Abstract
The use of open-source programming languages, such as R, in health decision sciences is growing and has the potential to facilitate model transparency, reproducibility, and shareability. However, realizing this potential can be challenging. Models are complex and primarily built to answer a research question, with model-sharing and transparency relegated to being secondary goals. Consequently, code is often neither well-documented nor systematically organized in a comprehensible and shareable approach. Moreover, many decision modelers are not formally trained in computer programming and may lack good coding practices, further compounding the problem of model transparency.
To address these challenges, we propose a high-level framework for model-based decision and cost-effectiveness analyses (CEA) in R. The proposed framework consists of a conceptual, modular structure and coding recommendations for the implementation of model-based decision analyses in R. This framework defines a set of common decision model elements divided into five components: (1) model inputs, (2) decision model implementation, (3) model calibration, (4) model validation, and (5) analysis. The first four components form the model development phase. The analysis component is the application of the fully developed decision model to answer the policy or the research question of interest, assess decision uncertainty, and/or to determine the value of future research through value of information (VOI) analysis.
In this framework, we also make recommendations for good coding practices specific to decision modeling, such as file organization and variable naming conventions. We showcase the framework through a fully functional, testbed decision model, which is hosted on GitHub for free download and easy adaptation to other applications. The use of this framework in decision modeling will improve code readability and model sharing, paving the way to an ideal, open-source world.
1. Introduction
Many journals now strongly encourage that the data and the code underlying an analysis be archived and made publicly available alongside the publication [1,2]. There are similar calls for making mathematical models that are the basis for health technology assessments (HTA) and cost-effectiveness analyses (CEAs) available to promote transparency, support reproducibility, and facilitate adaptation of existing models to new applications [3,4]. In formal HTA submissions, it is already expected that the model itself will be provided to clients and stakeholders for them to scrutinize and manipulate, necessitating a certain degree of model transparency and usability [5,6]. More broadly, the Open-Source Model Clearinghouse was recently launched as a database of open source models with a mandate to, in part, “facilitate adherence to standards calling for open disclosure of scientific software” [7]. Though it has been common wisdom that a detailed methods section and a lengthy appendix of equations should be sufficient to reproduce a mathematical model, this is not generally the case. Thus, to support the transparency of mathematical modeling, more and more emphasis is being placed on sharing the underlying model construction, be it implemented in a specific software platform or coded in a programming language [8–10].
For anyone who has ever looked under the hood of software source code, the naivety of transparency being achieved by sharing such code is obvious. Even for a well-trained and sophisticated programmer, coding entails a certain amount of personal style and preferences which may or may not be intuitive to the reader. Imagine, then, the even more extreme, yet still common, situation of releasing code that was never intended for public use to the public. If this is done as an after-thought, documentation may be lacking, and the code structure will likely be a byproduct of the complex decision history that it took to arrive at the final model structure rather than a pre-planned organizational structure. All these issues may be further obscured when proprietary software is required to view/operate the model, which may limit access to those with active licenses and installations. The appeal of proprietary software is often in the facilitation of model construction through a graphical interface that allows a user to point-and-click their way through an analysis. However, despite the initial user-friendliness of these software platforms, sharing the model alone still does not necessarily achieve transparency or reproducibility, as any manual point-and-click steps are not captured or recorded [11].
Health decision science and HTA are fields situated at the intersection of operations research, economics, statistics, medicine, and public health. Computer science and software development are generally not the major foci of decision-analytic training, as models are used to answer specific research questions, not necessarily as general tools for a client user-base. Thus, in order for the benefits of transparent and open model-sharing to be fully realized, guidance is needed on coding best practices as it relates to decision modeling so that these models can be read, scrutinized, and understood by their consumers.
The aim of this paper is to provide a high-level framework that sets a common structure for decision-model building for both model developers and model consumers. The development of this framework is the culmination of the research and pedagogical experiences of The Decision Analysis in R for Technologies in Health (DARTH) workgroup [12]. The DARTH framework modularizes decision models into a set of core components that are common across CEAs and HTA submissions, regardless of the type of mathematical model used. In this paper, we also provide a number of recommendations specific to decision-modeling applications relating to file organization, variable naming conventions, use of functions and data structures, and unit testing. However, these more detailed recommendations are suggestions only; the primary purpose of the DARTH framework is to outline a high-level organizational structure to code underlying a decision-modeling analysis. Given the diversity of applications and methodological needs of different analyses, we hope that the DARTH framework provides a scaffolding to facilitate readability, usability, and reproducibility of the analysis to others, without overly restricting the kinds of models and analyses that can be implemented in this framework. We showcase the DARTH framework through a fully functional, testbed decision model developed in R [11,13], implemented as an R package (darthpack) that is freely available for download via GitHub (https://github.com/DARTH-git/darthpack). The testbed model was designed to serve as a template for organizing and sharing model and analysis source code [14] that can be easily adapted to other applications and enhanced by other decision modelers. The adoption and promotion of this framework will create more readable, and thus more shareable models, paving the way to an open-source culture in health decision sciences.
2. Methods
2.1. Components of a decision model
DARTH framework is based on the premise that a comprehensive model-based decision and/or CEA will involve the same high-level model-development analysis components, regardless of the specific structure of the decision model being applied, be it a decision tree, Markov model, stochastic simulation model, and so on. In developing this framework, we strived to create a flexible framework that can successfully organize code relating to a diversity of model types and applications.
The framework we present here focuses on the organization of R code for the conduct of a decision analysis, but not on the specific content of the code within each component. We also assume that an analyst has already fully documented their biological, behavioral, and mathematical assumptions and decisions that went into their model and analysis in some kind of technical appendix. In our case study, we provide an example of how such documentation might look, but it is not the primary focus of this work. Thus, commentary in the code will primarily explain the functionality of that code, with the assumption that broader descriptions of the disease processes, interventions, and policy questions are provided alongside the code in a separate document.
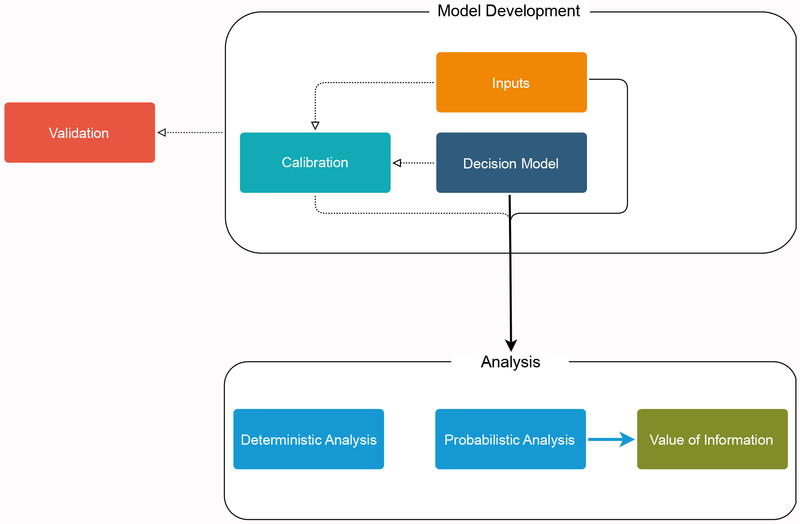
DARTH framework divides a decision analysis into five components: (1) model inputs, (2) decision model implementation, (3) model calibration, (4) model validation, and (5) analysis. The first four components form the model development phase, whereas the analysis component is the application of the final model to answer the policy or research question of interest, assess decision uncertainty, and/or to determine the value of future research through value of information (VOI) analysis. The same model from the development phase could be used to answer multiple research questions, which is why we make this distinction. The relationship between the five components is illustrated in Figure 1 and described in detail in the sections that follow.
Figure 1.
Schematic representation of the connectivity between the different components of the proposed DARTH coding framework.
2.1.1. Component 1: Define model inputs
In this component, all model input variables are declared and values are set. We broadly categorize input variables into three categories depending on how their values are informed: external, estimated, and calibrated. Parameters informed by external sources are set to a value either directly into an R script or read in from an external source, such as a .csv file or a data repository. These parameter values (and uncertainty ranges and distributions for probabilistic analyses) are derived from published literature or external data analyses not embedded into the analysis itself. Estimated parameters are those whose values are estimated through a primary data analysis conducted within the decision analysis. R has the advantage of being both a statistical and programming environment. This allows any necessary statistical analyses to be embedded directly within the decision analysis, further improving analysis transparency and reproducibility. The third type of model parameters are those that will be estimated via model calibration. In this first stage of the DARTH framework, we simply set these parameters to some valid but arbitrary “dummy” values that are compatible with the next phase of the analysis, model implementation, but are ultimately just placeholder values until we conduct the calibration phase. Not all models will utilize all three types of input variables or different models may rely more heavily on one input type than another. While we selected the three input parameter categories based on how models are typically parameterized, for any given application, it may make sense to organize input parameters according to a different set of categories. The point of this component is to group input variables together and organize them in a logical fashion that can be easily communicated to a user rather than rigidly prescribe a universal input parameter organizational structure.
2.1.2. Component 2: Decision model implementation
This implementation of the decision model component is the heart of the decision analysis. In this section of the DARTH framework, a function that maps model inputs to outputs is created, via the dynamic and/or stochastic processes that the decision model represents. The model itself could be a decision tree, Markov model, stochastic simulation, and so on. The output stored from the model at this stage should be sufficiently general and comprehensive to accommodate calibration, validation, and the main policy analysis. Constructing the model as a function at this stage facilitates subsequent components of model development and analysis, as these processes will all call the same model function but pass different parameter values and/or calculate different final outcomes from the model outputs. The model function also facilitates the use of parallel computing efforts for computationally intensive tasks, such as calibration and probabilistic sensitivity analysis (PSA).
We should note explicitly that the model function created here should have the capacity to capture the effect of any interventions or policy scenarios of interest on the outcomes of interest. The specific ways that intervention effects are incorporated into the model is a choice for the analyst. Interventions that reflect changing intensities of existing processes (e.g., increasing the frequency of screening) may be implemented by changing the values of relevant model input parameters. However, it is often the case that different interventions enable completely different pathways and processes in the model (e.g. medical management vs. surgery) and would be better captured by passing a categorical parameter value that indicates the intervention to be simulated. We do not recommend mixing these two cases and generally recommend using an explicit categorical intervention variable for generalizability. Ultimately, the analyst should decide how best to implement the functionality required for their application.
2.1.3. Component 3: Model calibration
In the model calibration component, unknown or highly uncertain model parameters are estimated by calibrating model outputs to match specified calibration targets [15–17]. This component involves both the setup of the calibration (specification of plausible ranges or prior distributions for input parameters to be calibrated, specification of calibration targets, calculation of corresponding values from model outputs and assessment of fit to targets) as well as the carrying out the calibration itself with a chosen algorithm. Once appropriate values, ranges, and/or distributions have been identified for calibrated parameters, these values will replace the placeholder values established in the model inputs component for the subsequent validation and analysis components. Though rare, not all models will have parameters that need to be calibrated. In such cases, the model calibration component can simply be omitted.
2.1.4. Component 4: Validation
Model validation should at the very least demonstrate the internal validity of the model. This means that the model reproduces outputs that correspond to its inputs [3,18]. For example, if an input parameter to the model was set to reflect a screening frequency of every 2 years, then the number of screenings conducted in the population over a given period of time should correspond to an average per-person screening frequency of every 2 years. Internal validity may also be demonstrated by plotting model-predicted outputs against calibration targets. Additionally, comparison of model outputs to other data sources not used in the model development (external validation) or to other models (comparative validity) may also be conducted here [19–23].
2.1.5. Component 5: Analysis
The analysis component is where the model developed in components 1-4 is applied to answer the question(s) of interest given current information. An analysis will generally be broken down into several subcomponents. As an example, we describe an analysis with three subcomponents: a probabilistic analysis (which includes the base case analysis), a deterministic scenario and sensitivity analysis (such as one- and two-way sensitivity analyses), and a value of information (VOI) analysis. However, in any given analysis, the analyst should create subcomponents as is relevant and appropriate for their application. Though the purposes and the structure of our example subcomponents vary, the general setup is similar. First, the analyst must specify the input parameter values that should be passed to the decision model function. Second, the analysis must setup calculation of the desired output values from the decision model outputs, which again are more comprehensive and detailed than may be necessary. For example, a Markov cohort model might output the cohort trace (distribution of the cohort across health states over the time horizon) or the transition dynamics array (proportion of the cohort that transitioned between any two health states in each cycle over the time horizon) [24], but in a given analysis, perhaps only the cohort’s survival over time is of interest. Within CEA in particular, there are many standard calculations, comparisons, and visualizations that are conducted based only on the total costs and quality-adjusted life-years (QALYs) calculated from model outputs for a set of strategies.
2.1.6. Subcomponent 5a: Probabilistic analysis
The probabilistic analysis subcomponent is the primary analysis component in the DARTH framework, which is typical for CEA following the recent guidance from the Second Panel on Cost-Effectiveness in Health and Medicine and satisfying the requirements of many health technology assessment agencies [6,25]. In a probabilistic analysis, also called a probabilistic sensitivity analysis (PSA), sets of input parameter values are randomly sampled from specified distributions. The model is then run for each set of parameter values, producing corresponding model outputs. Using analyst-specified functions that calculate outcomes of interest based on the model function output, means and standard deviations of these outcomes can be calculated from the PSA samples. For CEAs, primary outcomes of interest are generally total discounted costs and QALYs accrued over the analysis time horizon, though other intermediate outcomes may also be of interest. Interventions are then compared by calculating incremental cost-effectiveness ratios (ICERs) based on the expected cost and QALY outcomes from the PSA. We note that in the past, a primary analysis was often conducted using a single, deterministic set of base case parameter values but this practice is no longer recommended [6]. The distributions of outcomes produced from the PSA are also used to produce additional results regarding decision uncertainty, including cost-effectiveness acceptability curves (CEACs) and frontier (CEAF), expected loss curves (ELCs), and others [26]. For these common procedures, we rely on the decision-analytic modeling in R package, dampack, which is available for download here: https://github.com/DARTH-git/dampack. Instructions for installing dampack are described in the dampack GitHub repository.
2.1.7. Subcomponent 5b: Scenario and deterministic sensitivity analysis
The scenario and deterministic sensitivity analysis subcomponent is where the impact of individual or pairs of parameters on model outcomes can be assessed systematically through one-and two-way sensitivity analyses. An analyst may also wish to compare different scenarios (e.g. a high vs. low cost scenarios), either in a probabilistic or deterministic framing. Generally, these scenario analyses and sensitivity analyses would be secondary to the primary results presented in subcomponent 5a.
2.1.8. Subcomponent 5c: Value of information (VOI) analysis
In the VOI component, we determine whether further potential research is needed using the results from the PSA generated in the probabilistic analysis subcomponent. The most common VOI measures are the expected value of perfect information (EVPI), the expected value of partial perfect information (EVPPI), the expected value of sample information (EVSI) [27,28], and, more recently, the curve of optimal sample size (COSS) [29].
2.2. File structure and organization
A model implemented in the R programming environment will involve a series of scripts with the file extension “.R”. The analysis will also generally use and/or generate a number of data and output files, which may be either stored as internal R data files (using “.RData”, “.rda”, or “.rds” extensions) or as external data files, such as comma-separated-values (“.csv”) files. In the suggested organizational file structure of the DARTH framework, we use folders to delineate the different purposes that these files serve in the analysis. Within a folder, we append the relevant component number to the beginning of each file name to indicate where the file will be used or was created (in the case of outputs). Our suggested folder structure is summarized in Table 1. This structure is inspired by the organizational recommendations for an R package [30] and a simple reproducible workflow developed in the field of ecology and evolution [31]. As an example, consider the “data-raw” folder. The purpose of the data folder is to store the raw data files that will be cleaned, processed and/or analyzed to be used as inputs in the different components. The processed data would then be placed in the “data” folder, perhaps stored as the file “01_primary_data.RData” to indicate that it will inform model parameter values (the first framework component). Within this folder, we would likely also have a file named “01_inputs.csv” which would contain model parameter values derived externally from published literature. Finally, in addition to input data for the input generation component, an analyst might also have a “03_calibration_targets.RData” which stores the calibration target data that will be used to estimate unknown model parameters through calibration.
Table 1:
File folder structure for organizing model and analysis files used in the proposed DARTH coding framework.
| Folder name | Folder function |
|---|---|
| data-raw | This is where raw data is stored alongside “.R” scripts that read in raw data, process these data, and calls use_this::use_data(<processed data>) to save .rda formatted data files in the “data” folder. These data could include a “.csv” file with input parameters derived from the published literature, as well as internal R data files (with .RData, .rds, or .rda extensions) containing primary data from which model input values will be estimated through statistical models embedded into the analysis. |
| data | This is where input data is stored to be used in the different components of the CEA. These data could be generated from raw data stored in the “data-raw” folder. Essentially, this folder stores the cleaned or processed versions of raw data that has been gathered from elsewhere |
| R | This is where “.R” files that define functions to be used as part of the analysis are stored. These are functions that are specific to the analysis. The model will be one such function; however, other functions will likely be used, such as computing the fit of the model output to the specific calibration targets of the analysis. This folder also stores “.R” scripts that document the datasets in the “data” folder. |
| analysis | This is where interactive scripts of the analysis would be stored. These scripts control the overall flow of the analysis. This is also where many operations that ultimately become functions will be developed and debugged. |
| output | This is where output files of the analysis should be stored. These files may be internal R data files (“.RData”, “.rds”, “.rda”) or external data files (such as “.csv”). Examples of files stored here would be the output of the model calibration component or the PSA dataset generated in the uncertainty analysis component. These data files can then be loaded by other components without having to first rerun previous components (e.g. the calibrated model values can be loaded for a base case analysis without re-running the calibration). |
| figs | For analyses that will include figures, we generally create a separate figures folder. Though these could be stored in the output folder, it can be helpful to have a separate folder so that the images of the figure files can be easily previewed. This is particularly important for analyses that generate a large number of figures. |
| tables | This folder includes tables to be included in a publication or report, such as the table of intervention costs and effects and ICERs. |
| report | A report folder could be used to store R Markdown files to describe in detail the model-based CEA by using all the functions and data of the framework, run analyses and display figures. The R Markdown files can be compiled into .html, .doc or .pdf files to generate a report of the CEA. This report could be the document submitted to HTA agencies accompanying the R code of the model-based CEA. |
| vignettes | A vignettes folder could be used to describe the usage of the functions and data of each of some or all components of the framework through accompanying R Markdown files as documentation. The R Markdown file can use all the functions, outputs, and figures to integrate the R code into the Markdown text. |
| tests | A tests folder includes “.R” scripts that runs all the unit tests of the functions in the framework. A good practice is to have one file of tests for each complicated function or for each of the components of the framework. |
PSA probabilistic sensitivity analysis; ICERs incremental cost-effectiveness ratios; CEA cost-effectiveness analysis; HTA health technology assessment.
Our suggested file folder structure is fairly self-explanatory and certainly customizable. However, two folders that warrant further clarification are the “R” and “analysis” folders. The “R” folder is the traditional directory for storing functions for an R package. Here, we store a separate “.R” script with all the functions for each framework component as well as some auxiliary “.R” scripts, such as the description of the different data included in the R package. For example, the “data_init_params.R” script includes the description of the initial set of base-case parameters. To document the functions and processed data to be used as package data, we used roxygen2. roxygen2 is the recommended format to produce documentation for R packages. For a more detailed description on the different components and steps for building an R package, we refer the reader to the R package book by Hadley Wickham [30]. Formalizing operations into functions is especially advantageous for operations that will be repeated (e.g., calculation of total costs and QALYs from model output). A single function can replace multiple lines of code and modularizes operations, and any updates to these repeated operations will be propagated across all function calls. Using functions is considered a good programming practice [32]. Functions that are customized for the particular application, model, and/or analysis should be defined in the “.R” file corresponded to the component where they are first needed. For example, the decision model is implemented as a function, which is important since it will be called by so many other processes (calibration, validation, as well as the analysis components). Model calibration would also involve several custom functions, such as functions to derive outputs corresponding to the calibration targets from the model’s more generic, full output and functions to compare those model outputs to the calibration target values in terms of some kind of measure of “fit”. The analysis components will have many functions for calculating outputs of interests (e.g. aggregating costs and QALYs over a time horizon of interest), and running the model over different sets of input parameter values in deterministic and probabilistic sensitivity analyses.
The “analysis” folder is the traditional directory for storing the scripts with the code of R-based analyses. In this folder, we store a “.R” script for each framework component. These scripts are the overall control for these processes.
2.3. Naming conventions
Within the outlined file structures, we recommend that analysts use a consistent naming convention for variables and files throughout their code that balance readability and brevity. Different well thought out naming conventions have been proposed, including coding styles recommended by the tidyverse collection of R packages [33] and the Google R Style Guide [34]. We summarize our own naming convention, tailored to the specific types of parameters and files used in decision analytic modeling, in Table 2. In our naming convention, file names begin with the component number followed by some content descriptor, separated by underscores. User-defined functions are named starting with an action, followed by a descriptor, separated by underscores.
Table 2.
File and variable naming conventions in the proposed DARTH coding framework.
| Object type | Naming recommendation | Examples |
|---|---|---|
| Files | dir/<component number>_<description>.<ext> | •
analysis/θ1_model_inputs.R • R/θ2_simulation_model_functions.R |
| Functions | <action!>_<description> | •
generate_init_params() • generate_psa_params() |
| Variables | <x>_<y>_<var_name> where x = data type prefix y = variable type prefix var_name = brief descriptor |
• n_samp • hr_S1D • v_r_mort_by_age • a_M • l_params_all • df_out_ce |
Our variable naming conventions involve encoding certain features of the variable in the name. The suggested naming structure would be <x>_<y>_<var_name>, where x indicates the data type (e.g., scalar, vector, matrix, data frame, etc.), y is the variable type (e.g., probability, rate, relative risk, cost, utility, etc.), and var_name is some description of the variable presented separated by underscores. Suggested prefixes are summarized in Table 3.
Table 3.
Recommended prefixes in variable names that encode data and variable type.
| Prefix | Data type | Prefix | Variable type |
|---|---|---|---|
| <> (no prefix) | scalar | n | number |
| v | vector | p | probability |
| m | matrix | r | rate |
| a | array | u | utility |
| df | data frame | c | cost |
| dtb | data table | hr | hazard ratio |
| l | list | rr | relative risk |
| ly | life years | ||
| q | QALYs | ||
| se | standard error |
QALYs quality-adjusted life years
2.4. Unit testing
A full decision-analytic model, complete with all the modules outlined in the DARTH framework, will have complex interdependence between the various functions and processes. It is important to ensure that these functions behave as expected to maintain the integrity of the project. Thus, systematic testing is recommended alongside the development of the decision analysis source code. Testing increases confidence in the results and conclusions of the model and associated analyses and also allows the analyst to quickly identify whether modifications or additions to the analysis code impacts the behavior of the previously developed functions and processes [35]. A widely used testing method is unit testing, which tests a unit of code (often a function or a small process) to verify whether the code executes and generates outputs as intended.
For a comprehensive decision-analytic project, we suggest writing tests alongside the development of any new function or process or whenever a bug is found [30,35]. This practice results in a high level of test coverage of the analysis code, reducing the likelihood that unintended interactions or incompatibilities between functions and/or processes will go undetected. In practice, we suggest that each R script in the “R” folder have a corresponding testing “.R” script in a separate “tests” folder. The naming of a test file could begin with “test_”, followed by the file name of the source code that is the target of the testing. This file structure is also compatible with the R package structure.
In each test file, tests are organized by the functions or processes to be tested. A single function or process will likely be associated with multiple tests. For instance, unit testing of a function will involve testing that the function runs when inputs are of the right data type, that the function outputs of the right data type, and that the function outputs are correct in dimension and value for specific sets of input values. It is also important to test the error checking within a function, such that the function returns an error when invalid inputs are provided or unexpected results.
Comprehensive testing facilitates model sharing, as any downstream user wishing to modify the code can easily verify whether their changes to the original source code requires adjustments to be made to other parts of the code. To illustrate the use of unit testing, we provided examples of unit testing on two selected source code files in our case study using the R package testthat [30,36]. We only include a small number of tests so as to not overwhelm those new to testing; however, in practice, a comprehensive set of unit tests should be included.
2.5. Additional tools to support model transparency
A number of tools exist that can facilitate the decision modelers interaction with the R language. A useful and commonly used tool is RStudio, an open-source, integrated development environment (IDE) for R. RStudio offers functionality that facilitates R coding (e.g. syntax-highlighting). With RStudio it is possible to create projects, which are files with the “.Rproj” extension. An RStudio project creates a specific R session for the DARTH framework with its own working directory, workspace, history and source documents [41]. In other words, the RStudio project makes a standalone working environment without the trouble of having to specify where files are located when used in different computers. Additional functionality is embedded within the RStudio platform that allows the modeler to present the output of the analysis in a visually attractive and dynamic form. In particular, through the Shiny package, an interactive web app [37] can be developed to facilitate the usage of the decision model [38]. The Shiny app allows the user to modify the input parameters, rerun the model through the app’s interface and navigate through the updated results. Although Shiny has been developed to support web access to R models, it can also be downloaded and run locally. We have added the “Shiny_framework.R” file that generates the Shiny app in the GitHub repository, which can be executed locally once the darthpack repository is either downloaded or installed. An additional advantage of having the DARTH framework as an R package via darthpack is its integration with other packages to develop web applications with JavaScript, such as OpenCPU.
Once the analysis is completed, the user might be interested in generating a report of the findings. R Markdown is a functionality within RStudio that provides a dynamic solution to developing reports within an R environment. Once written, an R Markdown file can be “knitted” (transcribed) to a variety of different formats (.docx, .pdf, .html). There are a number of advantages associated with the use of R Markdown. The primary one is the integration of the report writing process with the data analysis or the simulation modeling. This allows for a better documented model-based CEA and a dynamic element to the report. For example, a report could be built in R Markdown while allowing for narrative that can be automatically updated conditional on the findings of the analysis. Another advantage of R Markdown is the ease of making a report publicly accessible because the ability for documents to be knitted in different web formats, allows them to be easily published on the web. With R Markdown, the description and reporting of the workflow of a CEA can be made more efficient with limited entry costs for those not already familiar with this functionality. Recently developed packages further enhance the functionality of R Markdown. For example, the bookdown facilitates the development of long reports [39]. We provide an example of how a report could be written in R Markdown with bookdown by describing the use of the functions of all the components of the DARTH framework using the case-study described below.
2.6. Case study: Sick-Sicker model
To showcase the DARTH framework, we performed a CEA of a hypothetical treatment using a state-transition cohort model on a hypothetical disease. For this CEA, we used the previously published Sick-Sicker model first described by Enns et al. [40]. Briefly, the Sick-Sicker model simulates a hypothetical cohort of 25-year-old healthy individuals with an age-specific background mortality that are at risk of developing a disease with two different stages of illness, “Sick” (S1) and “Sicker” (S2). Individuals in both the S1 and S2 states face an increased mortality and reduced quality of life (QoL) compared to healthy individuals. The hypothetical treatment improves QoL for individuals in the S1 state but has no effect on the QoL of those in the S2 state. While individuals who are afflicted with the illness can be identified through obvious symptoms, those in S1 cannot be easily distinguished from those in the S2 state. Thus, under the treatment strategy, all afflicted individuals are treated and accrue the costs of treatment, even though only those in S1 experience any benefit.
We assume that most parameters of the Sick-Sicker model and their uncertainty are known to the analyst and do not require any statistical estimation. However, because we cannot distinguish between S1 and S2, neither state-specific mortality hazard ratios nor the probability of progressing from S1 to S2 can be directly estimated. Therefore, we estimated these parameters by calibrating the model to epidemiological data. We internally validated the calibrated model by comparing the predicted outputs from the model, evaluated at the calibrated parameters, against the calibration targets.
As part of the CEA, we conducted different deterministic SA, including one-way and two-way SAs. To quantify the effect of parameter uncertainty on decision uncertainty, we conducted a PSA and reported our uncertainty analysis results with incremental costs and QALYs, ICERs, CEACs, CEAF, ELCs [26]. We also conducted a value of information (VOI) analysis to determine whether potential future research is needed to reduce parameter uncertainty.
The CEA of the Sick-Sicker model implemented in the DARTH framework may be downloaded from GitHub (https://github.com/DARTH-git/darthpack). We recommend either using the repository of this framework as a GitHub template or installing it as an R package. Using darthpack as a template allows users to easily modify any of the included files and is most appropriate for users wishing to adapt the DARTH framework to their own application model and analyses. To use darthpack as a template, users should first either clone the repository to their GitHub account or download it locally as a .zip file containing all files and folders. For users simply wishing to reproduce the existing analyses of the Sick-Sicker model in darthpack or conduct simple explorations using the included model and/or analysis functions, installing darthpack as a package is most appropriate. To install darthpack as a package, users should make use of the devtools package by typing devtools::install_github(“DARTH-git/darthpack”) . Detailed instructions on how to use and install the repository can be found in darthpack website (https://darth-git.github.io/darthpack/).
The DARTH framework is divided into different folders, described in Table 1, that could be accessed from the RStudio project “darthpack.Rproj”. A detailed description on how to install and use the DARTH framework on the Sick-Sicker mode can be found in the darthpack GitHub repository (https://github.com/DARTH-git/darthpack) and website (https://darth-git.github.io/darthpack). The framework of the case study is considered a finalized CEA so each of the components in the ‘analysis’ folder should be able to run independently of the rest of them. For example, if there is interest in reproducing the calibration component, the analyst or reviewer of the CEA can start by running the file “03_calibration.R” in the “analysis” folder, and so on. To reproduce the entire CEA, including all model development components and all analyses, the analyst should run the “_master.R” file in the “analysis” folder, which will execute the R scripts of each of the components. For a more detailed description of how the elements (functions, data and procedures) are interconnected within and between components for the Sick-Sicker model CEA case study, we recommend reading the vignettes of darthpack stored in the ‘vignettes’ folder of the repository. In addition, a detailed description of the CEA of Sick Sicker model can be found in the file “report.pdf’ stored in the “report/_book” folder and attached as supplementary material to this manuscript. This report could be used as a template for CEA that are submitted to HTA agencies for their approval. These documents describe the code in detail and will guide the reader on how to run code of the Sick-Sicker model implemented in the DARTH framework.
3. Discussion
We developed the DARTH framework as a way to support transparency, reproducibility, and model sharing in R-based decision analytic models and CEA. Adoption of this general framework will facilitate the sharing and readability of decision analytic models implemented in R as analysts adopting the framework will be familiar with the component structure and the specific choices and assumptions of each component can be easily scrutinized. The standardization of R code presented here may also support the broader use of R in formal HTA submissions, allowing for more complex modeling methods to be more transparently incorporated into decision-making regarding coverage of new health technologies.
As we illustrated in this paper, a traditional model-based decision analysis follows a well-defined conceptual structure. Despite this, our field lacks practical guidance for the implementation of decision modeling in programming languages. DARTH framework addresses this gap and will facilitate overall improvement in the quality, transparency, and reproducibility of decision models and analyses conducted in R. Frameworks like the one we propose have been adopted in other fields such as engineering, mathematics, and computer science to routinize frequently conducted analyses, leading to improvements in quality and efficiency in these methods [42]. There are additional benefits of using R as the platform to develop model-based CEA. One such benefit is that R has established packages that allow the evaluation of functions in parallel using different cores of computing systems. If components have processes that require the evaluation of the model multiple times (e.g., calibration, validation or PSA), the model evaluations can be carried out more efficiently by parallelizing these processes.
While a standardized framework can facilitate model sharing and readability, it must still be flexible enough to accommodate a wide variety of needs and applications. The framework we describe here is meant merely as the scaffolding for any given analysis; ultimately, the analyst should make design decisions that work for their particular use and that facilitate transparency to their audience, be it clients, stakeholders, a government agency, other academics, or the general public. Alongside the details of the DARTH framework, we have also attempted to provide the rationale behind our recommendations so that analysts may adapt the specific structures and recommendations to their needs while following the spirit of the framework.
DARTH framework is focused on the structure and organization of the source code underlying a decision model and analysis to support transparency and sharing. The DARTH framework facilitates dissemination by organizing all the code necessary to conduct a given set of analyses into a single directory that can be easily shared via a repository hosting service, such as GitHub, as we have done in our example model, or through open-source initiates, such as the Open-Source Model Clearinghouse [7]. The DARTH framework is built as an R package, which allows the model and analysis source code to be loaded directly into R. For a description of the steps involved in package development, see the R package book by Hadley Wickham [30]. A package has the advantage of generating a self-contained collection of code with explicit dependencies on other packages and versions with a standard downloading and installation process for users. A package is also advantageous if computationally-intensive functions have been compiled from C/C++ source code, as these functions will be available to the user as R functions. The C/C++ source code can be stored in a folder named “src” as part of an R package.
An R package makes it easier for others to use built-in functions, say for running a model with different input values or exactly reproducing the results of a set of pre-defined analyses. To modify the model structure or adapt it to a new application, the corresponding functions need to be modified and the package must be recompiled. This may be cumbersome in a model-development phase, when debugging and internal validation studies are being conducted. However, if RStudio is used for the package development or adaptation, compiling the package is an effortless task as long as all the R code is sound and well-implemented. If an analyst truly wants their model to be broadly used by practitioners, tools such as R shiny can make interacting with models more user-friendly. Documenting the model structure and different components in the CEA using R documentation and R Markdown also enhances the transparency of the decision models and associated analyses. The use of the DARTH framework alongside these complementary dissemination tools are the foundations for open, transparent and reproducible decision modeling, paving the way to an ideal, open-source world.
Supplementary Material
Key Points for Decision Makers.
The use of open-source software for model-based cost-effectiveness analyses is growing and has the potential to facilitate model transparency, reproducibility, and shareability. However, guidance as to how to structure the required components of such analyses is lacking.
A high-level coding framework can help standardize the construction of model-based decision and cost-effectiveness analyses, allowing the model code to be more easily read, scrutinized, and understood by others. The Decision Analysis in R for Technologies in Health (DARTH) framework modularizes decision models into a set of core components and provides guidance on how to structure and organize the implementation of these commons in R.
Adoption of this general framework will facilitate the sharing and readability of decision models implemented in R. It will also support the broader use of R in formal health technology assessment (HTA) submissions, allowing for more complex modeling methods to be more transparently incorporated into decision-making.
Acknowledgments
We thank Mr. Caleb Easterly for his helpful comments and suggestions on the code developed for this framework and Dr. Myriam Hunink for her overall contribution in the DARTH workgroup.
Funding/support
Dr. Alarid-Escudero was supported by a grant from the National Cancer Institute (U01- CA-199335) as part of the Cancer Intervention and Surveillance Modeling Network (CISNET). Dr. Enns was supported by a grant from the National Institute of Allergy and Infectious Disease of the National Institutes of Health under award no. K25AI118476. Dr. Jalal was supported by a grant from the National Institute of Health (KL2 TR0001856). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funding agencies had no role in the design of the study, interpretation of results, or writing of the manuscript. The funding agreement ensured the authors’ independence in designing the study, interpreting the data, writing, and publishing the report.
Footnotes
Data availability statement
Data and statistical code are provided in the GitHub repository (https://github.com/DARTH-git/darthpack) and the darthpack website (https://darth-git.github.io/darthpack). The version of darthpack released in this article is available at https://doi.org/10.5281/zenodo.3445451.
Conflict of interest
FAE reports no conflicts of interest. EK reports no conflicts of interest. PP reports no conflicts of interest. HJ reports no conflicts of interest. SYK reports no conflicts of interest. AY reports no conflicts of interest. EE reports no conflicts of interest.
Contributor Information
Fernando Alarid-Escudero, Drug Policy Program, Center for Research and Teaching in Economics (CIDE) - CONACyT, Aguascalientes, AGS, Mexico.
Eline Krijkamp, Epidemiology Department, Erasmus MC, Rotterdam, The Netherlands.
Petros Pechlivanoglou, The Hospital for Sick Children and University of Toronto, Toronto, Ontario, Canada.
Hawre Jalal, Department of Health Policy and Management, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, PA, USA.
Szu-Yu Zoe Kao, Division of Health Policy and Management, University of Minnesota School of Public Health, Minneapolis, MN, USA.
Alan Yang, The Hospital for Sick Children, Toronto, Ontario, Canada.
Eva A. Enns, Division of Health Policy and Management, University of Minnesota School of Public Health, Minneapolis, MN, USA.
References
- 1.Taichman DB. Data Sharing Statements for Clinical Trials: A Requirement of the International Committee of Medical Journal Editors. Ann Intern Med. 2017;14:e1002315–e1002315. [DOI] [PubMed] [Google Scholar]
- 2.Stanford. Data Availability Policies at Top Journals [Internet]. 2019. [cited 2019 Aug 2]. Available from: https://web.stanford.edu/~cy10/public/data/Data_Availability_Policies.pdf
- 3.Eddy DM, Hollingworth W, Caro JJ, Tsevat J, McDonald KM, Wong JB. Model transparency and validation: A report of the ISPOR-SMDM modeling good research practices task force-7. Med Decis Mak. 2012;32:733–43. [DOI] [PubMed] [Google Scholar]
- 4.Cohen JT, Neumann PJ, Wong JB. A Call for Open-Source Cost-Effectiveness Analysis. Ann Intern Med. 2017; 1—3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baio G, Heath A. When simple becomes complicated: why Excel should lose its place at the top table. Glob Reg Heal Technol Assess. 2017;4:0–0. [Google Scholar]
- 6.Canadian Agency for Drugs and Technologies in Health (CADTH). Procedure and Submission Guidelines for the CADTH Common Drug Review [Internet]. Ottawa, Canada; 2018. p. 1–113. Available from: https://www.cadth.ca/media/cdr/process/CDR_Submission_Guidelines.pdf [Google Scholar]
- 7.Center for the Evaluation of Value and Risk in Health. Open-Source Model Clearinghouse [Internet]. Tufts University Medical Center; 2019. [cited 2019 Feb 1]. Available from: http://healtheconomics.tuftsmedicalcenter.org/orchard/open-source-model-clearinghouse [Google Scholar]
- 8.Dunlop WCN, Mason N, Kenworthy J, Akehurst RL. Benefits, Challenges and Potential Strategies of Open Source Health Economic Models Pharmacoeconomics. Springer International Publishing; 2017;35:125–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sampson CJ, Wrightson T. Model Registration: A Call to Action PharmacoEconomics - Open. Springer International Publishing; 2017;1:73–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampson CJ, Arnold R, Bryan S, Clarke P, Ekins S, Hatswell A, et al. Transparency in Decision Modelling: What, Why, Who and How? Pharmacoeconomics. Springer International Publishing; 2019;June 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jalal H, Pechlivanoglou P, Krijkamp E, Alarid-Escudero F, Enns EA, Hunink MGM. An Overview of R in Health Decision Sciences. Med Decis Mak. 2017;37:735–46. [DOI] [PubMed] [Google Scholar]
- 12.Decision Analysis in R for Technologies in Health (DARTH) workgroup. Decision Analysis in R for Technologies in Health [Internet]. 2019. [cited 2019 Jan 1]. Available from: http://darthworkgroup.com
- 13.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 14.Marwick B, Boettiger C, Mullen L. Packaging Data Analytical Work Reproducibly Using R (and Friends). Am Stat. Taylor & Francis; 2018;72:80–8. [Google Scholar]
- 15.Stout NK, Knudsen AB, Kong CY (Joey), Mcmahon PM, Gazelle GS. Calibration Methods Used in Cancer Simulation Models and Suggested Reporting Guidelines. Pharmacoeconomics. 2009;27:533–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Briggs AH, Weinstein MC, Fenwick EAL, Karnon J, Sculpher MJ, Paltiel AD. Model Parameter Estimation and Uncertainty Analysis: A Report of the ISPOR-SMDM Modeling Good Research Practices Task Force Working Group-6. Med Decis Mak. 2012;32:722–32. [DOI] [PubMed] [Google Scholar]
- 17.Alarid-Escudero F, MacLehose RF, Peralta Y, Kuntz KM, Enns EA. Nonidentifiability in Model Calibration and Implications for Medical Decision Making. Med Decis Mak. 2018;38:810–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sargent RG. Verification and validation of simulation models. J Simul. Nature Publishing Group; 2013;7:12–24. [Google Scholar]
- 19.Goldhaber-Fiebert JD, Stout NK, Goldie SJ. Empirically evaluating decision-analytic models. Value Health. 2010;13:667–74. [DOI] [PubMed] [Google Scholar]
- 20.Rutter CM, Savarino JE. An evidence-based microsimulation model for colorectal cancer: validation and application. Cancer Epidemiol Biomarkers Prev. 2010;19:1992–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rutter CM, Knudsen AB, Marsh TL, Doria-Rose VP, Johnson E, Pabiniak C, et al. Validation of Models Used to Inform Colorectal Cancer Screening Guidelines: Accuracy and Implications. Med Decis Mak. 2016;36:604–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopec J a, Finès P, Manuel DG, Buckeridge DL, Flanagan WM, Oderkirk J, et al. Validation of population-based disease simulation models: a review of concepts and methods BMC Public Health. BioMed Central Ltd; 2010;10:710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Intervention and Surveillance Modelling Network (CISNET). About CISNET [Internet]. 2019. [cited 2019 Jul 16]. Available from: https://cisnet.cancer.gov/about/index.html
- 24.Krijkamp EM, Alarid-Escudero F, Enns E, Pechlivanoglou P, Hunink MM, Jalal H. A Multidimensional Array Representation of State-Transition Model Dynamics. bioRxiv 670612. 2019;June 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sculpher MJ, Basu A, Kuntz KM, Meltzer DO. Reflecting Uncertainty in Cost-Effectiveness Analysis In: Neumann PJ, Sanders GD, Russell LB, Siegel JE, Ganiats TG, editors. Cost-Effectiveness Heal Med. Second New York, NY: Oxford University Press; 2017. p. 289–318. [Google Scholar]
- 26.Alarid-Escudero F, Enns EA, Kuntz KM, Michaud TL, Jalal H. “Time Traveling Is Just Too Dangerous” But Some Methods Are Worth Revisiting: The Advantages of Expected Loss Curves Over Cost-Effectiveness Acceptability Curves and Frontier. Value Health. 2019;22:611–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raiffa H, Schlaifer RO. Applied Statistical Decision Theory. Cambridge, MA: Harvard Business School; 1961. [Google Scholar]
- 28.Claxton K, Posnett J. An economic approach to clinical trial design and research priority-setting. Health Econ. 1996;5:513–24. [DOI] [PubMed] [Google Scholar]
- 29.Jutkowitz E, Alarid-Escudero F, Kuntz KM, Jalal H. The Curve of Optimal Sample Size (COSS): A Graphical Representation of the Optimal Sample Size from a Value of Information Analysis Pharmacoeconomics. Springer International Publishing; 2019;37:871–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickham H R packages: Organize, test, document, and share your code. Spencer A, Marie Beaugureau, editors. Sebastopol, CA: O’Reilly Media; 2015. [Google Scholar]
- 31.Cooper N, Hsing P-Y, editors. A Guide to Reproducible Code in Ecology and Evolution. London, UK: British Ecology Society; 2017. [Google Scholar]
- 32.Kleijnen JPC. Verification and validation of simulation models. Eur J Oper Res. 1995;82:145–62. [Google Scholar]
- 33.Wickham H The tidyverse style guide [Internet]. 2019. [cited 2019 Jul 19]. Available from: https://style.tidyverse.org
- 34.Google. Google’s R Style Guide [Internet]. 2019. [cited 2019 Jul 24]. p. 1–6. Available from: https://google.github.io/styleguide/Rguide.xml
- 35.Martin RC. Clean code: a handbook of agile software craftsmanship. Boston, MA: Pearson Education; 2009. [Google Scholar]
- 36.Wickham H testthat: Get Started with Testing. R J. 2011;3:5. [Google Scholar]
- 37.Beeley C Web Application Development with R using Shiny. Birmingham, UK: Packt Publishing Ltd; 2013. [Google Scholar]
- 38.Incerti D, Curtis JR, Shafrin J, Lakdawalla DN, Jansen JP. A Flexible Open-Source Decision Model for Value Assessment of Biologic Treatment for Rheumatoid Arthritis Pharmacoeconomics. Springer International Publishing; 2019;37:829–43. [DOI] [PubMed] [Google Scholar]
- 39.Xie Y Bookdown: Authoring Books with R Markdown. 2016. [Google Scholar]
- 40.Enns EA, Cipriano LE, Simons CT, Kong CY. Identifying Best-Fitting Inputs in Health-Economic Model Calibration: A Pareto Frontier Approach. Med Decis Mak. 2015;35:170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.RStudio. Using projects [Internet]. 2019. [cited 2019 Feb 1]. Available from: https://support.rstudio.com/hc/en-us/articles/200526207-Using-Projects
- 42.David O, Ascough JC, Lloyd W, Green TR, Rojas KW, Leavesley GH, et al. A software engineering perspective on environmental modeling framework design: The Object Modeling System Environ Model Softw. Elsevier Ltd; 2013;39:201–13. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.