Transcriptomics combined with single-cell metabolomics give new information on the role of rays in lignification of developing xylem in Norway spruce.
Abstract
A comparative transcriptomic study and a single-cell metabolome analysis were combined to determine whether parenchymal ray cells contribute to the biosynthesis of monolignols in the lignifying xylem of Norway spruce (Picea abies). Ray parenchymal cells may function in the lignification of upright tracheids by supplying monolignols. To test this hypothesis, parenchymal ray cells and upright tracheids were dissected with laser-capture microdissection from tangential cryosections of developing xylem of spruce trees. The transcriptome analysis revealed that among the genes involved in processes typical for vascular tissues, genes encoding cell wall biogenesis-related enzymes were highly expressed in both developing tracheids and ray cells. Interestingly, most of the shikimate and monolignol biosynthesis pathway-related genes were equally expressed in both cell types. Nonetheless, 1,073 differentially expressed genes were detected between developing ray cells and tracheids, among which a set of genes expressed only in ray cells was identified. In situ single cell metabolomics of semi-intact plants by picoliter pressure probe-electrospray ionization-mass spectrometry detected monolignols and their glycoconjugates in both cell types, indicating that the biosynthetic route for monolignols is active in both upright tracheids and parenchymal ray cells. The data strongly support the hypothesis that in developing xylem, ray cells produce monolignols that contribute to lignification of tracheid cell walls.
In conifers such as Norway spruce (Picea abies), lignin is a major cell wall constituent of secondary xylem (wood), forming approximately 27% of the dry weight (Jaakkola et al., 2007), representing a major sink of carbon from photosynthesis in the form of secondary compounds derived from Phe (Pascual et al., 2016). In wood, the cell wall cellulose and hemicellulose network is impregnated with lignin, which sustains structural stability and supports water transport. The current view of the lignification process encompasses many steps, starting with the biosynthesis of monolignols in the cytosol and their transport across the plasma membrane. The monolignol transport mechanism is still only partly characterized, but the data obtained indicate that ATP-binding cassette (ABC) transporters have an important role (Miao and Liu, 2010; Alejandro et al., 2012). In the cell wall, monolignols are exposed to extracellular peroxidases and laccases, the enzymes initiating polymerization of lignin by oxidizing monolignols to phenolic radicals that couple nonenzymatically (Boerjan et al., 2003; Barros et al., 2015; Laitinen et al., 2017). Until now, it has not been clear whether monolignol biosynthesis in conifers is a cell-autonomous process in xylem tracheids, the main cell type in conifer wood, or whether neighboring cells (i.e. ray parenchymal cells) contribute to the production of monolignols. Cell culture studies with the angiosperm species Zinnia elegans indicate that nondifferentiating cells supply monolignols to differentiating tracheary elements (the good-neighbor hypothesis; Hosokawa et al., 2001; Tokunaga et al., 2005; Pesquet et al., 2013). Also, in inflorescence stems of Arabidopsis (Arabidopsis thaliana), parenchymal cells and developing fibers in the xylem produce monolignols for lignifying vessels, whereas in interfascicular fibers, lignification is a cell-autonomous process (Smith et al., 2013, 2017). In Norway spruce, the ray parenchymal cells do not seem to lignify during the growing season, based on staining with safranin and Alcian Blue for lignin and cellulose, respectively (see figure 1 in Marjamaa et al., 2003), providing a model system for testing the good-neighbor hypothesis. Also, in Scots pine (Pinus sylvestris), the cell walls of the present year’s ray cells do not lignify (Marjamaa et al., 2003), whereas the onset of lignification in ray parenchyma has been observed in a growth ring adjacent to the heartwood border in Scots pine (Bergström, 2003) and Pinus densiflora (Zheng et al., 2014). Therefore, the expression of lignin biosynthesis genes in the present year’s developing ray cells would strongly support the good-neighbor hypothesis in conifers.
Different transcriptomic approaches have been employed to reveal complex regulatory mechanisms and genes behind secondary xylem development (Carvalho et al., 2013; Nakano et al., 2015; Raherison et al., 2015; Zhong and Ye, 2015; Lamara et al., 2016; Barra-Jiménez and Ragni, 2017; Jokipii-Lukkari et al., 2017, 2018). Microarray and mRNA sequencing (RNA-Seq) approaches have identified a set of genes involved in cell wall polysaccharide and lignin synthesis as essential for cell wall morphology, composition, and wood quality. It has been shown that the processes are controlled by microRNA at the posttranscriptional level (Carvalho et al., 2013). Recently, an RNA-Seq approach conducted from mRNA isolated from xylem and cambium/phloem of Norway spruce collected over a year revealed a specific set of highly coexpressed monolignol biosynthesis genes with a high level of expression in developing xylem during the period of active lignification (Jokipii-Lukkari et al., 2018). Interactions in the transcriptional network controlling vascular development have also been studied (Duval et al., 2014; Raherison et al., 2015; Lamara et al., 2016; Jokipii-Lukkari et al., 2018). Identification of groups of transcription factors coexpressed with their potential target genes has revealed members of two transcription factor families that function as master switches in secondary cell wall development: NAC (NAM, ATAF1/2, and CUC2) and R2R3-MYB. They control gene expression of lower hierarchical level transcription factors as well as those encoding cellulose, hemicellulose, and lignin biosynthesis-related enzymes (Duval et al., 2014; Nakano et al., 2015; Zhong and Ye, 2015). Gene association analysis of wood properties in white spruce (Picea glauca) has linked the majority of wood trait-associated genes expressed in the secondary xylem with either NAC or MYB transcriptional regulators, suggesting that the regulatory mechanisms identified in model angiosperm systems are conserved in conifers (Lamara et al., 2016). For example, overexpression of two MYB genes of loblolly pine (Pinus taeda) in white spruce resulted in overlapping up-regulation of multiple phenylpropanoid pathway genes and enhanced lignin deposition (Bomal et al., 2008).
A recent gene expression Web resource, NorWood (Jokipii-Lukkari et al., 2017), was created by sequencing Norway spruce mRNA isolated from cryosections cut from the cambium and developing xylem into the previous year’s mature xylem. This database, together with the corresponding Populus tremula AspWoodresource, containing high-spatial-resolution data on gene expression over the corresponding sections (including phloem of European aspen [P. tremula]), have shown large-scale conservation in the transcriptome and coexpression networks of wood-forming tissues of softwood and hardwood species (Sundell et al., 2017). These resources represent a powerful community tool for generating evolutionary and developmental insights into the divergence of wood formation between angiosperms and gymnosperms and for advancing understanding on the regulation of wood development. However, these data do not allow for a separate study on gene expression of different cell types present in spruce xylem (i.e. upright tracheids, ray parenchymal cells [further referred to as tracheids and ray cells, respectively], ray tracheids, and epithelial cells of resin ducts; Fig. 1).
Figure 1.
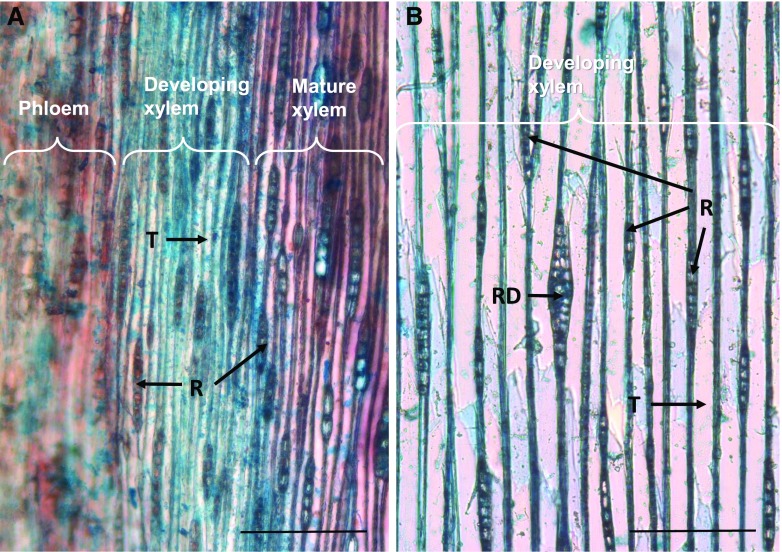
Photomicrographs of tangential cryosections of 4-year-old and 40-year-old Norway spruce stems stained with safranin and Alcian Blue. Note the smaller cell size in the young stem (A) in comparison with the old stem (B). Developing xylem (blue) and mature xylem (red) are stained with Alcian Blue and safranin, for cellulosic and lignified cell walls, respectively. In B, only the developing xylem is shown. Tracheid (T), resin duct (RD), and rays (R) are indicated. Bars = 200 μm.
An RNA-Seq study of individual cell types separated with laser-capture microdissection (LCM) from the bark of white spruce has revealed that analysis of the transcriptome at the whole-tissue level masks the cell type-specific expression of genes (Celedon et al., 2017). Hence, it is crucial to evaluate gene expression in specific cell types in order to understand the physiological phenomena occurring in those cells. Earlier, LCM was used successfully with other plant tissues (Ruel et al., 2009; Abbott et al., 2010; Hogekamp et al., 2011; Cañas et al., 2014; Nagy et al., 2014). To resolve the possible contribution of ray cells in lignification of tracheids in Norway spruce developing xylem, the LCM protocol for cryosections of developing xylem was first optimized, then ray cells and tracheids were collected separately with LCM (Blokhina et al., 2017). The isolated cells were then used for RNA extraction and transcriptome analysis. Since transcript abundances do not always translate into protein abundances, enzymatic activities, and hence, to metabolite levels, single-cell metabolome analysis was conducted separately from developing ray cells and tracheids using picoliter pressure probe-electrospray ionization-mass spectrometry (picoPPESI-MS; Nakashima et al., 2016) to investigate whether monolignol-related metabolites existed in developing ray cells.
Both the transcriptomic and metabolomic data strongly suggest that developing ray cells contribute to lignification of cell walls of developing tracheids. The analysis also identified a gene set that was expressed only in developing ray cells and not in tracheids. In our work, we have identified yet another role for the multifunctional ray parenchymal cells: participation in the lignification of adjacent tracheids.
RESULTS
The aim of this study was to evaluate whether ray cells contribute to lignification of tracheids according to the good-neighbor hypothesis suggested for angiosperms (Pesquet et al., 2013; Smith et al., 2013, 2017). To address this, global gene expression was investigated separately in upright tracheids and in ray parenchymal cells collected by LCM from developing xylem of mature Norway spruce trees grown in Finland. Due to size limitations in the experimental setup, young (approximately 4 years old) Norway spruce trees grown in Japan were used for single-cell metabolomics by picoPPESI-MS. Finally, the obtained results were compared with available Web resources to identify cell type-specific gene expression in ray parenchymal cells and xylem tracheids.
Tissue Anatomy of the Xylem of 4- and 40-Year Old Norway Spruce Trees
For practical reasons, spruce trees of different genotypes were used for transcriptomic and metabolomics analyses. Since trees of different maturity were utilized (approximately 40-year-old mature trees and approximately 4-year-old juvenile trees), the anatomy of xylem was investigated (Fig. 1). Xylem tracheids of a juvenile tree stem were narrower compared with those of a mature tree. The ratio of the number of rays to that of tracheids, however, was the same. In differentiating xylem of both juvenile and mature trees, developing resin ducts were present, but only occasionally.
Quality of RNA Extractions
Care was taken to exclude resin ducts and ray tracheids from the dissected material used for RNA isolation (Supplemental Fig. S1). Whole cryosections of developing xylem were also analyzed to address RNA quality issues during LCM (Blokhina et al., 2017) and to serve as a reference transcriptome. RNA of adequate to good quality was isolated from the tracheids and ray cells, with RNA integrity numbers in the range 6.1 to 9.2. Total RNA yield obtained from the microdissected cells was approximately 60 ng per sample for rays and tracheids and approximately 90 to 180 ng for the whole sections. The RNA yield per volume of the tissue was 0.5 ng mm−3 (whole sections), 3.4 ng mm−3 (tracheids), and 9.6 ng mm−3 (ray cells).
Gene Expression in Developing Tracheids and Ray Cells
In our RNA-Seq analysis, of 70,736 gene models present in the Norway spruce genome (version 1; Nystedt et al., 2013), 69.5% were expressed in at least some of the sample types analyzed. To evaluate whether differences existed in the most highly expressed genes in tracheids and ray cells, Gene Ontology (GO) category enrichment testing was conducted for the 500 most highly expressed genes of both cell types. This analysis revealed that similar cellular metabolism was active in developing tracheids and in developing ray cells (Fig. 2; Supplemental Table S1). For example, genes involved in response to various stimuli, and in phenylpropanoid, carbohydrate, protein, and amino acid metabolism, as well as in cell wall biogenesis were highly expressed in both cell types.
Figure 2.
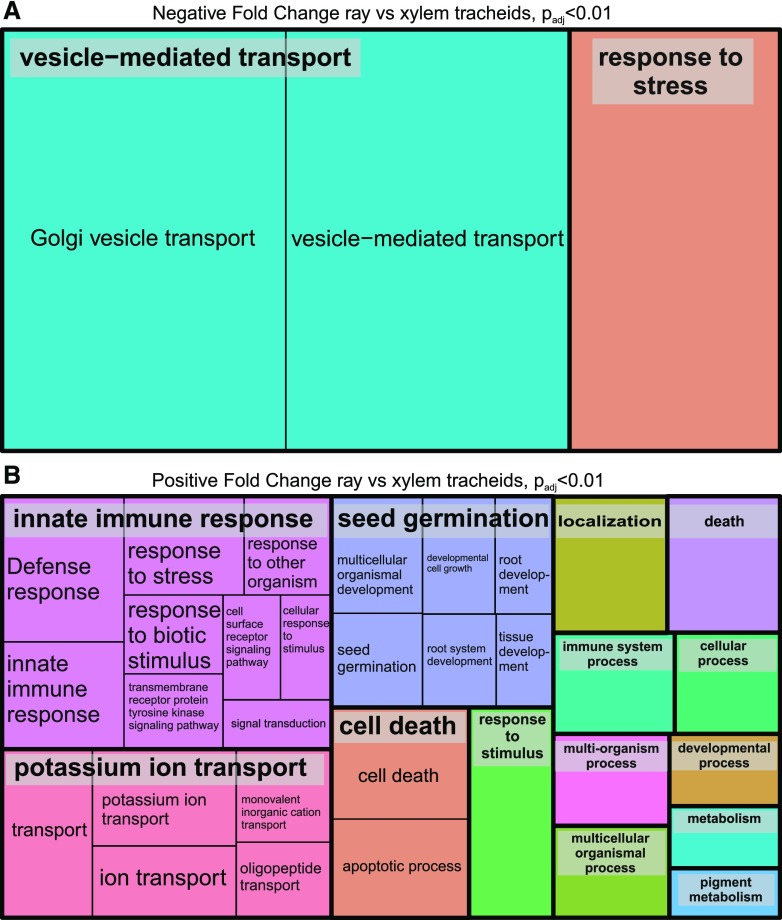
REVIGO Treemaps of 500 most highly expressed genes in developing xylem. The GO enrichment was realized using the Web-based tool agriGO (http://bioinfo.cau.edu.cn/agriGO/index.php; Du et al., 2010). Developing tracheids (A) and developing ray parenchymal cells (B) are shown. See Supplemental Table S1 for gene identifiers.
Some differences in transcript abundances were observed in the isolated xylem cell types. Of 1,073 differentially expressed genes, 541 genes were more highly expressed while expression of 532 genes was less abundant in developing ray cells than in developing tracheids (Padj < 0.01; Supplemental Table S2). Transcripts more abundant in ray cells contained, for example, genes related to immune response and signaling, ion transport, and cell death (Fig. 3B), whereas the GO terms vesicle transport and response to stress were enriched for the genes more highly expressed in tracheids (Fig. 3A).
Figure 3.
REVIGO Treemaps of differentially expressed genes. Genes more highly expressed in developing tracheids are shown in A, and those more highly expressed in developing ray parenchymal cells are shown in B (Padj < 0.01). The GO enrichment was realized using the Web-based tool agriGO (http://bioinfo.cau.edu.cn/agriGO/index.php; Du et al., 2010). See Supplemental Table S2 for gene identifiers.
Lignin Biosynthesis Genes: Shikimate, Phe/Tyr, Phenylpropanoid, and Monolignol Pathway Genes Were Highly Expressed in Both Ray Cells and Tracheids
To resolve whether developing ray cells synthesize monolignols, special focus was placed on the expression of the genes in the pathways generating monolignols. In the Norway spruce genome (version 1), there are over 100 shikimate and aromatic amino acid pathway-related genes leading to Phe and Tyr and over 200 phenylpropanoid and monolignol biosynthesis pathway-related genes leading to monolignols (Nystedt et al., 2013). Phenylpropanoid metabolism was active in both developing tracheids and ray cells, as was evident from the GO enrichment of the 500 most highly expressed genes (Fig. 2; Supplemental Table S1). Expression of the shikimate, aromatic amino acid, phenylpropanoid, and monolignol pathway-related genes was equally active in both cell types (Fig. 4; Supplemental Table S3). In general, enzymes encoded by the same genes were responsible for the phenylpropanoid metabolism in the cell types studied, since only a few genes were differentially expressed (Fig. 4; Supplemental Table S3). Of the 38 most highly expressed shikimate and Phe/Tyr pathway-related genes, transcripts of two (3-DEHYDROQUINATE SYNTHASE and 3-DEHYDROQUINATE DEHYDRATASE/SHIKIMATE DEHYDROGENASE; fold change 2.2 and 2.4, respectively) were less abundant, and one (AROGENATE DEHYDRATASE/PREPHENATE DEHYDRATASE; fold change 2.8) was more abundant in ray cells compared with tracheids (Padj < 0.05; Fig. 4; Supplemental Table S3). Eleven of the pathway genes detected in this study have been reported previously as induced twofold or greater under extracellular lignin-forming conditions in a cell culture of Norway spruce (Laitinen et al., 2017).
Figure 4.
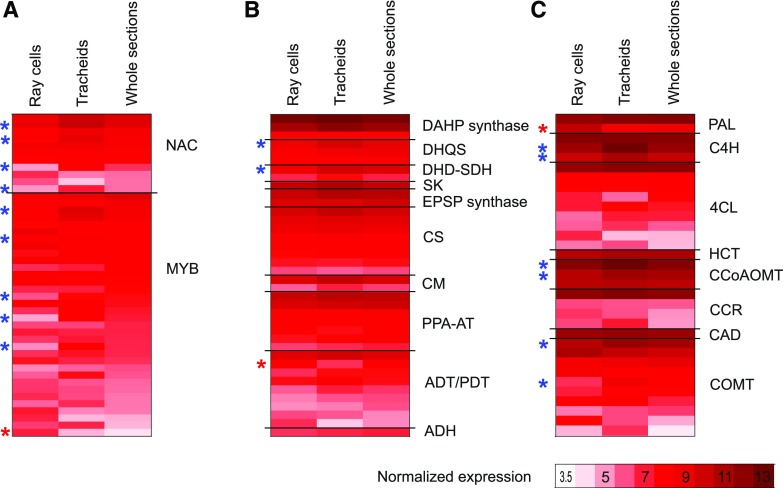
Average-normalized expression of genes important in lignification. NAC and MYB transcription factors (A), shikimate, Phe, and Tyr pathway genes leading to Phe and Tyr (B), and phenylpropanoid and monolignol biosynthesis pathway genes leading to monolignols (C) are shown in ray parenchymal cells, upright tracheids, and whole sections of developing xylem with expression levels higher than 5 (variance-stabilizing transformation [VST] > 5) at least in one of the sample types. Red stars count for significantly higher and blue for significantly lower levels of expression in ray cells than in tracheids (Padj ≤ 0.05). Shikimate, Phe, and Tyr pathway genes are according to Maeda and Dudareva (2012): ADH, arogenate dehydrogenase; ADT, arogenate dehydratase; CM, chorismate mutase; CS, chorismate synthase; DAHP synthase, 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase; DHD-SDH, 3-dehydroquinate dehydratase-shikimate dehydrogenase; DHQS, 3-dehydroquinate synthase; EPSP synthase, 5-enolpyruvylshikimate 3-phosphate synthase; PDT, prephenate dehydratase; PPA-AT, prephenate aminotransferase; SK, shikimate kinase. General phenylpropanoid pathway genes are as follows: CCoAOMT, caffeoyl-CoA 3-O-methyltransferase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate-CoA ligase; HCT, hydroxycinnamoyl-CoA shikimate/quinate hydroxycinnamoyl transferase; PAL, phenylalanine ammonia lyase. Monolignol pathway genes are as follows: CAD, cinnamyl alcohol dehydrogenase; CCR, cinnamoyl-CoA reductase; COMT, caffeate/5-hydroxyconiferaldehyde O-methyltransferase. p-Coumaroyl shikimate 3-hydroxylase is missing from the figure due to annotation difficulties related to cytochrome P450 genes, as enzyme activity cannot be inferred from the gene sequence. See Supplemental Table S3 for gene identifiers.
Most of the phenylpropanoid and monolignol pathway-related genes with expression greater than 5 (normalized using VST; scale 0–14) in at least one of the sample types analyzed here were equally expressed in both cell types (Fig. 4; Supplemental Table S3). One gene encoding Phe ammonia lyase (MA_10429279g0010) was more highly expressed in ray cells than in tracheids (Padj ≤ 0.05), whereas few genes had significantly higher expression in tracheids than in ray cells (i.e. two CINNAMIC ACID 4-HYDROXYLASE [C4H], two CAFFEOYL-CoA O-METHYLTRANSFERASE [CCoAOMT], and two CAFFEATE/5-HYDROXYCONIFERALDEHYDE O-METHYLTRANSFERASE [COMT]) genes. As syringyl units synthesized from sinapyl alcohol are not present in spruce lignin, the COMTs detected may belong to the flavonoid biosynthesis pathway.
Transcription Factors Regulating Cell Wall Development in Xylem
The NAC transcription factors VASCULAR-RELATED NAC-DOMAIN6 (VND6) and VND7 and MYB transcription factor family members MYB46 and MYB83 are known regulators of xylem differentiation and secondary cell wall formation in Arabidopsis (Nakano et al., 2015; Zhong and Ye, 2015; Heo et al., 2017). Of the 11 most highly expressed NAC genes in our data (Fig. 4; Supplemental Table S3), MA_6777g0010, a sequence homolog of white spruce PgNAC7 that is proposed to be the main master switch regulating secondary cell wall formation in white spruce (Duval et al., 2014), had no significant difference in expression in the sample types studied (normalized expression greater than 7.5). Four NAC genes (MA_95898g0010 [a sequence homolog of PgNAC8], MA_402393g0010, MA_27968g0010, and MA_18153g0010), on the other hand, were two to three times more highly expressed in tracheids than in ray cells (Padj < 0.05). From these NAC genes, MA_95898g0010 and MA_402393g0010 were detected in NorWood in the neighborhood of the secondary cell wall cellulose synthase genes (Jokipii-Lukkari et al., 2017), potentially suggesting a regulatory function. Five of the 34 most highly expressed MYB genes in this study were more highly expressed in tracheids than in ray cells (twofold to threefold), and one was more highly expressed in ray cells than in tracheids (2.5-fold; Padj < 0.05; Fig. 4; Supplemental Table S3). Additionally, two MYB genes (MA_962483g0010 and MA_16444g0030) with lower expression levels (VST < 5) were significantly more highly expressed in ray cells than in tracheids (fold change 2.4 and 5.7; Padj 0.04 and 0.014, respectively; Supplemental Table S3). MA_139238g0010 and MA_62361g0010, which are sequence homologs of loblolly pine PtMYB8 and PtMYB1 regulating secondary cell wall and/or monolignol biosynthesis (Bomal et al., 2008), on the other hand, were not differentially expressed in the cell types studied. The latter MYB gene, MA_62361g0010, was coexpressed with several secondary cell wall synthesis-related genes, including many monolignol biosynthesis genes in developing wood of Norway spruce (Jokipii-Lukkari et al., 2018), supporting its role in the regulation of monolignol biosynthesis (Bomal et al., 2008).
Expression of PEROXIDASE and LACCASE Genes in Developing Tracheids and Ray Cells
Unlike cell walls of ray tracheids, which are located on the top and the bottom of uniseriate rays, ray parenchymal cells do not lignify during the growing season of Norway spruce (Marjamaa et al., 2003). To investigate the expression of genes encoding oxidative enzymes involved in monolignol oxidation, the expression level of PEROXIDASEs and LACCASEs were analyzed separately in tracheids and ray cells. In the Norway spruce genome, class III secretory plant peroxidases occur as a large gene family containing 281 gene models (Nystedt et al., 2013). In the data, 201 putative PEROXIDASE genes had some expression, with 51 PEROXIDASEs having normalized expression over 1 (VST > 1) in any of the samples analyzed (Supplemental Table S4). Similar to the pathway genes leading to monolignols, most of the PEROXIDASE genes had no significant difference in expression between the cell types studied. Of all PEROXIDASEs, MA_91956g0010 had the highest expression in whole sections and in tracheids; furthermore, its expression was higher in tracheids than in ray cells (fold change 3.1; Padj < 0.05). MA_10432865g0020 (PaPX2; Marjamaa et al., 2006; Koutaniemi et al., 2007) and MA_10175544g0010, on the contrary, were the most highly expressed PEROXIDASE genes in ray cells and were significantly more expressed in ray cells than in tracheids (fold change 4 and 4.3; Padj < 0.001; Supplemental Table S4). In addition, the expression of four other PEROXIDASE genes was higher in ray cells than in tracheids, whereas one additional gene was more highly expressed in tracheids (Padj < 0.05). All of these PEROXIDASE genes had rather high expression levels in all sample types (normalized expression levels 3.4 to 10.5; Supplemental Table S4).
Out of 100 LACCASE genes expressed in the data set, 53 had normalized expression levels higher than 1 (Supplemental Table S4). Two genes (MA_10433259g0010 and MA_10431084g0010) were more highly expressed in tracheids than in ray cells (Padj < 0.05). The other LACCASE genes did not have statistically significant expression differences between these cell types.
LCM RNA-Seq Data in Comparison with the NorWood Database
To validate the data obtained in our microdissection analysis, gene expression was compared with that determined separately from cryomicroscopy sections of Norway spruce cut over the dividing cambium and developing xylem into the previous year’s mature xylem (Jokipii-Lukkari et al., 2017; norwood.congenie.org). More than half of the genes of the 500 genes with the highest expression values in the whole-section samples in this study (Supplemental Table S1) had expression in NorWood and could be found in the clustering analysis of Jokipii-Lukkari et al. (2017). One has to keep in mind, however, that only less than half of the expressed genes in our LCM RNA-Seq data were included in the clustering analysis represented in NorWood. Of genes having high expression in the whole-xylem sections, 145 genes belong to the NorWood data cluster related to secondary cell wall formation (Jokipii-Lukkari et al., 2017). In addition, 112 genes more expressed in tracheids than in ray cells (Supplemental Table S2) were mostly present in this same cluster. Genes more highly expressed in ray cells (Supplemental Table S2), on the other hand, had 46 matches to the cluster xylem expansion and 78 matches to the mature wood and the previous year’s latewood cluster. Many genes more abundantly expressed in ray cells, however, did not fall into any specific cluster. This observation suggests that there are differences in gene expression in developing ray cells as compared with those in the previous year’s mature wood, where the ray parenchymal cells and resin duct epithelial cells are the only living cells.
To investigate the behavior of the cell type-specific gene expression, genes expressed more highly in ray cells (541; Padj < 0.01) were used in NorWood to build a network having 191 members (threshold 5; Supplemental Table S2). Similarly, when all genes expressed more abundantly in tracheids (532; Padj < 0.01) were used, a network with 452 members was obtained (threshold 5; Supplemental Table S2).
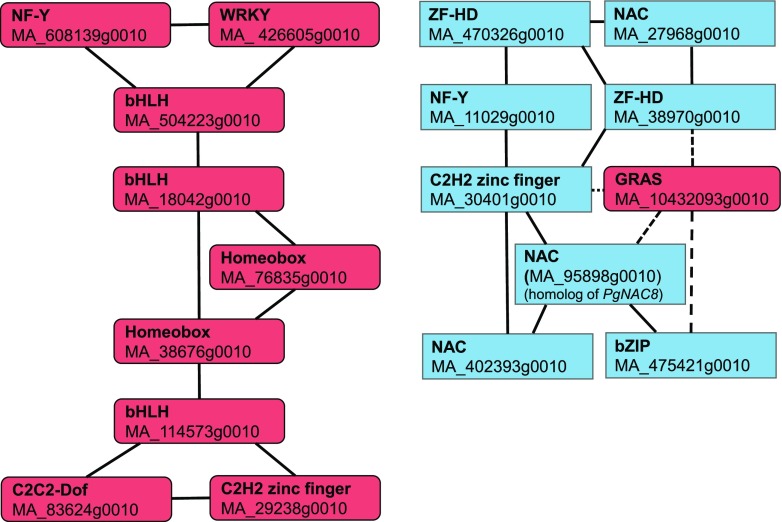
On the basis of the RNA-Seq data, 20 and 10 transcription factor genes were significantly more highly expressed in ray cells and tracheids, respectively (Padj < 0.01; Supplemental Table S2). These were used as an input in NorWood (threshold 3), and two coexpression subnetworks, both composed of nine members, were retrieved (Fig. 5; see Supplemental Table S8 for Arabidopsis homologs and description). Interestingly, one subnetwork was composed of only ray-enriched genes, whereas the other contained eight transcription factors whose genes were more highly expressed in tracheids, with negative correlation to one ray-enriched GRAS gene (Fig. 5). Next, these transcription factors and all pathway genes from the shikimate pathway down to monolignols with normalized expression over 5 were used as an input in NorWood (threshold 3). A network was created where the transcription factors were connected to 84 pathway genes (Supplemental Table S5).
Figure 5.
Coexpression subnetworks of transcription factors differentially expressed in developing ray cells and tracheids. Red color indicates transcription factors more highly expressed in ray cells, and blue indicates those more highly expressed in tracheids (Padj < 0.01). Solid lines indicate positive correlation, and dashed lines indicate negative correlation. Note that MA_95898g0010, which is more highly expressed in tracheids than in ray cells, is a sequence homolog of PgNAC8.
Developing Ray Cells Express a Set of Genes Not Expressed in Tracheids
Cell type-specific transcriptomes are known to be masked when an RNA-Seq analysis is conducted from the whole-complex tissue (Celedon et al., 2017). Our experimental setup using LCM to isolate specific cell types of developing xylem allowed the identification of genes that were expressed only in one cell type (i.e. in developing tracheids or in developing ray cells; Supplemental Table S6). None of the genes was solely expressed in tracheids, whereas expression of 69 genes was detected only in ray cells, with no detectable expression in tracheids (Table 1; Supplemental Table S6). The ray-only-expressed genes consisted of three major groups related to (1) RNA/DNA processing, transcription, and signaling (e.g. pentatricopeptide repeat-containing proteins, helicases); (2) carbohydrate metabolism (e.g. glycosyl transferases), and (3) transport. There was no annotation for one-third of these ray-only-expressed genes, suggesting novel and uncharacterized biological functions represented in developing ray cells. Furthermore, a number of genes with normalized expression levels below 1 (VST < 1) in tracheids (88 genes) were overrepresented in ray cells (Supplemental Table S6). Annotation of these genes revealed a pattern similar to that in the ray-only-expressed genes: RNA/DNA processing-, transcription-, signaling-, and transport-related genes were supplemented with genes encoding proteins related to disease resistance, ubiquitinylation, and lipid metabolism.
Table 1. Ray-only-expressed genes that have no expression, or have normalized expression level below 1 (VST < 1), in upright tracheids in developing xylem of Norway spruce.
NA, Not annotated.
| Gene Identifier | Uniprot Annotation | PFAM annotation |
|---|---|---|
| RNA/DNA processing | ||
| MA_10436812g0020 | Endoribonuclease Dicer homolog4 | Helicase_C, dsRNA_bind |
| MA_136489g0010 | ATP-dependent DNA helicase PcrA | AAA_11, 19, UvrD-helicase |
| MA_222324g0010 | ATP-dependent DNA helicase PcrA | UvrD_C, UvrD_C_2, Viral_helicase1 |
| MA_8906841g0010 | NA | UvrD-helicase |
| MA_953686g0010 | DNA replication ATP-dependent helicase/nuclease DNA2 | NA |
| MA_4430g0020 | Pentatricopeptide repeat-containing protein | PPR, TPR |
| MA_4430g0010 | Pentatricopeptide repeat-containing protein | DYW_deaminase, PPR, TPR |
| MA_500770g0010 | Pentatricopeptide repeat-containing protein | ATP13, PPR,RasGEF_N |
| MA_77707g0010 | Pentatricopeptide repeat-containing protein | Clathrin, DnaB, PPR, RPN7, TPR |
| MA_56436g0010 | Pentatricopeptide repeat-containing protein | DYW_deaminase, PPR, TPR |
| MA_62181g0010 | Pentatricopeptide repeat-containing protein | PPR, RPN7, TPR |
| MA_11619g0010 | Pentatricopeptide repeat-containing protein | DYW_deaminase, MRP-S27, PPR, RPN7, TPR |
| MA_108603g0010 | Pentatricopeptide repeat-containing protein | PPR |
| MA_257729g0010 | Pentatricopeptide repeat-containing protein | Ank, Apc3, Clathrin, DUF3368, ECSIT, MRP-S27, PPR, Rep_fac_C, TPR, TetR_C |
| MA_131437g0010 | Putative pentatricopeptide repeat-containing protein | DYW_deaminase, PPR, TPR |
| MA_10428974g0010 | tRNA modification GTPase MnmE | FeoB_N, GTP_EFTU, GTPase_Cys_C, MMR_HSR1 |
| MA_8617598g0010 | NA | HEPN, TPR |
| MA_165131g0010 | Nibrin | BRCT, FHA, PTCB-BRCT |
| General enzymes | ||
| MA_38431g0010 | Acetyl-CoA carboxylase1 | CPSase_L_D2, CPSase_L_chain |
| MA_103379g0010 | Biotin biosynthesis bifunctional protein BioWF | Aminotran_1_2 |
| MA_10435812g0010 | Imidazole glycerol phosphate synthase hisHF | GATase, GATase_3, GATase_5, Peptidase_C26, SNO |
| MA_89112g0010 | Violaxanthin deepoxidase | VDE |
| MA_193904g0010 | NA | Glyoxalase, Glyoxalase_2, Glyoxalase_4 |
| MA_157164g0010 | Rhodanese-like domain-containing protein4 | NA |
| MA_74391g0010 | NA | DUF607, Nudix_N |
| Redox | ||
| MA_17680g0010 | Cytochrome P450 | p450 |
| MA_10426917g0010 | Uncharacterized protein At3g17611 | DUF1412, Rhomboid, Rubredoxin |
| Carbohydrate related | ||
| MA_10435727g0010 | 1,4-α-Glucan-branching enzyme3 | CBM_48 |
| MA_863320g0010 | Endoglucanase1 | Glyco_hydro_9 |
| MA_619336g0010 | Hydroquinone glucosyltransferase | Glyco_trans_1_3, UDPGT |
| MA_407162g0010 | UDP-glycosyltransferase72B1 | Glyco_tran_28_C, Glyco_trans_1_3, UDPGT |
| MA_10259759g0010 | UDP-glycosyltransferase74E1 | UDPGT |
| MA_10433786g0010 | UDP-glycosyltransferase86A | UDPGT |
| Receptor-like kinases | ||
| MA_118627g0010 | LRR-like Ser/Thr-protein kinase BAM2 | LRRNT_2, LRR |
| MA_420984g0010 | Receptor protein kinase-like protein At4g34220 | LRRNT_2, LRR |
| MA_203596g0010 | Putative disease resistance protein At4g19050 | LRR |
| Transcription factors | ||
| MA_379291g0010 | NA | AKNA |
| MA_18230g0010 | NA | zf-CCHC, zf-GRF |
| Transport | ||
| MA_559373g0010 | Probable peptide/nitrate transporter | PTR2, TRAP_beta |
| MA_135402g0010 | Solute carrier family 15 member 1 | MFS_1, PTR2 |
| MA_9087020g0010 | Vacuolar protein sorting-associated protein18 homolog | Pep3_Vps18, TPR |
| MA_162910g0010 | WASH complex subunit 7 homolog | NA |
| MA_10430993g0010 | WASH complex subunit strumpellin | Strumpellin |
| MA_739378g0010 | HEAT repeat-containing protein6 | DUF4042, HEAT, IFRD |
| MA_5424g0010 | NA | HEAT |
| Protease | ||
| MA_141320g0010 | Protease Do-like8 | Trypsin_2 |
| MA_6458032g0010 | Subtilisin-like protease | Inhibitor_I9 |
| MA_10427774g0040 | NA | RINGv |
| Proteins not characterized or of unknown functions | ||
| MA_8138125g0010 | NA | NA |
| MA_211416g0010 | NA | NA |
| MA_362689g0010 | NA | NA |
| MA_10432337g0010 | NA | NA |
| MA_164263g0010 | NA | NA |
| MA_54857g0010 | NA | NA |
| MA_68192g0010 | NA | NA |
| MA_39199g0010 | NA | DUF3091 |
| MA_76111g0010 | NA | NA |
| MA_759716g0010 | NA | NA |
| MA_10430932g0010 | NA | NA |
| MA_2523991g0010 | NA | NA |
| MA_70653g0010 | NA | NA |
| MA_9999327g0010 | NA | NA |
| MA_99743g0030 | NA | NA |
| MA_10343198g0010 | NA | NA |
| MA_763224g0010 | NA | NA |
| MA_10110274g0010 | NA | NA |
| MA_70531g0010 | NA | DUF1645 |
| MA_8445490g0010 | Uncharacterized protein sll1770 | NA |
| MA_9440571g0010 | NA | NA |
In Situ Determination of Cellular Metabolites
In order to investigate whether the transcriptomic results translated to the metabolite level, picoPPESI-MS analysis was conducted separately on ray cells and tracheids of developing xylem of semi-intact plants of Norway spruce with a small part of bark tissue removed (Supplemental Fig. S2). The analysis showed that many lignin biosynthesis-related metabolites, including cinnamic acid or p-coumaraldehyde (mass-to-charge ratio [m/z] 147), p-coumaric acid (m/z 163), caffeic acid (m/z 179), coniferyl aldehyde (m/z 177), coniferyl alcohol (m/z 179), and p-coumaryl alcohol (m/z 149), were detected (as [M−H]−, M = molecular mass of chemical species) in both cell types, together with nine organic acids, sugars, most amino acids, glutathione, and abietic acid (Table 2; Fig. 6; Supplemental Fig. S3). Also, coniferin (a glycosylated form of coniferyl alcohol) and p-coumaryl alcohol 4-glucoside were detected as deprotonated forms (m/z 314 and 311, respectively) and as Cl− adduct ions ([M+Cl]−; m/z 377 and 347, respectively) in ray cells as well as in tracheids. The contents of coniferin, p-coumaryl alcohol 4-glucoside, and quinic acid (m/z 191) were overall greater in ray cells than in tracheids (see [coniferin−H]−, [coniferin+Cl]−, [p-coumaryl alcohol 4-glucoside−H]−, [p-coumaryl alcohol 4-glucoside+Cl]−, and a coniferin-sugar cluster ion {as [M′−H]−, M′ = summation of individual molecular mass of chemical cluster component species; [coniferin+Hex−H]−} in Table 2). Conversely, the contents of coniferyl aldehyde (m/z 177), malic acid ([malic acid−H]−; m/z 133), and their two malic acid-sugar clusters ([malic acid+Hex−H]− {m/z 313} and [malic acid+Hex2−H]− {m/z 475}), α-ketoglutaric acid (m/z 145), fumaric acid (m/z 115), and citric acid (m/z 191) were larger in tracheids than in ray cells (Table 2). Using the analytical method, caffeoylquinic acid, dihydroflavonol, leucodelphinidin, and Met were detected less frequently in both types of cells, whereas no detectable ionized signals were observed, for example, for any CoA conjugates, dihydroconiferin, and α-pinene in either of the cell types (Table 2).
Table 2. List of metabolites detected in ray cells and tracheids using picoPPESI-MS in negative ion mode.
Asterisks indicate significant differences between the ray cells (RC) and tracheids (TR) as follows: *, P < 0.05 and ***, P < 0.001.
| Metabolite | Molecular Formula | Ion Type Detected | Theoretical m/za | Signal Intensity | P | Relative Abundanceb | P | Frequency of Detectionc | MS/MS Results Shown in | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RC | TR | RC | TR | RC | TR | |||||||
| Monolignol pathways | ||||||||||||
| Cinnamic acid or p-Coumaraldehyde | C9H8O2 | [M−H]− | 147.045 | 3.9.E+04 | 1.4.E+05 | 0.359 | 0.019 | 0.034 | 0.416 | ++++ | ++++ | |
| p-Coumaric acid | C9H8O3 | [M−H]− | 163.040 | 4.1.E+04 | 5.0.E+04 | 0.749 | 0.015 | 0.024 | 0.416 | +++++ | ++++ | Supplemental Figure S3A.1 |
| 4-Coumaroyl-CoA | C30H38N7O18P3S | [M−H]− | 912.145 | – | – | – | – | – | – | – | – | |
| Caffeoyl-CoA | C30H42N7O19P3S | [M−H]− | 928.140 | – | – | – | – | – | – | – | – | |
| Feruloyl-CoA | C31H44N7O19P3S | [M−H]− | 942.155 | – | – | – | – | – | – | – | – | |
| Caffeic acid | C9H8O4 | [M−H]− | 179.035 | 1.0.E+05 | 2.7.E+05 | 0.392 | 0.045 | 0.168 | 0.353 | +++++ | ++++ | Supplemental Figure S3A.2 |
| Caffeoylshikimic acid | C16H16O8 | [M−H]− | 335.077 | – | – | – | – | – | – | – | – | |
| Caffeoylquinic acid | C16H18O9 | [M−H]− | 353.088 | 1.1.E+04 | 9.3.E+03 | 0.912 | 0.005 | 0.006 | 0.956 | ++ | ++ | |
| Coniferyl aldehyde | C10H10O3 | [M−H]− | 177.056 | 3.0.E+04 | 1.2.E+05 | 0.184 | 0.015 | 0.041 | 0.101 | ++++ | +++++ | Supplemental Figure S3A.3 |
| Coniferyl alcohol | C10H12O3 | [M−H]− | 179.071 | 1.1.E+06 | 1.5.E+06 | 0.705 | 0.527 | 0.446 | 0.717 | +++++ | +++++ | Supplemental Figure S3A.4 |
| p-Coumaryl alcohol | C9H10O2 | [M−H]− | 149.061 | 1.0.E+04 | 4.4.E+04 | 0.033 | 0.005 | 0.023 | 0.050 | +++ | ++++ | Supplemental Figure S3A.5 |
| Coniferin | C16H22O8 | [M−H]− | 341.124 | 1.5.E+05 | 1.3.E+05 | 0.872 | 0.055 | 0.029 | 0.451 | +++ | +++ | Supplemental Figure S3A.6 |
| Coniferin | C16H22O8 | [M+Cl]− | 377.101 | 5.9.E+06 | 2.6.E+06 | 0.296 | 2.415 | 1.239 | 0.368 | +++++ | ++++ | Supplemental Figure S3A.7 |
| p-Coumaryl alcohol 4-glucoside | C15H20O7 | [M−H]− | 311.114 | 1.3.E+04 | 2.7.E+03 | 0.480 | 0.004 | 0.002 | 0.656 | + | + | Supplemental Figure S3A.8 |
| p-Coumaryl alcohol 4-glucoside | C15H20O7 | [M+Cl]− | 347.090 | 1.0.E+05 | 5.3.E+04 | 0.494 | 0.043 | 0.027 | 0.611 | ++++ | +++ | Supplemental Figure S3A.9 |
| Dihydroconiferin | C16H24O8 | [M−H]− | 343.140 | – | – | – | – | – | – | – | – | |
| Dihydroconiferin | C16H24O8 | [M+Cl]− | 379.117 | – | – | – | – | – | – | – | – | |
| Shikimate and aromatic amino acid pathways | ||||||||||||
| Phosphoenolpyruvic acid | C3H5O6P | [M−H]− | 166.975 | 2.7.E+04 | 4.5.E+04 | 0.373 | 0.014 | 0.019 | 0.575 | ++++ | ++++ | |
| d-Erythrose 4-phosphoric acid | C4H9O7P | [M−H]− | 199.001 | 5.0.E+04 | 6.0.E+04 | 0.833 | 0.033 | 0.020 | 0.625 | ++++ | ++++ | Supplemental Figure S3A.10 |
| 3-Dehydroquinic acid | C7H10O6 | [M−H]− | 189.041 | 3.2.E+05 | 8.9.E+05 | 0.134 | 0.137 | 0.363 | 0.079 | +++++ | +++++ | Supplemental Figure S3A.11 |
| Shikimate-3-phosphoric acid | C7H11O8P | [M−H]− | 253.012 | – | – | – | – | – | – | – | – | |
| 5-Enolpyruvylshikimate | C10H13O10P | [M−H]− | 323.017 | – | – | – | – | – | – | – | – | |
| 3-Phosphoric acid (EPSP) | ||||||||||||
| Chorismic acid and prephenic acid | C10H10O6 | [M−H]− | 225.041 | 6.1.E+02 | 1.5.E+04 | 0.208 | 0.0004 | 0.004 | 0.172 | ++ | ++ | Supplemental Figure S3A.12 |
| Flavonoid pathways | ||||||||||||
| Flavanone | C15H12O2 | [M−H]− | 223.077 | – | – | – | – | – | – | – | – | |
| Sakuranetin or isosakuranetin | C16H14O5 | [M−H]− | 285.077 | – | – | – | – | – | – | – | – | |
| Dihydroflavonols | C15H12O3 | [M−H]− | 239.071 | 5.3.E+03 | 3.2.E+04 | 0.373 | 0.003 | 0.006 | 0.582 | +++ | ++ | |
| Leucocyanidin or melacacidin | C15H14O7 | [M−H]− | 305.067 | – | – | – | – | – | – | – | – | |
| Leucodelphinidin | C15H14O8 | [M−H]− | 321.062 | 7.2.E+03 | 1.4.E+04 | 0.571 | 0.003 | 0.009 | 0.448 | +++ | ++ | |
| Leucofisetinidin or leucopelargoni din | C15H14O6 | [M−H]− | 289.072 | – | – | – | – | – | – | – | – | |
| Leucopeonidin | C16H18O7 | [M−H]− | 321.098 | – | – | – | – | – | – | – | – | |
| Organic acids | ||||||||||||
| Malic acid | C4H6O5 | [M−H]− | 133.014 | 6.7.E+07 | 1.6.E+08 | 0.067 | 29.866 | 72.346 | 0.001*** | +++++ | +++++ | |
| Quinic acid | C7H12O6 | [M−H]− | 191.056 | 2.2.E+08 | 2.0.E+08 | 0.739 | 100.000 | 84.992 | 0.071 | +++++ | +++++ | |
| Oxaloacetic acid | C4H4O5 | [M−H]− | 130.999 | 7.5.E+03 | 5.6.E+05 | 0.152 | 0.002 | 0.332 | 0.152 | +++ | +++ | |
| Succinic acid | C4H6O4 | [M−H]− | 117.019 | 1.5.E+06 | 1.9.E+06 | 0.619 | 0.683 | 0.770 | 0.743 | +++++ | +++++ | |
| Ascorbic acid | C6H8O6 | [M−H]− | 175.025 | 4.7.E+06 | 5.8.E+06 | 0.679 | 2.385 | 2.654 | 0.828 | +++++ | +++++ | |
| 3-Phosphoglyceric acid | C3H7O7P | [M−H]− | 184.986 | 2.9.E+05 | 3.5.E+05 | 0.606 | 0.146 | 0.181 | 0.555 | +++++ | +++++ | |
| α-Ketoglutaric acid | C5H6O5 | [M−H]− | 145.014 | 4.1.E+05 | 6.8.E+05 | 0.197 | 0.170 | 0.354 | 0.084 | +++++ | +++++ | |
| Fumaric acid | C4H4O4 | [M−H]− | 115.004 | 1.7.E+06 | 3.5.E+06 | 0.160 | 0.751 | 1.525 | 0.029* | +++++ | +++++ | |
| Citric acid | C6H8O7 | [M−H]− | 191.020 | 4.1.E+06 | 2.4.E+07 | 0.021* | 2.052 | 12.433 | 0.040* | +++++ | +++++ | |
| Carbohydrate | ||||||||||||
| Hex | C6H12O6 | [M−H]− | 179.056 | 5.5.E+06 | 6.0.E+06 | 0.716 | 2.454 | 2.776 | 0.523 | +++++ | +++++ | |
| Hex | C6H12O6 | [M+Cl]− | 215.033 | 1.3.E+07 | 1.6.E+07 | 0.761 | 5.927 | 8.840 | 0.549 | +++++ | +++++ | |
| Hex2 | C12H22O11 | [M−H]− | 341.109 | 1.1.E+07 | 1.1.E+07 | 0.967 | 5.847 | 4.506 | 0.528 | +++++ | +++++ | |
| Hex2 | C12H22O11 | [M+Cl]− | 377.086 | 2.7.E+06 | 4.0.E+06 | 0.546 | 1.161 | 2.521 | 0.292 | +++++ | +++++ | |
| Pentose | C5H10O5 | [M−H]− | 149.046 | 5.4.E+06 | 6.0.E+06 | 0.812 | 2.394 | 2.382 | 0.982 | +++++ | +++++ | |
| Pentose | C5H10O5 | [M+Cl]− | 185.022 | 5.9.E+03 | 4.2.E+04 | 0.039 | 0.003 | 0.023 | 0.075 | + | ++++ | |
| Amino acids | ||||||||||||
| Ser | C3H7NO3 | [M−H]− | 104.035 | 5.4.E+04 | 5.1.E+04 | 0.911 | 0.033 | 0.021 | 0.499 | +++++ | ++++ | |
| Pro | C5H9NO2 | [M−H]− | 114.056 | 2.9.E+05 | 1.8.E+05 | 0.578 | 0.164 | 0.056 | 0.239 | +++++ | +++ | |
| Val | C5H11NO2 | [M−H]− | 116.072 | 1.6.E+05 | 1.4.E+05 | 0.793 | 0.087 | 0.057 | 0.500 | +++++ | ++++ | |
| Thr | C4H9NO3 | [M−H]− | 118.051 | 1.9.E+05 | 1.6.E+05 | 0.676 | 0.102 | 0.078 | 0.516 | +++++ | +++++ | |
| Leu, Ile | C6H13NO2 | [M−H]− | 130.087 | 3.0.E+05 | 2.1.E+06 | 0.335 | 0.156 | 1.911 | 0.334 | +++++ | ++++ | |
| Asp | C4H8N2O3 | [M−H]− | 131.046 | 1.2.E+05 | 1.2.E+05 | 0.978 | 0.073 | 0.036 | 0.453 | ++++ | ++++ | |
| Asp | C4H7NO4 | [M−H]− | 132.030 | 1.0.E+06 | 9.6.E+05 | 0.875 | 0.604 | 0.485 | 0.708 | +++++ | +++++ | |
| Gln | C5H10N2O3 | [M−H]− | 145.062 | 2.1.E+06 | 1.3.E+06 | 0.472 | 1.090 | 0.409 | 0.196 | +++++ | +++++ | |
| Glu | C5H9NO4 | [M−H]− | 146.046 | 9.9.E+05 | 1.1.E+06 | 0.752 | 0.513 | 0.537 | 0.920 | +++++ | +++++ | |
| Met | C5H11NO2S | [M−H]− | 148.044 | 1.0.E+04 | 3.6.E+03 | 0.567 | 0.005 | 0.001 | 0.430 | + | + | |
| His | C6H9N3O2 | [M−H]− | 154.062 | 8.3.E+04 | 7.7.E+04 | 0.916 | 0.041 | 0.025 | 0.448 | ++++ | +++++ | |
| Phe | C9H11NO2 | [M−H]− | 164.072 | 1.1.E+05 | 2.4.E+05 | 0.368 | 0.053 | 0.108 | 0.418 | +++++ | +++++ | |
| Arg | C6H14N4O2 | [M−H]− | 173.104 | 2.7.E+04 | 7.9.E+03 | 0.242 | 0.018 | 0.003 | 0.153 | +++ | ++ | |
| Tyr | C9H11NO3 | [M−H]− | 180.067 | 1.1.E+05 | 1.1.E+05 | 0.998 | 0.057 | 0.028 | 0.410 | ++++ | ++++ | |
| Trp | C11H12N2O2 | [M−H]− | 203.083 | 8.3.E+04 | 1.2.E+05 | 0.711 | 0.053 | 0.032 | 0.624 | ++++ | ++++ | |
| GABA | C4H9NO2 | [M−H]− | 102.056 | 7.5.E+04 | 4.1.E+04 | 0.405 | 0.040 | 0.015 | 0.171 | +++++ | ++++ | |
| Peptide | ||||||||||||
| Glutathione | C10H17N3O6S | [M−H]− | 306.077 | 6.1.E+05 | 8.1.E+05 | 0.645 | 0.313 | 0.317 | 0.979 | +++++ | +++++ | |
| Terpenoids | ||||||||||||
| Abietic acid | C20H30O2 | [M−H]− | 301.217 | 7.7.E+06 | 7.7.E+06 | 0.996 | 4.071 | 4.575 | 0.921 | +++++ | ++++ | |
| α-Pinene | C10H16 | [M−H]− | 135.118 | – | – | – | – | – | – | – | – | |
| Cluster ions | ||||||||||||
| Malic acid + Hex | C10H18O11 | [M−H]− | 313.078 | 6.6.E+05 | 9.0.E+06 | 0.219 | 0.290 | 2.258 | 0.069 | +++++ | +++++ | |
| Malic acid + Hex2 | C16H28O16 | [M−H]− | 475.130 | 2.4.E+06 | 4.1.E+06 | 0.286 | 1.093 | 2.474 | 0.152 | +++++ | +++++ | |
| Quinic acid + Hex | C13H24O12 | [M−H]− | 371.119 | 2.6.E+07 | 2.4.E+07 | 0.858 | 10.529 | 12.679 | 0.666 | +++++ | +++++ | |
| Quinic acid + Hex2 | C19H34O17 | [M−H]− | 533.172 | 3.1.E+07 | 1.7.E+07 | 0.298 | 11.558 | 9.478 | 0.686 | +++++ | +++++ | |
| Coniferin + Hex | C22H34O14 | [M−H]− | 521.188 | 1.3.E+05 | – | 0.337 | 0.039 | – | 0.323 | ++ | — | |
| Coniferin + Hex2 | C28H44O19 | [M−H]− | 683.240 | 1.3.E+05 | 1.0.E+05 | 0.852 | 0.041 | 0.019 | 0.528 | ++ | ++ | |
| Malic acid + coniferin | C20H28O13 | [M−H]− | 475.146 | 1.7.E+05 | 2.8.E+03 | 0.325 | 0.052 | 0.001 | 0.306 | ++ | + | |
| Quinic acid + coniferin | C23H34O14 | [M−H]− | 533.188 | 2.1.E+06 | 5.5.E+05 | 0.360 | 0.733 | 0.272 | 0.432 | +++ | ++ | |
All the theoretical values are quoted from Metlin (http://metlin.scripps.edu/index.php).
Values were calculated as a percentage of the base peak.
Signal detected (+); signal not detected (−). Frequencies of detection (x) in eight to nine individual measurements are indicated as follows: +++++, 80% < x < 100%; ++++, 60% < x < 80%; +++, 40% < x < 20%; ++, 20% < x < 40%; +, 0% < x < 20%.
Figure 6.
PicoPPESI-MS negative ion mode mass spectra directly obtained from ray cells (RC) and tracheids (TR) in semi-intact Norway spruce plants. Data are representative of similar experiments with eight to nine cells in each cell type.
DISCUSSION
Differentiation of xylem in conifers entails large changes in cellular biology, biochemistry, and transcriptome remodulation that lead to the formation of a thick secondary cell wall and lignification of xylem tracheids (Meents et al., 2018). In developing xylem of Norway spruce, the cell walls of ray parenchymal cells do not lignify (Marjamaa et al., 2003). They may lignify later during heartwood formation, as in Scots pine (Bergström 2003). The contribution of ray cells in monolignol biosynthesis was studied in this work. Samples for the LCM and metabolomic investigations were taken when secondary growth in Norway spruce was active (end of June in Finland, mid-April in Kumamoto, Japan). In southern Finland, xylem growth and lignification in Norway spruce starts in mid-May and continues until August (Marjamaa et al., 2003).
Developing Ray Cells Express Genes Encoding the Machinery for Lignin Biosynthesis and Contain Monolignols and Their Glucoconjugates
Ray cells have well-known roles in radial transport of water and nutrients and function as a storage compartment for starch, lipids, and proteins (Höll, 1975; Witt and Sauter, 1994). In Scots pine, up to 40% of nonstructural carbohydrates are stored in the sapwood parenchyma, making it an important reserve of carbohydrates (Jacquet et al., 2014). In angiosperms, ray parenchyma acts as an efficient symplastic route for xylem-phloem communication, forming specific conduits that support transport in longitudinal or radial directions (van Bel, 1990) and sustain xylem hydraulics and growth (Witt and Sauter, 1994). The transport function of rays is also essential for the cambium activity and for the survival of fusiform initials in the cambium (Lev-Yadun and Aloni, 1995).
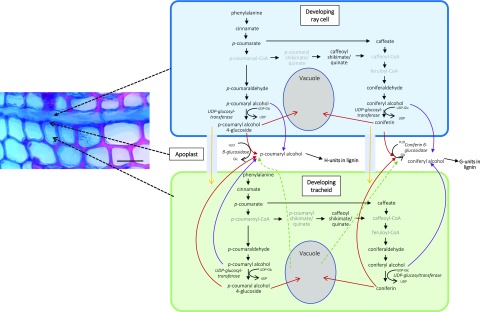
The RNA-Seq data obtained strongly suggested that in developing xylem of Norway spruce, ray cells synthesize monolignols, since most of the biosynthesis pathway genes were expressed equally highly in both tracheids and ray cells (Fig. 4). Furthermore, coniferyl and p-coumaryl alcohol, some intermediates in their biosynthetic pathway, and their glucoconjugates were detected by picoPPESI-MS analysis in both ray cells and tracheids (Table 2), confirming the transcriptomic results. As cell walls of ray parenchymal cells do not lignify in the growing season, it is probable that monolignols are transported to neighboring cells (upright and/or ray tracheids) and used there for cell wall lignification. Not only do ray cells possess the machinery for monolignol biosynthesis but also a number of genes that encode transporter proteins, including several ABC transporters (with no significant difference in expression between tracheids and ray cells). Some of these could encode transporter proteins in the plasma membrane, delivering monolignol alcohols to the apoplast of ray cells (Miao and Liu, 2010; Alejandro et al., 2012; Fig. 7). Monolignols would then need to diffuse through nonlignifying ray cell wall to the middle lamella, where lignification starts, and to adjacent lignifying tracheid cell walls. This is apparently occurring in postmortem lignification shown to exist in tracheary elements of Z. elegans and Arabidopsis (Pesquet et al., 2013; Smith et al., 2013, 2017). Alternatively, there is a possibility that monolignols produced by ray cells are transported to developing tracheids via a symplastic route (Fig. 7). Typically for conifers, rays in Norway spruce are one cell wide (uniseriate), and thus every ray cell is in contact with some tracheid cell (Fig. 1; Spicer, 2014). Pits are present in the cell walls of ray parenchymal cells that are shared with tracheids (Spicer, 2014). In tomato (Solanum lycopersicum), intracellular injections of a fluorescent dye revealed symplasmic connections between living fibers and adjacent ray parenchymal cells (van der Schoot and van Bel, 1990). Plasmodesmata also exist in pits located between developing, still-living tracheids and neighboring ray cells in conifers (Sokołowska, 2013). These would potentiate symplasmic transport of monolignols from ray cells to developing tracheids. There, the ray-synthesized monolignols could be transported to the cell wall via the same system(s) used for tracheid-self-generated monolignols. Although tempting to speculate, the data obtained in our experiments do not allow us to make any conclusion regarding the monolignol form (aglycone/glycoconjugate) that is transported into the apoplast in spruce. Expression of MA_8591669g0010, a sequence homolog for CONIFERIN β-GLUCOSIDASE of lodgepole pine (Pinus contorta; Dharmawardhana et al., 1995, 1999), was high in all samples studied in this work (normalized expression levels > 7); however, there was a significantly higher expression in tracheids than in ray cells (fold change 2.85; Padj < 0.05). In NorWood, this gene is highly expressed in developing xylem during secondary cell wall formation and programmed cell death (Jokipii-Lukkari et al., 2017; norwood.congenie.org), an observation supporting the idea that the enzyme has an important role for coniferyl alcohol liberation from coniferin for lignin biosynthesis.
Figure 7.
Putative lignin biosynthesis route in Norway spruce developing xylem based on single-cell metabolomics. Red arrows indicate transport of monolignol glucosides through the membrane via, for example, secondary active transport based on a proton gradient. Purple arrows indicate transport of monolignol alcohols through the membrane via, for example, ABC transporters (Miao and Liu, 2010; Alejandro et al., 2012). Yellow arrows indicate transport of monolignol glucosides to a neighboring cell via plasmodesmata. Green arrows indicate liberation of monolignol glucosides into the apoplast after vacuolar collapse. Monolignol names in gray were not detected. Bar = 50 μm.
In gymnosperms, coniferin has been shown to accumulate in the cambial zone and the developing xylem during cambium reactivation (Freudenberg and Harkin, 1963; Savidge, 1989; Aoki et al., 2016). In Japanese cypress (Chamaecyparis obtusa), coniferin was detected in vacuoles of xylem tracheids at the beginning and in the middle phase of lignification (i.e. during the active formation of secondary cell wall layers), after which it disappears (Morikawa et al., 2010; Tsuyama et al., 2013). Proton gradient-dependent transport of coniferin was detected in membrane vesicles prepared from differentiating xylem of hybrid poplar (Populus sieboldii × Populus grandidentata) and Japanese cypress, but this transport was across the tonoplast and endomembrane compartments (Tsuyama et al., 2013). It is possible that some glycoconjugates are stored in the vacuole, to be used for cell wall lignification later once the vacuole disrupts (Meents et al., 2018; Fig. 7). Alternatively, they are used as defense compounds in case of pathogen attack (Wang et al., 2013). In a feeding experiment with lodgepole pine seedlings, radiolabeled Phe was not incorporated into coniferin; however, the secondary cell walls of tracheids were strongly labeled (Kaneda et al., 2008). The data suggest a direct route from coniferyl alcohol to the lignin polymer. Also, ray cells had heavy labeling, but inhibitor experiments suggested that most label in ray cells incorporated into proteins. Some label, however, was also incorporated into phenylpropanoids in ray cells, indicating their participation in monolignol production (Kaneda et al., 2008). The effects of osmotic potential differences and/or wounding in the experiments by Kaneda et al. (2008), done with cut tissue sections immersed in 0.2 m Suc solution, could have affected the metabolome of the cells. In this respect, our in situ picoPPESI-MS approach, with careful consideration of the plant water status, minimizes the effect of tissue disturbance on the metabolome (Supplemental Fig. S4).
In addition to lignin formation, coniferyl alcohol, the main monolignol in Norway spruce, can be utilized for lignan synthesis. Lignans are optically active phenolic dimers and have various antioxidant effects (Hudgins et al., 2003). They are present in Norway spruce, especially in the knotwood (bases of branches), where they accumulate in large amounts and protect tissues against microbial attack (Willför et al., 2003, 2005; Piispanen et al., 2008). Small amounts of lignans have also been detected in the sapwood, but the amounts were clearly lower (less than 0.05% [w/w]) than in the knotwood (up to 15% [w/w]; Willför et al., 2005). In our analysis, no lignans (or other dilignols/oligolignols or their glycoconjugates) were reproducibly detected in either of the cell types. Hence, the ray cell-originated coniferyl alcohol seems to be used predominantly in the lignification of cell walls of upright tracheids and/or those of ray tracheids.
Transcription Factors Regulating Secondary Cell Wall Biosynthesis Have High Expression Levels in Both Upright Tracheids and in Ray Cells
Norway spruce sequence homologs of PgNAC7 (MA_6777g0010) and MYB transcription factors PtMYB8 and PtMYB1 (MA_139238g0010 and MA_62361g0010, respectively), suggested by earlier studies to be the main transcriptional regulators for secondary cell wall formation in loblolly pine (Bomal et al., 2008; Duval et al., 2014), had high expression in both ray cells and tracheids (Fig. 4; Supplemental Table S3). The transcription factor encoded by MA_62361g0010 was recently identified as a regulator likely to control seasonal lignification in Norway spruce together with another MYB (MA_9483804g0010) and a transcription factor, Asymmetric Leaves2/Lateral Organ Boundaries (MA_10434782g0020; Jokipii-Lukkari et al., 2018). In our study, all of these genes had high expression in all sample types (normalized expression level > 6.2), with no significant difference in expression between ray cells and tracheids. The observation that the genes encoding putative central regulators of secondary cell wall biosynthesis also have high expression in ray cells is interesting, as many of these transcription factors not only regulate lignin biosynthesis but also that of the cell wall polysaccharides. It is possible that repressors similar to VND-INTERACTING2 (VNI2; Yamaguchi et al., 2010) are preferentially present in developing ray cells. The absence of a clear VNI2 homolog in the spruce genome annotation (highest identity of 60% to At5g13180) prevented this hypothesis from being tested.
A previous study that utilized an extracellular lignin-forming cell culture of Norway spruce as a model for lignin biosynthesis had generated a gene coexpression network containing several transcription factors that were discussed as candidate regulators of monolignol biosynthesis (Laitinen et al., 2017). The present RNA-Seq data obtained using cells of developing xylem showed that out of the NAC and MYB candidate genes identified in the tissue culture study, only one NAC gene, MA_139896g0010, had a moderate expression level in rays and tracheids (normalized expression level of 2.4–2.8). Two genes encoding abscisic stress-ripening protein (MA_60383g0010 and MA_773806g0010), on the other hand, were quite highly expressed (normalized expression levels of 6.6–7.3 and 4.4–5.2, respectively), supporting that further studies are needed to resolve the role of this comparatively understudied transcription factor family in wood development. Additionally, two genes encoding (1) an ethylene-responsive transcription factor (MA_98464g0010; normalized expression level of 3.9–6) and (2) a TIFY transcription factor (MA_10427283g0030; normalized expression level of 1.5–2.3) warrant further attention in relation to wood development.
Many Genes Encoding Oxidative Enzymes Are Highly Expressed in Ray Cells
In Arabidopsis, both laccases and peroxidases have been shown to contribute to lignin biosynthesis by oxidizing monolignols to radicals that couple nonenzymatically (Berthet et al., 2011; Novo-Uzal et al., 2013; Zhao et al., 2013; Shigeto and Tsutsumi, 2016). In a Norway spruce cell culture that produces extracellular lignin (Kärkönen et al., 2002), however, peroxidases seem to have a major role in monolignol activation, as scavenging of apoplastic hydrogen peroxide efficiently hindered extracellular lignin formation (Laitinen et al., 2017). In a recent study in Norway spruce, several LACCASE genes and three PEROXIDASE genes were detected to coexpress with monolignol biosynthesis genes proposed as candidates for seasonal lignification (Jokipii-Lukkari et al., 2018). According to our study, most of the genes identified by Jokipii-Lukkari et al. (2018), and many other LACCASE and PEROXIDASE genes, had no significant difference in expression between developing ray cells and tracheids (Supplemental Table S4), suggesting that the proteins are synthesized in both cell types. PEROXIDASE genes MA_91956g0010 and MA_10430775g0010, however, had expression levels significantly higher in tracheids as compared with ray cells (2.6- to 3.1-fold; Padj < 0.05). The latter gene is strongly up-regulated (fold change 20.8) in the lignin-forming cell culture of spruce during lignin formation (Laitinen et al., 2017). Six PEROXIDASEs, on the other hand, were more highly expressed in ray cells than in tracheids (Padj < 0.05; Supplemental Table S4). Among these, MA_10432865g0020 (PaPX2; Marjamaa et al., 2006) was highly expressed in all sample types and was four times more highly expressed in ray cells than in tracheids (Supplemental Table S4). PaPX2 was observed to have the highest expression in the previous year’s late xylem (Jokipii-Lukkari et al., 2017; norwood.congenie.org). In an earlier study of a xylem sample collected in late summer, transcripts of PaPX2 were detected by in situ hybridization in tracheids but not in ray cells (Marjamaa et al., 2006). Additionally, PaPX2 expression is responsive to the pathogenic fungus Heterobasidium annosum and to developmental stress, being present, for example, during compression wood formation (Koutaniemi et al., 2007). Sequence homology comparisons of these Norway spruce peroxidases against Arabidopsis peroxidases with known association with lignin polymerization (AtPrx2/4/25/52/64/71/72; Herrero et al., 2013; Novo-Uzal et al., 2013; Shigeto et al., 2015; Zhong and Ye, 2015) did not identify homologs, as the nucleotide and amino acid sequences of the Norway spruce peroxidases are highly diverged from those of Arabidopsis, with similarities only in the active sites.
As genes encoding many peroxidases and laccases were equally highly expressed in both cell types, it is likely that products of LACCASE and PEROXIDASE genes expressed in ray cells are secreted into the cell wall of ray cells, from where some of them diffuse in the apoplastic fluid into the developing tracheid cell walls, where they function in the oxidative polymerization of coniferyl alcohol. However, a recent study in Arabidopsis showed that a lignification-related laccase, LAC4, was mobile in the primary cell wall but nonmobile in the secondary cell wall, suggesting that it was anchored to the secondary cell wall (Chou et al., 2018). Conifer laccases in native, glycosylated form are larger (70–120 kD; Koutaniemi et al., 2015) than peroxidases (35–60 kD; Koutaniemi et al., 2005). It is possible that peroxidases but not laccases are small enough to fit into the cell wall pores of partially lignified cell walls and can therefore move in the apoplastic fluid. Alternatively, laccases may be attached to some secondary cell wall components (Chou et al., 2018). Hence, it seems likely that the parenchymatous ray cells are not only participating in tracheid cell wall lignification by synthesizing monolignols but also by producing the enzymes (e.g. peroxidases) acting in the polymerization step.
Developing Ray Cells Express a Set of Genes Not Expressed in Tracheids
Ray cells contained a specific set of genes not expressed in tracheids (Table 1; Supplemental Table S6). These ray-cell-only-expressed genes included several putative PENTATRICOPEPTIDE REPEAT (PPR) protein genes. Additionally, there were six PPR genes that were expressed in both rays and tracheids, with a low expression level in tracheids (0.09–0.42; Supplemental Table S6). These nucleus-encoded proteins influence gene expression in chloroplasts and mitochondria by affecting RNA sequence, turnover, processing, or translation (Barkan and Small, 2014). Thus, they have vital effects on organelle biosynthesis and function. The enriched expression of these genes in ray cells suggests that active biogenesis of organelles occurs in developing ray cells. PPR-defective phenotypes are often associated with retarded growth, seedling lethality, or cell wall abnormalities, suggesting that the absence of the PPR-sustained regulation results in shortages in energy supply from mitochondria or chloroplasts (Barkan and Small, 2014). This has been verified with cotton (Gossypium spp.) transformants, where the absence of a mitochondria-targeting sequence in a PPR gene, Gh_A03G0489, resulted in diminished fiber wall thickening. The phenomenon is connected with the immature fiber locus, which is also associated with mitochondrial functioning (Thyssen et al., 2016).
Three UDP-GLYCOSYLTRANSFERASEs were detected among the ray-only-expressed genes (Table 1; Supplemental Table S6). In Arabidopsis, a protein encoded by the xylem-expressed UDP-GLYCOSYLTRANSFERASE72B1 has been shown to make monolignol glucoconjugates (Lin et al., 2016). A knockout of this gene led to strong up-regulation of genes involved in monolignol biosynthesis and polymerization, leading to ectopic and excessive lignification, suggesting that the enzyme is important for normal cell wall lignification (Lin et al., 2016). Since monolignol alcohols are toxic to plant cells (Väisänen et al., 2015), it is probable that monolignols produced by ray cells are glycosylated before transport into the cell wall of neighboring tracheids, especially if the transport to the neighboring cell occurs via the symplastic way (see above; Fig. 7). However, as coniferin and p-coumaryl alcohol glycoside were detected in both tracheids and ray cells (Table 2), it is likely that the same enzyme catalyzes the conjugation of monolignols to sugar moieties in both cell types.
CONCLUSION
In developing xylem of Norway spruce, ray parenchymal cells and developing upright tracheids show highly similar expression for most of the genes in the shikimate, Phe, phenylpropanoid, and monolignol pathways. Metabolome analysis of the cell sap of single tracheids and ray cells of semi-intact plants showed the presence of coniferyl and p-coumaryl alcohols and their glycoconjugates, as well as many of the monolignol biosynthesis pathway intermediates in both cell types. Only a few of the highly expressed genes encoding oxidative enzymes (peroxidases and laccases) differed significantly in expression between these cell types, whereas the majority of the PEROXIDASE and LACCASE genes had no significant difference in expression. These data suggest that, in addition to the well-described functions in storage and radial transport, developing ray cells contribute not only to monolignol biosynthesis but also to the machinery for lignin polymerization needed for cell wall lignification in tracheids. The analysis of transcription factor networks suggests that tracheids and ray cells have some differences in the regulation of expression of genes encoding enzymes leading to phenolic compounds. The LCM approach allowed identification of a set of transcripts in ray cells, with no detectable presence in tracheids. These encoded, for example, putative pentatricopeptide repeat proteins and proteins related to carbohydrate metabolism and transport, suggesting an active developing ray cell metabolism. These findings have advanced our understanding of the process of lignification, its regulatory mechanisms, and spatial organization.
MATERIALS AND METHODS
Plant Material and Cryosectioning for Transcriptomic Analysis
Cryosections of developing xylem of Norway spruce (Picea abies) were prepared from two approximately 40-year-old trees (clone no. E8504) felled during the active growth season (June 2014 and 2015) in Ruotsinkylä, southern Finland. Trunk discs were immediately frozen in liquid nitrogen and stored at −80°C. Cryosectioning was performed as described previously (Blokhina et al., 2017). Briefly, frozen stem discs were quickly (in approximately 1 min) sawn at room temperature into approximately 0.9- × 0.9- × 8- to 10-cm strips. Special care was taken to avoid thawing: the strip was briefly immersed in liquid nitrogen for 5 to 10 s and then sawn into 0.9- × 0.9- × 0.9-cm cubes. During sawing, the surface temperature of the blocks, as assessed with a surface thermometer, did not rise above −50°C. Sawn cubes were immediately mounted in a tissue-freezing medium, and 30- to 40-µm-thick tangential cryosections of developing xylem were cut in the cryotome (Leica, CM 3050 S) at −24°C and collected on glass slides (three to four sections per slide). After two consecutive fixation steps (70% [v/v] ethanol at −24°C and 100% ethanol at room temperature, in a laminar flow cabinet), sections were dried for 30 to 40 min in aseptic conditions and stored up to a maximum of 1 week in air-tight 50-mL Falcon tubes at −80°C prior to LCM. During fixation, the cryosections were gently flattened with a pipette tip to ensure successful laser dissection. The developing xylem was determined to be approximately 600 µm thick with 16 to 18 layers of lignifying cells as described by Blokhina et al. (2017). This parameter was determined separately from a 20-µm cross section stained with a mixture of 0.5% (w/v) Safranine O and 0.5% (w/v) Alcian Blue (1:1 in 50% [v/v] ethanol) for lignin (red) and cellulose (blue), respectively (Fig. 1). During collection of tangential sections for LCM, care was taken not to exceed the determined thickness of developing xylem, and routinely, after adjusting and trimming the block, four sections only (total thickness 120–160 µm) were collected from each cube.
LCM
Fixed cryosections were allowed to reach ambient temperature inside Falcon tubes to avoid condensation onto the slides. Dissection of developing ray cells and upright tracheids was performed by the Zeiss PALM MicroBeam LCM system with the following parameters: cut energy 60 to 65; LPC energy 10 to 30; cut speed 20 to 30; focus 65 to 70; and focus delta −2. The parameters are slide and cell type specific, and hence, small adjustments were performed when needed. The flatness of the section was crucial for successful cutting and catapulting (Blokhina et al., 2017). Ray cells and tracheids were collected separately into 500-µL adhesive cap tubes (Carl Zeiss), and the tubes were snap frozen in liquid nitrogen and stored at –80°C until RNA isolation. Special care was taken to exclude resin ducts and ray tracheids from the dissected cells used for RNA isolation (Supplemental Fig. S1). To avoid RNA degradation, each slide and the adhesive cap tube were used for microdissection for no more than 1.5 h, and RNase-free conditions were maintained throughout the cryosectioning and the LCM steps. Specific cell types collected during many LCM sessions were used for RNA isolation to achieve the adequate yield of RNA. The average area per each sample where RNA was isolated was 0.2 to 0.4 mm2. RNA from the following samples was prepared for the next-generation sequencing: ray parenchymal cells (excluding ray tracheids and resin ducts), upright tracheids, and whole sections of developing xylem including all cell types (tracheids, ray parenchymal cells, ray tracheids, and resin duct epithelial cells).
RNA Isolation and Next-Generation Sequencing
Total RNA was isolated with an RNeasy Plant Mini Kit (Qiagen, 74904) according to the manufacturer’s instructions. Final elution of RNA was performed with 30 µL of RNase-free water, after which RNA was quantified by Qubit Fluorometer v. 1.27. RNA quality was assessed by an Agilent 2100 Bioanalyzer using the Eukaryote Total RNA Pico kit with picogram sensitivity.
RNA from three replicates of ray cells and tracheids, and from four replicates of whole sections of developing xylem, was sequenced. Next-generation sequencing was performed in the Functional Genomics Unit, Biomedicum Helsinki, University of Helsinki in Illumina NextSeq Mid output 2 × 150 bp. The SMARTer Ultra Low Input RNA Kit for Sequencing v. 3 was used to generate high-quality cDNA compatible with the Illumina Nextera XT DNA Sample Preparation Kit.
Preprocessing of RNA-Seq Data and Differential Expression Analyses
The data preprocessing was performed as described at http://www.epigenesys.Eu/en/protocols/bio-informatics/1283-guidelines-for-rna-seq-data-analysis. Briefly, the quality of the raw sequence data was assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Residual rRNA contamination was assessed and filtered using SortMeRNA v. 1.9 (Kopylova et al., 2012; settings -n 8 -a 8 -v) using the rRNA sequences provided with SortMeRNA (rfam-5s-database-id98.fasta, rfam-5.8s-database-id98.fasta, silva-arc-16s-database-id95.fasta, silva-bac-16s-database-id85.fasta, silva-euk-18s-database-id95.fasta, silva-arc-23s-database-id98.fasta, silva-bac-23s-database-id98.fasta, and silva-euk-28s-database-id98.fasta). The data were then filtered to remove adapters and trimmed for quality using Trimmomatic v. 0.32 (Bolger et al., 2014; settings TruSeq3-PE-2.fa:2:30:10 LEADING:3 SLIDINGWINDOW:5:20 MINLEN:50). After both filtering steps, FastQC was run again to ensure that no technical artifacts were introduced. Filtered reads were aligned to the Norway spruce genome v. 1 (retrieved from the ConGenIE resource; Sundell et al., 2015) using STAR v. 2.4.0f1 (Dobin et al., 2013; nondefault settings: –outSAMstrandField intronMotif–readFilesCommand zcat–outSAMmapqUnique 254–quantMode TranscriptomeSAM–outFilterMultimapNmax 100–outReadsUnmapped Fastx–chimSegmentMin 1–outSAMtype BAM SortedByCoordinate–outWigType bedGraph–alignIntronMax 11000). Reasons for the use of the draft Norway spruce genome are explained in the next paragraph. The annotations obtained from the Norway spruce v. 1.0 GFF file contain only one transcript per gene model, and as such, did not need to be modified to generate synthetic gene models. This GFF file and the STAR read alignments were used as input to the HTSeq (Anders et al., 2015) htseq-count python utility to calculate exon-based read-count values. The htseq-count utility takes only uniquely mapping reads into account. Statistical analysis of single-gene differential expression between conditions was performed in R v. 3.2.3 using the Bioconductor v. 3.2 (Gentleman et al., 2004) DESeq2 package v. 1.10.1 (Love et al., 2014). False discovery rate-adjusted P values were used to assess significance; a common threshold of 1% was used throughout. For the data quality assessment and visualization, the read counts were normalized using a VST as implemented in DESeq2. The biological relevance of the data (e.g. biological replicate similarity) was assessed by principal component analysis and other visualizations (e.g. heat maps) using custom R scripts. One of the ray cell samples, S1-ray-cells, had very different characteristics from the other samples; this could be related to technical issues during the sample preparation. As a consequence, the sample was excluded from the downstream analyses. For all subsequent expression analyses, which were also performed in R, the normalized read counts obtained from DESeq2 were used. The differential expression analyses of the three conditions (ray cells, tracheids, and whole sections) were performed using DESeq2, and every comparison combination was conducted: ray cells versus tracheids, ray cells versus whole sections, and tracheids versus whole sections. An overview of the data, including raw and post-QC read counts and alignment rates, is given in Supplemental Table S7.
RNA-Seq analyses were performed by alignment to the draft Norway spruce genome assembly and annotation, the annotation of which has been shown to be comprehensive and to cover more than 90% of gene models expected to be present in conifers (Nystedt et al., 2013). Faced with a draft assembly, an alternative approach would be to perform a de novo transcript assembly. However, this has a number of inherent limitations, including incomplete and incorrect transcript reconstruction and fragmentation of the assembled transcript. These limitations must be weighed against those of using a draft genome assembly-based analysis, albeit one where the vast majority of protein-coding genes are expected to be represented. To explore this, we considered both the percentage of RNA-Seq reads aligning to the genome and, specifically, to annotated genes and contrasted this to a de novo transcriptome assembly using all RNA-Seq data generated in this study. On average, 83.5% of RNA-Seq reads aligned to the genome, with 59.5% aligning to annotated protein-coding genes. We frequently observed approximately 20% unaligned reads, which can vary substantially depending on the origin of the material. Unaligned reads can result from incomplete genome assembly but can also represent reads originating from other organisms such as fungi (Delhomme et al., 2015) and from technical sequencing artifacts. In contrast, the do novo transcriptome assembly (produced using default settings for the Trinity assembler) generated 4,100,427 transcripts, of which 361,899 were protein coding and 94% aligned to the genome. The alignment was performed using GMAP, and it reported 92,900, 35,910, and 950,912 unique, translocation, and multiple mappings, respectively. These overlapped (more than 95%) 12,848, 6,201, and 13,948 genes from the genome annotation of Nystedt et al. (2013), respectively, and originated from a total of 78,806 reconstructed transcripts (340,905 transcripts had an overlap of greater than 30%). The high number of noncoding transcripts that align to the genome combined with the extremely high number of transcripts in total highlight the extensive challenge faced in using a de novo-based approach for such analyses in Norway spruce. While there were a large number of protein-coding transcripts that aligned to the genome but did not overlap currently annotated genes, these included 60% and 32% with no or low expression in all samples [less than log2(5) reads], respectively, and hence that are of questionable biological relevance, with many representing incomplete genes (not spanning start-to-stop codons) and therefore potentially representing pseudogene fragments, which are prevalent in conifer genomes (Nystedt et al., 2013). As such, and in balance, the genome-based annotation is expected to yield biologically relevant expression results that capture the vast majority of informative genes while avoiding the many complications associated with using a de novo-based approach.
GO Enrichment Analysis
In order to increase coverage for GO enrichment, Arabidopsis (Arabidopsis thaliana) annotations were retrieved from ConGenIE.org (Sundell et al., 2015). Differentially expressed genes (Padj < 0.01) were analyzed with the agriGO Web tool (http://bioinfo.cau.edu.cn/agriGO/; Du et al., 2010) using hypergeometric testing (Yekutieli false discovery rate, significance level of 0.05). Results were visualized with REVIGO (http://revigo.irb.hr/; Supek et al., 2011) and R (r-project.org). The 500 most highly expressed genes from tracheids, ray cells, and whole sections were analyzed in the same way.
Plant Material for Single-Cell Metabolomic Analysis
To conduct in situ cell metabolomics, 4-year-old potted Norway spruce plants having a mean height of 41.5 cm were purchased from a nursery in Kamimashiki, Kumamoto, Japan, on April 13, 2018, and transported to Kyushu Okinawa Agricultural Research Center, Chikugo, Japan. The plants were stored in the growth chamber (18°C/16°C, 60%/70% relative humidity, and 350 µmol m−2 s−1 photosynthetically active radiation with a 13.5-h photoperiod). In April, the new xylem in the young trees was at the same developmental stage as in the mature trees used for LCM in June in Finland, justified by the number of newly formed xylem cell layers. The species identity of the trees was verified at the Botanical Herbarium of the University of Helsinki. It must be noted that trees used for transcriptomics and single-cell metabolomics were of different genotypes, different ages, and were grown in different environments.
In Situ Cell Metabolomics
Plants were transported to the laboratory set at 22°C to be used for the following in situ cell metabolomics. Prior to the analysis, plant water status, evaluated as stem water potential (SWP), was determined using a pressure chamber (Scholander et al., 1965) according to Turner (1988), taking care to avoid leaf water loss. Two twigs in the middle part of the stem were bagged with zip-top bags covered with aluminum film and allowed to equilibrate with the SWP for 20 min. When covering the twigs, some needles attached to the base of the branches, corresponding to the position of the pressure chamber lid, were removed, and the surface was covered with Vaseline prior to the covering. A preliminary experiment showed that leaf water potential equilibrated with the SWP in 20 min after the covering (Supplemental Fig. S4A). After 20 min, one of the covered twigs was removed and SWP was measured in the pressure chamber.
After the first SWP measurement, the plant was laid down and mounted on a laboratory jack-stand placed on a vibration-free table in the system of the on-site cell metabolome analysis using picoPPESI-MS (see Supplemental Fig. S1 in Wada et al., 2019). A part of the bark tissue (9.4 mm in length × 6.1 mm in width, the area was approximately 57.8 mm2) at the middle position of the stem was quickly removed using a small chisel under humid conditions to make a tangential section, and thereafter the stem was gently fixed to the holder in two positions (Supplemental Fig. S2, B and C).
Adopting the same system used by Wada et al. (2019), in situ cell metabolome analysis using picoPPESI-MS (Nakashima et al., 2016) was carried out individually for parenchymal ray cells and tracheids of developing xylem in semi-intact plants, where a small part of bark tissue was removed. Quartz microcapillary tips were prepared by a micropipette puller and were ground by using a microgrinder to prepare approximately 2-µm internal diameter tips. The orientation of the exposed tissue and the probe was set using a digital microscope (Supplemental Fig. S2). The cross section of the attached stems (and the exposed developing xylem tissues) was round, with the stem diameter of the cut position 8.3 ± 0.9 mm on average (n = 9). The quartz capillary tip was perpendicularly inserted into the tissue surface, while the location of the parenchymal ray cells and tracheids was identified with a microscope. This analytical method allowed cell metabolite profiling in real time at each target cell without any pretreatments under constant laboratory conditions. The quartz microcapillary tip was filled with a 0.01% (v/v) ionic liquid/silicone oil mixture (see below), and positive oil pressure of the mixture was maintained prior to tip insertion. Each punctured cell was located 50 to 75 μm below the cambium, where the stained sections indicated active lignin biosynthesis (Supplemental Fig. S2A). The microcapillary was moved with the aid of a motorized piezo-manipulator. After the picoliter-volume cell sap was collected by depressurizing in the microcapillary, the probe tip mounted on the 3D move/rotation micromanipulator was immediately oriented toward the orifice of an Orbitrap mass spectrometer (Q-Exactive; Thermo Fisher Scientific; Supplemental Fig. S2D) electrified with −4 kV using a high-voltage generator. The MS scan was performed in a negative ion mode with the instrumental settings of 200 ms as maximum injection time, inlet ion transfer tube temperature of 250°C, and resolution of 35,000. The picoPPESI-MS analysis was repeated with each cell type to collect technical replications (typically two to four), and the entire process of two types of cells, including positioning of the samples, was completed in 1 h per plant. All manipulations were conducted using the above-mentioned digital microscope, and the area where the bark was removed in the stem was humidified during all processes (Supplemental Fig. S2, A and B). For picoPPESI-MS, an ionic liquid, trihexyl (tetradecyl) phosphonium bis trifluoromethanesulfonyl amide (Cyphos IL109; Strem Chemicals), was suspended in phenyl methyl silicone oil (Wacker silicone fluid AS4) at a concentration of 0.01% (v/v) to enhance the electric conductivity of the silicone oil.
After completion of the cell metabolome analysis, another twig that had been covered with a zip-top bag was removed from the plant, and the second SWP measurement was conducted as described above. SWP at the first and second measurements was −0.42 and −0.4 MPa on average, respectively (Supplemental Fig. S4B). The values agree with the needle water potentials measured from the 4-year-old Norway spruce trees grown under nonstressed conditions (Ditmarová et al., 2010). The data indicate that there was no significant disturbance in plant water status after the bark removal, followed by picoPPESI-MS analysis, at least within 1 h (Supplemental Fig. S4B). All analyses were initiated between 9:30 am and 4:00 pm.
Identification of Metabolites
The list of monoisotopic exact m/z values for all the peaks on acquired mass spectra was extracted using the Qual Browser application in the Thermo Xcalibur software (Thermo Fisher Scientific). The observed peaks were then matched with theoretical masses of candidate metabolites in an online metabolomics database, Metlin (http://metlin.scripps.edu/index.php), allowing differences of less than 5 ppm. The putative metabolites were also confirmed with simulated isotopic ratios using the Qual Browser application. In addition to single-cell analyses, MS/MS analysis was conducted in the crude tissue extract. To do so, developing xylem tissues from the same plants were immediately frozen at −80°C for 2 h, and the tissues were freeze-dried. The tissue was ground to a fine powder with a mixer mill (MM400; Retch) and extracted with 50% (v/v) water/methanol. After centrifugation for 10 min at 10,000g at 4°C, the supernatant (crude tissue extract) was used for the MS/MS analysis. The collision-induced dissociation tandem MS analysis of the extract on putative monolignol metabolites was performed by using the same Orbitrap mass spectrometer coupled with the picoPPESI system in negative ion mode for further identification. For p-coumaric acid, caffeic acid, coniferyl aldehyde, coniferyl alcohol, p-coumaryl alcohol, coniferin, Cl− adduct ion of coniferin, p-coumaryl alcohol 4-glucoside, Cl− adduct ion of p-coumaryl alcohol 4-glucoside, d-erythrose 4-phosphate, 3-dehydroquinic acid, and chorismic acid/prephenic acid, the observed MS/MS fragmentation pattern was consulted with the PubChem library (https://pubchem.ncbi.nlm.nih.gov/). In the case of MS/MS identification for coniferin and coniferyl alcohol, 10 mm Glc, 10 mm Suc, plus 0.1 mm NaCl standard solutions were provided. All the standard chemicals and organic solvents used in the experiments were liquid chromatography-MS grade and purchased from Wako Pure Chemical Industries. Ultrapure water of 18.2 MΩ cm−1 was used throughout the experiment.
Statistical Analysis
Statistical analysis of cell metabolomics was conducted using Student’s t test in JMP software (version 12.1.0; SAS Institute).
Accession Numbers
The RNA-Seq data have been deposited to the European Nucleotide Archive ENA and are accessible with the accession number PRJEB10305/ERP011536.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. LCM of ray cells in developing xylem of spruce.
Supplemental Figure S2. Diagram showing the experimental flow of in situ cell metabolome analysis in developing xylem of Norway spruce plants using the picoPPESI-MS system.
Supplemental Figure S3. MS/MS analysis and isotope analysis performed by using an Orbitrap mass spectrometer coupled with the picoPPESI system in negative ion mode.
Supplemental Figure S4. Changes in leaf water potential measured after bagging the twigs with zip-top bags covered with aluminum film.
Supplemental Table S1. The 500 most highly expressed genes in whole sections, tracheids, and ray cells.
Supplemental Table S2. Differentially expressed genes in ray cells and tracheids.
Supplemental Table S3. Expression of genes encoding NAC and MYB transcription factors, enzymes of shikimate, aromatic amino acid, phenylpropanoid, and monolignol biosynthetic pathways.
Supplemental Table S4. Peroxidase and laccase gene expression.
Supplemental Table S5. Transcription factors and their neighbors that form a network in NorWood.
Supplemental Table S6. Genes expressed only in ray cells.
Supplemental Table S7. Summary of RNA-Seq sequencing reads and alignments.
Supplemental Table S8. Transcription factors differentially expressed in developing ray cells and tracheids.
ACKNOWLEDGMENTS
We thank Svetlana Kauppanen for microscopy of xylem. Next-generation sequencing was performed at the Functional Genomics Unit, Biomedicum Helsinki, University of Helsinki. We gratefully acknowledge Prof. Hiroshi Nonami, Dr. Hiroshi Nakano, and Prof. Rosa Erra-Balsells for helpful comments on the single-cell metabolomics.
Footnotes
This work was supported by the Academy of Finland (decisions 251390, 256174, 283245, and 317964 to A.K. and decision 316486 to O.B.) and the Faculty of Biological and Environmental Sciences of the University of Helsinki (to K.F.). T.L. was supported by the Finnish Cultural Foundation, Häme Regional Fund. N.D. was supported by the Knut and Alice Wallenberg Foundation and the UPSC Berzelii Centre. N.R.S. was supported by Trees and Crops for the Future.
Articles can be viewed without a subscription.
References
- Abbott E, Hall D, Hamberger B, Bohlmann J (2010) Laser microdissection of conifer stem tissues: Isolation and analysis of high quality RNA, terpene synthase enzyme activity and terpenoid metabolites from resin ducts and cambial zone tissue of white spruce (Picea glauca). BMC Plant Biol 10: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alejandro S, Lee Y, Tohge T, Sudre D, Osorio S, Park J, Bovet L, Lee Y, Geldner N, Fernie AR, et al. (2012) AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr Biol 22: 1207–1212 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki D, Hanaya Y, Akita T, Matsushita Y, Yoshida M, Kuroda K, Yagami S, Takama R, Fukushima K (2016) Distribution of coniferin in freeze-fixed stem of Ginkgo biloba L. by cryo-TOF-SIMS/SEM. Sci Rep 6: 31525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442 [DOI] [PubMed] [Google Scholar]
- Barra-Jiménez A, Ragni L (2017) Secondary development in the stem: When Arabidopsis and trees are closer than it seems. Curr Opin Plant Biol 35: 145–151 [DOI] [PubMed] [Google Scholar]
- Barros J, Serk H, Granlund I, Pesquet E (2015) The cell biology of lignification in higher plants. Ann Bot 115: 1053–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström B. (2003) Chemical and structural changes during heartwood formation in Pinus sylvestris. Forestry 76: 45–53 [Google Scholar]
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, et al. (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokhina O, Valerio C, Sokołowska K, Zhao L, Kärkönen A, Niittylä T, Fagerstedt K (2017) Laser capture microdissection protocol for xylem tissues of woody plants. Front Plant Sci 7: 1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomal C, Bedon F, Caron S, Mansfield SD, Levasseur C, Cooke JEK, Blais S, Tremblay L, Morency MJ, Pavy N, et al. (2008) Involvement of Pinus taeda MYB1 and MYB8 in phenylpropanoid metabolism and secondary cell wall biogenesis: A comparative in planta analysis. J Exp Bot 59: 3925–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañas RA, Canales J, Gómez-Maldonado J, Ávila C, Cánovas FM (2014) Transcriptome analysis in maritime pine using laser capture microdissection and 454 pyrosequencing. Tree Physiol 34: 1278–1288 [DOI] [PubMed] [Google Scholar]
- Carvalho A, Paiva J, Louzada J, Lima-Brito J (2013) The transcriptomics of secondary growth and wood formation in conifers. Mol Biol Int 2013: 974324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celedon JM, Yuen MMS, Chiang A, Henderson H, Reid KE, Bohlmann J (2017) Cell-type- and tissue-specific transcriptomes of the white spruce (Picea glauca) bark unmask fine-scale spatial patterns of constitutive and induced conifer defense. Plant J 92: 710–726 [DOI] [PubMed] [Google Scholar]
- Chou EY, Schuetz M, Hoffmann N, Watanabe Y, Sibout R, Samuels AL (2018) Distribution, mobility, and anchoring of lignin-related oxidative enzymes in Arabidopsis secondary cell walls. J Exp Bot 69: 1849–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhomme N, Sundström G, Zamani N, Lantz H, Lin YC, Hvidsten TR, Höppner MP, Jern P, Van de Peer Y, Lundeberg J, et al. (2015) Serendipitous meta-transcriptomics: The fungal community of Norway spruce (Picea abies). PLoS ONE 10: e0139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmawardhana DP, Ellis BE, Carlson JE (1995) A β-glucosidase from lodgepole pine xylem specific for the lignin precursor coniferin. Plant Physiol 107: 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmawardhana DP, Ellis BE, Carlson JE (1999) cDNA cloning and heterologous expression of coniferin β-glucosidase. Plant Mol Biol 40: 365–372 [DOI] [PubMed] [Google Scholar]
- Ditmarová L, Kurjak D, Palmroth S, Kmet J, Strelcová K (2010) Physiological responses of Norway spruce (Picea abies) seedlings to drought stress. Tree Physiol 30: 205–213 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z (2010) agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res 38: W64–W70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval I, Lachance D, Giguère I, Bomal C, Morency MJ, Pelletier G, Boyle B, MacKay JJ, Séguin A (2014) Large-scale screening of transcription factor-promoter interactions in spruce reveals a transcriptional network involved in vascular development. J Exp Bot 65: 2319–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenberg K, Harkin JM (1963) The glucosides of cambial sap of spruce. Phytochemistry 2: 189–193 [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. (2004) Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JO, Blob B, Helariutta Y (2017) Differentiation of conductive cells: A matter of life and death. Curr Opin Plant Biol 35: 23–29 [DOI] [PubMed] [Google Scholar]
- Herrero J, Fernández-Pérez F, Yebra T, Novo-Uzal E, Pomar F, Pedreño M, Cuello J, Guéra A, Esteban-Carrasco A, Zapata JM (2013) Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 237: 1599–1612 [DOI] [PubMed] [Google Scholar]
- Hogekamp C, Arndt D, Pereira PA, Becker JD, Hohnjec N, Küster H (2011) Laser microdissection unravels cell-type-specific transcription in arbuscular mycorrhizal roots, including CAAT-box transcription factor gene expression correlating with fungal contact and spread. Plant Physiol 157: 2023–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höll W. (1975) Radial transport in rays In Zimmermann MH, Milburn JA, eds, Transport in Plants I. Encyclopedia of Plant Physiology (New Series), Vol 1 Springer, Berlin, pp 432–450 [Google Scholar]
- Hosokawa M, Suzuki S, Umezawa T, Sato Y (2001) Progress of lignification mediated by intercellular transportation of monolignols during tracheary element differentiation of isolated Zinnia mesophyll cells. Plant Cell Physiol 42: 959–968 [DOI] [PubMed] [Google Scholar]
- Hudgins JW, Christiansen E, Franceschi VR (2003) Methyl jasmonate induces changes mimicking anatomical defenses in diverse members of the Pinaceae. Tree Physiol 23: 361–371 [DOI] [PubMed] [Google Scholar]
- Jaakkola T, Mäkinen H, Saranpää P (2007) Effects of thinning and fertilisation on tracheid dimensions and lignin content of Norway spruce. Holzforschung 61: 301–310 [Google Scholar]
- Jacquet JS, Bosc A, O’Grady A, Jactel H (2014) Combined effects of defoliation and water stress on pine growth and non-structural carbohydrates. Tree Physiol 34: 367–376 [DOI] [PubMed] [Google Scholar]
- Jokipii-Lukkari S, Delhomme N, Schiffthaler B, Mannapperuma C, Prestele J, Nilsson O, Street NR, Tuominen H (2018) Transcriptional roadmap to seasonal variation in wood formation of Norway spruce. Plant Physiol 176: 2851–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokipii-Lukkari S, Sundell D, Nilsson O, Hvidsten TR, Street NR, Tuominen H (2017) NorWood: A gene expression resource for evo-devo studies of conifer wood development. New Phytol 216: 482–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Rensing KH, Wong JCT, Banno B, Mansfield SD, Samuels AL (2008) Tracking monolignols during wood development in lodgepole pine. Plant Physiol 147: 1750–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärkönen A, Koutaniemi S, Mustonen M, Syrjänen K, Brunow G, Kilpeläinen I, Teeri TH, Simola LK (2002) Lignification related enzymes in Picea abies suspension cultures. Physiol Plant 114: 343–353 [DOI] [PubMed] [Google Scholar]
- Kopylova E, Noé L, Touzet H (2012) SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28: 3211–3217 [DOI] [PubMed] [Google Scholar]
- Koutaniemi S, Malmberg HAM, Simola LK, Teeri TH, Kärkönen A (2015) Norway spruce (Picea abies) laccases: Characterization of a laccase in a lignin-forming tissue culture. J Integr Plant Biol 57: 341–348 [DOI] [PubMed] [Google Scholar]
- Koutaniemi S, Toikka MM, Kärkönen A, Mustonen M, Lundell T, Simola LK, Kilpeläinen IA, Teeri TH (2005) Characterization of basic p-coumaryl and coniferyl alcohol oxidizing peroxidases from a lignin-forming Picea abies suspension culture. Plant Mol Biol 58: 141–157 [DOI] [PubMed] [Google Scholar]
- Koutaniemi S, Warinowski T, Kärkönen A, Alatalo E, Fossdal CG, Saranpää P, Laakso T, Fagerstedt KV, Simola LK, Paulin L, et al. (2007) Expression profiling of the lignin biosynthetic pathway in Norway spruce using EST sequencing and real-time RT-PCR. Plant Mol Biol 65: 311–328 [DOI] [PubMed] [Google Scholar]
- Laitinen T, Morreel K, Delhomme N, Gauthier A, Schiffthaler B, Nickolov K, Brader G, Lim KJ, Teeri TH, Street NR, et al. (2017) A key role for apoplastic H2O2 in Norway spruce phenolic metabolism. Plant Physiol 174: 1449–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamara M, Raherison E, Lenz P, Beaulieu J, Bousquet J, MacKay J (2016) Genetic architecture of wood properties based on association analysis and co-expression networks in white spruce. New Phytol 210: 240–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev-Yadun S, Aloni R (1995) Differentiation of the ray system in woody plants. Bot Rev 61: 45–88 [Google Scholar]
- Lin JS, Huang XX, Li Q, Cao Y, Bao Y, Meng XF, Li YJ, Fu C, Hou BK (2016) UDP-glycosyltransferase 72B1 catalyzes the glucose conjugation of monolignols and is essential for the normal cell wall lignification in Arabidopsis thaliana. Plant J 88: 26–42 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Dudareva N (2012) The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol 63: 73–105 [DOI] [PubMed] [Google Scholar]
- Marjamaa K, Hildén K, Kukkola E, Lehtonen M, Holkeri H, Haapaniemi P, Koutaniemi S, Teeri TH, Fagerstedt K, Lundell T (2006) Cloning, characterization and localization of three novel class III peroxidases in lignifying xylem of Norway spruce (Picea abies). Plant Mol Biol 61: 719–732 [DOI] [PubMed] [Google Scholar]
- Marjamaa K, Lehtonen M, Lundell T, Toikka M, Saranpää P, Fagerstedt KV (2003) Developmental lignification and seasonal variation in β-glucosidase and peroxidase activities in xylem of Scots pine, Norway spruce and silver birch. Tree Physiol 23: 977–986 [DOI] [PubMed] [Google Scholar]
- Meents MJ, Watanabe Y, Samuels AL (2018) The cell biology of secondary cell wall biosynthesis. Ann Bot 121: 1107–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao YC, Liu CJ (2010) ATP-binding cassette-like transporters are involved in the transport of lignin precursors across plasma and vacuolar membranes. Proc Natl Acad Sci USA 107: 22728–22733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa Y, Yoshinaga A, Kamitakahara H, Wada M, Takabe K (2010) Cellular distribution of coniferin in differentiating xylem of Chamaecyparis obtusa as revealed by Raman microscopy. Holzforschung 64: 61–67 [Google Scholar]
- Nagy NE, Sikora K, Krokene P, Hietala AM, Solheim H, Fossdal CG (2014) Using laser micro-dissection and qRT-PCR to analyze cell type-specific gene expression in Norway spruce phloem. PeerJ 2: e362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y, Yamaguchi M, Endo H, Rejab NA, Ohtani M (2015) NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front Plant Sci 6: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima T, Wada H, Morita S, Erra-Balsells R, Hiraoka K, Nonami H (2016) Single-cell metabolite profiling of stalk and glandular cells of intact trichomes with internal electrode capillary pressure probe electrospray ionization mass spectrometry. Anal Chem 88: 3049–3057 [DOI] [PubMed] [Google Scholar]
- Novo-Uzal E, Fernández-Pérez F, Herrero J, Gutiérrez J, Gómez-Ros LV, Bernal MA, Díaz J, Cuello J, Pomar F, Pedreño MA (2013) From Zinnia to Arabidopsis: Approaching the involvement of peroxidases in lignification. J Exp Bot 64: 3499–3518 [DOI] [PubMed] [Google Scholar]
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. (2013) The Norway spruce genome sequence and conifer genome evolution. Nature 497: 579–584 [DOI] [PubMed] [Google Scholar]
- Pascual MB, El-Azaz J, de la Torre FN, Cañas RA, Avila C, Cánovas FM (2016) Biosynthesis and metabolic fate of phenylalanine in conifers. Front Plant Sci 7: 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesquet E, Zhang B, Gorzsás A, Puhakainen T, Serk H, Escamez S, Barbier O, Gerber L, Courtois-Moreau C, Alatalo E, et al. (2013) Non-cell-autonomous postmortem lignification of tracheary elements in Zinnia elegans. Plant Cell 25: 1314–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piispanen R, Willför S, Saranpää P, Holmbom B (2008) Variation of lignans in Norway spruce (Picea abies [L.] Karst.) knotwood: Within-stem variation and the effect of fertilization at two experimental sites in Finland. Trees (Berl) 22: 317–328 [Google Scholar]
- Raherison ESM, Giguère I, Caron S, Lamara M, MacKay JJ (2015) Modular organization of the white spruce (Picea glauca) transcriptome reveals functional organization and evolutionary signatures. New Phytol 207: 172–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel K, Berrio-Sierra J, Derikvand MM, Pollet B, Thévenin J, Lapierre C, Jouanin L, Joseleau JP (2009) Impact of CCR1 silencing on the assembly of lignified secondary walls in Arabidopsis thaliana. New Phytol 184: 99–113 [DOI] [PubMed] [Google Scholar]
- Savidge RA. (1989) Coniferin, a biochemical indicator of commitment to tracheid differentiation in conifers. Can J Bot 67: 2663–2668 [Google Scholar]
- Scholander PF, Bradstreet ED, Hemmingsen EA, Hammel HT (1965) Sap pressure in vascular plants: Negative hydrostatic pressure can be measured in plants. Science 148: 339–346 [DOI] [PubMed] [Google Scholar]
- Shigeto J, Itoh Y, Hirao S, Ohira K, Fujita K, Tsutsumi Y (2015) Simultaneously disrupting AtPrx2, AtPrx25 and AtPrx71 alters lignin content and structure in Arabidopsis stem. J Integr Plant Biol 57: 349–356 [DOI] [PubMed] [Google Scholar]
- Shigeto J, Tsutsumi Y (2016) Diverse functions and reactions of class III peroxidases. New Phytol 209: 1395–1402 [DOI] [PubMed] [Google Scholar]
- Smith RA, Schuetz M, Karlen SD, Bird D, Tokunaga N, Sato Y, Mansfield SD, Ralph J, Samuels AL (2017) Defining the diverse cell populations contributing to lignification in Arabidopsis thaliana stems. Plant Physiol 174: 1028–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Schuetz M, Roach M, Mansfield SD, Ellis B, Samuels L (2013) Neighboring parenchyma cells contribute to Arabidopsis xylem lignification, while lignification of interfascicular fibers is cell autonomous. Plant Cell 25: 3988–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokołowska K. (2013) Symplasmic transport in wood: The importance of living xylem cells In Sokołowska K, Sowiński P, eds, Symplasmic Transport in Vascular Plants. Springer, New York, pp 101–132 [Google Scholar]
- Spicer R. (2014) Symplasmic networks in secondary vascular tissues: Parenchyma distribution and activity supporting long-distance transport. J Exp Bot 65: 1829–1848 [DOI] [PubMed] [Google Scholar]
- Sundell D, Mannapperuma C, Netotea S, Delhomme N, Lin YC, Sjödin A, Van de Peer Y, Jansson S, Hvidsten TR, Street NR (2015) The Plant Genome Integrative Explorer Resource:. New Phytol 208: 1149–1156. Available at: PlantGenIE.org [DOI] [PubMed] [Google Scholar]
- Sundell D, Street NR, Kumar M, Mellerowicz EJ, Kucukoglu M, Johnsson C, Kumar V, Mannapperuma C, Delhomme N, Nilsson O, et al. (2017) AspWood: High-spatial-resolution transcriptome profiles reveal uncharacterized modularity of wood formation in Populus tremula. Plant Cell 29: 1585–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T (2011) REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS ONE 6: e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyssen GN, Fang DD, Zeng L, Song X, Delhom CD, Condon TL, Li P, Kim HJ (2016) The immature fiber mutant phenotype of cotton (Gossypium hirsutum) is linked to a 22-bp frame-shift deletion in a mitochondria targeted pentatricopeptide repeat gene. G3 (Bethesda) 6: 1627–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga N, Sakakibara N, Umezawa T, Ito Y, Fukuda H, Sato Y (2005) Involvement of extracellular dilignols in lignification during tracheary element differentiation of isolated Zinnia mesophyll cells. Plant Cell Physiol 46: 224–232 [DOI] [PubMed] [Google Scholar]
- Tsuyama T, Kawai R, Shitan N, Matoh T, Sugiyama J, Yoshinaga A, Takabe K, Fujita M, Yazaki K (2013) Proton-dependent coniferin transport, a common major transport event in differentiating xylem tissue of woody plants. Plant Physiol 162: 918–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner NC. (1988) Measurement of plant water status by the pressure chamber technique. Irrig Sci 9: 289–308 [Google Scholar]
- Väisänen EE, Smeds AI, Fagerstedt KV, Teeri TH, Willför SM, Kärkönen A (2015) Coniferyl alcohol hinders the growth of tobacco BY-2 cells and Nicotiana benthamiana seedlings. Planta 242: 747–760 [DOI] [PubMed] [Google Scholar]
- van Bel AJE. (1990) Xylem-phloem exchange via the rays: The undervalued route of transport. J Exp Bot 41: 631–644 [Google Scholar]
- van der Schoot C, van Bel AJ (1990) Mapping membrane potential differences and dye-coupling in internodal tissues of tomato (Solanum lycopersicum L.). Planta 182: 9–21 [DOI] [PubMed] [Google Scholar]
- Wada H, Hatakeyama Y, Onda Y, Nonami H, Nakashima T, Erra-Balsells R, Morita S, Hiraoka K, Tanaka F, Nakano H (2019) Multiple strategies for heat adaptation to prevent chalkiness in the rice endosperm. J Exp Bot 70: 1299–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chantreau M, Sibout R, Hawkins S (2013) Plant cell wall lignification and monolignol metabolism. Front Plant Sci 4: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willför S, Hemming J, Reunanen M, Eckerman C, Holmbom B (2003) Lignans and lipophilic extractives in Norway spruce knots and stemwood. Holzforschung 57: 27–36 [Google Scholar]
- Willför SM, Sundberg AC, Rehn PW, Saranpää PT, Holmbom BR (2005) Distribution of lignans in knots and adjacent stemwood of Picea abies. Holz Roh Werkst 63: 353–357 [Google Scholar]
- Witt W, Sauter JJ (1994) Starch metabolism in poplar wood ray cells during spring mobilization and summer deposition. Physiol Plant 92: 9–16 [Google Scholar]
- Yamaguchi M, Ohtani M, Mitsuda N, Kubo M, Ohme-Takagi M, Fukuda H, Demura T (2010) VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in Arabidopsis. Plant Cell 22: 1249–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, Shao H, Wang X, Wang ZY, Dixon RA (2013) Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng P, Aoki D, Yoshida M, Matsushita Y, Imai T, Fukushima K (2014) Lignification of ray parenchyma cells in the xylem of Pinus densiflora. Part I. Microscopic investigation by POM, UV microscopy, and TOF-SIMS. Holzforschung 68: 897–905 [Google Scholar]
- Zhong R, Ye ZH (2015) Secondary cell walls: Biosynthesis, patterned deposition and transcriptional regulation. Plant Cell Physiol 56: 195–214 [DOI] [PubMed] [Google Scholar]