Abstract
The second WW domain (WW2) of the kidney and brain scaffolding protein, KIBRA, has an isoleucine (Ile-81) rather than a second conserved tryptophan and is primarily unstructured. However, it adopts the canonical triple-stranded antiparallel β-sheet structure of WW domains when bound to a two-PPXY motif peptide of the synaptic protein Dendrin. Here, using a series of biophysical experiments, we demonstrate that the WW2 domain remains largely disordered when bound to a 69-residue two-PPXY motif polypeptide of the synaptic and podocyte protein synaptopodin (SYNPO). Isothermal titration calorimetry and CD experiments revealed that the interactions of the disordered WW2 domain with SYNPO are significantly weaker than SYNPO's interactions with the well-folded WW1 domain and that an I81W substitution in the WW2 domain neither enhances binding affinity nor induces substantial WW2 domain folding. In the tandem polypeptide, the two WW domains synergized, enhancing the overall binding affinity with the I81W variant tandem polypeptide 2-fold compared with the WT polypeptide. Solution NMR results showed that SYNPO binding induces small but definite chemical shift perturbations in the WW2 domain, confirming the disordered state of the WW2 domain in this complex. These analyses also disclosed that SYNPO binds the tandem WW domain polypeptide in an antiparallel manner, that is, the WW1 domain binds the second PPXY motif of SYNPO. We propose a binding model consisting of a bipartite interaction mode in which the largely disordered WW2 forms a “fuzzy” complex with SYNPO. This binding mode may be important for specific cellular functions.
Keywords: isothermal titration calorimetry (ITC), nuclear magnetic resonance (NMR), circular dichroism (CD), protein-protein interaction, intrinsically disordered protein, PPXY motif, proline-rich motif, scaffolding protein, synaptopodin (SYNPO), WW domain
Introduction
KIBRA, also known as WWC1 (for WW and C2 domain containing), is a 125-kDa cytoplasmic scaffolding protein that is expressed mainly in the kidney and brain (1). It is a multifunctional protein implicated in the directional migration of podocytes (2–4), endosomal vesicle sorting (5), memory performance (6–8), cell growth and tumor suppression (9–11). Although these functional roles are well-documented, much less is known about how KIBRA engages partner proteins to facilitate these diverse functions.
The KIBRA polypeptide is organized into multiple protein-interacting modules. Of particular interest are two N-terminal WW domains, WW1 and WW2. The WW domain is a ∼35 residue globular domain that folds into a twisted, three-stranded anti-parallel β sheet and binds proline-rich motifs (12–14). Two conserved tryptophans (W), from which the WW name is derived, lie on opposite sides of the β sheet. The first tryptophan is part of a hydrophobic core essential for domain stability, and the second tryptophan, located in a solvent-exposed region, is a hydrogen bond donor important in ligand binding (15).
Human KIBRA WW1 and WW2 domains are separated by a 14-residue segment and recognize proline-rich motifs of the type PPXY (where P is proline, X is any amino acid, and Y is tyrosine). The domains share 61% sequence identity with one major difference; the WW2 domain has an isoleucine (Ile-81) at the position normally occupied by the second conserved tryptophan in most WW domain proteins. WW domain proteins with a substitution of the second conserved tryptophan are not uncommon; and a couple of these “atypical” WW domain proteins that have tyrosine or phenylalanine substitutions show reduced or complete loss of binding to specific targets (16, 17). Both human and mouse KIBRA WW2 domain display weak binding to PPXY-containing peptides (1, 18). Furthermore, recent extensive work by Zhang and co-workers (18) show that the mouse KIBRA WW2 domain is unstructured but adopts the canonical WW domain structure when bound to two of the three PPXY sites of the synaptic protein Dendrin. The KIBRA WW domains bind the two Dendrin PPXY sites, which are separated by two residues, with an unusually high nanomolar affinity. Given that the dissociation constants of WW domain-PPXY interactions are generally in the low micromolar range (17, 19), it is plausible that this high affinity binding induces folding of the WW2 domain.
Synaptopodin (SYNPO) is a 929-residue proline-rich actin cytoskeleton-associated protein, found in kidney podocytes and the dendrites of neurons (1, 20–22). In kidney podocytes, it is essential for the integrity of the podocyte actin cytoskeleton (23, 24) and forms a WW domain-PPXY–mediated complex with KIBRA implicated in cell migration (4). In dendrites, it plays an important role in regulating spine dynamics and synaptic plasticity (25). SYNPO has two PPXY motifs, PPTY (residues 562–565) and PPSY (residues 581–584) separated by 15 amino acids and located in a region with striking sequence similarity to the region harboring the Dendrin PPXY motifs (4). Thus SYNPO is a good candidate to provide additional insight into the structural heterogeneity of the KIBRA WW2 domain.
In this work, we characterize the interaction between a 69-residue recombinant SYNPO polypeptide and variants of the human KIBRA WW domain polypeptide by a series of biophysical techniques. We show that recombinant human KIBRA WW2 domain is primarily disordered, binds SYNPO with relatively weak affinity and remains largely disordered in the complex. The two proteins interact in an antiparallel manner with the well-folded WW1 domain making substantial contacts with the SYNPO polypeptide, whereas the WW2 domain forms a “fuzzy” complex. This structural heterogeneity of the WW2 domain is discussed in the context of specific functions of the KIBRA WW domains.
Results
SYNPO545–613 is primarily unstructured
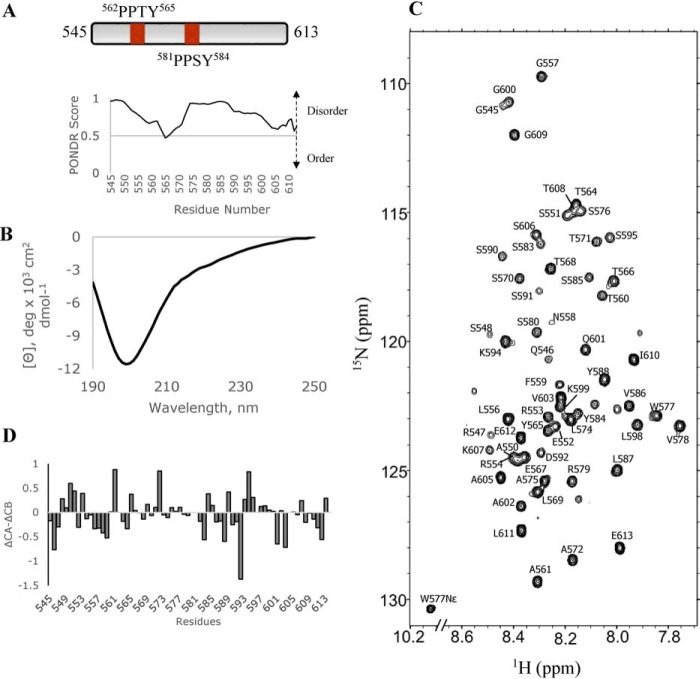
For this study, we used a 69-residue SYNPO polypeptide (SYNPO545–613) that includes both PPXY motifs, located at residues 562–565 and 581–584, and ∼28 flanking residues. The primary sequence of the SYNPO545–613 polypeptide was analyzed with PONDR, a computational tool for predicting unstructured regions in proteins (26, 27). The PONDR-generated disorder profile of SYNPO545–613 (Fig. 1) shows values greater than 0.5 indicative of a largely disordered polypeptide. To determine whether SYNPO545–613 is disordered as predicted, we used far UV CD and NMR spectroscopy to investigate the solution structure of the polypeptide. A strong negative signal at 200 nm in the far UV CD spectrum (Fig. 1B) and limited dispersion in the amide proton region of the 1H-15N HSQC2 spectrum (Fig. 1C) are consistent with a primarily disordered protein. To determine local structural propensities, backbone residue assignments were obtained for 57 of the 59 (Ser-596 and His-597 were not visible in the spectrum) nonproline residues. A plot of the deviations of the CA and CB chemical shift values (ΔCα–ΔCβ) from standard random coil values (28) in Fig. 1D, shows no strong propensity for this segment of SYNPO to form ordered structure. These results agree with the significant disorder reported for other members of the SYNPO family (29).
Figure 1.
SYNPO545–613 is primarily unstructured. A, schematic representation of the SYNPO545–613 construct showing the positions of the two PPXY (PPTY and PPSY) motifs. PONDR-generated disorder profile of SYNPO545–613. Values >0.5 indicate significant disorder in this segment of the protein. B, far UV CD spectrum of SYNPO545–613 collected in 10 mm sodium phosphate buffer, pH 8. SYNPO545–613 is predominantly unstructured as indicated by a strong ellipticity at 200 nm. C, 1H-15N HSQC spectrum of SYNPO545–613 showing backbone assignments for 57 of the 59 nonproline residues. The spectrum was recorded at 10 °C. D, a plot of the chemical shift differences (Δ13Cα–Δ13Cβ) as a function of residue number. ΔCA and ΔCB values were calculated by subtracting the random coil chemical shifts from experimentally determined chemical shift values.
Design of KIBRA constructs
Earlier studies assigned the KIBRA WW domain boundaries to residues 7–39 and 54–86, and identified P37A and P84A as mutations in the WW1 and WW2 domains, respectively, which abrogate binding to PPXY motifs (1). Guided by these findings and sequence-based secondary structure prediction, we designed the KWWTD, KWW1, and KWW2 constructs to include both, the first and the second WW domains, respectively, the P37ATD and P84ATD to probe binding to each WW domain in the context of the tandem WW domain construct, and the I81W mutants, I81WTD and KWW2-I81W, to restore the binding site tryptophan in the tandem and isolated WW domain constructs, respectively. A schematic of all the KIBRA constructs is shown in Fig. 2A.
Figure 2.

Schematic representation and far UV CD profiles of KIBRA WW domain constructs. A, The constructs used in this study were tandem WW domain (KWWTD), KWWTD variants with single point mutations in the WW1 (P37ATD) and WW2 (P84ATD and I81WTD) domains, individual WW domains (KWW1 or KWW2), and a KWW2 mutant (KWW2-I81W). Sequence-based secondary structure predictions are shown for the KWWTD construct. Two segments in the WW1 domain and a segment in the WW2 domain have β strand propensities (arrows). The rest of the protein is predicted to be disordered (dotted lines). B, far UV CD spectra of isolated WW domain constructs KWW1 (black), KWW2 (red), and KWW2-I81W (brown). C, far UV CD spectra of KWWTD (orange), P84ATD (green), I81WTD (magenta), and P37ATD (black). The P37A mutation decreases the signal at 230 nm. CD data were collected in 10 mm sodium phosphate, pH 6.8 or 8.0.
The KIBRA WW domains adopt different secondary structures
PSIPRED (30), a structure prediction algorithm that gives estimates of secondary structure based on a protein's primary sequence, predicts β strands for residues 22–26 and 32–35 in the WW1 domain, 70–72 in the WW2 domain (designated by arrows in Fig. 2A), and disorder for the rest of the KWWTD sequence. The CD spectrum of KWW1 shows a positive peak at 230 nm and a negative peak at 220 nm (black spectrum in Fig. 2B), which are signature WW domain peaks attributed to packing of the two tryptophans and β strands, respectively. Thus, the CD spectrum of KWW1 is consistent with a well-folded WW domain protein. In contrast, the spectra of KWW2 and KWW2-I81W lack the positive peak and show strong negative signals at 200 nm indicative of predominantly unstructured proteins (orange and brown spectra in Fig. 2B).
CD spectra for the tandem WW domain constructs, KWWTD, I81WTD, P84ATD, and P37ATD are shown in Fig. 2C. KWWTD, I81WTD, and P84ATD all show positive signals at 230 nm, albeit more reduced than the corresponding signal in the spectrum of the isolated WW1 domain in Fig. 2A, and negative signals at 220 and 200 nm. The similarities in the CD spectra suggest that the P84A and I81W point mutations do not significantly alter the secondary structure of the tandem WW domain polypeptide. The spectrum of the KWWTD polypeptide is qualitatively identical to the sum of the spectra for each WW domain (data not shown), which strongly implies that the domains retain their folded and unfolded structures in the tandem construct. In contrast, the CD spectrum of P37ATD (black spectrum in Fig. 2C) does not show a positive peak at 230 nm but only the negative signals at 220 and 200 nm, which clearly demonstrates that this polypeptide is predominantly unfolded. To summarize, the CD data demonstrate that the WW1 domain is folded, the WW2 domain is unfolded, the I81W and P84A mutations in the WW2 domain do not result in substantial folding of the WW2 domain, and a P37A mutation in the WW1 domain is enough to disrupt the folded structure.
Solution NMR studies of the KIBRA WW domains
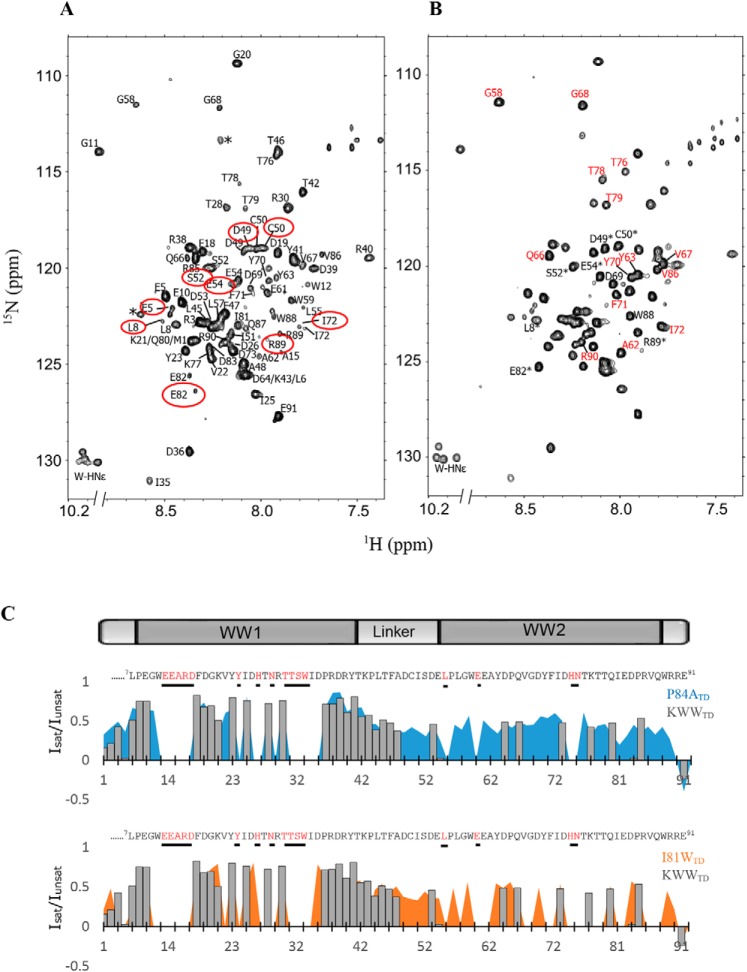
The 1H-15N HSQC spectrum of KWWTD with backbone assignments is shown in Fig. 3A. Heterogeneous peak intensities and substantial cross-peak overlap made it challenging to assign all the backbone resonances. Nonetheless we were able to unambiguously assign the 69 peaks (82 nonproline residues expected) visible in the spectrum. Missing peaks (Glu-13, Glu-14, Arg-16, Asp-17, Tyr-24, His-27, Asn-29, Thr-31, Thr-32, Ser-33, Trp-34, Glu-60, His-74, and Asn-75), mostly in the WW1 domain map to the 1st β strand and loop region (Glu-13, Glu-14, Arg-16 and -17), ligand-binding loop (His-27, Asn-29, Trp-34), and 3rd β strand (Thr-31, Thr-32, Ser-33) of the recent crystallographic structure of a ligand-bound mouse KIBRA WW domain (18). Chemical exchange with the solvent likely renders these signals undetectable. Two distinct peaks were assigned to N-terminal residues Glu-5 and Leu-8, which are adjacent to proline residues, inter-domain linker segment residues Asp-49, Cys-50, Ser-52, and Glu-54, and C-terminal residues Ile-72, Glu-82, and Arg-89 (circled in red, Fig. 3A). Five Trp indole NH signals rather than the four expected are observed in the indole region (1H 10.2 ppm; 15N 130 ppm) of the spectrum. Multiple peaks for the same NH are a strong indicator of multiple conformers in slow exchange. Peaks for the more folded KWW1 domain, example Gly-11 at 8.8 ppm and Arg-40 at 7.4 ppm, are more dispersed, whereas most peaks in the linker segment and the KWW2 domain are clustered in the middle of the spectrum.
Figure 3.
Resonance assignments and HetNOE plots of the KIBRA tandem WW domain constructs. A, 1H-15N HSQC spectrum of KWWTD showing backbone assignments for 69 of the 84 nonproline residues. Residues that display multiple conformations are circled in red. B, 1H-15N HSQC spectrum of P84ATD showing assignments for select peaks. Peaks designated with asterisks display multiple conformations in the WT KWWTD spectrum in A, but coalesce into single peaks in the P84ATD spectrum. Peaks labeled in red are more intense compared with equivalent peaks in A. C, plots of steady-state heteronuclear NOE (Isat/Iunsat) of individual residues in KWWTD (gray bar plot), P84ATD (blue area plot), and I81WTD (orange area plot). Residues for which assignments are not available are shown in red in the KWWTD sequence above each plot. Most assigned residues in the WW2 domain of KWWTD have HetNOEs close to zero, consistent with the disordered nature of the domain. Positive HetNOEs in the WW2 domain of P84ATD indicate transient structural rigidity. Error bars, ± 0.01- 0.06 are not shown for easier comparison.
The 1H-15N HSQC spectrum of P84ATD is shown in Fig. 3B. The spectrum is less heterogeneous than the KWWTD but is still missing the same peaks that were not visible in the spectrum of the WT protein. Several peaks (labeled in red, Fig. 3B) are significantly more intense than equivalent peaks in the WT spectrum. Peaks for residues Asp-49, Cys-50, Ser-52, Glu-54, Glu-82, and Arg-89 are denoted by asterisks. These peaks display multiple conformations in the WT spectrum but coalesce into single peaks in the P84ATD spectrum. The more homogeneous P84ATD spectrum may be due to reduced cis-trans isomerization. The 1H-15N HSQC spectrum of I81WTD was comparable with the KWWTD spectrum (see Fig. 6). However, the 1H-15N HSQC spectrum of P37ATD was less dispersed in the amide proton region, indicative of unfolding or aggregation of the polypeptide and consistent with the CD data (data not shown).
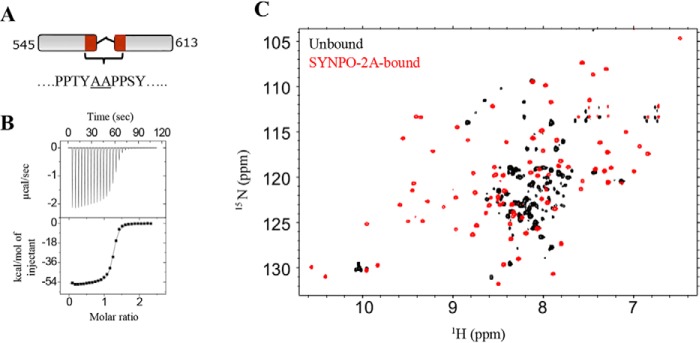
Figure 6.
SYNPO-2A binding induces changes in the KWWTD spectrum. A, schematic representation of the SYNPO-2A construct. The residues between the two PPXY motifs were replaced with two non-native alanine residues (underlined). B, ITC measurements for the KWWTD–SYNPO-2A interaction shows a binding affinity (Kd) of 0.1 μm. C, overlay of 1H-15N HSQC spectra of unbound (black) and SYNPO-2A–bound KWWTD (red) at a KWWTD:SYNPO-2A molar ratio of 1:1. Significant chemical shift dispersion in the SYNPO-2A–bound spectrum is indicative of binding-induced folding of the WW2 domain.
Backbone dynamics of the KIBRA tandem WW domain polypeptides
To gain insight into backbone dynamics of KWWTD, P84ATD, and I81WTD, we acquired steady-state heteronuclear NOE (HetNOE) data, which are sensitive to conformational dynamics in the picosecond to nanosecond time scale (31, 32). Generally, high positive (>0.6) HetNOE values suggest restricted internal motions, smaller positive values indicate substantial internal motions, and negative values are indicative of fully disordered regions. The HetNOE plots in Fig. 3C compare the WT KWWTD (gray bar plot) to P84ATD (blue area plot) and I81WTD (orange area plot). All three polypeptides show HetNOEs in the range of 0.5–0.9 for most residues in the WW1 domain. The relatively high values are indicative of restricted internal motions that support the conclusion that the WW1 domain is folded. For the KWWTD polypeptide, computed HetNOEs are close to zero for most residues immediately preceding the WW2 domain and within the WW2 domain. Equivalent residues in P84ATD (blue plot) and to a lesser extent I81WTD (orange plot) display relatively higher HetNOE values. These results are a clear indication of substantial internal motions in the WW2 domain of KWWTD.
Binding studies monitored by ITC
Binding energetics for SYNPO545–613-WW domain complexes were determined from isothermal titration calorimetry (ITC) experiments. Representative binding isotherms are shown in Fig. S1 and thermodynamic parameters are summarized in Table 1. All the interactions are enthalpically driven. Binding affinities are reported as dissociation constants (Kd), and range between 1.7 and 27 μm (Table 1). Binding of SYNPO545–613 to KWWTD is 2-fold weaker than I81WTD (Kd values of 2.2 and 0.9 μm, respectively) and ∼1.7-fold higher than interactions with P84ATD (compare 2.2 to 3.7 μm). The slight increase in the binding affinity of I81WTD relative to the WT KWWTD polypeptide suggests that the isoleucine (Ile-81) substitution in the WW2 domain reduces its affinity for SYNPO545–613. A stoichiometry of 1 is recorded for all interactions with the tandem WW domain polypeptides. As SYNPO545–613 is not expected to bind mutant domains in P37ATD and P84ATD a stoichiometry of 1 is interpreted as binding of a single WW domain to a single PPXY motif (1:1). For KWWTD and I81WTD, simultaneous interactions with both WW domains (2:2) could also result in a stoichiometry of 1.
Table 1.
Thermodynamics parameters for the SYNPO545–613 -KIBRA WW domain interaction
Changes in enthalpy (ΔH°), entropy (−TΔS°), and free energy of binding (ΔG°) are shown for interactions between KIBRA WW domains variants and SYNPO545–613. Values were determined at 25 °C from the average of two or three experiments using three different protein preparations. The error estimation is from experimental repeats.
| Titrant | n | Kd | ΔHo | −TΔSo | ΔGo |
|---|---|---|---|---|---|
| μm | kcal/mol | ||||
| KWWTD | 1.0 | 2.2 ± 0.1 | −38.3 ± 1.9 | 30.6 ± 1.9 | −7.71 ± 0.03 |
| P37ATD | 0.9 | 27 ± 6 | −17.5 ± 4.2 | 11.2 ± 4.3 | −6.23 ± 0.13 |
| P84ATD | 0.9 | 3.7 ± 0.1 | −29.3 ± 0.5 | 21.9 ± 0.5 | −7.41 ± 0.01 |
| I81WTD | 1.0 | 0.9 ± 0.1 | −49.8 ± 0.3 | 41.7 ± 0.3 | −8.24 ± 0.02 |
| KWW1 | 1.0 | 5.2 ± 0.4 | −23.4 ± 3.6 | 14.1 ± 3.6 | −7.21 ± 0.05 |
| KWW2 | 1.0a | >65 | NDb | ND | ND |
| KWW2 (I81W) | 1.0a | >60 | ND | ND | ND |
a Number of binding sites fixed to 1 in fitting procedure.
b ND, not determined.
Thermodynamic parameters for the interaction of SYNPO545–613 with the isolated KWW1, KWW2, and the WW2 domain mutant, KWW2-I81W are shown in Table 1. The Kd for the KWW1 domain-SYNPO545–613 interaction is (5.2 μm) ∼1.5–2–fold weaker than the P84ATD mutant (binding to the WW2 domain is abolished) or the KWWTD construct. This suggests that residues in the WW1 domain alone are not sufficient for optimum stability of the complex. Isotherms for the SYNPO545–613-KWW2 (Fig. S1F) and SYNPO545–613-KWW2-I81W (Fig. S1G) show low heat changes upon binding. The small magnitude of the heat changes make it difficult to fit the data and derive accurate thermodynamic parameters. Fixing the stoichiometry N, in the data analysis process to 1 (assuming binding to a single PPXY motif), rather than allowing it to be optimized as an additional fitting parameter, gave a minimum Kd value of 60 μm.
Solution NMR studies to map the KIBRA-SYNPO binding interface
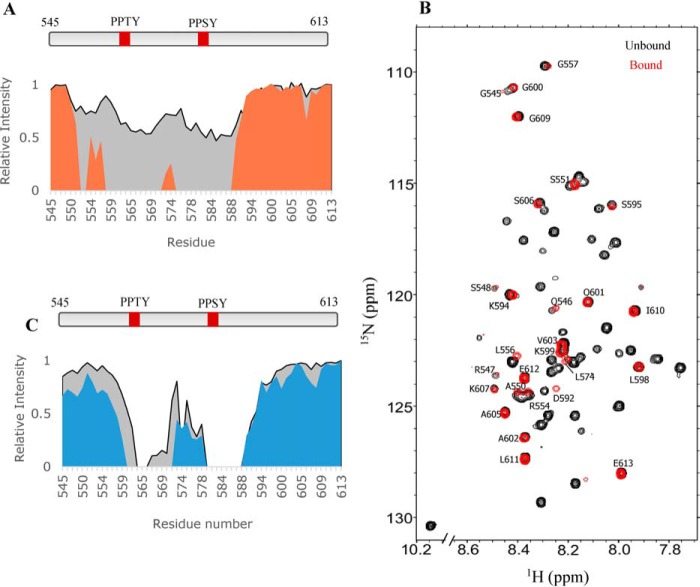
To map the binding interface of the SYNPO-KIBRA complex, we monitored chemical shift changes in the 1H-15N HSQC spectrum of SYNPO545–613, KWWTD, or mutant tandem polypeptides without or with the unlabeled binding partner. A change is defined as a shift and/or a decrease in peak intensity. Data for 15N-labeled SYNPO545–613 with unlabeled KWWTD added at molar ratios of 1:0.5 and 1:1 (SYNPO:KWWTD) are shown in Fig. 4. A plot of the normalized peak intensities of the 1:0.5 complex (Fig. 4A, gray plot) show a 43% decrease in peak intensities for residues 564–570 and 578–591. Increasing the molar ratio to 1:1 (orange plot in Fig. 4A and red spectrum in Fig. 4B) show complete loss of peaks corresponding to residues 558–571 and 575–588. Missing resonances are attributed to exchange broadening of peaks at the binding interface, as well as to slower tumbling of the complex. N-terminal residues 545–556, inter-PPXY motif linker segment residues 572–574 and C-terminal residues 592–613 are not perturbed (labeled peaks in Fig. 4B). A 1:2 complex did not show additional changes in chemical shift and intensities (data not shown) indicating that binding is saturated in the 1:1 complex. These observations are consistent with binding of both WW domains to both PPXY motifs.
Figure 4.
Residue-specific mapping of the KIBRA-binding site on SYNPO545–613. A, plots of relative peak intensity versus residue numbers of KIBRA-bound SYNPO545–613 at molar ratios of 0.5:1 (gray) and 1:1 (orange). B, overlay of 1H-15N HSQC spectra of unbound (black) and KWWTD-bound (red) 15N-labeled SYNPO545–613 at a 1:1 molar ratio. Peaks visible in the KIBRA-bound spectrum are labeled. C, plots of the relative intensity versus residue numbers of KWW1-bound SYNPO545–613 at molar ratios of 1:1 (gray) and 1:2 (blue). Binding is accompanied by complete loss of peaks in the vicinity of the PPXY motifs. Peak intensities are relative to the intensity of the same peak in unbound SYNPO545–613, which is taken as one.
To define the orientation of SYNPO545–613 in the complex, unlabeled KWW1 was titrated into 15N-labeled SYNPO545–613. Normalized peak intensities for the 1:1 (gray area plot) and 1:2 (blue area plot) complexes are shown in Fig. 4C. The 1:1 complex shows complete disappearance of peaks corresponding to residues 579–588, which include the second PPXY motif, and a few peaks corresponding to residues (564–566) in the vicinity of the first PPXY motif. More peaks in the 560–571 segment disappear when a higher concentration of unlabeled KWW1 is added (blue plot). The pattern of peak disappearance clearly shows that KWW1 first binds the second (PPSY) motif before it binds the first motif.
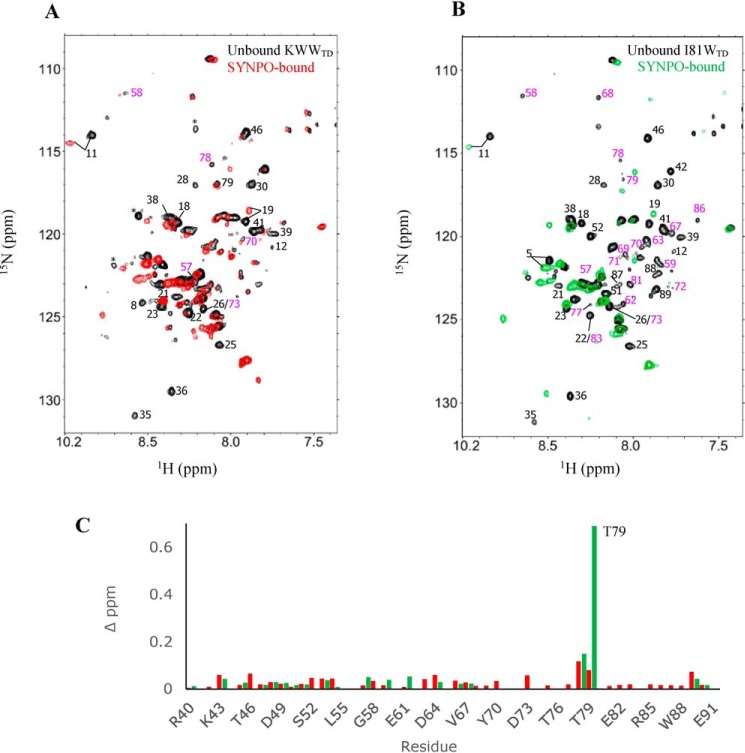
In a reciprocal NMR titration experiment, we monitored chemical shift changes in the 1H-15N HSQC spectrum of KWWTD or I81WTD when bound to saturating concentrations of unlabeled SYNPO545–613. Spectra of unbound (black) and SYNPO545–613-bound KWWTD (red) or I81WTD (green) at molar ratios of 1:1 are shown in Fig. 5, A and B. Peaks that are significantly attenuated or shifted in the SYNPO545–613-bound spectra are labeled. We reasoned that attenuated peaks are directly or indirectly involved in binding, and attribute the broadened resonances to exchange between unbound and bound conformations as well as to slower tumbling of the complex. Increasing the concentration of added SYNPO545–613 to a molar ratio of 1:2 did not result in additional changes in chemical shift or intensities (Fig. S2), a clear indication that binding is saturated in the 1:1 complex. Fewer WW2 domain peaks (labeled in magenta) are attenuated in the SYNPO545–613-bound KWWTD compared with the SYNPO545–613-bound I81WTD. This is consistent with the higher affinity of SYNPO545–613 for the I81W mutant. Fig. 5C show plots of the chemical shift changes in the SYNPO-bound WW2 domain of KWWTD (red) and I81WTD (green). Small but distinct changes in chemical shift are observed for most residues except Thr-79 in the SYNPO-bound I81WTD spectrum, which is significantly shifted. To summarize, SYNPO545–613 binding is accompanied by changes in chemical shift for select residues in both WW domains. This is clear indication that SYNPO binds both WW domains. The lack of chemical shift dispersion in the SYNPO-bound spectra of KWWTD and I81WTD is indication that the WW2 domain remains globally disordered in the SYNPO-bound complex.
Figure 5.
Residue-specific mapping of the SYNPO545–613-binding site on the KIBRA WW domains. Overlay of 2D 1H-15N HSQC spectra of 15N-labeled unbound (black) and SYNPO545–613-bound (A) KWWTD (red) and (B) I81WTD (green). Select peaks in the WW1 (black labels) and WW2 (magenta labels) domain that completely disappear or are significantly shifted in the SYNPO545–613-bound spectra are labeled with the residue number. These peaks are directly or indirectly involved in binding. Peaks designated with asterisks are part of the expression vector. The complexes were formed by mixing equal concentrations of unlabeled and 15N-labeled proteins. C, plots of chemical shift perturbation in the WW2 domain of SYNPO545–613-bound KWWTD (red) and SYNPO545–613-bound I81WTD (green). Binding induced small but definite changes in chemical shift for most residues. Thr-79 is significantly shifted in the SYNPO545–613-bound I81WTD spectrum.
Binding-induced folding of the KIBRA WW2 domain
Zhang and co-workers (18) reported that the mouse KIBRA WW2 domain adopts the canonical WW domain structure when bound to a two-PPXY motif-containing peptide of the neural and renal protein, Dendrin. The two PPXY sites in Dendrin are separated by only two residues, whereas the SYNPO545–613 PPXY sites are separated by a 15-residue linker. Thus we asked the question whether a SYNPO polypeptide with a two-residue linker would induce folding of the WW2 domain. To address this question, we engineered the SYNPO variant SYNPO-2A. The two PPXY sites in this variant are separated by two non-native alanine residues (Fig. 6). The ITC-measured dissociation constant of the SYNPO-2A–KWWTD interaction was 0.1 μm (Fig. 6B), which represents a 20-fold increase in binding affinity compared with the WT SYNPO545–613 (Kd of 2.2 μm in Table 1). Next, we monitored binding between 15N-labeled KWWTD and SYNPO-2A by NMR spectroscopy. The 1H-15N HSQC spectrum of SYNPO-2A-bound KWWTD is shown in Fig. 6C. The spectrum is well-dispersed, a clear indication that both WW domains are well-folded in the complex.
Discussion
Despite decades of well-documented studies on the functional roles of the KIBRA WW domains, how the domains, particularly the WW2 domain, bind partner PPXY proteins is unclear. A recent structural study of a mouse KIBRA tandem WW domain polypeptide bound to a peptide of the synaptic protein Dendrin, shows that the isolated WW2 domain is unfolded but adopts a well-folded structure in the complex (18). In contrast, the biophysical studies reported here clearly demonstrate that the WW2 domain remains largely disordered when bound to the actin-binding synaptic and podocyte protein Synaptopodin. The different folds of the WW2 domain in these independent studies clearly demonstrate that different PPXY partners induce varying degrees of folding of the WW2 domain. The extent of folding may be important for the diverse functions of KIBRA.
The isolated KIBRA WW2 domain binds PPXY peptides with weak affinity
In this study and previously reported studies (1, 18), the isolated WW2 domain displays weak binding to PPXY-containing peptides. Two unusual features of the KIBRA WW2 domain explain why it has a weak affinity for PPXY containing peptides: intrinsic disorder and the Ile-81 substitution of the conserved tryptophan. Disorder coupled to weak binding is inferred from observations that when the relatively well-folded WW1 domain is unfolded by introducing the P37A mutation, its affinity for the SYNPO polypeptide is also considerably weakened (Table 1). Pro-37 is a conserved WW domain residue and part of a hydrophobic core formed by the side chains of Trp-12 and Tyr-24. The hydrophobic core stabilizes the anti-parallel β sheet of the KIBRA WW1 domain (18), therefore it is not surprising that disrupting this core unfolds the WW1 domain. A mutation of the conserved proline in the WW domains of Yes-associated protein (YAP) and Formin-binding protein, FBP28, also unfold the domains (33, 34). Unexpectedly, alanine substitution in the equivalent position in the WW2 domain, Pro-84, leads to a substantial reduction in internal motions of the WW2 domain. This observation is intriguing because a stable hydrophobic core comprising Trp-59, Phe-71, and Pro-84 (equivalent to Trp-12, Tyr-24, and Pro-37 in the WW1 domain) is not expected to form in the primarily disordered WW2 domain. It is possible that Pro-84 is involved in cis-trans isomerization, which is abolished by the alanine mutation.
The second underlying cause of the weak affinity for PPXY peptides is the Ile-81 substitution in the WW2 domain. We find that as reported for the WW domains of Smad ubiquitination regulatory factor 2 and WW domain-containing oxidoreductase (16, 17), the loss of the binding site tryptophan reduces the affinity of the tandem WW domain polypeptide for SYNPO545–613. Isoleucine or leucine is the preferred amino acid in this position for KIBRA isoforms from Drosophila to humans, which suggests that this substitution is optimized for the functions of the KIBRA WW domains.
Similarities and differences between the KIBRA-SYNPO and KIBRA-Dendrin complexes
The solution studies reported here provide valuable molecular level insights into how the KIBRA WW domains, particularly, the WW2 domain, bind SYNPO. The pattern of peak disappearance in the NMR titration experiments (Fig. 4) strongly suggests that the second PPXY motif binds the N-terminal WW1 domain in the same anti-parallel orientation previously reported for the Dendrin polypeptide (18). The PPXY polypeptides may bind in a similar anti-parallel manner, but clearly there are distinct differences in how they influence the structure of the WW2 domain. The 1H-15N HSQC spectrum of the Dendrin-bound mouse KIBRA tandem WW domains shows significant peak dispersion consistent with global folding of the WW2 domain (18). In contrast, the limited dispersion in the 1H-15N HSQC spectrum of the 1:1 (Fig. 5) and 1:2 (Fig. S2) KIBRA-SYNPO complexes strongly argues against global folding of the WW2 domain. We do not rule out the possibility that in this dynamic complex, a population of the WW2 domain becomes folded upon binding to SYNPO545–613. However, this population is minimal compared with the population that remains disordered in the complex. Thus, whereas binding of Dendrin to the tandem WW domain of mouse KIBRA induces substantial folding of the WW2 domain, the data presented here clearly show that binding of SYNPO545–613 to the tandem WW domains of human KIBRA does not induce substantial folding of the WW2 domain.
A second major difference between the two binding partners is that whereas the dissociation constant of the SYNPO545–613-KIBRA complex is in the low micromolar range, a typical range for most WW domain-PPXY interactions (17, 35, 36), the interaction with Dendrin occurs with an unusually high nanomolar dissociation constant. The two PPXY sites in the Dendrin peptide are separated by only two amino acids; and increasing the number of amino acids between the two PPXY sites substantially reduced the binding affinity for the KIBRA WW domains (18). On the contrary, the two PPXY sites in the SYNPO545–613 polypeptide are separated by 15 residues. We demonstrate that a mutant SYNPO545–613 polypeptide, SYNPO-2A, with a two-residue linker binds the KWWTD polypeptide with a 20-fold higher affinity compared with the WT SYNPO545–613 polypeptide. Furthermore, initial NMR studies show that binding of SYNPO-2A to the KIBRA tandem WW domains induces substantial chemical shift changes in the 1H-15N HSQC spectrum (Fig. 6), similar to the KIBRA-Dendrin interaction (18). Clearly, whereas the WW2 domain remains largely disordered when bound to WT SYNPO, reducing the SYNPO PPXY linker to two residues induces substantial folding of the domain.
Model for the SYNPO-KIBRA interaction
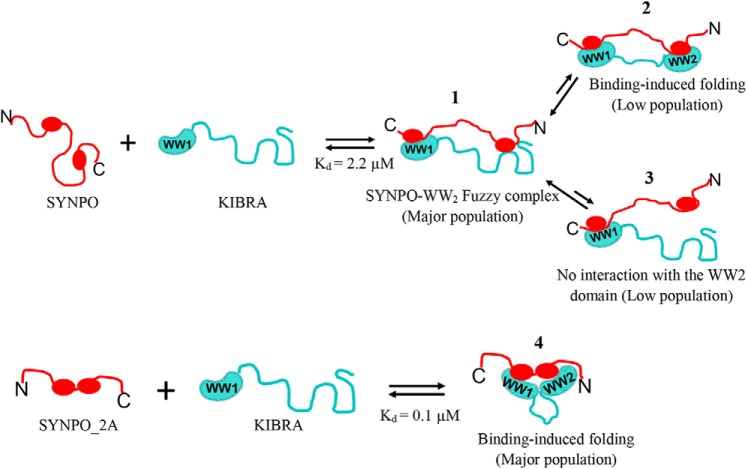
Fig. 7 depicts the proposed binding model for the KIBRA-SYNPO dynamic complex. In this bipartite mode of interaction, SYNPO first binds the WW1 domain via the second PPXY motif, inducing folding of a small percentage of WW2 domain (panel 2), whereas a substantial population remains largely disordered. A small fraction of the disordered WW2 domain remains unbound to SYNPO (panel 3), whereas a large proportion binds the first PPXY motif (panel 1). The bound WW2 domain is best described as a fuzzy complex, a term used to describe IDP complexes that defy the classic stable and well-folded binding interface (37–39) and retain conformational heterogeneity in the bound state. “Fuzziness” is reduced when the SYNPO PPXY sites are separated by two residues (panel 4). We speculate that partners in which the PPXY sites are separated by two residues will induce substantial folding of the WW2 domain, whereas partners with a longer linker will not. The structural variability of the WW2 domain may be critical for the diverse cellular functions of KIBRA.
Figure 7.
A model summarizing the interaction between the KIBRA WW domains and synaptopodin. The KIBRA WW domains (teal) comprise a well-folded WW1 domain and a disordered WW2 domain. SYNPO (red) has two PPXY motifs. The two proteins interact in an antiparallel manner, that is, the WW1 domain binds the second PPXY motif. The model depicts the populated conformations of the WW2 domain in the complex: 1) the WW2 domain remains largely unfolded forming a fuzzy complex with the first PPXY motif (most populated) and minor populations in which the WW2 domain; 2) undergoes a disorder-to-order transition; or 3) remains disordered and unbound to SYNPO; and 4) the SYNPO variant, SYNO-2A, binds the KIBRA WW domains with a higher affinity and induces substantial folding of the WW2 domain.
Experimental procedures
Cloning of constructs
All constructs were prepared using the PCR-based Gibson assembly cloning protocol (New England Biolabs). Briefly, a 69-residue Synaptopodin (Uniprot ID Q8N3V7) construct (hereafter referred to as SYNPO545–613), and KIBRA (Uniprot ID Q8IX03) WW domain constructs KWWTD (residues 1–91), KWW1 (residues 1–48), and KWW2 (residues 45–91) were generated by PCR. We also generated a SYNPO545–613 variant, SYNPO-2A, with a two-alanine residue linker between the two PPXY sites. PCR products were cloned into a modified pET24TM (Novagen) expression vector engineered with a tobacco etch virus protease recognition sequence to allow cleavage of the N-terminal poly-histidine tag. A Q5TM PCR-based mutagenesis protocol (New England Biolabs) was used to generate single site mutations of the tandem WW domain construct, P37ATD and P84ATD, which abrogate binding of PPXY motifs to the WW1 and WW2 domains, respectively (1), and I81W (I81WTD) and the isolated WW2 domain (KWW2-I81W). Primers for the PCR and mutagenesis reactions were purchased from Eurofins Genomics.
Recombinant protein production
Recombinant plasmids were transformed into Escherichia coli BL21 (DE3) cells. Cells were cultured at 37 °C in Lysogeny broth, or 15N-enriched MJ9 media supplemented with [12C]- or [13C]glucose to mid-log phase (optical density of 0.6–0.7). The temperature was reduced to 20 °C, and 0.1 mm isopropyl 1-thio-d-galactopyranoside was added to the culture to induce protein expression. Cultured cells were harvested after 12 h, lysed by sonication, and centrifuged to remove cell debris. The soluble His6-tagged proteins were first purified on a nickel-nitrilotriacetic acid affinity (Qiagen) column, followed by treatment with tobacco etch virus protease to remove the His6 tag, and finally purified on a Superdex-75 (GE Life Sciences) size exclusion chromatography column. Purity of recombinant proteins were assessed by SDS-PAGE. Protein concentrations were determined from the absorbance at 280 nm and computed molar extinction coefficients (http://web.expasy.org/protparam).
CD spectroscopy
Far UV CD data were recorded at 25 °C, on a JASCO 720 spectropolarimeter using a bandwidth of 1.0 nm and a path length of 1 mm. The experiments were collected on two independent samples prepared from two different protein preparations. Protein samples at concentrations of 10–20 μm were in buffer composed of 10 mm sodium phosphate, pH 6.8 or 8.0. Three scans were recorded on each sample. Reported data are the average of the experimental repeats.
Isothermal titration calorimetry
ITC data were collected on a VP-ITC instrument (Malvern instruments Inc.) set to 25 °C. 27 10-μl injections of 130–200 μm of the KIBRA constructs (KWWTD, KWW1, KWW2, P37ATD, P84ATD, KWW2-I81W and I81WTD) in buffer composed of 50 mm sodium phosphate, 50 mm NaCl, 5 mm β-mercaptoethanol, 1 mm sodium azide, pH 8, was titrated into 12–24 μm SYNPO545–613 in the same buffer. ITC data were analyzed with the Origin 7.0 software provided with the instrument. Data were fitted to a one-site model. Reported data are the average of two or three independent experiments performed under similar conditions using proteins from three different preparations. Three replicate titrations were carried out for each experiment. Standard deviations are computed from the average of the two experiments.
NMR data collection and analysis
NMR experiments were performed at 25 °C on a Bruker Avance III, 800 MHz spectrometer (Bruker BioSpin) equipped with a triple resonance cryogenic probe. NMR samples were prepared in 10 mm sodium phosphate buffer, pH 6.8, 10 mm sodium chloride, 5 mm tris(2-carboxyethyl)phosphine, and 10% D2O. A series of backbone assignments experiments (1H-15N HSQC, HNCACB, CBCA(CO)NH, and HBHA(CO)NH) were recorded on 13C-15N isotopically labeled KIBRA constructs (WWTD, P84ATD, P37ATD, and I81WTD) or SYNPO545–613 using BEST (band selective excitation short transient) (40) and/or TROSY (41) pulse sequences, and non-uniform sampling in the indirect dimensions. Non-uniform sampling data were reconstructed in NMRpipe (42). 1H chemical shifts were referenced to an internal 2,2-dimethylsilapentene-5-sulfonic acid. NMR data were visualized with the graphical program Sparky (47).
Titration data were collected on 15N-labeled SYNPO545–613 or 15N-labeled KIBRA constructs bound to the unlabeled partner. Unlabeled proteins were added at molar ratios of 1:0.5, 1:1, and 1:2 (labeled:unlabeled protein). The peak corresponding to the last residue (91 in isotopically labeled KWWTD/I81WTD and 613 in isotopically labeled SYNPO545–613) was used as an internal reference to correct for small changes in peak intensities across spectra. The relative intensity of each peak (measured as peak heights) was calculated as the ratio of the intensity of the peak in the spectrum of the bound protein to the intensity of the same peak in the spectrum of the unbound protein.
Steady-state 1H-15N heteronuclear NOEs data were collected as two two-dimensional interleaved spectra with and without proton saturation and a recycle delay of 8 s (43). NOE values were determined from the ratio of the peak intensities in the presence and absence of proton saturation. Errors associated with the data (σnoe) were estimated from the root mean square variation of the noise using Equation 1 (32).
| (Eq. 1) |
Where σsat and σunsat are the standard deviation values of peak intensities with and without saturation, respectively. HetNOE experiments for KWWTD were collected on two independent samples prepared from two different protein preparations.
Secondary structure propensity from NMR chemical shifts
Random coil values for NMR-based chemical shift propensities were compiled from the web-based algorithm of Poulsen et al. (44–46). Local structural propensities were calculated from the deviations of the experimental Cα and Cβ chemical shifts from expected random coil values (28). A negative difference between the Cα and the Cβ secondary chemical shifts (ΔCα–ΔCβ) is suggestive of β strand propensity; and a positive difference is attributed to helical propensity.
Chemical shift changes
Chemical shift for assigned residues in the C terminus (residue 45–91) of KWWTD and I81WTD were computed from the 1:1 titration point. The combined 1H and 15N chemical shift changes were calculated as follows,
| (Eq. 2) |
where HN and N are the 1H and 15N chemical shifts.
BMRB accession code
The chemical shifts for KIBRA KWWTD have been deposited in the Biological Magnetic Resonance Data Bank under accession code 27930.
Author contributions
E. K., D. J. R., and A. N. formal analysis; E. K., D. J. R., and A. N. investigation; J. K. and A. N. resources; A. N. conceptualization; A. N. supervision; A. N. funding acquisition; A. N. validation; A. N. writing-original draft with input from E. K.; A. N. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Professors Elisar Barbar and Andy Karplus for helpful discussions. NMR experiments were collected at the Oregon State University NMR Facility funded in part by National Institutes of Health HEI Grant 1S10OD018518 and M. J. Murdock Charitable Trust Grant 2014162.
This work was supported by start-up funds from the Oregon State University (OSU), OSU College of Science Disease Mechanism and Prevention Fund, and a New Investigator Award from the Medical Research Foundation of Oregon. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1 and S2.
The chemical shifts have been deposited in the Biological Magnetic Resonance Data Bank under accession code 27930.
- HSQC
- heteronuclear single quantum coherence
- HetNOE
- heteronuclear NOE
- ITC
- isothermal titration calorimetry.
References
- 1. Kremerskothen J., Plaas C., Büther K., Finger I., Veltel S., Matanis T., Liedtke T., and Barnekow A. (2003) Characterization of KIBRA, a novel WW domain-containing protein. Biochem. Biophys. Res. Commun. 300, 862–867 10.1016/S0006-291X(02)02945-5 [DOI] [PubMed] [Google Scholar]
- 2. Rosse C., Formstecher E., Boeckeler K., Zhao Y., Kremerskothen J., White M. D., Camonis J. H., and Parker P. J. (2009) An aPKC-exocyst complex controls paxillin phosphorylation and migration through localised JNK1 activation. PLos Biol. 7, e1000235 10.1371/journal.pbio.1000235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yoshihama Y., Sasaki K., Horikoshi Y., Suzuki A., Ohtsuka T., Hakuno F., Takahashi S., Ohno S., and Chida K. (2011) KIBRA suppresses apical exocytosis through inhibition of aPKC kinase activity in epithelial cells. Curr. Biol. 21, 705–711 10.1016/j.cub.2011.03.029 [DOI] [PubMed] [Google Scholar]
- 4. Duning K., Schurek E. M., Schlüter M., Bayer M., Reinhardt H. C., Schwab A., Schaefer L., Benzing T., Schermer B., Saleem M. A., Huber T. B., Bachmann S., Kremerskothen J., Weide T., and Pavenstädt H. (2008) KIBRA modulates directional migration of podocytes. J. Am. Soc. Nephrol. 19, 1891–1903 10.1681/ASN.2007080916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Traer C. J., Rutherford A. C., Palmer K. J., Wassmer T., Oakley J., Attar N., Carlton J. G., Kremerskothen J., Stephens D. J., and Cullen P. J. (2007) SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat. Cell Biol. 9, 1370–1380 10.1038/ncb1656 [DOI] [PubMed] [Google Scholar]
- 6. Vogt-Eisele A., Krüger C., Duning K., Weber D., Spoelgen R., Pitzer C., Plaas C., Eisenhardt G., Meyer A., Vogt G., Krieger M., Handwerker E., Wennmann D. O., Weide T., Skryabin B. V., et al. (2014) KIBRA (KIdney/BRAin protein) regulates learning and memory and stabilizes Protein kinase Mζ. J. Neurochem. 128, 686–700 10.1111/jnc.12480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Witte A. V., Kobe T., Kerti L., Rujescu D., and Floel A. (2016) Impact of KIBRA polymorphism on memory function and the hippocampus in older adults. Neuropsychopharmacology 41, 781–790 10.1038/npp.2015.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schneider A., Huentelman M. J., Kremerskothen J., Duning K., Spoelgen R., and Nikolich K. (2010) KIBRA: a new gateway to learning and memory? Front. Aging Neurosci. 2, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Genevet A., Wehr M. C., Brain R., Thompson B. J., and Tapon N. (2010) Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell 18, 300–308 10.1016/j.devcel.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stauffer S., Chen X., Zhang L., Chen Y., and Dong J. (2016) KIBRA promotes prostate cancer cell proliferation and motility. FEBS J. 283, 1800–1811 10.1111/febs.13718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anuj Arivazhagan L., Surabhi R. P., Kanakarajan A., Sundaram S., Pitani R. S., Mudduwa L., Kremerskothen J., Venkatraman G., and Rayala S. K. (2017) KIBRA attains oncogenic activity by repressing RASSF1A. Br. J. Cancer 117, 553–562 10.1038/bjc.2017.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Macias M. J., Hyvönen M., Baraldi E., Schultz J., Sudol M., Saraste M., and Oschkinat H. (1996) Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature 382, 646–649 10.1038/382646a0 [DOI] [PubMed] [Google Scholar]
- 13. Chen H. I., and Sudol M. (1995) The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc. Natl. Acad. Sci. U.S.A. 92, 7819–7823 10.1073/pnas.92.17.7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Martinez-Rodriguez S., Bacarizo J., Luque I., and Camara-Artigas A. (2015) Crystal structure of the first WW domain of human YAP2 isoform. J. Struct. Biol. 191, 381–387 10.1016/j.jsb.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 15. Andrè B., and Springael J. Y. (1994) WWP, a new amino acid motif present in single or multiple copies in various proteins including dystrophin and the SH3-binding Yes-associated protein YAP65. Biochem. Biophys. Res. Commun. 205, 1201–1205 10.1006/bbrc.1994.2793 [DOI] [PubMed] [Google Scholar]
- 16. Chong P. A., Lin H., Wrana J. L., and Forman-Kay J. D. (2006) An expanded WW domain recognition motif revealed by the interaction between Smad7 and the E3 ubiquitin ligase Smurf2. J. Biol. Chem. 281, 17069–17075 10.1074/jbc.M601493200 [DOI] [PubMed] [Google Scholar]
- 17. Schuchardt B. J., Bhat V., Mikles D. C., McDonald C. B., Sudol M., and Farooq A. (2013) Molecular origin of the binding of WWOX tumor suppressor to ErbB4 receptor tyrosine kinase. Biochemistry 52, 9223–9236 10.1021/bi400987k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ji Z., Li H., Yang Z., Huang X., Ke X., Ma S., Lin Z., Lu Y., and Zhang M. (2019) Kibra modulates learning and memory via binding to Dendrin. Cell Rep. 26, 2064–2077.e7 10.1016/j.celrep.2019.01.097 [DOI] [PubMed] [Google Scholar]
- 19. Iglesias-Bexiga M., Castillo F., Cobos E. S., Oka T., Sudol M., and Luque I. (2015) WW domains of the yes-kinase-associated-protein (YAP) transcriptional regulator behave as independent units with different binding preferences for PPxY motif-containing ligands. PLoS ONE 10, e0113828 10.1371/journal.pone.0113828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin T., Zhang L., Liu S., Chen Y., Zhang H., Zhao X., Li R., Zhang Q., Liao R., Huang Z., Zhang B., Wang W., Liang X., and Shi W. (2017) WWC1 promotes podocyte survival via stabilizing slit diaphragm protein dendrin. Mol. Med. Rep. 16, 8685–8690 10.3892/mmr.2017.7708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Asanuma K., Akiba-Takagi M., Kodama F., Asao R., Nagai Y., Lydia A., Fukuda H., Tanaka E., Shibata T., Takahara H., Hidaka T., Asanuma E., Kominami E., Ueno T., and Tomino Y. (2011) Dendrin location in podocytes is associated with disease progression in animal and human glomerulopathy. Am. J. Nephrol. 33, 537–549 10.1159/000327995 [DOI] [PubMed] [Google Scholar]
- 22. Kremerskothen J., Kindler S., Finger I., Veltel S., and Barnekow A. (2006) Postsynaptic recruitment of Dendrin depends on both dendritic mRNA transport and synaptic anchoring. J. Neurochem. 96, 1659–1666 10.1111/j.1471-4159.2006.03679.x [DOI] [PubMed] [Google Scholar]
- 23. Mundel P., Heid H. W., Mundel T. M., Krüger M., Reiser J., and Kriz W. (1997) Synaptopodin: an actin-associated protein in telencephalic dendrites and renal podocytes. J. Cell. Biol. 139, 193–204 10.1083/jcb.139.1.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Asanuma K., Yanagida-Asanuma E., Faul C., Tomino Y., Kim K., and Mundel P. (2006) Synaptopodin orchestrates actin organization and cell motility via regulation of RhoA signalling. Nat. Cell Biol. 8, 485–491 10.1038/ncb1400 [DOI] [PubMed] [Google Scholar]
- 25. Zhang X. L., Poschel B., Faul C., Upreti C., Stanton P. K., and Mundel P. (2013) Essential role for synaptopodin in dendritic spine plasticity of the developing hippocampus. J. Neurosci. 33, 12510–12518 10.1523/JNEUROSCI.2983-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Romero P., Obradovic Z., Kissinger C. R., Villafranca J. E., and Dunker A. K. (1997) Identifying disordered proteins from amino acid sequences. Proceedings of International Conference on Neural Networks, pp. 90–95, IEEE, Houston, TX [Google Scholar]
- 27. Garner E., Cannon P., Romero P., Obradovic Z., and Dunker A. K. (1998) Predicting disordered regions from amino acid sequence: common themes despite differing structural characterization. Genome Inform. Ser. Workshop Genome Inform. 9, 201–213 [PubMed] [Google Scholar]
- 28. Wishart D. S., and Case D. A. (2001) Use of chemical shifts in macromolecular structure determination. Methods Enzymol. 338, 3–34 [DOI] [PubMed] [Google Scholar]
- 29. Chalovich J. M., and Schroeter M. M. (2010) Synaptopodin family of natively unfolded, actin binding proteins: physical properties and potential biological functions. Biophys. Rev. 2, 181–189 10.1007/s12551-010-0040-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McGuffin L. J., Bryson K., and Jones D. T. (2000) The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405 10.1093/bioinformatics/16.4.404 [DOI] [PubMed] [Google Scholar]
- 31. Farrow N. A., Zhang O., Forman-Kay J. D., and Kay L. E. (1995) Comparison of the backbone dynamics of a folded and an unfolded SH3 domain existing in equilibrium in aqueous buffer. Biochemistry 34, 868–878 10.1021/bi00003a021 [DOI] [PubMed] [Google Scholar]
- 32. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., and Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 10.1021/bi00185a040 [DOI] [PubMed] [Google Scholar]
- 33. Koepf E. K., Petrassi H. M., Ratnaswamy G., Huff M. E., Sudol M., and Kelly J. W. (1999) Characterization of the structure and function of W → F WW domain variants: identification of a natively unfolded protein that folds upon ligand binding. Biochemistry 38, 14338–14351 10.1021/bi991105l [DOI] [PubMed] [Google Scholar]
- 34. Petrovich M., Jonsson A. L., Ferguson N., Daggett V., and Fersht A. R. (2006) Phi-analysis at the experimental limits: mechanism of β-hairpin formation. J. Mol. Biol. 360, 865–881 10.1016/j.jmb.2006.05.050 [DOI] [PubMed] [Google Scholar]
- 35. Schuchardt B. J., Bhat V., Mikles D. C., McDonald C. B., Sudol M., and Farooq A. (2014) Molecular basis of the binding of YAP transcriptional regulator to the ErbB4 receptor tyrosine kinase. Biochimie (Paris) 101, 192–202 10.1016/j.biochi.2014.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nyarko A. (2018) Differential binding affinities and allosteric conformational changes underlie interactions of Yorkie and a multivalent PPxY partner. Biochemistry 57, 547–556 10.1021/acs.biochem.7b00973 [DOI] [PubMed] [Google Scholar]
- 37. Sharma R., Raduly Z., Miskei M., and Fuxreiter M. (2015) Fuzzy complexes: specific binding without complete folding. FEBS Lett. 589, 2533–2542 10.1016/j.febslet.2015.07.022 [DOI] [PubMed] [Google Scholar]
- 38. Mittag T., Orlicky S., Choy W. Y., Tang X., Lin H., Sicheri F., Kay L. E., Tyers M., and Forman-Kay J. D. (2008) Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc. Natl. Acad. Sci. U.S.A. 105, 17772–17777 10.1073/pnas.0809222105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tompa P., and Fuxreiter M. (2008) Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 33, 2–8 10.1016/j.tibs.2007.10.003 [DOI] [PubMed] [Google Scholar]
- 40. Favier A., and Brutscher B. (2011) Recovering lost magnetization: polarization enhancement in biomolecular NMR. J. Biomol. NMR 49, 9–15 10.1007/s10858-010-9461-5 [DOI] [PubMed] [Google Scholar]
- 41. Salzmann M., Pervushin K., Wider G., Senn H., and Wüthrich K. (1998) TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc. Natl. Acad. Sci. U.S.A. 95, 13585–13590 10.1073/pnas.95.23.13585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 43. Farrow N. A., Zhang O., Forman-Kay J. D., and Kay L. E. (1994) A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J. Biomol. NMR 4, 727–734 10.1007/BF00404280 [DOI] [PubMed] [Google Scholar]
- 44. Kjaergaard M., Brander S., and Poulsen F. M. (2011) Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH. J. Biomol. NMR 49, 139–149 10.1007/s10858-011-9472-x [DOI] [PubMed] [Google Scholar]
- 45. Kjaergaard M., and Poulsen F. M. (2011) Sequence correction of random coil chemical shifts: correlation between neighbor correction factors and changes in the Ramachandran distribution. J. Biomol. NMR 50, 157–165 10.1007/s10858-011-9508-2 [DOI] [PubMed] [Google Scholar]
- 46. Schwarzinger S., Kroon G. J., Foss T. R., Chung J., Wright P. E., and Dyson H. J. (2001) Sequence-dependent correction of random coil NMR chemical shifts. J. Am. Chem. Soc. 123, 2970–2978 10.1021/ja003760i [DOI] [PubMed] [Google Scholar]
- 47. Goddard T. D., and Kneller D. G. (2005) Sparky 3, University of California, San Francisco [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.