Abstract
B cells are important modulators of immune responses both in autoimmunity and cancer. We have previously shown that B regulatory (Breg) cells promote pancreatic cancer via production of IL35, a heterodimeric cytokine comprised of the subunits p35 (Il12a) and Ebi3. However, it is not known how production of IL35 is regulated in vivo in the context of cancer-associated inflammation. To begin addressing this question, we have generated a knock-in mouse model, Il12aGFP, where an IRES-emGFP gene was inserted within the 3’ UTR of the Il12a locus. EmGFP signal in B cells from the Il12aGFP mice correlated with expression of p35 mRNA and protein. Using this model, we observed that in addition to Bregs, expression of GFP (p35) is upregulated in several other B cell subtypes in response to cancer. We assessed the expression of the other IL35 subunit, Ebi3, using a published tdTomato reporter model. We determined that Ebi3 expression was more tightly regulated in vivo and in vitro, suggesting that stimuli affecting Ebi3 upregulation are more likely to result in production of full IL35 heterodimer. We were also able to detect GFP and Tomato signal in myeloid & T cell lineages suggesting that these reporter models could also be used for tracking IL12-, IL27- and IL35-producing cells. Furthermore, using primary B cells isolated from reporter mice, we identified BCR, CD40 and TLR pathways as potential drivers of IL35 expression. These findings highlight the importance of pancreatic cancer-associated inflammatory processes as drivers of cytokine expression and provide a tool to dissect both disease-associated regulation of IL12- and IL35-competent lineage cells as well as establish assays for pharmacological targeting of individual subunits of heterodimeric IL12 family cytokines.
Keywords: IL35 cytokine, reporter model, regulatory B cell, pancreatic cancer
1. INTRODUCTION
The IL12 family of cytokines plays an important role in modulating disease-related immunological responses. These cytokines form via heterodimerization of p19, p28, p35, p40 and Ebi3 subunits, and include IL12 (p35/p40), IL35 (p35/Ebi3), IL23 (p19/p40) and IL27 (p28/Ebi3) (1). The IL12 family takes on a variety of roles to shape the fate of naïve T cells and modulate functions of effector T cells. Deregulation of effector functions of pro-inflammatory cytokines IL12 and IL23 as well as the immunosuppressive cytokines IL27 and IL35 have been associated with a slew of autoimmune conditions, such as EAE, uveitis, and MS, and has recently emerged as a prominent characteristic of cancer-associated inflammation, rendering these cytokines as crucial targets for therapy development (2–5).
Recently, we demonstrated that IL35+ B cells promote pancreatic cancer growth by eliciting immunosuppression (6, 7). Elevated levels of IL35 have been detected in cancer and can contribute to suppression of autoimmune conditions (2, 8–10). Several cell types, including naïve CD4+ T cells, Tregs, B cells, and dendritic cells have been shown to produce IL35 (2, 7, 9, 11, 12). Disease associated inflammation has been postulated to promote IL35 expression in immune cells, however, the precise mechanisms that regulate IL35 expression in cancer or other diseases remain unclear. Due to its heterodimeric nature, IL35 has proven difficult to study. It is not well understood how disease regulates endogenous expression of IL35 subunits, as typically cells are isolated and activated ex vivo in order to detect intracellular cytokine production. From a technical standpoint, it is not clear if detection of one of the subunits is sufficient to mark IL35-producing cells. Furthermore, there is accumulating evidence suggesting a role for mono- or homodimeric functions for p35 and/or Ebi3 subunits, making it important to determine endogenous sources of p35 and Ebi3 production and regulation (3, 13).
We sought to develop a murine model system that would allow us to study dynamic regulation of both IL35 subunits in the context of pancreatic cancer. To do this, we generated a novel IL12aGFP reporter strain, which allowed us to begin examination of Il12a expression patterns in healthy and tumor-bearing mice. To examine expression of Ebi3, we utilized a previously published TdTomato reporter strain (Ebi3Tom) (9).
2. MATERIALS AND METHODS
Mice
Il12aGFP mice were generated by the UNC Animal Models Core. A Cas9 guide RNA targeting the Il12a 3’UTR 70bp 3’ of the stop codon was produced by oligonucleotide-mediated cloning into a T7 promoter vector followed by in vitro transcription and spin column purification, with elution in microinjection buffer (protospacer sequence 5’- GATTCATAAGAGTCAGG −3’). The donor plasmid included a 1,397 bp 5’ homology arm, EMCV IRES, Emerald GFP coding sequence, Bovine Growth Hormone polyadenylation sequence and 1,436 bp 3’ homology arm in a pUC plasmid backbone. The donor plasmid was constructed by a modified Gibson assembly procedure using equimolar stoichiometry (1 picomole) of each DNA element and 20–40 bp overhangs with 2x assembly mix containing T5 flap endonuclease and Phusion (PMID: 21601685). The equimolar assembly reaction was thermocycled as follows: [37°C for 7.5 min, 50°C for 15 min, (55°C for 1 min decreasing by 1°C per cycle) where n = 10 cycles, 50°C for 35 min, and final soak 10°C]. Assembly mixes were purified over a silica minicolumn and quantitated by NanoDrop UV spectroscopy. Approximately 100 ng of purified assembly was transformed into 50 µl of commercially chemically competent Stellar cells. The final donor vector was Sanger-sequence verified. Donor plasmid was prepared by Qiagen High Speed Maxiprep protocol and resuspended in microinjection buffer. Recombinant Cas9 protein was expressed in E. coli and purified by the UNC Protein Expression and Purification Core Facility. C57BL/6J zygotes were microinjected with 400 nM Cas9 protein, 50 ng/µl guide RNA and 20 ng/µl donor plasmid in microinjection buffer (5 mM Tris pH7.5, 0.1 mM EDTA). Injected embryos were implanted in recipient pseudopregnant females. Resulting pups were screened by PCR for the presence of the knock-in event. Primers used to determine presence of Il12a-emGFP allele:
FWD 5’– AATGGGTCTAGGAGTGTGATGA –3’, REV 5’– AAATAACATATAGACAAACGCACACCG – 3’. Primers used to determine presence of Il12a- WT allele: FWD 5’– AATGGGTCTAGGAGTGTGATGA –3’, REV 5’–TGCTCTTCTGCTAACACATTGA –3’.
Ebi3Tom mice were obtained from D. Vignali (University of Pittsburgh) (9). In this mouse model a tdTomato-2A cassette was knocked-in after the transcriptional start site (ATG) in the Ebi3 locus. Six- to eight week-old wild-type (WT) C57Bl/6J mice were purchased from The Charles River Laboratories (strain #027). Leukocytes from spleens and tumors isolated from WT mice were used as negative controls for both GFP and Tomato fluorescence by flow cytometry. All mouse protocols were reviewed and approved by the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill.
Pancreatic Cancer Cell lines
The murine PDA cell line, KPC 4662, was originally derived from primary pancreatic tumor lesions of KPC mice by Dr. Robert Vonderheide’s lab (14). Cells were tested and confirmed to be mycoplasma and endotoxin free. Cells were maintained at 37°C and 5% CO2 in complete DMEM, which contains Dulbecco’s Modified Eagle medium (Gibco) supplemented with 10% FCS, 1% L-glutamine and 50 μg/ml gentamicin (Thermo Scientific).
Orthotopic Tumor Modeling Experiments
Mice were anesthetized using a ketamine (100 mg/kg)/Xylazine (10 mg/kg) cocktail administered via intraperitoneal injection. An incision in the left flank was made, and 75,000 KPC cells in ice-cold PBS mixed at 1:1 dilution with Matrigel (#354234, Corning) in a volume of 50 μL were injected using a 28-gauge needle. The incision was closed in two layers, with running 5–0 Vicryl RAPIDE sutures (Ethicon) for the body wall, and 5–0 PROLENE sutures (Ethicon) for the skin. All animals were given the pain reliever buprenorphine (0.1 mg/kg) subcutaneously once, directly after the conclusion of surgical procedure. Tumors and splenic tissues were harvested at 3 weeks post KPC cell injection.
Lymphocyte isolation
Single-cell suspensions were prepared from dissected tumors and spleens. Spleens were mechanically disrupted using a plunger end of a 5 mL syringe and resuspended in 1% FBS/PBS after passing through a 70-μm cell strainer (Falcon). Red blood cells were depleted from total splenocytes using 1x RBC Lysis Solution (eBioscience, 00–4333-57). For isolation of tumor-infiltrating lymphocytes, tumor tissue was minced into 1 to 2 mm pieces and digested with collagenase IV (1.25 mg/mL; #LS004188, Worthington), 0.1% trypsin inhibitor from soybean (# T9128, Sigma), hyaluronidase (1 mg/mL; # LS 002592, Worthington), and DNase I (100 mg/mL; # LS002007, Worthington) in complete DMEM for 30 minutes at 37°C. Cell suspensions were passed through a 70-μm cell strainer (Falcon) and resuspended in RPMI media (Gibco). Lymphocytes were isolated from processed tumor tissues by OptiPrep (Sigma) density gradient centrifugation. For experiments requiring ex vivo stimulation and intracellular cytokine staining, regulatory B cells (CD19+CD1dHiCD5+CD21Hi) and conventional B cells (CD19+CD1dLoCD5-CD21Lo) were sorted from single cell splenic suspensions using the BD FACS Aria III cell sorter according to manufacturer’s instructions and UNC flow cytometry core guidelines.
Flow cytometry profiling
Single cell splenic and tumor suspensions were blocked using TruStain FcX (Biolegend, 101319) at a concentration of 1µg/1×106 cells. Viability was assessed using the Zombie NIR or Zombie Violet (Biolegend, 423113 or 423105) according to manufacturer’s instructions. Cells were stained with fluorescent labelled antibodies (listed in Table 1) for 30 min on ice diluted in FACS buffer (PBS with 1% heat-inactivated FCS and 0.05% sodium azide). After staining, cells were washed twice with FACS buffer and re-suspended in FACS buffer (PBS with 1% FCS 0.05% sodium azide). Cells were fixed in IC Fixation buffer (eBioscience, 00–8222-49) for 10 minutes on ice after washing. Cells were washed twice in FACS buffer and then acquired on BD LSR Fortessa using manufacturer instructions and UNC flow cytometry core guidelines. Antibodies used in the study are listed in Supplementary Table 1.
Table 1
| Resource | Source | Identifier |
|---|---|---|
| Antibodies | ||
| anti-CD11c | Biolegend | N/A |
| anti-CD19 | Biolegend | N/A |
| anti-CD1d | BD Biosciences | N/A |
| anti-CD21 | Biolegend | N/A |
| anti-CD23 | Biolegend | N/A |
| anti-CD4 | Biolegend | N/A |
| anti-CD40 | Biolegend | N/A |
| anti-CD45 | Biolegend | N/A |
| anti-CD5 | Biolegend | N/A |
| anti-CD8 | Biolegend | N/A |
| anti-CD93 | BD Biosciences | N/A |
| anti-IgD | eBioscience | N/A |
| anti-IgM | Jackson Immune Research | N/A |
| anti-p35 | eBioscience | N/A |
| CD11b | BD Biosciences | N/A |
| CD3 | Biolegend | N/A |
| True Stain FCX | Biolegend | N/A |
| Bacterial Strains | ||
| Stellar competent cells | Takara Biosciences | N/A |
| Chemical | ||
| 2-mercaptoethanol | Thermo Scientific | N/A |
| Brefeldin A | Biolegend | N/A |
| buprenorphine | ||
| CO2 | ||
| CpG ODN1826 | Invivogen | N/A |
| cytofix/cytoperm buffer | BD | N/A |
| DMEM | Gibco | N/A |
| EDTA | Thermo Scientific | N/A |
| FCS | Corning | N/A |
| Gentamicin | Thermo Scientific | N/A |
| IC fixation buffer | eBioscience | N/A |
| Ionomycin | Cell Signalling | N/A |
| ketamine | ||
| L-glutamine | Thermo Scientific | N/A |
| LPS | Sigma | N/A |
| OpiPrep | Sigma | N/A |
| PBS | Thermo Scientific | N/A |
| Pen/strep | Thermo Scientific | N/A |
| Perfecta SYBR Green supermix | Quanta Biosciences | N/A |
| PMA | Sigma | N/A |
| R848 | Invivogen | N/A |
| RBC lysis solution | eBioscience | N/A |
| sodium Azide | sigma | N/A |
| Tris | Fisher Scientific | N/A |
| Xylazine | ||
| Critical Commercial Assay | ||
| B cell isolation kit | Stem Cell Technologies | N/A |
| Maxima First Strand cDNA synthesis kit | Thermo Scientific | N/A |
| Qiagen Hi Speed maxiprep kit | Qiagen | N/A |
| RNeasy minikit | Qiagen | N/A |
| Experimental models | ||
| mice | Charles River | C57Bl6/J |
| Oligonucleotides | ||
| b-actin fwd | Invitrogen | 5’-GGCTGTATTCCCCTCCATCG–3’ |
| b-actin rev | Invitrogen | 5’-CCAGTTGGTAACAATGCCATGT-3’ |
| emGFP fwd | Invitrogen | 5’-GACCACCTTGACCTACGGCG-3’ |
| emGFP rev | Invitrogen | 5’ –CCCTCGAACTTCACCTCGGC – 3’ |
| Il12a fwd | Invitrogen | 5’-CATCGATGAGCTGATGCAGT-3’ |
| Il12a rev | Invitrogen | 5’-CAGATAGCCCATCACCCTGT-3’ |
| ProteinPeptide | ||
| Collagenase IV | Worthington | N/A |
| DNAseI | Worthington | N/A |
| Flap endonuclease | NEB | N/A |
| Hyaluronidase | Worthington | N/A |
| Matrigel | Corning | N/A |
| Phusion polymerase | NEB | N/A |
| Soy Bean Trypsin Inhibitor | Sigma | N/A |
| Zombie NIR | Biolegend | N/A |
| Zombie Violet | Biolegend | N/A |
| Software | ||
| BD FACS DIVA | BD | N/A |
| FlowJo | TreeStar | N/A |
| Prism 8 | Graph Pad | N/A |
Intracellular cytokine staining
For ex vivo stimulation, tumor and spleen samples were processed as described above, single cell suspension was prepared, and sorted B cells were collected in complete RPMI media containing 10% FCS with 1x pen/strep. Sorted cells were cultured for 48 hours after the addition of LPS (1ug/ml, Sigma) and anti-CD40 (1ug/ml, Biolegend, Clone # HM40–3). For the final 5 hours of stimulation, cells were additionally incubated with 50 ng/ml PMA (Sigma, #P8139) and 200 ng/ml Ionomycin (Sigma, #I0634) in the presence of Golgistop Brefeldin A (1X, Biolegend) before subsequent staining. Cells were washed and blocked with TruStain FcX (Biolegend, #101319, 1µg/1×106 cells) for 5 minutes on ice. Viability was assessed using the Zombie NIR or Zombie Violet (Biolegend, #423113 or #423105) according to manufacturer’s instructions. Cells were then washed and stained with labeled antibodies against surface markers on ice for 30 min in FACS buffer (PBS with 1% FCS and 0.05% sodium azide). After surface staining, cells were washed, fixed and permeabilized using cytofix/cytoperm buffer (BD Biosciences, #554714) for 15 min at 4°C. Intracellular staining was performed using fluorophore-conjugated cytokine antibodies for 1 hour, at 4°C in the dark. After intracellular staining, cells were washed and resuspended in FACS buffer for acquisition by flow cytometry. After incubation, cells were washed, resuspended in FACS buffer, and acquired on LSR Fortessa (BD Bioscience) at the UNC Flow Cytometry Core Facility and analyzed by FlowJo version 10.2 (TreeStar, Inc.).
Quantitative PCR (qPCR) analysis
B cells were isolated from WT and Il12aGFP mouse spleens using a negative selection mouse B cell isolation kit (StemCell Technologies, #19854). RNA was purified from isolated B cells using the RNeasy mini kit (Qiagen, 74134). RNA was normalized to 1ug total RNA and cDNA were generated using the Maxima First Strand cDNA Synthesis Kit (Thermo Scientific, K1671). qPCR analysis was performed using PerfeCTa SYBR Green SuperMix (Quanta Biosciences) and the Applied Biosystems (ABS) 7500 real-time PCR system. Results were normalized to the expression of β-Actin and then calculated using ∆∆CT. All samples were run in triplicate. Primer sequences used for qPCR are β-Actin FWD 5’-GGCTGTATTCCCCTCCATCG–3’, β-Actin REV 5’-CCAGTTGGTAACAATGCCATGT-3’, Il12a FWD 5’-CATCGATGAGCTGATGCAGT-3’, Il12a REV 5’-CAGATAGCCCATCACCCTGT-3’, emGFP FWD 5’- GACCACCTTGACCTACGGCG - 3’, emGFP REV 5’ – CCCTCGAACTTCACCTCGGC – 3’.
In vitro reporter stimulation assay
B cells were isolated from Il12aGFP and Ebi3Tom mouse spleens using a negative selection mouse B cell isolation kit (StemCell Technologies, #19854). B cells were seeded in a 48-well plate (1e6/ml) in RPMI containing 10% heat-inactivated serum, 1x pen/strep, and B-mercaptoethanol. Cells were stimulated for 48 hours using the combinations of the following reagents: aIgM (10ug/ml; Jackson Immune Research #115–006-075), aCD40 (1ug/ml; Biolegend #102901), R848 (1ug/ml; Invivogen # tlrl-r848), LPS (1ug/ml; Sigma # 437627), CpG ODN1826 (1ug/ml; Invivogen # tlrl-1826).
Statistical analysis
6 mice were used in each group. Before analysis data were examined for quality and potential outliers. Significance in variations between two groups was determined by unpaired student t test (two-tailed). Statistical analysis were performed using GraphPad Prism software, Data is presented as mean ±s.e.m. p< 0.05 was considered statistically significant.
3. RESULTS
3.1. Generation of Il12aGFP mice.
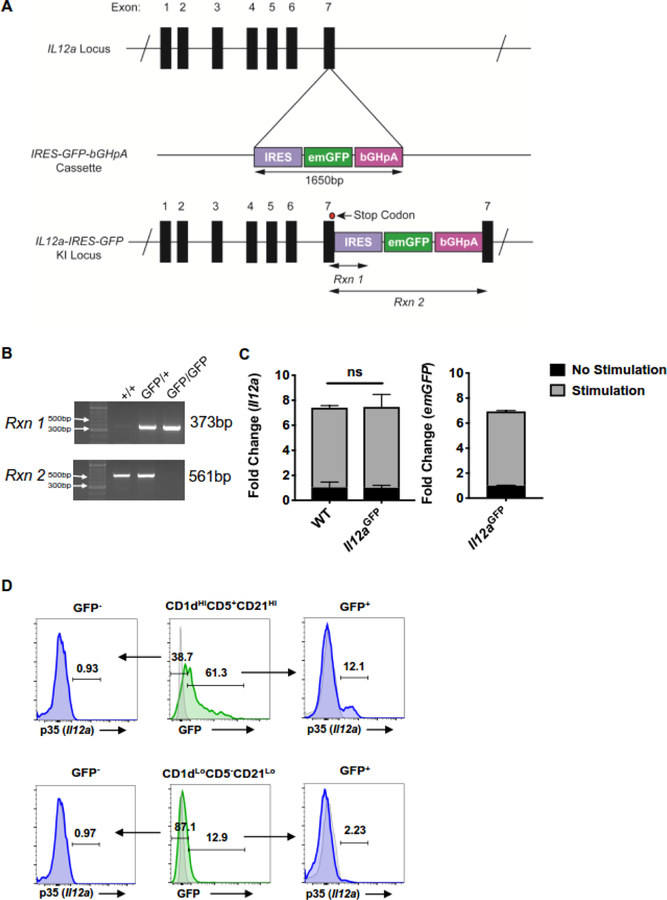
To establish a reporter mouse model for detection of endogenous Il12a expression, we engineered an emerald GFP (emGFP) expression vector containing an internal ribosomal entry site (IRES) and a bovine growth hormone polyadenylation sequence. The expression vector was inserted into C57Bl/6J zygotes using Cas9 and a guide RNA targeting a sequence 70bp downstream of the Il12a stop codon in the 3’ UTR (Figure 1A). Successful integration and zygosity was assessed by PCR (Figure 1B). The resulting mice did not display any defects in viability and fertility.
Figure 1. Generation of Il12aGFP KI mice and in vitro analysis of B cells.
(A) Schematic of IRES-emGFP insertion into the Il12a genomic locus in the 3’ UTR of exon 7.
(B) Genotyping of Il12aGFP mice by PCR. Rxn 1 determines insertion of IRES-emGFP cassette into Il12a exon 7 3’ UTR. Rxn 2 determines presence of WT Il12a allele without insertion of IRES-emGFP cassette.
(C) Splenic B cells (CD19+) isolated from healthy WT and Il12aGFP stimulated with LPS + αCD40 for 48 hours. Fold change of Il12a expression in healthy WT and Il12aGFP B cells (left) and emGFP (right) expression in IL12a-GFP B cells; n=3, ns = p>0.05.
(D) Splenic CD1dHiCD5+CD21Hi B cells (top) and CD1dLoCD5-CD21Lo B cells (bottom) from healthy Il12aGFP mice stimulated for 48 hours with LPS + αCD40. p35 (Il12a) and GFP protein expression were measured by flow cytometry.
To validate that expression of Il12a mRNA correlated with expression of emGFP mRNA, we used a previously established assay to stimulate splenic B cells from WT and Il12aGFP mice with LPS and anti-CD40, known activators of Il12a expression in cultured B cells (7). In comparison to unstimulated B cells we observed a significant increase in Il12a mRNA expression in both WT and Il12aGFP B cells. Furthermore, we did not see any significant differences in the fold change of Il12a expression between WT and Il12aGFP B cells upon stimulation, suggesting the IRES-emGFP cassette by itself does not alter endogenous basal Il12a expression (Figure 1C). We also observed that stimulated Il12aGFP B cells increase levels of Il12a and emGFP to a similar extent, demonstrating specific activation of emGFP expression under the Il12a promoter. To determine the relationship between GFP expression and p35 protein levels, we sorted regulatory (CD19+CD5+CD1dHiCD21Hi) and conventional (CD19+CD5-CD1dLoCD21Lo) B cells, and analyzed p35 and GFP expression in those subsets (Figure 1D). Consistent with our previous findings, only regulatory B cells exhibited GFP expression after stimulation and GFP+ regulatory B cells strongly correlated with production of the p35 protein in intracellular cytokine analysis assay. A large proportion of regulatory B cells and only a minor proportion of conventional B cells were GFP-positive, suggesting that GFP reporter readout might be more sensitive that p35 staining (Figure 1D). Furthermore, the GFP-negative populations of both B cell subsets were also negative for p35 production, suggesting that the Il12aGFP allele is a faithful reporter for p35 production (Figure 1D).
3.2. Modulation of Il12a expression in pancreatic cancer bearing mice.
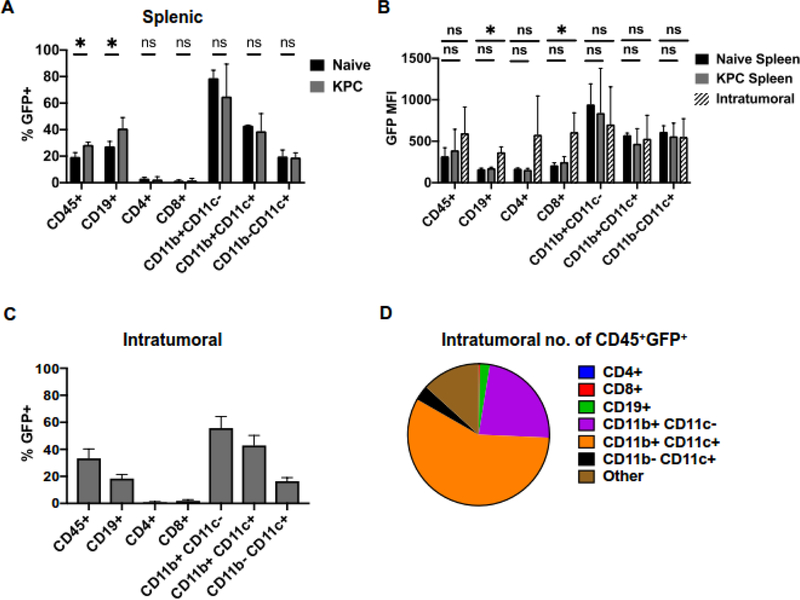
To understand how expression of p35 was modulated in the context of pancreatic cancer-induced inflammation in vivo, we performed orthotopic injections of primary syngeneic KrasG12D;Trp53R172H;p48Cre/+ (KPC) mouse pancreatic cancer cells into WT and Il12aGFP mice. Using the spleen as an indicator of a systemic immune response beyond the tumor itself, we noted an overall increase in the frequency of CD45+GFP+ immune cells compared to non-tumor bearing controls (Figure 2A, Sup. Figures 1 and 2). This increase primarily stemmed from CD19+GFP+ B cell compartment. We have previously shown that CD4+ T cells produced IL35 in pancreatic cancer (7). Consistent with this, we also observed that a subset of CD4+ cells was GFP positive, although the extent of reporter expression frequency in splenic CD4+ T cells did not appear to be regulated by cancer-associated inflammation (Figure 2A). Finally, a prominent fraction of myeloid cells expressed GFP (Figure 2A). Our prior observations showed that myeloid cells do not co-express p35 and Ebi3 to form IL35, but rather produce p40, suggesting that reporter activity might indicate formation of a IL12 heterodimer (7). We then asked whether pancreatic tumors increase the amount of p35 expression in B cells derived from spleens or tumors, by examining the mean fluorescent intensity (MFI) of GFP (Figure 2B). Splenic B cells from tumor bearing mice did not display increased GFP expression when compared to non-tumor controls, suggesting that pancreatic cancer associated inflammation modulates the number of p35 expressing B cells within the spleen, but not the amount of p35 per B cell. We did, however, note a significant increase of GFP MFI in the B cells found within the tumor compared to the spleen indicating that factors within the tumor microenvironment may direct the amount of p35 expression in B cells (Figure 2B). We also noted that intratumoral CD8+ T cells significantly increased GFP MFI, although the overall frequency of these cells within tumors was very low (Figure 2A, B).
Figure 2. Pancreatic cancer alters immune cell expression of IL12aGFP in vivo.
(A) Frequency of GFP+ immune cells isolated from spleens of KPC orthotopic tumor-bearing and non-tumor bearing (naïve) Il12aGFP mice measured by flow cytometry; n=6, *=p<0.05, ns=p>0.05.
(B) Mean fluorescent intensity (MFI) of GFP in GFP+ immune populations isolated from spleens and tumors of KPC orthotopic tumor-bearing and non-tumor bearing (naïve) Il12aGFP measured by flow cytometry mice; n=6, *=p<0.05, ns=p>0.05.
(C) Frequency of GFP+ immune cells isolated from orthotopic KPC pancreatic tumors of Il12aGFP mice measured by flow cytometry; n=6.
(D) Proportions of CD45+GFP+ cell populations isolated from Il12aGFP orthotopic KPC pancreatic tumors; n=6.
Frequencies of CD45+GFP+ immune cell subsets within the pancreatic tumors themselves largely mirrored the frequencies found in the tumor-bearing spleens (Figure 2C). To understand the extent of GFP-positive immune subset contribution to pancreatic TME, we analyzed the relative numbers of intratumoral GFP+ subsets. We observed that myeloid populations comprised the majority of GFP+ cells in the tumor (Figure 2D). While it appears that B cells comprise a minority of the GFP+ population within the tumor, pancreatic cancer has an effect on B cell Il12a expression systemically.
3.3. GFP expression marks multiple B cell subsets in pancreatic cancer.
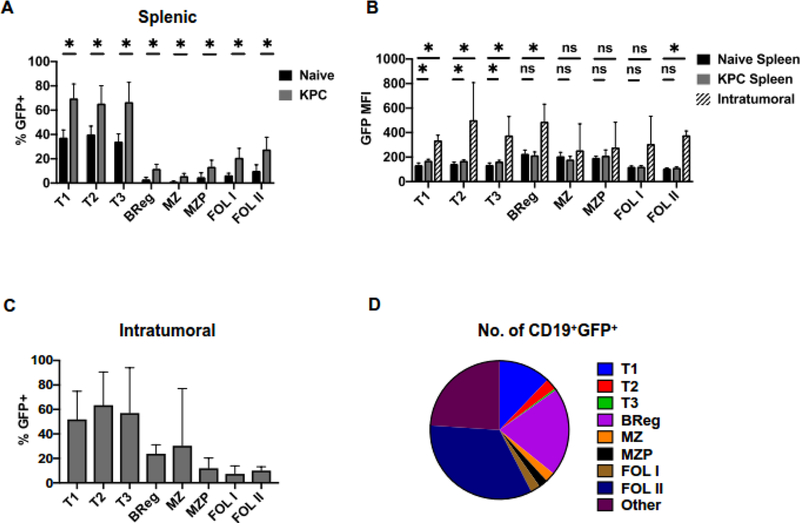
We have reported previously that IL35+ B cells promoted pancreatic cancer growth (6). We have thus far identified CD19+CD5+CD1dHiCD21Hi regulatory B cell subset (Breg) as expressing both p35 and Ebi3 subunits (Sup. Figures 3 and 4) (7). Using the Il12aGFP model, we wanted to understand which B cell subsets express p35 in vivo and upregulate its production in response to pancreatic cancer. We observed that all splenic B cell subsets that we examined expressed GFP basally and the frequency of GFP+ B cells in every population significantly increased in response to pancreatic cancer (Figure 3A). Interestingly, the majority of B cells within transitional populations (T1, T2, T3) had become GFP+ in response to pancreatic tumorigenesis, indicating that splenic B cells at the earliest developmental stages are highly susceptible to inflammatory signals. In addition to an increased frequency of GFP+ B cells in each subset, only the transitional B cell populations also displayed an increased amount of GFP expression per cell within the spleen (Figure 3B). We found that in addition to previously reported Bregs, several B cell subsets, including transitional, marginal zone, marginal zone progenitor and follicular B cells infiltrated pancreatic tumors (Figure 3B and C). P35 (GFP) signal was specifically induced in transitional, Breg and follicular type II cells within the tumor microenvironment, suggesting that localized cues might affect expression of p35 in restricted set of B cell populations (Figure 3B). Finally, we assessed the overall contribution of GFP+ B cell subsets to the make-up of the pancreatic TME. While transitional B cells displayed the highest frequency of GFP expression within the tumor, it is the follicular and Breg subsets that made up the majority of the GFP+ B cells within the tumor based on analysis of relative numbers (Figure 3C and D). Overall, our analysis suggests that pancreatic cancer-associated inflammatory cues contribute to increases in broad set of B cell populations that express p35, but that only select B cell subsets are able to upregulate the amount of p35 per cell specifically in response to cues at the tumor site.
Figure 3. Differential effect of inflammatory signals from pancreatic tumors on IL12aGFP expression in B cell subsets.
(A) Frequency of GFP+ B cell subsets isolated from spleens of KPC orthotopic tumor-bearing and non-tumor bearing (naïve) Il12aGFP mice measured by flow cytometry; n=6, *=p<0.05, ns=p>0.05.
(B) Mean fluorescent intensity (MFI) of GFP in GFP+ B cell subsets isolated from spleens and tumors of KPC orthotopic tumor-bearing and non-tumor bearing (naïve) Il12aGFP measured by flow cytometry mice; n=6, *=p<0.05, ns=p>0.05.
(C) Frequency of GFP+ B cell subsets isolated from orthotopic KPC pancreatic tumors of Il12aGFP mice measured by flow cytometry; n=6.
(D) Proportions of CD19+GFP+ cell populations isolated from Il12aGFP orthotopic KPC pancreatic tumors; n=6.
3.4. Ebi3-Tomato exhibits a restricted pattern of expression.
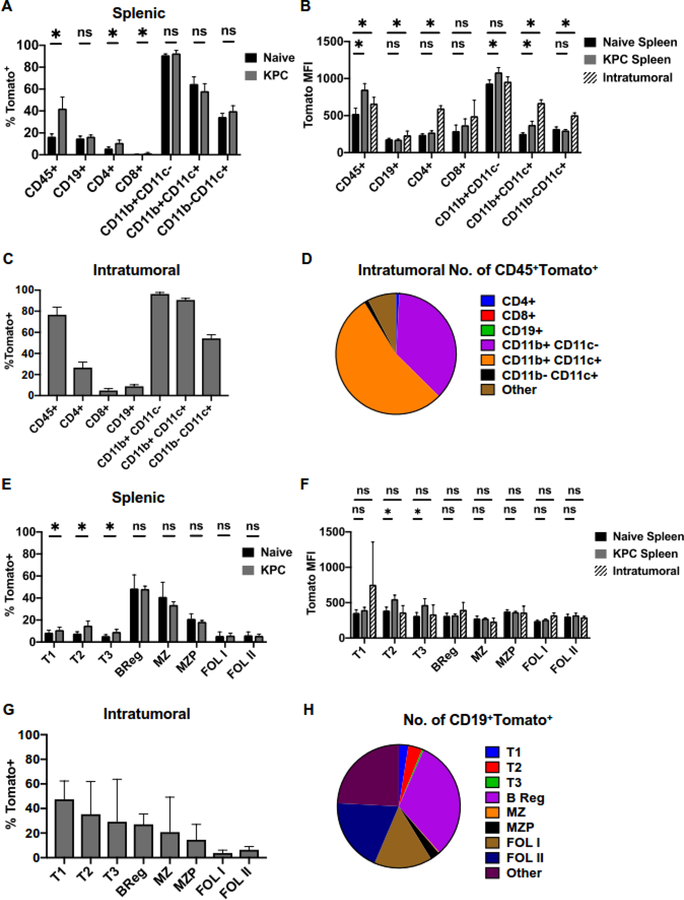
To understand how pancreatic cancer-associated inflammation regulates expression of Ebi3 subunit, we preformed orthotopic KPC injections into a previously published reporter strain Ebi3Tom (9). Presence of pancreatic tumors resulted in the overall increase in CD45+Tomato+ cells in the spleen (Figure 4A, Sup. Figure 5). We noted a specific increase in percent of CD4+ T cells that produces Ebi3, consistent with prior observations that CD4+ T cells upregulate IL35 in cancer (7, 9). However, this increase did not seem to account for almost doubling of frequency of CD45+Tomato+ cells in the spleen. To address this, we have analyzed the relative numbers of immune cell subsets in the spleens of tumor-naïve and tumor-bearing animals. We found that there was an increase in numbers of CD11b+CD11c-Tomato+ cells (2,719.8±1,119.7 in naïve spleens versus 11,111± 3,838.5 in spleens of tumor-bearing mice). Our prior work suggests that myeloid cells in pancreatic cancer can produce p28, a subunit, which can form cytokine IL27 upon binding to Ebi3, suggesting that myeloid cells may increase IL27 production in response to pancreatic inflammation (7).
Figure 4. Expression of EbI3Tom in immune subsets in response to pancreatic cancer.
(A) Frequency of Tomato+ immune cells isolated from spleens of KPC orthotopic tumor-bearing and non-tumor bearing (naïve) Ebi3Tom mice measured by flow cytometry; n=6, *=p<0.05, ns=p>0.05.
(B) Mean fluorescent intensity (MFI) of Tomato in Tomato+ immune populations isolated from spleens and tumors of KPC orthotopic tumor-bearing and non-tumor bearing (naïve) Ebi3Tom mice measured by flow cytometry mice; n=6, *=p<0.05, ns=p>0.05.
(C) Frequency of Tomato+ immune cells isolated from orthotopic KPC pancreatic tumors of Ebi3Tommice measured by flow cytometry; n=6.
(D) Proportions of CD45+Tomato+ cell populations isolated from Ebi3Tom orthotopic KPC pancreatic tumors; n=6.
(E) Frequency of Tomato+ B cell subsets isolated from spleens of KPC orthotopic tumor-bearing and non-tumor bearing (naïve) Ebi3Tom mice measured by flow cytometry; n=6, *=p<0.05, ns=p>0.05.
(F) Mean fluorescent intensity (MFI) of Tomato in Tomato+ B cell subsets isolated from spleens and tumors of KPC orthotopic tumor-bearing and non-tumor bearing (naïve) Ebi3Tom mice measured by flow cytometry mice; n=6, *=p<0.05, ns=p>0.05.
(G) Frequency of Tomato+ B cell subsets isolated from orthotopic KPC pancreatic tumors of Ebi3Tom mice measured by flow cytometry; n=6.
(H) Proportions of CD19+Tomato+ cell populations isolated from Ebi3Tom orthotopic KPC pancreatic tumors; n=6.
When we analyzed the amount of Tomato signal within immune cell subsets, we noted that distinct subsets responded to pancreatic cancer-associated inflammation in the manner that was location specific. Overall, Tomato MFI signal increased in CD45+ cells in both spleens and tumors (Figure 4B). Expression of Tomato signal was specifically enhanced in tumor-infiltrating B cells, CD4+ T cells and CD11b-CD11c+ dendritic cells, in intrasplenic CD11b+CD11c- myeloid cells and both intratumoral and intrasplenic CD11b+CD11c+ cells (Figure 4B). Within tumors, most CD45+ cells also expressed Tomato-reporter, and this was largely accounted for by Tomato+ myeloid cells, as judged both by frequency and relative numbers of cells (Figure 4C and D). Approximately 26% of CD4+ T cells and 8% of B cells produced Ebi3 as based on Tomato protein expression (Figure 4C). Overall, our analysis indicated that modulation of p35 largely accounted for changes in IL35 expression by B cells, and Bregs in particular, whereas modulation of Ebi3 may account for pancreatic cancer-associated changes in IL35 expression by CD4+ T cells.
Similar to modulation of Il12aGFP signal, we observed that splenic transitional B cell subsets significantly increased Tomato expression in response to pancreatic cancer, although the frequency of Tomato+ transitional subsets was overall quite low (Figure 4E, Sup. Figure 6). While intrasplenic Breg, MZ and MZP subsets expressed Tomato at frequencies of 20–48%, we did not observe any changes in the proportion of Tomato-expressing cells within these subsets in the presence of tumors (Figure 4E). Follicular B cell subsets in the spleens had overall low expression of Tomato protein, and this did not change in mice with pancreatic cancer (Figure 4E). We noted relatively few changes in the amount of Tomato signal per cell, as judged by Tomato MFI, only intrasplenic transitional B cell subsets upregulated Tomato MFI signal in response to cancer (Figure 4F). Finally, we assessed the overall contribution of Tomato+ B cell subsets to the pancreatic TME. In contrast with spleen, intratumoral transitional B cells had the highest frequency of Tomato expression, closely followed by Bregs and marginal zone B cells (Figure 4G). When we analyzed the relative numbers of cells comprising intratumoral B cell subsets, we observed that the follicular and Breg subsets made up the majority of the Tomato+ B cells (Figure 4G and H). Overall, our analysis suggests that B cell-specific expression of Ebi3 is more tightly regulated in vivo in response to pancreatic cancer-derived cues, as compared to expression of p35.
3.5. Stimulation of B cells reveals differential sensitivity of IL35 subunits to inflammatory signals.
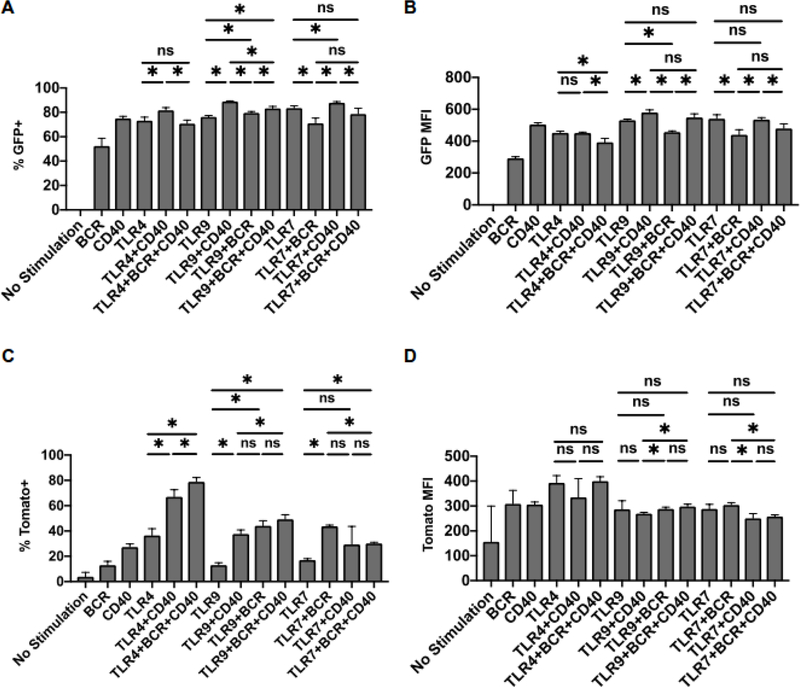
Multiple studies suggest that optimal activation and function of B cells relies on integration of various stimuli, such as BCR stimulation, CD40 activation and engagement of Toll like receptors (2, 15–22). To begin addressing the role of external signals in regulation of IL35 expression in B cells, we have cultured primary total splenic B cells from Il12aGFP and Ebi3Tom mice and stimulated B cells with agonists of BCR, CD40 and/or TLR pathways (Figure 5A–D). We found that frequency of GFP was robustly induced by most stimuli, closely followed by changes in GFP MFI (Figures 5A and B). We observed small, but statistically significant changes in proportion of GFP+ B cells when multiple stimuli were combined, although stimulation by CD40 agonist seemed to dampen this effect at least when used in combination with TLR4 and TLR7 agonists (Figure 5A). In contrast, robust changes in frequencies of Tomato+ B cells required coordinated activation of multiple stimuli (Figure 5C and D). These observations mirrored our analysis in vivo, and suggest that while expression of both p35 and Ebi3 subunits in B cells is directed by pancreatic cancer associated inflammation, regulation of p35 locus is more labile and can be triggered by a wide variety of inflammatory stimuli; whereas regulation of Ebi3 is significantly more constrained by the nature of stimulation.
Figure 5. In vitro stimulation of reporter B cells reveals differential regulation of Il12a and Ebi3 expression.
(A) Frequency of GFP+ B cells isolated from Il12aGFP spleens after stimulation of indicated receptors for 48 hours; n=3.
(B) Mean fluorescent intensity (MFI) of GFP in splenic Il12aGFP B cells after stimulation of indicated receptors for 48 hours in vitro; n=3.
(C) Frequency of Tomato+ B cells isolated from Ebi3Tom spleens after stimulation of indicated receptors for 48 hours; n=3.
(D) Mean fluorescent intensity (MFI) of Tomato in splenic Ebi3Tom B cells after stimulation of indicated receptors for 48 hours in vitro; n=3.
4. DISCUSSION
Cytokine IL35 is emerging as an important player in autoimmunity and cancer (2–5, 7, 8). In cancer, expression of IL35 has been postulated to come from both immune and epithelial compartments, although the precise cell type-specific and cancer type-specific pattern of expression remains to be elucidated (1, 7, 11, 23, 24). While increased IL35 levels in Tregs and B cells have so far been shown to contribute to suppression of productive T cell responses, IL35 in macrophages and cancer cells is an emerging regulator of angiogenesis and metastasis (1, 7, 24, 25). Furthermore, there is indication that IL35 may also be able to promote cancer cell growth by regulating proliferative capacity of cancer cells themselves (6, 23). It is not clear yet how cancer cell expression of IL35 affects immune responses and vice versa, whether IL35 produced by immune cells can directly regulate cancer cell properties.
One of the major difficulties in identifying IL35 producing cells has been lack of reagents and tracers that would allow to study modulation of this cytokine in a given disease setting. The Il12aGFP model allows for the elucidation of IL35 and/or IL12 producing cells, especially in vivo. In our study, we used mouse pancreatic cells that do not express IL35 (data not shown), thus elaborating on stromally sourced IL35. In the current work, we were able to utilize GFP analysis in combination with cell lineage markers in order to identify cell populations that make IL12a in vivo in the context of healthy and pancreatic tumor-bearing animals. We were also able to use an Ebi3Tom reporter mouse, to supplement our analysis of Ebi3-producing cells in vivo, in order to delineate IL35 producing immune subsets.
Since the GFP construct is located downstream of the stop codon of the Il12a gene, its expression should mirror activation of endogenous Il12a promoter. We found that expression of GFP mostly correlated with detection of p35 protein, but that not all GFP+ cells were positive for p35 staining, suggesting that the GFP signal is more sensitive than intracellular staining. This is possibly due to the longer half-life of GFP protein, or differential translational regulation of p35 versus GFP transcripts. Overall, our results suggest that Il12aGFP reporter is useful for tracking cell lineages committed to p35 production.
Using Il12aGFP and Ebi3Tom models, we observed that multiple immune cell lineages were committed to p35 and/or Ebi3 expression in vivo, as reflected by basal levels of GFP or Tomato signal. The greatest contributors to reporter signal were B cells and myeloid cells. In case of myeloid cells, our prior study utilizing intracellular cytokine staining for p40 and p28 proteins suggests that myeloid cells can express IL12 and/or IL27 (7). Injection of cancer cells into the pancreata of reporter mice has revealed overall increase in GFP and Tomato signal in immune cells, suggesting the presence of inflammatory pathways that may regulate cytokine production, although the extent to which immune subsets responded to pancreatic cancer was not equivalent. First, the splenic expression of Il12aGFP reporter was differentially regulated in B cell and myeloid cells, while expression of Ebi3Tom was altered in CD4+ T cells and myeloid cells. Second, additional and/or de novo changes in reporter expression by the immune cells isolated directly from tumors, suggested that localized signals may alter cytokine milieu. Finally, magnitude of reporter expression in B cell subsets in pancreatic cancer-bearing mice and in vitro shows that expression of p35 is more labile, while expression of the second IL35 subunit, Ebi3, is more tightly regulated in vivo and in vitro, suggesting that stimuli affecting Ebi3 upregulation are more likely to result in production of full heterodimer.
Specific triggers and determinants of IL35 upregulation in pancreatic and other cancer types remain unknown, and elucidating the cell type and cancer type specific expression pattern and function of IL35 are some of the vital next steps towards translating observations in mouse models to those most relevant for human cancer biology. Crossing reporter mouse models with spontaneous cancer models, such as KPC will allow for in vivo validation of cell-type specific expression pattern regulation. Increased expression of IL35 in pancreatic cancer has been correlated with worse survival in clinical patient cohorts (24), however, the source(s) of IL35, whether epithelial or stromal, that contributes to this correlation is not clear, since the tissue is analyzed by bulk RNA sequencing. Elucidating relevant IL35-producing cell populations in mouse and their counterparts in human cancers, along with generation of cell type specific signatures will allow us to deconvolute relative contribution of IL35 expression in cell type dependent manner and to further extend targeted therapeutic efforts.
To summarize, we have created a model to follow cells that produce p35 protein. Expression of the reporter enables tracing of both IL35 and IL12 producing cells, and its dynamic regulation by pancreatic cancer-associated inflammation provides rationale for future studies of p35 gene regulation in disease settings. While the baseline expression of either Il12aGFP or Ebi3Tom reporters were higher in vivo than in vitro, the reporter alleles responded to candidate stimuli in vitro, suggesting that the use of reporter models could be extended as a potential screening tool in identifying mechanisms of cytokine regulation. Finally, future single cell level analysis of mice that are double-positive for both Il12aGFP and Ebi3Tom will reveal which cells can co-express both proteins at the same time and allow for in depth studies on regulation of heterodimeric cytokine IL35.
Supplementary Material
Highlights.
A novel IL12a-GFP reporter mouse allows for detection of p35-producing cells in vitro and in vivo
Expression of p35 in immune cells is dynamically regulated by pancreatic cancer-associated inflammation
p35 expression marks multiple B cell subsets in pancreatic cancer
Model stimuli differentially alter p35 and Ebi3 expression profiles
ACKNOWLEDGMENTS
We thank N. Kren for discussions and Animal Studies Core for help with mouse colony maintenance. This work was supported in part by NCI R37 CA230786 (Y. P.-G.), University Cancer Research Fund at the University of North Carolina at Chapel Hill, United States (Y.P.-G.); AACR-PanCAN Pathway to Leadership Grant 13-70-25-PYLA (Y.P.-G.), V Foundation for Cancer Research grant DVP2019-016 (Y. P.-G.), Concern Foundation Conquer Cancer Now Award (Y.P.-G), the WUSTL SPORE Career Enhancement Award grant 1P50CA196510-01A1 from the NCI, NCI R01 CA203689 (D.A.A.V.), Cancer Cell Biology Training Program (CCBTP) grant 2T32CA071341-21 (D. M.) and NCI Comprehensive Cancer Center Support CORE grant (CA047904; D.A.A.V.). The UNC Flow Cytometry Core Facility, the UNC Animal Models Core and the UNC Lineberger Animal Studies Core are supported in part by P30 CA016086 Cancer Center Core Support Grant to the UNC Lineberger Comprehensive Cancer Center.
Footnotes
COMPETING FINANCIAL INTERESTS
D.A.A.V. has submitted patents covering IL-35 that are pending and are entitled to a share in net income generated from licensing of these patent rights for commercial development. D.O.C. is employed by, has equity ownership in and serves on the board of directors of TransViragen, the company which has been contracted by UNC-Chapel Hill to manage its Animal Models Core Facility.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
REFERENCES
- 1.Vignali DA, Kuchroo VK. IL-12 family cytokines: immunological playmakers. Nat Immunol 2012;13(8):722–8. Epub 2012/07/21. doi: 10.1038/ni.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen P, Roch T, Lampropoulou V, O’Connor RA, Stervbo U, Hilgenberg E, Ries S, Dang VD, Jaimes Y, Daridon C, Li R, Jouneau L, Boudinot P, Wilantri S, Sakwa I, Miyazaki Y, Leech MD, McPherson RC, Wirtz S, Neurath M, Hoehlig K, Meinl E, Grutzkau A, Grun JR, Horn K, Kuhl AA, Dorner T, Bar-Or A, Kaufmann SHE, Anderton SM, Fillatreau S. IL-35-producing B cells are critical regulators of immunity during autoimmune and infectious diseases. Nature 2014;507(7492):366–70. Epub 2014/02/28. doi: 10.1038/nature12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dambuza IM, He C, Choi JK, Yu CR, Wang R, Mattapallil MJ, Wingfield PT, Caspi RR, Egwuagu CE. IL-12p35 induces expansion of IL-10 and IL-35-expressing regulatory B cells and ameliorates autoimmune disease. Nat Commun 2017;8(1):719 Epub 2017/09/30. doi: 10.1038/s41467-017-00838-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang RX, Yu CR, Dambuza IM, Mahdi RM, Dolinska MB, Sergeev YV, Wingfield PT, Kim SH, Egwuagu CE. Interleukin-35 induces regulatory B cells that suppress autoimmune disease. Nat Med 2014;20(6):633–41. Epub 2014/04/20. doi: 10.1038/nm.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun T, Zhang D, Yang Y, Zhang X, Lv C, Fu R, Lv M, Liu W, Chen Y, Liu W, Huang Y, Xue F, Liu X, Zhang L, Li H, Yang R. Interleukin 35 may contribute to the loss of immunological self-tolerance in patients with primary immune thrombocytopenia. Br J Haematol 2015;169(2):278–85. Epub 2015/02/03. doi: 10.1111/bjh.13292. [DOI] [PubMed] [Google Scholar]
- 6.Pylayeva-Gupta Y, Das S, Handler JS, Hajdu CH, Coffre M, Koralov SB, Bar-Sagi D. IL35-Producing B Cells Promote the Development of Pancreatic Neoplasia. Cancer Discov 2016;6(3):247–55. Epub 2015/12/31. doi: 10.1158/2159-8290.CD-15-0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mirlekar B, Michaud D, Searcy R, Greene K, Pylayeva-Gupta Y. IL35 Hinders Endogenous Antitumor T-cell Immunity and Responsiveness to Immunotherapy in Pancreatic Cancer. Cancer Immunol Res 2018;6(9):1014–24. Epub 2018/07/08. doi: 10.1158/2326-6066.CIR-17-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pylayeva-Gupta Y Molecular Pathways: Interleukin-35 in Autoimmunity and Cancer. Clin Cancer Res 2016;22(20):4973–8. Epub 2016/09/02. doi: 10.1158/1078-0432.CCR-16-0743. [DOI] [PubMed] [Google Scholar]
- 9.Turnis ME, Sawant DV, Szymczak-Workman AL, Andrews LP, Delgoffe GM, Yano H, Beres AJ, Vogel P, Workman CJ, Vignali DA. Interleukin-35 Limits Anti-Tumor Immunity. Immunity 2016;44(2):316–29. Epub 2016/02/14. doi: 10.1016/j.immuni.2016.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee CC, Lin JC, Hwang WL, Kuo YJ, Chen HK, Tai SK, Lin CC, Yang MH. Macrophage-secreted interleukin-35 regulates cancer cell plasticity to facilitate metastatic colonization. Nat Commun 2018;9(1):3763 Epub 2018/09/16. doi: 10.1038/s41467-018-06268-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collison LW, Workman CJ, Kuo TT, Boyd K, Wang Y, Vignali KM, Cross R, Sehy D, Blumberg RS, Vignali DA. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature 2007;450(7169):566–9. Epub 2007/1½3. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- 12.Collison LW, Chaturvedi V, Henderson AL, Giacomin PR, Guy C, Bankoti J, Finkelstein D, Forbes K, Workman CJ, Brown SA, Rehg JE, Jones ML, Ni HT, Artis D, Turk MJ, Vignali DA. IL-35-mediated induction of a potent regulatory T cell population. Nat Immunol 2010;11(12):1093–101. Epub 2010/10/19. doi: 10.1038/ni.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chehboun S, Labrecque-Carbonneau J, Pasquin S, Meliani Y, Meddah B, Ferlin W, Sharma M, Tormo A, Masson JF, Gauchat JF. Epstein-Barr virus-induced gene 3 (EBI3) can mediate IL-6 trans-signaling. J Biol Chem 2017;292(16):6644–56. Epub 2017/03/11. doi: 10.1074/jbc.M116.762021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bayne LJ, Beatty GL, Jhala N, Clark CE, Rhim AD, Stanger BZ, Vonderheide RH. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell 2012;21(6):822–35. Epub 2012/06/16. doi: 10.1016/j.ccr.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schweighoffer E, Nys J, Vanes L, Smithers N, Tybulewicz VLJ. TLR4 signals in B lymphocytes are transduced via the B cell antigen receptor and SYK. J Exp Med 2017;214(5):1269–80. Epub 2017/03/31. doi: 10.1084/jem.20161117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haxhinasto SA, Bishop GA. Synergistic B cell activation by CD40 and the B cell antigen receptor: role of B lymphocyte antigen receptor-mediated kinase activation and tumor necrosis factor receptor-associated factor regulation. J Biol Chem 2004;279(4):2575–82. Epub 2003/11/08. doi: 10.1074/jbc.M310628200. [DOI] [PubMed] [Google Scholar]
- 17.Mizuno T, Rothstein TL. B cell receptor (BCR) cross-talk: CD40 engagement enhances BCR-induced ERK activation. J Immunol 2005;174(6):3369–76. [DOI] [PubMed] [Google Scholar]
- 18.Elgueta R, Benson MJ, de Vries VC, Wasiuk A, Guo Y, Noelle RJ. Molecular mechanism and function of CD40/CD40L engagement in the immune system. Immunol Rev 2009;229(1):152–72. Epub 2009/05/12. doi: 10.1111/j.1600-065X.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yanaba K, Bouaziz JD, Matsushita T, Tsubata T, Tedder TF. The development and function of regulatory B cells expressing IL-10 (B10 cells) requires antigen receptor diversity and TLR signals. J Immunol 2009;182(12):7459–72. Epub 2009/06/06. doi: 10.4049/jimmunol.0900270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pone EJ, Zhang J, Mai T, White CA, Li G, Sakakura JK, Patel PJ, Al-Qahtani A, Zan H, Xu Z, Casali P. BCR-signalling synergizes with TLR-signalling for induction of AID and immunoglobulin class-switching through the non-canonical NF-kappaB pathway. Nat Commun 2012;3:767 Epub 2012/04/05. doi: 10.1038/ncomms1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suthers AN, Sarantopoulos S. TLR7/TLR9- and B Cell Receptor-Signaling Crosstalk: Promotion of Potentially Dangerous B Cells. Front Immunol 2017;8:775 Epub 2017/07/29. doi: 10.3389/fimmu.2017.00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuraoka M, Snowden PB, Nojima T, Verkoczy L, Haynes BF, Kitamura D, Kelsoe G. BCR and Endosomal TLR Signals Synergize to Increase AID Expression and Establish Central B Cell Tolerance. Cell Rep 2017;18(7):1627–35. Epub 2017/02/16. doi: 10.1016/j.celrep.2017.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicholl MB, Ledgewood CL, Chen X, Bai Q, Qin C, Cook KM, Herrick EJ, Diaz-Arias A, Moore BJ, Fang Y. IL-35 promotes pancreas cancer growth through enhancement of proliferation and inhibition of apoptosis: evidence for a role as an autocrine growth factor. Cytokine 2014;70(2):126–33. Epub 2014/07/31. doi: 10.1016/j.cyto.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 24.Huang C, Li N, Li Z, Chang A, Chen Y, Zhao T, Li Y, Wang X, Zhang W, Wang Z, Luo L, Shi J, Yang S, Ren H, Hao J. Tumour-derived Interleukin 35 promotes pancreatic ductal adenocarcinoma cell extravasation and metastasis by inducing ICAM1 expression. Nat Commun 2017;8:14035 Epub 2017/0½0. doi: 10.1038/ncomms14035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang C, Li Z, Li N, Li Y, Chang A, Zhao T, Wang X, Wang H, Gao S, Yang S, Hao J, Ren H. Interleukin 35 Expression Correlates With Microvessel Density in Pancreatic Ductal Adenocarcinoma, Recruits Monocytes, and Promotes Growth and Angiogenesis of Xenograft Tumors in Mice. Gastroenterology 2018;154(3):675–88. Epub 2017/10/11. doi: 10.1053/j.gastro.2017.09.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.