Abstract
Racial/ethnic disparities have a significant impact on bladder cancer outcomes with African American patients demonstrating inferior survival over European-American patients. We hypothesized that epigenetic difference in methylation of tumor DNA is an underlying cause of this survival health disparity. We analyzed bladder tumors from African American and European-American patients using reduced representation bisulfite sequencing (RRBS) to annotate differentially methylated DNA regions. Liquid chromatography–mass spectrometry (LC-MS/MS) based metabolomics and flux studies were performed to examine metabolic pathways that showed significant association to the discovered DNA methylation patterns. RRBS analysis showed frequent hypermethylated CpG islands in African American patients. Further analysis showed that these hypermethylated CpG islands in patients are commonly located in the promoter regions of xenobiotic enzymes that are involved in bladder cancer progression. On follow-up, LC-MS/MS revealed accumulation of glucuronic acid, S-adenosylhomocysteine, and a decrease in S-adenosylmethionine, corroborating findings from the RRBS and mRNA expression analysis indicating increased glucuronidation and methylation capacities in African American patients. Flux analysis experiments with 13C-labeled glucose in cultured African American bladder cancer cells confirmed these findings. Collectively, our studies revealed robust differences in methylation-related metabolism and expression of enzymes regulating xenobiotic metabolism in African American patients indicate that race/ethnic differences in tumor biology may exist in bladder cancer.
Abnormal DNA methylation configurations are linked with adverse outcomes in bladder cancer. Here, we report racial disparities in specific DNA methylation patterns that could lead to silencing of xenobiotic genes. Downregulation of these genes may lead to bladder cancer progression in African American patients.
Introduction
Racial/ethnic disparities have a significant impact on bladder cancer (BLCA) outcomes with African American (AA) patients demonstrating inferior survival compared with European-American (EA) patients (1,2). Although access to quality health care is an important factor in this disparity, AAs experience worse outcomes even after receiving similar medical care provided to EA patients (1). Thus, the reasons behind the existing BLCA disparities in incidence, survival and mortality rates are complex and may include differences in tumor biology.
There is growing evidence that disparities in cancer may involve biological and environmental factors, as well as behavioral, socioeconomic, health care system and community influences. Furthermore, the efficacy of pharmacologic and other therapeutic interventions in the general population can depend on factors such as genetic ancestry, as shown for drug metabolism (3,4). The biological differences among populations, conjointly with environmental factors, may lead to discrepancies in incidence and progression of cancer. There have been limited research efforts to determine the role of biological determinants of BLCA racial disparities. Cancer is an epigenetic disease characterized by the dysregulation of DNA methylation patterns, which can modulate gene functions and malignant transformation (5,6). Various epigenetic mechanisms are influenced by environmental exposures (7). Social and environmental determinants, such as diet, sociocultural events and exposure to environmental chemicals are potential factors that can result in epigenetic changes (8). As histone and DNA methylations are affected by these factors, examining the role of epigenetics in BLCA disparities may lead to the discovery of novel molecular mechanisms of gene regulation and metabolism, that explains heterogeneity in tumor biology and disease outcome.
In this study, we reported the differences in the DNA methylation between AA and EA BLCA patients, which lead to dysregulation of various metabolic pathways, particularly perturbed phase II xenobiotic metabolism in AA BLCA. Metabolic flux analysis using 13C-labeled glucose in AA and EA BLCA cell lines further confirmed alterations in glucuronic acid metabolism, which provides the substrates for biotransformation reactions in phase II xenobiotic metabolism. The suppression of glucuronyl transferase leads to reduction in glucuronidation and an accumulation of glucuronic acid in AA BLCA.
Materials and methods
BLCA patient tissues
Frozen pathologically verified BLCA tissues for reduced-representation bisulfite sequencing (RRBS), metabolomics and protein analysis were obtained from the tumor banks of the University of Texas Southwestern Medical Center, TX, USA; Augusta University, GA, USA; and Baylor College of Medicine. All the samples were collected with informed consent and institutional review board approval. Tissues were reviewed for their tumor content by a genitourinary pathologist prior to analysis. Of the tissues used for this, 70% were ancestry verified. All the BLCA tissues’ clinical-pathologic characteristics are summarized in Supplementary Tables 1 and 2, available at Carcinogenesis Online.
Cell lines and culture methods
Human BLCA cell lines UMUC1, HTB-3, UMUC3 and UMUC5 were purchased from the American Type Culture Collection, a public repository. All cell lines were used within 6 months of their acquisition. American Type Culture Collection routinely verifies the identity of cell lines by short tandem repeat profiling. AA origin of HTB-3 and UMUC1 and EA origin of UMUC3 and UMUC5 cell lines were confirmed by ancestry typing.
DNA hydrolysis
Genomic DNA from ancestry-verified AA and EA BLCA tissues were isolated using Qiagen DNeasy blood and tissue kit by following the manufacturer’s protocol. The quality and quantity of DNA was determined by 260/280 ratio using BioTek microplate reader. DNA (1 μg) was denatured at 100°C for 3 min and subsequently chilled in ice. 1/10 vol of 0.1 M ammonium acetate (pH 5.3) and 2 U of nuclease P1 (Roche Molecular Biochemicals, Mannheim, Germany) were added and incubated at 45°C for 2 h. 1/10 vol of 1 M ammonium bicarbonate and 0.002 U venom phosphodiesterase I (Sigma) were added to this solution and incubation was continued for an additional 2 h at 37°C. The resulted mixture was further digested with 0.5 U alkaline phosphatase for 1 h at 37°C.
Measurements of methyl cytosine, cytosine and hydroxy methyl cytosine by liquid chromatography–mass spectrometry
The dried DNA samples were resuspended in 50 μL of methanol to water (50:50) containing 0.1% formic acid, and 10 µL of samples were injected into liquid chromatography–mass spectrometry (LC-MS/MS; 6495 QQQ Triple Quadrupole MS, Agilent Technologies). Analysis was carried out on high-performance liquid chromatography (HPLC) system equipped with a quaternary pump, a degasser, an auto sampler and a column compartment. Mass spectrometric detection was carried out using triple coupled quadrupole equipped with an electron spray ionization (ESI) source. The data acquisition was under the control of Mass Hunter software. The separation of methyl cytosine, cytosine and hydroxy methyl cytosine was achieved using an RRHD SB-CN column (1.8 μm, 3.0 × 100 mm, Agilent Technologies). The mobile phase consists of 0.1% formic acid (A) in water and acetonitrile (B) with a gradient elution at a flow rate of 0.3 ml/min. Gradient is spanning 2% B to 98% B over 15 min followed by 98% B to 2% B for 1 min. The typical operating source condition for MS scan in positive ESI mode was optimized. The list of single reaction monitoring transitions used to quantify metabolites is given in Table 1.
Table 1.
List of single reaction monitoring transitions for the metabolites measured in this study
| Metabolite name | Precursor ion | MRM/SRM (m/z) | Polarity | Retention time Rt (Min) |
|---|---|---|---|---|
| (m/z) | ||||
| Cytosine | 112 | 52/95/69 | Positive | 2.06 |
| Methyl cytosine | 126 | 54/81/109 | Positive | 2.1 |
| Hydroxy methyl cytosine | 142 | 81/124 | Positive | 2 |
| Glucoronic acid | 193 | 59/113 | Negative | 5.3 |
| S-(5'-Adenosyl)-l-methionine | 399.2 | 250.1/136.1 | Positive | 7 |
| SAH | 385.1 | 136/134 | Positive | 6.57 |
| SRM transitions of the metabolites measured for the flux | ||||
| SAH (M+0) | 385.1 | 134/136 | Positive | 6.57 |
| SAH (M+4) | 389.1 | 134/136 | Positive | 6.57 |
| S-(5'-Adenosyl)-l-methionine (M+0) | 399.2 | 136.1/250.1 | Positive | 7 |
| [13C]5 S-(5'-Adenosyl)-l-methionine (M+5) | 404.2 | 136.1/250.1 | Positive | 7 |
| Glucose Fructose (M+0) | 179 | 59 | Negative | 7.7 |
| Glucose Fructose (M+6) | 185.0762 | 61 | Negative | 7.7 |
| Glucoronic acid (M+0) | 193 | 59/113 | Negative | 5.3 |
| Glucoronic acid (M+6) | 199 | 59/113 | Negative | 5.3 |
SRM, single reaction monitoring.
RRBS sequencing
Genome-wide methylation analysis
Aberrant hypermethylation of CpG and transcriptional suppression is one of the most common genomic alterations in human cancers (9,10). To perform genome-wide analysis of DNA methylation, we used RRBS (11) to map methylated cytosines in AA and EA BLCA. In brief, 400 ng of genomic DNA is digested with MspI, end-repaired and A-tailed, and Illumina-compatible cytosine-methylated adaptor is ligated to the enzyme-digested DNA. Size-selected fragments representing sequences from 40 bp to 170 bp are bisulfite converted, and library preparation is carried out by PCR amplification and subsequently sequenced in a HiSeq4000 instrument (or higher).
RRBS data analysis
RRBS sequencing reads were aligned to UCSC Genome browser mm10 reference genome using Bowtie, and methylation status was classified using Bismark v0.7.11 (12). Differentially methylated regions (DMRs) between AA and EA BLCA were identified using DSS r package (13) implemented with Fisher’s exact test. In addition, we also analyzed the promotor methylations in The Cancer Genomic Atlas (TCGA), between AA and EA BLCA.
Pathway analysis
These DMRs were mapped to genes based on proximity to the promoter regions. The genes obtained from DMRs were subjected to online enrichment analysis platform ConsensusPathDB using kyoto encyclopedia of genes and genomes, Reactome and Wikipathways database pathway analysis (P < 0.05) to identify deregulated pathways in AA BLCA.
RNA-Seq analysis
The sequencing libraries were prepared with the mRNA HyperPrep kit (KAPA Biosystems), from 300 ng total RNA. Following manufacturer’s instructions, the first step selects for PolyA RNA using the mRNA Capture Beads (KAPA Biosystems). After PolyA selection, the remaining RNA is purified and fragmented then reverse transcribed into first-strand complementary DNA (cDNA) using random primers. The next step removes the RNA template and synthesizes a replacement strand, incorporating deoxyuridine triphosphate in place of dTTP to generate ds cDNA. Pure beads (KAPA Biosystems) are used to separate the ds cDNA from the second strand reaction mix resulting in blunt-ended cDNA. A single ‘A’ nucleotide is then added to the 3' ends of the blunt fragments. Multiple indexing adapters, containing a single ‘T’ nucleotide on the 3' end of the adapter, are ligated to the ends of the ds cDNA, preparing them for hybridization onto a flow cell. Adapter-ligated libraries are amplified by PCR, purified using pure beads and validated for appropriate size on a 4200 TapeStation D1000 Screentape (Agilent Technologies). The DNA libraries are quantitated using KAPA Biosystems qPCR kit and are pooled together in an equimolar fashion, following experimental design criteria. Each pool is denatured and diluted to 400 pM with 1% PhiX control library added. The resulting pool is then loaded into the appropriate NovaSeq Reagent cartridge for 100-cycle single-read sequencing on an S1 flowcell and sequenced on a NovaSeq6000 following the manufacturer’s recommended protocol (Illumina, unpublished data).
Measurement of the methylated and xenobiotic metabolites in AA and EA BLCA by LC-MS/MS
Total of 22 ancestry-verified BLCA tissues (13 AA and 9 EA) were used to measure the methylated and xenobiotic metabolites (Supplementary Table 2, available at Carcinogenesis Online). Ten milligrams of tissue was used for the metabolic extraction. The extraction step involved the addition of 750 µL ice-cold methanol to water (4:1) containing 20 µL spiked internal standards (l-tryptophan 15N2, l-Zeatin, l-glutamic acid d5, leucine 13C6, 15 N-glutamine) to each sample. Ice-cold chloroform and water were added in a 3:1 ratio for a final proportion of 4:3:2 methanol to chloroform to water. The organic (methanol and chloroform) and aqueous layers were mixed, dried and resuspended with methanol to water (50:50). The extract was deproteinized using a 3 kDa molecular filter (Amicon Ultracel-3K Membrane; Millipore Corporation) and the filtrate was dried under vacuum. Prior to LC-MS/MS, the dried extracts were resuspended in identical volumes of injection solvent composed of water to methanol (50:50). All the methylated and xenobiotic metabolites were separated on waters XBridge Amide column (3.5 µm, 4.6 × 100 mm). The mobile phase consists of 0.1% formic acid in water (A) and acetonitrile (B) at flow rate of 0.3 ml/min. The gradient program is as follows: 0 min (85% B); 3–12 min (85–10% B), 12–15 min (10% B), 16 min (85% B), followed by re-equilibration end of the gradient—the 23 min to the initial starting condition 85% B.
Measurements of S-adenosyl methionine and S-adenosylhomocysteine
S-adenosyl methionine (SAM) and S-adenosylhomocysteine (SAH) were measured in the extracted samples (tissues and cell lines) by LC-MS/MS. The separation of SAM and SAH were achieved on a Waters Xbridge Amide column (4.6 × 100 mm, 3.5 µm) using mobile phase of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) with 0.3 ml/min. The gradient used for the analysis is started with 85% B, at 3 min 30%B, at 10 min 2% B and re-equilibration till the end of the gradient 12 min. The single reaction monitoring transitions for SAM and SAH were depicted in Table 1. The operating source condition for MS scan in positive ESI mode was optimized.
Glucuronic acid measurements
To measure the glucuronic acid, samples were extracted with methanol to water (50:50) and deproteinized and analyzed by using LC-MS/MS. The glucuronic acid was retained on the Synergi 4u Max-RP 80A, Phenomenex column using mobile 1 mM ammonium acetate in water (A) and acetonitrile (B) with 0.5% ammonium hydroxide. The initial gradient started with 5% B, at 15 min 10% B, at 22 min 90% B, at 23 min 5% B till the end of the gradient 28 min. The flow rate used for the analysis is 0.3 ml/min. The single reaction monitoring transition for glucuronic acid was mentioned in Table 1.
Isotopic-labeled flux to measure the glucuronic acid
To quantify metabolic flux of glucose, cells were grown in 13C isotopic-labeled glucose (Cambridge Isotope Laboratories) media. Briefly, glucose-free media was supplemented with 12 mM 13C glucose for 6 h, and then cells were washed with ice-cold phosphate-buffered saline, immediately flash frozen and stored at −80ºC until metabolite extraction. Samples were extracted with methanol to water (50:50) followed by deproteinization and subjected to LC-MS/MS as mentioned earlier.
Immunoblotting analysis
Protein lysates were prepared using RIPA buffer (Sigma–Aldrich) with protease and phosphatase inhibitor cocktail (Thermo Scientific) and run on 4–20% Mini-PROTEAN® TGX™ gels using Tris/Glycine/SDS running buffer (Bio-Rad). Gels were blotted on polyvinylidene difluoride membranes. The membrane was blocked with 5% skim milk in TBST and probed with 1° and 2° antibodies. Horseradish-peroxidase-conjugated secondary antibodies were detected using the SuperSignal West Pico Chemiluminescent Substrate (Pierce Rockford)
Results
Identification of differences in DNA methylation between AA and EA patients
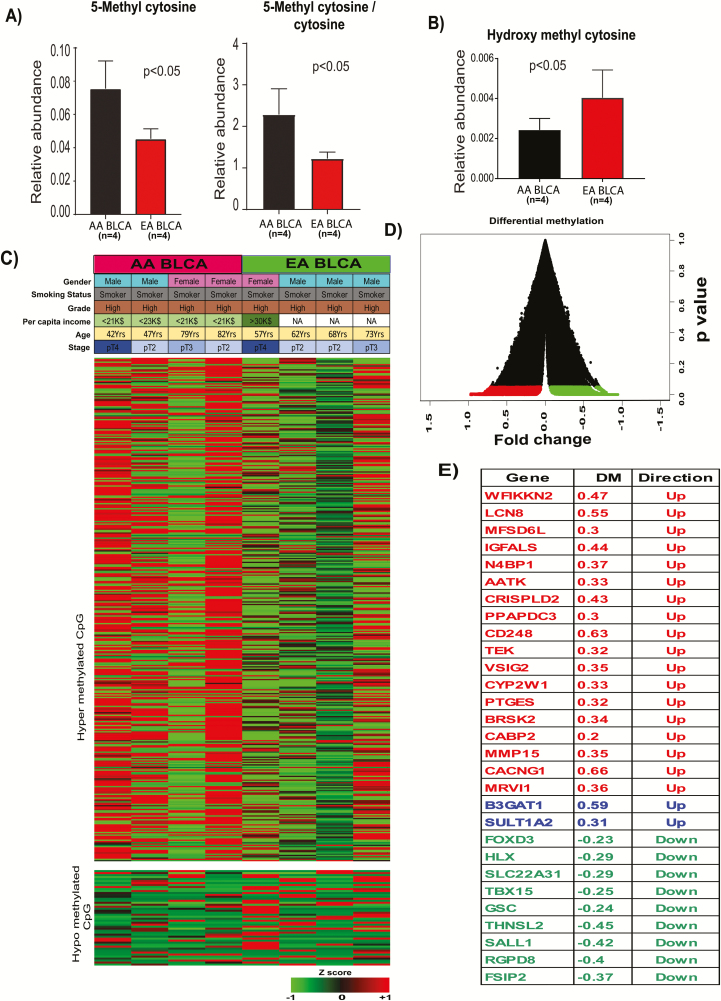
To understand the role of epigenetic-mediated DNA methylation in racial disparity, we measured 5-methylcytosine and ratio of 5-methylcytosine to cytosine, which are indicators of the DNA methylation status, using LC-MS/MS and observed higher levels of 5-methylcytosine in AA BLCA (Figure 1A). To further confirm the hypermethylation, we measured 5-hydroxymethylcytosine, which is an intermediate of the DNA demethylation process and observed low levels in AA BLCA (Figure 1B). To gain molecular insights into the epigenetic differences between AA and EA BLCA, we then performed genome-wide DNA methylation profiling of eight BLCA tissues (four AA and four EA) using RRBS followed by Illumina HiSeq3000 sequencing. This analysis identified 1071 DMRs (P < 0.01) in AA compared with EA BLCA (Figure 1C and D). The chromosomal location of these DMRs showed that their close proximity to 29 genes, which are known to be involved in various biological reactions (Figure 1E). To gain further strength in our findings, we validated our results in larger TCGA cohort with 20 AA and 290 EA BLCA patients. The TCGA methylation data were used for the analysis, and beta values were used to calculate the difference in methylation between AA and EA (AA minus EA). The TCGA analysis further supports the differential methylations in the promoters of the earlier observed genes in AA patients compared with EA patients (Supplementary Figure 1, available at Carcinogenesis Online).
Figure 1.
DMRs of AA and EA BLCA patient DNA. (A) Measurement of 5-Methyl cytosine and the ratio of methyl cytosine to cytosine in ancestry-verified AA (n = 4) and EA (n = 4) BLCA patients by LC-MS. (B) Measurement of hydroxy methyl cytosine in ancestry-verified AA (n = 4) and EA (n = 4) BLCA patients by LC-MS/MS. (C) Differential methylated regions in ancestry-verified AA (n = 4) and EA (n = 4) BLCA tissues. Red indicates hypermethylation and green indicates hypomethylation. (D) Volcano plot for DMRs in AA and EA BLCA. (E) Genes associated with hyper and hypomethylated CpG islands. DM, differential methylation AA minus EA.
Enrichment analysis of differentially methylated gene loci points to metabolism-related pathways in AA BLCA
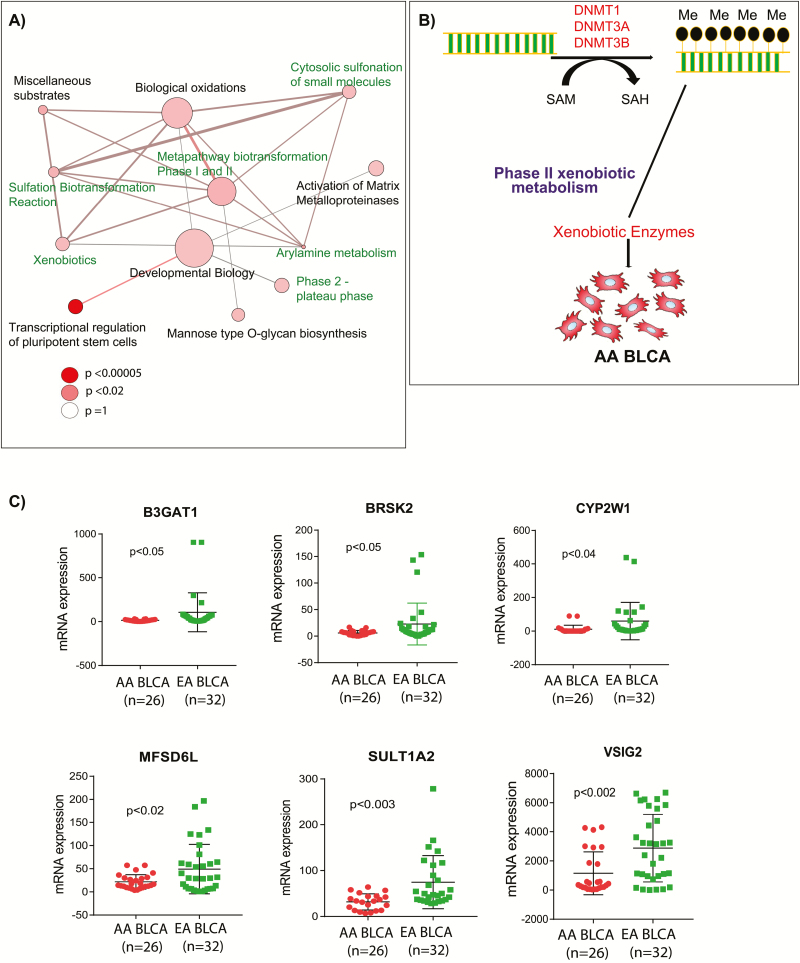
To understand the effects of these epigenetic differences on metabolic pathways in AA BLCA, we performed metabolic pathway analysis of the DMR-associated genes and identified several significantly enriched pathways (Figure 2A). Notably, these pathways cover xenobiotic pathways, particularly those of phase II biotransformation that plays an important role in the elimination of harmful xenobiotics. Glucuronidation and sulfonation are important biotransformation pathways that add glucuronic acid and sulfate moieties onto xenobiotics and render them more water-soluble, leading to their clearance. On the basis of this analysis, we hypothesized that hypermethylation at the promoters of the glucoronyltransferase leads to its suppression, impairing glucuronidation and accumulation of glucuronic acid in AA BLCA (Figure 2B). The expression of genes near the DMR-associated CpG islands was further validated using RNA Seq. Out of 29 genes, six genes were found to be significantly downregulated in AA compared with EA BLCA patients: B3GAT1 (Beta-1, 3-glucuronyltransferase), BRSK2 (BR serine/threonine kinase), CYP2W1 (cytochrome P450 family 2 subfamily W member), VSIG2 (V-set and immunoglobulin domain containing), MFSD6L (major facilitator superfamily domain containing 6 like) and SULT1A2 (sulfotransferase family 1A member; Figure 2C). The expression patterns for these genes were consistent with the observed differential methylations of CpG islands. Correlation analysis of promoter methylation and mRNA expression of VSIG2 and SULT1A2 in TCGA data corroborated the negative correlation specifically in AA tumors, further supporting altered xenobiotic metabolism in AA BLCA (Supplementary Figure 2, available at Carcinogenesis Online).
Figure 2.
Altered metabolic pathways and downregulation of key xenobiotic enzymes in AA BLCA. (A) Pathway analysis of the hyper- and hypomethylated genes from AA and EA BLCA. The node size is proportional to the number of genes in the pathway, and a colored node represents a statistically significant enrichment (P < 0.05). (B) Schematic overview of downregulation of xenobiotic enzymes by DNA methylation in AA BLCA. (C) mRNA expression of the genes BRSK2, B3GAT1, CYP2W1, MFSD6L, SULT1A2 and VSIG2 in ancestry-verified AA (n = 26) and EA BLCA (n = 32) tissues (P < 0.05, Student’s t test, data represent mean ± standard deviation).
Glucuronic acid accumulates in AA BLCA
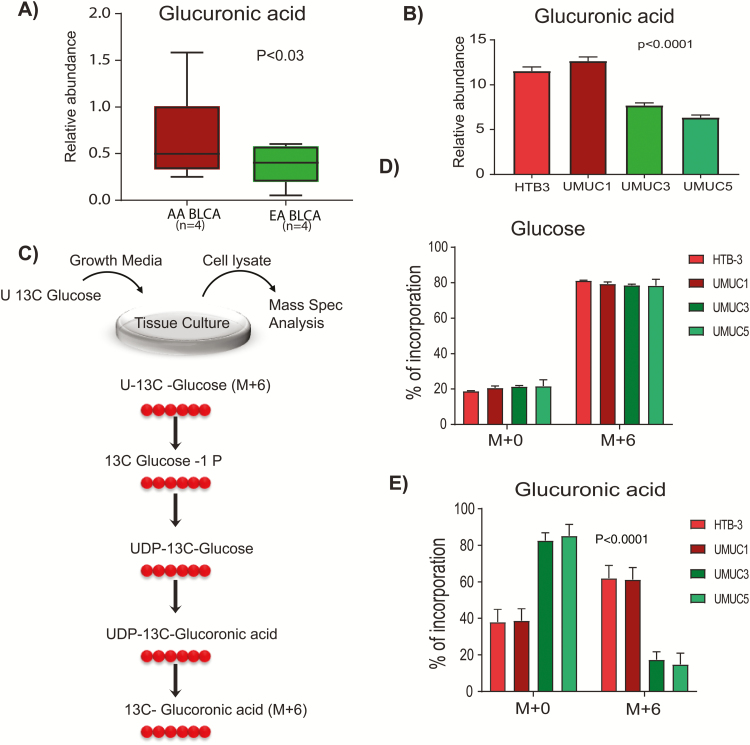
Glucuronidation is a phase II biotransformation reaction that involves the transfer of glucuronic acid from UDP glucuronic acid to xenobiotics to form polar conjugates that are easily excreted from the body (14). We measured glucuronic acid levels in tumors from AA and EA BLCA patients and found increased levels in AA patients (Figure 3A). We further measured glucuronic acid in AA BLCA cell lines (HTB3 and UMUC1), two EA BLCA cell lines (UMUC3 and UMUC5) and observed higher levels of glucuronic acid in AA BLCA compared with EA BLCA cell lines (Figure 3B). The glucuronic acid pathway is a minor route of glucose metabolism and provides uridine diphosphate (UDP) glucuronic acid for phase II xenobiotic biotransformation reactions and glycosaminoglycan metabolism (Figure 3C). To further confirm the deregulation of the glucuronic acid pathway, we performed metabolic flux analysis using 13C-labeled glucose in AA and EA BLCA cells lines. LC-MS analysis of flux confirmed the accumulation of glucuronic acid (m+6) in AA compared with EA BLCA, whereas glucose uptake was same in EA and AA BLCA cells (Figure 3D and E). Taken together, these results suggest that an altered glucuronic acid metabolism leads to high levels in AA compared with EA patients and cell lines.
Figure 3.
Accumulation of glucuronic acid in AA and EA BLCA. (A) Measurement of glucuronic acid levels in ancestry-verified AA (n = 4) and EA (n = 4) BLCA patients measured by LC-MS/MS (P < 0.03, Student’s t-test, data represent mean ± standard deviation). (B) Measurement of glucuronic acid levels in ancestry-verified AA (HTB3 and UMUC1) and EA (UMUC3 and UMUC5) BLCA cell lines measured by LC-MS/MS (n = 3 biological replicates; P < 0.05, Student’s t-test, data represent mean ± standard deviation). Peak area was normalized using 13C-labeled internal standards to represent relative abundance level. (C) Measurement of labelled glucoronic acid (M+6) using 13C glucose. (D) Measurement of 13C glucose (M+0 represents unlabeled glucose, and M+6 represents all six carbon-labeled glucose). (E) Glucuronic acid levels (M+0 represent unlabeled glucuronic acid, and M+6 represent all six carbon-labeled glucuronic acid) in ancestry verified AA (HTB3 and UMUC1) and EA (UMUC3 and UMUC5) BLCA cells measured by LC-MS/MS (n = 4 biological replicates)..
Reduced SAM and elevated methylated metabolites in AA BLCA
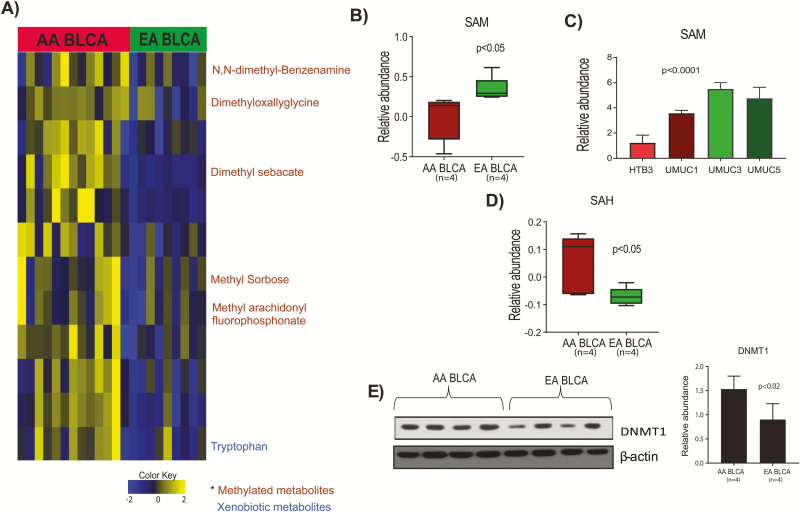
SAM is the universal methyl donor, and is used by methyltransferases to methylate DNA, RNA, proteins and metabolites (15). Once the methyl group is transferred from SAM to its substrate, SAH is produced. Methylated metabolites are also indicators of altered methylation metabolism. On the basis of the observed DNA hypermethylation in AA BLCA patients, we hypothesized that the methionine pathway is altered in AA BLCA. We measured methylated metabolites and observed elevated levels of N,N-dimethyl-benzenamine, dimethyloxalyglycine, dimethyl sebacate, methyl sorbose, in AA compared with EA BLCA (Figure 4A). We further measured levels of SAM in AA and EA BLCA and observed low levels of SAM in both patients and cell lines (Figure 4B and C). We extended the measurements of SAH in AA and EA BLCA and identified higher levels of SAH in AA compared with EA BLCA (Figure 4D), confirming the excessive utilization of SAM in AA compared with EA BLCA. DNMT1 is one of the methyltransferases that transfers the methyl group from SAM to cytosine of CpG islands. We measured DNMT 1 expression in AA and EA BLCA patients and identified elevated levels of DNMT1 in AA BLCA (Figure 4E). Overall these results revealed that SAM is the source of methyl group for DNA methylations process that influences expression of many genes during BLCA progression in AA BLCA.
Figure 4.
Methylated metabolites, SAM, SAH levels and DNMT1 expression in AA and EA BLCA. (A) Heat map of hierarchical clustering of differential metabolites from AA and EA BLCA patients. Columns represent individual tissue samples, and rows represent distinct metabolites. Shades of yellow and blue represent higher and lower levels of metabolites, respectively, relative to the median metabolite levels [false discovery rate (FDR) < 0.25s]. (B) Measurement of SAM levels in AA (n = 4) and EA (n = 4) BLCA patients by LC-MS/MS (P < 0.05, Student’s t-test, data represent mean ± standard deviation). (C) Measurement of SAM in AA (HTB3 and UMUC1) and EA (UMUC3 and UMUC5) BLCA cell lines by LC-MS/MS (P < 0.05, Student’s t-test, data represent mean ± standard deviation). (D) Measurement of SAH levels in AA and EA BLCA patients by LC-MS/MS. Peak area was normalized using 13C-labeled internal standards to represent relative abundance levels. (E) Immunoblot analysis of DNMT1 in AA (n = 4) and EA (n = 4) BLCA tissues and quantification.
Discussion
Disease-specific survival is worse for AA BLCA patients compared with other race/ethnic groups in the USA, even after accounting for differences in disease presentation (16). The disparity in BLCA is a complex problem due to the involvement of biological and environmental factors as well as socioeconomic and health care system (17). Social and environmental factors such as diet and exposure to environmental hazards can cause epigenetic changes that influence the expression of many genes (18). Here, we aimed to identify candidate biological determinants that contribute to the BLCA racial disparity by examining the DNA methylation differences and metabolic pathway alterations in AA over EA BLCA.
DNA methylation profiling enabled the identification of frequent hypermethylation regions in CpG islands of AA compared with EA BLCA tissues. DNA methylation, together with histone modifications, regulates the functioning of the genome by affecting chromatin architecture. Moreover, promoter hypermethylation plays a critical role in transcriptional regulation of important tumor suppressor and oncogenes during cancer progression (19–21). In this study, we observed and validated the hypermethylation at promoter regions of genes involved in xenobiotic metabolism, suggesting their potential role on disparity-driven BLCA progression among AA patients. Earlier studies from our group reported that altered xenobiotic metabolism play a pivotal role in BLCA progression (22).
Gene expression analysis of hyper- and hypomethylated CpG promoter regions helps to identify novel aberrantly expressed genes whose function might play an important role in AA BLCA progression. Altered metabolic pathways significantly associated with DMR genes, as the pathway-enrichment analysis identified deregulated metabolic pathways that may influence the tumor biology of AA patients than EA patients. Biotransformation reactions are commonly regulated by epigenetic regulation of genes in this pathway, potentially leading to accumulation of environmental carcinogens in AA BLCA that can contribute to carcinogenesis because increased urinary glucuronidation products can enhance DNA damage in BLCA (23). Another important deregulated pathway affected by the observed hypermethylation is the activation of matrix metalloproteinases (MMPs), indicated by hypermethylation of MMP15. These enzymes are responsible for the breakdown of the extracellular matrix during metastasis of cancer from primary tumors to distant organs (24,25). Our study also identified novel disparity-associated genes whose functions may play a crucial role in other pathways. For example, BRSK2, which is downregulated in AA BLCA, is a serine/threonine-protein kinase of the AMPK family that plays an important role in pancreatic ductal adenocarcinoma by suppression of mTORC1 activity via phosphorylation of tuberous sclerosis complex 2 (26). B3GAT1 is a glucanosyltransferase involved in glycosaminoglycan metabolism. Loss of B3GAT1 results in impairment of glucuronidation,which is involved in drug metabolism and clearance of xenobiotic compounds from the body, leading to the accumulation of hazardous compounds that may cause DNA instability and cancer. It also causes the accumulation of glucuronic acid, as we observed elevated levels in AA compared with EA BLCA. Of note, accumulated glucuronic acid was shown to have adverse effects in cancer (27). Therefore, the effects of the glucuronic acid pathway in racial disparity of AA BLCA progression will be an interesting area of research. CYP2W1 is a phase I xenobiotic monooxygenase known to metabolize various endogenous and exogenous substrates including lysophospholipids, procarcinogens and polycyclic aromatic hydrocarbons that are shown to have a crucial role in colorectal cancers (28). SULT1A1 and SULT1A2 are sulfotransferases that are inactivated by chromosomal aberrations in workers exposed to nitrotoluenes (29). Another study showed that the environmental pollutant 3-nitrobenzanthrone forms DNA adducts after reduction by NAD(P)H to quinone oxidoreductase and conjugation by acetyltransferases and sulfotransferases in human hepatic cytosols (30).
Glucuronic acid is an important component of hyaluronic acid synthesis, which plays an important role in extracellular matrix synthesis and also as a conjugating agent in xenobiotic metabolism, hence is gaining significant attention in cancer research. It has already been shown that glucuronic acid is elevated in breast cancer, and it is considered a biomarker for early detection of recurrent disease (27,31). Further studies on role of glucuronic acid in AA BLCA may improve understanding of molecular mechanisms associated with poor survival of AA BLCA. High levels of SAH and low levels of SAM infer the high consumption of SAM, which provides methyl donors for many epigenetic and physiological reactions such as the methylation of nucleic acids, histones, lipids, polyamines and proteins (32–34). Increased expression of DNMT1 in AA BLCA may utilize SAM for epigenetic modifications that in turn contribute to various processes of cancer development including tumor initiation, invasion, metastasis and chemotherapy resistance. The development of DNMT1 inhibitors as epigenetic drugs might prove useful as an epigenetic cancer therapy for AA BLCA patients.
In conclusion, our study reports higher DNA methylation in AA compared with EA BLCA patients, with the methylation-related metabolism being increased in BLCA patients, thereby specifically affecting xenobiotic metabolism pathways in these tumors. Metabolomic flux analysis further revealed the accumulation of glucuronic acid in AA compared with EA BLCA cell lines. Elevated levels of the methylated metabolite SAH and reduced levels of SAM corroborated the observed hypermethylations and altered methylation metabolism in AA BLCA. Further understanding and validation of these discoveries using large cohorts of BLCA specimens’ in vitro and in vivo studies will improve our knowledge on the development of disparity-specific therapeutic interventions to target BLCA in the future
Supplementary Material
Acknowledgements
We would like to thank Dr. Prashant Singh and Dr. Sean Glenn, Genomics Shared Resource at Roswell Park Comprehensive Cancer Center.
Glossary
Abbreviations
- AA
African American
- BLCA
Bladder Cancer
- cDNA
Complementary DNA
- DMR
Differentially methylated region
- EA
European American
- ESI
Electron spray ionization
- HPLC
High-performance liquid chromatography
- KEGG
Kyoto encyclopedia of genes and genomes
- LC-MS/MS
Liquid chromatography–mass spectrometry
- RRBS
Reduced-representation bisulfite sequencing
- TCGA
The cancer genomic atlas
- UDP
Uridine diphosphate
Funding
This research was supported by American Cancer Society (ACS) Award (127430-RSG-15-105-01-CNE to N.P., NIH/NCI R01CA220297 to N.P., NIH/NCI R01CA216426 to N.P. and NIH/NCI U01 CA167234 to A.S.K.). This project was also supported by the Agilent Technologies Center of Excellence (COE) and National Institute of Health (NIH) (P30 CA125123), CPRIT Proteomics and Metabolomics Core Facility (RP170005 to N.P.), and Dan L. Duncan Cancer Center. P.D.C. and M.M.I., were supported by Human Tissue Acquisition and Pathology at Baylor College of Medicine with funding from National Cancer Institute (NCI) (P30 CA125123). The Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health (NIH) (P30 CA142543 09).
Conflict of Interest Statement: None declared.
References
- 1. Mallin K., et al. (2011) Transitional cell carcinoma of the bladder: racial and gender disparities in survival (1993 to 2002), stage and grade (1993 to 2007). J. Urol., 185, 1631–1636. [DOI] [PubMed] [Google Scholar]
- 2. Schinkel J.K., et al. (2016) Overall and recurrence-free survival among black and white bladder cancer patients in an equal-access health system. Cancer Epidemiol., 42, 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Phan V.H., et al. (2009) Ethnic differences in drug metabolism and toxicity from chemotherapy. Expert Opin. Drug Metab. Toxicol., 5, 243–257. [DOI] [PubMed] [Google Scholar]
- 4. Ingelman-Sundberg M. (2008) Pharmacogenomic biomarkers for prediction of severe adverse drug reactions. N. Engl. J. Med., 358, 637–639. [DOI] [PubMed] [Google Scholar]
- 5. Jones P.A., et al. (2007) The epigenomics of cancer. Cell, 128, 683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Verma M., et al. (2002) Epigenetics in cancer: implications for early detection and prevention. Lancet. Oncol., 3, 755–763. [DOI] [PubMed] [Google Scholar]
- 7. Wolffe A.P., et al. (1999) Epigenetics: regulation through repression. Science, 286, 481–486. [DOI] [PubMed] [Google Scholar]
- 8. Mathers J.C., et al. (2010) Induction of epigenetic alterations by dietary and other environmental factors. Adv. Genet., 71, 3–39. [DOI] [PubMed] [Google Scholar]
- 9. Jones P.A., et al. (2002) The fundamental role of epigenetic events in cancer. Nat. Rev. Genet., 3, 415–428. [DOI] [PubMed] [Google Scholar]
- 10. Esteller M. (2005) DNA methylation and cancer therapy: new developments and expectations. Curr. Opin. Oncol., 17, 55–60. [DOI] [PubMed] [Google Scholar]
- 11. Akalin A., et al. (2012) methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol., 13, R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krueger F., et al. (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics, 27, 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Feng H., et al. (2014) A Bayesian hierarchical model to detect differentially methylated loci from single nucleotide resolution sequencing data. Nucleic Acids Res., 42, e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mulder G.J. (1992) Glucuronidation and its role in regulation of biological activity of drugs. Annu. Rev. Pharmacol. Toxicol., 32, 25–49. [DOI] [PubMed] [Google Scholar]
- 15. Finkelstein J.D. (1990) Methionine metabolism in mammals. J. Nutr. Biochem., 1, 228–237. [DOI] [PubMed] [Google Scholar]
- 16. Yee D.S., et al. (2011) Ethnic differences in bladder cancer survival. Urology, 78, 544–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hollenbeck B.K., et al. (2010) Racial differences in treatment and outcomes among patients with early stage bladder cancer. Cancer, 116, 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bachir B.G., et al. (2012) Cause-effect? Understanding the risk factors associated with bladder cancer. Expert Rev. Anticancer Ther., 12, 1499–1502. [DOI] [PubMed] [Google Scholar]
- 19. Estécio M.R., et al. (2011) Dissecting DNA hypermethylation in cancer. FEBS Lett., 585, 2078–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kulis M., et al. (2010) DNA methylation and cancer. Adv. Genet., 70, 27–56. [DOI] [PubMed] [Google Scholar]
- 21. Baylin S.B. (2005) DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol., 2 (suppl. 1), S4–11. [DOI] [PubMed] [Google Scholar]
- 22. Putluri N., et al. (2011) Metabolomic profiling reveals potential markers and bioprocesses altered in bladder cancer progression. Cancer Res., 71, 7376–7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Al-Zoughool M., et al. (2006) Effect of N-glucuronidation on urinary bladder genotoxicity of 4-aminobiphenyl in male and female mice. Environ. Toxicol. Pharmacol., 22, 153–159. [DOI] [PubMed] [Google Scholar]
- 24. Itoh Y., et al. (2002) Matrix metalloproteinases in cancer. Essays Biochem., 38, 21–36. [DOI] [PubMed] [Google Scholar]
- 25. Gialeli C., et al. (2011) Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J., 278, 16–27. [DOI] [PubMed] [Google Scholar]
- 26. Saiyin H., et al. (2017) BRSK2 induced by nutrient deprivation promotes Akt activity in pancreatic cancer via downregulation of mTOR activity. Oncotarget, 8, 44669–44681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Oikari S., et al. (2018) UDP-sugar accumulation drives hyaluronan synthesis in breast cancer. Matrix Biol., 67, 63–74. [DOI] [PubMed] [Google Scholar]
- 28. Chung F.F., et al. (2016) Cytochrome P450 2W1 (CYP2W1) in colorectal cancers. Curr. Cancer Drug Targets, 16, 71–78. [DOI] [PubMed] [Google Scholar]
- 29. Sabbioni G., et al. (2006) Biomarkers of exposure, effect, and susceptibility in workers exposed to nitrotoluenes. Cancer Epidemiol. Biomarkers Prev., 15, 559–566. [DOI] [PubMed] [Google Scholar]
- 30. Arlt V.M., et al. (2005) Environmental pollutant and potent mutagen 3-nitrobenzanthrone forms DNA adducts after reduction by NAD(P)H:quinone oxidoreductase and conjugation by acetyltransferases and sulfotransferases in human hepatic cytosols. Cancer Res., 65, 2644–2652. [DOI] [PubMed] [Google Scholar]
- 31. Yahya R.S., et al. (2014) Biochemical evaluation of hyaluronic acid in breast cancer. Clin. Lab., 60, 1115–1121. [DOI] [PubMed] [Google Scholar]
- 32. Shima H., et al. (2017) S-Adenosylmethionine synthesis is regulated by selective N6-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep., 21, 3354–3363. [DOI] [PubMed] [Google Scholar]
- 33. Hao X., et al. (2016) Immunoassay of S-adenosylmethionine and S-adenosylhomocysteine: the methylation index as a biomarker for disease and health status. BMC Res. Notes, 9, 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pegg A.E. (2016) Functions of polyamines in mammals. J. Biol. Chem., 291, 14904–14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.