Abstract
Phosphate starvation response (PSR) in nonmycorrhizal plants comprises transcriptional reprogramming resulting in severe physiological changes to the roots and shoots and repression of plant immunity. Thus, plant-colonizing microorganisms—the plant microbiota—are exposed to direct influence by the soil’s phosphorus (P) content itself as well as to the indirect effects of soil P on the microbial niches shaped by the plant. The individual contribution of these factors to plant microbiota assembly remains unknown. To disentangle these direct and indirect effects, we planted PSR-deficient Arabidopsis mutants in a long-term managed soil P gradient and compared the composition of their shoot and root microbiota to wild-type plants across different P concentrations. PSR-deficiency had a larger effect on the composition of both bacterial and fungal plant-associated microbiota than soil P concentrations in both roots and shoots.
To dissect plant–microbe interactions under variable P conditions, we conducted a microbiota reconstitution experiment. Using a 185-member bacterial synthetic community (SynCom) across a wide P concentration gradient in an agar matrix, we demonstrated a shift in the effect of bacteria on the plant from a neutral or positive interaction to a negative one, as measured by rosette size. This phenotypic shift was accompanied by changes in microbiota composition: the genus Burkholderia was specifically enriched in plant tissue under P starvation. Through a community drop-out experiment, we demonstrated that in the absence of Burkholderia from the SynCom, plant shoots accumulated higher ortophosphate (Pi) levels than shoots colonized with the full SynCom but only under Pi starvation conditions. Therefore, Pi-stressed plants are susceptible to colonization by latent opportunistic competitors found within their microbiome, thus exacerbating the plant’s Pi starvation.
This study shows that plants' response to phosphate starvation has more influence on plant microbiota than does the phosphate concentration itself; these changes are not necessarily beneficial for the plant, and can even exacerbate the phosphate starvation.
Introduction
Plants provide the primary energy source for terrestrial heterotrophs, most of which are microbial. The interaction of these microbial heterotrophs with plants ranges between the extremes of mutualistic symbiosis [1] and pathogenesis [2,3]. However, the vast majority of plant-associated microbial diversity, the plant microbiota, lies between these 2 extremes, inducing more subtle, context-dependent effects on plant health [4–6]. The microbiota consumes plant photosynthate [7–9], and it provides benefits via protection from pathogens [10–14] or abiotic stress [15,16] or by increasing nutrient bioavailability [4,17,18].
The plant microbiota is derived from the microbial community composition in soil [19–21], which is governed by its own set of ecological processes [22]. Correlations with soil microbial diversity, and by derivation, with plant microbiota composition and diversity, were observed for soil abiotic factors, such as pH [22–25], drought [25–30], and nutrient concentrations [22,25,31–35]. Soil nutrient concentrations, in particular orthophosphate (Pi)—the only form of phosphorus (P) that can be taken up by plants—produce comparatively modest changes in microbial community composition [35,36]. Nevertheless, available soil Pi concentrations influences where a plant–microbe interaction lies along the mutualism–pathogenicity continuum [17].
Nonmycorrhizal plants respond to phosphate limitation by employing a range of PSR mechanisms. These manifest as severe physiological and morphological changes to the root and shoot, such as lateral root growth prioritization, depletion of shoot Pi stores [37], and changes to root exudate profiles [38,39]; these changes can potentially affect both root and shoot microbiota. In Arabidopsis, most of the transcriptional PSR driving these physiological responses is controlled by the 2 partially redundant transcription factors PHOSPHATE STARVATION RESPONSE 1 (PHR1) and PHR1-LIKE (PHL1) [40]. As a result, the double mutant phr1 phl1 has an impaired PSR and accumulates a low level of Pi. Pi transport into roots relies on the PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1) gene, which is required for membrane localization of high-affinity phosphate transporters [41]. In axenic conditions, phf1 mutants constitutively express PSR and accumulate low levels of Pi [41]. In addition to inducing physiological changes, the plant’s response to its nutrient status is also linked to its immune system. PHR1 negatively regulates components of the plant immune system, which can lead to enhanced pathogen susceptibility but also to the alteration of the plant’s microbiota under phosphate starvation [4]. Arabidopsis microbiota are altered in phr1 phl1 and phf1 mutants [4,36] in experiments using both natural and synthetic microbial communities [4].
Here, we examined (i) the effect of soil P content on plant microbiota composition, (ii) how PSR modulates the plant microbiota, and (iii) the interplay between PSR and soil P content in shaping the plant microbiota composition. We used a combination of greenhouse experiments with differentially P-fertilized soils, Arabidopsis PSR mutants and laboratory microcosms utilizing tractable synthetic bacterial communities. Using PSR mutants planted in P-amended soil, we demonstrate that the plant PSR regulators have a profound effect on the composition of root and shoot microbiota, overshadowing the effect of the soil P content. We constructed an ecologically tractable system utilizing a complex bacterial synthetic community (SynCom) as a model of the plant root microbiome and used this system to study the interactions between microbiota assembly and abiotic stress. We demonstrate deterministic responses of the SynCom members to changes in Pi concentrations, and we identify Pi-dependent shifts in community composition along the mutualist–pathogen continuum.
Results
PSR is activated in soil
To better understand the effect of PSR genes on the plant microbiome under both Pi-limiting and Pi-replete conditions, we investigated how microbiota adapted to varying soil P levels interact with the plant’s PSR. We grew wild-type (wt) Arabidopsis and the PSR mutants phf1 and phr1 phl1 in soils collected from the “Halle long-term soil fertilization experiment,” ongoing since 1949 [42]. Throughout this long-term experiment, each transect of soil has received 1 of 3 P fertilization treatments: 0 (low), 15 (medium) and 45 (high) Kg[P].Ha−1.Year−1, resulting in a 3- to 5-fold difference in bioavailable P between the low and high treatments [43]. To differentiate the long-term adaptive effect of P limitation on the microbial community from the effect of short-term changes in P availability, we also fertilized a subset of the low P soil at the time of planting and designated this condition low+P.
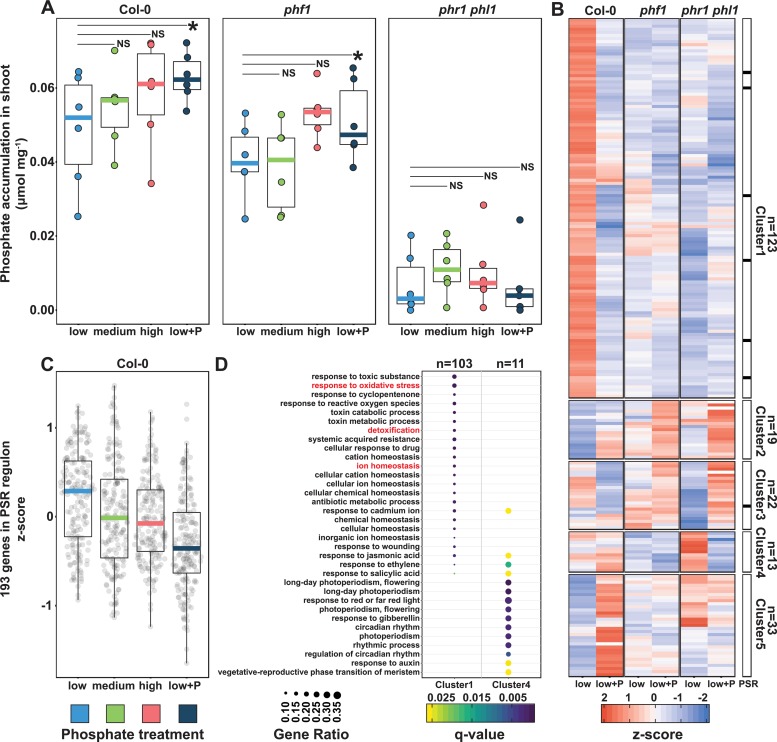
We examined whether PSR, defined and typically studied in axenic conditions, is active in our soil-based experimental system. We harvested 8-week-old plants grown in the different soils and quantified developmental and molecular phenotypes typically associated with PSR in both wt plants and mutants. We found a strong positive correlation among all developmental features analyzed: shoot area, shoot fresh weight, and shoot Pi accumulation across all soil conditions (Figs 1A and S1A–S1C and S1 Table and S1 Data). In wt plants, shoot Pi accumulation reflected soil P conditions (Fig 1A and S1 Data). As expected [44], phr1 phl1 showed a dramatic reduction in all phenotypic parameters (Figs 1A and S1B and S1C and S1 Data) and phf1 accumulated less shoot Pi than wt but did not display any obvious morphological effect (Figs 1A and S1B–S1D and S1 Data).
Fig 1. Plants respond to differential P conditions in soil.
(A) Free phosphate content normalized by shoot fresh weight (mmol·mg−1) across wt Col-0 plants and 2 PSR mutants, phf1 and phr1 phl1. Statistical significance between low P and low+P treatments was determined across each genotype independently by a paired t test (p < 0.05). (B) Heat map showing the average standardized expression of 210 DEGs across the low P and low+P samples in the Col-0, phf1 and phr1 phl1 genotypes. The black bar to the right highlights the distribution of 7 genes belonging to the in vitro defined PSR marker genes [4] across the 5 clusters in the heat map. (C) Average expression of 193 PSR marker genes [4] across the 4 phosphorus regimes in the Col-0 genotype. (D) GO enrichment for Clusters 1 and 4. Clusters 2, 3, and 6 did not show any statistically significant GO enrichment. The gene ratio is the proportion of genes per cluster that belong to a GO category. DEG, differentially expressed gene; GO, gene ontology; P, phosphorus; PSR, phosphate starvation response; wt, wild type.
To identify the transcriptomic signature of PSR in a low P soil, we compared the root transcriptomes of the 3 genotypes from the low P samples with those of the low+P samples (S2 Table). Using a likelihood ratio test, we identified 210 genes that were differentially expressed across genotypes and P conditions (q < 0.1). After hierarchical clustering, 123 (59%) of these genes fall into a single cluster (Cluster 1) of co-expressed genes that are exclusively highly expressed in wt under low P and not in either of the PSR mutants (Fig 1B and S1 Data). Thus, these genes represent a PSR under our experimental conditions. A gene ontology (GO) enrichment analysis (Fig 1D and S1 Data) illustrates that these genes are involved in processes such as ion homeostasis, detoxification, and response to oxidative stress. Interestingly, few PSR genes defined from in vitro experiments were significantly differentially expressed in our soil experiment. From a previously defined set of 193 PSR marker genes defined using 7-day-old seedlings exposed in vitro to P limitation for up to 2 days [4], only 7 were called as significant in our experiment using 8-week-old plants. Nevertheless, all 7 of these genes were enriched in wt in low P soil (Fig 1B and S2 Table). Surprisingly, despite the fact that phf1 and phr1 phl1 have contrasting transcriptional responses to Pi limitation in vitro [41], the 123 genes in Cluster 1 were not up-regulated in both mutants. To corroborate that the canonical in vitro defined PSR is also induced in wt plants, we compared the median expression of the set of 193 PSR marker genes [4] across the different soils (Figs 1C and S1E and S1 Data). As expected, shoot Pi content was significantly correlated with the induction of PSR marker genes (Figs 1C and S1F and S1 Data), with the highest median expression level in the low P conditions. We conclude that although the response of 8-week-old plants to low P conditions in natural soil is markedly different from in vitro defined PSR, wt plants indeed respond to low P conditions in the soils tested in a Pi concentration- and PHR1/PHL1-dependent manner.
Bacterial and fungal plant microbiota differ in plant recruitment patterns
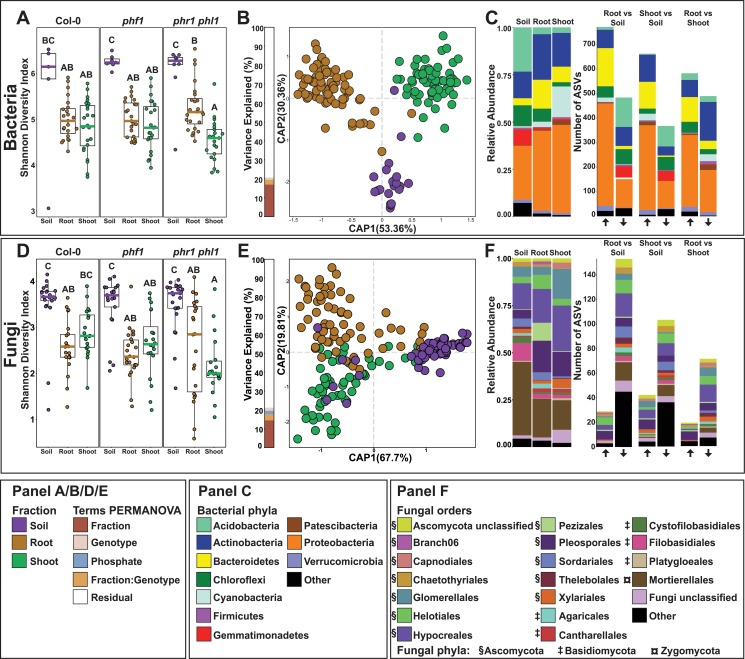
We studied the relationship between PSR and the plant microbiome in wt plants and the 2 PSR mutants grown in all 4 soils. Total DNA was extracted from shoots, roots, and soil, and the 16S rRNA (V3-V4) and ITS1 regions were amplified and sequenced to obtain bacterial and fungal community profiles, respectively. Sequences were collapsed into amplicon sequence variants (ASVs. Bacterial and fungal alpha- and beta-diversity measures conform to previously published data [19,36]: Microbial diversity decreased from the soil to the root and shoot compartments (Figs 2A and 2D and S2A and S2B and S3 Table and S1 Data), and roots and shoots harbor bacterial and fungal communities distinct from the surrounding soil community and from each other (Figs 2B and 2E and S2C–S2F and S4 Table and S1 Data). Plant-derived samples were primarily enriched in comparison to soil with members of the phyla Proteobacteria, Bacteroidetes, and Actinobacteria and depleted in members of Acidobacteria and Gemmatimonadetes (Figs 2C and S2E and S4 Table and S1 Data). Plant-enriched fungal ASVs belonged mainly to the phyla Ascomycota (orders Hypocreales and Pleosporales) and Basidiomycota (order Agaricales). Plant-depleted fungal ASVs belonged mainly to Filobasidiales (Basidiomycota) and Mortierellales (Zygomycota; Figs 2F and S2F and S5 Table and S1 Data).
Fig 2. Plant recruitment patterns of bacteria and fungi.
(A, D) Bacterial (A) and fungal (D) alpha diversity estimated using the Shannon Diversity Index. Letters represent post hoc test results, based on a full factorial ANOVA model. (B, E) CAP based on Bray-Curtis dissimilarities between bacterial (B) and fungal (E) communities across the soil, root, and shoot. The bar graph to the left of the CAP depicts the percentage of variance explained by statistically significant (p < 0.05) terms in a PERMANOVA model. (C) Left panel: Relative abundance profiles of the main bacterial phyla across the soil, root, and shoot fractions. Right panel: Number of statistically significant ASVs enriched in specific fractions. The arrows on the bottom of the panel denote the direction of the enrichment relative to the name of the contrast tested; the up arrow means enrichment in the left fraction of the contrast, whereas the down arrow means enrichment in the right fraction of the contrast (e.g., RootvsSoil, up arrow enriched in root relative to soil, bottom arrow enriched in soil relative to root). A detailed interactive visualization of the bacterial enrichment patterns across the multiple taxonomic levels can be found at https://itol.embl.de/tree/1522316254174701551987253. (F) Left panel: Relative abundance profiles of the main fungal orders across soil, root, and shoot fractions. Right Panel: Number of statistically significant ASVs enriched in specific fractions. The arrows on the bottom of the panel denote the direction of the enrichment relative to the name of the contrast tested; the up arrow signifies enrichment in the left fraction of the contrast, whereas the down arrow signifies enrichment in the right fraction of the contrast (e.g., RootvsSoil, up arrow enriched in root relative to soil, bottom arrow enriched in soil relative to root). Plot is colored by order. The symbols besides the colors in the legend denote phylum. A detailed interactive visualization of the fungal enrichment patterns across the multiple taxonomic levels can be found at https://itol.embl.de/tree/13656172137464831571097084. ASV, amplicon sequence variant; CAP, canonical analysis of principal coordinates; PERMANOVA, Permutational Multivariate Analysis of Variance.
To quantify the effect of soil community composition on the composition of root and shoot microbiota, we used Mantel tests to detect correlation between dissimilarity matrices of the 3 fractions (root, shoot, and soil). For bacteria, both root and shoot community dissimilarities were strongly correlated with soil community dissimilarity (S3A and S3B Fig and S1 Data), whereas for fungi, no correlation was measured between root and soil (S3D Fig and S1 Data), and only a weak correlation was measured between shoot and soil (S3E Fig and S1 Data). This observation indicates that both root and shoot bacterial communities are strongly dependent on soil community composition, despite the fact that bacterial microbiota are distinct from the soil community (Fig 2B and S1 Data). By contrast, the fungal microbiota composition both above and below ground is independent of relative abundances within the soil inoculum. This difference implies that the plant’s microbiota filtering mechanisms are fundamentally different for fungi and bacteria.
Shoot and root microbiota are both correlated and distinct
Shoot and root microbiomes are linked, and substantial crosstalk is expected to occur between these 2 niches [45,46]. We show here that roots and shoots harbor distinct communities from each other (Figs 2B and 2E and S2C–S2F and S4 Table and S1 Data). To further explore organ specificity in the plant microbiome composition, we compared root and shoot samples at the ASV level. Shoots were mainly enriched with the bacterial phyla Cyanobacteria and Patescibacteria compared to the root, whereas roots were enriched with Proteobacteria, Chloroflexi, and Bacteroidetes (Figs 2C and S2E and S4 Table and S1 Data). With regard to fungal orders, shoots were enriched with Capnodiales, Glomerellales, Pleosporales, and Hypocreales, whereas roots were enriched with Pezizales, Helotiales, and Mucorales (Figs 2F and S2F and S5 Table and S1 Data). The shoot enrichment of Cyanobacteria suggests that the availability of light is an important factor in niche differentiation within the plant [47–49]. We used Mantel tests to detect correlation between dissimilarity matrices of root and shoot samples. Despite the fact that they harbor distinct communities, roots and shoots were correlated with each other for both bacteria and fungi (S3C and S3F Fig and S1 Data). Thus, although roots and shoots form distinct bacterial and fungal niches, shifts in microbiota in both of these niches are correlated, suggesting that either one or both of these fractions serves as an inoculum source for the other.
The plant microbiome composition is driven by the plant PSR status
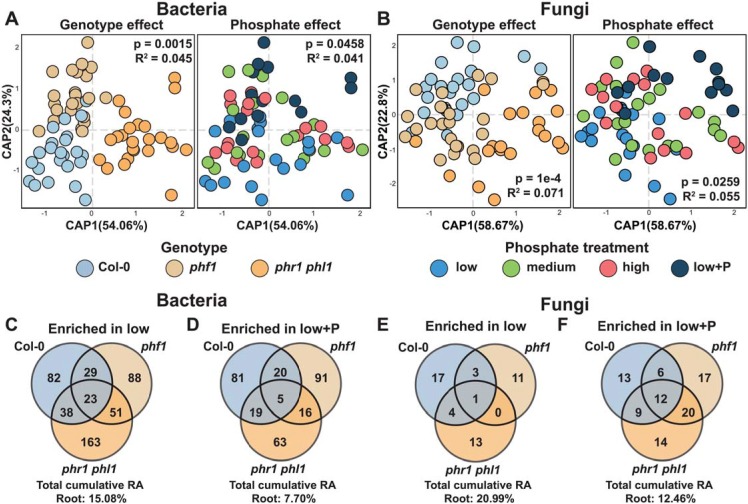
We investigated the influences of plant PSR signaling and the different soil P concentrations on microbial community composition. Constrained ordination showed significant differences, explaining a similar proportion of variance, in both bacterial and fungal community compositions. These differences persist across the P accumulation gradients represented by both the difference in soil P and PSR mutants (Figs 3A and 3B and S4A and S4B and S1 Data). This effect is maintained also when considering 13 soil edaphic factors that were measured in for the same soil samples [43] (S5A–S5D Fig and S1 Data). For both bacteria and fungi, the PSR genotype effect in roots was more consistent than the soil P effect (Fig 3A and 3B and S1 Data), which was mainly driven by the P-amended low+P samples. In shoots, both bacteria and fungi responded to PSR genotype but in this case did not respond to soil P (S4C and S4D Fig and S1 Data). We did not observe a significant soil P:genotype interaction effect for either bacteria or fungi (S4A and S4B Fig and S1 Data), confirming that phf1 and phr1 phl1 both had atypical bacterial microbiomes regardless of Pi status. As expected, we did not observe a PSR effect in the soil samples (S4E and S4F Fig and S1 Data). The notable genotype effect illustrates that the plant niche filtering (Fig 2B and 2E and S1 Data) is partly shaped by PSR.
Fig 3. Plant PSR controls the assembly of the plant microbiome.
(A, B) Canonical analysis of principal coordinates showing the influence of plant genotypes and soil P content over the (A) bacterial and (B) fungal communities in the root. The p-value and R2 values inside each plot are derived from a PERMANOVA model and correspond to the genotype and P term, respectively. (C, E) Venn diagrams showing the distribution of (C) bacterial and (E) fungal ASVs with statistically significant (q < 0.1) higher abundance in the low P treatment in comparison to the low+P treatment in the Col-0, phf1 and phr1 phl1 roots. (D, F) Venn diagrams showing the distribution of (D) bacterial and (F) fungal ASVs with statistically significant (q < 0.1) higher abundance in the low+P treatment in comparison to the low P treatment across the Col-0, phf1 and phr1 phl1 roots. ASV, amplicon sequence variant; P, phosphorus; PERMANOVA, Permutational Multivariate Analysis of Variance; PSR, phosphate starvation response; RA, relative abundance.
To define which taxa at the ASV-levels were influenced by soil P and/or plant PSR, we applied a generalized linear model (GLM) to the count datasets (S6 and S7 Tables). Contrasting the low P samples against the low+P samples, we detected 769 bacterial (S6 Table) and 140 fungal (S7 Table) ASVs, accounting for 23% and 33% of the bacterial and fungal abundance in the root, respectively, that were differentially abundant in at least 1 genotype (Fig 3C–3F and S1 Data). Of these, most (568 bacterial and 85 fungal ASVs) were genotype specific, suggesting that the Pi response of these taxa is not direct but is rather driven by Pi responses in the plant. Taken together, these results indicate that plant microbiota are relatively robust to differences in soil P content but are sensitive to the plant PSR status. Responses to soil P concentration are contingent on PSR regulatory elements under both low and high P conditions.
Bacterial synthetic communities modulate the plant PSR
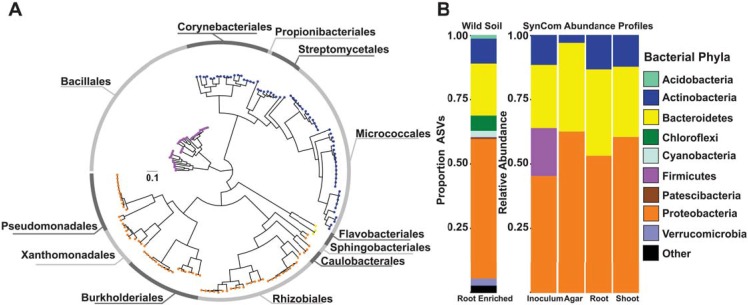
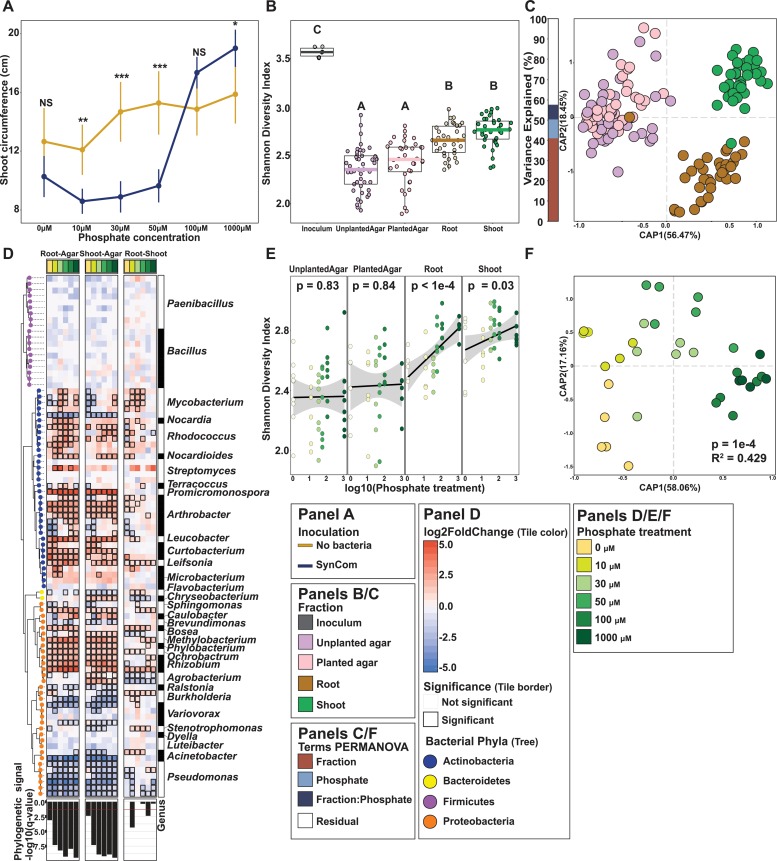
The results obtained from the soil experiment suggest that the community structure of the plant microbiome is not only determined by first-order interactions (plant–microbe, microbe–microbe, microbe–environment) but also by higher-order interactions, such as the effect of abiotic conditions on plant–microbe interactions. This is evident in the large proportion of ASVs that respond to soil P in a genotype-specific manner (Fig 3C–3F and S1 Data). To establish a system in which interactions of different orders of complexity can be studied reproducibly, we constructed a plant–microbe microcosm that can be deconstructed to its individual components while retaining a complexity that is comparable to natural ecological communities. We designed a representative bacterial SynCom from a culture collection composed of isolates derived from surface-sterilized Arabidopsis roots [50]. We selected 185 genome-sequenced isolates representing a typical plant-associated taxonomic distribution (Fig 4A and 4B and S1 Data). We grew each isolate separately and mixed the grown cultures to equal optical densities. We grew 7-day-old Arabidopsis seedlings in a Pi concentration gradient (0, 10, 30, 50, 100, 1000 μM KH2PO4) and concomitantly exposed them to the SynCom on vertical agar plates for 12 days.
Fig 4. Bacterial SynCom reproduces the typical plant-associated taxonomic distribution found in soil.
(A) Phylogenetic tree of 185 bacterial genomes included in the SynCom. The tree tips are colored according to the phylum classification of the genome in panel B; the outer ring shows the distribution of the 12 distinct bacterial orders present in the SynCom. (B) Left Panel: Proportion of ASVs enriched in the root in comparison to the natural soil across all treatments and genotypes based on a fitted GLM (q < 0.1). Each ASV is colored according to its phylum-level classification. Right Panel: Relative abundance profiles of bacterial isolates across the initial bacterial inoculum, planted agar, root, and shoot fractions. Each isolate is colored according to its phylum-level classification based on the genome-derived taxonomy. ASV, amplicon sequence variant; GLM, generalized linear model; SynCom, synthetic community.
First, we investigated whether PSR is induced in our experimental system. Similar to the natural soil-based experiment, we quantified developmental and transcriptional phenotypes associated with PSR in plants grown in different concentrations of Pi. In line with the work by Castrillo and colleagues [4], the presence of the SynCom consistently decreased primary root elongation across all Pi concentrations compared with the uninoculated control, but the Pi gradient did not affect this parameter (S6A Fig and S1 Data). Shoot size increased with Pi concentration, and the slope of this trend was affected by the presence of the SynCom: At high Pi, the SynCom tended to increase shoot size, whereas at low Pi the SynCom decreased it (Fig 5A and S1 Data), suggesting that the microbiome plays a role in shaping the plant’s response to different Pi concentrations.
Fig 5. Synthetic bacterial communities display deterministic community assembly in plants.
(A) Strip chart displaying the average shoot size of Col-0 Arabidopsis grown across a Pi gradient either in sterile conditions or with the SynCom. Each dot in the scatter plot represents the mean value for that particular treatment; the range crossing each dot represents the 95% confidence interval calculated. The lines are drawn to connect the means. (B) Alpha diversity across the fractions sampled was estimated using the Shannon Diversity index. An ANOVA model followed up by a Tukey HSD test were applied to estimate differences between inoculum, unplanted agar, planted agar, root, and shoot fractions. Letters represent the results of the post hoc test. (C) CAP based on Bray-Curtis dissimilarities between bacterial communities across the 4 fractions sampled. The bar graph to the left of the CAP depicts the percentage of variability explained by statistically significant (p < 0.05) terms in the PERMANOVA model. (D) Enrichment patterns of the SynCom. Each row along the different panels of the figure represents a USeq: a USeq encompasses a set of indistinguishable V3-V4 16S rRNA sequences present in the 185-member SynCom. Phylogenetic tree (on the left) is colored based on the phylum-level classification of the corresponding USeq. Each column in the heat maps represents a specific contrast in the enrichment model. We calculated root versus agar (left heat map), shoot versus agar (middle heat map), and root versus shoot (right heat map) enrichments within each Pi treatment (e.g., Root_0Pi versus Agar_0Pi). The heat maps are colored based on log2 fold changes derived from the fitted GLM. Positive fold changes (colored in red gradient) represent enrichments on the left side of the name of the contrast (e.g., Root-Agar, enriched in root in comparison to agar), whereas negative fold changes (colored in blue gradient) represent enrichments on the right side of the name of the contrast (e.g., Root-Agar, enriched in agar in comparison to agar). Boxed cells represent statistically significant enrichment/depletion. The bottom panel depicts the transformed (−log10) q-value derived from a phylogenetic signal Pagel’s λ test. Tests were performed per column in the heat map (e.g., Root0μM Pi versus Agar0μM Pi). (E) Bacterial alpha diversity estimated using the Shannon Diversity index. p-values derived from a linear model are shown for each fraction. Linear regression line is shown in black and the 95% confidence interval is shaded in gray. (F) CAP showing the influence of phosphate on the bacterial communities in the root. The bar graphs to the left of the CAP depict the percentage of variability explained by statistically significant (p < 0.05) variables based on a PERMANOVA model. CAP, canonical analysis of principal coordinates; GLM, generalized linear model; HSD, Honestly Significant difference; PERMANOVA, Permutational Multivariate Analysis of Variance; Pi, orthophosphate; SynCom, synthetic community; USeq, unique sequence.
We performed RNA sequencing (RNA-Seq) on inoculated and uninoculated seedlings exposed to high (1,000 μM) and low (50 μM) Pi. To confirm that our low Pi treatments induce PSR, we examined the expression of the 193 PSR markers defined in the work by Castrillo and colleagues [4]. We found that 168 of the 193 PSR markers genes were significantly induced in uninoculated plants at low Pi compared with high Pi conditions. In the presence of the SynCom, 184 out of 193 PSR marker genes were significantly induced, and the average fold change increased from 4.7 in uninoculated conditions to 11 in the presence of the SynCom (S6B Fig and S1 Data). We further examined whether the 123 low Pi-responsive genes from the soil experiment (Cluster 1 in Fig 1B) are overexpressed in the agar system as well. We found that 59 of the 123 genes (47.2%) were low Pi-enriched in uninoculated plants and 72 (58.5%) were low Pi-enriched in the presence of the SynCom. The average fold change for this set of 123 genes was 1.6 in uninoculated conditions and 2.0 in the presence of the SynCom (S6C Fig). These results confirm that (i) in both our systems, wild soils and axenic conditions, PSR is induced at low Pi, and (ii) the SynCom enhances this induction, similar to the results reported in the work by Castrillo and colleagues [4].
Bacterial SynComs display deterministic community assembly in plants
To quantify the establishment of the SynCom in the plants, we determined bacterial community composition after 12 days of co-inoculation in roots, shoots, and agar via 16S rRNA gene amplicon sequencing, mapping reads to 97 unique sequences (USeqs) representing the 185-strain SynCom (S8 Table). We found that plant roots and shoots sustained a higher bacterial alpha diversity than the surrounding agar (Fig 5B and S9 Table and S1 Data), an aspect in which our experimental system differs from a natural environment where species richness is higher in the surrounding soil than in the plant (Fig 2A and S1 Data). As in natural soil experimental systems, agar, roots, and shoots assembled distinct bacterial communities, and this difference among these 3 fractions explained most of the variance in community composition despite the different Pi concentrations (Figs 5C and S6D and S1 Data).
To study which strains are enriched in the roots and shoots under the different Pi concentrations, we utilized a GLM (S10 Table). Noticeably, plant (root and shoot) enrichment is strongly linked to phylogeny (Fig 5D and S1 Data) and is robust across the phosphate gradient assayed. In contrast, the root versus shoot comparison did not exhibit a significant phylogenetic signal, highlighting the fact that the ability to differentially colonize the shoot from the root under these conditions is phylogenetically scattered across the SynCom. As in the soil census, shoot, root, and agar beta diversities were significantly correlated (S6E–S6G Fig and S1 Data).
We hypothesized that by establishing a standardized protocol for producing the inoculum and controlling the growth conditions, we will have created a reproducible system in which most of the variance can be accounted for. To test this, we compared the number of ASVs and total relative abundance captured by the fraction (root/shoot versus soil/agar) in a GLM in the natural community experiment versus the SynCom experiment. Supporting our hypothesis, only 1,518 out of 3,874 measurable ASVs (32% of the total ASVs), accounting for 72% of the relative abundance in plant tissue, shift significantly between root and soil in the natural community survey, whereas 58 out of 97 USeqs (59%), accounting for 99% relative abundance in plant tissue, were significantly enriched or depleted in plant tissue in the SynCom experiment (Fig 5D and S1 Data).
These results indicate that plant colonization is largely deterministic in our SynCom system, in comparison to microbiomes in nature. The reproducibility of this system, coupled with our ability to edit it as a tool for hypothesis-testing, is crucial to bridge ecological observation with mechanistic understanding of plant–microbiota interactions.
P stress-induced changes in the root microbiome
The shifting role of the SynCom from increasing shoot size under replete Pi to decreasing shoot size and PSR induction under Pi limitation (Figs 5A and S6B and S1 Data) can be explained by either a shift in the lifestyle of individual bacteria along the mutualist–pathogen continuum or by changes in the microbiota composition along the Pi gradient. The latter would favor the proliferation of mutualist bacteria only when sufficient nutritional requirements are met. To measure the effect of the Pi concentration in the media on the SynCom composition in wt plants, we measured alpha and beta diversity along our Pi gradient (0, 10, 30, 50, 100, 1000 μM KH2PO4) in roots, shoots, and agar. We observed a positive correlation between alpha diversity and Pi concentrations, resembling a partial ecological diversity–productivity relationship—the prediction/observation of a bell-shaped response of ecological diversity to environmental productivity [51,52]—in roots and shoots but not in the surrounding agar (Fig 5E and S1 Data). As for beta diversity, the composition of the SynCom shifted significantly along the Pi concentration gradient (Figs 5F and S7A–S7E and S1 Data). Pi concentration therefore alters the plant microbiome, shifting from a net-positive outcome for the plant to a net-negative one as measured by shoot size (Fig 5A and S1 Data).
Burkholderia respond to Pi stress-induced changes in the plant
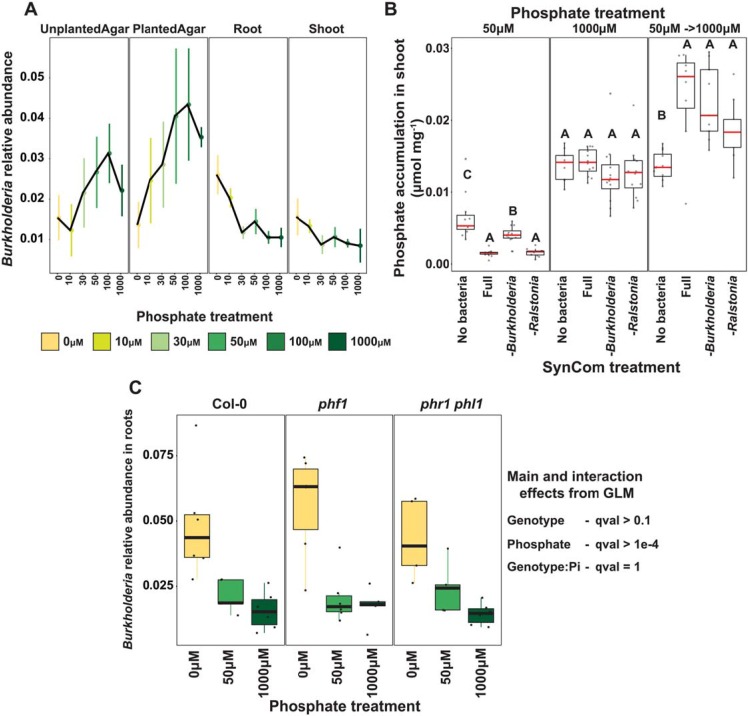
In a previous publication [4], we demonstrated that PHR1 negatively regulates defense-related genes under low Pi conditions. Suppression of plant defense and consequent alterations in colonization could account for some of the shift we observed from a beneficial to a detrimental community. We thus aimed to identify bacteria that respond to Pi stress-induced changes in the plant, rather than the Pi concentration itself. To do so, we searched for USeqs that displayed a strong Pi:fraction (shoot, root, agar) interaction in our GLM (S8A Fig and S11 Table and S1 Data). Two of the three USeqs displaying the strongest Pi:fraction interaction belonged to Burkholderiaceae, representing all 5 Burkholderia strains used in this experiment. The relative abundance of these USeqs is positively correlated with Pi concentration in the agar but is negatively correlated with Pi concentration in the root and shoot (Figs 6A and S8B and S8C and S1 Data). This pattern suggests that these strains are responding to physiological changes in the plant.
Fig 6. Bacterial strains respond to Pi-stress-induced physiological changes in the wt plants.
(A) Relative abundance of Burkholderia Useqs, both of which exhibit a statistically significant (q < 0.1) Pi-enrichment between the plant fractions and the agar fraction. The middle dot of each strip bar corresponds to the mean of that particular condition, the range of the strip bar corresponds to the 95% confidence interval of the mean. The lines are drawn connecting the means for each Pi concentration. (B) Box plots showing the phosphate accumulation in plants exposed to different SynComs across 3 phosphate treatments. Statistically significant differences among SynCom treatments were computed within each phosphate treatment separately using an ANOVA model. Letters represent the results of the post hoc test. (C) Box plot showing relative abundance of Burkholderia USeqs across 3 Pi concentrations and 3 plant genotypes. Summary of the NB-GLM for Burkholderia is shown on the right. GLM, generalized linear model; NB, no bacteria; Pi, orthophosphate; SynCom, synthetic community; USeq, unique sequence; wt, wild type.
To measure the physiological effect of the specific recruitment of Burkholderia under Pi stress on the plant, we conducted a drop-out experiment in which we compared plants inoculated with the full SynCom to plants inoculated with a SynCom excluding all 5 Burkholderia isolates. We also included a SynCom excluding all members of the neighboring Ralstonia clade (Fig 4A and S1 Data), which did not display any discernible Pi response. We measured shoot size and Pi concentrations in the shoots (a proxy for PSR) of plants grown in high (1,000 μM) and low (50 μM) Pi with the different SynComs. In addition, we measured shoot size and Pi content in a refeeding treatment with SynCom-inoculated plants grown in low (50 μM) Pi and then transferred to high Pi (1,000 μM) conditions. As seen before (Fig 5A and S1 Data), the SynCom decreased shoot sizes in low Pi and increased them in high Pi. However, the different taxon drop-outs did not affect shoot size compared with the full SynCom, except for a slight decrease in post-refeeding shoot size in the Ralstonia drop-out treatment (S9 Fig and S1 Data). All SynCom treatments decreased shoot Pi content in the low Pi conditions compared with the uninoculated plants but recovered to a higher shoot Pi level than the uninoculated treatments upon transferring to high Pi conditions, reproducing our previous report [4] (Fig 6B and S1 Data). Among inoculated treatments, plants colonized with the Burkholderia drop-out treatment (SynCom excluding all Burkholderia) had a higher Pi content than either plants colonized with the full SynCom or with the Ralstonia drop-out SynCom only in the low Pi conditions. There was no difference in shoot Pi among the SynCom treatments in either the high Pi treatment or following the refeeding treatment. This finding suggests that the enrichment of Burkholderia in plant tissue under Pi starvation can be considered a shift in the effect of bacteria on the plant from a positive interaction to a negative one.
To test whether Burkholderia are recruited to the plant under low Pi via a PSR-dependent mechanism, we inoculated 7-day-old wt, phf1, and phr1 phl1 seedlings with the SynCom and profiled the community composition after 12 days of growth in 3 Pi concentrations (0, 50, and 1,000 μM). In accordance with our soil experiment, and with the work by Castrillo and colleagues [4], community composition assembled in the roots of the 3 genotypes differed significantly (S10 Fig and S1 Data). However, Burkholderia sequences were enriched in low Pi in all 3 genotypes (Fig 6C and S1 Data), indicating that their recruitment to the root under low Pi is independent of PSR activation and of the immune dampening that accompanies it.
Discussion
This study shows that despite 60 years of differential fertilization, the plant’s PSR and accompanying changes to its microbiome composition between the low P and high P soils are subtle, possibly because Pi status of the plant is highly buffered by the plant ionomic regulatory network [53]. Only when comparing the low P versus the P-supplemented low+P samples is there a discernible difference in both plant transcriptome and shoot Pi accumulation, which correlates to a stronger effect on microbiota composition. This suggests that bioavailable Pi added to the soil is quickly consumed, and short-term amendments are needed in order to detect changes. A similar short-term fertilization approach was successfully applied in the work by Fabiańska and colleagues [36], using soils from a different long-term experimental site.
As opposed to in vitro defined transcriptomes [41], here, phf1 and phr1 phl1 plants, although each having a distinct transcriptional profile, were impaired in the response of a core set of genes (Cluster 1 in Fig 1B) to low P. This is despite the fact that phf1 plants were not impaired in growth (S1B and S1C Fig and S1 Data) and were only slightly impaired in shoot Pi accumulation (S1D Fig and S1 Data). A paralogous discrepancy was also observed in the work by Hiruma and colleagues [17], in which both phf1 and phr1 phl1 were impaired in their response to a plant growth promoting fungus, whereas only phr1 phl1 but not phf1 was impaired in P translocation to the shoot, raising the speculation that PHF1 is needed for fine, rather than bulk, P translocation within the plant. Our results support this idea and raise a similar speculation that PHF1-guided transporters are involved in fine grained phosphate translocation within the plant toward Pi sensing tissues.
An additional apparent departure from the axenically defined PSR transcriptional signature is that only 7 of the genes that were induced in wt plants under low Pi conditions overlapped with the list of 193 axenically defined PSR genes we used as a reference. It is notable, however, that despite the difference in plant age and environmental conditions between our soil and axenic agar systems, the agar-defined transcriptomic signature is detectable in the soil system (Fig 1C and S1 Data) and vice versa: the signature of the 123 genes identified in our soil system is detectable on agar (S6C Fig and S1 Data). Thus, despite the apparent differences, agar-defined PSR signatures are also maintained under complex real-life conditions.
Several studies link host physiological response to the soil phosphate status with the bacterial [4,38] and fungal [34,36] microbiome. A recent report of Arabidopsis planted in a 60-year-long annual P fertilization gradient (the same soil used in the current study) showed a modest P effect on plant microbiome composition [43]. Previously, we showed that PSR mutants in Arabidopsis have different bacterial microbiomes in Pi replete conditions [4], and a recent publication showed that PSR mutants had a slightly altered fungal microbiome in Pi-replete but not in Pi-depleted conditions [36]. Here, we analyzed fungi and bacteria side by side and demonstrated a pronounced effect of PSR impairment on both bacterial and fungal components of the plant microbiota. We noted an intriguing difference that emerged in the patterns of community assembly between bacteria and fungi (S3A, S3B, S3D and S3E Fig and S1 Data). The bacterial microbiota composition is strongly dependent on the soil bacterial community composition, whereas changes to the fungal microbiota are uncoupled from changes to the soil fungal community composition. This indicates that the plant is markedly more selective as to the fungi allowed to proliferate in its tissue than it is with bacteria. The observation that much of the modulation of the plant’s fungal microbiota is mediated by the bacterial microbiota itself [54] may also contribute to this complex pattern. Our results show that impairment of PSR genes profoundly affects the composition of the plant microbiota, independently of P conditions, and that observed shifts in root-derived microbial communities may not be a result of sensitivity to P concentrations but rather a response to PSR regulation in the hosts. The mechanism by which PSR regulation affects microbiota assembly is not fully understood. On one hand, PSR and plant immunity have been shown to be transcriptionally linked [4]. On the other, Pi depletion drastically changes the root’s exudate profiles [17,38,39,55], which have been shown to play a critical role in plant microbiota assembly [56,57]. It is likely that both factors contribute to shifts in microbiota composition. Evidently, however, the Burkholderia plant-enrichment in our SynCom occurs via a PSR-independent mechanism.
Our SynCom, comprising 185 genome-sequenced endophytic bacterial isolates, was designed to resemble a natural bacterial community (Fig 4B and S1 Data). The community assembly patterns shown for this system are highly reproducible, demonstrating that microbiome assembly is largely a deterministic process. The reproducibility and editability of this system is attractive for detailed mechanistic study of the processes that determine community assembly and its influence on plant phenotype and fitness. From within this SynCom, the genus Burkholderia emerges as a PSR-independent, low Pi-responsive taxon. We compared the effect of Burkholderia on shoot Pi accumulation from within a full SynCom (a realistic proxy for the bacterial community) to that of the full SynCom lacking Burkholderia, a strategy akin to knocking-out a gene of interest, also recently applied in the work by Durán and colleagues [54]. The control treatment for this type of approach is the full SynCom, whereas in a plant–bacterium binary association experiment it would typically be sterile conditions. Because both sterile conditions and binary association are strong deviations from conditions that may be encountered in the field, the results of binary association experiments may be correspondingly distorted. Using the drop-out approach, we expect to see more subtle differences, because the microbial load on the plant does not change much, but also that these differences be more relevant to the field—an expectation that is yet to be empirically tested. Our observation that dropping Burkholederia out of the SynCom increased shoot Pi in Pi-limiting conditions (50 μM Pi) but not in Pi-replete conditions (1,000 μM Pi) suggests that strains in this genus shift their relationship with the plant from a seeming commensal to a competitor/pathogen. Having ruled out PSR-dependent processes, another plausible explanation for Burkholderia enrichment patterns is that when Pi is limiting in the media, the plant becomes a source of Pi for the bacteria and strains with an enhanced ability to utilize plant-derived organic Pi or polyphosphate have an advantage under these conditions. Strains belonging to the genus Burkholderia have been shown to be particularly efficient at polyphosphate accumulation at pH 5.5, which is similar to the pH we used in our media [58].
Shifts in microbiota composition that accompany PSR are either adaptive to the plant or reflect opportunistic strategies by bacteria [4,17]. Under the former hypothesis, microbes recruited by the plant under Pi stress provide the plants with an advantage vis-a-vis coping with this stress, whereas under the latter, opportunistic microbes might be making a bad situation worse for the plant. In the case of Burkholderia in our SynCom, results support the latter hypothesis. Burkholderia contribute to depletion of shoot Pi stores, only under Pi-limiting conditions. However, plant-adaptive microbial recruitment under low Pi has been shown to occur as well [17]. The fact that bacteria responding to PSR genes are not a monophyletic group in soil indicates that multiple mechanisms are involved in community assembly. It is likely that these mechanisms encompass both plant-adaptive and opportunistic strategies.
Materials and methods
Soil P gradient experiment
Collection of soil from field site
Soil used in this experiment was collected from the long-term Pi fertilization field (“Field D”) trial at the Julius Kühn Experimental Station at Martin Luther University of Halle-Wittenberg (51°29′45.6′′N, 11°59′33.3′′E) [42,59]. Soil cores (10 cm diameter × 15 cm depth) were taken from 18 6 × 5 m unplanted plots belonging to 2 strips. These plots represent 3 P fertilization regimens: low, medium, and high P (0, 15, and 45 kg P ha−1 year−1, respectively). Differences in soil mineral content between strips and P fertilization regimens are reported in the work by Robbins and colleagues [41], showing that the different Pi regiments significantly differed only in P content. Soil cores were harvested in the middle of March 2016 (strip 1) and beginning of April 2016 (strip 2). Approximately 2 cm of the topsoil was discarded, and the remaining lower 13 cm of soil was stored at 4°C until use. Soils from each core were homogenized separately with a mesh sieve wire (5 × 5 m2). The sieved soil cores were stored at 4°C until use. About 300 g of soil were added to each pot (7 × 7 × 7 cm3).
Experimental design
Each of the 3 Arabidopsis genotypes was grown in soil from all 18 plots (6 plots per P treatment). In addition, a fourth P regimen designated “Low+P” was created by adding additional P to a set of pots with low P. The amount of P added to these pots is based on the difference in total P between low and high P plots. The average difference between low and high P over all the plots is 42 mg P per kg soil [43]. Per pot, this is 12.6 mg P (accounting for 300 g soil per pot). Thus, a 10 ml solution consisting of 4.2 mg P in the form of 20% K2HPO4 (MilliporeSigma, St. Louis, MO) and 80% KH2PO4 (MilliporeSigma, St. Louis, MO) was added to the pots in 3 applications (Weeks 2, 4, and 6) before watering (in order to distribute the P through the soil).
Thus, the experiment included 2 variables: soil treatment (low P, medium P, high P, low+P) and genotypes (Col-0, phf1, and phr1 phl1) with 6 independent replicates, amounting to 72 pots. Pot positions in the greenhouse were randomized.
Plant growth conditions
Arabidopsis thaliana ecotype Col-0 and mutants phf1 and phr1 phl1 (both in the Col-0 background) were used. Seeds were surface sterilized (20 min 70% EtOH (MilliporeSigma, St. Louis, MO), 10 s 100% EtOH) and planted directly onto moist soil. Sown seeds were stratified for 3 days at 4°C before being placed in a greenhouse under short-day conditions (6/18 day-night cycle; 19 to 21°C) for 8 weeks. Germinating seedlings were thinned to 4 plants per pot.
Sample harvest
After 8 weeks of growth, pots were photographed, and shoot size was quantified using WinRhizo software (Regent instruments Inc., Québec, Canada). Samples were harvested in random order to avoid any confounding circadian effect on the results. For DNA extraction, 2 roots, 2 shoots, and soil from each pot were harvested separately. Roots and shoots were rinsed in sterile water to remove soil particles, placed in 2 ml Eppendorf tubes (Eppendorf, Hamburg, Germany) with 3 sterile glass beads (MilliporeSigma, St. Louis, MO), then washed 3 times with sterile distilled water to remove soil particles and weakly associated microbes. Root and shoot tissue were then pulverized using a tissue homogenizer (TissueLyser II; Qiagen, Hilden, Germany) and stored at −80˚C until processing. Five ml of soil from each pot was suspended in 20 ml of sterile distilled water. The resulting slurry was sieved through a 100 μm sterile cell strainer (Fisher Scientific, Hampton, NH) and the flow-through was centrifuged twice at maximum speed for 20 minutes, removing the supernatant both times. The resulting pellet was stored at −80˚C until processing. For RNA extraction, one root system and one shoot were taken from 3 replicates of each treatment, washed lightly to remove soil particles, placed in 2 ml Eppendorf tubes with 3 glass beads and flash frozen with liquid nitrogen. Tubes were stored at −80˚C until processing. For shoot Pi measurement, 2 to 3 leaves from the remaining shoot in each pot were placed in an Eppendorf tube and weighed; 1% acetic acid (MilliporeSigma, St. Louis, MO) was then added, and samples were flash frozen and stored at −80°C until processing. The Ames method [60] was used to determine the phosphate concentration in these samples.
DNA extraction
DNA extractions were carried out on ground root and shoot tissue and soil pellets, using the 96-well-format MoBio PowerSoil Kit (MoBio Laboratories; Qiagen, Hilden, Germany) following the manufacturer’s instruction. Sample position in the DNA extraction plates was randomized, and this randomized distribution was maintained throughout library preparation and sequencing.
RNA extraction
RNA was purified from plant tissue using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions and stored at −80˚C.
Bacterial SynCom experiment
Bacterial isolation and culture
The 185-member bacterial SynCom contained genome-sequenced isolates obtained from Brassicaceae roots, nearly all Arabidopsis, planted in 2 North Carolina, US, soils. Because both bacteria and fungi responded similarly to PSR in our soil experiments, we only included bacteria, which are more compatible with our experimental system in our SynCom. A detailed description of this collection and isolation procedures can be found in the work by Levy and colleagues [50]. One week prior to each experiment, bacteria were inoculated from glycerol (MilliporeSigma, St. Louis, MO) stocks into 600 μL KB medium in a 96 deep well plate. Bacterial cultures were grown at 28°C, shaking at 250 rpm. After 5 days of growth, cultures were inoculated into fresh media and returned to the incubator for an additional 48 hours, resulting in 2 copies of each culture, 7 days old and 48 hours old. We adopted this procedure to account for variable growth rates of different SynCom members and to ensure that nonstationary cells from each strain were included in the inoculum. After growth, 48-hour-old and 7-day-old plates were combined and optical density (OD) of the culture was measured at 600 nm using an Infinite M200 Pro plate reader (TECAN, Männedorf, Switzerland). All cultures were then pooled while normalizing the volume of each culture according to the OD (we took a proportionally higher volume of culture from cultures with low OD). The mixed culture was then washed twice with 10 mM MgCl2 (MilliporeSigma, St. Louis, MO) to remove spent media and cell debris and vortexed vigorously with sterile glass beads to break up aggregates. OD of the mixed, washed culture was then measured and normalized to OD = 0.2. A total of 100 μL of this SynCom inoculum was spread on each agar plate prior to transferring seedlings.
Experimental design of agar experiments
We performed the Pi gradient experiment in 2 independent replicas (experiments performed at different times, with fresh bacterial inoculum and batch of plants), each containing 3 internal replications, amounting to 6 samples for each treatment. We had 2 SynCom treatments: no bacteria (NB) and SynCom; 6 Pi concentrations: 0, 10, 30, 50, 100, or 1,000 μM KH2PO4(henceforth, Pi); and 2 plant treatments: planted plates and unplanted plates (NP).
For the drop-out experiment, the entire SynCom, excluding all 5 Burkholderia and both Ralstonia isolates, was grown and prepared as described above. The Burkholderia and Ralstonia isolates were grown in separate tubes, washed, and added to the rest of the SynCom to a final OD of 0.001 (the calculated OD of each individual strain in a 185-Member SynCom at an OD of 0.2) to form the following 4 mixtures: (1) Full community—all Burkholderia and Ralstonia isolates added to the SynCom; (2) Burkholderia drop-out—only Ralstonia isolates added to the SynCom; (3) Ralstonia drop-out—only Burkholderia isolates added to the SynCom; (4) uninoculated plants—no SynCom. The experiment had 3 Pi conditions: low Pi (50 μM Pi), high Pi (1,000 μM Pi), and low→high Pi. Twelve days post-inoculation the low Pi and high Pi samples were harvested, and the low→high plants were transferred from 50 μM Pi plates to 1,000 μM Pi plates for an additional 3 days. The experiment was performed twice, and each rep consisted of 6 plates per SynCom mixture and Pi treatment, amounting to 72 samples. Upon harvest, shoot Pi accumulation was measured using the Ames method.
For the drop-out experiment with PSR mutants, the entire SynCom, excluding all 5 Burkholderia, was grown and prepared as described above. The Burkholderia isolates were grown in separate tubes, washed, and added to the SynCom to a final OD of 0.001 (the calculated OD of each individual strain in a 185-Member SynCom at an OD of 0.2) to form the following 2 mixtures: (1) Full community—all Burkholderia isolates added to the SynCom; (2) Burkholderia drop-out—no isolates added to the SynCom. For each SynCom, we inoculated 6 agar plates for each of 3 Pi conditions: 0, 50, and 1,000 μM Pi. Three 7-day-old seedlings from each of the 3 genotypes (wt Col-0, phf1, and phr1 phl1) were transferred to each plate. Roots were harvested 12 days post-inoculation, and bacterial DNA was extracted.
In vitro plant growth conditions
Arabidopsis thaliana accession Col-0 was used. All seeds were surface-sterilized with 70% bleach (Clorox, Oakland, CA), 0.2% Tween-20 (MilliporeSigma, St. Louis, MO) for 8 minutes, and rinsed 3 times with sterile distilled water to eliminate any seed-borne microbes on the seed surface. Seeds were stratified at 4°C in the dark for 2 days. Plants were germinated on vertical square 12 X 12 cm agar plates (Fisher Scientific, Hampton, NH) with Johnson medium (JM; [4]) containing 0.5% sucrose (MilliporeSigma, St. Louis, MO) and 1,000 μM Pi, for 7 days. Then, 10 plants were transferred to each vertical agar plate with amended JM lacking sucrose at one of the following experimental Pi concentrations: 0, 10, 30, 50, 100, or 1,000 μM Pi. The SynCom was spread on the agar prior to transferring plants. Each experiment included unplanted agar plates with SynCom for each media type (designated NP) and uninoculated plates with plants for each media type (designated NB). Plants were placed in randomized order in growth chambers and grown under a 16-hour dark/8-hour light regime at 21°C day/18°C night for 12 days (the period of time it takes roots to reach the bottom of the plate).
Sample harvest
Twelve days post-transferring, plates were imaged using a document scanner. For DNA extraction, roots, shoots, and agar were harvested separately, pooling 6 plants for each sample. Roots and shoots were placed in 2.0 ml Eppendorf tubes with 3 sterile glass beads. Samples were washed 3 times with sterile distilled water to remove agar particles and weakly associated microbes. Tubes were stored at −80˚C until processing. For RNA, samples were collected from a separate set of 2 independent experiments, using the same SynCom and conditions as above but with just 2 Pi concentrations: 1,000 μM Pi (high) and 50 μM Pi (low). Four seedlings were harvested from each sample, and samples were flash frozen and stored at −80˚C until processing.
DNA extraction
Root and shoot samples were lyophilized for 48 hours using a Freezone 6 freeze dry system (Labconco, Fisher Scientific, Hampton, NH) and pulverized using a tissue homogenizer (MP Biomedicals, Solon, OH). Agar from each plate was stored in a 30 ml syringe (Fisher Scientific, Hampton, NH) with a square of sterilized Miracloth (Millipore) at the bottom and kept at −20°C for a week. Syringes were then thawed at room temperature, and samples were squeezed gently into 50 ml tubes. Samples were centrifuged at maximum speed for 20 minutes, and most of the supernatant was discarded. The remaining 1 to 2 ml of supernatant containing the pellet was transferred into clean microfuge tubes. Samples were centrifuged again, supernatant was removed, and pellets were stored at −80°C until DNA extraction.
DNA extractions were carried out on ground root and shoot tissue and agar pellets using 96-well-format MoBio PowerSoil Kit (MOBIO Laboratories; Qiagen, Hilden, Germany) following the manufacturer’s instruction. Sample position in the DNA extraction plates was randomized, and this randomized distribution was maintained throughout library preparation and sequencing.
RNA extraction
RNA was extracted from Arabidopsis seedlings following the work by Ames [61]. Frozen seedlings were ground in liquid nitrogen, then homogenized in a buffer containing 400 μl of Z6-buffer; 8 M guanidinium-HCl (MilliporeSigma, St. Louis, MO), 20 mM MES, (MilliporeSigma, St. Louis, MO)20 mM EDTA (MilliporeSigma, St. Louis, MO) at pH 7.0; 400 μL phenol:chloroform:isoamylalcohol (25:24:1) (MilliporeSigma, St. Louis, MO) was added, and samples were vortexed and centrifuged (20,000g, 10 minutes) for phase separation. The aqueous phase was transferred to a new 1.5 ml tube, and 0.05 volumes of 1 N acetic acid (MilliporeSigma, St. Louis, MO) and 0.7 volumes 96% ethanol were added. The RNA was precipitated at −20°C overnight. Following centrifugation (20,000g, 10 minutes, 4°C), the pellet was washed with 200 μl sodium acetate (pH 5.2) (MilliporeSigma, St. Louis, MO) and 70% ethanol. The RNA was dried and dissolved in 30 μL of ultrapure water and stored at −80°C until use.
Quantification of plant phenotypes
The Ames method [60] was used to determine the phosphate concentration in the shoots of plants grown on different Pi regimens and treatments. Primary root length elongation was measured using ImageJ [62], and for shoot area and total root network measurement, WinRhizo software (Regent Instruments Inc., Quebec, Canada) was used.
DNA and RNA sequencing
Bacterial 16S sequencing
We amplified the V3-V4 regions of the bacterial 16S rRNA gene using primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). Two barcodes and 6 frames hifts were added to the 5’ end of 338F, and 6 frameshifts were added to the 806R primers, based on the protocol in the work by Lundberg and colleagues [63]. Each PCR reaction was performed in triplicate and included a unique mixture of 3 frameshifted primer combinations for each plate. PCR conditions were as follows: 5 μl Kapa Enhancer (Kapa Biosystems, Wilmington, MA), 5 μl Kapa Buffer A (Kapa Biosystems, Wilmington, MA), 1.25 μl of 5 μM 338F, 1.25 μl of 5 μM 806R, 0.375 μl mixed rRNA gene-blocking peptide nucleic acids (PNAs; 1:1 mix of 100 μM plastid PNA and 100 μM mitochondrial PNA; PNA Bio (Kapa Biosystems, Wilmington, MA), 0.5 μl Kapa dNTPs (Kapa Biosystems, Wilmington, MA), 0.2 μl Kapa Robust Taq (Kapa Biosystems, Wilmington, MA), 8 μl dH2O, 5 μl DNA; temperature cycling: 95°C for 60 seconds, 24 cycles of 95°C for 15 seconds, 78°C (PNA) for 10 seconds, 50°C for 30 seconds, 72°C for 30 seconds, 4°C until use. Following PCR cleanup, the PCR product was indexed using 96 indexed 806R primers with the same reaction mix as above and 9 cycles of the cycling conditions described in the work by Lundberg and colleagues [63]. PCR products were purified using AMPure XP magnetic beads (Beckman Coulter, Brea, CA) and quantified with a Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA). Amplicons were pooled in equal amounts and then diluted to 10 pM for sequencing. Sequencing was performed on an Illumina MiSeq instrument (Illumina, San Diego, CA) using a 600-cycle V3 chemistry kit. The raw data for the natural soil experiment is available in the NCBI SRA Sequence Read Archive (accession PRJNA531340). The raw data for the SynCom experiment is available in the NCBI SRA Sequence Read Archive (accession PRJNA531340).
Fungal/Oomycete ITS sequencing
We amplified the ITS1 region using primers ITS1-F (5′-CTTGGTCATTTAGAGGAAGTAA-3′; [64]) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′; [65]). Samples were diluted to concentrations of 3.5 ng μl−1 of DNA with nuclease-free water for the first PCR reaction to amplify the ITS1 region. Reactions were prepared in triplicate in 25 μl volumes consisting of 10 ng of DNA template, 1× incomplete buffer, 0.3% bovine serum albumin, 2 mM MgCl2, 200 μM dNTPs, 300 nM of each primer, and 2 U of DFS-Taq DNA polymerase (Bioron, Ludwigshafen, Germany); temperature cycling: 2 minutes at 94°C, 25 cycles: 30 seconds at 94°C, 30 seconds at 55°C, and 30 seconds at 72°C; and termination: 10 minutes at 72°C. PCR products were cleaned using an enzymatic cleanup (24.44 μl: 20 μl of template, 20 U of exonuclease I, 5 U of Antarctic phosphatase, 1× Antarctic phosphatase buffer; New England Biolabs, Frankfurt, Germany); incubation conditions were 30 minutes at 37°C, 15 minutes at 85°C; centrifuge 10 minutes at 4,000 rpm. A second PCR was then performed (2 minutes at 94°C; 10 cycles: 30 seconds at 94°C, 30 seconds at 55°C, and 30 seconds at 72°C; and termination: 10 minutes at 72°C), in triplicate using 3 μl of cleaned PCR product and sample-specific barcoded primers (5′- AATGATACGGCGACCACCGAGATCTACACTCACGCGCAGG-ITS1F-3′; 5′-CAAGCAGAAGACGGCATACGAGAT-BARCODE(12-NT)-CGTACTGTGGAGA-ITS2-3′). PCR reactions were purified using with Agencourt AMPure XP purification kit (Beckman Coulter, Krefeld, Germany). Amplicons were pooled in equal amounts and then diluted to 10 pM for sequencing. Sequencing was performed on an Illumina MiSeq instrument using a 600-cycle V3 chemistry kit. The raw data are available in the NCBI SRA Sequence Read Archive (Project Number PRJNA531340).
Plant RNA sequencing
Illumina-based mRNA-Seq libraries were prepared from 1 μg RNA following the work by Herrera Paredes and colleagues [38]. mRNA was purified from total RNA using Sera-mag oligo(dT) magnetic beads (GE Healthcare Life Sciences, Chicago, IL) and then fragmented in the presence of divalent cations (Mg2+) at 94°C for 6 minutes. The resulting fragmented mRNA was used for first-strand cDNA synthesis using random hexamers and reverse transcriptase (Enzymatics, Qiagen, Beverly, MA), followed by second-strand cDNA synthesis using DNA Polymerase I (Enzymatics, Qiagen, Beverly, MA) and RNAseH (Enzymatics, Qiagen, Beverly, MA). Double-stranded cDNA was end-repaired using T4 DNA polymerase (Enzymatics, Qiagen, Beverly, MA), T4 polynucleotide kinase (Enzymatics, Qiagen, Beverly, MA), and Klenow polymerase (Enzymatics, Qiagen, Beverly, MA). The DNA fragments were then adenylated using Klenow exo-polymerase (Enzymatics, Qiagen, Beverly, MA) to allow the ligation of Illumina Truseq HT adapters (D501–D508 and D701–D712; Illumina, San Diego, CA). Following library preparation, quality control and quantification were performed using a 2100 Bioanalyzer instrument (Agilent Technologies, Santa Clara, CA) and the Quant-iT PicoGreen dsDNA Reagent (Invitrogen, Carlsbad, CA), respectively. Libraries were sequenced using HiSeq4000 sequencers (Illumina, San Diego, CA) to generate 50-bp single-end reads.
Data processing and statistical analyses
Quantification of plant phenotypes—Soil experiment
To measure correlation between all measured plant phenotypes (shoot Pi, shoot weight, shoot size) we applied hierarchical clustering based on a matrix of Pearson correlation coefficients between all pairs of phenotypes. We used the R package corrplot version 0.84 [66] to visualize correlations. To compare shoot Pi accumulation, we treated the low P sample as the control, because this soil did not receive any treatment. We performed paired t tests between the different P-treated samples and the low P samples within each plant genotype independently (α < 0.05).
Amplicon sequence data processing—Soil experiments
Bacterial sequencing data were processed with MT-Toolbox [67]. Usable read output from MT-Toolbox (i.e., reads with 100% correct primer and primer sequences that successfully merged with their pair) were quality filtered using Sickle [68] by not allowing any window with a Q score under 20. After quality filtering, samples with low total reads recruited (<3,000 reads), amounting to 51 soil samples were discarded. Although this study focuses on the root microbiome, the relatively small number of remaining soil samples may have affected the results shown in S4E Fig. The remaining samples include at least 3 samples per genotype. The resulting sequences were collapsed into ASVs using the R package DADA2 version 1.8.1 [69]. Taxonomic assignment of each ASV was performed using the naïve Bayes kmer method implemented in the DADA2 package using the Silva 132 database as training reference [69].
Fungal ITS sequence data were processed using DADA2 [69] with default parameters using only the forward reads. Taxonomic assignment of each ASV was performed using the naïve Bayes kmer method implemented in the MOTHUR package [70] using the UNITE database [71] as training reference.
The resulting bacterial and fungal count tables were deposited at https://github.com/isaisg/hallepi.
Community analyses—Soil experiments
The resulting bacterial and fungal count tables were processed and analyzed with functions from the ohchibi package [72]. Both tables were rarefied to 3,000 reads per sample. An alpha diversity metric (Shannon diversity) was calculated using the diversity function from the vegan package version 2.5–3 [73]. We used ANOVA to test for differences in Shannon Diversity indices between groups. Tukey’s HSD post hoc tests here and elsewhere were performed using the cld function from the emmeans R package [74]. Beta-diversity analyses (Principal coordinate analysis and canonical analysis of principal coordinates [CAP]) were based on Bray-Curtis dissimilarity calculated from the rarefied abundance tables. We utilized the capscale function from the vegan R package v.2.5–3 [73] to compute a CAP. To analyze the full data set (all fraction, all genotypes, all phosphorus treatments), we constrained by fraction, plant genotype, and phosphorus fertilization treatment, while conditioning for the plot effect. We performed the genotype:phosphorus interaction analysis over each fraction independently, constraining for the plant genotype and phosphorus fertilization treatment while conditioning for the plot effect. In addition to CAP, we performed Permutational Multivariate Analysis of Variance (PERMANOVA) over the 2 data sets described above using the adonis function from the vegan package version 2.5–3 [73]. Finally, we used the function chibi.permanova from the ohchibi package to plot the R2 values for each significant term in the PERMANOVA model tested.
The relative abundance of bacterial phyla and fungal taxa were depicted using the stacked bar representation encoded in the function chibi.phylogram from the ohchibi package.
We used the R package DESeq2 version 1.22.1 [75] to compute the enrichment profiles for both bacterial and fungal ASVs. For the full data set model, we estimated main effects for each variable tested (Fraction, Plant genotype, and phosphorus fertilization) using the following design:
We delimited ASV fraction enrichments using the following contrasts: soil versus root, soil versus shoot, and root versus shoot. An ASV was considered statistically significant if it had q < 0.1.
We implemented a second statistical model in order to identify ASVs that exhibited statistically significant differential abundances depending on genotype. For this analysis, we utilized only root-derived low P and P-supplemented low P (low+P) treatments. We utilized a group design framework to facilitate the construction of specific contrasts. In the group variable we created, we merged the genotype and phosphate levels per sample (e.g., Col-0_lowP, phf1_low+P, or phr1 phl1_lowP). We controlled the paired structure of our design by adding a plot variable, resulting in the following model design:
We delimited 6 sets (S1, S2, S3, S4, S5, S6) of statistically significant (q < 0.1) ASVs from our model using the following contrasts:
S1 = {Samples from Col-0, higher abundance in low treatment in comparison to low+P treatment}
S2 = {Samples from phf1, higher abundance in low treatment in comparison to low+P treatment}
S3 = {Samples from phr1 phl1, higher abundance in low treatment in comparison to low+P treatment}
S4 = {Samples from Col-0, higher abundance in low+P treatment in comparison to low treatment}
S5 = {Samples from phf1, higher abundance in low+P treatment in comparison to low treatment}
S6 = {Samples from phr1 phl1, higher abundance in low+P treatment in comparison to low treatment}
The 6 sets described above were used to populate Fig 3C–3F.
The interactive visualization of the enrichment profiles was performed by converting the taxonomic assignment of each ASV into a cladogram with equidistant branch lengths using R. We used the interactive tree of life (iTOL) interface [76] to visualize this tree jointly with metadata files derived from the output of the statistical models described above. The cladograms for both bacteria and fungi can be downloaded from the links described above or via the iTOL user hallepi.
In order to compare beta-diversity patterns across samples, we only used samples coming from pots in which sequence data from all 3 fractions (soil, root, and shoot) passed quality filtering. Then, for each fraction, we estimated a distance structure between samples inside that fraction using the Bray-Curtis dissimilarity metric. Finally, we computed Mantel [77] correlations between pairs of distance objects (e.g., samples from root or samples from shoot) using the vegan package version 2.5–3 [73] implementation of the Mantel test.
All scripts and data sets required to reproduce the soil experiment analyses are deposited in the following GitHub repository: https://github.com/isaisg/hallepi/.
Inspection of other edaphic factors in the soil
To inspect whether the genotype or P effects that we observed are confounded by another edaphic factor in the soil, we cross-referenced our data set with the edaphic factors reported for the same soil plots in the work by Robbins and colleagues [43]. Because most of the edaphic factors are correlated (S5A Fig), we considered the first 3 principal components (PCs) derived from these edaphic factors. These 3 PCs encompass 87% of the cumulative variance in the edaphic factor matrix (S5B Fig). A PERMANOVA model of the root community composition that considers these 3 PCs assigns 21% of explained variance to the first PC, which is composed of 8 edaphic factors (S5C and S5B Fig). Nonetheless, the P and genotype variables explain a similar proportion of variance as in a model that did not account for the other edaphic factors, indicating that they are orthogonal to the other variables that can be accounted for and are not confounded by them.
Amplicon sequence data processing—SynCom experiments
SynCom sequencing data were processed with MT-Toolbox [67]. Usable read output from MT-Toolbox (i.e., reads with 100% correct primer and primer sequences that successfully merged with their pair) were quality filtered using Sickle [68] by not allowing any window with Q-score under 20. The resulting sequences were globally aligned to a reference set of 16S rRNA gene sequences extracted from genome assemblies of SynCom member strains. For strains that did not have an intact 16S rRNA gene sequence in their assembly, we generated the 16S rRNA gene using Sanger sequencing. The reference database also included sequences from known bacterial contaminants and Arabidopsis organellar 16S sequences (S12 Table). Sequence alignment was performed with USEARCH version 7.1090 [78] with the option ‘usearch_global’ at a 98% identity threshold. On average, 85% of sequences matched an expected isolate. Our 185 isolates could not all be distinguished from each other based on the V3-V4 sequence and were thus classified into 97 USeqs. A USeq encompasses a set of identical (clustered at 100%) 16S rRNA V3-V4 sequences coming from a single or multiple isolates.
Sequence mapping results were used to produce an isolate abundance table. The remaining unmapped sequences were clustered into Operational Taxonomic Units (OTUs) using UPARSE [79] implemented with USEARCH version 7.1090 at 97% identity. Representative OTU sequences were taxonomically annotated with the RDP classifier [80] trained on the Greengenes database [81] (4 February 2011). Matches to Arabidopsis organelles were discarded. The vast majority of the remaining unassigned OTUs belonged to the same families as isolates in the SynCom. We combined the assigned USeq and unassigned OTU count tables into a single table.
The resulting count table was processed and analyzed with functions from the ohchibi package. Samples were rarefied to 1,000 reads per sample. An alpha diversity metric (Shannon diversity) was calculated using the diversity function from the vegan package version 2.5–3 [73]. We used ANOVA to test for differences in alpha diversity between groups. Beta-diversity analyses (Principal coordinate analysis and CAP) were based on were based on Bray-Curtis dissimilarity calculated from the rarefied abundance tables. We used the capscale function from the vegan R package version 2.5–3 [73] to compute the CAP. To analyze the full data set (all fraction, all phosphate treatments), we constrained by fraction and phosphate concentration while conditioning for the replicate effect. We performed the Fraction:Phosphate interaction analysis within each fraction independently, constraining for the phosphate concentration while conditioning for the rep effect. In addition to CAP, we used PERMANOVA analysis over the 2 data sets described above using the adonis function from the vegan package version 2.5–3 [73]. Finally, we used the function chibi.permanova from the ohchibi package to plot the R2 values for each significant term in the PERMANOVA model tested.
We visualized the relative abundance of the bacterial phyla present in the SynCom using the stacked bar representation encoded in the chibi.phylogram from the ohchibi package.
We used the package DESeq2 version 1.22.1 [75] to compute the enrichment profiles for USeqs and OTUs present in the count table. For the full data set model, we estimated main effects for each variable tested (fraction and phosphate concentration) using the following model specification:
We calculated the USeqs/OTUs fraction enrichments using the following contrasts: agar versus root, agar versus shoot, and root versus shoot. A USeq/OTU was considered statistically significant if it had q < 0.1. In order to populate the heat maps shown in Fig 5C, we grouped the fraction and phosphate treatment variable into a new group variable that allowed us to fit the following model:
We used the fitted model to estimate the fraction effect inside each particular phosphate level (e.g., Root versus agar at 0Pi, or shoot versus agar at 1,000Pi).
Additionally, we utilized a third model for the identification of USeqs/OTUs that exhibited a significant Fraction:Phosphate interaction between the planted agar samples and the plant fractions (root and shoot). Based on the beta-diversity and alpha-diversity results, we only used samples that were treated with 0, 10, 100, and 1,000 μM of phosphate. We grouped the samples into 2 categories based on their phosphate concentration, low (0 μM and 10 μM) and high (100 μM and 1,000 μM). Then we used the following model specification to derive the desired interaction effect:
Finally, we subset USeqs that exhibited a significant interaction (Fraction:Category, q < 0.1) in the following 2 contrasts (planted agar versus root) and (planted agar versus shoot).
In order to compare beta-diversity patterns across samples, we only used samples coming from pots in which sequence data from all 3 fractions (soil root and shoot) passed quality filtering. Then, for each fraction, we estimated a distance structure between samples inside that fraction using the Bray-Curtis dissimilarity metric. Finally, we computed Mantel [77] correlations between pairs of distance objects (e.g., samples from root or samples from shoot) using the vegan package version 2.5–3 [73] implementation of the Mantel test.
For the drop-out experiment, we ran an ANOVA model inside each of the phosphate treatments tested (50 μM Pi, 1,000 μM Pi, and 50→1,000 μM Pi). We visualized the results of the ANOVA models using the compact letter display encoded in the CLD function from the emmeans package.
All scripts necessary to reproduce the synthetic community analyses are deposited in the following GitHub repository: https://github.com/isaisg/hallepi.
Phylogenetic inference of the SynCom isolates
To build the phylogenetic tree of the SynCom isolates, we utilized the supermatrix approach previously described in the work by Levy and colleagues [50]. Briefly, we scanned 120 previously defined marker genes across the 185 isolate genomes from the SynCom utilizing the hmmsearch tool from the hmmer version 3.1b2 [82]. Then, we selected 47 markers that were present as single copy genes in 100% of our isolates. Next, we aligned each individual marker using MAFFT [83] and filtered low quality columns in the alignment using trimAl [84]. Afterward, we concatenated all filtered alignments into a superalignment. Finally, FastTree version 2.1 [85] was used to infer the phylogeny utilizing the WAG model of evolution.
We utilized the inferred phylogeny along with the fraction fold change results of the main effect model to compute the phylogenetic signal (Pagel’s λ) [86] for each contrast (planted agar versus root, planted agar versus shoot, and root versus shoot) along each concentration of the phosphate gradient. The function phylosig from the R package phytools [87] was used to test for significance of the phylogenetic signal measured.
Multiple panel figures were constructed using the egg R package [88].
RNA-Seq read processing
Initial quality assessment of the Illumina RNA-Seq reads was performed using FastQC version 0.11.7 [89]. Trimmomatic version 0.36 [90] was used to identify and discard reads containing the Illumina adaptor sequence. The resulting high-quality reads were then mapped against the TAIR10 [91] Arabidopsis reference genome using HISAT2 version 2.1.0 [92] with default parameters. The featureCounts function from the Subread package [93] was then used to count reads that mapped to each one of the 27,206 nuclear protein-coding genes. Evaluation of the results of each step of the analysis was done with MultiQC version 1.1 [94]. Raw sequencing data and read counts are available at the NCBI Gene Expression Omnibus accession number GSE129396.
RNA-Seq statistical analysis—Soil experiment
To measure the transcriptional response to Pi limitation in soil, we used the package DESeq2 version 1.22.1 [75] to define differentially expressed genes (DEGs) using the raw count table described above. We used only samples from low P and P-supplemented low P (low+P) treatments along the 3 genotypes tested (Col-0, phf1, and phr1 phl1). We combined the genotype and P treatment variables into a new group variable (e.g., Col-0_lowP or phf1_low+P). Because we were interested in identifying DEGs among any pair of levels (6 levels) of the group variable (e.g., Col-0_lowP versus Col-0_low+P) we performed a likelihood ratio test (LRT) between a model containing the group variable and a reduced model containing only the intercept. Next, we defined DEGs as genes that had a q < 0.1.
For visualization purposes, we applied a variance stabilizing transformation to the raw count gene matrix. We then standardized (z-score) each gene along the samples. We subset DEGs from this standardized matrix and for each gene calculated the mean z-score expression value in a particular level of the group variable (e.g., Col-0_lowP); this resulted in a matrix of DEGs across the 6 levels in our design. Next, we created a dendrogram of DEGs by applying hierarchical clustering (method ward.D2, hclust R-base [95]) to a distance object based on the correlation (dissimilarity) of the expression profiles of the genes across the 6 levels in our design. Finally, we delimited the cluster of DEGs by cutting the output dendogram into 5 groups using the R-base cutree function [95]. GO enrichment was performed for each cluster of DEGs using the R package clusterProfiler [96].
For the PSR marker gene analysis, we downloaded the ID of 193 genes defined in the work by Castrillo and colleagues [4]. Then, we subset these genes from our standardized matrix and computed for each gene the mean z-score expression value in a particular level of the group variable. Finally, we visualized the average expression of this PSR regulon across our groups of interest utilizing the function chibi.boxplot from the ohchibi package.
All scripts necessary to reproduce the RNA-Seq analyses are deposited in the following GitHub repository: https://github.com/isaisg/hallepi.
RNA-Seq statistical analysis—SynCom experiment
To measure the transcriptional response to Pi limitation in the SynCom microcosm, we used the package DESeq2 version 1.22.1 [75] to define DEGs using the raw count gene table. We combined the bacteria (NB, Full SynCom) and P treatment variables into a new group variable (e.g., NB_50Pi or Full_1000Pi). Afterward we fitted the following model to our gene matrix:
Finally, utilizing the model fitted, we contrasted the phosphate treatment inside each level of the bacteria variable (e.g., NB_1000Pi versus NB_50Pi). Any gene with q < 0.1 was defined as differentially expressed.
For the PSR marker gene analysis, we downloaded the ID of 193 genes defined in the work by Castrillo and colleagues [4]. Then, we subset these genes from our standardized matrix and computed for each gene the mean z-score expression value in a particular level of the group variable. Finally, we visualized the average expression of the PSR regulon across our groups of interest utilizing the function chibi.boxplot from the ohchibi package.
All scripts necessary to reproduce the RNA-Seq analyses are deposited in the following GitHub repository: https://github.com/isaisg/hallepi.
Supporting information
(A) Heat map showing the all versus all pairwise Pearson correlation coefficient calculated between the quantified phenotypes associated with the PSR: shoot area, shoot fresh weight, and shoot free Pi accumulation. (B) Box plot showing the distribution of the shoot area measured across the P gradient within each of the 3 genotypes. (C) Boxplot showing the distribution of shoot fresh weight measured across the P gradient within each of the 3 genotypes. (D) Box plot showing the shoot Pi accumulation across the 3 genotypes. Letters represent the results of the post hoc test. (E) Box plots displaying the average expression of 193 PSR marker genes across the low and low+P samples in each of the 3 genotypes tested. (F) Scatter plot showing the relationship between the standardized average phosphate accumulation in leaves (x-axis) and the average standardized expression of 193 PSR marker genes (y-axis). The p-value and R value were calculated according to Pearson’s product moment correlation coefficient. P, phosphorus; Pi, orthophosphate; PSR, phosphate starvation response.
(TIF)
(A, B) Box plots showing the distribution of the alpha diversity (Shannon diversity index) across all levels of P in the soil for bacteria (A) and fungi (B). (C, D) PERMANOVA results in which the effect of the 3 variables (fraction, genotype, and soil P) and their interaction on the assembly of the bacterial (C) and fungal (D) communities were tested. (E) Relative abundance profiles of the main bacterial phyla in the 3 variables (fraction, genotype, and soil P) across all levels of P in the soils. (F) Relative abundance profiles of the main fungal orders in the 3 variables (fraction, genotype, and soil P) across all the levels of P in the soils. P, phosphorus; PERMANOVA, Permutational Multivariate Analysis of Variance.
(TIF)
(A, B, C) Correlation plots between Bray-Curtis distance matrices calculated for bacteria within soil treatments, root, and shoot fractions. The R and p-values were calculated using Mantel tests. (A) Correlation plot of soil versus root. (B) Correlation plot of soil versus shoot. (C) Correlation plot of root versus shoot. (D, E, F) Correlation plots between Bray-Curtis distance matrices calculated for fungi within soil treatments, root, and shoot fractions. The R and p-values were calculated using Mantel tests. R and p-values colored in red were calculated excluding the cloud of large distances appearing in graphs panels D and E. (D) Correlation plot of soil versus root. (E) Correlation plot of soil versus shoot. (F) Correlation plot of root versus shoot.
(TIF)
(A, B) PERMANOVA results showing the influence of the plant genotype and soil P concentration and their interaction on the assembly of the root (A) bacterial and (B) fungal communities. (C, D) CAP showing the effect of plant genotype and P content in the soil over the shoot (C) bacterial and (D) fungal communities. The p-value and R2 values in each plot are derived from a PERMANOVA model and correspond to the genotype and soil P term, respectively. (E, F) CAP showing the influence of genotype and P on the soil (E) bacterial and (F) fungal communities. Note smaller number of points in bacterial soil samples. The p-value and R2 values in each plot are derived from a PERMANOVA model and correspond to the genotype and soil P term, respectively. CAP, canonical analysis of principal coordinates; P, phosphorus; PERMANOVA, Permutational Multivariate Analysis of Variance.
(TIF)
(A) Correlation heat map of the 13 edaphic factors reported in the work by Robbins and colleagues [43]. (B) Bar plot displaying the amount of variance in the edaphic factor matrix explained by PC. (C) Bar plot depicting the contribution of the different edaphic factors to the first 3 PC. Colors denote the direction of the variable in PCA space. (D) Proportion of the variance explained for the different variables in models including (M2) and excluding (M1) edaphic factors for bacteria (left) and fungi (right). Only variables with a statistically significant effect are shown. P, phosphorus; PC, principal component; PCA, principal component analysis.
(TIF)
(A) Box plots displaying the primary root elongation of plants grown in a gradient of Pi concentrations in sterile conditions or with the SynCom. A t test was used for each Pi treatment to estimate differences between SynCom-treated and uninoculated plants. (B) Average expression of the 193 PSR markers genes in low (50 μM) and high (1,000 μM) Pi conditions within SynCom-treated and uninoculated plants. (C) Average expression of the 123 genes from Cluster 1 (Fig 1B) in low (50 μM) and high (1,000 μM) Pi conditions within SynCom-treated and uninoculated plants. (D) PERMANOVA model results showing the influence of the 2 variables (fraction and Pi concentration) and their interaction on the assembly of the bacterial community in the plant. (E, F, G) Correlation plots between Bray-Curtis distance matrices calculated for bacterial profile within agar, root, and shoot fractions. The R and p-values were calculated using Mantel tests. (E) Correlation plot of agar versus root. (F) Correlation plot of agar versus shoot. (G) Correlation plot of root versus shoot. PERMANOVA, Permutational Multivariate Analysis of Variance; Pi, orthophosphate; PSR, phosphate starvation response; SynCom, synthetic community.
(TIF)
(A, B) CAP showing the influence of Pi concentration in the media on the bacterial communities in the (A) plant shoot and (B) agar. The bar graphs to the left of each plot depict the percentage of variability explained by statistically significant (p < 0.05) variables based on a PERMANOVA model. (C, D, E) PERMANOVA model results showing the influence of Pi concentration on the assembly of the bacterial community in (C) roots, (D) shoot, and (E) agar. CAP, canonical analysis of principal coordinates; PERMANOVA, Permutational Multivariate Analysis of Variance; Pi, orthophosphate.
(TIF)
(A) Scatter plot (volcano plot) showing the results of the GLM interaction model between fraction and Pi concentration. The axes of the plot represent the output of the statistical test. The x-axis is the transformed q-value and the y-axis the log2 fold change. Each dot in the scatter plot represents a USeq. USeqs that showed a statistically significant fraction:Pi interaction are colored in red. USeqs genus and ID are displayed. The top right quadrant represents USeqs that are enriched in the plant tissues under low Pi conditions. (B, C) Relative abundance of Burkholderia Useqs 16 (B) and 30 (C) that exhibits a statistically significant (q < 0.1) Pi enrichment between the plant fractions and the agar fraction. The middle dot of each strip bar corresponds to the mean of that particular condition; the range of the strip bar corresponds to the 95% confidence interval of the mean. GLM, generalized linear model; Pi, orthophosphate; USeq, unique sequence.
(TIF)
Box plots showing shoot circumference in plants exposed to different SynComs across 3 phosphate treatments. Statistically significant differences among SynCom treatments were computed within each phosphate treatment separately using an ANOVA model. Letters represent the results of the post hoc test. SynCom, synthetic community.
(TIF)
CAP showing the influence of plant genotypes (A) and agar Pi concentration (B) over the bacterial SynCom in the root. The p-value and R2 values inside each plot are derived from a PERMANOVA model and correspond to the genotype and Pi terms, respectively. CAP, canonical analysis of principal coordinates; PERMANOVA, Permutational Multivariate Analysis of Variance; Pi, orthophosphate; SynCom, synthetic community.
(TIF)
Pi, orthophosphate.
(XLSX)
(XLSX)
ASV, amplicon sequence variant; GLM, generalized linear model.
(XLSX)
ASV, amplicon sequence variant; GLM, generalized linear model.
(XLSX)
ASV, amplicon sequence variant; GLM, generalized linear model; P, phosphorus; Pi, orthophosphate.
(XLSX)
ASV, amplicon sequence variant; GLM, generalized linear model; P, phosphorus; Pi, orthophosphate.
(XLSX)
USeq, unique sequence.
(XLSX)
(XLSX)
GLM, generalized linear model.
(XLSX)
GLM, generalized linear model; Pi, orthophosphate.
(XLSX)
(XLSX)
(ZIP)
Acknowledgments
We thank Dr. Wolfgang Gans and Prof. Edgar Peiter (Martin Luther University Halle-Wittenberg, Halle, Germany) for generously providing us with access to Halle soils; Prof. Paul Schulze-Lefert (Max Planck Institute for Plant Breeding Research, Cologne, Germany) for generously providing greenhouse and lab space; Stratton Barth, Julia Shen, Ellie Wilson, May Priegel, and Dilan Chudasma for technical assistance throughout the project; the Dangl lab microbiome group for useful discussions; Dr. Javier Paz-Ares, Dr. Antonio Leyva (National Centre for Biotechnology, Madrid, Spain), and Dr. Connor Fitzpatrick for critical comments on the manuscript.
Abbreviations
- ASV
amplicon sequence variant
- CAP
canonical analysis of principal coordinates
- DEG
differentially expressed gene
- GLM
generalized linear model
- GO
gene ontology
- HSD
honestly significant difference
- iTOL
interactive tree of life
- JM
Johnson medium
- LRT
likelihood ratio test
- NB
no bacteria
- NP
unplanted plates
- OD
optical density
- OTU
Operational Taxonomic Unit
- P
phosphorus
- PC
principal component
- PERMANOVA
Permutational Multivariate Analysis of Variance
- PHF1
PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1
- PHL1
PHR1-LIKE; PHR1, PHOSPHATE STARVATION RESPONSE 1
- Pi
orthophosphate
- PNA
peptide nucleic acid
- PSR
phosphate starvation response
- RNA-Seq
RNA sequencing
- SynCom
synthetic community
- USeq
unique sequence
- wt
wild type
Data Availability
The raw reads from the amplicon sequencing can be downloaded from the NCBI Sequence Read Archive (Project PRJNA531340). The raw reads for the RNA-Seq experiments can be downloaded from the omnibus accession GSE129396. In addition to the the raw reads, all the data structures and scripts used to analyze the data and generate figures can be downloaded from the following github repository: https://github.com/isaisg/hallepi An interactive visualization of the fraction enrichment profiles from the natural soil experiments can be accesed via the iTOL user hallepi or the following links: https://itol.embl.de/tree/1522316254174701551987253https://itol.embl.de/tree/13656172137464831571097084 Data underlying all figures in the manuscript is included within S1 Data.
Funding Statement
U.S. National Institute of Health NRSA (grant number F32-GM117758-02). Received by OMF. Office of Science (BER), U.S. Department of Energy (grant number DE-SC001043). Received by JLD. U.S. National Science Foundation (grant number IOS-1917270). Received by JLD. U.S. National Science Foundation INSPIRE (grant number IOS-1343020). Received by JLD. Howard Hughes Medical Institute. Received by JLD.The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Desbrosses GJ, Stougaard J. Root Nodulation: A Paradigm for How Plant-Microbe Symbiosis Influences Host Developmental Pathways. Cell Host Microbe. 2011;10: 348–358. 10.1016/j.chom.2011.09.005 [DOI] [PubMed] [Google Scholar]
- 2.Hacquard S, Kracher B, Hiruma K, Münch PC, Garrido-Oter R, Thon MR, et al. Survival trade-offs in plant roots during colonization by closely related beneficial and pathogenic fungi. Nat Commun. Nature Publishing Group; 2016;7: 11362 10.1038/ncomms11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411: 826–833. 10.1038/35081161 [DOI] [PubMed] [Google Scholar]
- 4.Castrillo G, Teixeira PJPL, Paredes SH, Law TF, de Lorenzo L, Feltcher ME, et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017;543: 513–518. 10.1038/nature21417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hacquard S, Spaepen S, Garrido-Oter R, Schulze-Lefert P. Interplay Between Innate Immunity and the Plant Microbiota. Annu Rev Phytopathol. Annual Reviews; 2017;55: 565–589. 10.1146/annurev-phyto-080516-035623 [DOI] [PubMed] [Google Scholar]
- 6.Haney CH, Samuel BS, Bush J, Ausubel FM, Gallo RL, Hooper LV., et al. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat Plants. Nature Publishing Group; 2015;1: 15051 10.1038/nplants.2015.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pickles BJ, Wilhelm R, Asay AK, Hahn AS, Simard SW, Mohn WW. Transfer of 13 C between paired Douglas-fir seedlings reveals plant kinship effects and uptake of exudates by ectomycorrhizas. New Phytol. 2016; 10.1111/nph.14325 [DOI] [PubMed] [Google Scholar]
- 8.Hernández M, Dumont MG, Yuan Q, Conrad R. Different bacterial populations associated with the roots and rhizosphere of rice incorporate plant-derived carbon. Appl Environ Microbiol. American Society for Microbiology; 2015;81: 2244–53. 10.1128/AEM.03209-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eissfeller V, Beyer F, Valtanen K, Hertel D, Maraun M, Polle A, et al. Incorporation of plant carbon and microbial nitrogen into the rhizosphere food web of beech and ash. Soil Biol Biochem. 2013;62: 76–81. 10.1016/j.soilbio.2013.03.002 [DOI] [Google Scholar]
- 10.Hu J, Wei Z, Friman V-P, Gu S-H, Wang X-F, Eisenhauer N, et al. Probiotic Diversity Enhances Rhizosphere Microbiome Function and Plant Disease Suppression. MBio. American Society for Microbiology; 2016;7: e01790–16. 10.1128/mBio.01790-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santhanam R, Luu VT, Weinhold A, Goldberg J, Oh Y, Baldwin IT. Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc Natl Acad Sci U S A. National Academy of Sciences; 2015;112: E5013–20. 10.1073/pnas.1505765112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM, et al. Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria. Science (80-). 2011;332. [DOI] [PubMed] [Google Scholar]
- 13.Shen Z, Ruan Y, Xue C, Zhong S, Li R, Shen Q. Soils naturally suppressive to banana Fusarium wilt disease harbor unique bacterial communities. Plant Soil. Springer International Publishing; 2015;393: 21–33. 10.1007/s11104-015-2474-9 [DOI] [Google Scholar]
- 14.Cha J-Y, Han S, Hong H-J, Cho H, Kim D, Kwon Y, et al. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J. Nature Publishing Group; 2016;10: 119–129. 10.1038/ismej.2015.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan Z, Druzhinina IS, Labbé J, Redman R, Qin Y, Rodriguez R, et al. Specialized Microbiome of a Halophyte and its Role in Helping Non-Host Plants to Withstand Salinity. Sci Rep. Nature Publishing Group; 2016;6: 32467 10.1038/srep32467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vargas L, Santa Brígida AB, Mota Filho JP, de Carvalho TG, Rojas CA, Vaneechoutte D, et al. Drought Tolerance Conferred to Sugarcane by Association with Gluconacetobacter diazotrophicus: A Transcriptomic View of Hormone Pathways. Yang H, editor. PLoS ONE. Public Library of Science; 2014;9: e114744 10.1371/journal.pone.0114744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiruma K, Gerlach N, Sacristán S, Nakano RT, Hacquard S, Kracher B, et al. Root Endophyte Colletotrichum tofieldiae Confers Plant Fitness Benefits that Are Phosphate Status Dependent. Cell. 2016;165: 464–474. 10.1016/j.cell.2016.02.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vessey JK. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil. Kluwer Academic Publishers; 2003;255: 571–586. 10.1023/A:1026037216893 [DOI] [Google Scholar]
- 19.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. Nature Research; 2012;488: 86–90. 10.1038/nature11237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeoh YK, Paungfoo-Lonhienne C, Dennis PG, Robinson N, Ragan MA, Schmidt S, et al. The core root microbiome of sugarcanes cultivated under varying nitrogen fertilizer application. Environ Microbiol. 2016;18: 1338–1351. 10.1111/1462-2920.12925 [DOI] [PubMed] [Google Scholar]
- 21.Edwards J, Johnson C, Santos-medellín C, Lurie E, Kumar N. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA. 2015; 10.1073/pnas.1414592112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci U S A. National Academy of Sciences; 2006;103: 626–31. 10.1073/pnas.0507535103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhalnina K, Dias R, de Quadros PD, Davis-Richardson A, Camargo FAO, Clark IM, et al. Soil pH Determines Microbial Diversity and Composition in the Park Grass Experiment. Microb Ecol. Springer US; 2015;69: 395–406. 10.1007/s00248-014-0530-2 [DOI] [PubMed] [Google Scholar]
- 24.Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. American Society for Microbiology; 2009;75: 5111–20. 10.1128/AEM.00335-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffiths RI, Thomson BC, James P, Bell T, Bailey M, Whiteley AS. The bacterial biogeography of British soils. Environ Microbiol. Blackwell Publishing Ltd; 2011;13: 1642–1654. 10.1111/j.1462-2920.2011.02480.x [DOI] [PubMed] [Google Scholar]
- 26.Santos-Medellín C, Edwards J, Liechty Z, Nguyen B, Sundaresan V. Drought Stress Results in a Compartment-Specific Restructuring of the Rice Root-Associated Microbiomes. MBio. American Society for Microbiology; 2017;8: e00764–17. 10.1128/mBio.00764-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naylor D, DeGraaf S, Purdom E, Coleman-Derr D. Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. Nature Publishing Group; 2017; 10.1038/ismej.2017.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angel R, Soares MIM, Ungar ED, Gillor O. Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME J. Nature Publishing Group; 2010;4: 553–563. 10.1038/ismej.2009.136 [DOI] [PubMed] [Google Scholar]
- 29.Xu L, Naylor D, Dong Z, Simmons T, Pierroz G, Hixson KK, et al. Drought delays development of the sorghum root microbiome and enriches for monoderm bacteria. Proc Natl Acad Sci U S A. National Academy of Sciences; 2018;115: E4284–E4293. 10.1073/pnas.1717308115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fitzpatrick CR, Copeland J, Wang PW, Guttman DS, Kotanen PM, Johnson MTJ. Assembly and ecological function of the root microbiome across angiosperm plant species. Proc Natl Acad Sci U S A. National Academy of Sciences; 2018;115: E1157–E1165. 10.1073/pnas.1717617115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramirez KS, Craine JM, Fierer N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob Chang Biol. 2012;18: 1918–1927. 10.1111/j.1365-2486.2012.02639.x [DOI] [Google Scholar]
- 32.Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. Nature Publishing Group; 2012;6: 1007–17. 10.1038/ismej.2011.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pantigoso HA, Manter DK, Vivanco JM. Phosphorus addition shifts the microbial community in the rhizosphere of blueberry (Vaccinium corymbosum L.). Rhizosphere. Elsevier; 2018;7: 1–7. 10.1016/J.RHISPH.2018.06.008 [DOI] [Google Scholar]
- 34.Yu P, Wang C, Baldauf JA, Tai H, Gutjahr C, Hochholdinger F, et al. Root type and soil phosphate determine the taxonomic landscape of colonizing fungi and the transcriptome of field-grown maize roots. New Phytol. Wiley/Blackwell (10.1111); 2018;217: 1240–1253. 10.1111/nph.14893 [DOI] [PubMed] [Google Scholar]
- 35.Leff JW, Jones SE, Prober SM, Barberán A, Borer ET, Firn JL, et al. Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc Natl Acad Sci U S A. National Academy of Sciences; 2015;112: 10967–72. 10.1073/pnas.1508382112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fabiańska I, Gerlach N, Almario J, Bucher M. Plant-mediated effects of soil phosphorus on the root-associated fungal microbiota in Arabidopsis thaliana. New Phytol. Wiley/Blackwell (10.1111); 2018; 10.1111/nph.15538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raghothama KG. PHOSPHATE ACQUISITION. Annu Rev Plant Physiol Plant Mol Biol. Annual Reviews; 1999;50: 665–693. 10.1146/annurev.arplant.50.1.665 [DOI] [PubMed] [Google Scholar]
- 38.Herrera Paredes S, Gao T, Law TF, Finkel OM, Mucyn T, Teixeira PJPL, et al. Design of synthetic bacterial communities for predictable plant phenotypes. Kemen E, editor. PLoS Biol. Public Library of Science; 2018;16: e2003962 10.1371/journal.pbio.2003962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziegler J, Schmidt S, Chutia R, Müller J, Böttcher C, Strehmel N, et al. Non-targeted profiling of semi-polar metabolites in Arabidopsis root exudates uncovers a role for coumarin secretion and lignification during the local response to phosphate limitation. J Exp Bot. Oxford University Press; 2016;67: 1421 10.1093/jxb/erv539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, et al. A Central Regulatory System Largely Controls Transcriptional Activation and Repression Responses to Phosphate Starvation in Arabidopsis. Ecker JR, editor. PLoS Genet. Public Library of Science; 2010;6: e1001102 10.1371/journal.pgen.1001102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez E, Solano R, Rubio V, Leyva A, Paz-Ares J. PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 Is a Plant-Specific SEC12-Related Protein That Enables the Endoplasmic Reticulum Exit of a High-Affinity Phosphate Transporter in Arabidopsis. PLANT CELL ONLINE. 2005;17: 3500–3512. 10.1105/tpc.105.036640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gransee A, Merbach W. Phosphorus dynamics in a long-term P fertilization trial on Luvic Phaeozem at Halle. J Plant Nutr Soil Sci. Wiley-Blackwell; 2000;163: 353–357. [DOI] [Google Scholar]
- 43.Robbins C, Thiergart T, Hacquard S, Garrido-Oter R, Gans W, Peiter E, et al. Root-Associated Bacterial and Fungal Community Profiles of Arabidopsis thaliana Are Robust Across Contrasting Soil P Levels. Phytobiomes. Phytobiomes; 2018;2: PBIOMES-09-17-0. 10.1094/PBIOMES-09-17-0042-R [DOI] [Google Scholar]
- 44.Phillipson BA. PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 Is a Plant-Specific SEC12-Related Protein That Enables the Endoplasmic Reticulum Exit of a High-Affinity Phosphate Transporter in Arabidopsis. Plant Cell Online. American Society of Plant Biologists; 2005;17: 3500–3512. 10.1105/tpc.105.036640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, Wassmann R, et al. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. Nature Publishing Group; 2012;6: 1378–1390. 10.1038/ismej.2011.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature. Nature Publishing Group; 2015;528: 364–369. 10.1038/nature16192 [DOI] [PubMed] [Google Scholar]
- 47.Atamna-Ismaeel N, Finkel OM, Glaser F, Sharon I, Schneider R, Post AF, et al. Microbial rhodopsins on leaf surfaces of terrestrial plants. Environ Microbiol. 2012;14 10.1111/j.1462-2920.2011.02554.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atamna-Ismaeel N, Finkel O, Glaser F, von Mering C, Vorholt JA, Koblížek M, et al. Bacterial anoxygenic photosynthesis on plant leaf surfaces. Environ Microbiol Rep. 2012;4 10.1111/j.1758-2229.2011.00323.x [DOI] [PubMed] [Google Scholar]
- 49.Finkel OM, Delmont TO, Post AF, Belkin S. Metagenomic signatures of bacterial adaptation to life in the phyllosphere of a salt-secreting desert tree. Appl Environ Microbiol. 2016;82 10.1128/AEM.00483-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levy A, Salas Gonzalez I, Mittelviefhaus M, Clingenpeel S, Herrera Paredes S, Miao J, et al. Genomic features of bacterial adaptation to plants. Nat Genet. Nature Publishing Group; 2018;50: 138–150. 10.1038/s41588-017-0012-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kassen R, Buckling A, Bell G, Rainey PB. Diversity peaks at intermediate productivity in a laboratory microcosm. Nature. Nature Publishing Group; 2000;406: 508–512. 10.1038/35020060 [DOI] [PubMed] [Google Scholar]
- 52.Tilman D. Resource competition and community structure. Princeton University Press; 1982. [PubMed] [Google Scholar]
- 53.Baxter IR, Vitek O, Lahner B, Muthukumar B, Borghi M, Morrissey J, et al. The leaf ionome as a multivariable system to detect a plant’s physiological status. Proc Natl Acad Sci. 2008;105: 12081–12086. 10.1073/pnas.0804175105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Durán P, Thiergart T, Garrido-Oter R, Agler M, Kemen E, Schulze-Lefert P, et al. Microbial Interkingdom Interactions in Roots Promote Arabidopsis Survival. Cell. Elsevier; 2018;175: 973-983.e14. 10.1016/j.cell.2018.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pant B-D, Pant P, Erban A, Huhman D, Kopka J, Scheible W-R. Identification of primary and secondary metabolites with phosphorus status-dependent abundance in A rabidopsis, and of the transcription factor PHR1 as a major regulator of metabolic changes during phosphorus limitation. Plant Cell Environ. John Wiley & Sons, Ltd (10.1111); 2015;38: 172–187. 10.1111/pce.12378 [DOI] [PubMed] [Google Scholar]
- 56.Huang AC, Jiang T, Liu Y-X, Bai Y-C, Reed J, Qu B, et al. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science. American Association for the Advancement of Science; 2019;364: eaau6389. 10.1126/science.aau6389 [DOI] [PubMed] [Google Scholar]
- 57.Stringlis IA, Yu K, Feussner K, de Jonge R, Van Bentum S, Van Verk MC, et al. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc Natl Acad Sci U S A. National Academy of Sciences; 2018;115: E5213–E5222. 10.1073/pnas.1722335115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mullan A, Quinn JP, McGrath JW. Enhanced phosphate uptake and polyphosphate accumulation in Burkholderia cepacia grown under low-pH conditions. Microb Ecol. Springer-Verlag; 2002;44: 69–77. 10.1007/s00248-002-3004-x [DOI] [PubMed] [Google Scholar]
- 59.Merbach W, Garz J, Schliephake W, Stumpe H, Schmidt L. The long-term fertilization experiments in Halle (Saale), Germany—Introduction and survey. J Plant Nutr Soil Sci. Wiley-Blackwell; 2000;163: 629–638. [DOI] [Google Scholar]
- 60.Ames BN. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. Academic Press; 1966;8: 115–118. 10.1016/0076-6879(66)08014-5 [DOI] [Google Scholar]
- 61.Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal Biochem. Academic Press; 1987;163: 16–20. 10.1016/0003-2697(87)90086-8 [DOI] [PubMed] [Google Scholar]
- 62.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. Nature Publishing Group; 2012;9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL. Practical innovations for high-throughput amplicon sequencing. Nat Methods. Nature Publishing Group; 2013;10: 999–1002. 10.1038/nmeth.2634 [DOI] [PubMed] [Google Scholar]
- 64.Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol Ecol. Wiley/Blackwell (10.1111); 1993;2: 113–118. 10.1111/j.1365-294x.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- 65.White T.J., Bruns T.D, Lee S.B., Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics PCR protocols: A Guide to Methods and Applications. Academic Press; 1990. pp. 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- 66.Taiyun Wei M, Taiyun Wei cre A, Simko aut V, Levy ctb M, Xie ctb Y, Jin ctb Y, et al. Package “corrplot” Title Visualization of a Correlation Matrix. 2017.
- 67.Yourstone SM, Lundberg DS, Dangl JL, Jones CD. MT-Toolbox: improved amplicon sequencing using molecule tags. BMC Bioinformatics. BioMed Central; 2014;15: 284 10.1186/1471-2105-15-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joshi N, Fass J. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.33) [Software]. Available at https://github.com/najoshi/sickle. 2011.
- 69.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. Nature Publishing Group; 2016;13: 581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. American Society for Microbiology; 2009;75: 7537–41. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nilsson RH, Larsson K-H, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, et al. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. Narnia; 2019;47: D259–D264. 10.1093/nar/gky1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gonzalez IS. isaisg/ohchibi: iskali. 2019; 10.5281/ZENODO.2593691 [DOI] [Google Scholar]
- 73.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’hara RB, et al. Package “vegan.” 2015.
- 74.Package “emmeans” Type Package Title Estimated Marginal Means, aka Least-Squares Means. 2019; 10.1080/00031305.1980.10483031 [DOI]
- 75.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. BioMed Central; 2014;15: 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44: W242–W245. 10.1093/nar/gkw290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. American Association for Cancer Research; 1967;27: 209–20. [PubMed] [Google Scholar]
- 78.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. Oxford University Press; 2010;26: 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 79.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10: 996–998. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- 80.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl Environ Microbiol. 2007;73: 5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. American Society for Microbiology; 2006;72: 5069–72. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wheeler TJ, Eddy SR. nhmmer: DNA homology search with profile HMMs. Bioinformatics. Oxford University Press; 2013;29: 2487–2489. 10.1093/bioinformatics/btt403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. Oxford University Press; 2013;30: 772–80. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. Oxford University Press; 2009;25: 1972–3. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Price MN, Dehal PS, Arkin AP. FastTree 2 –Approximately Maximum-Likelihood Trees for Large Alignments. Poon AFY, editor. PLoS ONE. Public Library of Science; 2010;5: e9490 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harvey, Pagel M. The comparative method in evolutionary biology. 1991;
- 87.Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. John Wiley & Sons, Ltd (10.1111); 2012;3: 217–223. 10.1111/j.2041-210X.2011.00169.x [DOI] [Google Scholar]
- 88.Package “egg” Type Package Title Extensions for “ggplot2”: Custom Geom, Plot Alignment, Symmetrised Scale, and Fixed Panel Size. 2018.
- 89.Andrews S. Babraham Bioinformatics—FastQC A Quality Control tool for High Throughput Sequence Data [Internet]. 2018. pp. 3–5. doi:v.0.11.5 [Google Scholar]
- 90.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. Oxford University Press; 2014;30: 2114–20. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, et al. The arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. genesis. John Wiley & Sons, Ltd; 2015;53: 474–485. 10.1002/dvg.22877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. NIH Public Access; 2015;12: 357–60. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. Oxford University Press; 2013;41: e108 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. Narnia; 2016;32: 3047–3048. 10.1093/bioinformatics/btw354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Team RDC. R: A language and environment for statistical computing. 2011; [Google Scholar]
- 96.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS. Mary Ann Liebert, Inc.; 2012;16: 284 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Heat map showing the all versus all pairwise Pearson correlation coefficient calculated between the quantified phenotypes associated with the PSR: shoot area, shoot fresh weight, and shoot free Pi accumulation. (B) Box plot showing the distribution of the shoot area measured across the P gradient within each of the 3 genotypes. (C) Boxplot showing the distribution of shoot fresh weight measured across the P gradient within each of the 3 genotypes. (D) Box plot showing the shoot Pi accumulation across the 3 genotypes. Letters represent the results of the post hoc test. (E) Box plots displaying the average expression of 193 PSR marker genes across the low and low+P samples in each of the 3 genotypes tested. (F) Scatter plot showing the relationship between the standardized average phosphate accumulation in leaves (x-axis) and the average standardized expression of 193 PSR marker genes (y-axis). The p-value and R value were calculated according to Pearson’s product moment correlation coefficient. P, phosphorus; Pi, orthophosphate; PSR, phosphate starvation response.
(TIF)
(A, B) Box plots showing the distribution of the alpha diversity (Shannon diversity index) across all levels of P in the soil for bacteria (A) and fungi (B). (C, D) PERMANOVA results in which the effect of the 3 variables (fraction, genotype, and soil P) and their interaction on the assembly of the bacterial (C) and fungal (D) communities were tested. (E) Relative abundance profiles of the main bacterial phyla in the 3 variables (fraction, genotype, and soil P) across all levels of P in the soils. (F) Relative abundance profiles of the main fungal orders in the 3 variables (fraction, genotype, and soil P) across all the levels of P in the soils. P, phosphorus; PERMANOVA, Permutational Multivariate Analysis of Variance.
(TIF)
(A, B, C) Correlation plots between Bray-Curtis distance matrices calculated for bacteria within soil treatments, root, and shoot fractions. The R and p-values were calculated using Mantel tests. (A) Correlation plot of soil versus root. (B) Correlation plot of soil versus shoot. (C) Correlation plot of root versus shoot. (D, E, F) Correlation plots between Bray-Curtis distance matrices calculated for fungi within soil treatments, root, and shoot fractions. The R and p-values were calculated using Mantel tests. R and p-values colored in red were calculated excluding the cloud of large distances appearing in graphs panels D and E. (D) Correlation plot of soil versus root. (E) Correlation plot of soil versus shoot. (F) Correlation plot of root versus shoot.
(TIF)
(A, B) PERMANOVA results showing the influence of the plant genotype and soil P concentration and their interaction on the assembly of the root (A) bacterial and (B) fungal communities. (C, D) CAP showing the effect of plant genotype and P content in the soil over the shoot (C) bacterial and (D) fungal communities. The p-value and R2 values in each plot are derived from a PERMANOVA model and correspond to the genotype and soil P term, respectively. (E, F) CAP showing the influence of genotype and P on the soil (E) bacterial and (F) fungal communities. Note smaller number of points in bacterial soil samples. The p-value and R2 values in each plot are derived from a PERMANOVA model and correspond to the genotype and soil P term, respectively. CAP, canonical analysis of principal coordinates; P, phosphorus; PERMANOVA, Permutational Multivariate Analysis of Variance.
(TIF)
(A) Correlation heat map of the 13 edaphic factors reported in the work by Robbins and colleagues [43]. (B) Bar plot displaying the amount of variance in the edaphic factor matrix explained by PC. (C) Bar plot depicting the contribution of the different edaphic factors to the first 3 PC. Colors denote the direction of the variable in PCA space. (D) Proportion of the variance explained for the different variables in models including (M2) and excluding (M1) edaphic factors for bacteria (left) and fungi (right). Only variables with a statistically significant effect are shown. P, phosphorus; PC, principal component; PCA, principal component analysis.
(TIF)
(A) Box plots displaying the primary root elongation of plants grown in a gradient of Pi concentrations in sterile conditions or with the SynCom. A t test was used for each Pi treatment to estimate differences between SynCom-treated and uninoculated plants. (B) Average expression of the 193 PSR markers genes in low (50 μM) and high (1,000 μM) Pi conditions within SynCom-treated and uninoculated plants. (C) Average expression of the 123 genes from Cluster 1 (Fig 1B) in low (50 μM) and high (1,000 μM) Pi conditions within SynCom-treated and uninoculated plants. (D) PERMANOVA model results showing the influence of the 2 variables (fraction and Pi concentration) and their interaction on the assembly of the bacterial community in the plant. (E, F, G) Correlation plots between Bray-Curtis distance matrices calculated for bacterial profile within agar, root, and shoot fractions. The R and p-values were calculated using Mantel tests. (E) Correlation plot of agar versus root. (F) Correlation plot of agar versus shoot. (G) Correlation plot of root versus shoot. PERMANOVA, Permutational Multivariate Analysis of Variance; Pi, orthophosphate; PSR, phosphate starvation response; SynCom, synthetic community.
(TIF)
(A, B) CAP showing the influence of Pi concentration in the media on the bacterial communities in the (A) plant shoot and (B) agar. The bar graphs to the left of each plot depict the percentage of variability explained by statistically significant (p < 0.05) variables based on a PERMANOVA model. (C, D, E) PERMANOVA model results showing the influence of Pi concentration on the assembly of the bacterial community in (C) roots, (D) shoot, and (E) agar. CAP, canonical analysis of principal coordinates; PERMANOVA, Permutational Multivariate Analysis of Variance; Pi, orthophosphate.
(TIF)
(A) Scatter plot (volcano plot) showing the results of the GLM interaction model between fraction and Pi concentration. The axes of the plot represent the output of the statistical test. The x-axis is the transformed q-value and the y-axis the log2 fold change. Each dot in the scatter plot represents a USeq. USeqs that showed a statistically significant fraction:Pi interaction are colored in red. USeqs genus and ID are displayed. The top right quadrant represents USeqs that are enriched in the plant tissues under low Pi conditions. (B, C) Relative abundance of Burkholderia Useqs 16 (B) and 30 (C) that exhibits a statistically significant (q < 0.1) Pi enrichment between the plant fractions and the agar fraction. The middle dot of each strip bar corresponds to the mean of that particular condition; the range of the strip bar corresponds to the 95% confidence interval of the mean. GLM, generalized linear model; Pi, orthophosphate; USeq, unique sequence.
(TIF)
Box plots showing shoot circumference in plants exposed to different SynComs across 3 phosphate treatments. Statistically significant differences among SynCom treatments were computed within each phosphate treatment separately using an ANOVA model. Letters represent the results of the post hoc test. SynCom, synthetic community.
(TIF)
CAP showing the influence of plant genotypes (A) and agar Pi concentration (B) over the bacterial SynCom in the root. The p-value and R2 values inside each plot are derived from a PERMANOVA model and correspond to the genotype and Pi terms, respectively. CAP, canonical analysis of principal coordinates; PERMANOVA, Permutational Multivariate Analysis of Variance; Pi, orthophosphate; SynCom, synthetic community.
(TIF)
Pi, orthophosphate.
(XLSX)
(XLSX)
ASV, amplicon sequence variant; GLM, generalized linear model.
(XLSX)
ASV, amplicon sequence variant; GLM, generalized linear model.
(XLSX)
ASV, amplicon sequence variant; GLM, generalized linear model; P, phosphorus; Pi, orthophosphate.
(XLSX)
ASV, amplicon sequence variant; GLM, generalized linear model; P, phosphorus; Pi, orthophosphate.
(XLSX)
USeq, unique sequence.
(XLSX)
(XLSX)
GLM, generalized linear model.
(XLSX)
GLM, generalized linear model; Pi, orthophosphate.
(XLSX)
(XLSX)
(ZIP)
Data Availability Statement
The raw reads from the amplicon sequencing can be downloaded from the NCBI Sequence Read Archive (Project PRJNA531340). The raw reads for the RNA-Seq experiments can be downloaded from the omnibus accession GSE129396. In addition to the the raw reads, all the data structures and scripts used to analyze the data and generate figures can be downloaded from the following github repository: https://github.com/isaisg/hallepi An interactive visualization of the fraction enrichment profiles from the natural soil experiments can be accesed via the iTOL user hallepi or the following links: https://itol.embl.de/tree/1522316254174701551987253https://itol.embl.de/tree/13656172137464831571097084 Data underlying all figures in the manuscript is included within S1 Data.