Abstract
This study introduces a new approach for assessing the effects of pediatric epilepsy on a language connectome. Two novel data‐driven network construction approaches are presented. These methods rely on connecting different brain regions using either extent or intensity of language related activations as identified by independent component analysis of fMRI. An auditory word definition decision task paradigm was used to activate the language network for 29 patients and 30 controls. Evaluations illustrated that pediatric epilepsy is associated with a network efficiency reduction. Patients showed a propensity to inefficiently use the whole brain network to perform the language task; whereas, controls seemed to efficiently use smaller segregated network components to achieve the same task. To explain the causes of the decreased efficiency, graph theoretical analysis was performed. The analysis revealed substantial global network feature differences between the patients and controls for the extent of activation network. It also showed that for both subject groups the language network exhibited small‐world characteristics; however, the patient's extent of activation network showed a tendency toward randomness. It was also shown that the intensity of activation network displayed ipsilateral hub reorganization on the local level. We finally showed that a clustering scheme was able to fairly separate the subjects into their respective patient or control groups. The clustering was initiated using local and global nodal measurements. Compared to the intensity of activation network, the extent of activation network clustering demonstrated better precision. This ascertained that the network differences presented by the networks were associated with pediatric epilepsy. Hum Brain Mapp 35:5996–6010, 2014. © 2014 Wiley Periodicals, Inc.
Keywords: functional network, language network, graph theory, functional magnetic resonance imaging, independent component analysis
INTRODUCTION
Recent studies have focused on exploring the morphology as well the functional activities of the brain as intricate and often subtle networks of interconnected elements [Essen et al., 2013; Fornito et al., 2013; Hosseini and Kesler, 2013; Sporns, 2011, 2013; Varoquaux and Craddock, 2013]. Such elements might be structural or functional depending on the study's objectives. Graph theory [Chartrand, 1985] has played an essential role in offering a formal framework to study such networks. For example, language activation patterns, as captured by functional MRI (fMRI), have been studied extensively in healthy subjects [Gaillard, 2004; Wu et al., 2013]. Yet, new insights are gained when looking at these activation patterns from a graph perspective.
Additionally, other studies have highlighted the possible effects of different neurological disorders on different brain networks. For example, epilepsy [Killory et al., 2011; Liao et al., 2010a, 2010b; Wang et al., 2012], Parkinson's disease [Wu et al., 2009], aphasia [Sonty et al., 2007], attention deficit hyperactivity disorder [Fair, 2012; Yu‐Feng et al., 2007], Alzheimer's disease [Greicius et al., 2004], and depression [Zeng et al., 2013, 2012]. Epilepsy is a “network disease” [Bonilha et al., 2012], and its effects have consequently been investigated by functional connectivity networks [Vlooswijk et al., 2010; Waites et al., 2006; Zhang et al., 2009], morphological or structural networks [Bernhardt et al., 2011], or a combination of them [Zhang et al., 2011].
Most functional networks studies rely on resting state fMRI [Bettus et al., 2010; Chen et al., 2011; Doucet et al., 2012; Hosseini and Kesler, 2013]. Functional connectivity within the language network is reduced in adults with epilepsy and is associated with worse language performance [Besseling et al., 2013; Pravatà et al., 2011; Vlooswijk et al., 2010; Waites et al., 2006]. Less is known about functional connectivity within language areas in pediatric epilepsy populations. Language networks are commonly disrupted in patients with epilepsy; this disruption is present early in the course of the disease; furthermore, most young adult focal epilepsy has onset in late childhood and adolescence [Berl et al., 2014; Gaillard et al., 2007]. Functional connectivity during language tasks has not been investigated in children [Parkinson 2002; Steinberg et al., 2013], nor has graph theory been applied to these functional analyses. Therefore, the focus in this study is directed at language network connectivity in older children and adolescents, during a language task, from a graph theoretical perspective. The goal was threefold: (1) constructing a data‐driven functional brain network by connecting different spatially independent units generated by independent component analysis (ICA) [Comon, 1994]; (2) investigating the functional network characteristics using graph theory by identifying and extracting relevant features for the language task at hand; and (3) studying the network topology and changes induced by pediatric epilepsy on the global and local levels.
The fMRI ICs were used to construct two distinct networks for each subject. The networks were constructed, capturing the extent and intensity of language related activation separately. These large‐scale whole brain networks were used to study the effects of epilepsy on the network reorganization and topology. Afterward, certain network features were used to blindly cluster the subjects into two groups representing the patients and controls. The unsupervised clustering was to ascertain that the disease induced, or was associated with, the changes observed in the network features.
METHODS
Participants and Data Collection
The data used in this study was collected by a multisite consortium and repository for pediatric epilepsy. This repository is hosted at our institution (http://mri-cate.fiu.edu) to study the effects of pediatric epilepsy on the brain structure and function [Lahlou et al., 2006]. The datasets selected for this study came from three leading pediatric hospitals: British Columbia Children's Hospital, Children's Healthcare of Atlanta, and Children's National Medical Center. All three locations used a 3‐Tesla Siemens Trio MRI scanner. Institutional review board requirements were followed where the parents gave written informed consent and children gave assent. The datasets were deidentified to insure confidentiality.
All subjects underwent fMRI data acquisition while performing an Auditory Descriptive Decision Task (ADDT) devised to stimulate the temporal and the inferior frontal cortex as described in [Berl et al., 2012; Gaillard et al., 2007]. The subjects were shown an object and then were subjected to an auditory stimulus describing the object. If the description matched the object the subjects were instructed to press the “True” button, otherwise they were instructed to press the “False” button. The description was repeated every 3‐sec period where a “True” pair appeared pseudorandomly with a 70% chance. At rest the subjects listened to description in reverse speech and were coached to press a button on beeps generated following the audio. Furthermore, the difficulty of the paradigm was adjusted appropriately to match the subject's age group.
A total of 29 pediatric epilepsy patients and 30 age and sex matched control subjects were recruited. The patients were between 9.5 and 18.5 years with an average age of 13.5 ± 2.45 years, 14 females, 21 right‐handed, 17 with a remote symptomatic seizure etiology, 5.5 ± 4.70 years average age of first seizure, 9 ± 4.12 years average age at habitual seizure onset, 24 left hemispheric focus, four right hemispheric focus, one bilateral focus; all patients went through presurgical evaluation, and 13 patients underwent epilepsy surgery. None of the patients had any seizure for 24 h prior to the fMRI study. The controls were between 10 and 20 years of age with an average of 13.5 ± 2.98 years, 14 females, 18 right‐handed, and 12 with unknown handedness. All controls were native English speakers free of any current or past neurological or psychiatric disease. Furthermore, there were six patients with right language laterality whereas all controls had left language laterality. Table 1 summarizes the demographics.
Table 1.
Summary of the demographics for patients and controls
| Age (years) | Females | Handedness | Language laterality | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Min | Max | Average | STD | Right | Left | Unknown | Left | Right | ||
| Patients (29) | 9.5 | 18.5 | 13.5 | 2.45 | 14 | 21 | 6 | 2 | 23 | 6 |
| Controls (30) | 10 | 20 | 13.5 | 2.98 | 14 | 18 | 0 | 12 | 30 | 0 |
To ascertain that the chosen demographics do not have an effect on the study, statistical tests were performed on the age, gender, and language laterality. There was no significant age different between the subject groups with P = 0.614. Similarly, no significant gender difference was found between the subject groups as tested with Fisher's exact test (two‐sided P = 1, one‐sided P = 0.554). There was slightly significant language laterality difference between the subject groups using Fisher's exact text (two‐sided P = 0.011, one‐sided P = 0.011).
The language laterality index was calculated by combining a bootstrap procedure with a histogram analysis [Wang et al., 2013a; Wilke and Schmithorst, 2006]. The masked areas were thresholded, and then data was obtained for the left and right sides. The data was converted into a vector whose elements were used in a bootstrapped resampling method to compute the lateralization index as given in Eq. (1):
| (1) |
All the possible LI values were plotted in a histogram, from which the mean of the 50% central values was used as the selected LI value. The threshold was obtained by the mean intensity of the voxels in the image. The toolbox used for these calculations is described in [Wilke and Lidzba, 2007].
In addition, a high resolution isotropic structural T1 MRI scan was acquired for each subject. These T1 scans aid in registering each subject's fMRI space to a common stereotaxic space, which is defined by a brain template.
fMRI Preprocessing
Each subject's fMRI dataset was preprocessed using the FMRIB Software Library [Jenkinson et al., 2012; Smith et al., 2004] as follows: temporally high pass filtered with a cutoff frequency of 0.01 Hz to remove the MRI scanner's baseline wandering effect, head motion corrected using MCFLIRT [Jenkinson et al., 2002], slice time corrected, deskulled to remove nonbrain tissues using brain extraction tool (BET) [Smith, 2002], and spatially smoothed with a 5 mm full width at half maximum to increase the signal to noise ratio. Datasets were inspected to ascertain that head motion did not exceed 1 mm in any of the major axes directions. Afterward, each of the preprocessed datasets was passed to the probabilistic IC analysis (PICA) algorithm to get its spatially independent latent sources as implemented in MELODIC [Beckmann, 2012; Beckmann and Smith, 2004].
Coregistering the AAL90 to the fMRI Space
The Automated Anatomical Labeling (AAL90) atlas [Tzourio‐Mazoyer et al., 2002], which includes 90 cortical and subcortical regions in the MNI152 space, was registered to each subject's fMRI space; afterward, it was superimposed over each of the subject's spatially ICs. For each subject the registration was as follows: deskulling of the fMRI dataset, deskulling of its corresponding T1 both using BET, registering the deskulled fMRI to the deskulled T1 using an affine transformation (12 degrees of freedom), registering the deskulled T1 to the MNI152 brain also using an affine transformation. The two aforementioned registration steps were concatenated into a single transformation matrix. This single matrix was then inverted and applied to register the AAL90 atlas to the subject's fMRI space. Each registration step was performed using the FMRIB's Linear Image Registration Tool (FLIRT) [Jenkinson et al., 2002].
Functional Network Construction
Networks or graphs consist of a group of nodes connected by edges. The goal in this implementation step was to determine a functional brain network, which is essentially defining the nodes and connecting them by valid edges. The node selection process has an impact on the nature of the resulting networks; however, when comparing groups of subjects using the same selection process the effect is counteracted [Zalesky et al., 2010]. Consequently, the nodes were defined as the different 90 regions of the AAL90 atlas. These nodes will be comparable across subjects. In this study, two novel methods to connect these nodes based on the extent and the intensity of activation are proposed.
Extent of activation network
After coregistering the AAL90 atlas to the subject's fMRI space, the atlas is superimposed over each spatial IC. Hence, each of the 90 atlas regions will have several accompanying ICs. For each of these regions we count the number of activated voxels at each IC. The temporal profile of the IC with the highest activated voxel count is associated with the particular atlas region. Eventually, every region in the atlas will be associated with a single temporal signal. A 90 × 90 correlation matrix is thus constructed using Pearson's correlation between the temporal signals for each subject. The resulting matrix is a graph adjacency matrix representing the functional brain network of the subject at hand while capturing its extent of activation.
Intensity of activation network
Similarly, an adjacency matrix capturing the intensity of activation can be constructed by repeating the same steps as in section Extent of Activation Network, but instead of counting the number of activated voxel, the average absolute z values of the activated voxels is computed within each region at each IC. The resulting graph adjacency matrix represents the functional brain network of the subject at hand while capturing its intensity of activation.
Thresholding the Adjacency Matrix
All graphs constructed using either of the connectivity methods, extent or intensity, are undirected weighted graphs. In this study, the focus was placed on connectivity whether two nodes were connected or not; therefore, the absolute value of the adjacency matrix was thresholded to an unweighted form. In this section, a thresholding scheme based on graph density was used to facilitate the selection of an objective threshold comparable across all subjects. A graph density can be defined as:
| (2) |
For a fully connected graph, where all nodes are connected directly to all other nodes, D = 1. On the other extreme, a disconnected graph will yield D = 0. Density is thus a measure of a network's wiring cost. Therefore, thresholding using this measure would facilitate comparison across networks. For example, two brain networks with the same density will have the same number of nodes and the same number of edges (same wiring cost). However, the edges are shifted reflecting the state of the subject's network.
Figure 1 shows the relation between thresholding using Pearson's correlation coefficient and the graph density. Clearly, higher correlation threshold values lead to lower density graphs. Thus, a bidisectional algorithm is used to determine the corresponding correlation threshold value given a selected density.
Figure 1.
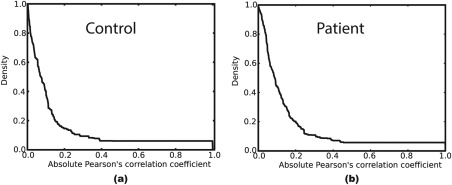
The thresholding relationship between the absolute value of Pearson's correlation coefficient and its corresponding graph density applied to a (a) control's and (b) patient's adjacency matrix. The figure clearly shows that higher correlation thresholds lead to a less dense graph.
To justify using the graph density as opposed to the correlation values for thresholding, raw histogram correlation values of the patients and the controls are used to gauge both extent and intensity of the activation distributions in terms of Pearson's correlation versus number of edges. Figure 2 shows these histograms for the ICA‐based methods in contrast to a traditional network construction method, which uses the blood‐oxygen‐level dependent (BOLD) average within each region of the atlas as the representative temporal profile.
Figure 2.
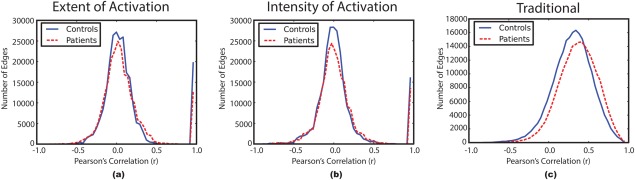
Histograms of the combined correlation values for the patients and controls groups. Controls: solid blue, Patients: dashed red. (a) Extent of activation network, (b) intensity of activation network, and (c) traditional network. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Figure 2 illustrates a slight distinction between the patients and controls for the traditional method, which agrees with [Fornito et al., 2013] in schizophrenia; whereas, our ICA‐based methods produced correlation values that are comparable between the subject groups. This makes the density threshold selection more neutral toward the correlation value. In other words, selecting a specific density threshold for all subjects will produce correlation thresholds that are similar between the groups; whereas, the traditional method produces different correlation values for the same density value in each subject group. Using this approach, the ambiguity as to which threshold is to be selected (Pearson's correlation or density) is resolved.
A density threshold that guarantees a connected graph with the cheapest wiring cost was selected for our study. A connected graph is a network where every node will have a path to any other node in the network, directly or indirectly. To measure a graph connectedness, we use the normalized size of the largest connected component. A connected component in a graph is a group of nodes that can reach each other within the component; that is, in a connected graph all nodes can reach each other and the largest connected component contains all nodes, hence its normalized size is 1. Similarly, if there are several connected components within the network, which do not have any connecting links, then the size of the largest connected component will be less than 1.
For each network construction method, the normalized size of the largest connected components is computed for every subject across a density range (from 10 to 70%) assumed on the basis of the results in Figure 3. The figure shows that density thresholds of 65 and 55% will guarantee a connected graph (normalized size of the largest connected component = 1) for all subjects in the extent of activation network and intensity of activation network, respectively.
Figure 3.
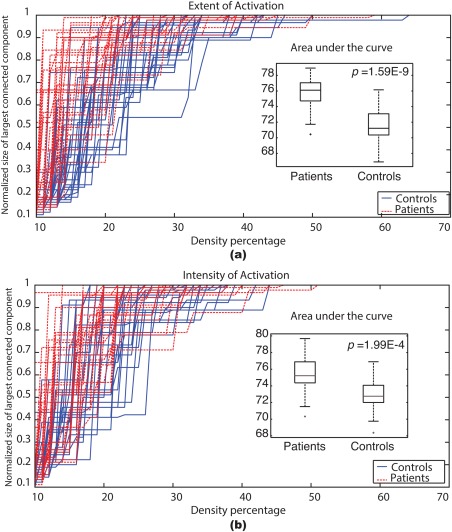
The normalized size of the largest connected component across a range of graph densities (10–70%) for each subject. Controls: solid blue, Patients: dashed red. The insets show box plots of the areas under the curve for the patients and the controls after excluding the outliers. (a) Extent of activation network, (b) intensity of activation network. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Networks with a density higher than 50% are not biological and tend to be random [Hosseini et al. 2012; Kaiser and Hilgetag, 2006]. Figure 3 shows that only two patients and one control subject exceed the 50% density threshold; therefore, these subjects are considered outliers and are excluded from further analyses. Consequently, all networks, regardless of the method, are thresholded using a 50% density threshold and the resulting binary graphs are used for further studies. Moreover, this threshold is in the range suggested in [Reus and van den Heuvel, 2013] for structural networks. Finally, the areas under the curves in Figure 3 are determined to investigate any differences associated with epilepsy.
Graph Measures
Graph theoretical analysis was performed on both functional network methods. Graph theory provides many global and local quantitative measures to analyze the brain network dynamics. The following succinctly summarizes the measures used in this study. Most of these measures were implemented using functions from the Python NetworkX library [Hagberg et al., 2008 2008].
Clustering coefficient
The clustering coefficient ci of a node ni captures the cliquishness within its neighborhood ki. The clustering coefficient of a node in a binary undirected graph is calculated as the fraction of the number of edges Ei to the total number of possible edges within the node's neighborhood [Watts and Strogatz, 1998]:
| (3) |
Thereafter, the clustering coefficient of the whole network was calculated as the average clustering coefficient of all its constituent nodes n:
| (4) |
Characteristic path length
The shortest path length d(ni, nj) between all ni and nj where i ≠ j was calculated. Thereafter, these path lengths were averaged to get the characteristic path length as expressed in Eq. (5)
| (5) |
Small‐world index
Each subjects' functional brain network was compared to 100 null networks [Hosseini et al. 2012; Maslov and Sneppen, 2002]; these null networks had the same degree and degree distribution of the compared network. The averages of the 100 clustering coefficients C random and the characteristic path lengths L random were compared to the subject's brain clustering coefficient C net and characteristic path length L net. Compared to random networks, a small‐world network has greater clustering coefficient γ = C net/C random > 1 and similar characteristic path length λ = L net/L random ≈ 1 [Watts and Strogatz, 1998]. The two small‐world features can be combined into a single scalar index σ = γ/λ which is typically greater than 1 for small‐world networks.
Degree centrality
The normalized degree centrality C D of a node ni in a graph with n nodes is simply the fraction of the number of connection ki the node has over the total number of possible connections (n − 1)
| (6) |
Eigenvector centrality
Eigenvector centrality measures the importance of a node in a graph; it is a referential measure that gives higher values to nodes that connect to higher value nodes. In other words, it assigns high values to nodes communicating with central nodes in the network [Bonacich, 1987].
Unsupervised Clustering
This step aims at classifying the subjects into distinct groups in an unsupervised and data‐driven approach. A dual‐level clustering scheme was used, the first level consisted of three self‐organizing map (SOM) classifiers; whereas, the second level, which aggregates and clusters the outputs of the first level, consisted of a single k‐means classifier with k = 2. Figure 4 describes the structure of this classification system. Three feature vectors were computed for each subject based on the degree centrality, clustering coefficient, and eigenvector centrality; each of these feature vectors was set up as 1 × 90‐dimentional vector on the basis of the AAL90 atlas. Next, each feature vector was used as an input for one of the SOM classifiers. SOM is a type of artificial neural network. In this study, 500 training steps were used and an initial neighborhood size of 3 was assumed.
Figure 4.
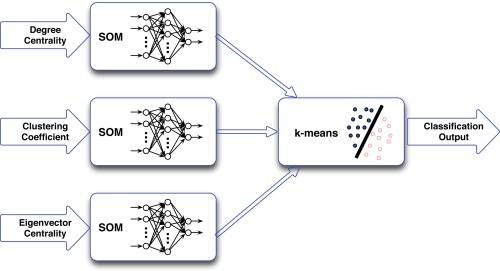
Schematic diagram of the dual‐level classification system. The system consists of three SOM classifiers followed by a single k‐means classifier. The input feature vectors of each patient were: degree centrality, clustering coefficient, and eigenvector centrality. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
RESULTS
Density and Thresholding Analyses
Figure 3 shown earlier provided the normalized size of the largest connected component as a function of graph density. It illustrates that both the extent of activation networks and the intensity of activation networks were able to delineate the patients from the controls. Furthermore, for both networks, most patients had a very large connected component with relatively small density values. This observation implies that patients' brain tend to use more regions to perform the language task; whereas, the controls tend to compartmentalize the brain into separate smaller connected components when performing the ADDT task. To confirm these observations statistically, we calculated the area under the curve for each subject then we compared the patient population to the control population using a boxplot and a t‐test. The t‐test confirmed that both networks could separate the patients/controls groups with a P = 1.59E‐9 for the extent of activation networks and a P = 1.99E‐4 for the intensity of activation networks. The boxplots provided as insets in Figure 3 show that patients have a greater area as compared to controls.
Global Network Features
To study the brain language network general dynamics and its topology, the global network features were assessed. The average clustering coefficients and characteristic path length were calculated for both networks. The extent of activation network showed a significant clustering difference between the patient and control groups P = 7.258E‐5; whereas, the intensity of activation network did not yield a significant clustering difference P = 0.47. Similarly, the extent of activation network showed a significant characteristic path length difference between the patients and controls P = 0.004; whereas, the intensity of activation network showed no significant path length difference P = 0.22. Figure 5 summarizes these findings as box plots.
Figure 5.
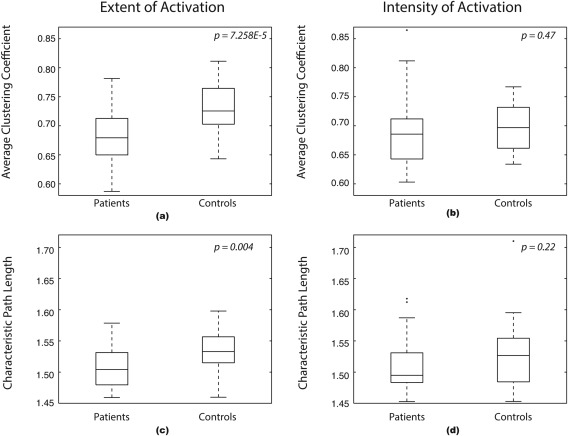
Global network features box plots. (a) Average clustering coefficient of the extent of activation network, (b) average clustering coefficient of the intensity of activation network, (c) Characteristic path length of the extent of activation network, and (d) characteristic path length of the intensity of activation network.
The functional network topology of each subject was investigated by comparing it to 100 null random networks as explained earlier. The average γ = C net/C random for all subjects was 1.71 ± 0.11 for the extent of activation and 1.68 ± 0.09 for the intensity of activation networks. Similarly, the average λ = L net/L random of all subjects for the extent of activation and intensity of activation was 0.94 ± 0.01 and 0.93 ± 0.02, respectively. Subsequently, each network method thresholded with the 50% density threshold resulted in small‐world indices σ = γ/λ greater than 1 with P = 1.0E‐13 and averaged 1.82 ± 0.1 and 1.79 ± 0.08 for the extent of activation and intensity of activation, respectively.
We found out that each of the network construction methods generated small‐world networks. To distinguish the topology differences between the subject groups, we compared their small‐world network parameters using t‐tests. For the extent of activation network the clustering parameter γ was higher for controls as compared to patients with P = 4.21E‐6; similarly, the path length parameter λ was higher for controls as compared to patients with P = 0.007. Consequently, the small‐world index σ was higher for controls compared to patients P = 1.21E‐5. On the contrary, there was no significant difference between the groups for any of the small‐world parameter in the intensity of activation network P = 0.019 for γ, P = 0.284 for λ, and P = 0.021 for σ. Recall that σ = γ/λ.
Local Network Features
After investigating the global network features, the focus was shifted to the individual node features and the association of epilepsy with such features. Two local network features were used: degree centrality and eigenvector centrality. Each feature was calculated for every node, and then the subjects were separated into patients and controls groups. A t‐test was used to highlight nodes that were different between the groups. To control for the multiple testing error rate (90 t‐tests), the Benjamini–Hochberg (BH) method [Benjamini and Hochberg, 1995] was used at 10% false discovery rate (FDR) for each network.
Figure 6 shows the local network differences between the subject groups on the nodal level. The figure illustrates that for the extent of activation network only the eigenvector centrality of the right inferior temporal gyrus was significantly higher for patients compared to controls. Similarly, Figure 6 shows that for the intensity of activation network the degree centrality of the left anterior cingulate gyrus, left fusiform gyrus, and left thalamus was higher for patients compared to controls.
Figure 6.
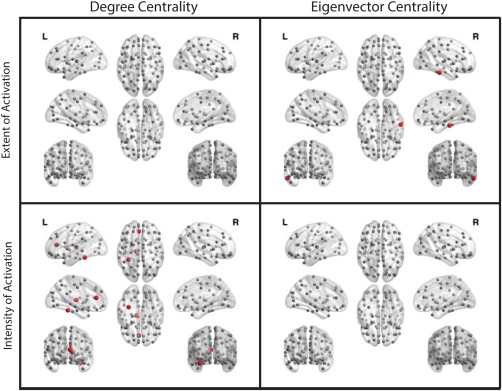
Local network differences at 10% FDR. For the extent of activation networks, first row, and for the intensity of activation networks, second row. Red nodes indicate patients have greater centrality values compared to controls; while, gray nodes indicate no significant difference between the groups. The figure was generated using BrainNet Viewer [Xia et al., 2013]. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Moreover, Figure 6 shows that except for the right inferior temporal gyrus in the eigenvector centrality measure of the extent of activation network, all other significant nodes were left hemispheric. Additionally, Figure 6 illustrates that all significant nodes had greater values for patients compared to controls.
To examine if the local nodal differences or the hemispheric disparity observed above can be explained by atypical language laterality, the effect of the laterality index, as defined in Eq. (1), on the centrality measures was tested for all significant nodes. No effect was found for either of the networks at 5% FDR using the BH method [Benjamini and Hochberg, 1995]. In other word, atypical language laterality does not seem associated with local hub shifts. It is noted, however, that only six patients had right language laterality, whereas the rest of the patients and all controls had typical left language laterality. Additionally, the epileptogenic focus was left hemispheric for 24 patients, right hemispheric for four patients, and one patient had a bilateral focus.
Clustering Results
Several studies showed that epilepsy affects brain language networks [Campo et al., 2013; Vlooswijk et al., 2010, 2011; Wang et al., 2013a; You et al., 2013, 2011]. In this study global and local graph measures were used as feature vectors in a dual‐level clustering scheme. Figure 7a,b illustrate the results of the unsupervised clustering for the extent of activation network and intensity of activation network, respectively. These results show that the extent of activation network produced better clustering outcomes compared to those produced from the intensity of activation network. Table 2 enumerates the confusion matrices of both networks and confirms this observation. The table shows that the extent of activation network clustering has 70.00% precision, 77.78% sensitivity, 68.97% specificity, and 73.21% accuracy. Conversely, the intensity of activation network clustering has 62.07% precision, 66.67% sensitivity, 62.07% specificity, and 64.29% accuracy.
Figure 7.
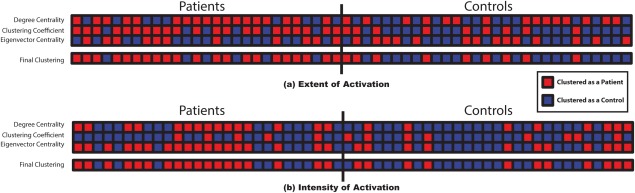
Clustering results for (a) the extent of activation network and (b) the intensity of activation network. The first three lines of each subfigure illustrate the results of the SOMs for the: degree centrality, clustering coefficent, and eigenvector centrality feature vectors in order. The last line shows the final and second level k‐means clustering results. The subjects on the left side are patients and the subjects on the right side are controls. A red square represents a subject clustered as a patient; whereas, a blue square represents a subject clustered as a control. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Table 2.
Clustering confusion matrix for the extent of activation and intensity of activation networks
| Extent of activation | Intensity of activation | ||||||
|---|---|---|---|---|---|---|---|
| Clustered as | Clustered as | ||||||
| Patients | Controls | Patients | Controls | ||||
| Patients | 21 | 6 | Patients | 18 | 9 | ||
| Actual | Controls | 9 | 20 | Actual | Controls | 11 | 18 |
| Precision | 70.00% | Precision | 62.07% | ||||
| Sensitivity | 77.78% | Sensitivity | 66.67% | ||||
| Specificity | 68.97% | Specificity | 62.07% | ||||
| Accuracy | 73.21% | Accuracy | 64.29% | ||||
DISCUSSION
In this study, two novel data‐driven network construction methods were introduced. We used these methods to study the association of epilepsy with the changes in the whole brain language network during a language task. The results showed that epilepsy is associated with network changes where the patients showed less efficient networks compared to controls. These network changes manifested on the global and topological levels for the extent of activation network but not for the intensity of activation networks. Conversely, the intensity of activation network revealed changes on the local level. Topologically, all networks were shown to have small‐world network architecture with subtle differences between the subject groups. On the local level, the extent of activation based networks showed ipsilateral nodal centrality reorganization. An unsupervised clustering system was able to objectively separate the patients and controls validating the correlation of epilepsy with the observed network changes.
The thresholding scheme used in this study showed a difference between the patient and control groups. Patients had greater area under the curve for both network construction methods. The greater area under the curve for the patients implies that they can achieve the language task with less dense graphs. In other words, the patients reach a connected graph with smaller densities than controls. Hence, the whole brain language network is less efficient in patients compared to controls. Controls tend to compartmentalize the brain into several smaller connected components as opposed to using the whole brain to achieve a single language task. This decrease in efficiency agrees with other studies that demonstrated effects of epilepsy on memory networks [Campo et al., 2013; Vlooswijk et al., 2011; Voets et al., 2009], on resting state networks [Bettus et al., 2009, 2010; Doucet et al., 2012; Liao et al., 2010a; Mankinen et al., 2012; Morgan et al., 2010; Waites et al., 2006; Zhang et al., 2010], and on language networks [Karunanayaka et al., 2011; Vlooswijk et al., 2010].
Karunanayaka et al. [2011] used a semantic/tone decision fMRI task [Binder et al., 1997] to study the different semantic networks, detected by ICA, and their correlation with the performed task. They concluded that epilepsy negatively affects the left hemispheric language network in patients with left focused temporal lobe epilepsy; furthermore, they showed that epilepsy also altered other nodes in the network in both left and right temporal lobe epilepsy. Likewise, Vlooswijk et al. [2010] used a covert word‐generation and text reading paradigms to study the effect of epilepsy on language networks. They constructed their networks by connecting highly active regions identified by the model‐dependent generalized linear model (GLM) analysis. Similar to our study, they reported a decrease in functional connectivity in the language areas, and general reduction in language performance for patients. Unlike these studies, the findings presented here are based on whole brain functional networks constructed using an auditory word definition decision task. Our networks were data‐driven and proved to be useful at detecting brain dynamics while preserving its temporal characteristics throughout task performance.
To explain the difference that was observed between the patients and controls, global network features were assessed to understand the network dynamics and to define its topology. The extent of activation networks showed significant global differences; whereas, the intensity of activation networks showed no such differences between the subject groups. Consequently, the networks express different information because they were constructed on different bases, namely the extent of activation and intensity of activation.
Moreover, we found, for both network construction methods, the patient and control networks showed a small‐world network topology. The extent of activation network showed a lower small‐world clustering parameter compared to controls indicating the tendency of the patients' network toward a more random network. Conversely, the intensity of activation network did not show any difference between the subject groups. The small‐world network topology has higher information transfer efficiency and better synchronization when compared to random networks; hence, the extent of activation network showed that patients have reduced information transfer efficiency matching the inefficiency we observed through thresholding and on the global level. These findings match other studies that find typical and atypical brain networks have a small‐world architecture and that neurological disorders introduce changes to the network while maintaining the overall small‐world network architecture [Bernhardt et al., 2011; Hosseini and Kesler, 2013; Zhang et al., 2011].
Regardless of the graph global features and topology, it is unrealistic to assume that neurons are optimally and intricately connected knowing the whole brain network topology and structure. It is fair, however, to assume that neurons act on a local information optimization to achieve a certain task as explained in [Fornito et al., 2013]. Therefore, we studied two centrality measures to identify the local differences in language networks between children with epilepsy and normal controls.
A node centrality emphasizes its importance as a hub in the brain information highway. The different centrality measures used in this study convey certain common, yet nonredundant information [Zuo et al., 2012]. In this study, the extent of activation networks showed a single significant node between the centrality measures. Conversely, the intensity of activation networks highlighted several significant nodes located in the left hemisphere.
Moreover, the significant nodes showed an interesting phenomenon where the left hemispheric nodes had greater centrality values for the patients. This observation might be attributed to the effect of pediatric epilepsy on the reorganization of the language network [You et al., 2013, 2011]. It was also shown that language laterality was not correlated with this nodal reorganization; however, the small number of subjects with atypical right language laterality might have biased our observation.
To verify that the global and local network changes were associated with pediatric epilepsy, the two centrality measures in addition to a global nodal feature were used as input vectors to an unsupervised clustering system. The data‐driven clustering system was able to fairly group the subjects into patients and controls for both networks with better results for the extent of activation networks compared to the intensity of activation network. The intensity of activation network yielded clusters with fair precision, which is close to other studies that reported precision values around 75% in: epilepsy [Zhang et al., 2012], attention‐deficit hyperactivity disorder [Colby et al., 2012; Dai et al., 2012], autism [Anderson et al., 2011], Alzheimer's disease [Chen et al., 2011; Wang et al., 2013b], schizophrenia [Bassett et al., 2012], and major depression [Lord et al., 2012; Zeng et al., 2012]. Therefore, as in previous studies [You et al., 2013, 2011], we concur that pediatric epilepsy can cause, or is associated with, local hub shifts in the language network, similar to the shifts shown in the intensity of activation network.
There are certain limitations in our study. Some clinical data such as comorbid conditions was not uniformly assessed and recorded; hence, the effects of comorbidities, such as disorders of executive function or language measures could not be studied. All study patients had focal epilepsy, however, the epilepsy focus, underlying cause, and use of antiepileptic medications differed among our study population. Previous work demonstrated common effects of different seizure focus location and pathology on language network expression [Berl et al., 2014; Gaillard et al., 2007; Stewart et al., 2014]. Future studies should focus on more homogeneous populations to explore further the findings presented here. Some medications, such as Topiramate, may have an effect on the BOLD response [Szaflarski and Allendorfer, 2012]; the effect of antiepileptic medications on network connectivity is not known and remains a subject of additional investigation. The findings here represent associations with epilepsy. It is not clear whether epilepsy, its underlying cause or its treatment, is the primary force underlying the differences observed in this study.
In addition, many computational and statistical methods were implemented in this study. Bayesian implementation could have been used in the multiple comparisons analysis performed in section Local Network Features. However, we elected to adopt the BH method [Benjamini and Hochberg, 1995], which is considered a gold standard in theory and application for controlling the multiple testing error rate, the so‐called FDR. Based on our knowledge, there are several Bayesian methods of multiple comparisons. Yet, they all ought to control or minimize other types of error rate, such as Bayesian FDR (BFDR). As a result, it is unfair to compare the Bayesian method to the BH methods in theory. Empirically, however, simulations in various cases, including the statistical model considered in Section Local Network Features, showed very similar results to BH, except for some rare cases were it showed slightly better results. Several real data applications in gene expression microarrays analysis support these observations [Muller et al., 2004; Sarkar et al., 2008; Wu and Pena, 2013]. The BH method has become widely adopted because of its ease of computation and implementation. It would be very interesting to compare BH method's results shown in this study to the aforementioned Bayesian methods' results, and to develop a computational package that can be used for other similar applications. This could yield a new methodology in the field of applied statistics in the way similar studies could be gauged in future research.
In conclusion, this study has explored the association of epilepsy with changes in a language connectome. Two complementary network construction methods were introduced and useed. Patients with epilepsy showed reduced network efficiency compared to controls. Patients useed the whole brain network to achieve a language task whereas controls used segregated network components to achieve the same task. These network alterations were on the global level for the extent of activation networks and on the local level for the intensity of activation networks. Furthermore, all networks showed a small‐world topology. These observations were confirmed by an unsupervised classification system, which was able to fairly cluster the subjects into their corresponding group. Some clinical data such as, comorbid conditions, IQ, and the effect of antiepileptic medications were not uniformly recorded and evaluated, which pointed out certain limitations. Additionally, the epilepsy type and seizure location of the studied population was inhomogeneous, future studies should direct the focus on a more homogenous population to narrow down our findings.
ACKNOWLEDGMENT
The authors thank all of the children and families who participated in our study.
REFERENCES
- Anderson JS, Nielsen JA, Froehlich AL, Dubray MB, Druzgal TJ, Cariello AN, Cooperrider JR, Zielinski BA, Ravichandran C, Fletcher PT, Alexander AL, Bigler ED, Lange N, Lainhart JE (2011): Functional connectivity magnetic resonance imaging classification of autism. Brain 134:3742–3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Nelson BG, Mueller BA, Camchong J, Lim KO (2012): Altered resting state complexity in schizophrenia. NeuroImage 59:2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann C, Smith S (2004): Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans Med Imag 23:137–152. [DOI] [PubMed] [Google Scholar]
- Beckmann CF (2012): Modelling with independent components. NeuroImage 62:1–11. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995): Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
- Berl MM, Mayo J, Parks EN, Rosenberger LR, Vanmeter J, Ratner NB, Vaidya CJ, Gaillard WD (2012): Regional differences in the developmental trajectory of lateralization of the language network. Hum Brain Mapp 35:270–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berl MM, Zimmaro LA, Khan OI, Dustin I, Ritzl E, Duke ES, Sepeta LN, Sato S, Theodore WH, Gaillard WD (2014): Characterization of atypical language activation patterns in focal epilepsy. Ann Neurol 75:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt BC, Chen Z, He Y, Evans AC, Bernasconi N (2011): Graph‐theoretical analysis reveals disrupted small‐world organization of cortical thickness correlation networks in temporal lobe epilepsy. Cerebral Cortex 21:2147–2157. [DOI] [PubMed] [Google Scholar]
- Besseling RMH, Overvliet GM, Jansen JFA, van der Kruijs SJM, Vles JSH, Ebus, SCM. , Hofman PAM, de Louw AJA, Aldenkamp, AP , Backes WH (2013): Aberrant functional connectivity between motor and language networks in rolandic epilepsy. Epilepsy Res 107:253–262. [DOI] [PubMed] [Google Scholar]
- Bettus G, Guedj E, Joyeux F, Confort‐Gouny S, Soulier E, Laguitton V, Cozzone PJ, Chauvel P, Ranjeva J‐P, Bartolomei F, Guye M (2009): Decreased basal fMRI functional connectivity in epileptogenic networks and contralateral compensatory mechanisms. Hum Brain Mapp 30:1580–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettus G, Bartolomei F, Confort‐Gouny S, Guedj E, Chauvel P, Cozzone PJ, Ranjeva J‐P, Guye M (2010): Role of resting state functional connectivity MRI in presurgical investigation of mesial temporal lobe epilepsy. J Neurol Neurosurg Psychiatry 81:1147–1154. [DOI] [PubMed] [Google Scholar]
- Binder JR, Frost JA, Hammeke TA, Cox RW, Rao SM, Prieto T (1997): Human brain language areas identified by functional magnetic resonance imaging. The J Neurosci 17:353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacich P (1987): Power and centrality: A family of measures. Am J Sociol 92:1170–1182. [Google Scholar]
- Bonilha L, Nesland T, Martz GU, Joseph JE, Spampinato MV, Edwards JC, Tabesh A (2012): Medial temporal lobe epilepsy is associated with neuronal fibre loss and paradoxical increase in structural connectivity of limbic structures. J Neurol Neurosurg Psychiatry 83:903–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo P, Garrido MI, Moran RJ, García‐Morales I, Poch C, Toledano R, Gil‐Nagel A, Dolan RJ, Friston KJ (2013): Network reconfiguration and working memory impairment in mesial temporal lobe epilepsy. NeuroImage 72C:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand G (1985): Introductory graph theory. New York: Dover.
- Chen G, Ward B, Xie C, Li W, Wu Z, Jones J, Franczak M, Antuono P, Li S‐J (2011): Classification of Alzheimer disease, mild cognitive impairment, and normal cognitive status with large‐scale network analysis based on resting‐state functional MR imaging. Radiology 259:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JB Colby, JD Rudie, JA Brown, PK Douglas, MS Cohen, et al.. (2012) Insights into multimodal imaging classification of ADHD. Front Syst Neurosci 6:59 Doi: 10.3389/fnsys.2012.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comon P (1994): Independent component analysis, a new concept? Signal Process 36:287–314. [Google Scholar]
- Dai D, Wang J, Hua J, He H (2012): Classification of ADHD children through multimodal magnetic resonance imaging. Front Syst Neurosci 6:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet G, Osipowicz K, Sharan A, Sperling MR, Tracy JI (2012): Extratemporal functional connectivity impairments at rest are related to memory performance in mesial temporal epilepsy. Hum Brain Mapp 34:2202–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essen DCV, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K, Consortium FTW‐MH (2013): The WU‐Minn human connectome project: An overview. NeuroImage 80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fair DA, Nigg JT, Iyer S, Bathula D, Mills KL, Dosenbach NUF, Schlaggar BL, Mennes M, Gutman D, Bangaru S, Buitelaar JK, Dickstein DP, Di Martino A, Kennedy DN, Kelly C, Luna B, Schweitzer JB, Velanova K, Wang YF, Mostofsky S, Castellanos FX, Milham MP (2012): Distinct neural signatures detected for ADHD subtypes after controlling for micro‐movements in resting state functional connectivity MRI data. Front Syst Neurosci, 6:80. Doi: 10.3389/fnsys.2012.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Breakspear M (2013): Graph analysis of the human connectome: Promise, progress, and pitfalls. NeuroImage 80:426–444. [DOI] [PubMed] [Google Scholar]
- Gaillard WD (2004): Functional MR imaging of language, memory, and sensorimotor cortex. Neuroimaging Clin N Am 14:471–485. [DOI] [PubMed] [Google Scholar]
- Gaillard WD, Berl MM, Moore EN, Ritzl EK, Rosenberger LR, Weinstein SL, Conry JA, Pearl PL, Ritter FF, Sato S, Vezina LG, Vaidya CJ, Wiggs E, Fratalli C, Risse G, Ratner NB, Gioia G, Theodore WH (2007): Atypical language in lesional and nonlesional complex partial epilepsy. Neurology 69:1761–1771. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V (2004): Default‐mode network activity distinguishes Alzheimer's disease from healthy aging: Evidence from functional MRI. Proc Natl Acad Sci USA 101:4637–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AA Hagberg, DA Schult, PJ Swart (2008): Exploring network structure, dynamics, and function using NetworkX. In G. Varoquaux, T. Vaught, J. Millman (Eds.) SciPy 2008: Proceedings of the 7th Python in Science Conference pp 11–15.
- Hosseini SMH, Kesler SR (2013): Comparing connectivity pattern and small‐world organization between structural correlation and resting‐state networks in healthy adults. NeuroImage 78C:402–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini SMH, Hoeft F, Kesler SR (2012): GAT: A graph‐theoretical analysis toolbox for analyzing between‐group differences in large‐scale structural and functional brain networks. PLoS ONE 7:e40709 Doi: 10.1371/journal.pone.0040709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S (2002): Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage 17:825–841. [DOI] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM (2012): Fsl. NeuroImage 62:782–790. [DOI] [PubMed] [Google Scholar]
- Kaiser M, Hilgetag CC (2006): Nonoptimal component placement, but short processing paths, due to long‐distance projections in neural systems. PLoS Comput Biol 2:e95 Doi: 10.1371/journal.pcbi.0020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunanayaka P, Kim KK, Holland SK, Szaflarski JP (2011): The effects of left or right hemispheric epilepsy on language networks investigated with semantic decision fMRI task and independent component analysis. Epilepsy Behav 20:623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killory BD, Bai X, Negishi M, Vega C, Spann MN, Vestal M, Guo J, Berman R, Danielson N, Trejo J, Shisler D, Novotny EJ Jr, Constable RT, Blumenfeld H (2011): Impaired attention and network connectivity in childhood absence epilepsy. Neuroimage 56:2209–2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M Lahlou, MR Guillen, M Adjouadi, WD Gaillard (2006): An online web‐based repository site of fMRI medical images and clinical data for childhood epilepsy. The 11th world congress on internet in medicine Ontario, Canada: Mednet; pp 120–127 [Google Scholar]
- Liao W, Zhang Z, Pan Z, Mantini D, Ding J, Duan X, Luo C, Lu G, Chen H (2010a): Altered functional connectivity and small‐world in mesial temporal lobe epilepsy. PLoS ONE 5:e8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Zhang Z, Pan Z, Mantini D, Ding J, Duan X, Luo C, Wang Z, Tan Q, Lu, G , Chen H (2010b): Default mode network abnormalities in mesial temporal lobe epilepsy: A study combining fMRI and DTI. Hum Brain Mapp 32:883–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord A, Horn D, Breakspear M, Walter M (2012): Changes in community structure of resting state functional connectivity in unipolar depression. PLoS ONE 7:e41282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankinen K, Jalovaara P, Paakki J‐J, Harila M, Rytky S, Tervonen O, Nikkinen J, Starck T, Remes J, Rantala H, Kiviniemi V (2012): Connectivity disruptions in resting‐state functional brain networks in children with temporal lobe epilepsy. Epilepsy Res 100:168–178. [DOI] [PubMed] [Google Scholar]
- Maslov S, Sneppen K (2002): Specificity and stability in topology of protein networks. Science 296:910–913. [DOI] [PubMed] [Google Scholar]
- Morgan V, Gore J, Abou‐Khalil B (2010): Functional epileptic network in left mesial temporal lobe epilepsy detected using resting fMRI. Epilepsy Res 88:168–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Parmigiani G, Robert C, Rousseau J (2004): Optimal sample size for multiple testing: the case of gene expression microarrays. J Am Stat Assoc 99:990–1001. [Google Scholar]
- Parkinson GM (2002): High incidence of language disorder in children with focal epilepsies. Dev Med Child Neurol, 44:533–537. Doi: 10.1111/j.1469-8749.2002.tb00325.x. [DOI] [PubMed] [Google Scholar]
- Pravatà E, Sestieri C, Mantini D, Briganti C, Colicchio G, Marra C, Colosimo C, Tartaro A, Romani GL, Caulo M (2011): Functional connectivity MR imaging of the language network in patients with drug‐resistant epilepsy. Am J Neuroradiol 32:532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reus MA, van den Heuvel MP (2013): Estimating false positives and negatives in brain networks. NeuroImage 70:402–409. [DOI] [PubMed] [Google Scholar]
- Sarkar SK, Zhou T, Ghosh D (2008): A general decision theoretic formulation of procedures controlling FDR and FNR from a Bayesian perspective. Stat Sin 18:925–945. [Google Scholar]
- Smith SM (2002): Fast robust automated brain extraction. Hum Brain Mapp 17:143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S, Jenkinson M, Woolrich M, Beckmann C, Behrens T, Johansen‐Berg H, Bannister P, De Luca M, Drobnjak I, Flitney D, Niazy R, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady J, Matthews P (2004): Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage 23:S208–S219. [DOI] [PubMed] [Google Scholar]
- Sonty SP, Mesulam MM, Weintraub S, Johnson NA, Parrish TB, Gitelman DR (2007): Altered effective connectivity within the language network in primary progressive aphasia. J Neurosci 27:1334–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O (2011): The human connectome: a complex network. Ann N Y Acad Sci 1224:109–125. [DOI] [PubMed] [Google Scholar]
- O Sporns (2013): The human connectome: Origins and challenges. NeuroImage 80:53–61. [DOI] [PubMed] [Google Scholar]
- ME Steinberg, NB Ratner, W Gaillard, M Berl (2013): Fluency patterns in narratives from children with localization related epilepsy. Journal of Fluency Disorders 38:193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CC, Swanson SJ, Sabsevitz DS, Rozman ME, Janecek JK, Binder JR (2014): Predictors of language lateralization in temporal lobe epilepsy. Neurophsychologia 60:93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaflarski JP, Allendorfer JB (2012): Topiramate and its effect on fMRI of language in patients with right or left temporal lobe epilepsy. Epilepsy Behav 24:74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio‐Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M (2002): Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single‐subject brain. NeuroImage 15:273–289. [DOI] [PubMed] [Google Scholar]
- Varoquaux G, Craddock RC (2013): Learning and comparing functional connectomes across subjects. NeuroImage 80:405–415. [DOI] [PubMed] [Google Scholar]
- Vlooswijk MCG, Jansen JFA, Majoie HJM, Hofman PAM, de Krom MCTFM, Aldenkamp AP, Backes WH (2010): Functional connectivity and language impairment in cryptogenic localization‐related epilepsy. Neurology 75:395–402. [DOI] [PubMed] [Google Scholar]
- Vlooswijk MCG, Vaessen MJ, Jansen JFA, de Krom MCFTM, Majoie HJM, Hofman PAM, Aldenkamp AP, Backes WH (2011): Loss of network efficiency associated with cognitive decline in chronic epilepsy. Neurology 77:938–944. [DOI] [PubMed] [Google Scholar]
- Voets NL, Adcock JE, Stacey R, Hart Y, Carpenter K, Matthews PM, Beckmann CF (2009): Functional and structural changes in the memory network associated with left temporal lobe epilepsy. Hum Brain Mapp 30:4070–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites AB, Briellmann RS, Saling MM, Abbott DF, Jackson GD (2006): Functional connectivity networks are disrupted in left temporal lobe epilepsy. Ann Neurol 59:335–343. [DOI] [PubMed] [Google Scholar]
- Wang J, You X, Wu W, Guillen MR, Cabrerizo M, Sullivan J, Donner E, Bjornson B, Gaillard WD, Adjouadi M (2013a): Classification of fMRI patterns‐A study of the language network segregation in pediatric localization related epilepsy. Hum Brain Mapp 35:1446–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zuo X, Dai Z, Xia M, Zhao Z, Zhao X, Jia J, Han Y, He Y (2013b): Disrupted functional brain connectome in individuals at risk for Alzheimer's Disease. Biol Psychiatry 73:472–481. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang Z, Jiao Q, Liao W, Chen G, Sun K, Shen L, Wang M, Li K, Liu Y, Lu G (2012): Impairments of thalamic nuclei in idiopathic generalized epilepsy revealed by a study combining morphological and functional connectivity MRI. PLoS ONE 7:e39701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts D, Strogatz S (1998): Collective dynamics of “small‐world” networks. Nature 393:440–442. [DOI] [PubMed] [Google Scholar]
- Wilke M, Lidzba K (2007): LI‐tool: A new toolbox to assess lateralization in functional MR‐data. J Neurosci Methods 163:128–136. [DOI] [PubMed] [Google Scholar]
- Wilke M, Schmithorst VJ (2006): A combined bootstrap/histogram analysis approach for computing a lateralization index from neuroimaging data. NeuroImage 33:522–530. [DOI] [PubMed] [Google Scholar]
- Wu K, Taki Y, Sato K, Hashizume H, Sassa Y, Takeuchi H, Thyreau B, He Y, Evans AC, Li X, Kawashima R, Fukuda H (2013): Topological organization of functional brain networks in healthy children: differences in relation to age, sex, and intelligence. PLoS ONE 8: e55347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Wang L, Chen Y, Zhao, C. , Li K, Chan P (2009): Changes of functional connectivity of the motor network in the resting state in Parkinson's disease. Neurosci Lett 460:6–10. [DOI] [PubMed] [Google Scholar]
- Wu W, Pena E (2013): Bayes multiple decision functions. Electron J Stat 7:1272–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Wang J, He Y (2013): BrainNet viewer: A network visualization tool for human brain connectomics. PLoS ONE 8:e68910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Adjouadi M, Wang J, Guillen MR, Bernal B, Sullivan J, Donner E, Bjornson B, Berl M, Gaillard WD (2013): A decisional space for fMRI pattern separation using the principal component analysis‐a comparative study of language networks in pediatric epilepsy. Hum Brain Mapp 34:2330–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Adjouadi M, Guillen MR, Ayala M, Barreto A, Rishe N, Sullivan J, Dlugos, D , Vanmeter J, Morris D, Donner E, Bjornson B, Smith ML, Bernal B, Berl M, Gaillard WD (2011): Sub‐patterns of language network reorganization in pediatric localization related epilepsy: A multisite study. Hum Brain Mapp 32:784–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu‐Feng Z, Yong H, Chao‐Zhe Z, Qing‐Jiu C, Man‐Qiu S, Meng L, Li‐Xia T, Tian‐Zi J, Yu‐Feng W (2007): Altered baseline brain activity in children with ADHD revealed by resting‐state functional MRI. Brain Dev 29:83–91. [DOI] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Bullmore ET (2010): Network‐based statistic: Identifying differences in brain networks, NeuroImage 53:1197–1207, ISSN 1053–8119, http://dx.doi.org/10.1016/j.neuroimage.2010.06.041. [DOI] [PubMed] [Google Scholar]
- Zeng L‐L, Shen H, Liu L, Hu D (2013): Unsupervised classification of major depression using functional connectivity MRI. Hum Brain Mapp 35:1630–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L‐L, Shen H, Liu L, Wang L, Li B, Fang P, Zhou Z, Li Y, Hu D (2012): Identifying major depression using whole‐brain functional connectivity: A multivariate pattern analysis. Brain 135:1498–1507. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lu G, Zhong Y, Tan Q, Yang Z, Liao W, Chen Z, Shi J, Liu Y (2009): Impaired attention network in temporal lobe epilepsy: A resting FMRI study. Neurosci Lett 458:97–101. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lu G, Zhong Y, Tan Q, Liao W, Wang Z, Wang Z, Li K, Chen H, Liu Y (2010): Altered spontaneous neuronal activity of the default‐mode network in mesial temporal lobe epilepsy. Brain Res 1323:152–160. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Liao W, Chen H, Mantini D, Ding J‐R, Xu Q, Wang Z, Yuan C, Chen G, Jiao Q, Lu G (2011): Altered functional‐structural coupling of large‐scale brain networks in idiopathic generalized epilepsy. Brain 134:2912–2928. [DOI] [PubMed] [Google Scholar]
- Zhang J, Cheng W, Wang Z, Zhang Z, Lu W, Lu G, Feng J (2012): Pattern classification of large‐scale functional brain networks: Identification of informative neuroimaging markers for epilepsy. PLoS ONE 7:e36733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X‐N, Ehmke R, Mennes M, Imperati D, Castellanos FX, Sporns O, Milham, MP (2012): Network centrality in the human functional connectome. Cereb Cortex 22:1862–1875. [DOI] [PubMed] [Google Scholar]


