Abstract
Pausing by RNA polymerase (RNAP) during transcription regulates gene expression in all domains of life. In this review, we recap the history of transcriptional pausing discovery, summarize advances in our understanding of the underlying causes of pausing since then, and describe new insights into the pausing mechanisms and pause modulation by transcription factors gained from structural and biochemical experiments. The accumulated evidence to date suggests that upon encountering a pause signal in the nucleic-acid sequence being transcribed, RNAP rearranges into an elemental, catalytically inactive conformer unable to load NTP substrate. The conformation, and as a consequence lifetime, of an elemental paused RNAP is modulated by backtracking, nascent RNA structure, binding of transcription regulators, or a combination of these mechanisms. We conclude the review by outlining open questions and directions for future research in the field of transcriptional pausing.
Keywords: Elemental pause, RNA hairpin pause, backtrack pause, NusA, NusG, RfaH
I. Introduction
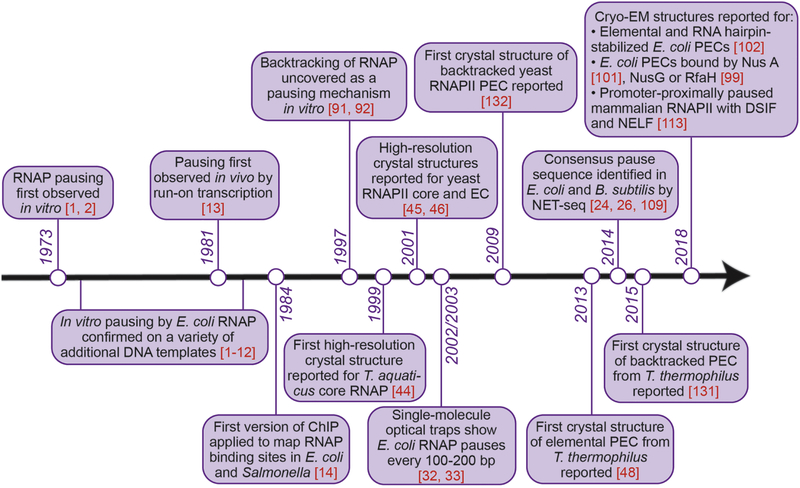
In 1973, Nancy Maizels observed that Escherichia coli RNA polymerase (RNAP) transiently paused at discrete sites during transcription of the lactose operon in vitro. This was indicated by the accumulation and disappearance of RNA transcripts of discrete intermediate lengths over the course of the transcription reaction [1]. That same year, Dahlberg and Blattner reported a similar phenomenon, this time during RNA synthesis on bacteriophage λ DNA [2]. These two seminal studies marked the start of an entire field of research on transcriptional pausing (Figure 1), the breadth of which increased immensely over the past 4.5 decades to encompass not only basic research into the fundamental nature of a paused RNAP, but also its modulation by transcription factors across all domains of life, genome-wide sequence dependence of pausing, and the ever-growing body of important roles transcriptional pausing plays in fine-tuning gene expression.
Figure 1.
Milestones in the field of transcriptional pausing research, with emphasis on pausing by bacterial RNA polymerase.
The initially observed in vitro pausing of bacterial RNAP during transcription was confirmed on a variety of DNA templates in the 1970s and early 1980s, suggesting that pausing is ubiquitous and generating some insight into its underlying causes. In many cases, these pauses took place immediately following synthesis of an RNA hairpin structure: E. coli trp (tryptophan operon) leader and terminator [3–5]; E. coli thr (threonine operon) leader [6]; some sites in the E. coli rrnB (one of the seven ribosomal RNA operons in E. coli) leader [7]; some sites in bacteriophage λ tR1 terminator, located downstream of the structural gene for the lytic repressor cro [8]; and some sites in SV40 virus DNA F1 region [9]. At other sites, however, RNAP paused following synthesis of an RNA apparently lacking a hairpin structure: the initially reported E. coli lacZ [1, 10, 11] and bacteriophage λ 6S RNA [2]; phage T7 early DNA region [12]; other sites in the E. coli rrnB leader [7]; and other sites in the SV40 DNA F1 region [9]. Thus, although not all in vitro pauses observed in early studies correlated with a predicted secondary structure in the nascent RNA, formation of a stable RNA hairpin did appear to signal RNAP to pause.
The phenomenon of transcriptional pausing was first demonstrated in vivo in 1981 by the Chambon laboratory in hen erythrocytes, where a transcription run-on technique (Box 1) revealed accumulation of eukaryotic RNAPII, or Pol II (we will use “RNAPII” throughout this review to avoid confusion with DNA polymerase II) at the 5’-end of the β-globin gene – an indication of RNAPII pausing proximally to the gene promoter [13]. The next milestone in the field arrived with the advent of methods to map RNAP binding to specific DNA locations in vivo (later termed chromatin immunoprecipitation, ChIP; Box 1), first successfully applied by Gilmour and Lis in 1984 to map bacterial RNAP binding sites across several genes in E. coli and Salmonella [14]. Using ChIP in eukaryotes, transcription start site-proximal accumulation of RNAPII was demonstrated on a handful of additional eukaryotic genes in mid-1980s, in Drosophila and mammalian cells [15–17]. Importantly, by complementing ChIP with nuclear run-on experiments, the Lis laboratory showed these accumulated RNAPII molecules to be transcriptionally engaged but paused, as opposed to simply being tightly associated with the promoter [16]. In the mid-2000s, approaches that coupled ChIP with either microarray hybridization (ChIP-chip) or with high-throughput sequencing (ChIP-seq) revealed that RNAP pausing in vivo is not restricted to the promoter regions of specific genes but occurs globally at 5’ and 3’ ends of genes [18–22]. More recently, techniques mapping RNAP position at nucleotide resolution, including native elongating transcript sequencing (NET-seq, Box 1) and precision nuclear run-on and sequencing (PRO-seq, Box 1), established that transcriptional pauses are universal: they take place throughout the gene body across the genomes of different organisms, ranging from bacteria to human cells, and occur on average every 20–100 base pairs (bp) of DNA transcribed [23–26]. Altogether, these studies suggested that pausing is a general feature of transcription and is not limited to specific genes or specific locations within the genes.
Box 1. Methods to map paused RNA polymerase (RNAP) in vivo.
Refer to [58], [59] and [74] for more detailed description of the methods, including limitations of each, and relevant literature resources.
Transcription run-on technique determines in vivo location of an actively transcribing RNAP on a gene from the analysis of nascent RNA product of that gene. In a transcription run-on experiment, cells are lysed and transcription is halted. Transcription by RNAPs that are still bound to DNA and nascent RNA is then briefly re-started in vitro in the presence of radiolabeled or modified NTPs. The amount of labeled RNA produced at a given location of a gene reports on the abundance of RNAP at that location and thus serves as a measure of RNAP occupancy at the genes of interest. Global run-on sequencing (GRO-seq) [75] using brominated NTPs or 4-thioUTP, and its nucleotide-resolution variation, precision nuclear run-on and sequencing (PRO-seq) [76] using chain-terminating biotinylated NTPs, are genome-wide transcription run-on methods, in which labeled RNA is purified, converted into a DNA sequencing library, and sequenced.
Chromatin immunoprecipitation (ChIP) determines in vivo location of RNAP on genes from the analysis of DNA crosslinked to RNAP and immunoprecipitated with an RNAP-specific antibody. In a ChIP experiment, RNAP is reversibly cross-linked to the chromatin DNA, typically with formaldehyde, although in the seminal versions of this approach intact cells were irradiated with UV light to crosslink DNA to the protein irreversibly [14]. Protein-DNA complexes are then fragmented and solubilized, and DNA cross-linked to RNAP is purified by immunoprecipitation, followed by reversal of the crosslinks. The relative amount of DNA captured this way is quantified by microarray hydridization (ChIP-chip), high-throughput sequencing (ChIP-seq), or quantitative PCR (ChIP-qPCR). Versions of ChIP-seq that introduce exonuclease digestion step to eliminate DNA not directly bound to RNAP (ChIP-exo and ChIP-nexus) achieve higher resolution and sensitivity.
Native elongating transcript sequencing (NET-seq) determines both the genomic location and abundance of RNAP from the analysis of 3’-ends of nascent RNA transcripts. Nascent RNA associated with the extremely stable RNAP-RNA-DNA transcription complex is purified by either immunoprecipitation or cell fractionation. The 3’-ends of these purified RNAs are converted into a DNA library, which is sequenced to reveal the identities and abundance of each 3’-end. Mapping the sequencing reads to the genome being analyzed reports DNA strand-specific genomic position and the RNAP active site at single-nucleotide resolution. NET-seq has been given other names (e.g., mNET-seq, NET-prism) when other pull-down targets are used [26, 77, 78], but it would lessen confusion to use NET-seq (without modifiers) for all methods that sequence nascent transcripts 3’-ends from the RNAP active site.
Sequencing- and microarray-based approaches to detect transcriptional pauses in vivo have been complemented by imaging real-time transcription in live cells. In 2007, the Singer laboratory measured mammalian RNAPII transcription kinetics in living cells by using a combination of RNAPII fluorescent fusion and fluorescent phage MS2-labeled mRNA transcripts, generated from an engineered lacO gene array. Here, computational modeling of fluorescence recovery after photobleaching (FRAP) of tagged α-amanitin-resistant RNAPII revealed at least three kinetically different populations of RNAPII, representing enzyme engaged in transcription initiation, elongation and pausing [27]. By implementing a version of this approach a decade later, this time with endogenously expressed mammalian GFP-RNAPII, Steurer et al. assigned the paused kinetic population to the promoter-proximally paused RNAPII, with calculated residence time of only 42 s – in sharp contrast to much longer 20 min RNAPII spends bound to the chromatin [28]. These findings suggested a rapid turnover of RNAPII at promoters due to termination of promoter-proximally paused RNAPII, which could contribute to regulation of gene expression by, for instance, keeping promoters of active genes free of nucleosomes.
As the tools to map and image RNAP pauses in vivo became more and more sophisticated, so did the tools to observe transcriptional pausing in purified in vitro systems – beyond the traditional workhorse, gel electrophoresis. Optical traps, single-molecule Förster resonance energy transfer (smFRET) microscopy, and fluorescence co-localization microscopy approaches allowed detection and monitoring of single molecules of RNAP in action [29–31]. Optical trapping studies in early 2000s, for instance, revealed that individual bacterial RNAP molecules pause every 100–200 bp of transcribed DNA – a number confirmed by in vivo RNAP mapping methods years later – for brief durations of 1–6 s on average at saturating 1 mM NTPs (Figure 1) [32, 33]. Additionally, smFRET studies reported in 2005 and Co-localization Single-Molecule Spectroscopy (CoSMoS) studies reported in 2016 demonstrated at the single-molecule level that σ70, which is historically thought of solely as a transcription initiation protein that aids in promoter recognition, can remain bound to a fraction of elongating RNAPs in vitro well past initiation [34, 35], confirming an observation made in ensemble measurements earlier [36, 37]. This retained σ factor then appears to modulate the behavior of RNAP during productive RNA synthesis, e.g., by inducing pauses at promoter-like sequences and by blocking binding of competing transcription regulators to RNAP [34, 38, 39], although the question of σ retention vs. re-binding in vivo remains open [40]. By monitoring initial transcription, i.e. synthesis of the early short RNAs by RNAP, in real time with smFRET, the Kapanidis lab captured a long pause (~20 s) during the transition from a 6-nucleotide (nt) to a 7-nt RNA transcript at a lac promoter [41]. At around the time of the Kapanidis report, the Weiss group observed the “initiation pause” in their smFRET and single-molecule magnetic tweezers experiments [42]. The initiation pause may serve as a decision point between promoter escape to productive elongation and aborting RNA synthesis, and is largely controlled by a specific region of the bacterial initiation factor σ70 [41, 43]. Here, single-molecule studies revealed features of transcriptional pausing often hidden in ensemble measurements.
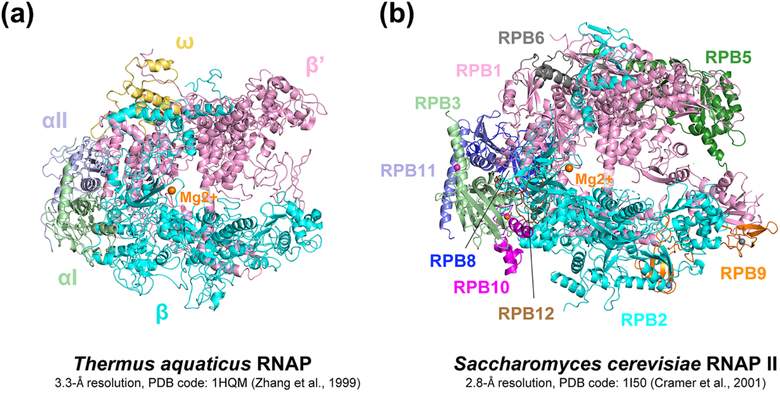
Along with the improvements in genomic analyses and single-molecule assays described above, structural studies of RNAPs both in prokaryotes and eukaryotes has progressed apace since 1999, and the new structural information greatly aided the design and interpretation of the biochemical assays. The first high-resolution crystal structure of a multi-subunit cellular RNAP, of a thermostable RNAP from Thermus aquaticus (T. aquaticus) [44] (Figure 2A) provided a long-awaited structural framework to interpret decades of biochemical and genetic results. A Saccharomyces cerevisiae (S. cerevisiae) RNAPII crystal structure followed in 2001, confirming high structural conservation of RNAP from prokaryotes to eukaryotes [45, 46] (Figure 2B). Many crystal structures containing thermophilic bacterial RNAPs from T. aquaticus or T. thermophilus followed, including structures of an elongation complex (EC) and an elemental paused complex (ePEC) [47, 48]. More than a decade passed until E. coli RNAP structures were determined in 2013 by three different groups independently [49–51]. During this time of great advancement in our structural understanding of the bacterial transcription cycle, X-ray crystallography was the only structural tool available, limiting structural studies to transcription complexes that could be crystallized and sometimes complicating structural analyses due to the confounding effects of crystal packing forces. Fortunately, thanks to the recent advances in cryo-electron microscopy (cryo-EM), structure determination of challenging targets, including RNAPs, at atomic resolution without crystallization became possible [52]. As a result, dozens of structures of transcription complexes without the influence of crystal packing forces have been reported in the last few years and deepened our understanding of transcription, including transcriptional pausing control, which we cover in the Sections II and III.
Figure 2. First high-resolution structures of RNAP.
A. First high resolution structure of a bacterial RNAP, T. aquaticus RNAP core at 3.3 Å resolution (Zhang et al., 1999). B. First high resolution structure of a eukaryotic RNAP, S. cerevisiae RNAP II core at 2.8 Å resolution (Cramer et al., 2001). All subunits are labeled on the figure and catalytic magnesium is drawn as an orange sphere in the center of the structures.
Pausing of RNAP during transcription is broadly involved in regulating gene expression in both prokaryotes and eukaryotes. Generally speaking, transcriptional pauses define windows of time and space for co-transcriptional regulatory events to occur. In some cases, pausing of RNAP at specific positions on the DNA template provides a time window to allow the interaction of the transcribing complex with small molecules, regulatory proteins or RNAs [53–55]. In other cases, pauses make transcribing complexes susceptible to termination, thus prematurely terminating RNA synthesis [56, 57] and decreasing RNA abundance in the cell – unless regulatory proteins stabilize these complexes for continued transcription (“antitermination”). The body of specific examples of the established and emerging roles transcriptional pauses play to regulate gene expression continues to grow. In eukaryotes, promoter-proximally paused RNAPII physically blocks nucleosome re-assembly (“nucleosome occlusion”) to keep promoters open and accessible to activator and transcription factor binding; allows rapid or synchronous gene activation; and couples elongation and co-transcriptional RNA processing (5’-capping, 3’-end processing, splicing), such that nascent RNA is protected from degradation and efficiently matures into a functional mRNA [58, 59]. In bacteria, transcriptional pauses control co-transcriptional folding of nascent RNA into its biologically functional forms, such as catalytic RNAs or alternative structures of riboswitch or attenuator RNAs [60–64]. Paused bacterial RNAP gives the translating ribosome time to catch up and release RNAP from the pause, thereby synchronizing transcription with translation [65, 66]. Transcriptional pauses at the intrinsic (factor-independent) termination sites provide time for a terminator hairpin to form [67]. Pausing also serves as a key first step in Rho-dependent termination of transcription by stalling RNAP at terminators long enough for Rho protein to bind nascent RNA and dissociate it from the transcribing complex [68]. Finally, RNAP pausing precedes excision of misincorporated nucleotides, thus playing an important role in proofreading and maintaining fidelity of transcription [69–71].
While paused RNAP regulates co-transcriptional events like RNA folding and regulator binding, these events and regulators in turn modulate pausing behavior of RNAP [53, 60, 72, 73]. Pausing of RNAP in vivo is fine-tuned by a plethora of cellular regulators, a few of which are discussed in this review. Although the knowledge of intricacies of pausing regulation in vivo might make it an attractive target for applied research, before this complex regulation can be fully exploited, we must first understand the basic molecular mechanisms by which RNAP enters and escapes a transient pause state, alone and in the presence of regulatory proteins. Accordingly, this review focuses on the accumulated biochemical and structural insights into the structural and mechanistic basis of transcription pausing.
II. Mechanisms of pausing by RNAPs
A. Types of transcriptional pauses
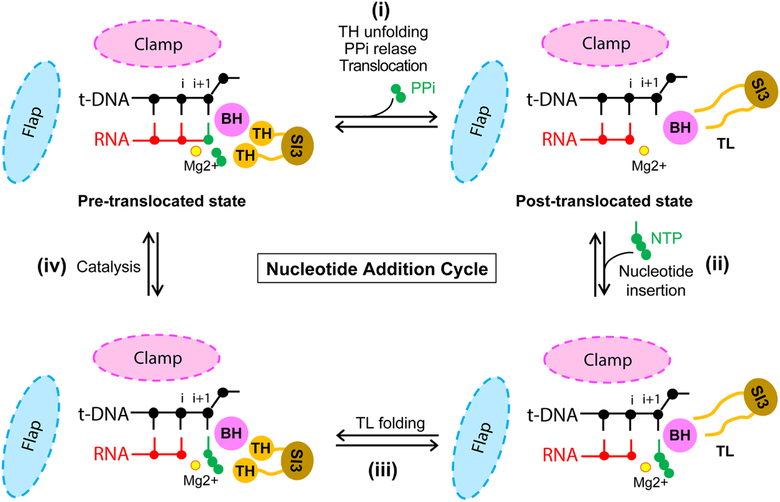
RNA synthesis during productive elongation takes place inside an elongation complex (EC), composed of RNAP and the nucleic acids (DNA and nascent RNA). Transcript extension within the EC occurs one nucleotide at a time through a repeating sequence of steps termed the nucleotide addition cycle (NAC) (Figure 3). The NAC is comprised of (i) nucleic-acid translocation from pre- to post-translocated register, which vacates the binding site for the incoming nucleoside triphosphate (NTP) and aligns the next template DNA base for base-pairing; (ii) NTP binding; (iii) folding of the RNAP active site-proximal trigger loop (TL) motif into the trigger helices (TH); and (iv) formation of the phosphodiester bond [79]. Unfolding of the TH and release of pyrophosphate accompany the translocation step to prepare active site for the next nucleotide addition cycle. RNAPs from many Gram-negative bacterial lineages, including E. coli, contain a large amino-acid sequence insertion (sequence insertion 3, or SI3) in the middle of their TL [80], which has to move during TL-TH transition, thereby affecting RNAP function during elongation, pausing, and proofreading hydrolysis [81–83]. Transcriptional pauses interrupt the NAC at one or more of the above steps in a paused elongation complex (PEC). Pauses characterized to date represent off-pathway events, meaning that they branch off of the main nucleotide addition pathway and kinetically compete with it [12, 84] (Figure 4). A corollary of this definition of pausing is that formation of the offline pause state involves a change in the structure of the EC that disrupts the NAC. Thus, a key feature of an off-pathway pause is that only a fraction of the ECs transcribing through the site enter the pause state, with the rest of the complexes moving past the site without pausing. An on-pathway, so-called pre-translocated pause has also been proposed to result from slow translocation from pre- to post-translocated register following nucleotide addition [85, 86]. The phenomenon resulting from translocation bias toward the pre-translocated state, however, is best defined as a slow nucleotide addition step rather than a “pre-translocated pause” because the EC is not structurally rearranged during this slow step, and the pre- and post-translocated registers likely still equilibrate rapidly relative to the rate of nucleotide addition (see accompanying review of the mechanism of translocation by Belogurov and Artsimovitch). Such slow nucleotide addition steps may promote pausing at a site by allowing time for formation of an off-pathway pause state [87, 88]. Regardless of the detailed mechanism, pauses are caused by sequence-specific interactions of RNAP with DNA and nascent RNA rather than by stochastic fluctuations in the structure of EC independent of nucleic-acid sequence.
Figure 3. Schematic diagram of nucleotide addition cycle.
An active-site view of the nucleotide addition cycle by RNAP. A part of RNA-DNA hybrid in the proximity of the catalytic magnesium is drawn with the neighboring protein components, BH, TL and SI3 connected to the TL. Clamp domain of RNAP forms the main channel that accomodates the RNA-DNA hybrid, and the flap domain is located at the upstream end of RNA-DNA hybrid. The locations of clamp and flap domains are marked by colored ovals and their sizes are not to scale. Each round of nucletide addition occurs in the following steps: (i) nucleic-acid translocation from pre- to post-translocated register, which vacates the binding site for the incoming nucleoside triphosphate (NTP) and aligns the next template DNA base for base-pairing with NTP; (ii) NTP binding; (iii) TL folding into the trigger helices (TH); and (iv) formation of the phosphodiester bond. Unfolding of the TH and release of pyrophosphate accompany the translocation step (i).
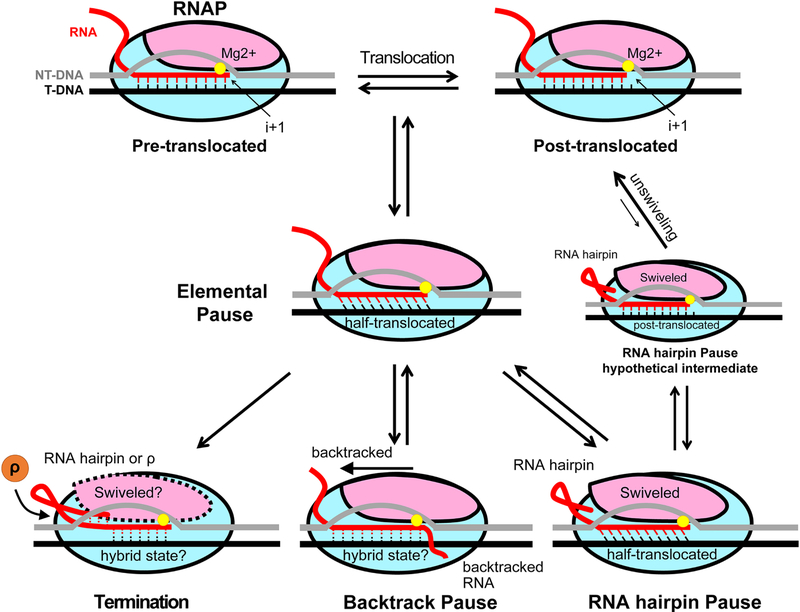
Figure 4. Schematic diagram of transcriptional pauses.
In the on-pathway process of nucleotide addition, RNAP translocates from pre-translocated to post-translocated register to vacate the i+1 site for the next nucleotide to bind as shown in the top part of Figure 3. During this translocation, RNAP can isomerize into an off-pathway conformation that blocks catalysis for a few seconds in response to the nucleic acid sequence interacting with the enzyme. This isomerized state of an elongation complex (EC) is termed the ‘elemental pause’. Elemental paused EC samples multiple RNA-DNA hybrid registers, including the half-translocated register (see the main text). Elemental paused EC can further isomerize into longer-lived paused states, such as backtrack and RNA hairpin-stabilized pauses, or precede transcription termination. In an RNA hairpin-stabilized paused EC, RNA hairpin formation in the RNA exit channel induces conformational change of RNAP (swiveling), while RNA-DNA hybrid remains half-translocated. To escape a hairpin-stabilized pause, the minor population of swiveled RNAP with post-translocated RNA-DNA hybrid may be unswiveled, returning to the on-pathway. In a backtrack paused EC, RNAP moves backward on the nucleic acids, extruding the 3′-end of RNA into the secondary channel. Hybrid conformation here is drawn with dotted lines because both canonical pre-translocated and the half-translocated hybrid conformations have been observed in backtracked ECs. During intrinsic termination, RNA hairpin in a paused EC invades RNA-DNA hybrid, while Rho-dependent termination is likely preceded by an elemental pause. The conformation of RNA-DNA hybrid in the termination process is unknown.
Transcriptional pauses are believed to initiate with an isomerization of the EC into an off-pathway, transient (lifetime of a few seconds), catalytically inactive elemental pause state, producing an elemental paused elongation complex (ePEC) [61, 84, 88] (Figure 4). The ePEC can then further re-arrange into long-lived pause states by backtracking (i.e., a backward movement of RNAP that disengages the 3’ RNA end from the active site and extrudes it into the secondary channel); by the formation of RNA secondary structures, such as hairpins, in the RNA exit channel that modify ePEC conformation [60]; by the action of transcription regulators, or a combination of several of these mechanisms. Backtracking appears to be the most common mechanism of stabilizing the initial elemental pause [89–92] and is favored when the RNA-DNA hybrid is destabilized by a UA-rich RNA-DNA hybrid or by nucleotide misincorporation into the transcript [71, 93]. The two well-characterized pause examples in bacteria are accompanied by backtracking: the ops (operon polarity suppressor) pause, which occurs in the early transcribed region of E. coli operons that encode or affect biosynthesis of extracytoplasmic macromolecules (e.g., hemolysin) [89]; and promoter-proximal pauses caused by σ70 factor failing to disengage from the transcription complex after initiation, thus allowing the σ70-associated ECs pause at –10-like promoter sequences downstream of the promoter [94–98]. Backtracking at the ops assists loading of the transcription factor RfaH onto the EC [53, 99].
Another means of stabilizing the elemental pause is via formation of an RNA hairpin in the RNA exit channel of an ePEC. Formation of an RNA duplex in the RNA exit channel of an ePEC 11–12 nt upstream from a paused RNA 3′ nt prolongs the initial elemental pause ~10-fold [100]. An RNA hairpin-stabilized pause can be prolonged even further, up to 100-fold, by the transcription factor NusA [100, 101], as described in later sections of this review. The spacing between the RNA duplex in the RNA exit channel of RNAP and 3’ end of the transcript determines the interactions of the duplex with the flap domain of RNAP [102, 103]. Because of the specific spacing requirement between the 3’ nt of the elemental paused RNA and the upstream RNA duplex, hairpin-stabilized pauses likely occur infrequently, although genome-wide mapping of such pauses in any organism remains an important experimental challenge due to the complexities of predicting RNA structures. Hairpin-stabilized pauses are prevalent in attenuation control of enterobacterial amino-acid biosynthetic operons (e.g., the his pause in the leader region of histidine biosynthetic operon), where they synchronize transcription of the attenuation control regions with translation of the leader peptide-coding regions. A hairpin-stabilized pause governs folding of a regulatory leader RNA in mgtA operon, which encodes an Mg2+ transporter in enterobacteria, into one of two mutually exclusive conformations, one of which serves as a substrate for Rho-dependent transcription termination at high intracellular Mg2+ [104, 105]. Finally, RNA hairpin formation serves as a precursor to transcription termination, a platform for recruitment of anti-termination proteins (e.g., λN), and plays a role in reiterative transcription, or transcript slippage [60].
B. Sequence specificity of transcriptional pausing
Multiple lines of evidence over the years suggested that RNAP pausing is programmed by specific elements in the nucleic-acid sequence. In vitro work on the his pause in E. coli showed that certain base substitutions in the template DNA increased pausing, whereas others decreased it, and altering the 3’-terminal nucleotide at the pause site changed not only pause strength but also the location of the pause on the transcribed template [106]. Biochemical experiments with E. coli RNAP pausing in the early transcribed region of the bacteriophage T7 D111 deletion variant also pointed to the importance of the nature of the incoming NTP [107]. Single-molecule studies on E. coli RNAP transcribing an rpoB gene demonstrated a statistically significant variation in pausing as a function of template position [33]. A burning question became: is there a common nucleic acid sequence shared between different genes that drives RNAP to pause?
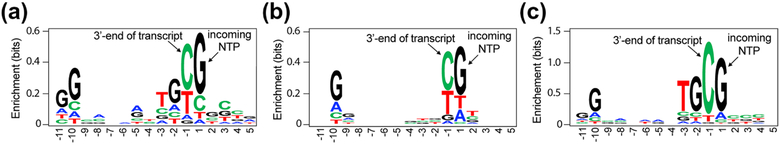
The efforts to identify the consensus sequence that causes transcriptional pausing date almost 4 decades back. In 1981, based on the analyses of several E. coli RNAP pause sites in the early transcribed region of bacteriophage T7 and its D111 deletion mutant mentioned above, Aivazashvili and colleagues suggested that the rate of nucleotide addition was influenced by the identity of the nucleobase at the 3’ end of the RNA transcript, the identity of the next NTP to be incorporated into the paused RNA, and on the nucleic-acid sequence context in the immediate proximity of these two RNA elements [108]. Sequencing of nascent RNAs from bacteria by NET-seq by several labs in 2014 and 2015, followed by sequence alignments of the observed pause sites, finally revealed a 16-nt consensus elemental pause sequence conserved among different bacterial RNAPs [24, 26, 109] (Figure 5). The consensus pause signal is multipartite, as discussed in a later section of this review, where elements that have most pronounced effect on pausing propensity are the G−10 in the nascent RNA of the upstream fork junction and Y−1G+1, where −1 refers to the 3’ end of nascent RNA +1 represents the incoming NTP [24, 84], although it is not known whether the sequence specificity comes from template DNA, non-template DNA, nascent RNA, or a combination of thereof. In addition to these sequence elements independently identified by Larson et al. [24], Vvedenskaya et al. [26] and Imashimizu et al. [109], the consensus determined by Larson et al. also contains a G−11 in the non-template DNA. The difference could potentially stem from the difference in PEC trapping methods between the studies, where relatively slow cooling of cells by Vvedenskaya et al. and Imashimizu et al. (vs. flash-freezing in liquid nitrogen by Larson et al.) may have captured only the strongest contributors to pausing. Imashimizu et al. work further showed that PECs positioned on the consensus nucleic-acid sequence sample multiple translocation registers including post-, pre-translocated and 1-nt backtracked hybrid conformations [109].
Figure 5. Consensus elemental pause sequence.
A. Consensus sequence logo obtained by using NET-seq from B. subtilis and E. coli. Cells were harvested by filtration and flash-frozen in liquid nitrogen before RNA isolation. Native elongating transcripts were harvested by immunoprecipitating Flag-tagged RNAP and the transcripts were convered to DNA for sequencing (Larson, 2014). B. Consensus sequence obtained by NET-seq in E. coli under conditions similar to those in panel A, except the cells were not flash-frozen in liquid nitrogen but instead cooled on dry ice. (Vvedenskaya, 2014) C. Consensus sequence logo obtained by RNET-seq (read-length-specific NET-seq) (Imashimizu, 2015). Contrast to the experiments performed in above panel B and C, only the RNA region buried in RNAP was isolated and analyzed.
In E. coli, the consensus pause sequence accounted not only for the known regulatory pause sites, such as attenuator pauses in thr, leu and his leader regions, but also allowed identification of ~20,000 additional previously undocumented in vivo sites [24]. In the case of expressed genes, these transcriptional pauses were enriched at translation start sites and occurred within the first 100 nt, in line with transcriptional pausing playing an important role in synchronizing transcription and translation in bacteria.
C. The elemental pause
As backtrack- and RNA hairpin-stabilized pauses were studied in more detail, a transient intermediate that connects these off-pathway long-lived pauses to the on-pathway efficient elongation was proposed and its existence supported by single-molecule optical assays and cross-linking experiments [33, 89, 110, 111]. Newman et al. found that 95% of RNAP pausing on the rpoB template exhibited transient pausing a few seconds long while the remaining 5% pausing lasted much longer, in excess of 25 s, probably aided by RNAP backtracking or nascent RNA hairpin formation. The major transient pausing was observed throughout the rpoB DNA template and was not affected by assisting vs. opposing applied force, suggesting that this frequent pausing was rate-limited by neither backtracking nor hyper-translocation [33, 110]. Based on realization that these non-backtracked pauses appeared to also precede the hairpin-stabilized and possibly the backtrack-stabilized paused states, this pause was named ‘elemental’ [61].
Initial structural studies of elemental paused transcription complexes used X-ray crystallography [48]. Three separate crystal structures of Thermus RNAP assembled on a minimal nucleic acid scaffold comprising the E. coli his pause sequence but without the RNA pause hairpin (so an elemental pause scaffold) were determined: one crystal form with T. aquaticus RNAP at 7.8 Å resolution, and two with T. thermophilus RNAP at 4.5 and 3.6 Å resolution. All three structures shared two common features, an open clamp and a kinked bridge helix (BH) that blocked substrate NTP binding. The higher resolution structure (3.6 Å) revealed the kinked BH trapped the template DNA base from entering the active site; thus, the complex was in a state of partial translocation. This active-site conformation resembled that of the active site structure in the α-amanitin-stalled S. cerevisiae RNAPII EC [112]. In addition, the RNA exit channel was widened, suggesting an RNA hairpin could form within the RNA exit channel.
Recently, however, an ePEC structure was determined by single-particle cryo-EM suggesting different features for an elemental pause [102]. The cryo-EM sample was prepared with E. coli RNAP on a full nucleic acid scaffold comprising the E. coli his pause sequence including the RNA pause hairpin (see below). Thanks to the ability to separate distinct conformational states through cryo-EM analysis, a minor population (~12%) of the particles missing the RNA pause hairpin was detected (it is not known if the RNA hairpin was degraded or simply unformed in these particles). RNA hairpin-less ECs in the cryo-EM preparation led to a 5.5-Å electron density map with a 4.2 Å local resolution around the active site and showed the following preliminary features of ePEC. First, the clamp was closed in contrast to the crystal structures. Second, the active site exhibited a half-translocated state in which the RNA was post-translocated while the template DNA was pre-translocated. In this conformation, the NTP substrate-binding site was empty but the template DNA base for the substrate was still base-paired with its non-template DNA partner, thereby inhibiting binding of the incoming NTP, consistent with the result that elemental pause escape was not dependent on the NTP concentration [84]. This half (or partial)-translocated state may be a common feature of paused transcription complexes, as it was also observed in an RNA hairpin-stabilized paused state, backtracked RNAPII and in promoter-proximally paused RNAPII structures [101, 102, 113, 114] as discussed in a later section.
On the basis of the reported consensus sequence (see section IIB above) and structures of the ePEC discussed in this section, Saba et al. recently performed a battery of biochemical assays to probe the molecular mechanism of elemental pause [84]. The authors showed that the elemental pause signal is multipartite, similar to an RNA hairpin-stabilized pause [115]. The elemental pause dwell time was affected by mutations in the upstream fork junction, downstream fork, RNA-DNA hybrid, and downstream DNA duplex, implying that the elemental pause mechanism is complex, requiring orchestration of multiple interactions between RNAP and nucleic acids. In addition, the authors showed that the ePEC samples the 1-nt backtracked state in addition to the pre-translocated [24, 26] and half-translocated [102] states, in agreement with a previous report [109], although backtracking did not limit the rate of pause escape on the consensus pause sequence. Instead, based on the results of the fluorescence-based translocation assays with the fluorescent guanine analog 6-methylisoxanthopterin (6-MI) at either the upstream fork of RNA-DNA hybrid or downstream DNA duplex of the transcription bubble, the authors proposed that progressing past the half-translocated state rate-limits escape from the pause, although the route by which this barrier is overcome remains to be determined. Interestingly, the rate of RNA-DNA hybrid translocation at the upstream fork was similar in non-paused and elemental paused ECs. This study triggers additional questions on the details of the elemental pause mechanism. For example, how the hybrid translocation at the upstream fork is uncoupled with downstream duplex translocation, and what interactions between RNAP and consensus DNA or RNA sequence induce the elemental pause.
In summary, from the biochemical and structural studies of the elemental pause, we can conclude the following.
The elemental pause signal is multipartite: sequences at the upstream fork, hybrid, downstream fork and downstream duplex DNA affect pause duration [24, 26, 84, 110]. Among the elements of the sequence, the upstream-fork G at −10 and the downstream-fork YG at −1 and +1 are most crucial for the pause, although the effect from other nucleic-acid scaffold areas might have averaged out in the analysis of biochemical pause-escape data.
An ePEC samples multiple translocation registers during its lifetime, including the half-translocated, pre-translocated, and 1-nt backtracked states. Post-translocated state cannot exist in an elemental paused state because ePEC does not bind NTP substrate while post-translocated state has an empty active site for the substrate binding. An ePEC ensemble likely consists mostly of pre-translocated and half-translocated populations, whereas a backtracked state exists only transiently, since backtracking did not affect pause life times although it was observed indirectly via GreA/B cleavage [84]. The equilibrium between pre- and half-translocated states might be determined by the DNA sequence.
Complete downstream DNA translocation to form the fully post-translocated state is the rate-limiting step for elemental pause escape. Nevertheless, as the elemental pause is caused by multiple signals spanning the upstream fork to the downstream DNA duplex, a more careful explanation of how multiple signals orchestrate a block to downstream DNA translocation is needed [84]. Determination of higher resolution ePEC structures by cryo-EM, avoiding crystal packing issues, will deepen our understanding of the molecular mechanism of elemental pausing in transcription.
D. RNA hairpin-stabilized pausing
Since the discovery of RNA hairpin-stabilized pausing, studies have been directed towards uncovering the mechanistic basis for exactly how an RNA hairpin prolongs the lifetime of a transcriptional pause. In an early hypothesis, the RNAP was viewed as a rigid body that could not accommodate an RNA hairpin within the RNA exit channel [116]. Consequently, the folding of the RNA hairpin might ‘pull’ the RNA transcript out of the RNA exit channel in the upstream direction, thus altering the nucleic-acid structure in the active site and inhibiting addition of the next nucleotide. This model was refuted by the finding that a 1-nt insertion between the RNA hairpin and RNA-DNA hybrid increased pause duration, instead of releasing the tension of RNA pulling and shortening the pause as predicted by the model [72].
Another general hypothesis for an RNA hairpin-stabilized pausing mechanism is an allosteric model in which global conformational changes in RNAP induced by RNA hairpin formation in the RNA exit channel modulate catalysis by altering the active site architecture allosterically. Note that the RNA exit channel where the RNA hairpin forms is more than 50 Å away from the RNAP active site Mg2+. To probe the active site architecture at an RNA hairpin-stabilized pause, Toulokhonov et al. performed cross-linking experiments between RNAP and RNA, where photosensitive cross-linkable nucleotide analogs were placed at −11 position of RNA and the 3’-end of RNA. These cross-linking experiments revealed that RNA duplex formation in the RNA exit channel did not change the cross-linking pattern at −11 position of RNA at the entrance of the exit channel but altered the crosslinking ratio between β and β’ subunits at the 3’-end of RNA near the active site, supporting the allosteric model [72]. A nucleotide cross-linkable analog introduced into the loop region of RNA hairpin cross-linked to β flap-tip helix region of RNAP indicating the interaction between RNA hairpin and β flap-tip [103]. Interestingly, an RNAP mutant lacking the entire flap tip or flap-tip-helix abolished or decreased the RNA hairpin-stabilized pausing on the his pause sequence without altering RNA hairpin formation or RNA-DNA hybrid register [103]. Furthermore, the pause suppressing RNAP mutants, β’ F773V and β T563I, where mutations are located adjacent to the active site, exhibited slower RNA duplex formation in the exit channel, supporting the idea that the exit channel and active site are energetically linked [100].
In accordance with the crucial role of the TL in the NAC, TL dynamics has been shown to affect pausing as well [81, 117–121]. Deletion of the TL reduced the fold-change in catalysis rates both in the paused and non-paused states, but diminished the effect of hairpin formation on pause duration [120]. A cysteine-pair reporter assay revealed that in the RNA hairpin paused RNAP TL movement is restricted. Thus, even if a hairpin-stabilized PEC samples the post-translocated register, which can bind NTP, the TL probably does not fold properly in response to NTP binding in the active site [119]. Complete deletion or small deletions in SI3 abrogate the RNA pause hairpin effect without affecting the elemental pause [81, 83, 122].
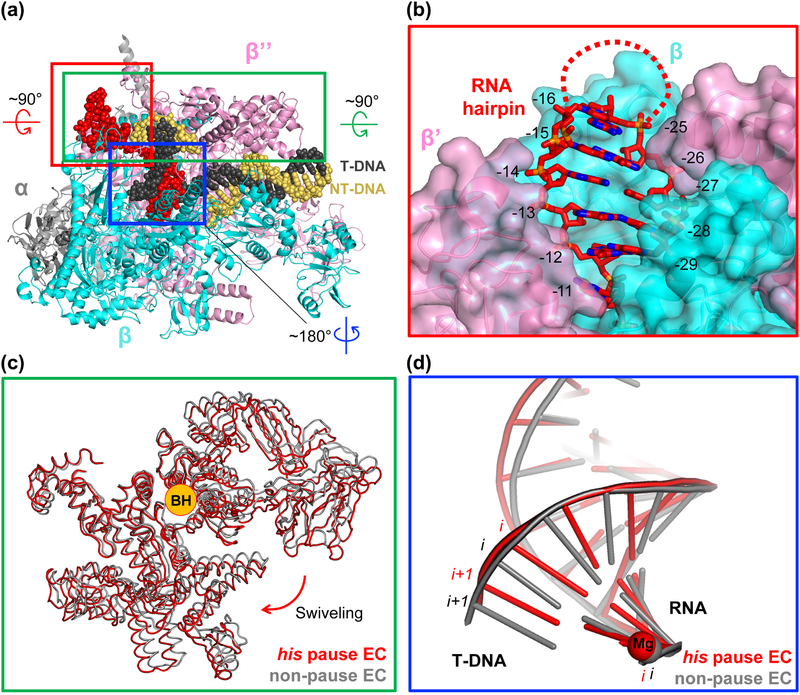
Aided by the recent technical advances in cryo-EM, structures of his pause elongation complexes containing his RNA pause hairpin (hisPEC) with and without NusA were independently determined by two groups [101, 102]. The most significant structural features of the hisPEC were consistent in both structures regardless of the presence of NusA. The interesting features observed in these structures are as follows (Figure 6).
Figure 6. Structural features of an RNA hairpin-stabilized pause elongation complex (hisPEC).
A. The overall structure of E. coli hisPEC. Specific areas are boxed, rotated, and zoomed for detailed description. B. Red boxed region from A rotated by 90° and magnified to show RNA exit channel with his pause RNA hairpin stem duplex in it. RNA hairpin stem duplex formed fully and resided in the RNA exit channel while the loop region of the hairpin was disordered (marked by a dotted line). The opening of the channel widened compared to that of non-paused EC. The residues in RNA are numbered by their positions relative to the active site, where the RNA 3’-end nucleotide is at −1 position. The −11 RNA base connects RNA-DNA hybrid and RNA hairpin. C. Green boxed region from panel A rotated by 90° and zoomed in. Comparison of Cα traces of swivel modules from hisPEC and non-pause EC (PDB code: 6ALF). Compared to the swivel module of a non-pause EC, swivel module in hisPEC rotated about 3° around the axis overlapping BH. D. Blue boxed region from panel A rotated by 180° and zoomed in. Only nucleic acid structures were drawn for clarity. Non-pause EC exhibits a post-translocated hybrid while hisPEC has a half-translocated hybrid, harboring DNA base in i+1 site base-paired with RNA base in i site.
The RNA hairpin stem resided within the RNA exit channel while the loop is exposed to the solvent and not resolved in the cryo-EM structure (Figure 6A, B). The inner wall of the RNA exit channel is lined with positively charged amino acid residues, presumably assisting formation of an RNA hairpin within the channel.
Contrary to the expectation from crystallographic work, the RNAP clamp was closed, but not in the position seen in a non-pause EC. Instead, a set of RNAP domains including the clamp, jaw, shelf, and SI3 domains (mostly in β’ subunit) was rotated (‘swiveled’) by about 3° around an axis almost overlapping the BH (Figure 6A, C). Swiveling explains the results of the cysteine-pair reporter assay with hisPEC mentioned above: the swiveled clamp also favors the disulfide bond formation designed to promote open clamp conformation in the assay. The swiveling movement inhibits nucleotide addition because the swiveled SI3 would clash with the β lobe upon TL folding, which is a crucial step for catalysis, explaining the pause-reducing effect from SI3 deletion [81, 83, 122].
The RNA-DNA hybrid was observed in a half-translocated state, as described for the ePEC in section IIC (Figure 6D). This is consistent with the idea that the RNA hairpin pause derives from the ePEC.
Based on the accumulated evidence so far, we envision the following model for the formation of an RNA hairpin-stabilized pause. First, RNAP pauses at a pause site via the elemental pause mechanism, providing a time window for RNA hairpin formation in the RNA exit channel. At this stage, the RNA-DNA hybrid assumes a half-translocated conformation, at least in a subpopulation of the complexes. Second, RNA hairpin formation allosterically stabilizes the swiveled conformer, thereby relocating SI3. This relocation inhibits proper folding of the TL for the NAC and blocks nucleotide addition for minutes. Unfortunately, the TL was disordered in both cryo-EM structures of hairpin-stabilized PECs and flap-tip helix was invisible in hisPEC without NusA, complicating understanding of the roles of TL and flap-tip in RNA hairpin-stabilized PEC. This may suggest that TL, flap-tip, or both stabilize RNA-hairpin pause in a way that cryo-EM cannot reveal due to the dynamic conformational change, or play roles during the initial stages of entry into an RNA hairpin-stabilized pause rather than stabilizing the final paused state. Escape from a hairpin-stabilized pause requires both the reversal of swiveling and a shift of RNA-DNA hybrid to a post-translocated state. It is possible that a minor population, unresolved by single-particle cryo-EM, of the swiveled hairpin-stabilized PECs occupies the post-translocated register, which is capable of binding NTP but not catalysis due to inhibited TL folding (Box 2).
Box 2. The motions of biomolecular complexes differ from those of macroscopic machines.
It is important to remember that structural states (i.e., conformations) of macromolecules are in constant thermal motion such that all available conformations are stochastically sampled with varying probabilities. For example, ECs and PECs sample different states, with the relative stabilities of the array of states determining the longevity of events such as pausing. In a paused complex, a non-paused state may be sampled transiently but not readily lead to pause escape because, for example, NTP substrate fails to bind while the state exists. It is tempting to describe functional states of biomolecular machines, like RNAP, in terms reminiscent of discrete, long-lived states of a macroscopic machine that cycle in precisely timed intervals, but the consequences of thermal activity create a much different reality at the molecular scale. Side chains and domains of RNAP are in constant movement relative to one another driven by thermal energy (e.g., see simulations in [135]). As a consequence, ECs fluctuate to transiently attain PEC-like conformations (and vice versa) even though the alternative state may be occupied for only a small fraction of the time. Structures of ECs or PECs, whether crystal structures or cryo-electron microscopy structures, should not be interpreted as snapshots of a rigid machine, but as the most probable representations of a dynamic distribution of structures.
E. Parallels between bacterial RNAP and eukaryotic RNAPII pausing
Transcriptional pausing in eukaryotes plays critical roles in maintenance of nucleosome-free regions at promoters, activation of gene expression, RNA splicing, polyadenylation, and cellular differentiation and development [58, 59, 123, 124]. In particular, promoter-proximal pausing by eukaryotic RNAPII is a rate-determining step for transcription elongation and serves as a checkpoint for transcript and RNAPII modification [74]. Promoter-proximal pausing has not been observed in yeast but is widespread in other eukaryotes, such as Drosophila and humans [19, 125–128]. The recently reported cryo-EM structure of a promoter-proximally paused Sus scrofa (pig) RNAPII contains Homo sapiens DSIF (DRB sensitivity-inducing factor) and NELF (negative elongation factor) [113]. DSIF is a heterodimer of Spt4 and Spt5. Spt5 is an ortholog of bacterial transcription elongation factor NusG, and the NusG family of regulators is the only transcription factor family conserved in all domains of life [129]. NELF contains four subunits, NELF-A, -B, -C/D, and -E. In the structure, NELF binds to the RNAPII funnel region, which is equivalent to the secondary channel in prokaryotic RNAP, bridges the core and shelf modules, and contacts the TL. Thus, NELF restrains mobility of part of RNAPII required for pause release. In addition, NELF prevents binding of the anti-pausing transcription elongation factor IIS (TFIIS) by occupying its binding site [130]. Interestingly, the RNA-DNA hybrid in the promoter-proximal paused RNAPII structure is half-translocated, superimposable with the elemental and RNA hairpin-stabilized PECs of E. coli RNAP near the active site, suggesting that the half-translocated conformation of the hybrid might be a universal mechanism for interrupting RNAP catalysis. In addition, Spt5 binds to the same site of RNAPII as NusG binds on E. coli RNAP [99].
A half-translocated RNA-DNA hybrid has also been observed in a structure of paused S. cerevisiae RNAPII EC stabilized by backtracking. Structures of backtracked PECs from multiple species and with various length of backtracked RNA have been reported [114, 131, 132], but only the S. cerevisiae RNAPII complex with a 9-mer backtracked RNA exhibited the half-translocated RNA-DNA hybrid, also called a “tilted” hybrid [114]. Why only this particular backtracked complex has a tilted hybrid is unclear, but neither the length of RNA nor the species origin of RNAP would explain this observation since other backtracked complexes that either contain RNAP from the same species or have backtracked RNAs of various lengths did not exhibit the half-translocated hybrid [132]. The nucleic-acid sequence used to reconstitute backtracked RNAP complexes, the way of assembling the backtracked complexes or the different crystal packing forces experiences by each complex might have influenced the observed hybrid conformation.
A half-translocated RNA-DNA hybrid was also observed in the structures of initial transcribing complex (ITC) of S. cerevisiae RNAPII [133, 134], where the initial RNA-DNA hybrids shorter than 8-bp presented as half-translocated. Interestingly, addition of NTP substrate to a half-translocated ITC changed its register to post-translocated, with NTP bound in the i+1 site, suggesting that the half-translocated hybrid might compensate for instability of a short hybrid to help transcription with short RNA. This might indicate that the half-translocated state is not only a catalytically inactive paused intermediate in elongation but also an initiation intermediate that tolerates short RNA-DNA hybrids.
The structures described above demonstrate that RNAPII exhibits the half-translocated hybrid, just as prokaryotic RNAPs, and all observed RNAPII half-translocated hybrids are either in a paused state or in an initially transcribing state, which are not on-line elongating states. Combined with the prokaryotic RNAP structures, this finding suggests that the half-translocated, or tilted, hybrid structure could be a universal conformation accompanying the paused state.
F. Structural features of backtrack pauses in prokaryotes and eukaryotes
The first backtracked RNAP complex structure was solved with S. cerevisiae RNAPII in 2009 by X-ray crystallography [132]. In this study, multiple backtracked complex structures were determined by using RNA transcripts containing mismatched nucleotides at the RNA 3’-end or by using a template DNA strand bearing DNA damage downstream of the 3’-end of the RNA. The overall crystal structures of the RNAPII backtracked complexes were similar regardless of the cause of backtracking or backtracked RNA lengths. In the structures, the first backtracked nucleotide (3’ to the i+1 site) was bound in a pocket created by the BH, TL and other RNAPII residues, termed the “P” site based on its potential role in proofreading. The rest of the backtracked RNA appeared highly mobile and had no clear electron density in the secondary channel.
Another S. cerevisiae RNAPII backtracked complex structure containing a 15-mer polyC RNA revealed better electron density for the backtracked RNA, showing that it makes interactions with the conserved residues lining the inside of the secondary channel and suggesting that backtracked RNA might have a preferred conformation [114]. A TFIIS-bound backtracked complex structure showed that TFIIS rearranges the backtracked RNA location within the secondary channel to perform RNA cleavage, explaining why TFIIS-catalyzed cleavage of long backtracked RNAs is slower than that of short backtracked RNAs. As commented in the previous section, the RNA-DNA hybrid in the backtracked RNAPII was tilted about ~25° similarly to the half-translocated state observed in E. coli hisPEC, although this altered hybrid conformation was not observed in the previously reported S. cerevisiae RNAPII backtracked complex [132].
Backtracked complex structures were also determined with bacterial RNAPs. Sekine et al. solved crystal structures of T. thermus RNAP backtracked complex with and without GreA/Gfh1 chimeric protein [131]. Consistent with the earlier reported backtracked RNAPII structures [114, 132], first backtracked nucleotide was located in the P site and the clamp was closed in T. thermus RNAP backtracked complex. GreA/Gfh1 chimeric protein binding to backtracked T. thermus RNAP induced ‘ratcheting’ opening the clamp and rotating the shelf module whereas TFIIS binding to S. cerevisiae RNAPII did not change the clamp position.
In summary, the crystal structures of backtracked ECs from eukaryotes and prokaryotes showed few conformational changes in RNAP compared to non-paused EC whereas the hybrid conformation varied in different complexes. The first backtracked RNA base was located in the ‘P site’ at the end of the secondary channel and the remainder of the backtracked RNA was extruded into the secondary channel. To avoid the possibility of having crystal-packing biased structures, obtaining cryo-EM structures of backtracked ECs is necessary.
III. Regulation of RNAP pausing by transcription factors
To tune gene expression, every step of the transcription cycle is modulated by a multiplicity of transcription factors, and the regulation of transcriptional pausing during elongation is no exception. In this section, we describe bacterial transcription factors known to enhance or attenuate RNAP pausing and summarize what is known about them mechanistically.
A. Stabilization of RNA hairpin-dependent pausing by NusA
NusA (N utilization substance protein A) is a multi-functional transcription elongation factor that is universally conserved among eubacteria, and plays diverse roles in a context-dependent manner [136]. In this section, our discussion will be limited to E. coli NusA because the function of NusA in other species is not yet known clearly. NusA was first discovered (and named, N-utilizing substance) as a factor required for λ phage protein N-mediated antitermination, along with other factors such as NusG, B, and E [137]. NusA was also shown to be necessary for the antitermination of ribosomal RNA synthesis [138] and for the efficient expression of endogenous genes like β-galactosidase [139]. In contrast to its antitermination activity in the context of λN antitermination complex, NusA also enhances intrinsic termination [140] and extends the lifetime of pauses at sites where pausing is prolonged by nascent RNA structures [89, 141]. In addition, NusA facilitates RNA folding, transcription-translation coupling, and DNA repair, probably partially by modulating transcriptional pausing [63, 142, 143]. Out of the many roles of NusA, here we will focus on its RNA hairpin-stabilized pause enhancing activity [89].
Suitable to its multiple functionalities, NusA contains multiple domains: NTD (N-terminal domain), S1, KH1 (K-homology), KH2, AR1 (acidic-rich), and AR2 [144, 145]. NusA-NTD is necessary and sufficient to enhance pausing of RNAP [146]. S1, KH1 and KH2 bind RNA [147, 148] and the two AR domains modulate binding affinity of NusA to RNAP [146]. In particular, AR2 auto-inhibits NusA by blocking RNA binding to it in the absence of RNAP, and this auto-inhibition is relieved in the presence of RNAP by AR2 binding to the C-terminal domain of the RNAP α subunit [149]. The contributions of NusA to stimulation of intrinsic termination are partially from its pause-enhancing activity, but not exclusively dependent on it [146].
Recently, two independent structures of NusA-bound E. coli RNAP elongation complexes – the hisPEC and the λN anti-termination complex – were reported in contrasting biological contexts [101, 150].
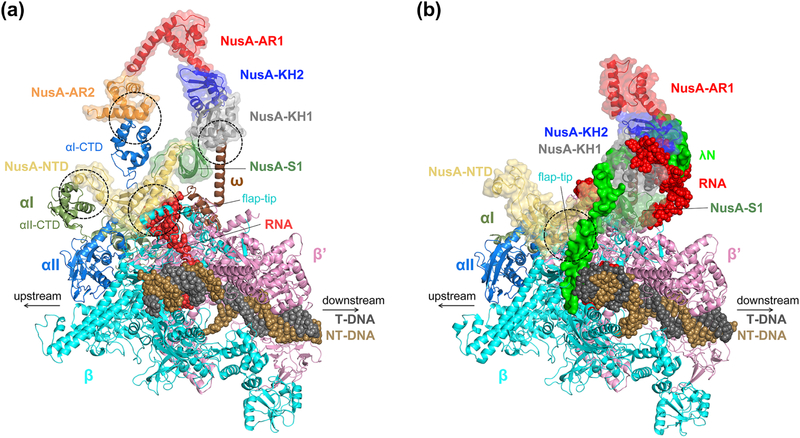
The 3.6 Å cryo-EM structure of a NusA-bound hisPEC structure revealed four main interactions of NusA to the TEC to stabilize the RNA hairpin pause (Figure 7A). First, NusA-NTD was bound to the FTH of RNAP, as predicted from previous biochemical and NMR studies [72, 151, 152]. The observation that deletion of the FTH abolished the pause-enhancing effect of the NusA-NTD also emphasizes the role of this interaction in hairpin pause stabilization [72, 146]. Second, NusA-AR2 was bound to the α1-CTD of RNAP. This interaction relieves the auto-inhibition of NusA, as proposed from gel-shift assays and NMR study [146, 153]. Third, NusA-NTD was bound to the α2-CTD. Deletion of the α-CTDs eliminated the pause-enhancing activity of the NusA-NTD, implying that the interaction between NusA-NTD and α2-CTD is required for this activity. This interaction likely increases the binding affinity of NusA-NTD to RNAP rather than playing a functional role since the α2-CTD is highly mobile without NusA and does not have a defined interaction with other RNAP subunits [101]. Fourth, the NusA-KH domains were bound to the RNAP ω subunit. A KH deletion did not abolish pause-enhancing activity of NusA, but higher concentrations of NusA were required for maximum activity, indicating that the KH domain contributes to NusA affinity for the EC [146]. The interaction between the KH domains and the RNAP ω subunit explains the positive effect of the KH domains on the binding affinity of NusA to EC.
Figure 7. Comparison of NusA-bound hisPEC and λN anti-termination complex.
Protein region is drawn in cartoon format and nucleic acids are drawn as spheres. NusA is marked by a transparent surface and each domain of NusA is colored differently. A. hisPEC-NusA complex (PDB code: 6FLQ). The main contact points between RNAP and NusA are marked with dotted circles. B. λN anti-termination complex (PDB code: 6GOV) drawn from the same vantage point as in panel A. λN is drawn as a non-transparent green surface. The main contact point between RNAP and NusA is marked with a dotted circle. NusA-AR2 was disordered. NusB and NusE are not shown for clarity.
In the 3.7 Å cryo-EM structure of the λN anti-termination complex, NusA exhibited a largely distinct conformation compared to NusA in the hisPEC-NusA complex due to extensive interaction with λN (Figure 7B) [150]. The λN anti-termination complex comprises NusA, NusB, NusE and NusG in addition to λN and the RNAP EC. In the structure, λN remodeled the Nus factors to inhibit their pause stabilizing or termination-promoting functions, as well as penetrated the EC to stabilize an active RNAP conformation. Superimposition of the hisPEC-NusA complex with the λN anti-termination complex revealed that the angle between the Cα of D344 at the end of the KH2 domains from both structures and the Cα of NusA D103, a residue that contact with RNAP at the same position in both structures, is ~ 40°. Contrary to the hisPEC-bound NusA that relocated the β flap-tip to stabilize the RNA hairpin and enhance pausing allosterically, NusA in the λN anti-termination complex was displaced from the RNA exit channel and so was prevented from stabilizing a pause RNA hairpin. The β flap-tip was also moved away with NusA-NTD from the RNA exit channel. The RNAP α-CTDs were not visualized in the λN anti-termination complex, suggesting that the αCTD-NusA interactions were broken during λN-mediated repositioning of NusA.
In summary, the two structures of NusA-bound elongation complexes demonstrated that:
NusA promotes the RNA hairpin-stabilized pause by situating the RNAP flap-tip to stabilize the RNA hairpin or to enhance the allosteric effect of the RNA hairpin.
λN eliminates the pause-enhancing activity of NusA by relocating NusA and altering the interactions between NusA and RNAP.
B. Pause suppression by NusG and RfaH
Although pausing aids timely recruitment of transcription regulators, guides nascent RNA folding, opposes promoter occlusion by nucleosomes, and permits termination, excessive pausing can lead to premature transcription termination or genome instability [59]. NusG and RfaH both are members of the NusG/Spt5 family, and their most prominent function is to suppress pausing in order to prevent these deleterious effects. Both NusG and RfaH consist of an N-terminal domain NGN (NusG-like N-terminal domain) and a C-terminal domain KOW (named after its discoverers, Kyrpides, Ouzounis, Woese [154]). NusG reduces backtrack pauses whereas RfaH inhibits both backtrack and RNA hairpin-stabilized pauses [155]. Other than their anti-pausing properties, NusG and RfaH have a few differences. RfaH, whose RNAP-binding surface is autoinhibited by the CTD, can be loaded onto RNAP only at a specific sequence ops exposed in the non-template DNA of a PEC that can displace the CTD. In contrast, E. coli NusG does not exhibit sequence specificity [89] although B. subtilis and M. tuberculosis NusGs exhibit some sequence-specific stimulation of pausing or termination [73, 156]. Both NusG and RfaH KOW domains are known to bind to ribosomal proteins such as S10 [157, 158], whereas NusG KOW interacts with termination factor Rho to favor its action at suboptimal termination sequences [159, 160].
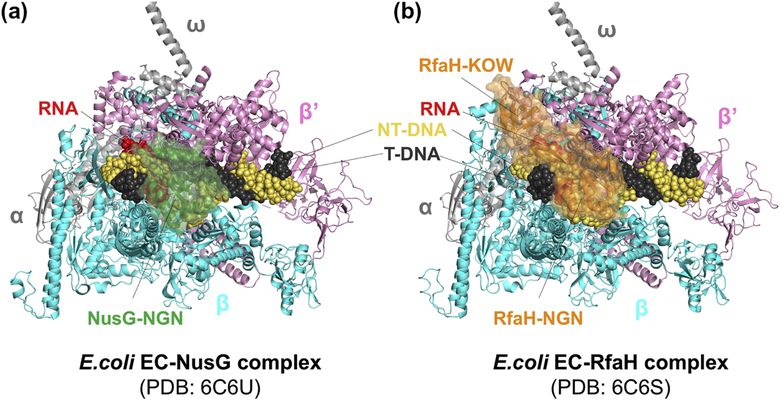
Cryo-EM structures of NusG- and RfaH-bound ECs in combination with biochemical assays revealed the anti-pausing mechanism of NusG and RfaH in the work by Kang et al. [99] (Figure 8). Both NusG and RfaH bind to the same site on RNAP and contact the β protrusion, β lobe domain, and β’ clamp helices. Although RNAP structures in NusG- and RfaH-bound ECs were similar to that of a non-paused EC, the angle between upstream and downstream duplex DNAs decreased upon binding of NusG and RfaH, implying NusG and RfaH rearrange the upstream duplex DNA. Based on psoralen cross-linking at the upstream DNA duplex near transcription bubble fork, the authors suggested that NusG and RfaH inhibit backtracking by stabilizing upstream duplex DNA, which must be melted during backtracking [161]. Additionally, biochemical assays revealed that RfaH could inhibit RNA hairpin-stabilized pausing by tightly binding to EC and preventing swiveling caused by RNA hairpin formation. In contrast, NusG showed much lower affinity to the EC explaining why NusG could not block swiveling.
Figure 8. NusG- and RfaH-bound EC structures.
RNAP subunits and nucleic acids are labelled. A. E. coli NusG (colored in green) binds between β and β’ subunits, contacting β protrusion, β lobe, and β’ clamp helices. NusG KOW domain was disordered in the cryo-EM structure. B. E. coli RfaH (colored in orange) binds to the same site as NusG in panel A. In addition, RfaH recognizes a short-hairpin formed by ops sequence in the non-template DNA (not visible in the figure) and RfaH KOW domain binds flap tip of RNAP, covering upstream duplex DNA near the transcriptpion bubble.
C. Roles of the NusG-family transcription factors in prokaryotes: RfaH and others
RfaH is a paralog of NusG and a poster child showing how NusG family transcription factors can be utilized for the expression of specific genes. RfaH was first discovered as a component in lipopolysaccharide synthesis [162]. It was first thought to be an enzyme but was later shown to be a positive regulator of rfa operon [163]. Afterward, the list of gene operons regulated by RfaH, or RfaH regulons, has increased to include tra (F factor synthesis), hly (hemolysin synthesis), and kps (exopolysaccharide synthesis) [164]. As mentioned above, RfaH needs to be recruited to the ops site exposed on the non-template single-stranded DNA in the EC [53, 165]. Deletion of either RfaH or the ops sequence abrogated full expression of the genes in the operon, showing a consistent pattern of moderate transcription decrease in early genes and of almost complete abolition of transcription of the distal genes [164, 166–170].
A number of biochemical experiments, combined with multiple structural studies [99, 158, 171], demonstrated that RfaH assists the full expression of long operons that contain the ops site in their leader sequence by inhibiting both backtracking and RNA hairpin pause. The polarity-suppressing activity of RfaH is likely common in NusG-like proteins from diverse bacterial species. Myxococcus xanthus taA is a NusG-like protein and located first in the type I polyketide synthase (PKS) gene cluster for the antibiotic TA (Myxovirescin) [172]. Deletion of taA does not affect the normal growth and development of the cells but abrogates antibiotic production. Bacteroides fragilis NCTC9343 synthesizes eight distinct capsular polysaccharides (PS) from separate biosynthetic loci, and each locus contains a NusG-like proteins, named UpxY where x varies from a to h. Deletion of UpxY abolished transcription of the operon, whereas swapping a short NGN peptide between UpxYs altered the specificity of UpxYs toward the gene cluster. The swapped peptide was located at the homologous region to the ops recognition site in RfaH, implying that UpxY might recognize a specific sequence in the operon through this region in order to activate transcription in a similar way to RfaH [99]. Bacillus amyloliquefaciens LoaP (Long operon-associated Protein) is another example of a NusG-like protein that regulates transcription elongation of a biosynthetic gene cluster [173]. In contrast to taA and UpxY that control one operon, LoaP controls two type I PKS gene clusters, difficidin and macrolactin synthetic clusters. Systematic NusG-like protein sequence alignment in the study also revealed that most LoaP homologues in NusG-like proteins are located adjacent to large gene clusters, implying universal usage of NusG-like proteins as polarity-suppressing biosynthesis regulators.
IV. Concluding remarks
Since its inception in the early 1970s, the field of transcriptional pausing research has evolved and expanded. Our understanding of pausing mechanisms has been deepened, largely thanks to advances in experimental methods: ensemble and single-molecule in vitro approaches, improved structural biology methods, and in vivo genome-wide sequencing and imaging techniques. Nevertheless, open questions still remain (Box 3), making this active field of research an exciting avenue for the future.
Box 3. Open questions in transcriptional pausing research.
What is the mechanistic basis of sequence specificity of transcriptional pausing; i.e., why does RNA polymerase pause more frequently on some sequences and not others?
How general is the bacterial consensus pause sequence?
Does the consensus pause sequence change in response to environmental changes or in different species?
What could adjust the pause efficiency and duration other than the DNA sequence?
What is the structural basis of hairpin-stabilized pausing in bacterial RNAPs lacking SI3?
How can we apply the understanding of pausing mechanisms to practical purposes, e.g., bacterial engineering for natural product production or clinical diagnostic and therapeutic application?
Although we now know the sequence elements associated with paused RNAP and that these sequences cause pausing by inducing the half-translocated state, at least in some bacteria, we do not fully understand how interactions of these pause sequences with RNAP interfere with translocation. One future direction that could shed light on this issue is determination of high-resolution structures of ePECs formed by nucleotide addition. Because of the transient nature of ePECs, it seems likely that complexes that arrive at the pause by NTP incorporation, as opposed to direct reconstitution at the pause site, would better represent physiologically relevant species. These structural outputs could then be used as a starting point for theoretical work. Computational analyses of the PECs in different nucleic-acid contexts in turn could complement structural efforts to address the mechanistic basis for the sequence specificity of transcriptional pausing.
The consensus pause sequence (Figure 5) described in this review was determined by sequencing nascent RNA from E. coli and B. subtilis grown in a rich medium [24, 26]. Although purified RNAPs from several additional organisms (Rhodobacter sphaeroides, Mycobacteria bovis, Thermus thermophiles, and mammalian RNAPII from B. taurus) responded to the consensus pause sequence in vitro [24], it seems important to verify that the consensus holds across prokaryotes and eukaryotes by NET-seq, or determine if other pause sequences exist. Another interesting open question is whether changing growth conditions will alter the consensus pause sequence. For example, the small-molecule alarmone (p)ppGpp, which rapidly accumulates in bacterial cells exposed to environmental or nutritional stress (so-called stringent response), enhances in vitro pausing of E. coli RNAP during elongation on genes under stringent control [174, 175]. Furthermore, different solutes (e.g., chaotropic vs. kosmotropic) have been demonstrated to differentially affect RNAP conformation and both its interactions with nucleic acids and its pausing behavior in vitro [115]. Increased pressure has also been shown to influence elongation behavior of RNAP in vitro [176, 177]. Finally, altering pH changes transcriptional pausing both in vitro and in vivo [178, 179]. Thus, it would be interesting to see if subjecting bacteria to stress or changing osmolarity, pressure, or pH within the cells alters the consensus pause sequence in vivo. These effects may be of particular importance in industrial applications of synthetic biology where growth conditions may be suboptimal or highly variable in addition to insights into fundamental mechanisms that may be gained by defining the effects.
From the structures of RNA hairpin-stabilized PECs from E. coli, it is clear that the SI3 domain of RNAP plays a key role in inhibiting catalysis at the pause site by not allowing TL folding as a consequence of swiveling. Many bacterial RNAPs, however, lack the SI3 domain yet are capable of sensing RNA structure as a pause signal, raising the question of whether these bacteria employ a different mechanism for hairpin-stabilized pausing. For example, Bacillus subtilis RNAP, which does not contain SI3, stalled at a site which has a hairpin structure similar to his pause hairpin but shifted away from the RNA 3’-end by one nt [180]. Determination of PEC structures from these organisms will shed light on this question and inform us on how conserved the allosteric mode of hairpin action is.
Heterologous expression of biosynthetic gene clusters for natural product synthesis is often unsuccessful for poorly understood reasons. One possible reason for these failures could be our inability to predict pausing in a given organism to avoid deleterious pauses or programming the EC to override them (e.g., using regulators like RfaH). If pauses could be predicted accurately, in order to remove negative effects from the synthetic sequence (analogously to codon optimization for recombinant protein expression) or optimally maintain RNA folding and transcription–translation coupling, then genetic design will be more powerful. Furthermore, our growing understanding of NusG-family factors modulating transcription of biosynthetic gene clusters in different bacteria is likely to boost our ability to generate natural products heterologously. Another area that will benefit from the ability to predict, and control, transcriptional pauses is bacterial engineering for clinical diagnostic and therapeutic applications [181].
Highlights.
RNA polymerase pausing is widespread and plays key roles in fine-tuning gene expression
Most pauses initiate via an elemental pause state unable to load the NTP substrate
RNA structure prolongs elemental pause by stimulating RNAP conformational changes
Regulators either promote or inhibit RNAP rearrangements associated with pausing
Abbreviations:
- RNAP
RNA polymerase
- RNAPII
eukaryotic Pol II RNA polymerase
- EC
elongation complex
- PEC
paused elongation complex
- ePEC
elemental paused elongation complex
- bp
base pair
- nt
nucleotide
- NTP
nucleoside triphosphate
- NAC
nucleotide addition cycle
- TL
trigger loop
- BH
bridge helix
- FT
flap tip
- FTH
flap-tip helix
- SI3
sequence insertion 3
- CTD
carboxy-terminal domain
- NTD
amino-terminal domain
- NGN
NusG-like N-terminal domain
- NET-seq
native elongating transcript sequencing
- CPX
cysteine-pair cross-linking
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Maizels NM. The Nucleotide Sequence of the Lactose Messenger Ribonucleic Acid Transcribed from the UV5 Promoter Mutant of Escherichia coli. Proceedings of the National Academy of Sciences. 1973;70:3585–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Dahlberg JE, Blattner FR. Virus Res, ICN-UCLA Symp Mol Biol Proc, 2nd. 1973:533. [Google Scholar]
- [3].Winkler ME, Yanofsky C. Pausing of RNA polymerase during in vitro transcription of the tryptophan operon leader region. Biochemistry. 1981;20:3738–44. [DOI] [PubMed] [Google Scholar]
- [4].Farnham PJ, Platt T. Rho-independent termination: dyad symmetry in DNA causes RNA polymerase to pause during transcription in vitro. Nucleic Acids Res. 1981;9:563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lee F, Squires CL, Squires C, Yanofsky C. Termination of transcription in vitro in the escherichia coli tryptophan operon leader region. Journal of Molecular Biology. 1976;103:383–93. [DOI] [PubMed] [Google Scholar]
- [6].Gardner JF. Initiation, pausing, and termination of transcription in the threonine operon regulatory region of Escherichia coli. J Biol Chem. 1982;257:3896–904. [PubMed] [Google Scholar]
- [7].Kingston Chamberlin MJ. Pausing and attenuation of in vitro transcription in the rrnB operon of E. coli. Cell. 1981;27:523–31. [DOI] [PubMed] [Google Scholar]
- [8].Adhya S, Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–96. [DOI] [PubMed] [Google Scholar]
- [9].Reisbig RR, Hearst JE. Escherichia coli deoxyribonucleic acid dependent ribonucleic acid polymerase transcriptional pause sites on SV40 DNA F1. Biochemistry. 1981;20:1907–18. [DOI] [PubMed] [Google Scholar]
- [10].Gilbert W, Maizels N, Maxam A. Sequences of controlling regions of the lactose operon. Cold Spring Harbor Symp Quant Biol. 1974;38:845–55. [DOI] [PubMed] [Google Scholar]
- [11].Gilbert WJ. Starting and stopping sequences of the RNA polymerase In: Losick R, Chamberlin MJ, editors. RNA Polymerase. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1976. p. 193–205. [Google Scholar]
- [12].Kassavetis GA, Chamberlin MJ. Pausing and termination of transcription within the early region of bacteriophage T7 DNA in vitro. J Biol Chem. 1981;256:2777–86. [PubMed] [Google Scholar]
- [13].Gariglio P, Bellard M, Chambon P. Clustering of RNA polymerase B molecules in the 5′ moiety of the adult β-globin gene of hen erythrocytes. Nucleic Acids Research. 1981;9:2589–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gilmour DS, Lis JT. Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:4275–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gilmour DS, Lis JT. RNA polymerase II interacts with the promoter region of the noninduced hsp70 gene in Drosophila melanogaster cells. Molecular and Cellular Biology. 1986;6:3984–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. [DOI] [PubMed] [Google Scholar]
- [17].Spencer CA, Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990;5:777–85. [PubMed] [Google Scholar]
- [18].Buck MJ, Lieb JD. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics. 2004;83:349–60. [DOI] [PubMed] [Google Scholar]
- [19].Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, et al. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nature Structural &Amp; Molecular Biology. 2007;15:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Reppas NB, Wade JT, Church George M, Struhl K. The Transition between Transcriptional Initiation and Elongation in E. coli Is Highly Variable and Often Rate Limiting. Molecular Cell. 2006;24:747–57. [DOI] [PubMed] [Google Scholar]
- [22].Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Larson MH, Mooney RA, Peters JM, Windgassen T, Nayak D, Gross CA, et al. A pause sequence enriched at translation start sites drives transcription dynamics in vivo. Science (New York, NY). 2014;344:1042–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, et al. Native Elongating Transcript Sequencing Reveals Human Transcriptional Activity at Nucleotide Resolution. Cell. 2015;161:541–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Vvedenskaya IO, Vahedian-Movahed H, Bird JG, Knoblauch JG, Goldman SR, Zhang Y, et al. Interactions between RNA polymerase and the “core recognition element” counteract pausing. Science (New York, NY). 2014;344:1285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, et al. In vivo dynamics of RNA polymerase II transcription. Nature structural & molecular biology. 2007;14:796–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Steurer B, Janssens RC, Geverts B, Geijer ME, Wienholz F, Theil AF, et al. Live-cell analysis of endogenous GFP-RPB1 uncovers rapid turnover of initiating and promoter-paused RNA Polymerase II. Proceedings of the National Academy of Sciences. 2018;115:E4368–E76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Davenport RJ, Wuite GJ, Landick R, Bustamante C. Single-molecule study of transcriptional pausing and arrest by E. coli RNA polymerase. Science (New York, NY). 2000;287:2497–500. [DOI] [PubMed] [Google Scholar]
- [30].Larson MH, Landick R, Block SM. Single-molecule studies of RNA polymerase: one singular sensation, every little step it takes. Mol Cell. 2011;41:249–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dangkulwanich M, Ishibashi T, Bintu L, Bustamante C. Molecular Mechanisms of Transcription through Single-Molecule Experiments. Chemical Reviews. 2014;114:3203–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Adelman K, La Porta A, Santangelo TJ, Lis JT, Roberts JW, Wang MD. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:13538–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–47. [DOI] [PubMed] [Google Scholar]
- [34].Harden TT, Wells CD, Friedman LJ, Landick R, Hochschild A, Kondev J, et al. Bacterial RNA polymerase can retain σ70 throughout transcription. Proceedings of the National Academy of Sciences. 2016;113:602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kapanidis AN, Margeat E, Laurence TA, Doose S, Ho SO, Mukhopadhyay J, et al. Retention of Transcription Initiation Factor σ70 in Transcription Elongation: Single-Molecule Analysis. Molecular Cell. 2005;20:347–56. [DOI] [PubMed] [Google Scholar]
- [36].Bar-Nahum G, Nudler E. Isolation and characterization of sigma(70)-retaining transcription elongation complexes from Escherichia coli. Cell. 2001;106:443–51. [DOI] [PubMed] [Google Scholar]
- [37].Mukhopadhyay J, Kapanidis AN, Mekler V, Kortkhonjia E, Ebright YW, Ebright RH. Translocation of sigma(70) with RNA polymerase during transcription: fluorescence resonance energy transfer assay for movement relative to DNA. Cell. 2001;106:453–63. [DOI] [PubMed] [Google Scholar]
- [38].Ring BZ, Roberts JW. Function of a nontranscribed DNA strand site in transcription elongation. Cell. 1994;78:317–24. [DOI] [PubMed] [Google Scholar]
- [39].Mooney RA, Landick R. Tethering s70 to RNA polymerase reveals high in vivo activity of s factors and s70-dependent pausing at promoter-distal locations. Genes Dev. 2003;17:2839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mooney RA, Darst SA, Landick R. Sigma and RNA polymerase: an on-again, off-again relationship? Mol Cell. 2005;20:335–45. [DOI] [PubMed] [Google Scholar]
- [41].Duchi D, Bauer DL, Fernandez L, Evans G, Robb N, Hwang LC, et al. RNA Polymerase Pausing during Initial Transcription. Mol Cell. 2016;63:939–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lerner E, Chung S, Allen BL, Wang S, Lee J, Lu SW, et al. Backtracked and paused transcription initiation intermediate of Escherichia coli RNA polymerase. Proceedings of the National Academy of Sciences. 2016;113:E6562–E71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dulin D, Bauer DLV, Malinen AM, Bakermans JJW, Kaller M, Morichaud Z, et al. Pausing controls branching between productive and non-productive pathways during initial transcription in bacteria. Nature Communications. 2018;9:1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–24. [DOI] [PubMed] [Google Scholar]
- [45].Cramer P, Bushnell D, Kornberg R. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science (New York, NY). 2001;292:1863–76. [DOI] [PubMed] [Google Scholar]
- [46].Gnatt A, Cramer P, Fu J, Bushnell D, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science (New York, NY). 2001;292:1876–82. [DOI] [PubMed] [Google Scholar]
- [47].Vassylyev D, Vassylyeva M, Perederina A, Tahirov T, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–62. [DOI] [PubMed] [Google Scholar]
- [48].Weixlbaumer A, Leon K, Landick R, Darst SA. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152:431–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zuo Y, Wang Y, Steitz TA. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell. 2013;50:430–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Murakami KS. X-ray crystal structure of Escherichia coli RNA polymerase sigma70 holoenzyme. The Journal of biological chemistry. 2013;288:9126–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Bae B, Davis E, Brown D, Campbell EA, Wigneshweraraj S, Darst SA. Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma70 domain 1.1. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:19772–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bai XC, McMullan G, Scheres SH. How cryo-EM is revolutionizing structural biology. Trends in biochemical sciences. 2015;40:49–57. [DOI] [PubMed] [Google Scholar]
- [53].Artsimovitch I, Landick R. The Transcriptional Regulator RfaH Stimulates RNA Chain Synthesis after Recruitment to Elongation Complexes by the Exposed Nontemplate DNA Strand. Cell. 2002;109:193–203. [DOI] [PubMed] [Google Scholar]
- [54].Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The Speed of RNA Transcription and Metabolite Binding Kinetics Operate an FMN Riboswitch. Molecular Cell. 2005;18:49–60. [DOI] [PubMed] [Google Scholar]
- [55].Widom JR, Nedialkov YA, Rai V, Hayes RL, Brooks CL, Artsimovitch I, et al. Ligand Modulates Cross-Coupling between Riboswitch Folding and Transcriptional Pausing. Molecular Cell. 2018;72:541–52.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495–504. [DOI] [PubMed] [Google Scholar]
- [57].Proudfoot NJ. Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science (New York, NY). 2016;352:aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature Reviews Genetics. 2012;13:720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Mayer A, Landry HM, Churchman LS. Pause & go: from the discovery of RNA polymerase pausing to its functional implications. Current opinion in cell biology. 2017;46:72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zhang J, Landick R. A Two-Way Street: Regulatory Interplay between RNA Polymerase and Nascent RNA Structure. Trends in biochemical sciences. 2016;41:293–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Landick R The regulatory roles and mechanism of transcriptional pausing. Biochem Soc Trans. 2006;34:1062–6. [DOI] [PubMed] [Google Scholar]
- [62].Steinert H, Sochor F, Wacker A, Buck J, Helmling C, Hiller F, et al. Pausing guides RNA folding to populate transiently stable RNA structures for riboswitch-based transcription regulation. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pan T, Sosnick T. RNA FOLDING DURING TRANSCRIPTION. Annual Review of Biophysics and Biomolecular Structure. 2006;35:161–75. [DOI] [PubMed] [Google Scholar]
- [65].Landick R, Carey J, Yanofsky C. Translation activates the paused transcription complex and restores transcription of the trp operon leader region. Proceedings of the National Academy of Sciences. 1985;82:4663–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation Between Translating Ribosomes and RNA Polymerase in Transcription Elongation. Science (New York, NY). 2010;328:504–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ray-Soni A, Bellecourt MJ, Landick R. Mechanisms of Bacterial Transcription Termination: All Good Things Must End. Annu Rev Biochem. 2016;85:319–47. [DOI] [PubMed] [Google Scholar]
- [68].Richardson J Rho-dependent termination and ATPases in transcript termination. Biochim Biophys Acta. 2002;1577:251. [DOI] [PubMed] [Google Scholar]
- [69].Erie D, Hajiseyedjavadi O, Young M, von Hippel P. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science (New York, NY). 1993;262:867–73. [DOI] [PubMed] [Google Scholar]
- [70].Sydow JF, Cramer P. RNA polymerase fidelity and transcriptional proofreading. Current Opinion in Structural Biology. 2009;19:732–9. [DOI] [PubMed] [Google Scholar]
- [71].Gamba P, James K, Zenkin N. A link between transcription fidelity and pausing in vivo. Transcription. 2017;8:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Toulokhonov I, Artsimovitch I, Landick R. Allosteric Control of RNA Polymerase by a Site That Contacts Nascent RNA Hairpins. Science (New York, NY). 2001;292:730–3. [DOI] [PubMed] [Google Scholar]
- [73].Yakhnin AV, Murakami KS, Babitzke P. NusG Is a Sequence-specific RNA Polymerase Pause Factor That Binds to the Non-template DNA within the Paused Transcription Bubble. Journal of Biological Chemistry. 2016;291:5299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nature Reviews Molecular Cell Biology. 2015;16:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Core LJ, Waterfall JJ, Lis JT. Nascent RNA Sequencing Reveals Widespread Pausing and Divergent Initiation at Human Promoters. Science (New York, NY). 2008;322:1845–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kwak H, Fuda NJ, Core LJ, Lis JT. Precise Maps of RNA Polymerase Reveal How Promoters Direct Initiation and Pausing. Science (New York, NY). 2013;339:950–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Mylonas C, Tessarz P. NET-prism enables RNA polymerase-dedicated transcriptional interrogation at nucleotide resolution. RNA biology. 2019:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Nojima T, Gomes T, Grosso ARF, Kimura H, Dye MJ, Dhir S, et al. Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing. Cell. 2015;161:526–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Mishanina TV, Palo MZ, Nayak D, Mooney RA, Landick R. Trigger loop of RNA polymerase is a positional, not acid–base, catalyst for both transcription and proofreading. Proceedings of the National Academy of Sciences. 2017;114:E5103–E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: structural analysis. J Mol Biol. 2010;395:686–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Windgassen T, Mooney RA, Nayak D, Palangat M, Zhang J, Landick R. Trigger-helix folding pathway and SI3 mediate catalysis and hairpin-stabilized pausing by Escherichia coli RNA polymerase. Nucleic Acids Res. 2014;pii: gku997:epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Zakharova N, Bass I, Arsenieva E, Nikiforov V, Severinov K. Mutations in and monoclonal antibody binding to evolutionary hypervariable region of Escherichia coli RNA polymerase beta’ subunit inhibit transcript cleavage and transcript elongation. The Journal of biological chemistry. 1998;273:24912–20. [DOI] [PubMed] [Google Scholar]
- [83].Artsimovitch I, Svetlov V, Murakami K, Landick R. Co-overexpression of E. coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J Biol Chem. 2003;278:12344–55. [DOI] [PubMed] [Google Scholar]
- [84].Saba J, Chua XY, Mishanina TV, Nayak D, Windgassen TA, Mooney RA, et al. The elemental mechanism of transcriptional pausing. eLife. 2019;8:e40981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Bai L, Shundrovsky A, Wang MD. Sequence-dependent Kinetic Model for Transcription Elongation by RNA Polymerase. Journal of Molecular Biology. 2004;344:335–49. [DOI] [PubMed] [Google Scholar]
- [86].Bochkareva A, Yuzenkova Y, Tadigotla VR, Zenkin N. Factor-independent transcription pausing caused by recognition of the RNA-DNA hybrid sequence. The EMBO journal. 2012;31:630–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Imashimizu M, Kireeva ML, Lubkowska L, Gotte D, Parks AR, Strathern JN, et al. Intrinsic translocation barrier as an initial step in pausing by RNA polymerase II. J Mol Biol. 2013;425:697–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Gabizon R, Lee A, Vahedian-Movahed H, Ebright RH, Bustamante CJ. Pause sequences facilitate entry into long-lived paused states by reducing RNA polymerase transcription rates. Nat Commun. 2018;9:2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci U S A. 2000;97:7090–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Reeder TC, Hawley DK. Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell. 1996;87:767–77. [DOI] [PubMed] [Google Scholar]
- [91].Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA:DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. [DOI] [PubMed] [Google Scholar]
- [92].Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J Biol Chem. 1997;272:15329–38. [DOI] [PubMed] [Google Scholar]
- [93].Nudler E RNA Polymerase Backtracking in Gene Regulation and Genome Instability. Cell. 2012;149:1438–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Ring B, Yarnell W, Roberts J. Function of E. coli RNA polymerase sigma factor sigma70 in promoter-proximal pausing. Cell. 1996;86:485–93. [DOI] [PubMed] [Google Scholar]
- [95].Brodolin K, Zenkin N, Mustaev A, Mamaeva D, Heumann H. The sigma 70 subunit of RNA polymerase induces lacUV5 promoter-proximal pausing of transcription. Nature structural & molecular biology. 2004;11:551–7. [DOI] [PubMed] [Google Scholar]
- [96].Nickels BE, Mukhopadhyay J, Garrity SJ, Ebright RH, Hochschild A. The σ70 subunit of RNA polymerase mediates a promoter-proximal pause at the lac promoter. Nature Structural &Amp; Molecular Biology. 2004;11:544. [DOI] [PubMed] [Google Scholar]
- [97].Perdue S, Roberts J. A backtrack-inducing sequence is an essential component of Escherichia coli sigma70-dependent promoter-proximal pausing. Mol Microbiol. 2010;78:636–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Marr MT, Roberts JW. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol Cell. 2000;6:1275–85. [DOI] [PubMed] [Google Scholar]
- [99].Kang JY, Mooney RA, Nedialkov Y, Saba J, Mishanina TV, Artsimovitch I, et al. Structural Basis for Transcript Elongation Control by NusG Family Universal Regulators. Cell. 2018;173:1650–62.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Hein PP, Kolb KE, Windgassen T, Bellecourt MJ, Darst SA, Mooney RA, et al. RNA polymerase pausing and nascent-RNA structure formation are linked through clamp-domain movement. Nature structural & molecular biology. 2014;21:794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Guo X, Myasnikov AG, Chen J, Crucifix C, Papai G, Takacs M, et al. Structural Basis for NusA Stabilized Transcriptional Pausing. Mol Cell. 2018;69:816–27.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Kang JY, Mishanina TV, Bellecourt MJ, Mooney RA, Darst SA, Landick R. RNA Polymerase Accommodates a Pause RNA Hairpin by Global Conformational Rearrangements that Prolong Pausing. Mol Cell. 2018;69:802–15.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol Cell. 2003;12:1125–36. [DOI] [PubMed] [Google Scholar]
- [104].Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, et al. Riboswitch control of Rho-dependent transcription termination. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:5376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Hollands K, Sevostiyanova A, Groisman EA. Unusually long-lived pause required for regulation of a Rho-dependent transcription terminator. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E1999–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Chan CL, Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol. 1993;233:25–42. [DOI] [PubMed] [Google Scholar]
- [107].Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8900–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Aivazashvili VA, Bibilashvili R, Vartikian RM, Kutateladze TA. [Effect of the primary structure of RNA on the pulse character of RNA elongation in vitro by Escherichia coli RNA polymerase: a model]. Mol Biol (Mosk). 1981;15:915–29. [PubMed] [Google Scholar]
- [109].Imashimizu M, Takahashi H, Oshima T, McIntosh C, Bubunenko M, Court DL, et al. Visualizing translocation dynamics and nascent transcript errors in paused RNA polymerases in vivo. Genome Biology. 2015;16:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Herbert R, La Porta A, Wong B, Mooney R, Neuman K, Landick R, et al. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Markovtsov V, Mustaev A, Goldfarb A. Protein-RNA interactions in the active center of the transcription elongation complex. Proc Natl Acad Sci USA. 1996;93:3221–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nature structural & molecular biology. 2008;15:811–8. [DOI] [PubMed] [Google Scholar]
- [113].Vos SM, Farnung L, Urlaub H, Cramer P. Structure of paused transcription complex Pol II-DSIF-NELF. Nature. 2018;560:601–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–53. [DOI] [PubMed] [Google Scholar]
- [115].Chan C, Landick R. Effects of neutral salts of transcript elongation and pausing suggest the his leader pause RNA hairpin interacts with an easily disordered region of RNA polymerase. J Mol Biol. 1997;268:37–53. [DOI] [PubMed] [Google Scholar]
- [116].Yarnell WS, Roberts JW. Mechanism of instrinsic termination and antitermination. Science (New York, NY). 1999;284:611–5. [DOI] [PubMed] [Google Scholar]
- [117].Bar-Nahum G, Epshtein V, Ruckenstein A, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–93. [DOI] [PubMed] [Google Scholar]
- [118].Epshtein V, Mustaev A, Markovtsov V, Bereshchenko O, Nikiforov V, Goldfarb A. Swing-gate model of nucleotide entry into the RNA polymerase active center. Mol Cell. 2002;10:623–34. [DOI] [PubMed] [Google Scholar]
- [119].Nayak D, Voss M, Windgassen T, Mooney RA, Landick R. Cys-pair reporters detect a constrained trigger loop in a paused RNA polymerase. Mol Cell. 2013;50:882–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–19. [DOI] [PubMed] [Google Scholar]
- [121].Weilbaecher R, Hebron C, Feng G, Landick R. Termination-altering amino acid substitutions in the ß’ subunit of Escherichia coli RNA polymerase identify regions involved in RNA chain elongation. Genes Dev. 1994;8:2913–7. [DOI] [PubMed] [Google Scholar]
- [122].Conrad T, Frazier M, Joyce A, Cho B, Knight E, Lewis N, et al. RNA polymerase mutants found through adaptive evolution re-program Escherichia coli for optimal growth in minimal media. Proc Natl Acad Sci U S A. 2010;107:20500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Carrillo Oesterreich F, Bieberstein N, Neugebauer KM. Pause locally, splice globally. Trends in cell biology. 2011;21:328–35. [DOI] [PubMed] [Google Scholar]
- [124].Levine M Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, et al. RNA polymerase is poised for activation across the genome. Nature genetics. 2007;39:1507–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Stargell LA, Struhl K. Mechanisms of transcriptional activation in vivo: two steps forward. Trends in genetics : TIG. 1996;12:311–5. [DOI] [PubMed] [Google Scholar]
- [128].Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, et al. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nature genetics. 2007;39:1512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Werner F A nexus for gene expression-molecular mechanisms of Spt5 and NusG in the three domains of life. J Mol Biol. 2012;417:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Palangat M, Renner DD, Price DH, Landick R. DSIF/NELF, a negative elongation factor for human RNA polymerase II, inhibits the anti-arrest, transcript cleavage factor TFIIS. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15036–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Sekine S, Murayama Y, Svetlov V, Nudler E, Yokoyama S. The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions. Mol Cell. 2015;57:408–21. [DOI] [PubMed] [Google Scholar]
- [132].Wang D, Bushnell DA, Huang X, Westover KD, Levitt M, Kornberg RD. Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution. Science (New York, NY). 2009;324:1203–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Cheung AC, Sainsbury S, Cramer P. Structural basis of initial RNA polymerase II transcription. The EMBO journal. 2011;30:4755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Liu X, Bushnell DA, Silva DA, Huang X, Kornberg RD. Initiation complex structure and promoter proofreading. Science (New York, NY). 2011;333:633–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Silva D-A, Weiss DR, Pardo Avila F, Da L-T, Levitt M, Wang D, et al. Millisecond dynamics of RNA polymerase II translocation at atomic resolution. Proceedings of the National Academy of Sciences. 2014;111:7665–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Nudler E, Gottesman ME. Transcription termination and anti-termination in E. coli. Genes Cells. 2002;7:755–68. [DOI] [PubMed] [Google Scholar]
- [137].Friedman DI, Baron LS. Genetic characterization of a bacterial locus involved in the activity of the N function of phage lambda. Virology. 1974;58:141–8. [DOI] [PubMed] [Google Scholar]
- [138].Vogel U, Jensen KF. NusA is required for ribosomal antitermination and for modulation of the transcription elongation rate of both antiterminated RNA and mRNA. The Journal of biological chemistry. 1997;272:12265–71. [DOI] [PubMed] [Google Scholar]
- [139].Kung H, Spears C, Weissbach H. Purification and properties of a soluble factor required for the deoxyribonucleic acid-directed in vitro synthesis of beta-galactosidase. The Journal of biological chemistry. 1975;250:1556–62. [PubMed] [Google Scholar]
- [140].Greenblatt J, McLimont M, Hanly S. Termination of transcription by nusA gene protein of Escherichia coli. Nature. 1981;292:215–20. [DOI] [PubMed] [Google Scholar]
- [141].Farnham PJ, Greenblatt J, Platt T. Effects of NusA protein on transcription termination of the tryptophan operon of Escherichia coli. Cell. 1982;29:945–51. [DOI] [PubMed] [Google Scholar]
- [142].Cohen SE, Godoy VG, Walker GC. Transcriptional modulator NusA interacts with translesion DNA polymerases in Escherichia coli. Journal of bacteriology. 2009;191:665–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Squires CL, Zaporojets D. Proteins shared by the transcription and translation machines. Annual review of microbiology. 2000;54:775–98. [DOI] [PubMed] [Google Scholar]
- [144].Beuth B, Pennell S, Arnvig KB, Martin SR, Taylor IA. Structure of a Mycobacterium tuberculosis NusA-RNA complex. The EMBO journal. 2005;24:3576–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Worbs M, Bourenkov GP, Bartunik HD, Huber R, Wahl MC. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol Cell. 2001;7:1177–89. [DOI] [PubMed] [Google Scholar]
- [146].Ha KS, Toulokhonov I, Vassylyev DG, Landick R. The NusA N-terminal domain is necessary and sufficient for enhancement of transcriptional pausing via interaction with the RNA exit channel of RNA polymerase. J Mol Biol. 2010;401:708–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Bycroft M, Hubbard TJ, Proctor M, Freund SM, Murzin AG. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–42. [DOI] [PubMed] [Google Scholar]
- [148].Gibson TJ, Thompson JD, Heringa J. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS letters. 1993;324:361–6. [DOI] [PubMed] [Google Scholar]
- [149].Mah TF, Kuznedelov K, Mushegian A, Severinov K, Greenblatt J. The alpha subunit of E. coli RNA polymerase activates RNA binding by NusA. Genes Dev. 2000;14:2664–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Krupp F, Said N, Huang YH, Loll B, Burger J, Mielke T, et al. Structural Basis for the Action of an All-Purpose Transcription Anti-termination Factor. Mol Cell. 2019;74:143–57.e5. [DOI] [PubMed] [Google Scholar]
- [151].Ma C, Mobli M, Yang X, Keller AN, King GF, Lewis PJ. RNA polymerase-induced remodelling of NusA produces a pause enhancement complex. Nucleic Acids Res. 2015;43:2829–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Yang X, Molimau S, Doherty GP, Johnston EB, Marles-Wright J, Rothnagel R, et al. The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA. EMBO reports. 2009;10:997–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Schweimer K, Prasch S, Sujatha PS, Bubunenko M, Gottesman ME, Rosch P. NusA interaction with the alpha subunit of E. coli RNA polymerase is via the UP element site and releases autoinhibition. Structure (London, England : 1993). 2011;19:945–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Kyrpides NC, Woese CR, Ouzounis CA. KOW: a novel motif linking a bacterial transcription factor with ribosomal proteins. Trends in biochemical sciences. 1996;21:425–6. [DOI] [PubMed] [Google Scholar]
- [155].Kolb KE, Hein PP, Landick R. Antisense oligonucleotide-stimulated transcriptional pausing reveals RNA exit channel specificity of RNA polymerase and mechanistic contributions of NusA and RfaH. The Journal of biological chemistry. 2014;289:1151–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Kalyani BS, Kunamneni R, Wal M, Ranjan A, Sen R. A NusG paralogue from Mycobacterium tuberculosis, Rv0639, has evolved to interact with ribosomal protein S10 (Rv0700) but not to function as a transcription elongation-termination factor. Microbiology (Reading, England). 2015;161:67–83. [DOI] [PubMed] [Google Scholar]
- [157].Burmann BM, Schweimer K, Luo X, Wahl MC, Stitt BL, Gottesman ME, et al. A NusE:NusG complex links transcription and translation. Science (New York, NY). 2010;328:501–4. [DOI] [PubMed] [Google Scholar]
- [158].Burmann BM, Knauer SH, Sevostyanova A, Schweimer K, Mooney RA, Landick R, et al. An alpha helix to beta barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell. 2012;150:291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Peters JM, Mooney RA, Grass JA, Jessen ED, Tran F, Landick R. Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 2012;26:2621–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Lawson MR, Ma W, Bellecourt MJ, Artsimovitch I, Martin A, Landick R, et al. Mechanism for the Regulated Control of Bacterial Transcription Termination by a Universal Adaptor Protein. Mol Cell. 2018;71:911–22.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Turtola M, Belogurov GA. NusG inhibits RNA polymerase backtracking by stabilizing the minimal transcription bubble. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Wilkinson RG, Stocker BAD. Genetics and Cultural Properties of Mutants of Salmonella typhimurium lacking Glucosyl or Galactosyl Lipopolysaccharide Transferases. Nature. 1968;217:955–7. [DOI] [PubMed] [Google Scholar]
- [163].Lindberg AA, Hellerqvist C-G. Rough Mutants of Salmonella typhimurium: Immunochemical and Structural Analysis of Lipopolysaccharides from rfaH Mutants. Microbiology (Reading, England). 1980;116:25–32. [DOI] [PubMed] [Google Scholar]
- [164].Bailey MJ, Hughes C, Koronakis V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol Microbiol. 1997;26:845–51. [DOI] [PubMed] [Google Scholar]
- [165].Belogurov GA, Mooney RA, Svetlov V, Landick R, Artsimovitch I. Functional specialization of transcription elongation factors. The EMBO journal. 2009;28:112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Bailey MJ, Koronakis V, Schmoll T, Hughes C. Escherichia coli HlyT protein, a transcriptional activator of haemolysin synthesis and secretion, is encoded by the rfaH (sfrB) locus required for expression of sex factor and lipopolysaccharide genes. Mol Microbiol. 1992;6:1003–12. [DOI] [PubMed] [Google Scholar]
- [167].Beutin L, Manning PA, Achtman M, Willetts N. sfrA and sfrB products of Escherichia coli K-12 are transcriptional control factors. Journal of bacteriology. 1981;145:840–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Stevens MP, Clarke BR, Roberts IS. Regulation of the Escherichia coli K5 capsule gene cluster by transcription antitermination. Mol Microbiol. 1997;24:1001–12. [DOI] [PubMed] [Google Scholar]
- [169].Nieto JM, Bailey MJA, Hughes C, Koronakis V. Suppression of transcription polarity in the Escherichia coli haemolysin operon by a short upstream element shared by polysaccharide and DNA transfer determinants. Molecular Microbiology. 1996;19:705–13. [DOI] [PubMed] [Google Scholar]
- [170].Zuber PK, Artsimovitch I, NandyMazumdar M, Liu Z, Nedialkov Y, Schweimer K, et al. The universally-conserved transcription factor RfaH is recruited to a hairpin structure of the non-template DNA strand. eLife. 2018;7:e36349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, et al. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell. 2007;26:117–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Paitan Y, Orr E, Ron EZ, Rosenberg E. A nonessential signal peptidase II (Lsp) of Myxococcus xanthus might be involved in biosynthesis of the polyketide antibiotic TA. Journal of bacteriology. 1999;181:5644–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Goodson JR, Klupt S, Zhang C, Straight P, Winkler WC. LoaP is a broadly conserved antiterminator protein that regulates antibiotic gene clusters in Bacillus amyloliquefaciens. Nature microbiology. 2017;2:17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Krohn M, Wagner R. Transcriptional pausing of RNA polymerase in the presence of guanosine tetraphosphate depends on the promoter and gene sequence. The Journal of biological chemistry. 1996;271:23884–94. [DOI] [PubMed] [Google Scholar]
- [175].Kingston RE, Nierman WC, Chamberlin MJ. A direct effect of guanosine tetraphosphate on pausing of Esherichia coli RNA polymerase during RNA chain elongation. J Biol Chem. 1981;256:2787–97. [PubMed] [Google Scholar]
- [176].Erijman L, Clegg M. Heterogeneity of E. coli RNA polymerase revealed by high pressure. J Mol Biol. 1995;253:259–65. [DOI] [PubMed] [Google Scholar]
- [177].Erijman L, Clegg RM. Reversible stalling of transcription elongation complexes by high pressure. Biophys J. 1998;75:453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. A pH-responsive riboregulator. Genes & Development. 2009;23:2650–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Nechooshtan G, Elgrably-Weiss M, Altuvia S. Changes in transcriptional pausing modify the folding dynamics of the pH-responsive RNA element. Nucleic Acids Res. 2014;42:622–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Yakhnin AV, Yakhnin H, Babitzke P. RNA polymerase pausing regulates translation initiation by providing additional time for TRAP-RNA interaction. Mol Cell. 2006;24:547–57. [DOI] [PubMed] [Google Scholar]
- [181].Riglar DT, Silver PA. Engineering bacteria for diagnostic and therapeutic applications. Nature reviews Microbiology. 2018;16:214–25. [DOI] [PubMed] [Google Scholar]