Abstract
Objectives
We proposed the application of a multivariate cross‐sectional framework based on a combination of a variable selection method and a multiple factor analysis (MFA) in order to identify complex meaningful biological signals related to attention‐deficit/hyperactivity disorder (ADHD) symptoms and hyperactivity/inattention domains.
Methods
The study included 135 children from the general population with genomic and neuroimaging data. ADHD symptoms were assessed using a questionnaire based on ADHD‐DSM‐IV criteria. In all analyses, the raw sum scores of the hyperactivity and inattention domains and total ADHD were used. The analytical framework comprised two steps. First, zero‐inflated negative binomial linear model via penalized maximum likelihood (LASSO‐ZINB) was performed. Second, the most predictive features obtained with LASSO‐ZINB were used as input for the MFA.
Results
We observed significant relationships between ADHD symptoms and hyperactivity and inattention domains with white matter, gray matter regions, and cerebellum, as well as with loci within chromosome 1.
Conclusions
Multivariate methods can be used to advance the neurobiological characterization of complex diseases, improving the statistical power with respect to univariate methods, allowing the identification of meaningful biological signals in Imaging Genetic studies.
Keywords: ADHD, Imaging Genetics, LASSO‐ZINB, multiple factor analysis, neurogenetics
1. INTRODUCTION
Attention‐deficit/hyperactivity disorder (ADHD) is characterized by hyperactivity and inattention symptoms with a clinical prevalence estimated between 5% and 7% (Cardo, Servera, & Llobera, 2017). These symptoms have a complex and polygenic etiology where multiple genes of small effect, altered brain development, and exposure to environmental risk factors play a role in the development of the disorder (Arcos‐Burgos, Vélez, Solomon, & Muenke, 2012; Polanczyk, de Lima, Horta, Biederman, & Rohde, 2007). Neuroimaging is commonly used to quantify brain structure and function, being a useful tool to unravel the complex etiology of ADHD symptoms. Neuroimaging data interpretation and understanding greatly improves with genetic studies. Imaging Genetics (IG) studies seek to analyze the combined effect of multiple genetic and brain volume variations as potential biomarkers of disease, providing a better understanding of mechanisms of neurodevelopmental domains (Bogdan et al., 2017).
However, the dimensionality of the data in IG studies represents the principal handicap in the acquisition of analytical perspectives on neurodevelopmental domains and complex neurological diseases, amplifying existing issues of reliability and interpretability of the results. Conventional strategies often test millions of single nucleotide polymorphisms (SNPs) for association with a given neuroimaging‐based measure, or, conversely, multiple neuroimaging‐based measures are tested for association with candidate genetic variants. These strategies ignore potential combined effects between genetic and neuroimaging correlates and reduces statistical power due to multiple testing correction. Several pioneering studies have proposed different analytical procedures to assess neurological and biological patterns of ADHD symptoms, by combining SNPs and magnetic resonance imaging (MRI)‐derived features, such as gene‐set analysis (Mous et al., 2015; Vilor‐Tejedor, Gonzalez, & Calle, 2015) and parallel independent component analysis (Khadka et al., 2016). These methods have the advantage of integrating and capturing underlying multivariate relationships but commonly perform complex dimensionality reduction procedures for which it is difficult to discern the individual information patterns related to the phenotype of interest.
The extraction of informative genetic and imaging‐based features for in IG studies often relies on variable selection. This selection is based on an initial identification of the most informative variables, and the integration of different sources of data through multivariate multiscale techniques, such as multiple factor analysis (MFA). The main benefits of this process are twofold: first, it removes any redundant features, which may improve prediction accuracy in addition to supporting the interpretability of results. Second, it helps in the generation of post hoc inferences.
The use of variable selection methods can also be seen as an alternative strategy of genome‐wide association studies for the identification of informative genetic markers related to neurodevelopmental domains, reducing the dimensionality of the data. In this context, in psychiatric research, it is uncommon the use of dichotomous status—one that indicates whether the disorder is present or absent. However, such dichotomous classification does not capture accurately the variability observed in the general population, because individuals who present extreme score values are grouped together with individuals whose symptoms are just above the diagnosis threshold. This dichotomous approach contrasts with the notion of a continuum distribution of the symptoms describing a dimensional spectrum, ranging from normal to dysfunctional. In this regard, when analyzing ADHD symptoms as a continuous score, the phenotype often presents skewed and overdispersed distribution, and, to date, little attention has been paid to their efficient statistical modeling. Moreover, still with the development of newly advanced methodologies that allow novel analytical possibilities, the vast majority of these methods have not yet been used in the IG field of ADHD (Vilor‐Tejedor, Cáceres, Pujol, Sunyer, & González, 2016).
In order to overcome this methodological scarcity, we proposed an analytical framework, which consists of a two‐step analysis combining a variable selection strategy designed for a count data distribution, along with a dimensionality reduction approach. In the first step, we performed separate selection of most relevant SNPs and brain regions of interest (ROIs) for ADHD via penalized zero‐inflated negative binomial regression analysis (LASSO‐ZINB; Mallick & Tiwari, 2016). In the second step, we jointly evaluated preselected ROIs and SNPs by applying multiple factor analysis (MFA) to determine significant genetic and imaging correlates of ADHD variability (Escofier & Pagès, 1990).
The main aim of the application of our proposed analytical framework was to identify the most informative genetic and brain structure factors, which in turn, had better explain the underlying structure of total ADHD symptoms and hyperactivity and inattention domains.
2. MATERIALS AND METHODS
2.1. Participants
This study is based on a subsample of the BRrain dEvelopment and Air pollution ultrafine particles in school childrEn (BREATHE) project. This population‐based cohort of primary schoolchildren was designed to analyze the adverse effects of traffic‐related air pollution on neurodevelopment. As part of the BREATHE project design, 38 schools located in the metropolitan area of Barcelona were selected based on modeled air pollution levels to achieve maximum exposure contrast. Further details on study design and sociodemographic descriptive for low and high polluted schools can be found elsewhere (Sunyer et al., 2015). From those schools, a total of 2,875 children aged 7 to 10 years and enrolled in the 2nd, 3rd, and 4th primary grades were recruited. Genotypic, neuroimaging, and behavioural data were available for a subset of 135 individuals. All the parents or legal guardians of the study subjects provided signed informed consent as approved by the Ethics Committees of the centers involved in the study.
2.2. Total ADHD symptoms and hyperactivity and inattention domains
ADHD symptoms were collected using a questionnaire based on the ADHD criteria of the Diagnostic and Statistical Manual of Mental Disorders, fourth edition (4th ed., DSM–IV–TR; American Psychiatric Association, 1994), completed by teachers. ADHD‐DSM–IV consists of a list of 18 symptoms, assessing two separate symptom groups: inattention (nine symptoms) and hyperactivity/impulsivity (nine symptoms). Each ADHD symptom was rated in a 4‐point scale of frequency from never or rarely (0) to very often (3). The continuous measure of total ADHD symptoms was calculated as the sum of the score of each item. This measure ranges from 0 to 54. Hyperactivity and inattention domains were calculated as the sum of the scores of the nine items corresponding to each dimension. In all analyses, total ADHD symptoms and hyperactivity and inattention domains were treated as continuous outcome variables modeled by zero‐inflated negative‐binomial distributions, previously shown to improve the statistical modeling of ADHD studies (Vilor‐Tejedor, Alemany, et al., 2016).
2.3. Genomic assessment
Genome‐wide genotyping was performed using the HumanCore BeadChip WG‐330‐1101 (Illumina) at the Spanish National Genotyping Centre (CEGEN) ‐ Spanish National Cancer Research Centre (CNIO). Genotype calling was done using the GeneTrain2.0 algorithm (with a default threshold of 0.15) based on HapMap clusters implemented in the GenomeStudio software. PLINK was used for the genetic data quality control (Purcell et al., 2007). We applied the following sample quality control thresholds: sample call rate > 97% (N = 3 exclusions) and heterozygosity 4 SD (N = 5 exclusions). Then, we checked sex discordances (N = 18 exclusions, 1%) and relatedness (N = 80 exclusions: 1 twin, 32 siblings, 39 cousins, 8 incongruent sibling's couples). In total, we excluded 106 subjects (6%). Genetic variants were filtered by SNP call rate > 95%, Minor alelle frequency (MAF) > 1%, and Hardy‐Weinberg equilibrium. (HWE) pvalue > 1.10E‐6 (N = 58,827 exclusions, 19.68%). We additionally used principal component analysis to identify population structure patterns. The two first components showed a homogeneous cluster indicating that it is not necessary to exclude any sample based on this criterium. The final genetic data set included 246,103 SNPs. A full description of the genotyping and quality control procedures can be find elsewhere (Alemany et al., 2016). Moreover, a priori linkage disequilibrium‐based SNP pruning was performed. The pruning procedure consisted on calculating linkage disequilibrium (LD) between each pair of SNPs considering a windows of 50 SNPs. One of a pair of SNPs with an LD greater than 0.2 were removed, shifting then the windows 5 SNPs forward and repeating the procedure. This procedure is extensively documented in Purcell et al. (2007). The pruned genetic data set included 70,707 SNPs.
2.4. Neuroimaging assessment
MRI of brain anatomy was performed using a 1.5 Tesla Signa Excite system (General Electric, Milwaukee, WI, USA) equipped with an eight‐channel phased‐array head coil and single‐shot echo planar imaging software. High‐resolution three‐dimensional anatomical images were obtained using an axial T1‐weighted three‐dimensional fast spoiled gradient recalled acquisition in the steady state inversion recovery‐prepared sequence. A total of 134 contiguous slices were acquired with repetition time of 11.9 ms; echo time of 4.2 ms; flip angle of 15°; field of view of 30 cm; 256 × 256 pixel matrixes; and slice thickness of 1.2 mm. All the anatomical images were visually inspected, and subjects with poor quality images were discarded. Cortical reconstruction and volumetric segmentation were carried out using FreeSurfer tool (http://surfer.nmr.mgh.harvard.edu/). Specifically in this study, measures of 42 subcortical structures obtained from FreeSurfer automatic segmentation were used. For each isolated structure, the program estimates the absolute volume in mm3. No adjustment for global volumes (e.g., total brain or intracranial volume) was used. Processing steps included removal of non‐brain tissue, segmentation of the subcortical white matter (WM) and deep gray matter (GM) volumetric structures in native space, tessellation of the GM and WM boundary, registration to a spherical atlas which is based on individual cortical folding patterns to match cortical geometry across subjects and creation of a variety of surface based data. More details can be found in Mortamais et al. (2017) and Pujol et al. (2016).
2.5. Statistical analysis
A two‐step procedure was conducted (Figure 1). In the first step, we performed variable selection based on a count data distribution to select SNPs associated with each ADHD dimension (total ADHD symptoms and hyperactivity and inattention domains) fitting penalized zero inflated negative binomial regression analysis (LASSO‐ZINB) models. LASSO is a modeling method that shrinks the coefficients of uninformative variables towards zero, thus, performing variable selection on strongest effects. Cross‐validated log‐likelihood values for the shrinkage parameter (λ) were computed though the mpath R package (Mallick & Tiwari, 2016). These parameters are those used to control the amount of shrinkage that is applied to the estimates in the penalized zero‐inflated negative binomial regression models. We, therefore, selected SNPs with regression coefficients different from zero. LASSO‐ZINB models were computed with mpath package of R. At the second step of analysis, we computed MFA to determine the main genetic and brain structures influencing ADHD domains variability. All SNPs selected in the previous step were included in the multivariate analysis. MFA was used to estimate the relationships among genetic and neuroimaging correlates in order to identify concurrent changes in total ADHD, inattention, and hyperactivity dimensions. Specifically, MFA standardizes variables in each predefined block of data and calculates the global axes, which are the linear combination of original parameters that maximize the global data variance. Hence, MFA highlights the most significant features and data structures. MFA was computed using the FactoMineR R package. All models at both steps were adjusted for age, sex, school, and the first 10 principal components.
Figure 1.
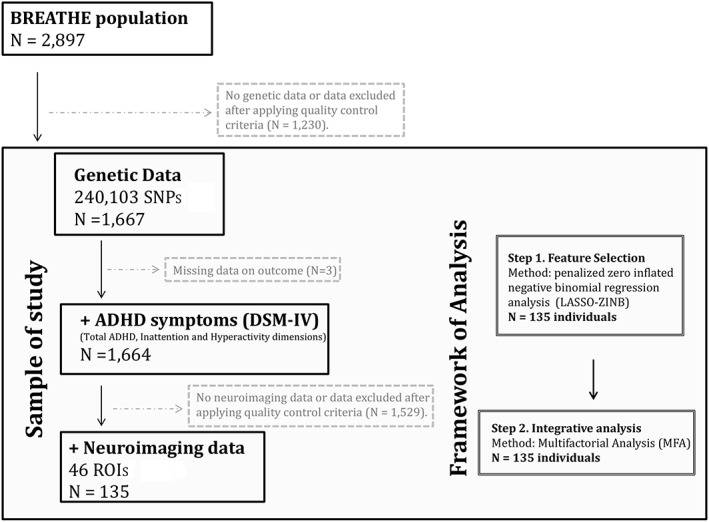
Flow chart depicting the final sample size for each outcome analyzed and the analytical procedure. Solid lines and boxes represent individuals remaining in the study. Dashed lines and boxes represent individuals excluded. Reason and number of individuals excluded are indicated in dashed boxes. Double boxes indicate the analytical procedure performed. SNPs: single nucleotide polymorphisms; ROIs: brain regions of interest; ADHD: attention‐deficit/hyperactivity disorder
3. RESULTS
3.1. Descriptives
Table 1 shows descriptive of age, sex, and the mean, standard deviation, and range scores for total ADHD symptoms, hyperactivity domain, and inattention domain, for the whole sample and the neuroimaging subsample. Samples were balanced for age and gender. We also showed correlation patterns (Pearson correlation statistics) between total ADHD symptoms and inattention and hyperactivity dimensions. The three outcomes analyzed are highly correlated (r > 0.6; p < 2e‐16). Figure 2 shows the empirical distributions of ADHD symptoms and hyperactivity and inattention domains. We can see that the distribution of the data is positively skewed because a high proportion of the individuals have zero count scores. Table S1 summarizes the cerebral structures analyzed in the study. Figure 3 shows the pattern of correlations (Pearson correlation statistics) among the volumes for all the selected anatomical structures. Brain structures highly correlated in size across subjects (i.e., structural covariance) included perivascular spaces with basal ganglia and WM with cingulate cortex. In addition, we specifically identified a negative correlation between perivascular spaces and cerebral ventricles. Descriptive results are in general agreement with previous studies (Lukoshe, White, Schmidt, van der Lugt, & Hokken‐Koelega, 2013).
Table 1.
Descriptive data of the sample
| N = 135 (IG sample) | |
|---|---|
| Age in years, mean (SD), range | 9.07 (0.82) 7.6–11.0 |
| Sex distribution (F/M) | 65/70 (48.15%) |
| DSM‐IV ADHD symptoms | |
| Raw score, mean (SD) | 7.78 (9.02) |
| Range | 0–42 |
| Wilcoxon test sex differences, pvalue | W = 2,727; p = 1.43e‐3 |
| DSM‐IV inattention domain | |
| Raw score, mean (SD) | 4.56 (4.53) |
| Range | 0–24 |
| Wilcoxon test sex differences, pvalue | W = 2,849; p = 0.043 |
| DSM‐IV hyperactivity domain | |
| Raw score, mean (SD) | 3.22 (5.76) |
| Range | 0–23 |
| Wilcoxon test sex differences, pvalue | W = 2,751; p = 0.0014 |
Note. SDQ: Strengths and Difficulties Questionnaire; ADHD: attention‐deficit/hyperactivity disorder; SD: standard deviation; IG: Imaging Genetic sample.
Figure 2.
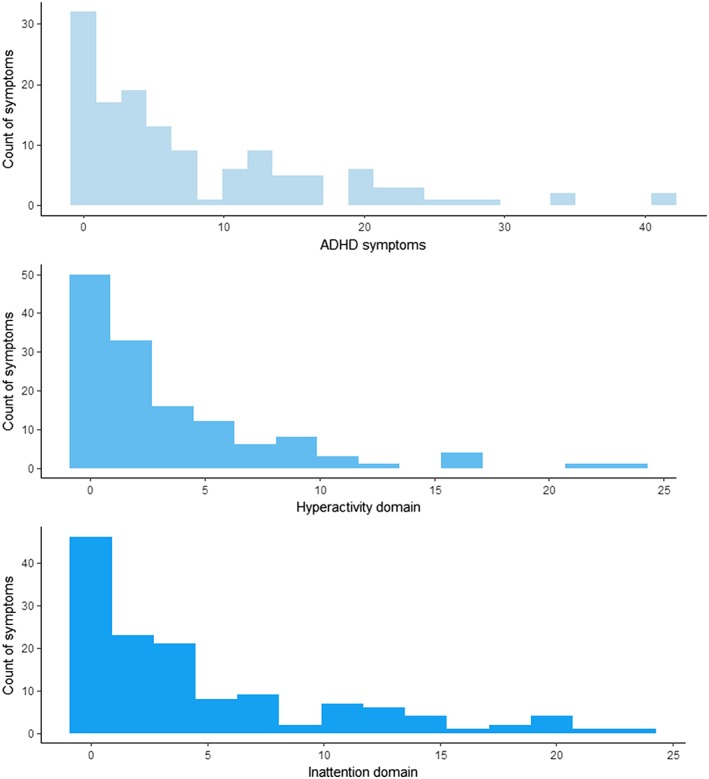
Observed frequency distribution of attention‐deficit/hyperactivity disorder (ADHD) symptoms and hyperactivity and inattention domains
Figure 3.
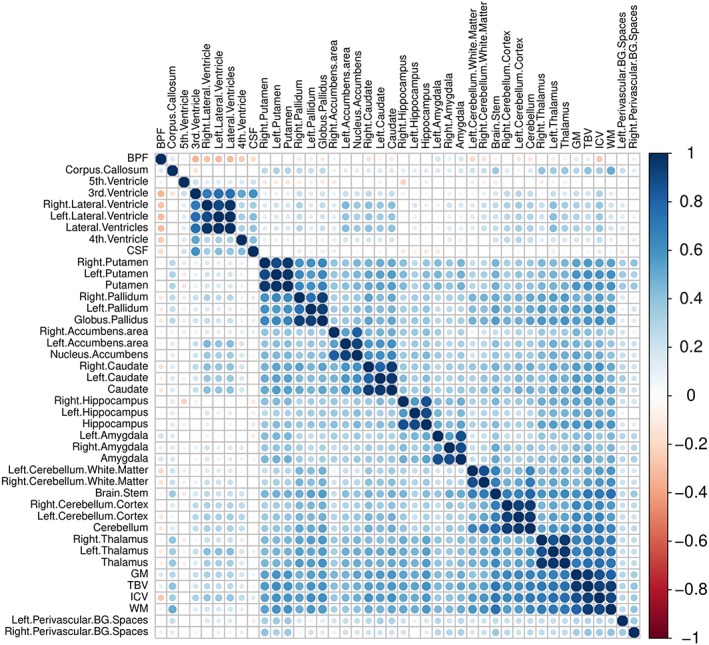
Correlation heatmap showing pair correlations across brain structures of BREATHE project subsample, with blue color indicating positive correlations and red color indicating negative correlations. 3rd.Ventricle: third ventricle; 4th.Ventricle: fourth ventricle; CSF: cerebrospinal fluid; 5th.Ventricle: septum pellucidum; ICV: total intracranial volume; WM: white matter; TBV: total brain volume; BPF: brain parenchymal fraction; GM: gray matter; Perivascular.BG.Spaces: basal ganglia perivascular spaces. Abbreviations for all brain structures are shown in Table S5
3.2. Step 1. Feature selection
LASSO‐ZINB selected 694 SNPs and 17 ROIs for total ADHD symptoms, 1,849 SNPs and 8 ROIs for hyperactivity domain, and 683 SNPs and 10 ROIs for inattention dimension.
3.3. Step 2. Multiple factor analysis
By applying multiple factor analysis (MFA), we found that the total variability explained by the three first principal components is 6.7% for total ADHD symptoms, 36.9% for hyperactivity domain, and 7.4% for inattention domain. Cumulative explained variance considering the whole number of components is reported in detail in Tables S2–S4.
For total ADHD symptoms, the first dimension of the MFA is mainly positively correlated to specific brain structures, whereas the second and third dimensions were mainly correlated with genetic components. The first dimension was correlated to WM and GM volumes (adj_pvalue = 1.65E‐57; adj_pvalue = 4.71E‐52; Figure S1), while the rs763173 and the rs12563394 within the CAMTA1 (Calmodulin Binding Transcription Activator 1), and the rs301806 within the RERE (Arginine/Glutamic Acid Dipeptide Repeats) genes, respectively were the top SNPs negatively correlated to the second dimension of the MFA (adj_pvalue = 3.28E‐07; adj_pvalue = 3.28E‐07, adj_pvalue = 1.25E‐06). Moreover, the rs11808641 and rs6683453 were the top intergenic SNPs positively correlated with the third dimension of the MFA (adj_pvalue = 1.45E‐07; adj_pvalue = 7.84E‐07), and the rs4654432, the top intronic variant negatively correlated (adj_pvalue = 7.84E‐07; Table 2). For hyperactivity domain, the first dimension was mainly positively correlated to GM and WM (adj_pvalue = 5.71E‐64, adj_pvalue = 3.46E‐49; Figure S3). The second dimension was positively correlated to the intergenic SNPs, rs4912036 and rs609506 (adj_pvalue = 4.20E‐06, adj_pvalue = 7.76E‐06) and negatively correlated to the intergenic SNP rs10799708, and the rs7531434 within the FBXO44 (F‐Box Protein 44) gene (adj_pvalue = 1.58E‐05, adj_pvalue = 1.66E‐05), whereas the third dimension was positively correlated to the rs4614226 and rs11808641 (adj_pvalue = 3.99E‐08, adj_pvalue = 1.83E‐07) and negatively correlated to rs9988443 (adj_pvalue = 8.36E‐08; Table 3). Also for inattention domain, the first dimension of the MFA was positively correlated to WM, in addition to cerebellum (adj_pvalue = 6.42E‐77, adj_pvalue = 2.05E‐16; Figure S3). The second dimension was positively correlated to the intergenic SNPs (rs11808641) and the intragenic SNP (rs301806) within the RERE gene (adj_pvalue = 3.1E‐09, adj_pvalue = 7.2E‐09) and negatively correlated to the intragenic SNP (rs299499; adj_pvalue = 1.02E‐08). Finally, the top SNP correlated with the third dimension was the rs10864315 within the PER3 (Period circadian regulator 3) gene (adj_pvalue = 1.12E‐09; Table 4).
Table 2.
Correlation coefficients between each variable and the first and second dimensions of the MFA for ADHD symptoms
| Correlation | pvalue | adj_pvalue | |
|---|---|---|---|
| Dimension 1 | |||
| GM | 0.927232688564944 | 1.38E−44 | 1.65E−43 |
| WM | 0.910424492643076 | 7.85E−39 | 4.71E−38 |
| Thalamus_total | 0.717095025829273 | 1.34E−08 | 5.38E−08 |
| Cerebellum_total | 0.683429156162821 | 6.67E−06 | 2.00E−05 |
| VentralDC_total | 0.676995448028593 | 1.99E−05 | 4.77E−05 |
| Putamen_total | 0.634838447442277 | 1.36E−02 | 2.72E−02 |
| Pallidum_total | 0.593448654612394 | 3.32E+00 | 5.70E+00 |
| Caudate_total | 0.585407440089544 | 8.85E+00 | 1.33E+01 |
| Hippocampus_total | 0.574935685894525 | 3.04E+01 | 4.06E+01 |
| Amygdala_total | 0.490218128831766 | 1.59E+05 | 1.91E+05 |
| Accumbens_total | 0.45703786625667 | 2.52E+06 | 2.75E+06 |
| CorpusCallosum_total | 0.36853162551687 | 1.09E+09 | 1.09E+09 |
| Dimension 2 | |||
| rs299499 | 0.405688972338025 | 1.05E+08 | 4.48E+08 |
| rs6577561 | 0.396601916550131 | 1.92E+08 | 5.43E+06 |
| rs241239 | 0.389752000367744 | 2.98E+08 | 7.21E+08 |
| rs11121354 | 0.38768509204982 | 3.39E+08 | 7.21E+08 |
| rs9434723 | 0.382335061401637 | 4.74E+08 | 8.95E+08 |
| rs2095904 | 0.380289200809443 | 5.37E+08 | 9.14E+06 |
| rs2289731 | 0.378160998010948 | 6.12E+08 | 9.41E+08 |
| rs1541318 | 0.376829383381258 | 6.64E+08 | 9.41E+08 |
| rs4926481 | 0.369494028743745 | 1.03E+09 | 1.33E+09 |
| rs12041403 | 0.368486221566673 | 1.10E+09 | 1.33E+09 |
| rs6678792 | 0.357626908188188 | 2.06E+09 | 2.33E+09 |
| rs1009941 | 0.34279496000648 | 4.70E+09 | 4.70E+09 |
| rs428180 | −0.347986047251672 | 3.54E+09 | 3.76E+09 |
| rs7517675 | −0.399011088637171 | 1.64E+08 | 5.43E+06 |
| rs12563394 | −0.428266644675448 | 2.20E+07 | 1.25E+08 |
| rs301806 | −0.451582291220506 | 3.86E+06 | 3.28E+07 |
| rs763173 | −0.452099796238842 | 3.70E+06 | 3.28E+07 |
| Dimension 3 | |||
| rs11808641 | 0.467209943606731 | 1.11E+06 | 1.45E+07 |
| rs6683453 | 0.433795581928581 | 1.47E+07 | 7.84E+07 |
| rs4614226 | 0.403942736141467 | 1.18E+08 | 3.85E+08 |
| rs228648 | 0.399816246208454 | 1.55E+08 | 4.04E+08 |
| rs504560 | 0.358303096603233 | 1.98E+09 | 2.86E+09 |
| rs428180 | 0.346584675383812 | 3.82E+09 | 4.76E+09 |
| rs1040397 | 0.345633822436828 | 4.03E+09 | 4.76E+09 |
| rs7523335 | 0.340903344883599 | 5.21E+09 | 5.56E+09 |
| rs1556691 | 0.339692021868727 | 5.56E+09 | 5.56E+09 |
| rs9988443 | −0.363767401067916 | 1.45E+09 | 2.35E+09 |
| rs4654512 | −0.370752418497216 | 9.58E+08 | 1.78E+09 |
| rs10864315 | −0.391665962623566 | 2.63E+08 | 5.71E+08 |
| rs4654432 | −0.430969435191676 | 1.81E+07 | 7.84E+07 |
Note. Variables with correlation coefficients significantly different from zero are shown. ADHD: attention‐deficit/hyperactivity disorder; GM: gray matter; MFA: multiple factor analysis; WM: white matter.
Table 3.
Correlation coefficients between each variable and the first and second dimensions of the MFA for hyperactivity domain
| Correlation | pvalue | adj_pvalue | |
|---|---|---|---|
| Dimension 1 | |||
| GM | 0.941771297833597 | 8.16E−51 | 5.71E−50 |
| WM | 0.899728612092088 | 9.90E−36 | 3.46E−35 |
| Putamen_total | 0.580555543615691 | 1.58E+01 | 3.68E+01 |
| Pallidum_total | 0.574558798033791 | 3.18E+01 | 5.57E+01 |
| Amygdala_total | 0.47812901810156 | 4.50E+04 | 6.30E+05 |
| Accumbens_total | 0.455283404150271 | 2.89E+06 | 3.37E+06 |
| CorpusCallosum_total | 0.416646105322838 | 5.00E+07 | 5.00E+07 |
| Dimension 2 | |||
| rs4912036 | 0.423924740764028 | 3.00E+07 | 4.20E+08 |
| rs609506 | 0.404918666989798 | 1.11E+07 | 7.76E+08 |
| rs2744750 | 0.383141786757075 | 4.51E+08 | 1.58E+09 |
| rs780568 | 0.375125352977279 | 7.37E+08 | 1.66E+09 |
| rs705690 | 0.370867760135604 | 9.51E+08 | 1.66E+09 |
| rs1878052 | 0.36668839763096 | 1.22E+09 | 1.77E+08 |
| rs3117048 | 0.361143153479151 | 1.68E+09 | 2.14E+08 |
| rs241278 | 0.356522374059979 | 2.19E+09 | 2.24E+09 |
| rs1891215 | 0.356183232911795 | 2.24E+09 | 2.24E+09 |
| rs1541318 | −0.35841355236352 | 1.97E+09 | 2.24E+09 |
| rs12122426 | −0.36602000730949 | 1.27E+09 | 1.77E+08 |
| rs1556691 | −0.371358957099761 | 9.24E+08 | 1.66E+09 |
| rs7531434 | −0.374452951530456 | 7.67E+08 | 1.66E+09 |
| rs10799708 | −0.38344428926564 | 4.42E+08 | 1.58E+09 |
| Dimension 3 | |||
| rs4614226 | 0.481704454191441 | 3.32E+05 | 3.99E+04 |
| rs11808641 | 0.447828604469368 | 5.15E+06 | 1.83E+07 |
| rs6683453 | 0.432701939651229 | 1.60E+07 | 3.19E+07 |
| rs12743431 | 0.393084777654286 | 2.40E+08 | 4.12E+08 |
| rs10915271 | 0.374844959387008 | 7.49E+08 | 8.99E+08 |
| rs17472583 | 0.359573890812884 | 1.84E+09 | 2.01E+09 |
| rs11579829 | −0.35618121321993 | 2.24E+09 | 2.24E+09 |
| rs6426389 | −0.37649430455298 | 6.78E+08 | 8.99E+08 |
| rs4233264 | −0.387189378180583 | 3.50E+08 | 5.25E+08 |
| rs1149048 | −0.43800798109189 | 1.08E+07 | 2.59E+07 |
| rs4654512 | −0.445591798677868 | 6.11E+06 | 1.83E+07 |
| rs9988443 | −0.464441181119758 | 1.39E+06 | 8.36E+06 |
Note. Variables with correlation coefficients significantly different from zero are shown. GM: gray matter; MFA: multiple factor analysis; WM: white matter.
Table 4.
Correlation coefficients between each variable and the first and second dimensions of the MFA for inattention domain
| Correlation | pvalue | adj_pvalue | |
|---|---|---|---|
| Dimension 1 | |||
| WM | 0.963279880355951 | 8.03E−65 | 6.42E−63 |
| Thalamus_total | 0.695059811866848 | 8.61E−07 | 2.79E−06 |
| VentralDC_total | 0.693975730674085 | 1.05E−06 | 2.79E−06 |
| Cerebellum_total | 0.636810693363686 | 1.02E−02 | 2.05E−02 |
| Putamen_total | 0.591831551865405 | 4.06E+00 | 6.49E+00 |
| Pallidum_total | 0.565833525203952 | 8.60E+01 | 1.15E+01 |
| CorpusCallosum_total | 0.448392856194293 | 4.93E+06 | 5.64E+06 |
| Accumbens_total | 0.382375347484038 | 4.73E+08 | 4.73E+08 |
| Dimension 2 | |||
| rs11808641 | 0.514582053857106 | 1.72E+04 | 3.10E+05 |
| rs301806 | 0.495818211403101 | 9.68E+04 | 7.16E+05 |
| rs7517675 | 0.493210871845464 | 1.22E+05 | 7.16E+05 |
| rs428180 | 0.490213056002427 | 1.59E+05 | 7.16E+05 |
| rs4614226 | 0.422198150192695 | 3.39E+07 | 7.63E+07 |
| rs7523335 | 0.41216712378504 | 6.80E+07 | 1.22E+08 |
| rs12743431 | 0.410291940545789 | 7.72E+07 | 1.26E+08 |
| rs6663882 | 0.406457031793968 | 1.00E+08 | 1.50E+08 |
| rs7520373 | 0.403339362344629 | 1.23E+08 | 1.71E+08 |
| rs6683453 | 0.368771374849255 | 1.08E+09 | 1.39E+09 |
| rs10910053 | 0.362807560536103 | 1.53E+09 | 1.83E+09 |
| rs2840528 | 0.346905564303549 | 3.76E+09 | 3.98E+09 |
| rs12137865 | −0.337123488336764 | 6.38E+09 | 6.38E+09 |
| rs9988443 | −0.358567451741447 | 1.95E+09 | 2.20E+09 |
| rs4654512 | −0.416181426154807 | 5.16E+07 | 1.03E+08 |
| rs2784739 | −0.424406276455231 | 2.90E+07 | 7.45E+07 |
| rs2289731 | −0.429919659475771 | 1.95E+07 | 5.86E+07 |
| rs299499 | −0.483472232523089 | 2.86E+05 | 1.03E+06 |
| Dimension 3 | |||
| rs1556691 | 0.454934732975396 | 2.97E+06 | 1.26E+07 |
| rs6678792 | 0.440289134782256 | 9.11E+05 | 2.58E+07 |
| rs12756299 | 0.388778218453741 | 3.16E+08 | 5.38E+08 |
| rs4926481 | 0.378742253879418 | 5.91E+08 | 8.37E+08 |
| rs707476 | 0.358118137794679 | 2.00E+09 | 2.43E+09 |
| rs2071986 | 0.342546478920176 | 4.77E+09 | 4.77E+09 |
| rs641473 | −0.347691436835545 | 3.60E+08 | 3.82E+09 |
| rs227163 | −0.350051522358633 | 3.16E+09 | 3.58E+09 |
| rs1878052 | −0.372165545241512 | 8.80E+08 | 1.15E+09 |
| rs863171 | −0.380437393089976 | 5.33E+08 | 8.23E+08 |
| rs10864330 | −0.389215346206817 | 3.08E+08 | 5.38E+08 |
| rs609506 | −0.416057780048898 | 5.20E+07 | 1.11E+08 |
| rs2032563 | −0.431401893712581 | 1.75E+07 | 4.26E+07 |
| rs780568 | −0.441299552357602 | 8.45E+06 | 2.58E+07 |
| rs705690 | −0.469364014327263 | 9.33E+05 | 5.29E+06 |
| rs4654432 | −0.491056687206438 | 1.48E+05 | 1.26E+06 |
| rs10864315 | −0.524520764781794 | 6.59E+03 | 1.12E+05 |
Note. Variables with correlation coefficients significantly different from zero are shown. MFA: multiple factor analysis; WM: white matter.
We also showed the quality of representation of each group of data (neuroimaging, genetics, and ADHD domains). For total ADHD symptoms, both groups have different coordinates on the MFA dimensions, which mean that their contribution is dissimilar to each factor. For instance, neuroimaging information is correlated to the second dimension whereas genetic data in both (Figure S4). Similar patterns were observed for hyperactivity and inattention domains (Figures S5–S6).
4. DISCUSSION
This study proposed the application of a two‐step analysis combining a variable selection strategy designed for a count data distribution along with a MFA approach. The aim was to increase the validity of the results in IG studies and ultimately provide further insights into ADHD. This proposed strategy was inspired by the conclusions of earlier research, which indicated that the use of zero‐inflated negative binomial regression models for count data outcomes provides better parameter estimations (Vilor‐Tejedor, Alemany, et al., 2016). In the proposed study, the application of variable selection based on a LASSO‐ZINB distribution constitutes a novel aspect of the treatment of these data, which accounts for the extra variability associated with the overabundant zero observations in ADHD dimensions. Furthermore, LASSO‐ZINB provides an alternative to marginal feature selection, in which multiple genetic signals and brain structures possibly correlated, can be analyzed in a single model (Szymczak et al., 2009). Although this method has provided a successful performance in a genetic association study (Mallick & Tiwari, 2016) to our knowledge, this is the first time that LASSO‐ZINB is applied in an IG study.
On the other hand, the application of MFA was also novel in IG studies of ADHD. MFA allows determining different correlation patterns between genetic and imaging correlates and the symptomatology. MFA can also deal with large numbers of predictor variables even in the presence of complex interactions, which is not allowed in common association analysis (de Tayrac, Lê, Aubry, Mosser, & Husson, 2009). Hence, MFA could be an attractive strategy in IG studies given its simplicity and structure, which allows the detection of complex relationships between genetic variants and neuroimaging data. Moreover, prior variable selection provides high accuracy on the MFA, which, in turn, represents a measure of importance reflecting the impact of each variable of the model with the outcome of interest.
Results obtained after applying MFA indicate that WM and GM were the most important brain‐MRI measurements in terms of variability explained associated with total ADHD symptoms and hyperactivity domain, in addition to cerebellum structure for inattention domains. Interestingly, an abnormal increase of WM may be related to non‐optimal brain remodeling (deficient axonal pruning), resulting in excessive anatomical connections (Fields, 2010; Tau & Peterson, 2010). However, larger WM volumes in ADHD could also relate to significantly accelerated myelination, as measurable WM in conventional (T1‐weighted) anatomical MRI scans correspond to myelinated WM (Paus et al., 2001). It is important to note that myelination is an active process throughout life (Narayan, Kass, & Thomas, 2007; Yakovlev & Lecours, 1967), which is physiologically accelerated in early postnatal years and during adolescence (Paus et al., 2001; Pujol et al., 2004), and enhanced by repetitive use or skill learning (McKenzie et al., 2014; Pujol et al., 2006). Patholsogical acceleration of myelination has been suggested, for example, in melancholic depression (Soriano‐Mas et al., 2011), childhood obesity (Ou, Andres, Pivik, Cleves, & Badger, 2015), heavy cannabis use (Matochik, Eldreth, Cadet, & Bolla, 2005), and pathological lying (Yang et al., 2005). In addition, for inattention domain, the MFA procedure determined that the cerebellum was also an important brain structure influencing the variability explained. Although cerebellum structure has not been directly related to total ADHD symptoms, some studies have already suggested the involvement of the cerebellum structure in ADHD. For instance, Goetz, Schwabova, Hlavka, Ptacek, and Surman (2017) suggest a cerebellar involvement in dopaminergic neurotransmission which, in turn, affects ADHD symptoms development. Other studies found cerebellar structural abnormalities, which explained a significant amount of the variance of ADHD symptoms (Bledsoe, Semrud‐Clikeman, & Pliszka, 2011).
Moreover, MFA results associated these findings with two intragenic SNPs (rs301806 and rs12563394) within the CAMTA1 gene, which has been related to episodic tremor and motor impairment (Monies et al., 2017; Shinawi, Coorg, Shimony, Grange, & Al‐Kateb, 2015). Interestingly, it has been demonstrated that ADHD children usually present difficulties in activities that require motor coordination (Goulardins, Marques, & De Oliveira, 2017; Kaiser, Schoemaker, Albaret, & Geuze, 2015). Thus, the results obtained could suggest the mediation of specific genetic factors in total ADHD symptoms and motor impairment. MFA also found an intragenic SNP (rs7531434) within the FBXO44 gene, associated with hyperactivity dimension. Although this gene has not been directly implicated in the hyperactivity domain, it has been potentially related with phosphorylation‐dependent ubiquitination processes, which in turn could play an important role in mechanisms that regulate accumulation at synaptic site in the brain (Hunter, 2007). Finally, for the inattention domain, MFA found two intragenic SNPs, the rs10864315 within the PER3 gene and the rs301806 within the CAMTA1 gene. Interestingly, the rs301806 was previously found in the MFA for total ADHD symptoms, suggesting the shared contribution of genetic mechanisms between total ADHD symptoms and inattention domains. Furthermore, the involvement of PER1 gene in the influence of sleep duration and circadian regulation (Hida et al., 2015; Husse, Hintze, Eichele, Lehnert, & Oster, 2012) could be seen as an interesting finding due to the prevalence of sleep problems in ADHD individuals (Bogdan & Reeves, 2018; Cassoff, Wiebe, & Gruber, 2012), especially for inattention domains (Gruber et al., 2012).
However, the current results should be interpreted in the context of some limitations and strengths. First, the main limitation of the study is the modest sample size. Second, the use of a cross‐sectional design prevents us from addressing issues such as reverse‐causation. Third, although the proposed analytical procedure minimizes the internal validation error of prediction, the lack of a replication sample requires the present findings to be considered with caution. Fourth, neurodevelopmental information related to ADHD domains were reported by only one informant (teachers), which provides insufficient information results in a lack of information about the occurrence of these symptoms, and constitutes a subjective measure of the score of these symptoms. Moreover, we examined a statistical framework that, in turn, provide greater complexity in the interpretation of the results. The optimal manner in which to interpret and visualize such results and the suitability of applied statistical techniques (e.g., multiple testing corrections) to such data analysis require further exploration. Finally, results from the analysis of total ADHD symptoms as outcome may be led by common effects that arise from the combination of hyperactivity and inattention domains into a total score. We could hypothesize that it is necessary to study the hyperactivity and inattention domains separately because they may be related to specific causal pathways that are not associated with the total ADHD symptoms. However, the analysis of total ADHD symptoms allows to capturing individuals presenting combined symptoms of hyperactivity and inattention domains, suggesting a representation of the most serious cases of ADHD symptoms, given the combination of both types of domains at the same time.
The strengths of the study include several aspects to overcome these limitations. First, we used a population‐based sample of children, increasing the generalization of the results. Second, the use of a continuous score for total ADHD symptoms and hyperactivity and inattention domains increase statistical power and allows the application of a novel framework and advanced statistical methods, which in turn, contribute to a better characterization and understanding of ADHD symptoms (Goodkind et al., 2015; Hudziak, Achenbach, Althoff, & Pine, 2007; Lubke, Hudziak, Derks, van Bijsterveldt, & Boomsma, 2009; MacCallum, Zhang, Preacher, & Rucker, 2002). Third, the proposed variable selection strategy used in the first analytical step (LASSO‐ZINB) is able to conduct simultaneous model selection and strable effect estimation in the presence of multicollinearity, which, in addition reduces the burden of multiple testing. Because multicollinearity can result in large and opposite signed estimator values for correlated predictors, a penalty function is imposed to keep the value of predictors below a pre‐specified value and so select them efficiently and also taking into account the existence of multicollinearity (Mallick & Tiwari, 2016).
To conclude, the application and development of novel analytical frameworks and advanced statistical methods allow determining the combined effects of genetics and neuroimaging in the context of complex disorders such as ADHD. Therefore, future research may benefit from the application of multivariate strategies.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest and financial disclosures.
Supporting information
Table S1 Brain region description for the Imaging Genetic sample (N = 135; mean Age = 8.4 [0.82])
Table S2. Proportion of variances retained by the different components for ADHD symptoms
Table S3. Proportion of variances retained by the different components for hyperactivity domain.
Table S4. Proportion of variances retained by the different components for inattention domain.
Table S5. Labels of Brain Regions of Interest included in the analysis.
Figure S1. Multiple factor Analysis. Representation of variables on the first plane for total ADHD symptoms.
Figure S2. Multiple factor Analysis. Representation of variables on the first plane for hyperactivity domain.
Figure S3. Multiple factor Analysis. Representation of variables on the first plane for inattention domain.
Figure S4. Representations of groups/blocks of variables for total ADHD symptoms. Each group/block of variables is represented by a single point.
Figure S5. Representations of groups/blocks of variables for hyperactivity domain. Each group/block of variables is represented by a single point.
Figure S6. Representations of groups/blocks of variables for inattention domain. Each group/block of variables is represented by a single point.
ACKNOWLEDGMENTS
Natalia Vilor‐Tejedor is funded by a pre‐doctoral grant from the Agència de Gestió d'Ajuts Universitaris i de Recerca (2017 FI_B 00636), Generalitat de Catalunya – Fons Social Europeu. Silvia Alemany thanks the Institute of Health Carlos III for her Sara Borrell postdoctoral grant (CD14/00214). The research leading to these results has also received funding from the European Research Council under the ERC Grant Agreement 268479—the BREATHE project and by Grant MTM2015‐68140‐R from the Ministerio de Economía e Innovación (Spain). ISGlobal is a member of the CERCA Programme, Generalitat de Catalunya.
Vilor‐Tejedor N, Alemany S, Cáceres A, et al. Sparse multiple factor analysis to integrate genetic data, neuroimaging features, and attention‐deficit/hyperactivity disorder domains. Int J Methods Psychiatr Res. 2018;27:e1738 10.1002/mpr.1738
REFERENCES
- Alemany, S. , Vilor‐Tejedor, N. , Bustamante, M. , Pujol, J. , Macià, D. , Martínez‐Vilavella, G. , … Sunyer, J. (2016). A genome‐wide association study of attention function in a population‐based sample of children. PLoS One, 11(9), e0163048 10.1371/journal.pone.0163048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcos‐Burgos, M. , Vélez, J. I. , Solomon, B. D. , & Muenke, M. (2012). A common genetic network underlies substance use disorders and disruptive or externalizing disorders. Human Genetics, 131(6), 917–929. 10.1007/s00439-012-1164-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Association, A. P (1994). Diagnostic and statistical manual of mental disorders: DSM‐IV [Internet] (4th ed.). ( p. 866). Washington (DC): American Psychiatric Association. [Google Scholar]
- Bledsoe, J. C. , Semrud‐Clikeman, M. , & Pliszka, S. R. (2011). Neuroanatomical and neuropsychological correlates of the cerebellum in children with attention‐deficit/hyperactivity disorder–combined type. Journal of the American Academy of Child & Adolescent Psychiatry, 50(6), 593–601. 10.1016/j.jaac.2011.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan, A. R. , & Reeves, K. W. (2018). Sleep duration in relation to attention deficit hyperactivity disorder in American adults. Behavioral Sleep Medicine, 16(3), 235–243. 10.1080/15402002.2016.1188391 [DOI] [PubMed] [Google Scholar]
- Bogdan, R. , Salmeron, B. J. , Carey, C. E. , Agrawal, A. , Calhoun, V. D. , Garavan, H. , … Goldman, D. (2017). Imaging genetics and genomics in psychiatry: A critical review of progress and potential. Biological Psychiatry, 82(3), 165–175. 10.1016/j.biopsych.2016.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardo, E. , Servera, M. , & Llobera, J. (2017). Estimation of the prevalence of attention deficit hyperactivity disorder among the standard population on the island of Majorca. Revista de Neurologia, 44(1), 10–14. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/17199223 [PubMed] [Google Scholar]
- Cassoff, J. , Wiebe, S. T. , & Gruber, R. (2012). Sleep patterns and the risk for ADHD: A review. Nature and Science of Sleep, 4, 73–80. 10.2147/NSS.S31269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escofier, B. , & Pagès, J. (1990). Analyses factorielles simples et multiples: Objectifs, méthodes, interprétation. Paris: Dunod. [Google Scholar]
- Fields, R. D. (2010). Neuroscience. Change in the brain's white matter. Science (New York, N.Y.), 330(6005), 768–769. 10.1126/science.1199139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz, M. , Schwabova, J. P. , Hlavka, Z. , Ptacek, R. , & Surman, C. B. (2017). Dynamic balance in children with attention‐deficit hyperactivity disorder and its relationship with cognitive functions and cerebellum. Neuropsychiatric Disease and Treatment, 13, 873–880. 10.2147/NDT.S125169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodkind, M. , Eickhoff, S. B. , Oathes, D. J. , Jiang, Y. , Chang, A. , Jones‐Hagata, L. B. , … Etkin, A. (2015). Identification of a common neurobiological substrate for mental illness. JAMA Psychiatry, 72(4), 305 10.1001/jamapsychiatry.2014.2206–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulardins, J. B. , Marques, J. C. B. , & De Oliveira, J. A. (2017). Attention deficit hyperactivity disorder and motor impairment. Perceptual and Motor Skills, 124(2), 425–440. 10.1177/0031512517690607 [DOI] [PubMed] [Google Scholar]
- Gruber, R. , Michaelsen, S. , Bergmame, L. , Frenette, S. , Bruni, O. , Fontil, L. , & Carrier, J. (2012). Short sleep duration is associated with teacher‐reported inattention and cognitive problems in healthy school‐aged children. Nature and Science of Sleep, 4, 33–40. 10.2147/NSS.S24607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida, A. , Kitamura, S. , Katayose, Y. , Kato, M. , Ono, H. , Kadotani, H. , … Mishima, K. (2015). Screening of clock gene polymorphisms demonstrates association of a PER3 polymorphism with morningness–eveningness preference and circadian rhythm sleep disorder. Scientific Reports, 4(1), 6309 10.1038/srep06309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudziak, J. J. , Achenbach, T. M. , Althoff, R. R. , & Pine, D. S. (2007). A dimensional approach to developmental psychopathology. International Journal of Methods in Psychiatric Research, 16 Suppl, 1(S1), S16–S23. 10.1002/mpr.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter, T. (2007). The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Molecular Cell, 28(5), 730–738. 10.1016/J.MOLCEL.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Husse, J. , Hintze, S. C. , Eichele, G. , Lehnert, H. , & Oster, H. (2012). Circadian clock genes Per1 and Per2 regulate the response of metabolism‐associated transcripts to sleep disruption. PLoS One, 7(12), e52983 10.1371/journal.pone.0052983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, M.‐L. , Schoemaker, M. M. , Albaret, J.‐M. , & Geuze, R. H. (2015). What is the evidence of impaired motor skills and motor control among children with attention deficit hyperactivity disorder (ADHD)? Systematic review of the literature. Research in Developmental Disabilities, 36, 338–357. 10.1016/j.ridd.2014.09.023 [DOI] [PubMed] [Google Scholar]
- Khadka, S. , Pearlson, G. D. , Calhoun, V. D. , Liu, J. , Gelernter, J. , Bessette, K. L. , & Stevens, M. C. (2016). Multivariate imaging genetics study of MRI gray matter volume and SNPs reveals biological pathways correlated with brain structural differences in attention deficit hyperactivity disorder. Frontiers in Psychiatry, 7 10.3389/fpsyt.2016.00128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubke, G. H. , Hudziak, J. J. , Derks, E. M. , van Bijsterveldt, T. C. E. M. , & Boomsma, D. I. (2009). Maternal ratings of attention problems in ADHD: Evidence for the existence of a continuum. Journal of the American Academy of Child and Adolescent Psychiatry, 48(11), 1085–1093. 10.1097/CHI.0b013e3181ba3dbb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukoshe, A. , White, T. , Schmidt, M. N. , van der Lugt, A. , & Hokken‐Koelega, A. C. (2013). Divergent structural brain abnormalities between different genetic subtypes of children with Prader‐Willi syndrome. Journal of Neurodevelopmental Disorders, 5(1), 31 10.1186/1866-1955-5-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum, R. C. , Zhang, S. , Preacher, K. J. , & Rucker, D. D. (2002). On the practice of dichotomization of quantitative variables. Psychological Methods, 7(1), 19–40. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11928888 [DOI] [PubMed] [Google Scholar]
- Mallick, H. , & Tiwari, H. K. (2016). EM adaptive LASSO—A multilocus modeling strategy for detecting SNPs associated with zero‐inflated count phenotypes. Frontiers in Genetics, 7, 32 10.3389/fgene.2016.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matochik, J. A. , Eldreth, D. A. , Cadet, J.‐L. , & Bolla, K. I. (2005). Altered brain tissue composition in heavy marijuana users. Drug and Alcohol Dependence, 77(1), 23–30. 10.1016/j.drugalcdep.2004.06.011 [DOI] [PubMed] [Google Scholar]
- McKenzie, I. A. , Ohayon, D. , Li, H. , Paes de Faria, J. , Emery, B. , Tohyama, K. , & Richardson, W. D. (2014). Motor skill learning requires active central myelination. Science, 346(6207), 318–322. 10.1126/science.1254960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monies, D. , Abou Al‐Shaar, H. , Goljan, E. A. , Al‐Younes, B. , Al‐Breacan, M. M. A. , Al‐Saif, M. M. , … Bohlega, S. (2017). Identification of a novel genetic locus underlying tremor and dystonia. Human Genomics, 11(1), 25 10.1186/s40246-017-0123-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortamais, M. , Pujol, J. , van Drooge, B. L. , Macià, D. , Martínez‐Vilavella, G. , Reynes, C. , … Sunyer, J. (2017). Effect of exposure to polycyclic aromatic hydrocarbons on basal ganglia and attention‐deficit hyperactivity disorder symptoms in primary school children. Environment International, 105, 12–19. 10.1016/j.envint.2017.04.011 [DOI] [PubMed] [Google Scholar]
- Mous, S. E. , Hammerschlag, A. R. , Polderman, T. J. C. , Verhulst, F. C. , Tiemeier, H. , van der Lugt, A. , … Posthuma, D. (2015). A population‐based imaging genetics study of inattention/hyperactivity: Basal ganglia and genetic pathways. Journal of the American Academy of Child and Adolescent Psychiatry, 54(9), 745–752. 10.1016/j.jaac.2015.05.018 [DOI] [PubMed] [Google Scholar]
- Narayan, S. , Kass, K. E. , & Thomas, E. A. (2007). Chronic haloperidol treatment results in a decrease in the expression of myelin/oligodendrocyte‐related genes in the mouse brain. Journal of Neuroscience Research, 85(4), 757–765. 10.1002/jnr.21161 [DOI] [PubMed] [Google Scholar]
- Ou, X. , Andres, A. , Pivik, R. T. , Cleves, M. A. , & Badger, T. M. (2015). Brain gray and white matter differences in healthy normal weight and obese children. Journal of Magnetic Resonance Imaging, 42(5), 1205–1213. 10.1002/jmri.24912 [DOI] [PubMed] [Google Scholar]
- Paus, T. , Collins, D. L. , Evans, A. C. , Leonard, G. , Pike, B. , & Zijdenbos, A. (2001). Maturation of white matter in the human brain: A review of magnetic resonance studies. Brain Research Bulletin, 54(3), 255–266. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/11287130 [DOI] [PubMed] [Google Scholar]
- Polanczyk, G. , de Lima, M. S. , Horta, B. L. , Biederman, J. , & Rohde, L. A. (2007). The worldwide prevalence of ADHD: A systematic review and metaregression analysis. The American Journal of Psychiatry, 164(6), 942–948. 10.1176/ajp.2007.164.6.942 [DOI] [PubMed] [Google Scholar]
- Pujol, J. , López‐Sala, A. , Sebastián‐Gallés, N. , Deus, J. , Cardoner, N. , Soriano‐Mas, C. , … Sans, A. (2004). Delayed myelination in children with developmental delay detected by volumetric MRI. NeuroImage, 22(2), 897–903. 10.1016/j.neuroimage.2004.01.029 [DOI] [PubMed] [Google Scholar]
- Pujol, J. , Martínez‐Vilavella, G. , Macià, D. , Fenoll, R. , Alvarez‐Pedrerol, M. , Rivas, I. , … Sunyer, J. (2016). Traffic pollution exposure is associated with altered brain connectivity in school children. NeuroImage, 129, 175–184. 10.1016/j.neuroimage.2016.01.036 [DOI] [PubMed] [Google Scholar]
- Pujol, J. , Soriano‐Mas, C. , Ortiz, H. , Sebastian‐Galles, N. , Losilla, J. M. , & Deus, J. (2006). Myelination of language‐related areas in the developing brain. Neurology, 66(3), 339–343. 10.1212/01.wnl.0000201049.66073.8d [DOI] [PubMed] [Google Scholar]
- Purcell, S. , Neale, B. , Todd‐Brown, K. , Thomas, L. , Ferreira, M. A. R. , Bender, D. , … Sham, P. C. (2007). PLINK: A tool set for whole‐genome association and population‐based linkage analyses. The American Journal of Human Genetics, 81(3), 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinawi, M. , Coorg, R. , Shimony, J. S. , Grange, D. K. , & Al‐Kateb, H. (2015). Intragenic CAMTA1 deletions are associated with a spectrum of neurobehavioral phenotypes. Clinical Genetics, 87(5), 478–482. 10.1111/cge.12407 [DOI] [PubMed] [Google Scholar]
- Soriano‐Mas, C. , Hernández‐Ribas, R. , Pujol, J. , Urretavizcaya, M. , Deus, J. , Harrison, B. J. , … Cardoner, N. (2011). Cross‐sectional and longitudinal assessment of structural brain alterations in melancholic depression. Biological Psychiatry, 69(4), 318–325. 10.1016/j.biopsych.2010.07.029 [DOI] [PubMed] [Google Scholar]
- Sunyer, J. , Esnaola, M. , Alvarez‐Pedrerol, M. , Forns, J. , Rivas, I. , López‐Vicente, M. , … Querol, X. (2015). Association between traffic‐related air pollution in schools and cognitive development in primary school children: A prospective cohort study. PLoS Medicine, 12(3), e1001792 10.1371/journal.pmed.1001792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczak, S. , Biernacka, J. M. , Cordell, H. J. , González‐Recio, O. , König, I. R. , Zhang, H. , & Sun, Y. V. (2009). Machine learning in genome‐wide association studies. Genetic Epidemiology, 33(S1), S51–S57. 10.1002/gepi.20473 [DOI] [PubMed] [Google Scholar]
- Tau, G. Z. , & Peterson, B. S. (2010). Normal development of brain circuits. Neuropsychopharmacology: Official Publication of the American College of Neuropsychopharmacology, 35(1), 147–168. 10.1038/npp.2009.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Tayrac, M. , Lê, S. , Aubry, M. , Mosser, J. , & Husson, F. (2009). Simultaneous analysis of distinct Omics data sets with integration of biological knowledge: Multiple factor analysis approach. BMC Genomics, 10, 32 10.1186/1471-2164-10-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilor‐Tejedor, N. , Alemany, S. , Forns, J. , Cáceres, A. , Murcia, M. , Macià, D. , … González, J. R. (2016). Assessment of susceptibility risk factors for ADHD in imaging genetic studies. Journal of Attention Disorders.. 10.1177/1087054716664408 [DOI] [PubMed] [Google Scholar]
- Vilor‐Tejedor, N. , Cáceres, A. , Pujol, J. , Sunyer, J. , & González, J. R. (2016). Imaging genetics in attention‐deficit/hyperactivity disorder and related neurodevelopmental domains: State of the art. Brain Imaging and Behavior. 10.1007/s11682-016-9663-x, 11, 1922–1931. [DOI] [PubMed] [Google Scholar]
- Vilor‐Tejedor, N. , Gonzalez, J. R. , & Calle, M. L. (2015). Efficient and powerful method for combining p‐values in Genome‐wide Association Studies. IEEE/ACM Transactions on Computational Biology and Bioinformatics/IEEE, ACM. 10.1109/TCBB.2015.2509977, 13, 1100–1106. [DOI] [PubMed] [Google Scholar]
- Yakovlev, P. , & Lecours, A. (1967), January 1). The myelogenetic cycles of regional maturation of the brain In Regional development of the brain in early life. Retrieved from https://www.scienceopen.com/document?vid=1c16c21a-8793-4f48-bfcc-87c6f65e1bf9 [Google Scholar]
- Yang, Y. , Raine, A. , Lencz, T. , Bihrle, S. , Lacasse, L. , & Colletti, P. (2005). Prefrontal white matter in pathological liars. British Journal of Psychiatry, 187(4), 320–325. 10.1192/bjp.187.4.320 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Brain region description for the Imaging Genetic sample (N = 135; mean Age = 8.4 [0.82])
Table S2. Proportion of variances retained by the different components for ADHD symptoms
Table S3. Proportion of variances retained by the different components for hyperactivity domain.
Table S4. Proportion of variances retained by the different components for inattention domain.
Table S5. Labels of Brain Regions of Interest included in the analysis.
Figure S1. Multiple factor Analysis. Representation of variables on the first plane for total ADHD symptoms.
Figure S2. Multiple factor Analysis. Representation of variables on the first plane for hyperactivity domain.
Figure S3. Multiple factor Analysis. Representation of variables on the first plane for inattention domain.
Figure S4. Representations of groups/blocks of variables for total ADHD symptoms. Each group/block of variables is represented by a single point.
Figure S5. Representations of groups/blocks of variables for hyperactivity domain. Each group/block of variables is represented by a single point.
Figure S6. Representations of groups/blocks of variables for inattention domain. Each group/block of variables is represented by a single point.


