Abstract
The parathyroid hormone 1 receptor (PTH1R) mediates the biologic actions of parathyroid hormone (PTH) and parathyroid hormone–related protein (PTHrP). Here, we showed that salt-inducible kinases (SIKs) are key kinases that control the skeletal actions downstream of PTH1R and that this GPCR, when activated, inhibited cellular SIK activity. Sik gene deletion led to phenotypic changes that were remarkably similar to models of increased PTH1R signaling. In growth plate chondrocytes, PTHrP inhibited SIK3, and ablation of this kinase in proliferating chondrocytes rescued perinatal lethality of PTHrP-null mice. Combined deletion of Sik2 and Sik3 in osteoblasts and osteocytes led to a dramatic increase in bone mass that closely resembled the skeletal and molecular phenotypes observed when these bone cells express a constitutively active PTH1R that causes Jansen’s metaphyseal chondrodysplasia. Finally, genetic evidence demonstrated that class IIa histone deacetylases were key PTH1R-regulated SIK substrates in both chondrocytes and osteocytes. Taken together, our findings establish that SIK inhibition is central to PTH1R action in bone development and remodeling. Furthermore, this work highlights the key role of cAMP-regulated SIKs downstream of GPCR action.
Keywords: Bone Biology, Endocrinology
Keywords: Bone development, Bone disease, Osteoclast/osteoblast biology
Introduction
The parathyroid hormone 1 receptor (PTH1R) is a class B, 7-transmembrane G protein–coupled receptor (GPCR) (1) whose biologic actions control mineral ion homeostasis (2), bone development (3, 4), and bone remodeling (5). Two endogenous peptide ligands, parathyroid hormone (PTH) and parathyroid hormone–related protein (PTHrP), activate this widely expressed receptor (6). Highlighting the importance of this GPCR, synthetic PTH1R peptide agonists are currently used for treatment of osteoporosis (7) and hypoparathyroidism (8). Despite these advances, the downstream signaling steps linking PTH1R activation and the resulting formation of cAMP levels to changes in gene expression in vivo remain incompletely understood.
Salt-inducible kinases (SIKs) represent a subfamily of AMP-activated protein kinase (AMPK) family kinases (9, 10). High dietary salt intake increases Sik1 expression in the adrenal glands (11). In contrast, Sik2 and Sik3 are generally expressed at a constitutive level in multiple tissues (12). SIK cellular activity is regulated predominantly by opposing activities of 2 upstream kinases. Liver kinase B1 (LKB1, encoded by the Stk11 gene) phosphorylates the activation loop of all AMPK family kinases, including SIKs, and therefore stimulates SIK cellular activity (13, 14). In contrast, cAMP-dependent protein kinase A (PKA) phosphorylates SIKs at C-terminal residues outside of the kinase domain, leading to SIK inhibition through an allosteric mechanism involving 14-3-3 binding and altered substrate availability (15–18).
Class IIa histone deacetylases (HDACs) and CREB-regulated transcription coactivators (CRTCs) (19, 20) are key SIK substrates (9, 10, 21, 22). When phosphorylated, class IIa HDACs and CRTC proteins are retained in the cytoplasm by 14-3-3 proteins. When SIK activity is inhibited by PKA phosphorylation, class IIa HDAC and CRTC phosphorylation levels are reduced, leading to nuclear translocation where they regulate target gene expression. Nuclear class IIa HDACs predominantly block MEF2-driven gene expression, while nuclear CRTC proteins coactivate CREB-driven target genes. Therefore, PKA-dependent SIK inhibition serves as a key link between GPCR activation and gene expression changes. This model has been proposed in diverse biologic systems, including myocytes (22, 23), macrophages (downstream of prostaglandin E2) (24–27), hepatocytes (downstream of glucagon) (16), and melanocytes (downstream of melanocyte stimulating hormone) (9, 28). In addition, we recently described SIK2 as an important PKA-dependent substrate downstream of PTH1R action in osteocytes (29). The aforementioned studies have mainly used in vitro cell culture systems and small-molecule kinase inhibitors to reach these conclusions. Moreover, the relative role of PKA-dependent SIK phosphorylation (relative to other PKA substrates) in the biologic actions of PTH1R remains poorly understood. Therefore, the goal of the current study was to use genetic approaches to determine the role of salt-inducible kinases downstream of PTH1R action in vivo. Based on the signaling model in which PTH1R action inhibits SIK cellular function, we predicted that SIK gene deletion might mimic the actions of excessive PTH1R signaling in target cells.
Here, we report genetic evidence demonstrating central roles for SIKs downstream of PTH1R action. During endochondral bone formation, PTHrP signaling leads to PKA-dependent phosphorylation and inactivation of SIK3. Mice with universal Sik3 knockout (KO) display delayed chondrocyte hypertrophy (30), similar to what is seen with transgenic overexpression of PTHrP in chondrocytes (31); in these growth plates, class IIa HDAC phosphorylation at 14-3-3 binding sites is reduced (32). We show that Sik3 deficiency rescues the perinatal lethality observed in Pthrp-KO mice, which is due to accelerated chondrocyte differentiation and thus premature skeletal mineralization. To complement these studies on the PTH1R/SIK pathway in growth plate chondrocytes, we studied the consequences of deleting SIKs in mature osteoblasts and osteocytes. Combined ablation of SIK2 and SIK3 dramatically increased bone mass and bone turnover, a phenotype highly reminiscent of what is seen with constitutive PTH1R activation in these cells. Bone transcriptomic profiling by RNA-Seq confirmed striking molecular concordance between the effects of SIK2/3 deletion and ligand-independent PTH1R signaling. Finally, we asked which SIK substrates contribute to these phenotypic changes. In both growth plate chondrocytes and osteoblasts/osteocytes, class IIa HDACs are essential for the skeletal phenotypes seen with SIK deletion. Taken together, these results support the importance of a cAMP/PKA/SIK/class IIa HDAC signaling axis for the physiologic skeletal actions downstream of the PTH1R.
Results
SIK3 is a key mediator of PTHrP action in growth plate chondrocytes.
Previous studies have suggested that SIK3, a kinase expressed in growth plate chondrocytes, controls chondrocyte hypertrophy in mice and humans (30, 33). PTHrP, secreted from chondrocytes near the ends of bone, acts on its receptor on proliferating chondrocytes to block their differentiation into post-proliferative hypertrophic chondrocytes, thereby allowing them to continue proliferating (4). PTHrP signaling in chondrocytes reduces class IIa HDAC (HDAC4 and HDAC5) phosphorylation at 14-3-3 binding sites and promotes their nuclear translocation (32, 34). In the nucleus, class IIa HDACs block chondrocyte hypertrophy by inhibiting the actions of the hypertrophy-promoting transcription factors MEF2C and RUNX2 (35, 36). Despite these advances, the precise link between PTHrP signaling in growth plate chondrocytes and class IIa HDAC phosphorylation has remained obscure. To investigate the hypothesis that SIK3 is a target of PTHrP signaling, we treated primary rib chondrocytes with PTH and examined SIK3 phosphorylation. As shown in Figure 1A, in chondrocytes, PTH1R activation led to reduced HDAC4/5 phosphorylation at the first 14-3-3 binding site on these comigrating proteins and concomitant SIK3 phosphorylation at T441, one of SIK3’s 3 PKA phosphorylation sites (37).
Figure 1. Sik3 deletion rescues perinatal lethality of Pthrp-deficient mice.
(A) Primary rib chondrocytes isolated from newborn WT mice were treated with vehicle or PTH (1–34, 100 nM) for 30 minutes, followed by immunoblotting as indicated. Phosphorylated HDAC4-Ser245 (p-HDAC4-Ser245), p-HDAC5-Ser250, and p-HDAC7-Ser178 represent the residues in mouse HDAC4 protein. Here, contemporaneous immunoblots were run in parallel. This experiment was performed twice, and representative results are shown. (B, top) H&E staining of proximal tibia at birth (original magnification, ×100) demonstrates that the lethal phenotype of the Pthrp-KO mouse is rescued by Sik3 gene deletion. Each mouse genotype shown is defined as follows: Sik3fl/+, Pthrp-KO (universal Pthrp–/–), Sik3-cHET (Sik3fl/+ Col2a1-Cre), Sik3-cKO Pthrp-KO (Sik3fl/fl Pthrp–/– Col2a1-Cre), and Sik3-cKO (Sik3fl/fl Col2a1-Cre). Numbers represent the average length of the proliferating chondrocyte region (black lines) (mean ± SEM, n = 3, biological triplicates; we measured the average length using 6–9 sections per mouse). *P < 0.01, **P < 0.001 by 1-way ANOVA followed by Dunnett’s test for multiple comparisons, when the Pthrp-KO measurement is control. (B, bottom) Col10a1 mRNA in situ hybridization of the anterior rib cage at birth (original magnification, ×40). Abnormal Col10a1 mRNA expression in the Pthrp-KO mice is reduced or absent with combined Sik3 gene deletion (red arrowheads). Normal Col10a1 mRNA expression in the sternum is missing in the Pthrp and Sik3 double-KO mouse and the Sik3-KO mouse (black arrowheads). S indicates the lower end of the sternum. (C) H&E staining of proximal tibia (top) and anterior rib cage (bottom) at P26 (original magnification, ×40) from surviving postnatal mice. Bone formation is severely affected in the Sik3 and Pthrp double-KO mouse and the Sik3-cKO mouse: secondary ossification center (black arrowheads in the top panels) and bone formation in the sternum (black arrowheads in the bottom panels) are missing. Abnormal chondrocyte hypertrophy in the anterior rib is not seen in the Sik3 and Pthrp double-KO mouse (red arrowheads). Scale bars (red lines): 500 μm.
To identify in vivo correlates of the link between PTH1R signaling and SIK3 phosphorylation, we asked whether deletion of the Sik3 gene in chondrocytes, which is known to delay chondrocyte hypertrophy (30), might rescue the phenotype of mice lacking the Pthrp gene. For this, Sik3fl/fl mice were mated with Col2a1-Cre mice (38) to produce chondrocyte-specific Sik3 gene deletion. Mice lacking Pthrp die immediately after birth from respiratory failure caused by a small circumference of the rib cage in association with extensive chondrocyte hypertrophy and mineralization that are not seen in normal rib cartilage (39). Strikingly, the lethal phenotype of the Pthrp-KO mouse is completely rescued by homozygous ablation of Sik3 in chondrocytes. In Pthrp-KO mice, the distance from the top of the tibial growth plate to the beginning of the hypertrophic layer was extremely short as a result of the accelerated chondrocyte hypertrophy (Figure 1B, top panel). As shown in Figure 1B (top), the accelerated chondrocyte hypertrophy in the Pthrp-KO mouse was completely abrogated when Sik3 was deleted simultaneously in chondrocytes. Unlike Pthrp-KO mice, which showed abnormal Col10a1 mRNA expression (a marker of hypertrophic chondrocytes) in the anterior ribs, Pthrp/Sik3 double-KO mice exhibited no Col10a1 mRNA expression in the anterior ribs (Figure 1B, bottom, red arrowheads). Notably, normal Col10a1 expression on sternum (Sik3fl/+ in Figure 1B, bottom) was missing in the Sik3 and Pthrp double-KO mouse as well as in the Sik3-KO mouse (Figure 1B, bottom, black arrowheads). Partial suppression of Col10a1 mRNA expression was noted in the Sik3-cHET (Sik3fl/+ Col2a1-Cre) Pthrp-KO mice (Figure 1B, bottom, red arrowheads), although this was insufficient to rescue perinatal lethality due to Pthrp deletion.
Consistent with rescued rib chondrocyte hypertrophy, we observed rescue of perinatal lethality of Pthrp-KO mice in compound Pthrp/Sik3 mutants. While mice lacking Pthrp uniformly died just after birth, Pthrp and Sik3 double-KO mice survived up to 1 month after birth. The cause of death in compound Pthrp/Sik3 mutants was not abnormal rib chondrocyte hypertrophy and mineralization (Figure 1C, bottom, red arrowheads). Rather, these mice exhibited other phenotypes, such as a complete failure of tooth eruption (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI130126DS1), a phenotype identical to what is observed when Pth1r is deleted in dental mesenchymal progenitors (38, 40–42). Taken together, these results establish a key role for SIK3 inhibition downstream of PTHrP action in chondrocytes in the growth plate.
SIK1 and SIK2 support the actions of SIK3 on chondrocyte differentiation.
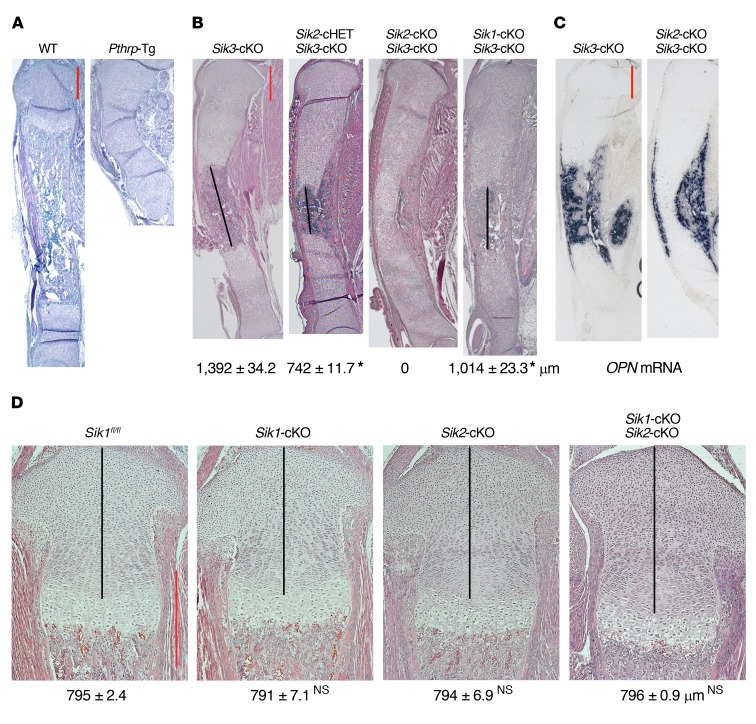
The universal Sik3-KO mice and the chondrocyte-specific Sik3 conditional KO (cKO) mice showed delayed chondrocyte hypertrophy compared with WT controls (ref. 30, Figure 1B, and Figure 2A). However, the phenotype of Sik3-KO mice was not as severe as the delay of chondrocyte hypertrophy in chondrocyte-specific Pthrp transgenic (Tg) mice (Figure 2A). Pthrp-Tg mice (Pthrp-Tg/+) exhibit only round chondrocytes at birth, and therefore subsequent bone formation is severely delayed after birth (31). The more severe phenotype with Pthrp overexpression suggests that other SIK family proteins (SIK1 or SIK2) may control chondrocyte hypertrophy in addition to SIK3. To confirm this hypothesis, we generated the double-KO mice of Sik1 and Sik3 or Sik2 and Sik3. Sik2 and Sik3 are linked by 5 Mb on mouse chromosome 9; therefore, a large number of progeny (>120) were screened in order to ultimately identify mice with both floxed alleles present in cis (Supplemental Figure 2). By additional deletion of the Sik1 or Sik2 gene in the Sik3-cKO mouse, we observed longer growth plate and delayed separation of proximal and distal growth plates at birth (Figure 2B, black lines). These results suggest that SIK1 and SIK2 also regulate chondrocyte differentiation when SIK3 is absent, but that Sik2 gene deletion exhibits more obvious effects. We found a stepwise delay of chondrocyte differentiation when the Sik3-cKO deletion was combined with the Sik2-cHET and then the Sik2-cKO deletion (Figure 2, A and B). In newborn Sik2 and Sik3 double-cKO mice, bone formation started from both perichondria, but the proximal and distal growth plates were not separated by bone invasion, judging by osteopontin (OPN; marker for late hypertrophic chondrocytes and bone) mRNA expression (Figure 2C). The Sik1-cKO, Sik2-cKO, and Sik1/Sik2 double-cKO exhibited normal growth plate phenotypes (Figure 2D), highlighting that SIK3 is the predominant SIK isoform in regulating chondrocyte differentiation.
Figure 2. SIK1 and SIK2 control chondrocyte hypertrophy in addition to SIK3.
(A and B) H&E staining of whole tibia at birth (original magnification, ×20) shows that additional homozygous deletion of Sik1 or Sik2 in the Sik3-cKO mouse delays chondrocyte hypertrophy. Each mouse genotype shown is defined as follows: Sik3-cKO (Sik3fl/fl Col2a1-Cre), Sik2-cHET Sik3-cKO (Sik2fl/+ Sik3fl/fl Col2a1-Cre), Sik2-cKO Sik3-cKO (Sik2fl/fl Sik3fl/fl Col2a1-Cre), Sik1-cKO Sik3-cKO (Sik1fl/fl Sik3fl/fl Col2a1-Cre). Numbers represent average length of the bone region between proximal and distal growth plates (black lines) (mean ± SEM, n = 3, biological triplicates; we measured the average length using 6–9 sections for each mouse). *P < 0.001 by 1-way ANOVA followed by Dunnett’s test, when the Sik3-cKO measurement is control. P values less than 0.05 were considered significant. (C) In situ hybridization for osteopontin (OPN) mRNA on the whole tibia at birth (original magnification, ×20). (D) H&E staining of proximal tibial growth plate at birth (original magnification, ×100). Numbers represent the average length of the proliferating chondrocyte region (black lines) (mean ± SEM, n = 3, biological triplicates; we measured the average length using 6–9 sections for each mouse). By 1-way ANOVA followed by Dunnett’s test when the corresponding WT (Sik1fl/fl) measurement is control, we found no significant (NS) differences by knocking out Sik1 and/or Sik2. P values less than 0.05 were considered significant. SIK3 is the major mediator. SIK3 alone can exhibit normal phenotype without SIK1 and SIK2. Scale bars (red lines): 500 μm.
Combined SIK2/3 deletion in osteoblasts and osteocytes causes high bone mass with accelerated bone turnover.
Having established a key role for SIK3 downstream of PTH1R action in growth plate chondrocytes, we next asked whether SIK deletion in osteoblasts and osteocytes might mimic the skeletal effects of constitutive PTH1R signaling. Jansen’s metaphyseal chondrodysplasia is caused by rare activating Pth1r mutations, most commonly Pth1rH223R (43, 44). When this Pth1r variant is expressed in osteoblasts or osteocytes via transgene, mice show very high trabecular bone mass associated with increased bone remodeling (45, 46). PTH signaling in osteoblasts leads to PKA-mediated inhibition of SIK2, and small-molecule pan-SIK inhibitors mimic skeletal PTH action (29). Therefore, we predicted that deletion of SIKs in osteoblasts and osteocytes might mimic the high turnover phenotype seen with PTH1RH223R expression in these cells. Analysis of existing cortical bone RNA-Seq data sets indicates that all 3 SIKs are expressed at moderate levels in bone (Supplemental Figure 3A). Moreover, recent single-cell RNA-Seq data sets demonstrate coexpression of all 3 SIKs in Pth1r-expressing osteoblasts (47, 48). For these studies, Dmp1-Cre (9.6-kb promoter element; ref. 49) was used to achieve deletion in mature osteoblasts and osteocytes. “Single” mutants lacking Sik1, Sik2, or Sik3 showed no obvious skeletal phenotype as assessed by micro-CT and analysis of serum bone turnover markers procollagen type 1 N-terminal propeptide (P1NP) and C-terminal telopeptide (CTX) (Supplemental Figure 3, B and C, and Supplemental Table 1 for all micro-CT data).
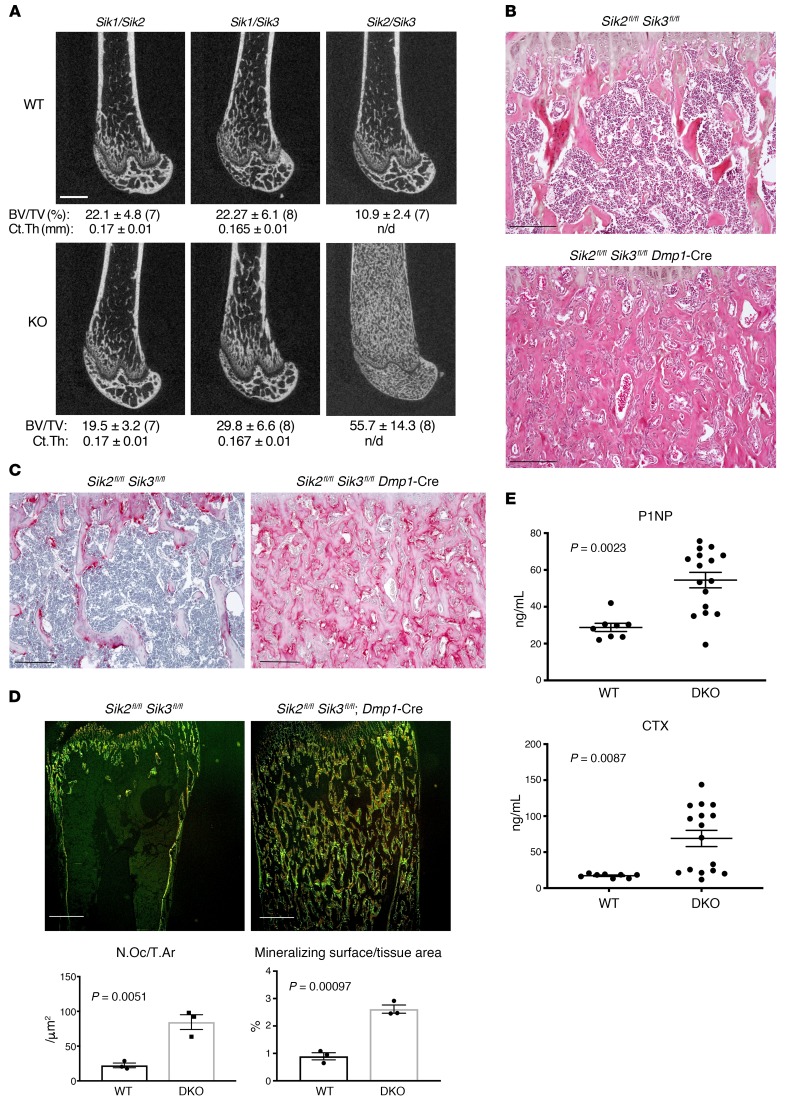
However, using the same strategy, we generated mice lacking Stk11, the upstream kinase required for activation of all 3 SIK isoforms, and observed markedly increased trabecular bone mass (Supplemental Figure 3D). Therefore, given the potential for functional redundancy among SIK isoforms, we generated all possibilities of combined double Sik mutants using Dmp1-Cre. While Sik1/Sik2 and Sik1/Sik3 compound mutant mice showed no dramatic skeletal phenotype when bred to Dmp1-Cre transgenic mice, Sik2/Sik3 compound mutant animals (Sik2fl/fl Sik3fl/fl Dmp1-Cre) showed a dramatic high bone mass phenotype associated with expansion of trabecular bone throughout the marrow cavity (Figure 3A and Supplemental Figure 4). Histologic analysis of Sik2fl/fl Sik3fl/fl Dmp1-Cre long bones demonstrated increased trabecular bone mass with an expansion of marrow stromal cells (Figure 3B and Supplemental Figure 5; here we define bone marrow stromal cells as non-hematopoietic cells located away from bone surfaces) and increased tartrate-resistant acid phosphatase–positive (TRAP-positive) osteoclasts lining trabecular surfaces (Figure 3, C and D). Dynamic histomorphometry demonstrated high bone formation throughout trabecular and cortical surfaces in Sik2fl/fl Sik3fl/fl Dmp1-Cre animals (Figure 3D and Supplemental Figure 6). Consistent with TRAP staining and dynamic histomorphometry results, serum P1NP (a marker of bone formation) and CTX (a marker of bone resorption) analysis from these mice confirmed a state of accelerated bone turnover (Figure 3E) despite normal circulating levels of calcium, phosphate, and PTH (Supplemental Figure 7).
Figure 3. Combined SIK2/3 deletion in osteoblasts and osteocytes causes high bone mass with accelerated bone turnover.
(A) Representative femur micro-CT images from littermate control (top) and compound SIK mutant (bottom) 8-week-old male mice of the indicated genotypes. Mean trabecular bone volume fraction (± SD) in the distal metaphysis (BV/TV) is listed below each representative image, with sample size analyzed of each group following in parentheses. Below trabecular BV/TV, the midshaft cortical thickness (Ct.Th) is listed. All WT mice studied bear the indicated loxP-flanked allele but are negative for the Dmp1-Cre transgene (which on its own does not cause a discernible skeletal phenotype). n/d, not determined. Scale bar: 1 mm. (B) H&E-stained proximal tibia histology from 8-week-old WT and SIK2/3 double-mutant mice from A shown at ×100 original magnification (results shown are representative of n = 6 mice per group). (C) TRAP-stained (pink) proximal tibia histology from WT and SIK2/3 double-mutant mice. (D) Top: WT and SIK2/3 double-mutant mice were labeled with calcein at P26, demeclocycline at P27, then sacrificed 1 day later for dynamic histomorphometry on non-decalcified, plastic-embedded sections. Representative images at ×4 original magnification showing fluorescent signals from dual calcein/demeclocycline labeling are shown; scale bars: 500 μm. Bottom: Quantification of osteoclast and trabecular mineralizing surface (normalized to tissue area) is shown; n = 3 mice per genotype were analyzed. (E) Fasting serum from 8-week-old WT (n = 8) and SIK2/3 double-mutant (DKO, n = 16) mice was collected and analyzed for P1NP (bone formation marker) and CTX (bone resorption marker). Student’s t tests were used for comparison between WT and SIK2/3-DKO mice, with P values listed in the figure panels. Scale bars: 100 μm for all histology panels in this figure unless otherwise indicated. See also Supplemental Figures 1–6.
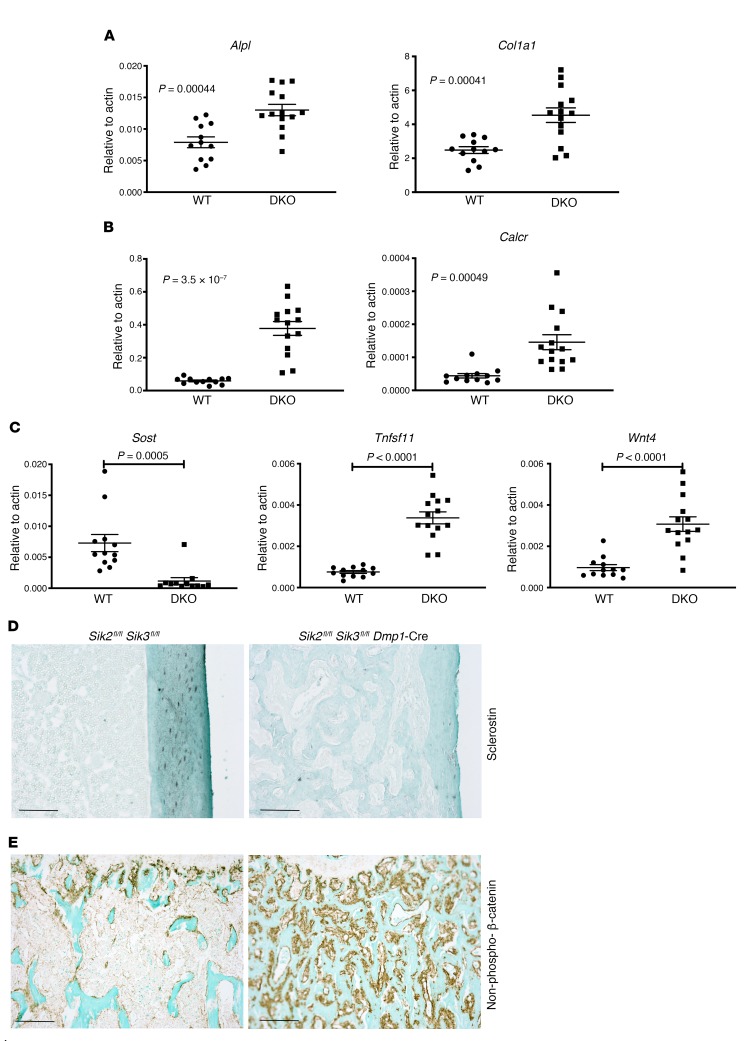
Next, we isolated cortical bone RNA from WT and Sik2fl/fl Sik3fl/fl Dmp1-Cre mice in order to determine molecular correlates associated with this dramatic high bone mass phenotype. As expected, expression of osteoblast markers (Alpl and Col1a1) and osteoclast markers (Acp5 [the gene encoding TRAP] and Calcr) was elevated compared with control (Figure 4, A and B). We previously defined a core group of 142 genes whose expression is acutely regulated by both PTH and small-molecule SIK inhibitors in a transformed osteoblastic/osteocytic cell line (29). As shown in Figure 4C (and below in Figure 5), Sik2fl/fl Sik3fl/fl Dmp1-Cre mice showed differential expression of PTH/SIK target genes (Sost, Tnfsf11, and Wnt4) in the expected direction. Sik2fl/fl Sik3fl/fl Dmp1-Cre mice showed reduced osteocytic sclerostin expression in situ (Figure 4D). Consistent with reduced sclerostin mRNA and protein levels, Sik2fl/fl Sik3fl/fl Dmp1-Cre mice showed increased numbers of osteoblastic cells on bone surfaces staining positive for nonphosphorylated (activated) β-catenin (Figure 4E).
Figure 4. Molecular correlates of high bone mass phenotype in Sik2fl/fl Sik3fl/fl Dmp1-Cre mice.
(A–C) Marrow-flushed cortical bone RNA was isolated from WT (n = 12) and SIK2/3 mutant (n = 14) 6-week-old mice, followed by quantitative reverse transcriptase PCR analysis for the indicated osteoblast marker (A), osteoclast marker (B), and PTH/SIK inhibitor–responsive genes (C). Student’s t tests were used for comparison between WT and Sik2/3-DKO mice, with P values listed in the figure panels. (D) Sclerostin immunohistochemistry (brown; original magnification, ×100) from WT and Sik2fl/fl Sik3fl/fl Dmp1-Cre mice. Osteocytes in mutant trabecular and cortical bone stain poorly for sclerostin. (E) Immunohistochemistry for nonphosphorylated (S33/S37/T41) β-catenin in the primary and secondary spongiosa. Immunostaining results show representative data from n = 5 mice per genotype. Scale bars: 100 μm for all panels in this figure.
Figure 5. Global molecular concordance between Sik2fl/fl Sik3fl/fl Dmp1-Cre and C1HR mice.
(A) Left: H&E-stained images (original magnification, ×100) in the primary and secondary spongiosa of 6-week-old Col1a1-PTH1RH223R (C1HR) mice showing increased bone mass and increased marrow stromal cells. Middle: TRAP-stained images (original magnification, ×20) revealing increased osteoclasts on trabecular surfaces of C1HR mice. Right: Immunostaining for activated β-catenin reveals increased WNT pathway activity in C1HR animals. See Figure 3, B and C, and Figure 4E for similarities to Sik2fl/fl Sik3fl/fl Dmp1-Cre mice. Scale bars: 100 μm for left and right panels, 1 mm for middle panels. (B) RNA-Seq was performed on bone RNA isolated from Sik2fl/fl Sik3fl/fl Dmp1-Cre mice (n = 6) with n = 8 littermate controls, and C1HR mice (n = 12) with n = 11 littermate controls. Volcano plots show the relationship between fold change and statistical significance across these 2 data sets. (C) Venn diagram analysis of lists of differentially expressed genes (DEGs) (log2 FC > 2, FDR < 0.05) between the 2 comparisons. Hypergeometric P values for all overlap analyses are less than 0.05. (D) Heatmap showing extensive coregulation of gene expression in SIK2/3 mutant and C1HR mice. Each row represents a distinct DEG (encompassing all genes found in the first Venn diagram in C), and each column represents a mouse of the indicated genotype. The log2 fold change of each gene is expressed versus the mean of the control mice. (E) Scatterplot demonstrating extensive coregulation between SIK2/3 mutants and C1HR mice. Each dot represents the average log2 fold change for all genes detected by RNA-Seq comparing mutant versus control. Linear regression analysis between the 2 data sets reveals P < 0.0001, slope = 0.9112, R2 = 0.5914. (F) Similar scatterplot demonstrating relative lack of concordance between effects of SIK2/3 deletion in osteoblasts/osteocytes and effects of 2 weeks of sclerostin antibody treatment. Linear regression analysis between the 2 data sets reveals P < 0.0001, slope = 0.1155, R2 = 0.051. See also Supplemental Figures 6–8.
Molecular concordance between Sik2fl/fl Sik3fl/fl Dmp1-Cre and PTH1RH223R-expressing mice.
We noted clear phenotypic similarities between the skeletal phenotypes of Sik2fl/fl Sik3fl/fl Dmp1-Cre mice and mice expressing a constitutively activated PTH1R (H223R, associated with Jansen’s metaphyseal chondrodysplasia) in osteoblasts or osteocytes (45, 46). Like Sik2fl/fl Sik3fl/fl Dmp1-Cre mice, Col1a1-PTH1RH223R (hereafter referred to as C1HR) animals also showed increased trabecular bone mass with marrow stromal cells (50, 51), increased TRAP-positive osteoclasts on trabecular surfaces, and increased nonphosphorylated (activated) β-catenin–positive cells lining bone (Figure 5A). To further explore the molecular concordance between these 2 models of apparent hyperactive PTH1R signaling, we performed bulk RNA-Seq on bone RNA isolated from both mutant strains along with matched WT littermate controls (see Methods). Initial analysis of both comparisons revealed a large number of differentially expressed genes (DEGs; fold change > 2, FDR < 0.05) versus respective control RNA samples (Figure 5B and Supplemental Table 2 for RNA-Seq data). Sik2fl/fl Sik3fl/fl Dmp1-Cre mice showed 3600 DEGs (1327 upregulated, 2273 downregulated), while C1HR mice showed 5909 DEGs (3385 upregulated, 2524 downregulated).
Comparing lists of DEGs, 71.3% of the genes regulated in Sik2fl/fl Sik3fl/fl Dmp1-Cre mice were also differentially expressed in C1HR animals (Figure 5C). This striking overlap was not due to random chance (hypergeometric P < 10–1066). A similar degree of overlap was noted when directionality of gene expression changes (vs. WT controls) was accounted for: 74.6% of upregulated DEGs in Sik2fl/fl Sik3fl/fl Dmp1-Cre mice were upregulated in C1HR animals (hypergeometric P < 10–942), and 65.8% of downregulated DEGs in Sik2fl/fl Sik3fl/fl Dmp1-Cre mice were downregulated in C1HR animals (hypergeometric P < 10–1807). Nonrandom overlap also existed between genes acutely coregulated by PTH and the small-molecule SIK inhibitor YKL-05-093 in cultured Ocy454 cells (29) and genes regulated in both Sik2fl/fl Sik3fl/fl Dmp1-Cre and C1HR mice in vivo (hypergeometric P = 0.016). Visualization of all quantified genes by heatmap (Figure 5D) revealed groups of coregulated genes, as did a scatterplot showing the average fold change for each gene in each RNA-Seq comparison (Figure 5E). Gene ontology analysis of co-upregulated genes revealed enrichment in expected terms such as osteoclast differentiation, ECM organization, WNT signaling, and endochondral bone morphogenesis (Supplemental Figure 8). Bone cell WNT signaling output (52) was clearly upregulated in both Sik2fl/fl Sik3fl/fl Dmp1-Cre and C1HR mice (Supplemental Figure 9A), and it is clear that multiple mechanisms contributed to this effect. As shown in Supplemental Figure 9B, expression of the paracrine-acting WNT inhibitor Sost (53) was not downregulated in C1HR animals (likely because of low activity of the Col1a1 promoter in Sost-expressing osteocytes). However, expression of multiple canonical and noncanonical WNT ligands was upregulated in Sik2fl/fl Sik3fl/fl Dmp1-Cre and C1HR mice (Supplemental Figure 9C), suggesting that multiple mechanisms contribute to increased WNT activity downstream of PTH1R signaling in bone.
Molecular concordance between Sik2fl/fl Sik3fl/fl Dmp1-Cre and C1HR mice could be due to specific engagement of common signaling pathways, or it could be due to shared expression of common genes nonspecifically associated with increased bone formation, increased osteoblast number, and increased trabecular bone mass. Therefore, we compared DEGs seen in Sik2fl/fl Sik3fl/fl Dmp1-Cre mice with those seen after treatment with neutralizing anti-sclerostin antibody (Scl-Ab; ref. 54), another distinct model of high bone formation and increased trabecular bone mass. Two weeks of Scl-Ab treatment caused differential expression of 1030 genes (649 upregulated, 381 downregulated; Supplemental Figure 10A). Scatterplot and Venn diagram analysis demonstrated minimal concordance between Scl-DEGs compared with Sik2fl/fl Sik3fl/fl Dmp1-Cre and C1HR mice. For instance, 74.6% of upregulated genes in Sik2fl/fl Sik3fl/fl Dmp1-Cre mice were also upregulated in C1HR mice, while only 7.4% of those same genes were also upregulated by Scl-Ab treatment (Figure 5F and Supplemental Figure 10, B and C). Taken together, these results suggest that common engagement of a shared specific signaling pathway drives the striking phenotypic and molecular concordance observed between Sik2fl/fl Sik3fl/fl Dmp1-Cre and C1HR mice.
Class IIa HDACs are key downstream mediators of PTH1R/SIK action in the growth plate and in osteoblasts/osteocytes.
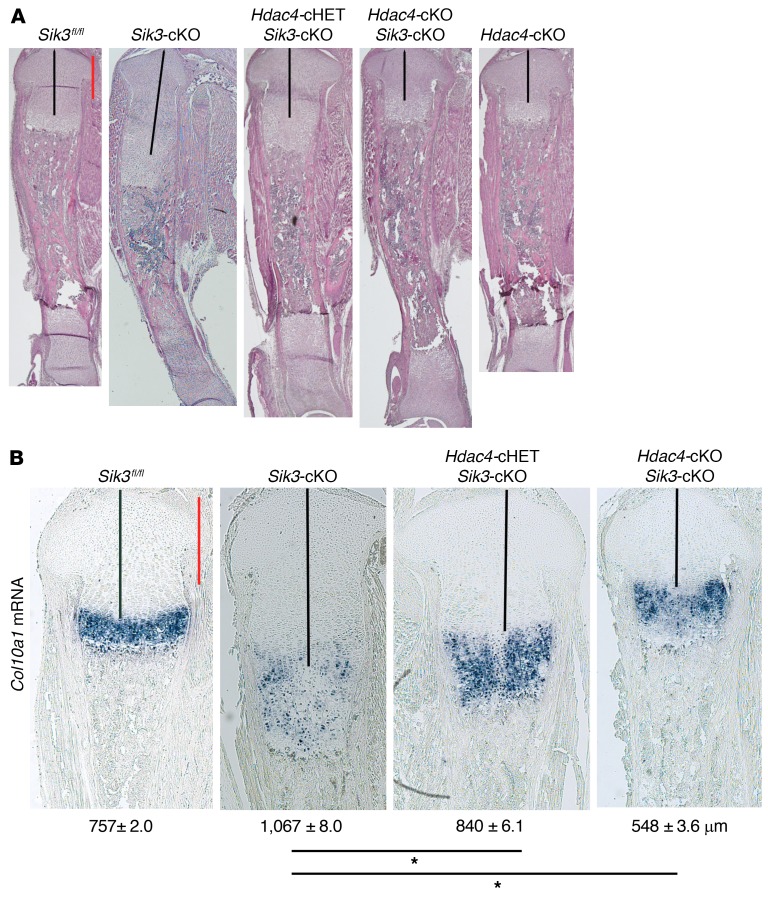
While multiple SIK substrates have been reported (55, 56), work from many biologic systems supports a model in which class IIa HDACs and CRTC proteins are key mediators of SIK-dependent cAMP-regulated changes in gene expression (9, 10). We recently reported a key role for class IIa HDAC proteins in suppressing MEF2C-driven chondrocyte hypertrophy downstream of PTHrP action (32). Therefore, we tested the role of HDAC4 as a mediator of SIK3 action in the growth plate by generating chondrocyte-specific compound Sik3/Hdac4 mutant mice. As shown in Figure 6A, by deleting the Hdac4 gene in the Sik3-cKO mouse, stepwise shorter proliferating chondrocyte regions were observed at birth, supporting the idea that HDAC4 acts downstream of SIK3 to delay chondrocyte hypertrophy. Delayed chondrocyte hypertrophy observed in the chondrocyte-specific Sik3-cKO mice was completely abrogated with concomitant Hdac4 gene deletion. Consistent with these histologic data, deleting Hdac4 in Sik3-cKO mice resulted in accelerated Col10a1 expression similar to what is seen in Hdac4-cKO (Hdac4fl/fl Col2a1-Cre) mice (ref. 35 and Figure 6B). Sik3/Hdac4 double-cKO mice appeared phenotypically similar to Hdac4-cKO mice (Figure 6A), indicating that HDAC4 functions downstream as a key mediator of SIK3 signaling.
Figure 6. Class IIa HDACs are key downstream mediators of PTH1R/SIK action in the growth plate.
(A) H&E staining of the whole tibia at birth (original magnification, ×20) reveals that the expanded growth plate in the Sik3-cKO mouse is abrogated when the Hdac4 gene is simultaneously deleted. Each mouse genotype shown is defined as follows: Sik3fl/fl, Sik3-cKO (Sik3fl/fl Col2a1-Cre), Hdac4-cHET Sik3-cKO (Hdac4fl/+ Sik3fl/fl Col2a1-Cre), Hdac4-cKO Sik3-cKO (Hdac4fl/fl Sik3fl/fl Col2a1-Cre), Hdac4-cKO (Hdac4fl/fl Col2a1-Cre). (B) In situ hybridization for Col10a1 mRNA on the proximal tibial growth plate at birth (original magnification, ×40). Numbers represent the average length of the proliferating chondrocyte region (black lines) (mean ± SEM, n = 3, biological triplicates; we measured the average length using 6–9 sections for each mouse). *P < 0.001 by 1-way ANOVA followed by Dunnett’s test, when the Sik3-cKO measurement is control. P values less than 0.05 were considered significant. Scale bars (red lines): 500 µm.
We next asked whether class IIa HDACs might also be required for the skeletal phenotype seen in Sik2fl/fl Sik3fl/fl Dmp1-Cre mice. Here, a similar compound mutant strategy was used in which Hdac4fl/fl Hdac5–/– Dmp1-Cre mice were intercrossed to Sik2fl/fl Sik3fl/fl Dmp1-Cre animals. As we previously reported (29), Hdac4fl/fl Hdac5–/– Dmp1-Cre mice showed cortical and trabecular osteopenia. As detailed above, Sik2fl/fl Sik3fl/fl Dmp1-Cre mice showed massive increases in trabecular bone mass through the marrow cavity. In contrast, compound Hdac4fl/fl Hdac5–/– Sik2fl/fl Sik3fl/fl Dmp1-Cre animals showed osteopenia similar to what was observed in Hdac4fl/fl Hdac5–/– Dmp1-Cre mice (Figure 7A). At the histologic level, compound Hdac4fl/fl Hdac5–/– Sik2fl/fl Sik3fl/fl Dmp1-Cre quadruple-KO mice showed osteopenia similar to what was observed in Hdac4fl/fl Hdac5–/– Dmp1-Cre double-KO animals (Figure 7B). Sclerostin immunohistochemistry revealed, as expected (29), high sclerostin levels in mice lacking Hdac4/Hdac5, low osteocytic sclerostin expression in Sik2fl/fl Sik3fl/fl Dmp1-Cre animals, and high sclerostin in quadruple mutants (Figure 7C). In sum, these data reveal that class IIa HDACs are key downstream mediators of the biologic effects of SIK deletion in growth plate chondrocytes and osteoblasts/osteocytes. Since SIK-deficient phenotypes are completely reversed in the presence of concomitant class IIa HDAC deletion, class IIa HDACs likely function as vital downstream mediators of SIK action in bone in vivo.
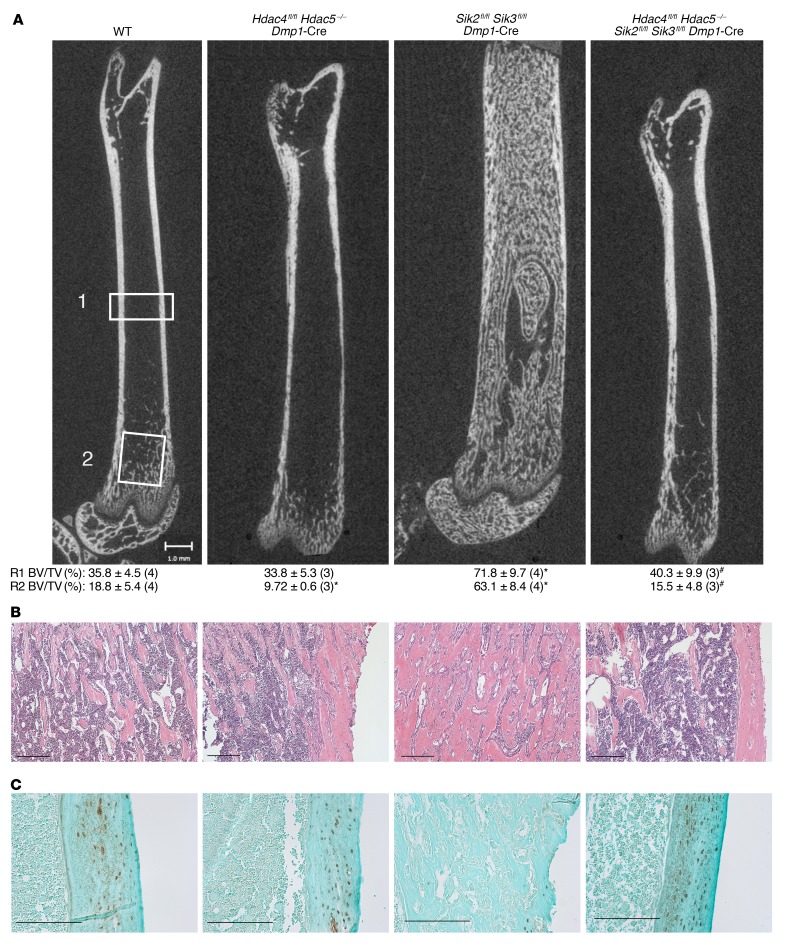
Figure 7. Class IIa HDACs are key downstream mediators of PTH1R/SIK action in osteoblasts/osteocytes.
(A) Femurs from 6-week-old mice of the indicated genotypes were analyzed by micro-CT. Full-length images are representative of at least n = 3 mice per genotype; littermate controls were used for all experiments. Mean R1 (midshaft cortical) bone volume fraction (BV/TV, determined from region “1” above including cortical bone) and R2 (trabecular) bone volume fraction (BV/TV, determined from region “2” above excluding cortical bone) (± SD) are shown for each genotype, with sample size analyzed in parentheses. *P < 0.05 vs. WT, #P < 0.05 vs. Sik2/3-DKO. For R1 midshaft whole bone BV/TV comparisons: WT vs. Sik2/3-DKO, P = 0.00053; Sik2/3-DKO vs. Sik2/3 Hdac4/5 quadruple-KO, P = 0.0083. For R2 trabecular BV/TV comparisons: WT vs. Hdac4/5-DKO, P = 0.037; WT vs. Sik2/3-DKO, P = 0.000114; Sik2/3-DKO vs. Sik2/3 Hdac4/5 quadruple-KO, P = 0.000336. One-way ANOVA followed by Dunnett’s multiple-comparisons test was used to determine P values. (B) Representative H&E stain (sample size analyzed of each group is shown in A) of the primary and secondary spongiosa in the proximal tibia at ×100 original magnification. (C) Sclerostin immunohistochemistry from sections in B shown at the level of midshaft cortical bone. Sclerostin immunoreactivity levels are high in HDAC4/5-deficient mice, low in Sik2fl/fl Sik3fl/fl Dmp1-Cre mice, and high in compound HDAC4/5 SIK2/3 mutants. Scale bars: 100 μm for histology panels and 1 mm for micro-CT panels.
Discussion
Intracellular signaling downstream of PTH1R has dramatic effects on growth plate chondrocytes, osteoblasts, and osteocytes in bone development and remodeling. Here, using in vivo genetic evidence, we demonstrate that salt-inducible kinases control the biologic actions of PTH1R. In growth plate chondrocytes, we show that PTH1R activation leads to PKA-mediated SIK3 T441 phosphorylation, a signaling event known to block the cellular action of SIK3 (15, 37). In osteocytes, PTH1R signaling triggers PKA-dependent phosphorylation of SIK2 at multiple sites. In osteoblasts, PTH1R signaling targets both SIK2 and SIK3 cellular activity. In chondrocytes, PTH in vitro similarly triggers phosphorylation of SIK3. SIK3 deletion phenocopies the effects of constitutive PTH1R signaling, and rescues lethality observed in the Pthrp-KO mice. In osteoblasts/osteocytes, combined SIK2/3 deletion mimics the effects of constitutive PTH1R signaling at the phenotypic and molecular levels. Therefore, we conclude that SIK inhibition is a pivotal intracellular step in these downstream biologic actions of PTH1R.
While multiple SIK substrates have been reported in diverse biologic systems (55, 56), a consensus model has emerged in which class IIa HDACs and CRTC proteins are key mediators of the biologic actions of these kinases in normal physiology (9, 10). Hdac4-KO mice exhibit limb deformities due to exuberant RUNX2- and MEF2C-driven chondrocyte hypertrophy (35, 36). We recently reported that HDAC4 is required for PTHrP to inhibit chondrocyte hypertrophy (32), and nuclear HDAC4 accumulation is noted in SIK3-deficient chondrocytes (30). Here we show that HDAC4 is also required for the growth plate phenotype associated with Sik3 deficiency. These genetic results place HDAC4 “downstream” of PTHrP/SIK3 signaling in the control of chondrocyte hypertrophy. While the role of CRTC proteins in chondrocyte maturation remains to be determined, our results clearly suggest a dominant role of class IIa HDACs as physiologically important SIK substrates in the growth plate.
The specific role of PKA-mediated SIK2/3 phosphorylation in blocking the cellular activity of these kinases in bone development and homeostasis remains to be tested in vivo. Recently, a mouse ethylnitrosurea (ENU) mutagenesis screen for phenotypes related to sleep abnormalities identified the Sleepy SIK3 mutant as a strain with markedly prolonged sleep time. This SIK3 DNA change leads to an in-frame loss of the 52–amino acid exon 13, which includes a PKA phosphorylation site at S551 (57). Phosphoproteomic analysis of Sik3Sleepy CNS tissue revealed a potential neuron-specific role for SIK3 in phosphorylating several synaptic proteins (58). More recently, the specific role of one PKA site within exon 13, serine 551, was tested in vivo. SIK3 S551A mice show hypersomnia similar to that of the exon 13–skipped Sleepy mutants (59). Therefore, PKA-mediated SIK3 S551 phosphorylation plays a key role in control of sleep-related neurotransmission by SIK3. However, the role of S551 in controlling the growth plate actions of SIK3 downstream of PTH1R remains to be determined. Furthermore, like SIK2, there are multiple PKA phosphorylation sites of SIK3 that are likely to work in conjunction to promote signal-dependent 14-3-3 association (15).
Recently, humans with homozygous Sik3 missense mutation have been reported to show a Jansen’s metaphyseal chondrodysplasia–like growth plate phenotype (33). These findings are entirely consistent with our data demonstrating that PTHrP inhibits SIK3 action in the growth plate, and that homozygous Sik3 gene deletion rescues perinatal lethality in mice lacking PTHrP. Krakow and colleagues proposed that altered mTOR signaling (via SIK3-mediated regulation of DEPTOR activity) provides the mechanistic link explaining this phenotype. Our studies with compound SIK3/HDAC4 mutants suggest that class IIa HDACs are likely the SIK3 substrates that dominate in regulation of the differentiation of chondrocytes in the growth plate. The role of DEPTOR in chondrocyte maturation and its regulation by SIKs remains to be studied using mouse genetics. Our current work suggests that small-molecule SIK agonists, if identified, or class IIa HDAC inhibitors (60, 61) might be of therapeutic utility for growth plate and bone abnormalities in individuals with Jansen’s metaphyseal chondrodysplasia.
In both chondrocytes and osteoblasts/osteocytes, class IIa HDACs are required for the biologic phenotypes associated with SIK gene deletion. These results suggest that class IIa HDACs are crucial downstream mediators of cellular SIK action. However, the relative role of CRTC proteins in the cellular actions of SIKs remains to be determined. Many genes regulated by PTH1R signaling are thought to be direct CREB targets (62), and it is likely that CRTC proteins synergize with direct PKA-mediated CREB phosphorylation to induce targets such as RANKL. We acknowledge the possibility that the strong phenotypic consequences of class IIa HDAC deficiency may obscure important roles for CRTC proteins and/or other SIK substrates. Therefore, future studies will be needed to better understand how CRTC proteins contribute to the cellular actions of SIKs in vivo.
In osteoblasts and osteocytes, PTH1R signaling reduces cellular SIK activity, as manifested by dephosphorylation and nuclear translocation of class IIa HDACs and CRTC proteins (29, 63). We previously observed PTH-stimulated PKA-dependent phosphorylation of SIK2 (but not SIK3) in a transformed osteocyte-like cell line. While no overt bone phenotype is present when SIK2 is deleted with Dmp1-Cre, compound Sik2 and Sik3 deletion leads to a dramatic high bone mass phenotype similar to what is observed with PTH1RH223R expression in osteoblasts and/or osteocytes. Therefore, it is likely that PTH1R signaling acutely inhibits the cellular activity of both related kinases (SIK2 and SIK3) in vivo in osteoblasts and osteocytes. Further studies are needed to better understand the compensatory relationships between these closely related kinases. For example, compound SIK1/SIK3 mutant mice show increased cortical bone area and trends (not statistically significant) toward increased trabecular bone mass (Figure 3A) and increased bone turnover. Pharmacologic inhibitors that target all 3 SIK isoforms mimic PTH action in vivo (29). Our current results suggest that specific inhibitors of only SIK2 and SIK3 may be sufficient to boost bone formation and bone mass in therapeutic settings.
Transcriptomic analysis of Sik2fl/fl Sik3fl/fl Dmp1-Cre and C1HR mice revealed a common signature associated with WNT pathway upregulation. Previous studies have linked PTH1R signaling in osteoblasts/osteocytes with increased WNT pathway output (5, 64), and intact canonical WNT signaling is required for intermittent PTH treatment to increase bone mass (65, 66). Multiple mechanisms have been proposed to explain cross-talk between PTH1R and WNT signaling pathways, including direct interactions between PTH1R and the WNT coreceptor LRP6, suppression of the WNT inhibitors SOST and DKK1, and induction of WNT ligands by bone marrow T cells (67–70). Expression of multiple canonical and noncanonical WNT ligands is stimulated by PTH1R activation via SIK2/3 inhibition. Therefore, these results add to our understanding of how WNT signaling is stimulated by PTH1R action.
While SIK gene deletion may not perfectly mimic the effects of PKA-dependent kinase inhibition, we do note that T163A SIK3 kinase–deficient mice show short bone length similar to that in mice completely lacking SIK3 (71). Therefore, it is likely that the major function of SIK3 is to serve as a protein kinase. Small-molecule SIK inhibitors have been studied as research tools and potential therapeutic agents for cancer (72–75), inflammatory bowel disease (76), skin pigmentation (28), and osteoporosis (29). Our current in vivo results indicate that a major biologic action of SIKs is to mediate multiple biologic actions of PTH1R signaling.
Our initial work to identify SIKs downstream of PTH1R action in bone emanated from studies designed to understand how PTH signaling suppresses SOST expression (29). Our current findings demonstrate that the ability of PTH1R to inhibit SIKs accounts for many of the dominant biologic aspects of PTH1R action in bone development and remodeling. Future studies are needed to determine whether SIK inhibition plays a similar dominant role in any of the extraskeletal actions downstream of PTH1R, or in other GPCR/Gsα/cAMP-linked signaling systems.
Methods
Genetically modified mice.
The following published genetically modified strains were used: germline Pthrp-deficient mice (RRID: MGI:2154515) (39), Hdac4-floxed mice (RRID: MGI:4418117) (77), germline Hdac5-deficient mice (RRID: MGI:3056065) (78), Col2a1-Cre mice (RRID: MGI:2176070) (38), Dmp1-Cre mice (RRID: MGI:3784520) (49), Stk11-floxed mice (RRID: MGI:4440829) (79) (The Jackson Laboratory, JAX 014143), Sik1-floxed mice (RRID: MGI:5648544) (80), and Sik2-floxed mice (RRID: MGI:5905012) (16). Sik3tm1a(EUCOMM)Hmgu mice (RRID: MGI:5085429) were purchased from the European Conditional Mouse Mutagenesis Program (EUCOMM) and bred to PGK1-FLPo mice (The Jackson Laboratory, JAX 011065) to generate mice bearing a loxP-flanked SIK3 allele. See Supplemental Figure 2 for details on generation of compound SIK2/SIK3-floxed mice in cis. For all other compound mutant strains, single-mutant mice were intercrossed and then F1 progeny bred to obtain compound mutants along with WT (Cre-negative) littermate controls. All strains were backcrossed to C57BL/6 for at least 4 generations. WT (Cre-negative) littermate controls were used for all studies to account for potential influence of genetic background.
Primary rib chondrocyte culture.
Primary rib chondrocytes were isolated from WT newborn mice using the protocol in ref. 81 with modification. The front part of the rib cage was dissected by fine forceps under a dissecting microscope. Hypertrophic chondrocytes and the bone region on the posterior rib cage were removed. The anterior rib cages were incubated in a digestion medium containing 0.2% (approximately 600 U/mL) Type 2 Collagenase (Worthington, LS004176), DMEM, and 10% FBS at room temperature. Muscle, soft tissues, and sternal bones were removed by fine forceps. The anterior rib cartilages were further digested in a 6-well plate with the fresh digestion medium at 37°C overnight (1 well for ribs from 2 pups). Cells were released from the cartilage matrix by gentle pipetting with a p1000 tip 10 times, and then passed through nylon mesh cell strainers (BD Falcon). We mixed the flow through and spun down the cells to remove collagenase. We cultured the cells in DMEM/10% FBS in a 10-cm dish until evening. We collected and counted cells for frozen stock using BAMBANKER media (FUJIFILM Wako Chemicals, 302-14681). PTH treatments (1–34 peptide courtesy of Ashok Khatri in the Massachusetts General Hospital Peptide Core) were performed when cells were 50%–60% confluent.
Histology and immunohistochemistry.
Formalin-fixed paraffin-embedded decalcified tibia sections from newborn, 6-week-old, and 8-week-old mice were obtained. H&E staining was performed on some sections using standard protocols. For immunohistochemistry, antigen retrieval was performed using proteinase K (20 μg/mL) for 15 minutes. Endogenous peroxidases were quenched, and slides were blocked in TNB buffer (Perkin-Elmer), then stained with primary antibody for 1 hour at room temperature. Anti-sclerostin antibody (Scl-Ab; R&D, BAF1589) was used at a 1:200 dilution; anti–β-catenin (active; Cell Signaling Technology, 9561) was used at a 1:1000 dilution. Sections were washed and incubated with HRP-coupled secondary antibodies, signals were amplified using tyramide signal amplification, and HRP detection was performed using DAB (Vector) for 2–3 minutes. Slides were briefly counterstained with fast green before mounting. For sclerostin immunostains, all photomicrographs were taken 3 mm below the growth plate on the lateral side of the tibia. Sections from at least 4 mice per experimental group were analyzed. Analysis of immunostaining was done based on coded sample numbers in a completely blinded manner.
In situ hybridization.
Digoxigenin-labeled (DIG-labeled) complementary RNA (cRNA) probes were synthesized using DIG RNA-labeling mix (Roche, 11277073910) with T7 RNA polymerase (Roche, 10881767001) or SP6 RNA polymerase (Roche, 10810274001) according to the manufacturer’s protocol. After DNase (Promega, M6101) treatment and subsequent ethanol precipitation using LiCl (MilliporeSigma, L9650), we resuspended the cRNA pellet in 100 μL RNase-free water. The target regions of the cRNA probes for Col10a1 and osteopontin are listed in Supplemental Table 3.
After baking, deparaffinization, and rehydration, the sections were subjected to fixation using 4% paraformaldehyde (PFA; Thermo Fisher Scientific, 416785000) for 10 minutes at room temperature. Thereafter, sections were subjected to Proteinase K treatment (Thermo Fisher Scientific, 17916; 3 μg/mL in PBS, pH 7.4) for 10 minutes at room temperature, followed by postfixation in 4% PFA for 5 minutes at room temperature, and then acetylation for 10 minutes at room temperature using 0.25% acetic acid and 0.1 M triethanolamine (MilliporeSigma, A6404 and T1377) in Milli-Q water (MilliporeSigma). Each step was followed by 2 PBS washes for 5 minutes. After the last PBS wash, the slides were immersed in 80% ethanol for 3 minutes and then air-dried briefly. We used 0.5 μL DIG-labeled cRNA probe per slide with 300 μL hybridization buffer containing 50% formamide (MilliporeSigma, F9037), 10 mM Tris (pH 8.0), 200 μg/mL yeast transfer RNA (MilliporeSigma, R8759), 1× Denhardt’s solution (MilliporeSigma, D2532), 10% dextran sulfate (MilliporeSigma, D8906), 600 mM NaCl, 0.25% SDS, and 1 mM EDTA (pH 8.0). We incubated the slides with glass coverslips in a moist slide box (with 50% formamide in 5× saline–sodium citrate [SSC] at the bottom) at 58°C overnight.
After overnight incubation, samples were washed sequentially with SSC (2× SSC at room temperature for 30 minutes, 2× SSC at 60°C for 1 hour, and then 0.2× SSC at 60°C for 1 hour). We blocked the slides at room temperature for 1 hour using 5% heat-inactivated sheep serum (MilliporeSigma, S3772) and 5% Blocking Reagent (Roche, 11096176001) in 100 mM Tris (pH 7.5)/150 mM NaCl buffer. Then, we incubated the slides with anti-DIG–alkaline phosphatase antibody (Roche, 11093274910) (1:2500 dilution by the blocking buffer) at room temperature for 2 hours. After 2 washes with 100 mM Tris (pH 7.5)/150 mM NaCl buffer for 15 minutes and a subsequent wash with 100 mM Tris (pH 9.5)/100 mM NaCl/50 mM MgCl2 buffer for 10 minutes, we incubated the slides with BM-Purple (Roche, 11442074001) at room temperature for 1–4 days until visible signals could be observed. We mounted the slides with Aqua-Mount Slide Mounting Media (Thermo Fisher Scientific, 13800) after fixing the slides with 4% PFA in PBS (pH 7.4) at room temperature for 30 minutes and washing subsequently by PBS.
Immunoblotting.
Whole cell lysates were collected by 200 μL of 4× Laemmli SDS sample buffer (Boston Bioproduct, BP-110R). Twenty microliters of lysates were separated by 10% SDS-PAGE (Expedeon, NXG01027), and proteins were transferred to Hybond ECL nitrocellulose membrane (Amersham, RPN303D). Membranes were blocked with 5% BSA (Boston Bioproduct, P-753) in TBS-T, and incubated with primary antibody overnight at 4°C. All antibodies (SIK3 pT441, HDAC4 pS245, and β-actin) were used at 1:1000 dilutions by the blocking solution. The next day, membranes were washed 3 times in TBS-T, incubated with HRP-coupled secondary antibodies (Santa Cruz Biotechnology, sc-2004) at 1:5000 dilution by TBS-T, and washed, and signals were detected with enhanced chemiluminescence (ECL; Bio-Rad, Clarity Western ECL Substrate, 1705060). All immunoblots were repeated at least twice with comparable results obtained. See complete unedited blots in the supplemental material.
Histomorphometry.
Right tibiae from 4-week-old mice were subjected to bone histomorphometric analysis. The mice were given calcein (20 mg/kg by i.p. injection) and demeclocycline (40 mg/kg by i.p. injection) 2 days and 1 day before necropsy, respectively. The tibia was dissected and fixed in 70% ethanol for 3 days. Fixed bones were dehydrated in graded ethanol, then infiltrated and embedded in methylmethacrylate without demineralization. Undecalcified 5- and 10-μm-thick longitudinal sections were obtained using a microtome (Leica Biosystems, RM2255). The 5-μm sections were stained with Goldner trichrome, and at least 2 nonconsecutive sections per sample were examined for measurement of cellular parameters. The 10-μm sections were left unstained for measurement of dynamic parameters, and only double labels were measured, avoiding nonspecific fluorochrome labeling. A standard dynamic bone histomorphometric analysis of the tibial metaphysis was done using the Osteomeasure analyzing system (Osteometrics Inc.). Measurements were performed in the area of secondary spongiosa, 200 μm below the proximal growth plate. The observer was blinded to the experimental genotype at the time of measurement. The structural, dynamic, and cellular parameters were calculated and expressed according to the standardized nomenclature (82). Because of overt differences in trabecular bone content comparing WT and SIK2/3 mutant mice, we chose to normalize parameters to tissue area rather than bone surfaces.
Micro-CT.
Assessment of bone morphology and microarchitecture was performed with high-resolution micro–computed tomography (μCT40; Scanco Medical) in 8-week-old male mice. Femurs and vertebrae were dissected, fixed overnight in neutral-buffered formalin, then stored in 70% EtOH until the time of scanning. In brief, the distal femoral metaphysis and mid-diaphysis were scanned using 70 kVp peak x-ray tube potential, 113 mAs x-ray tube current, 200 milliseconds integration time, and 10 μm isotropic voxel size. Cancellous (trabecular) bone was assessed in the distal metaphysis, and cortical bone was assessed in the mid-diaphysis. The femoral metaphysis region began 1700 μm proximal to the distal growth plate and extended 1500 μm distally. Cancellous bone was separated from cortical bone with a semiautomated contouring program. For the cancellous bone region we assessed bone volume fraction (BV/TV, percent), trabecular thickness (Tb.Th, mm), trabecular separation (Tb.Sp, mm), trabecular number (Tb.N, 1/mm), connectivity density (Conn.D, 1/mm), and structure model index. Transverse CT slices were also acquired in a 500-μm-long region at the femoral mid-diaphysis to assess total cross-sectional area, cortical bone area, and medullary area (Tt.Ar, Ct.Ar, and Ma.Ar, respectively, all mm2); bone area fraction (Ct.Ar/Tt.Ar, percent), cortical thickness (Ct.Th, mm), porosity (Ct.Po, percent), and minimum (Imin, mm4), maximum (Imax, mm4), and polar (J, mm4) moments of inertia. Bone was segmented from soft tissue using the same threshold, 300 mg hydroxyapatite/cm3 for trabecular and 700 mg hydroxyapatite/cm3 for cortical bone. Scanning and analyses adhered to the guidelines for the use of micro-CT for the assessment of bone architecture in rodents (83). Micro-CT analysis was done in a completely blinded manner with all mice assigned to coded sample numbers. Since Sik2/3 mutant bones show abnormal trabecular bone throughout the length of the femur, standard midshaft cortical parameters could not be determined for these mice. Therefore, for analysis of compound mutants shown in Figure 7A, we analyzed the bone volume fraction (BV/TV) through a fixed region in the femur midshaft that includes the cortical bone shell. This “R1” BV/TV parameter allows us to quantify the extent of abnormal medullary bone in the Sik2/3 mutants, which is absent in all other strains analyzed, including the Sik2/3 Hdac4/5 compound mutant mice.
Serum analysis.
Three-hour fasting serum was collected from mice just before sacrifice by retro-orbital bleed. Serum was isolated and analyte levels were determined using the following commercially available detection kits: P1NP from Immunodiagnostic Systems (AC-33F1), CTX from Immunodiagnostic Systems (AC-06F1), PTH from Immunotopics (60-2305), phosphate from BioVision (K410-500), and total calcium from Stanbio (0150). All absolute concentrations were determined based on interpolation from standard curves provided by the manufacturer.
Gene expression studies.
For long-bone RNA isolation from control and Sik2/3 double-KO animals, mice were sacrificed and both femurs were rapidly dissected on ice. Soft tissue was removed and epiphyses cut. Bone marrow cells were then removed by serial flushing of the marrow cavity with ice-cold PBS until bone appeared completely white. TRIzol (Life Technologies) was added to remaining bone tissue, and samples were frozen at –80°C and then homogenized. RNA was then extracted per the manufacturer’s instructions, and further purified on RNeasy microcolumns (Qiagen). RNA with an A260/280 ratio less than 1.7 was not used for downstream analysis; this workflow allows us to avoid use of samples with excessive bone matrix protein contamination. For cDNA synthesis, 1 μg RNA was used in synthesis reactions according to the instructions of the manufacturer (Primescript RT). SYBR Green–based quantitative PCR detection was performed using FastStart Universal SYBR Green (Roche) on a StepOne Plus (Applied Biosystems) thermocycler. All PCR primer sequences are listed in Supplemental Table 3. Transcript levels for genes of interest were determined relative to a housekeeping gene (β-actin) using standard methods (84). A similar protocol for cortical bone isolation was used for the comparison of effects of sclerostin antibody treatment versus vehicle. For bone RNA isolation from control and C1HR mice, the same protocol was used with the exception that the marrow flushing step was not performed. In light of overt phenotypic differences between control and experimental (Sik2/3 double-KO, C1HR-transgenic, Scl-Ab–treated) mice, we acknowledge that differential expression of many genes may be due to changes in cellular tissue composition. Therefore, we focus on similarities and differences across comparisons (WT/Sik2/3 double-KO vs. WT/C1HR, WT/Sik2/3 double-KO vs. control/Scl-Ab, WT/C1HR vs. control/Scl-Ab), to determine the overall molecular concordance between the 3 different models of increased trabecular bone mass.
RNA-Seq.
RNA sequencing from bone RNA samples for Figure 4 was conducted using a BGISEQ500 platform (BGI) (85). Briefly, RNA samples with RNA integrity number values greater than 8.0 were used for downstream library construction. mRNAs were isolated by PAGE, followed by adaptor ligation and reverse transcription with PCR amplification. PCR products were again purified by PAGE and dissolved in elution buffer solution. Double-stranded PCR products were heat-denatured and circularized by the splint oligo sequence. The single-stranded circle DNA was formatted as the final sequencing library, and validated on a bioanalyzer (Agilent 2100) before sequencing. The library was amplified with phi29 to generate DNA nanoballs, which were loaded into the patterned nanoarray followed by SE50 sequencing. On average we obtained 20 million reads per bone RNA sample. Sequencing reads were mapped by the STAR aligner (86) to the mm9 reference genome using Ensembl annotation. Gene expression counts (cpm) were calculated using HTSeq version 0.6.0 (87). Differential expression analysis was performed using the edgeR package (88) based on the criteria of more than 2-fold change in expression value versus control and FDRs less than 0.05. The RNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE133988).
Cortical bone RNA-Seq data from control and sclerostin antibody–treated mice were as previously described (54). Briefly, cortical bone RNA was isolated and sequencing libraries were generated using the TruSeq RNA Sample Preparation Kit version 2 (Illumina). Nine barcoded libraries were pooled per lane of PE50 sequencing on an Illumina HiSeq 2000. Reads were mapped to the mouse genome with RUM (http://www.cbil.upenn.edu/RUM/userguide.php); then differential expression analysis was performed with edgeR. The RNA-Seq data were deposited to ArrayExpress under accession number E-MTAB-8113.
Bioinformatics.
Venn diagrams of differentially expressed gene sets were generated using genes with more than 2-fold change in expression values and FDR less than 0.05. Gene ontology enrichment analyses were performed using Enrichr (89). Statistical testing for the significance of gene set overlaps was performed by hypergeometric test with a P value cutoff of 0.05. Venn diagrams of DEG lists were constructed using the Venny tool (http://bioinfogp.cnb.csic.es/tools/venny/).
Study approval.
Animals were housed in the Center for Comparative Medicine at Massachusetts General Hospital, and all experiments were approved by the hospital’s Subcommittee on Research Animal Care.
Quantification and statistics.
When 2 groups were compared, statistical analyses were performed using unpaired 2-tailed Student’s t test. When multiple simultaneous comparisons were performed between WT and mutant strains, the method of Benjamini, Krieger, and Yekutieli was used to determine the FDR. When more than 2 experimental groups were compared, 1-way ANOVA (GraphPad Prism 8.0) with Dunnett’s correction was performed. P values less than 0.05 were considered to be significant. The numbers of mice studied in all experiments are described in the figure legends, and in all figures data points represent individual mice. Data are expressed as mean ± SEM.
Author contributions
SN, TJG, HJ, HMK, MLB, and MNW designed research studies. SN, MJO, CDC, HN, CJJ, and MNW conducted the experiments. SN, MJO, CDC, JDSM, DJB, MB, MN, WO, MLB, and MNW acquired and analyzed data. MC and RIS performed bioinformatics analyses. MF and RB contributed reagents. UA contributed an RNA-Seq data set. MNW, SN, and HMK wrote the original manuscript, which was edited and approved by all coauthors. MNW and HMK acquired funding and supervised the study.
Supplementary Material
Acknowledgments
We thank Eric Olson, Jerry Feng, and Kei Sakamoto for providing reagents, David Fisher, Nathanael Gray, Michael Mannstadt, Lauren Surface, and Tatsuya Kobayashi for helpful discussions, members of the Wein laboratory for fruitful discussions, and Elizabeth Williams for assistance with animal husbandry. MNW acknowledges funding support from the American Society of Bone and Mineral Research, the Harrington Discovery Institute, and the NIH (DK116716, AR066261, and AR067285). HMK acknowledges funding support from the NIH (AR066261 and DK011794). RIS acknowledges funding support from the NIH (DK040561). MF acknowledges funding support from CNRS, the Société Francophone du Diabète (SFD), and the Fondation pour la Recherche Médicale (FRM). TJG and HJ acknowledge funding from the NIH (DK113039 and DK011794). RB acknowledges funding support from the NIH (DK092590 and AR059847). WO acknowledges funding support from the NIH (DE027421).
Version 1. 08/20/2019
In-Press Preview
Version 2. 10/22/2019
Electronic publication
Version 3. 12/02/2019
Print issue publication
Footnotes
Conflict of interest: MNW and HMK are coinventors on a pending patent (US patent application 16/333,546) regarding the use of SIK inhibitors for osteoporosis. MNW and TJG receive research support from Radius Health. MNW and HMK receive research support from Galapagos NV.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(12):5187–5203.https://doi.org/10.1172/JCI130126.
Contributor Information
Shigeki Nishimori, Email: snishimori@mgh.harvard.edu.
Hiroshi Noda, Email: h.noda470@gmail.com.
Murat Cetinbas, Email: cetinbas@molbio.mgh.harvard.edu.
Janaina da Silva Martins, Email: JDASILVAMARTINS@mgh.harvard.edu.
Ugur Ayturk, Email: ayturku@HSS.EDU.
Daniel J. Brooks, Email: djbrooks@bidmc.harvard.edu.
Michael Bruce, Email: MBRUCE3@mgh.harvard.edu.
Mizuki Nagata, Email: mizukin@umich.edu.
Wanida Ono, Email: wono@umich.edu.
Christopher J. Janton, Email: CJANTON@mgh.harvard.edu.
Mary L. Bouxsein, Email: mbouxsei@bidmc.harvard.edu.
Marc Foretz, Email: marc.foretz@inserm.fr.
Rebecca Berdeaux, Email: rebecca.berdeaux@uth.tmc.edu.
Ruslan I. Sadreyev, Email: sadreyev@molbio.mgh.harvard.edu.
Thomas J. Gardella, Email: Gardella@helix.MGH.Harvard.edu.
Harald Jüppner, Email: hjueppner@partners.org.
Henry M. Kronenberg, Email: hkronenberg@partners.org.
Marc N. Wein, Email: MNWEIN@partners.org.
References
- 1. Gardella T, Jüppner H, Bringhurst F, Potts J Jr. Receptors for parathyroid hormone (PTH) and PTH-related protein. In: Bilezikian J, Raisz L, Martin T, eds. Principles of Bone Biology. 3rd ed. Amsterdam, Netherlands: Elsevier; 2008:555–576. [Google Scholar]
- 2.Bergwitz C, Jüppner H. Regulation of phosphate homeostasis by PTH, vitamin D, and FGF23. Annu Rev Med. 2010;61:91–104. doi: 10.1146/annurev.med.051308.111339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Crombrugghe B, Lefebvre V, Nakashima K. Regulatory mechanisms in the pathways of cartilage and bone formation. Curr Opin Cell Biol. 2001;13(6):721–727. doi: 10.1016/S0955-0674(00)00276-3. [DOI] [PubMed] [Google Scholar]
- 4.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 5.Wein MN, Kronenberg HM. Regulation of bone remodeling by parathyroid hormone. Cold Spring Harb Perspect Med. 2018;8(8):a031237. doi: 10.1101/cshperspect.a031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheloha RW, Gellman SH, Vilardaga JP, Gardella TJ. PTH receptor-1 signalling—mechanistic insights and therapeutic prospects. Nat Rev Endocrinol. 2015;11(12):712–724. doi: 10.1038/nrendo.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neer RM, et al. Effect of parathyroid hormone (1-34) on fractures and bone mineral density in postmenopausal women with osteoporosis. N Engl J Med. 2001;344(19):1434–1441. doi: 10.1056/NEJM200105103441904. [DOI] [PubMed] [Google Scholar]
- 8.Mannstadt M, et al. Efficacy and safety of recombinant human parathyroid hormone (1-84) in hypoparathyroidism (REPLACE): a double-blind, placebo-controlled, randomised, phase 3 study. Lancet Diabetes Endocrinol. 2013;1(4):275–283. doi: 10.1016/S2213-8587(13)70106-2. [DOI] [PubMed] [Google Scholar]
- 9.Wein MN, Foretz M, Fisher DE, Xavier RJ, Kronenberg HM. Salt-inducible kinases: physiology, regulation by cAMP, and therapeutic potential. Trends Endocrinol Metab. 2018;29(10):723–735. doi: 10.1016/j.tem.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakamoto K, Bultot L, Göransson O. The salt-inducible kinases: emerging metabolic regulators. Trends Endocrinol Metab. 2018;29(12):827–840. doi: 10.1016/j.tem.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Takemori H, Halder SK, Nonaka Y, Okamoto M. Cloning of a novel kinase (SIK) of the SNF1/AMPK family from high salt diet-treated rat adrenal. FEBS Lett. 1999;453(1–2):135–139. doi: 10.1016/s0014-5793(99)00708-5. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto M, Takemori H, Katoh Y. Salt-inducible kinase in steroidogenesis and adipogenesis. Trends Endocrinol Metab. 2004;15(1):21–26. doi: 10.1016/j.tem.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- 14.Lizcano JM, et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 2004;23(4):833–843. doi: 10.1038/sj.emboj.7600110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonntag T, Vaughan JM, Montminy M. 14-3-3 proteins mediate inhibitory effects of cAMP on salt-inducible kinases (SIKs) FEBS J. 2018;285(3):467–480. doi: 10.1111/febs.14351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Patel K, et al. The LKB1-salt-inducible kinase pathway functions as a key gluconeogenic suppressor in the liver. Nat Commun. 2014;5:4535. doi: 10.1038/ncomms5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henriksson E, et al. SIK2 regulates CRTCs, HDAC4 and glucose uptake in adipocytes. J Cell Sci. 2015;128(3):472–486. doi: 10.1242/jcs.153932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henriksson E, et al. The AMPK-related kinase SIK2 is regulated by cAMP via phosphorylation at Ser358 in adipocytes. Biochem J. 2012;444(3):503–514. doi: 10.1042/BJ20111932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat Rev Mol Cell Biol. 2011;12(3):141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Screaton RA, et al. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119(1):61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 22.Berdeaux R, et al. SIK1 is a class II HDAC kinase that promotes survival of skeletal myocytes. Nat Med. 2007;13(5):597–603. doi: 10.1038/nm1573. [DOI] [PubMed] [Google Scholar]
- 23.Stewart R, Akhmedov D, Robb C, Leiter C, Berdeaux R. Regulation of SIK1 abundance and stability is critical for myogenesis. Proc Natl Acad Sci U S A. 2013;110(1):117–122. doi: 10.1073/pnas.1212676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clark K, et al. Phosphorylation of CRTC3 by the salt-inducible kinases controls the interconversion of classically activated and regulatory macrophages. Proc Natl Acad Sci U S A. 2012;109(42):16986–16991. doi: 10.1073/pnas.1215450109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundberg TB, et al. Small-molecule screening identifies inhibition of salt-inducible kinases as a therapeutic strategy to enhance immunoregulatory functions of dendritic cells. Proc Natl Acad Sci U S A. 2014;111(34):12468–12473. doi: 10.1073/pnas.1412308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacKenzie KF, et al. PGE(2) induces macrophage IL-10 production and a regulatory-like phenotype via a protein kinase A-SIK-CRTC3 pathway. J Immunol. 2013;190(2):565–577. doi: 10.4049/jimmunol.1202462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luan B, Yoon YS, Le Lay J, Kaestner KH, Hedrick S, Montminy M. CREB pathway links PGE2 signaling with macrophage polarization. Proc Natl Acad Sci U S A. 2015;112(51):15642–15647. doi: 10.1073/pnas.1519644112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mujahid N, et al. A UV-independent topical small-molecule approach for melanin production in human skin. Cell Rep. 2017;19(11):2177–2184. doi: 10.1016/j.celrep.2017.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wein MN, et al. SIKs control osteocyte responses to parathyroid hormone. Nat Commun. 2016;7:13176. doi: 10.1038/ncomms13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasagawa S, et al. SIK3 is essential for chondrocyte hypertrophy during skeletal development in mice. Development. 2012;139(6):1153–1163. doi: 10.1242/dev.072652. [DOI] [PubMed] [Google Scholar]
- 31.Weir EC, Philbrick WM, Amling M, Neff LA, Baron R, Broadus AE. Targeted overexpression of parathyroid hormone-related peptide in chondrocytes causes chondrodysplasia and delayed endochondral bone formation. Proc Natl Acad Sci U S A. 1996;93(19):10240–10245. doi: 10.1073/pnas.93.19.10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimori S, et al. PTHrP targets HDAC4 and HDAC5 to repress chondrocyte hypertrophy. JCI Insight. 2019;4(5):97903. doi: 10.1172/jci.insight.97903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Csukasi F, et al. The PTH/PTHrP-SIK3 pathway affects skeletogenesis through altered mTOR signaling. Sci Transl Med. 2018;10(459):eaat9356. doi: 10.1126/scitranslmed.aat9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozhemyakina E, Cohen T, Yao TP, Lassar AB. Parathyroid hormone-related peptide represses chondrocyte hypertrophy through a protein phosphatase 2A/histone deacetylase 4/MEF2 pathway. Mol Cell Biol. 2009;29(21):5751–5762. doi: 10.1128/MCB.00415-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vega RB, et al. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119(4):555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 36.Arnold MA, et al. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12(3):377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Berggreen C, Henriksson E, Jones HA, Morrice N, Göransson O. cAMP-elevation mediated by β-adrenergic stimulation inhibits salt-inducible kinase (SIK) 3 activity in adipocytes. Cell Signal. 2012;24(9):1863–1871. doi: 10.1016/j.cellsig.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26(2):145–146. doi: 10.1002/(SICI)1526-968X(200002)26:2<145::AID-GENE14>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 39.Karaplis AC, et al. Lethal skeletal dysplasia from targeted disruption of the parathyroid hormone-related peptide gene. Genes Dev. 1994;8(3):277–289. doi: 10.1101/gad.8.3.277. [DOI] [PubMed] [Google Scholar]
- 40.Ono W, Sakagami N, Nishimori S, Ono N, Kronenberg HM. Parathyroid hormone receptor signalling in osterix-expressing mesenchymal progenitors is essential for tooth root formation. Nat Commun. 2016;7:11277. doi: 10.1038/ncomms11277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schipani E, et al. Targeted expression of constitutively active receptors for parathyroid hormone and parathyroid hormone-related peptide delays endochondral bone formation and rescues mice that lack parathyroid hormone-related peptide. Proc Natl Acad Sci U S A. 1997;94(25):13689–13694. doi: 10.1073/pnas.94.25.13689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Philbrick WM, Dreyer BE, Nakchbandi IA, Karaplis AC. Parathyroid hormone-related protein is required for tooth eruption. Proc Natl Acad Sci U S A. 1998;95(20):11846–11851. doi: 10.1073/pnas.95.20.11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schipani E, Kruse K, Jüppner H. A constitutively active mutant PTH-PTHrP receptor in Jansen-type metaphyseal chondrodysplasia. Science. 1995;268(5207):98–100. doi: 10.1126/science.7701349. [DOI] [PubMed] [Google Scholar]
- 44.Schipani E, et al. Constitutively activated receptors for parathyroid hormone and parathyroid hormone-related peptide in Jansen’s metaphyseal chondrodysplasia. N Engl J Med. 1996;335(10):708–714. doi: 10.1056/NEJM199609053351004. [DOI] [PubMed] [Google Scholar]
- 45.Calvi LM, et al. Activated parathyroid hormone/parathyroid hormone-related protein receptor in osteoblastic cells differentially affects cortical and trabecular bone. J Clin Invest. 2001;107(3):277–286. doi: 10.1172/JCI11296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Brien CA, et al. Control of bone mass and remodeling by PTH receptor signaling in osteocytes. PLoS One. 2008;3(8):e2942. doi: 10.1371/journal.pone.0002942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baryawno N, et al. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177(7):1915–1932.e16. doi: 10.1016/j.cell.2019.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tikhonova AN, et al. The bone marrow microenvironment at single-cell resolution. Nature. 2019;569(7755):222–228. doi: 10.1038/s41586-019-1104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu Y, Xie Y, Zhang S, Dusevich V, Bonewald LF, Feng JQ. DMP1-targeted Cre expression in odontoblasts and osteocytes. J Dent Res. 2007;86(4):320–325. doi: 10.1177/154405910708600404. [DOI] [PubMed] [Google Scholar]
- 50.Calvi LM, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425(6960):841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 51.Ono N, et al. Osteopontin negatively regulates parathyroid hormone receptor signaling in osteoblasts. J Biol Chem. 2008;283(28):19400–19409. doi: 10.1074/jbc.M800005200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nioi P, et al. Transcriptional profiling of laser capture microdissected subpopulations of the osteoblast lineage provides insight into the early response to sclerostin antibody in rats. J Bone Miner Res. 2015;30(8):1457–1467. doi: 10.1002/jbmr.2482. [DOI] [PubMed] [Google Scholar]
- 53.Baron R, Kneissel M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat Med. 2013;19(2):179–192. doi: 10.1038/nm.3074. [DOI] [PubMed] [Google Scholar]
- 54.Kedlaya R, et al. Sclerostin inhibition reverses skeletal fragility in an Lrp5-deficient mouse model of OPPG syndrome. Sci Transl Med. 2013;5(211):211ra158. doi: 10.1126/scitranslmed.3006627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahmed AA, et al. SIK2 is a centrosome kinase required for bipolar mitotic spindle formation that provides a potential target for therapy in ovarian cancer. Cancer Cell. 2010;18(2):109–121. doi: 10.1016/j.ccr.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakamaki J, et al. Role of the SIK2-p35-PJA2 complex in pancreatic β-cell functional compensation. Nat Cell Biol. 2014;16(3):234–244. doi: 10.1038/ncb2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Funato H, et al. Forward-genetics analysis of sleep in randomly mutagenized mice. Nature. 2016;539(7629):378–383. doi: 10.1038/nature20142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Z, et al. Quantitative phosphoproteomic analysis of the molecular substrates of sleep need. Nature. 2018;558(7710):435–439. doi: 10.1038/s41586-018-0218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Honda T, et al. A single phosphorylation site of SIK3 regulates daily sleep amounts and sleep need in mice. Proc Natl Acad Sci U S A. 2018;115(41):10458–10463. doi: 10.1073/pnas.1810823115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guerriero JL, et al. Class IIa HDAC inhibition reduces breast tumours and metastases through anti-tumour macrophages. Nature. 2017;543(7645):428–432. doi: 10.1038/nature21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lobera M, et al. Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat Chem Biol. 2013;9(5):319–325. doi: 10.1038/nchembio.1223. [DOI] [PubMed] [Google Scholar]
- 62.Onal M, St John HC, Danielson AL, Pike JW. Deletion of the distal Tnfsf11 RL-D2 enhancer that contributes to PTH-mediated RANKL expression in osteoblast lineage cells results in a high bone mass phenotype in mice. J Bone Miner Res. 2016;31(2):416–429. doi: 10.1002/jbmr.2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ricarte FR, Le Henaff C, Kolupaeva VG, Gardella TJ, Partridge NC. Parathyroid hormone(1-34) and its analogs differentially modulate osteoblastic Rankl expression via PKA/SIK2/SIK3 and PP1/PP2A-CRTC3 signaling. J Biol Chem. 2018;293(52):20200–20213. doi: 10.1074/jbc.RA118.004751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Delgado-Calle J, Sato AY, Bellido T. Role and mechanism of action of sclerostin in bone. Bone. 2017;96:29–37. doi: 10.1016/j.bone.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kedlaya R, et al. Adult-onset deletion of β-catenin in (10kb)Dmp1-expressing cells prevents intermittent PTH-induced bone gain. Endocrinology. 2016;157(8):3047–3057. doi: 10.1210/en.2015-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu C, et al. Postnatal deletion of β-catenin in osterix-expressing cells is necessary for bone growth and intermittent PTH-induced bone gain. J Bone Miner Metab. 2018;36(5):560–572. doi: 10.1007/s00774-017-0873-0. [DOI] [PubMed] [Google Scholar]
- 67.Wan M, et al. Parathyroid hormone signaling through low-density lipoprotein-related protein 6. Genes Dev. 2008;22(21):2968–2979. doi: 10.1101/gad.1702708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo J, et al. Suppression of Wnt signaling by Dkk1 attenuates PTH-mediated stromal cell response and new bone formation. Cell Metab. 2010;11(2):161–171. doi: 10.1016/j.cmet.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Terauchi M, et al. T lymphocytes amplify the anabolic activity of parathyroid hormone through Wnt10b signaling. Cell Metab. 2009;10(3):229–240. doi: 10.1016/j.cmet.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bellido T. Downregulation of SOST/sclerostin by PTH: a novel mechanism of hormonal control of bone formation mediated by osteocytes. J Musculoskelet Neuronal Interact. 2006;6(4):358–359. [PubMed] [Google Scholar]
- 71.Darling NJ, Toth R, Arthur JS, Clark K. Inhibition of SIK2 and SIK3 during differentiation enhances the anti-inflammatory phenotype of macrophages. Biochem J. 2017;474(4):521–537. doi: 10.1042/BCJ20160646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miranda F, et al. Salt-inducible kinase 2 couples ovarian cancer cell metabolism with survival at the adipocyte-rich metastatic niche. Cancer Cell. 2016;30(2):273–289. doi: 10.1016/j.ccell.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 73.Zhou J, et al. A novel compound ARN-3236 inhibits salt-inducible kinase 2 and sensitizes ovarian cancer cell lines and xenografts to paclitaxel. Clin Cancer Res. 2017;23(8):1945–1954. doi: 10.1158/1078-0432.CCR-16-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tarumoto Y, et al. LKB1, salt-inducible kinases, and MEF2C are linked dependencies in acute myeloid leukemia. Mol Cell. 2018;69(6):1017–1027.e6. doi: 10.1016/j.molcel.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maxfield KE, Macion J, Vankayalapati H, Whitehurst AW. SIK2 restricts autophagic flux to support triple-negative breast cancer survival. Mol Cell Biol. 2016;36(24):3048–3057. doi: 10.1128/MCB.00380-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sundberg TB, et al. Development of chemical probes for investigation of salt-inducible kinase function in vivo. ACS Chem Biol. 2016;11(8):2105–2111. doi: 10.1021/acschembio.6b00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Potthoff MJ, et al. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J Clin Invest. 2007;117(9):2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chang S, McKinsey TA, Zhang CL, Richardson JA, Hill JA, Olson EN. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol Cell Biol. 2004;24(19):8467–8476. doi: 10.1128/MCB.24.19.8467-8476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468(7324):653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nixon M, et al. Skeletal muscle salt inducible kinase 1 promotes insulin resistance in obesity. Mol Metab. 2016;5(1):34–46. doi: 10.1016/j.molmet.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakamura Y, Inloes JB, Katagiri T, Kobayashi T. Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol Cell Biol. 2011;31(14):3019–3028. doi: 10.1128/MCB.05178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dempster DW, et al. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 2013;28(1):2–17. doi: 10.1002/jbmr.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Müller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res. 2010;25(7):1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- 84.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 85.Fehlmann T, et al. cPAS-based sequencing on the BGISEQ-500 to explore small non-coding RNAs. Clin Epigenetics. 2016;8:123. doi: 10.1186/s13148-016-0287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Anders S, Pyl PT, Huber W. HTSeq — a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.