Abstract
Hypertrophic cardiomyopathy (HCM) is the most common inherited human heart disease. The same disease has a high prevalence in cats, where it is also suspected to be inherited. More than 1500 variants in MYBPC3, MYH7 and other sarcomeric genes are associated with human HCM, while in cats, only two causative variants in MYBPC3 are currently known. Here, we describe an adult Domestic Shorthair cat with arterial thromboembolism and heart failure that was diagnosed with HCM on necropsy. Sequencing of the coding regions of MYBPC3 and MYH7 revealed 21 variants, of which the MYH7 c.5647G>A (p.(Glu1883Lys)) variant was further analysed, because its orthologous variant had already been reported in a human patient with HCM, but with limited causal evidence. This variant affects the highly conserved assembly competence domain, is predicted in silico to be damaging and was found only once in population databases. Recently, functional studies have confirmed its predicted damaging effect and a paralogous variant in MYH6 has been associated with cardiac disease in humans as well. This report of an orthologous variant in a cat with HCM and its absence in 200 additional cats provides further evidence for its disease-causing nature. As the first report of feline HCM caused by a variant in MYH7, this study also emphasises this gene as a candidate gene for future studies in cats and highlights the similarity between human and feline HCM.
Subject terms: Cardiac hypertrophy, Disease genetics
Introduction
Hypertrophic cardiomyopathy (HCM; Phenotype MIM number: 192600) is the most common inherited human heart disease, affecting at least 1 in 500 people [1]. It is characterised by concentric hypertrophy of the left ventricle that cannot be attributed to secondary causes [2]. The clinical expression is widely variable, ranging from longevity without symptoms to congestive heart failure or sudden death [2]. More than 1500 variants are associated with human HCM, most of them limited to a single patient family [3]. They mainly reside in sarcomeric genes, of which MYBPC3 and MYH7 are the most important, containing 70% of the variants [1].
MYBPC3 (human geneID: 4607) consists of 33 exons and encodes the cardiac myosin-binding protein C of 1274 amino acids. The protein seems to play a key role in the regulation of sarcomeric contractility and may also influence sarcomere structure [4, 5]. Known causative variants in MYBPC3 are either missense or truncating variants [6]. MYH7 (human geneID: 4625) comprises 40 exons and encodes myosin-7 of 1935 amino acids, the dominant myosin heavy chain protein in cardiac and type I skeletal muscle fibres [7, 8]. This protein generates contractile force through hydrolisation of ATP and interactions with actin at the N-terminal head and channels this force through its C-terminal tail to the cytoskeleton [8]. HCM-causing variants in MYH7 are principally missense variants [6]. Missense variants in MYH7 have also been associated with dilated cardiomyopathy, left ventricular non-compaction and skeletal muscle diseases [2, 9]. One such skeletal muscle disease is myosin storage myopathy (MSM), characterised by slowly progressive muscle weakness and hyaline bodies in type I muscle fibres, that is caused by variants in exons 37–40 (exons are numbered like in NG_007884.1) [9].
HCM (OMIA 000515-9685) is the most common heart disease in cats, affecting almost 15% of the feline population [10]. HCM is often associated with certain breeds, for example the Maine Coon, Ragdoll and British Shorthair breeds [11]. Nevertheless, most patients are non-pedigree Domestic Shorthairs [12]. Feline HCM is considered the best spontaneous animal model for human HCM for its similarity in morphology, histopathology and clinical course [13, 14]. A genetic aetiology is suspected in several breeds as well as in Domestic Shorthairs [12]. However, only two causative variants in cats have yet been identified in MYBPC3, XM_019812396.1:c.91G>C in Maine Coons [15] and c.2455C>T in Ragdolls [11]. An orthologue of the latter variant also causes HCM in humans [16].
The aim of this study was to identify the causative variant in cats affected with HCM. Based on the hypothesis that human and feline HCM have a similar genetic cause, we sequenced the coding regions of MYBPC3 (geneID: 101094698) and MYH7 (geneID: 101096736). In this report, we describe the case in which a causal variant was identified. The results from eight other cases that were sequenced, but where no causal variant was found, are described in Supplementary Table 6.
Materials and methods
Clinical examination
A male castrated Domestic Shorthair, estimated to be six years old, was presented to the emergency department of Ghent University’s small animal clinic for acute paraplegia, pain and severe dyspnoea with cyanosis. A physical examination and thoracic and cardiac focused assessment sonography scan were performed [17]. Due to the severe clinical signs and poor prognosis, the cat was euthanised.
Pathological examination
A complete necropsy was performed, including examination and weighing of the heart, and the heart was frozen at −80 °C thereafter. A transverse slice of frozen ventricular tissue was fixated in 4% buffered formaldehyde, paraffin embedded and routinely processed for histopathological examination. Five micrometre sections were stained with haematoxylin-eosin, Von Giesson, Masson’s Trichrome, Prussian blue and Alcian Blue (pH 2.5). Immunohistochemical stainings included elastin (Novocastra lyophilised monoclonal mouse antibody elastin, Cat No. NCL-ELASTIN, Leica biosystems, Newcastle, UK) and smooth muscle actin (monoclonal mouse anti-human smooth muscle actin clone 1A4, Cat No. M085101-2, Dako, Glostrup, Denmark).
Genetic analysis
Genomic DNA was isolated from 100 mg frozen heart tissue from the case by proteinase K digestion and subsequent phenol/chloroform extraction and ethanol precipitation [18]. The purity and quantity of the DNA were determined with the NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The gDNA was first tested for the two known feline HCM-causing MYBPC3 c.91G>C and c.2455C>T variants using an in-house developed probe-based qPCR assay.
Total RNA was isolated from 100 mg frozen heart tissue from the case using the Aurum Total RNA Fatty and Fibrous Tissue Kit, including an on-column DNase digest (Bio-Rad Laboratories, Hercules, CA, USA). The purity and quantity of the RNA were determined with the NanoDrop ND-1000 spectrophotometer, the integrity was assessed by evaluating the 28S/18S rRNA bands on agarose gel and possible DNA contamination was detected with minus-RT-PCR using an in-house developed UBC integrity assay [19]. Reverse transcription was performed on 1 μg high-quality, DNA-free RNA using the ImProm-II Reverse Transcription System (Promega Corporation, Madison, WI, USA) with oligo(dT) and random hexamer primers. Complementary DNA was 10 times diluted with water and 2 μl was used as a template for PCR. The integrity and amplificability of the cDNA were tested with the UBC integrity assay.
Primer pairs were designed to generate amplicons covering the complete coding regions of MYBPC3 (Acc. No.: XM_019812396.1) and MYH7 (Acc. No.: XM_006932746.4) on cDNA using the NCBI Primer-BLAST software [20], checking for primer specificity and avoiding known SNPs. Regions that form secondary structures, as predicted by Mfold [21], were excluded as primer binding site. PCR was performed with Tempase Hot Start polymerase (VWR International, Leuven, Belgium) and amplicons were analysed via agarose gel electrophoresis. Sequencing reactions were performed with the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA) with individual PCR primers as sequencing primers. The sequences were run at Eurofins Genomics (Ebersberg, Germany) and the results were analysed with BioEdit v7.2.5 [22]. For missense variants, the evolutionary conservation of the affected amino acid was checked via ClustalW and quantified with the ConSurf web tool [23] and the functional effect of the substitution was predicted in silico with PROVEAN [24] and PolyPhen-2 [25]. The Exome Variant Server [26], Exome Aggregation Consortium [27] and Genome Aggregation Database (GnomAD) [27] databases were searched for orthologous variants in humans.
A BseRI PCR-RFLP assay was developed to genotype exon 38 of MYH7 (Acc. No.: NC_018728.3; exon 38 spans positions 76166287 to 76166382) for the c.5647G>A (p.(Glu1883Lys)) variant on gDNA and validated by sequencing. The assay was performed on DNA extracted from either whole blood using proteinase K digestion as described in Van Poucke et al. [28]. or from heart tissue as outlined above in 200 additional cats. These were 125 Domestic Shorthairs and 25 cats each from the Ragdoll, Maine Coon and British Shorthair breeds. The resulting fragments were evaluated via gel electrophoresis.
The sequences of the primer pairs and details on the (q)PCR, sequencing and assay reactions are given in Supplementary Tables 1–4.
Results
Clinical features
The six-year-old Domestic Shorthair cat was presented to the university hospital with acute paralysis of first one and then also the other pelvic leg. Three weeks previously, he had developed a cough that did not respond to antibiotics. The owner had noted no other clinical signs. The only information about family members that the owner could provide was that the only littermate, a female, had been found dead suddenly at the age of two years.
On inspection and physical examination the cat had tachypnoea, severe dyspnoea and cyanosis. Femoral pulses were absent and the hind legs were paralysed, cold and extremely painful when manipulated. Lung sounds were muffled ventrally and the dyspnoea made cardiac auscultation very difficult. The cat was hypothermic with a rectal temperature of 36 °C.
Focused thoracic sonography showed multiple B-lines and focused cardiac sonography showed subjective thickening of the left ventricular free wall. The cat was diagnosed with thromboembolism of the distal aorta most likely due to HCM, with a suspicion of pulmonary oedema. No further examinations were performed because of the poor general condition of the cat. Because of the severity of the clinical signs and the poor prognosis, the cat was euthanized.
Pathological features
Pathological examination revealed subjectively hypertrophic skeletal muscles and an enlarged heart that weighed 27.2 gram (normal weight: <20 g) [29]. The left ventricle felt firm on palpation and the left atrium was dilated. Transverse sectioning revealed concentric hypertrophy of the left ventricular wall with narrowing of the ventricular lumen. The thicknesses of the left ventricular free wall, interventricular septum and right ventricular free wall were 12, 9 and 3 mm, respectively. The lungs were congested and oedematous. Serohaemorrhagic fluid was present in the pleural cavity, trachea and nasal cavity. The kidneys showed multiple chronic infarctions.
Histopathological examination of the left ventricle myocardium revealed a diffuse hypertrophy and karyomegaly of the cardiomyocytes with multifocal areas of increased branching. There were also numerous small fibrotic septa intersecting the myocardium (interstitial fibrosis) with adjacent myofibre disarray. The subendocardial region of the left ventricle showed a non-circumferential extensive fibrosis with few scattered small areas of chondroid metaplasia and areas with large aggregates of hemosiderin-laden macrophages (iron-positive on Prussian blue). This thick subendocardial layer stained red on Von Giesson and blue on both Masson’s Trichrome and Alcian blue and showed a gradual increase in tissue density from luminal (loosely arranged collagen fibres) to myocardial (compact collagenous connective tissue). Immunohistochemistry for elastin revealed multifocal large areas of increased elastin deposition within the subendocardial layer. The gross and histopathological lesions are consistent with a HCM with areas of substantial subendocardial replacement fibrosis. Histopathological images of the heart are given in Supplementary Figs. 1–5.
Genetic analysis
The qPCR assay was negative for both known MYBPC3 c.91G>C and c.2455C>T variants. By sequencing the coding region of the cDNA, seven silent and four missense variants in MYBPC3 and nine silent and one missense variant in MYH7 were found. They were all archived in the EVA database (https://www.ebi.ac.uk/eva/?Study-Browser&browserType=sgv; project ID: PRJEB30318; analysis ID: ERZ795310) and are listed in Supplementary Table 5.
Only one variant, MYH7 c.5647G>A (p.(Glu1883Lys)), was found to change an amino acid that is highly conserved, both in its metazoan orthologues and in its feline and human paralogues, and predicted to be deleterious by PROVEAN (score: −2.613) and PolyPhen-2 (HumDiv score: 1.000, HumVar score: 0.978). This variant was absent in the 200 additionally screened cats and its human orthologue was counted once in the 31,396 alleles in the GnomAD database and not found in the other human databases.
Discussion
The physical examination abnormalities in this cat were highly suggestive of thromboembolism of the distal aorta. Most cases of feline arterial thromboembolism are caused by underlying cardiac disease, most commonly HCM [30]. The respiratory signs were compatible with acute pain and with acute, severe congestive heart failure. The history of coughing might also be related to the heart disease, although cardiogenic coughing is rare in cats [31]. Ultrasound examination was limited by the critical condition of the patient, but was indicative for HCM.
In cases of cardiomyopathy with severe fibrosis, there can be a thin line between HCM and restrictive cardiomyopathy, and there is evidence that the two conditions are different phenotypical expressions of the same genetic disease [32]. Restrictive cardiomyopathy can also occur secondary to radiation fibrosis, nutritional deficiencies or infiltrative diseases such as amyloidosis, hypereosinophilia, sarcoidosis or endomyocarditis [33]. In cats, it has also been suggested that it is the result of an end-stage HCM with myocardial failure or infarction [34]. Our case is compatible with a primary HCM because of the heart’s increased weight, size, wall thickness and firmness. It also displayed all histological features predominant for HCM such as cardiomyocytes displaying hypertrophy, karyomegaly, branching and myofibre disarray, admixed with myocardial interstitial fibrosis [35, 36]. The (sub)endocardial fibrosis was classified as a replacement fibrosis (secondary to ischaemic infarction) as the histological features are consistent with chronic granulation/scar tissue and the presence of large aggregates of hemosiderin-laden macrophages is an indication of an old haemorrhage or infarct. The littermate’s history of sudden death is also compatible with HCM, although no necropsy had been performed to identify the cause of death in that cat [37].
Of the 21 variants that were found, only the five missense variants were further investigated, as silent variants are not associated with HCM [2]. The MYBPC3 c.772G>A (p.(Val258Ile)), c.2765C>T (p.(Pro922Leu)) and c.3109G>A (p.(Ala1037Thr)) variants were not considered to be causal, as the effects of the amino acid substitutions were predicted to be benign and each variant was found in the reference sequence of the mouse, rat, horse and at least one species of the Felidae family. The MYBPC3 c.220G>A (p.(Ala74Thr)) variant has been proposed earlier as a causative variant for HCM, but subsequent studies found that this variant is very common and not significantly associated with HCM [38].
This left only the MYH7 c.5647G>A variant as a possible causative variant, which has a human orthologue, NM_000257.3:c.5647G>A (p.(Glu1883Lys); rs121913652). This orthologue was described by Tajsharghi et al. [39] in a consanguineous human family in which three of four siblings were diagnosed with both HCM and MSM as adults. Two patients died of heart failure and the coding regions of MYH7 in the third patient were sequenced. This patient was a homozygote for the variant, but its causality has been disputed because of limited evidence (see the discussion on ClinVar, variation ID: 14121).
The human and feline cases had comparable HCM, but apparently different degrees of skeletal muscle disease. The human patients displayed a short stature, thoracic scoliosis, calf hypertrophy in one case and progressive muscle weakness [39]. In contrast, the feline case had no musculoskeletal abnormalities other than the generalised muscle hypertrophy observed on necropsy. The owner had not observed muscle weakness. This comparative lack of muscle disease might be explained by a species difference in muscle fibre types. As type I muscle fibres, that express MYH7, make up a larger proportion of muscles in large animals such as humans than in smaller cats [8], a defect in MYH7 may have more severe consequences in humans. The muscular hypertrophy observed in this cat might have compensated the defect to an extent that allowed normal locomotor function. However, it is also possible that some degree of muscle weakness was present, but not recognised, as even severe myopathy in cats can go unnoticed by the owner and veterinarian [40, 41]. Histopathology of skeletal muscles was not available to compare to the MSM phenotype in humans.
The human patient carrying the MYH7 c.5647G>A variant was a homozygote and Tajsharghi et al. [39] suggested an autosomal recessive pattern of inheritance for this variant. However, both HCM and MSM generally show an autosomal dominant pattern of inheritance and this might also have been the case in this human family [2, 9]. The two affected siblings that were not genotyped could be heterozygotes and it is not clear whether cardiomyopathy was ruled out in the parents, especially the father who died of stroke at the age of 58 [39]. The cat described here was a heterozygote, consistent with an autosomal dominant pattern of inheritance. Because of the suspicion of HCM in the littermate, it is considered unlikely that the causative variant was a de novo variant. The MYH7 c.5647G>A variant is extremely rare in humans and the absence of this variant in the 200 additional cats suggests that it is also rare in cats.
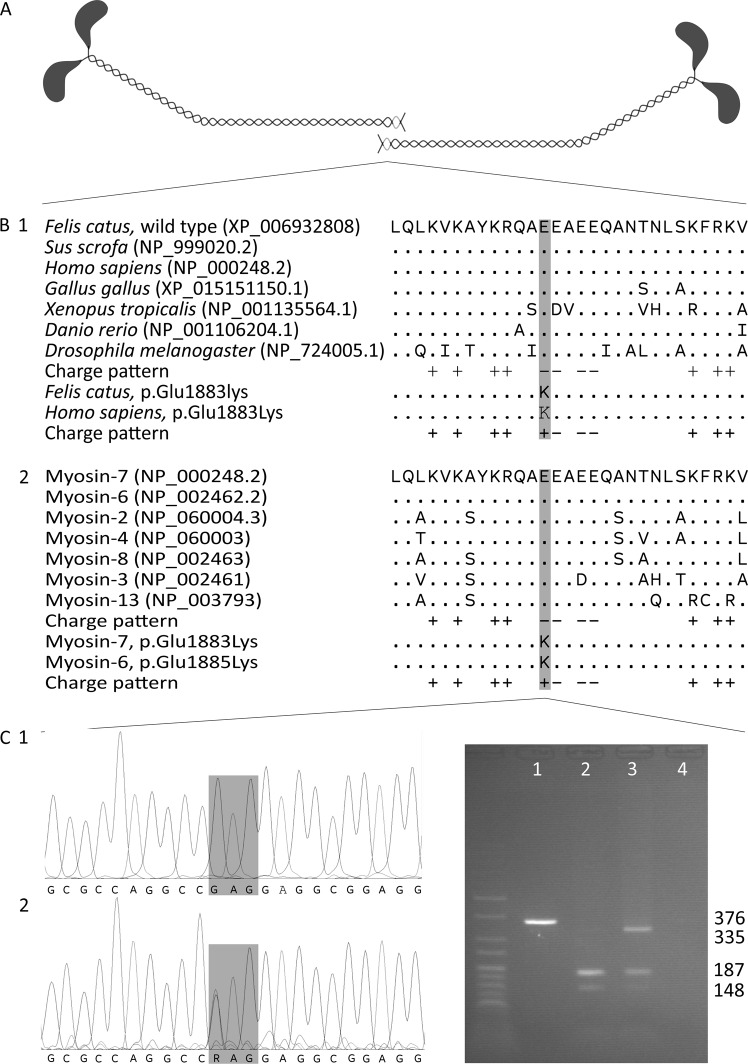
The affected glutamic acid residue is part of the assembly competence domain (ACD), a C-terminal 29 amino acid region that has a distinct pattern of four negative charges surrounded by positive charges and is essential for sarcomeric thick filament assembly [42]. During the assembly process, the tails of myosin heavy chains form coiled coils, the coils dimerise tail-to-tail and then aggregate into filaments [43]. The tail-to-tail dimerisation requires the correct functioning of the ACD, which is highly conserved in myosin-7 orthologues across species as well as in the striated muscle myosin heavy chain paralogues of cats and humans (Fig. 1) [42, 43].
Fig. 1.
Position and evolutionary conservation of the ACD. a Tail-to-tail dimerisation of the coiled coils formed by myosin heavy chains. The ACD regions are shaded light-grey. b The amino acid sequence and charge pattern of the ACD in myosin-7 (myosin heavy chain for Drosophila) orthologues across species (1) and skeletal and cardiac muscle myosin heavy chain paralogues in humans (2). Accession numbers are given in parentheses and identical amino acids are depicted as “.”. The variants cause the substitution of a conserved, negatively charged glutamic acid residue by a positively charged lysine residue. c Chromatograms of a homozygous wild-type cat (1) and the heterozygous case (2). The variant changes codon 1883 from GAG to AAG. d Agarose gel electrophoresis of an uncleaved amplicon (1), 376 bp long, the cleaved amplicons of a homozygous wild-type cat (2) and the heterozygote case (3) and a negative control (4). The amplicon is always cleaved at an internal cleavage site in a large fragment of 335 bp and a small fragment of 41 bp. The wild-type large fragment contains a second recognition site and is cleaved in fragments of 187 and 148 bp, while the variant large fragment does not contain a recognition site and remains intact. Two percent agarose gel with HyperLadder V
The p.(Glu1883Lys) variant substitutes one of the four negative charges by a positive charge. In vitro assessment of this substitution shows a lower stability and higher solubility of the protein, indicating a reduced assembling capacity [44]. For a similar substitution (Glu1886Lys), it was shown that the extent of filament assembly is reduced to 60% compared to the wild type [45]. Expression of Glu1883Lys mutant protein in a Drosophila animal model causes ultrastructural thick filament misalignment and disrupted sarcomere structure in pupae. In adults, these abnormalities worsen and are complemented with hyaline bodies reminiscent of human MSM. These adults show a compromised ability to fly and jump, indicating severe functional effects of this variant, as predicted by computational and predictive data [44].
In addition, a paralogous variant in MYH6, the major myosin heavy chain gene expressed in the cardiac atria [7], causes an identical charge pattern disruption of the ACD with deleterious phenotypical effects in humans. This variant, NM_002471.3:c.5653G>A (p.(Glu1885Lys); rs760353963), is considered to be the causative variant in a family where Wolff–Parkinson–White syndrome segregates in an autosomal dominant pattern with incomplete penetrance [46]. It is also considered to have contributed to lethal congenital heart disease in a compound heterozygote who also carried another MYH6 variant [47].
By adding the data of the orthologous feline case to that of the already described human family, together with the updated population data, the recently described functional studies and the paralogous human MYH6 variant, there is now enough evidence to classify the NM_000257.3:c.5647G>A (p.(Glu1883Lys)) variant as HCM-causing, according to the standards and guidelines for the interpretation of sequence variants [48].
This is the first report of HCM caused by a variant in MYH7 in a cat. An earlier study sequencing sarcomeric candidate genes, including MYH7, in 14 HCM-affected cats did not identify any disease-causing variant [49]. A recent, more extensive study identified several candidate variants, but not in MYH7 [50]. It might be interesting to investigate if this variant is also the causal variant in other Domestic Shorthair HCM cases.
Variants that cause HCM in humans are generally rare and the clinical significance of a variant found in human patients is not always clear [1, 2]. The identification of an orthologous variant in a spontaneous animal model of HCM can support its causality in humans. The involvement of MYH7, in addition to MYBPC3, in feline HCM suggests that its similarity to human HCM extends to the genetic level. As most cases of feline HCM are idiopathic, further investigations on its aetiology are needed to confirm this genetic similarity. In addition to the phenotypic similarity, this similarity would make cats with HCM a suitable animal model for the development of preventive and therapeutic strategies for HCM.
Supplementary information
Acknowledgements
The authors wish to thank Linda Impe, Ruben Van Gansbeke, Dominique Vander Donckt, Delphine Ameye, Sarah Loomans, Joachim Christiaens and Christian Puttevils for their excellent technical assistance. TS's PhD is funded by the Ghent University’s Special Research Fund.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-019-0431-4) contains supplementary material, which is available to authorised users.
References
- 1.Sabater-Molina M, Perez-Sanchez I, Hernandez Del Rincon JP, Gimeno JR. Genetics of hypertrophic cardiomyopathy: a review of current state. Clin Genet. 2018;93:3–14. doi: 10.1111/cge.13027. [DOI] [PubMed] [Google Scholar]
- 2.Marian AJ, Braunwald E. Hypertrophic cardiomyopathy. Genetics, pathogenesis, clinical manifestations, diagnosis, and therapy. Circ Res. 2017;121:749–70. doi: 10.1161/CIRCRESAHA.117.311059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maron BJ, Ommen SR, Semsarian C, Spirito P, Olivotto I, Maron MS. Hypertrophic cardiomyopathy: present and future, with translation into contemporary cardiovascular medicine. J Am Coll Cardiol. 2014;64:83–99. doi: 10.1016/j.jacc.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Nyland LR, Palmer BM, Chen Z, Maughan DW, Seidman SE, Seidman JG, et al. Cardiac myosin binding protein-C is essential for thick-filament stability and flexural rigidity. Biophys J. 2009;96:3273–80. doi: 10.1016/j.bpj.2008.12.3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta MK, Robbins J. Post-translational control of cardiac hemodynamics through myosin binding protein C. Pflug Arch. 2014;466:231–6. doi: 10.1007/s00424-013-1377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19:192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mascarello F, Toniolo L, Cancellara P, Reggiani C, Maccatrozzo L. Expression and identification of 10 sarcomeric MyHC isoforms in human skeletal muscles of different embryological origin. Diversity and similarity in mammalian species. Ann Anat. 2016;207:9–20. doi: 10.1016/j.aanat.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91:1447–531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 9.Tajsharghi H, Oldfors A. Myosinopathies: pathology and mechanisms. Acta Neuropathol. 2013;125:3–18. doi: 10.1007/s00401-012-1024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payne JR, Brodbelt DC, Luis Fuentes V. Cardiomyopathy prevalence in 780 apparently healthy cats in rehoming centres (the CatScan study) J Vet Cardiol. 2015;17(Suppl 1):S244–S257. doi: 10.1016/j.jvc.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Meurs KM, Norgard MM, Ederer MM, Hendrix KP, Kittleson MD. A substitution mutation in the myosin binding protein C gene in ragdoll hypertrophic cardiomyopathy. Genomics. 2007;90:261–4. doi: 10.1016/j.ygeno.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Côté E, MacDonald KA, Meurs KM, Sleeper MM Hypertrophic Cardiomyopathy. In: Feline cardiology. Chichester, UK: John Wiley & Sons; 2011. p. 103–75.
- 13.Freeman LM, Rush JE, Stern JA, Huggins GS, Maron MS. Feline Hypertrophic Cardiomyopathy: a spontaneous large animal model of human HCM. Cardiol Res. 2017;8:139–42. doi: 10.14740/cr578w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maron BJ, Fox PR. Hypertrophic cardiomyopathy in man and cats. J Vet Cardiol. 2015;17(Suppl 1):S6–S9. doi: 10.1016/j.jvc.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Meurs KM, Sanchez X, David RM, Bowles NE, Towbin JA, Reiser PJ, et al. A cardiac myosin binding protein C mutation in the Maine Coon cat with familial hypertrophic cardiomyopathy. Hum Mol Genet. 2005;14:3587–93. doi: 10.1093/hmg/ddi386. [DOI] [PubMed] [Google Scholar]
- 16.Ripoll Vera T, Monserrat Iglesias L, Hermida Prieto M, Ortiz M, Rodriguez Garcia I, Govea Callizo N, et al. The R820W mutation in the MYBPC3 gene, associated with hypertrophic cardiomyopathy in cats, causes hypertrophic cardiomyopathy and left ventricular non-compaction in humans. Int J Cardiol. 2010;145:405–7. doi: 10.1016/j.ijcard.2010.04.032. [DOI] [PubMed] [Google Scholar]
- 17.Lisciandro GR. Abdominal and thoracic focused assessment with sonography for trauma, triage, and monitoring in small animals. J Vet Emerg Crit Care. 2011;21:104–22. doi: 10.1111/j.1476-4431.2011.00626.x. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch EF, Maniatis T. Commonly used techniques in molecular cloning. In: Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. p. E.1–E.39.
- 19.Van Poucke M, Peelman L Flexible, multi-use, PCR-based nucleic acid integrity assays based on the ubiquitin C gene. 2017. https://www.biorxiv.org/content/10.1101/168195v1.
- 20.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 23.Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nhlbi Go Exome Sequencing Project (ESP). Exome variant server [Internet]. Seattle, WA, USA [Cited 13 Dec 2018]. http://evs.gs.washington.edu/EVS/.
- 27.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Poucke M, Vandesompele J, Mattheeuws M, Van Zeveren A, Peelman LJ. A dual fluorescent multiprobe assay for prion protein genotyping in sheep. BMC Infect Dis. 2005;5:13. doi: 10.1186/1471-2334-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kittleson MD, Meurs KM, Munro MJ, Kittleson JA, Liu S-K, Pion PD, et al. Familial hypertrophic cardiomyopathy in Maine Coon cats: an animal model of human disease. Circulation. 1999;99:3172–80. doi: 10.1161/01.CIR.99.24.3172. [DOI] [PubMed] [Google Scholar]
- 30.Luis Fuentes V. Arterial thromboembolism: risks, realities and a rational first-line approach. J Feline Med Surg. 2012;14:459–70. doi: 10.1177/1098612X12451547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gompf RE. The history and physical examination. In: Tilley LP, Smith FWK, Oyama MA, Sleeper MM, editors. Manual of canine and feline cardiology. 4th ed. St. Louis, MO, USA: Elsevier Saunders; 2008, p. 2–23.
- 32.Kubo T, Gimeno JR, Bahl A, Steffensen U, Steffensen M, Osman E, et al. Prevalence, clinical significance and genetic basis of hypertrophic cardiomyopathy with restrictive phenotype. J Am Coll Cardiol. 2007;49:2419–26. doi: 10.1016/j.jacc.2007.02.061. [DOI] [PubMed] [Google Scholar]
- 33.Mitchell, RN Heart. In: Kumar V, Abbas AK, Aster JC, editors. Robbins basic pathology, 9th ed. Philadelphia, PA, USA: Elsevier Saunders; 2013, p. 365–406.
- 34.Côté E, MacDonald KA, Meurs KM, Sleeper MM. Restrictive/unclassified cardiomyopathy. In: Feline cardiology. Chichester, UK: John Wiley & Sons; 2011, p. 177–81.
- 35.Fox PR. Hypertrophic cardiomyopathy. Clinical pathologic correlates. J Vet Cardiol. 2003;5:39–45. doi: 10.1016/S1760-2734(06)70051-0. [DOI] [PubMed] [Google Scholar]
- 36.Biasato I, Francescone L, La Rosa G, Tursi M. Anatomopathological staging of feline hypertrophic cardiomyopathy through quantitative evaluation based on morphometric and histopathological data. Res Vet Sci. 2015;102:136–41. doi: 10.1016/j.rvsc.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 37.Wilkie LJ, Smith K, Luis Fuentes V. Cardiac pathology findings in 252 cats presented for necropsy; a comparison of cats with unexpected death versus other deaths. J Vet Cardiol. 2015;17(Suppl 1):S329–S340. doi: 10.1016/j.jvc.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 38.Longeri M, Ferrari P, Knafelz P, Mezzelani A, Marabotti A, Milanesi L, et al. Myosin-binding protein C DNA variants in domestic cats (A31P, A74T, R820W) and their association with hypertrophic cardiomyopathy. J Vet Intern Med. 2013;27:275–85. doi: 10.1111/jvim.12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tajsharghi H, Oldfors A, Macleod DP, Swash M. Homozygous mutation in MYH7 in myosin storage myopathy and cardiomyopathy. Neurology. 2007;68:962. doi: 10.1212/01.wnl.0000257131.13438.2c. [DOI] [PubMed] [Google Scholar]
- 40.Blunden AS, Gower S. Hypertrophic feline muscular dystrophy: diagnostic overview and a novel immunohistochemical diagnostic method using formalin-fixed tissue. Vet Rec. 2011;168:510. doi: 10.1136/vr.d1119. [DOI] [PubMed] [Google Scholar]
- 41.Remmers G, Hayden DW, Jaeger MA, Ervasti JM, Valberg SJ. Postanesthetic death in a cat with myopathy. Vet Pathol. 2015;52:186–8. doi: 10.1177/0300985814524797. [DOI] [PubMed] [Google Scholar]
- 42.Sohn RL, Vikstrom KL, Strauss M, Cohen C, Szent-Gyorgyi AG, Leinwand LA. A 29 residue region of the sarcomeric myosin rod is necessary for filament formation. J Mol Biol. 1997;266:317–30. doi: 10.1006/jmbi.1996.0790. [DOI] [PubMed] [Google Scholar]
- 43.Kachur TM, Pilgrim DB. Myosin assembly, maintenance and degradation in muscle: role of the chaperone UNC-45 in myosin thick filament dynamics. Int J Mol Sci. 2008;9:1863–75. doi: 10.3390/ijms9091863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viswanathan MC, Tham RC, Kronert WA, Sarsoza F, Trujillo AS, Cammarato A, et al. Myosin storage myopathy mutations yield defective myosin filament assembly in vitro and disrupted myofibrillar structure and function in vivo. Hum Mol Genet. 2017;26:4799–813. doi: 10.1093/hmg/ddx359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Armel TZ, Leinwand LA. Mutations in the beta-myosin rod cause myosin storage myopathy via multiple mechanisms. Proc Natl Acad Sci USA. 2009;106:6291–6. doi: 10.1073/pnas.0900107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bowles NE, Jou CJ, Arrington CB, Kennedy BJ, Earl A, Matsunami N, et al. Exome analysis of a family with Wolff-Parkinson-White syndrome identifies a novel disease locus. Am J Med Genet. 2015;167A:2975–84. doi: 10.1002/ajmg.a.37297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Preuss C, Capredon M, Wünnemann F, Chetaille P, Prince A, Godard B, et al. Family based whole exome sequencing reveals the multifaceted role of notch signaling in congenital heart disease. PLoS Genet. 2016;12:e1006335. doi: 10.1371/journal.pgen.1006335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meurs KM, Norgard MM, Kuan M, Haggstrom J, Kittleson M. Analysis of 8 sarcomeric candidate genes for feline hypertrophic cardiomyopathy mutations in cats with hypertrophic cardiomyopathy. J Vet Intern Med. 2009;23:840–3. doi: 10.1111/j.1939-1676.2009.0341.x. [DOI] [PubMed] [Google Scholar]
- 50.Matos J, Psifidi A, Connolly D, Wilkie L, Piercy R, Fuentes V. Candidate genes for feline hypertrophic cardiomyopathy: analysis of 18 sarcomeric and non-sarcomeric genes. Ir Vet J. 2018;71:17. doi: 10.1186/s13620-018-0126-0. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.