Abstract
Staphylococcus epidermidis is one of the most prevalent prokaryotic species on human skin and mucosal membranes that constitute the commensal flora. S. epidermidis has become one of the most common causes of primary bacteremia. Infections are difficult to diagnose because the pathogen has natural niches on human skin and the ability to adhere to inanimate surfaces via biofilms. Alarmingly, S. epidermidis has acquired resistance to many antibiotics, which presents a danger to human health. Known as methicillin-resistant S. epidermidis (MRSE), most clinical isolates of MRSE in North America exhibit β-lactam resistance primarily due to the presence of mecA, a gene that bestows β-lactam antibiotic resistance in a manner similar to methicillin-resistant Staphylococcus aureus (MRSA). MecA encodes for expression of penicillin-binding protein 2a (PBP2a), which is absent in β-lactam susceptible strains of S. epidermidis. We can disable this resistance factor in MRSE with 600-Da branched polyethylenimine (BPEI). Cationic BPEI targets anionic wall teichoic acid (WTA), an essential cofactor for proper functioning of PBP2a. We found that BPEI synergizes the activity of β-lactam antibiotics against MRSE. Growth curves suggest that the combination of BPEI and oxacillin is bactericidal. Electron micrographs indicate abnormalities in the cellular septa and cell walls of treated samples. Therefore, first-line clinical treatments can be effective against MRSE when used in combination with BPEI.
Keywords: antibiotic resistance, branched polyethylenimine, Staphylococcus epidermidis, wall teichoic acid, β-lactams
Introduction
Previously known as a harmless commensal species on human skin, Staphylococcus epidermidis is a causative agent of nosocomial infections, along with Staphylococcus aureus.[1] With the widespread use of indwelling medical devices and implanted foreign bodies, as well as the rising number of immunocompromised patients,.[2] S. epidermidis has become a significant threat to the public health. In the US, S. epidermidis vascular-catheter-related bloodstream infections create an economic burden of approximately $2 billion/year.[3] Unlike the more aggressively virulent and coagulase-positive Staphylococcus aureus, S. epidermidis belongs to the coagulase-negative staphylococci and is rarely a life-threatening infection outside of hospital settings. However, its prevalence in hospitals and the commensal lifestyle on human skin have granted S. epidermidis resilience against treatments. Complications in treatment are often due to the presence of specific antibiotic resistance genes and the production of biofilms. S. epidermidis, an opportunistic pathogen, colonizes abiotic surfaces, such as medical devices, for weeks to months.[4] Polysaccharide intercellular adhesin, also known as poly-N-acetyl glucosamine (PIA/PNAG), was found to have a major function in S. epidermidis biofilm formation. PIA/PNAG is governed by the icaADBC operon, which appears more frequently in isolates of S. epidermidis from hospital-related infections than isolates from healthy individuals. As a permanent skin-colonizer, S. epidermidis may also have elaborate mechanisms that allow it to attach to cardiac devices, surgical sites, central nervous system shunts, prosthetic joints, and vascular grafts.[3–5] S. epidermidis is the second-most common cause of prosthetic valve endocarditis with 24% mortality, surpassed in frequency only by S. aureus.[5] Hospital isolates of S. epidermidis are methicillin resistant at significantly higher rates (75–90%) than S. aureus (40–60%).[3] The emergence of methicillin-resistant S. epidermidis (MRSE) infections emphasizes the need for antimicrobial drug design to yield new treatments before the pathogens become resistant to the last-resort antibiotics.
β-Lactam antibiotics, such as amoxicillin and oxacillin, inhibit bacterial cell wall synthesis by irreversibly binding to the penicillin-binding proteins (PBPs), a subgroup of enzyme transpeptidases. β-lactam binding to PBPs prevents them from cross-linking the cell wall peptidoglycan. Without cell wall synthesis, bacteria are unable to replicate and consequently die. However, MRSE has the staphylococcal cassette chromosome mec (SCCmec), which contains the gene mecA. MecA is ultimately translated to create PBP2a, a unique penicillin-binding protein that has a low affinity for β-lactam antibiotics relative to other PBPs, thereby making these first-choice antibiotics ineffective.[3] In our study, we disabled this resistance factor using 600-Da branched polyethylenimine (BPEI), a low-molecular-weight cationic polymer. Data show that a combination of BPEI and β-lactam antibiotics kills MRSE. The proposed mechanism of action is that BPEI electrostatically binds to wall teichoic acid (WTA), which is an anionic polymer in the cell wall of S. epidermidis and other Gram-positive bacteria. WTA has been suggested to play an important role in the localization of penicillin binding proteins, especially PBP2a.[6] Foxley et al. successfully potentiated β-lactams against MRSA by targeting WTA with BPEI.[7] We believe a similar mechanism of action exists between BPEI and β-lactams against MRSE. Thus, when BPEI binds to WTA, PBP2a loses its ability to function, and MRSE’s susceptibility to β-lactams is restored.
Results and Discussion
From a comparative epidemiology study in Belgium[8] S. epidermidis isolates from catheter-related bloodstream infections (CRBSI) were found to have significant resistance to more antibiotics than the commensal isolates. More than 75% of the CRBSI isolates contained the resistance gene mecA, while only 3% of the S. epidermidis isolates from healthy individuals did. Additionally, the majority of the MRSE isolates are resistant to multiple antibiotics including oxacillin, erythromycin, and ciprofloxacin.[8] S. epidermidis is involved in hospital-acquired infections[9] which pose a great danger to public health. We were motivated to better understand the pathogenesis of MRSE as well as the pharmacology of BPEI and antibiotics against S. epidermidis laboratory strains and clinical isolates using multiple analytical methods.
Checkerboard assays indicate synergistic effects between BPEI and β-lactams
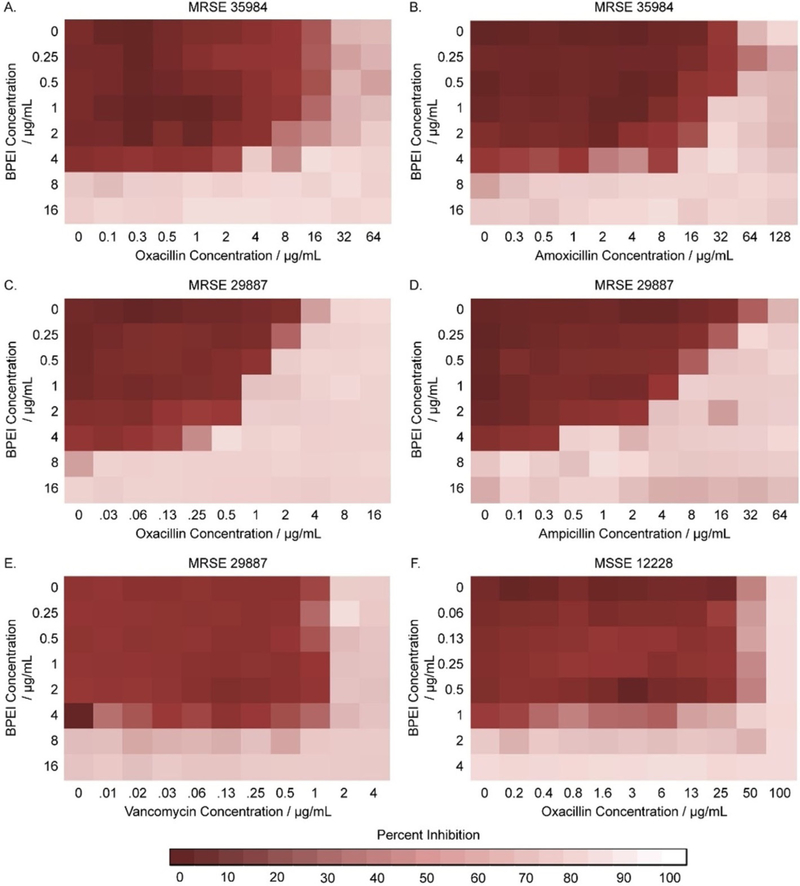
To investigate the synergy between 600-Da BPEI and antibiotics against MRSE, checkerboard assays were conducted. Synergistic effects were observed between BPEI and β-lactam antibiotics (oxacillin, ampicillin, and amoxicillin), but not between BPEI and vancomycin (Table 1). Synergy occurred when their FICI was less than 0.5, while additive effects were indicated by an FICI between 0.5 to 1.[10] The MICs of BPEI and several antibiotics were tested against two laboratory strains of methicillin-resistant Staphylococcus epidermidis, MRSE ATCC® 29887™[11] and MRSE ATCC® 35984™,[12] and one methicillin-susceptible strain, MSSE ATCC® 12228™.[12] As a result, the BPEI MICs for both MRSE 35984 (Figure 1 A,B) and MRSE 29887 (Figure 1 C–E) were 8 μgmL−1, while the BPEI MIC for MSSE 12228 (Figure 1 F) was 2 μgmL−1. The oxacillin MICs for MRSE 35984, MRSE 29887, and MSSE 12228 were 32 μgmL−1, 8 μgmL−1, and 0.1 μgmL−1, respectively. Checkerboard assays showed synergy between BPEI and oxacillin against both MRSE 35984 (FICI = 0.5) and MRSE 29887 (FICI = 0.375). In the presence of 4 μgmL−1 600-Da BPEI, the oxacillin MICs for MRSE 35984 and MRSE 29887 were reduced from 32 to 4 μg mL−1 and from 8 to 0.5 μgmL−1, respectively. Additionally, BPEI had synergy with amoxicillin (FICI = 0.5) against MRSE 35984 and ampicillin (FICI = 0.31) against MRSE 29887. Linezolid was also found to have synergy or additivity with BPEI (FICI = 0.38/0.75 against MRSE 35984/MRSE 29887). No synergy was seen between BPEI and vancomycin (FICI = 1, Figure 1E). BPEI did not potentiate β-lactam activity against the susceptible strain, MSSE 12228, which lacks PBP2a (FICI = 1, Figure 1 F). The MIC of each antibiotic was lowered by a factor ranging from 4-fold (linezolid from 1 to 0.25 μgmL−1) to 256-fold (amoxicillin from 64 to 0.25 μgmL−1) when combined with 4 μgmL−1 BPEI (Table 1). To evaluate our approach further, two clinical MRSE isolates from patients at the University of Oklahoma College of Medicine were also tested. In Table 1, both clinical isolates had strong antibiotic resistance with high oxacillin MICs (64 and 128 μgmL−1). When combined with 4 μgmL−1 BPEI, the MIC of oxacillin in both cases dramatically dropped to 2 μgmL−1, making the bacteria no longer resistant. These data show that BPEI can effectively potentiate traditional antibiotics against MRSE.
Table 1.
Synergy of 600-Da BPEI and antibiotics against two MRSE laboratory reference strains and two MRSE clinical isolates.
| Strain | Antibiotic | BPEI | MIC [μgmL−1][a] Antibiotic |
Antibiotic + BPEI (4 μg mL−1) |
FICI[b] | Outcome |
|---|---|---|---|---|---|---|
| MRSE ATCC 35984 | Oxacillin | 8 ± 0.090 | 32 ± 0.060 | 4 ± 0.006 | 0.5 | Synergy |
| Amoxicillin | 8 ± 0.016 | 64 ± 0.013 | 0.25 ± 0.021 | 0.5 | Synergy | |
| Linezolid | 8 ± 0.026 | 1 ± 0.014 | 0.25 ± 0.011 | 0.38 | Synergy | |
| Vancomycin | 8 ± 0.003 | 2 ± 0.003 | 2 ± 0.024 | 1 | No synergy | |
| MRSE ATCC 29887 | Oxacillin | 8 ± 0.059 | 4 ± 0.001 | 0.5 ± 0.0003 | 0.375 | Synergy |
| Ampicillin | 8 ± 0.006 | 32 ± 0.042 | 0.5 ± 0.007 | 0.31 | Synergy | |
| Linezolid | 8 ± 0.012 | 1 ± 0.005 | 0.25 ± 0.013 | 0.75 | Additivity | |
| Vancomycin | 8 ± 0.028 | 2 ± 0.001 | 2 ± 0.015 | 1 | No synergy | |
| MRSE OU26 | Oxacillin | 64 ± 0.002 | 32 ± 0.008 | 2 ± 0.071 | 0.17 | Synergy |
| MRSE OU29 | Oxacillin | 128 ± 0.037 | 16 ± 0.001 | 2 ± 0.022 | 0.27 | Synergy |
Minimum inhibitory concentration; values are the mean ±SD of n = 3 determinations.
Minimum fractional inhibitory concentration index.
Figure 1.
Checkerboard assays testing the synergy of BPEI and antibiotics on MRSE 35984, MRSE 29887, and MSSE 12228. Synergy between BPEI and β-lactams was observed on MRSE 35984 (A, B) and MRSE 29887 (C, D), but not on MSSE 12228 (F). No synergy was found between BPEI and vancomycin on MRSE 29887 (E). Each heat map is the average of three trials in the change of OD600.
Synergistic effects were observed between BPEI and β-lactams but not vancomycin because the targets of β-lactams and vancomycin are distinctly different. Vancomycin binds to the D-Ala-D-Ala stems of bacterial cell wall peptidoglycan, whereas β-lactams bind to PBPs.[13] BPEI binds to WTA, a cofactor of PBP2a, while WTA is not connected with vancomycin activity. BPEI and β-lactams target the PBPs resulting in synergy: BPEI-WTA interaction inhibits PBP2a, while β-lactams inhibit the other PBPs.[13b] A similar explanation can be used to explain the data with methicillin-susceptible S. epidermidis (MSSE) that does not have PBP2a. Likewise, WTA is not involved in the regulation of other PBPs in MSSE. Therefore, disabling WTA does not potentiate antibiotics against MSSE.
Wall teichoic acid (WTA) plays an important role in BPEI potentiation of β-lactams against the Gram-positive MRSE. Studies have shown that WTA is required for antibiotic resistance and biofilm formation in MRSE.[14] By using NMR spectroscopy, Foxley et al. demonstrated that BPEI and WTA interacted via electrostatic binding of positively charged amine groups on BPEI and negatively charged phosphate groups on WTA.[15] We believe a similar mechanism explains the results obtained in this study. Our hypothesis is that the BPEI-WTA attractions cause steric hindrance to prevent WTA-PBP2a interaction, thereby disabling the resistance factor PBP2a. The β-lactams prevent the other PBPs from cross-linking the cell wall; consequently, the MRSE cells are killed. Synergistic and additive effects of the BPEI + antibiotic combinations signify enormous potential benefits for patient health because these first-line antibiotics are FDA-approved. Also, the low molecular weight 600-Da BPEI has been tested to have very low cytotoxicity and nephrotoxicity.[7] Thus, BPEI could be a drug-candidate to treat MRSE infections.
Growth curves confirm antimicrobial activity of oxacillin was restored
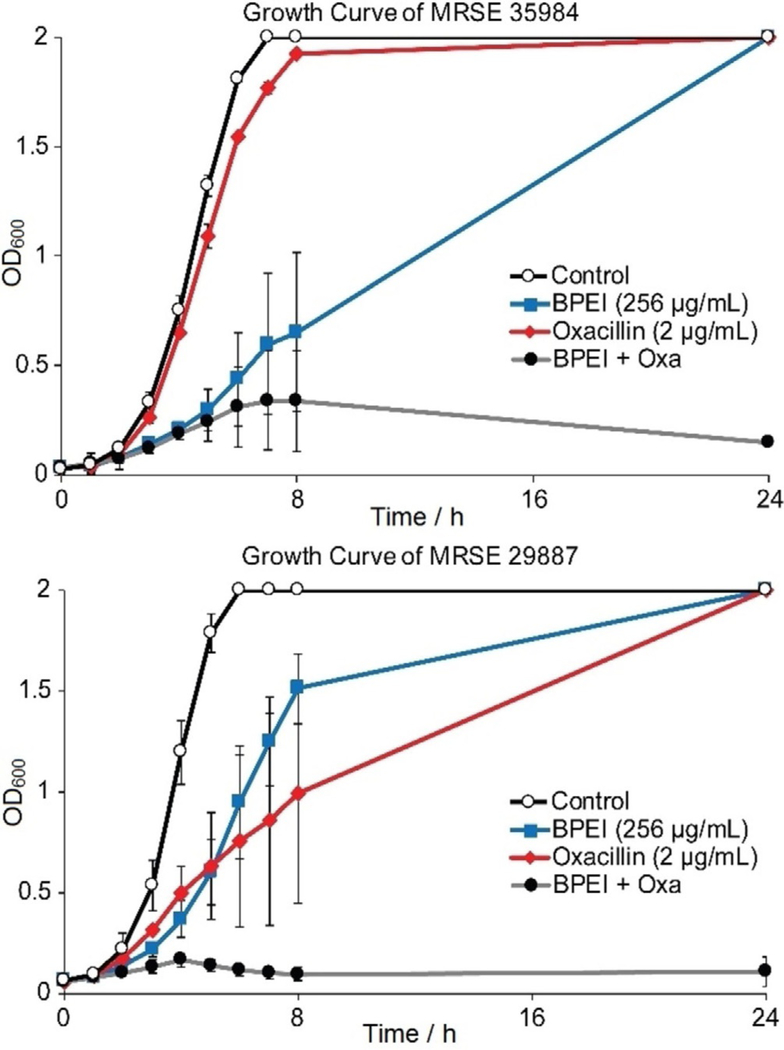
Antimicrobial activity of oxacillin against MRSE 35984 and MRSE 29887 was restored by 600-Da BPEI. However, by comparing data in Figure 1, it is harder to disable resistance in MRSE 35984. This was confirmed by evaluating the time-dependence of antimicrobial activity. Growth curves of the two resistant strains were monitored by recording their OD600 for 24 hours and are shown as the averages of three separate trials (Figure 2). The bacteria exposed to either BPEI alone or oxacillin alone reached stationary phase. However, each strain failed to do so in the combination treatment of BPEI + oxacillin. The OD600 increased for all of the samples except for those treated with the BPEI + oxacillin. In the BPEI + oxacillin treated samples, the OD600 dropped after 4 hours for MRSE 29887 and after 8 hours for MRSE 35984. This indicates a strong antimicrobial activity and also suggests bactericidal activity of our BPEI + β-lactam combination.
Figure 2.
BPEI restores bactericidal activity of oxacillin in the combination treatment (grey curve) against both MRSE 29887 and MRSE 35984. Error bars denote standard deviation (n = 3).
Scanning and transmission electron microscopies confirm BPEI’s mechanism of action
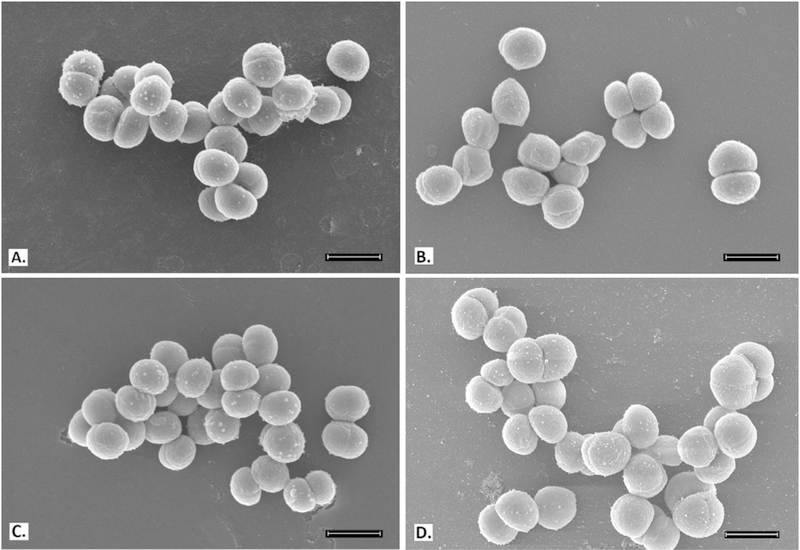
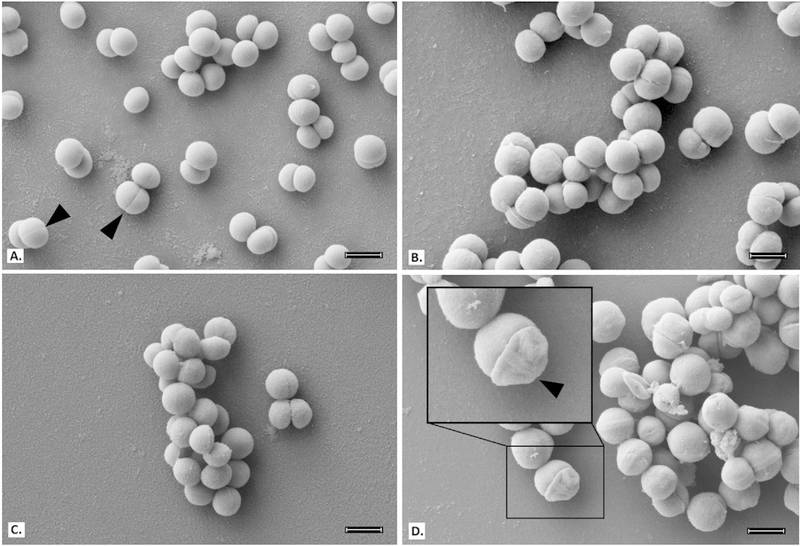
To better understand and confirm BPEI’s mechanism of action against MRSE, electron microscopy was used to examine treated samples of MRSE 35984 and MSSE 12228. Bacterial cells were collected at mid-log phase. SEM images of MRSE 35984 include the untreated control, BPEI-treated, (Figure 4) oxacillin-treated, and combination BPEI + oxacillin-treated samples (Figure 3). The cells in the control sample have smooth, rounded shapes with very clear division septa (Figure 3A). BPEI-treated (Figure 3B) and oxacillin-treated (Figure 3C) cells show no noticeable differences in appearance. However, many of the cells exposed to the BPEI + oxacillin combination have an abnormal appearance, including concave distortion and thicker division septa (Figure 3D). These cellular morphology changes were not observed on the susceptible strain, MSSE 12228 (Figure 4).
Figure 4.
Scanning electron micrographs of MSSE 12228; scale bar = 1 μm. Untreated control (A), BPEI (32 μg mL−1) treated (B), oxacillin (0.003 μg mL−1) treated (C), and combination of BPEI + oxacillin (32 μg mL−1 + 0.003 μg mL−1) treated cells (D) show no noticeable differences.
Figure 3.
Scanning electron micrographs of MRSE 35984; scale bar = 1 μm. Untreated control cells appear smooth and rounded with clear division septa (arrows) (A). BPEI (256 μgmL−1) treated cells (B) and oxacillin (2 μgmL−1) treated cells (C) show no significant difference. Combination of BPEI + oxacillin (256 μg mL−1 + 2 μg mL−1) treated cells (D) show extreme distortions and thicker division septa (inset).
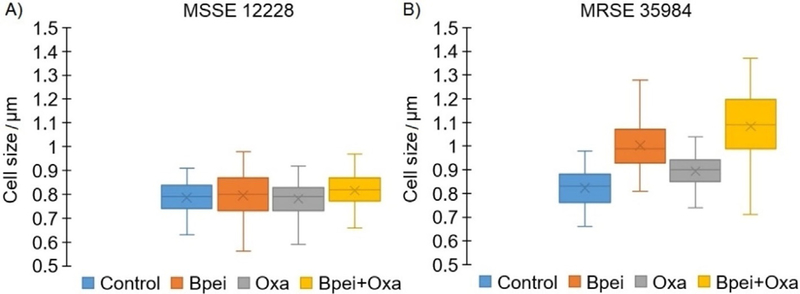
The majority of cells from BPEI-treated and the BPEI + oxacillin treated samples (Figure 3B,D) appear qualitatively larger than the control sample. To confirm the difference in cell size, cell diameters were measured for both MSSE 12228 and MRSE 35984. As shown in Figure 5A, MSSE 12228 did not show a statistically significant difference (ANOVA, p-value >0.001) in cell size among the four treated groups: control (0.79± 0.06 μm), BPEI-treated (0.81 ±0.09 μm), oxacillin-treated (0.79± 0.08 μm), and combination BPEI + oxacillin-treated (0.82 ± 0.07 μm). The cell sizes of MRSE 35984 (Figure 5B) were found to be significantly different (ANOVA, p-value<0.001) among the four treated groups: control (0.82±0.07 μm), BPEI-treated (1.00±0.10 μm), oxacillin-treated (0.89±0.07 μm), and combination BPEI + oxacillin-treated (1.08 ±0.15 μm).
Figure 5.
Cell size measurements of MSSE 12228 and MRSE 35984 from SEM images among four treated groups: untreated control, BPEI (256 μgmL−1) treated, oxacillin (2 μg mL−1) treated, and combination BPEI + oxacillin treated. N = 100 cells per group. Mean value, X-marker; median value, center line; lower quartile and upper quartile, lower and upper ends of the box; minimum and maximum values, whiskers outside the box. No significant difference in cell size among treated groups in MSSE 12228 (ANOVA, p-value >0.001) (A). Significant difference in cell size among treated groups in MRSE 35984 (ANOVA, p-value < 0.001) (B).
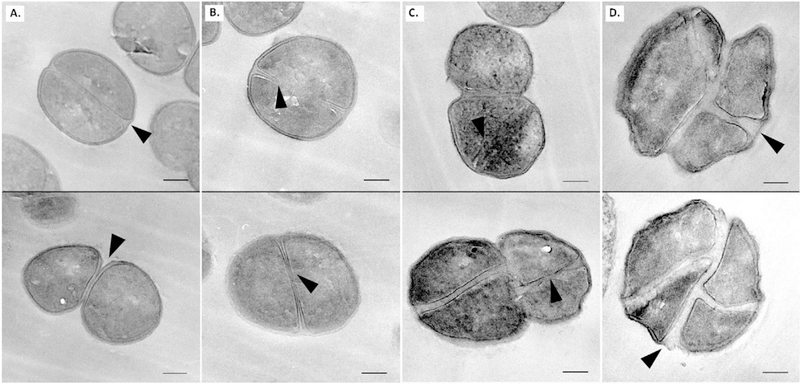
To further elucidate the mechanism of action for our BPEI + β-lactam treatment against MRSE 35984, cross-sections of the four treated groups were imaged with TEM. TEM allows higher magnification and observation of intracellular structures of the cells. The untreated control cells (Figure 6A) have well-rounded morphologies and complete division septa, while BPEI-treated (Figure 6B) and oxacillin-treated (Figure 6C) samples display many incomplete division septa. The combination BPEI + oxacillin-treated cells (Figure 6D) have the largest sizes, abnormal multi-division septa, and unrounded shapes that the other treated groups do not possess. Each division septum appears thicker than in the other groups, supporting the SEM data (Figure 3D). Exposure to the BPEI + oxacillin combination makes MRSE cells unable to divide and ultimately causes them to burst, resulting in cell death.
Figure 6.
Transmission electron micrographs of MRSE 35984; scale bar = 200 nm; arrows show division septa. Untreated control cells appear rounded with complete division septa (A). BPEI (256 μgmL−1) treated cells (B) and oxacillin treated (2 μgmL−1) cells (C) show incomplete division septa. Combination of BPEI + oxacillin (256 μg mL−1 + 2 μg mL−1) treated cells (D) appear to be the largest with abnormal division septa and concave morphology.
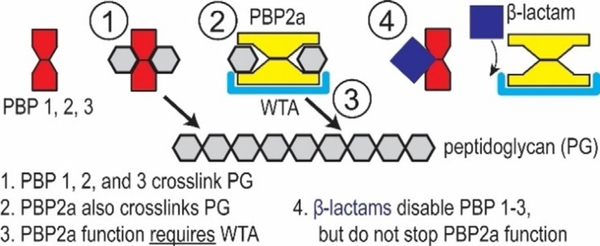
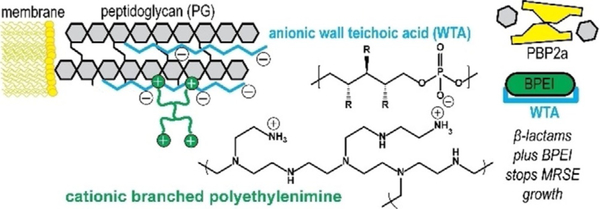
The cell envelope of Gram-positive bacteria is a strong framework whose masonry-like architecture protects against internal turgor pressure and external attack by antimicrobial agents. The cell wall is maintained in a robust state by dynamic turnover that removes weakened portions while constructing new cell wall layers. Autolysins break down the cell wall by hydrolyzing the bond between N-acetylmuramic acid and N-acetylglucosamine groups of peptidoglycan. Synthesis of new cell wall layers involves assembly of precursors in the cytoplasm prior to transport across the membrane. Next, transgly-cosylase and transpeptidase enzymes assemble the new components in their proper configuration. These processes establish the current strategy of interrupting cell wall construction as a target for many antibacterial drugs. β-lactam antibiotics bind to transpeptidase penicillin binding proteins (PBPs) to prevent crosslinking of nascent peptidoglycan (Figure 7). Degradation by autolysins coupled with the inability to replace peptidoglycan leads to thinning of the cell wall, loss of integrity, and rupture from internal pressure. However, MRSE endures by expressing PBP2a, whose enzymatic functions proceed in the presence of β-lactams. Cationic BPEI binds to anionic WTA (figure 8) to prevent WTA function, one of which is to serve as a scaffold for PBP2a enzymatic activity. In this manner, BPEI disables resistance from PBP2a.
Figure 7.
Illustrative description of crosslinking of cell wall peptidoglycan in MRSE. The PBPs complete the crosslinking process unless inhibited by β-lactam antibiotics. One mechanism of β-lactam resistance is the expression PBP2a for cell wall crosslinking in the presence of β-lactams.
Figure 8.
Instead of targeting WTA biosynthesis, BPEI attacks mature WTA in the cell wall. WTA is an anionic phosphodiester biomolecule and we use cationic BPEI to disable WTA via an electrostatic interaction. In this manner, we can disable resistance from PBP2a and restore MRSE susceptibility to β-lactam antibiotics. For WTA, the side chains, R, can be either N-acetylglucosamine, D-alanine, or hydroxy groups.
Conclusion
Antimicrobial resistance is a critical and increasing world-wide threat to the public health. The CDC estimates that at least million people in the US alone are infected by antibiotic-resistant bacteria annually, leading to 23000 deaths and $20 billion/year in US healthcare costs.[16] The threat from MRSE appears lower than MRSA because of reduced virulence and fewer toxins. However, with its ubiquitous niche on the human skin, MRSE cannot be overlooked. S. epidermidis bacteria can be found on nearly every medical device.[17] Fortunately, WTA in these bacteria is a weakness in antimicrobial resistance mechanisms.[6,14b,18] Previous attempts to disable PBP2a by stopping WTA biosynthesis have not survived preclinical trials. Both MRSA and MRSE employ PBP2a, whose functionality requires the presence of WTA. Their resistance can be overcome by disabling WTA, as demonstrated with WTA-deficient strains of MRSA (ΔtarO mutants),[6] by inhibiting the WTA synthesis protein TarO with tunicamycin,[19] or by inhibiting another bio-synthesis protein, TarG, with Targocil®.[14b,20] The work in these reports is strong. Targocil® and related compounds suffer from protein binding that reduces in vivo efficacy[21] but new compounds have mitigated this drawback.[22] Compounds that inhibit WTA and potentiate β-lactams against MRSA also lead to potentiation against MRSE.[14b]
Nevertheless, WTA remains an under exploited weakness, and our data show that cationic 600-Da BPEI targets this weakness.[7,15] We depart from the status quo of stopping WTA bio-synthesis[14b,19] and instead target mature WTA. WTA is an anionic phosphodiester, and cationic 600-Da BPEI disables WTA via an electrostatic interaction. This, in turn, disables PBP2a. In combination, 600-Da BPEI and β-lactam antibiotics inhibit MRSE growth (this work) and MRSA growth.[7,15] The effect is characterized as synergistic from the FICI. Measuring MRSE growth reveals that bacteria exposed to sub-lethal concentrations of BPEI and oxacillin fail to reach exponential phase when the two compounds are combined. This data indicates that the mechanism by which BPEI + oxacillin prevents growth of MRSE is bactericidal. Additional checkerboard assays demonstrate anti-MRSE potency of amoxicillin and ampicillin mixed with 600-Da BPEI. Likewise, higher 600-Da BPEI concentrations decrease further the antibiotic MIC values. Yet, potentiation of linezolid challenges the premise that our discovery applies to WTA-based resistance mechanisms, such as the function of PBP2a. From calculation of the FICI, the effect is a characterized as additive rather than synergy, and thus BPEI and linezolid affect different bacterial components. We know that BPEI binds to anionic sites of the cell wall envelope[7,15] whereas linezolid acts in the cytoplasm to interfere with protein synthesis via the ribosomal 50S subunit. The potentiation of linezolid is most likely to occur by increasing its permeation into the cytoplasm. Therefore, BPEI reduces barriers from the cell membrane and/or the extracellular biofilm of MRSE. Yet, 600-Da BPEI does not alter the MIC of vancomycin, whose mode-of-action is constrained to the cell wall peptidoglycan. Work to evaluate increased cross-membrane permeation of antibiotics due to the presence of 600-Da BPEI is underway.
The overuse of antibiotics is reflected in the pervasiveness of MRSE. From previous studies, the staphylococcal cassette chromosome mec, which codes for methicillin resistance, is suggested to be a one-way horizontal gene transfer from S. epidermidis to the well-known super-bug, MRSA.[23] As a ubiquitous commensal species of human microflora, S. epidermidis contains an optimum reservoir for antibiotic resistance genes, making it an implicit danger. Catheter-related bloodstream infections (CRBSI) are caused primarily by coagulase-negative Staphylococci, which include S. epidermidis, and they can usually be treated with glycopeptides (such as vancomycin) without catheter removal. However, the chances of bacteremia recurring could be as high as 20%,[24] and the effectiveness of the last-resort antibiotics (linezolid, vancomycin, and daptomycin) has been reduced significantly against MRSE.[25] Some MRSE clinical isolates have intermediate resistance to vancomycin and caused complications in patients with sepsis and peritonitis.[26] Thus, a need exists to evaluate methods of combating these infections. Our strategy of combining traditional β-lactams with BPEI holds promise in the ongoing battle against MRSE.
Experimental Section
Materials:
In this work, the Staphylococcus epidermidis bacteria were purchased from the American Type Culture Collection (ATCC 29887: methicillin-resistant, ATCC 35984: methicillin-resistant, and ATCC 12228: methicillin-susceptible). Two clinical isolates of MRSE were provided by Cindy McCloskey, M.D. at the University of Oklahoma College of Medicine. Chemicals were purchased from Sigma-Aldrich (DMSO, growth media, and electron microscopy fixatives). Antibiotics were purchased from Gold Biotechnology. 600-Da BPEI was purchased from Polysciences.
Checkerboard assay:
Checkerboard assays were conducted to identify synergy between BPEI and antibiotics against bacteria. The full procedure is outlined in Foxley et al.[7] Briefly, stock solutions of oxacillin, ampicillin, amoxicillin, linezolid, and vancomycin were made in DMSO. Antibiotic solutions were serial diluted and added to pre-sterilized 96-well plates so that the final DMSO concentration was less than 1 %. A stock culture was made of one colony per mL and added 1 % v/v to cation-adjusted Mueller-Hinton broth (MHB) in the 96-well plates. Optical density readings at 600 nm (OD600) were made using a Tecan Infinite M20 plate reader immediately after inoculation. The plates were incubated for 24 h at 35 °C, and a final OD600 reading was measured. The change in OD600 was found by subtracting the initial OD600 from the final OD600 reading. A change in OD600 greater than 0.05 indicated positive growth. The minimum inhibitory concentration (MIC) was assigned as the lowest concentration of antibiotic or BPEI that inhibited growth. The fractional inhibitory concentration index (FICI) was calculated using the previously established equation.[10] An FICI lower than 0.5 indicated synergy, an FICI between 0.5 and 1 indicated additivity, and an FICI equal to 1 indicated no synergy. Each assay was done in triplicate.
Growth curve:
Tryptic Soy Broth (TSB) growth media augmented with various amounts of BPEI and/or oxacillin was inoculated 0.5% from overnight cultures of MRSE 29887 and MRSE 35984. Cells were grown at 35 °C with shaking. The OD600 was recorded every hour for each sample. Each study was done in triplicate.
Scanning electron microscopy:
MRSE 35984 and MSSE 12228 cells were inoculated 0.5% from an overnight culture and grown at 35 °C with shaking. Each strain of bacteria was grown in four separate conditions: BPEI, oxacillin, combination (BPEI + oxacillin), and control. The OD600 was monitored, and growth was stopped at late-lag phase (OD600 = 0.20–0.25). Samples were fixed with Karnovsky fixative (2% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer) for 30 min. The cells were then fixed with 1% OsO4 for 30 min in the dark. The cells were washed with water three times. A couple drops of each sample were placed on clean, poly-L-lysine coated coverslips and air-dried for 30 min. To dehydrate, the samples went through a series of ethanol solutions (20%, 35%, 50%, 70%, and 95%), spending 15 min in each solution. Afterward, the samples were dried with hexamethyldisilazane (HMDS) and sputter-coated with AuPd. A Zeiss NEON SEM was used to image the samples at 5 kV accelerating voltage. Size analysis was performed on ImageJ.
Transmission electron microscopy:
MRSE 35984 cells were inoculated 0.5% from an overnight culture and grown at 35 °C with shaking in four separate conditions: BPEI, oxacillin, combination (BPEI + oxacillin), and control. The OD600 was monitored, and growth was stopped at late-lag phase (OD600 = 0.20–0.25). Samples were fixed with Karnovsky fixative (2% glutaraldehyde and 2% paraformaldehyde in 0.1 M cacodylate buffer) for 2 h. The cells were washed three times with 0.2 M cacodylate buffer and then stained with 1 % OsO4 in 1.5% potassium ferrocyanide for 2 h in the dark. Samples were washed three times with water and stained with 1 % uranyl acetate for 30 min. After three more washes with water, the cells were dehydrated with 50%, 70%, 95%, 95%, and 100% ethanol (20 min in each solution). The samples were immersed in propylene oxide (PO) for 1 h to remove residual ethanol. The cell samples were then infiltrated with 2:1 of PO: Epon for 1 h, 1:1 of PO: Epon for 1 h, 2:1 of PO: Epon for 2 h, and pure Epon overnight. The cells were transferred into embedding molds with fresh Epon and allowed to sink to the bottom for 24–48 h at 60°C. The sample blocks were ultra-thin-sectioned by an ultramicrotome to 80–100 nm thickness. The thin sections were placed on TEM copper grids and then stained with 1 % uranyl acetate for 30 min. A JEOL2000FX TEM was used to image the samples at 200 kV accelerating voltage. Size analysis was performed on ImageJ.
Acknowledgements
This work was possible due to the kindness and contributions of Daniel Glatzhofer, PhD, Robert Cichewicz, PhD, Scott Russel, PhD, and Preston Larson, PhD. We also thank Cindy McCloskey, M.D. for the clinical isolates. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. Funding was provided by the Oklahoma Center of Advancement of Science and Technology and The University of Oklahoma.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- [1].Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA, Science 2009, 324, 1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Percival SL, Suleman L, Vuotto C, Donelli G, J. Med. Microbiol 2015, 64, 323–334. [DOI] [PubMed] [Google Scholar]
- [3].Otto M, Nat. Rev. Microbiol. 2009, 7, 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Neely AN, Maley MP, J. Clin. Microbiol. 2000, 38, 724–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chu VH, Miro JM, Hoen B, Cabell CH, Pappas PA, Jones P, Stryjewski ME, Anguera I, Braun S, Munoz P, Commerford P, Tornos P, Francis J, Oyonarte M, Selton-Suty C, Morris AJ, Habib G, Almirante B, Sexton DJ, Corey GR, Fowler VG, Heart 2008, 95, 570–576.18952633 [Google Scholar]
- [6].Farha MA, Leung A, Sewell EW, D’Elia MA, Allison SE, Ejim L, Pereira PM, Pinho MG, Wright GD, Brown ED, ACS Chem. Biol. 2013, 8, 226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Foxley MA, Wright SN, Lam AK, Friedline AW, Strange SJ, Xiao MT, Moen EL, Rice CV, ACS Med. Chem. Lett. 2017, 8, 1083–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cherifi S, Byl B, Deplano A, Nonhoff C, Denis O, Hallin M, J. Clin. MicroBiol. 2013, 51, 1541–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].a) Snowden JN, Beaver M, Smeltzer MS, Kielian T, Infect. Immun. 2012, 80, 3206–3214; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Heim CE, Hanke ML, Kielian T, in Methods in Molecular Biology, Humana Press, 2013, pp. 183–191. [DOI] [PubMed] [Google Scholar]
- [10].Odds FC, J. Antimicrob. Chemother. 2003, 52, 1–1. [DOI] [PubMed] [Google Scholar]
- [11].Lowdin E, Odenholt I, Cars O, Antimicrob. Agents Chemother. 1998, 42, 2739–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Busetti A, Crawford DE, Earle MJ, Gilea MA, Gilmore BF, Gorman SP, Laverty G, Lowry AF, McLaughlin M, Seddon KR, Green Chem. 2010, 12, 420. [Google Scholar]
- [13].a) Knox JR, Pratt RF, Antimicrob. Agents Chemother. 1990, 34, 1342–1347; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hartman BJ, Tomasz A, J. Bacteriol. 1984, 158, 513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].a) Mann PA, Müller A, Wolff KA, Fischmann T, Wang H, Reed P, Hou Y, Li W, Müller CE, Xiao J, Murgolo N, Sher X, Mayhood T, Sheth PR, Mirza A, Labroli M, Xiao L, McCoy M, Gill CJ, Pinho MG, Schneider T, Roemer T, PLoS Pathog. 2016, 12, e1005585; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang H, Gill CJ, Lee SH, Mann P, Zuck P, Meredith TC, Murgolo N, She X, Kales S, Liang L, Liu J, Wu J, Santa Maria J., Su J, Pan J, Hailey J, McGuinness D, Tan CM, Flattery A, Walker S, Black T, Roemer T, Chem. Biol. 2013, 20, 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Foxley MA, Friedline AW, Jensen JM, Nimmo SL, Scull EM, King JB, Strange S, Xiao MT, Smith BE, Thomas KJ III, Glatzhofer DT, Cichewicz RH, Rice CV, J. Antibiot. 2016, 69, 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Antibiotic/Antimicrobial Resistance (AR/AMR), Biggest Threats and Data, Centers for Disease Control and Prevention (CDC), US Department of Health and Human Services, 2013: http://www.cdc.gov/drugresistance/threat-report-2013/. [Google Scholar]
- [17].Vuong C, Otto M, Microbes Infect. 2002, 4, 481–489. [DOI] [PubMed] [Google Scholar]
- [18].Pasquina LW, Santa Maria J. P., Walker S, Curr. Opin. MicroBiol. 2013, 16, 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Campbell J, Singh AK, Santa Maria J. P., Kim Y, Brown S, Swoboda JG, Mylonakis E, Wilkinson BJ, Walker S, ACS Chem. Biol. 2011, 6, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].a) Swoboda JG, Meredith TC, Campbell J, Brown S, Suzuki T, Bollenbach T, Malhowski AJ, Kishony R, Gilmore MS, Walker S, ACS Chem. Biol. 2009, 4, 875–883; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) D’Elia MA, Millar KE, Beveridge TJ, Brown ED, Bacteriol J.. 2006, 188, 8313–8316; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Bhavsar AP, Erdman LK, Schertzer JW, Brown ED, J. Bacteriol. 2004, 186, 7865 – 7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lee SH, Wang H, Labroli M, Koseoglu S, Zuck P, Mayhood T, Gill C, Mann P, Sher X, Ha S, Yang S-W, Mandal M, Yang C, Liang L, Tan Z, Tawa P, Hou Y, Kuvelkar R, DeVito K, Wen X, Xiao J, Batchlett M, Balibar CJ, Liu J, Xiao J, Murgolo N, Garlisi CG, Sheth PR, Flattery A, Su J, Tan C, Roemer T, Sci. Transl. Med. 2016, 8, 329–332. [DOI] [PubMed] [Google Scholar]
- [22].Mandal M, Tan Z, Madsen-Duggan C, Buevich AV, Caldwell JP, Dejesus R, Flattery A, Garlisi CG, Gill C, Ha SN, Ho G, Koseoglu S, Labroli M, Basu K, Lee SH, Liang L, Liu J, Mayhood T, McGuinness D, McLaren DG, Wen X, Parmee E, Rindgen D, Roemer T, Sheth P, Tawa P, Tata J, Yang C, Yang S-W, Xiao L, Wang H, Tan C, Tang H, Walsh P, Walsh E, Wu J, Su J, J. Med. Chem. 2017, 60, 3851–3865. [DOI] [PubMed] [Google Scholar]
- [23].a) Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F, Lancet 2006, 367, 731–739; [DOI] [PubMed] [Google Scholar]; b) Hanssen AM, Kjeldsen G, Sollid JUE, Antimicrob. Agents Chemother. 2003, 48, 285–296; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Marraffini LA, Sontheimer EJ, Science 2008, 322, 1843–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Raad I, Hanna H, Maki D, Lancet Infect. Dis. 2007, 7, 645–657. [DOI] [PubMed] [Google Scholar]
- [25].a) Gagnon RF, Richards GK, Subang R, ASAIO J. 1992, 38, M596– M599; [DOI] [PubMed] [Google Scholar]; b) Raad I, Hanna H, Jiang Y, Dvorak T, Reitzel R, Chaiban G, Sherertz R, Hachem R, Antimicrob. Agents Chemother. 2007, 51, 1656–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].a) Sieradzki K, Roberts RB, Serur D, Hargrave J, Tomasz A, J. Clin. MicroBiol. 1999, 37, 39–44; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Schwalbe RS, Stapleton JT, Gilligan PH, New Engl. J. Med. 1987, 316, 927–931. [DOI] [PubMed] [Google Scholar]