Abstract
The blood-brain barrier (BBB) is composed of brain microvascular endothelial cells (BMECs) that regulate brain homeostasis, and astrocytes within the brain are involved in the maintenance of the BBB or modulation of its integrity in disease states via secreted factors. A major challenge in modeling the normal or diseased BBB is that conventional in vitro models lack either the physiological complexity of the BBB or key functional features such as formation of a sufficiently tight barrier. In this study, we utilized human induced pluripotent stem cell (hiPSC)-derived BMECs in a BBB-on-a-chip device that supports flow and coculture with an astrocyte-laden 3D hydrogel. The BMECs are separated from the hydrogel by a porous membrane with either 0.4 or 8.0 μm pore size, making the device suitable for studying the transport of molecules or cells, respectively, across the BBB. In addition, all cells seeded in the device are differentiated from the same hiPSC line, which could enable genetic and rare disease modeling. Formation of a confluent BMEC barrier was confirmed by immunocytochemistry of tight junction proteins and measurement of fluorescein permeability. Integrity of the barrier was further assessed by performing impedance spectroscopy in the device. Finally, the ability of this device to recapitulate a disease model of BBB disruption was demonstrated, with apical addition of TGF-β1 leading to transendothelial electrical resistance reduction and indicators of astrocyte activation. These results demonstrate the utility of the fabricated device for a broad range of applications such as drug screening and mechanistic studies of BBB disruption.
INTRODUCTION
In recent years, there has been increasing interest in the development of physiologically relevant in vitro models of human tissues and organs for drug development and disease modeling, mainly due to the failure of animal models to recapitulate the physiology of the human body. In fact, 90% of drugs fail in early clinical trials because they elicit toxic responses in humans or they do not demonstrate the same efficacy observed in animal models.1 Different individuals may also exhibit heterogeneous responses to certain therapeutics as a result of genetic variability.1,2 The use of human induced pluripotent stem cells (hiPSCs) to develop in vitro models of human tissues is a promising approach to overcome the aforementioned issues. Harvesting cells from the skin or blood of patients and reprogramming them into pluripotent stem cells, which have a high self-renewal capacity and ability to differentiate into most human cell types, enables the establishment of patient-specific in vitro models of various organs. However, the conventional static, 2D cell cultures lack certain important characteristics of the human body such as biomechanical forces, three-dimensionality, and relevant surface-to-volume ratios.3,4
Utilizing microfluidic platforms for cell culture can address these limitations. Hence, integration of hiPSC-derived cells and microfluidic platforms to create in vitro models of human tissues is a promising alternative to animal models and traditional 2D in vitro models for human disease modeling and drug discovery. The blood-brain barrier (BBB) has been a particularly challenging part of the body to model in vitro since none of the available immortalized human brain-specific microvascular endothelial cell (BMEC) lines or primary BMECs are able to mimic the in vivo barrier properties of the BBB.5,6 hiPSC-derived BMECs, on the other hand, have been shown to better mimic the characteristics of the in vivo BBB, such as possessing high transendothelial electrical resistance (TEER) and expressing all of the crucial tight junction proteins, efflux transporters, and solute carriers that are found in the BMECs lining human brain microvessels.7–10 Further, it is not fully understood whether BBB disruption is the cause or consequence of various neurodegenerative diseases,11 and isogenic hiPSC-based models of the neurovascular unit can be used to address this question, as protocols have been established to differentiate multiple cell types of the neurovascular unit from hiPSCs, including astrocytes, microglia, and, recently, pericytes.12,13 For instance, it is reported that hiPSC-derived astrocytes from patients with Alzheimer's disease14–16 or multiple sclerosis17 exhibit genetic and molecular differences compared to those derived from healthy individuals. Isogenic models that combine multiple patient-derived cells of the neurovascular unit could enable studies to determine whether these changes affect barrier properties of the BBB.
The BBB-on-a-chip platforms that have been developed can be categorized into extracellular matrix (ECM)-based and membrane-based devices.18 In one type of ECM-based device, BMECs are cultured on a hydrogel, with the BMEC monolayer perpendicular to the bottom surface of device. While it is possible to culture the other cell types, such as astrocytes and neurons, in 3D in these types of devices, the orientation of the BMECs makes imaging of the cells and tight junction proteins as well as measuring TEER challenging. In addition, a recent study has shown that the direct seeding of hiPSC-derived BMECs onto hydrogels significantly reduces their barrier tightness.19 In another type of ECM-based device, a cylindrical lumen structure is fabricated in a hydrogel to better mimic the geometry of human vessels. However, since previous studies have reported a lack of response by the BMECs to flow and curvature,20–22 geometry might not be a crucial factor in improving the functionality of these cells. Membrane-based devices, in which the BMECs are cultured on a membrane sandwiched between two layers, provide better control over the surface area of the BMEC monolayer and enable measurement of TEER as well as easy visualization of the BMECs. However, in most membrane-based devices, cells are cultured in 2D on each side of the membrane. Recently, models constructed using self-organized microvessels in ECM have been developed that possess in vivo-like geometry.23,24 Nevertheless, it is challenging to quantify the barrier properties of the BMECs in these devices. While primary and immortalized human cell lines have been used in the majority of BBB-on-a-chip devices,25–32 in recent years, a few studies have reported the use of hiPSC-derived BMECs in fabrication of their devices. However, these systems use either BMECs alone19,33,34 or BMECs cocultured in 2D with primary rat astrocytes35 or primary human astrocytes and pericytes.36,37 Astrocytes are the most abundant cells in the brain38 that are located in close proximity to the brain microvasculature. Covering 99% of the microvasculature surface in the brain by their endfeet,39 astrocytes play a key role in controlling the permeability of the BBB by secreting various factors during BBB homeostasis or upon stimulation in different disease conditions.38 Therefore, incorporation of human astrocytes into BBB-on-a-chip devices is essential to appropriately model regulation of the healthy and diseased BBB. Recently, prerosette neural progenitor cells derived from hiPSCs were used in a device in coculture with BMECs,37 but they were maintained in 2D and contained a mixture of multiple cell types.
Here, we report an isogenic hiPSC-derived BBB-on-a-chip platform consisting of a BMEC monolayer and astrocytes cultured in 3D. Our device design incorporates elements of both membrane-based and ECM-based devices, enabling us to make use of the advantages of both types of devices. Three-dimensional culture of astrocytes has been shown to be more physiologically relevant than the 2D culture, thereby improving functionality of these cells.40,41 The 3D culture of astrocytes has also been shown to differentially modulate BBB integrity compared to the 2D culture of astrocytes.42 Hence, in this study, the astrocytes were cultured in a 3D hydrogel with nutrients to support their culture provided by medium in adjacent channels. BMECs were cultured on a membrane located on top of the hydrogel channel, enabling easy visualization and control over the surface area of the BMEC monolayer. Membranes with 0.4 μm or 8.0 μm pore diameters were used to fabricate the devices. Use of membranes with 8.0 μm pore diameter allows for the study of transmigration of cells into the brain, while membranes with 0.4 μm pore diameters are commonly used to investigate molecule transport across the BBB. We also demonstrated the ability to evaluate the electrical properties of the BMEC monolayer in our model using impedance spectroscopy. Finally, as an example of utilizing this device for disease modeling, the effects of apical delivery of TGF-β1 on BBB integrity and astrocyte activation were shown, as high levels of TGF-β1 in the plasma are implicated in BBB disruption in various diseases, including acute liver failure and melanoma.43–45 This hiPSC-derived model recapitulates key characteristics of the in vivo BBB and thus is suitable for various applications including disease modeling, mechanistic studies of BBB disruption, and therapeutic discovery.
MATERIALS AND METHODS
Microfluidic device design and fabrication
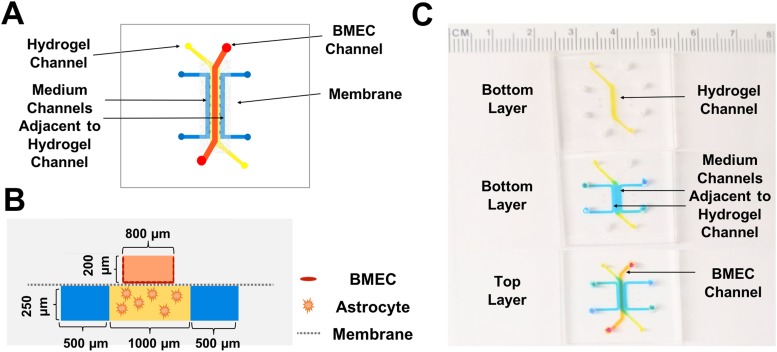
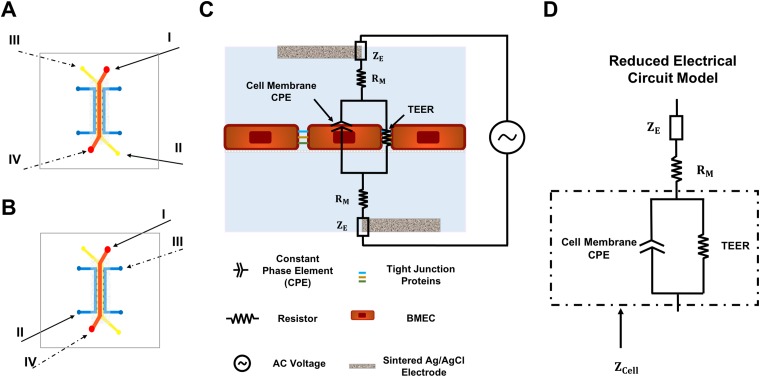
Top and front view schematics of the fabricated BBB-on-a-chip device are shown in Figs. 1(a) and 1(b), respectively. The device used in this study consists of a track-etched 0.4 μm or 8.0 μm polyethylene terephthalate (PET) membrane sandwiched between two polydimethylsiloxane (PDMS) layers. The bottom layer (basolateral or “brain” side) includes three channels: a middle channel composed of a hydrogel and two adjacent channels containing medium that are separated from the hydrogel channel by PDMS posts. The PDMS posts secure the hydrogel in the middle channel while allowing it to maintain contact with the cell culture medium. This prevents dehydration of the hydrogel and provides the astrocytes encapsulated in the hydrogel with the required nutrients. Additionally, electrodes can be inserted into the medium-containing channels for TEER measurements across the BMEC monolayer. The medium in these channels can also be used to measure the levels of cytokines and growth factors secreted by astrocytes upon stimulation. The large size of the astrocytes cultured in the hydrogel channel and the effect of channel height on current distribution in impedance analysis46 were taken into account when designing the channel heights [Fig. 1(b)]. The top layer (apical or “blood” side) has a single rectangular channel in which the differentiated BMECs are cultured on a collagen IV and fibronectin-coated PET membrane. As components of the basement membrane in the brain,47 these proteins enhance the adhesion of BMECs to the membrane by providing an in vivo-like surface for the cells. The length of the straight part of this channel is 7 mm, and the surface area of the membrane in contact with both the basolateral and apical sides of the device is 0.065 cm2. Figure 1(c) contains images of the fabricated device showing the position of each channel.
FIG. 1.
Schematics and images of the fabricated BBB-on-a-chip device. (a) Top view schematic of the device and the positions of the channels. (b) Cross-sectional view schematic of the fabricated device and the channel dimensions. (c) Images of the fabricated device, with different colored dyes used to visualize the various channels. The hydrogel channel is shown in yellow, the BMEC channel in orange, and the medium channels adjacent to the hydrogel channel in blue.
Two different masters for the top and bottom layers of the device were fabricated using SU-8 2100 (MicroChem Corporation) following the manufacturer's procedures to achieve structures with the desired thickness. In order to fabricate the top and bottom layers, Sylgard 184 (Corning, USA) prepolymer and cross-linker were mixed at a 1:10 ratio and subsequently defoamed using a THINKY planetary centrifugal mixer. Next, this mixture was cast on the fabricated masters and degassed for more than 1 h in a desiccator prior to baking in a 75 °C oven for at least 2 h to cross-link the prepolymer. The cross-linked PDMS was peeled off of the masters and cut into rectangular pieces. Holes were punched at the ends of each channel using biopsy punches.
Polyethylene terephthalate (PET) membranes with pore sizes of either 0.4 μm or 8.0 μm were sandwiched between the fabricated PDMS layers using the method developed by Aran et al.48 In brief, membranes were first cut out with a scalpel from BD Falcon Cell Culture Inserts (6-well plate inserts) and then oxygen plasma-treated, followed by wet chemical silanization in the 3-amino-propyltriethoxysilane (APTES) solution. Next, PDMS pieces were plasma oxygen-activated, and the silanized membrane was directly bonded to the top layer, followed by bonding of the top PDMS layer-membrane complex to the bottom PDMS layer. Finally, the assembled devices were attached to No. 1 microscope coverslips by oxygen plasma treatment of both the devices and the coverslips. For devices with hydrogels, the hydrogel channel in the bottom layer was coated with poly-d-lysine in order to enhance the adhesion of the hydrogel to the PDMS posts by establishing stronger physical interactions between the hydrogel and the PDMS surface.
BMEC differentiation and culture
The protocol developed by Hollmann et al.10 was used to differentiate hiPSCs to BMECs. In summary, IMR90-4 hiPSCs were maintained on 6-well plates coated with hESC-qualified Matrigel (Corning) in the TeSR-E8 medium (STEMCELL Technologies) until they reached about 80% confluency. At this point (day-1), cells were detached and singularized using Accutase (STEMCELL Technologies) and seeded at 160 000 cells per well in Matrigel-coated 6-well plates using the TeSR-E8 medium supplemented with 10 μM Y-27632 (STEMCELL Technologies). The next day (day 0), the medium was switched to TeSR-E6 (STEMCELL Technologies), and the medium was changed every day for the next 3 days (days 1–3). On day 4 of differentiation, the medium was switched to human endothelial serum-free medium (Thermo Fisher Scientific) containing 1% bovine platelet poor plasma-derived serum (Fisher Scientific), which is referred to as the EC medium throughout the text, supplemented with 10 μM retinoic acid (MilliporeSigma) and 20 ng/ml human basic fibroblast growth factor (Peprotech). On day 6, cells were subcultured onto 6-well plates that were coated with collagen IV and fibronectin to purify BMECs through selective adhesion. One day after subculture, BMECs were ready to be seeded in the devices.
Astrocyte culture
Differentiated astrocytes were a kind gift from Dr. Ethan S. Lippmann (Vanderbilt University). Astrocytes were also differentiated from IMR90-4 iPSCs using the protocol developed by Hollmann et al.10 This differentiation protocol produces a mixture of astrocytes and glial progenitors. Since the majority of these cells express glial fibrillary acidic protein (GFAP), an astrocyte marker, they are referred to as astrocytes throughout the text. A vial of differentiated astrocytes was thawed and cultured on Matrigel-coated 6-well plates in the TeSR-E6 medium supplemented with recombinant human epidermal growth factor (Peprotech) and recombinant human ciliary neurotrophic factor (Peprotech). The medium was changed every other day until the cells were seeded into the fabricated device.
Seeding cells in the device
Devices with or without hydrogels in the middle bottom channel were used to study the biocompatibility of the chemically modified membrane for culture of BMECs and to confirm the formation of a barrier on the membrane. Devices with hydrogels either contained astrocyte-laden hydrogels (referred to as coculture with astrocytes) or blank hydrogels without cells (referred to as monoculture). Fabricated devices were sterilized by adding 70% ethanol to the channels, followed by aspirating the ethanol and drying in a biosafety cabinet under laminar flow. The membrane in the top channel was coated with collagen IV (0.8 mg/ml, MilliporeSigma) and fibronectin (0.2 mg/ml, MilliporeSigma) for 4 h. For devices with a hydrogel in the bottom channel, the astrocyte-laden or blank hydrogel was then introduced into the hydrogel channel and incubated at 37 °C for 30 min to cross-link the hydrogel. The hydrogel consisted of collagen type I (4 mg/ml, Corning), growth factor reduced Matrigel (2 mg/ml, Corning), and Glycosil hyaluronic acid (2 mg/ml, MilliporeSigma). This optimized hydrogel was previously shown to support the physiologically relevant culture of human astrocytes and minimize their activation in vitro.39 Next, the EC medium was introduced into the two channels adjacent to the hydrogel. For devices without hydrogels, the EC medium was placed in the middle bottom channel.
Purified BMECs were treated with Accutase until about 90% of cells were visually detached from the surface. The detached cells were centrifuged and seeded in the device at a ratio of 1 well of a 6-well plate to 3 microfluidic devices (around 500 000 cells per device) by directly adding 100 μl of cells in the EC medium to the reservoirs in the top channel. These reservoirs consisted of 1000 μl pipette tips inserted into the inlet and outlet of the channel, which facilitated the cell seeding process and allowed for culture of cells under both static and dynamic conditions. Cells were allowed to attach to the coated membrane for at least 1 h, followed by removal of nonadherent cells by introducing the EC medium to the reservoirs to apply flow and collecting the nonadherent cell solution from the outlet reservoir. At this point, the devices could be connected to a syringe pump for dynamic culture or they could remain under static culture. For experiments that involved flow, namely, the addition of TGF-β1 to the apical channel to determine its effect on barrier properties and astrocyte activation, the BMECs were kept in static culture prior to initiating the experiment. BMEC barrier formation was confirmed by TEER measurements under both dynamic and static cultures in the device. Differences were not expected, as it has been previously shown that the hiPSC-derived BMECs do not align and elongate and their barrier properties do not change under dynamic culture.20–22,33,35
Immunostaining and imaging of cells
BMECs were stained with calcein AM (Thermo Fisher Scientific) following the manufacturer's protocol to visualize live cells. BMECs that were cultured in the fabricated devices were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min. Then, the cells were permeabilized with 0.2% Triton X-100 in PBS for an additional 5 min. Next, the cells were blocked in 10% normal goat serum for 1 h before they were incubated with primary antibodies, namely, occludin (1:200, mouse monoclonal OC-3F10, Invitrogen), claudin-5 (1:200, mouse monoclonal 4C3C2, Invitrogen), VE-cadherin (1:50, mouse monoclonal BV9, Santa Cruz), or ZO-1 (1:200, rabbit polyclonal, Invitrogen) at 4 °C overnight. The next day, the cells were washed three times with PBS prior to adding the secondary antibodies. Goat antirabbit Alexa Fluor 594 (1:200, Invitrogen) or goat antimouse Alexa Fluor 488 (1:200, Invitrogen) were incubated for 1 h at room temperature. Finally, the cells were washed three times with PBS before imaging. F-actin staining was conducted using Alexa Fluor 488-conjugated phalloidin (Invitrogen) following the manufacturer's protocol for staining of fixed cells. Astrocytes were either prestained with membrane dyes, Cellbrite Green or Texas Red-conjugated Wheat Germ Agglutinin (WGA), or stained after fixation following the manufacturer's protocol. In order to stain for GFAP, astrocytes were fixed with 4% paraformaldehyde for 15 min and then permeabilized with 0.2% Triton X-100 in PBS for 7 min. The blocking and primary antibody incubation steps were the same as for the BMEC staining. GFAP antibody (1:500, rabbit monoclonal EPR1034Y, Abcam) and goat antirabbit Alexa Fluor 488 (1:500, Abcam) were the primary and secondary antibodies used for astrocyte staining. The cells were incubated with secondary antibodies for 2 h followed by three PBS washes. All steps for astrocytes staining in the device were conducted using the pipette tip reservoirs connected to the medium channels adjacent to the hydrogel channel, while all solutions for BMEC staining were added to the apical channel. Calcein AM and phase contrast imaging in the device and 2D astrocyte imaging were performed using the EVOS FL Auto microscope (Thermo Fisher Scientific). All the other imaging in the device was performed with the Nikon A1Rsi Confocal microscope with SIM super resolution.
Efflux transporter activity assay
Two days after seeding the BMECs in the devices, the medium was removed and the cells were washed once with HBSS (Thermo Fisher Scientific). Next, the devices were treated for 15 min with 100 μM verapamil (Abcam) in HBSS with 25 mM HEPES (Thermo Fisher Scientific). The control devices received HBSS with 25 mM HEPES. Subsequently, devices were incubated with 2 μM of calcein AM for 20 min. Then, calcein AM was removed and the cells were washed with HBSS prior to imaging with the EVOS Fl Auto microscope. Three devices were tested per condition, and three images were taken for each device using the 20× objective. The fluorescence intensity was evaluated by calculating average pixel intensity for each image using ImageJ.
Impedance spectroscopy
Two sintered Ag/AgCl electrode pellets (1 mm diameter, Harvard Apparatus) were used to measure the electrical impedance across the BMEC monolayer. These electrodes were connected to an LCR meter (Keysight Technologies, E4980AL/032), and impedance magnitude and phase angle were recorded in the range of 20 Hz to 100 kHz. In order to account for the electrode impedance, first the electrode impedance across a membrane coated with collagen IV and fibronectin (blank device) was measured by immersing the electrodes in the medium reservoirs. Based on the type of the device (with hydrogel or without hydrogel), the appropriate pair of reservoirs was chosen, with one reservoir connected to the apical channel and the other connected to the basolateral part of the device. For devices with hydrogels, the basolateral reservoir was connected to one of the adjacent medium channels, and for devices without hydrogels, the reservoir was connected to the middle channel, which was filled with the medium. After cell seeding and formation of a confluent monolayer, the same procedure was repeated for devices with BMECs. Based on an electrical circuit model, the electrode impedance was extracted. Next, by knowing the electrode impedance, the values of other elements were determined by fitting the measured impedance magnitude and phase angle to the proposed model using a least-squares optimization method. The detailed procedure can be found in the supplementary material (Supplementary Methods and Fig. S1).
Fluorescein permeability measurement
In order to determine the permeability of fluorescein molecules across the BMEC monolayer, sodium fluorescein powder was dissolved in the EC medium and filter sterilized. A 10 μM fluorescein solution was introduced to the top channel using the medium reservoirs, while the bottom channel was filled with the EC medium without sodium fluorescein. The amount of the medium in the reservoirs was adjusted such that the height of the medium in each reservoir was approximately equal in order to minimize the effect of fluorescein transfer across the channel due to pressure differences. After 1 h, the medium from the bottom channel was removed, and the concentration of sodium fluorescein was determined using a Synergy H1 hybrid multimode plate reader (BioTek). Approximating that the concentration throughout the top channel remains relatively constant, the permeability of the cell monolayer can be found using Eq. (1). In this equation, is the concentration of fluorescein in the basolateral part of the device after 1 h (found from a calibration curve), is the volume of the EC medium in the basolateral channel, is the concentration of fluorescein that was introduced in the apical channel, A is the common surface area between apical and basolateral channel, and t is the incubation time. After determining the permeability of the devices with BMECs and blank devices (without BMECs), the permeability of the BMEC layer can be found using Eq. (2),
| (1) |
| (2) |
TGF-β1 treatment of BMECs
For the devices that were treated with TGF-β1, the EC medium containing 25 ng/ml recombinant human TGF-β1 (Abcam) was added to the apical part of the device under 4 μl/min flow, while the same flow rate of the EC medium was applied to the control devices. The flow was stopped after 6 h, and the reservoirs were connected to the devices for impedance spectroscopy. After impedance spectroscopy, the devices were reconnected to the syringe pump until the next day (32 h after introducing TGF-β1), when the cells were fixed and permeabilized for immunostaining.
RESULTS AND DISCUSSION
Culture of BMECs in the device
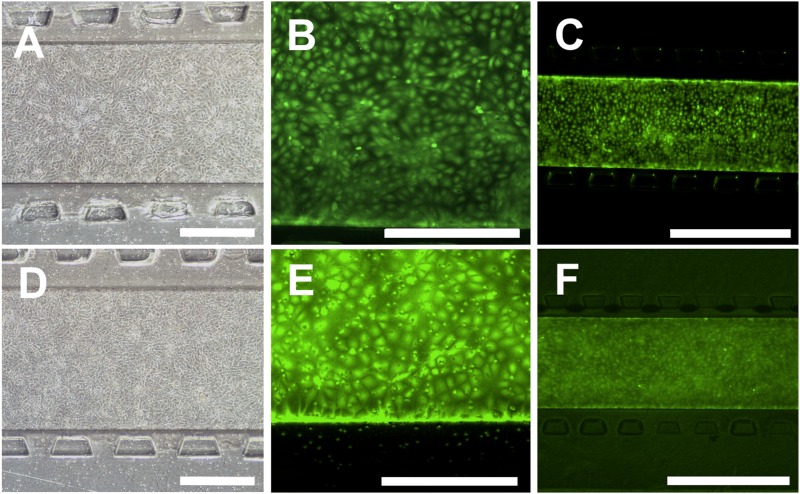
Due to the difference in the surface chemistry of the membrane in the device compared to the standard cell culture PET membrane and the changes in culture conditions in the microfluidic device as compared to the standard 2D systems, the compatibility of the fabricated devices for the culture of BMECs and formation of a confluent BMEC monolayer was assessed. Purified BMECs were attached to the collagen IV- and fibronectin-coated membranes and formed a confluent layer 1 day after seeding [Figs. 2(a) and 2(d)], and the cells remained viable and confluent 2 days postseeding on the 0.4 μm [Figs. 2(b) and 2(c)] and 8.0 μm [Figs. 2(e) and 2(f)] membranes. For the 0.4 μm membrane, monolayer formation was observed in both devices with and without a hydrogel in the bottom channel. However, for devices with the 8.0 μm membrane, having a hydrogel in the bottom channel was necessary to form a confluent layer. In the absence of a hydrogel, the BMECs migrated through the membrane pores and either detached from the membrane or in some locations formed a double BMEC layer. To our knowledge, this is the first report demonstrating culture of hiPSC-derived BMECs on a membrane with 8.0 μm pore size that results in a successful formation of a cellular monolayer with requisite BMEC barrier properties, which is essential for studies concerning transport of cells such as cancer cells and immune cells across the BBB.
FIG. 2.
BMECs cultured in the device remain viable and form a confluent monolayer in devices with a hydrogel. (a)–(c) The phase contrast image (a) and calcein AM staining [(b) and (c)] of BMECs in devices with a 0.4 μm membrane. (d)–(f) The phase contrast image (d) and calcein AM staining [(e) and (f)] of BMECs in devices with an 8.0 μm membrane. All images show full coverage of the membrane with live cells. Phase contrast imaging was performed 1 day after seeding, and live cell staining was performed 2 days after seeding. Scale bars for (a), (b), (d), and (e) indicate 400 μm, and scale bars for (c) and (f) indicate 1000 μm.
Characterization of hiPSC-derived astrocytes and their culture in the device
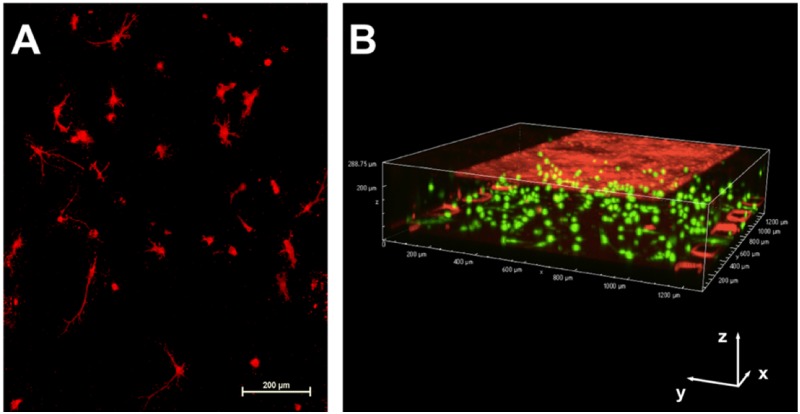
Immunofluorescence analysis of differentiated astrocytes and glial progenitors grown in the 2D culture was performed to evaluate expression of GFAP [supplementary material, Fig. S2(a)]. Quantification of the images by evaluating the ratio of GFAP-expressing cells to total cells showed that 75.8 ± 5.7% of the cells were GFAP-positive, which was expected as the differentiation protocol used to derive these cells produces a mixed population of astrocytes and glial progenitors.49 The astrocytes were cultured in a hydrogel that was introduced to the basolateral channel of the device. Imaging of the astrocyte-laden hydrogel demonstrated that it remained confined to the middle channel in the bottom layer of the device [supplementary material, Figs. S2(b) and S2(c)], which was in contact with the membrane on which the BMECs are seeded. Partial contact between astrocyte endfeet and BMECs was observed in 8.0 μm membrane devices, as shown in the supplementary material, Fig. S2(d). Astrocytes cultured in the device developed expected morphologies for astrocytes cultured in 3D (mostly stellate, but others bipolar or round50,51) and remained alive for at least 3 days in the device [Fig. 3(a)]. A 3D image of the device containing differentiated astrocytes and BMECs is shown in Fig. 3(b), demonstrating the localization of each cell type in the isogenic BBB-on-a-chip device.
FIG. 3.
Visualization of astrocyte morphology and localization in the device. (a) The Z-stack image of the astrocytes cultured in the device and stained with WGA Texas Red-X conjugate demonstrates development of physiological morphology of astrocytes in the hydrogel. Scale bar indicates 200 μm. (b) The 3D image of the device with astrocytes stained with Cellbrite Green (membrane dye) and BMECs stained with Cell Tracker Red. Scale bars in the x, y, and z directions indicate 200 μm.
Efflux transporter activity
Calcein AM diffuses into the cells through the plasma membrane. After being metabolized by esterases inside the cells, it becomes fluorescent and hydrophilic and thus is unable to diffuse out of the cells.52 Calcein AM is a substrate of the efflux pumps multidrug resistance-associated protein 1 (MRP1) and P-glycoprotein 1 (P-gp or MDR1), which can actively pump this molecule out of the cells.53 As a result, calcein AM can be used to study the efflux transporter activity. Verapamil, which inhibits the activity of both MRP1 and P-gp,54 was used to evaluate the efflux transporter activity within the device. The cells that were treated with verapamil showed 53% higher fluorescence intensity, indicating higher calcein AM retention in the cells when the efflux transporter activity was inhibited [supplementary material, Figs. S3(a) and S3(b)]. These results demonstrate that the BMECs retain the functional efflux activity when cultured in the device.
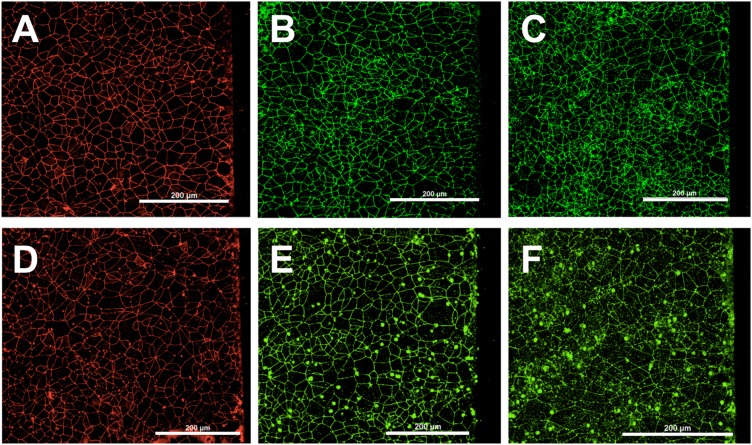
Expression of tight junction proteins
Immunocytochemistry of tight junction proteins is commonly used to visualize and quantify the expression of these molecules as well as to determine their localization.55,56 The horizontal placement of the BMEC layer on the membrane in our device enables us to utilize this approach to visualize the tight junction molecules and to study alterations in their expression or localization. Two days after seeding the device, the confluent BMEC monolayer was fixed and stained for ZO-1, occludin, and claudin-5, the tight junction proteins that are responsible for the low permeability of the BBB.57 Figures 4(a)–4(f) show the requisite expression and continuous junctional localization of these proteins on BMECs throughout the membrane in devices with 0.4 μm and 8.0 μm membranes.
FIG. 4.
BMECs seeded in devices with a hydrogel show proper tight protein expression. Immunostaining of the BMEC layer on a 0.4 μm (a)–(c) or 8 μm (d)–(f) membrane for ZO-1 [(a) and (d)], occludin [(b) and (e)], and claudin-5 [(c) and (f)]. Scale bars indicate 200 μm.
Evaluating permeability of the BMEC monolayer within the device
In order to demonstrate the formation of a tight barrier in the fabricated device seeded with BMECs, we quantified the permeability of sodium fluorescein across the cell layer by introducing a solution of this molecule to the apical channel of the devices without hydrogels and measuring the fluorescence intensity of the solution in the bottom channel. As shown in the supplementary material, Fig. S5(a), the permeability of the cell monolayer remains below 10−6 cm/s for at least 2 days, which is within the same order of magnitude as hiPSC-derived BMECs cultured on Transwells.10 This shows the suitability of the fabricated device for culture of BMECs, and its ability to support the formation of a cellular barrier that mimics the in vivo characteristics of the BBB. The ability to quantify permeability of the BMEC monolayer to different molecules of interest is important for drug delivery applications. It is also possible to determine the permeability of the BMEC monolayer in a device with a hydrogel using confocal laser scanning microscopy if the ECM diffusivity is known, and there is no pressure difference across the BMECs monolayer.58 However, this assay might not be ideal for characterizing barrier properties of the BMEC layer at various time points because it is invasive, time consuming, and prone to error due to possible batch to batch variability in hydrogel properties, limitations of the confocal optical section thickness, and photobleaching of fluorescent molecules.
Impedance spectroscopy to determine TEER and membrane capacitance
Evaluating the electrical resistance across a monolayer of cells is a standard and noninvasive alternative assay to assess the integrity of cellular barriers. Two approaches are commonly used to determine the TEER in microfluidic devices: (1) measuring the difference in impedance “magnitude” at two selected frequencies59 to compare the high- vs low-frequency response or (2) measuring the complex impedance (magnitude and phase) at a constant frequency for a device without cells and devices with cells and calculating the difference due to the addition of cells to the system.30,35,60–64 These approaches, however, can produce inaccurate results because small geometric variabilities between the devices at the microscale inherent to the fabrication process can cause large differences in the measured impedance values. The impedance of different components of the device (e.g., electrodes vs cell membrane) will have different frequency responses, so selecting only one or two frequencies may not provide an accurate measurement of the TEER and cell membrane capacitance. Furthermore, impedance analyzers often exhibit lower accuracy at very low or very high frequencies, so simply selecting the largest possible frequency range will often produce more error in the TEER calculation. An alternative approach is to use impedance spectroscopy, which measures the magnitude and phase of the complex impedance at many frequencies, typically covering several orders of magnitude in the frequency response of the device. Impedance spectroscopy is able to determine the electrical properties of a given device in a more accurate way because it accounts for device variability and the frequency response of each component.
There are several studies that have reported employing impedance spectroscopy to find TEER.65–68 In this study, a 2-probe impedance spectroscopy method was applied using two sintered Ag/AgCl electrodes. Different inlets, one located on the top layer and the other on the bottom layer, were used to insert the electrodes and measure the impedance across the cellular barrier. Two measurements were conducted for each device and based on the type of the device used for the experiment, electrodes were inserted to the appropriate pair of reservoirs connected to the inlets, I and II or III and IV, as shown in Figs. 5(a) and 5(b). Our method offers several advantages over previous studies. First, inserting electrodes into the channel inlets eliminates the need for printing or embedding electrodes into the device, which simplifies the device fabrication process without sacrificing accurate determination of both cell membrane capacitance and TEER. Second, as opposed to previous applications of impedance spectroscopy to these systems65,68 that neglect phase information, both impedance magnitude and phase angle are used to calculate TEER and cell membrane capacitance in this study, which improves the precision of calculated parameters. The phase angle of the complex impedance is related to energy storage capacity, so including the phase angle in this analysis is especially important when calculating cell membrane capacitance.69 Finally, error due to nonuniformity of current distribution in our device is negligible due to the high TEER values of the BMECs incorporated in our system and the relatively large height of the channels, as discussed elsewhere.66
FIG. 5.
Electrode placement for 2-point impedance spectroscopy and the electrical circuit model of the device. (a) and (b) Schematics showing the inlets used for placement of electrodes for devices without a hydrogel (a) and with a hydrogel (b). For the two-probe measurement, measurements are made by inserting the electrodes into reservoirs I and II (solid arrows) or III and IV (dashed arrows). (c) The electrical circuit diagram for the device with BMECs showing the various elements that contribute to the overall impedance. (d) The reduced electrical circuit model that was used for fitting the measured data and finding the electrical properties of the cellular barrier.
The various elements of the device that contribute to the total impedance and the reduced circuit model are shown in Figs. 5(c) and 5(d), respectively. The same pair of electrodes and the same electrolyte (EC medium) were used to record the impedance values for all the devices, so the impedance of the electrodes is theoretically the same for all samples. The cell monolayer behaves as a capacitor due to the very thin plasma membrane of cells composed of an insulating phospholipid bilayer that separates the ions located inside and outside the cells. This can be modeled as a nonideal capacitor, commonly known as a constant phase element (CPE).65,66 The junctional proteins that firmly link the BMECs together act like an electrical resistor that impedes the movement of ions across the BMECs monolayer. This electrical resistor is in parallel with the CPE of the cell membrane, while other electrical components of the device such as medium resistance and electrode impedance are in series with the cell layer components. First, the effect of culturing BMECs in the device on the impedance between the two points, one located in the apical reservoir and the other in the basolateral reservoir, was evaluated by measuring impedance magnitude and phase angle as a function of frequency before and after seeding BMECs in the device. As shown in the supplementary material, Figs. S4(a) and S4(b), the formation of a cellular barrier by BMECs cultured on the membrane causes an increase in the measured impedance magnitude, as well as a negative, frequency-dependent phase angle.
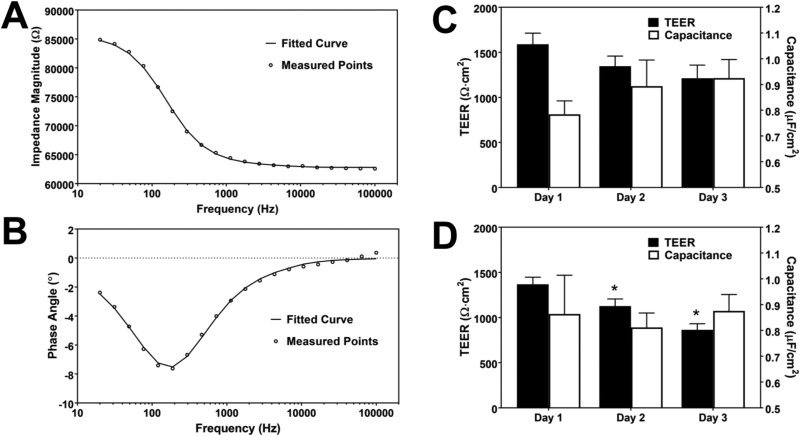
Subsequently, in order to determine the important electrical parameters of the monolayer—TEER and cell membrane capacitance—the data were fit to a circuit model using the least-squares fitting method in MATLAB. The detailed procedure for isolating the electrical components of the cell layer can be found in the supplementary material. In summary, the impedance of the electrodes was first determined in a blank device, which refers to the same type of device without BMECs for each group. Next, using the same electrodes in the same electrolyte, the impedance over frequency for the devices with BMECs was recorded. By subtracting the impedance of the electrodes and fitting the real and imaginary parts of the measured impedance to the equivalent electrical circuit, TEER, membrane capacitance, and medium resistance could be found. An example of the fitted data is demonstrated in Figs. 6(a) and 6(b). The normalized TEER and membrane capacitance for this device are 1401 Ω cm2 and 0.79 μF/cm2, respectively. TEER and capacitance were evaluated for devices with hydrogels [Figs. 6(c) and 6(d)] and without hydrogels [supplementary material, Fig. S5(b)]. The average TEER values on day 1 for BMECs cultured in devices with a hydrogel using the 0.4 μm or 8.0 μm membrane were 1590 ± 214 Ω cm2 and 1369 ± 134 Ω cm2, respectively, showing the formation of a tight barrier as early as 1 day after cell seeding. The TEER value remained in an acceptable range for studying transport of small and large drug molecules70 for at least 3 days, after which the TEER typically drops but remains above 400 Ω cm2 (data not shown). The effect of coculture with astrocytes on the TEER value is shown in the supplementary material, Fig. S6, 1 day after initiating culture of cells in the device. While there is an increase in the average TEER value for devices that are cocultivated with astrocytes, the difference between two groups is not statistically significant. This is in agreement with reported TEER results using the same cell types in a Transwell system for the first 3 days of coculture.10 The calculated normalized cell membrane capacitance in the device [Figs. 6(c) and 6(d)] is within the range of reported cell membrane capacitance values for different cell types (approximately 0.5–2 μF/cm2).71,72 While cell membrane capacitance has not traditionally been quantified in BBB-on-a-chip devices, evaluating capacitance of the cellular layer could potentially be used to study physiological phenomena involving alteration of the cell membrane surface area, such as endocytosis and exocytosis.73,74 Impedance spectroscopy can also be used to distinguish between paracellular and transcellular resistance and the difference between apical and basolateral membrane capacitance upon stimulation by various factors.75,76
FIG. 6.
Impedance spectroscopy results and the calculated electrical properties of the BMEC layer. (a) and (b) Plot of experimentally measured data points and the model (indicated by the fitted curve) based on the fitted values for impedance magnitude (a) and phase angle (b) as a function of frequency for devices with BMECs. (c) and (d) TEER and the normalized membrane capacitance of the BMEC layer in devices with a hydrogel in the basolateral channel with 0.4 μm (c) and 8.0 μm (d) membrane evaluated 1, 2, and 3 days after seeding BMECs in the devices. Each group consists of 3 devices (* indicates P < 0.01 compared to the previous day).
Effect of TGF-β1 on TEER in the BBB model
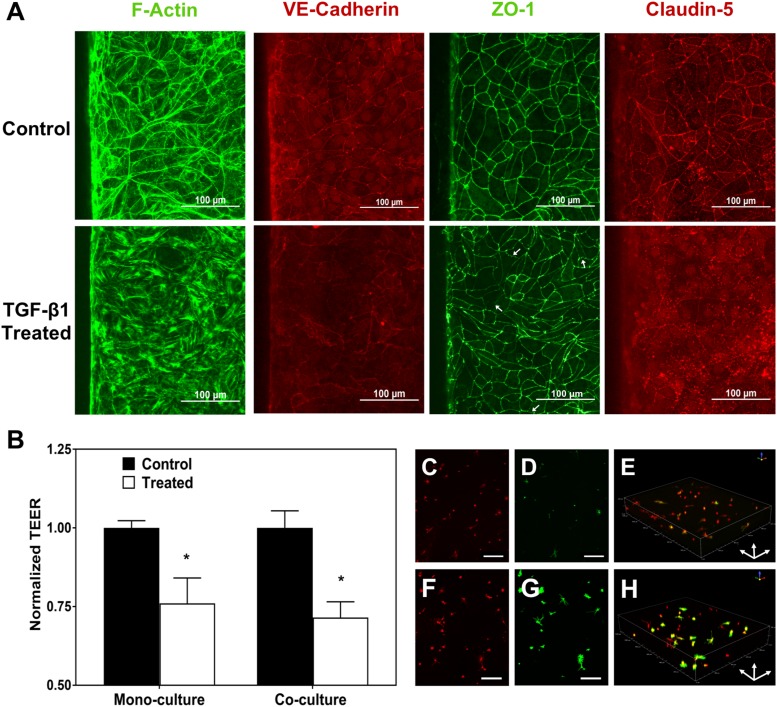
TGF-β1 is known to be present at higher levels in the plasma of patients suffering from acute liver failure as well as melanoma patients. The high level of TGF-β1 in the plasma results in increased permeability of the BBB in the case of liver failure45,77 and is thought to be involved in extravasation of melanoma cells to the brain by causing BMECs to undergo an endothelial to mesenchymal transition.44 In order to recapitulate the effect of TGF-β1 on BBB permeability, 25 ng/ml TGF-β1 was added to the apical part of the device at a flow rate of 4 μl/min, and both TEER and expression of F-actin and junctional proteins were evaluated 6 h after TGF-β1 administration [Figs. 7(a) and 7(b)]. Similar to other reports,78 F-actin staining indicated formation of stress fibers in the treated samples, which commonly results in increase in permeability of endothelial layers79 [Fig. 7(a)]. In addition, staining for VE-cadherin and claudin-5 showed decreased junctional localization of these proteins upon TGF-β1 treatment [Fig. 7(a)]. Finally, even though ZO-1 expression remained junctional, more regions of discontinuity and frayed junctions (shown with white arrows) were evident in the treated samples [Fig. 7(a)]. The addition of TGF-β1 to the apical channel decreased the TEER in devices with BMECs alone as well as in devices containing cocultures of astrocytes and BMECs by 24% and 28%, respectively [Fig. 7(b)], further confirming disruption of the BBB after treatment with TGF-β1.
FIG. 7.
Studying changes in barrier properties and junctional protein expression of BMECs and activation of astrocytes after TGF-β1 treatment. (a) The effect of TGF-β1 on the formation of stress fibers and junctional protein expression. White arrows show frayed junctions in ZO-1 staining. (b) The effect of TGF-β1 on TEER of the BMEC layer for devices with only BMECs (monoculture) and coculture of BMECs and astrocytes (* indicates P < 0.05 compared to control). (c)–(e) Astrocytes cultured in an untreated device fixed and stained with WGA (c) and for GFAP (d). (e) The 3D image of the astrocytes stained for GFAP (green) and cell membrane (WGA; red) in an untreated device. (f)–(h) Astrocytes cultured in devices treated with TGF-β1 in the apical channel fixed and stained with WGA (f) and for GFAP (g). (h) The 3D image of astrocytes stained for GFAP (green) and cell membrane (red) in a treated device showing upregulation of GFAP expression in the astrocytes. Scale bars indicate 200 μm.
Evaluation of astrocyte activation in the basolateral channel
Astrocyte activation or reactive gliosis is observed in various pathologies of the central nervous system80 and is thought to be a promising therapeutic target.40 Astrocyte activation is characterized by hypertrophy of astrocyte processes and upregulation of multiple markers including GFAP, vimentin, and s100β.40,80–84 Staining astrocytes for GFAP expression in order to detect their activation during various brain pathologies is a standard in vivo assay. However, using this assay in conventional in vitro platforms is challenging since astrocytes cultured in 2D are already in a highly reactive state,40,85,86 and thus it is difficult to monitor changes in their activation. Moreover, reactive gliosis is a heterogeneous state and depending on the stimulus causing activation, the response from astrocytes is different.87 Therefore, starting from an already reactive population of astrocytes complicates the analysis. The use of a previously optimized hydrogel for culturing human astrocytes in this study enables evaluation of changes in GFAP expression and astrocyte morphology in order to study astrocyte activation due to different factors, since basal astrocyte reactivity markedly decreases when cultured in 3D in the hydrogel.40
As TGF-β1 is unable to cross an intact BBB,88 we hypothesized that the disruption of the BMEC barrier observed upon TGF-β1 addition to the apical channel of the devices would facilitate the transport of TGF-β1 to the hydrogel channel due to the increase in permeability. In order to test this hypothesis, devices that were treated with 25 ng/ml of TGF-β1 were fixed, and the astrocytes in the basolateral channel were stained for GFAP and labeled with WGA TX-Red®-X conjugate after 32 h of treatment. Using confocal microscopy, an increase in expression of GFAP by astrocytes in the devices that were treated with TGF-β1 was observed, indicating activation of the astrocytes and consistent with previous reports that TGF-β1 induces upregulation of GFAP in astrocytes [Figs. 7(c)–7(h)].40,80,89–92 These results are also in agreement with the mechanism proposed by McMillin et al. in which high levels of TGF-β1 on the blood side of the BBB due to acute liver failure result in the penetration of TGF-β1 and other toxins into the brain through disruption of the BBB integrity.43,45 These results indicate the utility of this model for screening for astrocyte activation, and future studies could utilize activated astrocytes in the model to understand their role in the BBB modulation.
CONCLUSIONS
Incorporation of hiPSC-derived cells in a microfluidic device is a promising approach toward the establishment of tissue-on-chip platforms that can better resolve the current limitations associated with animal studies. Here, we report a BBB-on-a-chip platform that is composed of BMECs that form a highly selective barrier and astrocytes, the most abundant cells in the brain and active players in central nervous system diseases. The BMECs and astrocytes that were utilized in our device were differentiated from the same hiPSC cell line to establish an isogenic in vitro model of the BBB. This is particularly important for personalized medicine applications since it demonstrates the possibility of creating patient-specific BBB-on-a-chip platforms. BMECs can be cultured and form a monolayer barrier on membranes with 8.0 μm pore size, which can be used to study the interaction of immune cells and cancer cells with the BMECs and astrocytes during their adhesion to the BMEC layer and their transmigration into the brain. We also report the ability to measure TEER in the device using impedance spectroscopy via a 2-probe method. Furthermore, capacitance of the cell monolayer, another useful characteristic of the cellular barrier, can be determined using this approach. As an example of modeling a disease condition, we demonstrate that high levels of TGF-β1 in the apical channel of the device decrease the TEER and can activate the astrocytes in the basolateral part of the device, which are seeded in 3D in a hydrogel for physiologically relevant culture. Further molecular assays to study the role of astrocytes in different diseases could be conducted in our fabricated platform. The use of hiPSC-derived cells also enables the study of rare diseases and the examination of potential therapeutic strategies in a dynamic system. Other hiPSC-derived cell types found in the neurovascular unit such as pericytes, neurons, and microglia that play a vital role in certain diseases could also be added to the basolateral part of the device if desired. Overall, we believe this device has the potential to be incorporated in many in vitro studies due to its ability to support various standard in vitro assays such as immunocytochemistry, TEER and permeability measurement, and ELISA assays on the conditioned medium, while the cells are cultured in 3D in the presence of flow.
SUPPLEMENTARY MATERIAL
See the supplementary material for supplementary methods describing calculation of TEER and cell membrane capacitance from impedance spectroscopy and for Figs. S1–S5. Figure S1 summarizes the approach used to find TEER and capacitance. Figure S2 provides images of astrocytes in both 2D and 3D culture in the device and visualization of partial contact between astrocyte endfeet and BMECs. Figure S3 shows the efflux transporter activity of BMECs in the device. Figure S4(a) shows the effect of BMECs on the measured impedance of the device. Figure S5 includes quantification of sodium fluorescein permeability as well as TEER and capacitance of the BMEC monolayer in devices without a hydrogel. Figure S6 demonstrates the effect of coculture with astrocytes on TEER values of the BMEC monolayer in the device.
ACKNOWLEDGMENTS
This work was supported by the University of Minnesota. Portions of this work were conducted in the Minnesota Nano Center, which is supported by the National Science Foundation through the National Nano Coordinated Infrastructure Network (NNCI) under Award No. ECCS-1542202. Confocal microscopy imaging and analysis was performed at the University Imaging Centers, University of Minnesota. Differentiated astrocytes used in this work were a kind gift from Dr. Ethan S. Lippmann (Vanderbilt University).
REFERENCES
- 1.Low L. A. and Tagle D. A., Expert Rev. Precis. Med. Drug Dev. 3, 137 (2018). 10.1080/23808993.2018.1456333 [DOI] [Google Scholar]
- 2.Nelson M. R., Johnson T., Warren L., Hughes A. R., Chissoe S. L., Xu C.-F., and Waterworth D. M., Nat. Rev. Genet. 17, 197 (2016). 10.1038/nrg.2016.12 [DOI] [PubMed] [Google Scholar]
- 3.Liu C., Oikonomopoulos A., Sayed N., and Wu J. C., Development 145, dev156166 (2018). 10.1242/dev.156166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker G. M., Zeringue C., and Beebe D. J., Lab Chip 4, 91 (2004). 10.1039/b311214d [DOI] [PubMed] [Google Scholar]
- 5.Srinivasan B., Kolli A. R., Esch M. B., Abaci H. E., Shuler M. L., and Hickman J. J., J. Lab. Autom. 20, 107 (2015). 10.1177/2211068214561025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helms H. C., Abbott N. J., Burek M., Cecchelli R., Couraud P., Deli M. A., Galla H. J., Romero I. A., Shusta E. V., Stebbins M. J., Vandenhaute E., Weksler B., and Brodin B., J. Cereb. Blood Flow Metab. 36, 862 (2016). 10.1177/0271678X16630991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lippmann E. S., Al-Ahmad A., Azarin S. M., Palecek S. P., and Shusta E. V., Sci. Rep. 4, 4160 (2014). 10.1038/srep04160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lippmann E. S., Azarin S. M., Kay J. E., Nessler R. A., Wilson H. K., Al-Ahmad A., Palecek S. P., and Shusta E. V., Nat. Biotechnol. 30, 783 (2012). 10.1038/nbt.2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian T., Maguire S. E., Canfield S. G., Bao X., Olson W. R., Shusta E. V., and Palecek S. P., Sci. Adv. 3, 48 (2017). 10.1126/sciadv.1701679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollmann E. K., Bailey A. K., Potharazu A. V., Neely M. D., Bowman A. B., and Lippmann E. S., Fluids Barriers CNS 14, 1 (2017). 10.1186/s12987-017-0059-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y., Zhang L., Pu H., Mao L., Hu X., Jiang X., Xu N., Stetler R. A., Zhang F., Liu X., Leak R. K., Keep R. F., Ji X., and Chen J., Nat. Commun. 7, 10523 (2016). 10.1038/ncomms10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamieson J. J., Linville R. M., Ding Y. Y., Gerecht S., and Searson P. C., Fluids Barriers CNS 16, 1 (2019). 10.1186/s12987-019-0136-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stebbins M. J., Gastfriend B. D., Canfield S. G., Lee M. S., Richards D., Faubion M. G., Li W. J., Daneman R., Palecek S. P., and Shusta E. V., Sci. Adv. 5, eaau7375 (2019). 10.1126/sciadv.aau7375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J., Davis M. D., Martens Y. A., Shinohara M., Graff-Radford N. R., Younkin S. G., Wszolek Z. K., Kanekiyo T., and Bu G., Hum. Mol. Genet. 26, 2690 (2017). 10.1093/hmg/ddx155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones V. C., Atkinson-Dell R., Verkhratsky A., and Mohamet L., Cell Death Dis. 8, 1 (2017). 10.1038/cddis.2017.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oksanen M., Petersen A. J., Naumenko N., Puttonen K., Lehtonen Š, Gubert Olivé M., Shakirzyanova A., Leskelä S., Sarajärvi T., Viitanen M., Rinne J. O., Hiltunen M., Haapasalo A., Giniatullin R., Tavi P., Zhang S. C., Kanninen K. M., Hämäläinen R. H., and Koistinaho J., Stem Cell Rep. 9, 1885 (2017). 10.1016/j.stemcr.2017.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perriot S., Mathias A., Perriard G., Canales M., Jonkmans N., Merienne N., Meunier C., El Kassar L., Perrier A. L., Laplaud D. A., Schluep M., Déglon N., and Du Pasquier R., Stem Cell Rep. 11, 1199 (2018). 10.1016/j.stemcr.2018.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogorad M. I., DeStefano J., Karlsson J., Wong A. D., Gerecht S., and Searson P. C., Lab Chip 15, 4242 (2015). 10.1039/C5LC00832H [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katt M. E., Linville R. M., Mayo L. N., Xu Z. S., and Searson P. C., Fluids Barriers CNS 15, 7 (2018). 10.1186/s12987-018-0092-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Destefano J. G., Xu Z. S., Williams A. J., Yimam N., and Searson P. C., Fluids Barriers CNS 14, 1 (2017). 10.1186/s12987-017-0068-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinitz A., DeStefano J., Ye M., Wong A. D., and Searson P. C., Microvasc. Res. 99, 8 (2015). 10.1016/j.mvr.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye M., Sanchez H. M., Hultz M., Yang Z., Bogorad M., Wong A. D., and Searson P. C., Sci. Rep. 4, 4681 (2014). 10.1038/srep04681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campisi M., Shin Y., Osaki T., Hajal C., Chiono V., and Kamm R. D., Biomaterials 180, 117 (2018). 10.1016/j.biomaterials.2018.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phan D. T., Bender R. H. F., Andrejecsk J. W., Sobrino A., Hachey S. J., George S. C., and Hughes C. C., Exp. Biol. Med. 242, 1669 (2017). 10.1177/1535370217694100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adriani G., Ma D., Pavesi A., Kamm R. D., and Goh E. L. K., Lab Chip 17, 448 (2016). 10.1039/C6LC00638H [DOI] [PubMed] [Google Scholar]
- 26.Ugolini G. S., Occhetta P., Saccani A., Re F., Krol S., Rasponi M., and Redaelli A., J. Micromech. Microeng. 28, 044001 (2018). 10.1088/1361-6439/aaa816 [DOI] [Google Scholar]
- 27.Marino A., Tricinci O., Battaglini M., Filippeschi C., Mattoli V., Sinibaldi E., and Ciofani G., Small 14, 1702959 (2018). 10.1002/smll.201702959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griep L. M., Wolbers F., de Wagenaar B., Ter Braak P. M., Weksler B. B., Romero I. A., Couraud P. O., Vermes I., van der Meer A. D., and van den Berg A., Biomed. Microdevices 15, 145 (2013). 10.1007/s10544-012-9699-7 [DOI] [PubMed] [Google Scholar]
- 29.Bang S., Lee S., Ko J., Son K., Tahk D., Ahn J., Im C., and Jeon N. L., Sci. Rep. 7, 1 (2017). 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu H., Li Z., Yu Y., Sizdahkhani S., Ho W. S., and Yin F., Sci. Rep. 6, 1 (2016). 10.1038/s41598-016-0001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herland A., Van Der Meer A. D., Fitzgerald E. A., Park T., and Jelle J., PLoS One 11, e0150360 (2016). 10.1371/journal.pone.0150360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Booth R. and Kim H., Lab Chip 12, 1784 (2012). 10.1039/c2lc40094d [DOI] [PubMed] [Google Scholar]
- 33.Linville R. M., Destefano J. G., Sklar M. B., Xu Z., Farrell A. M., Bogorad M. I., Chu C., Walczak P., Cheng L., Mahairaki V., Whartenby K. A., Calabresi P. A., and Searson P. C., Biomaterials 190–191, 24 (2019). 10.1016/j.biomaterials.2018.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faley S. L., Neal E. H., Wang J. X., Bosworth A. M., Weber C. M., Balotin K. M., Lippmann E. S., and Bellan L. M., Stem Cell Rep. 12, 474 (2019). 10.1016/j.stemcr.2019.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y. I., Abaci H. E., and Shuler M. L., Biotechnol. Bioeng. 114, 184 (2017). 10.1002/bit.26045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park T. E., Mustafaoglu N., Herland A., Hasselkus R., Mannix R., FitzGerald E. A., Prantil-Baun R., Watters A., Henry O., Benz M., Sanchez H., McCrea H. J., Goumnerova L. C., Song H. W., Palecek S. P., Shusta E., and Ingber D. E., Nat. Commun. 10, 1 (2019). 10.1038/s41467-018-07882-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vatine G. D., Barrile R., Workman M. J., Sances S., Barriga B. K., Rahnama M., Barthakur S., Kasendra M., Lucchesi C., Kerns J., Wen N., Spivia W. R., Chen Z., Van Eyk J., and Svendsen C. N., Cell Stem Cell 24, 995 (2019). 10.1016/j.stem.2019.05.011 [DOI] [PubMed] [Google Scholar]
- 38.Cheslow L. and Alvarez J. I., Curr. Opin. Pharmacol. 26, 39 (2016). 10.1016/j.coph.2015.09.010 [DOI] [PubMed] [Google Scholar]
- 39.Mathiisen T. M., Lehre K. P., Danbolt N. C., and Ottersen O. L. E. P., Glia 58, 1094 (2010). 10.1002/glia.20990 [DOI] [PubMed] [Google Scholar]
- 40.East E., Golding J. P., and Phillips J. B., J. Tissue Eng. Regener. Med. 3, 634 (2009). 10.1002/term.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Puschmann T. B., Zanden C., De Pablo Y., Kirchhoff F., Pekna M., Liu J., and Pekny M., Glia 440, 432 (2013). 10.1002/glia.22446 [DOI] [PubMed] [Google Scholar]
- 42.Hawkins B. T., Grego S., and Sellgren K. L., Brain Res. 1608, 167 (2015). 10.1016/j.brainres.2015.02.025 [DOI] [PubMed] [Google Scholar]
- 43.McMillin M., Grant S., Frampton G., Petrescu A. D., Williams E., Jefferson B., Thomas A., Brahmaroutu A., and Demorrow S., J. Neuroinflammation 16, 69 (2019). 10.1186/s12974-019-1455-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krizbai I. A., Gasparics Á, Nagyőszi P., Fazakas C., Molnár J., Wilhelm I., Bencs R., Rosivall L., and Sebe A., PLoS One 10, e0119655 (2015). 10.1371/journal.pone.0119655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mcmillin M. A., Frampton G. A., Seiwell A. P., Patel N. S., Jacobs A. N., and Demorrow S., Lab. Investig. 95, 903 (2015). 10.1038/labinvest.2015.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeste J., Illa X., Gutiérrez C., Solé M., Guimerà A., and Villa R., J. Phys. D Appl. Phys. 49, 375401 (2016). 10.1088/0022-3727/49/37/375401 [DOI] [Google Scholar]
- 47.Wong A. D., Ye M., Levy A. F., Rothstein J. D., Bergles D. E., and Searson P. C., Front. Neuroeng. 6, 7 (2013). 10.3389/fneng.2013.00007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aran K., Sasso L. A., Kamdar N., and Zahn J. D., Lab Chip 10, 548 (2010). 10.1039/b924816a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lippmann E. S., Estevez-Silva M. C., and Ashton R. S., Stem Cells 32, 1032 (2014). 10.1002/stem.1622 [DOI] [PubMed] [Google Scholar]
- 50.Zuidema J. M., Gilbert R. J., and Gottipati M. K., Cells Tissues Organs 205, 372 (2019). 10.1159/000494667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balasubramanian S., Packard J. A., Leach J. B., and Powell E. M., Tissue Eng. A 22, 885 (2016). 10.1089/ten.tea.2016.0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vellonen K. S., Honkakoski P., and Urtti A., Eur. J. Pharm. Sci. 23, 181 (2004). 10.1016/j.ejps.2004.07.006 [DOI] [PubMed] [Google Scholar]
- 53.Essodaïgui M., Broxterman H. J., and Garnier-Suillerot A., Biochemistry 37, 2243 (1998). 10.1021/bi9718043 [DOI] [PubMed] [Google Scholar]
- 54.Goh L. B., Spears K. J., Yao D., Ayrton A., Morgan P., Roland Wolf C., and Friedberg T., Biochem. Pharmacol. 64, 1569 (2002). 10.1016/S0006-2952(02)01355-2 [DOI] [PubMed] [Google Scholar]
- 55.Buckley A. G., Looi K., Iosifidis T., Ling K., Sutanto E. N., Martinovich K. M., Kicic-starcevich E., Garratt L. W., Shaw N. C., Lannigan F. J., Larcombe A. N., Zosky G., Knight D. A., Rigby P. J., Kicic A., and Stick S. M., Biol. Proced. Online 20, 3 (2018). 10.1186/s12575-018-0070-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson L. G., Adv. Drug Deliv. Rev. 57, 111 (2005). 10.1016/j.addr.2004.08.004 [DOI] [PubMed] [Google Scholar]
- 57.Luissint A., Artus C., Glacial F., and Ganeshamoorthy K., Fluids Barriers CNS 9, 23 (2012). 10.1186/2045-8118-9-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kwak B., Ozcelikkale A., Shin C. S., Park K., and Han B., J. Control. Release 194, 157 (2014). 10.1016/j.jconrel.2014.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Partyka P. P., Godsey G. A., Galie J. R., Kosciuk M. C., Acharya N. K., Nagele R. G., and Galie P. A., Biomaterials 115, 30 (2017). 10.1016/j.biomaterials.2016.11.012 [DOI] [PubMed] [Google Scholar]
- 60.Deosarkar S. P., Prabhakarpandian B., Wang B., Sheffield J. B., Krynska B., and Kiani M. F., PLoS One 10, e0142725 (2015). 10.1371/journal.pone.0142725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim H. J., Huh D., Hamilton G., and Ingber D. E., Lab Chip 12, 2165 (2012). 10.1039/c2lc40074j [DOI] [PubMed] [Google Scholar]
- 62.Walter F. R., Valkai S., Kincses A., Petneházi A., Czeller T., Veszelka S., Ormos P., Deli M. A., and Dér A., Sens. Actuators B 222, 1209 (2016). 10.1016/j.snb.2015.07.110 [DOI] [Google Scholar]
- 63.Van Der Helm M. W., Odijk M., Frimat J., Van Der Meer A. D., Eijkel J. C. T., Van Den Berg A., and Segerink L. I., Biosens. Bioelectron. 85, 924 (2016). 10.1016/j.bios.2016.06.014 [DOI] [PubMed] [Google Scholar]
- 64.Ooth R. B. and Im H. K., Annu. Rev. Biomed. Eng. 42, 2379 (2014). 10.1007/s10439-014-1086-5 [DOI] [Google Scholar]
- 65.Henry O. Y. F., Villenave R., Cronce M. J., Leineweber W. D., Benz A., and Ingber D. E., Lab Chip 17, 2264 (2017). 10.1039/C7LC00155J [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeste J., Illa X., Gutiérrez C., Solé M., Guimerà A., and Villa R., J. Phys. D Appl. Phys. 49, 375401 (2016). 10.1088/0022-3727/49/37/375401 [DOI] [Google Scholar]
- 67.Douville N. J., Tung Y., Li R., Wang J. D., El-sayed M. E. H., and Takayama S., Anal. Chem. 82, 2505 (2010). 10.1021/ac9029345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van der Helm M. W., Henry O. Y. F., Bein A., Hamkins-Indik T., Cronce M. J., Leineweber W. D., Odijk M., van der Meer A. D., Segerink L. I., and Ingber D. E., Lab Chip 19, 452 (2019). 10.1039/C8LC00129D [DOI] [PubMed] [Google Scholar]
- 69.Swisher S. L., Lin M. C., Liao A., Leeflang E. J., Khan Y., Pavinatto F. J., Mann K., Naujokas A., Young D., Roy S., Harrison M. R., Arias A. C., Subramanian V., and Maharbiz M. M., Nat. Commun. 6, 6575 (2015). 10.1038/ncomms7575 [DOI] [PubMed] [Google Scholar]
- 70.Mantle J. L., Min L., and Lee K. H., Mol. Pharm. 13, 4191 (2016). 10.1021/acs.molpharmaceut.6b00818 [DOI] [PubMed] [Google Scholar]
- 71.Golowasch J., Thomas G., Taylor A. L., Patel A., Pineda A., Khalil C., Nadim F., Golowasch J., Thomas G., Al T., Patel A., Pineda A., and Khalil C., J. Neurophysiol. 102, 2161 (2009). 10.1152/jn.00160.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Asami K., Takahashit Y., and Takashima S., Biophys. J. 58, 143 (1990). 10.1016/S0006-3495(90)82360-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee R., Kim J., Kim S. Y., Jang S. M., Lee S.-M., Choi I.-H., Park S. W., Shin J.-S., and Yoo K.-H., Lab Chip 12, 2377 (2012). 10.1039/c2lc21236f [DOI] [PubMed] [Google Scholar]
- 74.Rituper B., Guc A., Jorgac J., Flašker A., Kreft M., and Zorec R., Nat. Protoc. 8, 1169 (2013). 10.1038/nprot.2013.069 [DOI] [PubMed] [Google Scholar]
- 75.Krug S. M., Fromm M., and Gu D., Biophys. J. 97, 2202 (2009). 10.1016/j.bpj.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmid T., Bogdan M., and Gu D., PLoS One 8, e62913 (2013). 10.1371/journal.pone.0062913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dhanda S. and Sandhir R., Mol. Neurobiol. 55, 3642 (2018). 10.1007/s12035-017-0521-7 [DOI] [PubMed] [Google Scholar]
- 78.Birukova A. A., Adyshev D., Gorshkov B., Birukov K. G., and Verin A. D., FEBS Lett. 579, 4031 (2005). 10.1016/j.febslet.2005.06.018 [DOI] [PubMed] [Google Scholar]
- 79.Vandenbroucke E., Mehta D., Minshall R., and Malik A. B., Ann. N. Y. Acad. Sci. 1123, 134 (2008). 10.1196/annals.1420.016 [DOI] [PubMed] [Google Scholar]
- 80.Pekny M., Wilhelmsson U., and Pekna M., Neurosci. Lett. 565, 30 (2014). 10.1016/j.neulet.2013.12.071 [DOI] [PubMed] [Google Scholar]
- 81.Pekny M. and Nilsson M., Glia 50, 427 (2005). 10.1002/glia.20207 [DOI] [PubMed] [Google Scholar]
- 82.Hamby M. E. and Sofroniew M. V., Neurotherapeutics 7, 494 (2010). 10.1016/j.nurt.2010.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liddelow S. A. and Barres B. A., Immunity 46, 957 (2017). 10.1016/j.immuni.2017.06.006 [DOI] [PubMed] [Google Scholar]
- 84.Gomes F. C. A., Paulin D., and Moura Neto V., Braz. J. Med. Biol. Res. 32, 619 (1999). 10.1590/S0100-879X1999000500016 [DOI] [PubMed] [Google Scholar]
- 85.Placone A. L., Mcguiggan P. M., Bergles D. E., Guerrero-cazares H., Quinones-Hinojosa A., and Searson P. C., Biomaterials 42, 134 (2015). 10.1016/j.biomaterials.2014.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li S., Uno Y., Rudolph U., Cobb J., Liu J., Anderson T., Levy D., Balu D. T., and Coyle J. T., Biochem. Pharmacol. 151, 245 (2018). 10.1016/j.bcp.2017.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anderson M. A., Ao Y., and Sofroniew M. V., Neurosci. Lett. 565, 23 (2014). 10.1016/j.neulet.2013.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kastin A. J., Akerstrom V., and Pan W., J. Mol. Neurosci. 21, 43 (2003). 10.1385/JMN:21:1:43 [DOI] [PubMed] [Google Scholar]
- 89.Reilly J. F., Maher P. A., and Kumari V. G., Glia 22, 202 (1998). [DOI] [PubMed] [Google Scholar]
- 90.Laping N. J., Morgan T. E., Nichols N. R., Rozovsicy I., Zarow C., Finch C. E., and Angeles L., Neuroscience 58, 563 (1994). 10.1016/0306-4522(94)90081-7 [DOI] [PubMed] [Google Scholar]
- 91.Schachtrup C., Ryu J. K., Helmrick M. J., Vagena E., Galanakis D. K., Degen J. L., Margolis R. U., and Akassoglou K., J. Neurosci. 30, 5843 (2010). 10.1523/JNEUROSCI.0137-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cullen D. K., Simon C. M., and Laplaca M. C., Brain Res. 8, 103 (2007). 10.1016/j.brainres.2007.04.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See the supplementary material for supplementary methods describing calculation of TEER and cell membrane capacitance from impedance spectroscopy and for Figs. S1–S5. Figure S1 summarizes the approach used to find TEER and capacitance. Figure S2 provides images of astrocytes in both 2D and 3D culture in the device and visualization of partial contact between astrocyte endfeet and BMECs. Figure S3 shows the efflux transporter activity of BMECs in the device. Figure S4(a) shows the effect of BMECs on the measured impedance of the device. Figure S5 includes quantification of sodium fluorescein permeability as well as TEER and capacitance of the BMEC monolayer in devices without a hydrogel. Figure S6 demonstrates the effect of coculture with astrocytes on TEER values of the BMEC monolayer in the device.