Abstract
Background:
Epigenetic alterations comprise key regulatory events that dynamically alter gene expression and their deregulation is commonly linked to the pathogenesis of various diseases, including cancer. Unlike DNA mutations, epigenetic alterations involve modifications to proteins and nucleic acids that regulate chromatin structure without affecting the underlying DNA sequence, altering the accessibility of the transcriptional machinery to the DNA, thus modulating gene expression. In cancer cells, this often involves the silencing of tumor suppressor genes or the increased expression of genes involved in oncogenesis. Advances in laboratory medicine have made it possible to map critical epigenetic events, including histone modifications and DNA methylation, on a genome-wide scale. Like the identification of genetic mutations, mapping of changes to the epigenetic landscape has increased our understanding of cancer progression. However, in contrast to irreversible genetic mutations, epigenetic modifications are flexible and dynamic, thereby making them promising therapeutic targets. Ongoing studies are evaluating the use of epigenetic drugs in chemotherapy sensitization and immune system modulation. With the preclinical success of drugs that modify epigenetics, along with the FDA approval of epigenetic drugs including the DNA methyltransferase 1 (DNMT1) inhibitor 5-azacitidine and the histone deacetylase (HDAC) inhibitor vorinostat, there has been a rise in the number of drugs that target epigenetic modulators over recent years.
Conclusion:
We provide an overview of epigenetic modulations, particularly those involved in cancer, and discuss the recent advances in drug development that target these chromatin-modifying events, primarily focusing on novel strategies to regulate the epigenome.
Keywords: Epigenetics, Chromatin, Cancer, Methylation, Acetylation, Histones, DNA
1. INTRODUCTION
Conrad Waddington coined the term ‘epigenetics’ to describe heritable changes in phenotype that were independent of the changes in DNA sequence. More recently, epigenetics refers to chromatin-based events that regulate gene expression through biological mechanisms that involve changes to chromatin structure. Deregulation of these chromatin-modifying processes is central to the genesis of various diseases, including cancer. The eukaryotic genome is highly complex and is composed of 23 pairs of chromosomes that encode approximately 20,000 genes. The extended length of eukaryotic DNA measures about 2 meters in length. In order for it to fit inside the nucleus of human cells, the diameter of which is merely 5-10 µm, the DNA needs to be tightly and efficiently packaged. Histones are positively charged scaffolding proteins that are required for DNA packaging within the nucleus [1]. Histones are highly abundant in the basic amino acids lysine and arginine, which facilitate their binding and interaction with the negatively charged DNA. Eukaryotic DNA and the histone proteins together form the macromolecular chromatin complex in higher organisms. In 1974, Roger Kornberg described the basic structural unit of chromatin and termed it the nucleosome. The nucleosome core is comprised of 147 base pairs of DNA wrapped 1.65 times around four core histone proteins (H2A, H2B, H3 and H4) [2]. Histone H1, the linker histone, facilitates DNA compaction by stringing the bead-like nucleosomes together, leading to higher-order chromatin structures [3]. Histone H5 is another variant form of linker histone that assists in condensation of nucleosome chains [4]. The protruding N-terminal tails of histones are subject to several post-translational modifications that can significantly alter chromatin architecture by affecting their interaction with DNA [5].
DNA and histone modifications impart dynamic plasticity to the epigenome and are controlled by chromatin modifying enzymes. (Table 1) describes the various DNA and histone modifications known to date. Biological processes like DNA replication and transcription require DNA binding factors to access the appropriate regions on DNA to faithfully carry out their functions. This access is extremely limited due to the tightly-packed nano-architecture of chromatin. DNA and histone modifications can dictate the chromatin assembly by altering the non-covalent interactions between the nucleosomes. The altered chromatin structure can then either serve as a docking site for transcription factors or can hinder the binding of these factors to the chromatin. Histone post-translational modifications and DNA methylation are thus important regulators of nucleosome dynamics, thereby impacting gene regulation [6]. Epigenetic repression, in general, is attributed to a more compact topology of chromatin and is directly governed by DNA and histone methylation and nucleosome aggregation by the cohesin and polycomb complexes. Conversely, epigenetic activation is a result of a more open conformation of chromatin structure, also called a nucleosome free region (NFR), that is facilitated by histone post-translational modifications such as acetylation or the incorporation of histone variants [7].
Table 1. DNA and Histone modifications and their functions.
| Chromatin Modification | Residues Modified | Enzymes | Function |
|---|---|---|---|
| DNA Modifications | |||
| 5-methylcytosine | Cytosine | DNA methyltransferases | Transcription [30] |
| 5-hydroxymethylcytosine | Cytosine | Ten-eleven translocation enzymes | Transcription, self-renewal of stem cells [32] |
| 5-formylcytosine | Cytosine | Ten-eleven translocation enzymes | Transcription [30] |
| 5-carboxylcytosine | Cytosine | Ten-eleven translocation enzymes | Transcription [30] |
| Histone Modifications | |||
| Acetylation | K-ac | HAT1, CBP/p300, PCAF/GCN5, TIP60 HB01, ScSAS3 ScSAS2 (SpMST2) ScRTT109 |
Transcription, repair, replication, condensation [65] |
| Methylation | K-me1, K-me2, K-me3, R-me1 R-me2a R-me2s | SirT2 (ScSir2), SUV39H1, SUV39H2 G9a, ESET/SETDB1 EuHMTase/GLP, CLL8 SpClr4, MLL1, MLL2, MLL3, MLL4, MLL5, SET1A, SET1B, ASH1, Sc/Sp SET1, SET2, (Sc/Sp SET2), NSD1, SYMD2, DOT1, Sc/Sp DOT1, Pr-SET 7/8, SUV4 20H1, SUV420H2, SpSet 9, EZH2, RIZ1, LSD1/BHC110, JHDM1a, JHDM1b, JHDM2a, JHDM2b, JMJD2A/JHDM3A, JMJD2B, JMJD2C/GASC1, JMJD2D, CARM1, PRMT4, PRMT5 |
Transcription, repair [81] |
| Phosphorylation | S-p, T-p, Y-p | Haspin, MSK1, MSK2, CKII Mst1, JAK2, EGFR |
Transcription, repair, condensation [91, 164, 165] |
| Ubiquitylation | K-ub | Bmi/Ring1A, RNF20/RNF40 | Transcription, repair [99] |
| Sumoylation | K-su | SAE1/SAE2, UBC9, PIAS family proteins, RanBP2, Pc2 | Transcriptional repression, repair [103] |
| Neddylation | K-ned | UBA3/NAE1, UBE2F, RNF111 | Transcription, repair [112] |
| ADP ribosylation | E-ar | PARP1, SIRT4 | Transcription, repair [92] |
| Deimination | R > Cit | Protein arginine deiminases | Transcription and decondensation [92] |
| Proline isomerization | P-cis > P-trans | ScFPR4 | Transcription [92] |
| Formylation | K-fo | DNA binding [163] | |
| Hydroxylation | Y-OH | JMJD6 | Chromatin demethylation [163] |
| O-GlcNAcylation | S-GlcNAc, T-GlcNAc | O-Linked N-acetylglucosamine transferase | Transcription [163] |
| Butyrylation | K-bu | p300, CBP | Transcription [163] |
| Propionylation | K-pr | MYST family | Not known [163] |
| Crotonylation | K-cr | p300 | Transcription [163] |
Epigenetic modifiers are broadly classified into three groups. Epigenetic “writers” are a dedicated group of enzymes that add modifications to histones or DNA. Methylation (DNA methylation, protein arginine methylation, histone lysine methylation) and acetylation (histone acetylation) are among the most widely studied modifications catalyzed by these enzymes. The chemical modifications added by epigenetic writers are recognized by a set of proteins called epigenetic “readers” that mediate the effects of these modifications on to the transcription machinery. Epigenetic readers (DNA methylation readers, histone methylation readers and histone acetylation readers) contain specialized domains that can efficiently detect and bind to the modifications added onto the DNA and histones and can assist in the recruitment of transcriptional activators or repressors. The post-translational modifications laid down by epigenetic writers can be reversed by another set of enzymes called epigenetic “erasers” (DNA demethylases, histone demethylases and histone deacetylases) [8]. The addition and removal of the chemical tags can be dictated by the cell’s response to its environment or internal homeostasis, as per its requirement, and results in modification of gene expression through changes in DNA accessibility and the recruitment of proteins that activate or repress gene transcription. While these three classes of proteins are involved in gene regulation under normal cellular conditions, deregulation of the expression and activity of these epigenetic regulators has been frequently implicated in human disease, including the development and progression of cancer. The dynamic and reversible nature of these enzymatic epigenetic modifications makes them ideal targets for drug discovery and cancer therapeutics (Fig. 1).
Fig. (1).
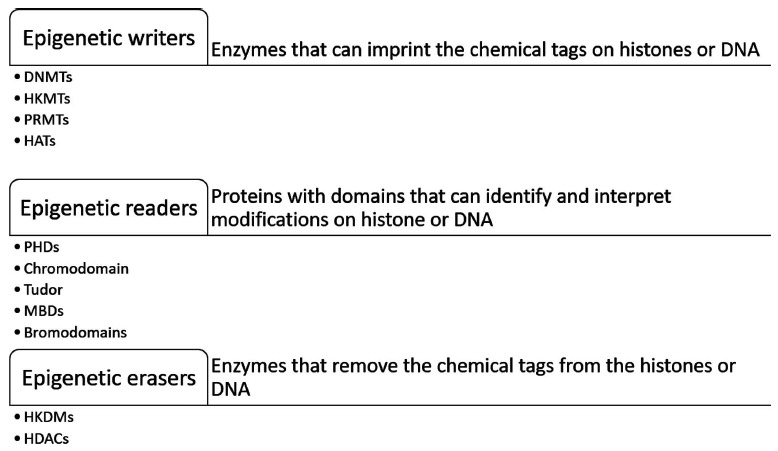
Epigenetic readers, writers and erasers.
The epigenetic machinery is extremely complex and heterogeneous, particularly in disease states, where changes in expression and activity of chromatin modifying enzymes can greatly influence changes in chromatin structure and gene expression. Due to advances in laboratory technology (see below), we can now precisely profile epigenetic mechanisms involved in the progression of cancer and can develop epigenetic inhibitors to combat the disease. Expanding literature has clearly established the prevalence of mutations in readers, writers and erasers apart from the ones found in chromatin remodeling proteins. Almost 20% of all cancers display a mutation in the mammalian SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin-remodeling complex [9]. Such a high mutation rate allows cancer cells to re-wire the transcriptional machinery in order to adapt to environmental changes. A variety of epigenetic drugs have been approved by the FDA against cancer. Methylation inhibitors such as Azacytidine and Decitabine were approved in 2004 and 2006, respectively, for leukemia and myelodysplastic syndromes and histone deacetylase inhibitors like Vorinostat, Romidepsin, Panobinostat and Belinostat were approved for lymphoma [10]. The last decade has witnessed the development of novel targeted therapies specifically designed against the epigenetic components, i.e. readers, writers and erasers. The ones that made it to clinical trials include JQ1 (BET bromodomain protein inhibitor), LSD1 (lysine-specific histone demethylase–1) inhibitors, mutant IDH1 and IDH2 inhibitors, EZH2 (enhancer of zeste 2) inhibitors, PRMT5 (protein arginine methyltransferase 5) and DOT1L (disruptor of telomeric silencing–1–like) inhibitors [11].
Having highlighted the significant impact of chromatin remodeling on the regulation of molecular activities within a cell, here we discuss various epigenetic factors and mutations responsible for modifying the topology of chromatin and the role of these factors during progression of the diseased condition, especially, but not limited to, cancer. We further discuss the progress achieved in the development of pharmacological inhibitors against the deregulated epigenetic factors.
2. MUTATIONS IN EPIGENETIC MEDIATORS
Regulated changes in chromatin structure and gene expression act to maintain cellular homeostasis. Perturbations in normal gene expression form the basis of many human diseases, including cancer. Epigenetics is a major determinant of the normal genetic program and its dysregulation plays a pivotal role in the development and progression of tumorigenesis, allowing for suppression of tumor suppressor genes and the increased expression of oncogenes. Additionally, changes in chromatin structure have a multifaceted function in regulating the enzymes involved in carcinogenesis. Comprehensive analysis of the genetic mutations in chromatin reveals that they are sufficient to fuel cancer progression. Findings from cancer genome sequencing projects suggest that 50% of cancers harbor mutations in the chromatin or chromatin-associated proteins [12]. Such mutations can reprogram epigenetic signaling and result in abnormal gene expression, which results in diseases like cancer, neuropsychiatric disorders and autoimmune diseases [13].
The frequent occurrence of mutations in chromatin regulatory proteins establishes that pervasive epigenetic dysregulation promotes cancer initiation and development. The Polycomb (PcG) and Trithorax (Trx) complexes are amongst the most commonly mutated epigenetic proteins in cancer [14]. PcG and Trx essentially act opposite to each other. PcG aids in tri-methylation at histone H3 lysine 27 (H3K27), a marker of repressed genes, whereas Trx catalyzes methylation of histone H3 Lys4 (H3K4), which is a modification that is strongly associated with active promoters [15]. Mutation in the TrxG member MLL1 by chromosomal translocation has been documented in childhood mixed-lineage leukemias (MLLs) [16]. Point mutations in EZH2 (Y641), the enzymatic subunit of the PcG complex, are frequently observed in non-Hodgkin lymphoma [17]. EZH2 Y641 expressing cells show an increase in the methylation levels at H3K27me3 [18-19]. In addition to Y641, other EZH2 activating mutations such as A677G [20] and A687V [21] have also been reported. These studies encouraged the development of EZH2 inhibitors for cancer therapy. GSK126, a small molecule inhibitor of EZH2, has been shown to specifically inhibit the proliferation of B-cell lymphomas that contain activating EZH2 mutations [22]. Pre-clinical studies with CPI-1205, a potent selective and reversible EZH2 inhibitor, showed advanced anti-proliferative effects in lymphoma and prostate cancer cell models and in patients with non-Hodgkin lymphoma [23]. Another first-in-class selective EZH2 inhibitor, tazemetostat, has been investigated for safety and clinical activity in patients with relapsed or refractory B-cell non-Hodgkin lymphoma and advanced solid tumors. Tazemetostat was found to exhibit a suitable safety profile and significant anti-cancer activity in patients and is now being tested in phase 2 studies [24].
Alterations or mutations in histones themselves can act to reprogram the epigenome and promote cancer progression. For instance, in pediatric glioma, mutations at two critical positions in histone H3.3, K27M and G34R/V, have been shown to be strongly associated with glioblastoma (GBM) pathogenesis [25]. Tumors which possess H3K27M mutation exhibit lower levels of H3K27me3 [26]. Use of a H3K27M transgene or peptide showed a reduction of endogenous H3K27me2/3 through a mechanism involving an inhibition of the PRC2 complex via interaction with EZH2 [27], whereas H3G34R/V mutations do not necessarily affect H3K27me2/3 levels but they do inhibit H3K36me exclusive to the HA-tagged H3.2/3 purified oligonucleosomes [27]. Apart from histone H3, histone H1 can also be mutated in cancer conditions. Histone H1 mutant H1S102F shows reduced capacity to associate with chromatin [28], suggesting that this mutant histone could result in local changes in chromatin structure, although this mutation requires further investigation. These reports collectively establish the association between the mutations in regulatory components of chromatin and the potential for cancer development.
3. EPIGENETIC MODIFICATIONS TO DNA
DNA methylation is one of the best-described epigenetic events that controls gene expression and is dysregulated in diseases like cancer. It is generally considered to be a mediator of silencing gene expression and usually occurs at cytosine residues, forming 5-methylcytosine. Methylation of cytosine residues occurs predominantly in the CpG islands within the genome. DNA methyltransferases (DNMTs) are the group of enzymes that establish and maintain methylation patterns in the CpG islands throughout the genome by catalyzing the transfer of a methyl group to a cytosine residue within DNA, with additional methylation of RNA molecules. DNMTs 1 and 3 localize to the nucleus, while DNMT2 localizes to the cytoplasm [29]. DNMT3a and 3b (also known as de novo methyltransferases) can methylate an unmethylated DNA and are particularly important in germ cells and embryonic development. DNMT1 maintains methylation patterns after DNA replication, showing a preference for hemimethylated DNA, helping to maintain methylation at most targeted CpG sites in dividing cells [30]. DNMT2 acts as an RNA methyltransferase [29]. DNA methylation can go beyond the methyl-additive DNMTs. Ten-eleven translocation (TET) proteins have been shown to convert 5-methylcytosine (5mC) to 5-hydroxymethylcy- tosine (5hmC) in cultured cells [31], which may serve as a means of demethylation of cytosine residues or may be involved in other functional roles [32]. Moreover, hydrolytic deamination of cytosine or 5-methylcytosine by cytidine deaminases results in mutation to thymidine, changing the genetic code if not repaired. DNA methylation at sites other than CpG sites has also been reported [33]. Cells with methylated non-CpG sites show lower DNA methylation at protein binding sites and enhancers.
DNA methylation is frequently observed to be deregulated in various cancer types and is an important mechanism for gene silencing, particularly of tumor suppressor genes, genes involved in DNA repair and those that help to maintain the epithelial phenotype. Suppression of gene expression due to changes in methylation patterns can occur in cancer types within the same tissue. For example, different subtypes of breast cancer display different patterns of DNA methylation. This abnormal hypermethylation of DNA is more prominent in luminal ERα positive breast cancer compared to ERα negative subtypes [34]. Notably, various genes that are essential for DNA repair are hypermethylated in colon cancer. Promoter methylation of the DNA mismatch repair gene MutL homolog 1 (MLH1) in sporadic primary colorectal cancers results in loss of protein expression and a loss of mismatch repair function [35-36]. In prostate cancer, hypermethylation of promoters of epithelium-specific genes and hypomethylation of epithelial to mesenchymal transition (EMT) markers are observed, resulting in EMT and stemness [37]. In these cells, methylation of CpG islands by DNMT3A was shown to play an important role in the observed phenotype. The expression of various tumor suppressor genes is repressed by promoter hypermethylation, which contributes to tumor initiation and progression. Hypermethylation of the cyclin-dependent kinase inhibitor 2A (CDKN2A) gene, which codes for p16-INK4a, was one of the earlier observations of a tumor-promoting role of DNA hypermethylation [38]. Other genes encoding CDK inhibitory proteins, including p15/CdkN2B [39] and p57KIP1 [40], have also been shown to be repressed through hypermethylation. Hypermethylation of the BRCA1 gene, which promotes homologous recombination and is considered to play a pivotal role in DNA repair by maintaining genome integrity, has been demonstrated in multiple cases of breast and ovarian cancers [41, 42]. Another methyltransferase, EHMT2 (Euchromatic Histone Lysine Methyltransferase 2), is involved in the methylation of histone H3 on Lysine 9. This methylation event is also associated with transcriptional repression and, like DNMTs, EHMT2 protein expression and activity is increased in human tumors [43-45]. Inhibition of EHMT2 expression inhibits cancer cell proliferation and tumor metastasis, demonstrating the importance of Histone H3 methylation in cancer progression [43, 44, 46].
Given the reversible nature of methylation patterns in the repression of genes that act to inhibit cancer development and progression, various DNMT1 inhibitors/ hypomethylating drugs have been developed and tested for cancer treatment (Table 2). 5-Azacytidine, a potent DNMT inhibitor, and its derivatives are the most successful epigenetic drugs to date and are currently used to treat leukemia and myelodysplastic syndromes (MDS). Initially considered as a cytotoxic agent, azacytidine was later found to possess hypomethylation activity as it incorporates into the DNA of rapidly dividing cancer cells [47]. 5-Aza is transported into the cells by the human concentrative nucleoside transporter 1 (hCNT1) [48], where it is phosphorylated by kinases to convert it into its active triphosphate form, which is then incorporated in DNA or RNA. Incorporated azacytidine restricts the methylation of DNA by DNMT due to the replacement of C5 of cytosine with N5 of the modified pyrimidine of azacytidine and directs DNMT towards proteasomal degradation [49]. Apart from DNMT disruption through DNA incorporation, 5-Aza can also incorporate within RNA and induce ribosomal disassembly, which then hinders the translation of tumor promoting proteins [50]. Decitabine (5-Aza-2’deoxycytadine), the deoxy form of 5-Aza, incorporates more efficiently into DNA than does 5-Aza. Decitabine was recently shown to restore the levels of the transcription factor Sox2 accompanied by a decrease in invasive and migratory potential of various head and neck cancer cell lines [51]. Instability of azacytidine and decitabine in the human physiological environment is the major limitation of these inhibitors. Guadecitabine, decitabine linked to a deoxyguanosine, is a more stable analog and shows improved bioavailability and stability [52]. Guadecitabine clinical trials involving 93 patients with acute myelogenous leukemia (AML) or MDS showed favorable results [53]. A recent advance has come in the identification of a compound that inhibits both DNA and histone methylation. A first in class dual DNMT/EHMT2 inhibitor, CM272, promotes apoptosis and hinders cell proliferation in vitro and in xenografts of hematological neoplasias, including AML, acute lymphoblastic leukemia (ALL) and diffuse large B-cell lymphoma (DLCBL) [54].
Table 2. DNMT inhibitors.
| Chemical Name | Mechanism of Action | Cancer Type | Clinical Stage |
|---|---|---|---|
| 5-Azacytidine | Cytosine analog | High-risk myelodysplastic syndromes (MDS) | FDA approved for treatment of MDS [48] |
| 5-Aza-2’-deoxycytidine | Cytosine analog | (MDS) | FDA approved for treatment of MDS [48] |
| SGI-110 | Cytosine analog | Acute myelogenous leukemia (AML) and MDS | Phase III clinical trial in AML [53] |
| 5-Fluro-2’-deoxycytidine | Cytosine analog | Refractory Solid Tumors | Phase II clinical trial in refractory solid tumors [161] |
| Zebularine | Cytosine analog | Human bladder cancer, AML cell lines and primary AML samples | Preclinical [161] |
| CP-4200 | Cytosine analog | MDS | Preclinical [161] |
| RG108 | Small molecule inhibitor | Endometrial cancer | Preclinical [161] |
| Nanaomycin A | Small molecule inhibitor | A549, HL60, and HCT116 cancer cell lines | Preclinical [161] |
| Procainamide | Antiarrythmic drug | Cloned T-cell lines | Preclinical [55] |
| Hydralazine | Antidiuretic | Cloned T-cell lines | Preclinical [55] |
| Valproic acid | Antiepileptic and mood stabilizer | AML or MDS | Preclinical [76] |
Hydralazine and procainamide are two non-nucleoside based DNMT inhibitors that have been tested for their potential therapeutic effect in solid tumors. Hydralazine, a widely-used vasodilator for the treatment of high blood pressure, blocks DNA methyltransferase activity by interaction of its nitrogen residues with the Lys-162 and Arg-240 residues of DNMT [55]. Procainamide, a sodium channel blocker used for the treatment of arrhythmias, acts as a competitive inhibitor of methyltransferase activity by binding to DNMTs. Although these non-nucleoside DNMT inhibitors exhibit lower cytotoxicity, they are not as potent as hypomethylating agents as is 5-Aza [56]. Other strategies used for DNMT inhibition are briefly described in (Table 2).
Although genomic hypermethylation in cancer has gathered immense attention, little is known about cancer associated global hypomethylation. Human cancers, including prostate metastatic tumors, leukocytes from B-cell chronic lymphocytic leukemia, hepatocellular carcinomas and cervical cancer exhibit global genomic hypomethylation as compared to their respective normal tissues [57-60]. Recently PRMT6, a protein arginine methyltransferase that demethylates histone H3 arginine 2 (H3R2me2a), has been shown to induce global hypomethylation in breast cancer cells [61]. The chromatin signature of DNA hypomethylated sequences is more profound at genomic regions that are marked with a repressive H3K9me3 histone mark [62]. Thus it is reasonable to propose that both localized hypermethylation and global hypomethylation play an important role in cancer progression.
4. HISTONE MODIFICATIONS
Proteins of the histone family are some of the most abundant and conserved alkaline proteins in eukaryotes and play pivotal roles in DNA packaging and gene regulation. The DNA strand is wrapped around the histone octamer, which is composed of two copies of each of the core histones (H3, H2A, H2B and H4). Histone H1 is a linker histone and helps secure the DNA wound around the nucleosome [63]. The N-termini of histone is amenable to various post-translational modifications including, but not limited to, methylation, acetylation, sumoylation, ubiquitylation, neddylation, phosphorylation and ADP ribosylation. Post-translational modifications to histones can alter the accessibility of DNA to proteins and transcription factors. An open euchromatin structure is more amenable to binding of transcription factors and is casually linked with gene activation, whereas a closed heterochromatin structure is more restrictive and is a marker of repressed genes. This circuitry involving post-translational modifications of histones is thus considered crucial for the regulation of gene expression. Apart from the canonical histones, the epigenetic landscape also consists of histone variants which determine nucleosome stability. Histone variants are more commonly descendants of H1, H2A and H3 families. Their sequences and structures vary from the core histones and are subjected to distinct post-translational modifications. Emerging evidence suggests a role for core histones, histone variants and their PTMs as potential drivers of cancer [64]. Thus multiple small molecule inhibitors of these epigenetic events are currently being evaluated in the clinic for cancer treatment and are discussed below.
4.1. Histone Acetylation
Histone acetylation is largely associated with active transcription and is mainly localized at enhancer regions, promoters and the gene body. Addition of a negatively charged acetyl group to a histone tail results in charge neutralization of the positively charged histone. This positive charge neutralization weakens the electrostatic interaction between histones and the negatively charged DNA, resulting in an open conformation of chromatin that is more accommodating to transcription factors. Histone acetylation is catalyzed by histone acetyltransferases (HATs) which include the members of the GNAT family (Gcn5-related N-acetyltransferases) that specifically target H3K9, H3K14, H3K36 histones, the MYST family that acetylate H4K5, H4K8, H4K12, H4K16, H3K14, H3K23 histones and the CBP/p300 (cAMP response element binding protein) family that is responsible for acetylating H2AK5, H3K9, H3K23, H3K56 residues [65]. The acetylation reaction mainly involves the transfer of an acetyl group from the acetyl-coenzyme A (acetylCoA) cofactor to the e-amino nitrogen of lysine residues. Besides charge neutralization, lysine acetylation also supports the binding of proteins with bromodomains (readers), which identify this modification and aid in the recruitment of transcriptional activators [66]. Thus acetylated lysines can serve as docking sites for regulatory factors to bind to their target sequences and guide the recruitment of additional transcription factors that stimulate gene expression.
Given the diverse role of histone acetylation in regulation of various downstream biological processes, it is not surprising that deregulated histone modification is commonly linked with the genesis of various diseases. A number of reports describe the link between over-expressed histone deacetylases (HDACs) and cancer progression or clinical poor outcome [67]. HDAC inhibitors have been successfully used in combination with DNA-damaging agents, taxanes, death receptor agonists and hormonal therapies [68]. Recently, it was established that paclitaxel-resistant non-small cell lung cancer (NSCLC) cells exhibited increased HDAC expression and activity, which augmented the rate of cell proliferation. HDAC inhibition in these cells led to paclitaxel sensitization and the induction of apoptosis [69]. In epidermal growth factor receptor (EGFR)-mutant lung cancer cells, the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA, vorinostat) markedly reduced cell proliferation and reduced tumor growth in a xenograft model by causing cell cycle arrest in the G2/M phase [70]. Similarly, in multidrug resistant colorectal cancer, inhibition of HDAC7 using trichostatin A (TSA) relieved the histone hypoacetylation at CNT2, which is a transporter of natural nucleosides and nucleoside‐derived drugs [71]. HDAC inhibitors have also been shown to restore the expression of tumor suppressor genes, such as tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and Death Receptor 5 (DR5), which are frequently involved in the induction of cellular apoptosis. Alternatively, they have also been shown to inhibit the expression of pro-survival genes such as B-cell lymphoma 2 (BCL2). HDAC inhibitors can also enhance immune responses and upregulate major histocompatibility complex class (MHC) I and II proteins and co-stimulatory molecules such as CD80 and CD86 [72]. To date, three HDAC inhibitors, vorinostat, romidepsin and panobinostat (LBH-589, PS), have been FDA approved and are used for the treatment of T-cell lymphoma and multiple myeloma.
TSA, an antifungal agent, has been shown to possess reversible HDAC inhibitory activity that regulates apoptosis, angiogenesis and cell differentiation. TSA promotes histone H4 acetylation and results in increased expression of the cyclin-dependent kinase inhibitor p21, which is associated with G1 phase arrest [73]. The mechanism of anti-cancer activity of vorinostat is quite similar to TSA. Vorinostat represses telomerase activity by up-regulating p21. Valproic acid (VPA), long-used as an anti-epileptic agent due to its blockage of sodium channels in the brain, also acts as an HDAC inhibitor [74] and has been shown to enhance the sensitivity of lung cancer cells to cisplatin [75]. Combination therapy of VPA with the CDK inhibitor P276-00 demonstrated an enhanced therapeutic effect on NSCLC cell lines [76]. Belinostat, a pan-histone deacetylase inhibitor, has been FDA approved for the treatment of relapsed or refractory peripheral T-cell lymphoma (PTCL). Yet another HDAC inhibitor, Panobinostat, was recently shown to sensitize EGFR-mutated and wild-type NSCLC cells to the anti-proliferative activity of the EGFR inhibitor erlotinib by enhancing the acetylation of histone H3 [77].
Apart from targeting the aberrant expression of HDACs, various strategies targeting bromodomain (BRD) and extra-terminal domain (BET) proteins have been actively explored over the past several years. BRD-containing proteins are the readers of acetyl marks on histone tails which on interaction with acetylated chromatin results in recruitment of regulatory factors that influence gene expression. Their druggable nature has encouraged the development of targeted therapies in recent years. Two small-molecule inhibitors of BET proteins, JQ1 and I-BET, have shown favorable results in pre-clinical studies and are currently being tested in clinical trials [78]. JQ1 binds particularly well to BRD4 and inhibit the transcription of the MYC oncogene [79], which is known to promote hematological cancers such as acute myeloid leukemia, B cell acute lymphoblastic leukemia and diffuse large B cell lymphoma. A comprehensive review describing the discovery and uses of bromodomain inhibitors was recently published [80].
4.2. Histone Methylation
Methylation is one of the most complex and well-studied PTMs of the histone. Histone tails can be methylated at lysine and arginine residues, but the fact that these amino acids can be mono-, di- or tri-methylated adds additional layers of complexity to the functional responses to these modifications. Histone methylation is catalyzed by enzymes which can be classified into three distinct families of proteins —the SET-DOMAIN-containing protein family, the non-SET-domain proteins DOT1/DOT1L and the protein arginine methyltransferases (PRMT1) family. The SET-domain containing protein methyltransferase superfamily includes members that can methylate lysines in histone tails. Alternatively, DOT1 family members methylate K79 in the globular region of histone H3 and are structurally different from SET-domain proteins. PRMTs catalyze the transfer of methyl groups from S-adenosyl-L-methionine (SAM) to the guanidino nitrogens of arginine residues.
Histone lysine methylation has been reported to occur at H3 and H4. In contrast to lysine acetylation, which is usually a mark of gene activation, lysine methylation can result in either gene activation or repression, depending upon the site being methylated [81]. Classically, H3K27me and H3K9me are considered to depict a silent or compressed chromatin whereas H3K4me, H3K36 and H3K79 are implicated in activation of transcription. H3K27me is the most well characterized modification and is catalyzed by the PcG protein and SET-domain containing histone methyltransferase EZH2. EZH2 forms a complex with EED and SUZ12 in order to methylate H3K27. The tri-methylation mark imparted by EZH2 represents transcriptional repression; however, the mechanism underlying this transcriptional downregulation is still poorly understood. In contrast to acetylation, histone methylation does not result in charge alteration, but instead promotes the recruitment of regulatory factors which ultimately results in gene suppression. Studies have shown that EZH2 is required for recruitment of DNMT to its target gene promoters and subsequent methylation of DNA [82]. These studies also highlight the interconnectivity between DNA and histone methylation. EZH2 has been frequently found to be overexpressed in breast, prostate, lung and blood cancers [83]. GSK126 is an EZH2 inhibitor with a Ki value of 0.5–3 nM against wild-type EZH2 and all mutant forms. GSK126 has shown potent anti-proliferative activities in diffuse large B-cell lymphomas that contain EZH2 activating mutations [83]. Various other EZH2 inhibitors are currently under clinical trials and many other inhibitors of mutant EZH2 are currently being developed.
DOT1L (disruptor of telomeric silencing 1-like) methylates H3K79, which can initiate an active transcriptional state. Various reports have suggested a direct correlation between DOT1L and breast cancer malignancy. Apart from breast cancer, DOT1L plays an important role in the progression of lung cancer. Knockdown of DOT1L using siRNA blocked the proliferation of both A549 and NCI-H1299 cells [84]. DOT1L is also a target for mixed lineage leukemia (MLL). EPZ004777 is a DOT1L inhibitor which exhibits selective antitumor activity against MLL [85].
Similar to lysine methylation, arginine methylation can either represent active or repressive chromatin. Arginine methylation is difficult to detect in vivo; nevertheless, the prevalence of a number of PRMTs suggests that this is a relatively widespread modification. Extended literature demonstrates increased levels of PRMTs in cancer progression. Recently, higher expression of PRMT5 was reported in lymphoma, leukemia and solid tumors [86]. A selective inhibitor of PRMT5 enzymatic activity, EPZ015666 (GSK3235025), exhibits potent anti-proliferative effects in both in vitro and in vivo models of MCL [87]. In NSCLC patients, PRMT1 and CARM1 have been observed to be overexpressed. PRMT1 promotes NSCLC progression and metastasis by methylating the transcription factor Twist1, which further represses CDH1 (E-cadherin), promoting EMT [88]. In breast cancer, PRMT7 supports EMT and metastasis by inhibiting CDH1 expression [89]. Together, these reports suggest that the development of inhibitors to arginine methylation could have an impact on several cancer types.
4.3. Histone Phosphorylation
Apart from acetylation and methylation, histone phosphorylation has also been long known to be involved in cellular responses, especially after DNA damage and during cell division. Serine, threonine and tyrosine are the amino acids on histone tails that are prone to phosphorylation. Phosphorylation of the histone variant H2A.X at Ser 139 after DNA damage identifies the site of impaired DNA in the chromatin [69, 90]. Phosphorylation of histone H3 at S10 is an exclusive histone mark associated with cellular transformation [91]. In mammalians, MSK1/2 and RSK2 are the two kinases that have been shown to target H3S10. This modification has been shown to be important for the activation of NFKB-regulated genes [92]. H3S10 phosphorylation is also widely known to aid transcriptional activation by promoting H3K14 acetylation and altering the conformation of chromatin. During mitosis, H3S10 can be phosphorylated by Aurora B kinase which results in displacement of heterochromatin protein 1 (HP1) from H3K9me, which is commonly associated with compressed chromatin [93]. Recently, Aurora B kinase has also been shown to phosphorylate S31 of histone H3.3 [94]. Other sites of phosphorylation linked to gene activation are S28 and T11 [65]. Threonine phosphorylation has not been demonstrated rigorously; nevertheless, H2A threonine 119 phosphorylation by nucleosomal histone kinase-1 has been shown to play a pivotal role in cell cycle progression [95]. Some studies suggest a plausible link between histone phosphorylation and acetylation. One school of thought suggests that both these PTMs are spatially linked but are independent of each other, whereas another presumes that the two PTMs co-exist due to simultaneous recruitment of kinases and histone acetyltransferases [96].
4.4. Histone Ubiquitination
The 76 amino acid protein Ubiquitin (Ub) is ubiquitously distributed and highly conserved throughout eukaryotic organisms. Ub is known to regulate diverse molecular processes including the targeting of proteins for degradation by the 26S proteasome, activation of stress responses, cell-cycle regulation, protein trafficking, endocytosis signaling and transcriptional regulation [97]. Ub conjugates to its substrate proteins via a 3 step enzymatic cascade involving a ubiquitin activating enzyme (E1), which is followed by its conjugation to a ubiquitin-conjugating enzyme (E2) through a thioester bond. The final step involves transferring ubiquitin from the E2 enzyme to a target lysine residue in the substrate protein by a RING (Really Interesting New Gene) E3 ubiquitin ligase, or through direct transfer of the ubiquitin to a catalytic cysteine in the E3 ubiquitin ligase, the latter occurring with HECT (Homologous to E6 AP Carboxyl Terminus) and RBR (RING-Between-RING) ligases. These latter E3 ubiquitin ligases then directly transfer the Ub onto the target substrate. Protein targets can either be monoubiquitinated with a single Ub on a lysine, or polyubiquitinated through the stepwise addition of a ubiquitin chain. Histone H2A was the first protein identified as a target of ubiquitination, with modification at lysine 119 [98]. In mammals the majority of H2A and H2B is monoubiquitinated; Lys-119 (UbH2A) and H2B at Lys-120 (UbH2B) [99]. Histone ubiquitination is associated with cellular processes such as DNA repair, transcriptional regulation and genome stability. Histone ubH2A mediates transcriptional silencing via polycomb proteins. Other core histones H3 and H4 can also be monoubiquitinated in response to DNA damage [100]. H2B ubiquitination plays an essential role in both transcriptional activation and tumor suppression [101]. A number of studies have shown lower H2Bub1 expression in gastric, parathyroid and colorectal tumors [102].
4.5. Histone Sumoylation
Small ubiquitin-related modifier (SUMO) has been well-characterized as a ubiquitin-like protein frequently involved in post-translational modifications which impinges on various biological processes [103]. Three members of the SUMO family have been characterized: SUMO-1, SUMO-2 and SUMO-3. A fourth member of this family was later identified (SUMO-4) which, like the other members, regulates subcellular localization and stability of target proteins. Conjugation of SUMO to target proteins involves a similar set of enzymatic reactions as ubiquitination (E1 activating enzyme, E2 conjugating enzyme, E3 protein ligase). Various sumoylation substrates have been described to date, some of which are: p53 [104], MDM2 [105], PML [106], etc. Histone H4 has also been reported to be modified by SUMO. Histone H4 sumoylation (suH4) results in gene repression mediated by histone deacetylases and HP1 [107]. Recently it was shown that suH4 stimulated the activity of lysine-specific demethylase 1 (LSD1) which represses eukaryotic gene expression by demethylating mono- and dimethylated Lys4 in histone H3 [108]. Apart from histones, SUMO can modify histone modifying enzymes such as HDACs, EZH2 and KDM5B [109]. Sumoylated proteins have also been shown to be important for the DNA damage repair response. In human cells, histone variant H2A.Z-2 is SUMOylated by PIAS4, which is essential for its exchange at the DNA damage sites [110]. Increased expression of SUMO proteins has been linked with cancer development; therefore, SUMO proteins could be novel targets for cancer therapeutics [111].
4.6. Histone Neddylation
Protein neddylation is carried out by addition of the NEDD8 protein, which shares a high degree of structural similarity to Ub. NEDD8 addition to proteins utilizes similar chemical reactions as ubiquitination; however, NEDD8 conjugated proteins are more prevalent in the nucleus than in the cytoplasm [112]. Cullin, a vital subunit in the SCF E3 Ub ligase complex, is the most widely-studied neddylation substrate. NEDD8 conjugates to and introduces a conformational change in cullin which promotes its binding to an E2 Ub conjugase that ultimately results in protein ubiquitination and degradation [113]. In neuroendocrine tumors, increased neddylation of cullin has been positively correlated with tumor progression [114]. Elevated levels of neddylation enzymes (e.g. NEDD8 E1, NAE1/UBA3 and NEDD8 E2, UBE2M/UBE2F) have been reported in lung cancer, liver cancer, colorectal cancer, glioblastoma, nasopharyngeal carcinoma and esophageal squamous cell carcinoma, as compared to normal tissues [115]. Higher expression of these enzymes has also been associated with lower overall survival in patients [116]. Histone H4 and H2A have recently been identified as NEDD8 target proteins. Interestingly, neddylation of H2A suppresses its ubiquitination and thus interferes with DNA damage repair [117]. On the contrary, NEDD8 conjugation to N-terminal lysine residues of H4 in response to DNA damage results in recruitment of factors that facilitate the amplification of the Ub cascade and thus assist in DNA damage repair [118]. Given the role of NEDD in cancer progression, neddylation appears to be a promising therapeutic target. MLN4924 (pevonedistat), developed by Millennium Pharmaceuticals, is a small molecule inhibitor of the E1 NEDD8-activating enzyme and is currently in phase I/II clinical trials. MLN4924 inactivates the first step of the neddylation cascade [119]. MLN4924 interferes with cullin neddylation and blocks CRL activation, which results in accumulation of various CRLs substrates. This further triggers various cell death pathways in cancer cells. MLN4924 has also been used in combination with other anti-cancer drugs. Combination of MLN4924 and azacytidine in patients with AML (NCT01814826) exhibited promising clinical effects in phase 1b. The combination therapy resulted in 33% and 22% complete and partial responses, respectively, which was superior to azacitidine or MLN4924 alone response [120].
5. EPIGENETICS OF RNA
Modifications to RNA molecules occur rather frequently and are critical for molecular regulation of biological processes. In contrast to DNA, RNA is known to consist of more than 100 types of modifications resulting in alternative nucleotide forms [121]. This large concoction of RNA nucleotides allows for the diverse catalytic and regulatory functions, apart from its conventional role of translating genetic code to functional proteins. These nucleotides can alter the structure and complementarity of RNA and can also render the RNA molecules more amenable to binding to various proteins within the cellular machinery. Despite the multi-functional prowess of RNA molecules, very little is known about epigenetic modification of RNA and its impact on biological processes. Here we discuss in brief the recent developments in epigenetic modifications to RNA.
5.1. RNA Methylation
RNA is composed of purine and pyrimidine rings that can be chemically altered by addition of various chemical groups such as acetyl, isopentenyl and threonylcarbamoyl, but the most frequently found and extensively studied modification is the addition of methyl groups. Methylated nucleotides have been identified and studied expansively in rRNAs and tRNAs, but are lesser known in mRNAs. rRNA modifications mostly regulate the quality control checkpoints during the assembly of ribosomes, whereas tRNA modifications are associated with tRNA folding and stability [122-123]. Methylation of adenosine at the N6 position (m6A) is considered to be the predominant mRNA modification and is also detected in rRNA and tRNA [124-125]. Other known modifications of mRNA include m5C and 2’O methylation [126].
m6A modifications reportedly play a role in biological processes such as mRNA splicing and stability [127]. Similar to other epigenetic modifications to histone and DNA, m6A is dynamic and reversible. Lin et al. demonstrated the functional importance of the m6A mRNA modification by knocking out METTL3 RNA methyltransferase, the enzyme responsible for N6 adenosine methylation, which resulted in apoptosis in human lung cancer cells [128]. They showed that METTL3 promotes translation by interacting with the translation initiation machinery and is required for growth and invasion of lung cancer cells. The role of m6A RNA modification as a positive regulator for cell programming to pluripotency was also described recently. Other studies have shown that m6A modification is essential for sustaining the ground state of human and mouse embryonic stem cells (ESCs) [129-130]. Increased m6A has been found to have significant effects on cancer initiation and progression. Post-translation modifications also play a pivotal role in the repair of damaged DNA by regulating the access to chromatin and the recruitment of DNA repair proteins to the damaged sites. Recently it was shown that UV-induced DNA damage resulted in the accumulation of m6A at the sites of DNA damage. Xiang et al. showed that the catalytic activity of MTTL3 and MTTL14 rapidly increases after UV irradiation, which further results in polymerase κ localization near the damaged sites. The authors suggest that m6A methylation may be important for pol κ mediated DNA repair [131].
5.2. Non-Coding RNA (ncRNA)
A ncRNA is a functional RNA molecule that is not translated into polypeptides or protein sequence, but instead regulates the expression of genes at transcriptional or post-transcriptional level. ncRNAs can be classified as short ncRNAs (<30 nucleotides) and long ncRNAs (>200 nucleotides). Short ncRNAs can be further grouped as microRNA (miRNAs), short interfering (siRNA) and piwi-interacting (piRNA). Aberrant expression of miRNAs has been frequently associated with cancer development. MiRNAs can function as tumor suppressors (miR-15a and miR-16-1) as well as oncogenes (miR-155 or members of the miR-17–92 cluster) [132]. MiRNA can target multiple protein coding regions and regulate the rate of translation of about 60% of protein-coding genes [133]. Let7 is one of the most studied tumor suppressive miRNA and is implicated in tumorigenesis of the head and neck, lung, colon, rectum and ovary [134]. Additionally, some reports suggest that miRNAs can control the expression of epigenetic regulatory enzymes such as DNMT, HATs and HMTs [135]. A recent review describes in detail various small ncRNAs identified as drivers of oncogenesis in various cancer types [136].
A regulatory role for long ncRNAs in various biological processes has surfaced over the past decade. The mechanisms underlying the regulatory roles of lnc RNA is heterogeneous and primarily depends upon their localization and interacting proteins. LncRNAs can orchestrate chromatin folding by directing the epigenetic regulators to specific promoters of protein coding genes. Deregulated lncRNA can have a significant effect on the process of tumorigenesis [137]. Various lncRNAs have been shown to functionally interact with epigenetic components and control the deposition of histone marks on the chromatin. X-inactive specific transcript (Xist), a lncRNA involved in X chromosome inactivation (XCI) in early female embryonic development, is one of the first known lncRNAs involved in the formation of a repressive complex on the chromatin. Xist requires two proteins, hnRNPK (heterogeneous nuclear ribonucleoprotein K) and SHARP (SMRT/histone deacetylase 1 (HDAC1)-associated repressor protein), for its interaction with PRC2 to maintain the silenced state during the imprinted phase of X-chromosome inactivation [138]. HOX transcript antisense RNA (HOTAIR) lncRNA has also been known to repress gene expression and its deregulation was shown to promote the epigenetic alterations favorable for tumor growth and metastasis, particularly in breast cancer. HOTAIR can acquire secondary structure that favors its association with PRC2 proteins, which can then deposit H3K27m3 repressive marks and inhibit transcription [139]. Various other lncRNAs involved in the pathogenesis of cancer have been discussed recently [140]. Taken together, these reports suggest lncRNAs as a lucrative therapeutic target for cancer therapy.
6. TOOLS TO STUDY THE EPIGENOME
The most common methods used for assessing epigenetic changes include microarray-based techniques and next-generation sequencing (NGS). These approaches have been widely applied to analyze DNA methylation, DNA-protein interactions and the non-coding RNAs such as miRNAs and lncRNAs.
6.1. DNA Methylation Analysis
DNA methylation can be assessed in a variety of ways. A commonly explored method involves the use of sodium bisulfite to differentially convert unmethylated cytosines to uracil, while methylated residues are unchanged. The detection of methylated cytosine residues is then checked either by PCR or Sanger sequencing to map differentially-methylated regions in the genome. The sodium bisulfite approach can also be followed by NGS technology, which allows for robust methylation profiling of the entire genome-hence the name whole genome bisulfite sequencing (WGBS) [141]. For qualitative analysis of the methylation pattern when using sodium bisulfite, high-resolution melt (HRM) analysis can be carried out. Using this method, uracil-containing DNA fragments are amplified by PCR and then subjected to decreasing temperatures. To determine changes in the methylation profile of a test DNA sample both the melt temperature and melt curve are compared to a standard DNA sample [142]. Methylated DNA can also be measured using commercially available microarrays for specific regions of interest. This represents a fast and cost-effective technique; it relies on the use of immunoprecipitation beads coupled to antibodies against 5-methylcytosine to determine the content of methylated DNA fragments. This approach is known as Methylated DNA immunoprecipitation or MeDIP. However, there are several commercial kits that enable the measurement of methylated DNA in the cell lysate without prior immunoprecipitation using an enzyme-linked immunosorbent assay (ELISA). It’s a quick, easy, and suitable technique for the identification of wide changes in global DNA methylation. Utilizing this method, methylated cytosine content can be accurately assessed using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) [141].
6.2. DNA-Protein Interaction Analysis
Beyond DNA methylation, another widely-explored area in the epigenetic field involves the study of histones modifications. The interaction between these proteins and DNA not only impacts the expression of genes, but also affects the accessibility and packaging of DNA itself. The gold standard method for investigating histone-DNA binding is chromatin immunoprecipitation (ChIP). This technique relies on specific antibodies directed against the modified histones (usually methylated or acetylated) followed by DNA analysis using conventional PCR, qPCR, microarray hybridization or sequencing. ChIP followed by massive parallel sequencing, also referred to as ChIP–Seq, is the most recent and commonly applied approach to characterize histone-associated DNA fragments. The objective of ChIP-Seq is to determine the association between histones or histone modifications and their specific regions in the genome (for example, transcription factor binding sites). ChIP-Seq also enables the assessment of the effect of chromatin changes on gene expression that can be correlated with key regulatory events important for numerous biological functions [143].
6.3. Chromatin Accessibility Analysis
Chromatin plays a major role in directing gene expression by controlling the accessibility of DNA to transcription factors and other regulating proteins. The conformation of chromatin can be assessed using nucleases that digest the chromatin in its open state (euchromatin) only while the inaccessible form (heterochromatin) remains intact. qPCR can then be performed to detect any regions in the genome that are epigenetically silenced by comparing the shift in the quantity of the PCR product between nuclease treated and untreated samples. A high throughput approach, such as NGS, may be applied after the nuclease’s treatment, (example: DNase I-Seq) which enables the analysis of the whole genome chromatin accessibility and the comparison of DNA-structural differences between samples on a genome-wide large scale [144].
6.4. Non-Coding RNAs Analysis
Non-coding RNA measurement can be achieved using different methods including qPCR, microarrays and sequencing. There are a variety of kits that enable extracting small RNAs while preserving their delicate structures. Following isolation, RNA molecules are reverse transcribed into cDNA and analyzed by qPCR. More commonly, the cDNA product is sequenced using an NGS based technology known as RNA-Seq. This also involves the use of computational approaches to measure the RNA content in biological samples. This method offers a significant advantage over hybridization-based techniques due to its ability to identify structural variants and discover new transcript products of gene fusion and alternative splicing, especially when it is accompanied by deep RNA-Seq [145]. Alternatively, hybridization-based technologies allow for the detection of ncRNAs present at low levels that are not accurately assessed by high throughput sequencing.
7. EPIGENETIC DRUGS USED FOR TREATING OTHER DISEASES
Epigenetic deregulation has been reported to play a significant role in health conditions other than cancer. In mice, histone acetylation plays a significant role in memory formation, such that deregulation of histone H4 lysine12 (H4K12) acetylation results in failure of hippocampal gene expression that is required for memory consolidation. Aged mice treated with vorinostat show improved memory and re-establish learning-induced gene expression [146]. In mouse and rat models, vorinostat has been shown to exhibit anti-rheumatic activity [147]. Moreover, vorinostat reportedly slows down the progression of Huntington-like syndrome in mice [148]. Additionally, mice that were deficient in the CBP gene showed signs of impaired memory [149]. Several reports highlight the anti-inflammatory and specific immune modulatory activity of HDACi [150-151]. Valproic acid has been tested for treating schizophrenia and Alzheimer's Disease (AD). VPA has been shown to reduce Aβ plaque in AD transgenic mice. VPA inhibits GSK-3β-mediated γ-secretase cleavage of APP which decreases Aβ production and improves the memory in AD mouse model [152]. HDACi have also been shown to possess immune suppressive activity. Regulatory T-cells (Tregs) control transplant tolerance by suppressing immune reactivity. HDACi regulates the transcription of FOXP3, a transcription factor that plays a pivotal role in the immunosuppressive function of Treg cells [153]. HDACi have also been implicated in type 2 diabetes. HDAC5 regulates a crucial glucose transporter, GLUT4, which is frequently deregulated in diabetes [154]. Recently it was shown that overexpression of HDAC7 in pancreatic islets and clonal beta cells causes beta cell dysfunction and treating HDAC7 overexpressing cells with TSA (an inhibitor of class I and class II HDACs) and MC1568 (an inhibitor of class II HDACs) reversed this effect [155]. Another independent study describes the effect of HDAC3 on dual specificity phosphatase 5 (DUSP5) which results in the initiation of diabetic cardiomyopathy (DCM) [156].
Additional roles for DNA and histone methylation in non-oncogenic diseases are also emerging. Increased DNMT expression is associated with hypermethylation of REELIN, an extracellular protein involved in neuronal migration and positioning during brain development and which might be involved in the development of schizophrenia [157]. The DNMT inhibitor zebularine disrupts neuro inflammation in LPS administered mice [158]. Recently it was shown that DNMT inhibition using 5‐aza‐2′‐deoxycytidine (dAZA) and zebularine alters the cocaine self-administration capability in rats [159]. A potential role of DNA methylation in systemic lupus erythematosus (SLE), an autoimmune disease, has also been discussed lately [160, 161]. Epigenetic modifications also play an important role in the pathology of multiple sclerosis (MS) [162].
CONCLUSION
In the last two decades, we have witnessed great progress in understanding the impact of epigenome on pathological conditions like cancer. Advancements in technological capabilities have allowed us to identify various new histone posttranslational modifications [163] and to visualize high-resolution maps for the epigenetic modifications on chromatin that can drive cancer progression. In spite of this impressive progress, there are still a lot of ‘not-so-well understood’ epigenetic phenomena. Our current understanding of the complexity of human epigenetics is still preliminary and further studies are required to translate the exciting epigenetic discoveries from the lab into the clinic.
ACKNOWLEDGEMENTS
Declared none.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
J.K., A.D. and S.T.E. wrote the paper and J.K. made the figures and tables. This work was supported by grant number 5R01CA187342 from the National Cancer Institute.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Morgan M.A., Shilatifard A. Chromatin signatures of cancer. Genes Dev. 2015;29:238–249. doi: 10.1101/gad.255182.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornberg R.D. Chromatin structure: A repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 3.Laybourn P.J., Kadonaga J.T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991;254:238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- 4.Cutter A.R., Hayes J.J. Linker histones: Novel insights into structure-specific recognition of the nucleosome. Biochem. Cell Biol. 2016;95:171–178. doi: 10.1139/bcb-2016-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos E.I., Reinberg D. Histones: Annotating chromatin. Annu. Rev. Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 6.Campos E.I., Reinberg D. New chaps in the histone chaperone arena. Genes Dev. 2010;24:1334–1338. doi: 10.1101/gad.1946810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai W.K., Pugh B.F. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat. Rev. Mol. Cell Biol. 2017;18:548. doi: 10.1038/nrm.2017.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswas S., Rao C.M. Epigenetic tools (the writers, the readers and the erasers) and their implications in cancer therapy. Eur. J. Pharmacol. 2018;837:8–24. doi: 10.1016/j.ejphar.2018.08.021. [DOI] [PubMed] [Google Scholar]
- 9.Kadoch C., Crabtree G.R. Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci. Adv. 2015;1:e1500447. doi: 10.1126/sciadv.1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verma M., Kumar V. Epigenetics Aging Longevity. Elsevier; 2018. Epigenetic drugs for cancer and precision medicine. pp. 439–451. [Google Scholar]
- 11.Dawson M.A. The cancer epigenome: Concepts, challenges, and therapeutic opportunities. Science. 2017;355:1147–1152. doi: 10.1126/science.aam7304. [DOI] [PubMed] [Google Scholar]
- 12.Shen H., Laird P.W. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suvà M.L., Riggi N., Bernstein B.E. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piunti A., Shilatifard A. Epigenetic balance of gene expression by polycomb and compass families. Science. 2016;352(6290):aad9780. doi: 10.1126/science.aad9780. [DOI] [PubMed] [Google Scholar]
- 15.Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 16.Ziemin-van der Poel S., McCabe N.R., Gill H.J., Espinosa R., Patel Y., Harden A., Rubinelli P., Smith S.D., LeBeau M.M., Rowley J.D. Identification of a gene, MLL, that spans the breakpoint in 11q23 translocations associated with human leukemias. Proc. Natl. Acad. Sci. USA. 1991;88:10735–10739. doi: 10.1073/pnas.88.23.10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morin R.D., Mendez-Lago M., Mungall A.J., Goya R., Mungall K.L., Corbett R.D., Johnson N.A., Severson T.M., Chiu R., Field M. Frequent mutation of histone-modifying genes in non-hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sneeringer C.J., Scott M.P., Kuntz K.W., Knutson S.K., Pollock R.M., Richon V.M., Copeland R.A. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl. Acad. Sci. USA. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yap D.B., Chu J., Berg T., Schapira M., Cheng S.W., Moradian A., Morin R.D., Mungall A.J., Meissner B., Boyle M., Marquez V.E., Marra M.A., Gascoyne R.D., Humphries R.K., Arrowsmith C.H., Morin G.B., Aparicio S.A. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117:2451–2459. doi: 10.1182/blood-2010-11-321208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCabe M.T., Graves A.P., Ganji G., Diaz E., Halsey W.S., Jiang Y., Smitheman K.N., Ott H.M., Pappalardi M.B., Allen K.E. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc. Natl. Acad. Sci. USA. 2012;109:2989–2994. doi: 10.1073/pnas.1116418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ott H.M., Graves A., Pappalardi M.B., Huddleston M., Halsey W.S., Hughes A., Groy A., Dul E., Jiang Y., Bai Y. A687V EZH2 is a driver of histone H3 lysine 27 (H3K27) hyper-trimethylation. Mol. Cancer Ther. 2014;13:3062–3073. doi: 10.1158/1535-7163.MCT-13-0876. [DOI] [PubMed] [Google Scholar]
- 22.McCabe M.T., Ott H.M., Ganji G., Korenchuk S., Thompson C., Van Aller G.S., Liu Y., Graves A.P., Diaz E., LaFrance L.V. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 23.Taplin M-E., Hussain A., Shore N.D., Bradley B., Trojer P., Lebedinsky C., Senderowicz A.M., Antonarakis E.S. A phase 1b/2 study of CPI-1205, a small molecule inhibitor of EZH2, combined with enzalutamide (E) or abiraterone/prednisone (A/P) in patients with metastatic castration resistant prostate cancer (mCRPC). J. Clin. Oncol. 2018;36:s6. [Google Scholar]
- 24.Italiano A., Soria J-C., Toulmonde M., Michot J-M., Lucchesi C., Varga A., Coindre J-M., Blakemore S.J., Clawson A., Suttle B. Tazemetostat, an EZH2 inhibitor, in relapsed or refractory B-cell non-hodgkin lymphoma and advanced solid tumours: A first-in-human, open-label, phase 1 study. Lancet Oncol. 2018;19:649–659. doi: 10.1016/S1470-2045(18)30145-1. [DOI] [PubMed] [Google Scholar]
- 25.Schwartzentruber J., Korshunov A., Liu X-Y., Jones D.T., Pfaff E., Jacob K., Sturm D., Fontebasso A.M., Quang D-A.K., Tönjes M. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 26.Venneti S., Garimella M.T., Sullivan L.M., Martinez D., Huse J.T., Heguy A., Santi M., Thompson C.B., Judkins A.R. Evaluation of h istone 3 lysine 27 trimethylation (H3K27me3) and enhancer of zest 2 (EZH 2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3a K27M mutant glioblastomas. Brain Pathol. 2013;23:558–564. doi: 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis P.W., Müller M. Koletsky, M.; Cordero, F.; Lin, S.; Banaszynski, L.; Garcia, B. A.; Muir, T.; Becher, O.; Allis, C. D. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H., Kaminski M.S., Li Y., Yildiz M., Ouillette P., Jones S., Fox H., Jacobi K., Saiya-Cork K., Bixby D., Lebovic D., Roulston D., Shedden K., Sabel M., Marentette L., Cimmino V., Chang A.E., Malek S.N. Mutations in linker histone genes HIST1H1 B, C, D, and E; OCT2 (POU2F2); IRF8; and ARID1A underlying the pathogenesis of follicular lymphoma. Blood. 2014;123:1487–1498. doi: 10.1182/blood-2013-05-500264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goll M.G., Kirpekar F., Maggert K.A., Yoder J.A., Hsieh C-L., Zhang X., Golic K.G., Jacobsen S.E., Bestor T.H. Methylation of tRNAAsp by the DNA methyltransferase homolog DNMT2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. [DOI] [PubMed] [Google Scholar]
- 30.Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. doi: 10.1038/nature14192. [DOI] [PubMed] [Google Scholar]
- 31.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Branco M.R., Ficz G., Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 2011;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 33.Lister R., Pelizzola M., Dowen R.H., Hawkins R.D., Hon G., Tonti-Filippini J., Nery J.R., Lee L., Ye Z., Ngo Q-M. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stefansson O.A., Moran S., Gomez A., Sayols S., Arribas-Jorba C., Sandoval J., Hilmarsdottir H., Olafsdottir E., Tryggvadottir L., Jonasson J.G. A DNA methylation‐based definition of biologically distinct breast cancer subtypes. Mol. Oncol. 2015;9:555–568. doi: 10.1016/j.molonc.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herman J.G., Umar A., Polyak K., Graff J.R., Ahuja N., Issa J.P., Markowitz S., Willson J.K., Hamilton S.R., Kinzler K.W., Kane M.F., Kolodner R.D., Vogelstein B., Kunkel T.A., Baylin S.B. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc. Natl. Acad. Sci. USA. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wajed S.A., Laird P.W., DeMeester T.R. DNA methylation: An alternative pathway to cancer. Ann. Surg. 2001;234:10–20. doi: 10.1097/00000658-200107000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pistore C., Giannoni E., Colangelo T., Rizzo F., Magnani E., Muccillo L., Giurato G., Mancini M., Rizzo S., Riccardi M. DNA methylation variations are required for epithelial-to-mesenchymal transition induced by cancer-associated fibroblasts in prostate cancer cells. Oncogene. 2017;36:5551–5566. doi: 10.1038/onc.2017.159. [DOI] [PubMed] [Google Scholar]
- 38.Merlo A., Herman J.G., Mao L., Lee D.J., Gabrielson E., Burger P.C., Baylin S.B., Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat. Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 39.Cameron E.E., Bachman K.E., Myohanen S., Herman J.G., Baylin S.B. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 40.Shin J.Y., Kim H.S., Park J., Park J.B., Lee J.Y. Mechanism for inactivation of the KIP family cyclin-dependent kinase inhibitor genes in gastric cancer cells. Cancer Res. 2000;60:262–265. [PubMed] [Google Scholar]
- 41.Dobrovic A., Simpfendorfer D. Methylation of the BRCA1 gene in sporadic breast cancer. Cancer Res. 1997;57:3347–3350. [PubMed] [Google Scholar]
- 42.Catteau A., Harris W.H., Xu C-F., Solomon E. Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: Correlation with disease characteristics. Oncogene. 1999;18:1957–1965. doi: 10.1038/sj.onc.1202509. [DOI] [PubMed] [Google Scholar]
- 43.Hua K.T., Wang M.Y., Chen M.W., Wei L.H., Chen C.K., Ko C.H., Jeng Y.M., Sung P.L., Jan Y.H., Hsiao M., Kuo M.L., Yen M.L. The H3K9 methyltransferase G9a is a marker of aggressive ovarian cancer that promotes peritoneal metastasis. Mol. Cancer. 2014;13:189–201. doi: 10.1186/1476-4598-13-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen M.W., Hua K.T., Kao H.J., Chi C.C., Wei L.H., Johansson G., Shiah S.G., Chen P.S., Jeng Y.M., Cheng T.Y., Lai T.C., Chang J.S., Jan Y.H., Chien M.H., Yang C.J., Huang M.S., Hsiao M., Kuo M.L. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010;70:7830–7840. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- 45.Zhong X., Chen X., Guan X., Zhang H., Ma Y., Zhang S., Wang E., Zhang L., Han Y. Overexpression of G9a and MCM7 in oesophageal squamous cell carcinoma is associated with poor prognosis. Histopathology. 2015;66:192–200. doi: 10.1111/his.12456. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J., He P., Xi Y., Geng M., Chen Y., Ding J. Down-regulation of G9a triggers DNA damage response and inhibits colorectal cancer cells proliferation. Oncotarget. 2015;6:2917–2927. doi: 10.18632/oncotarget.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones P.A., Taylor S.M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 48.Rius M., Stresemann C., Keller D., Brom M., Schirrmacher E., Keppler D., Lyko F. Human concentrative nucleoside transporter 1-mediated uptake of 5-azacytidine enhances DNA demethylation. Mol. Cancer Ther. 2009;8:225–231. doi: 10.1158/1535-7163.MCT-08-0743. [DOI] [PubMed] [Google Scholar]
- 49.Ghoshal K., Datta J., Majumder S., Bai S., Kutay H., Motiwala T., Jacob S.T. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol. Cell. Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Stresemann C., Lyko F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer. 2008;123:8–13. doi: 10.1002/ijc.23607. [DOI] [PubMed] [Google Scholar]
- 51.Hess J., Homann S., Laureano N.K., Tawk B., Bieg M., Hostenech X.P., Freier K., Weichert W., Zaoui K. Tumor cell plasticity in the pathogenesis and prognosis of head and neck cancer. Laryngorhinootologie. 2018;97:S94–S95. [Google Scholar]
- 52.Griffiths E., Choy G., Redkar S., Taverna P., Azab M., Karpf A.R. Sgi-110: DNA methyltransferase inhibitor oncolytic. Drugs Future. 2013;38:535–543. [PMC free article] [PubMed] [Google Scholar]
- 53.Issa J-P.J., Roboz G., Rizzieri D., Jabbour E., Stock W., O’Connell C., Yee K., Tibes R., Griffiths E.A., Walsh K. Safety and tolerability of guadecitabine (SGI-110) in patients with myelodysplastic syndrome and acute myeloid leukaemia: A multicentre, randomised, dose-escalation phase 1 study. Lancet Oncol. 2015;16:1099–1110. doi: 10.1016/S1470-2045(15)00038-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.San José-Enériz E., Agirre X., Rabal O., Vilas-Zornoza A., Sanchez-Arias J.A., Miranda E., Ugarte A., Roa S., Paiva B., de Mendoza A.E-H. Discovery of first-in-class reversible dual small molecule inhibitors against G9a andDNMTs in hematological malignancies. Nat. Commun. 2017;8:15424. doi: 10.1038/ncomms15424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh N., Dueñas‐González A., Lyko F., Medina‐Franco J.L. Molecular modeling and molecular dynamics studies of hydralazine with human DNA methyltransferase 1. ChemMedChem. 2009;4:792–799. doi: 10.1002/cmdc.200900017. [DOI] [PubMed] [Google Scholar]
- 56.Chuang J.C., Yoo C.B., Kwan J.M., Li T.W., Liang G., Yang A.S., Jones P.A. Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2′-deoxycytidine. Mol. Cancer Ther. 2005;4:1515–1520. doi: 10.1158/1535-7163.MCT-05-0172. [DOI] [PubMed] [Google Scholar]
- 57.Bedford M.T., Van Helden P.D. Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res. 1987;47:5274–5276. [PubMed] [Google Scholar]
- 58.Wahlfors J., Hiltunen H., Heinonen K., Hamalainen E., Alhonen L., Janne J. Genomic hypomethylation in human chronic lymphocytic leukemia. Blood. 1992;80:2074–2080. [PubMed] [Google Scholar]
- 59.Lin C-H., Hsieh S-Y., Sheen I-S., Lee W-C., Chen T-C., Shyu W-C., Liaw Y-F. Genome-wide hypomethylation in hepatocellular carcinogenesis. Cancer Res. 2001;61:4238–4243. [PubMed] [Google Scholar]
- 60.Kim Y.I., Giuliano A., Hatch K.D., Schneider A., Nour M.A., Dallal G.E., Selhub J., Mason J.B. Global DNA hypomethylation increases progressively in cervical dysplasia and carcinoma. Cancer. 1994;74:893–899. doi: 10.1002/1097-0142(19940801)74:3<893::aid-cncr2820740316>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 61.Veland N., Hardikar S., Zhong Y., Gayatri S., Dan J., Strahl B.D., Rothbart S.B., Bedford M.T., Chen T. The arginine methyltransferase PRMT6 regulates DNA methylation and contributes to global DNA hypomethylation in cancer. Cell Rep. 2017;21:3390–3397. doi: 10.1016/j.celrep.2017.11.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Perez R.F., Tejedor J.R., Bayon G.F., Fernández A.F., Fraga M.F. Distinct chromatin signatures of DNA hypomethylation in aging and cancer. Aging Cell. 2018;17:e12744. doi: 10.1111/acel.12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 64.Buschbeck M., Hake S.B. Variants of core histones and their roles in cell fate decisions, development and cancer. Nat. Rev. Mol. Cell Biol. 2017;18:299–314. doi: 10.1038/nrm.2016.166. [DOI] [PubMed] [Google Scholar]
- 65.Biswas S., Rao C.M. Epigenetics in cancer: Fundamentals and beyond. Pharmacol. Ther. 2017;173:118–134. doi: 10.1016/j.pharmthera.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 66.Taverna S.D., Li H., Ruthenburg A.J., Allis C.D., Patel D.J. How chromatin-binding modules interpret histone modifications: Lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Falkenberg K.J., Johnstone R.W. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 2014;13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 68.Perego P., Zuco V., Gatti L., Zunino F. Sensitization of tumor cells by targeting histone deacetylases. Biochem. Pharmacol. 2012;83:987–994. doi: 10.1016/j.bcp.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 69.Wang L., Li H., Ren Y., Zou S., Fang W., Jiang X., Jia L., Li M., Liu X., Yuan X. Targeting HDAC with a novel inhibitor effectively reverses paclitaxel resistance in non-small cell lung cancer via multiple mechanisms. Cell Death Dis. 2017;7:e2063. doi: 10.1038/cddis.2015.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wei Y., Zhou F., Lin Z., Shi L., Huang A., Liu T., Yu D., Wu G. Antitumor effects of histone deacetylase inhibitor suberoylanilide hydroxamic acid in epidermal growth factor receptor-mutant non-small-cell lung cancer lines in vitro and in vivo. Anticancer Drugs. 2018;29:262–270. doi: 10.1097/CAD.0000000000000597. [DOI] [PubMed] [Google Scholar]
- 71.Ye C., Han K., Lei J., Zeng K., Zeng S., Ju H., Yu L. Inhibition of HDAC7 reverses CNT2 repression in colorectal cancer by up‐regulating histone acetylation state. Br. J. Pharmacol. 2018;175(22):4209–4217. doi: 10.1111/bph.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ansari J., Shackelford R.E., El-Osta H. Epigenetics in non-small cell lung cancer: From basics to therapeutics. Transl. Lung Cancer Res. 2016;5:155–171. doi: 10.21037/tlcr.2016.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mukhopadhyay N.K., Weisberg E., Gilchrist D., Bueno R., Sugarbaker D.J., Jaklitsch M.T. Effectiveness of trichostatin A as a potential candidate for anticancer therapy in non–small-cell lung cancer. Ann. Thorac. Surg. 2006;81:1034–1042. doi: 10.1016/j.athoracsur.2005.06.059. [DOI] [PubMed] [Google Scholar]
- 74.Gottlicher M., Minucci S., Zhu P., Kramer O.H., Schimpf A., Giavara S., Sleeman J.P., Lo Coco F., Nervi C., Pelicci P.G., Heinzel T. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gavrilov V., Lavrenkov K., Ariad S., Shany S. Sodium valproate, a histone deacetylase inhibitor, enhances the efficacy of vinorelbine-cisplatin-based chemoradiation in non-small cell lung cancer cells. Anticancer Res. 2014;34:6565–6572. [PubMed] [Google Scholar]
- 76.Shirsath N., Rathos M., Chaudhari U., Sivaramakrishnan H., Joshi K. Potentiation of anticancer effect of valproic acid, an antiepileptic agent with histone deacetylase inhibitory activity, by the cyclin-dependent kinase inhibitor P276-00 in human non-small-cell lung cancer cell lines. Lung Cancer. 2013;82:214–221. doi: 10.1016/j.lungcan.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 77.Greve G., Schiffmann I., Pfeifer D., Pantic M., Schüler J., Lübbert M. The pan-HDAC inhibitor panobinostat acts as a sensitizer for erlotinib activity in EGFR-mutated and-wildtype non-small cell lung cancer cells. BMC Cancer. 2015;15:947–956. doi: 10.1186/s12885-015-1967-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sakaguchi T., Yoshino H., Sugita S., Osako Y., Yonemori M., Miyamoto K., Nakagawa M., Enokida H. Bromodomain protein BRD4 inhibition as a novel therapeutic approach in sunitinib-resistant renal cell carcinoma. Eur. Urol. Suppl. 2018;17:e60 [Google Scholar]
- 80.Pérez-Salvia M., Esteller M. Bromodomain inhibitors and cancer therapy: From structures to applications. Epigenetics. 2017;12:323–339. doi: 10.1080/15592294.2016.1265710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Y., Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 82.Viré E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J-M. The polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 83.Song Y., Wu F., Wu J. Targeting histone methylation for cancer therapy: Enzymes, inhibitors, biological activity and perspectives. J. Hematol. Oncol. 2016;9:49. doi: 10.1186/s13045-016-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim W., Kim R., Park G., Park J-W., Kim J-E. Deficiency of H3K79 histone methyltransferase Dot1-like protein (DOT1L) inhibits cell proliferation. J. Biol. Chem. 2012;287:5588–5599. doi: 10.1074/jbc.M111.328138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Daigle S.R., Olhava E.J., Therkelsen C.A., Majer C.R., Sneeringer C.J., Song J., Johnston L.D., Scott M.P., Smith J.J., Xiao Y. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Blanc R.S., Richard S. Arginine methylation: The coming of age. Mol. Cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 87.Chan-Penebre E., Kuplast K.G., Majer C.R., Boriack-Sjodin P.A., Wigle T.J., Johnston L.D., Rioux N., Munchhof M.J., Jin L., Jacques S.L. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol. 2015;11:432–437. doi: 10.1038/nchembio.1810. [DOI] [PubMed] [Google Scholar]
- 88.Avasarala S., Van Scoyk M., Rathinam M.K.K., Zerayesus S., Zhao X., Zhang W., Pergande M.R., Borgia J.A., DeGregori J., Port J.D. PRMT1 is a novel regulator of epithelial-mesenchymal-transition in non-small cell lung cancer. J. Biol. Chem. 2015;290:13479–13489. doi: 10.1074/jbc.M114.636050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yao R., Jiang H., Ma Y., Wang L., Wang L., Du J., Hou P., Gao Y., Zhao L., Wang G. PRMT7 induces epithelial-to-mesenchymal transition and promotes metastasis in breast cancer. Cancer Res. 2014;74:5656–5667. doi: 10.1158/0008-5472.CAN-14-0800. [DOI] [PubMed] [Google Scholar]
- 90.Rossetto D., Avvakumov N., Côté J. Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics. 2012;7:1098–1108. doi: 10.4161/epi.21975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li B., Huang G., Zhang X., Li R., Wang J., Dong Z., He Z. Increased phosphorylation of histone H3 at serine 10 is involved in Epstein-Barr virus latent membrane protein-1-induced carcinogenesis of nasopharyngeal carcinoma. BMC Cancer. 2013;13:124. doi: 10.1186/1471-2407-13-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 93.Fischle W., Tseng B.S., Dormann H.L., Ueberheide B.M., Garcia B.A., Shabanowitz J., Hunt D.F., Funabiki H., Allis C.D. Regulation of HP1–chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 94.Li M., Dong Q., Zhu B. Aurora Kinase B phosphorylates histone H3.3 at serine 31 during mitosis in mammalian cells. J. Mol. Biol. 2017;429:2042–2045. doi: 10.1016/j.jmb.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 95.Aihara H., Nakagawa T., Yasui K., Ohta T., Hirose S., Dhomae N., Takio K., Kaneko M., Takeshima Y., Muramatsu M. Nucleosomal histone kinase-1 phosphorylates H2A Thr 119 during mitosis in the early Drosophila embryo. Genes Dev. 2004;18:877–888. doi: 10.1101/gad.1184604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sawicka A., Seiser C. Sensing core histone phosphorylation-a matter of perfect timing. Biochim. Biophys. Acta. Gene Regul. Mech. 2014;1839:711–718. doi: 10.1016/j.bbagrm.2014.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pickart C.M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 98.Goldknopf I., Taylor C.W., Baum R.M., Yeoman L.C., Olson M., Prestayko A., Busch H. Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. J. Biol. Chem. 1975;250:7182–7187. [PubMed] [Google Scholar]
- 99.Osley M.A. Regulation of histone H2A and H2B ubiquitylation. Brief. Funct. Genomics. 2006;5:179–189. doi: 10.1093/bfgp/ell022. [DOI] [PubMed] [Google Scholar]
- 100.McClurg U.L., Robson C.N. Deubiquitinating enzymes as oncotargets. Oncotarget. 2015;6:9657–9668. doi: 10.18632/oncotarget.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Davie J.R., Murphy L.C. Inhibition of transcription selectively reduces the level of ubiquitinated histone H2B in chromatin. Biochem. Biophys. Res. Commun. 1994;203:344–350. doi: 10.1006/bbrc.1994.2188. [DOI] [PubMed] [Google Scholar]
- 102.Dwane L., Gallagher W.M., Chonghaile T.N., O’Connor D.P. The emerging role of non-traditional ubiquitination in oncogenic pathways. J. Biol. Chem. 2017;292:3543–3551. doi: 10.1074/jbc.R116.755694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Melchior F. SUMO-nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 104.Gostissa M., Hengstermann A., Fogal V., Sandy P., Schwarz S.E., Scheffner M., Del Sal G. Activation of p53 by conjugation to the ubiquitin‐like protein SUMO‐1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Buschmann T., Fuchs S.Y., Lee C-G., Pan Z-Q., Ronai Z. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell. 2000;101:753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- 106.Müller S., Matunis M.J., Dejean A. Conjugation with the ubiquitin‐related modifier SUMO‐1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shiio Y., Eisenman R.N. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dhall A., Weller C.E., Chu A., Shelton P.M., Chatterjee C. Chemically sumoylated histone H4 stimulates intranucleosomal demethylation by the LSD1-CoREST complex. ACS Chem. Biol. 2017;12:2275–2280. doi: 10.1021/acschembio.7b00716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shanmugam M.K., Arfuso F., Arumugam S., Chinnathambi A., Jinsong B., Warrier S., Wang L.Z., Kumar A.P., Ahn K.S., Sethi G. Role of novel histone modifications in cancer. Oncotarget. 2018;9:11414–11426. doi: 10.18632/oncotarget.23356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fukuto A., Ikura M., Ikura T., Sun J., Horikoshi Y., Shima H., Igarashi K., Kusakabe M., Harata M., Horikoshi N. SUMO modification system facilitates the exchange of histone variant H2A. Z-2 at DNA damage sites. Nucleus. 2018;9:87–94. doi: 10.1080/19491034.2017.1395543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Han Z-J., Feng Y-H., Gu B-H., Li Y-M., Chen H. The post-translational modification, sumoylation, and cancer. Int. J. Oncol. 2018;52:1081–1094. doi: 10.3892/ijo.2018.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kamitani T., Kito K., Nguyen H.P., Yeh E.T. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 113.Duda D.M., Borg L.A., Scott D.C., Hunt H.W., Hammel M., Schulman B.A. Structural insights into NEDD8 activation of cullin-RING ligases: Conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Salon C., Brambilla E., Brambilla C., Lantuejoul S., Gazzeri S., Eymin B. Altered pattern of Cul‐1 protein expression and neddylation in human lung tumours: Relationships with CAND1 and cyclin E protein levels. J. Pathol. 2007;213:303–310. doi: 10.1002/path.2223. [DOI] [PubMed] [Google Scholar]
- 115.Zhou L., Zhang W., Sun Y., Jia L. Protein neddylation and its alterations in human cancers for targeted therapy. Cell. Signal. 2018;•••:92–102. doi: 10.1016/j.cellsig.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li L., Wang M., Yu G., Chen P., Li H., Wei D., Zhu J., Xie L., Jia H., Shi J. Overactivated neddylation pathway as a therapeutic target in lung cancer. J. Natl. Cancer Inst. 2014;106:dju083. doi: 10.1093/jnci/dju083. [DOI] [PubMed] [Google Scholar]
- 117.Li T., Guan J., Huang Z., Hu X., Zheng X. RNF168-mediated H2A neddylation antagonizes ubiquitylation of H2A and regulates DNA damage repair. J. Cell Sci. 2014;127:2238–2248. doi: 10.1242/jcs.138891. [DOI] [PubMed] [Google Scholar]
- 118.Ma T., Chen Y., Zhang F., Yang C-Y., Wang S., Yu X. RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Mol. Cell. 2013;49:897–907. doi: 10.1016/j.molcel.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Soucy T.A., Smith P.G., Milhollen M.A., Berger A.J., Gavin J.M., Adhikari S., Brownell J.E., Burke K.E., Cardin D.P., Critchley S. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 120.Swords R.T., Savona M.R., Maris M.B., Erba H.P., Berdeja J.G., Foran J.M., Hua Z., Faessel H.M., Dash A.B., Sedarati F. Pevonedistat (MLN4924), an investigational, first-in-class NAE inhibitor, in combination with azacitidine in elderly patients with acute myeloid leukemia (AML) considered unfit for conventional chemotherapy: Updated results from the phase 1 C15009 trial. Blood. 2014;124:2313. [Google Scholar]
- 121.Liu N., Pan T. RNA epigenetics. Transl. Res. 2015;165:28–35. doi: 10.1016/j.trsl.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Song X., Nazar R.N. Modification of rRNA as a ‘quality control mechanism’in ribosome biogenesis. FEBS Lett. 2002;523:182–186. doi: 10.1016/s0014-5793(02)02986-1. [DOI] [PubMed] [Google Scholar]
- 123.Agris P.F. Decoding the genome: A modified view. Nucleic Acids Res. 2004;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wei C-M., Gershowitz A., Moss B. N6, O2′-dimethyladenosine a novel methylated ribonucleoside next to the 5′ terminal of animal cell and virus mRNAs. Nature. 1975;257:251–253. doi: 10.1038/257251a0. [DOI] [PubMed] [Google Scholar]
- 125.Narayan P., Rottman F.M. An in vitro system for accurate methylation of internal adenosine residues in messenger RNA. Science. 1988;242:1159–1162. doi: 10.1126/science.3187541. [DOI] [PubMed] [Google Scholar]
- 126.Squires J.E., Patel H.R., Nousch M., Sibbritt T., Humphreys D.T., Parker B.J., Suter C.M., Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bokar J.A. Fine-tuning of RNA functions by modification and editing. Springer; 2005. The biosynthesis and functional roles of methylated nucleosides in eukaryotic mRNA. pp. 141–177. [Google Scholar]
- 128.Lin S., Choe J., Du P., Triboulet R., Gregory R.I. The m 6 A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Batista P.J., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley D.M., Lujan E., Haddad B., Daneshvar K. M6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y.S. M6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 131.Xiang Y., Laurent B., Hsu C-H., Nachtergaele S., Lu Z., Sheng W., Xu C., Chen H., Ouyang J., Wang S. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573–576. doi: 10.1038/nature21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 133.Friedman R.C., Farh K.K-H., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lopez-Serra P., Esteller M. DNA methylation-associated silencing of tumor-suppressor microRNAs in cancer. Oncogene. 2012;31:1609–1622. doi: 10.1038/onc.2011.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Guil S., Esteller M. DNA methylomes, histone codes and miRNAs: Tying it all together. Int. J. Biochem. Cell Biol. 2009;41:87–95. doi: 10.1016/j.biocel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 136.Anastasiadou E., Jacob L.S., Slack F.J. Non-coding RNA networks in cancer. Nat. Rev. Cancer. 2018;18:5–18. doi: 10.1038/nrc.2017.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schmitt A.M., Chang H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chu C., Zhang Q.C., Da Rocha S.T., Flynn R.A., Bharadwaj M., Calabrese J.M., Magnuson T., Heard E., Chang H.Y. Systematic discovery of Xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M-C., Hung T., Argani P., Rinn J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Morlando M., Fatica A. Alteration of epigenetic regulation by long noncoding RNAs in cancer. Int. J. Mol. Sci. 2018;19:E570. doi: 10.3390/ijms19020570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kurdyukov S., Bullock M. DNA methylation analysis: Choosing the right method. Biology (Basel) 2016;5:3–24. doi: 10.3390/biology5010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Smith E., Jones M.E., Drew P.A. Quantitation of DNA methylation by melt curve analysis. BMC Cancer. 2009;9:123. doi: 10.1186/1471-2407-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mundade R., Ozer H.G., Wei H., Prabhu L., Lu T. Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond. Cell Cycle. 2014;13:2847–2852. doi: 10.4161/15384101.2014.949201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Natarajan A., Yardımcı G.G., Sheffield N.C., Crawford G.E., Ohler U. Predicting cell-type–specific gene expression from regions of open chromatin. Genome Res. 2012;22:1711–1722. doi: 10.1101/gr.135129.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tripathi R., Chakraborty P., Varadwaj P.K. Unraveling long non-coding RNAs through analysis of high-throughput RNA-sequencing data. Noncoding RNA Res. 2017;2:111–118. doi: 10.1016/j.ncrna.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Peleg S., Sananbenesi F., Zovoilis A., Burkhardt S., Bahari-Javan S., Agis-Balboa R.C., Cota P., Wittnam J.L., Gogol-Doering A., Opitz L. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 147.Lin H.S., Hu C.Y., Chan H.Y., Liew Y.Y., Huang H.P., Lepescheux L., Bastianelli E., Baron R., Rawadi G., Clément‐Lacroix P. Anti‐rheumatic activities of histone deacetylase (HDAC) inhibitors in vivo in collagen‐induced arthritis in rodents. Br. J. Pharmacol. 2007;150:862–872. doi: 10.1038/sj.bjp.0707165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hockly E., Richon V.M., Woodman B., Smith D.L., Zhou X., Rosa E., Sathasivam K., Ghazi-Noori S., Mahal A., Lowden P.A. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington’s disease. Proc. Natl. Acad. Sci. USA. 2003;100:2041–2046. doi: 10.1073/pnas.0437870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Alarcón J.M., Malleret G., Touzani K., Vronskaya S., Ishii S., Kandel E.R., Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: A model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 150.Brogdon J.L., Xu Y., Szabo S.J., An S., Buxton F., Cohen D., Huang Q. Histone deacetylase activities are required for innate immune cell control of Th1 but not Th2 effector cell function. Blood. 2007;109:1123–1130. doi: 10.1182/blood-2006-04-019711. [DOI] [PubMed] [Google Scholar]
- 151.Adcock I. Hdac inhibitors as anti‐inflammatory agents. Br. J. Pharmacol. 2007;150:829–831. doi: 10.1038/sj.bjp.0707166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Qing H., He G., Ly P.T., Fox C.J., Staufenbiel M., Cai F., Zhang Z., Wei S., Sun X., Chen C-H. Valproic acid inhibits Aβ production, neuritic plaque formation, and behavioral deficits in Alzheimer’s disease mouse models. J. Exp. Med. 2008;205:2781–2789. doi: 10.1084/jem.20081588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tao R., De Zoeten E.F., Özkaynak E., Chen C., Wang L., Porrett P.M., Li B., Turka L.A., Olson E.N., Greene M.I. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 154.McGee S.L., Hargreaves M. Exercise and skeletal muscle glucose transporter 4 expression: Molecular mechanisms. Clin. Exp. Pharmacol. Physiol. 2006;33:395–399. doi: 10.1111/j.1440-1681.2006.04362.x. [DOI] [PubMed] [Google Scholar]
- 155.Daneshpajooh M., Bacos K., Bysani M., Bagge A., Laakso E.O., Vikman P., Eliasson L., Mulder H., Ling C. HDAC7 is overexpressed in human diabetic islets and impairs insulin secretion in rat islets and clonal beta cells. Diabetologia. 2017;60:116–125. doi: 10.1007/s00125-016-4113-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Xu Z., Tong Q., Zhang Z., Wang S., Zheng Y., Liu Q., Qian L., Chen S-y., Sun J., Cai L. Inhibition of HDAC3 prevents diabetic cardiomyopathy in OVE26 mice via epigenetic regulation of DUSP5-ERK1/2 pathway. Clin. Sci. (Lond.) 2017;131:1841–1857. doi: 10.1042/CS20170064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dong E., Grayson D.R., Guidotti A., Ruzicka W., Veldic M., Costa E. Reviewing the role of DNA (cytosine-5) methyltransferase overexpression in the cortical GABAergic dysfunction associated with psychosis vulnerability. Epigenetics. 2007;2:29–36. doi: 10.4161/epi.2.1.4063. [DOI] [PubMed] [Google Scholar]
- 158.Matt S.M., Zimmerman J.D., Lawson M.A., Bustamante A.C., Uddin M., Johnson R.W. Inhibition of DNA methylation with zebularine alters lipopolysaccharide-induced sickness behavior and neuroinflammation in mice. Front. Neurosci. 2018;12:636. doi: 10.3389/fnins.2018.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fonteneau M., Filliol D., Anglard P., Befort K., Romieu P., Zwiller J. Inhibition of DNA methyltransferases regulates cocaine self‐administration by rats: A genome‐wide DNA methylation study. Genes Brain Behav. 2017;16:313–327. doi: 10.1111/gbb.12354. [DOI] [PubMed] [Google Scholar]
- 160.Hedrich C.M., Mäbert K., Rauen T., Tsokos G.C. DNA methylation in systemic lupus erythematosus. Epigenomics. 2017;9:505–525. doi: 10.2217/epi-2016-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sato T., Issa J., Kropf P., Hypomethylating Drugs D.N.A. in Cancer Therapy. Cold Spring Harb. Perspect. Med. 2017;7:a026948. doi: 10.1101/cshperspect.a026948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Castro K., Casaccia P. Epigenetic modifications in brain and immune cells of multiple sclerosis patients. Mult. Scler. J. 2018;24:69–74. doi: 10.1177/1352458517737389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhao Y. Garcia, B.A. Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harb. Perspect. Biol. 2015;7:a025064. doi: 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dawson M.A. Bannister; A.J.; Göttgens B.; Foster, S.D.; Bartke, T.; Green, A.R.; Kouzarides, T. JAK2 phosphorylates histone H3Y41 and excludes HP1α from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chou R.H. Wang, Y.N.; Hsieh, Y.H.; Li, L.Y.; Xia, W.; Chang, W.C.; Chang, L.C.; Cheng, C. C.; Lai, C.C.; Hsu, J.L.; Chang, W.J.; Chiang, S.Y.; Lee, H. J.; Liao, H. W.; Chuang, P. H.; Chen, H.Y.; Wang, H.L.; Kuo, S.C.; Chen, C.H.; Yu, Y.L.; Hung, M.C. EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev. Cell. 2014;30:224–237. doi: 10.1016/j.devcel.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]


