Abstract
Ribosome driven protein biosynthesis comprises four phases: initiation, elongation, termination and recycling. In bacteria, ribosome recycling requires ribosome recycling factor and elongation factor G, and several structures of bacterial recycling complexes have been determined. In the eukaryotic and archaeal kingdoms, however, recycling involves the ABC-type ATPase ABCE1 and little is known about its structural basis. Here we present cryo-EM reconstructions of eukaryotic and archaeal ribosome recycling complexes containing ABCE1 and the termination factor paralog Pelota. These structures reveal the overall binding mode of ABCE1 to be similar to canonical translation factors. Moreover, the iron-sulfur cluster domain of ABCE1 interacts with and stabilizes Pelota in a conformation that reaches towards the peptidyl transferase center, thus explaining how ABCE1 may stimulate peptide-release activity of canonical termination factors. Employing the mechanochemical properties of ABCE1, a conserved mechanism in archaea and eukaryotes is suggested that couples translation termination to recycling, and eventually to re-initiation.
Introduction
Recycling of ribosomes for a new round of translation initiation is an essential part of protein synthesis. In archaea and eukaryotes recycling has been shown to require the highly conserved and essential ABC-type ATPase ABCE1 (Rli1p in Saccharomyces cerevisiae with 46.7% identity to aABCE1 in Pyrococcus furiosus)1–4. ABCE1 can dissociate ribosomes into subunits either after canonical termination by release factors (RFs)4 or after recognition of stalled ribosomes by mRNA surveillance factors such as Pelota (Dom34p in S. cerevisiae, aPelota in P. furiosus)5. Crystal structures of archaeal ABCE1 (aABCE1) revealed two nucleotide binding domains (NBDs) in a typical head-to-tail orientation as observed for most of the other members of the ABC protein family6–8. Additional unique structural features of ABCE1 proteins are a helix-loop-helix (HLH) motif, a highly conserved hinge domain and an iron-sulfur cluster domain (FeS) containing two [4Fe-4S]2+ clusters6,9.
In eukaryotes ABCE1 can be found associated with ribosomes and small ribosomal subunits, but also with RFs and initiation factors (eRF1, eIF2, eIF3 and eIF5)10,11. Notably, ABCE1 physically interacts with eRF1 and directly influences its function in stop-codon recognition and peptidyl-tRNA hydrolysis12,13. During recycling ABCE1 can split post termination complexes (post-TCs) obtained with eRF1 and eRF3 into free 60S subunits and tRNA-and mRNA-bound 40S subunits4. A similar role for ABCE1 was found in an archaeal translation system in which ABCE1 together with aRF1 was shown to dissociate ribosomes into subunits upon ATP-binding8.
ABCE1 also acts together with the eRF1 paralog Pelota5. In S. cerevisiae, Dom34 and the eRF3 paralog Hbs1 were described as mRNA surveillance factors recognizing stalled elongating ribosomes14. Such stalls can occur on mRNAs with stable secondary structures, truncations or lacking a stop codon, so that further elongation or canonical termination is prevented. In the so-called no-go mRNA decay (NGD) or non-stop mRNA decay (NSD) pathways, such stalled ribosomes are recognized by Dom34 and Hbs1 (NGD, NSD)14 or by another eRF3 paralog Ski7 (NSD) eventually triggering mRNA degradation14–18. A cryo-EM structure of a stalled ribosome bound to Dom34-Hbs1 shows that Dom34 occupies the ribosomal A site, whereas Hbs1 binds the ribosome similar to other translational GTPases, such as EF-Tu19. Dom34-Hbs1 alone displays ribosome dissociation activity and splits stalled reconstituted ribosomes that contain P-site peptidyl-tRNA20. In a mammalian system, however, ABCE1 is strictly required for ribosome disassembly of both programmed and also vacant ribosomes by Pelota and Hbs15. Taken together ABCE1 is likely the general ribosome recycling factor in archaea and eukaryotes. In contrast to the analogous bacterial system, however, ABCE1 acts not only after canonical RF-dependent termination but also after Pelota-dependent recognition of stalled ribosomes.
It is not known how ABCE1 functions on the ribosome in concert with Pelota or RFs, and how the mechanochemical properties of ABCE1 are employed for ribosome recycling. To address these questions, we determined cryo-EM structures of eukaryotic and archaeal recycling complexes containing Pelota and ABCE1.
Cryo-EM reconstruction and model of Pelota-ABCE1-ribosome complexes
Recycling complexes were obtained by in vitro reconstitution of the 70S and 80S ribosomes with purified Pelota and ABCE1 orthologs. For the generation of a yeast (S. cerevisiae) 80S ribosome-Dom34-Rli1 complex we used ribosome nascent chain complexes (RNCs) stalled by an mRNA with a synthetic stem-loop (SL)14,21, a complex used previously for an 80S-Dom34-Hbs1 cryo-EM reconstruction19. For archaeal (P. furiosus) 70S-aPelota-aABCE1 complexes, 70S ribosomes were purified from a translation extract22. Simultaneous ribosome binding of Pelota and ABCE1 in the presence of non-hydrolysable ADPNP was shown by pelleting assays in the yeast and the archaeal system (Suppl. Fig. 1a, b). Notably, aABCE1-dependent splitting of archaeal ribosomes was not detectable with ADPNP, but strictly required ATP (Suppl. Fig. 1c, d).
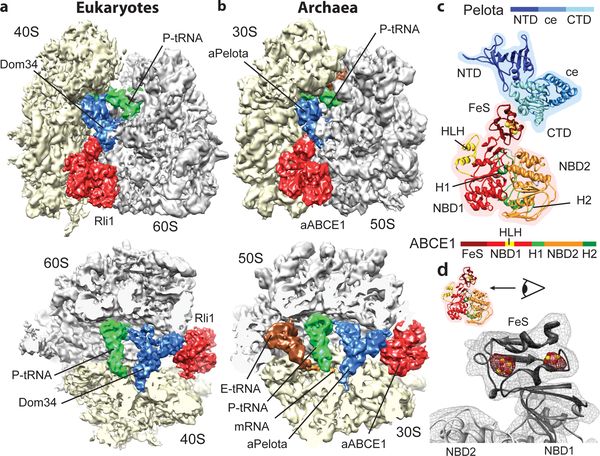
Using cryo-EM in combination with single particle analysis we determined the structures of the SL-RNC-Dom34-Rli1 complex from yeast and the 70S-aPelota-aABCE1 complex from P. furiosus. Computational sorting was performed to generate homogeneous populations of ribosomal complexes containing Pelota, ABCE1 and P-site tRNA. The resolution of the final maps was determined to be 7.2 Å for the yeast complex and 6.6 Å for the archaeal complex (Suppl. Fig. 2). In both reconstructions we observed density for Pelota in the ribosomal A site, for ABCE1 in the GTPase translation factor binding site, and for tRNA in the P site (Fig. 1a, 1b). Additional E-site tRNA density is present in the archaeal ribosome. Both reconstructions are remarkably similar to each other with respect to conformation and the ribosomal interaction patterns of the ABCE1 and Pelota orthologs. Using available crystal structures we could unambiguously assign and position the individual domains of Pelota - divided into N-terminal domain (NTD), central domain (ce) and C-terminal domain (CTD) - and ABCE1 - divided into the N-terminal iron sulfur cluster domain (FeS), NBD1 containing a HLH motif, NBD2 and the hinge domain (Fig. 1c; Suppl. Fig. 3).
Fig. 1: The ribosome-bound Pelota-ABCE1 complex.
(a, b) Cryo-EM reconstructions of the eukaryotic SL-RNC-Dom34-Rli1 and the archaeal 70S-aPelota-aABCE1 complex at 7.2 Å and 6.6 Å resolution, respectively. Extra densities were observed for Dom34/aPelota and Rli1/aABCE1 in the canonical factor binding site as well as for P-site tRNA, E-site tRNA and mRNA. The upper section represents side views, the lower section top views, where large and small subunits were cut. (c) Homology model for ribosome-bound Pelota and ABCE1 in transparent density. The individual domains are color-coded as in the schematic representation of domain organization. N-terminal Domain (NTD), central domain (ce) and C-terminal domain (CTD) are indicated; FeS indicates iron-sulfur cluster domain; NBD1 and 2 indicate nucleotide binding domain 1 and 2; HLH indicates helix-loop-helix motif; H1 and H2 indicates hinge 1 and hinge 2 domain. (d) Zoom on the FeS domain of aABCE1. The density for the two [4Fe-4S]2+ is displayed in red mesh at high contour level.
Strikingly, the two electron dense [4Fe-4S]2+ clusters of ABCE1 can be clearly resolved as distinct spheres at high contour levels in both maps validating the positioning of crystal structures in the EM maps (Fig. 1d). For molecular analysis we used the crystal structure of the yeast 40S subunit23, the model of the yeast 80S ribosome24,25 and, in addition, we built a homology-based molecular model of the archaeal 70S ribosome.
Ribosome-ABCE1 interaction
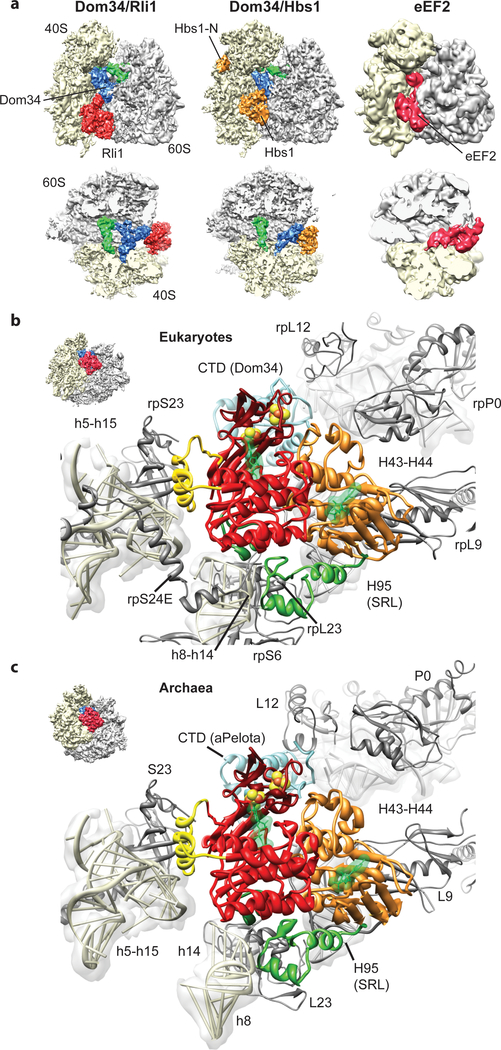
ABCE1 binds to ribosomes in the intersubunit space, where canonical translational GTPases such as EF-Tu, EF-G/eEF2 or Hbs1 also interact with the ribosome (Fig. 2a)19,26–29. ABCE1 excludes these factors from binding at the same time, and we thus conclude that dissociation of Hbs1 or aEF1α, or in case of termination, eRF3 or aEF1α, has to precede ABCE1 binding. Similar to these GTPases, the ATPase ABCE1 contacts the small ribosomal subunit, specifically rRNA helices h5, h8, h14 and h15 (Suppl. Table 1, 2). The h5-h15 region interacts with domain II of the translational GTPases, whereas the h8-h14 junction is the most proximal region to the GTPase switch regions30,31 (Suppl. Fig. 4). Interestingly, the same regions are contacted by ABCE1 via two specific, up to now unexplained, structural features of ABCE1-type ABC-ATPases. The HLH motif of ABCE1 contacts the h5-h15 junction, whereas the hinge region establishes extensive contacts to the h8-h14 junction. In contrast to translational GTPases that engage in close interaction with the sarcin-ricin-loop of H95 (SRL), contacts of ABCE1 to the large subunit are essentially limited to L9 in both species. Despite the overall striking similarity between Rli1 and aABCE1 in their ribosome interaction mode, additional minor contacts are present in the yeast complex: Rli1 contacts rpS6e and rpS24e on the small subunit, and, on the large subunit, rpP0 and a small region of the SRL (H95) which is different from the binding region of translational GTPases (Fig. 2b, c). Unexpectedly, the FeS cluster domain of ABCE1 does not directly bind the ribosome but instead interacts with Pelota only. These interactions are conserved between yeast and archaea. In summary, ABCE1 establishes multiple contacts with both small and large ribosomal subunits as well as with the release factors (and their paralogs) and these interactions involve all domains of ABCE1. Such tight recognition provides a rationale for direct mechanochemical coupling of ATP driven conformational changes in ABCE1 with structural changes in the ribosome that are critical for termination and release.
Fig. 2: Interaction of Pelota and ABCE1 with the ribosome.
(a) Comparison of the SL-RNC-Dom34-Rli1 cryo-EM map with the SL-RNC-Dom34-Hbs1 and the 80S-eEF2 maps. Views are as in Fig. 1a and 1b. (b, c) Interactions of ABCE1 with the eukaryotic (b) and the archaeal (c) ribosome. The view is indicated by a thumbnail. The domain color code is as in Fig. 1c.
ABCE1 stabilized conformational switch of Pelota
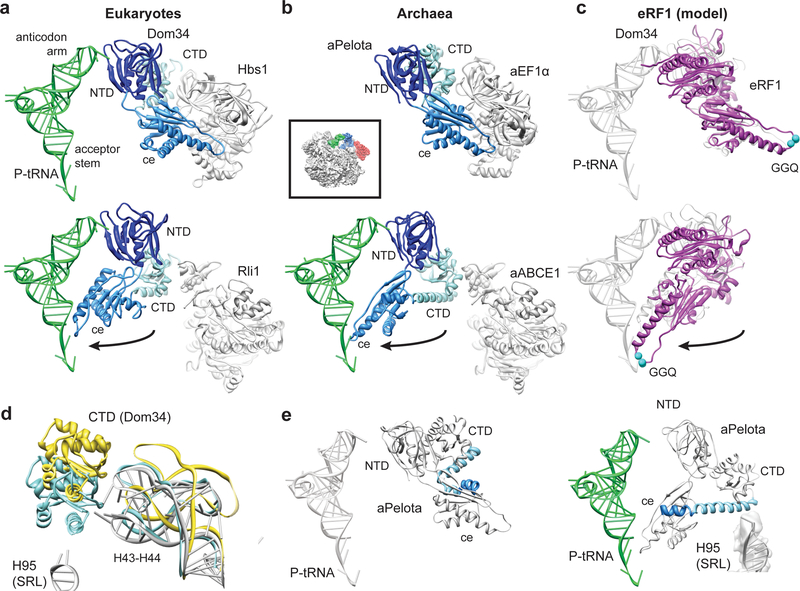
The FeS domain of ABCE1 binds to the CTD of Pelota and we observe a large-scale conformational change in the central domain and CTD compared to the Dom34 structure in the Hbs1-bound state19 (Fig. 3). By contrast, the NTD of Pelota is essentially unchanged in these two structures where they are located in the A site contacting rRNA helices h18, h28, h31, h34, and h44 and additionally the ribosomal protein rpS30 in yeast. The β3-β4 loop reaches deeply into the A site and at the current resolution we observe additional contacts of this loop of aPelota to the ribosomal protein S5 (Suppl. Fig. 3c). The most dramatic rearrangements, however, occur in the central domain of both Pelota orthologs: the central domain of Dom34 bound to Hbs1 in the yeast ribosome is tightly packed against Hbs119, very similar to the domain arrangement in a crystal structure of an aPelota-aEF1α complex32 (Fig. 3b); in the presence of ABCE1, however, the central domain of Dom34 or aPelota is rotated by approximately 140 degrees towards the P-site tRNA (Suppl. Movie 1, 2). In this conformation it establishes numerous new contacts to rRNA in domain IV and V of the large subunit (Suppl. Table 1, 2). The positively charged loop β10-α3 directly contacts the P-site tRNA acceptor stem and the ribosomal protein L10e in archaea. In the closely related RF1 proteins, this loop contains the GGQ motif that is essential for catalyzing the hydrolysis of peptide from peptidyl-tRNA33,34. When modeling ribosome-bound eRF1 on the basis of the Pelota conformation observed in the presence of ABCE1, the GGQ motif of eRF1 would be ideally positioned to interact with the CCA-end of the P-site peptidyl-tRNA in the peptidyl transferase center (Fig. 3c). This may explain how ABCE1 can stimulate termination activity in vivo and in vitro12,13.
Fig. 3: Domain movements in Pelota and eRF1.
(a) Comparison of the ribosome-bound Dom34 conformation in complex with Hbs1 (top section) and Rli1 (lower section). (b) Comparison of the aPelota-aEF1α crystal structure32 with the ribosome-bound aPelota in complex with aABCE1. The central domain of Pelota swings out towards the P-site tRNA. (c) Models for eRF1 before and after the suggested movement of the central domain. (d) Conformation of the Dom34 CTD and the stalk base rRNA (H43-H44) when bound to Hbs1 (yellow) and to Rli1 (blue). rRNA conformation without factors bound is shown in grey. (e) In aPelota three separate small helices refold into a long α-helix during movement of the central domain bridging the CTD and the central domain.
Since the CTD of Pelota establishes the only contact site to ABCE1 via the FeS cluster domain, the interaction surface is rather small (440 Å2) compared to that between ribosome-bound Dom34 and Hbs1 trapped in the GTP state (1940 Å2)19. In the ABCE1-bound conformation, the CTD is rotated downwards by approximately 15 degrees together with a movement of the ribosomal stalk base (H43-H44, rpL12), similar to that induced by eEF2 binding (Suppl. Fig. 5)28,29. Both the stalk base and the CTD of Dom34 move closer towards the SRL (H95) and a strong contact between the CTD of Dom34 (helix α7) and the SRL is established (Fig. 3d). Very similar conformations of the stalk base, as well as of the central domain and CTD, were observed for aPelota on the archaeal ribosome, although some molecular details of domain fold and ribosome interaction pattern also differ between Dom34 and aPelota. Interestingly, helices α5, α6 and α7 that link the central domain and the CTD of aPelota establish one long alpha helix with a kink between α5 and α6 in the presence of aABCE1 that reaches from the SRL (H95) deeply into the A site (Fig. 3e).
In summary, in both species the presence of ABCE1 stabilizes an alternative conformation of Pelota on the ribosome, primarily affecting the central domain that reaches through the A site to contact the P-site tRNA. An analogous behavior of the closely related RFs would ideally position the conserved GGQ-loop for catalyzing peptidyl-tRNA hydrolysis.
Mechanochemical activity of ABCE1 on the ribosome
Typically, ABC proteins generate mechanochemical work by nucleotide driven clamp like motions of the two NBDs: in the apo- or ADP-bound state, NBDs adopt an open conformation often linked to a higher affinity for the given substrate of the ABC enzyme. ATP binding triggers a closed state, by binding to Walker A and B motifs of one NBD and the opposing conserved LSGGQ-loop (signature motif) of the other NBD that coordinates the γ-phosphate of ATP for subsequent hydrolysis35. ATP-binding or subsequent ATP-hydrolysis lead to a “power stroke” that usually causes concomitant conformational changes in connected domains or binding partners.
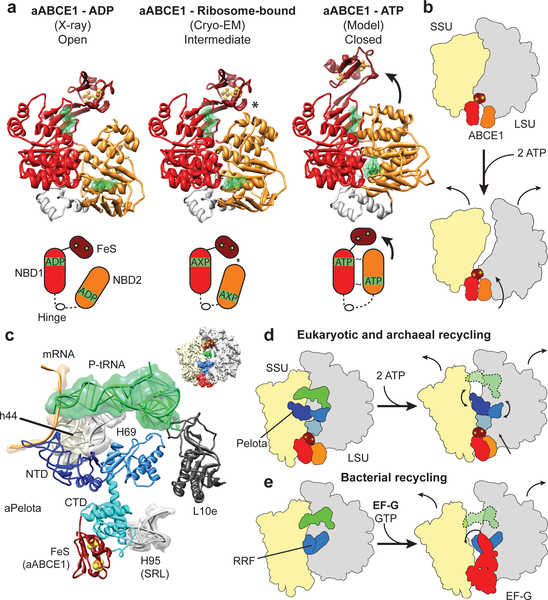
To analyze the mechanochemical function of ABCE1 in ribosome splitting we compared the ribosome-bound conformation of ABCE1 with the open ADP-bound form as observed in the crystal6–8 and with a model for the closed ATP-bound state (Fig. 4a). The model for the closed state is derived by individually superimposing the NBDs of ABCE1 to NBDs crystallized in the ATP bound state36. Interestingly, neither the open nor the closed model can be easily modeled into the electron density in the reconstructions. In both reconstructions, we observe an intermediate, half-open state of the two NBDs: NBD2 rotates by approximately 17 degrees towards NBD1 and the FeS cluster domain. However, an additional upward movement by 8 Å of NBD2 would be required in order to obtain the fully closed conformation where the signature motif of one NBD domain contacts the nucleotide binding domain pocket of the other NBD domain (Suppl. Movie 3). Notably, in the observed half-open state of ABCE1 we find a contact between the NBD2 domain and the FeS cluster domain, which is not seen in the crystal structures of the open state. Adoption of the fully closed ATP-bound conformation would therefore require a substantial shift of the FeS cluster domain, also of about 8 Å (Fig. 4a) to avoid a steric clash. While the limited resolution of the reconstruction doesn’t allow for any conclusions regarding the nature of the bound nucleotide, the conformations of the individual lobes within the NBD1 and NBD2 domains more closely resemble those of the ADP-bound crystal structures. The similarity of the “intermediate” conformation in both reconstructions suggests that binding to the ribosome induces an allosteric change in ABCE1, perhaps related to allosteric control of ABC transporter by substrate binding37.
Fig. 4: Mechanochemical activity of ABCE1 on the ribosome.
(a) Crystal structure of the open (ADP-bound) aABCE1, the cryo-EM structure of the ribosome-bound aABCE1 and homology model of the closed (ATP-bound) aABCE1 including schematic drawings. An asterisk indicates a contact between NBD2 and the FeS domain of aABCE1. (b) Ribosomal subunits may be dissociated by following the trajectory of aABCE1 domain closure upon ATP-binding. (c) Interactions of the aPelota NTD and central domain within the ribosome. (d) ABCE1 domain closure could lead to an allosteric cascade with the FeS domain acting as a bolt on the CTD of Pelota to rearrange the NTD and central domain of Pelota. This mechanism would be analogous to the splitting reaction in bacteria by RRF and EF-G as depicted in (e).
The finding that ATP hydrolysis4,5 is required for full splitting activity strongly suggests that ABCE1 indeed has to undergo a conversion from the observed half-open pre-splitting conformation to the fully closed ATP state in order to efficiently dissociate ribosomes. Therefore, we analyzed the effect of ATP-dependent NBD domain closure by superimposing the half-open ribosome-bound state of ABCE1 with the model for the closed-state. ABCE1 in the closed conformation would not sterically clash with the ribosomal subunits to induce splitting. One possibility is that the small and large ribosomal subunits follow the trajectory of NBD1 and NBD2 of ABCE1, respectively. In this case the ribosomal subunits would sufficiently rotate away from each other so as to affect the inter-subunit bridges and, thus, the overall ribosome stability (Fig. 4b).
It is more likely, however, that the transition of ABCE1 through the closed conformation triggers an allosteric cascade affecting Pelota: The FeS cluster domain of ABCE1 contacts the NBD2 domain already in the half-open state and has to follow the movement of the NBD2 during closure. This conformational change of the FeS cluster domain towards the intersubunit space is likely to be transmitted to Pelota via the close interaction with its CTD. A shift of the CTD would in turn be transmitted to both the NTD and the central domain of Pelota. These Pelota domains establish a network of contacts with the small and the large ribosomal subunit as well as with the P-site tRNA (Fig. 4c). Numerous mutations indeed underline the functional importance of these domains for the activity of Pelota (Suppl. Table 1, 2). A conformational shift can be easily envisaged to cause dissociation of the ribosome by destabilizing inter-subunit bridges and the P-site tRNA. A function of the FeS cluster domain of ABCE1 as a structural bolt to remodel Pelota by transmitting ATP induced changes from the NBDs is in good agreement with the finding that deletion of this domain abolishes splitting activity8. Enhanced stability of the domain provided by the FeS cluster may be required in the transmission of ABCE1’s mechanochemical power for ribosome splitting.
Although employing an entirely different cast of characters, this scenario is structurally reminiscent of bacterial ribosome recycling by ribosome recycling factor (RRF) and EF-G. In this case, an EF-G-based GTP-dependent conformational switch positions RRF to clash with the small ribosomal subunit, inevitably promoting subunit dissociation8 (Fig.4d, e).
Conclusion
Taken together we provide a structural basis and a universal mechanistic model for eukaryotic and archaeal ribosome recycling in which ABCE1 actively coordinates rescue (or translation termination) with recycling and re-initiation (Fig. 5):
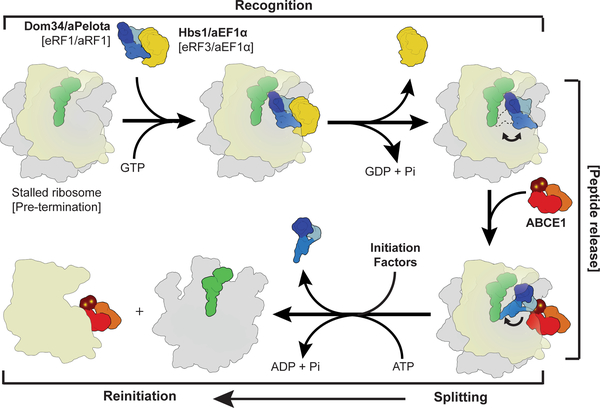
Fig. 5: Scheme of archaeal and eukaryotic ribosome recycling bridging termination with initiation.
A translational GTPase (Hbs1/aEF1α/eRF3) delivers the factor, which recognizes stalled ribosomes (Pelota) or pre-termination complexes (eRF1/aRF1). After GTP hydrolysis, the GTPase dissociates and ABCE1 can bind. ABCE1 induces or stabilizes the swung-out conformation of Pelota (or RF1), which would lead to peptide release in case of termination. Ribosome splitting is induced after ATP-binding to ABCE1 and hydrolysis. In eukaryotes initiation factors can bind during the splitting reaction coupling ribosome recycling with re-initiation. After splitting ABCE1 stays associated with the small ribosomal subunit.
In the first stage, the recognition stage, the sensing factors Pelota (for rescue) or RF1 (for termination) are delivered to stalled ribosomes or pre-termination complexes by EF-Tu like GTPases. In the next step, the GTPase dissociates to allow ABCE1 binding to the ribosome. ABCE1 interacts with the C-terminal domain of Pelota (or of RF1s in termination) to stabilize the extended conformation of the central domain. In the case of translation termination, the GGQ motif of RF1 will be positioned proximal to the CCA-end of the P-site tRNA to catalyze peptide release; in the case of ribosome rescue, the central domain will be tightly accommodated proximal to the peptidyl transferase center. Subsequently, in both cases, ABCE1 triggers ribosome disassembly into subunits by a power stroke upon NBD domain closure and ATP hydrolysis5. Our biochemical and structural data suggest a universal role of ATP hydrolysis in the mechanism of ABCE1-driven recycling. The conformational switch of ABCE1 could cause either a direct disruption of the ribosomal intersubunit bridges or, more likely, further conformational changes via an allosteric cascade from the FeS cluster domain of ABCE1 to the central domain and NTD of Pelota. In the archaeal system ABCE1 remains bound to the small ribosomal subunit after splitting8 and it has been also found on the small subunit in eukaryotes10,11. Notably, ribosome recycling is coupled in eukaryotes with re-initiation when initiation factors such as eIF3, eIF1 and eIF1A bind the small ribosomal subunit as recycling is completed38. An initial recruitment of eIF3 to the 80S ribosome may even occur directly via ABCE1 interaction with the eIF3 subunit eIF3j (Hcr1p in yeast), even before recycling is completed13,39,40. In contrast, the analogous bacterial recycling system consisting of RRF and EF-G acts only after termination is completed and the participation of initiation factors is less clear41. In conclusion, the archaeal and eukaryotic kingdoms have maintained an extremely conserved general ribosome recycling system with an ABC-type ATPase at the core: the mechanochemical properties of ABCE1 are employed through a still somewhat enigmatic FeS cluster domain. This domain triggers an allosteric cascade that actively coordinates translation termination or rescue with recycling12, and eventually with re-initiation. It remains a puzzle as to why a complex FeS cluster domain is apparently employed for a structural role only and has not been replaced by a simpler structure over billions of years of evolution. Thus, it is highly desirable to seek deeper insight into additional functions of ABCE1 in processes such as translation initiation and ribosome assembly.
Methods Summary
Programmed yeast SL-RNCs were prepared from cell-free extracts as described19,42. Archaeal ribosomes were purified from cell free extracts22 by sucrose density centrifugation. Ribosome binding partners (Dom34, aPelota, ABCE1, aRF1 and aIF6) were expressed in E. coli or S. cerevisiae (Rli1) and purified using affinity chromatography. Ligands were reconstituted in vitro with SL-RNCs or 70S ribosomes, and binding was analyzed by SDS-PAGE after pelleting of ribosome-bound fractions. Splitting activity was monitored in sucrose gradients using UV-profiles. For cryo-EM, yeast and archaeal recycling complexes were vitrified and data were collected on a Titan Krios electron microscope (FEI Company). Single particle analysis and 3D reconstruction was done using the SPIDER software package43. Homology models were generated using HHPRED44 and MODELLER45.
Supplementary Material
Acknowledgements
We thank Alexandra Scheele and Andrea Gilmozzi for excellent technical assistance, and Daniel Wilson for critical discussions. This work was supported by grants of the DFG, SFB594 (to R.B.), SFB646 (to T.B., R.B. and K.-P. H.), NIH U19 AI083025 (to K.-P. H.) and by the Fonds der chemischen Industrie (to S.F.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Full methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Winzeler EA et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999). [DOI] [PubMed] [Google Scholar]
- 2.Coelho CM et al. Growth and cell survival are unevenly impaired in pixie mutant wing discs. Development 132, 5411–5424 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Estevez AM, Haile S, Steinbuchel M, Quijada L & Clayton C. Effects of depletion and overexpression of the Trypanosoma brucei ribonuclease L inhibitor homologue. Mol. Biochem. Parasitol 133, 137–141 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Pisarev AV et al. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell 37, 196–210 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pisareva VP, Skabkin MA, Hellen CU, Pestova TV & Pisarev AV Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. Embo J. (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karcher A, Schele A & Hopfner K-P X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi. J. Biol. Chem 283, 7962–7971 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Karcher A, Buttner K, Martens B, Jansen RP & Hopfner KP X-ray structure of RLI, an essential twin cassette ABC ATPase involved in ribosome biogenesis and HIV capsid assembly. Structure 13, 649–659 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Barthelme D. et al. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc. Natl. Acad. Sci (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barthelme D. et al. Structural organization of essential iron-sulfur clusters in the evolutionarily highly conserved ATP-binding cassette protein ABCE1. J. Biol. Chem 282, 14598–14607 (2007). [DOI] [PubMed] [Google Scholar]
- 10.Dong J. et al. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J. Biol. Chem 279, 42157–42168 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Andersen D & Leevers S. The essential Drosophila ATP-binding cassette domain protein, Pixie, binds the 40S ribosome in an ATP-dependent manner and is required for translation initiation. J. Biol. Chem 282, 14752–14760 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Shoemaker CJ & Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc. Natl. Acad. Sci (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khoshnevis S et al. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep 11, 214–219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doma M & Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440, 561–564 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atkinson G, Baldauf S & Hauryliuk V. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evolutionary Biology 8, 290 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee HH et al. Structural and functional insights into Dom34, a key component of no-go mRNA decay. Mol. Cell 27 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Frischmeyer P et al. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295, 2258–2261 (2002). [DOI] [PubMed] [Google Scholar]
- 18.van Hoof A, Frischmeyer PA, Dietz HC & Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295, 2262–2264 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Becker T et al. Structure of the no-go mRNA decay complex Dom34–Hbs1 bound to a stalled 80S ribosome. Nat. Struct. Mol. Biol 18, 715–720 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Shoemaker CJ, Eyler DE & Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330, 369–372, (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosoda N et al. Translation termination factor eRF3 mediates mRNA decay through the regulation of deadenylation. J. Biol. Chem 278, 38287–38291 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Endoh T, Kanai T & Imanaka T A highly productive system for cell-free protein synthesis using a lysate of the hyperthermophilic archaeon, Thermococcus kodakaraensis. Appl. Microbiol. Biotechnol 74, 1153–1161 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Rabl J, Leibundgut M, Ataide SF, Haag A & Ban N Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331, 730–736 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Armache JP et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-Å cryo-EM map of the 80S eukaryotic ribosome. Proc. Natl. Acad. Sci 107, 19754–19759 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armache JP et al. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-Å resolution. Proc. Natl. Acad. Sci 107, 19748–19753 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmeing TM et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326, 688–694 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao YG et al. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326, 694–699 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Taylor DJ et al. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 26, 2421–2431, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spahn CM et al. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J. 23, 1008–1019 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villa E et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc. Natl. Acad. Sci 106, 1063–1068 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Connell SR et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol. Cell 25, 751–764 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi K et al. Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1α complex. Proc. Natl. Acad. Sci 107, 17575–17579 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frolova L et al. Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5, 1014–1020 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song H et al. The crystal structure of human eukaryotic release factor eRF1 - mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100, 311–321 (2000). [DOI] [PubMed] [Google Scholar]
- 35.Hopfner KP & Tainer JA Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr. Opin. Struct. Biol 13, 249–255 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Smith PC et al. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol. Cell 10, 139–149 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Locher KP Review. Structure and mechanism of ATP-binding cassette transporters. Philos. Trans. R. Soc. Lond. B. Biol. Sci 364, 239–245 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pisarev AV, Hellen CU & Pestova TV Recycling of eukaryotic posttermination ribosomal complexes. Cell 131, 286–299 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kispal G et al. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J. 24, 589–598 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yarunin A et al. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 24, 580–588 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pavlov MY, Antoun A, Lovmar M & Ehrenberg M Complementary roles of initiation factor 1 and ribosome recycling factor in 70S ribosome splitting. EMBO J. 27, 1706–1717 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beckmann R et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107, 361–372 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Frank J et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol 116, 190–199 (1996). [DOI] [PubMed] [Google Scholar]
- 44.Soding J, Biegert A & Lupas AN The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33, W244–248 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eswar N, Eramian D, Webb B, Shen MY & Sali A Protein structure modeling with MODELLER. Methods Mol. Biol 426, 145–159 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.