Abstract
As MDM4 and HPV16 E6 oncoproteins play important roles in inhibition of p53 activity, a functional polymorphism (rs4245739) in the 3’ UTRs of MDM4 targeted by miRNA-191 may alter its expression level or functional efficiency, thus affecting tumor status and survival in HPV-positive squamous cell carcinoma of oropharynx (SCCOP). A total of 564 incident SCCOP patients with definitive radiotherapy were included for determination of tumor HPV16 status and genotypes of the polymorphism. Univariate and multivariable Cox models were performed to assess the associations between the polymorphism and outcomes. We found that MDM4 rs4245739 had statistically significant associations with tumor HPV-positivity and survival of SCCOP patients. Patients with AC/CC variant genotypes of MDM4 rs4245739 were approximately 3-fold more likely to be HPV16-positive tumors among SCCOP patients compared with common homozygous AA genotype (aOR, 3.2, 95%CI, 1.9–5.5). Moreover, patients with MDM4 rs4245739 AC/CC variant genotypes had significantly better overall, disease-specific, and disease-free survival compared with those with the corresponding common homozygous AA genotype (all log-rank: P < 0.05); and these genotypes were significantly associated with an approximately 3 to 4 times reduced risk of overall death, death owing to disease, and recurrence after multivariable adjustment. Finally, the significant effects of MDM4 rs4245739 polymorphism on survival were found among HPV16-positive SCCOP patients only after the stratified analyses by tumor HPV status. We concluded that MDM4 rs4245739 polymorphism is significantly associated with tumor HPV status and survival of SCCOP, especially in HPV16-positive SCCOP patients treated with definitive radiotherapy; nevertheless, prospective larger studies are warranted.
Keywords: MDM4 3’UTR variant, miRNA, HPV status, oropharyngeal cancer, biomarkers
Introduction
Owing to screening and HPV prophylactic vaccination programs, rates of cervical cancer in developed countries have plateaued or declined. Meanwhile, the incidence of squamous cell carcinoma of the oropharynx (SCCOP), a distinct subgroup of head and neck cancers, has increased rapidly in recent years, despite the decrease in tobacco use in the U.S. The rising incidence of SCCOP in the U.S. is likely a consequence of persistent infection with HPV, predominantly high-risk type 16 (HPV16) [1–3]. In the U.S., HPV accounts for approximately 70–80% of SCCOP (primarily tonsils and base of tongue) [4]. HPV16 is the most common high-risk type, found in 90–95% of tumor HPV-positive [HPV(+)] SCCOP patients as reported by others [3,5]. Unfortunately, there is currently no effective screening method to identify patients with tumor HPV(+) SCCOP. Moreover, even among patients with HPV(+) SCCOP, there remains heterogeneity in clinical outcomes. Identification of patients with SCCOP needing treatment intensification and those able to benefit from reduction of treatment intensity is critical to more effective and less morbid treatment. Thus, due to the increasing incidence of HPV(+) SCCOP, novel biomarkers for screening at risk of and prognosis for HPV-related SCCOP patients are greatly needed.
The survival of patients with HPV(+) SCCOP is markedly improved compared to patients with HPV(−) SCCOP, which carries an overall 5-year relative survival rate of approximately 50% [6–8]. The prognosis of patients with SCCOP depends on clinical features, epidemiologic variables, and lifestyle factors [9–18], but these current prognostic markers remain imperfect in predicting clinical outcomes. HPV status has emerged as the dominant prognostic marker. However, even SCCOP patients with the same tumor HPV status can have significantly heterogeneous clinical outcomes. Alongside clinical and epidemiologic prognostic factors, molecular biomarkers may play a key role by increasing the accuracy and validity of outcome prediction. At present, no significant molecular biomarkers are available for prognostication in HPV(+) SCCOP patients. Thus, the identification of individual SCCOP patients at high risk of recurrence or death is clinically relevant, and new biomarkers for identifying and further stratifying HPV(+) SCCOP patients may ensure optimal therapy. Despite advances in diagnostic techniques, the majority of SCCOP patients are still diagnosed with late-stage SCCOP, and consequently, leading to low survival [19–23]. Thus, identifying molecular biomarkers which may lead to improved survival for patients with SCCOP is urgently needed.
p53 as an important tumor suppressor plays a critical role in genome integrity and acts as “the guardian of genome in many human cancers, especially in SCCHN [24,25], while mouse double minute 2 (MDM2) and 4 (MDM4) inhibit the tumor suppressor activity of p53 through the interaction between p53 and the two negative modulators [26–28]. MDM4 is an oncoprotein which negatively regulates the p53. It is well documented that overexpression of this protein leads to many types of tumors, including SCCHN[24,28]. Therefore, MDM4 either directly binds to p53 and inhibits its transcriptional activity [29,30] or binds to MDM2 to regulate its role in inhibition of p53 activity[31,32]. Additionally, MDM4 plays an essential role in MDM2-MDM4–p53 regulatory circuit to enhance the function of the E3 ubiquitin ligase of MDM2 and promoting degradation of p53[33]. Therefore, either down-expression or over-expression of MDM4 gene may affect cancer risk and prognosis[34,35]. The translational potential of MDM4 for predicting survival has become a novel therapeutic target for p53 reactivation in cancer treatment [26].
Recent studies suggested that the variation in the 3’-UTR of MDM4 can lead to a decreased risk of various malignancies [36]. Although we have previously reported the associations of several genetic variants of MDM4, including MDM4 rs11801299 and rs10900598 in the 3′-untranslated region (3′-UTR) and rs1380576 in the first intron, with cancer risk and prognosis [37–41], their exact functional changes remain unknown. A functional polymorphism of MDM4 rs4245739 in the 3’UTR of this gene has been identified, and creates a target binding site of miRNA-191, leading to a decreased level of MDM4 protein. The occurrence of the C minor allele of rs4245739 (A>C) in the 3’-UTR of MDM4 has been found to be associated with reduced cancer risk, progression of metastasis, and cancer-related death[36]. Furthermore, a recent larger meta-analysis showed that MDM4 rs4245739 is associated with a reduced overall risk of cancer[36].
To date, no studies have investigated whether MDM4 rs4245739 is associated with tumor HPV status and survival of SCCOP patients, particularly HPV16-positive SCCOP patients after radiation or chemoradiation. In the present study, we aimed to assess the distribution of genotypes of this polymorphism involved in these molecular pathways in HPV16-positive and HPV16-negative tumors among patients with SCCOP and evaluate whether this genetic variant affects HPV16 tumor status and survival of SCCOP patients.
Materials and methods
Study patient population
In this study, there are a total of 564 patients with incident SCCOP from an ongoing molecular epidemiology study of squamous cell carcinoma of the head and neck at The University of Texas MD Anderson Cancer Center from January 2000 to May 2013. All patients were recruited consecutively with newly diagnosed, histopathologically confirmed, and untreated SCCOP without restrictions on age, sex, ethnicity, cancer stage, or histology. At recruitment, a written informed consent was obtained from each patient and the study was approved by the Institutional Review Board of MD Anderson Cancer Center (Protocol #: LAB00–062). At enrollment the patients were asked to conduct an epidemiological questionnaire for information on demographic and risk factors such as smoking and alcohol status. Each patient also provided a total of 30 mL of blood sample as required by the parent study for genotyping/phenotype biomarker study. The detailed data regarding study patients on enrollment, epidemiological, and clinical information have been stated in our previously study[42].
HPV16 determination in tumor specimens
Genomic DNA samples of 474 patients were extracted from paraffin-embedded tumor tissues of SCCOP patients and determined for HPV16 tumor status using polymerase chain reaction and in situ hybridization methods described previously[42,43]. For the rest of 90 SCCOP patients, the presence of tumor HPV16 in tumor specimens was determined using in situ hybridization and p16 immunohistochemistry from patients’ HPV16 data in their clinical records, as the pathology laboratory in our institution had begun classifying all SCCOP specimens for HPV16 status as a standard clinical practice and routine laboratory assay.
MDM4 genotyping
The genomic DNA extracted from blood samples was used for MDM4 genotyping. The genotyping of 481 patients was performed by the PCR-based restriction fragment length polymorphism (PCR-RFLP) as previously reported [44]. Moreover, a 10% random sample of PCR-RFLP genotyping was repeated with a 100% reproducibility. Additionally, the rest of genotyping of 83 cases was performed using Illumina HumanOmniExpress-12v1 BeadChip with a genotyping call rate > 95%.
Determination of tumor MDM4 protein expression
In this patient cohort, 61 SCCOP patients, whose tumor specimens available, were selected for MDM4 determination by the immunohistochemistry (IHC) as previously described (45). Briefly, the sections were deparafinized with xylene and dehydrated with ethanol. Antigen retrieval was performed by microwave heating methods. Samples were blocked in 2% horse serum in PBS-Tween 20 for 30 min. After blocking, the sections were then incubated with primary antibody for MDM4 (AB112, 1:150, Santa Cruz Biotechnology) for 2 hours at 37oC. MDM4 protein was detected using the Vectastain ABC Elite kit. The sections were then reviewed and categorized under a microscope as positively- or negatively-stained (section with ≥ 10% positively stained tumor cells were considered to be positively-stained)(45).
Patient follow-up
We followed up and monitored the patients throughout their treatment and post-treatment courses from scheduled routine clinical and radiographic examinations. Patients were classified as disease free if they had no disease recorded on the date of the last visit at our institution. Clinical data on stage of the index tumor at presentation, site of the index tumor, and treatment, were documented at initial presentation and from follow-up examinations. Index cancer stage was classified as either clinical stage I/II (early-stage) or clinical stage III/IV (late-stage) disease. We classified treatment into the two categories: radiotherapy only or radiochemotherapy. Medical comorbidities were classified as several categorizes including none, mild, moderate, or severe comorbidities according to a modification of the Kaplan-Feinstein comorbidity index (Adult Comorbidity Evaluation 27).
Statistical analysis
Statistical Analysis System software (Version 9.4; SAS Institute, Cary, NC) was used for all of the statistical analyses. The primary endpoints of this study included overall deaths, deaths due to disease, and disease recurrence. The date of the end of treatment to the date of last follow-up or clinical detection of recurrent cancer (local, regional, or distant) was defined as the disease-specific survival (DFS). Patients who had recurrence free or lost to follow-up were censored. The time from first appointment to death from any cause or date of last follow-up was defined as the overall survival (OS), and the time from first appointment to death from disease or date of last follow-up was defined as the disease-specific survival (DSS). The patients alive at the end of the study or lost to follow-up were censored for either OS or DSS determination. In the univariate analysis, we assessed epidemiological variables at the time of diagnosis, including age in years, ethnicity, sex, smoking and alcohol status, HPV status, and clinical characteristics including as index tumor stage, comorbidity, and treatment. For association of the variant with HPV16 tumor status, the variables including age, ethnicity, sex, smoking and alcohol status were included for multivariable analysis as the univariate analysis did not indicate significant associations of HPV16 tumor status with these clinical characteristics including as index tumor stage, comorbidity, and treatment. The univariate prognostic analysis was not statistically significant for some variables, while these variables were retained in the main-effects and final multivariable model because of biological, epidemiological and clinical significance considerations. We employed the dominant mode of genotype coding a priori for the SNP analysis as done in our previous analyses and publications (37, 40, 41). The Kaplan–Meier method was used to compare survival between patients with different genotypes. We also calculated the log-rank statistic for testing the hypothesis of a difference in survival between genotyping groups. Moreover, we assessed whether the genotypes were statistically associated with survival among SCCOP patients using a Cox proportional hazards model after adjustment with above important confounders. Finally, the stratification analysis by tumor HPV status was also performed in a similar multivariable analysis. A two-sided statistical significance was set at p < 0.05.
Results
Characteristics of study patients
The characteristics of the 564 patients are summarized in Table 1. As we previously reported [46], the patients had a median age of 55 years at diagnosis ranged from 28 to 82 years. Approximately 518 (92%) of patients were non-Hispanic white, and approximately 491(87.1%) of the cases were males. About 284 (50.3%) patients had a reported history of smoking, and 377 (66.8%) cases were reported a history of alcohol drinking. Among these patients, approximately 482 (85%) cases were tumor HPV16 positive, 93.6% had late-stage disease. For treatment, approximately 431 (76.4%) patients received radiation plus chemotherapy and 133 (23.6%) cases received radiation alone, while all patients received radiotherapy. A total of 78 patients died from any cause (44 died from SCCOP), and 64 patients had disease recurrence after a 34.5 months median follow-up. Additionally, in this SCCOP patient cohort, 19 cases were African American, of whom 10 cases were tumor HPV16 positive.
Table 1.
Characteristics of study patients with SCCOP (N = 564)
| Characteristics | All patients (N, %) | HPV(+) SCCOP (N, %) | HPV(−) SCCOP (N, %) |
|---|---|---|---|
| Age | |||
| ≤ 57 years | 343(60.8) | 293 (60.8) | 50 (61.0) |
| > 57 years | 221(39.2) | 189 (39.2) | 32 (39.0) |
| Sex | |||
| Male | 491(87.1) | 428 (88.8) | 63 (76.8) |
| Female | 73(12.9) | 54 (11.2) | 19 (23.2) |
| Ethnicity | |||
| Non-Hispanic white | 518(91.8) | 447 (92.7) | 71 (86.6) |
| Other | 46(8.2) | 35 (7.3) | 11 (13.4) |
| Smoking | |||
| Never | 280(49.7) | 244 (50.6) | 36 (43.9) |
| Ever | 284(50.3) | 238 (49.4) | 46 (56.1) |
| Alcohol use | |||
| Never | 187(33.2) | 158 (32.8) | 29 (35.4) |
| Ever | 377(66.8) | 324 (67.2) | 53 (64.6) |
| Index cancer stage | |||
| I or II | 36(6.4) | 33 (6.9) | 3 (3.7) |
| III or IV | 528(93.6) | 449 (93.1) | 79 (96.3) |
| Comorbidity | |||
| None or mild | 511(90.6) | 438 (90.9) | 73 (89.0) |
| Moderate to severe | 53(9.4) | 44 (9.1) | 9 (11.0) |
| Treatment | |||
| Radiotherapy only | 133(23.6) | 117 (24.3) | 16 (19.5) |
| Radiochemotherapy | 431(76.4) | 365 (75.7) | 66 (80.5) |
| Death, all causes | |||
| Yes | 78(14.0) | 66 (13.7) | 12 (14.8) |
| No | 485(86.0) | 416 (86.3) | 69 (85.2) |
| Death, owing to disease | |||
| Yes | 44(8.0) | 37 (7.7) | 7 (8.6) |
| No | 519(92.0) | 445 (92.3) | 74 (91.4) |
| Recurrence | |||
| Yes | 64(11.4) | 55 (11.4) | 9 (11.0) |
| No | 500(88.6) | 427 (88.6) | 73 (89.0) |
| Follow up (Months) | |||
| Median | 34.5 | 35.2 | 31.2 |
| Range | 0.1–207.0 | 1.3–207.0 | 0.1–77.2 |
Association of MDM4 rs4245739 genotypes with tumor HPV16-positive SCCOP patients
The genotype distributions for MDM4 rs4245739 polymorphism are shown in Table 2. HPV16-positive patients were more likely to have the variant genotypes for MDM4 rs4245739 compared with the HPV16-negative patients (P < 0.05). The patients with the variant AC+CC genotypes of MDM4 rs4245739 had an approximately 3.2-fold higher risk, of having an HPV16-positive tumor compared with the patients with the corresponding common AA homozygous genotype (aOR, 3.2, 95% CI, 1.9–5.5).
Table 2.
Association of MDM4 variant with tumor HPV16-positive SCCOP patients
| Genotypes | HPV16-positive tumors (n, %) | HPV16-negative tumors (n, %) | cOR†, 95% CI | aOR*, 95% CI |
|---|---|---|---|---|
| MDM4rs4245739 | ||||
| AAb | 270 (92.1) | 23 (7.9) | 1.0 | 1.0 |
| AC + CC | 212 (78.2) | 59 (21.8) | 3.3 (2.0–5.5) | 3.2 (1.9–5.5) |
Reference group.
Crude OR.
Adjusted for sex, age, ethnicity, smoking and alcohol drinking status.
MDM4 rs4245739 genotypes are associated with survival of SCCOP patients
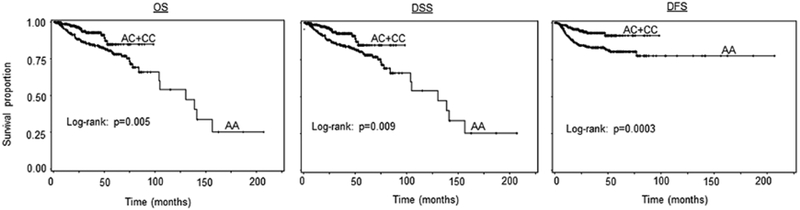
The genotype distributions for MDM4 rs4245739 polymorphism and associations with survival in 564 SCCOP patients are shown in Table 3. The AC/CC variant genotypes of MDM4 rs4245739 were significantly associated with better OS, DSS, and DFS than the corresponding common homozygous AA genotype (log-rank P = 0.005, 0.009, and 0.0003 for OS, DSS, and DFS, respectively, Fig. 1). The multivariable Cox regression analysis was adjusted with other important confounders, including age, sex, ethnicity, smoking, alcohol, disease stage, comorbidity, and HPV status and treatment (Table 3). The significant associations of OS with treatment (aHR, 1.7, 95%CI, 1.1–3.2) and tumor HPV status (aHR, 0.5, 95%CI, 0.3–0.9) were observed, while such significant associations were not found for other prognostic factors (aHR, 2.6, 95%CI, 0.9–7.4 for sex, 1.5, 0.9–2.4 for age, 1.9, 0.9–3.8 for ethnicity, 1.0, 0.5–1.4 for smoking, 0.6, 0.3–1.1 for alcohol drinking, 0.9, 0.3–3.1 for disease stage, and 0.9, 0.4–2.1 for comorbidity, respectively. The similar trend in associations of these confounding factors with DSS and DFS was found. After adjustment with these covariates, we found that after chemoradiation the patients with AC/CC variant genotypes of MDM4 rs4245739 had reduced risk of overall death (HR, 0.4, 95% CI, 0.3–0.8), death due to disease (HR, 0.4, 95% CI, 0.2–0.8), and disease recurrence (HR, 0.4, 95% CI, 0.2–0.7) compared with the patients with AA genotype.
Table 3.
Association of MDM4 variant with OS, DSS, and DFS of SCCOP patients (N = 564)
| Genotypes | OS | DSS | DFS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Overall death/Total | P | cHRa (95% CI) | aHR* (95% CI) | Death, owing to disease/Total | P | cHRa (95% CI) | aHR* (95% CI) | Recc./Total | P | cHRa (95% CI) | aHR* (95% CI) | |
| MDM4rs4245739 | 0.005 | 0.009 | 0.0003 | |||||||||
| AAb | 60/293 | 1.0 | 1.0 | 34/293 | 1.0 | 1.0 | 48/293 | 1.0 | 1.0 | |||
| AC + CC | 18/271 | 0.5(0.3–0.8) | 0.4(0.3–0.8) | 10/271 | 0.4(0.1–0.8) | 0.4(0.2–0.8) | 16/271 | 0.4(0.2–0.8) | 0.4(0.2–0.7) | |||
P: log-rank test.
Crude HR.
Hazard ratio adjusted for age, sex, ethnicity, smoking status, alcohol use status, stage, comorbidity, HPV16 status, and treatment.
Reference group.
Rec.: Recurrence
Figure 1.
Survival analysis by MDM4 rs4245739 genotypes among all SCCOP patients (N = 564).
Association between MDM4 rs4245739 genotypes and survival of tumor HPV-positive and HPV-negative SCCOP patients
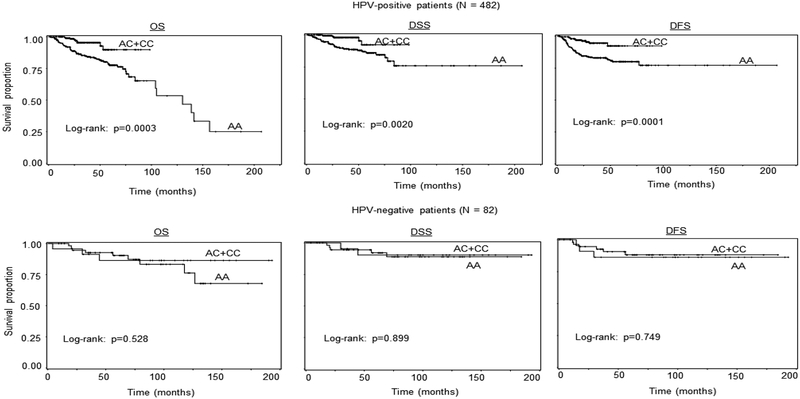
As shown in Table 2, MDM4 rs4245739 AC+CC genotypes were more likely to be tumor HPV-positive SCCOP patients compared with AA genotype (OR, 3.2, 95%CI, 2.0–5.5). Because approximately 85% of SCCOP patients were tumor HPV16-positive SCCOP cases in this study patient cohort, we assessed the effect of MDM4 rs4245739 polymorphism on survival in HPV16-positive cases only. As shown in Figure 2, the Kaplan–Meier univariate survival analysis showed that SCCOP patients with the MDM4 rs4245739 AC+CC variant genotypes had significantly better OS, DSS, and DFS than those with the corresponding common homozygous AA genotype (log-rank: all P < 0.01). Moreover, the multivariable Cox regression analysis demonstrated significantly associations of this MDM4 genetic variant with OS, DSS, and DFS among 482 HPV16-positive SCCOP patients and 82 HPV16-negative cases (Table 4). With adjustment of other important confounding variables, we found that the patients carrying the MDM4 rs4245739 AC+CC variant genotypes had significantly reduced risk of overall death, death from SCCOP, and recurrence (HR, 0.3, 95% CI, 0.1–0.6 for OS; HR, 0.3, 95% CI, 0.1–0.7 for DSS; and HR, 0.3, 95% CI, 0.1–0.6 for DFS, respectively) than those with MDM4 rs4245739 AA genotype as shown in Table 4. Additionally, in this study, we also performed survival analysis in tumor HPV-negative SCCOP patients, and we did not find the similarly significant associations partially due to either small sample size or few outcome events in this small subgroup.
Figure 2.
Survival analysis by MDM4 rs4245739 genotypes among 482 tumor HPV16-positive and 82 HPV16-negative SCCOP patients.
Table 4.
Association of MDM4 variant with OS, DSS, and DFS of SCCOP patients stratified by tumor HPV16 status
| Genotypes | OS | DSS | DFS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Overall death/Total | Log-rank P value | aHRa (95% CI) | Death, owing to disease/Total | Log-rank P value | aHRa (95% CI) | Recc./Total | Log-rank P value | aHRa (95% CI) | |
| Tumor HPV16-positive SCCOP patients (N = 482) | |||||||||
| MDM4rs4245739 | 0.0003 | 0.002 | 0.0001 | ||||||
| AAb | 57/270 | 1.0 | 32/270 | 1.0 | 45/270 | 1.0 | |||
| AC + CC | 9/212 | 0.3(0.1–0.6) | 5/212 | 0.3 (0.1–0.7) | 10/212 | 0.3(0.1–0.6) | |||
| Tumor HPV16-negative SCCOP patients (N = 82) | |||||||||
| MDM4rs4245739 | 0.528 | 0.899 | 0.749 | ||||||
| AAb | 3/23 | 1.0 | 2/23 | 1.0 | 3/23 | 1.0 | |||
| AC + CC | 9/59 | 2.0 (0.4–8.9) | 5/59 | 4.1(0.5–36.5) | 6/59 | 1.2(0.2–6.4) | |||
Hazard ratio adjusted for age, sex, ethnicity, alcohol use status, stage, comorbidity, and treatment.
Reference group.
Rec.: Recurrence
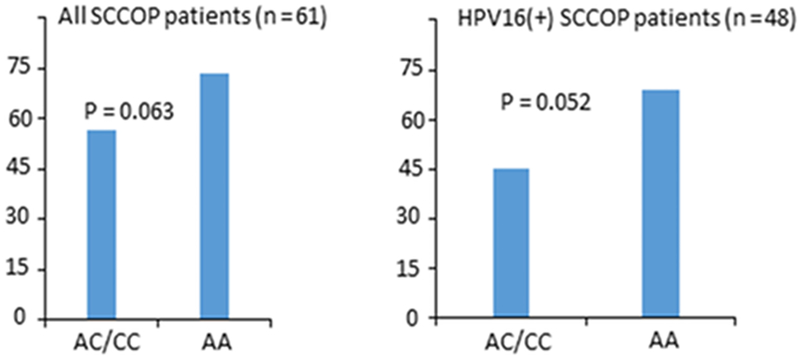
To further characterize the potentially functional relevance of this polymorphism, we performed correlation analyses between tumor MDM4 protein expression by IHC and genotypes of this polymorphism among a subset of 61 SCCOP patients. Among 61 SCCOP patients, 48 cases had HPV16 positive tumors and 13 were tumor HPV16-negative. Overall, MDM4 rs4245739 AC/CC variant genotypes had a lower MDM4 protein expression than MDM4 rs4245739 AA genotype among all 61 SCCOP patients although the difference did not reach the statistically significant level (P = 0.063). However, after we stratified the patients by tumor HPV16 status, we found that HPV16 positive SCCOP patients with AC/CC genotypes had a borderline significantly lower MDM4 protein expression than those with MDM4 rs4245739 AA genotype (P = 0.052) as shown in Fig 3. Additionally, we also assessed correlation between MDM4 and miR-191 gene expression levels in the Framingham Heart Study (FHS) Offspring Exam 8 cohort (n=1902). Our analysis showed that the MDM4 expression is negatively correlated with miR-191 (cor. = −0.04; P = 0.0699, N = 1902). Furthermore, the expression level of MDM4 and miR-191 by AC/CC genotype was weakly negatively correlated with each other in the dominant genotypic group (cor. = −0.05; P = 0.12; N = 909). These results indicated a moderate correlation between MDM4 and miR-191 expression levels, which might partially support our biological assumption that increased binding of miR-191 on 3’-UTR of MDM4 with AC/CC genotype may result in its degradation and thereby increases p53 activity.
Figure 3.
Comparison between MDM4 expression levels and MDM4 rs4245739 genotypes among all 61 SCCOP patients and 48 tumor HPV16(+) SCCOP patients.
Discussion
In this study, we have identified MDM4 rs4245739 variant to be associated with tumor HPV16 status and survival of SCCOP, particularly for HPV(+) SCCOP. Thus, this polymorphism likely alters the regulation of MDM4 and its related molecular activities such as apoptosis, and such changes may affect HPV16 clearance and escape from immune surveillance and clinical outcome. Our findings support that MDM4 rs4245739 genetic polymorphism is more likely biologically functional and might affect the individual differences in tumor HPV16 status and survival of SCCOP patients after definitive radiotherapy.
MDM4, a MDM2 homologue, is located on chromosome1q32 and is a p53-interacting protein [47,48]. It shares a NH2-terminal p53-binding domain with MDM2 and negatively affects expression and activity of p53 in different human tumors [47]. Because of lack of appreciable ubiquitin ligase activity for MDM4, it is biologically different that MDM4 majorly contributes to inhibition of p53-mediated transcriptional transactivation, but MDM2 largely contributes to degradation of p53 [48]. Moreover, MDM4 may bind to MDM2 to enhance the ability of MDM2 for regulation of p53. Thus, it is likely that future work can develop novel molecules that target cancers that contain inactive wild-type p53 through blockage of p53-MDM2/MDM4 interactions.
While tumor HPV status remains one of the strongest biomarkers for predicting outcome of SCCOP, identification of novel and robust biomarkers for predicting HPV status in SCCOP is clinically useful. As HPV-positive SCCOP patients have heterogeneous clinical outcomes[25], it is possible for physicians to optimize patient stratification for HPV-positive SCCOP patients by this MDM4 variant for better personalized treatment.
This current study found that MDM4 rs4245739 was associated with tumor HPV16 status in SCCOP. Although we and others have reported associations of some MDM4 genetic variants with risk and outcomes of head and neck cancers or HPV-associated head and neck cancers [37,40,41,49,50], no studies have been focused on this functional polymorphism. Additionally, most of previous studies either had different HPV16 status of study patients or had heterogeneous tumor sites [43,51–53]. Thus, it is likely that additional confounders may bias the estimates of the associations. Our results from the current study demonstrated that MDM4 rs4245739 might serve as a biomarker for HPV16-positive tumors of SCCOP.
To date, we do not know how this polymorphism affects the function of MDM4 and how it exactly affects tumor HPV16 status and survival of SCCOP, whereas it is biologically plausible that HPV16 E6 and MDM4 may jointly affect these outcomes of SCCOP through biological activities via regulation of p53-dependent pathway. The reason why this MDM4 polymorphism modifies these outcomes of SCCOP patients could be that the functional rs4245739 SNP A>C locating in the 3’-untrasnlated (3’-UTR) region of MDM4 forms a miR-191 targeting site [54]. MiR-191 may bind to 3’-UTR of MDM4 in favor of rs4245739 C allele; and such changes might lead to a reduced expression of MDM4, thus subsequently increasing the activities of p53.
MDM4 as an oncoprotein plays a negatively role in the regulation of p53; and its overexpression can result in cancer progression and development[36]. However, whether the MDM4-dependent pathway in p53 degradation is active in HPV positive tumors or whether degradation of p53 depends entirely on the HPV16 E6-dependent pathway in such tumors remains unclear. In HPV positive cells, the HPV16 E6-dependent pathway of p53 degradation was not only active but was required for p53 degradation, while MDM2-dependent pathway was actually inactive or less active [55, 56]. In another study, HPV16 E6 mediated degradation was dominant over MDM2 in cervical cancer [57]. The E6 might differently induce p53 degradation from the way as MDM2 to promote p53 degradation, indicating important differences between these two pathway of MDM2 and HPV16 E6 of p53 degradation [58]. As MDM4 is a closely related protein to MDM2 in regulation of the p53 pathway, we assumed that MDM4 might be in a process analogous to these MDM2-mediated p53 degradation in HPV positive SCCOP. It is also likely that MDM4 may interact with HPV16 E6 in p53 degradation in other different pathways, which require further investigation. Recent evidence supported that the variation in the 3’-UTR of MDM4 can lead to a decreased risk of many types of cancer. For example, the rs4245739 C minor allele in the 3’-UTR of MDM4 has been demonstrated to reduce the risk of cancer, progression and improve clinical outcomes[36]. Moreover, many functional studies have demonstrated that the C minor allele creates a new binding site for miR-191[36], which results in a reduced expression of MDM4. A more recent meta-analysis with a large sample size supported as association of this polymorphism with a decreased risk of cancer[36].
Biologically, HPV E6 can inhibit p53 through proteasomal degradation[59], and MDM4 can directly bind to the p53 transactivation domain to inhibit activities of p53. Therefore, the down-expression of MDM4 could increase the p53 activity, and functionally influence different p53-related pathways, thus leading to changes of several p53-dependent cellular activities (e.g., cell growth suppression, apoptosis induction) by affecting the interaction between p53 and HPV16 through degradation or inactivation of HPV16 E6 oncoprotein [60]. Thus, it is biologically likely that this specific MDM4 variant could affect association with tumor HPV16 status and survival in SCCOP through the interplay between p53 tumor suppressor and HPV16 E6 oncogene.
While HPVs can bypass immune systems; and MDM4 rs4245739 polymorphism can affect the apoptotic capacity though p53-dependent pathway, these two molecular pathways thus may jointly control the HPV clearance through escape of immune surveillance, thus eventually affecting the SCCOP patient’s tumor HPV status and outcome[61]. Thus, we speculate the AC+CC variant genotypes of MDM4 rs4245739 may cause the reduced MDM4 expression to affect the regulation of p53-dependent pathways and p53-dependent activities, such as apoptotic response. Thus, MDM4 rs4245739 AC+CC genotypes might alter regulation in these pathways not to enable many HPV-infected cells to escape or counterattack against the immune system, resulting in more likely HPV-positive tumors and better clinical outcomes. In the current study, we found that patients with MDM4 rs4245739 AC+CC variant genotypes had favorable prognosis in HPV16 positive SCCOP patients, which is consistent with our current finding that these AC+CC variant genotypes are significantly associated with HPV16 positive SCCOP tumors. HPV positive SCCOP patients typically have few somatic genetic changes (e.g., intact p53), compared to those with HPV negative SCCOP, whose tumors are majorly caused by smoking and commonly have somatic mutations such as p53 mutations. Thus, these patients with p53 mutations likely have a poor response to radiotherapy, partially due to inactivation of the p53-mediated apoptotic pathway, while HPV-positive SCCOP patients harboring intact p53 have better response to chemoradiotherapy or radiation treatment, due to induction of apoptosis for tumor cells. Thus, it is our speculation that HPV-positive SCCOP patients carrying MDM4 rs4245739 AC+CC variant genotypes will have higher apoptotic efficacy than patients with the corresponding common AA genotype; and subsequently have a better response to chemoradiotherapy, leading to a better survival. Nevertheless, these findings are warranted for further validation in well-designed prospective studies.
In this study, we also found that the AC+CC genotype patients had follow-up that was only half as long as follow-up for AA genotype patients as shown in Fig 1 and Fig 2. We found that the patients with AC/CC genotypes did have shorter follow up time than those with AA genotype. Moreover, the last censored patient with AC/CC genotypes was tumor HPV(+), and had shorter follow up (approximately 8.5 years) due to lost to follow up, thus resulting in that the Kaplan-Meier curves appeared to end quite differently.
Some potential strengths and limitations of this study should be considered. This study has several strengths including: 1) a homogenous group of patients with SCCOP tumors only; 2) determination of tumor HPV16 status instead of serological status; and 3) well controlled quality for MDM4 genotyping. Hopefully these strengths may help prevent the bias in associations likely from the confounding factors and may also help improve accuracy of the associations in the current study. Despite of these several strengths, we should interpret our findings with caution due to several limitations. These limitations include: 1) no frequency matching on several variables between HPV16-positive and HPV16-negative patients; 2) a possible selection bias due to inclusion of a relative small sample size; 3) certain degree of potential misclassification for tumor HPV16 status with the limitation of current determination methods available for HPV; 4) not counting for other unknown potential confounders due to the hospital-based cohort; and 5) no similar effect of this polymorphism on survival in HPV-negative patients due to either no significant association between them or small sample size, misclassification of HPV status by other high-risk HPV types, and few events of outcome among this HPV-negative subgroup. Therefore, future larger well-designed and prospective, or multicenter studies are necessary to verify our findings.
In conclusion, this study may indicate that MDM4 rs4245739 could be a biomarker for tumor HPV16-positivity of patients with SCCOP. Moreover, this variant might be a useful prognostic biomarker of SCCOP patients, particularly HPV16-positive SCCOP patients. Nevertheless, future larger, well-designed prospective studies are warranted for more accurate evaluation of clinical validity and utility of this biomarker before clinical implementation.
Acknowledgments:
The authors gratefully thank Mr. Joseph A Munch from Scientific Publication Department at the UT MD Anderson Cancer Center for article editing and Ms. Yingdong Li for laboratory support.
Funding: This work was supported by the National Institutes of Health R01 ES-11740 (to Q.W.), CA133099 (to G.L.), and CA186261–01A1 (to G.L.)
Abbreviations:
- MDM4
mouse double minute 4
- miRNA
microRNA
- SCCOP
squamous cell carcinoma of the oropharynx
- SCCHN
squamous cell carcinomas of the head and neck
- HPV
human papillomavirus
- OR
odds ratio
- CI
confidence intervals
- HR
hazard ratio
- 3’ UTRs
3’ untranslated regions
Footnotes
Conflict of Interest: None declared.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
References
- 1.Chaturvedi AK, Engels EA, Pfeiffer RM et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. Journal of clinical oncology 2011;29(32):4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mork J, Lie AK, Glattre E et al. Human papillomavirus infection as a risk factor for squamous-cell carcinoma of the head and neck. New England Journal of Medicine 2001;344(15):1125–1131. [DOI] [PubMed] [Google Scholar]
- 3.Gillison ML, Koch WM, Capone RB et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. Journal of the National Cancer Institute 2000;92(9):709–720. [DOI] [PubMed] [Google Scholar]
- 4.Viens LJ. Human papillomavirus–associated cancers—United States, 2008–2012. MMWR Morbidity and mortality weekly report 2016;65. [DOI] [PubMed] [Google Scholar]
- 5.Herrero R, Castellsagué X, Pawlita M et al. Human papillomavirus and oral cancer: the International Agency for Research on Cancer multicenter study. Journal of the National Cancer Institute 2003;95(23):1772–1783. [DOI] [PubMed] [Google Scholar]
- 6.Pintos J, Franco EL, Black MJ, Bergeron J, Arella M. Human papillomavirus and prognoses of patients with cancers of the upper aerodigestive tract. Cancer: Interdisciplinary International Journal of the American Cancer Society 1999;85(9):1903–1909. [DOI] [PubMed] [Google Scholar]
- 7.Báez A, Almodovar JI, Cantor A et al. High frequency of HPV16‐associated head and neck squamous cell carcinoma in the Puerto Rican population. Head & Neck: Journal for the Sciences and Specialties of the Head and Neck 2004;26(9):778–784. [DOI] [PubMed] [Google Scholar]
- 8.Koskinen WJ, Chen RW, Leivo I et al. Prevalence and physical status of human papillomavirus in squamous cell carcinomas of the head and neck. International Journal of Cancer 2003;107(3):401–406. [DOI] [PubMed] [Google Scholar]
- 9.Rischin D, Young RJ, Fisher R et al. Prognostic significance of p16INK4A and human papillomavirus in patients with oropharyngeal cancer treated on TROG 02.02 phase III trial. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2010;28(27):4142–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bansal D, Vineetha R, Pai KM. Postradiation trismus and its impact on quality of life in patients with head and neck cancer: a commentary. Oral surgery, oral medicine, oral pathology and oral radiology 2016;121(2):196–197. [DOI] [PubMed] [Google Scholar]
- 11.SEER(2007). Surveillance, Epidemiology and End Results stat fact sheets: National Cancer Institute. 2007.
- 12.Alho OP HK ea. Clinical Oncology, 3rd ed ed.: Elsevier Churchill Livingston; 2004. [Google Scholar]
- 13.Alho OP, Hannula K, Luokkala A, Teppo H, Koivunen P, Kantola S. Differential prognostic impact of comorbidity in head and neck cancer. Head & neck 2007;29(10):913–918. [DOI] [PubMed] [Google Scholar]
- 14.Baatenburg de Jong RJ, Hermans J, Molenaar J, Briaire JJ, le Cessie S. Prediction of survival in patients with head and neck cancer. Head & neck 2001;23(9):718–724. [DOI] [PubMed] [Google Scholar]
- 15.Piccirillo JF, Lacy PD, Basu A, Spitznagel EL. Development of a new head and neck cancer-specific comorbidity index. Archives of otolaryngology--head & neck surgery 2002;128(10):1172–1179. [DOI] [PubMed] [Google Scholar]
- 16.Christensen AJ, Moran PJ, Ehlers SL, Raichle K, Karnell L, Funk G. Smoking and drinking behavior in patients with head and neck cancer: effects of behavioral self-blame and perceived control. Journal of behavioral medicine 1999;22(5):407–418. [DOI] [PubMed] [Google Scholar]
- 17.Matthias C, Harreus U, Strange R. Influential factors on tumor recurrence in head and neck cancer patients. European archives of oto-rhino-laryngology : official journal of the European Federation of Oto-Rhino-Laryngological Societies (EUFOS) : affiliated with the German Society for Oto-Rhino-Laryngology - Head and Neck Surgery 2006;263(1):37–42. [DOI] [PubMed] [Google Scholar]
- 18.Browman GP, Mohide EA, Willan A et al. Association between smoking during radiotherapy and prognosis in head and neck cancer: a follow-up study. Head & neck 2002;24(12):1031–1037. [DOI] [PubMed] [Google Scholar]
- 19.Carvalho AL, Nishimoto IN, Califano JA, Kowalski LP. Trends in incidence and prognosis for head and neck cancer in the United States: a site-specific analysis of the SEER database. Int J Cancer 2005;114(5):806–816. [DOI] [PubMed] [Google Scholar]
- 20.Marur S, Forastiere AA. Head and neck cancer: changing epidemiology, diagnosis, and treatment. Mayo Clinic proceedings 2008;83(4):489–501. [DOI] [PubMed] [Google Scholar]
- 21.Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. The New England journal of medicine 2001;345(26):1890–1900. [DOI] [PubMed] [Google Scholar]
- 22.Goy J, Hall SF, Feldman-Stewart D, Groome PA. Diagnostic delay and disease stage in head and neck cancer: a systematic review. The Laryngoscope 2009;119(5):889–898. [DOI] [PubMed] [Google Scholar]
- 23.Cooper JS, Porter K, Mallin K et al. National Cancer Database report on cancer of the head and neck: 10-year update. Head & neck 2009;31(6):748–758. [DOI] [PubMed] [Google Scholar]
- 24.Burgess A, Chia KM, Haupt S, Thomas D, Haupt Y, Lim E. Clinical Overview of MDM2/X-Targeted Therapies. Frontiers in oncology 2016;6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou G, Liu Z, Myers JN. TP53 Mutations in Head and Neck Squamous Cell Carcinoma and Their Impact on Disease Progression and Treatment Response. Journal of cellular biochemistry 2016;117(12):2682–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wasylishen AR, Lozano G. Attenuating the p53 Pathway in Human Cancers: Many Means to the Same End. Cold Spring Harbor perspectives in medicine 2016;6(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends in cell biology 2010;20(5):299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nature reviews Cancer 2013;13(2):83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marine JC, Jochemsen AG. Mdmx as an essential regulator of p53 activity. Biochemical and biophysical research communications 2005;331(3):750–760. [DOI] [PubMed] [Google Scholar]
- 30.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nature reviews Cancer 2006;6(12):909–923. [DOI] [PubMed] [Google Scholar]
- 31.Sharp DA, Kratowicz SA, Sank MJ, George DL. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. The Journal of biological chemistry 1999;274(53):38189–38196. [DOI] [PubMed] [Google Scholar]
- 32.Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. MDM2 interacts with MDMX through their RING finger domains. FEBS letters 1999;447(1):5–9. [DOI] [PubMed] [Google Scholar]
- 33.Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell death and differentiation 2006;13(6):927–934. [DOI] [PubMed] [Google Scholar]
- 34.Bao J, Nanding A, Song H, Xu R, Qu G, Xue Y. The overexpression of MDM4: an effective and novel predictor of gastric adenocarcinoma lymph node metastasis. Oncotarget 2016;7(41):67212–67222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salvi S, Calistri D, Gurioli G et al. Copy number analysis of 24 oncogenes: MDM4 identified as a putative marker for low recurrence risk in non muscle invasive bladder cancer. International journal of molecular sciences 2014;15(7):12458–12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moszynska A, Gebert M, Collawn JF, Bartoszewski R. SNPs in microRNA target sites and their potential role in human disease. 2017;7(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu H, Wang LE, Liu Z et al. Polymorphisms of MDM4 and risk of squamous cell carcinoma of the head and neck. Pharmacogenetics and genomics 2011;21(7):388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang MY, Jia M, He J et al. MDM4 genetic variants and risk of gastric cancer in an Eastern Chinese population. Oncotarget 2017;8(12):19547–19555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun T, Lee GS, Oh WK et al. Single-nucleotide polymorphisms in p53 pathway and aggressiveness of prostate cancer in a Caucasian population. Clinical cancer research : an official journal of the American Association for Cancer Research 2010;16(21):5244–5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu H, Sturgis EM, Liu Z, Wang LE, Wei Q, Li G. Modifying effect of MDM4 variants on risk of HPV16-associated squamous cell carcinoma of oropharynx. Cancer 2012;118(6):1684–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu Z, Sturgis EM, Zhu L et al. Mouse double minute 4 variants modify susceptibility to risk of recurrence in patients with squamous cell carcinoma of the oropharynx. 2018;57(3):361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guan X, Sturgis EM, Lei D et al. Association of TGF-beta1 genetic variants with HPV16-positive oropharyngeal cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2010;16(5):1416–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji X, Sturgis EM, Zhao C, Etzel CJ, Wei Q, Li G. Association of p73 G4C14-to-A4T14 polymorphism with human papillomavirus type 16 status in squamous cell carcinoma of the head and neck in non-Hispanic whites. Cancer 2009;115(8):1660–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu J, Tang X, Li M et al. Functional MDM4 rs4245739 genetic variant, alone and in combination with P53 Arg72Pro polymorphism, contributes to breast cancer susceptibility. Breast cancer research and treatment 2013;140(1):151–157. [DOI] [PubMed] [Google Scholar]
- 45.Liang ZD, Lippman SM, Wu TT, et al. RRIG1 mediates effects of retinoic acid receptor beta2 on tumor cell growth and gene expression through binding to and inhibition of RhoA. Cancer Res 2006;66(2):7111–7118. [DOI] [PubMed] [Google Scholar]
- 46.Tao Y, Sturgis EM, Huang Z et al. TGFbeta1 Genetic Variants Predict Clinical Outcomes of HPV-Positive Oropharyngeal Cancer Patients after Definitive Radiotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research 2018;24(9):2225–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barboza JA, Iwakuma T, Terzian T, El-Naggar AK, Lozano G. Mdm2 and Mdm4 loss regulates distinct p53 activities. Molecular cancer research : MCR 2008;6(6):947–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riemenschneider MJ, Knobbe CB, Reifenberger G. Refined mapping of 1q32 amplicons in malignant gliomas confirms MDM4 as the main amplification target. Int J Cancer 2003;104(6):752–757. [DOI] [PubMed] [Google Scholar]
- 49.Jin L, Sturgis EM, Zhang Y et al. Genetic variants in p53-related genes confer susceptibility to second primary malignancy in patients with index squamous cell carcinoma of head and neck. Carcinogenesis 2013;34(7):1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Z, Sturgis EM, Zhang Y et al. Combined p53-related genetic variants together with HPV infection increase oral cancer risk. Int J Cancer 2012;131(3):E251–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ji X, Neumann AS, Sturgis EM et al. p53 codon 72 polymorphism associated with risk of human papillomavirus-associated squamous cell carcinoma of the oropharynx in never-smokers. Carcinogenesis 2008;29(4):875–879. [DOI] [PubMed] [Google Scholar]
- 52.Chen X, Sturgis EM, Etzel CJ, Wei Q, Li G. p73 G4C14-to-A4T14 polymorphism and risk of human papillomavirus-associated squamous cell carcinoma of the oropharynx in never smokers and never drinkers. Cancer 2008;113(12):3307–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X, Sturgis EM, El-Naggar AK, Wei Q, Li G. Combined effects of the p53 codon 72 and p73 G4C14-to-A4T14 polymorphisms on the risk of HPV16-associated oral cancer in never-smokers. Carcinogenesis 2008;29(11):2120–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang MJ, Luo YJ, Shi ZY et al. The associations between MDM4 gene polymorphisms and cancer risk. Oncotarget 2016;7(34):55611–55623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hengstermann A, Linares LK, Ciechanover A, Whitaker NJ, Scheffner M. Complete switch from Mdm2 to human papillomavirus E6-mediated degradation of p53 in cervical cancer cells. Proc Natl Acad Sci U S A 2001; 98(3):1218–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hietanen S, Lain S, Krausz E, Blattner C, Lane DP. Activation of p53 in cervical carcinoma cells by small molecules. Proc Natl Acad Sci U S A 2000;97(15):8501–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Traidej M, Chen L, Yu D, Agrawal S, Chen J. The roles of E6-AP and MDM2 in p53 regulation in human papillomavirus-positive cervical cancer cells. Antisense Nucleic Acid Drug Dev 2000; 10(1):17–27. [DOI] [PubMed] [Google Scholar]
- 58.Camus S, Higgins M, Lane DP, Lain S. Differences in the ubiquitination of p53 by Mdm2 and the HPV protein E6. FEBS Lett 2003;;536(1–3):220–224. [DOI] [PubMed] [Google Scholar]
- 59.Thomas M, Pim D, Banks L. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene 1999;18(53):7690–7700. [DOI] [PubMed] [Google Scholar]
- 60.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell 1993;75(3):495–505. [DOI] [PubMed] [Google Scholar]
- 61.Favre A, Paoli D, Poletti M et al. The human palatine tonsil studied from surgical specimens at all ages and in various pathological conditions. 1. Morphological and structural analyses. Zeitschrift fur mikroskopisch-anatomische Forschung 1986;100(1):7–33. [PubMed] [Google Scholar]