Abstract
Alternative splicing of pre-mRNA is an essential post- and co-transcriptional mechanism of gene expression regulation that produces multiple mature mRNA transcripts from a single gene. Genetic mutations that affect splicing underlie numerous devastating diseases. The complexity of splicing regulation allows for multiple therapeutic approaches to correct disease-associated mis-splicing events. In this review, we first highlight recent findings from therapeutic strategies that have used splice switching antisense oligonucleotides and small molecules that bind directly to RNA. Second, we summarize different genetic and chemical approaches to target components of the spliceosome to correct splicing defects in pathological conditions. Finally, we present an overview of compounds that target kinases and accessory pathways that intersect with the splicing machinery. Advancements in the understanding of disease-specific defects caused by mis-regulation of alternative splicing will certainly increase the development of therapeutic options for the clinic.
Keywords: alternative splicing, antisense oligonucleotides, RNA therapeutics, spliceosome, RNA binding proteins, RNA binding drugs
1. Introduction
The initial product of gene transcription is an immature pre-mRNA that contains non-coding intronic sequences interspersed between coding exonic sequences. The maturation of mRNA requires removal of intronic sequences and splicing together of adjacent exons prior to translation into protein. This process is termed constitutive splicing and occurs at every intron-exon boundary [1]. In the late 1970’s and early 1980’s several groups unexpectedly observed multiple species of mRNAs that contained non-intervening sequences from a single gene [2–4]. The most plausible explanation suggested that a previously unknown pre-mRNA processing step exists that functions to excise specific exons and splice together non-adjacent exons. This process, now ubiquitously known as alternative splicing, is an important evolutionarily conserved mechanism that increases proteome complexity from a limited genome.
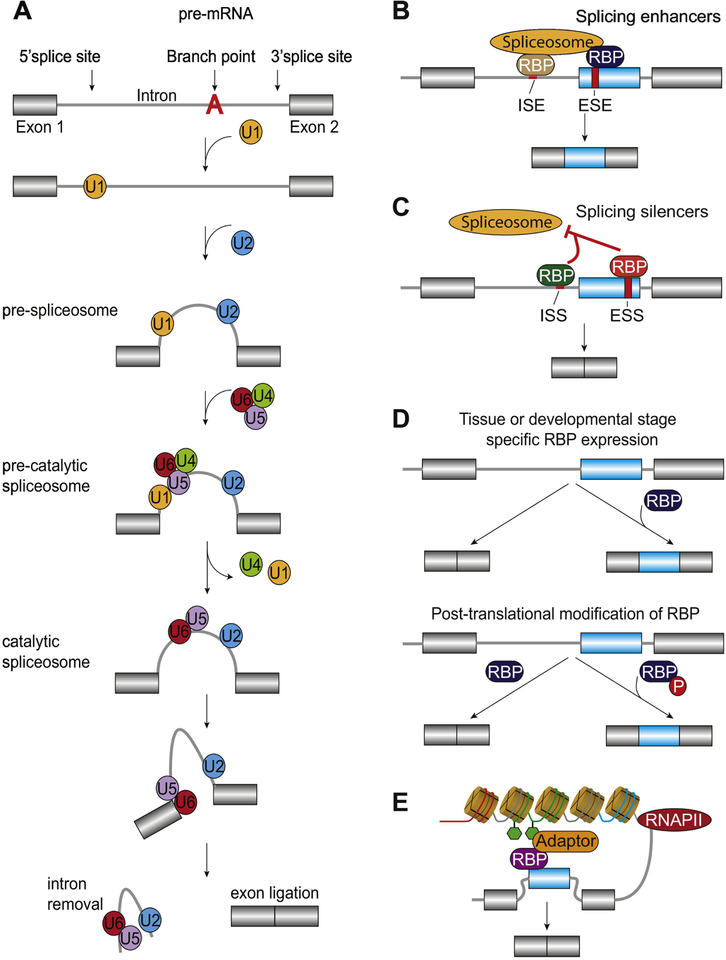
Constitutive splicing relies on the recognition of exon boundaries and is controlled by the spliceosome, a complex ribonucleoprotein molecular machine. The spliceosome is composed of over 150 proteins, in addition to the catalytic five small-nuclear ribonucleoproteins (snRNPs) U1, U2, U4, U5, and U6. Consensus sequences in the pre-mRNA ensure that spliceosome assembly occurs properly. The branch point sequence along with the 5’ and 3’ splice sites (5’SS and 3’SS, respectively) are the core sequences necessary for spliceosome recognition and assembly. The branch point sequence is located 18–40 nucleotides upstream of the 3’SS followed by a polypyrimidine tract [5]. Assembly of the molecular machinery occurs through a series of reactions, the first is the recognition of the 5’SS by the U1 snRNP creating the early complex. The prespliceosomal complex is subsequently formed by recognition of the U2 snRNP onto the branch point sequence. The final step is completed through the recruitment and exchange of the tri-snRNP U4/U6–U5 to excise the intron and ligate remaining exonic sequences through transesterification reactions (Fig. 1A).
Figure 1.
Regulation of constitutive and alternative splicing. A. Assembly of the spliceosome begins with the recognition of the 5’splice site by the U1 snRNP. The pre-spliceosome complex is formed by recognition of U2 snRNP onto the branch point sequence. The pre-catalytic spliceosome is created by recruitment of the complex formed of snRNP of U4, U5 and U6. The catalytic spliceosome is completed through dissociation of U4 and U1. The final step is to excise the intronic regions and ligate remaining exonic sequences through transesterification reactions. Adapted from Shi [154]. B-C. Binding of RNA binding proteins (RBPs) to intronic or exonic splicing enhancers (ISE and ESE, respectively) or silencers (ISS and ESS, respectively) promotes exon retention (B) and skipping (C), respectively [155]. D. RBP expression is regulated in a tissue and developmental specific manner and post-translational modifications such as phosphorylation (shown as P in red) impact their ability to recognize cis-regulatory elements [155]. E. Alternative splicing is coupled to RNA polymerase II (RNAPII) kinetics and epigenetic modifications through adaptor proteins that concurrently recognize histone modifications and RBPs. Adapted from Luco et al. [6].
Alternative splicing reactions occur in a manner similar to constitutive splicing; however, several factors influence the regulation of splice site selection [6–8]. Alternatively spliced exons often have weaker splice sites, i.e. different than the consensus sequences, and therefore are not as well recognized by the spliceosome, compared to constitutive exons [9,10]. Additionally, other sequences inherent to the pre-mRNA (cis-regulatory elements) recruit trans-acting RNA binding proteins (RBPs) and splicing factors in a sequence specific manner [11–14]. The cis-regulatory elements are classically grouped by their location and regulatory function. Regulation of splicing can occur through RBPs bound to splice enhancing motifs found within an intron or exon (ISE and ESE, respectively). This stabilizes the formation of the spliceosome and promotes the recognition and retention of an exon (Fig. 1B). Contrastingly, RBPs bound to splice silencing motifs within an intron or exon (ISS and ESS, respectively) prevents spliceosomal formation and promotes removal of an exon [9,15–17] (Fig. 1C). Expression patterns of RBPs differ by tissue type and developmental stage (Fig. 1D). Furthermore, numerous RBPs are rich in serine and threonine residues, making them attractive targets for phosphorylation. Phosphorylation of RBPs can alter their subcellular localization, splice site specificity, stability, or their ability to interact with the spliceosome and other accessory proteins [18] (Fig. 1D).
The contribution of cis-regulatory elements, RBPs, and splicing factors in deciding splice site selection does not fully explain the regulatory mechanisms governing alternative splicing [6]. This suggests that there must be other post- and/or co-transcriptional mechanisms that explain the regulation. Indeed, RNA secondary structure impacts the ability of RBPs to bind mRNA and cooperate in alternative splicing regulation [19–23]. Removal of introns and alternative exons has also been observed prior to polyadenylation of an mRNA [24], suggesting that transcriptional kinetics plays an important role in regulating alternative splicing. In support of this, RNA polymerase II pausing alters splice site selection [25] and chromatin architecture and histone modifications are also major players in alternative splicing regulation [6,26–28]. Current models of alternative splicing suggest that each of these layers of regulation likely act in concert (Fig. 1E).
While the physiological significance of alternative splice forms remain understudied, it is known that harmful human diseases can result when alternative splicing is misregulated [29] due to: (i) alterations in trans-acting factors in diseases such as myotonic dystrophy, and amyotrophic lateral sclerosis (ALS) or (ii) mutations in cis-elements that regulate splicing in diseases such as Hutchinson-Gilford syndrome, and retina pigmentosa. It has been previously shown that 303 genes and 370 diseases are associated with the disruption of alternative splicing and that 2,337 splicing mutations are linked to diseases [30]. One of the most well described alternative splicing-linked diseases associated with altered trans-acting factors is myotonic dystrophy, a multisystemic disorder resulting in muscle weakness and loss, cardiac defects, and neurological features. Myotonic dystrophies are partially caused by the deregulation of the RBPs muscleblind like splicing regulator 1 (MBNL1) and CUGBP Elav-like family member 1 (CELF1) [31,32]. The RBP TDP-43 (TDPBP) is the major disease related protein in ALS [33]. In motor neurons of humans with ALS, TDP-43 is hyperphosphorylated and ubiquitinated. This results in TDP-43 being inappropriately redistributed from the nucleus to the cytoplasm [33]. Splicing alterations are also observed in ALS-linked mutations in TDP-43 [34]. On the other hand, when genetic mutations in cis-elements cause the mis-activation of splice sites, diseases such as Hutchinson-Gilford progeria syndrome are seen [35]. Similarly, retinitis pigmentosa, a common cause of blindness from retinal degradation, has been linked to mutations in the splice sites involved in pre-mRNA processing [36].
As the understanding of the mechanisms behind alternative splicing become clearer, promising new therapeutics are being designed to combat human diseases. Here we review the current knowledge on approaches to correct mis-splicing associated with specific diseases. First, we briefly describe several classes of molecules that bind directly to RNA to re-direct alternative splicing. Next, we describe how dysregulation of the spliceosome contributes to disease and the strategies using small molecules and genetic methods being utilized to correct the splicing defects. We then discuss the development of small molecules that correct inappropriate activity of kinases that regulate alternative splicing reactions. Lastly, we summarize several non-canonical pathways that have been shown to intersect with alternative splicing and the compounds that alter their activity.
2. RNA binding molecules
2.1. Splice-switching oligonucleotides
Several groups have utilized antisense oligonucleotides (ASOs) to therapeutically manipulate alternative splicing. ASOs are small and synthetically made molecules that bind through Watson-Crick base-pairing to a complementary pre-mRNA sequence [37]. ASOs can act in several manners which primarily depend on the location of their binding. Endogenous mRNAs can be targeted for RNase degradation after a RNA-DNA hybrid is created by ASO binding [38]. ASOs can also be used to drive alternative splicing reactions by binding to well characterized intronic or exonic splicing enhancers or silencers in pathologies where aberrant splicing is the underlying cause of the disease. This class of ASOs is known as splice-switching oligonucleotides (SSOs). A recent review in 2016 comprehensively characterized the approaches to therapeutically target alternative splicing using SSOs [39]. We will highlight a few of the latest findings after that review and will focus on spinal muscle atrophy (SMA) and Duchenne’s muscular dystrophy as SSOs therapies for these diseases have made significant progress in clinical trials.
SMA is a genetic disease that in severe cases is fatal within the first two years after birth [40]. The root cause of SMA stems from a marked reduction in survival motor protein (SMN) expression which also correlates with disease severity [41]. Humans have two highly homologous genes, SMN1 and SMN2, that encode for an identical SMN protein [40,42,43]. However, in SMA two different mechanisms account for reduced SMN protein expression. First, humans with SMA have homozygous loss of SMN1 which results in reduced SMN protein expression [40,44] (Fig. 2A, top). Furthermore, SMN2 is unable to compensate for the loss of SMN1 due to a cytosine to thymine transition at the +6 position (C6T) in exon 7 [43,45]. This transition is sufficient to reduce the inclusion of exon 7 [43,45] and thus, the SMN protein generated from SMN2 is shorter and less functional than the one generated from the SMN1 gene [46–48]. The C6T transition in exon 7 of the SMN2 gene creates a novel ESS which promotes binding of heterogeneous nuclear ribonucleoprotein-A1 (HNRNPA1), a known splicing repressor [49]. Binding of HNRNPA1 to the ESS in SMN2 exon 7 prevents proper formation of the U2 snRNP complex and promotes splicing of exon 7 [49] (Fig. 2A, bottom). One therapeutic strategy to treat SMA is to target trans-acting splicing activators and repressors of SMN2 exon 7. A few studies have accomplished this in vitro [50–52] and at least 40 RBPs have been implicated in splicing of exon 7 of the SMN2 gene [53]. While those studies provided crucial information about the molecular mechanisms contributing to SMA development and progression, targeting trans-acting factors has not been an effective strategy in treating SMA. Another approach is to target cis-regulatory elements within the SMN2 pre-mRNA to promote exon 7 inclusion. This was first successfully achieved in 2001 using SSOs directed against the 3’SS of exon 8 [54]. Numerous studies since then have been focused on targeting other cis-regulatory elements using SSOs and have generated successful therapies for SMA. Most notably is the discovery of the intronic splicing silencer N1 (ISS-N1) element in intron 7 downstream of the 5’SS of SMN2 exon 7 [55]. SMN2 exon 7 inclusion is promoted when ISS-N1 is deleted and in the presence of ASOs targeted against this element [55]. These findings led to the development of a highly specific ASO based therapy aiming to promote SMN2 exon 7 inclusion. In late 2016, the United States Food and Drug Administration (FDA) approved Spinraza (nusinersen), an 18-mer SSO that promotes retention of SMN2 exon 7 and increased SMN protein expression, for the treatment of SMA in humans [56,57]. The complex mechanism of action of ISS-N1-targeting SSOs is thoroughly described by Singh et al. [58]. Briefly, binding of Spinraza to ISS-N1 induces secondary structural changes in SMN2 pre-mRNA which blocks HNRNPA1/A2 binding sites and thus prevents removal of SMN2 exon 7 [58,59] (Fig. 2B). Additional structural rearrangements increase the accessibility of binding sites for TIA1 cytotoxic granule associated RNA binding protein (TIA1) which in turn increase U1 recruitment and exon 7 retention [58]. The phosphate linkages and 2’-hydroxyl groups of the ribofuranosyl rings of Spinraza have been chemically modified to phosphorothioate linkages and 2’-O-2-methoxyethyl groups, respectively [39,60,61]. Modifications to SSOs at the 2’-position in the sugar moiety provide resistance against RNase-H degradation [39] and are common in SSOs to extend their biological availability. Spinraza is the first and only FDA approved therapy for humans suffering from SMA.
Figure 2.
RNA binding molecules that modulate alternative splicing. A. Patients with spinal muscular atrophy (SMA) have reduced survival motor neuron (SMN) protein expression as a result of a homozygous deletion of the survival motor neuron-1 gene (SMN1). SMN2 gene is a homolog of SMN1 gene, however exon 7 is often skipped in SMN2 which results in the expression of a less functional SMN protein. The asterisk (*) indicates the presence of a cytosine to thymine transition at the +6 position (C6T) in exon 7 of SMN2 B. SMN protein expression can be restored in humans suffering of SMA using Spinraza. Spinraza is a splice switching oligonucleotide (SSO) that binds to the intronic splicing silencer N1 element (ISS-N1) in SMN2 (red portion of the intron between exon 7 and 8). Binding of Spinraza to ISS-N1 promotes SMN2 exon 7 retention by blocking the binding of a heterogeneous nuclear ribonucleoprotein (HNRNP) to the exonic splicing silencer (ESS) created by the C6T mutation in exon 7. C. Mutations or deletions in the shown exons of the DMD gene are a hallmark of Duchenne’s muscular dystrophy. This causes a frame shift and results in a non-functional dystrophin protein. SSO therapies can induce targeted exonic skipping in this region and realigns the reading frame to produce a more functional dystrophin protein. D. Structural differences between exondys 51 and kyndrisa backbones. Diagrams are based on backbones commonly used in antisense oligonucleotide therapies [39]. E. Proposed mechanisms of action of SMN2 exon 7 retention by small molecules binding to RNA. LMI070 binds the 5’ splice site (5’SS) of exon 7 and increases the affinity of the U1 complex to the 5’SS (left). Binding of SMN-C to two sites in exon 7 recruits the U1 complex and prevents HNRNPG-mediated removal of exon 7 (middle). Binding of SMN-C to a single AGGAAG motif in SMN2 exon 7 recruits far-upstream element binding protein-1 (FUBP1) and the KH-type splicing regulatory protein (KHSRP) to the exon while preventing HNRNPG-mediated removal of exon 7 (right). ex: exon.
Duchenne’s muscular dystrophy is a severe neuromuscular disease caused by mutations in the DMD gene. These mutations perturb the normal reading frame and cause premature stop codons in the mRNA which result in truncated, reduced, or non-functional dystrophin protein [39,62–64]. Dystrophin links the myofibrillar cytoskeleton to the surrounding extracellular matrix [62]. Thus, altered dystrophin levels disrupt structural components necessary for normal muscle architecture. The pathology of Duchenne’s muscular dystrophy is characterized by progressive muscle weakness and atrophy, inflammation, cardiomyopathies, and an early death [62,65]. Numerous mutations in the DMD gene occur between exons 45 and 63 [62]. SSO targeted therapies have focused on promoting exon skipping in this region which restores the open reading frame and produces partially functional dystrophin proteins [62] (Fig. 2C). Targeting exon 51 was selected as a candidate because the frameshift induced by its skipping produces a longer, more functional dystrophin protein compared to transcripts containing exon 51. Furthermore, this strategy has the potential to have clinical benefit for the largest subpopulation (~13%) of all Duchenne’s muscular dystrophy patients [37,62,63,66–68]. In 2016, the FDA approved the use of eteplirsen (exondys 51), a 30-mer phosphomorpholidate oligonucleotide (morpholino), for the treatment of Duchenne’s muscular dystrophy [63,67,69]. Approval was granted based on findings demonstrating that eteplirsen increases the percentage of muscle fibers expressing the dystrophin protein in humans with mutations within exon 51 [70,71]. There were no FDA approved therapies for Duchenne’s muscular dystrophy until this point in time.
Kyndrisa (also known as drisapersen, PRO051, and GSK2402968), sponsored by BioMarin, is another SSO related to eteplirsen and targets a similar region within exon 51 [39]. Phase-II trials demonstrated improvements in a six-minute walk test in boys with Duchenne’s muscular dystrophy that received injections of kyndrisa (clinicaltrials.gov codes and ). Kyndrisa progressed to Phase-III randomized, double blind, placebo-controlled study with larger enrollments and longer study timelines (). Though the treatments were well tolerated by the patients, the results failed to show any significant benefit on walking ability compared to placebo, likely due to increased variation in the data [72] and thus regulatory approval was not given to kyndrisa. Interestingly, a post-hoc analysis of the data from the 48-week Phase-III trial showed improvements in six-minute walking distance in a subpopulation of the patients that have baseline six-minute walking distances of 300–400 meters [72]. This suggests that kyndrisa may in fact be efficacious in Duchenne’s muscular dystrophy patients with a less severe disease progression. Though initially rejected for approval, we are currently unaware of whether or not kyndrisa is being pursued any further as an SSO therapy for Duchenne’s muscular dystrophy after these findings were published.
Kyndrisa and exondys 51 target a similar sequence in the DMD gene to promote exon 51 skipping, however kyndrisa is a 20-mer nucleotide and is shortened at the 5’ and 3’ ends by eight and two nucleotides, respectively, compared to exondys 51 [39]. Additionally, kyndrisa is modified to be a 2’-O-methyl (2’OMe) phosphorothioate oligonucleotide and is not a morpholino like exondys 51 [39,68]. Other SSO-mediated therapies that promote exon skipping between exons 45–63 of the DMD gene have also progressed to clinical trials. The PRO044 compound targets DMD exon 44 and entered several clinical trials (, , and ), however its sponsor BioMarin stopped pursuing SSOs for the treatment of Duchenne’s muscular dystrophy when kyndrisa failed to obtain FDA approval. Sarepta Therapeutics is currently recruiting for the ESSENCE trial () of two morpholinos, SRP-4045 and SRP-4053 (also known as casimersen and golodirsen, respectively), that target exons 45 and 53, respectively. Results have not been posted for the ESSENCE trial as it is estimated to be completed in May 2023.
In summary, even though SSOs can target similar sequences in the genome, as in the case of exondys 51 and kyndrisa, subtle differences in the SSO chemistries may underlie the differing outcomes observed in their clinical trials [68] (Fig. 2D). Similarly, even a single nucleotide difference in an ASO target can significantly impact a splicing outcome. The cytosine residue at the 10th position of SMN2 intron 7 is the first residue of ISS-N1 and is necessary for the ASO-induced inclusion of exon 7 [73]. An identically sized ASO that starts at the eleventh residue of SMN2 intron 7 produces an opposite effect on SMN2 splicing [73]. These studies highlighted the challenges faced when optimizing SSO-based therapeutics and the importance in understanding how minor chemical modifications to SSOs and target sequences can alter their pharmacologic properties. Ongoing studies with SSO-targeted therapies are laying the groundwork to optimize future therapies in diseases that could benefit from corrected splicing patterns.
2.2. Small molecules
Though SSOs are showing promising results in treating SMA, Duchenne’s muscular dystrophy, and other diseases, there is always a need to develop drugs with improved clinical outcomes and pharmacologic traits. While fewer examples exist, recent advances in high-throughput drug screening are identifying small molecules that directly bind RNA to modulate alternative splicing decisions; several of which have shown promising efficacy thus far. Initial in vitro screens identified the antineoplastic antibiotic aclarubicin (aclacinomycin A) as a potential candidate. Treatment of cells with aclarubicin induced retention of SMN2 exon 7 and increased SMN protein expression [74]. The mechanism of aclarubcin-induced SMN2 splicing was never fully elucidated because it was found to be toxic and not likely useful in the clinic. Several other classes of compounds have been identified as potential candidates to promote retention of SMN2 exon 7 [75]. Among them, three orally available compounds, SMN-C1, SMN-C2, and SMN-C3 were shown to be potent within the nanomolar range, to increase SMN protein expression, and to prevent the neuromuscular pathology classically observed in a mouse model of SMA [75]. These results suggested that SMN-C1, SMN-C2, and SMN-C3 may have a therapeutic benefit in humans with SMA. Several academic and pharmaceutical laboratories have developed other small molecules to target exon 7 of SMN2 since the initial study identifying SMN-C compounds. Subsequent fine tuning of side chains to optimize the pharmacokinetic and pharmacodymanic properties have led to favorable outcomes in achieving optimal potency and safety [76–80]. Notably, two small molecules have progressed to clinical trials. First, LMI070 (also known as NVS-SM1 and branaplan) is currently sponsored by Novartis for treating Type 1 SMA (). In preclinical models of SMA, LMI070 showed beneficial outcomes in survival and SMN protein expression [76,80]. Studies using analogs of LMI070 revealed that their mechanism of action is via sequence-selective binding to RNA to induce stabilization of a double-stranded RNA structure [76]. In the presence of LMI070 analogs the U1 snRNP complex has enhanced affinity for the 5′SS of SMN2 exon 7 (Fig. 2E, left). This promotes the inclusion of SMN2 exon 7 and the production of full length SMN protein [76,80].
A second small molecule that binds SMN2 pre-mRNA to modulate splicing and SMN protein expression is risdiplam (also known as RG7916 and RO7034067). Risdiplam is currently sponsored by Roche in the FIREFISH, SUNFISH, and JEWLEFISH clinical trials (, , and , respectively) to study the efficacy of the drug in infants and adults with either type 1, 2, or 3 SMA. The structure of risdiplam has not been disclosed; however, the previously identified compounds SMN-C2 and SMN-C3 are closely related analogs of RG7916 [75,81]. A proposed model of SMN-C mechanism of action suggests that exon 7 inclusion is increased when the splicing modifier binds to two sites present within SMN2 exon 7 [82]. In the presence of various SMN-C compounds, HNRNPG disassociates from the pre-mRNA allowing the U1 snRNP complex to promote the inclusion of exon 7 [82] (Fig. 2E, middle). Extending those studies, Wang et al. demonstrated that SMN-C2 and SMN-C3 bind to an AGGAAG motif located at the +24 to +29 region of SMN2 exon 7 [81] inducing conformational changes in the pre-mRNA. These conformational changes displace HNRNPG and promote pre-mRNA recognition by the Far-upstream element binding protein-1 (FUBP1) and the KH-type splicing regulatory protein (KHSRP) [81] (Fig. 2E, right). Interestingly, the ESE in exon 7 contains an AGGAAG motif and provides complimentary support to each of the aforementioned mechanistic studies on SMN-C compounds.
3. Therapeutically targeting the spliceosome in cancer
Several cancers result from point mutations within genes encoding members of the spliceosome. The subcomplex splicing factor-3B (SF3B) is located within the U2 snRNP and contains seven subunits, one of them being SF3B1 [83]. SF3B contributes to the activation of the first catalytic step of the splicing reaction and nuclear retention of pre-mRNA for normal gene expression [84] and is involved in RNA splice site selection [85]. One of the most prevalent mutations in alternative splicing-associated cancers occurs within the SF3B1 gene, specifically within the HEAT (Huntingtin, EF3, PP2A, and TOR1) domains of this protein [86]. SF3B1 mutations result in breast [87] and lung [88] cancers, as well as chronic myelomonocytic [86] and lymphocytic [89] leukemias. Taken together, this evidence suggests that targeting components of the spliceosome and, specifically those within the SF3B subcomplex, may be a viable option as a therapeutic strategy.
3.1. Small molecules targeting SF3B
Since 1996 when the initial inhibitor of SF3B was developed, the list of small molecules used to target the SF3B spliceosomal subcomplex has grown significantly. The first reported small molecule was FR901464, a natural product harvested from the fermentation broth of pseudomonas [90]. FR901464 causes cell cycle arrest and, thus, halted proliferation of cancerous cells in mouse models of solid tumors and leukemias [84]. In vitro treatment of HeLa cells with FR901464 or its methylated derivative, spliceostatin A, increased the presence of unspliced pre-mRNAs out of the nucleus [84]. This is consistent with the role of SF3B in nuclear retention of pre-mRNAs [84]. For example, FR901464 treatment produced unspliced cyclin-dependent kinase inhibitor 1B (CDKN1B, also known as p27) pre-mRNA with a premature termination codon that produces a truncated protein named p27* [84]. It is thought that p27* inhibits cancer cell growth by reducing the activity of the cyclin-dependent protein kinases (CDK). Additionally, p27* may stabilize endogenous p27 which also reduces CDK activity and arrests the cell cycle [84].
As spliceostatin A and FR901464 grew in popularity as therapeutic compounds that target the SF3B complex, numerous other molecules were designed as anti-tumor drugs. Sudemycins are a class of compounds that maintain the FR901464 pharmacophore but are less structurally complex [91]. Current studies are disclosing the functional differences between these compounds and demonstrate that there are variable alternative splicing patterns triggered by spliceostatin A and sudemycin K [92]. While spliceostatin A modulates intron retention, sudemycins strongly influence exon skipping [92]. The variability in alternative splicing observed between compounds suggests that sensitivity to molecular targets result in differences in splicing modulation.
Despite the continued development of antitumor compounds targeting the SF3B complex, there are still only a few orally available drugs in the clinics. Seiler et al. have identified an orally-available small molecule H3B-8800 that directly interacts with the SF3B complex [93]. H3B-8800 is a potent inhibitor of canonical and aberrant splicing caused by mutations in the SF3B1 gene and can kill tumor cells harboring this mutation [93]. The Phase-I clinical trial using H3B-8800 has begun and is expected to be completed by the end of 2019 (). It is hopeful that this compound will be the first SF3B1 inhibitor to be clinically used.
The number of SF3B inhibitors is growing; however, the downstream effects of these compounds remain to be elucidated. The use of SF3B inhibitors results in altered mRNA biogenesis and accumulation of unspliced mRNA within nuclear speckles [94]. The resulting nuclear speckles predominantly contain the spliceosome components which were unused and are located proximal to the transcription sites [94]. Compounds that target SF3B1 also activate an alternative export pathway such that intron-containing pre-mRNAs bypass splicing and are transported into the cytoplasm from the nuclear speckle [95]. Under normal conditions, unspliced mRNA is degraded by a standard nonsense-mediated pathway initiated by recognition of the exon junction complex. However, the exon junction complex is unable to stably form around the unspliced mRNA upon treatment with SF3B1 inhibitors [95]. Therefore, SF3B1 inhibitors trigger an alternate non-sense mediated mRNA decay pathway independent of the exon junction complex.
In summary, several cancers are associated with mis-splicing induced by mutations in the SF3B1 gene. Small molecule inhibitors that target the SF3B spliceosomal subcomplex are currently showing beneficial effects on killing tumorigenic cells and reducing cancer cell proliferative capacity (Fig. 3). The most well-characterized small molecules include spliceostatin A (FR901464) and sudemycins; however, similar and clinically relevant compounds are currently under development. While no compounds have made it through Phase-I of clinical trials, it is hopeful that continuing studies will build upon this knowledge and will lead to the development of targeted therapies to inhibit SF3B activity.
Figure 3.
Representation of normal cell conditions with canonical splicing, followed by cancerous cells, and finally cancerous cells treated with SF3B1 inhibitors. Red regions represent mis-splicing, such as intron retention or exon skipping. Cancerous cells depend on canonical splicing patterns within the cell to continue replication, resulting in aberrant proteins. When cancerous cells are exposed to SF3B1 inhibitors (such as sudemycins, spliceostatin A, or H3B-8800), they do not contain sufficient canonical splicing products to continue proliferation, therefore resulting in apoptosis.
3.2. Genetically targeting the spliceosome
Overexpression of human oncogenes drives pro-tumorigenic functions within susceptible cells, increasing total RNA and protein production [96,97]. This increase in RNA levels leads to oncogenic stress on the spliceosome for greater mRNA processing. Therefore, it is no surprise that inhibition of the spliceosome within tumorigenic cells leads to global defects in RNA processing and cellular dysfunction.
In the previous section, we described therapeutics that have direct interactions with the U2 snRNP (i.e. SF3B). Here we focus on indirect therapeutics to genetically target the spliceosome. The bud31 homolog protein (BUD31) is a core spliceosomal component and is responsible for the catalytic activity and assembly of the spliceosome through interactions with SF3B [98]. BUD31 was identified as a lethal gene in cancerous cells and has been utilized as an antitumor target because its deletion induces apoptosis and halts proliferation within tumorigenic cells [99].
Similar to the discovery of BUD31, several studies implicated the ubiquitin-specific peptidase 39 (USP39) as a genetic anticancer target [100,101]. USP39 is an integral part of pre-mRNA splicing catalysis, specifically by recruiting the tri-snRNP U4/U6–U5. Depletion of USP39 by lentiviral short hairpin RNAs results in cell cycle arrest and apoptosis within tumorigenic cells in vitro [100,101] and in vivo [101].
Overall, genetically targeting the spliceosome is a therapeutic option for cancer treatment. While these types of therapies remain to be used in the clinical setting, they are a candidate against pharmacologic inhibitors.
4. Targeting kinases and phosphatases involved in splicing
Post-translational modifications of proteins are an effective method to fine tune cellular processes. Protein phosphorylation is quite possibly the most well studied post-translational modification. Environmental cues and disease states trigger changes in intracellular signaling pathways which results in activation or suppression of protein kinases. These signaling cascades can intersect with RBPs involved in splice site selection and, furthermore, several kinases known to phosphorylate RBPs are dysregulated in diseases [102,103]. Recent work has focused on understanding context and disease specific RBP phosphorylation. The hope behind these studies is to develop targeted therapies to modulate alternative splicing by acting on the kinases and phosphatases that regulate RBP activity.
4.1. Serine-arginine protein kinase family (SRPKs)
The serine/arginine (SR) proteins are an important family of RBPs involved in splice site recognition and contain amino acid residues that are substrates for a number of kinases [104,105]. The serine-arginine protein kinase family (SRPK) specifically targets serine residues located within arginine rich motifs [106]. In a viral model known to hijack a cell’s RNA processing machinery SRp75 phosphorylation was increased by 20-fold compared to controls [107]. This evidence suggested that targeting SRPKs could have therapeutic benefit as antivirals. The group screened a chemical library for SPRK-selective inhibitors and identified an isonicotinamide compound, SRPIN340, that acts as an ATP competitive inhibitor to attenuate kinase activity [107]. SRPIN340 reduced SR protein phosphorylation and inhibited viral propagation without adverse toxicity when given orally to rats at very high doses (2,000 mg/kg) [107].
Elevated SPRK levels are also observed in several cancers. Addition of SRPIN340 caused inconsistent effects on splicing in several in vitro models of lymphoid and myeloid leukemia with elevated SRPK levels [102,105]. However, in multiple models of leukemia, SRIPN340 uniformly induced significant cell death and reduced phosphorylation of the SPRK substrates known as SR splice factor-2 (SRSF2), SRSF4, SRSF5, and SRSF6 [105]. Many solid tumors, including melanoma, rely on the pro-angiogenic effects of the vascular endothelial growth factor (VEGF) for tumor progression. SRSF1 drives VEGF splice form expression toward a pro-angiogenic splice form [102]. SRPIN340 treatment in vivo reduced melanoma tumor size and the expression of the pro-angiogenic VEGF splice form [102]. Lastly, SPHINX, an isonicotinamide analog of SRPIN340, also reduced the expression of the pro-angiogenic VEGF splice forms and SRSF1 phosphorylation in a model of age-related macular degeneration [108].
These studies provide promising evidence that SRPKs can be effectively targeted in several diseases. It is possible that in diseases for which there is no therapeutic benefit of SRPK inhibition, like HIV [107], there are more dominant mechanisms that contribute to the pathology. Additionally, other disease-related mechanisms may exist to overcome SPRK inhibition. Further investigation is certainly needed to thoroughly understand the mechanisms by which mis-regulation of SRPKs contribute to mis-splicing and furthermore, if correcting mis-splicing can improve disease outcomes.
4.2. CDC2-like family of kinases (CLKs)
Members of the SR protein family are also substrates of the CDC2-like family of kinases (CLKs). Several CLK inhibitors have been identified and found to elicit effects on splicing. The first CLK inhibitor to be discovered was the cell permeable benzothiazole, TG003 [109], which is an ATP competitive inhibitor with selective potency toward CLK1, CLK2, and CLK4. TG003 affects splicing through the reduction of phosphorylation of several SR proteins, including the splicing factor SRSF1 (also known as SF2/ASF) [109]. TG003 has also been used to restore the reading frame of the dystrophin protein induced by a point mutation in exon 31 of the DMD gene. The point mutation generates a premature stop codon and reduced dystrophin protein expression [110]. SRSF9 (also known as SRp30c) is thought to be involved in the recognition of DMD exon 31 for splice selection [110], suggesting that the mutation in exon 31 disrupts an ESE or creates an ESS. Interestingly, addition of TG003 induced exon 31 skipping in muscles of humans with Duchenne’s muscular dystrophy carrying the mutation in exon 31 [110]. It is not clear yet whether or not TG003 restored dystrophin protein expression to a functional level.
CLKs are also important contributors to splicing regulation during cell cycle progression [111]. Of the alternative splicing events observed during cell cycle, 94% are blocked by either TG003 or another CLK inhibitor, KH-CB19. Interestingly, only 65% of the splicing events during cell cycle progression were blocked by both TG003 and KH-CB19 [111]. This suggests that there is a significant overlap in the inhibition of CLKs by TG-003 and KH-CB19, but also that there is likely some degree of differential selectivity toward CLK family members and SR protein phosphorylation between those two molecules. Consistent with this hypothesis, KH-CB19 co-crystalized with the ATP binding site of several CLKs, reduced SR protein phosphorylation, and selectively inhibited CLK1 and CLK4 with reduced affinity for CLK2 and CLK3 [112]. Another series of small molecules (Cpd1, Cpd2, Cpd3) developed as CLK inhibitors reduced SR protein phosphorylation and induced concomitant large scale changes in alternative splicing of genes involved in growth and survival [113].
Two analogs of the CDK inhibitors named CGP-74514A and aminopurvalanol A have selectivity for CLKs and potency in the nanomolar range while reducing off target inhibition of CDKs [103]. Lastly, two other compounds, leucettine L41 and T-025, were initially identified as CLK specific inhibitors that modulate alternative splicing through reductions in SR protein phosphorylation [114,115]. Both molecules co-crystalize with the ATP binding pocket of CLKs, however upon further investigation they were found to be effective inhibitors of dual-specificity, tyrosine phosphorylation regulated kinases (DYRKs) with potencies in the low nanomolar range [114,115].
To our knowledge, SRPKs and CLKs are currently the only kinases to which small molecules have been specifically developed to re-direct splicing decisions in diseases. The SRPK and CLK families of proteins are not particularly large, however they do have overlapping substrates and are quite promiscuous in their cellular functions [106,116]. This presents a significant challenge in developing targeted therapies against kinases involved in alternative splicing. Therefore, future studies should focus on understanding the substrate specificity of each of the SRPK and CLK family members and which splicing events are under their control. This will help develop more effective inhibitors and will narrow the window of off-target effects on splicing.
4.3. Protein phosphatases
Shi et al. [117] provided the first direct evidence of the role that protein phosphatases play in phosphorylation-dephosphorylation cycles during splicing regulation. Authors have demonstrated that the U2 and U5 snRNP spliceosomal components are substrates of protein phosphatase 1 (PP1) and PP2A [117]. Furthermore, dephosphorylation of U2 and U5 by PP1 and PP2A causes structural rearrangements in the spliceosome. This is essential for the transition from the first to the second catalytic step in pre-mRNA splicing that generates a lariat intron and ligates exons when the phosphate in the 3’SS is ‘attacked’ by the 3’ hydroxyl group of the 5’ exon [117,118].
Targeting protein phosphatases may also have applicability to human diseases of mis-splicing. For example, the transformer-2-beta homolog 1 (TRA2B, also known as splicing factor serine/arginine-rich 10 (SFRS10)) is a member of the SR protein family and promotes retention of exon 7 of SMN2 by binding to an AG-rich ESE in exon 7 [50]. The RNA recognition motif of TRA2B interacts with PP1 [119]. Increasing PP1 expression reduces the inclusion of TRA2B dependent exons, including SMN2 exon 7. Similarly, PP1 inhibitors such as tautomycin and cantharidin promote SMN2 exon 7 accumulation in fibroblasts from children with type I SMA and also increase SMN protein expression in the spinal cord and liver of transgenic mice that contain human SMN2 [119]. In a related study, Zhang et al. synthesized analogs of cantharidin in an effort to identify novel compounds to modulate alternative splicing and promote SMN2 exon 7 retention [120]. Five novels compounds, isocantharidin and pseudocantharidins A-D, increased the inclusion of SMN2 exon 7 and expression of SMN protein in fibroblasts isolated from a patient with type I SMA [120]. Though each of the compounds elicited similar effects on SMN2 pre-mRNA splicing and SMN protein expression, their ability to modulate the activity of PP1 and PP2A differed. Isocantharidin and pseudocantharidin A reduced the activity of PP1 and PP2A whereas pseudocantharidin D had no effect on their activity. Intriguingly, pseudocantharidin B and pseudocantharidin C increased PP2A activity and had no effect on PP1 [120]. Activation of PP2A with pseudocantharidin B and pseudocantharidin C induced the dephosphorylation of threonine-33 on TRA2B and promoted SMN2 exon 7 inclusion. The precise mechanism of how phosphorylation of TRA2B controls splice site selection and how these five cantharidin analogs elicit positive effects on SMN2 exon 7 inclusion is still unknown.
High-throughput screens have identified other phosphatase inhibitors belonging to a group of 1,4-naphthoquinones that inhibit pre-mRNA splicing [121,122]. Formation of the catalytic spliceosome is inhibited in the presence of naphthoquinones, such as NSC659999 and NSC95397 [122]. Additional effects on splicing may also occur because naphthoquinones generate reactive oxygen species that can oxidize and indirectly inactivate PP2A [122]. These studies provide clear data supporting the role of phosphatases in the regulation of alternative splicing. Overall, the activity of several kinases involved in splicing reactions is antagonized by protein phosphatases and, thus, the functional role of phosphatases can aid in the understanding of splicing mechanisms.
5. Therapeutic potential to disrupt splicing through associated pathways
Numerous other kinases and phosphatases that are canonically involved in growth and metabolism also regulate components of the splicing machinery. Furthermore, the contribution of epigenetics to alternative splicing regulation is becoming more readily apparent [6,8,123–125]. There is an abundance of compounds available to chemically alter these pathways; however, they were not initially developed to modulate alternative splicing decisions. Table 1 highlights a few of lesser well studied pathways and modifications involved in splicing regulation and compounds to chemically alter their function. Several of the compounds are in clinical trials, but none of them specifically investigate changes in alternative splicing as an endpoint. Because less is known about the precise mechanisms of how these post-translational signals regulate alternative splicing, we envision that future studies carefully interpret data generated using chemical inhibitors and consider their impact on alternative splicing regulation.
Table 1.
Signaling pathways and epigenetic marks associated with alternative splicing regulation along with the respective compounds that alter their activity. It is important to note that not every study mentioned in this table measured alternative splicing as an outcome.
| Target selectivity |
Compounds | Function | References to target effects on splicing |
|---|---|---|---|
| mTOR | Rapamycin and analogs (Sirolimus, everolimus, temsirolimus) Torin |
Inhibitor | [130,131] |
| AMPK | AICAR metformin |
Activator | [132,133] |
| Compound C (dorsomorphin) |
Inhibitor | ||
| JNK | SP600125 CC-401 JNK-IN-8 |
Inhibitor | [134,135] |
| GSK3 | CHIR-99021 SB216763 CHIR-98014 |
Inhibitor | [136–138] |
| ERK | VTX-11e SCH772984 XMD8–92 GDC-0994 |
Inhibitor | [139,140] |
| PKC | Enzastaurin Sotrastaurin Go 6983 Staurosporine Quercetin |
Inhibitor | [141,142] |
| PI3K / AKT | LY294002 Rigosertib Wortmannin |
Inhibitor | [126,143–145] |
| AKT specific | A-443654 AR-42 AR-67 AZD5363 GSK6900693 GSK795 MK-2206 Perifosine Tricirbine |
||
| STK38 | BAPTA-AM | Activator | [18] |
| Protein phosphatases |
Okadaic acid Tautomyinc Microcystin-LR Sodium orthovanadate Sodium fluoride Pseudocantharidins NSC659999 NSC95397 |
Inhibitor | [89,121,122,146,147] |
| Histone acetylation | BA3 Anacardic acid Garcinol C646 |
Inhibit acetyltransferase |
[148–151] |
| SAHA MS-275 Scriptad Trichostatin A DHC Nicotine amide Sirtinol Splitomicin |
Inhibit deacetylase |
||
| Histone methylation |
Sudemycin E EPZ5657 EPZ005687 BIX 01294 MM-012 3-deazaneplanocin |
Inhibit methyltransferase |
[26–28,152,153] |
| GSK J4 OG-L002 JIB-04 Tranylcypromine |
Inhibit demethylase |
An example of one such study used an AKT serine/threonine kinase (AKT) inhibitor to demonstrate that control of the splicing machinery is intimately coupled to phosphorylation events and epigenetic marks [126]. Data from a phosphoproteomic screen indicated that the three AKT isoforms differentially affect proteins involved in RNA processing. One specific RNA processing protein IWS1 (named “interacts with SUPT6H, CTD assembly factor 1”) is phosphorylated by AKT1 and AKT3, but not AKT2. Phosphorylation of IWS1 occurs when it is part of a complex with the histone chaperone suppressor of Ty6 (SUPT6H) and Aly proteins. Upon IWS1 phosphorylation, the histone methyltransferase SET-domain containing 2 (SETD2) is recruited to the IWS1 protein complex and trimethylates histone 3 (H3) at the lysine 36 (H3K36me3). This methylation mark is recognized by the chromatin adaptor protein MORF-related gene on chromosome 15 (MORF4L1, also known as MRG15). MORF4L1/MRG15 then provides a docking site for the polypyrimidine tract binding protein-1 (PTBP1) splicing factor where it promotes skipping of exon IIIb in the fibroblast growth factor receptor 2 gene (FGFR-2) [126]. Splicing decisions are a complex interplay between epigenetics and chromatin architecture and the signaling pathways that regulate cellular localization and activation status of RNA processing factors.
In summary, post-translational modifications of proteins contribute significantly to alternative splicing decisions. Additionally, this further increases the number of potential therapeutic targets in diseases caused by mis-splicing. One concern in the field is the broad number of splicing events under the control of phosphorylation signaling cascades or epigenetic changes. While some compounds have shown efficacy in changing splicing outcomes, it seems that many of them are merely just tools to examine the mechanisms of alternative splicing regulation. As such, significant work is needed to overcome this hurdle and to develop targeted compounds with the specificity to correct mis-splicing events while limiting off-target effects.
6. Conclusion
Alternative splicing is intimately coupled to multiple mechanisms that act in concert. It is evident that defects in any of these regulatory processes can be the foundation of disease development or progression. As knowledge of alternative splicing regulation continues to evolve it is likely that even more diseases will be associated with mis-splicing. The complexity of regulation of splicing opens the possibility that it is permissive to multiple avenues of therapeutic development. Indeed, in recent years significant milestones have been achieved in the field and targeted therapeutics are being developed that effectively re-direct alternative splicing with beneficial therapeutic outcomes.
There is no doubt that SSOs are currently showing the most promise in the clinical setting. To our knowledge Spinraza and Exondys 51 are the only FDA approved drugs marketed to alter disease-associated mis-splicing events. Several traits of SSOs make them attractive molecules to redirect alternative splicing and may reduce the barriers for future development. The precision with which SSOs base-pair to their target sequence reduces the chances of off-target binding. Likewise, because the target sequences in the genome are known ahead of time, SSOs are generally easy to design for splicing events in other genes [39,69]. SSOs are also quite stable and are synthetically prepared so they do not show variability between batch preparations [69]. Determining the optimal chemical modifications and routes of administration are important things to consider in SSO development because minor changes in these can alter their pharmacologic efficacy [39]. Though SSOs may currently have an advantage over other therapies in the clinic, the advent of new technologies is shedding light on new ways to specifically target splicing modulation.
Disease-associated proteins have long been the major targets of drug development. However, continued research in RNA biology, alternative splicing regulation, and improved understanding of RNA secondary and tertiary structures now provide the ability to identify small molecule ligands for specific RNA structures [69,127]. The Inforna 2.0 and selective 2’-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) platforms combine chemo- and bioinformatics and are pioneering the recent advances in the ability to identify small molecules that specifically target RNA structures [128,129]. SHAPE-MaP was designed to identify RNA motifs using algorithms to model RNA structure based on massively parallel sequencing data generated from mutational profiles and SHAPE reactivity data [129]. Data from SHAPE-MaP studies identified the RNA binding mechanism of action behind the SMN-C compounds used to treat SMA. To our knowledge this is the only small molecule that interacts directly with RNA structures to induce conformational changes capable of modulating alternative splicing. Inforna 2.0 was designed to identify specific interactions between RNA motifs and small molecules. First, RNA motifs from a desired RNA target are mined from sequence information. Inforna then generates a list of candidate molecules by comparing the previously generated RNA-motifs to a database that contains all known RNA motif-small molecule interactions [128]. These two platforms generate data that complement one another and have the potential to greatly expand the number of druggable targets. For example, transcription factors are often considered to be undruggable [127]. In the future, it may be possible to develop small molecules that target and induce the degradation of mRNAs that encode for transcription factors. This may be a way to circumvent the previously thought undruggabliity of transcription factors. We are certain that the utilization of technologies like SHAPE-MaP and Inforna 2.0 [81,129] in splicing studies will greatly expand the list of druggable targets in diseases associated with defects in splicing.
Highlights.
Numerous diseases are associated with mis-splicing of pre-mRNA
ASOs were the first FDA approved therapies to re-direct splicing
RNA binding small molecules alter splicing and show promise in clinical trials
Genetic and chemical modification of the spliceosome are therapies for cancer
Ongoing splicing studies target epigenetics, accessory proteins, and RNA structure
Acknowledgements
The authors would like to acknowledge the following funding sources: the National Institutes of Health (1R01GM130866) (to JG), the American Heart Association (19CDA34660248) (to JG), an IBM/Junior Faculty Development Award from UNC-Chapel Hill (to JG), a Pilot & Feasibility Research Grant from the Nutrition and Obesity Research Center at UNC-Chapel Hill (UNC NORC) (P30DK056350) (to JG), startup funds from UNC-Chapel Hill (to JG), a NCTraCs Pilot Grant (550KR181805) from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health, through Grant Award Number UL1TR002489 (to JG), the Basil O’Connor Starter Scholar Award from the March of Dimes Foundation (5-FY18–36) (to JG), and the Postbaccalaureate Research Education Program at the University of North Carolina at Chapel Hill (UNC-PREP) which is funded by the NIH-NIGMS (R25-GM089569) grant (funding to JRG). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interests
None
References
- [1].Faustino NA, Cooper TA, Pre-mRNA splicing and human disease, Genes Dev, 17 (2003) 419–437. [DOI] [PubMed] [Google Scholar]
- [2].Early P, Rogers J, Davis M, Calame K, Bond M, Wall R, Hood L, Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways, Cell, 20 (1980) 313–319. [DOI] [PubMed] [Google Scholar]
- [3].Knapp G, Beckmann JS, Johnson PF, Fuhrman SA, Abelson J, Transcription and processing of intervening sequences in yeast tRNA genes, Cell, 14 (1978) 221–236. [DOI] [PubMed] [Google Scholar]
- [4].Berget SM, Moore C, Sharp PA, Spliced segments at the 5’ terminus of adenovirus 2 late mRNA, Biochemistry, 74 (1977) 3171–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Will CL, Lührmann R, Spliceosome structure and function, Cold Spring Harb. Perspect. Biol, 3 (2011) a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T, Epigenetics in alternative pre-mRNA splicing, Cell, 144 (2011) 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Baralle FE, Giudice J, Alternative splicing as a regulator of development and tissue identity, Nat. Rev. Mol. Cell Biol, 18 (2017) 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Naftelberg S, Schor IE, Ast G, Kornblihtt AR, Regulation of alternative splicing through coupling with transcription and chromatin structure, Annu. Rev. Biochem, 84 (2015) 165–198. [DOI] [PubMed] [Google Scholar]
- [9].Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, Stamm S, Function of alternative splicing, Gene, 514 (2013) 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Garg K, Green P, Differing patterns of selection in alternative and constitutive splice sites, Genome Res, 17 (2007) 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pedrotti S, Giudice J, Dagnino-Acosta A, Knoblauch M, Singh RK, Hanna A, Mo Q, Hicks J, Hamilton S, Cooper TA, The RNA-binding protein Rbfox1 regulates splicing required for skeletal muscle structure and function, Hum. Mol. Genet, 24 (2015) 2360–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Li Q, Zheng S, Han A, Lin CH, Stoilov P, Fu XD, Black DL, The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation, Elife, 3 (2014) e01201. doi: 10.7554/eLife.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Han A, Stoilov P, Linares AJ, Zhou Y, Fu XD, Black DL, De novo prediction of PTBP1 binding and splicing targets reveals unexpected features of its RNA recognition and function, PLoS Comput. Biol, 10 (2014) e1003442. doi: 10.1371/journal.pcbi.1003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yeo G, Burge CB, Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals, J. Comput. Biol, 11 (2004) 377–394. [DOI] [PubMed] [Google Scholar]
- [15].Yang J, Hung LH, Licht T, Kostin S, Looso M, Khrameeva E, Bindereif A, Schneider A, Braun T, RBM24 is a major regulator of muscle-specific alternative splicing, Dev. Cell, 31 (2014) 87–99. [DOI] [PubMed] [Google Scholar]
- [16].Goren A, Ram O, Amit M, Keren H, Lev-Maor G, Vig I, Pupko T, Ast G, Comparative analysis identifies exonic splicing regulatory sequences—the complex definition of enhancers and silencers, Mol. Cell, 22 (2006) 769–781. [DOI] [PubMed] [Google Scholar]
- [17].Echeverria GV, Cooper TA, Muscleblind-like 1 activates insulin receptor exon 11 inclusion by enhancing U2AF65 binding and splicing of the upstream intron, Nucleic Acids Res, 42 (2014) 1893–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Liu J, Kong X, Mun Lee Y, Kai Zhang M, Yan Guo L, Lin Y, Kwang Lim T, Lin Q, Qin Xu X, Stk38 modulates Rbm24 protein stability to regulate sarcomere assembly in cardiomyocytes, Sci. Rep, 7 (2017) 44870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kroun Damgaard C, Østergaard Tange T, Kjems J, hnRNP A1 controls HIV-1 mRNA splicing through cooperative binding to intron and exon splicing silencers in the context of a conserved secondary structure, RNA, 8 (2002) 1401–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Buckanovich RJ, Darnell RB, The neuronal RNA binding protein Nova-1 recognizes specific RNA targets in vitro and in vivo, Mol. Cell. Biol, 17 (1997) 3194–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Buratti E, Baralle FE, Influence of RNA secondary structure on the pre-mRNA splicing process, Mol. Cell. Biol, 24 (2004) 10505–10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Singh NN, Singh RN, Androphy EJ, Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes, Nucleic Acids Res, 35 (2007) 371–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Singh NN, Lawler MN, Ottesen EW, Upreti D, Kaczynski JR, Singh RN, An intronic structure enabled by a long-distance interaction serves as a novel target for splicing correction in spinal muscular atrophy, Nucleic Acids Res, 41 (2013) 8144–8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Beyer AL, Osheim YN, Splice site selection, rate of splicing, and alternative splicing on nascent transcripts, Genes Dev, 2 (1988) 754–765. [DOI] [PubMed] [Google Scholar]
- [25].Roberts G, Gooding C, Mak HY, Smith CWJ, Proudfoot NJ, Co-transcriptional commitment to alternative splice site selection, Nucleic Acids Res, 26 (1998) 5568–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T, Regulation of alternative splicing by histone modifications, Science, 327 (2010) 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gelfman S, Cohen N, Yearim A, Ast G, DNA-methylation effect on cotranscriptional splicing is dependent on GC architecture of the exon-intron structure, Genome Res, 23 (2013) 789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gutierrez-Arcelus M, Ongen H, Lappalainen T, Montgomery SB, Buil A, Yurovsky A, Bryois J, Padioleau I, Romano L, Planchon A, Falconnet E, Bielser D, Gagnebin M, Giger T, Borel C, Letourneau A, Makrythanasis P, Guipponi M, Gehrig C, Antonarakis SE, et al. , Tissue-specific effects of genetic and epigenetic variation on gene regulation and splicing, PLoS Genet, 11 (2015) e1004958. doi: 10.1371/journal.pgen.1004958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tazi J, Bakkour N, Stamm S, Alternative splicing and disease, Biochim. Biophys. Acta, 1792 (2009) 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang J, Zhang J, Li K, Zhao W, Cui Q, SpliceDisease database: linking RNA splicing and disease, Nucleic Acids Res, 40 (2012) D1055–D1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Cieply B, Carstens RP, Functional roles of alternative splicing factors in human disease, Wiley Interdiscip. Rev. RNA, 6 (2015) 311–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pistoni M, Ghigna C, Gabellini D, Alternative splicing and muscular dystrophy, RNA Biol, 7 (2010) 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, a Kretzschmar H, Trojanowski JQ, Lee VM-Y, Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis, Science, 314 (2006) 130–133. [DOI] [PubMed] [Google Scholar]
- [34].Arnold ES, Ling S-C, Huelga SC, Lagier-Tourenne C, Polymenidou M, Ditsworth D, Kordasiewicz HB, McAlonis-Downes M, Platoshyn O, Parone PA, Da Cruz S, Clutario KM, Swing D, Tessarollo L, Marsala M, Shaw CE, Yeo GW, Cleveland DW, ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43, Proc. Natl. Acad. Sci. U. S. A, 110 (2013) E736–E745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Pollex R, Hegele R, Hutchinson-Gilford progeria syndrome, Clin. Genet, 66 (2004) 375–381. [DOI] [PubMed] [Google Scholar]
- [36].Mordes D, Luo X, Kar A, Kuo D, Xu L, Fushimi K, Yu G, Sternberg P, Wu JY, Wu JY, Pre-mRNA splicing and retinitis pigmentosa, Mol. Vis, 12 (2006) 1259–1271. [PMC free article] [PubMed] [Google Scholar]
- [37].Rinaldi C, Wood MJA, Antisense oligonucleotides: the next frontier for treatment of neurological disorders, Nat. Rev. Neurol, 14 (2017) 9–21. [DOI] [PubMed] [Google Scholar]
- [38].Wu H, Lima WF, Zhang H, Fan A, Sun H, Crooke ST, Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs, J. Biol. Chem, 279 (2004) 17181–17189. [DOI] [PubMed] [Google Scholar]
- [39].Havens MA, Hastings ML, Splice-switching antisense oligonucleotides as therapeutic drugs, Nucleic Acids Res, 44 (2016) 6549–6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lefebvre S, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, Denis Le Paslier L, Fr J, Cohen D, Weissenbach J, Munnich A, Melki J, Identification and characterization of a spinal muscular atrophy-determining gene, Cell, 80 (1995) 155–165. [DOI] [PubMed] [Google Scholar]
- [41].Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J, Correlation between severity and SMN protein level in spinal muscular atrophy, Nat. Genet, 16 (1997) 265–269. [DOI] [PubMed] [Google Scholar]
- [42].Bürglen L, Lefebvre S, Clermont O, Burlet P, Viollet L, Cruaud C, Munnich A, Melki J, Structure and organization of the human survival motor neurone (SMN) gene, Genomics, 32 (1996) 479–482. [DOI] [PubMed] [Google Scholar]
- [43].Lorson CL, Hahnen E, Androphy EJ, Wirth B, A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy, Genetics, 96 (1999) 6307–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mailman MD, Heinz JW, Papp AC, Snyder PJ, Sedra MS, Wirth B, Burghes AHM, Prior TW, Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2, Genet. Med, 4 (2002) 20–26. [DOI] [PubMed] [Google Scholar]
- [45].Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AHM, Mcpherson JD, A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2, Hum. Mol. Genet, 8 (1999) 1177–1183. [DOI] [PubMed] [Google Scholar]
- [46].Vitte J, Fassier C, Tiziano FD, Dalard C, Soave S, Roblot N, Brahe C, Saugier-Veber P, Paul Bonnefont J, Melki J, Refined characterization of the expression and stability of the SMN gene products, Am J Pathol, 171 (2007) 1269–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Burnett BG, Muñoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH, Regulation of SMN protein stability, Mol. Cell. Biol, 29 (2009) 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Cho S, Dreyfuss G, A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity, Genes Dev, 24 (2010) 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kashima T, Manley JL, A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy, Nat. Genet, 34 (2003) 460–463. [DOI] [PubMed] [Google Scholar]
- [50].Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B, Htra2-B1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2), Proc. Natl. Acad. Sci, 97 (2000) 9618–9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cartegni L, Hastings ML, Calarco JA, De Stanchina E, Krainer AR, Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2, Am. J. Hum. Genet, 78 (2006) 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Cartegni L, Krainer AR, Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1, Nat. Genet, 30 (2002) 377–384. [DOI] [PubMed] [Google Scholar]
- [53].Singh RN, Singh NN, Mechanism of splicing regulation of spinal muscular atrophy genes, Adv. Neurobiol, 20 (2018) 31–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lim SR, Hertel KJ, Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3’ splice site pairing, J. Biol. Chem, 276 (2001) 45476–45483. [DOI] [PubMed] [Google Scholar]
- [55].Singh NK, Singh NN, Androphy EJ, Singh RN, Splicing of a critical exon of human survival motor neuron is regulated by a unique silencer element located in the last intron, Mol. Cell. Biol, 26 (2006) 1333–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].FDA US, Spinraza (nusinersen) Injection, (2016). [Google Scholar]
- [57].Corey DR, Nusinersen, an antisense oligonucleotide drug for spinal muscular atrophy, Nat. Neurosci, 20 (2017) 497–499. [DOI] [PubMed] [Google Scholar]
- [58].Singh NN, Howell MD, Androphy EJ, Singh RN, How the discovery of ISS-N1 led to the first medical therapy for spinal muscular atrophy, Gene Ther, 24 (2017) 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR, Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice, Am. J. Hum. Genet, 82 (2008) 834–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, Kirschner J, Chiriboga CA, Saito K, Servais L, Tizzano E, Topaloglu H, Tulinius M, Montes J, Glanzman AM, Bishop K, Zhong ZJ, Gheuens S, Bennett CF, Schneider E, Farwell W, et al. , Nusinersen versus sham control in infantile-onset spinal muscular atrophy, N. Engl. J. Med, 377 (2017) 1723–1732. [DOI] [PubMed] [Google Scholar]
- [61].Finkel RS, Chiriboga CA, Vajsar J, Day JW, Montes J, De Vivo DC, Yamashita M, Rigo F, Hung G, Schneider E, Norris DA, Xia S, Bennett CF, Bishop KM, Treatment of infantile-onset spinal muscular atrophy with nusinersen: a phase 2, open-label, dose-escalation study, Lancet, 388 (2016) 3017–3026. [DOI] [PubMed] [Google Scholar]
- [62].Robinson-Hamm JN, Gersbach CA, Gene therapies that restore dystrophin expression for the treatment of Duchenne muscular dystrophy, Hum. Genet, 135 (2016) 1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Stein CA, Eteplirsen approved for Duchenne muscular dystrophy: the FDA faces a difficult choice, Mol. Ther, 24 (2016) 1884–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Tuffery-Giraud S, Miro J, Koenig M, Claustres M, Normal and altered pre-mRNA processing in the DMD gene, Hum. Genet, 136 (2017) 1155–1172. [DOI] [PubMed] [Google Scholar]
- [65].Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K, Abbs S, Garralda ME, Bourke J, Wells DJ, Dickson G, Wood MJ, Wilton SD, Straub V, Kole R, Shrewsbury SB, Sewry C, Morgan JE, Bushby K, Muntoni F, Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study, Lancet, 378 (2011) 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Kinali M, Arechavala-Gomeza V, Feng L, Cirak S, Hunt D, Adkin C, Guglieri M, Ashton E, Abbs S, Nihoyannopoulos P, Garralda ME, Rutherford M, Mcculley C, Popplewell L, Graham IR, Dickson G, Wood MJ, Wells DJ, Wilton SD, Kole R, et al. , Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy: a single-blind, placebo-controlled, dose-escalation, proof-of-concept study, Lancet Neurol, 8 (2009) 918–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Stein CA, Castanotto D, FDA-approved oligonucleotide therapies in 2017, Mol. Ther, 25 (2017) 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Mendell JR, Sahenk Z, Rodino-Klapac LR, Clinical trials of exon skipping in Duchenne muscular dystrophy, Expert Opin. Orphan Drugs, 5 (2017) 683–690. [Google Scholar]
- [69].Lieberman J, Tapping the RNA world for therapeutics, Nat. Struct. Mol. Biol, 25 (2018) 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Aartsma-Rus A, Krieg AM, FDA approves eteplirsen for Duchenne muscular dystrophy: The next chapter in the eteplirsen saga, Nucleic Acid Ther, 27 (2017) 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].FDA US, Press Announcements - FDA grants accelerated approval to first drug for Duchenne muscular dystrophy, FDA News Release, (2016). [Google Scholar]
- [72].Goemans N, Mercuri E, Belousova E, Komaki H, Dubrovsky A, McDonald CM, Kraus JE, Lourbakos A, Lin Z, Campion G, Wang SX, Campbell C, Araujo A, Bertini E, Born P, Cances C, Chabrol B, Chae J-H, Colomer Oferil J, Comi GP, et al. , A randomized placebo-controlled phase 3 trial of an antisense oligonucleotide, drisapersen, in Duchenne muscular dystrophy, Neuromuscul. Disord, 28 (2018) 4–15. [DOI] [PubMed] [Google Scholar]
- [73].Singh NN, Hollinger K, Bhattacharya D, Singh RN, An antisense microwalk reveals critical role of an intronic position linked to a unique long-distance interaction in pre-mRNA splicing, RNA, 16 (2010) 1167–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Andreassi C, Jarecki J, Zhou J, Coovert DD, Monani UR, Chen X, Whitney M, Pollok B, Zhang M, Androphy E, Burghes AHM, Aclarubicin treatment restores SMN levels to cells derived from type I spinal muscular atrophy patients, Hum. Mol. Genet, 10 (2001) 2841–2849. [DOI] [PubMed] [Google Scholar]
- [75].Naryshkin NA, Weetall M, Dakka A, Narasimhan J, Zhao X, Feng Z, Ling KKY, Karp GM, Qi H, Woll MG, Chen G, Zhang N, Gabbeta V, Vazirani P, Bhattacharyya A, Furia B, Risher N, Sheedy J, Kong R, Ma J, et al. , SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy, Science, 345 (2014) 688–693. [DOI] [PubMed] [Google Scholar]
- [76].Palacino J, Swalley SE, Song C, Cheung AK, Shu L, Zhang X, Van Hoosear M, Shin Y, Chin DN, Keller CG, Beibel M, Renaud NA, Smith TM, Salcius M, Shi X, Hild M, Servais R, Jain M, Deng L, Bullock C, et al. , SMN2 splice modulators enhance U1–pre-mRNA association and rescue SMA mice, Nat. Chem. Biol, 11 (2015) 511–517. [DOI] [PubMed] [Google Scholar]
- [77].Woll MG, Qi H, Turpoff A, Zhang N, Zhang X, Chen G, Li C, Huang S, Yang T, Moon Y-C, Lee C-S, Choi S, Almstead NG, Naryshkin NA, Dakka A, Narasimhan J, Gabbeta V, Welch E, Zhao X, Risher N, et al. , Discovery and optimization of small molecule splicing modifiers of survival motor neuron 2 as a treatment for spinal muscular atrophy, J. Med. Chem, 59 (2016) 6070–6085. [DOI] [PubMed] [Google Scholar]
- [78].Ratni H, Karp GM, Weetall M, Naryshkin NA, V Paushkin S, Chen KS, Mccarthy KD, Qi H, Turpoff A, Woll MG, Zhang X, Zhang N, Yang T, Dakka A, Vazirani P, Zhao X, Pinard E, Green L, David-Pierson P, Tuerck D, et al. , Specific correction of alternative survival motor neuron 2 splicing by small molecules: discovery of a potential novel medicine to treat spinal muscular atrophy, J. Med. Chem, 59 (2016) 6086–6100. [DOI] [PubMed] [Google Scholar]
- [79].Pinard E, Green L, Reutlinger M, Weetall M, Naryshkin NA, Baird J, Chen KS, V Paushkin S, Metzger F, Ratni H, Hoffmann F, Discovery of a novel class of survival motor neuron 2 splicing modifiers for the treatment of spinal muscular atrophy, J. Med. Chem, 60 (2017) 4444–4457. [DOI] [PubMed] [Google Scholar]
- [80].Cheung AK, Hurley B, Kerrigan R, Shu L, Chin DN, Shen Y, Je Sung M, Hou Y, Axford J, Cody E, Sun R, Fazal A, Fridrich C, Sanchez CC, Tomlinson RC, Jain M, Deng L, Hoffmaster K, Song C, Van Hoosear M, et al. , Discovery of small molecule splicing modulators of survival motor neuron-2 (SMN2) for the treatment of spinal muscular atrophy (SMA), J. Med. Chem, 61 (2018) 11021–11036. [DOI] [PubMed] [Google Scholar]
- [81].Wang J, Schultz PG, Johnson KA, Mechanistic studies of a small-molecule modulator of SMN2 splicing, Proc. Natl. Acad. Sci, 115 (2018) E4604–E4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Sivaramakrishnan M, Mccarthy KD, Campagne S, Huber S, Meier S, Augustin A, Heckel T, Meistermann H, Hug MN, Birrer P, Moursy A, Khawaja S, Schmucki R, Berntenis N, Giroud N, Golling S, Tzouros M, Banfai B, Duran-Pacheco G, Lamerz J, et al. , Binding to SMN2 pre-mRNA-protein complex elicits specificity for small molecule splicing modifiers, Nat. Commun, 8 (2017) doi: 10.1038/s41467-017-01559-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Cretu C, Schmitzová J, Ponce-Salvatierra A, Dybkov O, De Laurentiis EI, Sharma K, Will CL, Urlaub H, Lührmann R, Pena V, Molecular architecture of SF3b and structural consequences of its cancer-related mutations, Mol. Cell, 64 (2016) 307–319. [DOI] [PubMed] [Google Scholar]
- [84].Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, Tani T, Horinouchi S, Yoshida M, Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA, Nat. Chem. Biol, 3 (2007) 576–583. [DOI] [PubMed] [Google Scholar]
- [85].Webb TR, Joyner AS, Potter PM, The development and application of small molecule modulators of SF3b as therapeutic agents for cancer, Drug Discov. Today, 18 (2013) 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Ishiyama K, Mori H, et al. , Frequent pathway mutations of splicing machinery in myelodysplasia, Nature, 478 (2011) 64–69. [DOI] [PubMed] [Google Scholar]
- [87].Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, Bowlby R, Shen H, Hayat S, Fieldhouse R, Lester SC, Tse GMK, Factor RE, Collins LC, Allison KH, Chen Y-Y, et al. , Comprehensive molecular portraits of invasive lobular breast cancer, Cell, 163 (2015) 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H, et al. , Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing, Cell, 150 (2012) 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, Zhang W, Vartanov AR, Fernandes SM, Goldstein NR, Folco EG, Cibulskis K, Tesar B, Sievers QL, Shefler E, Gabriel S, et al. , SF3B1 and other novel cancer genes in chronic lymphocytic leukemia, N. Engl. J. Med, 365 (2011) 2497–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Nakajima H, Hori Y, Terano H, Okuhara M, Manda T, Matsumoto S, Shimomura K, New antitumor substances, FR901463, FR901464 and FR901465, J. Antibiot. (Tokyo), 49 (1996) 1204–1211. [DOI] [PubMed] [Google Scholar]
- [91].Fan L, Lagisetti C, Edwards CC, Webb TR, Potter PM, Sudemycins, novel small molecule analogues of FR901464, induce alternative gene splicing, ACS Chem. Biol, 6 (2011) 582–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Vigevani L, Gohr A, Webb T, Irimia M, Valcárcel J, Molecular basis of differential 3′ splice site sensitivity to anti-tumor drugs targeting U2 snRNP, Nat. Commun, 8 (2017) doi: 10.1038/s41467-017-02007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Seiler M, Yoshimi A, Darman R, Chan B, Keaney G, Thomas M, Agrawal AA, Caleb B, Csibi A, Sean E, Fekkes P, Karr C, Klimek V, Lai G, Lee L, Kumar P, Lee SC-W, Liu X, Mackenzie C, Meeske C, et al. , H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers, Nat. Med, 24 (2018) 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Lamond AI, Spector DL, Nuclear speckles: a model for nuclear organelles, Nat. Rev. Mol. Cell Biol, 4 (2003) 605–612. [DOI] [PubMed] [Google Scholar]
- [95].Carvalho T, Martins S, Rino J, Marinho S, Carmo-Fonseca M, Pharmacological inhibition of the spliceosome subunit SF3b triggers EJC-independent NMD, J. Cell Sci, 130 (2017) 1519–1531. [DOI] [PubMed] [Google Scholar]
- [96].Lin CY, Lovén J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA, Transcriptional amplification in tumor cells with elevated c-Myc, Cell, 151 (2012) 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ruggero D, The role of Myc-induced protein synthesis in cancer, Cancer Res, 69 (2009) 8839–8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Saha D, Khandelia P, O’Keefe RT, Vijayraghavan U, Saccharomyces cerevisiae NineTeen complex (NTC)-associated factor Bud31/Ycr063w assembles on precatalytic spliceosomes and improves first and second step pre-mRNA splicing efficiency, J. Biol. Chem, 287 (2012) 5390–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hsu TY-T, Simon LM, Neill NJ, Marcotte R, Sayad A, Bland CS, Echeverria GV, Sun T, Kurley SJ, Tyagi S, Karlin KL, Dominguez-Vidaña R, Hartman JD, Renwick A, Scorsone K, Bernardi RJ, Skinner SO, Jain A, Orellana M, Lagisetti C, et al. , The spliceosome is a therapeutic vulnerability in MYC-driven cancer, Nature, 525 (2015) 384–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Gan Z, Han K, Lin S, Hu H, Shen Z, Min D, Knockdown of ubiquitin-specific peptidase 39 inhibited the growth of osteosarcoma cells and induced apoptosis in vitro, Biol. Res, 50 (2017) doi: 10.1186/s40659-017-0121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zhao Y, Zhang B, Lei Y, Sun J, Zhang Y, Yang S, Zhang X, Knockdown of USP39 induces cell cycle arrest and apoptosis in melanoma, Tumor Biol, 37 (2016) 13167–13176. [DOI] [PubMed] [Google Scholar]
- [102].V Gammons M, Lucas R, Dean R, Coupland SE, Oltean S, Bates DO, Targeting SRPK1 to control VEGF-mediated tumour angiogenesis in metastatic melanoma, Br. J. Cancer, 111 (2014) 477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Shi Y, Park J, Lagisetti C, Zhou W, Sambucetti LC, Webb TR, A triple exon-skipping luciferase reporter assay identifies a new CLK inhibitor pharmacophore, Bioorg Med Chem Lett, 27 (2017) 406–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Ohe K, Hagiwara M, Modulation of alternative splicing with chemical compounds in new therapeutics for human diseases, ACS Chem. Biol, 10 (2015) 914–924. [DOI] [PubMed] [Google Scholar]
- [105].Siqueira R. Pais, de Almeida Alves Barbosa É, Polêto M. Depólo, Righetto G. Lima, Seraphim T. Vargas, Salgado R. Locatelli, Ferreira J. Gasperazzo, de Andrade Barros M. Vinícius, de Oliveira L. Licursi, Albertoni Laranjeira A. Brunelli, Almeida M. Rogéria, Júnior A. Silva, Rangel Fietto J. Lopes, Kobarg J, de Oliveira E. Basílio, Teixeira R. Ricardo, Borges J. César, Yunes J. Andrés, Bressan G. Costa, Potential antileukemia effect and structural analyses of SRPK inhibition by N-(2-(Piperidin-1-yl)-5-(trifluoromethyl)phenyl) isonicotinamide (SRPIN340), PLoS One, 10 (2015) e0134882. doi: 10.1371/journal.pone.0134882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Giannakouros T, Nikolakaki E, Mylonis I, Georgatsou E, Serine-arginine protein kinases: a small protein kinase family with a large cellular presence, FEBS J, 278 (2011) 570–586. [DOI] [PubMed] [Google Scholar]
- [107].Fukuhara T, Hosoya T, Shimizu S, Sumi K, Oshiro T, Yoshinaka Y, Suzuki M, Yamamoto N, Herzenberg LA, Herzenberg LA, Hagiwara M, Utilization of host SR protein kinases and RNA-splicing machinery during viral replication, Proc. Natl. Acad. Sci, 103 (2006) 11329–11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].V Gammons M, Fedorov O, Ivison D, Du C, Clark T, Hopkins C, Hagiwara M, Dick AD, Cox R, Harper SJ, Hancox JC, Knapp S, Bates DO, Topical antiangiogenic SRPK1 inhibitors reduce choroidal neovascularization in rodent models of exudative AMD, Investig. Opthalmology Vis. Sci, 54 (2013) 6052–6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Muraki M, Ohkawara B, Hosoya T, Onogi H, Koizumi J, Koizumi T, Sumi K, Yomoda J, V Murray M, Kimura H, Furuichi K, Shibuya H, Krainer AR, Suzuki M, Hagiwara M, Manipulation of alternative splicing by a newly developed inhibitor of Clks, J. Biol. Chem, 279 (2004) 24246–24254. [DOI] [PubMed] [Google Scholar]
- [110].Nishida A, Kataoka N, Takeshima Y, Yagi M, Awano H, Ota M, Itoh K, Hagiwara M, Matsuo M, Chemical treatment enhances skipping of a mutated exon in the dystrophin gene, Nat. Commun, 2 (2011) doi: 10.1038/ncomms1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Dominguez D, Tsai Y-H, Weatheritt R, Wang Y, Blencowe BJ, Wang Z, An extensive program of periodic alternative splicing linked to cell cycle progression, Elife, 5 (2016) e10288. doi: 10.7554/eLife.10288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Fedorov O, Huber K, Eisenreich A, Filippakopoulos P, King O, Bullock AN, Szklarczyk D, Jensen LJ, Fabbro D, Trappe J, Rauch U, Bracher F, Knapp S, Specific CLK inhibitors from a novel chemotype for regulation of alternative splicing, Chem. Biol, 18 (2011) 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Araki S, Dairiki R, Nakayama Y, Murai A, Miyashita R, Iwatani M, Nomura T, Nakanishi O, Inhibitors of CLK protein kinases suppress cell growth and induce apoptosis by modulating pre-mRNA splicing, PLoS One, 10 (2015) e0116929. doi: 10.1371/journal.pone.0116929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Debdab M, Carreaux F-O, Renault S, Soundararajan M, Fedorov O, Filippakopoulos P, Lozach O, Babault L, Tahtouh T, Baratte B, Ogawa Y, Hagiwara M, Eisenreich A, Rauch U, Knapp S, Meijer L, Bazureau J-P, Leucettines, a class of potent inhibitors of cdc2-like kinases and dual specificity, tyrosine phosphorylation regulated kinases derived from the marine sponge leucettamine B: modulation of alternative pre-RNA splicing, J. Med. Chem, 54 (2011) 4172–4186. [DOI] [PubMed] [Google Scholar]
- [115].Iwai K, Yaguchi M, Nishimura K, Yamamoto Y, Tamura T, Nakata D, Dairiki R, Kawakita Y, Mizojiri R, Ito Y, Asano M, Maezaki H, Nakayama Y, Kaishima M, Hayashi K, Teratani M, Miyakawa S, Iwatani M, Miyamoto M, Klein MG, et al. , Anti-tumor efficacy of a novel CLK inhibitor via targeting RNA splicing and MYC-dependent vulnerability, EMBO Mol. Med, 10 (2018) e8289. doi: 10.15252/emmm.201708289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Colwill K, Feng LL, Yeakley JM, Gish GD, Cáceres JF, Pawson T, Fu XD, SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors, J. Biol. Chem, 271 (1996) 24569–24575. [DOI] [PubMed] [Google Scholar]
- [117].Shi Y, Reddy B, Manley JL, PP1/PP2A phosphatases are required for the second step of pre-mRNA splicing and target specific snRNP proteins, Mol. Cell, 23 (2006) 819–829. [DOI] [PubMed] [Google Scholar]
- [118].Staley JP, Guthrie C, Mechanical devices of the spliceosome: motors, clocks, springs, and things, Cell, 92 (1998) 315–326. [DOI] [PubMed] [Google Scholar]
- [119].Novoyatleva T, Heinrich B, Tang Y, Benderska N, Butchbach MER, Lorson CL, Lorson MA, Ben-Dov C, Fehlbaum P, Bracco L, Burghes AHM, Bollen M, Stamm S, Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing, Hum. Mol. Genet, 17 (2008) 52–70. [DOI] [PubMed] [Google Scholar]
- [120].Zhang Z, Kelemen O, van Santen MA, Yelton SM, Wendlandt AE, Sviripa VM, Bollen M, Beullens M, Urlaub H, Lührmann R, Watt DS, Stamm S, Synthesis and characterization of pseudocantharidins, novel phosphatase modulators that promote the inclusion of exon 7 into the SMN (survival of motoneuron) pre-mRNA, J. Biol. Chem, 286 (2011) 10126–10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Berg MG, Wan L, Younis I, Diem MD, Soo M, Wang C, Dreyfuss G, A quantitative high-throughput in vitro splicing assay identifies inhibitors of spliceosome catalysis, Mol. Cell. Biol, 32 (2012) 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Effenberger KA, Perriman RJ, Bray WM, Lokey RS, Ares M, Jurica MS, Jurica MS, A high-throughput splicing assay identifies new classes of inhibitors of human and yeast spliceosomes, J. Biomol. Screen, 18 (2013) 1110–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Pacini C, Koziol MJ, Bioinformatics challenges and perspectives when studying the effect of epigenetic modifications on alternative splicing, Phil. Trans. R. Soc. B, 373 (2018) 20170073. doi: 10.1098/rstb.2017.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Khan DH, Jahan S, Davie JR, Pre-mRNA splicing: role of epigenetics and implications in disease, Adv. Biol. Regul, 52 (2012) 377–388. [DOI] [PubMed] [Google Scholar]
- [125].Lev Maor G, Yearim A, Ast G, The alternative role of DNA methylation in splicing regulation, Trends Genet, 31 (2015) 274–280. [DOI] [PubMed] [Google Scholar]
- [126].Sanidas I, Polytarchou C, Hatziapostolou M, Ezell SA, Kottakis F, Hu L, Guo A, Xie J, Comb MJ, Iliopoulos D, Tsichlis PN, Phosphoproteomics screen reveals Akt isoform-specific signals linking RNA processing to lung cancer, Mol. Cell, 53 (2014) 577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Connelly CM, Moon MH, Schneekloth JS, The emerging role of RNA as a therapeutic target for small molecules, Cell Chem. Biol, 23 (2016) 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Disney MD, Winkelsas AM, Pradeep Velagapudi S, Southern M, Fallahi M, Childs-Disney JL, Inforna 2.0: a platform for the sequence-based design of small molecules targeting structured RNAs, ACS Chem Biol, 11 (2016) 1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Siegfried NA, Busan S, Rice GM, Nelson JAE, Weeks KM, RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP), Nat. Methods, 11 (2014) 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Passacantilli I, Frisone P, De Paola E, Fidaleo M, Paronetto MP, hnRNPM guides an alternative splicing program in response to inhibition of the PI3K/AKT/mTOR pathway in Ewing sarcoma cells, Nucleic Acids Res, 45 (2017) 12270–12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Lee G, Zheng Y, Cho S, Jang C, England C, Dempsey JM, Yu Y, Liu X, He L, Cavaliere PM, Chavez A, Zhang E, Isik M, Couvillon A, Dephoure NE, Blackwell TK, Yu JJ, Rabinowitz JD, Cantley LC, Blenis J, Post-transcriptional regulation of de novo lipogenesis by mTORC1-S6K1-SRPK2 signaling, Cell, 171 (2017) 1545–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Laustriat D, Gide J, Barrault L, Chautard E, Benoit C, Auboeuf D, Boland A, Battail C, Artiguenave F, Deleuze J-F, Bénit P, Rustin P, Franc S, Charpentier G, Furling D, Bassez G, Nissan X, Martinat C, Peschanski M, Baghdoyan S, In vitro and in vivo modulation of alternative splicing by the biguanide metformin, Mol. Ther. - Nucleic Acids, 4 (2015) e262. doi: 10.1038/mtna.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Brockhoff M, Rion N, Chojnowska K, Wiktorowicz T, Eickhorst C, Erne B, Frank S, Angelini C, Furling D, Ruegg MA, Sinnreich M, Castets P, Targeting deregulated AMPK/mTORC1 pathways improves muscle function in myotonic dystrophy type I, J. Clin. Invest, 127 (2017) 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Martinez NM, Agosto L, Qiu J, Mallory MJ, Gazzara MR, Barash Y, Fu X-D, Lynch KW, Widespread JNK-dependent alternative splicing induces a positive feedback loop through CELF2-mediated regulation of MKK7 during T-cell activation, Genes Dev, 29 (2015) 2054–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Pelisch F, Blaustein M, Kornblihtt AR, Srebrow A, Cross-talk between signaling pathways regulates alternative splicing: a novel role for JNK, J. Biol. Chem, 280 (2005) 25461–25469. [DOI] [PubMed] [Google Scholar]
- [136].Shinde MY, Sidoli S, Kulej K, Mallory MJ, Radens CM, Reicherter AL, Myers RL, Barash Y, Lynch KW, Garcia BA, Klein PS, Phosphoproteomics reveals that glycogen synthase kinase-3 phosphorylates multiple splicing factors and is associated with alternative splicing, J. Biol. Chem, 292 (2017) 18240–18255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Hernandez F, Perez M, Lucas JJ, Mata AM, Bhat R, Avila J, Glycogen synthase kinase-3 plays a crucial role in tau exon 10 splicing and intranuclear distribution of SC35: implications for Alzheimers disease, J. Biol. Chem, 279 (2004) 3801–3806. [DOI] [PubMed] [Google Scholar]
- [138].Heyd F, Lynch KW, Phosphorylation-dependent regulation of PSF by GSK3 controls CD45 alternative splicing, Mol. Cell, 40 (2010) 126–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Al-Ayoubi AM, Zheng H, Liu Y, Bai T, Eblen ST, Mitogen-activated protein kinase phosphorylation of splicing factor 45 (SPF45) regulates SPF45 alternative splicing site utilization, proliferation, and cell adhesion, Mol. Cell. Biol, 32 (2012) 2880–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Tisserant A, König H, Signal-regulated pre-mRNA occupancy by the general splicing factor U2AF, PLoS One, 3 (2008) e1418. doi: 10.1371/journal.pone.0001418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Gascoigne N, Zal T, Kaye J, Rao S, Duncan McCuaig R, Dunn J, Li J, Masch A, Knaute T, Schutkowski M, Zerweck J, PKC-theta is a novel SC35 splicing factor regulator in response to T cell activation, Front. Immunol, 6 (2015) 562. doi: 10.3389/fimmu.2015.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Verma SK, Deshmukh V, Liu P, Nutter CA, Espejo R, Hung ML, Wang GS, Yeo GW, Kuyumcu-Martinez MN, Reactivation of fetal splicing programs in diabetic hearts is mediated by protein kinase C signaling, J. Biol. Chem, 288 (2013) 35372–35386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Zhou Z, Qiu J, Liu W, Zhou Y, Plocinik RM, Li H, Hu Q, Ghosh G, Adams JA, Rosenfeld MG, Fu X-D, The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus, Mol. Cell, 47 (2012) 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Blaustein M, Pelisch F, Tanos T, Muñoz MJ, Wengier D, Quadrana L, Sanford JR, Muschietti JP, Kornblihtt AR, Cáceres JF, Coso OA, Srebrow A, Concerted regulation of nuclear and cytoplasmic activities of SR proteins by AKT, Nat. Struct. Mol. Biol, 12 (2005) 1037–1044. [DOI] [PubMed] [Google Scholar]
- [145].Li P, Carter G, Romero J, Gower KM, Watson J, Clk/STY (cdc2-like kinase 1) and Akt regulate alternative splicing and adipogenesis in 3T3-L1 pre-adipocytes, PLoS One, 8 (2013) e53268. doi: 10.1371/journal.pone.0053268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Black AJ, Schilder RJ, Kimball SR, Palmitate-and C6 ceramide-induced Tnnt3 pre-mRNA alternative splicing occurs in a PP2A dependent manner, Nutr. Metab, 15 (2018) doi: 10.1186/s12986-018-0326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Mermoud JE, Cohen P ’, A.L. Lamond, Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing, Nucleic Acids Res, 20 (1992) 5263–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Kuhn AN, Van Santen MA, Schwienhorst A, Urlaub H, Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation, RNA, 15 (2009) 153–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Dušková E, Hnilicová J, Staněk D, CRE promoter sites modulate alternative splicing via p300-mediated histone acetylation, RNA Biol, 11 (2014) 865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Xu Y, Zhao W, Olson SD, Prabhakara KS, Zhou X, Alternative splicing links histone modifications to stem cell fate decision, Genome Biol, 19 (2018) doi: 10.1186/s13059-018-1512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].da Gloria VG, de Araujo MM, Santos AM, Leal R, de Almeida SF, Carmo AM, Moreira A, T cell activation regulates CD6 alternative splicing by transcription dynamics and SRSF1, J. Immunol, 193 (2014) 391–399. [DOI] [PubMed] [Google Scholar]
- [152].Convertini P, Shen M, Potter PM, Palacios G, Lagisetti C, De La Grange P, Horbinski C, Fondufe-Mittendorf YN, Webb TR, Stamm S, Sudemycin E influences alternative splicing and changes chromatin modifications, Nucleic Acids Res, 42 (2014) 4947–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Sims RJ III, Millhouse S, Chen C-F, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D, Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing, Mol. Cell, 28 (2007) 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Shi Y, Mechanistic insights into precursor messenger RNA splicing by the spliceosome, Nat Rev Mol Cell Biol, 18 (2017) 655–670. [DOI] [PubMed] [Google Scholar]
- [155].Fu X-D, Ares M, Context-dependent control of alternative splicing by RNA-binding proteins, Nat. Rev. Genet, 15 (2014) 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]