Abstract
The colonisation of specific body sites in contact with the external environment by microorganisms is both well-described and universally accepted, whereas, the existence of microbial evidence in other “classically sterile” locations including the blood, synovial space, and lungs, is a relatively new concept. Increasingly, a role for the microbiome in disease is being considered, and it is therefore necessary to increase our understanding of these. To date, little data support the existence of a “synovial fluid microbiome”.
Methods
The presence and identity of bacterial and fungal DNA in the synovial fluid of rheumatoid arthritis (RA) patients and healthy control subjects was investigated through amplification and sequencing of the bacterial 16S rRNA gene and fungal internal transcribed spacer region 2 respectively. Synovial fluid concentrations of the cytokines IL-6, IL-17A, IL22 and IL-23 were determined by ELISA.
Results
Bacterial 16S rRNA genes were detected in 87.5% RA patients, and all healthy control subjects. At the phylum level, the microbiome was predominated by Proteobacteria (Control = 83.5%, RA = 79.3%) and Firmicutes (Control = 16.1%, RA = 20.3%), and to a much lesser extent, Actinobacteria (Control = 0.2%, RA = 0.3%) and Bacteroidetes (Control = 0.1%, RA = 0.1%). Fungal DNA was identified in 75% RA samples, and 88.8% healthy controls. At the phylum level, synovial fluid was predominated by members of the Basidiomycota (Control = 53.9%, RA = 46.9%) and Ascomycota (Control = 35.1%, RA = 50.8%) phyla. Statistical analysis revealed key taxa that were differentially present or abundant dependent on disease status.
Conclusions
This study reports the presence of a synovial fluid microbiome, and determines that this is modulated by disease status (RA) as are other classical microbiome niches.
Introduction
The microbiome
The term “microbiome” describes the entire habitat, including the microorganisms (bacteria, archaea, lower and higher eukaryotes, and viruses), their genomes, and the surrounding environmental conditions. Herein, we utilise this term to denote the existence of a microbial community evidenced through the survey of 16S rRNA genes. In contrast, the term “microbiota” refers to the assemblage of viable microorganisms that comprise these communities [1]. Whilst previous estimates stated that the number of microbial cells present in our microbiota exceeded our own by approximately one order of magnitude, current estimates are more conservative and suggest that the number of bacterial cells in the human body is roughly equal to the number of human cells, representing a mass of approximately 0.2 kg [2]. In either case, the microbiota represents a significant source of non-host biological material. The microbiota undertakes essential biological processes and thus it is unsurprising that a number of disease states are associated with changes in microbiome composition, termed “dysbiosis”. Whilst the colonisation of specific body sites in contact with the external environment (such as the gastrointestinal tract, skin and vagina) by microorganisms is both well-described and universally accepted [3], the existence of microorganisms and or microbial DNA in other “classically sterile” locations including the blood [4], synovial space [5], and lungs [6–9], is a relatively new concept.
Rheumatoid arthritis and dysbiosis
Rheumatoid arthritis (RA) is a chronic autoimmune disease that causes synovial joint inflammation leading to significant joint pain, swelling, and significant disability with increased morbidity and mortality. RA is estimated to affect 0.5–1% of the population worldwide [10]. RA aetiology still remains unclear. Although the autoimmune hypothesis is well established the causes generating this self-directed immune reaction are still poorly understood. Although convention still suggests otherwise, an overwhelming volume of epidemiological and experimental data indicates a microbial origin for RA (reviewed extensively by Pretorius and colleagues [11]) and therefore increasingly, a role for the altered microbiome composition in disease initiation and progression is considered. To date, RA has been associated with dysbiosis of the oral [12, 13], gastrointestinal [12, 14–22], and respiratory [23–25] microbiomes and associated with the presence of specific organisms (most commonly P. mirabilis) within the urinary tract [26–29], suggesting that RA results in (or from) pan-microbiome dysbiosis. Studies have further begun to associate RA, or stages thereof, with the presence or absence of specific bacteria, including the expansion of rare bacterial linages [16]. Such findings have the potential to increase our disease understanding, and further suggest that the microbiome may afford a valuable source of novel biomarkers and or novel targets for therapeutic modulation.
The classical assertion that RA lacks a microbial component stems from the generalised inability of clinicians and scientists to observe microbial colonies on synthetic media when samples from RA patients are subjected to routine culture. Such assessments crudely characterise organisms as “alive” or “dead” (or perhaps more accurately, present or not present) based upon this colony forming ability, yet do not routinely recognise the potential for the presence of viable-non culturable bacteria. Furthermore, extensive evidence now provides for the existence of bacterial DNA and RNA in classically sterile areas leaving the possibility of; (a) microbial translocation from classical niches (e.g. the gastrointestinal tract) followed by opsonisation leaving only the nucleic acid, (b) the passage of cell-free microbial nucleic acid through the circulatory system, (c) microbial translocation followed by a state of dormancy due to unfavourable environmental conditions. With relevance to the first two scenarios, it is well-established that the immune system is able to differentiate self from non-self-nucleic acid (DNA and RNA) through specific pattern recognition receptions [30], and that foreign DNA is recognised by TLR9, and RNA is recognised by TLR3 on immune cells, resulting in the upregulation of type I interferons and various pro-inflammatory cytokines [31] (also associated with rheumatoid arthritis) [32]. TLR9 is found in the endosome of peripheral blood mononuclear cells, such as monocytes, macrophages, T, B, and NK cells, whereas TLR3 is expressed only in the endosomes of myeloid derived cells such as dendritic cells and monocytes [33]. With relevance to the latter scenario, dormant or VBNC bacterial cells may persist within the joints of arthritic patients, undetected by routine culture, whilst retaining the ability to shed inflammatory agents such as lipopolysaccharide (LPS) and other antigenic components [11], yet such samples may present as culture-negative. Indeed, bacterial LPS is able to simulate many of the common RA-associated cytokines [34].
Evidence of bacteria in the synovial fluid: Origin and transport
Under normal physiological conditions, one expects the synovial space to be sterile. Indeed, the presence of viable bacteria results in a diagnosis of septic arthritis, a condition regarded as a true medical emergency [35]. Nevertheless, numerous studies report the presence of microorganisms, or microorganism-derived genetic material in the synovial fluid of RA patients and healthy control subjects [36–42]. Interestingly, these studies overwhelmingly identify bacteria most commonly found to reside within the oral cavity, and these findings are accompanied by evidence demonstrating the presence of these DNAs and or antibodies directed against the originating organism in the blood [36, 37, 43–46]. On the basis of these findings, we suggest that bacteria / bacterial nucleic acid may reach the synovial space via the blood (as evidenced by their concurrent presence in both fluids), and further, that the inflammatory environment of the synovial tissues may facilitate the trapping of these bacterial DNAs, increasing their apparent concentration in this location [37].
Despite extensive epidemiological and experimental evidence supporting a microbial cause in RA, mounting evidence linking infection of the oral tissues with RA [23, 47–58], and data demonstrating the appearance of microbial DNA from distant microbiome niches in the synovial space (see above), there has been limited study of the bacterial community in this compartment [5] and no global characterisation of the fungal DNA present in this location to date. To this end, we investigated the presence of bacterial (via sequencing of the 16S rRNA gene) and fungal (via sequencing of the ITS2 region) DNA, in the synovial fluid of human RA patients and healthy control subjects using cutting-edge next generation sequencing and bioinformatic techniques. Furthermore, we evaluated any apparent changes in the bacterial or fungal communities detected, in the context of key markers of synovial inflammation (IL-6, IL-17A, IL-22 and IL-23).
Materials and methods
Donor population
This study investigated the presence of bacterial and fungal DNA in synovial fluid samples from sixteen rheumatoid arthritis patients, and nine sex and BMI-matched healthy control (disease-free) individuals who donated a sample of their synovial fluid for research purposes. Synovial fluid samples were aspirated from the knee joint using aseptic technique, transferred to a sterile micro-centrifuge tube, and stored at -80°C prior to further analysis. Samples were procured from Sera Laboratories Limited.
The Independent Investigational Review Board Inc. (BRI-0722) ethically approved sample collection by Sera Laboratories Limited from human donors giving informed written consent. Furthermore, the authors obtained ethical approval from Keele University Ethical Review Panel 3 for the study reported herein. All experiments were performed in accordance with relevant guidelines and regulations.
Measuring synovial fluid cytokines
Markers of inflammation were measured in synovial fluid utilising the Human Magnetic Luminex Screening Assay following the manufacturer’s instructions (R&D Systems, Minneapolis, USA). Levels of IL-6, IL-17A, IL-22, and IL-23 were analysed by Luminex kit LXSAHM-04. Prior to analysis, synovial fluid samples were centrifuged at 16,000 x g for four minutes and diluted in a 1:2 ratio by adding 25 μl of sample to 25 μl of Assay Buffer prior to analysis.
Microbiome characterisation
Amplification and sequencing of the bacterial 16S rRNA variable region 4 and fungal ITS region was used to characterise the bacterial and fungal DNA present in the synovial fluid samples.
The bacterial 16S rRNA and fungal ITS2 genes were amplified by direct PCR utilising the oligonucleotide primers detailed in (Table 1). Such approaches lack the need for a separate DNA extraction phase, often associated with the introduction of contaminants that impact upon all downstream applications. A first round of PCR using oligonucleotide primers 16SV4 F/R and ITS2 F/R was carried out with four microliters of each synovial fluid sample as template in a final volume of 20μl. Reactions comprised 10μl Phusion blood PCR buffer (Thermofisher), 0.4μl (2 U) Phusion blood DNA polymerase, 1μl of each primer (10μM), and 3.6μl of nuclease-free water that had been subject to 15 minutes UV-irradiation. A negative control reaction in which synovial fluid was substituted with UV-irradiated nuclease-free water was run in parallel with our experimental samples to confirm that none of the reagents were contaminated by target DNA. Cycling parameters comprised an initial denaturation step performed at 98°C for 5 minutes, followed by 33 cycles of denaturation (98°C, 10 seconds), annealing (55°C, 5 seconds) and extension (72°C, 15 seconds). A final elongation of 7 minutes at 72°C completed the reaction. Following gel electrophoresis of 5μl of each resulting PCR product to confirm successful amplification, the remainder (15μl) was purified of excess primer and PCR reagents using the QIAquick PCR Purification kit.
Table 1. Oligonucleotide primers used in this study.
| Primer Name | Primer Sequence (5’– 3’) | Length |
|---|---|---|
| 16SV4_F | GTGCCAGCMGCCGCGGTAA | 19 |
| 16SV4_R | GGACTACHVGGGTWTCTAAT | 20 |
| 16SV4_XT_F | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGTGCCAGCMGCCGCGGTAA | 52 |
| 16SV4_XT_R | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGGACTACHVGGGTWTCTAAT | 54 |
| ITS2_F | GCA TCG ATG AAG AAC GCAGC | 20 |
| ITS2_R | TCCTCCGCTTATTGATATGC | 20 |
| ITS2_XT_F | TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG GCATCGATGAAGAACGCAGC | 53 |
| ITS2_XT_R | GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG TCCTCCGCTTATTGATATGC | 54 |
In order to add sequencing adapters to facilitate MiSeq library preparation, a second round of PCR amplification was conducted in a total volume of 50μl comprising 10μl 5X Platinum Super-Fi Buffer, 1μl 10mM dNTP mixture, 0.5μl Platinum Super-Fi polymerase, 2.5μl of each XT_tagged primer (16SV4_XT F/R or ITS2_XT F/R), 5μL from the successful first round PCR reaction and 38.5 μL of UV-irradiated nuclease-free water. Cycling parameters comprised an initial denaturation step performed at 98°C for 2 minutes, followed by 7 cycles of denaturation (98°C, 10 seconds), and annealing / extension (72°C, 20 seconds) extension (72°C, 15 seconds). A final elongation of 5 minutes at 72°C completed the reaction. Two controls were run in parallel with this process and these comprised a standard PCR negative control (in which template was replaced with UV irradiated molecular biology grade water) and the first round PCR control which was progressed alongside our experimental samples. All PCR products and our two control reactions were purified using AMPure XP magnetic beads (Agencourt) at a ratio of 0.8 beads to sample (v/v), eluted in 20μl of UV-irradiated nuclease-free water, and visualised by agarose gel electrophoresis and ehidium bromide staining as above. All PCR negative reactions were subject to high sensitivity DNA quantification using the Qubit 3.0 hsDNA kit (Invitrogen) to quantitate any DNA in these samples.
Amplicons were barcoded using the Nextera DNA library kit, multiplexed for efficiency, and sequenced using the Ilumina MiSeq system with a 250bp paired-end read metric. Bioinformatic analysis was performed using QIIME implemented as part of the Nephele 16S / ITS paired-end QIIME pipeline using open reference clustering against the SILVA database for bacteria and the ITS database for fungi at a sequence identity of 99%. All other parameters remained as default. The statistical significance of differences in the abundance of individual bacteria and fungi between the RA and control donors was determined by applying a two-tailed, Mann Whitney test using GraphPad Prism V8.0.
Results
Clinical features of donors and results of 16S rRNA and ITS2 PCR amplification
Synovial fluid samples were obtained from 25 human donors. Sixteen patients were diagnosed with RA, and of these, 8 were male and 8 were female. The RA patients’ ages ranged from 52 to 74 years, with a mean of 65.3 years. Nine control synovial fluid samples were obtained from 5 males and 4 females. Their ages ranged from 50 to 68 years, with a mean of 61.0 years. There were no significant differences in the ages of the two cohorts (Unpaired T-test; P = > 0.05) (Table 2).
Table 2. Characteristics of the patient population.
| Patient ID# | Age | Diagnosis | 16S rRNA PCR | ITS2 PCR |
|---|---|---|---|---|
| 1339 | 61–65 | RA | + | − |
| 1340 | 66–70 | RA | + | + |
| 1341 | 66–70 | RA | + | + |
| 1342 | 66–70 | RA | + | − |
| 1343 | 66–70 | RA | + | − |
| 1344 | 66–70 | RA | + | − |
| 1345 | 56–60 | RA | + | + |
| 1346 | 50–55 | RA | + | + |
| 1347 | 50–55 | RA | + | + |
| 1348 | 66–70 | RA | + | + |
| 1349 | 71–75 | RA | + | + |
| 1350 | 66–70 | RA | + | + |
| BRH1095336 | 66–70 | RA | + | + |
| BRH1095337 | 66–70 | RA | − | + |
| BRH1095338 | 66–70 | RA | − | + |
| BRH1095339 | 66–70 | RA | + | + |
| 1351 | 56–60 | Healthy | + | + |
| 1352 | 61–65 | Healthy | + | − |
| 1353 | 61–65 | Healthy | + | + |
| 1354 | 50–55 | Healthy | + | + |
| 1355 | 50–55 | Healthy | + | + |
| 1356 | 71–75 | Healthy | + | + |
| 1357 | 61–65 | Healthy | + | + |
| 1358 | 50–55 | Healthy | + | + |
| 1359 | 66–70 | Healthy | + | + |
Utilising PCR amplification, bacterial 16S DNA was detected in the synovial fluid of 14 of out 16 patients with RA (87.5%), and in 9 out of 9 (100%) healthy control subjects. ITS2 amplification, indicative of the presence of fungi, was detected in 12 of 16 (75%) RA samples, and 8 out of 9 (89%) healthy control samples. Our various experimental controls designed to identify target contamination amongst the molecular reagents and plasticware used consistently failed to produce a visible band following PCR and agarose gel electrophoresis. Furthermore, these samples were subjected to DNA quantification using the Qubit 3.0 high-sensitivity DNA kit (Invitrogen) and were found to be below the limit of detection in all cases.
Assessment of inflammatory markers in synovial fluid
Synovial fluid concentrations of interleukin 6 (IL-6), 17A (IL-17A), 22 (IL-22) and 23 (IL-23) were measured using the Luminex system as described earlier. Mean interleukin levels in healthy control and RA patient synovial fluid were significantly different, with cytokines being present at higher levels in the RA synovial fluid in all cases (Unpaired T- test) (Table 3).
Table 3. Differences in cytokine levels among.
| Cytokine | Control mean (SD) (pg/mL) | RA mean (SD) (pg/mL) | P value (pg/mL) |
|---|---|---|---|
| IL-6 | 156.43 (100.30) | 1257.80 (905.66) | <0.005 |
| IL-17A | 11.68 (3.92) | 65.44 (16.51) | <0.005 |
| IL-22 | 220.14 (85.02) | 873.00 (376.46) | <0.005 |
| IL-23 | 145.64 (64.43) | 296.21 (97.07) | <0.005 |
Characterisation of Bacterial populations via 16S rDNA sequencing of synovial fluid
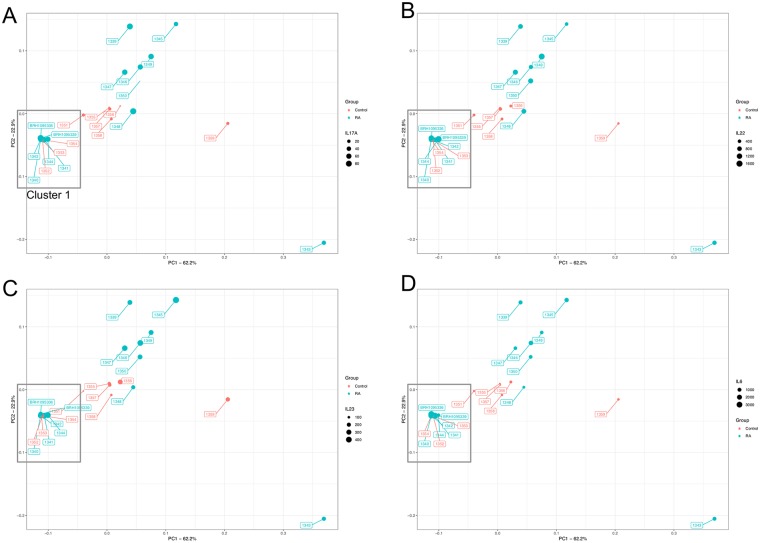
The presence of bacterial DNA in synovial fluid was evaluated via PCR amplification and sequencing of the bacterial 16S rRNA gene, variable region 4. An average of 75,000 reads were generated for each of the samples; 70,658 reads in the RA samples, and 80,533 reads in the control samples. Although the control samples generated more reads on average, this difference was not statistically significant (Unpaired T test; P = > 0.05), and nevertheless, rarefaction was used prior to differential abundance analysis to control for differences in sequencing depth. Following taxonomic classification using the Nephele platform as described above, our first analytical approach utilised Principal Coordinates Analysis (PCoA) to reduce the complexity of the data obtained, and to visualise any obvious separation between the various experimental samples (Fig 1).
Fig 1. Principal coordinate analysis plot generated from a Bray Curtis distance matrix of synovial fluid bacterial community structure for control subjects (red) and arthritic patients (blue) as determined via amplification and sequencing of the 16S rRNA gene variable region 4 (V4).
Proportions of variation explained by the principal coordinates are designated on the axes. PCoA found that the maximal variation were 62.2% (PC1), and 22.9% (PC2). The microbiome of samples that appear in close proximity to each other are more similar in composition. The data points in each pane are sized according to the synovial concentration of IL-17A (A), IL-22 (B), IL-23 (C), and IL-6 (D).
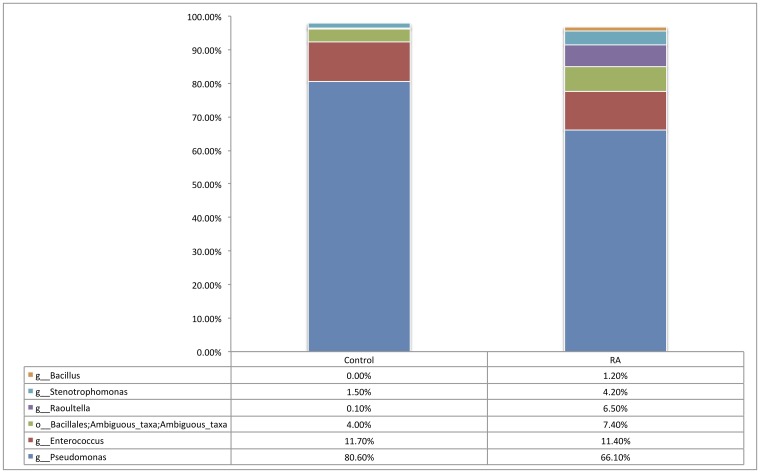
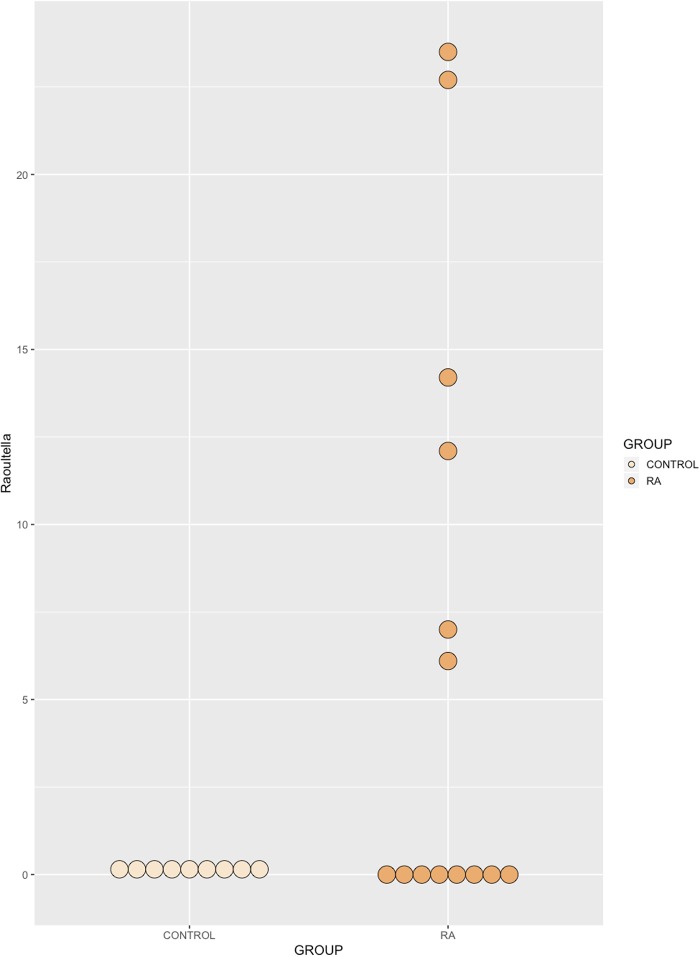
At the phylum level, the synovial fluid microbiome was predominated by Proteobacteria (Control = 83.5%, RA = 79.3%) and Firmicutes (Control = 16.1%, RA = 20.3%), and to a much lesser extent, Actinobacteria (Control = 0.2%, RA = 0.3%) and Bacteroidetes (Control = 0.1%, RA = 0.1%). At the genus level, our synovial fluid samples were predominated by the genus Pseudomonas (Control = 80.6%, RA = 66.1%, P = 0.30), followed by the genus Enterococcus (Control = 11.7%, RA = 11.4%, P = 0.95). To a lesser extent, synovial fluid samples contained members of the Stenotrophomonas (Control = 1.5%, RA = 4.2%, P = 0.97), Raoultella (Control = <0.1%, RA = 6.5%, P = 0.051) and Bacillus (Control = 0%, RA = 1.2%, P = 0.99) genera, the latter two being detected almost exclusively in the synovial fluid of our RA patients (Fig 2). Finally, unidentified taxa of the Bacillales order (Control = 4%, RA = 7.4%, P = 0.97) were also present in relatively high abundance yet could not be further taxonomically classified. Statistical analysis revealed that whilst the abundance of most bacterial DNA, at different taxonomic levels, was unaffected by disease status, the abundance of Raoultella was borderline significant (P = 0.051) and observed as the almost exclusive presence of this organism’s DNA in the synovial fluid of RA patients only (Fig 3).
Fig 2. Relative abundance of genera representing >1% of the total bacterial population in synovial fluid samples of rheumatoid arthritis (RA, n = 14) and control (Control, n = 9) samples as determined by amplification and sequencing of the 16S rRNA gene variable region 4.
Data are mean abundance expressed as a percentage of the total bacterial sequence count.
Fig 3. Relative abundance of the genus Raoultella, identified in the synovial fluid of healthy donors and RA patients by amplification and sequencing of the 16S rRNA gene variable region.
A borderline significant (p = 0.051) increase in the mean abundance of Raoultella was identified in RA synovial fluid relative to healthy control synovial fluid. Data are individual sample abundances expressed as a percentage of the total bacterial sequence count.
Inflammatory markers and modulated by bacterial microbiome PCoA cluster
Based upon the clustering of samples observed following PCoA (Fig 1), we investigated whether the samples in “cluster 1”, had significantly different inflammatory marker levels to the remaining samples, in addition to different microbial populations. Interestingly, IL-6 was significantly higher in PCoA cluster 1 compared with the remaining samples (Two Way ANOVA; P = 0.030) yet all other inflammatory markers were unaffected by PCoA cluster (P > = 0.05).
Characterisation of fungal populations via ITS2 sequencing of synovial fluid
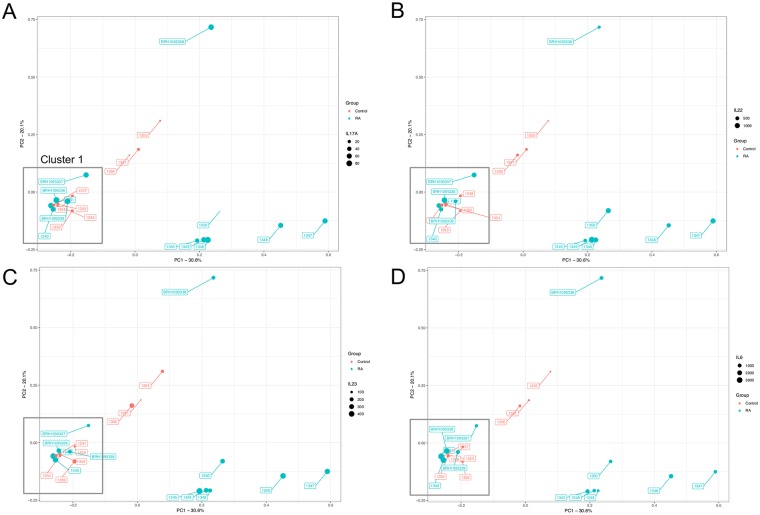
The presence of fungal DNA in synovial fluid was evaluated via PCR amplification and sequencing of the fungal ITS2 gene. An average of 50,000 reads were generated for each of the samples; 52,113 reads in the RA samples, and 49,929 reads in the Control samples. Although the RA samples generated more reads on average, this difference was not statistically significant (P = > 0.05). Following taxonomic classification using the Nephele platform as described above, our first approach utilised Principal Coordinates Analysis (PCoA) to reduce the complexity of the data obtained, and to visualise any obvious separation between the two experimental groups (Fig 4).
Fig 4. Principal coordinate analysis plot generated from a Bray Curtis distance matrix of synovial fluid fungal community structure for control subjects (red) and arthritic patients (blue) as determined by amplification and sequencing of the ITS2 gene.
Proportions of variation explained by the principal coordinates are designated on the axes. PCoA found that the maximal variation were 30.6% (PC1), and 20.1%. The microbiome of samples that appear in close proximity to each other are more similar in composition. The data points in each pane are sized according to the synovial concentration of IL-17A (A), IL-22 (B), IL-23 (C), and IL-6 (D).
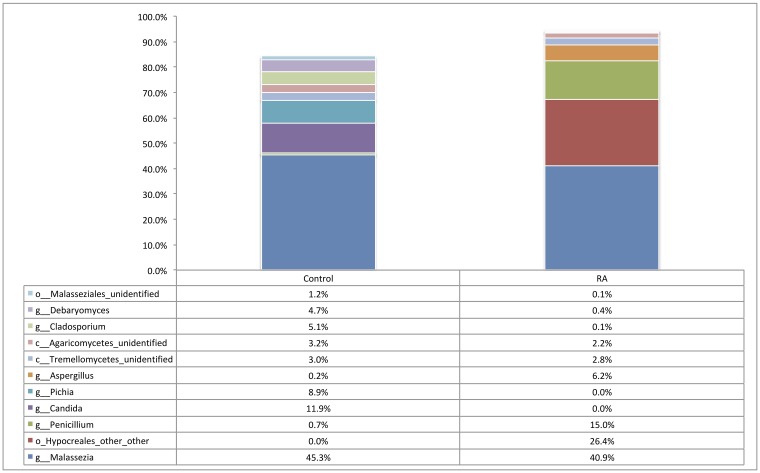
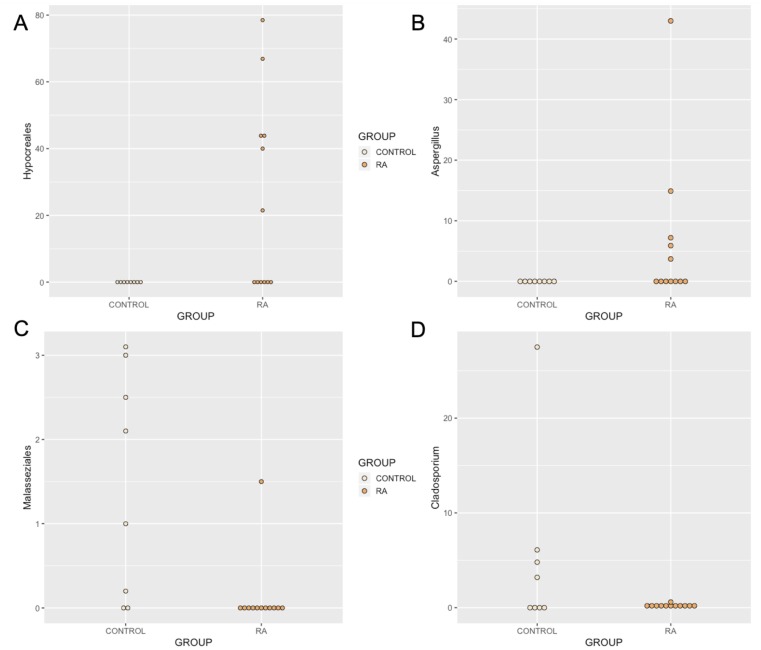
At the phylum level, synovial fluid was found to be predominated by members of the Basidiomycota (Control = 53.9%, RA = 46.9%) and Ascomycota (Control = 35.1%, RA = 50.8%) phyla. At the genus level, synovial fluid samples were predominated by the genus Malassezia, which accounted for 45.3% and 40.9% of the total number of fungal sequences identified in the Control and RA donors respectively (P = 0.8). To a lesser extent, synovial fluid samples also contained organisms from the genera Penicillium (Control = 0.7%, RA = 15%, P = 0.6), Candida (Control = 11.9%, RA = 0.00%, P = 0.4), Pichia (Control = 8.9%, RA = 0.00%, P = 0.4), Aspergillus (Control = 0.2%, RA = 6.2%, P = 0.14), Cladosporium (Control = 5.1%, RA = 0.1%, P = 0.019), and Debaryomyces (Control = 4.7%, RA = 0.4%, P = 0.7). Furthermore, genus level classification revealed the presence of specific unclassified organisms belonging to the classes Tremellomycetes (Control = 3%, RA = 2.8%, P = 0.98), and Agaricomycetes (Control = 3.2%, RA = 2.2%, P = 0.3), and the orders Hypocreales (Control = 0.00%, RA = 26.4%, P = 0.012) and Malasseziales (Control = 1.2%, RA = 0.1%, P = 0.002) (Fig 5). Statistical analysis revealed that whilst the abundance of most fungal DNA was unaffected by disease state, select organisms were differentially abundant or present (Fig 6). An unclassified member of the Hypocreales order (P = 0.012) and a member of the genus Aspergillus was detected almost exclusively in RA synovial fluid samples only (P = 0.14 including all data, P = 0.057 excluding samples where <1% of reads mapped to this taxa). Conversely, a member of the order Malasseziales (P = 0.002), and the genus Cladosporium (P = 0.019) were observed almost exclusively in control synovial fluid.
Fig 5. Relative abundance of taxa representing >1% of the total fungal population in each synovial fluid sample (RA, n = 12) and (Control, n = 9) as determined by amplification and sequencing of the fungal ITS2 gene.
Data are mean abundance expressed as a percentage of the total fungal sequence count.
Fig 6. Relative abundance of (A) Hypocreales, (B), Aspergillus, (C) Malasseziales, and (D) Cladosporium identified in the synovial fluid of RA patients and healthy control subjects.
Data determined by the amplification and sequencing of the ITS2 gene. (A) The relative abundance of order Hypocreales was increased in synovial fluid of RA patients relative to healthy control synovial fluid. (B) The relative abundance of genus Aspergillus was increased in RA synovial fluid samples compared with healthy subjects (borderline significant). (C), and (D) The abundance of the order Malasseziales and genus Cladosporium were significantly decreased in the synovial fluid of RA patients in comparison to the synovial fluid of healthy control subjects. Data are individual expression values expressed as a percentage of the total fungal read count in each sample. The statistical significance between groups was determined by unpaired t test where P ≤ 0.05.
Inflammatory markers and fungal microbiome PCoA cluster
Based solely upon the clustering of samples observed following PCoA, we investigated whether samples in PCoA “cluster 1” had significantly different inflammatory marker profiles, in addition to different microbial populations, than the remaining samples. IL-6 was significantly higher in PCoA cluster 1 compared with the remaining samples (Two Way ANOVA; P = 0.016). Further, the interaction between PCoA cluster and disease status (control or RA) was also significant (P = 0.048). All other inflammatory markers were unaffected by PCoA cluster (P > = 0.05).
Discussion
Mounting evidence suggests pan-microbiome dysbiosis in patients with RA. Moreover, various studies spanning many decades link the development of RA with infections of the oral tissues or urinary system, and report the presence of microorganisms or microorganism-derived genetic material in the synovial fluid; a compartment classically considered to be sterile. To our surprise, only one study has investigated the potential for a “synovial fluid microbiome” using next generation sequencing [5]. To this end, we investigated the presence of bacterial and fungal DNA in the synovial fluid of rheumatoid arthritis (RA) patients and healthy control subjects through amplification and sequencing of the 16S rRNA and ITS2 genes.
Evidence of bacteria
Synovial fluid samples from 25 subjects were analysed in this study and our analyses revealed the presence of a complex synovial microbiome community in health and disease. Bacterial 16S rRNA was detected in fourteen out of sixteen (87.5%) RA patients, and nine out of nine (100%) healthy control subjects. At the phylum level, the synovial fluid microbiome was predominated by Proteobacteria (81.1%) and Firmicutes (18.5%), and to a much lesser extent, Actinobacteria (0.3%) and Bacteroidetes (0.1%). These phyla were also identified as the most abundant in the only other comparable study [5], albeit in slightly different proportions. Whilst the major phyla identified appear similar to those of the blood microbiome [59], they are present in the synovial fluid at different levels, suggesting a distinct community structure.
At the genus level, our synovial fluid samples were predominated by DNA of the genus Pseudomonas and Enterococcus, to a lesser extent, unidentified taxa of the Bacillales order and Stenotrophomonas genus. The genera Raoultella and Bacillus were detected almost exclusively in the synovial fluid of our RA patients (Figs 2 and 3). DNA of Pseudomonas species, which was the most common genus to be identified in the study cohort, has previously been identified in the synovial fluid of patients with RA [42, 60].
Whilst the bacterial communities characterised in the disease and control states were largely consistent, the presence of the genus Raoultella was largely restricted to the RA synovial fluid in our study (present only in 4 of our control samples with read counts < 0.3% yet detected in 6 of our RA patient samples with read abundances in the range 6.1–23.5%). Interestingly, when present, Raoultella often occupied a large proportion of the total bacterial read count. This is the first study to describe the presence of DNA from the genus Raoultella in RA synovial fluid samples, however, two major species are capable of causing infections in man: Raoultella ornithinolytica and Raoultella planticola, and have been shown to be capable of reaching the articular joint compartment [61–63]. Moreover, Raoultella are characterised as histamine generating bacteria [64], a key pro-inflammatory molecule found in RA blood, articular cartilage, synovial fluid and synovial tissue [65]. It is plausible that the presence and or indeed abundance of Raoultella DNA may correlate with specific disease features (for example; disease stage, severity, current disease activity, treatment regimen), and we therefore plan to explore this relationship further in the future, particularly with the aim of identifying the origin of the DNA found.
With regards the numerous reports detailing a link between oral and urinary tract associated bacteria and RA, we reviewed the results of our taxonomic classification looking specifically for the presence and abundance of these organisms. Bacteria of the genus Porphyromonas were largely absent with the exception of three synovial fluid samples (Control = 2, RA = 1), which returned a small number of reads mapping to this genus. These results differ from those of Zhao and colleagues who report the presence of Porphyromonas in all of the samples they sequenced [5], irrespective of disease state [5]. Moreover, no reads mapping to the genus Proteus were identified in any sample analysed, however numerous other members of the order Enterobacteriales to which Proteus belongs were identified, including Raoultella. The presence of Porphyromonas (gingivalis) has been previously associated with increased levels of protein citrullination as a result of its bacterial PPAD enzyme, and increased anti-citrullinated protein antibodies (ACPAs). Although we could not determine ACPA titres in our samples due to sample availability, one possible explanation for the absence of Porphyromonas in our study is that our patients presented with lower anti-citrullinated protein antibody levels, relative to the Zhao study. In considering the presence additional disease-associated organisms, it is important to note here that none of our RA patients reported oral or urinary conditions, and it is therefore further plausible that the above bacteria are present only during active oral or urinary disease. A separate study is therefore warranted to further investigate these phenomena including RA patients (without co-morbidities), RA patients with poor oral health, and patients with periodontitis alone (without RA).
Evidence of fungi
Fungal DNA was identified in 12 out of 16 (75%) RA samples, and 8 out of 9 (88.8%) healthy controls. Our various experimental controls consistently failed to produce a visible band following PCR and agarose gel electrophoresis. Furthermore, DNA quantification using the Qubit 3.0 high-sensitivity DNA kit (Invitrogen) was consistently below the limit of detection. These indicate that the fungal DNA identified originates from within the samples rather than from contaminants introduced. At the phylum level, synovial fluid was predominated by members of the Basidiomycota and Ascomycota phyla. At the genus level, synovial fluid samples were predominated by the genus Malassezia, which accounted for 45.3% and 40.9% of the total number of fungal sequences identified in the Control and RA donors respectively. To a lesser extent, synovial fluid samples also contained organisms from the genera Penicillium, Candida, Pichia, Aspergillus, Cladosporium, and Debaryomyces. Furthermore, genus level classification revealed the presence of specific unclassified organisms belonging to the classes Tremellomycetes and Agaricomycetes, and the orders Hypocreales and Malasseziales (Fig 6).
Again we identified key taxa that appeared to be differentially present or abundant dependent on disease status. The order Hypocreales was selectively abundant in RA patients (7 / 12) and absent from healthy control synovial fluid. Interestingly, when present, members of the Hypocreales order occupied a large proportion of the total fungal read count (~ 40%). Evidence for the presence of members of this order have been previously identified in the healthy human oral cavity [66] and found to be ubiquitous in human plasma [67], however no reports directly link this order with RA.
Further, the abundance of genus Aspergillus was borderline significant; detected more commonly in the synovial fluid of RA patients, and occupying an average of ~ 10.0% of the read population when present. Aspergillus has been identified in the healthy human gut [68] and oral cavity [66] in various studies, and is a prevalent opportunistic pathogen of the lungs, particularly affecting immunocompromised individuals, where it causes a range of pulmonary diseases [69]. Furthermore, Aspergillus galactomannan antigen level has been shown to elevate in a non-specific manner among patients with RA [70], further hinting towards a possible association.
The presence members of the genus Cladosporium were largely absent in the synovial fluid of all RA patients in comparison to the synovial fluid of healthy control subjects (present in 5 / 8 healthy control patients in comparison with 2 / 12 RA samples). Cladosporium has been shown to colonise the healthy human gut [68] and oral cavity [66], and is encountered commonly in human clinical samples [71], thus evidence of Cladosporium DNA in the healthy patient is perhaps not unexpected. That said, it’s absence from the synovial fluid of the majority of RA patients may suggest an altered fungal community in some distant microbial niche, that presents as a reduction in translocated Cladosporium DNA. These findings all warrant further investigation.
In-vivo studies have demonstrated that mycotoxins may play a role in the pathogenesis of RA [72]. Furthermore, cells such as Th17 that are involved in the immunity against fungi are also implicated in the pathogenesis of RA [73]. These indicate that fungal presence in synovial fluid of patients with RA may contribute to pathogenesis. However, as fungal DNA was also found in healthy controls, further studies are needed, with quantification of the load of fungal DNA and taxa specific components.
Changes in microbiome with synovial fluid cytokine levels
Numerous cytokines have been implicated in the pathogenesis of RA. Levels of IL-17-A, IL-22, IL23, and IL-6 have been identified to be higher in diseased joints than in healthy [74, 75], and IL-6 has been identified to play a key role in the pathogenesis of RA. It is implicated in pathogenesis of local joint disease as well as systemic manifestations [76, 77]. Serum and synovial fluid IL-6 levels correlate with disease severity and activity [78, 79]. We found that IL-6 levels were higher among a sub-cluster of the RA population defined according to the fungal and bacterial microbiome. As mentioned earlier, Th17 cells are important in the immunity against fungal pathogens, and IL-6 is involved in the differentiation of lymphocytes to Th17 cells [77]. This could indicate that the differences in the fungal microbiome within patients with RA might be related to disease severity through an IL-6 –Th17 mediated pathway.
Limitations
Bacterial DNA was identified in 14/16 patients with RA, and in all disease free controls whereas fungal DNA was identified in 12 out of 16 RA samples, and 8 out of 9 healthy controls. These results are consistent with other published works that have previously reported the presence of bacterial DNA in the majority, but not necessarily all, samples investigated [80, 81]. A potential limitation of this study is that we did not elect to sequence of experimental controls, and relied on the absence of a detectable signal following agarose gel electrophoresis and QuBit analysis using the high sensitivity DNA kit. Whilst is plausible that sequencing could have revealed the presence of low abundance target DNA introduced during our experimental procedures we suggest that this (potential) DNA would be (1) present at a much lower concentration (given our experimental samples were clearly visible following gel electrophoresis relative to the absence of any bands for our experimental controls), and (2) consistent given that all samples were processed in parallel. Whilst we acknowledge this potential limitation, it does not detract from our finding of statistically different levels of specific bacteria between the two experimental groups, despite all samples being processed at the same time utilising the same specific batches of reagents.
Conclusions
This is one of the first studies to report the existence of a synovial fluid microbiome, and to determine that this is modulated by disease status (RA) as are other classical microbiome niches. Our recent work on the blood microbiome [4], and that of others [82–92], characterised a complex community of organisms (or the genetic material thereof, more specifically) that likely originate in one of the classical microbiome niches (the gut, mouth, urogenital tract, skin) and reach the circulation through a process termed atopobiosis. We hypothesise that the same process may be true of the community we characterised here in the synovial fluid, with the circulatory system acting as a conduit between their usual place of habitation and the synovial compartment. Further studies incorporating pan-microbiome analysis are required to support these data.
Preprint
This manuscript has been published as a preprint bioRxiv [93].
Data Availability
Data deposited at OSF, DOI 10.17605/OSF.IO/HK4F6.
Funding Statement
DBMH/DPT R35 Higher Committe for Education Development http://www.hcediraq.org/HCED_english_website/homeen.html The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Marchesi JR, Ravel J. The vocabulary of microbiome research: a proposal. Microbiome. 2015;3:31 Epub 2015/07/30. 10.1186/s40168-015-0094-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016;14(8):e1002533 Epub 2016/08/19. 10.1371/journal.pbio.1002533 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markova ND. L-form bacteria cohabitants in human blood: significance for health and diseases. Discov Med. 2017;23(128):305–13. . [PubMed] [Google Scholar]
- 4.Whittle E, Leonard M, Harrison R, Gant T, Tonge D. Multi-Method Characterisation of the Human Circulating Microbiome. BioRxiv. 2018. Epub 2018. 10.1101/359760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Y, Chen B, Li S, Yang L, Zhu D, Wang Y, et al. Detection and characterization of bacterial nucleic acids in culture-negative synovial tissue and fluid samples from rheumatoid arthritis or osteoarthritis patients. Sci Rep. 2018;8(1):14305 Epub 2018/09/24. 10.1038/s41598-018-32675-w . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao Q, Jiang F, Yin R, Wang J, Xia W, Dong G, et al. Interplay between the lung microbiome and lung cancer. Cancer Lett. 2018;415:40–8. Epub 2017/12/02. 10.1016/j.canlet.2017.11.036 . [DOI] [PubMed] [Google Scholar]
- 7.Moffatt MF, Cookson WO. The lung microbiome in health and disease. Clin Med (Lond). 2017;17(6):525–9. 10.7861/clinmedicine.17-6-525 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Dwyer DN, Dickson RP, Moore BB. The Lung Microbiome, Immunity, and the Pathogenesis of Chronic Lung Disease. J Immunol. 2016;196(12):4839–47. 10.4049/jimmunol.1600279 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Hao K, Yang T, Wang C. Role of the Lung Microbiome in the Pathogenesis of Chronic Obstructive Pulmonary Disease. Chin Med J (Engl). 2017;130(17):2107–11. 10.4103/0366-6999.211452 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gibofsky A. Overview of epidemiology, pathophysiology, and diagnosis of rheumatoid arthritis. Am J Manag Care. 2012;18(13 Suppl):S295–302. . [PubMed] [Google Scholar]
- 11.Pretorius E, Akeredolu OO, Soma P, Kell DB. Major involvement of bacterial components in rheumatoid arthritis and its accompanying oxidative stress, systemic inflammation and hypercoagulability. Exp Biol Med (Maywood). 2017;242(4):355–73. Epub 2016/11/26. 10.1177/1535370216681549 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21(8):895–905. Epub 2015/07/27. 10.1038/nm.3914 . [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Oliva I, Paropkari AD, Saraswat S, Serban S, Yonel Z, Sharma P, et al. Dysbiotic Subgingival Microbial Communities in Periodontally Healthy Patients With Rheumatoid Arthritis. Arthritis Rheumatol. 2018;70(7):1008–13. Epub 2018/05/21. 10.1002/art.40485 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202 10.7554/eLife.01202 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taneja V. Arthritis susceptibility and the gut microbiome. FEBS Lett. 2014;588(22):4244–9. 10.1016/j.febslet.2014.05.034 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 2016;8(1):43 Epub 2016/04/21. 10.1186/s13073-016-0299-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Zeng B, Zhang J, Li W, Mou F, Wang H, et al. Role of the Gut Microbiome in Modulating Arthritis Progression in Mice. Sci Rep. 2016;6:30594 10.1038/srep30594 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scher JU, Littman DR, Abramson SB. Microbiome in Inflammatory Arthritis and Human Rheumatic Diseases. Arthritis Rheumatol. 2016;68(1):35–45. 10.1002/art.39259 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 2016. 10.1002/art.39783 . [DOI] [PubMed] [Google Scholar]
- 20.Vaahtovuo J, Munukka E, Korkeamäki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol. 2008;35(8):1500–5. Epub 2008/06/01. . [PubMed] [Google Scholar]
- 21.Liu X, Zou Q, Zeng B, Fang Y, Wei H. Analysis of fecal Lactobacillus community structure in patients with early rheumatoid arthritis. Curr Microbiol. 2013;67(2):170–6. Epub 2013/03/13. 10.1007/s00284-013-0338-1 . [DOI] [PubMed] [Google Scholar]
- 22.Wu X, Liu J, Xiao L, Lu A, Zhang G. Alterations of Gut Microbiome in Rheumatoid Arthritis. Osteoarthritis and Cartilage. 2017;25:S287–S8. [Google Scholar]
- 23.Brusca SB, Abramson SB, Scher JU. Microbiome and mucosal inflammation as extra-articular triggers for rheumatoid arthritis and autoimmunity. Curr Opin Rheumatol. 2014;26(1):101–7. 10.1097/BOR.0000000000000008 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demoruelle MK, Solomon JJ, Fischer A, Deane KD. The lung may play a role in the pathogenesis of rheumatoid arthritis. Int J Clin Rheumtol. 2014;9(3):295–309 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scher JU, Joshua V, Artacho A, Abdollahi-Roodsaz S, Öckinger J, Kullberg S, et al. The lung microbiota in early rheumatoid arthritis and autoimmunity. Microbiome. 2016;4(1):60 Epub 2016/11/17. 10.1186/s40168-016-0206-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebringer A, Rashid T. Rheumatoid arthritis is an autoimmune disease triggered by Proteus urinary tract infection. Clin Dev Immunol. 2006;13(1):41–8. 10.1080/17402520600576578 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebringer A, Rashid T. Rheumatoid arthritis is caused by a Proteus urinary tract infection. APMIS. 2014;122(5):363–8. Epub 2013/08/29. 10.1111/apm.12154 . [DOI] [PubMed] [Google Scholar]
- 28.Tani Y, Tiwana H, Hukuda S, Nishioka J, Fielder M, Wilson C, et al. Antibodies to Klebsiella, Proteus, and HLA-B27 peptides in Japanese patients with ankylosing spondylitis and rheumatoid arthritis. J Rheumatol. 1997;24(1):109–14. . [PubMed] [Google Scholar]
- 29.Wilson C, Thakore A, Isenberg D, Ebringer A. Correlation between anti-Proteus antibodies and isolation rates of P. mirabilis in rheumatoid arthritis. Rheumatol Int. 1997;16(5):187–9. 10.1007/bf01330294 . [DOI] [PubMed] [Google Scholar]
- 30.Chi H, Flavell RA. Innate recognition of non-self nucleic acids. Genome Biol. 2008;9(3):211 Epub 2008/03/10. 10.1186/gb-2008-9-3-211 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Atianand MK, Fitzgerald KA. Molecular basis of DNA recognition in the immune system. J Immunol. 2013;190(5):1911–8. 10.4049/jimmunol.1203162 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castañeda-Delgado JE, Bastián-Hernandez Y, Macias-Segura N, Santiago-Algarra D, Castillo-Ortiz JD, Alemán-Navarro AL, et al. Type I Interferon Gene Response Is Increased in Early and Established Rheumatoid Arthritis and Correlates with Autoantibody Production. Front Immunol. 2017;8:285 Epub 2017/03/20. 10.3389/fimmu.2017.00285 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enninga EA, Nevala WK, Holtan SG, Markovic SN. Immune Reactivation by Cell-Free Fetal DNA in Healthy Pregnancies Re-Purposed to Target Tumors: Novel Checkpoint Inhibition in Cancer Therapeutics. Front Immunol. 2015;6:424 Epub 2015/08/26. 10.3389/fimmu.2015.00424 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossol M, Heine H, Meusch U, Quandt D, Klein C, Sweet MJ, et al. LPS-induced cytokine production in human monocytes and macrophages. Crit Rev Immunol. 2011;31(5):379–446. . [DOI] [PubMed] [Google Scholar]
- 35.Ross JJ. Septic Arthritis of Native Joints. Infect Dis Clin North Am. 2017;31(2):203–18. Epub 2017/03/30. 10.1016/j.idc.2017.01.001 . [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Martinez RE, Abud-Mendoza C, Patiño-Marin N, Rizo-Rodríguez JC, Little JW, Loyola-Rodríguez JP. Detection of periodontal bacterial DNA in serum and synovial fluid in refractory rheumatoid arthritis patients. J Clin Periodontol. 2009;36(12):1004–10. 10.1111/j.1600-051X.2009.01496.x . [DOI] [PubMed] [Google Scholar]
- 37.Moen K, Brun JG, Valen M, Skartveit L, Eribe EK, Olsen I, et al. Synovial inflammation in active rheumatoid arthritis and psoriatic arthritis facilitates trapping of a variety of oral bacterial DNAs. Clin Exp Rheumatol. 2006;24(6):656–63. . [PubMed] [Google Scholar]
- 38.Ogrendik M. Rheumatoid arthritis is linked to oral bacteria: etiological association. Modern rheumatology. 2009;19(5):453–6. 10.1007/s10165-009-0194-9 [DOI] [PubMed] [Google Scholar]
- 39.Reichert S, Haffner M, Keyßer G, Schäfer C, Stein JM, Schaller HG, et al. Detection of oral bacterial DNA in synovial fluid. Journal of clinical periodontology. 2013;40(6):591–8. 10.1111/jcpe.12102 [DOI] [PubMed] [Google Scholar]
- 40.Témoin S, Chakaki A, Askari A, El-Halaby A, Fitzgerald S, Marcus RE, et al. Identification of oral bacterial DNA in synovial fluid of arthritis patients with native and failed prosthetic joints. Journal of clinical rheumatology: practical reports on rheumatic & musculoskeletal diseases. 2012;18(3):117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gérard HC, Wang Z, Wang GF, El-Gabalawy H, Goldbach-Mansky R, Li Y, et al. Chromosomal DNA from a variety of bacterial species is present in synovial tissue from patients with various forms of arthritis. Arthritis Rheum. 2001;44(7):1689–97. . [DOI] [PubMed] [Google Scholar]
- 42.Kempsell KE, Cox CJ, Hurle M, Wong A, Wilkie S, Zanders ED, et al. Reverse transcriptase-PCR analysis of bacterial rRNA for detection and characterization of bacterial species in arthritis synovial tissue. Infect Immun. 2000;68(10):6012–26. 10.1128/iai.68.10.6012-6026.2000 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hitchon CA, Chandad F, Ferucci ED, Willemze A, Ioan-Facsinay A, van der Woude D, et al. Antibodies to porphyromonas gingivalis are associated with anticitrullinated protein antibodies in patients with rheumatoid arthritis and their relatives. J Rheumatol. 2010;37(6):1105–12. Epub 2010/05/01. 10.3899/jrheum.091323 . [DOI] [PubMed] [Google Scholar]
- 44.Lundberg K, Kinloch A, Fisher BA, Wegner N, Wait R, Charles P, et al. Antibodies to citrullinated alpha-enolase peptide 1 are specific for rheumatoid arthritis and cross-react with bacterial enolase. Arthritis Rheum. 2008;58(10):3009–19. 10.1002/art.23936 . [DOI] [PubMed] [Google Scholar]
- 45.Ogrendik M, Kokino S, Ozdemir F, Bird PS, Hamlet S. Serum antibodies to oral anaerobic bacteria in patients with rheumatoid arthritis. MedGenMed. 2005;7(2):2 Epub 2005/06/16. . [PMC free article] [PubMed] [Google Scholar]
- 46.Moen K, Brun JG, Madland TM, Tynning T, Jonsson R. Immunoglobulin G and A antibody responses to Bacteroides forsythus and Prevotella intermedia in sera and synovial fluids of arthritis patients. Clin Diagn Lab Immunol. 2003;10(6):1043–50. 10.1128/CDLI.10.6.1043-1050.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartold PM, Marshall RI, Haynes DR. Periodontitis and Rheumatoid Arthritis: A Review. J Periodontol. 2005;76 Suppl 11S:2066–74. 10.1902/jop.2005.76.11-S.2066 . [DOI] [PubMed] [Google Scholar]
- 48.Chen HH, Huang N, Chen YM, Chen TJ, Chou P, Lee YL, et al. Association between a history of periodontitis and the risk of rheumatoid arthritis: a nationwide, population-based, case-control study. Ann Rheum Dis. 2013;72(7):1206–11. Epub 2012/08/31. 10.1136/annrheumdis-2012-201593 . [DOI] [PubMed] [Google Scholar]
- 49.de Pablo P, Dietrich T, McAlindon TE. Association of periodontal disease and tooth loss with rheumatoid arthritis in the US population. J Rheumatol. 2008;35(1):70–6. Epub 2007/11/15. . [PubMed] [Google Scholar]
- 50.Forner L, Larsen T, Kilian M, Holmstrup P. Incidence of bacteremia after chewing, tooth brushing and scaling in individuals with periodontal inflammation. J Clin Periodontol. 2006;33(6):401–7. 10.1111/j.1600-051X.2006.00924.x . [DOI] [PubMed] [Google Scholar]
- 51.Horliana AC, Chambrone L, Foz AM, Artese HP, Rabelo MeS, Pannuti CM, et al. Dissemination of periodontal pathogens in the bloodstream after periodontal procedures: a systematic review. PLoS One. 2014;9(5):e98271 Epub 2014/05/28. 10.1371/journal.pone.0098271 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koziel J, Mydel P, Potempa J. The link between periodontal disease and rheumatoid arthritis: an updated review. Curr Rheumatol Rep. 2014;16(3):408 10.1007/s11926-014-0408-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ortiz P, Bissada NF, Palomo L, Han YW, Al-Zahrani MS, Panneerselvam A, et al. Periodontal therapy reduces the severity of active rheumatoid arthritis in patients treated with or without tumor necrosis factor inhibitors. J Periodontol. 2009;80(4):535–40. 10.1902/jop.2009.080447 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scher JU, Ubeda C, Equinda M, Khanin R, Buischi Y, Viale A, et al. Periodontal disease and the oral microbiota in new-onset rheumatoid arthritis. Arthritis Rheum. 2012;64(10):3083–94. 10.1002/art.34539 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao X, Liu Z, Shu D, Xiong Y, He M, Xu S, et al. Association of Periodontitis with Rheumatoid Arthritis and the Effect of Non-Surgical Periodontal Treatment on Disease Activity in Patients with Rheumatoid Arthritis. Med Sci Monit. 2018;24:5802–10. Epub 2018/08/20. 10.12659/MSM.909117 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li R, Tian C, Postlethwaite A, Jiao Y, Garcia-Godoy F, Pattanaik D, et al. Rheumatoid arthritis and periodontal disease: What are the similarities and differences? Int J Rheum Dis. 2017;20(12):1887–901. Epub 2018/01/09. 10.1111/1756-185X.13240 . [DOI] [PubMed] [Google Scholar]
- 57.Cheng Z, Meade J, Mankia K, Emery P, Devine DA. Periodontal disease and periodontal bacteria as triggers for rheumatoid arthritis. Best Pract Res Clin Rheumatol. 2017;31(1):19–30. Epub 2017/09/01. 10.1016/j.berh.2017.08.001 . [DOI] [PubMed] [Google Scholar]
- 58.Potempa J, Mydel P, Koziel J. The case for periodontitis in the pathogenesis of rheumatoid arthritis. Nat Rev Rheumatol. 2017;13(10):606–20. Epub 2017/08/24. 10.1038/nrrheum.2017.132 . [DOI] [PubMed] [Google Scholar]
- 59.Whittle E, Leonard MO, Harrison R, Gant TW, Tonge DP. Multi-Method Characterization of the Human Circulating Microbiome. Frontiers in Microbiology. 2019;9(3266). 10.3389/fmicb.2018.03266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olsen-Bergem H, Kristoffersen AK, Bjørnland T, Reseland JE, Aas JA. Juvenile idiopathic arthritis and rheumatoid arthritis: bacterial diversity in temporomandibular joint synovial fluid in comparison with immunological and clinical findings. Int J Oral Maxillofac Surg. 2016;45(3):318–22. Epub 2015/11/06. 10.1016/j.ijom.2015.08.986 . [DOI] [PubMed] [Google Scholar]
- 61.Bonnet E, Julia F, Giordano G, Lourtet-Hascoet J. Joint infection due to Raoultella planticola: first report. Infection. 2017;45(5):703–4. Epub 2017/03/23. 10.1007/s15010-017-1006-3 . [DOI] [PubMed] [Google Scholar]
- 62.Levorova J, Machon V, Guha A, Foltan R. Septic arthritis of the temporomandibular joint caused by rare bacteria Raoultella ornithinolytica. Int J Oral Maxillofac Surg. 2017;46(1):111–5. Epub 2016/10/07. 10.1016/j.ijom.2016.09.008 . [DOI] [PubMed] [Google Scholar]
- 63.Venus K, Vaithilingam S, Bogoch II. Septic arthritis of the knee due to Raoultella ornithinolytica. Infection. 2016;44(5):691–2. Epub 2016/08/03. 10.1007/s15010-016-0930-y . [DOI] [PubMed] [Google Scholar]
- 64.Kanki M, Yoda T, Tsukamoto T, Shibata T. Klebsiella pneumoniae produces no histamine: Raoultella planticola and Raoultella ornithinolytica strains are histamine producers. Appl Environ Microbiol. 2002;68(7):3462–6. 10.1128/AEM.68.7.3462-3466.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim KW, Kim BM, Lee KA, Lee SH, Firestein GS, Kim HR. Histamine and Histamine H4 Receptor Promotes Osteoclastogenesis in Rheumatoid Arthritis. Sci Rep. 2017;7(1):1197 Epub 2017/04/26. 10.1038/s41598-017-01101-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peters BA, Wu J, Hayes RB, Ahn J. The oral fungal mycobiome: characteristics and relation to periodontitis in a pilot study. BMC Microbiol. 2017;17(1):157 Epub 2017/07/12. 10.1186/s12866-017-1064-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Beatty M, Guduric-Fuchs J, Brown E, Bridgett S, Chakravarthy U, Hogg RE, et al. Small RNAs from plants, bacteria and fungi within the order Hypocreales are ubiquitous in human plasma. BMC Genomics. 2014;15:933 Epub 2014/10/25. 10.1186/1471-2164-15-933 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hallen-Adams HE, Suhr MJ. Fungi in the healthy human gastrointestinal tract. Virulence. 2017;8(3):352–8. Epub 2016/10/13. 10.1080/21505594.2016.1247140 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Singh H, Joshi P, Khanna V, Gupta SG, Arora S, Maurya V. Pulmonary Aspergilloma in Rheumatoid Arthritis. Med J Armed Forces India. 2003;59(3):254–6. Epub 2011/07/21. 10.1016/S0377-1237(03)80024-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Horie M, Tamiya H, Goto Y, Suzuki M, Matsuzaki H, Hasegawa WT, et al. Nonspecific elevation of serum Aspergillus galactomannan antigen levels in patients with rheumatoid arthritis. Respir Investig. 2016;54(1):44–9. Epub 2015/09/26. 10.1016/j.resinv.2015.08.002 . [DOI] [PubMed] [Google Scholar]
- 71.Sandoval-Denis M, Sutton DA, Martin-Vicente A, Cano-Lira JF, Wiederhold N, Guarro J, et al. Cladosporium Species Recovered from Clinical Samples in the United States. J Clin Microbiol. 2015;53(9):2990–3000. Epub 2015/07/15. 10.1128/JCM.01482-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jahreis S, Kuhn S, Madaj AM, Bauer M, Polte T. Mold metabolites drive rheumatoid arthritis in mice via promotion of IFN-gamma- and IL-17-producing T cells. Food Chem Toxicol. 2017;109(Pt 1):405–13. Epub 2017/09/19. 10.1016/j.fct.2017.09.027 . [DOI] [PubMed] [Google Scholar]
- 73.van Hamburg JP, Tas SW. Molecular mechanisms underpinning T helper 17 cell heterogeneity and functions in rheumatoid arthritis. J Autoimmun. 2018;87:69–81. Epub 2017/12/16. 10.1016/j.jaut.2017.12.006 . [DOI] [PubMed] [Google Scholar]
- 74.Kim K-W, Kim H-R, Park J-Y, Park J-S, Oh H-J, Woo Y-J, et al. Interleukin-22 promotes osteoclastogenesis in rheumatoid arthritis through induction of RANKL in human synovial fibroblasts. Arthritis & Rheumatism. 2012;64(4):1015–23. 10.1002/art.33446 [DOI] [PubMed] [Google Scholar]
- 75.Moura RA, Cascão R, Perpétuo I, Canhão H, Sousa E, Mourão AF, et al. Cytokine profile in serum and synovial fluid of patients with established rheumatoid arthritis. Annals of the Rheumatic Diseases. 2010;69(Suppl 2):A51–A. 10.1136/ard.2010.129643g [DOI] [Google Scholar]
- 76.Yoshida Y, Tanaka T. Interleukin 6 and rheumatoid arthritis. Biomed Res Int. 2014;2014:698313 Epub 2014/01/12. 10.1155/2014/698313 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Srirangan S, Choy EH. The role of interleukin 6 in the pathophysiology of rheumatoid arthritis. Ther Adv Musculoskelet Dis. 2010;2(5):247–56. 10.1177/1759720X10378372 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rajaei E, Mowla K, Hayati Q, Ghorbani A, Dargahi-Malamir M, Hesam S, et al. Evaluating the relationship between serum level of interleukin-6 and rheumatoid arthritis severity and disease activity. Curr Rheumatol Rev. 2019. Epub 2019/02/06. . [DOI] [PubMed] [Google Scholar]
- 79.Matsumoto T, Tsurumoto T, Shindo H. Interleukin-6 levels in synovial fluids of patients with rheumatoid arthritis correlated with the infiltration of inflammatory cells in synovial membrane. Rheumatol Int. 2006;26(12):1096–100. 10.1007/s00296-006-0143-2 . [DOI] [PubMed] [Google Scholar]
- 80.Sun W, Dong L, Kaneyama K, Takegami T, Segami N. Bacterial diversity in synovial fluids of patients with TMD determined by cloning and sequencing analysis of the 16S ribosomal RNA gene. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2008;105(5):566–71. Epub 2008/02/21. 10.1016/j.tripleo.2007.08.015 . [DOI] [PubMed] [Google Scholar]
- 81.Siala M, Jaulhac B, Gdoura R, Sibilia J, Fourati H, Younes M, et al. Analysis of bacterial DNA in synovial tissue of Tunisian patients with reactive and undifferentiated arthritis by broad-range PCR, cloning and sequencing. Arthritis Res Ther. 2008;10(2):R40 Epub 2008/04/14. 10.1186/ar2398 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kell DB, Pretorius E. On the translocation of bacteria and their lipopolysaccharides between blood and peripheral locations in chronic, inflammatory diseases: the central roles of LPS and LPS-induced cell death. Integr Biol (Camb). 2015;7(11):1339–77. 10.1039/c5ib00158g . [DOI] [PubMed] [Google Scholar]
- 83.Bhattacharyya M, Ghosh T, Shankar S, Tomar N. The conserved phylogeny of blood microbiome. Mol Phylogenet Evol. 2017;109:404–8. Epub 2017/02/12. 10.1016/j.ympev.2017.02.001 . [DOI] [PubMed] [Google Scholar]
- 84.Mangul S, M Olde Loohuis L, Ori A, Jospin G, Koslicki D, Yang HT, et al. Total RNA Sequencing reveals microbial communities in human blood and disease specific effects. bioRxiv. 2016. [Google Scholar]
- 85.Païssé S, Valle C, Servant F, Courtney M, Burcelin R, Amar J, et al. Comprehensive description of blood microbiome from healthy donors assessed by 16S targeted metagenomic sequencing. Transfusion. 2016;56(5):1138–47. 10.1111/trf.13477 . [DOI] [PubMed] [Google Scholar]
- 86.Potgieter M, Bester J, Kell DB, Pretorius E. The dormant blood microbiome in chronic, inflammatory diseases. FEMS Microbiol Rev. 2015;39(4):567–91. 10.1093/femsre/fuv013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Amar J, Lange C, Payros G, Garret C, Chabo C, Lantieri O, et al. Blood microbiota dysbiosis is associated with the onset of cardiovascular events in a large general population: the D.E.S.I.R. study. PLoS One. 2013;8(1):e54461 10.1371/journal.pone.0054461 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rajendhran J, Shankar M, Dinakaran V, Rathinavel A, Gunasekaran P. Contrasting circulating microbiome in cardiovascular disease patients and healthy individuals. Int J Cardiol. 2013;168(5):5118–20. Epub 2013/08/02. 10.1016/j.ijcard.2013.07.232 . [DOI] [PubMed] [Google Scholar]
- 89.Dinakaran V, Rathinavel A, Pushpanathan M, Sivakumar R, Gunasekaran P, Rajendhran J. Elevated levels of circulating DNA in cardiovascular disease patients: metagenomic profiling of microbiome in the circulation. PLoS One. 2014;9(8):e105221 Epub 2014/08/18. 10.1371/journal.pone.0105221 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sze MA, Tsuruta M, Yang SW, Oh Y, Man SF, Hogg JC, et al. Changes in the bacterial microbiota in gut, blood, and lungs following acute LPS instillation into mice lungs. PLoS One. 2014;9(10):e111228 Epub 2014/10/21. 10.1371/journal.pone.0111228 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jeon SJ, Cunha F, Vieira-Neto A, Bicalho RC, Lima S, Bicalho ML, et al. Blood as a route of transmission of uterine pathogens from the gut to the uterus in cows. Microbiome. 2017;5(1):109 Epub 2017/08/25. 10.1186/s40168-017-0328-9 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Martel J, Wu CY, Huang PR, Cheng WY, Young JD. Pleomorphic bacteria-like structures in human blood represent non-living membrane vesicles and protein particles. Sci Rep. 2017;7(1):10650 Epub 2017/09/06. 10.1038/s41598-017-10479-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hammad DB, Tonge DP. Molecular Characterisation of the Synovial Fluid Microbiome. bioRxiv. 2018. 10.1101/405613 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data deposited at OSF, DOI 10.17605/OSF.IO/HK4F6.