Abstract
Forward genetic screens have successfully identified many genes and continue to be powerful tools for dissecting biological processes in Arabidopsis and other model species. Next-generation sequencing technologies have revolutionized the time-consuming process of identifying the mutations that cause a phenotype of interest. However, due to the cost of such mapping-by-sequencing experiments, special attention should be paid to experimental design and technical decisions so that the read data allows to map the desired mutation. Here, we simulated different mapping-by-sequencing scenarios. We first evaluated which short-read technology was best suited for analyzing gene-rich genomic regions in Arabidopsis and determined the minimum sequencing depth required to confidently call single nucleotide variants. We also designed ways to discriminate mutagenesis-induced mutations from background Single Nucleotide Polymorphisms in mutants isolated in Arabidopsis non-reference lines. In addition, we simulated bulked segregant mapping populations for identifying point mutations and monitored how the size of the mapping population and the sequencing depth affect mapping precision. Finally, we provide the computational basis of a protocol that we already used to map T-DNA insertions with paired-end Illumina-like reads, using very low sequencing depths and pooling several mutants together; this approach can also be used with single-end reads as well as to map any other insertional mutagen. All these simulations proved useful for designing experiments that allowed us to map several mutations in Arabidopsis.
INTRODUCTION
Whole-genome massive sequencing (WGS) has opened a new pathway for mutation mapping in organisms both with and without sequenced genomes (1). Most types of naturally occurring and induced mutations have been successfully mapped using WGS: Single Nucleotide Polymorphisms (SNPs), small insertions and deletions (indels), insertional elements and structural mutations (2–5). However, WGS experiments are relatively expensive and require considerable effort to obtain the samples required, to create and sequence libraries, and to analyze the data produced to obtain meaningful information. Therefore, optimizing the experimental design is highly important for obtaining the desired biological information on the first attempt. This process could involve reviewing the literature for an equivalent experimental design or performing pilot experiments (6). However, the former option depends on availability, while the latter requires an extra investment.
A third option is simulating the experiment in silico (7,8). For example, a computer text file representing a mutated genome can be created and WGS reads can be simulated and analyzed in the same manner as real reads to predict, for instance, whether a certain read depth (RD) will be suitable for a particular purpose. With this information in hand, the actual experiment can be performed more efficiently. Other customizable parameters that can be simulated include the mutagenesis density (point and insertional mutations), type of sequencing library (single-end versus paired-end reads), insert size in paired-end libraries, read length, base calling error rate, RD distribution genome-wide, and so on (9). Furthermore, rather difficult questions might be raised during the experimental design using particular mapping strategies: for example, the accuracy of mapping ethyl methanesulfonate (EMS)-induced mutations via bulk F2 segregant analysis largely depends on the size of the F2 mapping population, as well as the RD used (10,11). A researcher can perform multiple rounds of a simulation experiment, progressively modify the values of different parameters as desired, and analyze the outcome.
Simulations can also be used to validate an analysis workflow (12). For instance, a FASTA genome with a transposon inserted in a known position can be created and massively sequenced in silico, and the resulting reads can be used as a substrate to compare the ability of different analysis workflows to determine the insertion position (the so-called known-truth approach; 13). This approach expands the usefulness of simulated data to test if a certain experimental design will be suitable for answering a research question. Finally, benchmarking of computational tools such as variant-calling software also depends on the use of simulated data (14–16).
The rapid development of novel software tools has increasingly facilitated the simulation of Next Generation Sequencing (NGS) experiments (reviewed in 17). Simulated experiments performed to date include those simulating DNA structural variation (18), RNA-sequencing (RNA-seq) differential-expression studies (19,20), bisulfite sequencing (21), studies based on tumor sequencing data (22) and data from Quantitative Trait Loci (QTL) analysis (23), sequencing of heterogeneous populations (7), and de novo genome assembly (8). A combination of simulations and pilot experiments can also be performed. Such a two-step approach has been successfully used to improve the power of RNA-seq experiments for assessing differential gene expression (24–26).
The motivation for performing the current study emerged from questions that arose daily in our laboratory while designing WGS experiments to identify mutations induced in wild-type genetic backgrounds of the model plant Arabidopsis thaliana (hereafter, Arabidopsis). Here, we describe how we used simulations to aid in the design of actual WGS experiments, in which we successfully identified several genes causal for phenotypes of interest, which carried mutations induced by EMS. We also describe simulations for mutant genomes carrying multiple insertional mutations.
MATERIALS AND METHODS
Simulations
To simulate mutant genomes, FASTA files containing reference Arabidopsis sequences (27,28) were obtained from the National Center for Biotechnology Information (NCBI) and modified by making single base changes or by inserting long sequences at random positions. The density of natural SNPs used was that observed between the Arabidopsis accessions Columbia-0 (Col-0) and Landsberg erecta (Ler). The density of EMS-induced SNPs used was 4–14 per Mb of reference sequence, as described previously (29,30). To simulate meiotic recombination and artificial selection, two FASTA files representing homologous chromosomes were used as input to generate sets of recombinant sequences. Crossover frequency distribution in Arabidopsis was obtained from Salomé et al. (31), and crossover positions were randomly distributed. Short reads were simulated by taking substrings at random positions from the input sequence, and GC content bias in libraries was created by counter-selecting reads with a probability proportional to their distance from neutral GC content and a strength parameter ranging from 0 to 1, as in the following formula: strength × 2 × (|GC% – 50|). Library fragment sizes in paired-end libraries and read lengths in Ion Proton-like libraries were assumed to follow a normal distribution. The base calling error frequency used was 0–1% (32,33). In all scripts, random numbers were simulated using the Mersenne Twister algorithm (34). These procedures were implemented in Python2 and are available under the GPL-3.0 license (Scripts S1–S3). The arguments required by the programs and the values used in all simulations are detailed in Supplementary Table S1.
Plant materials and preparation of DNA for sequencing
Seeds of the wild-type accessions Col-0 and Ler of Arabidopsis thaliana L. Heynh. were first obtained from the Nottingham Arabidopsis Stock Center (NASC) and then propagated by self pollination in our laboratory. The EMS-induced mutants angulata1-1 (anu1-1), anu12-1, denticulata3 (den3), den6-1, ondulata4 (ond4) and scabra1-1 (sca1-1), as well as 19 other mutants derived from the same mutagenesis used to create background SNP masks, belong to the Micol collection of leaf mutants, which were obtained previously in our laboratory in the Ler background (35). The T-DNA mutants were obtained from the SALK collection (36), and the specific lines were previously analyzed and described in Wilson-Sánchez et al. (37). Isolation of the ago1-25 mutant (EMS; Col-0) was described in Morel et al. (38). Plants were grown and crossed as described previously (37). To create mapping populations, the mutants were either backcrossed or pseudo-backcrossed (see Results) and the F2 progeny of the selfing of a single F1 plant was used. Leaf tissue from F2 individuals exhibiting the phenotype of interest was combined in approximately equal amounts (by weight) to obtain approximately equimolar DNA populations. Total genomic DNA was purified using Mini or Midi DNA Plant Kits (Qiagen, Venlo, The Netherlands). DNA concentration was determined with a Qubit dsDNA HS Assay Kit in a Qubit 2.0 fluorometer (Thermo Fisher Scientific, Waltham, MA, USA), and its integrity was confirmed by gel electrophoresis.
Library preparation and sequencing
Short read data were generated in-house with an Ion Proton sequencer or externally on the HiSeq2000 and HiSeq2500 platforms at BGI (Beijing Genomics Institute, Hong Kong) and STAB-VIDA (Caparica, Portugal), respectively. For the samples sequenced with the Ion Proton, all devices and reagents were obtained from Life Technologies (now Thermo Fisher Scientific). DNA libraries were prepared with Ion Shear Plus and Ion Xpress Plus kits, amplified via five PCR cycles, and assessed for quality with a Bioanalyzer 2100 using a DNA High Sensitivity Chip (Agilent Technologies, Santa Clara, CA, USA). Sequencing templates were created with an Ion OneTouch and an Ion ES instrument using an Ion PI OT2 200 Kit v3 and analyzed with a Qubit 2.0 fluorometer using an Ion Sphere Quality Control Kit. Sequencing was performed with an Ion PI 200 Kit v3 in an Ion PI Chip v2.
Data analysis
DNA sequence complexity was measured using SeqComplex (http://caballero.github.io/SeqComplex/). Read data in FASTQ format obtained from simulations and from the HiSeq and Ion Proton sequencers were analyzed identically (Supplementary Figure S1). Assessment of raw read quality was performed with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Nucleotide calls at the 3′ ends of reads with Phred scores < 15 were trimmed.
Alignments were performed with Bowtie2 (39) or BWA-MEM (40), most often using default parameters for single- and paired-end reads. Analyses and routine tasks with SAM/BAM alignments were performed with tools from the SAMtools (41), BAMtools (42), BEDtools (43), and QualiMap (44) suites. SNP calling was performed with GATK Unified Genotyper (45) or SAMtools mpileup and BCFtools call (41,46). SNP datasets were filtered by the QUAL field (47) according to the needs of each experiment. To create the masks of natural SNPs (Supplementary Tables S2–S4), the VCF files from the individual mutants were filtered on RD (≥7) and alternative allele frequency (≥0.4) and merged using custom scripts developed for Easymap, a mapping-by-sequencing program for which a manuscript is in preparation. Finally, variants were filtered according to the number of samples in which they appeared.
Availability of read and variant data
The short read data from some samples analyzed in this work are available from the NCBI Sequence Read Archive database (http://www.ncbi.nlm.nih.gov/sra) or from the authors upon request: the anu1-1 and anu12-1 mutants (accession SRP043639; 48); den3 mutant (upon request); sca1-1 mutant (SRP050297; 49); two libraries, one created from the pooled DNA of 100 phenotypically mutant plants of the F2 progeny of an ago1-25 × Col-0 cross (SRX3510107) and the other from the Col-0 parental line (SRX3510106); a library created from the pooled DNA of 109 double mutant plants selected from the F2 progeny of a den6-1 × ond4 cross (SRX3510108); and two libraries from pools of five SALK T-DNA mutant lines (SRX473258; 37).
The short read data from the mutants used to create SNP masks (35) were not deposited at NCBI. However, the consensus lists of Col-0-Ler SNPs that were created are shown in Supplementary Tables S2–S4.
RESULTS
The first decision in WGS: choosing a sequencing platform and RD for SNP analysis
During the course of a forward genetics project to study leaf development, we wanted to use WGS to identify the causal genes for several Arabidopsis leaf-shape mutants isolated in a forward genetics screen of a population derived from EMS mutagenesis. Two sequencing platforms were available to us: HiSeq2000 (Illumina) and Ion Proton (Life Technologies, now Thermo Fisher Scientific), which yield ∼100-bp paired-end reads and ∼200-bp single-end reads, respectively. Using a custom Python script (Script S1 and Supplementary Figure S1), we simulated reads from these two platforms (Simulation 1, in Supplementary Table S1) and tested the ability of two common aligner programs, Bowtie2 (39) and BWA-MEM (40), to align the reads to a reference sequence. We used a 5-Mb sequence from gene-rich regions of the Arabidopsis Col-0 accession genome (complexity ∼10; 50) as a template reference sequence, as such regions are the usual targets of gene cloning experiments, and repetitive low-complexity regions reduce the performance of the aligners (51). The percentage of unaligned reads from the two platforms was similar regardless of the aligner used (Table 1). The number of reads that could not be mapped unambiguously was also comparable. Since we concluded that these two types of reads are equally suitable for analyzing gene-rich regions from Arabidopsis, we considered both sequencing platforms in subsequent cloning experiments.
Table 1.
Percentage of simulated HiSeq2000-like and Ion proton-like reads aligned
| Sequencing platform simulated | Aligner | Reads | ||
|---|---|---|---|---|
| Not aligned | Aligned once | Aligned more than once | ||
| HiSeq2000-likea | Bowtie2 | 0.01 | 96.65 | 3.33 |
| BWA-MEM | 0.50 | 98.08 | 1.41 | |
| Ion Proton-likeb | Bowtie2 | 0.01 | 97.03 | 2.96 |
| BWA-MEM | 0.60 | 97.79 | 1.62 | |
aFixed-length (100 nt), paired-end library.
bVariable length (200 nt average), single-end library.
Next, we wanted to know how mean RD affects coverage (CN), i.e. the percentage of the template sequence with RD ≥ N (52); the latter condition is critical for reliable genome-wide base calling. A chosen mean RD cannot ensure that all bases are read at that depth (Supplementary Figure S2), as fragment libraries are largely random, and several biases commonly arise during library preparation (such as the preference for DNA fragments with neutral GC content during library PCR amplification) (53–56), which in turn further increases the RD variance of a given sample. We simulated libraries with increasing degrees of GC bias strength (Simulation 2, in Supplementary Table S1) and found that a 100% strength yielded an RD distribution that closely matched that of real Illumina libraries (Supplementary Figure S2). Consequently, in all subsequent simulations involving RD as a variable, we used a 100% GC bias strength.
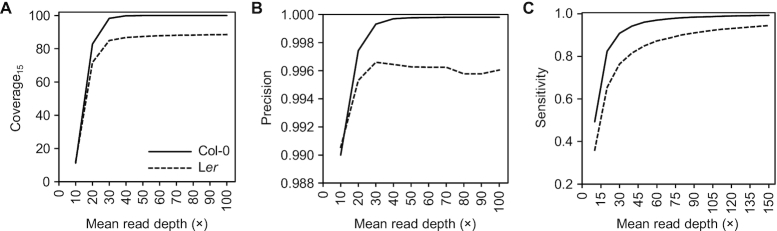
We then simulated several datasets with increasing mean RD (10× to 100×) and analyzed the resulting C15, a threshold that we established arbitrarily, because RD ≥ 15 is expected to allow confident base calling and variant analysis (Simulation 3, in Supplementary Table S1). Increasing the mean RD also increased C15 (solid line in Figure 1A). When using ∼40× mean RD, >99.7% of the template sequence passed the C15 threshold, and further reduction of the number of under-read nucleotides required a substantial increase in RD and costs. To test how RD would affect C15 in a non-reference genome, we repeated this experiment with reads simulated from Ler (28) and aligned to Col-0. Saturation was observed at ∼40–50× RD (dashed line in Figure 1A), although the maximum was at 88.6%, likely due to large chromosomic stretches present only in Col-0.
Figure 1.
Effect of read depth on variant calling in gene-rich regions in Arabidopsis. (A) Effect of mean read depth (RD) on template coverage (C15, percentage of template sequence read with RD ≥ 15×). (B, C) Effect of mean RD on the (B) precision and (C) sensitivity of the analysis. Precision is calculated as TP/(TP + FP), and sensitivity as TP/(TP + FN). TP: True positive SNPs. FP: False positive SNPs. FN: False negative SNPs.
Accurate base determination and variant detection also require a minimum RD to minimize the impact of errors generated during library preparation, sequencer base calling errors (0.1–1% for different sequencing platforms), or reads misaligned to the reference sequence (32,33), which when combined interfere with the statistical models that variant callers employ to determine DNA bases (57,58). First, we sought to determine the minimum RD needed to call SNPs systematically with a standard variant calling workflow. We simulated a 10 Mb high-complexity template from a Col-0 gene-rich region and a polymorphic version with 10 000 (0.1%) random SNPs (Script S2 and Supplementary Figure S1). Next, we simulated read datasets from 10× to 150× mean RD and a 1% base calling error rate (Simulation 4, in Supplementary Table S1), and investigated how RD affects the precision and sensitivity of SNP calling (14) using Samtools with default parameters (41). Precision (fraction of reported SNPs that are not false positives) and sensitivity (fraction of SNPs in the sample that are detected) showed an asymptotic response to RD increase, reaching their maximum (∼100% and ∼99%, respectively) at ∼40× and ∼80× RD, respectively (solid lines in Figure 1B, C). To study how RD affects SNP calling in a non-reference genome, we repeated the previous simulation but generated reads from a 10-Mb high-complexity Ler template with 0.1% random SNPs (28) and aligned them to Col-0. Natural variants were subtracted using both experimental and simulated control data (Supplementary Table S2, 600× RD Ler reads aligned to Col-0). Precision increased with RD up to 30× and then did not respond further. At any given RD, it was always lower than in the reference genome, due to false positives derived from misalignments in divergent regions. Sensitivity again showed an asymptotic pattern but approached the maximum later than in the reference genome, again likely due to the higher rate of misalignments and unaligned reads.
False negatives, which reduce sensitivity, are mainly due to the presence of stretches without aligned reads. However, shallowly read stretches coupled with sequencing or alignment errors can cause both false negatives and false positives, which reduce precision. This issue can be mitigated by filtering out low-quality SNPs, but at the expense of missing some true positives (13). Our results show that increasing the mean RD is effective to reduce both false positives and false negatives in gene-rich regions of Arabidopsis. Based on the results of C15 analysis (Figure 1A) and SNP calling (Figure 1B, C), we decided that 40–50× RD provided the best compromise between cost and accuracy for our WGS genetic analyses in gene-rich genomic regions in Arabidopsis.
Mapping EMS-induced mutations in non-reference backgrounds with prior linkage analysis information
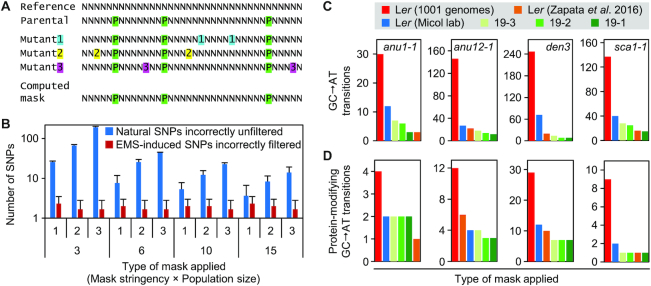
We previously isolated 255 Arabidopsis mutants after EMS mutagenesis (35). Linkage analysis using molecular markers allowed us to delimit candidate intervals for the mutations that cause the phenotypes of these mutants (59). Since these mutants are in the Ler genetic background, their genome sequences have hundreds of thousands of SNPs with the reference accession Col-0 (60, and our unpublished data), making it difficult to discriminate between natural and EMS-derived SNPs. A Ler genome sequence is available (28), but whether its quality is high enough for reliable SNP calling has not been tested. Moreover, wild-type lines used in any laboratory harbor up to a few thousand SNPs compared to other lines considered to be the same accession (61–65). Therefore, the parental line of the mutants should always be sequenced to create an SNP mask. However, it is difficult to capture all SNPs in a single sequencing run (Figure 1C; 13,57). Alternatively, if several mutants obtained from the same parental line must be analyzed, their SNPs can be combined to create more comprehensive multi-sample lists of natural SNPs. These datasets can subsequently be used as SNP masks to analyze individual mutants (Figure 2A; 48,66,67).
Figure 2.
SNP calling in non-reference genomes. (A) Procedure used to create a natural (parental) SNP mask by integrating the data from different mutants obtained from the same mutagenesis (see also Supplementary Figure S3). N indicates any nucleotide. Nucleotides represented by a P shaded in green are SNPs between the reference sequence (Reference) and the sequence of the line that was subjected to mutagenesis (Parental). Nucleotides shown as 1, 2 and 3 (shaded in blue, yellow and magenta, respectively) are specific of each mutant line and are absent from the genomes of other mutants or the reference and parental lines. The computed mask only includes the SNPs shaded in green. (B) Effect of masking natural SNPs with different population sizes (X-axis, bottom) and SNP selection stringencies (minimum number of samples in which the SNPs were detected; X-axis, top) (n = 3). Error bars indicate standard deviation. (C, D) Number of remaining SNPs in the candidate intervals of the anu1-1 (0.216 Mb), anu12-1 (0.754 Mb), den3 (2.539 Mb) and sca1-1 (0.760 Mb) mutants after filtering with different single-sample (red and blue) and multi-sample (green) SNP masks, or direct alignment of the reads to the Ler genome assembly (orange) (28). The Ler (Micol lab) data correspond to the parental line of the mutants analyzed. The multi-sample masks were constructed from 19 sibling mutants and include all SNPs present in at least 3, 2 or 1 samples. (C) Number of all remaining GC→AT transitions. (D) Number of remaining protein-modifying GC→AT transitions.
To assess how this approach could be applied to our mutants in the Ler background, we simulated mutant populations (Supplementary Figure S1) with an increasing number of individuals to create different SNP masks (masking populations), as well as three additional mutants representing problem individuals to be analyzed. Each mutant consisted of a 5-Mb gene-rich sequence carrying 50 000 parental SNPs (common to all three samples, introduced to simulate natural SNPs between the reference accession and the mutated lines) and 60 GA→CT random transitions (unique to each sample, to simulate EMS-induced SNPs; 68,69) (Simulation 5, in Supplementary Table S1). We simulated reads with 40× RD and called SNPs. We then created SNP masks using masking populations of varying sizes and stringencies (i.e. the minimum number of mutants in the masking population where a given SNP has to be detected in order to include it in the mask) (Supplementary Figure S3; 67). Finally, we used these masks to filter the SNPs of the three simulated problem individuals and to evaluate their performance.
As shown in Figure 2B, we observed contaminating natural SNPs, but increasing the size of the masking population reduced its number. On the other hand, increasing the population size did not significantly increase the number of incorrectly filtered EMS-induced SNPs. For each population size, we created masks of three different stringency levels (Supplementary Figure S3; 67). The least stringent masks (those that included all SNPs detected in at least one individual of the masking population) were best able to filter out contaminating natural SNPs but did not significantly increase the number of incorrectly filtered EMS-induced SNPs (Figure 2B). Therefore, the larger the masking population and the less stringent the SNP selection criteria, the greater the ability of the resulting SNP mask to filter out natural SNPs; at the same time, such a mask will very rarely filter out an EMS-induced SNP.
Using the previous results as a benchmark, we set out to create an SNP mask between our current Ler laboratory line and Col-0 (TAIR10). We used WGS data from 19 sibling non-allelic mutants derived from the same mutagenesis (35), created SNP masks with low stringency (SNPs detected in ≥1, ≥2 or ≥3 of the 19 samples; Supplementary Tables S2–S4), and applied the masks to four mutants that we previously isolated from the same mutagenesis (48,49). We compared our simulation-derived approach (multi-sample masks) to simply subtracting SNPs from the Ler 1001 genome data (70) or from a single sequencing run of the Ler parental line (single-sample masks). We also compared the masks with direct alignment to the Ler assembly by Zapata et al. (28). As shown in Figure 2C, D, multi-sample masks outperformed single-sample masks for filtering out natural SNPs, and they did not incorrectly filter out the causal mutations. Alignment to the Ler assembly (28) gave comparable results (Figure 2C, D; Table 2). However, for the vast majority of non-Col-0 accessions no assembly information is available. Overall, our results suggest that multi-sample masking is a powerful, background-independent approach for analyzing mutants in all non-reference backgrounds. The masks developed in this experiment are currently being used to filter out natural SNPs in mutants whose causal genes are in the process of being identified.
Table 2.
Number of reliable SNPs in chromosome 2 called using the multi-mask approach and the Ler genome assembly of Zapata et al. (2016)
| Mutant | Multi-sample masking (19–1 mask) | Alignments to the Ler assembly of Zapata et al. (2016) |
|---|---|---|
| anu1-1 | 254 | 233 |
| anu12-1 | 72 | 189 |
| den3 | 118 | 121 |
| sca1-1 | 223 | 207 |
| Total | 667 | 750 |
Reliable SNPs are those with an allele frequency >0.75 in a high-complexity genomic region: Col-0 chromosome 2 segment ranging from 8 000 000 to 18 585 056 bp in TAIR10, which corresponds to 7 348 758 to 19 037 554 bp in the Ler assembly of Zapata et al. (28).
Backcross mapping-by-sequencing to detect recessive EMS-induced mutations without prior linkage analysis information
In recent years, we have had to identify the genes underlying several novel Arabidopsis mutants for which there was no previous knowledge of the position of the causal mutation. WGS can replace the time-consuming process of map-based cloning (linkage analysis coupled to candidate gene sequencing) when applied to F2 populations derived from a cross between a mutant and a polymorphic line (mapping-by-sequencing; 71–74). In addition, a single backcross and the use of EMS-induced SNPs as mapping markers has been successfully employed to map causal mutations in plants (10,62,75–79). This option makes it easier to detect the SNP causing the phenotype of interest (the causal mutation), since the number of natural SNPs is typically reduced from several hundred thousand to 500–1500 (30,80).
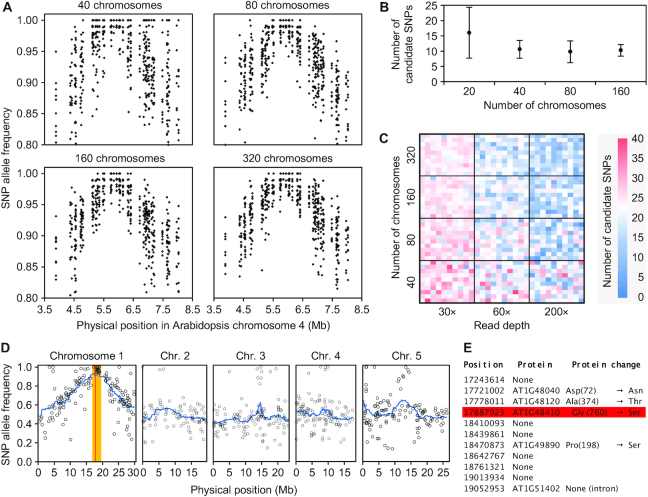
We decided to use backcross mapping-by-sequencing to accelerate mutation identification. First, to assess how the size of the F2 mapping population affects mapping precision, we simulated a version of Arabidopsis chromosome 4 with 109 mutations representing the natural divergence between the Col-0 reference sequence and a laboratory-grown Col-0 line (61–65), together with 232 EMS GC→AT transitions. We then obtained sets of 40, 80, 160 and 320 backcross F2 recombinant chromosomes (10 replicates) artificially selected based on their harboring a single transition (at position 5 845 220 bp; Script S3 and Supplementary Figure S1). We then simulated 100× RD reads from each set, called SNP variants, and characterized the candidate regions obtained (Simulation 6, in Supplementary Table S1). A plot of the variant allele frequencies (AFs) against their position on the chromosome forms a concave curve, with its maximum near the position of the causal mutation (Figure 3A). The number of candidate mutations (arbitrarily established as all SNPs with an AF ≥ 0.98) decreased when we increased the number of chromosomes from 40 to 80 but remained constant when we further increased the number of chromosomes from 160 and 320 (Figure 3A, B). This result suggests that if the number of reads at a given locus (RD) is lower than the number of recombinant chromosomes (i.e. the mapping population size; MPS), read sampling acts as a limiting factor when assessing the allelic frequencies in the population.
Figure 3.
Mapping-by-sequencing EMS-induced mutations using backcross-derived F2 populations. (A) Variation of SNP allele frequency around the causal mutation after simulating 100× RD reads from mapping populations of different sizes. Each SNP is represented by 10 dots, each corresponding to an experimental replicate. (B) Effect of the number of chromosomes analyzed on the number of candidate SNPs (with an allele frequency ≥ 0.98). Data were computed from 10 simulated replicates of each population size. Error bars indicate standard deviation. (C) Combined effect of mapping population size and sequencing read depth on the number of candidate SNPs. Each heatmap unit represents a single simulated experiment. (D, E) Identification of the ago1-25 mutation using mapping-by-sequencing. (D) Positions and allele frequencies of the SNPs detected in the mapping population. The candidate interval is shown in orange; it is defined based on a cluster of SNPs with allele frequencies ≥0.95. The red line indicates the position of the ago1-25 mutation. (E) Effect of the GC→AT substitutions found in the candidate interval, including the ago1-25 mutation (red square).
To test this hypothesis, we simulated different combinations of MPS × RD in sets of 100 replicates (Simulation 7, in Supplementary Table S1). As shown in Figure 3C, increasing the MPS from 40 to 320 chromosomes while using a low RD (30×) throughout had a significant but mild effect on the number of candidate mutations (24% reduction; P = 0.0001 in a Student's t-test). Similarly, keeping the MPS low (40 chromosomes) and increasing RD had a limited impact on the number of candidate mutations, although this number was greater than in the previous case (37% reduction, P = 0.0001). The greatest effect was observed when we simultaneously increased MPS and RD (63% reduction, P = 0.0001). These results suggest that both parameters can act as bottlenecks in a mapping-by-sequencing experiment, as indicated in previous studies (11). Visual inspection of the results, which is often how mapping-by-sequencing results are analyzed, revealed that higher MPS renders a smoother AF curve, while higher RD reduces the noise in AF sampling (Supplementary Figure S4). The availability of smooth, low noise curves makes it easier to identify the candidate mutations.
Achieving RDs such as 200× in genomes of hundreds of Mb requires large amounts of short read data, which are normally not acceptable or available for a cloning experiment. Therefore, RD will be the limiting parameter in mapping-by-sequencing experiments in which it is easy to obtain large F2 populations. Still, lower RDs can be sufficient if SNP density is not very high. To confirm this notion, we backcrossed the EMS-induced argonaute1-25 (ago1-25) mutant, which was isolated in the Col-0 genetic background and has a characteristic recessive morphological phenotype (38), and pooled the DNA from 100 phenotypically mutant F2 plants (200 chromosomes). We sequenced at 60× RD and determined the positions and AF of all SNPs detected. We observed clear linkage of the phenotype to a narrow region in chromosome 1 (Figure 3D), which contained only 11 SNPs with AF ≥ 0.95. Five of these SNPs altered protein sequences, including a C→T transition at position 17 887 923 bp (Gly760→Ser), which is known to be responsible for the mutant phenotype of ago1-25 (Figure 3E). We used these simulations and a real-data experiment as the foundation for other mapping-by-sequencing projects.
M2 populations derived from a single M1 individual have also been used to map causal mutations in EMS-induced Arabidopsis mutants (78). An M2 population contains twice as many EMS mutations as an F2 population derived from a mutant that has been selfed several times (Supplementary Figure S5A). Only half of these mutations within the candidate interval will cosegregate with the causal mutation (Supplementary Figure S5A). To compare the outcome of M2 vs. backcross-F2 mapping-by-sequencing experiments, we simulated both corresponding populations (Simulation 8, in Supplementary Table S1) and counted the number of candidate mutations in the candidate intervals We found no significant differences in the number of candidate mutations obtained using these two approaches (Supplementary Figure S5B, C), suggesting that they reach similar mapping accuracy. However, while M2 mapping populations can be obtained faster, F2 mapping populations can still be preferred to perform complementation analysis prior to mapping-by-sequencing.
Pseudo-backcrossing coupled to high RD sequencing
To reduce the sizes of candidate intervals without increasing the cost per mutant, we designed a novel approach that we termed pseudo-backcrossing coupled to high RD sequencing: first, two mutants exhibiting different, monogenic phenotypes caused by recessive mutations in non-linked loci, both arising from the same mutagenesis, are crossed to obtain the F2 progeny; a mapping population of double mutants isolated because of their additive phenotypes is then constructed, and genomic DNA from these double mutants is sequenced at a high RD (Figure 4A). The advantage of this technique is that the higher RD allows for narrower candidate intervals compared to the conventional technique (Figure 3C). The cost per mutant does not increase because only one sequencing library has to be generated (at a total cost of only a few hundred dollars), and the increased number of reads analyzed, although more expensive, should be useful to identify the underlying mutation for two separate mutants. These approaches will only be useful for mutations affecting unlinked genes and causing additive and clearly distinguishable phenotypes.
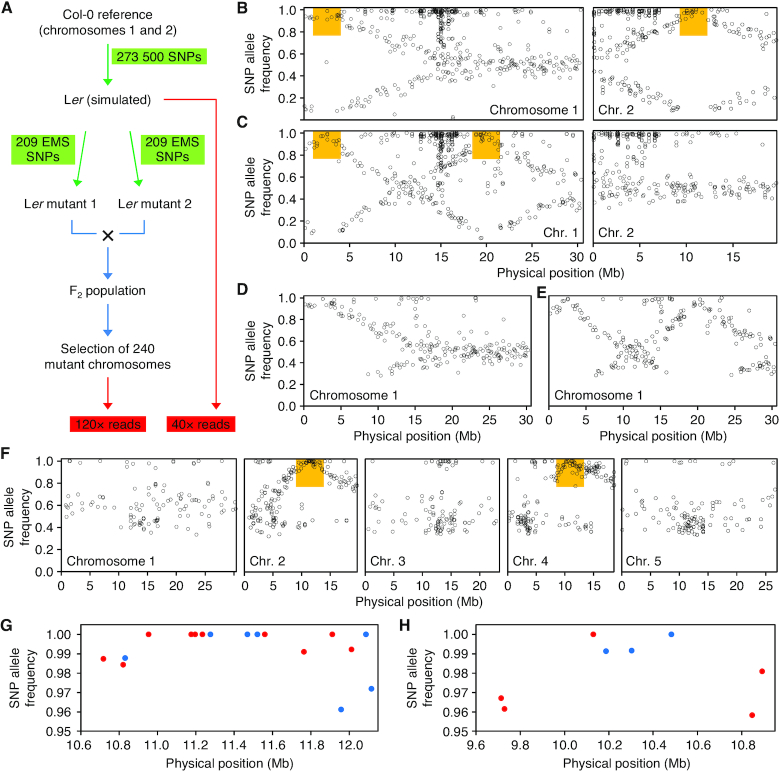
Figure 4.
Mapping-by-sequencing EMS-induced mutations using pseudo-backcrossing-derived F2 populations. (A) Overview of reads simulation from a pseudo-backcross. Green arrows and labels indicate mutagenesis. Blue arrows denote recombination and selection. Red arrows and labels indicate massive sequencing. (B–E) Simultaneous identification of two candidate intervals in simulated pseudo-backcross-derived F2 mapping populations. (B) Position and allele frequency of the SNPs detected in a simulated mapping population derived from a cross of two mutants carrying (B) unlinked and (C) linked recessive mutations. Orange boxes indicate the locations of the candidate SNPs. (D, E) Overview of chromosome 1 showing only SNPs with a QUAL value > 200. Note that the application of this filter to remove background SNPs results in the unintentional removal of variants with low allele frequency. (F–H) Simultaneous identification of two candidate intervals in an F2 mapping population derived from a ond4 × den6-1 cross. (F) Genome-wide positions and allele frequencies of the SNPs detected in the mapping population after SNP quality filtering. The orange boxes indicate the locations of the candidate SNPs. (G, H) Positions and allele frequencies of the candidate SNPs on chromosomes (G) 2 and (H) 4. Blue dots: protein-modifying SNPs. Red dots: non-protein modifying SNPs.
To determine whether the use of a pseudo-backcross-derived F2 mapping population would allow us to simultaneously map two of our EMS-induced mutants in the Ler background, we performed the following simulation: we generated 273 000 random SNPs on Col-0 chromosomes 1 and 2 to simulate the Ler strain, which we then used as a template to create two sibling mutants, each with 209 random EMS-type SNPs; we then generated 240 F2 double-mutant genomes for positions chr1:2 500 000 and chr2:11 000 000; finally, we simulated 40× RD reads from the Ler parental line and high-depth reads (120×) for the F2 population (Simulation 9, in Supplementary Table S1). Following read alignment and variant calling, we subtracted the SNPs detected in the parental line from the F2 SNP list. As shown in Figure 4B, the AFs of the SNPs from each simulated mutant described complementary traces along the chromosomes and diverged around the positions of the two causal mutations, allowing us to distinguish two candidate intervals at the expected positions. This experiment was repeated 10 times, yielding comparable results. The detection of a significant number of SNPs with AF near 1 is due to the presence of unfiltered background SNPs and to sequencing and alignment errors in the F2 population.
Next, we investigated whether this approach can also be used to define candidate regions for partially linked mutations. We simulated F2 double-mutant genomes selected for positions chr1:2 500 000 and chr1:20 000 000, which rendered only 1/24 (<1/16) double mutants. Again, visualizing the AF of the SNPs present in the double mutant population allowed us to determine the locations of the two candidate regions (Figure 4C). Comparable results were obtained in the 10 replicates performed. Filtering out the SNPs with a QUAL value < 200 (see Materials and Methods) removed many pericentromeric SNPs; many SNPs with low AF around the candidate intervals were also removed in this way (Figure 4D, E).
Taking these results into account, we applied this strategy to simultaneously map two unlinked Ler mutations: ond4 and den6-1 (35). After crossing the ond4 and den6-1 mutants, we isolated 109 individuals (218 chromosomes) from the F2 progeny that unequivocally showed both mutant phenotypes in an additive manner (Supplementary Figure S6), extracted and pooled DNA from these plants, and sequenced it at 120× RD. After removing background SNPs, plotting the frequencies of all EMS-type SNPs allowed us to profile two candidate regions, including a region on chromosomes 2 and 4 around positions 11.2 and 10.3 Mb, respectively (Figure 4F). These two candidate regions contained seven and three GC→AT transitions, respectively, that were predicted to modify protein sequences (blue dots in Figure 4G, H), as well as ten and five, respectively, that were conservative (red dots in Figure 4G, H). This result shows that pseudo-backcrossing offers the advantages of high RD without an increase in sequencing cost.
Mapping T-DNA insertions in pooled samples
The annotated T-DNA insertions from mutant collections such as SALK (36) and SAIL (81) are not always responsible for an observed phenotype due to the presence of multiple insertions per line. Indeed, we previously faced this situation with several Arabidopsis mutants from the SALK collection, which we subjected to mapping-by-sequencing (37), since we aimed to analyze by WGS small groups of T-DNA mutant lines using a simple method compatible with DNA pooling that takes advantage of paired-end read properties and prior knowledge of the insertion sequence (37,82; Figure 5A). We reasoned that aligning paired-end reads from these mutants to the T-DNA sequence in paired-end mode would result in a portion of the read pairs, i.e. those spanning the junctions between the plant DNA and the T-DNA insertion, with only one of the two mates aligned. Thus, selecting these read pairs and aligning them to the Arabidopsis reference sequence would generate output comprising clusters of alignments around the T-DNA positions (blue rectangles in Figure 5A). A similar approach could be used with single reads aligned in local mode, in which the nucleotide positions immediately adjacent to the T-DNA insertions would show a signature consisting of clipped alignments (red rectangles in Figure 5A).
Figure 5.
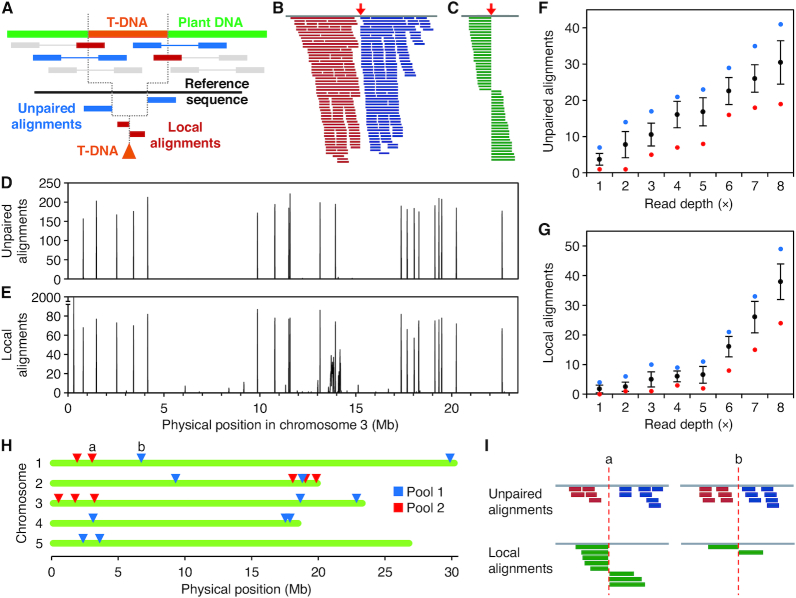
Mapping T-DNA insertions with WGS paired-end reads. (A) Procedure devised to map T-DNA insertions with massive paired-end reads. Total reads are aligned to the T-DNA sequence. Pairs with only one mate aligned (blue) or clipped sequences (red) are then aligned to the genome sequence, delimiting the position of the insertion. Reads and template are not drawn to scale. (B, C) Output of the alignment of simulated reads selected for mapping to the genomic sequence. The red arrows indicate the positions of the insertions simulated. (B) Unpaired alignments from a population of paired-end reads. (C) Local alignments (30–70 bp long) from a population of 90-mer reads. (D, E) Profile of the accumulation of (D) unpaired and (E) clipped alignments from simulated reads on Arabidopsis chromosome 3. Bin size is 10 kb. (E) Note that alignments peak at the same bins as in (D). Also note that false positives appear at positions 0.3 and 13.5–14.5 Mb. (F,G) Average number of (F) unpaired and (G) local alignments per insertion at different average read depths. Black dots and bars represent mean and standard deviation, respectively. Blue and red dots represent the maximum and minimum values, respectively. n = 100 insertions. (H, I) Insertions found in two pools of SALK mutants sequenced with low (5×) read depth from an experiment described in a published work (37). (I) Read signatures from two insertions labeled as a and b in (H).
To validate this protocol prior to sequencing our mutants we performed simulations that we did not describe in (37); these simulations are provided here. We simulated a 1-Mb genome from the Arabidopsis reference sequence and inserted a 4.2 kb T-DNA sequence from the pBIN-pROK2 vector (36). We simulated 100× paired-end reads (Simulation 11, in Supplementary Table S1) and analyzed them as described above. As predicted, a very small portion of the total read pairs was discordantly aligned at the ends of the T-DNA sequence. In addition, the unaligned mates, when filtered and realigned to the 1-Mb Arabidopsis reference sequence, clustered around the position of the in silico inserted T-DNA (Figure 5B). Forward reads were clustered together on the left flank and reverse reads on the right flank. After the local alignment, we identified a set of alignments that unambiguously pinpointed the insertion bp coordinate (Figure 5C).
To test the reliability of this workflow on an Arabidopsis genome-wide scale, we simulated a whole genome with 100 randomly positioned T-DNA insertions and attempted to map them with paired-end reads at a common RD (40×; Simulation 12, in Supplementary Table S1). We classified the unpaired alignments to the Arabidopsis sequence in bins of 10 kb and managed to obtain one signal peak for every T-DNA insertion simulated (Figure 5D). The number of reads supporting each insertion was homogenous (190.1 ± 18.5 reads per insertion), suggesting that this method is highly reliable at 40× RD. Moreover, 100% of the insertion positions accumulated locally aligned reads that precisely indicated the insertion position (72.2 ± 11.4 reads per insertion; Figure 5E). However, local alignments occurred in other parts of the genome as well, due to sequence similarity between different regions of the genome (e.g., the leftmost peak in Figure 5E), preventing us from using them without prior paired-end read information. One way to overcome this problem in the absence of paired-end reads would be to use >100 nt reads and only consider rather long local alignments.
For our previous study (37), we found it necessary to determine the minimum RD needed to safely map all insertions in a single sample, consisting of a pool of T-DNA lines. To this end, we made a preliminary simulation of our workflow, which is extended here (Simulation 13, in Supplementary Table S1), of sets of paired-end reads with decreasing RD down to 1× and characterized the number of supporting reads for each insertion. As shown in Figure 5F–G, in our simulations the number of supporting reads depends on the RD used, and very low RD values are sufficient to map insertions in a pool of T-DNA lines. From the results shown in Figure 5G and H, we arbitrarily set 5× as the lowest RD advisable for our purposes. The knowledge gained in this simulation experiment allowed us to design the actual experiment that we described in reference 37: we pooled the genomic DNA from 10 T-DNA mutants in two samples, which were sequenced on an Illumina HiSeq2000 at 25× RD per sample (∼5× per individual genome). We were able to unambiguously map 11 previously annotated insertions in these lines, together with 8 additional, non-annotated insertions, suggesting that most or all T-DNAs had been detected in our analysis (37 and Figure 5H–I).
DISCUSSION
In this study, we showed how simulated data could be used to help optimize the experimental design of mutation cloning projects that make use of WGS. We made useful predictions using relatively simple programs because the processes simulated, namely DNA mutagenesis, chromosomal recombination, and short-read sequencing, were assumed to be random for our purposes (31,36,69,83). We successfully accomplished three tasks: comparing short read technologies, evaluating experimental designs, and testing our analysis workflows. Our approach allowed us to obtain highly meaningful results for some experiments while quickly discarding dead-end approaches, saving laboratory resources (for example, regarding dominant mutations, we discarded the use of phenotypically mutant individuals from an F2 mapping population to define a sharp candidate interval).
Benchmarking
The use of simulated data is sometimes the only option available when performing a benchmarking experiment. For example, we assessed the percentage of HiSeq2000-like and Ion Proton-like reads that mapped unambiguously to gene-rich chromosomal regions. This experiment could not have been performed with experimental (real) reads, as these are obtained from whole nuclear DNA and it is therefore impossible to determine which reads originated from these regions. Similarly, we calculated the precision and sensitivity of an SNP calling workflow at different RDs. This analysis cannot easily be performed with real read data because it is impossible to know the positions of all SNPs in a given sample prior to sequencing (15).
Discriminating EMS-induced SNPs in backgrounds with high SNP density
For robust analysis of mutants derived from EMS mutagenesis, it is critical to obtain SNP call sets with the minimum number of false positives and false negatives. We found that in our typical analysis target, Arabidopsis gene-rich regions, a value of 50× RD is convenient for calling SNPs, since at this depth, both precision and sensitivity reached a plateau level.
Analyzing EMS mutants obtained from a non-reference accession leads to the detection of hundreds of thousands of background SNPs (28), making it more difficult to identify the mutation that causes the phenotype of interest. We demonstrated that using a background mask that integrates data from different samples rather than simply sequencing the parental line of a given mutant not only makes it unnecessary to sequence the parental DNA, but it also facilitates the elimination of background SNPs (Figure 2). This is because a single sequencing run on the background line at a commonly used RD never uncovers all variant positions, as revealed by our simulations. We also showed that the higher the number of samples used to create the mask and the lower the stringency to include SNPs in the mask, the greater its capacity to filter out background SNPs. However, the following tradeoff should always be considered: very lenient masks may include false SNPs (read errors or misalignments), whereas very stringent masks may discard some natural SNPs. As a result, very lenient masks may wrongly filter out EMS-induced SNPs in the problem sample, and very stringent masks may not filter out many natural SNPs (67). The high density of background SNPs in a typical analysis involving two different accessions (28) stresses the need for a powerful method for eliminating these SNPs. For example, not eliminating 1% of the background SNPs in a sample containing 500 000 such SNPs would result in retaining 5000 undesired variants in the analysis. We have shown that our SNP masking approach gives similar results to the direct variant calling using non-reference genome assemblies as a template (Figure 2). However, it should be taken into account that high-quality genome assemblies for non-reference lines are generally not available, and that the multi-mask approach is background independent.
Mapping EMS-induced causal mutations with an F2 recombinant mapping population
The ultimate goal of mapping-by-sequencing experiments is to define a candidate interval containing the phenotype-causing mutation as narrow as possible (71). In agreement with previously published results (11), we found that in the ranges normally used in actual experiments, both the number of chromosomes in a mapping population and the sequencing RD act as limiting factors for narrowing down a candidate interval. The effect of the first factor is obvious, since the number of recombination events near the causal mutation depends on the total number of chromosomes present. We also found that large mapping populations, independently of RD, produced lower variability in the number of candidate SNPs between simulation replicates, contributing to more predictable results (Figure 3B). We found that sequencing RD is a limiting factor to narrowing down a candidate interval when it is below the number of unique recombinant chromosomes, as parts of the chromosomes are never read under these conditions (first column in Figure 3C). For example, considering a mapping population of 200 chromosomes with only 20 reads overlapping a given polymorphic marker, most of the information available for this marker in the population is never extracted. On the contrary, increasing RD over the size of the mapping population did reduce the number of candidate mutations (bottom row in Figure 3C). The most likely explanation for this, given that sequencing is a random sampling process (83), is that not all chromosomes are read an equivalent number of times at a given locus. For example, for a particular marker in a population with 30 chromosomes read at 30× RD, some chromosomes could be read more than once, while others remain unread, whereas at 300×, these differences would likely be neutralized due to deeper sampling.
Also, increasing RD produces more accurate AF values and results in less noisy AF graphs (Supplementary Figure S4), which help delimit a candidate interval. In traditional linkage analysis, hundreds or even thousands of individuals are genotyped for markers close to the causal mutation, allowing even very low-frequency recombinant markers to be detected. Reminiscent of this approach, Hartwig et al. (10) used very high RD to resolve the candidate interval of an EMS-induced mutant: after 40× RD sequencing of a mapping population of 540 chromosomes, they detected apparent complete linkage between three candidate mutations (AF = 1). Next, they performed 20 000× RD targeted sequencing and found a recombination rate of >2% for two of the loci, which led them to establish the remaining mutation as the causal one.
Sequencing a whole genome with high RD and performing targeted sequencing are not always possible due to cost constraints. Using simulated data, we showed that two EMS-derived mutants could be mapped using a single mapping population obtained from a pseudo-backcross. This method produced twice the usual number of reads without increasing the cost per mutant. This approach is feasible only if the two mutants are not totally linked and if the double mutant is easy to score. By crossing the ond4 and den6-1 recessive mutants, we successfully defined two candidate intervals with very few candidate mutations (Figure 4G, H). To our knowledge, this is the first report of this approach, which could be useful for other researchers who wish to identify several mutated genes derived from a single EMS mutagenesis.
Mapping large DNA insertions
While attempting to map T-DNA insertions in Arabidopsis, we demonstrated how performing simulations allowed us to easily validate and optimize a simple pipeline for mapping the insertions using high-throughput read data. Although novel tools to map insertions are constantly being released (84–86), our approach is very simple and requires only a standard read aligner that supports single- and paired-end reads and both end-to-end and local alignments. The method used here is suitable for use with low RD reads and pooled DNA, as demonstrated using both simulated and real data. One drawback of this method is that it requires prior knowledge of the sequence being mapped, which might not always be available. Overall, the results presented here suggest that simulating WGS mapping experiments is a useful procedure to design better real experiments.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank J.M. Serrano, M.J. Ñíguez and J. Castelló for their excellent technical assistance.
Notes
Present address: David Wilson-Sánchez, Max Planck Institute for Plant Breeding Research, Carl-von-Linné-Weg 10, Köln, 50829, Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Ciencia, Investigación y Universidades of Spain [BIO2014-53063-P and PGC2018-093445-B-I00, to J.L.M.]; Generalitat Valenciana [Prometeo/2019/117 to J.L.M.]. Funding for open access charge: Ministerio de Ciencia, Investigación y Universidades of Spain [PGC2018-093445-B-I00].
Conflict of interest statement. None declared.
REFERENCES
- 1. Schneeberger K., Weigel D.. Fast-forward genetics enabled by new sequencing technologies. Trends Plant Sci. 2011; 16:282–288. [DOI] [PubMed] [Google Scholar]
- 2. Korbel J.O., Urban A.E., Affourtit J.P., Godwin B., Grubert F., Simons J.F., Kim P.M., Palejev D., Carriero N.J., Du L. et al.. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007; 318:420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doitsidou M., Poole R.J., Sarin S., Bigelow H., Hobert O.. C. elegans mutant identification with a one-step whole-genome-sequencing and SNP mapping strategy. PLoS One. 2010; 5:e15435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karakoc E., Alkan C., O’Roak B.J., Dennis M.Y., Vives L., Mark K., Rieder M.J., Nickerson D.A., Eichler E.E.. Detection of structural variants and indels within exome data. Nat. Methods. 2011; 9:176–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Williams-Carrier R., Stiffler N., Belcher S., Kroeger T., Stern D.B., Monde R.A., Coalter R., Barkan A.. Use of Illumina sequencing to identify transposon insertions underlying mutant phenotypes in high-copy Mutator lines of maize. Plant J. 2010; 63:167–177. [DOI] [PubMed] [Google Scholar]
- 6. Smith H.E., Fabritius A.S., Jaramillo-Lambert A., Golden A.. Mapping challenging mutations by whole-genome sequencing. G3. 2016; 6:1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Killcoyne S., del Sol A.. FIGG: simulating populations of whole genome sequences for heterogeneous data analyses. BMC Bioinformatics. 2014; 15:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou X., Peris D., Kominek J., Kurtzman C.P., Hittinger C.T., Rokas A.. in silico Whole Genome Sequencer & Analyzer (iWGS): a computational pipeline to guide the design and analysis of de novo genome sequencing studies. G3. 2016; 6:3655–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pratas D., Pinho A.J., Rodrigues J.M.. XS: a FASTQ read simulator. BMC Research Notes. 2014; 7:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hartwig B., James G.V., Konrad K., Schneeberger K., Turck F.. Fast isogenic mapping-by-sequencing of ethyl methanesulfonate-induced mutant bulks. Plant Physiol. 2012; 160:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. James G.V., Patel V., Nordstrom K.J., Klasen J.R., Salome P.A., Weigel D., Schneeberger K.. User guide for mapping-by-sequencing in Arabidopsis. Genome Biol. 2013; 14:R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robert C., Watson M.. Errors in RNA-Seq quantification affect genes of relevance to human disease. Genome Biol. 2015; 16:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brockman W., Alvarez P., Young S., Garber M., Giannoukos G., Lee W.L., Russ C., Lander E.S., Nusbaum C., Jaffe D.B.. Quality scores and SNP detection in sequencing-by-synthesis systems. Genome Res. 2008; 18:763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Highnam G., Wang J.J., Kusler D., Zook J., Vijayan V., Leibovich N., Mittelman D.. An analytical framework for optimizing variant discovery from personal genomes. Nat. Commun. 2015; 6:6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Talwalkar A., Liptrap J., Newcomb J., Hartl C., Terhorst J., Curtis K., Bresler M., Song Y.S., Jordan M.I., Patterson D.. SMaSH: a benchmarking toolkit for human genome variant calling. Bioinformatics. 2014; 30:2787–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clevenger J.P., Ozias-Akins P.. SWEEP: a tool for filtering high-quality SNPs in polyploid crops. G3. 2015; 5:1797–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Escalona M., Rocha S., Posada D.. A comparison of tools for the simulation of genomic next-generation sequencing data. Nat. Rev. Genet. 2016; 17:459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qin M., Liu B., Conroy J.M., Morrison C.D., Hu Q., Cheng Y., Murakami M., Odunsi A.O., Johnson C.S., Wei L. et al.. SCNVSim: somatic copy number variation and structure variation simulator. BMC Bioinformatics. 2015; 16:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Benidt S., Nettleton D.. SimSeq: a nonparametric approach to simulation of RNA-sequence datasets. Bioinformatics. 2015; 31:2131–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Frazee A.C., Jaffe A.E., Langmead B., Leek J.T.. Polyester: simulating RNA-seq datasets with differential transcript expression. Bioinformatics. 2015; 31:2778–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rackham O.J., Dellaportas P., Petretto E., Bottolo L.. WGBSSuite: simulating whole-genome bisulphite sequencing data and benchmarking differential DNA methylation analysis tools. Bioinformatics. 2015; 31:2371–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yuan X., Zhang J., Yang L.. IntSIM: an integrated simulator of Next-Generation Sequencing data. IEEE Trans. Biomed. Eng. 2016; 64:441–451. [DOI] [PubMed] [Google Scholar]
- 23. Kessner D., Novembre J.. Power analysis of artificial selection experiments using efficient whole genome simulation of quantitative traits. Genetics. 2015; 199:991–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Busby M.A., Stewart C., Miller C.A., Grzeda K.R., Marth G.T.. Scotty: a web tool for designing RNA-Seq experiments to measure differential gene expression. Bioinformatics. 2013; 29:656–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luo H., Li J., Chia B.K., Robson P., Nagarajan N.. The importance of study design for detecting differentially abundant features in high-throughput experiments. Genome Biol. 2014; 15:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guo Y., Zhao S., Li C.I., Sheng Q., Shyr Y.. RNAseqPS: a web tool for estimating sample size and power for RNAseq experiment. Cancer Informatics. 2014; 13:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000; 408:796–815. [DOI] [PubMed] [Google Scholar]
- 28. Zapata L., Ding J., Willing E.M., Hartwig B., Bezdan D., Jiao W.B., Patel V., Velikkakam James G., Koornneef M., Ossowski S. et al.. Chromosome-level assembly of Arabidopsis thaliana Ler reveals the extent of translocation and inversion polymorphisms. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:4052–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Colbert T., Till B.J., Tompa R., Reynolds S., Steine M.N., Yeung A.T., McCallum C.M., Comai L., Henikoff S.. High-throughput screening for induced point mutations. Plant Physiol. 2001; 126:480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jander G., Baerson S.R., Hudak J.A., Gonzalez K.A., Gruys K.J., Last R.L.. Ethylmethanesulfonate saturation mutagenesis in Arabidopsis to determine frequency of herbicide resistance. Plant Physiol. 2003; 131:139–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Salomé P.A., Bomblies K., Fitz J., Laitinen R.A., Warthmann N., Yant L., Weigel D.. The recombination landscape in Arabidopsis thaliana F2 populations. Heredity. 2012; 108:447–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ledergerber C., Dessimoz C.. Base-calling for next-generation sequencing platforms. Brief. Bioinform. 2011; 12:489–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fox E.J., Reid-Bayliss K.S., Emond M.J., Loeb L.A.. Accuracy of next generation sequencing platforms. J. Next Gener. Sequenc. Applic. 2014; 1:1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Matsumoto M., Nishimura T.. Mersenne twister: a 623-dimensionally equidistributed uniform pseudorandom number generator. ACM Trans. Model. Comput. Simul. 1998; 8:3–30. [Google Scholar]
- 35. Berná G., Robles P., Micol J.L.. A mutational analysis of leaf morphogenesis in Arabidopsis thaliana. Genetics. 1999; 152:729–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R. et al.. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003; 301:653–657. [DOI] [PubMed] [Google Scholar]
- 37. Wilson-Sánchez D., Rubio-Díaz S., Muñoz-Viana R., Pérez-Pérez J.M., Jover-Gil S., Ponce M.R., Micol J.L.. Leaf phenomics: a systematic reverse genetic screen for Arabidopsis leaf mutants. Plant J. 2014; 79:878–891. [DOI] [PubMed] [Google Scholar]
- 38. Morel J.B., Godon C., Mourrain P., Beclin C., Boutet S., Feuerbach F., Proux F., Vaucheret H.. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002; 14:629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013; 16 March 2013, preprint: not peer reviewedhttps://arxiv.org/abs/1303.3997.
- 41. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 Genome Project Data Processing Subgroup . The Sequence alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Barnett D.W., Garrison E.K., Quinlan A.R., Stromberg M.P., Marth G.T.. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011; 27:1691–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. García-Alcalde F., Okonechnikov K., Carbonell J., Cruz L.M., Gotz S., Tarazona S., Dopazo J., Meyer T.F., Conesa A.. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 2012; 28:2678–2679. [DOI] [PubMed] [Google Scholar]
- 45. DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. et al.. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011; 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011; 27:2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T. et al.. The variant call format and VCFtools. Bioinformatics. 2011; 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mateo-Bonmatí E., Casanova-Sáez R., Candela H., Micol J.L.. Rapid identification of angulata leaf mutations using next-generation sequencing. Planta. 2014; 240:1113–1122. [DOI] [PubMed] [Google Scholar]
- 49. Mateo-Bonmatí E., Casanova-Sáez R., Quesada V., Hricova A., Candela H., Micol J.L.. Plastid control of abaxial-adaxial patterning. Sci. Rep. 2015; 5:15975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wootton J.C., Federhen S.. Statistics of local complexity in amino acid sequences and sequence databases. Computers & Chemistry. 1993; 17:149–163. [Google Scholar]
- 51. Phan V., Gao S., Tran Q., Vo N.S.. How genome complexity can explain the difficulty of aligning reads to genomes. BMC Bioinformatics. 2015; 16(Suppl. 17):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sims D., Sudbery I., Ilott N.E., Heger A., Ponting C.P.. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 2014; 15:121–132. [DOI] [PubMed] [Google Scholar]
- 53. Poptsova M.S., Il’icheva I.A., Nechipurenko D.Y., Panchenko L.A., Khodikov M.V., Oparina N.Y., Polozov R.V., Nechipurenko Y.D., Grokhovsky S.L.. Non-random DNA fragmentation in next-generation sequencing. Sci. Rep. 2014; 4:4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ossowski S., Schneeberger K., Clark R.M., Lanz C., Warthmann N., Weigel D.. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008; 18:2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Aird D., Ross M.G., Chen W.S., Danielsson M., Fennell T., Russ C., Jaffe D.B., Nusbaum C., Gnirke A.. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011; 12:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van Dijk E.L., Jaszczyszyn Y., Thermes C.. Library preparation methods for next-generation sequencing: tone down the bias. Exp. Cell Res. 2014; 322:12–20. [DOI] [PubMed] [Google Scholar]
- 57. Ajay S.S., Parker S.C., Abaan H.O., Fajardo K.V., Margulies E.H.. Accurate and comprehensive sequencing of personal genomes. Genome Res. 2011; 21:1498–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nielsen R., Paul J.S., Albrechtsen A., Song Y.S.. Genotype and SNP calling from next-generation sequencing data. Nat. Rev. Genet. 2011; 12:443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Robles P., Micol J.L.. Genome-wide linkage analysis of Arabidopsis genes required for leaf development. Mol. Genet. Genomics. 2001; 266:12–19. [DOI] [PubMed] [Google Scholar]
- 60. Schneeberger K., Ossowski S., Ott F., Klein J.D., Wang X., Lanz C., Smith L.M., Cao J., Fitz J., Warthmann N. et al.. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:10249–10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Uchida N., Sakamoto T., Kurata T., Tasaka M.. Identification of EMS-induced causal mutations in a non-reference Arabidopsis thaliana accession by whole genome sequencing. Plant Cell Physiol. 2011; 52:716–722. [DOI] [PubMed] [Google Scholar]
- 62. Allen R.S., Nakasugi K., Doran R.L., Millar A.A., Waterhouse P.M.. Facile mutant identification via a single parental backcross method and application of whole genome sequencing based mapping pipelines. Front. Plant Sci. 2013; 4:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shao M.R., Shedge V., Kundariya H., Lehle F.R., Mackenzie S.A.. Ws-2 introgression in a proportion of Arabidopsis thaliana Col-0 stock seed produces specific phenotypes and highlights the importance of routine genetic verification. Plant Cell. 2016; 28:603–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jiang C., Mithani A., Belfield E.J., Mott R., Hurst L.D., Harberd N.P.. Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res. 2014; 24:1821–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Exposito-Alonso M., Becker C., Schuenemann V.J., Reitter E., Setzer C., Slovak R., Brachi B., Hagmann J., Grimm D.G., Jiahui C. et al.. The rate and potential relevance of new mutations in a colonizing plant lineage. PLOS Genetics. 2018; 14:e1007155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Flowers E.B., Poole R.J., Tursun B., Bashllari E., Pe’er I., Hobert O.. The Groucho ortholog UNC-37 interacts with the short Groucho-like protein LSY-22 to control developmental decisions in C. elegans. Development. 2010; 137:1799–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Minevich G., Park D.S., Blankenberg D., Poole R.J., Hobert O.. CloudMap: a cloud-based pipeline for analysis of mutant genome sequences. Genetics. 2012; 192:1249–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Krieg D.R. Ethyl methanesulfonate-induced reversion of bacteriophage T4rII mutants. Genetics. 1963; 48:561–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Greene E.A., Codomo C.A., Taylor N.E., Henikoff J.G., Till B.J., Reynolds S.H., Enns L.C., Burtner C., Johnson J.E., Odden A.R. et al.. Spectrum of chemically induced mutations from a large-scale reverse-genetic screen in Arabidopsis. Genetics. 2003; 164:731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. The 1001 Genomes Consortium 1,135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016; 166:481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A.H., Nielsen K.L., Jorgensen J.E., Weigel D., Andersen S.U.. SHOREmap: simultaneous mapping and mutation identification by deep sequencing. Nat. Methods. 2009; 6:550–551. [DOI] [PubMed] [Google Scholar]
- 72. Cuperus J.T., Montgomery T.A., Fahlgren N., Burke R.T., Townsend T., Sullivan C.M., Carrington J.C.. Identification of MIR390a precursor processing-defective mutants in Arabidopsis by direct genome sequencing. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Austin R.S., Vidaurre D., Stamatiou G., Breit R., Provart N.J., Bonetta D., Zhang J., Fung P., Gong Y., Wang P.W. et al.. Next-generation mapping of Arabidopsis genes. Plant J. 2011; 67:715–725. [DOI] [PubMed] [Google Scholar]
- 74. Rishmawi L., Sun H., Schneeberger K., Hulskamp M., Schrader A.. Rapid identification of a natural knockout allele of ARMADILLO REPEAT-CONTAINING KINESIN1 that causes root hair branching by mapping-by-sequencing. Plant Physiol. 2014; 166:1280–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Abe A., Kosugi S., Yoshida K., Natsume S., Takagi H., Kanzaki H., Matsumura H., Yoshida K., Mitsuoka C., Tamiru M. et al.. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012; 30:174–178. [DOI] [PubMed] [Google Scholar]
- 76. Petit J., Bres C., Mauxion J.P., Tai F.W., Martin L.B., Fich E.A., Joubes J., Rose J.K., Domergue F., Rothan C.. The Glycerol-3-Phosphate Acyltransferase GPAT6 from tomato plays a central role in fruit cutin biosynthesis. Plant Physiol. 2016; 171:894–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zuryn S., Le Gras S., Jamet K., Jarriault S.. A strategy for direct mapping and identification of mutations by whole-genome sequencing. Genetics. 2010; 186:427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wachsman G., Modliszewski J.L., Valdes M., Benfey P.N.. A simple pipeline for mapping point mutations. Plant Physiol. 2017; 174:1307–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Thole J.M., Strader L.C.. Next-generation sequencing as a tool to quickly identify causative EMS-generated mutations. Plant Signal. Behav. 2015; 10:e1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ashelford K., Eriksson M.E., Allen C.M., D’Amore R., Johansson M., Gould P., Kay S., Millar A.J., Hall N., Hall A.. Full genome re-sequencing reveals a novel circadian clock mutation in Arabidopsis. Genome Biol. 2011; 12:R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Sessions A., Burke E., Presting G., Aux G., McElver J., Patton D., Dietrich B., Ho P., Bacwaden J., Ko C. et al.. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002; 14:2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lambirth K.C., Whaley A.M., Schlueter J.A., Bost K.L., Piller K.J.. CONTRAILS: a tool for rapid identification of transgene integration sites in complex, repetitive genomes using low-coverage paired-end sequencing. Genomics Data. 2015; 6:175–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Reuter J.A., Spacek D.V., Snyder M.P.. High-throughput sequencing technologies. Mol. Cell. 2015; 58:586–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Henaff E., Zapata L., Casacuberta J.M., Ossowski S.. Jitterbug: somatic and germline transposon insertion detection at single-nucleotide resolution. BMC Genomics. 2015; 16:768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Jiang C., Chen C., Huang Z., Liu R., Verdier J.. ITIS, a bioinformatics tool for accurate identification of transposon insertion sites using next-generation sequencing data. BMC Bioinformatics. 2015; 16:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ecovoiu A.A., Ghionoiu I.C., Ciuca A.M., Ratiu A.C.. Genome ARTIST: a robust, high-accuracy aligner tool for mapping transposon insertions and self-insertions. Mobile DNA. 2016; 7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.