Abstract
Background and Aims
Tree rings, as archives of the past and biosensors of the present, offer unique opportunities to study influences of the fluctuating environment over decades to centuries. As such, tree-ring-based wood traits are capital input for global vegetation models. To contribute to earth system sciences, however, sufficient spatial coverage is required of detailed individual-based measurements, necessitating large amounts of data. X-ray computed tomography (CT) scanning is one of the few techniques that can deliver such data sets.
Methods
Increment cores of four different temperate tree species were scanned with a state-of-the-art X-ray CT system at resolutions ranging from 60 μm down to 4.5 μm, with an additional scan at a resolution of 0.8 μm of a splinter-sized sample using a second X-ray CT system to highlight the potential of cell-level scanning. Calibration-free densitometry, based on full scanner simulation of a third X-ray CT system, is illustrated on increment cores of a tropical tree species.
Key Results
We show how multiscale scanning offers unprecedented potential for mapping tree rings and wood traits without sample manipulation and with limited operator intervention. Custom-designed sample holders enable simultaneous scanning of multiple increment cores at resolutions sufficient for tree ring analysis and densitometry as well as single core scanning enabling quantitative wood anatomy, thereby approaching the conventional thin section approach. Standardized X-ray CT volumes are, furthermore, ideal input imagery for automated pipelines with neural-based learning for tree ring detection and measurements of wood traits.
Conclusions
Advanced X-ray CT scanning for high-throughput processing of increment cores is within reach, generating pith-to-bark ring width series, density profiles and wood trait data. This would allow contribution to large-scale monitoring and modelling efforts with sufficient global coverage.
Keywords: X-ray CT, computed tomography, tree ring analysis, multiscale imaging, earth system sciences, increment cores, wood traits, densitometry, deep learning, scanner simulation
INTRODUCTION
Tree rings, as archives of the past and biosensors of the present, offer unique opportunities to study influences of the fluctuating environment, including climate- or disturbance-induced tree growth anomalies over decades to centuries (Babst et al., 2017), and deliver essential input for global vegetation modelling (Beeckman, 2016; Zuidema et al., 2018) within land surface models. In addition to conventional tree ring width (TRW) series, wood traits are of special interest. For instance, X-ray densitometry has been used for a long time, and it has been shown that maximum latewood density (MXD) (Schweingruber et al., 1978) is a temperature proxy superior to that of TRW for certain coniferous wood species (Björklund et al., 2014). Quantitative wood anatomy, i.e. quantifying the number and size of anatomical components of which a tree ring consists, is another promising approach (e.g. Fonti et al., 2008; Cuny et al., 2015; García-González et al., 2016; Carrer et al., 2017) to build long-term records of wood traits, documenting tree performance and tree stress (Beeckman, 2016), while mapping of long-term wood chemistry variations can also be of use (e.g. Hevia et al., 2018). To contribute to earth system sciences, however, sufficient spatial coverage is required, thus necessitating large data sets, since it is known that sampling design is a consequential issue, influencing the outcome of tree ring studies (Brienen et al., 2012; Nehrbass-Ahles et al., 2014).
The above-mentioned issues point to the need for high-throughput mapping of the tree ring (at the intra-annual scale), especially when tree ring data series are to be coupled with earth observation satellite data and used in land surface models. Rather than merely relying on databases reporting on general averages, the latter models should be fed with detailed individual-based (Scheiter et al., 2013) pith to bark trait data to properly account for spatial and temporal inter- and intraspecies variation in performance.
Different techniques have been reported in the literature (see, for example, Liang et al., 2013; Scharnweber et al., 2016; Jacquin et al., 2017). In particular, progress in preparation and analysis of long thin sections has significantly advanced the field of quantitative wood anatomy (Gärtner and Nievergelt, 2010; Gärtner et al., 2015). Although TRW and wood trait measurements are often labour intensive and time consuming, limiting the number of samples that can be reasonably analysed, progress in efficient thin sectioning and imaging (using, for example, a slide scanner) is further reducing the time needed for processing. In most cases, however, sample treatment is still necessary for proper analysis, by sub-sampling of the increment cores (or stem discs), sawing to an appropriate thickness, polishing of the surface or sectioning. These procedures call for skilled and trained staff. Moreover, standardization is challenging for most methods, yet is essential to guarantee comparability.
X-ray computed tomography (CT) is one of the few candidate techniques to take tree ring research on increment cores to the next level and is complementary to existing techniques. It is non-invasive, requires limited sample preparation, enables 3-D tree ring analysis (Van den Bulcke et al., 2014) and quantitative wood anatomy, and has the potential for semi-automated or guided ring indication. In all, it is a promising technique for mass processing of increment cores (e.g. Steffenrem et al., 2014; De Mil et al., 2016; Jacquin et al., 2019). Additionally, thanks to recent developments in scanner simulation (Dhaene, 2017), calibration-free X-ray densitometry is within reach.
We mainly report here on a series of scans of increment cores, performed on state-of-the-art X-ray CT systems, developed by the Ghent University Centre for X-ray Tomography (UGCT, www.ugct.ugent.be) and TESCAN XRE (www.XRE.be, part of the TESCAN ORSAY HOLDING a.s.), formerly known as XRE, a UGCT spin-off company. We show how automated multiscale scanning and reconstruction can be used to handle large physical increment core sets in different configurations for archiving, TRW measurements and densitometry. Additionally, volume of interest (VOI) scanning allows for zooming in on specific sections of interest at high resolution without sample manipulation. Dedicated sub-sampling and high-resolution scanning can even resolve cell-level details, as such complementing the thin section approach. We furthermore illustrate how accurate simulation of an X-ray CT scanner enables estimation of wood density without the use of calibration material and highlight the potential of neural-based learning techniques for tree ring indications and quantitative wood anatomy on such scans.
MATERIALS AND METHODS
X-ray CT scan systems
The CoreTOM scan system has been developed by TESCAN XRE and is designed for multiscale imaging of elongated core-like objects up to 1 m in length. It consists of a mechanical set-up with ten motorized axes mounted on high-precision granite and is equipped with a high-power 180 kV microfocus X-ray source (true spatial resolution down to 3 μm) and multiple high-speed flat panel detectors for high sample throughput. In this study, both a 43 × 43 cm2 a-Si flat panel (2800 × 2800 pixels) with a pixel size of 150 μm and a 14.5 × 11.4 cm2 CMOS flat panel (1900 × 1500 pixels) with a pixel size of 75 μm are used. The VOI scanning feature allows for automated scanning of a specific region of an object at a higher resolution. The scanner is controlled via the LabView®-based control software platform Acquila (currently further developed and licensed by TESCAN XRE), which is based on the acquisition software originally developed at UGCT (Dierick et al., 2010).
The HECTOR scan system is developed and built by the Radiation Physics group of the UGCT in collaboration with XRE. It consists of a mechanical set-up with nine motorized axes and has a 240 kV XWT 240-SE directional microfocus source from X-RAY WorX with a large 40 × 40 cm2 PerkinElmer 1620 CN3 CS flat panel detector (2000 × 2000 pixels) with a pixel size of 200 μm. The two translation stages of the detector allow for tiled tomography, thus resulting in a large field of view (FOV; 80 × 80 cm2). The scanner is controlled via a LabView®-based control software platform (Dierick et al., 2010). More information can be found in Masschaele et al. (2013).
The Nanowood scan system has been developed and built by the Radiation Physics group of the UGCT and is equipped with two separate X-ray tubes and two different X-ray detectors to allow for optimal scanning for a very wide range of samples. It consists of a mechanical set-up with seven motorized axes, has an open-type Hamamatsu transmission tube for extra-high-resolution scans and a closed-type Hamamatsu directional tube used for larger samples. An 11 megapixel Photonic Science VHR CCD camera with a pixel size of approx. 7 μm is complemented with a large area Varian flat-panel detector. The scanner is controlled via the same LabView®-based control software platform as HECTOR (Dierick et al., 2010). More information can be found in Dierick et al. (2014).
The operational details of all scanners are summarized in Table 1. Increment cores and sample holders
Table 1.
Operational details of the different X-ray CT scan systems
| Maximum object diameter (cm) | Maximum object height (m) | Maximum object weight (kg) | Maximum resolution (μm) | |
|---|---|---|---|---|
| CoreTOM | 60 | 1 | 50 | 3 |
| HECTOR | 60 | 1 | 80 | 3 |
| Nanowood | 30 | 0.3 | 5 | 0.4 |
The maximum height and diameter reported here are related to the maximum field of view (FOV) of the scanner without sample manipulation.
Increment cores of different wood species were selected, including three temperate broad-leaved species, namely oak (Quercus spp.), beech (Fagus sylvatica) and European white birch (Betula pendula), the temperate coniferous Scots pine (Pinus silvestris) and the brevi-deciduous tropical tree species limba (Terminalia superba). All but the latter species were scanned with the CoreTOM scan system at TESCAN XRE, whereas the limba samples were scanned with the HECTOR scan system at UGCT. HECTOR is currently fully implemented in the Arion software toolbox (Dhaene et al., 2015) and therefore well suited for illustrating calibration-free densitometry (see further).
Increment cores, stored in paper straws, were mounted in three differently sized sample holders. For details on the exact configuration and mounting procedure we refer to Van den Bulcke et al. (2014) and De Mil et al. (2016). Sample holder 1, with a diameter of 10 cm and a variable length (currently up to 60 cm), can hold 33 increment cores and was designed for high-throughput processing purposes. Sample holder 2, with a diameter of 3 cm and a length of 16 cm, contains six increment cores, allows scanning at higher resolution and was specifically designed for accurate densitometry. Sample holder 3 is suited for the highest resolution scans on intact increment cores to resolve as much anatomical detail as possible and can hold up to 1 m of core, whether stacked on top of each other or not.
Finally, to highlight the potential of smaller sample sizes and cell-level resolution, an additional series of scans was performed using the Nanowood scan system to image a splinter-sized specimen of European larch (Larix decidua).
X-ray CT scanning and calibration-free densitometry
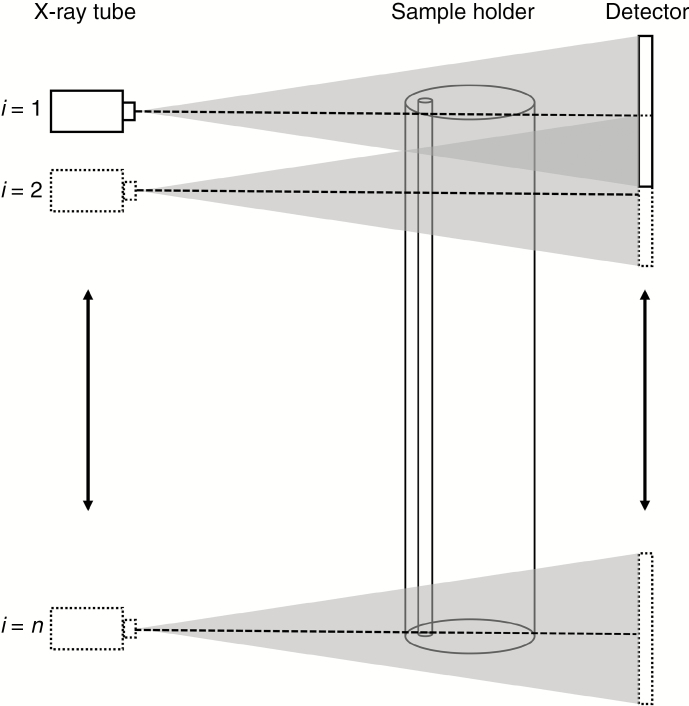
Overlapping smooth stacked scans were performed per sample holder with the X-ray CT systems described above. The principle of stacked scanning is schematically visualized in Fig. 1.
Fig. 1.
Principle of stacked scanning: tube and detector move simultaneously and, at each position, i, a full tomography scan is made of the samples in the sample holder, ensuring a pre-defined overlap with the previous scan. The total number of scans is n. Note that instead of moving the tube and detector, the sample could move upwards (or downwards) as well. The dotted line in the centre of the beam represents the central plane.
The high-resolution stacked scans of the larch sample on Nanowood were done by moving the sample upwards instead of the tube and detector since Nanowood has a fixed height for both components. Reconstructions were performed using the XRE reconstruction tools in Acquila or the Octopus Reconstruction package (Vlassenbroeck et al., 2007) on a workstation with 16 CPU cores, 128 GB RAM and a 1080 Ti Nvidia GPU. Stacked scans per sample holder were stitched and merged afterwards using the XRE reconstruction software or a custom-written routine in Fiji (Schindelin et al., 2012) based on the stitching algorithm of Preibisch et al. (2009).
Density estimates of the samples were based on two different procedures: the method described in De Ridder et al. (2011) using a reference material with a chemical composition similar to that of wood on one hand and simulated on the other hand using the Arion software (Dhaene et al., 2015). For the latter, the following equation was solved to obtain density ρ:
The reconstructed mass attenuation coefficient in each voxel of a 3-D volume is represented by μ. μ/ρ(Ei)z is the mass attenuation coefficient of the material z present in the sample at energy Ei. Si’’ represents the number of photons emitted by the X-ray source and Dinc,i represents the deposited energy in the detector per incident photon. For more details on the method, the reader is referred to Dhaene et al. (2015).
RESULTS
An overview of the scanned samples, the acquisition parameters, resolution, number of scans per stack and resulting reconstructed data volume is given in Table 2.
Table 2.
Details of the scans
| Scanner | Samples | Resolution (μm) | No of projections per scan | Exposure time per projection (ms) | No of scans per stack | Stitched 16-bit reconstructed data volume (GB) |
|---|---|---|---|---|---|---|
| CoreTOM | 33 cores (oak) | 60.0 | 2160 | 1400 | 8 | Approx. 28 |
| VOI single core (oak) | 13.5 | 1800 | 1000 | 13 | Approx. 68 | |
| 6 cores (oak, beech, birch, pine)* | 23.7 | 2250 | 100 | 6 | Approx. 21 | |
| 1 core (pine)* | 8.0 | 1600 | 150 | 15 | Approx. 55 | |
| 1 core (oak)* ,† | 4.5 | 2400 | 75 | 73 | Approx. 140 | |
| 1 core (pine)* | 4.0 | 3000 | 70 | 70 | Approx. 275 | |
| HECTOR | 6 cores | 35.0 | 2000 | 1000 | 1 | Approx. 2 |
| Nanowood | 1 splinter-sized specimen (larch) | 0.8 | 2000 | 1400 | 10 | Approx. 8 |
*Faster CMOS detector mounted on the CoreTOM system.
†Eight-bit reconstruction instead of 16-bit
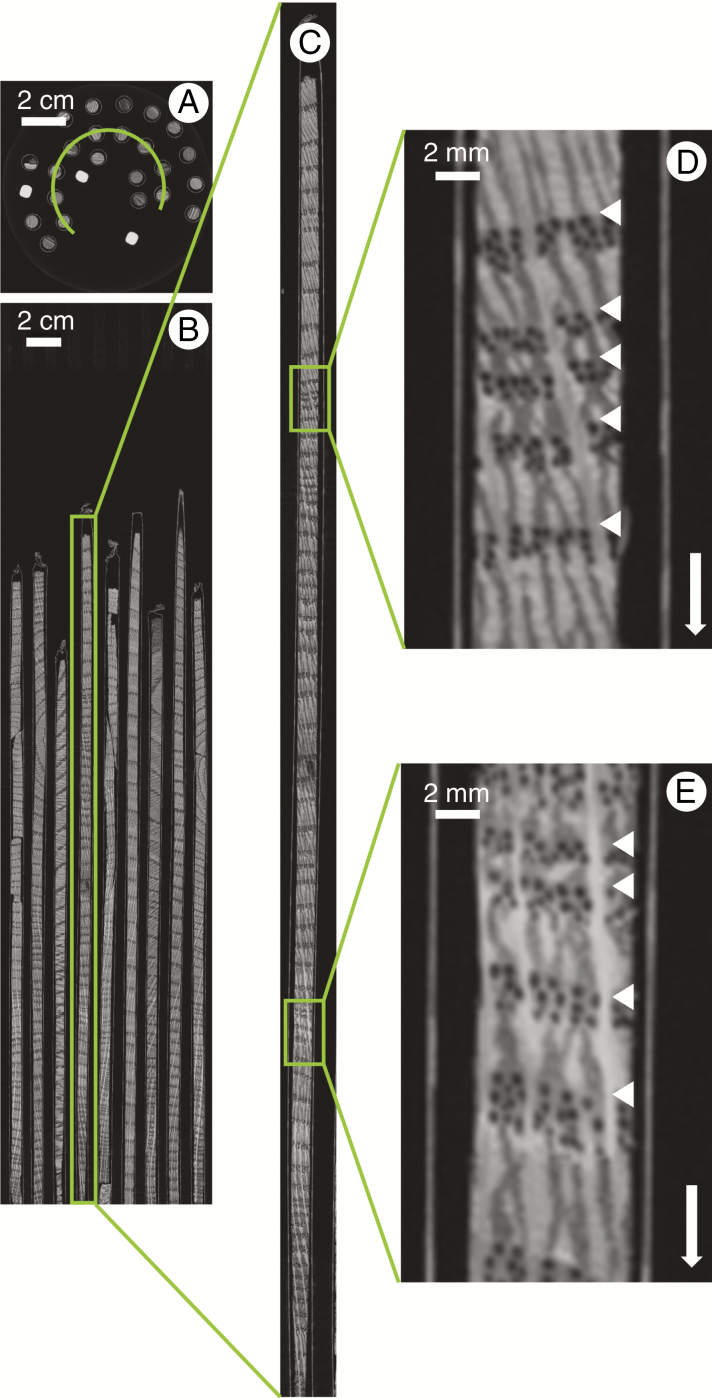
Figure 2 shows the result of stacked scanning and stitching of a sample holder with a series of Quercus spp. increment cores at 60 μm resolution. The scan resolution allows the tree ring boundaries to be indicated rather easily and is in most cases sufficient for TRW measurements, yet lacks detail for further analysis, such as, for instance, earlywood vessel measurements (Fonti et al., 2008).
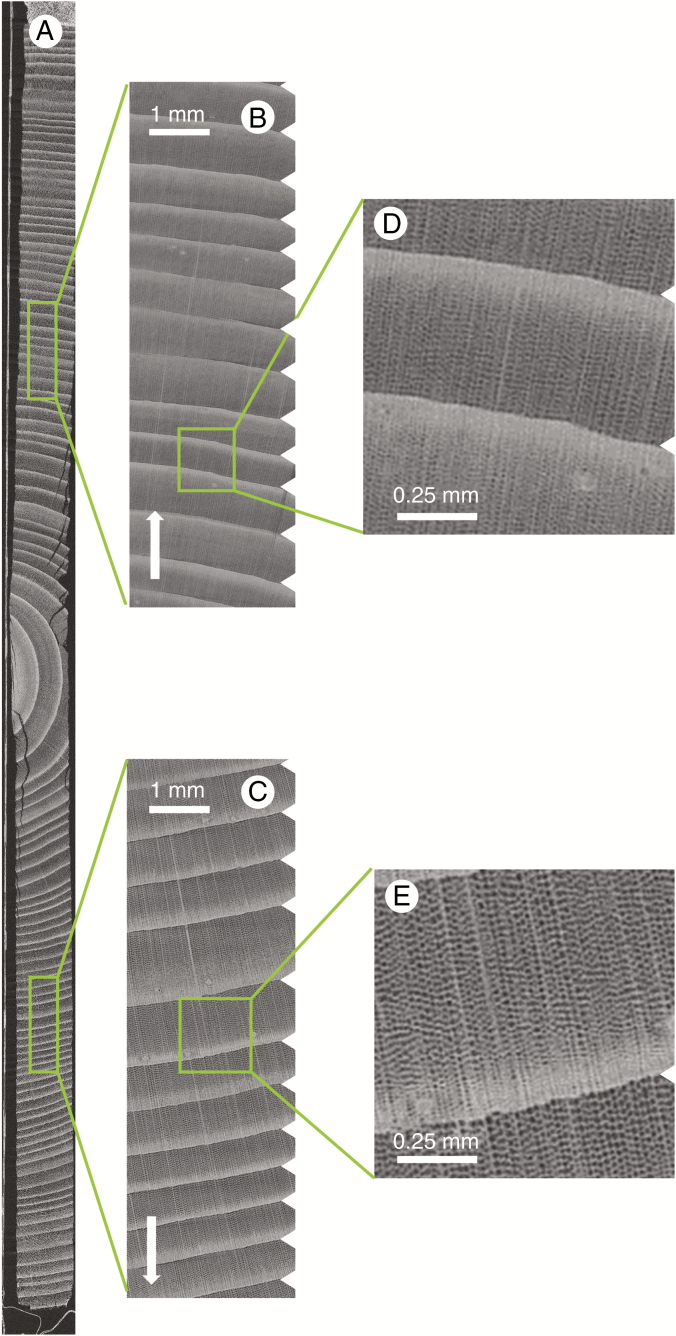
Fig. 2.
Cross-section through sample holder 1 (A), with a longitudinal section along the green arc depicted in A (B). A single core of 32.8 cm length was selected (C) to illustrate the level of detail (D and E). The white arrow indicates the pith to bark direction, and the white triangles demarcate ring boundaries.
To image a single core at much higher resolution, zoomed projection images of the selected core are obtained. The result of such a VOI scan is shown in Fig. 3, which has been acquired using the automated VOI scanning protocol implemented in the CoreTOM acquisition software Acquila. A single oak core is positioned in the FOV and is fully scanned at 13.5 μm resolution. Obviously, the scan resolution allows tree ring boundaries to be indicated and has clear potential for the visualization and analysis of larger anatomical features such as earlywood vessels, but can also be sufficient for quantification of other tissue proportions such as parenchyma and rays.
Fig. 3.
VOI scan of an oak increment core of 25.6 cm length (A), with a zoomed-in section illustrating the clear growth ring boundaries and earlywood vessels (B and C), and further zoomed-in sections highlighting that even smaller vessels can be discerned properly (D and E). The white arrow indicates the pith to bark direction, and the white triangles demarcate ring boundaries.
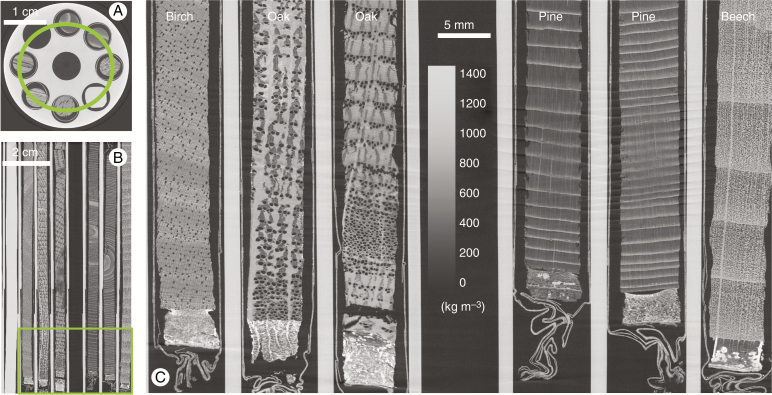
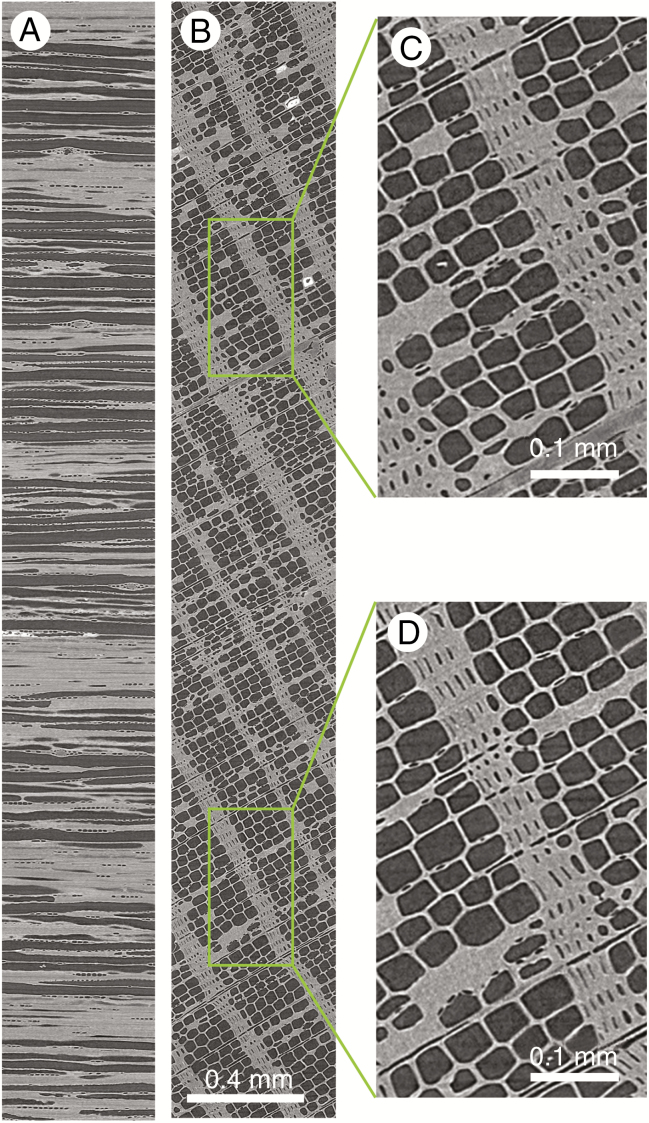
Six increment cores of different wood species have been mounted in sample holder 2 (16 cm in length) and scanned at a resolution of 23.7 μm. Figure 4 shows a cross-section through the holder and illustrates the level of detail that can be noted for a series of wood species. Furthermore, it shows a clear view of the bark and even the different paper layers of the straws containing the increment cores.
Fig. 4.
Cross-section through sample holder 2 with six increment cores (A), with a longitudinal section along the green circle depicted in A (B). Zoomed-in section at the bottom shows the bark and wood anatomical details that can be discerned (C). The scale bar is given in the middle as well as a density greyscale in kg m–3.
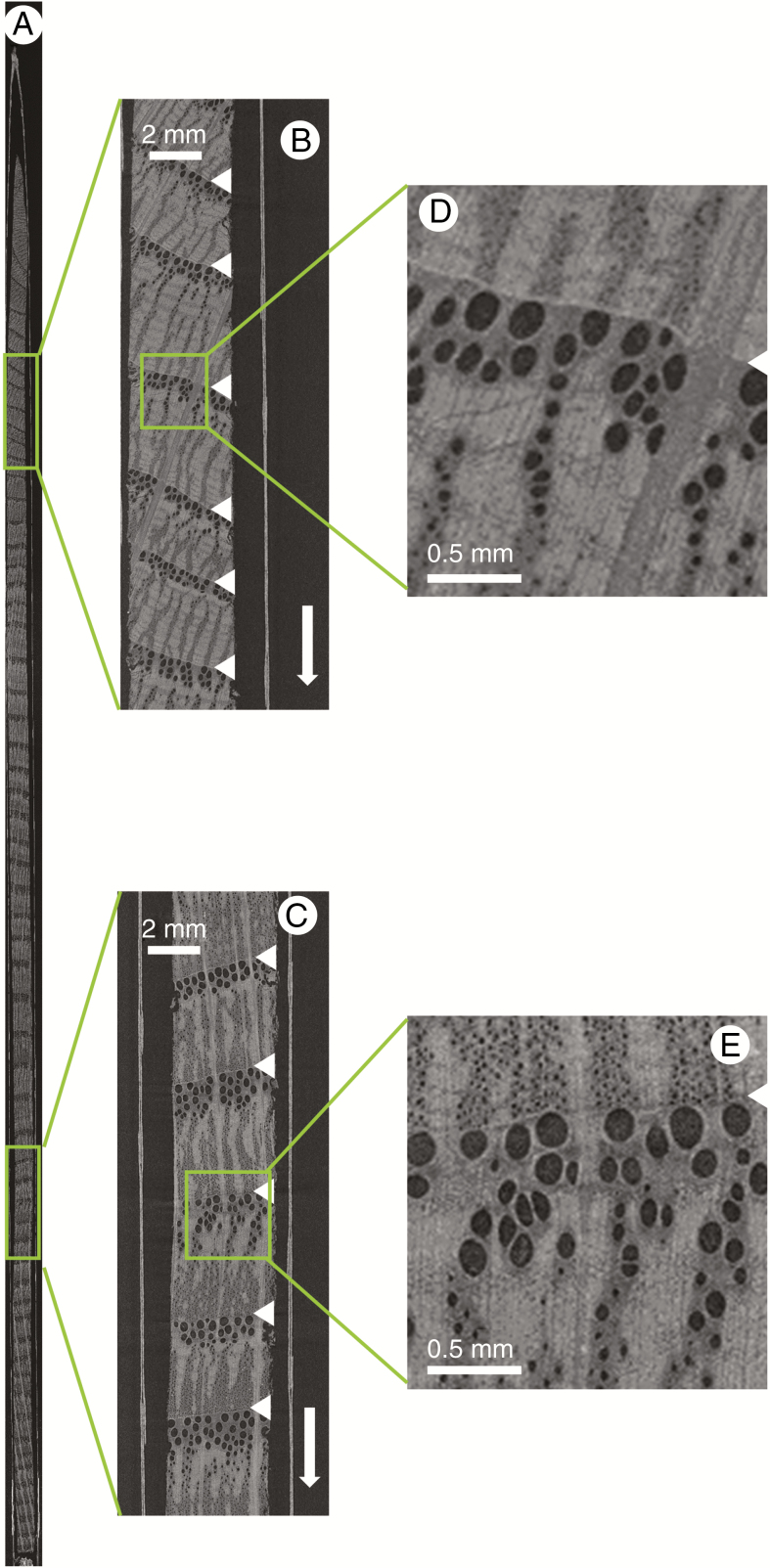
Only single increment cores (sample holder 3) fit within the FOV when scanning at a higher resolution. Figure 5 shows the result of scanning a single Scots pine core of 11.5 cm length. The resolution is 8 μm and individual earlywood cells can be seen, yet not quantified in terms of shape and size.
Fig. 5.
Scan of a Scots pine increment core of 11.5 cm length in sample holder 3 (A), with zoomed-in sections illustrating the clear growth ring boundaries (B and C), and further zoomed-in sections highlighting that individual earlywood cells can be discerned (D and E). The white arrow indicates the pith to bark direction, and the white indentations demarcate ring boundaries.
A small section of the best possible high-resolution scan of an entire increment core of oak and Scots pine is given in Fig. 6. Anatomical details are well resolved, yet cell-level details such as cell walls and measurable sizes of earlywood cells are not yet within reach at this resolution.
Fig. 6.
Small section of a high-resolution scan of an oak (A) and Scots pine (B) increment core in sample holder 3. The white arrow indicates the pith to bark direction, and the white indentations demarcate ring boundaries.
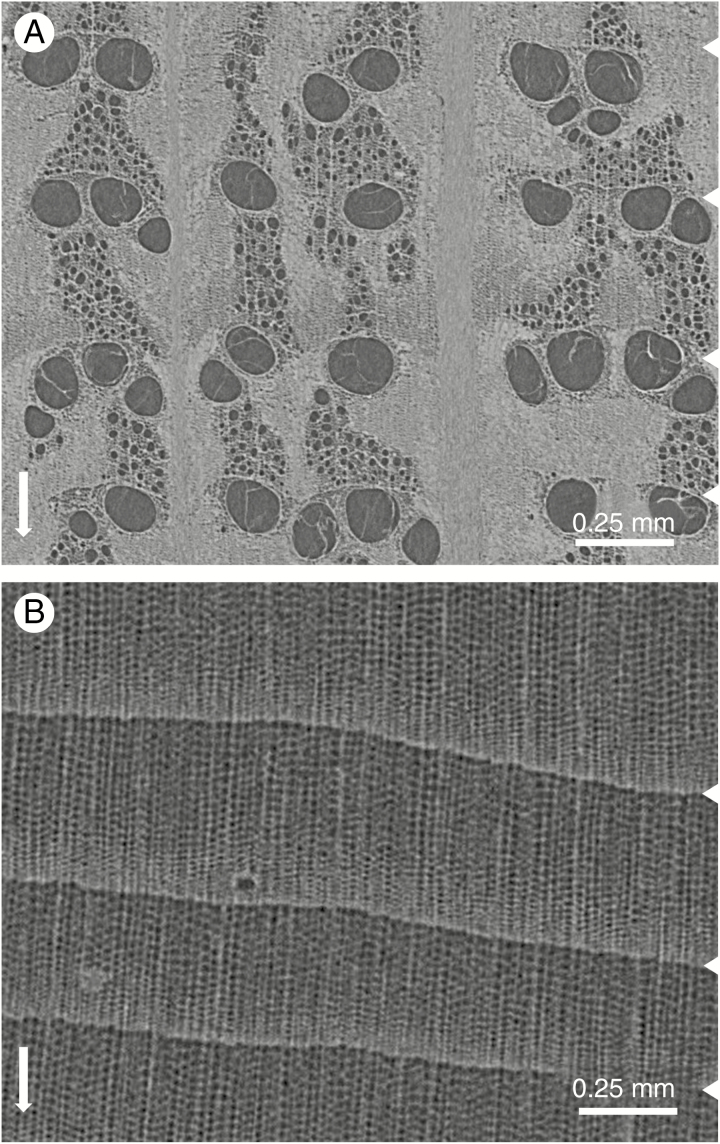
Scans at sub-micron resolution are needed to get a proper view of cell-level details. Such an example is given in Fig. 7, showing a cross-section through a splinter-sized larch stick scanned with the Nanowood scan system.
Fig. 7.
Sub-micron scan of a needle-shaped larch stick of approx. 4 mm length (A, quasi-tangential; B, transversal), with zoomed-in sections (C and D) illustrating the clear cell-level details that can be obtained.
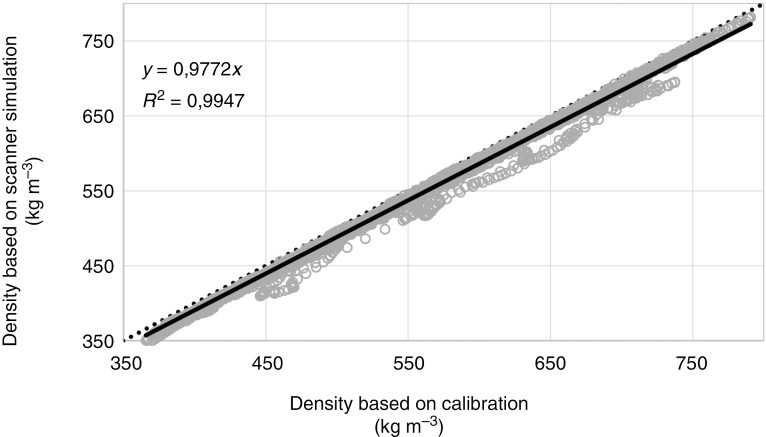
For densitometry (see, for example, Bastin et al., 2015), six increment cores of limba were scanned in sample holder 2. The holder, similar to the one illustrated in Fig. 4, consists of reference material with a chemical composition close to that of wood and is used to convert attenuation coefficients to density estimates. Figure 8 illustrates the density profiles of a sub-section of the six limba cores plotted against the results of the scanner simulation approach. The density profiles are obtained according to the procedure described in De Ridder et al. (2010) using the software reported in Van den Bulcke et al. (2014) and De Mil et al. (2016). The correlation between both methods is very high.
Fig. 8.
Scatterplot of densities calculated according to the procedure reported in De Ridder et al. (2010) and values derived from scanner simulation for a series of subsections from limba increment cores. The grey circles are the data points, the solid black line represents the linear fit (R2 and equation given in the upper left-hand corner) and the black dotted line indicates the 1:1 line.
DISCUSSION
The potential use of X-ray CT scanning for tree ring analysis was reported in the early 1980s (Onoe et al., 1984) and was explicitly mentioned as a technique needed for tree ring research by Grabner et al. (2009). Several researchers such as Okochi et al. (2007), Bil et al. (2012) and Stelzner and Million (2015) used X-ray CT scanning with the scope of characterizing cultural and archaeological objects, yet these studies are mostly limited to the measurement of TRWs and wood identification.
High-throughput processing of increment cores has only been accomplished recently (Maes et al., 2017; Vannoppen et al., 2017), showing the extraction of both TRW series as well as density profiles (De Mil et al., 2016; Vannoppen et al., 2018).
Sample preparation
One of the disadvantages of many existing techniques for analysis on increment cores is the need for sample manipulation and preparation, depending on skilled staff; see, for example, the protocol for sectioning for quantitative wood anatomy by von Arx et al. (2016). In contrast, increment core manipulation is limited for X-ray CT scanning. Furthermore, both scanning and reconstruction can be automated, limiting operator intervention for data acquisition. A well-designed sample holder for multiple cores (see, for example, Steffenrem et al., 2014; Van den Bulcke et al., 2014; Jacquin et al., 2019) also enables automation of part of the processing, limiting the need for further intervention along the processing pipeline. The number of cores that can be scanned simultaneously and the maximum length of the cores are merely determined by (1) sample holder design and (2) hardware limits of the scanning system, and both can be optimized.
Scanning and reconstruction
Exposure time is a major consideration when scanning since it directly affects total scan time. The exposure time will have a major influence on the reconstruction quality that needs to be achieved. Longer scans will result in a better signal to noise ratio for a given set-up, yet this is also influenced by the X-ray source (voltage, power and spot size), the composition of the sample and detector hardware (sensitivity and dynamic range). The effect of different detectors on exposure time can be seen in Table 2. To obtain the highest possible scan quality, the full dynamic range of the detector should be explored (du Plessis et al., 2017), yet, more importantly, scan time should also be fine-tuned to the research question at hand. Automated overnight scanning is an attractive option especially for stacked scanning at high resolution. It should be mentioned that X-ray imaging beamlines at synchrotron facilities allow for very fast, high-throughput scanning at high resolution, yet they are less accessible for frequent use and, in most cases, they have limited options for scanning large and long objects, and are thus less suited for multiscale scanning, as shown here.
Within the scope of tree-ring-related analyses, an essential point of discussion is the resolution needed to extract reliable quantitative data. The multiscale approach shown here illustrates that, depending on the research question at hand, a different scale can be of interest. It should be noted that CT is a digital imaging technique, discretizing a volume in a stack of 3-D voxels. As such, increasing the resolution for a given volume by a factor of 2 will lead to an increase of the resulting data by a factor of 8. Additionally, for this increase in resolution, more stringent requirements are imposed on the measurement system, resulting in an even larger increase of acquisition time. The choice of the imaging resolution to be used is, therefore, a very difficult trade-off between the detail and the total volume that needs to be imaged, notably between medical CT and micro-CT (Jacquin et al., 2019).
Multiscale imaging enables scanning of large numbers of increment cores at a moderate resolution (Fig. 2) for archiving purposes. In certain cases, it is also sufficiently good for TRW measurements (De Groote et al., 2018; 110 μm resolution) and density profiling (Vannoppen et al., 2018; 110 μm resolution). Furthermore, analysis of increment cores scanned at this resolution can guide scanning at higher resolutions (Fig. 3), focusing, for instance, on narrower rings (Fig. 4, oak and pine sample) or most interesting cores. At higher resolutions (Figs 5 and 6), discerning tree ring boundaries does not only depend on density differences but they can be derived from anatomical features directly. Virtual sectioning of scans at cell-level resolution is also within reach (Fig. 7), although long thin sections are currently setting the bar for quantitative wood anatomy (e.g. Saß and Eckstein, 1994; Gärtner et al., 2015). The use of staining agents could furthermore unlock new methods of visualization and analysis (Pauwels et al., 2013; Staedler et al., 2013; Van Loo et al., 2014).
Nevertheless, many micro-CT systems are built with a directional-type X-ray source, and, as such, they are limited to a resolution of a few microns, similar to the HECTOR and CoreTOM systems used in this study. When higher resolutions are desired, a different hardware configuration is needed. The latter is based on either a different type of X-ray source (see, for example, Masschaele et al., 2007; Dierick et al., 2014; Busse et al., 2018), optical magnification at the detector side (see, for example, Busse et al., 2018) or even X-ray optics (see, for example, Wong et al., 2014). Although a hardware configuration such as that of Nanowood enables both sub-micron resolutions and scans of larger objects, it makes the system mechanically very complex and halves the available measurement time. Having two separate scanners specifically tuned to their purpose and resolution is thus far more ideal. The larch sample clearly shows that sub-micron resolution scans enable cell-level detail due to the specific combination of a transmission tube and a sensitive detector with small pixel size. Although sub-sampling was needed, VOI imaging of certain sections of an increment core could also be possible at an even higher resolution than presented in Fig. 6.
Finally, given that the FOV is limited by the size of the detector for most scanners, the stacked approach illustrated in Fig. 1 is key to covering the entire increment core length, subsequently followed by automated stitching of the separate volumes. The latter is a non-trivial task and needs proper evaluation when stitching several tens of scans (Figs 5 and 6). Furthermore, for the standard cone beam circular geometry, information on the transitions along the Z-axis is lost to some extent due to cone beam artefacts (De Witte, 2010). Sufficient overlap between consecutive scans is mandatory to reduce this. An alternative approach is helical scanning and related reconstruction (e.g. Van den Bulcke et al., 2014) based on either the patented Katsevich approach (Katsevich, 2002) or on iterative reconstruction (De Schryver, 2017). Helical scanning requires clever computing if used to reconstruct large data sets (cf. full increment cores of which a small sub-sample is visualized in Fig. 6) and needs motion compensation strategies for long scans (Latham et al., 2018).
Finally, at sub-micron resolution, phase contrast is a prominent effect. When left unprocessed, the edge enhancement effect is particularly disturbing for quantitative image analysis. However, several processing algorithms, of which the Paganin algorithm (Paganin et al., 2002) is currently the most used solution, exist to cope with this issue (Burvall et al., 2011; Boone et al., 2012).
Data analysis
A number of packages exist to handle 2-D imagery and extract ring width, density and wood traits. Well-known commercial tree ring software such as WinDendro™ and CooRecorder are used frequently for TRW measurements on flatbed scans of polished wood surfaces and the analysis of conventional densitometry images. The commercial packages WinCELL™, ROXAS (depending on Image-Pro Plus; von Arx and Carrer, 2014) and the open-source software ImageJ (e.g. Schuldt et al., 2013) are powerful software tools for building vessel and lumen chronologies, mainly using thin cross-sections. All these packages could handle the 2-D X-ray-based cross-sections generated here. To exploit the 3-D nature of the images, however, the MATLAB®-based software toolboxes described in Van den Bulcke et al. (2014) and De Mil et al. (2016) could be used for TRW and density profiling. The process of the latter is clearly facilitated when density values are reconstructed directly from scanner simulation rather than the conversion of attenuation coefficients to density values based on a reference material (Fig. 8).
Furthermore, given that wood is a highly structured and organized material, conventional machine learning techniques for pixel-based classification can be used in the context of image segmentation, object detection and boundary detection. A well-known example of this is the WEKA toolbox for ImageJ (developed by Arganda-Carreras et al., 2017), which facilitates the integration of machine learning schemes with image processing modules into the pipeline and allows for users to provide feedback by correcting or adding labels. Consequently, with the WEKA toolbox, one can efficiently delineate wood vessels and parenchyma (De Mil et al., 2018).
Conventional machine learning techniques are, however, limited in their ability to process data in their raw form. On the other hand, deep learning methods can result in multiple levels of representation by composing simple non-linear modules that each transform the representation at one level (starting with the raw input) into a representation at a higher, slightly more abstract level (LeCun et al., 2015). The image data sets from De Mil et al. (2016) and those presented herein can be of significant value for testing the latest deep learning architecture, potentially assisting in much of the manual measurement work still done today, similar to the work reported by Fabijańska and Danek (2018) on optical images. Deep learning to extract ring boundaries but also for anatomical feature detection could potentially revolutionize the field. X-ray CT scanning can generate such large amounts of data (see, for instance, the www.dendrochronomics.ugent.be website where we keep track of the number of cores scanned), needed for the training of a deep neural net. Moreover, data augmentation, which is beneficial for the training of a deep neural net, can be implemented in a clever way, given the species-specific prior knowledge of the anatomical structure and tree ring boundaries of wood.
Furthermore, time series of TRW, density and quantitative wood anatomy could be used in predictive modelling, adopting the principle of investigating the range of responses among individual sample trees (Carrer, 2011) rather than relying on mean chronologies. Therefore, the large data sets lend themselves to machine learning methods (see De Gooijer et al., 2006; Voyant et al., 2017 for reviews on this topic). In order to capture time-dependent information, time-aware methods such as recurrent neural networks could be used to analyse even undated series properly in terms of the environmental factor under study. For example, reservoir computing has proven to give good results on time series modelling tasks without being too computationally demanding (Jaeger and Haas, 2004; wyffels and Schrauwen, 2010).
Finally, although not illustrated in this study, the latest developments in X-ray CT scanning in combination with X-ray fluorescence (e.g. Laforce et al., 2017) could also lead to extracting chemical information from increment cores, similarly to what is currently already accomplished in 2-D using, for example, the ITRAX scanner (e.g. Hevia et al., 2018).
Data handling
Large amounts of data are generated (Table 2) and need to be stored efficiently, as either 16-bit or 8-bit images. Well-known formats are multipage TIFF, HDF5 and JP2 with different compression alternatives. Exploiting the 3-D nature of the data of increment cores can be of interest (see, for example, Van den Bulcke et al., 2014), yet, in other cases, storing a sub-set of transversal sections, preferentially filtered in the longitudinal direction to increase the signal-to-noise ratio, is adequate. Furthermore, although stitching conveniently results in single increment core volumes, analysing volumes separately is also an option. Sufficient data storage and secure back-up are essential, as well as up-to-date computer infrastructure for reconstruction and analysis. Cloud-based solutions are under development, especially for medical imaging and synchrotron beamlines (see, for example, Meng et al., 2011; Bednarz et al., 2015), compelling automated data management.
Several initiatives have been launched to make X-ray CT data available via open repositories, such as, for example, for digital morphology (Davies et al., 2017), forensic analysis (Rowe et al., 2016) or mathematical purposes (De Carlo et al., 2018). The complexity of information stored in the increment cores calls for online storage as well, which would enable the research community to exploit the data, similar to existing tree ring databases such as the International Tree-Ring Data Bank (ITRDB).
CONCLUSIONS
X-ray CT scanning, accelerated by an increase in computing power and significant progress in both hardware and software, has become an eminent 3-D microscopy technique in many research disciplines. Its potential in tree ring research is promising since large data sets are needed to couple tree ring series to global monitoring and modelling efforts. In all, a dedicated X-ray CT scanning system could be used optimally to scan large core sets and analyse them semi-automatically with custom-written software. The multiscale scanning approach illustrated here allows for optimal selection of increment cores, enabling scanning of selected cores at high resolution without any sample manipulation. Although optimal results for cell-level scans are currently only obtained when sub-sampling cores, they do not yet allow for the same workflow as conventional thin section approaches recently developed. Nevertheless, further improvements are expected to allow sub-micron VOI scanning of intact increment cores. Multiscale imaging will also help to exploit maximally the information stored in wood at different microscopic resolutions from the tissue level (e.g. wood density) to the sub-cellular level such as cell walls and pits. Hence, it is complementary to the thin section approach. Calibration-free densitometry and novel staining procedures can also open up new avenues for analysis. X-ray CT data are also very well suited for calculation of MXD and are potentially an important tool for climate reconstructions.
Standardized X-ray CT scanning would also allow for re-analysis of existing core databases, whereas new online databases of the generated CT volumes would unlock the data for other researchers to explore and use the images, similar to existing international tree ring databases. Establishing a dedicated X-ray CT facility servicing the tree ring and land surface modelling community and even forest inventory campaigns could be a final aim. Not only would this allow a standard for scanning to be set, but it would also allow optimal use of resources and access to the latest developments, and complement the existing suite of techniques.
FUNDING
The Special Research Fund of Ghent University (BOF-UGent) is acknowledged for the funding under grant number BOF17-GOA-015, the financial support of the UGCT Center of Expertise (BOF.EXP.2017.0007) and the funding of the XINCAST project.
ACKNOWLEDGEMENTS
We would like to thank Stijn Willen for his help with the preparation of sample holders and samples.
LITERATURE CITED
- Arganda-Carreras I, Kaynig V, Rueden C, et al. . 2017. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics 33: 2424–2426. [DOI] [PubMed] [Google Scholar]
- von Arx G, Carrer M. 2014. ROXAS – a new tool to build centuries-long tracheid-lumen chronologies in conifers. Dendrochronologia 32: 290–293. [Google Scholar]
- von Arx G, Crivellaro A, Prendin AL, Čufar K, Carrer M. 2016. Quantitative wood anatomy – practical guidelines. Frontiers in Plant Science 7: 781. doi: 10.3389/fpls.2016.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst F, Poulter B, Bodesheim P, Mahecha MD, Frank DC. 2017. Improved tree-ring archives will support earth-system science. Nature Ecology & Evolution 1: 8. doi: 10.1038/s41559-016-0008. [DOI] [PubMed] [Google Scholar]
- Bastin J-F, Fayolle A, Tarelkin Y, Van den Bulcke J, de Haulleville T, Mortier F, et al. 2015. Wood specific gravity variations and biomass of central african tree species: The simple choice of the outer wood. PLoS ONE 10: e0142146. doi:10.1371/journal.pone.0142146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarz T, Wang D, Arzhaeva Y, et al. . 2015. Cloud based toolbox for image analysis, processing and reconstruction tasks. In: Sun C, Bednarz T, Pham TD, Vallotton P, Wang D, eds. Signal and image analysis for biomedical and life sciences. Cham: Springer, 191–205. [DOI] [PubMed] [Google Scholar]
- Beeckman H. 2016. Wood anatomy and trait-based ecology. IAWA Journal 37: 127–151. [Google Scholar]
- Bill J, Daly A, Johnsen Ø, Dalen KS. 2012. DendroCT – dendrochronology without damage. Dendrochronologia 30: 223–230. [Google Scholar]
- Björklund JA, Gunnarson BE, Seftigen K, Esper J, Linderholm HW. 2014. Blue intensity and density from northern Fennoscandian tree rings, exploring the potential to improve summer temperature reconstructions with earlywood information. Climate of the Past 10: 877–885. [Google Scholar]
- Boone MN, Devulder W, Dierick M, Brabant L, Pauwels E, Van Hoorebeke L. 2012. Comparison of two single-image phase-retrieval algorithms for in-line x-ray phase-contrast imaging. Journal of the Optical Society of America A 29: 2667–2672. [DOI] [PubMed] [Google Scholar]
- Brienen RJ, Helle G, Pons TL, Guyot JL, Gloor M. 2012. Oxygen isotopes in tree rings are a good proxy for Amazon precipitation and El Nino-Southern Oscillation variability. Proceedings of the National Academy of Sciences, USA 109: 16957–16962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burvall A, Lundström U, Takman PA, Larsson DH, Hertz HM. 2011. Phase retrieval in X-ray phase-contrast imaging suitable for tomography. Optics Express 19: 10359–10376. [DOI] [PubMed] [Google Scholar]
- Busse M, Müller M, Kimm MA, et al. . 2018. Three-dimensional virtual histology enabled through cytoplasm-specific X-ray stain for microscopic and nanoscopic computed tomography. Proceedings of the National Academy of Sciences, USA 115: 2293–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer M. 2011. Individualistic and time-varying tree-ring growth to climate sensitivity. PLoS One 6: e22813. doi: 10.1371/journal.pone.0022813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer M, Castagneri D, Prendin AL, Petit G, von Arx G. 2017. Retrospective analysis of wood anatomical traits reveals a recent extension in tree cambial activity in two high-elevation conifers. Frontiers in Plant Science 8: 737. doi: 10.3389/fpls.2017.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuny HE, Rathgeber CB, Frank D, et al. . 2015. Woody biomass production lags stem-girth increase by over one month in coniferous forests. Nature Plants 1: 15160. doi: 10.1038/nplants.2015.160. [DOI] [PubMed] [Google Scholar]
- Davies TG, Rahman IA, Lautenschlager S, et al. . 2017. Open data and digital morphology. Proceedings of the Royal Society B: Biological Sciences 284: 20170194. doi: 10.1098/rspb.2017.0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carlo F, Gürsoy D, Ching DJ, et al. . 2018. TomoBank: a tomographic data repository for computational x-ray science. Measurement Science and Technology 29: 034004. [Google Scholar]
- De Gooijer JG, Hyndman RJ. 2006. 25 years of time series forecasting. International Journal of Forecasting 22: 443–473. [Google Scholar]
- De Groote SR, Vanhellemont M, Baeten L, et al. . 2018. Competition, tree age and size drive the productivity of mixed forests of pedunculate oak, beech and red oak. Forest Ecology and Management 430: 609–617. [Google Scholar]
- De Mil T, Vannoppen A, Beeckman H, Van Acker J, Van den Bulcke J. 2016. A field-to-desktop toolchain for X-ray CT densitometry enables tree ring analysis. Annals of Botany 117: 1187–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mil T, Tarelkin Y, Hahn S, et al. . 2018. Wood density profiles and their corresponding tissue fractions in tropical angiosperm trees. Forests 9: 763. doi: 10.3390/f9120763. [DOI] [Google Scholar]
- De Ridder M, Van den Bulcke J, Vansteenkiste D, et al. . 2010. High-resolution proxies for wood density variations in Terminalia superba. Annals of Botany 107: 293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schryver T. 2017. Fast imaging in non-standard X-ray computed tomography geometries. PhD Thesis, Ghent University. [Google Scholar]
- De Witte Y. 2010. Improved and practically feasible reconstruction methods for high resolution X-ray tomography. PhD Thesis, Ghent University. [Google Scholar]
- Dhaene J. 2017. Development and application of a highly accurate polychromatic X-ray microtomography simulator. PhD Thesis, Ghent University. [Google Scholar]
- Dhaene J, Pauwels E, De Schryver T, De Muynck A, Dierick M, Van Hoorebeke L. 2015. A realistic projection simulator for laboratory based X-ray micro-CT. Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms 342: 170–178. [Google Scholar]
- Dierick M, Van Loo D, Masschaele B, Boone M, Van Hoorebeke L. 2010. A LabVIEW® based generic CT scanner control software platform. Journal of X-ray Science and Technology 18: 451–461. [DOI] [PubMed] [Google Scholar]
- Dierick M, Van Loo D, Masschaele B, et al. . 2014. Recent micro-CT scanner developments at UGCT. Nuclear Instruments and Methods in Physics Research Section B: Beam Interactions with Materials and Atoms 324: 35–40. [Google Scholar]
- Du Plessis A, Broeckhoven C, Guelpa A, Le Roux SG. 2017. Laboratory x-ray micro-computed tomography: a user guideline for biological samples. GigaScience 6: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabijańska A, Danek M. 2018. DeepDendro – a tree rings detector based on a deep convolutional neural network. Computers and Electronics in Agriculture 150: 353–363. [Google Scholar]
- Fonti P, García-González I. 2008. Earlywood vessel size of oak as a potential proxy for spring precipitation in mesic sites. Journal of Biogeography 35: 2249–2257. [Google Scholar]
- García-González I, Souto-Herrero M, Campelo F. 2016. Ring-porosity and earlywood vessels: a review on extracting environmental information through time. IAWA Journal 37: 295–314. [Google Scholar]
- Gärtner H, Nievergelt D. 2010. The core-microtome: a new tool for surface preparation on cores and time series analysis of varying cell parameters. Dendrochronologia 28: 85–92. [Google Scholar]
- Gärtner H, Cherubini P, Fonti P, et al. . 2015. A technical perspective in modern tree-ring research – how to overcome dendroecological and wood anatomical challenges. Journal of Visualized Experiments 97: doi: 10.3791/52337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabner M, Salaberger D, Okochi T. 2009. The need of high resolution μ-X-ray CT in dendrochronology and in wood identification. In: Proceedings of 6th International Symposium on Image and Signal Processing and Analysis, 2009. IEEE, 349–352. [Google Scholar]
- Hevia A, Sánchez-Salguero R, Camarero JJ, et al. . 2018. Towards a better understanding of long-term wood-chemistry variations in old-growth forests: a case study on ancient Pinus uncinata trees from the Pyrenees. Science of the Total Environment 625: 220–232. [DOI] [PubMed] [Google Scholar]
- Jacquin P, Longuetaud F, Leban J.M, Mothe F. 2017. X-ray microdensitometry of wood: a review of existing principles and devices. Dendrochronologia 42: 42–50. [Google Scholar]
- Jacquin P, Mothe F, Longuetaud F, Billard A, Kerfriden B, Leban JM. 2019. CarDen: a software for fast measurement of wood density on increment cores by CT scanning. Computers and Electronics in Agriculture 156: 606–617. [Google Scholar]
- Jaeger H, Haas H. 2004. Harnessing nonlinearity: predicting chaotic systems and saving energy in wireless communication. Science 304: 78–80. [DOI] [PubMed] [Google Scholar]
- Katsevich A. 2002. Theoretically exact filtered backprojection-type inversion algorithm for spiral CT. SIAM Journal on Applied Mathematics 62: 2012–2026. [Google Scholar]
- Laforce B, Masschaele B, Boone MN, et al. . 2017. Integrated three-dimensional microanalysis combining X-ray microtomography and X-ray fluorescence methodologies. Analytical Chemistry 89: 10617–10624. [DOI] [PubMed] [Google Scholar]
- Latham S, Kingston A, Recur B et al. 2018. Reprojection alignment for trajectory perturbation estimation in IEEE Transactions on Computational Imaging 4: 271–283. [Google Scholar]
- LeCun Y, Bengio Y, Hinton G. 2015. Deep learning. Nature 521: 436–444. [DOI] [PubMed] [Google Scholar]
- Liang W, Heinrich I, Helle G, Liñán ID, Heinken T. 2013. Applying CLSM to increment core surfaces for histometric analyses: a novel advance in quantitative wood anatomy. Dendrochronologia 31: 140–145. [Google Scholar]
- Maes SL, Vannoppen A, Altman J, et al. . 2017. Evaluating the robustness of three ring-width measurement methods for growth release reconstruction. Dendrochronologia 46: 67–76. [Google Scholar]
- Masschaele BC, Cnudde V, Dierick M, Jacobs P, Van Hoorebeke L, Vlassenbroeck J. 2007. UGCT: new X-ray radiography and tomography facility. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 580: 266–269. [Google Scholar]
- Masschaele B, Dierick M, Van Loo D, et al. . 2013. HECTOR: a 240kV micro-CT setup optimized for research. Journal of Physics: Conference Series 463: 012012. [Google Scholar]
- Meng B, Pratx G, Xing L. 2011. Ultrafast and scalable cone-beam CT reconstruction using MapReduce in a cloud computing environment. Medical Physics 38: 6603–6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass-Ahles C, Babst F, Klesse S, et al. . 2014. The influence of sampling design on tree-ring-based quantification of forest growth. Global Change Biology 20: 2867–2685. [DOI] [PubMed] [Google Scholar]
- Okochi T, Hoshino Y, Fujii H, Mitsutani T. 2007. Nondestructive tree-ring measurements for Japanese oak and Japanese beech using micro-focus X-ray computed tomography. Dendrochronologia 24: 155–164. [Google Scholar]
- Onoe M, Tsao JW, Yamada H, et al. . 1984. Computed tomography for measuring the annual rings of a live tree. Nuclear Instruments and Methods in Physics Research 221: 213–220. [Google Scholar]
- Paganin D, Mayo SC, Gureyev TE, Miller PR, Wilkins SW. 2002. Simultaneous phase and amplitude extraction from a single defocused image of a homogeneous object. Journal of Microscopy 206: 33–40. [DOI] [PubMed] [Google Scholar]
- Pauwels E, Van Loo D, Cornillie P, Brabant L, d Van Hoorebeke L. 2013. An exploratory study of contrast agents for soft tissue visualization by means of high resolution X-ray computed tomography imaging. Journal of Microscopy 250: 21–31. [DOI] [PubMed] [Google Scholar]
- Preibisch S, Saalfeld S, Tomancak P. 2009. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25: 1463–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe TB, Luo ZX, Ketcham RA, Maisano JA, Colbert MW. 2016. X-ray computed tomography datasets for forensic analysis of vertebrate fossils. Scientific Data 3: 160040. doi: 10.1038/sdata.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saß U, Eckstein D. 1994. Preparation of large thin sections and surfaces of wood for automatic image analysis. Holzforschung 48: 117–118. [Google Scholar]
- Scharnweber T, Hevia A, Buras A, van der Maaten E, Wilmking M. 2016. Common trends in elements? Within- and between-tree variations of wood-chemistry measured by X-ray fluorescence – a dendrochemical study. Science of the Total Environment 566: 1245–1253. [DOI] [PubMed] [Google Scholar]
- Scheiter S, Langan L, Higgins SI. 2013. Next-generation dynamic global vegetation models: learning from community ecology. New Phytologist 198: 957–969. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nature Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldt B, Leuschner C, Brock N, Horna V. 2013. Changes in wood density, wood anatomy and hydraulic properties of the xylem along the root-to-shoot flow path in tropical rainforest trees. Tree Physiology 33: 161–174. [DOI] [PubMed] [Google Scholar]
- Schweingruber FH, Fritts HC, Bräker OU, Drew LG, Schär E. 1978. The X-ray technique as applied to dendroclimatology. Tree-Ring Bulletin 38. [Google Scholar]
- Staedler YM, Masson D, Schönenberger J. 2013. Plant tissues in 3D via X-ray tomography: simple contrasting methods allow high resolution imaging. PLoS One 8: e75295. doi: 10.1371/journal.pone.0075295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffenrem A, Kvaalen H, Dalen KS, Høibø OA. 2014. A high-throughput X-ray-based method for measurements of relative wood density from unprepared increment cores from Picea abies. Scandinavian Journal of Forest Research 29: 506–514. [Google Scholar]
- Stelzner J, Million S. 2015. X-ray computed tomography for the anatomical and dendrochronological analysis of archaeological wood. Journal of Archaeological Science 55: 188–196. [Google Scholar]
- Van den Bulcke J, Wernersson EL, Dierick M, et al. . 2014. 3D tree-ring analysis using helical X-ray tomography. Dendrochronologia 32: 39–46. [Google Scholar]
- Van Loo D, Bouckaert L, Leroux O, et al. . 2014. Contrast agents for soil investigation with X-ray computed tomography. Geoderma 213: 485–491. [Google Scholar]
- Vannoppen A, Maes S, Kint V, et al. . 2017. Using X-ray CT based tree-ring width data for tree growth trend analysis. Dendrochronologia 44: 66–75. [Google Scholar]
- Vannoppen A, Boeckx P, De Mil T, et al. . 2018. Climate driven trends in tree biomass increment show asynchronous dependence on tree-ring width and wood density variation. Dendrochronologia 48: 40–51. [Google Scholar]
- Vlassenbroeck J, Dierick M, Masschaele B, Cnudde V, Van Hoorebeke L, Jacobs P. 2007. Software tools for quantification of X-ray microtomography at the UGCT. Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 580: 442–445. [Google Scholar]
- Voyant C, Notton G, Kalogirou S, et al. . 2017. Machine learning methods for solar radiation forecasting: a review. Renewable Energy 105: 569–582. [Google Scholar]
- Wong J, D’Sa D, Foley M, Chan JGY, Chan HK. 2014. NanoXCT: a novel technique to probe the internal architecture of pharmaceutical particles. Pharmaceutical Research 31: 3085–3094. [DOI] [PubMed] [Google Scholar]
- wyffels F, Schrauwen B. 2010. A comparative study of reservoir computing strategies for monthly time series prediction. Neurocomputing 73: 1958–1964. [Google Scholar]
- Zuidema PA, Poulter B, Frank DC. 2018. A wood biology agenda to support global vegetation modelling. Trends in Plant Science 23: 1006–1015. [DOI] [PubMed] [Google Scholar]