Extraintestinal pathogenic Escherichia coli (ExPEC) is an important human and animal pathogen. Despite the apparent similarities in their known virulence attributes, some ExPEC strains can cross the host species barrier and present a zoonotic potential, whereas other strains exhibit host specificity, suggesting the existence of unknown mechanisms that remain to be identified.
KEYWORDS: ExPEC, host adaptation, pathogenicity, septicemia, TraDIS
ABSTRACT
Extraintestinal pathogenic Escherichia coli (ExPEC) is an important human and animal pathogen. Despite the apparent similarities in their known virulence attributes, some ExPEC strains can cross the host species barrier and present a zoonotic potential, whereas other strains exhibit host specificity, suggesting the existence of unknown mechanisms that remain to be identified. We applied a transposon-directed insertion site sequencing (TraDIS) strategy to investigate the ExPEC XM strain, which is capable of crossing the host species barrier, and to screen for virulence-essential genes in both mammalian (mouse) and avian (duck) models of E. coli-related septicemia. We identified 151 genes essential for systemic infection in both mammalian and avian models, 97 required only in the mammalian model, and 280 required only in the avian model. Ten genes/gene clusters were selected for further validation, and their contributions to ExPEC virulence in both mammalian and avian models or mammalian- or avian-only models were confirmed by animal tests. This represents the first comprehensive genome-wide analysis of virulence-essential genes required for systemic infections in two different host species and provides a further comprehensive understanding of ExPEC-related virulence, host specificity, and adaptation.
INTRODUCTION
Extraintestinal pathogenic Escherichia coli (ExPEC) is an important bacterial pathogen that causes severe extraintestinal infections in humans and animals (1–3). ExPEC is among the most common pathogens in the urinary tract, causing 75% of urinary tract infections (UTIs) (4, 5), and the second most common bacterial pathogen associated with neonatal meningitis, causing high mortality in newborns (6). Additionally, ExPEC represents the leading cause of bloodstream infections in nursing homes, hospitals, and young children. In 2001, 40,000 deaths were attributed to ExPEC-associated sepsis, which is 500-fold greater than the number of deaths caused by E. coli O157:H7 (7). Moreover, ExPEC causes extraintestinal infections in many farm animals, companion animals, and wild animals, leading to respiratory or systemic diseases, with ExPEC strains of avian origin being the best studied (8–11).
ExPEC strains of human and animal origin share a genetic background and are not distinguishable by genotype, serotype, virulence-associated genes, or genome content, despite their isolation from various hosts and tissues (12–15). Additionally, E. coli isolates from retail foods, including milk, meat, and eggs, are indistinguishable from human ExPEC isolates (10, 16, 17). Such observations have led to the hypothesis that ExPEC exhibits a zoonotic potential and might represent a foodborne source of infection leading to human diseases. Indeed, some ExPEC strains of avian origin cause meningitis in a rat model of human neonatal meningitis (18), and some human ExPEC strains cause colisepticemia in avian models (19, 20). These data provide compelling evidence that ExPEC can cross the host species barrier to cause disease in both mammals and avian species and demonstrate the zoonotic potential of ExPEC. However, these results should not be overinterpreted to presume that all or even most ExPEC strains possess a zoonotic potential, as other studies indicate that many ExPEC strains exhibit degrees of host specificity, with only a subset of ExPEC strains, including those of both human and avian origins, exhibiting zoonotic potential (21).
Many virulence genes have been identified, and their underlying mechanisms have been elucidated in ExPEC (22–24); however, little is understood regarding the mechanisms associated with host-specific infections caused by ExPEC. Despite apparent similarities in their known virulence attributes, some ExPEC strains harbor zoonotic potential, while others do not, suggesting that unknown mechanisms underlie host specificity in some ExPEC strains. In this study, we applied a transposon-directed insertion site sequencing (TraDIS) (25) strategy to investigate a highly virulent ExPEC strain of avian origin in a duck model of avian colibacillosis (avian model) and a mouse model of human E. coli-related septicemia (mammalian model). We successfully identified genes essential for systemic infection of both mammals and avian species, those essential only in mammals, and those essential only in avian species. Moreover, we further confirmed these findings using animal virulence tests. Our results significantly advance our understanding of the molecular mechanisms underlying ExPEC-related host specificity/adaptation.
RESULTS
Selection of an appropriate ExPEC strain with zoonotic potential and determination of the input dose for TraDIS.
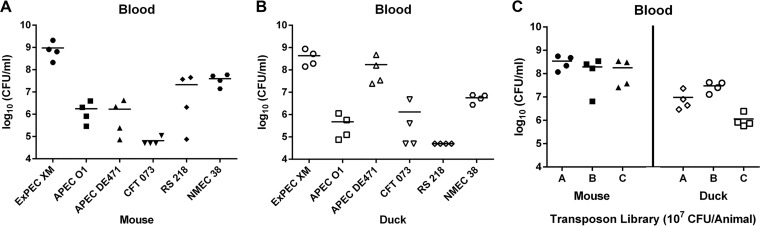
To select an appropriate ExPEC strain with zoonotic potential for TraDIS, six ExPEC strains, including three strains of avian origin (ExPEC XM, APEC O1, and APEC DE471) and three strains of human origin (CFT073, RS218, and NMEC 38), were tested in a duck model of avian E. coli-related septicemia (avian model) and in a mouse model of human E. coli-related septicemia (mammalian model). The animals were inoculated with 5 × 107 CFU of ExPEC via intraperitoneal injection, and the blood bacterial load was determined 12 h after infection. The results showed that ExPEC strains RS218 and APEC DE471 exhibited a certain degree of host specificity (Fig. 1A and B), with the human ExPEC strain RS218 being significantly more virulent in the mouse model than in the duck model, whereas the avian ExPEC strain APEC DE471 was significantly more virulent in the duck model than in the mouse model. Additionally, the other four ExPEC strains demonstrated no significant difference in virulence in the two animal models. Strain ExPEC XM was the most virulent strain in both the mammalian and avian models, suggesting its ability to cross the host species barrier. Therefore, this strain with zoonotic potential was used for subsequent TraDIS screening.
FIG 1.
Blood bacterial load of ExPEC strains during systemic infection in mice and ducks. (A) Bacterial loads of different ExPEC strains in the blood of the mammalian model at 12 h postinoculation. Four BALB/c mice were inoculated with 5 × 107 cells via intraperitoneal injection, and each data point indicates the CFU count obtained from an individual mouse. (B) Bacterial loads of different ExPEC strains in the blood of the avian model at 12 h postinoculation. Four Pekin ducks were inoculated with 5 × 107 cells via intraperitoneal injection, and each data point indicates the CFU count obtained from an individual duck. (C) Every four mice or ducks served as one group, with three biological replicates included. Twelve mice and 12 ducks were inoculated with the mini-Tn5 transposon mutant library. The blood bacterial load is indicated. These colonies were harvested for genomic DNA extraction and used for TraDIS.
To determine the appropriate inoculation dose of ExPEC XM for the screening of host adaptation genes, four inoculation doses (1 × 105, 1 × 106, 1 × 107, and 1 × 108 CFU/animal) were compared in the mouse and duck models. Inoculation of 105 and 106 CFU of ExPEC XM resulted in a low percentage of animals developing septicemia, whereas inoculation of 108 CFU resulted in the sudden death of the animals prior to the reisolation of bacteria. An inoculation dose of 107 CFU led to reproducible disease progression and allowed bacterial reisolation from each infected animal.
Construction of the transposon mutant library and screening in animal models.
To provide a sufficient saturation density for the identification of ExPEC XM genes essential for the systemic infection of different hosts, a genome-saturating mini-Tn5 transposon mutant library containing 100,000 kanamycin-resistant transformants was generated. An estimated 52,503 transposon mutants are required to generate a 99.99% saturation of the ExPEC XM genome, which is 5.4 Mbp in length (GenBank accession numbers NZ_CP025328.1 and NZ_CP025329.1). This library was submitted to three rounds of growth in lysogeny broth (LB) to enrich for mutants that did not exhibit fitness in vitro (26). The final library was used as the input for the infection experiments.
Every four mice or ducks served as one group, and three biological replicates were included. Therefore, a total of 12 mice and 12 ducks were inoculated with 107 CFU of transposon mutants (input), and bacteria were reisolated from blood after 12 h of infection (Fig. 1C). Genomic DNA was isolated from the recovered bacteria, and the transposon insertion sites in the input (inoculum) and in the output (collected bacteria) were determined using TraDIS. Each sample yielded 60 million single-end reads that were tagged with a transposon-specific sequence, and approximately 70% of these reads mapped to the ExPEC XM genome using Bowtie software (27) in the FASTQ format (Table 1). In total, 112,234 and 110,861 unique insertion sites were identified in mammal and avian input pools, respectively, equating to an average of one insertion site every 48.41 bp or 49.01 bp, respectively. Reproducibility was high among the three groups for each model, with Spearman correlation coefficients (R2) being >0.99 (see Fig. S1 in the supplemental material). In addition, a Bland-Altman plot further showed that approximately 94% of the reads were within the limits of agreement (LOA) (Fig. S2).
TABLE 1.
Summary of sequencing and mapping results of TraDIS
| Sample name | No. of tagged reads | No. (%) of reads mapped to ExPEC XM chromosome | No. of unique insertion sites in ExPEC |
|---|---|---|---|
| Mammalian input | 62,393,529 | 43,784,685 (70.18) | 112,234 |
| Mammalian output A | 57,925,375 | 40,873,130 (70.56) | 81,910 |
| Mammalian output B | 66,796,639 | 48,850,553 (73.13) | 83,336 |
| Mammalian output C | 57,934,293 | 40,127,493 (69.26) | 82,061 |
| Avian input | 59,394,008 | 41,267,083 (69.48) | 110,861 |
| Avian output A | 59,136,332 | 39,465,856 (66.72) | 79,079 |
| Avian output B | 50,696,348 | 36,852,275 (72.69) | 76,392 |
| Avian output C | 55,513,958 | 36,507,790 (65.76) | 74,345 |
Identification of essential genes in ExPEC XM involved in systemic infection.
A fitness factor was calculated for each gene, as previously reported (26, 28). Briefly, reads for each transposon insertion site were normalized by dividing the total number of reads obtained from each sample, and a fitness factor for each site was calculated by dividing the normalized output reads by the normalized input reads. The fitness factor for a given gene was obtained from the average fitness factors of all insertion sites in this gene. A fitness factor of ≤1 indicated that the given mutant strain was outcompeted and that the interrupted gene might contribute to virulence.
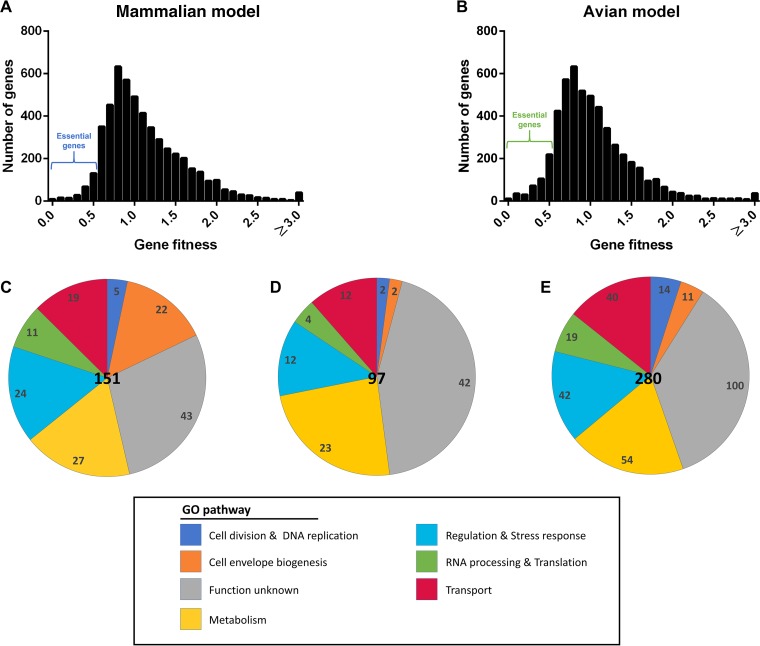
The insertion sites were evenly distributed throughout the genome of ExPEC XM (Fig. 2A and B and Fig. S3A). Genes with fitness factors of ≤0.5 (i.e., less than a 2-fold change in frequency) in either model were considered candidates of essential genes. In total, 248 and 431 genes important in the mammalian model or avian model were identified as candidates of essential genes (Table S1 and Fig. S3B). The reads of every gene from input and output samples were analyzed by use of the Bioconductor package edgeR to provide a reference for the essential genes (Table S2). The functional category of each gene was identified based on the Gene Ontology from the ExPEC XM annotation (Fig. 2C to E). Among them, several essential genes located in the same operon or in the same pathway are shown in Fig. 3. Several essential gene candidates were previously identified to be virulence-associated genes in ExPEC, including the capsular synthetic gene (in the neu-kps gene cluster) (29, 30), the lipopolysaccharide (LPS) synthetic gene (in the rml and waa gene clusters) (31, 32), two-component regulatory system genes (envZ-ompR) (33), an antioxidant gene regulator (oxyR) (34), as well as genes coding for enterobacterial common antigen (ECA) (wzzE, wecF, wzyE, and wecG) (35) and siderophore transport (iro gene cluster) (36, 37).
FIG 2.
Summary of in vivo TraDIS screening results. (A and B) An overall fitness factor was determined for each gene as described in the text. The regions of the essential genes for the mammalian model and the avian model are shown. (C to E) Classification of essential genes. The number of shared virulence-essential genes necessary for systemic infection of both mammalian and avian models (C), mammalian models (D), and avian models (E) in each Gene Ontology functional category.
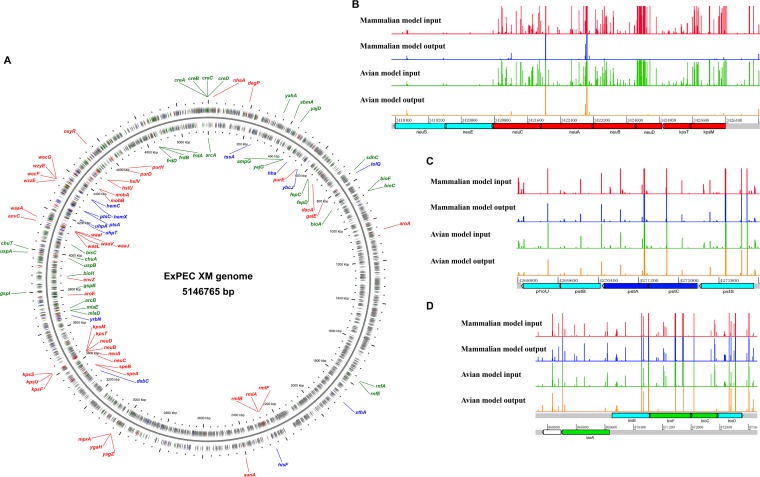
FIG 3.
Overview of the candidate essential genes in the ExPEC XM genome involved in systemic infection. (A) The circular diagram shows the locations of 85 essential gene candidates on the ExPEC XM genome. Several essential genes located in the same operon or in the same pathway are shown. The two rings containing gray, red, blue, and green arrows illustrate the coding sequences, shared virulence-essential genes for systemic infection of both mammalian and avian models, virulence-essential genes identified only in the mammalian model, and virulence-essential genes identified only in the avian model, respectively, on the forward and reverse strands of the genome. (B to D) The three insets represent a close-up look at three regions on the genome, with the graphs showing the location and the relative number of each mutant found in the samples.
Shared virulence-essential genes associated with systemic infection in both mammalian and avian models of E. coli-related septicemia.
The disruption of some ExPEC XM genes led to significant virulence attenuation in both the mammalian and the avian models, indicating shared virulence mechanisms. We identified, by TraDIS screening, a total of 151 genes that were essential in both the mammalian and the avian models (Table S1). Genes involved in metabolism accounted for 18% of the shared essential genes. They featured prominently, were significantly enriched in the entire genome, and included nucleotide metabolism genes (purDH) (38), polyamine synthesis genes (speAB) involved in putrescine synthesis (39), and molybdenum cofactor biosynthesis genes (mobAB) (40), identified in both the mammalian and the avian models in our primary screen. Approximately 15% of the essential genes were involved in the biogenesis of bacterial surface structures, including capsule biosynthesis genes (in the neu-kps gene cluster) and LPS biosynthesis genes (in the rml and waa gene clusters). The third most abundant category contained genes encoding outer and inner membrane proteins and transporters (13%), including metal ion transporters, such as iron uptake systems, peptide and amino acid transporters, Na+/H+ antiporters, and phospholipid transporters. Additionally, many genes involved in regulation, RNA processing, translation, the stress response, cell division, and DNA replication were identified in our primary screen of both the mammalian and the avian models and included genes related to two-component regulatory systems (envZ-ompR), the antioxidant stress response (oxyR), and mprA, a suppressor of the multidrug resistance gene cluster emrAB (41) (Fig. 2C).
Six genes/gene clusters, including the neu-kps gene cluster, the rml gene cluster, mprA, nhaA, yga, and sanA, were selected for further validation in both the mammalian and the avian models. These genes/gene clusters were deleted, and the mutant strains were coinfected with the wild-type strain (ratio, 1:1). The deletion of the neu-kps (encoding the capsule) and rml (encoding the O antigen) gene clusters (Table S3) resulted in the significant attenuation of ExPEC virulence (by 10,000-fold) (Fig. 4A), which was consistent with previous reports and which validated our screening results. Surprisingly, deletion of nhaA or mprA, each of which is encoded by both commensal and pathogenic E. coli strains, led to at least a 10,000-fold attenuation of virulence upon coinfection with the wild type in both the mammalian and the avian models (Fig. 4A). The nhaA and mprA mutant strains were further tested individually in the mammalian model separately from the wild-type strain, and both mutants showed at least a 100,000-fold virulence attenuation compared with the virulence of the wild-type strain (Fig. 4B). Furthermore, mice infected with either mutant strain presented no symptoms and showed a phenotype similar to that of control mice inoculated with only phosphate-buffered saline (PBS). Furthermore, the ygaZ and sanA mutants showed 1,000-fold virulence attenuation relative to the virulence of the wild-type strain in the coinfection study (Fig. 4A).
FIG 4.
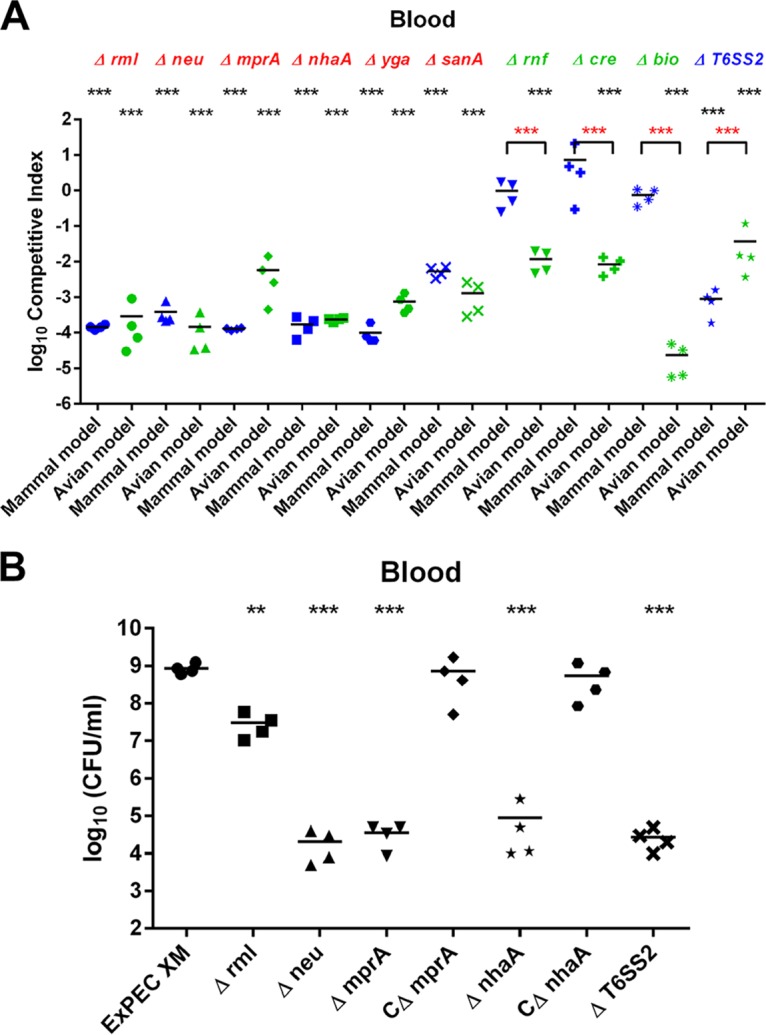
(A) In vivo competition assays in the mammalian and the avian models. Values less than 0 indicate reduced fitness compared to that of the wild-type strain. (B) Virulence tests of individual mutant strains in the mammalian model. P values were determined by Student’s t test. **, P < 0.01; ***, P < 0.001.
Virulence-essential genes identified only in the avian model of E. coli-related septicemia.
There were 280 virulence-essential genes identified only in the avian model by TraDIS screening (Table S1). These included genes involved in regulatory functions, the synthesis of cell surface structures and membranes, and metabolism (Fig. 2E). The most striking finding was the identification of multiple transposon insertions within bioA, bioF, bioC, and bioH, resulting in a significant attenuation of their levels in the primary screening of the avian model, whereas interruption of these genes had only a minor effect on the mammalian model, with fitness factors ranging from 0.17 to 0.44. These results suggest that biotin synthesis is involved in specific host adaptation. Therefore, the operon bioABFCD was selected for further study.
The gene cluster for biotin synthesis was deleted, and the resulting mutant strain was coinfected with the wild type in order to validate the virulence attenuation. The results indicated that the mutant was outcompeted by the wild-type strain in the avian model, with a competitive index (CI) of 2.3 × 10−5, indicating 100,000-fold virulence attenuation. In contrast, we observed no significant difference in virulence from that of the wild-type strain in the mammalian model (CI, 0.74), demonstrating that biotin biosynthetic genes are important for ExPEC virulence only in avian species and not in mammals (Fig. 4A). Furthermore, expression of these genes in ExPEC XM revealed that the bioABFC genes were significantly upregulated by 8- to 16-fold during culture in avian serum compared with their level of expression in LB (42).
Another interesting finding was the Rnf electron transport complex. The genes rnfA and rnfE and the cotranscribed gene nth, encoding endonuclease III, were identified by TraDIS screening only in the avian model. The fitness factors for these genes were less than 0.5 in the avian model and ranged from 0.78 to 1.05 in the mammalian model, suggesting their involvement in host specificity. Coinfection experiments showed that the mutant strain lacking the rnfABCDEG-nth genes was significantly outcompeted (100-fold; P < 0.001) by the wild-type strain in the avian model (Fig. 4A). As expected, this effect was host specific and either not observed or observed to a very limited degree in the mammalian model. These data confirm the involvement of the rnf subunit genes in host adaptation during systemic infection in the avian model but not in the mammalian model.
Additionally, transposon insertions in the creABCD genes, encoding a two-component system, led to a significant decrease in the fitness factor in the avian model (fitness factor range, 0.24 to 0.4). CI values in the mammalian and the avian models of 7.3 and 8.3 × 10−3, respectively, confirmed that mutant virulence was specifically attenuated during systemic infection of the avian model but not of the mammalian model (Fig. 4A).
Virulence-essential genes identified only in the mammalian model of E. coli-related septicemia.
A total of 97 virulence-essential genes were exclusively identified in the mouse model by TraDIS screening (Table S1). Similar to those identified in both the mammalian and the avian models and those identified only in the avian model, these genes were classified as being involved in metabolism, bacterial cell surface synthesis, membrane transport activity, and regulatory and other functions (Fig. 2D).
Interestingly, a mutant strain with transposon insertions in tssA, which is involved in the biosynthesis of the type VI secretion system (T6SS), was identified only in the mammalian model. In the mammalian model, the fitness factor for tssA was 0.46, whereas in the avian model, it was 0.73, suggesting that this gene is likely more important for systemic infection in the mammalian model than in the avian model. Therefore, the gene cluster of the type VI secretion system was selected for further investigation. The type VI secretion system mutant strain was generated and coinfected with the wild-type strain in both the mammalian and the avian models. The results showed CIs of 8.8 × 10−4 in the mammalian model and 3.7 × 10−2 in the avian model, suggesting that the type VI secretion system is more important for ExPEC-specific systemic infection in the mammalian model than in the avian model (Fig. 4A). Furthermore, this mutant strain showed a 100,000-fold attenuation of virulence in the mouse model in the absence of the wild-type strain (Fig. 4B).
DISCUSSION
ExPEC is a major cause of extraintestinal diseases in humans and animals, including bacteremia, urinary tract infections (UTIs), and meningitis. The accumulated evidence suggests that some human and animal ExPEC strains are genotypically and phenotypically indistinguishable and harbor zoonotic potential, whereas other ExPEC strains exhibit host specificity, despite the apparent similarities in their known virulence attributes, thereby suggesting unknown mechanisms underlying the associated differences. Here, we screened transposon mutants of ExPEC XM in mammalian (mouse) and avian (duck) models of systemic infection, representing human and avian E. coli-related septicemia, respectively. For the first time, ExPEC genes that are important for systemic infection of both animal models and that contribute to host specificity/adaptation were successfully identified. Understanding the genetic and molecular basis of host specificity/adaptation by ExPEC is important to elucidate its pathogenic mechanisms, develop better animal models, and design new strategies and therapeutics for the control of ExPEC-related diseases.
It is unsurprising that the neu and rml gene clusters were identified to be genes important for systemic infection of both mammals and avian species, given that they encode the K1 capsule and the O antigen, respectively, and are associated with ExPEC virulence (30, 32). In addition to the genes responsible for K1-capsule and O-antigen synthesis, mprA and nhaA were identified, and deletion of either gene significantly attenuated ExPEC virulence by >10,000-fold in both coinfection assays with the wild-type strain and infection assays with the mutant strain alone. The mprA gene was previously identified to be a regulator controlling the expression of E. coli microcins and the multidrug resistance pump EmrAB (41). Additionally, several recent studies showed that mprA is required for capsule production in ExPEC and Klebsiella pneumoniae (43, 44), is essential for ExPEC virulence, and can be inhibited by a small molecule, DU011 (45). The gene nhaA encodes an E. coli integral membrane protein, which functions as the Na+/H+ antiporter, exchanging Na+ for H+ across the cytoplasmic membrane and other intracellular membranes (46). It is essential for Na+, pH, and volume homeostasis, the processes crucial for cell viability (47). In addition, the NhaA homolog in Yersinia pestis is reportedly essential for virulence (48). Our results further confirm that mprA and nhaA are genes essential for ExPEC virulence. Furthermore, there is an urgent need to develop an efficacious, broadly cross-protective vaccine against most E. coli pathotypes, given the high cost associated with ExPEC infections in humans and animals and the increasing antibiotic resistance. Both mprA and nhaA are conserved in all E. coli pathotypes, and deletion of either gene significantly attenuates ExPEC virulence in the bloodstreams of both mammals and avian but does not affect mutant growth in vitro. If deletion of either gene would not affect E. coli’s growth in the host intestine, they could be ideal targets for the development of broadly conserved vaccine antigens and novel antimicrobial drugs.
Among the virulence-essential genes identified only in the avian model, each gene in the biotin synthesis bioABFCDH operon was identified. Biotin functions as an essential cofactor for carboxylases and decarboxylases in all organisms and is part of key metabolic pathways, including fatty acid biosynthesis, replenishment of the tricarboxylic acid cycle, and amino acid metabolism (49). Bacteria, plants, and some fungi can synthesize biotin de novo from a pimeloyl coenzyme A precursor, whereas avian species and mammals can obtain biotin only from external sources (50). Previous studies have shown that biotin synthesis is necessary in all stages of the Mycobacterium tuberculosis life cycle due to the lack of a transporter necessary for scavenging exogenous biotin, whereas it is required for the rapid escape of Francisella from phagosomes and its intracellular replication in the cytosol (51). In the present study, we found that biotin synthesis was required only during systemic infection in avian species and not in mammals, with the genes associated with biotin synthesis being significantly upregulated during infection only in avian species. This is possibly due to the following: (i) the biotin concentration in duck (avian) tissues is lower than that in mouse (mammalian) tissues, (ii) ExPEC infection of ducks (avian species) requires more biotin than infection of mice (mammals), and (iii) both case (i) and case (ii) are simultaneously true.
Type VI secretion systems have been characterized in many Gram-negative bacteria, including bacterial pathogens (52). Bacteria take advantage of the type VI secretion system by directly injecting protein effectors across the cellular envelope into target cells. This system functions in interbacterial competition, signaling, and environmental fitness and contributes to the virulence of bacterial pathogens (53–55). At least two type VI secretion systems (T6SS1 and T6SS2) have been reported in ExPEC and are involved in bacterial competition (56, 57). However, type VI secretion systems have thus far not been associated with bacterial host specificity or adaptation in ExPEC. In the present study, T6SS2 was identified in the primary screen from the mammalian model but not in the primary screen from the avian model. Moreover, the T6SS2 deletion strain showed 100-fold attenuated virulence in the mammalian model relative to its virulence in the avian model, suggesting that this system is involved in host specificity and/or adaptation, although its underlying mechanisms remain unknown and require further study. Ma et al. tested T6SS2 of the same strain in both mammalian and avian models; however, no significant difference in virulence was observed between wild-type and mutant strains in either model (56). This may be because only 1 gene, either vgrG or clpV, was deleted in previous study, while the whole 17-gene cluster of T6SS2 was deleted in our study (56).
In summary, we used a TraDIS strategy to successfully identify 151 ExPEC genes important for the systemic infection of both mammals and avian species, 97 genes important for the systemic infection of mammals, and 280 genes important for the systemic infection of avian species, with several genes being confirmed to contribute to ExPEC host specificity/adaptation. This represents the first comprehensive genome-wide survey of ExPEC genes required for systemic infections in two different host species. The identification of bacterial genes involved in human adaptation presents a special challenge for experimentalists. In this study, we used a mouse model of human E. coli-related septicemia to screen for ExPEC genes that are important during human infection, an approach with obvious limitations due to the fundamental differences in human and mouse anatomies and functional elements. Recent technological advances in creating humanized chimeric mice might overcome such limits and provide unprecedented insights into the mechanisms of the host interaction with human pathogens. Additionally, understanding the mechanisms associated with ExPEC-related host specificity/adaptation is the ultimate goal. Although a large number of candidate genes were identified in this study, the roles of only some of them were confirmed; however, their underlying mechanisms remain unknown. Future studies will, it is hoped, provide a more comprehensive understanding of the mechanisms involved in ExPEC-specific host specificity/adaptation.
MATERIALS AND METHODS
Ethics statement.
All animal experimental procedures were conducted according to the guidelines of the Experimental Animal Management Measures of Jiangsu Province and approved by the Laboratory Animal Monitoring Committee of Jiangsu Province (Nanjing, China).
Bacteria and culture conditions.
ExPEC XM (O2:K1:H7), which is of the B2 and sequence type 95 (ST95) phylogenetic groups and which was isolated from the blood of a duck with colibacillosis, was used for the infection studies and library construction (56). The strains and plasmids used in this study are listed in Table S2 in the supplemental material. The bacterial strains were routinely cultured at 37°C on solid LB agar or in liquid LB medium supplemented, when appropriate, with the following antibiotics: ampicillin (Amp; 100 μg/ml), kanamycin (Kan; 50 μg/ml), nalidixic acid (Nal; 30 μg/ml), or chloramphenicol (Cm; 25 μg/ml).
Generation of the transposon mutant library.
Tn5 insertion mutants were generated in ExPEC XM using the pUTmini-Tn5km2 plasmid (Ampr Kmr). Briefly, pUTmini-Tn5km2 plasmids were purified from clones and individually transformed into the donor strain E. coli S17-1 λpir. Conjugation experiments were performed by growing each E. coli S17-1 λpir clone and the ExPEC Nalr strain to an optical density at 600 nm of ∼1. To construct transposon mutants of ExPEC, 1 ml of donor cells and 1 ml of recipient cells were harvested and washed three times in PBS. After discarding the supernatant, the cells were resuspended in 100 μl PBS, plated onto LB agar plates, and incubated at 37°C for 4 h. Colonies were collected, resuspended in PBS, and plated onto LB agar containing Kan and Nal. After incubation overnight at 37°C, the total number of colonies was estimated by counting a proportion from each of multiple plates. Kan- and Nal-resistant colonies were resuspended in sterilized LB broth using a bacteriological spreader before adding sterile PBS. Each batch contained an estimated 10,000 mutants, and a total of 100,000 transformants was generated by pooling 10 mutant batches. The entire mini-Tn5 mutant collection was grown three times in LB for 12 h at 37°C, and the resulting pool was used as the inoculum for experimental infections.
Mouse and duck infection experiments.
BALB/c mice (6 weeks old) and Pekin ducks (2 weeks old) were purchased from the Comparative Medicine Center of Yangzhou University (Nanjing, China). All animal experiments included populations with 50% males and 50% females, and all animals had access to sterile water and food ad libitum.
For virulence comparison experiments, mice and ducks (n = 4/each) were inoculated with 5 × 107 CFU of selected strains via intraperitoneal injection, followed by euthanization after 12 h and blood harvesting for plating on LB agar and determining the bacterial load. For transposon library screening, mice and ducks were inoculated with 105 (n = 4), 106 (n = 4), 107 (n = 4), and 108 (n = 4) CFU of ExPEC XM bacteria to determine the appropriate dose. Upon determination of 107 CFU as the appropriate dose, animals (n = 12 each) were administered the transposon library (input), and after 12 h, mice and ducks were euthanized to collect blood, which was plated on LB agar for 8 h at 37°C to reduce the proportion of host components. Colonies from blood cultures were harvested, and bacterial pellets were stored at −80°C. For coinfection experiments, wild-type and mutant strains were cultured to the late log phase and resuspended in PBS to yield 5 × 108 CFU/ml containing an equal number of wild-type and mutant strains. The inoculum (100 μl) was injected into the enterocoelia, and then the mice and ducks were euthanized at 12 h postinoculation and the bacterial load was calculated as described above. CIs were calculated as the ratio of the mutant strain versus the wild-type strain in the blood to the mutant strain versus the wild-type strain in the inoculum.
TraDIS.
Genomic DNA was isolated from the inoculum used for infections and from blood-derived cultures using a bacterial genomic DNA extraction kit (TaKaRa, Shiga, Japan). Genomic DNA (5 μg) was sheared to yield ∼500-bp fragments (Covaris, Woburn, MA, USA). The subsequent steps were performed according to the Illumina TruSeq nano DNA library preparation reference guide (revision D) for DNA end repair, DNA end adenylation, and adapter ligation, and the PCR enrichment step was performed at 98°C for 30 s, followed by 22 cycles of 98°C for 10 s, 60°C for 30 s, and 72°C for 1 min using a custom transposon-specific primer (TraDIS-F for enrichment of transposon insertion sites) and an index primer (one index per sample to allow for multiplex sequencing). Libraries from the input and output samples were sequenced in lanes using paired sequencing and an Illumina HiSeq sequencer 2000 (Illumina, San Diego, CA, USA) at Wuhan NextOmics (Wuhan, China).
Analysis of TraDIS data.
The transposon end (P5 end) raw sequencing reads from FASTQ files were picked and filtered, and split index sequences were combined with the transposon-specific sequences using NGSQCToolkit (version 2.3.3; http://www.nipgr.res.in/ngsqctoolkit.html). These reads were aligned to the ExPEC XM genome (GenBank accession number for the chromosome, NZ_CP025328.1; GenBank accession number for the plasmid, NZ_CP025329.1) using Bowtie software (27). Subsequent analysis steps were performed as previously described (26, 28) to calculate the number of sequencing reads (raw read counts) and the number of different insertion sites for each gene, which were then used to estimate the fitness and identify essential genes. The normalized read counts from three biological replicates of each model were loaded into the edgeR package (version 3.26.5) using the R environment (version 3.6.0) (58). The read counts and insertion sites were visualized using the Artemis (version 18.0.2) genome browser (59). The circular genome diagram was generated by the CGView server (60).
Recombinant DNA techniques.
PCR, electroporation, and DNA gel electrophoresis were performed as described by Sambrook and Russell (61) unless otherwise indicated. Recombinant plasmids and PCR products were purified using a MiniBEST DNA fragment purification kit (TaKaRa) or a MiniBEST plasmid purification kit (TaKaRa). Deletion mutants were constructed using the bacteriophage lambda Red recombinase system described by Datsenko and Wanner (62). All oligonucleotide primers used in this study were purchased from GenScript (Nanjing, China) and are listed in Table S3. All mutants were created using the ExPEC XM strain, and homologous recombination constructions were generated using PCR-purified products harboring a selectable antibiotic resistance gene and 60-nucleotide homology extensions. The antibiotic resistance gene was removed by transforming the pCP20 plasmid carrying a flippase (63). Mutants were confirmed by PCR and sequencing. For complementation, the coding sequences of the genes plus their putative promoter regions were amplified (the primers used are listed in Table S4 in the supplemental material) from the ExPEC XM strain and independently cloned into pGEN-MCS (42) using the BamHI and EcoRI restriction sites.
Accession number(s).
The sequences for the ExPEC XM chromosome and ExPEC XM plasmid have been deposited in the NCBI GenBank database under accession numbers NZ_CP025328.1 and NZ_CP025329.1, respectively. The raw PacBio sequence reads have been deposited in the Sequence Read Archive (SRA) under accession number SRX3460479. The TraDIS reads have been deposited in SRA under accession numbers SRX5318349, SRX5319314, SRX5319611, SRX5322355, SRX5322360, SRX5322748, SRX5324829, and SRX5325696.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yinli Bao, Jiale Ma, Wentong Cai, Ying Zheng, and Fengwei Jiang for their detailed and constructive comments, as well as Li Chen and Haiyan Ren for their assistance with the animal experiments.
This work was funded by the Natural Science Foundation of China (grant no. 31470243).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00666-19.
REFERENCES
- 1.Wiles TJ, Kulesus RR, Mulvey MA. 2008. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol 85:11–19. doi: 10.1016/j.yexmp.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Köhler C-D, Dobrindt U. 2011. What defines extraintestinal pathogenic Escherichia coli? Int J Med Microbiol 301:642–647. doi: 10.1016/j.ijmm.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Teng C-H, Cai M, Shin S, Xie Y, Kim K-J, Khan NA, Di Cello F, Kim KS. 2005. Escherichia coli K1 RS218 interacts with human brain microvascular endothelial cells via type 1 fimbria bacteria in the fimbriated state. Infect Immun 73:2923–2931. doi: 10.1128/IAI.73.5.2923-2931.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lane M, Mobley H. 2007. Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int 72:19–25. doi: 10.1038/sj.ki.5002230. [DOI] [PubMed] [Google Scholar]
- 5.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 6.Wijetunge D, Gongati S, DebRoy C, Kim K, Couraud P, Romero I, Weksler B, Kariyawasam S. 2015. Characterizing the pathotype of neonatal meningitis causing Escherichia coli (NMEC). BMC Microbiol 15:211. doi: 10.1186/s12866-015-0547-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mellata M. 2013. Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog Dis 10:916–932. doi: 10.1089/fpd.2013.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dho-Moulin M, Fairbrother JM. 1999. Avian pathogenic Escherichia coli (APEC). Vet Res 30:299–316. [PubMed] [Google Scholar]
- 9.Johnson JR, Kuskowski MA, Owens K, Clabots C, Singer RS. 2009. Virulence genotypes and phylogenetic background of fluoroquinolone-resistant and susceptible Escherichia coli urine isolates from dogs with urinary tract infection. Vet Microbiol 136:108–114. doi: 10.1016/j.vetmic.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Jakobsen L, Spangholm DJ, Pedersen K, Jensen LB, Emborg H-D, Agersø Y, Aarestrup FM, Hammerum AM, Frimodt-Møller N. 2010. Broiler chickens, broiler chicken meat, pigs and pork as sources of ExPEC related virulence genes and resistance in Escherichia coli isolates from community-dwelling humans and UTI patients. Int J Food Microbiol 142:264–272. doi: 10.1016/j.ijfoodmicro.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 11.Guenther S, Grobbel M, Lübke-Becker A, Goedecke A, Friedrich ND, Wieler LH, Ewers C. 2010. Antimicrobial resistance profiles of Escherichia coli from common European wild bird species. Vet Microbiol 144:219–225. doi: 10.1016/j.vetmic.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 12.Ewers C, Li G, Wilking H, Kiebling S, Alt K, Antao E, Laturnus C, Diehl I, Glodde S, Homeier T. 2007. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int J Med Microbiol 297:163–176. doi: 10.1016/j.ijmm.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Johnson TJ, Wannemuehler Y, Johnson SJ, Stell AL, Doetkott C, Johnson JR, Kim KS, Spanjaard L, Nolan LK. 2008. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl Environ Microbiol 74:7043–7050. doi: 10.1128/AEM.01395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clermont O, Olier M, Hoede C, Diancourt L, Brisse S, Keroudean M, Glodt J, Picard B, Oswald E, Denamur E. 2011. Animal and human pathogenic Escherichia coli strains share common genetic backgrounds. Infect Genet Evol 11:654–662. doi: 10.1016/j.meegid.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Johnson TJ, Kariyawasam S, Wannemuehler Y, Mangiamele P, Johnson SJ, Doetkott C, Skyberg JA, Lynne AM, Johnson JR, Nolan LK. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J Bacteriol 189:3228–3236. doi: 10.1128/JB.01726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maluta RP, Logue CM, Casas MRT, Meng T, Guastalli EAL, Rojas TCG, Montelli AC, Sadatsune T, de Carvalho Ramos M, Nolan LK, da Silveira WD. 2014. Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS One 9:e105016. doi: 10.1371/journal.pone.0105016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nandanwar N, Janssen T, Kühl M, Ahmed N, Ewers C, Wieler LH. 2014. Extraintestinal pathogenic Escherichia coli (ExPEC) of human and avian origin belonging to sequence type complex 95 (STC95) portray indistinguishable virulence features. Int J Med Microbiol 304:835–842. doi: 10.1016/j.ijmm.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 18.Tivendale KA, Logue CM, Kariyawasam S, Jordan D, Hussein A, Li G, Wannemuehler Y, Nolan LK. 2010. Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect Immun 78:3412–3419. doi: 10.1128/IAI.00347-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moulin-Schouleur M, Répérant M, Laurent S, Brée A, Mignon-Grasteau S, Germon P, Rasschaert D, Schouler C. 2007. Extraintestinal pathogenic Escherichia coli strains of avian and human origin: link between phylogenetic relationships and common virulence patterns. J Clin Microbiol 45:3366–3376. doi: 10.1128/JCM.00037-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skyberg JA, Johnson TJ, Johnson JR, Clabots C, Logue CM, Nolan LK. 2006. Acquisition of avian pathogenic Escherichia coli plasmids by a commensal E. coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect Immun 74:6287–6292. doi: 10.1128/IAI.00363-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ron EZ. 2006. Host specificity of septicemic Escherichia coli: human and avian pathogens. Curr Opin Microbiol 9:28–32. doi: 10.1016/j.mib.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Maddux JT, Stromberg ZR, Curtiss R III, Mellata M. 2017. Evaluation of recombinant attenuated Salmonella vaccine strains for broad protection against extraintestinal pathogenic Escherichia coli. Front Immunol 8:1280. doi: 10.3389/fimmu.2017.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Germon P, Chen Y-H, He L, Blanco JE, Bree A, Schouler C, Huang S-H, Moulin-Schouleur M. 2005. ibeA, a virulence factor of avian pathogenic Escherichia coli. Microbiology 151:1179–1186. doi: 10.1099/mic.0.27809-0. [DOI] [PubMed] [Google Scholar]
- 24.Schubert S, Picard B, Gouriou S, Heesemann J, Denamur E. 2002. Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infect Immun 70:5335–5337. doi: 10.1128/IAI.70.9.5335-5337.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langridge GC, Phan M-D, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res 19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subashchandrabose S, Smith SN, Spurbeck RR, Kole MM, Mobley HL. 2013. Genome-wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog 9:e1003788. doi: 10.1371/journal.ppat.1003788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramsey ME, Hyde JA, Medina-Perez DN, Lin T, Gao L, Lundt ME, Li X, Norris SJ, Skare JT, Hu LT. 2017. A high-throughput genetic screen identifies previously uncharacterized Borrelia burgdorferi genes important for resistance against reactive oxygen and nitrogen species. PLoS Pathog 13:e1006225. doi: 10.1371/journal.ppat.1006225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buckles EL, Wang X, Lane MC, Lockatell CV, Johnson DE, Rasko DA, Mobley HL, Donnenberg MS. 2009. Role of the K2 capsule in Escherichia coli urinary tract infection and serum resistance. J Infect Dis 199:1689–1697. doi: 10.1086/598524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma J, An C, Jiang F, Yao H, Logue C, Nolan LK, Li G. 2018. Extraintestinal pathogenic Escherichia coli increase extracytoplasmic polysaccharide biosynthesis for serum resistance in response to bloodstream signals. Mol Microbiol 110:689–706. doi: 10.1111/mmi.13987. [DOI] [PubMed] [Google Scholar]
- 31.Johnson JR. 2003. Microbial virulence determinants and the pathogenesis of urinary tract infection. Infect Dis Clin North Am 17:261–278. doi: 10.1016/S0891-5520(03)00027-8. [DOI] [PubMed] [Google Scholar]
- 32.Bao Y, Zhang H, Huang X, Ma J, Logue CM, Nolan LK, Li G. 2018. O-specific polysaccharide confers lysozyme resistance to extraintestinal pathogenic Escherichia coli. Virulence 9:666–680. doi: 10.1080/21505594.2018.1433979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai SJ, Inouye M. 2002. EnvZ-OmpR interaction and osmoregulation in Escherichia coli. J Biol Chem 277:24155–24161. doi: 10.1074/jbc.M110715200. [DOI] [PubMed] [Google Scholar]
- 34.Johnson JR, Clabots C, Rosen H. 2006. Effect of inactivation of the global oxidative stress regulator oxyR on the colonization ability of Escherichia coli O1:K1:H7 in a mouse model of ascending urinary tract infection. Infect Immun 74:461–468. doi: 10.1128/IAI.74.1.461-468.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phan M-D, Peters KM, Sarkar S, Lukowski SW, Allsopp LP, Moriel DG, Achard ME, Totsika M, Marshall VM, Upton M, Scott BA, Mark SA. 2013. The serum resistome of a globally disseminated multidrug resistant uropathogenic Escherichia coli clone. PLoS Genet 9:e1003834. doi: 10.1371/journal.pgen.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magistro G, Hoffmann C, Schubert S. 2015. The salmochelin receptor IroN itself, but not salmochelin-mediated iron uptake promotes biofilm formation in extraintestinal pathogenic Escherichia coli (ExPEC). Int J Med Microbiol 305:435–445. doi: 10.1016/j.ijmm.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Henderson IR, Nataro JP. 2001. Virulence functions of autotransporter proteins. Infect Immun 69:1231–1243. doi: 10.1128/IAI.69.3.1231-1243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma J, Cai X, Bao Y, Yao H, Li G. 2018. Uropathogenic Escherichia coli preferentially utilize metabolites in urine for nucleotide biosynthesis through salvage pathways. Int J Med Microbiol 308:990–999. doi: 10.1016/j.ijmm.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Shaibe E, Metzer E, Halpern YS. 1985. Metabolic pathway for the utilization of l-arginine, l-ornithine, agmatine, and putrescine as nitrogen sources in Escherichia coli K-12. J Bacteriol 163:933–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ninfa AJ, Magasanik B. 1986. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc Natl Acad Sci U S A 83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lomovskaya O, Lewis K, Matin A. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J Bacteriol 177:2328–2334. doi: 10.1128/jb.177.9.2328-2334.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang F, An C, Bao Y, Zhao X, Jernigan RL, Lithio A, Nettleton D, Li L, Wurtele ES, Nolan LK. 2015. ArcA controls metabolism, chemotaxis, and motility contributing to the pathogenicity of avian pathogenic Escherichia coli. Infect Immun 83:3545–3554. doi: 10.1128/IAI.00312-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goh KG, Phan M-D, Forde BM, Chong TM, Yin W-F, Chan K-G, Ulett GC, Sweet MJ, Beatson SA, Schembri MA. 2017. Genome-wide discovery of genes required for capsule production by uropathogenic Escherichia coli. mBio 8:e01558-17. doi: 10.1128/mbio.01558-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dorman MJ, Feltwell T, Goulding DA, Parkhill J, Short FL. 2018. The capsule regulatory network of Klebsiella pneumoniae defined by density-TraDISort. mBio 9:e01863-18. doi: 10.1128/mbio.01863-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arshad M, Goller CC, Pilla D, Schoenen FJ, Seed PC. 2016. Threading the needle: small-molecule targeting of a xenobiotic receptor to ablate Escherichia coli polysaccharide capsule expression without altering antibiotic resistance. J Infect Dis 213:1330–1339. doi: 10.1093/infdis/jiv584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Padan E, Tzubery T, Herz K, Kozachkov L, Rimon A, Galili L. 2004. NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+ antiporter. Biochim Biophys Acta 1658:2–13. doi: 10.1016/j.bbabio.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 47.Padan E. 2008. The enlightening encounter between structure and function in the NhaA Na+-H+ antiporter. Trends Biochem Sci 33:435–443. doi: 10.1016/j.tibs.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 48.Minato Y, Ghosh A, Faulkner WJ, Lind EJ, Schesser Bartra S, Plano GV, Jarrett CO, Hinnebusch BJ, Winogrodzki J, Dibrov P, Häse CC. 2013. Na+/H+ antiport is essential for Yersinia pestis virulence. Infect Immun 81:3163–3172. doi: 10.1128/IAI.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tong L. 2013. Structure and function of biotin-dependent carboxylases. Cell Mol Life Sci 70:863–891. doi: 10.1007/s00018-012-1096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zempleni J, Wijeratne SS, Hassan YI. 2009. Biotin. BioFactors 35:36–46. doi: 10.1002/biof.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Napier BA, Meyer L, Bina JE, Miller MA, Sjöstedt A, Weiss DS. 2012. Link between intraphagosomal biotin and rapid phagosomal escape in Francisella. Proc Natl Acad Sci U S A 109:18084–18089. doi: 10.1073/pnas.1206411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ho BT, Dong TG, Mekalanos JJ. 2014. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 15:9–21. doi: 10.1016/j.chom.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RRS, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37. doi: 10.1016/j.chom.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin J, Zhang W, Cheng J, Yang X, Zhu K, Wang Y, Wei G, Qian P-Y, Luo Z-Q, Shen X. 2017. A Pseudomonas T6SS effector recruits PQS-containing outer membrane vesicles for iron acquisition. Nat Commun 8:14888. doi: 10.1038/ncomms14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schwarz S, Hood RD, Mougous JD. 2010. What is type VI secretion doing in all those bugs? Trends Microbiol 18:531–537. doi: 10.1016/j.tim.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma J, Bao Y, Sun M, Dong W, Pan Z, Zhang W, Lu C, Yao H. 2014. Two functional type VI secretion systems in avian pathogenic Escherichia coli are involved in different pathogenic pathways. Infect Immun 82:3867–3879. doi: 10.1128/IAI.01769-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma J, Sun M, Bao Y, Pan Z, Zhang W, Lu C, Yao H. 2013. Genetic diversity and features analysis of type VI secretion systems loci in avian pathogenic Escherichia coli by wide genomic scanning. Infect Genet Evol 20:454–464. doi: 10.1016/j.meegid.2013.09.031. [DOI] [PubMed] [Google Scholar]
- 58.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M-A, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 60.Stothard P, Wishart DS. 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 61.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 62.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14. doi: 10.1016/0378-1119(95)00193-A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.