitive control and discourse comprehension. NeuroImage 58:675–686. [DOI] [PubMed] [Google Scholar]
- Newman SD, Malaia E, Seo R, Cheng H (2013): The effect of individual differences in working memory capacity on sentence comprehension: An fMRI study. Brain Topogr 26:458–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand WM (1971): Objective criteria for the evaluation of clustering methods. J Am Stat Assoc 66:846–850. [Google Scholar]
- Raz G, Winetraub Y, Jacob Y, Kinreich S, Maron‐Katz A, Shaham G, Podlipsky I, Gilam G, Soreq E, Hendler T (2012): Portraying emotions at their unfolding: A multilayered approach for probing dynamics of neural networks. NeuroImage 60:1448–1461. [DOI] [PubMed] [Google Scholar]
- Raz G, Jacob Y, Gonen T, Winetraub Y, Flash T, Soreq E, Hendler T (2014): Cry for her or cry with her: Context‐dependent dissociation of two modes of cinematic empathy reflected in network cohesion dynamics. Soc Cogn Affect Neurosci 9:30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K, Rowe JB, Passingham RE (2002): Parahippocampal reactivation signal at retrieval after interruption of rehearsal. J Neurosci 22:6315–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoğlu Ü, Pearlson GD, Kiehl KA, Wang YM, Michael AM, Calhoun VD (2010): A method for evaluating dynamic functional network connectivity and task‐modulation: Application to schizophrenia. Magn Reson Mater Phys Biol Med 23:351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurz M, Radua J, Aichhorn M, Richlan F, Perner J (2014): Fractionating theory of mind: A meta‐analysis of functional brain imaging studies. Neurosci Biobehav Rev 42:9–34. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD (2007): Dissociable intrinsic connectivity networks for salience processing and executive control. J Neurosci off J Soc Neurosci 27:2349–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Li Z, Zeng L‐L, Yuan L, Chen F, Liu Z, Hu D (2014): Internetwork dynamic connectivity effectively differentiates schizophrenic patients from healthy controls. Neuroreport 25:1344–1349. [DOI] [PubMed] [Google Scholar]
- Shibata M, Toyomura A, Motoyama H, Itoh H, Kawabata Y, Abe J (2012): Does simile comprehension differ from metaphor comprehension? A functional MRI study. Brain Lang 121:254–260. [DOI] [PubMed] [Google Scholar]
- Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD (2012): Decoding subject‐driven cognitive states with whole‐brain connectivity patterns. Cereb Cortex 22:158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Moeller S, Xu J, Auerbach EJ, Woolrich MW, Beckmann CF, Jenkinson M, Andersson J, Glasser MF, Essen DCV, Feinberg DA, Yacoub ES, Ugurbil K (2012): Temporally‐independent functional modes of spontaneous brain activity. Proc Natl Acad Sci 109:3131–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaniol J, Davidson PSR, Kim ASN, Han H, Moscovitch M, Grady CL (2009): Event‐related fMRI studies of episodic encoding and retrieval: Meta‐analyses using activation likelihood estimation. Neuropsychologia 47:1765–1779. [DOI] [PubMed] [Google Scholar]
- Spreng RN, Sepulcre J, Turner GR, Stevens WD, Schacter DL (2013): Intrinsic architecture underlying the relations among the default, dorsal attention, and frontoparietal control networks of the human brain. J Cogn Neurosci 25:74–86. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3816715/. Last accessed on 27/07/2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sripada C, Angstadt M, Kessler D, Phan KL, Liberzon I, Evans GW, Welsh RC, Kim P, Swain JE (2014): Volitional regulation of emotions produces distributed alterations in connectivity between visual, attention control, and default networks. NeuroImage 89:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KS, Seminowicz DA, Davis KD (2009): Two systems of resting state connectivity between the insula and cingulate cortex. Hum Brain Mapp 30:2731–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossel S, Geng JJ, Fink GR (2014): Dorsal and ventral attention systems distinct neural circuits but collaborative roles. Neuroscientist 20:150–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni T, Speer NK, Zacks JM (2008): Neural substrates of narrative comprehension and memory. NeuroImage 41:1408–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD (2011): Large‐scale automated synthesis of human functional neuroimaging data. Nat Methods 8:665–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki J, Ochsner K (2012): The neuroscience of empathy: Progress, pitfalls and promise. Nat Neurosci 15:675–680. Available at: http://www.ncbi.nlm.nih.gov/pubmed/22504346. Last accessed on 27/07/2016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information Table 1.


