Abstract
Non-invasive approaches for early detection of bladder cancer are actively being investigated. We recently developed a urine based molecular assay for the detection and surveillance of bladder neoplasms (UroSEEK). UroSEEK is designed to detect alterations in 11 genes that includes most common genetic alterations in bladder cancer. In this study we analyzed 527 cases including 373 non-invasive and 154 invasive urothelial carcinomas of bladder from trans-urethral resections or cystectomies performed at 4 institutions (1991–2016). Two different mutational analysis assays of a representative tumor area were performed: first, a singleplex PCR assay for evaluation of the TERT promoter region (TERTSeqS) and second, a multiplex PCR assay using primers designed to amplify regions of interest of 10 (FGFR3, PIK3CA, TP53, HRAS, KRAS, ERBB2, CDKN2A, MET, MLL, and VHL) genes (UroSeqS). Overall 92% of all bladder tumors were positive for at least one genetic alteration in the UroSEEK panel. We found TERT promoter mutations in 77% of low-grade non-invasive papillary carcinomas with relatively lower incidence of 65% in high-grade non-invasive papillary carcinomas and carcinomas in situ; p=0.017. Seventy-two percent of pT1 and 63% of muscle-invasive bladder tumors harbored TERT promoter mutations with g.1295228C>T alteration being the most common in all groups. FGFR3 and PIK3CA mutations were more frequent in low-grade non-invasive papillary carcinomas compared to high-grade non-invasive papillary carcinomas and carcinomas in situ (p<0.0001), while the opposite was true for TP53 (p<0.0001). Significantly higher rates of TP53 and CDKN2A mutation rates (p=0.005 and 0.035, respectively) were encountered in muscle-invasive bladder tumors compared to those of pT1 stage. The overwhelming majority of all investigated tumors showed at least one mutation among UroSEEK assay genes confirming the comprehensive coverage of the panel and supporting its potential utility as a non-invasive urine based assay.
Keywords: bladder cancer, non-invasive assay, urine, molecular diagnostics, FFPE
Introduction
Bladder cancer is the fourth most common cancer in men and the most common malignancy of the urinary tract in both male and female. In 2019, 80,470 new cases are being estimated in the USA leading to 17,670 deaths [1].
Invasive urothelial carcinoma evolves through two distinct pathways: low-and high-grade non-invasive papillary urothelial carcinoma and “flat” carcinoma in situ [2]. Due to high recurrence rates and likelihood of progression to muscle-invasive bladder cancer, periodical follow-up with cystoscopy and cytology for patients diagnosed with non-invasive tumors is required [3–6]. These procedures entail an estimated three billion dollar burden to the health care system every year [7]. Alternative approaches for surveillance are therefore needed.
Early detection of bladder cancer remains a challenge in clinical practice. Hematuria, with or without lower urinary tract symptoms, is the most common presenting symptom. While asymptomatic microscopic hematuria is prevalent (up to 31%) [8, 9], only a small fraction of patients will ultimately be diagnosed with bladder cancer (3–5%) [10]. Therefore, risk stratification with a non-invasive method to avoid unneeded cystoscopy is of great utility. Desquamated urothelial cells in urine have long been used as a valuable source for non-invasive detection of bladder cancer. Beside cystoscopy, urine cytology remains the gold standard for bladder cancer detection. However, its overall low sensitivity (11–76%) especially in low-grade tumors tampers its utility. Several FDA approved urine-based non-invasive assays are currently available (e.g. UroVysion, ImmunoCyt/uCyt+™, NMP22®, and BTA®) for both early detection and surveillance of bladder cancer. Sensitivity and specificity for these assays range from 56% to 78% and 74% to 88%, respectively [11–18]. We recently developed a non-invasive bladder cancer assay with promising performance characteristics in both early detection and surveillance setting. When combined with cytology, a sensitivity of 95% and a specificity of 93% were reached in early detection cohort [19]. The assay, termed “UroSEEK”, consists of three components (TERTSeqS, UroSeqS and Fast SeqS). It covers molecular alterations that are frequently encountered in bladder cancer in addition to aneuploidy (FastSeqS). The alterations include TERT promoter mutations (TERTSeqS) that occur in up to 80% of bladder cancers [20, 21] and 10 additional genes: FGFR3, PIK3CA, HRAS, KRAS, TP53, CDKN2A, ERBB2, MLL, MET, and VHL (UroSeqS) [22–24].
Evidently, a non-invasive mutation based approach can only be effective if its genes are closely matched to the tumors it aims to detect. Therefore, in the current study we sought to identify the distribution of the UroSEEK gene panel in archival tumor tissues from a multi-institutional international cohort of bladder urothelial carcinoma. Relationship with tumor grade and stage was also assessed.
Material and Methods
Patient samples and clinical data
The study was approved by the Institutional Board Review of participating institutions. Required material transfer agreements were obtained. The 527 formalin-fixed, paraffin-embedded bladder urothelial carcinoma specimens were collected between 1991 and 2016 from four international academic institutions (Johns Hopkins Hospital, Baltimore, MD, United States; A.C. Camargo Cancer Center, Sao Paulo, Brazil; Osaka University Hospital, Osaka, Japan; and Hacettepe University Hospital, Ankara, Turkey). The mutational findings of the UroSEEK gene panel in a subset (102 tumors) were described in our previously reported study describing the non-invasive multigene assay (UroSEEK) [19]. All histologic sections from transurethral resection of bladder tumor and cystectomy specimens were reviewed by a genitourinary pathologist to confirm the diagnosis and select a representative tumor area. Corresponding formalin-fixed, paraffin-embedded blocks were cored for DNA purification as previously described [20]. Clinicopathologic data was obtained from electronic medical record. Only cases with a minimum follow-up time of 3 month were included in outcome analysis. Disease recurrence was defined as the development of histologically documented tumor occurrence. Progression was defined as the occurrence of histologically documented upgrade or upstage of disease.
Mutation analysis
Mutation and data analysis was performed as previously described [19, 20, 25, 26]. In brief, purified DNA was submitted for SafeSeqS analysis, a sequencing error-reduction technique capable of discriminating mutations from artifactual sequencing variants introduced during the sequencing process [27, 28]. Two different mutational analysis assays were performed: first, a singleplex PCR assay for evaluation of TERT promoter region (TERTSeqS) and a second multiplex PCR assay [20] using primers designed to amplify regions of interest of 10 (FGFR3, PIK3CA, TP53, HRAS, KRAS, ERBB2, CDKN2A, MET, MLL, and VHL) genes (UroSeqS). Primers are listed in Supplementary Table S1 [19].
In order to evaluate the statistical significance of observed mutations, DNA from white blood cells of 188 unrelated healthy individuals was also assessed. A variant was scored as a mutation only if the mutant allele frequency was much higher than that observed in normal white blood cells. As previously described [19], the classification of a sample’s DNA status was based on two complementary criteria applied to each mutation: 1) the difference between the average mutant allele frequency in the sample of interest and the corresponding maximum mutant allele frequency observed for that same mutation in a set of controls; and 2) the Stouffer’s Z-score obtained by comparing the mutant allele frequency in the sample of interest to a distribution of normal controls.
Statistical analysis
Statistical analysis was performed using R version 3.5.1 (2018–07-02) from the R Foundation for Statistical Computing (Vienna, Austria). For hypothesis testing, statistical significance was established at p<0.05 for 2 tails of distribution. The relationship between mutation status and pathological and outcome variables was analyzed by Chi-square test with Yates’ continuity correction.
Results
Clinicopathologic features
Five hundred twenty-seven tumors from 484 patients were included in the study. One-hundred and thirteen patients were female and 371 were male. The median age was 68 years (range 28–96). Three hundred twenty-nine patients were Caucasian, 68 were Asian, 46 were African-American with the race in the remaining 41 patients being undetermined or from other category. The tumor included 188 low-grade non-invasive papillary carcinomas, 129 high-grade non-invasive papillary carcinomas, 56 carcinomas in situ, and 154 high grade invasive urothelial carcinoma including 111 pT1 and 43 ≥pT2 (muscle-invasive) tumors.
Mutation Analysis
The frequency of mutations of all genes of the UroSEEK gene panel across histopathological categories is summarized in Figure 1. The mutation variants of analyzed genes are listed in Supplementary Table S2. Overall 93% of non-invasive and 92% of invasive tumors were positive for at least one mutation in genes included in UroSEEK assay.
Figure 1.
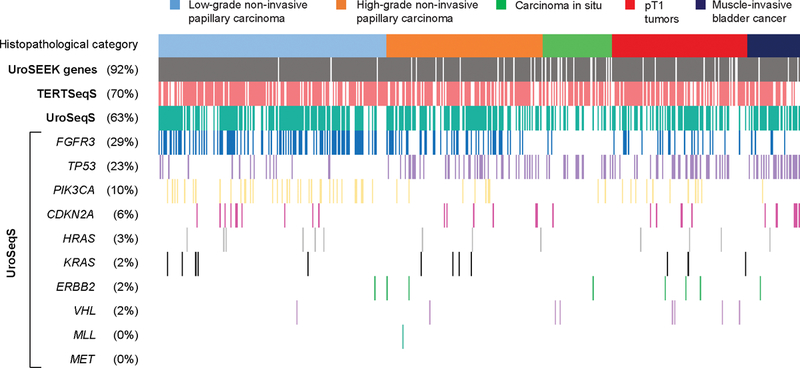
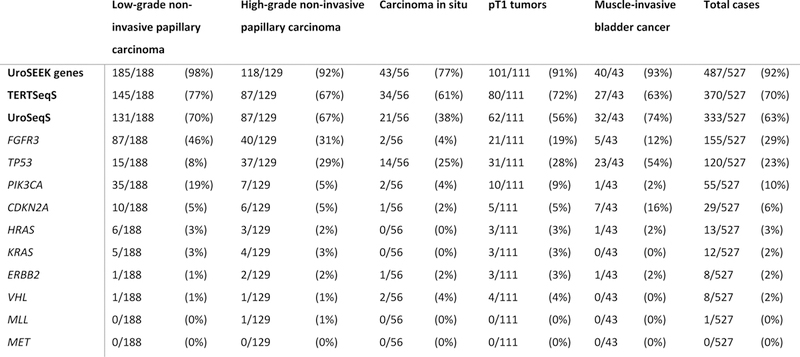
Distribution of mutations in UroSEEK gene panel and its TERTSeqS and UroSeqS components across 527 bladder carcinomas. A) Oncoplot graphic representation of data. Each column represents one tumor. The top line colored boxes indicate the histopathological category. Rows indicate affected gene(s) and their rate of mutations. Detailed listing of the absolute numbers of mutations is shown in B).
TERT promoter mutations were identified in 70% of all cases with the most common alteration being g.1295228C>T, followed by g.1295250C>T. A previously unreported variant (g.1295223G>T; in 1 case) was detected. A higher incidence of TERT promoter mutation was identified in low-grade non-invasive papillary carcinomas compared to high-grade non-invasive papillary carcinomas and carcinomas in situ (77% vs 65%; p=0.017; see Table 1). Although higher frequency of TERT promoter mutations occurred in pT1 cases (72%) compared to muscle-invasive bladder cancer (63%), the difference was not statistically significant.
Table 1.
Distribution of mutations of UroSEEK gene panel in non-invasive bladder cancer
| Low-grade non-invasive papillary carcinoma | High-grade non-invasive papillary carcinoma + Carcinoma in situ | p-value | |||
|---|---|---|---|---|---|
| UroSEEK genes | 185/188 | (98%) | 161/185 | (87%) | <0.0001 |
| TERTSeqS | 145/188 | (77%) | 121/185 | (65%) | 0.017 |
| UroSeqS | 131/188 | (70%) | 108/185 | (58%) | 0.030 |
| FGFR3 | 87/188 | (46%) | 42/185 | (23%) | <0.0001 |
| TP53 | 15/188 | (8%) | 51/185 | (28%) | <0.0001 |
| PIK3CA | 35/188 | (19%) | 9/185 | (5%) | <0.0001 |
| CDKN2A | 10/188 | (5%) | 7/185 | (4%) | 0.644 |
| HRAS | 6/188 | (3%) | 3/185 | (2%) | 0.515 |
| KRAS | 5/188 | (3%) | 4/185 | (2%) | 1.0 |
| ERBB2 | 1/188 | (1%) | 3/185 | (2%) | 0.604 |
| VHL | 1/188 | (1%) | 3/185 | (2%) | 0.604 |
| MLL | 0/188 | (0%) | 1/185 | (1%) | 0.994 |
| MET | 0/188 | (0%) | 0/185 | (0.0%) | . |
Among the 10 genes included in the UroSeqS assay, FGFR3 and PIK3CA mutations occurred significantly more often in low-grade non-invasive papillary carcinoma tumors compared to high-grade non-invasive papillary carcinomas and carcinomas in situ (p<0.0001), while the reverse was true for TP53 (p<0.0001; see Table 1). In invasive bladder cancer CDKN2A and TP53 mutations were more commonly observed in muscle-invasive bladder cancer compared to pT1 tumors (P= 0.035 and 0.005, respectively; see Table 2). Regarding mutation variants, p.S249C was the most frequent FGFR3 mutation (67%), while p.E545K and p.H66P were the most common PIK3CA and CDKN2A mutations, (49% and 79%, respectively; see Supplementary Table S2).
Table 2.
Distribution of mutations of UroSEEK gene panel in invasive bladder cancer
| PT1 TUMORS | Muscle-invasive bladder cancer | p-value | |||
|---|---|---|---|---|---|
| UroSEEK genes | 101/111 | (91%) | 40/43 | (93%) | 0.933 |
| TERTSeqS | 80/111 | (72%) | 27/43 | (63%) | 0.354 |
| UroSeqS | 62/111 | (56%) | 32/43 | (74%) | 0.053 |
| FGFR3 | 21/111 | (19%) | 5/43 | (12%) | 0.399 |
| TP53 | 31/111 | (28%) | 23/43 | (54%) | 0.005 |
| PIK3CA | 10/111 | (9%) | 1/43 | (2%) | 0.273 |
| CDKN2A | 5/111 | (5%) | 7/43 | (16%) | 0.035 |
| HRAS | 3/111 | (3%) | 1/43 | (2%) | 1.0 |
| KRAS | 3/111 | (3%) | 0/43 | (0%) | 0.661 |
| ERBB2 | 3/111 | (3%) | 1/43 | (2%) | 1.0 |
| VHL | 4/111 | (4%) | 0/43 | (0%) | 0.486 |
| MLL | 0/111 | (0%) | 0/43 | (0%) | . |
| MET | 0/111 | (0%) | 0/43 | (0%) | . |
Two hundred and nineteen tumors demonstrated co-occurrence of mutations in both assay components with 59% of tumors harboring TERT promoter mutation also showing a mutation in at least one UroSeqS gene (p=0.001). Finally, occasional cases demonstrated multiple variants of TERT promoter (13 tumors) and TP53 (14 tumors) mutations and one case displayed two FGFR3 variant mutations.
Mutational analysis in patients with sequential tumors
Tissues from sequential tumors were available for mutational analysis in 36 patients. As illustrated in Figure 2, TERT promoter mutations were absent in all samples in 7 of the 36 patients. In the remaining 29 patients, the same TERT promoter mutation was persistently present across tumors in 22 and variably found in 7 patients. Only 2 of 15 patients with observed FGFR3 mutations had the same mutation present across tumors.
Figure 2.
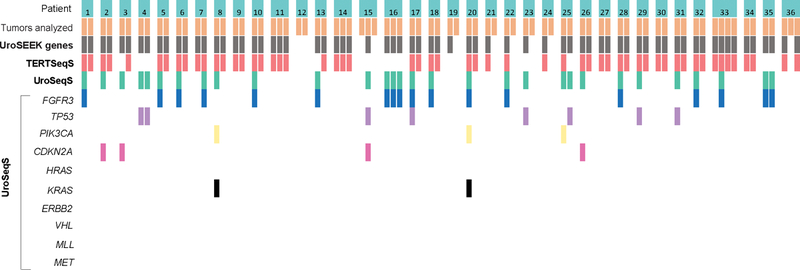
Oncoplot representation of mutations in UroSEEK gene panel and its TERTSeqS and UroSeqS components in 36 patients with sequential tumors. Each broad column represents one patient. The individual tumor per patient (orange box) and their mutated gene(s) are indicated in each row.
Association of UroSEEK assay with outcome
The distribution of the three hundred-three cases that met the minimum follow-up requirement for outcome analysis was as follows: 124 low-grade non-invasive papillary carcinomas, 78 high-grade non-invasive papillary carcinomas, 24 carcinomas in situ, 58 pT1 and 19 muscle-invasive bladder tumors.
Association of tumor UroSEEK, TERTSeqS and UroSeqS findings with recurrence and progression are summarized in Table 3. As shown, 94% of tumors with subsequent recurrence had mutation in at least one of the 11 genes included in UroSEEK. A comparable rate of 95% was found in tumors without recurrence (p=0.784). Only 21 patients developed progression during follow-up. Tumors with progression were less likely to harbor a mutation in one or more UroSEEK assay genes (81% vs. 96%; p=0.016).
Table 3.
Association of UroSEEK gene panel with outcome
| Recurrence | p-value | Progression | p-value | |||
|---|---|---|---|---|---|---|
| Yes N=132 |
No N=171 |
Yes N=21 |
No N=282 |
|||
| Mutation in UroSEEK genes | 124/132 (94%) |
163/171 (95%) |
0.784 | 17/21 (81%) |
270/282 (96%) |
0.016 |
| Mutation in TERTSeqS | 101/132 (77%) |
115/171 (67%) |
0.101 | 12/21 (57%) |
204/282 (72%) |
0.217 |
| Mutation in UroSeqS | 87/132 (66%) |
120/171 (70%) |
0.505 | 12/21 (57%) |
195/282 (69%) |
0.369 |
Discussion
In our current multi-center study we investigated the comprehensive coverage of the UroSEEK gene assay in archival bladder cancer tissues. The TERTSeqS component detected mutations in the TERT promoter region in 70% of all tumors. Mutations in UroSeqS gene panel were found in 63% of all cases. Combing these two components led to a capture rate for alteration of the UroSEEK assay in 92% of all tumors. More specifically, in the subset of low-grade non-invasive papillary carcinoma cases, we found at least one gene alteration in 98% of the cases with 77% being positive for TERTSeqS and 70% positive for UroSeqS. This is especially important given the low sensitivity of routine cytology for the diagnosis of low grade non-invasive tumors. As evidenced in our recent study, urine cytology was negative in all 49 low-grade non-invasive papillary carcinoma cases, while urine UroSEEK was positive in 2/3 of these cases [19].
The current observation of 54% of TP53 mutation in muscle-invasive bladder cancer is in line with findings of recent studies such as The Cancer Genome Atlas [23] and Kim et al. [29] where TP53 was altered in 48% and 57% of cases, respectively. We observed a lower frequency for PIK3CA, ERBB2 and MLL in muscle-invasive bladder cancer compared to these two studies.
Our study represents one of the largest assessment of a multigene assay in low-grade non-invasive papillary carcinoma tumors to date where 188 cases were analyzed. In this group, FGFR3 mutations were the most frequent (46%) among the 10 genes included in UroSeqS followed by PIK3CA mutation in 19%. These rates are lower than those obtained by Hurst et al. [30] in their study of whole exome and targeted sequencing of 82 Ta tumors (79% and 54% for FGFR3 and PIK3CA mutations, respectively). This could be due to our hotspot focused mutational analysis approach given that Hurst et al. have identified many non-hotspot mutations in their analysis.
TERT promoter mutations are one of the most frequent alteration in bladder cancer and its variants [25, 26, 31–33]. In the current analysis, we found TERT promoter mutations in 70% of all cases with the highest rate observed in low-grade non-invasive papillary carcinomas (77%). The here found distribution rates of TERT promoter mutations in non-invasive lesions is in line with our originally reported rates in conventional urothelial carcinoma in Kinde et al. where the low-grade non-invasive papillary carcinoma group had the highest rate of TERT promoter mutations of 86% [20]. In contrast, Pietzak et al. found TERT promoter mutations to be more frequent in their high-grade non-invasive papillary carcinoma cases compared to low-grade non-invasive papillary carcinomas (88% and 61 %, respectively) [34].
Our analysis of sequential tumors in 36 patients unveiled an identical TERT promoter mutation across tumors in 22 of the 29 patients harboring TERT promoter mutations. In surveillance setting, UroSEEK is envisioned to be use for follow-up in patients whose index tumor is positive for at least one alteration in the panel. This consistency of TERT promoter alteration across tumors, if proven in a larger cohort of sequential tumors, suggests that assessment of the index tumor might be sufficient and could obviate the need for repeat sequential tumor testing. The same approach would not apply to cases where the index tumor is only positive for one of the ten genes in UroSeqS where we found inconsistency in mutation detection across tumors from the same patient. This might in part be due to tumor molecular heterogeneity that was not captured by our adopted technique.
Given the nature of the cohort in the original study describing the non-invasive multigene assay (UroSEEK) [19], analysis of association with outcome could only be performed in a subset of patients. This is due to the fact that in a proportion of cases tumors that are contemporaneous with analyzed urine samples were selected. Strengths of the study include the large number of cases in the multi-institutional cohort (527 tumors) and the advantageous error proof nature of the SafeSeqS technique.
In conclusion, the overwhelming majority of all investigated tumors showed at least one mutation among genes included in the recently reported UroSEEK assay, across tumor grade and stage. This confirms the comprehensive coverage of UroSEEK panel and support its potential utility as a non-invasive urine based assay. Our findings are especially reassuring in the subset of low-grade non-invasive papillary carcinoma where routine cytology lacks sensitivity.
Supplementary Material
Acknowledgements
We are grateful for the generous support provided by the Johns Hopkins Greenberg Bladder Cancer Institute, Henry and Marsha Laufer, the Virginia and DK Ludwig Fund for Cancer Research, the Commonwealth Foundation, the John Templeton Foundation, and the Conrad R Hilton Foundation. All sequencing was performed at the Sol Goldman Sequencing Facility at Johns Hopkins. This work was also supported by grants from the NIH (Grants CA-77598, CA 06973, GM 07309, and ES019564).
Footnotes
Disclosure/Conflict of Interest
Nickolas Papadopoulos, Ken W Kinzler, Bert Vogelstein: Founder of Personal Genome Diagnostics and PapGene and advise Sysmex-Inostics. Kinzler and Vogelstein also advise Eisai. Vogelstein is also an advisor to Camden Partners. These companies and others have licensed technologies from Johns Hopkins that are related to the work described in this paper. These licenses are associated with equity or royalty payments to Papadopoulos, Kinzler, Netto, and Vogelstein. Additional patent applications on the work described in this paper may be filed by Johns Hopkins University. The terms of these arrangements are managed by the university in accordance with its conflict of interest policies. The other authors declare that no competing interests exist.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 2.Netto GJ. Clinical applications of recent molecular advances in urologic malignancies: no longer chasing a “mirage”? Adv in Anat Pathol 2013;20:175–203. [DOI] [PubMed] [Google Scholar]
- 3.Chamie K, Litwin MS, Bassett JC, et al. Recurrence of high-risk bladder cancer: a population-based analysis. Cancer 2013;119:3219–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Millan-Rodriguez F, Chechile-Toniolo G, Salvador-Bayarri J, et al. Primary superficial bladder cancer risk groups according to progression, mortality and recurrence. J Urol 2000;164:680–4. [DOI] [PubMed] [Google Scholar]
- 5.Chang SS, Boorjian SA, Chou R, et al. Diagnosis and Treatment of Non-Muscle Invasive Bladder Cancer: AUA/SUO Guideline. J Urol 2016;196:1021–9. [DOI] [PubMed] [Google Scholar]
- 6.Babjuk M, Bohle A, Burger M, et al. EAU Guidelines on Non-Muscle-invasive Urothelial Carcinoma of the Bladder: Update 2016. Eur Urol 2017;71:447–61. [DOI] [PubMed] [Google Scholar]
- 7.Netto GJ, Epstein JI. Theranostic and prognostic biomarkers: genomic applications in urological malignancies. Pathology 2010;42:384–94. [DOI] [PubMed] [Google Scholar]
- 8.Wood PW. Urothelial Tumors of the Bladder. In: Wein AJ, Kavoussi LR, Novick AC, Partin AW, Peters CA. Campbell-Walsh Urology 10th ed. Philadelphia, PA: Elsevier Saunders, 2012. p.2309–2334 [Google Scholar]
- 9.Davis R, Jones JS, Barocas DA, et al. Diagnosis, evaluation and follow-up of asymptomatic microhematuria (AMH) in adults: AUA guideline. J Urol 2012;188:2473–81. [DOI] [PubMed] [Google Scholar]
- 10.Davis R, Jones JS, Barocas DA et al. Diagnosis, Evaluation and Follow-up of Asymptomatic Microhematuria (AMH) in Adults, 2016, [cited 10/10/2018]. Available from https://www.auanet.org/guidelines/asymptomatic-microhematuria-(amh)-guideline [DOI] [PubMed]
- 11.Smith ZL, Guzzo TJ. Urinary markers for bladder cancer. F1000Prime Rep 2013;5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Sullivan P, Sharples K, Dalphin M, et al. A multigene urine test for the detection and stratification of bladder cancer in patients presenting with hematuria. J Urol 2012;188:741–7. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Que H, Suo C, et al. Evaluation of the NMP22 BladderChek test for detecting bladder cancer: a systematic review and meta-analysis. Oncotarget 2017;8:100648–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He H, Han C, Hao L, Zang G. ImmunoCyt test compared to cytology in the diagnosis of bladder cancer: A meta-analysis. Oncol Lett 2016;12:83–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lavery HJ, Zaharieva B, McFaddin A, Heerema N, Pohar KS. A prospective comparison of UroVysion FISH and urine cytology in bladder cancer detection. BMC Cancer 2017;17:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lokeshwar VB, Habuchi T, Grossman HB, et al. Bladder tumor markers beyond cytology: International Consensus Panel on bladder tumor markers. Urology 2005;66:35–63. [DOI] [PubMed] [Google Scholar]
- 17.Guo A, Wang X, Gao L, et al. Bladder tumour antigen (BTA stat) test compared to the urine cytology in the diagnosis of bladder cancer: A meta-analysis. Can Urol Assoc J 2014;8:E347–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou R, Gore JL, Buckley D, et al. Urinary Biomarkers for Diagnosis of Bladder Cancer: A Systematic Review and Meta-analysis. Ann Intern Med 2015;163:922–31. [DOI] [PubMed] [Google Scholar]
- 19.Springer SU, Chen CH, Rodriguez Pena MDC, et al. Non-invasive detection of urothelial cancer through the analysis of driver gene mutations and aneuploidy. Elife 2018;7. [DOI] [PMC free article] [PubMed]
- 20.Kinde I, Munari E, Faraj SF, et al. TERT Promoter Mutations Occur Early in Urothelial Neoplasia and are Biomarkers of Early Disease and Disease Recurrence in Urine. Cancer Res 2013;73:7162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A 2013;110:6021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hedegaard J, Lamy P, Nordentoft I, et al. Comprehensive Transcriptional Analysis of Early-Stage Urothelial Carcinoma. Cancer Cell 2016;30:27–42. [DOI] [PubMed] [Google Scholar]
- 23.Robertson AG, Kim J, Al-Ahmadie H, et al. Comprehensive Molecular Characterization of Muscle-Invasive Bladder Cancer. Cell 2017;171:540–56 e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan TZ, Rouanne M, Tan KT, Huang RY, Thiery JP. Molecular Subtypes of Urothelial Bladder Cancer: Results from a Meta-cohort Analysis of 2411 Tumors. Eur Urol 2019; 75(3):423–432 [DOI] [PubMed] [Google Scholar]
- 25.Rodriguez Pena MDC, Tregnago AC, Eich ML, et al. Spectrum of genetic mutations in de novo PUNLMP of the urinary bladder. Virchows Arch 2017;471:761–7. [DOI] [PubMed] [Google Scholar]
- 26.Nguyen D, Taheri D, Springer S, et al. High prevalence of TERT promoter mutations in micropapillary urothelial carcinoma. Virchows Arch 2016;469:427–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinde I, Bettegowda C, Wang Y, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med 2013;5:167ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A 2011;108:9530–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim PH, Cha EK, Sfakianos JP, et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur Urol 2015;67:198–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurst CD, Alder O, Platt FM, et al. Genomic Subtypes of Non-invasive Bladder Cancer with Distinct Metabolic Profile and Female Gender Bias in KDM6A Mutation Frequency. Cancer Cell 2017;32:701–15 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cowan M, Springer S, Nguyen D, et al. High prevalence of TERT promoter mutations in primary squamous cell carcinoma of the urinary bladder. Mod Pathol 2016;29:511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowan ML, Springer S, Nguyen D, et al. Detection of TERT promoter mutations in primary adenocarcinoma of the urinary bladder. Hum Pathol 2016;53:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palsgrove DN, Taheri D, Springer SU, et al. Targeted sequencing of Plasmacytoid urothelial carcinoma reveals frequent TERT promoter mutations. Hum Pathol 2018. [DOI] [PMC free article] [PubMed]
- 34.Pietzak EJ, Bagrodia A, Cha EK, et al. Next-generation Sequencing of Nonmuscle Invasive Bladder Cancer Reveals Potential Biomarkers and Rational Therapeutic Targets. Eur Urol 2017;72:952–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


